Abstract
The obligate intracellular bacterial pathogen, Chlamydia trachomatis, develops within a membrane-bound vacuole termed the inclusion. Affinity purification-mass spectrometry (AP-MS) experiments to study the interactions that occur at the chlamydial inclusion membrane have been performed and, more recently, combined with advances in C. trachomatis genetics. However, each of the four AP-MS published reports used either different experimental approaches or statistical tools to identify proteins that localize at the inclusion. We critically analyzed each experimental approach and performed a meta-analysis of the reported statistically significant proteins for each study, finding that only a few eukaryotic proteins were commonly identified between all four experimental approaches. The two similarly conducted in vivo labeling studies were compared using the same statistical analysis tool, Significance Analysis of INTeractome (SAINT), which revealed a disparity in the number of significant proteins identified by the original analysis. We further examined methods to identify potential background contaminant proteins that remain after statistical analysis. Overall, this meta-analysis highlights the importance of carefully controlling and analyzing the AP-MS data so that pertinent information can be obtained from these various AP-MS experimental approaches. This study provides important guidelines and considerations for using this methodology to study intracellular pathogens residing within a membrane-bound compartment.
INTRODUCTION
Chlamydia trachomatis, a developmentally regulated obligate intracellular bacterium, causes the most common bacterial sexually transmitted disease [1]. C. trachomatis is among several pathogenic intracellular bacteria that develop within a host-derived vacuole, typically referred to as a Bacteria Containing Vacuole (BCV) [2-4]. For Chlamydia, the BCV is referred to as an inclusion. Throughout its biphasic developmental cycle, C. trachomatis resides within the inclusion, which is modified by secreted C. trachomatis inclusion membrane (Inc) proteins [5-7]. Incs are characterized as having two or more hydrophobic transmembrane domains [6, 8], with the N- and C-termini exposed to the host cytosol [9]. Although there are over 50 predicted Inc proteins [10], the roles and binding partners of Incs remain largely unknown due to the difficulties in purifying Incs and maintaining interacting partners by traditional affinity purification methods [11-13]. Before the development of a genetic transformation system in C. trachomatis, various in vitro methods were used to identify potential protein-protein interacting partners [14-16]. More recently, four large-scale proteomics experiments to identify inclusion-associated proteins at the C. trachomatis inclusion membrane have been published with two 2019 studies leveraging advances in genetic manipulation of C. trachomatis [12, 17-20]. This has allowed more direct experimental approaches to be implemented to identify chlamydial Inc-binding partners or inclusion-associated proteins. Each approach affinity-purified tagged proteins or whole inclusions and then the purified proteins were identified by mass spectrometry. However, each of these AP-MS studies featured different experimental approaches and/or utilized different statistical analysis tools to assign significance to the identified proteins [12, 18-20]. We compare these experimental approaches, critically analyzing the limitations of each.
In one of these large scale AP-MS studies, Mirrashidi et al. transiently transfected uninfected host cells with epitope-tagged chlamydial Incs (full length and/or cytoplasmic domains), using the Strep-tag® system, and identified by mass spectrometry putative host cell binding partners [18]. Their approach led to the identification of the IncE binding partners, sorting nexin (SNX) 5 and SNX6 [18, 21]. One limitation of this experimental methodology is that hydrophobic Incs or only the cytosolic domains of Incs were ectopically expressed in eukaryotic host cells out of their normal spatial context [18] rather than anchored within the inclusion membrane [22]. Note that, by design, this approach does not allow for Inc-Inc interactions. Ectopically expressed Incs aggregate in micelle-type structures [22], which could increase the possibility of detecting false interactions or, conversely, excluding true interacting proteins. Another group, Aeberhard et al., purified chlamydial inclusions from infected host cells and identified inclusion-associated proteins by mass spectrometry (i.e., inclusion-MS) [19]. In agreement with Mirrashidi et al. [18], the inclusion-MS study also identified both SNX5 and SNX6 at the inclusion [19]. Although this method also validated the localization of additional eukaryotic proteins at the inclusion membrane, the purification of inclusions is labor-intensive and resulted in a reported final total recovery rate of only eight percent of all inclusions [19]. Inclusion-MS is also not sufficient to detect Incs, which eliminates the possibility of understanding Inc-Inc interactions [19]. Importantly, no other group has attempted to replicate these experimental approaches, which makes it difficult to directly compare the datasets to eliminate false-positive protein identifications.
In vivo proximity labeling systems used in the context of chlamydial genetic approaches have recently been implemented as molecular tools to overcome the limitations of studying the chlamydial inclusion membrane with the experimental approaches described above [11, 23-25]. The ascorbate peroxidase (APEX2) proximity labeling system covalently modifies proximal or interacting proteins in vivo, in the context of infection. This system yields a significant advantage over in vitro experiments to detect protein-protein interactions between hydrophobic membrane proteins that are otherwise difficult to study. Specifically, this methodology circumvents issues with detergents disrupting protein-protein interactions because proximal or interacting proteins are covalently modified (e.g. with a biotin molecule) so protein-protein interactions do not need to be maintained during lysis. The biotin-modified proteins are affinity-purified (e.g., using streptavidin) and identified by mass spectrometry. The APEX2 system is likely to prove useful for understanding how intracellular bacteria modify their BCV membranes to interact with the host cell in ways that facilitate pathogen growth. In the context of understanding chlamydial-host interactions, this approach utilizes C. trachomatis transformants that are induced to express Inc-APEX2 fusion proteins, which facilitates correct protein folding and localization to the inclusion membrane compared to ectopic expression of tagged Incs [11, 12, 20]. Two recent reports utilized the APEX2 proximity labeling system to identify potential binding partners of Incs in the C. trachomatis L2 inclusion [12, 20]. These two proximity labeling studies were methodologically very similar but differed in the statistical analysis of the AP-MS datasets as one study used a G-test or t-test to identify significant proteins [20], while the other study used Significance Analysis of INTeractome (SAINT) to identify significant proteins [12]. Nevertheless, these data can be directly compared to better contextualize each study’s results and render greater confidence in the proteins identified from both studies.
In this meta-analysis, we first compared the statistically significant proteins reported in the four AP-MS C. trachomatis inclusion membrane interaction studies to identify common proteins, while noting the differences in processing and identification. Next, we directly compared the two proteomics experiments that used the APEX2 proximity labeling system [12, 20]. We processed the Mascot data reported by Dickinson et al. [20] using the SAINT statistical analysis tool [26] described in Olson et al. [12] to determine the effect of statistical analysis tools on the identification of significant proteins. We found that there were notable differences in the number of significant proteins found using SAINT compared to the G- and t-test as reported by Dickinson et al. [20]. Finally, we examined our Inc-APEX2 AP-MS datasets [12] using a more rigorous minimum peptide threshold to determine how these parameters affected the SAINT statistically significant eukaryotic and chlamydial proteins. The results suggest that a rigorous statistical analysis is critical to eliminate likely false-positive hits from these datasets.
MATERIALS AND METHODS
Source of AP-MS data and how these data were used
The statistically significant proteins reported by Mirrashidi et al. [18], Aeberhard et al. [19], Dickinson et al. [20], and Olson et al. [12] with data source are listed in Table 1. The statistically significant protein identifiers from each source (listed in Table 1) were uploaded into UniProt (www.uniprot.org) using the “retrieve/mapping” tool to obtain the most current UniProt KB annotation. These UniProt KB protein annotations (listed in Table S1) were used for the Venny [27] comparison of these experimental datasets. The complete UniProt mapped input list of proteins from each of the experimental datasets are found in Table S1. Using the current version of UniProt, some entries mapped to more than one identifier (Table S1). Venny 2.0 [27], a Venn diagram tool (https://bioinfogp.cnb.csic.es/tools/venny/index.html), was used to compare the proteins identified in Mirrashidi et al. [18], Aeberhard et al. [19], Olson et al. [12], and Dickinson et al. [20].
Table 1.
Comparison of large-scale AP-MS C. trachomatis L2 studies
| Experimental procedure |
Source of data used in this study |
Statistical test used for data analysis |
Statistical significance cut-off reported |
Statistically significant proteins# |
|---|---|---|---|---|
| Strep-Tag® AP-MSa | Table S1; "prey.entry.name" column | CompPASS and MiST | Top 1% ComPASS; MiST ≥ 0.7 | 335 (N=3) |
| Inclusion-MSb | Table S1; “Protein ID” column | SILAC/Two-sided Wilcoxon test with Benjamini-Hochberg | p≤ 0.05 | 352 (N=3) |
| IncB-APEX2 c | S1 Table; Complete proteomic data | G-test and T-test | p≤ 0.05 | 399 (N=6) |
| IncF-APEX2, IncA-APEX2, IncATM-APEX2d | Table 1, Table S1. Significant C. trachomatis L2 proteins; Table S2. statistically significant eukaryotic proteins | Significance Analysis of INTeractome (SAINT) | BFDR ≤ 0.05 | 199 (N=5) |
The statistically significant proteins at 24 hpi for all but the Strep-tag experiments by Mirrashidi et al. which transiently transfected (uninfected) HEK293T cells
Mirrashidi et al.; 331 protein entries reported by Mirrashidi et al. mapped to 335 proteins (Uniprot)
Aeberhard et al.; 350 protein entries reported by Aeberhard et al. mapped to 352 proteins (Uniprot)
Dickinson et al.; 396 protein entries reported by Dickinson et al. mapped to 399 proteins (Uniprot)
Olson et al.; 199 unique protein entries combined using each IncF-APEX2, IncA-APEX2, and IncATM-APEX2
SAINT analysis of Dickinson et al. [20]
To analyze the results of Dickinson et al. [20] using Significance Analysis of INTeractome (SAINT), the Mascot results files were downloaded from the PRIDE consortium (PRIDE via Proteomexchange; PXD012494). These results files were input into Scaffold Viewer (http://www.proteomesoftware.com/products/free-viewer) to visualize spectral count data for each protein. In Scaffold, the parameters were set to 95% protein threshold, one-peptide minimum threshold, and 95% peptide threshold. The generated “samples report” file was exported as an excel file, which contains the protein identification and spectral counts for each biological replicate and bait protein (e.g., IncB-APEX2) at 8, 16, and 24 hours post-infection, and used to create the SAINT input files (i.e., bait, prey, interaction files) (Table S2). The SAINT input files were analyzed using the GUI Significance Analysis of INTeractome (SAINT) interface available via the APOSTL Galaxy Server (http://apostl.moffitt.org/)_(Table S2). SAINT is a statistical tool which accounts for protein length and uses Bayesian statistics to calculate a Bayesian False Discovery Rate (BFDR) for each prey-bait interaction indicated [26, 28]. A Venny comparison of the SAINT data with Dickinson et al. reported data is provided in Table S3 and Table S4.
Analysis of Dickinson et al. statistically significant proteins using the CRAPome [29]
The Contaminant Repository for Affinity Purification (CRAPome; www.crapome.org) [29], was used to further analyze the reported data from Dickinson et al. for potential contamination with background proteins. The list of statistically significant proteins reported by Dickinson et al. was input into the CRAPome and the results for each prey identified are reported in Table S5. Each sample was normalized by dividing the average spectral counts (out of the 411 experiments available in the CRAPome) by the amino acid length. The “percent of experiments identified” column was calculated by dividing the average spectral counts by the 411 total AP-MS experiments in the CRAPome that contain the spectral count information.
Identification of mitochondrial proteins that have homology to C. trachomatis proteins
AP-MS peptide samples from uninfected HeLa cells supplemented with biotin, which were prepared as previously described [12], were analyzed using the Mascot server, which searched the Swiss-Prot database selected for C. trachomatis strain 434/Bu. Proteins with spectral counts in three or more out of six total replicates were then BLAST searched against Homo sapiens using NCBI Protein BLAST. The resulting human proteins were then identified by UniProtKB. Subcellular localization, query cover, Expect value and percent identity from the BLAST search are provided. These data are listed in Table S6.
Analysis of two-peptide minimum threshold on Olson et al. SAINT significant eukaryotic and C. trachomatis L2 proteins
To determine the SAINT significant proteins using a two-peptide minimum threshold in Scaffold, the filtering parameters reported by Olson et al. [12] for the eukaryotic (PRIDE accession number: PXD015890) and C. trachomatis L2 proteins (PRIDE accession number: PXD015883) identified were changed to a two-peptide minimum, and the generated “samples report” file was exported as an excel file. The SAINT input files were created from this output [26] and analyzed using SAINT v3.6.1 through the APOSTL Galaxy Server (http://apostl.moffitt.org/). The samples report and SAINT results for eukaryotic proteins are in Table S8 and C. trachomatis L2 proteins in Table S9.
RESULTS AND DISCUSSION
A Meta-Analysis of C. trachomatis AP-MS and Inclusion-MS Experiments to Identify Protein-Protein Interactions at the Chlamydial Inclusion Membrane
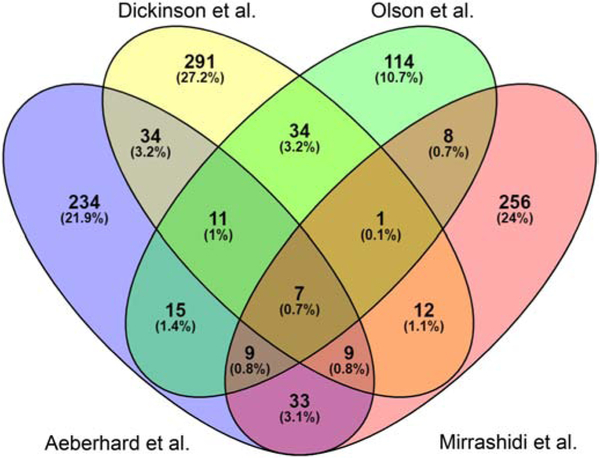
Given the complex nature of the protein-protein interactions (PPIs) that occur at the C. trachomatis L2 inclusion membrane, we sought to compare previously identified inclusion-associated proteins from four AP-MS datasets. Briefly, Mirrashidi et al. [18] transfected uninfected HEK293T cells with tagged Incs, which were purified by AP-MS; Aeberhard et al. [19] purified C. trachomatis L2 inclusions from eukaryotic cells; both Dickinson et al. [20] and Olson et al. [12] used the APEX2 proximity labeling system to tag interacting proteins in vivo, followed by AP-MS to identify interacting or proximal proteins. To compare the proteins that were identified using each of these experimental approaches, we analyzed the statistically significant proteins that were reported from each experiment (summarized in Table 1). Aeberhard et al. identified 350 (p≤0.05) statistically significant proteins for purified inclusions [19], Mirrashidi et al. identified 331 (MiST score (1 = perfect score)) statistically significant proteins by transfecting uninfected cells with epitope-tagged Incs [18], Dickinson et al. (24 hpi time point only) identified 396 (p≤0.05) statistically significant proteins using C. trachomatis L2 IncB-APEX2 transformants [20], and Olson et al. identified 199 (p≤0.05) statistically significant proteins combined from C. trachomatis L2 IncF-APEX2, IncA-APEX2, and IncATM-APEX2 transformants [12]. We used Venny [27], to compare these lists with Venn diagrams (http://bioinfogp.cnb.csic.es/tools/venny/) to detect the commonly identified, statistically significant proteins from each AP-MS and inclusion-MS experimental dataset (Fig. 1; Table S1) [12, 18-20].
Fig. 1.
Venny comparison of eukaryotic proteins at the C. trachomatis inclusion reported by Olson et al., Aeberhard et al., Mirrashidi et al., and Dickinson et al. The significant proteins reported from each study were input into Venny to determine commonly identified proteins. The total number of proteins as well as the percentage of total is indicated within each overlapping section.
Highlighting the numerous differences in each of these experimental protocols, only 0.7% (7 proteins) of the total input for Venny were commonly identified as statistically significant in all four inclusion-MS or AP-MS experimental datasets (Table 1, Table S1; Fig. 1). These proteins were: Leucine-Rich Repeat Flightless-Interacting Protein 1 (LRRF1, LRRFIP1), Myosin Phosphatase-Targeting Subunit 1 (MYPT1, PPP1R12A), Reticulon-4 (RTN4), Sorting Nexin-1 (SNX1), Tropomodulin-3 (TMOD3), 14-3-3 protein beta (YWHAB), and 14-3-3 protein eta (YWHAH)(Table S1. In agreement with the current literature, several of the statistically significant eukaryotic proteins identified by all four studies have been reported to localize at the inclusion membrane (Table S1). These include 14-3-3 Beta [14], MYPT1 [30, 31], SNX1 [19], and LRRF1 [12]. This highlights the potential for the three other commonly identified proteins, Reticulon-4, Tropomodulin-3, and 14-3-3 eta, to localize at the chlamydial inclusion during infection of a host cell.
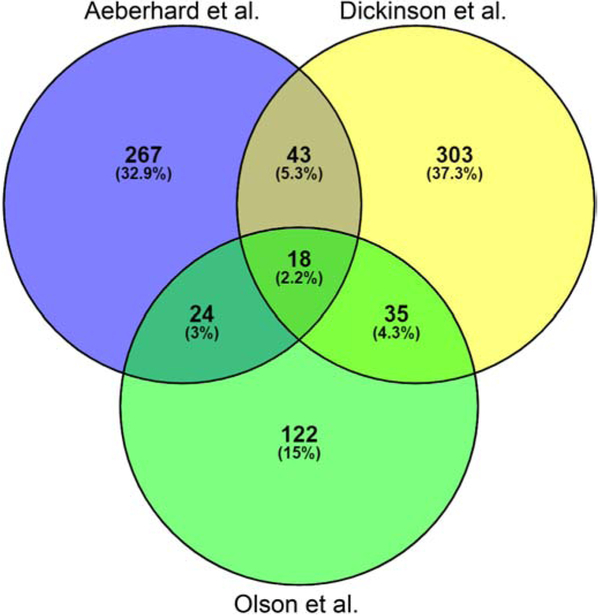
Comparison of Aeberhard et al. [19], Dickinson et al. [20], and Olson et al. [12] reported statistically significant eukaryotic proteins
We next compared the reported significant proteins from each of the three studies that were performed using chlamydial infected cells and found that 18 proteins or 2.2% of the total proteins are common between these three experiments (Fig. 2, Table S1). In addition to the seven commonly identified proteins from all four AP-MS experiments indicated above, there were 11 additional commonly identified proteins from the Aeberhard et al., Dickinson et al., and Olson et al. studies. The 11 additional commonly identified proteins were: Alpha-actinin-4 (ACTN4_HUMAN), Brain acid soluble protein 1 (BASP1_HUMAN), Caprin-1 (CAPR1_HUMAN), Elongation factor 1-delta, EF-1-delta (EF1D_HUMAN), Membrane-associated progesterone receptor component 1 (PGRC1_HUMAN), Stress-induced-phosphoprotein 1, (STIP1_HUMAN), Tropomyosin alpha-3 chain (TPM3_HUMAN), Tropomyosin alpha-4 chain (TPM4_HUMAN), Vinculin (VINC_HUMAN), Nuclease-sensitive element-binding protein 1 (YBOX1_HUMAN), and 14-3-3 protein theta (1433T_HUMAN). This may represent a small but biologically relevant increase due to these experiments being carried out in the context of C. trachomatis infection [12, 19, 20], rather than ectopically expressing Incs in uninfected host cells [18]. Fewer of these proteins have been validated for their localization thus far, but the inclusion has been shown to be surrounded by an actin and intermediate filament (e.g., vinculin) cytoskeleton network [32]. This would explain the identification of such factors as TPM3 and TPM4, for example [33, 34].
Fig. 2.
Venny comparison of eukaryotic proteins identified at 24 hours post-infection in Olson et al. compared to Dickinson et al. and Aeberhard et al. experimental approaches. The reported significant proteins from each study using C. trachomatis L2 infected eukaryotic cells were input into Venny to determine commonly identified proteins. The total number of proteins as well as the percentage of total is indicated within each overlapping section.
Comparison of Dickinson et al. and Olson et al. reported statistically significant eukaryotic proteins
Finally, we directly compared the statistically significant proteins reported from the published C. trachomatis L2 APEX2 proximity labeling system experiments [12, 20]. This is the first time that two large-scale AP-MS experiments using the same APEX2 proximity labeling system tool to identify interactions at the C. trachomatis inclusion membrane have been reported [12, 20]. A comparison of the experimental parameters used in each APEX2 experiment is summarized in Table 2. Both experiments fused APEX2 to the C-terminus of an Inc to detect protein-protein interactions at the inclusion membrane in vivo.
Table 2.
Affinity purification-mass spectrometry experimental parameters reported in Dickinson et al. and Olson et al. &
| Dickinson et al.* | Olson et al.& | |
|---|---|---|
| Tissue type | HeLa 229 cells | HeLa 229 cells |
| Inc-APEX2 construct | IncB-APEX2 | IncF-APEX2, IncA-APEX2, IncATM-APEX2 |
| Affinity Purification | Streptavidin-agarose resin | Streptavidin Magnetic beads |
| Elution | On resin trypsin digestion | SDS-PAGE and sectioned |
| Digestion Enzyme(s) | Trypsin | Trypsin and AspN |
| Mass Spectrometer | Thermo Fisher Velos Orbitrap | Thermo Fisher Orbitrap Lumos |
| Software | MS-GF+ Release (v2016.10.24) | Mascot and Scaffold |
| Protein modifications | Carbamidomethyl and Oxidation | Carbamidomethyl and Oxidation |
| Fasta search Homo sapiens | UniProt SPROT accessed 20170412 | UniProt Human accessed 20180927 |
| Fasta search C. trachomatis | C. trachomatis L2 434/Bu pL2 Plasmid accessed 20180105 | C. trachomatis L2 434/Bu accessed 20180330 |
| Peptide/protein Filtering | False Discovery Rate ≤ 1%, Unique peptides, requiring a minimum of six amino acids in length, were filtered using an MS-GF threshold of ≤ 1×10−9, corresponding to an estimated false-discovery rate (FDR) <1% at a peptide level. | Scaffold filtering: 95% protein threshold, 1-peptide minimum, 95% peptide threshold |
| Additional data processing | Relative peptide abundances were log-transformed. Elimination of statistical outliers was confirmed using a standard Pearson correlation at a sample level | |
| Statistical Test | G-test and T-test | SAINT# |
previously published in Dickinson et al., PLoS Pathog. 2019 15(4):e1007698, doi: 10.1371/journal.ppat.1007698
previously published in Olson et al., Infection and Immunity. 2019
Significance Analysis of INTeractome (SAINT)
The major differences between these studies include the specific Inc(s) used for each of the Inc-APEX2 constructs, the protein digestion enzymes used in mass spectrometry sample preparation, and statistical analysis of mass spectrometry data. The specifics of each study are listed in Table 2. Of note, Dickinson et al. used C. trachomatis L2 transformed with IncB-APEX2 that uniformly decorated the inclusion [20] (Table 2). The localization of IncB in this context is different than the reports of endogenous IncB, which has been shown to localize in microdomains within the inclusion membrane [35]. Olson et al. used C. trachomatis L2 transformed with IncA-APEX2, IncF-APEX2, or IncATM-APEX2 (a truncated IncA construct), all of which uniformly decorated the inclusion similar to endogenous IncA or IncF, respectively [6-8].
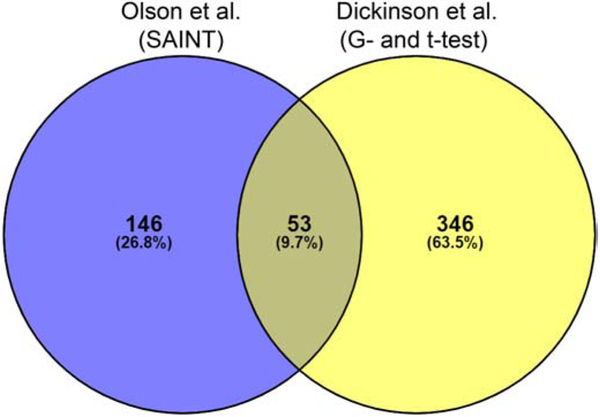
Venny [27] was used to compare the statistically significant proteins reported at 24 hpi by both Dickinson et al. [20] and Olson et al. [12]. At 24 hpi, 399 statistically significant proteins were reported by Dickinson et al. using the G and t-test [20], while 199 statistically significant proteins were reported by Olson et al. using Significance Analysis of INTeractome (SAINT) [12] (Fig. 3, Table S1). Fifty-three proteins (9.7% of the total protein input) were commonly identified in both studies; given that different Inc proteins were used for these proximity labeling experiments, some differences may be expected. However, each Inc-APEX2 construct, IncB-APEX2 used by Dickinson et al. [20] and IncF-APEX2, IncA-APEX2, and IncATM-APEX2 used by Olson et al. [12], uniformly labeled the inclusion when the expression of the constructs was induced. Therefore, it is surprising that at 24 hpi, only approximately 13% of the statistically significant proteins reported by Dickinson et al. (using the G and t-test) [20] were commonly identified by Olson et al. (using SAINT) [12] (Fig. 3, Table S1). In addition, greater than 88% of Dickinson et al. [20] and 72% of Olson et al. [12] datasets contained unique proteins (i.e., proteins not commonly identified by both studies) (Fig. 3, Table S1).
Fig. 3.
Venny comparison of reported eukaryotic proteins at 24 hours post-infection by Olson et al. (SAINT analysis) and Dickinson et al. (G- and t-test analysis) using the in vivo ascorbate peroxidase proximity labeling system (APEX2) combined with AP-MS. The reported significant proteins from each study were input into Venny to determine commonly identified proteins. The total number of proteins as well as the percentage of total is indicated within each overlapping section.
SAINT analysis of Dickinson et al. identified eukaryotic proteins from APEX2 proximity labeling AP-MS data
One possible explanation for the differences in the reported significant proteins is the statistical analysis tools used for each experiment. For example, Olson et al. [12] used SAINT, which takes into account the protein length when calculating the probability that a protein is a true interacting protein and not a false-positive [26]. The probability is reported as Bayesian False Discovery Rate (BFDR) [26]. Dickinson et al. used an unspecified G- or t-test to assign significance to their mass spectrometry data. Because both Dickinson et al. and Olson et al. used the APEX2 proximity labeling system, it is possible for the first time to directly compare the proteins identified in these experiments. Although it is not possible to change the experimental conditions prior to mass spectrometry (Table 2), the raw mass spectrometry data can be reanalyzed. To directly compare the results from Dickinson et al. [20] with the SAINT statistically significant proteins reported by Olson et al. [12] and to minimize differences in the AP-MS data processing between these two datasets, we analyzed the Dickinson et al. datasets [20] using SAINT as described in the Materials and Methods. By the SAINT analysis tool, at 24 hpi, 76 eukaryotic proteins were found to be statistically significant (Table 3, Table S2-S3).
Table 3.
Comparison of Dickinson et al. results relative to the indicated statistical tests
| Time (hours post-infection) |
Number of statistically significant& proteins |
Number of proteins identified by both statistical tests |
|
|---|---|---|---|
| G/t-test* | SAINT# | ||
| 8 hpi | 90 | 0 | n/a |
| 16 hpi | 180 | 17 | 13 |
| 24 hpi | 399 | 76 | 70 |
previously published in Dickinson et al., PLoS Pathog. 2019 15(4):e1007698, doi:10.1371/journal.ppat.1007698
Analyzed for this study (BFDR ≤0.05)
p ≤0.05
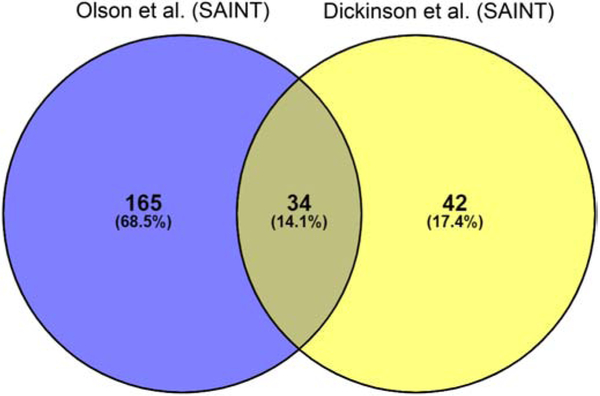
To determine if these SAINT analyzed data appeared more similar to the data reported by Olson et al., we compared the 24 hpi SAINT analyzed Dickinson et al. datasets with the 24 hpi SAINT analyzed Olson et al. datasets. When the same tool was used to calculate statistical significance, the number of commonly identified statistically significant proteins increased from 9.7% (Fig. 3) to 14.1% (Fig. 4). In addition, 45% (34 of 76 proteins) of the Dickinson et al. SAINT significant proteins were commonly identified by Olson et al.. Furthermore, now only 55% (42 of 76 proteins) of the Dickinson et al. SAINT significant proteins were unique (Fig. 4), compared to 88% (354 of 399 proteins) of proteins from the G-test and t-test results (Fig. 3). The different Inc-APEX2 (IncB vs. IncA and IncF) constructs and overexpression of each construct used to identify proximal or interacting proteins likely contribute to the remaining proteins that are unique in each dataset.
Fig. 4.
Venny comparison of SAINT analyzed Dickinson et al. eukaryotic proteins at 24 hpi. The raw data reported by Dickinson et al. at 24 hpi were analyzed by SAINT then compared to Olson et al. reported 24 hpi SAINT significant proteins. The total number of proteins as well as the percentage of total is indicated within each overlapping section.
While the specific implications for the mislocalization of Incs and “flooding” the inclusion membrane with additional Incs is not well understood at this time [13], the phenotypic changes in IncF-APEX2 localization upon overexpression support the concept that overexpression may impact the organization of the inclusion membrane [11]. It is also likely that overexpression of other Inc proteins will subsequently influence or change the recruitment of certain host proteins to the inclusion. In support, Rucks et al. showed that reducing the expression levels of IncF-APEX2 rescued normal inclusion development [11], indicating that carefully assessing induction time and amount of the inducing agent used (i.e., overexpression levels) will influence the interaction data. It is important to note, that while IncB-APEX2 did not localize in the inclusion in a manner that is consistent with reports of endogenous IncB [35], Dickinson et al. did not report any abnormalities in inclusion size or bacterial morphology upon IncB-APEX2 overexpression [20].
Comparison of SAINT significant Dickinson et al. eukaryotic protein datasets with the previously reported G- and t-test significant proteins at 24 hpi
Because the SAINT analyzed Dickinson et al. datasets drastically decreased the overall number of statistically significant proteins compared to those reported by Dickinson et al. using the G- and t-test at 24 hpi, we further examined which proteins were statistically significant using each analysis tool. The statistically significant proteins reported by Dickinson et al. using the G- and t-test (p ≤ 0.05) [25] were compared to the SAINT analyzed significant proteins (BFDR ≤ 0.05) (Table 3, Table S2-S3). At 24 hpi, the 399 statistically significant eukaryotic proteins were identified using the G-test and t-test compared to only 76 statistically significant eukaryotic proteins identified by SAINT (Table 3; Table S5). We used Venny [27] to determine which proteins were commonly identified by each statistical analysis tool (i.e., SAINT and the G- and t-test). Most SAINT significant proteins (70 of 76 proteins) were also identified as significant using the G- and t-test (Table 3; Table S3). Six proteins were unique to the SAINT statistical analysis tool (i.e., not determined to be statistically significant by G- and t-test) at 24 hpi: Hsc70-interacting protein, Hip (F10A1_HUMAN), Protein transport protein Sec16A (SC16A_HUMAN), Src substrate cortactin (SRC8_HUMAN), Tropomyosin beta chain (TPM2_HUMAN), HLA class I histocompatibility antigen (1A30_HUMAN), and Ataxin-2-like protein (ATX2L_HUMAN) (Table 3, Table S3). In support of these SAINT data, cortactin has been previously shown to localize with the chlamydial inclusion [36].
These results indicate that despite using permissive Scaffold parameters (95% protein, 1-peptide minimum, 95% peptide identification), SAINT provides a more rigorous analysis than the G-test and t-test alone (Table 3, Table S3). Most of the proteins identified by SAINT were also identified by the G-test and T-test which indicates that SAINT has more stringent parameters to calculate statistical significance (Table 3) and that amino acid length of each prey protein is an important aspect in these statistical calculations
Comparison of SAINT significant Dickinson et al. eukaryotic protein datasets with the reported G- and t-test significant proteins at 16 and 8 hpi
A stated goal of the Dickinson et al. study was to use the APEX2 system to understand how the proteome around the chlamydial inclusion changes over the course of the developmental cycle, and AP-MS data were taken from 8 hpi (early developmental cycle), 16 hpi (mid-developmental cycle), and 24 hpi (mid-late developmental cycle). As we have already re-analyzed the 24 hpi dataset above, we performed a similar re-analysis of the 16 and 8 hpi datasets and again compared SAINT significant proteins with the G- and t-test reported statistically significant proteins.
At 16 hpi, Dickinson et al. reported 180 statistically significant proteins using the G- and t-test method, while only 17 statistically significant proteins were identified by SAINT (Table 3, Table S3). Consistent with the re-analysis of the 24 hpi dataset, at 16 hpi, most of the G- and t-test significant proteins were also detected using SAINT (13 of 17 total proteins) (Table S3). The four unique proteins identified only by the SAINT analysis included: Caprin-1 (CAPR1_HUMAN), Microtubule-associated protein 4 (MAP4_HUMAN), Src substrate cortactin (SRC8_HUMAN), and Vimentin (VIME_HUMAN) (Table 3, Table S3). Consistent with previous studies, cortactin has been shown to be associated with the inclusion [36], and both vimentin [32] and microtubules [37] have been extensively studied for their roles near the inclusion during infection.
Finally, the SAINT analysis was applied to the 8 hpi Dickinson et al. dataset [20]. At 8 hpi, using the G-test and t-test, Dickinson et al. reported 90 statistically significant proteins [20]. In contrast, the SAINT analyzed Dickinson et al. dataset did not identify any statistically significant proteins (Table 3). These SAINT data are further supported by the findings published by Dickinson et al. in which they used RNAi specific for the genes corresponding to 64 of the 90 statistically significant proteins at 8 hpi to validate eukaryotic proteins that were recruited to the inclusion [20]. Of the 64 proteins that underwent further testing by RNAi, only silencing of two genes (Stress-induced-phosphoprotein 1 (STIP1) and Myosin light polypeptide 6 (MYL6) yielded a 2-fold decrease in the production of infectious progeny [20]. By the G- and t-test, STIP1, and MYL6 were also statistically significant at 16 and 24 hpi, but only SAINT significant at 24 hpi (Table S3, Table S4). The proteins that were silenced by RNAi in Dickinson et al. [20] are also directly compared to the SAINT calculated BFDR in Table S4.
Analysis of Dickinson et al. and Olson et al. reported statistically significant eukaryotic proteins using the CRAPome [29]
The most striking difference between the original dataset reported by Dickinson et al. and the SAINT analyzed datasets was with the 8 hpi samples, where, by SAINT, no proteins were identified as statistically significant. We asked if there was another metric of eliminating background proteins from a list of statistically significant proteins. Hence, we analyzed the G- and t-test reported statistically significant Dickinson et al. datasets using the CRAPome [29] to determine if their original list of statistically significant proteins was contaminated with background proteins. The CRAPome is a repository for background, contaminant proteins identified from AP-MS experiments [29]. A list of eukaryotic proteins is queried, and the CRAPome reports the number of experiments that identified the protein including the average spectral counts for each experiment that identified the queried protein. The Dickinson et al. G- and t-test reported statistically significant proteins from each 8, 16, and 24 hpi were queried into the CRAPome. The CRAPome reported average spectral count data for each queried protein was normalized to the queried protein length. We then used a two-pronged approach to defining contaminant proteins as those with spectral counts in greater than 30% of the experiments (i.e., 411 total AP-MS experiments uploaded to the CRAPome) and being above an arbitrary cut-off of greater than 0.02 for the average spectral counts normalized to protein length (Table S5).
Using these two criteria, at 8 hpi, 15 of the 90 (16.7%) reported statistically significant (by G- and t-test) proteins are potentially background contaminant proteins (Table 4; Table S5). At 16 hpi, 19 of 180 (10.6%) significant proteins (by G- and t-test), and at 24 hpi 33 of 399 (8.3%) significant proteins (by the G- and t-test) are potential background proteins. We then queried the SAINT analyzed Dickinson et al. datasets at each timepoint into the CRAPome to determine if the SAINT analysis reduced the number of contaminant proteins. While SAINT decreased the number of statistically significant proteins detected (Table 3), a similar percentage of the SAINT significant proteins identified fit the criteria for probable contaminant proteins except for 8 hpi, where no proteins were identified as SAINT significant (Table 4). The proteins that were flagged as potential contaminants from the SAINT analysis were also common to the G- and t-test CRAPome analysis, indicating that regardless of the statistical tool used, some contaminant proteins will be identified as statistically significant (Table 4).
Table 4.
CRAPome analysis of Dickinson et al. reported G- and t-test datasets, SAINT analyzed Dickinson et al. and Olson et al. SAINT datasets
| Dickinson et al. datasets |
Statistical analysis tool used |
Statistically significant proteins queried |
Identified contaminant proteins |
Commonly identified proteins |
Proteins identified as contaminants (%) |
|---|---|---|---|---|---|
| 8 hpi | G- and t-test | 90 | 15 | 16.67 | |
| 8 hpi | SAINT# | 0 | n/a | n/a | n/a |
| 16 hpi | G- and t-test | 180 | 19 | 10.56 | |
| 16 hpi | SAINT# | 17 | 3 | (2)* | 17.65 |
| 24 hpi | G- and t-test | 399 | 33 | 8.27 | |
| 24 hpi | SAINT# | 76 | 11 | (11)* | 14.47 |
| Olson et al. datasets |
Statistical analysis tool |
Total proteins input |
Identified contaminant proteins |
Commonly identified proteins |
Proteins identified as contaminants (%) |
| 24 hpi 1-peptide | SAINT | 199 | 13 | 6.53 | |
| 24 hpi 2-peptide | SAINT | 101 | 7 | (7)& | 6.93 |
SAINT statistical significance calculated using 1-peptide minimum threshold in Scaffold
denotes the commonly identified proteins using G- and t-test and SAINT
denotes commonly identified proteins using SAINT 1-peptide and 2-peptide
We also ran our 24 hpi SAINT analyzed AP-MS data [12] through the CRAPome to identify potential contaminant proteins for our datasets. At 24 hpi, 13 of 199 (6.5%) proteins were identified as potential contaminants using a one-peptide minimum threshold as reported by Olson et al. [12]. When the threshold was increased to the two-peptides minimum (see “Intra-experimental analysis of Olson et al. AP-MS” section below), then 7 of 101 (6.9%) statistically significant proteins were identified as contaminants (Table 4 and Table S5). The same seven proteins identified as contaminants from the two-peptide minimum dataset were also identified as contaminants in the one-peptide dataset (Table 4 and Table S5), indicating that some contaminant proteins will be identified regardless of the stringency of the minimum peptide threshold. Overall, to reduce the number of contaminant proteins in future AP-MS studies and to identify potential contaminant proteins, it is important to evaluate statistically significant proteins using the CRAPome. We have compiled a list of the eukaryotic proteins that were commonly identified as contaminants in these APEX2 studies (Table S5).
To help further distinguish contaminant proteins from true positive interacting partners, we have also compiled a list of eukaryotic proteins that were identified in uninfected cells that share sequence homology with chlamydial proteins (Table S6). At 8 hpi, C. trachomatis is in the early stages of the developmental cycle, the inclusions are very small, and chlamydial protein content will be orders of magnitude less than the eukaryotic host background. Further, the host cell produces naturally biotinylated host proteins, some of which have high homology to bacterial proteins, including C. trachomatis proteins. These facts can lead to false identification of chlamydial proteins by mass spectrometry at early time points post-infection. For these early time points a secondary method of labeling chlamydial proteins to differentiate from host proteins may be necessary. The use of both the CRAPome [29] and Table S6 containing chlamydial proteins with homology to eukaryotic cells may provide the best insight into background proteins.
Analysis of temporal recruitment of eukaryotic proteins identified by Dickinson et al. at the inclusion membrane
An enticing utility of the APEX2 proximity labeling system is the possibility of identifying “snapshots” of interactions or chlamydial inclusion proteomes throughout the developmental cycle. As noted above, Dickinson et al. obtained AP-MS data at each 8, 16, and 24 hpi to identify temporal changes in eukaryotic protein recruitment to the C. trachomatis L2 inclusion. We used Venny to examine which SAINT-identified statistically significant proteins remained associated with the inclusion from 16 to 24 hpi to distinguish the eukaryotic proteins that are potentially temporally recruited to the inclusion membrane from those proteins that remain associated with the inclusion throughout the developmental cycle.
We first compared the commonly identified statistically significant proteins as originally reported by Dickinson et al. using the G- and t-test. From these analyses, 85.5% (154 of 180) of Dickinson et al. 16hpi protein hits (i.e., G-test and t-test significant proteins) were commonly detected at 24 hpi (Table 3 and Table S3) [20]. Consistent with the G- and t-test datasets results, the SAINT analyzed Dickinson et al. datasets have a high percent of commonly identified proteins at 16 and 24 hpi. 94.1% (16 of 17 proteins) of the statistically significant proteins at 16 hpi were also significant at 24 hpi (Table 3 and Table S3). The unique protein at 16 hpi (i.e., not significant at 24 hpi) was aspartyl/asparaginyl beta-hydroxylase (ASPH_HUMAN). These results are not unexpected because both 16 hpi and 24 hpi are mid-developmental cycle, and the requirements for C. trachomatis L2 development at these times are likely not very different. This analysis also suggests the chlamydial inclusion proteome may be quite stable during the mid-developmental cycle period.
SAINT analysis of Dickinson et al. chlamydial proteins identified by APEX2 proximity labeling AP-MS
In contrast to the experiments in Mirrashidi et al. [18] and Aeberhard et al. [19], the APEX2 in vivo proximity labeling system allows for the detection of proximal or interacting C. trachomatis L2 Inc proteins [12, 20]. Again, to minimize differences in the datasets, we analyzed the chlamydial protein datasets from Dickinson et al. using SAINT (see Materials and Methods) (Table S2). We then compared the statistically significant proteins detected in the Dickinson et al. and Olson et al. APEX2 proximity labeling experiments. Both APEX2 studies identified four statistically significant C. trachomatis L2 inclusion membrane proteins (Incs) with the Inc, CT223, being identified in both studies (Table 5) [12, 20].
Table 5.
Comparison of SAINT statistically significant C. trachomatis L2 proteinsa identified in two APEX2 proximity labeling studies
| Dickinson et al. (SAINT) | ||||
| IncB-APEX2 | 24 hpi | BFDR* | 16 hpi | BFDR* |
| IncG | 0 | IncG | 0 | |
| CT223 | 0.01 | |||
| CT228 | 0.01 | |||
| CT813 | 0.02 | |||
| Olson et al. (SAINT) | ||||
| 24 hpi | BFDR* | |||
| IncF-APEX2 | CT223 | 0 | ||
| IncD | 0.02 | |||
| IncF | 0.03 | |||
| IncAtm-APEX2 | IncA | 0 | ||
| CT223 | 0.02 | |||
| IncA-APEX2 | OmcB | 0 | ||
| CT223 | 0 | |||
| IncA | 0 | |||
Protein name indicated using C. trachomatis serovar D naming convention
SAINT calculated Bayesian False Discovery Rate (BFDR)
Unique to individual experimental datasets, the SAINT analyzed Dickinson et al. dataset identified IncG, CT228, and CT813 as statistically significant at 24 hpi (Table 5; Table S2). However, only IncG was SAINT significant at 16 hpi, and no Incs were significant at 8 hpi (Table 5; Table S2) [20]. The lack of statistically significant Incs identified at the 8hpi is consistent with the smaller inclusions and few early Inc proteins being localized to the inclusion membrane at this time [38]. In contrast, the Olson et al. study identified IncD, IncF, and IncA as SAINT statistically significant (Table 5). The differences in statistically significant Incs identified in each dataset may reflect the use of different Inc-APEX2 fusion proteins in each study, but the organization of Incs in the inclusion membrane is currently not well defined [13].
Intra-experimental analysis of Olson et al. AP-MS identified eukaryotic and C. trachomatis L2 proteins
After comparing the two APEX2 proximity labeling studies, we aimed to further examine how various parameters such as minimum peptide threshold would impact our own APEX2 AP-MS datasets. We have previously published the SAINT statistically significant proteins identified using a one-peptide minimum threshold [12]. A two-peptide minimum (e.g., two unique peptides per parent protein) is generally accepted in the proteomics field as more rigorous than a one-peptide minimum threshold for mass spectrometry data. However, conflicting reports in the field suggest that the two-peptide minimum may not significantly change the overall number of proteins identified [39]. We decided to examine how the proposed, more rigorous, peptide threshold cut-off would affect the SAINT identified statistically significant eukaryotic and C. trachomatis L2 proteins within our own data set [12].
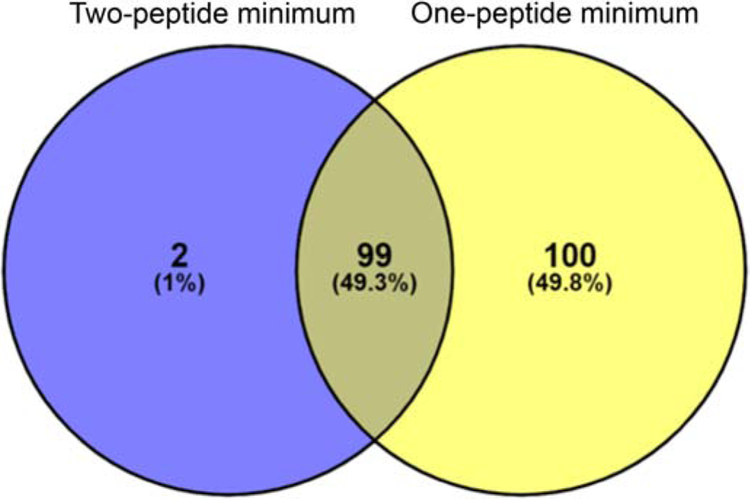
As reported by Olson et al. [12], the one-peptide minimum threshold identified 199 SAINT statistically significant proteins. To re-analyze these data under the new parameters, we applied the two-peptide minimum threshold filtering parameters in Scaffold to each of the eukaryotic (Table S7) and C. trachomatis L2 (Table S8) protein datasets (see Materials and Methods). After applying the two-peptide minimum (i.e., including hits from each IncF-APEX2, IncATM-APEX2, and IncA-APEX2), 101 unique eukaryotic proteins were identified by SAINT as statistically significant (Table S7; Fig.5). All but two of the proteins from the two-peptide minimum analysis were also identified in the one-peptide minimum dataset. For the eukaryotic protein datasets, the increased stringency of using a two-peptide minimum may be beneficial to decrease the overall false-discovery rate and reduce potential false-positive identifications. This is speculation as only the localization of some of these SAINT significant eukaryotic proteins have been validated thus far.
Fig. 5.
The effect of minimum peptide threshold on identification of SAINT significant proteins. Olson et al. The SAINT significant eukaryotic proteins were calculated using a two-peptide minimum threshold and compared to a one-peptide minimum threshold as reported in Olson et al. The total number of proteins as well as the percentage of total is indicated within each overlapping section.
Next, to determine if the minimum number of peptides would affect statistically significant chlamydial proteins identified, SAINT was applied to spectral count data obtained from the two-peptide minimum threshold (Table S8). We thought the two-peptide minimum might negatively impact the identification of C. trachomatis L2 Inc proteins because proteomics studies have found that hydrophobic proteins are often underrepresented in mass spectrometry experiments [40]. For example, proteins that contain large transmembrane regions (e.g., Incs) typically have fewer tryptic peptides, thus are less frequently detected by the mass spectrometer. Surprisingly, the two-peptide minimum did not affect the number of SAINT statistically significant chlamydial Inc proteins in our dataset (Table 6). In fact, the same proteins were found to be statistically significant, with minimal change in BFDR, regardless of a one-peptide or two-peptide minimum (Table 6). These data suggest that, for identifying chlamydial proteins, a one-peptide threshold is sufficient.
Table 6.
Comparison of minimum peptide threshold on SAINT calculated statistically significant C. trachomatis L2 proteins
| Sample | Protein (Uniprot ID) | Proteina | 1 peptide minimum BFDR b |
2 peptide minimum BFDR b |
|---|---|---|---|---|
| IncF-APEX2 | A0A0H3MKT3_CHLT2 | CT223 | 0 | 0 |
| INCD_CHLT2 | IncD | 0.02 | 0.01 | |
| INCF_CHLT2 | IncF | 0.03 | 0.02 | |
| IncATM-APEX2 | A0A0H3MD02_CHLT2 | IncA | 0 | 0 |
| A0A0H3MKT3_CHLT2 | CT223 | 0.02 | 0 | |
| IncA-APEX2 | OMCB_CHLT2 | OmcB | 0 | 0 |
| A0A0H3MKT3_CHLT2 | CT223 | 0 | 0 | |
| A0A0H3MD02_CHLT2 | IncA | 0 | 0 | |
| MOMP_CHLT2 | MOMP | ns | 0.05 |
Protein name indicated using C. trachomatis serovar D naming convention
Bayesian False Discovery Rate (SAINT) ns; not significant
Determination of availability of APEX2 amino acid targets of chlamydial Inc proteins
After determining the peptide threshold had a minimal effect on the identification of chlamydial Inc proteins, we aimed to better understand the ability of APEX2 to covalently tag different proteins. This is because the use of both AspN and trypsin for protein digestion to enhance peptide sequence coverage and identification [12, 41] resulted in the identification of only four Inc proteins as significant by SAINT analysis. These results were surprising as there are 50+ predicted Incs expressed on the inclusion membrane [10], and a previous study indicated that both IncF and IncA could potentially interact with at least 8 and 4 additional Inc proteins, respectively [42]. The specific amino acid residues that APEX2 covalently modifies with a biotin molecule are cysteine, tyrosine, tryptophan, and histidine [11, 25, 43], but we have not mapped how prevalent these amino acids are outside of the large hydrophobic transmembrane regions [40]. It is plausible that Incs that have fewer APEX2 modifiable residues will have decreased total biotinylation and may not be as efficiently enriched during the affinity purification steps as the proteins that have numerous biotin modifiable residues. Overall, this would result in these proteins being more difficult to detect by mass spectrometry. We analyzed our chlamydial datasets [12] to understand how these intrinsic differences in the amino acid composition of Inc proteins might influence our analysis (Table 7; SFig.1). We found in general that chlamydial Inc proteins with 11 or more biotin modifiable residues are identified with statistical significance (BFDR ≤0.05) [12].
Table 7.
Analysis of APEX2 modifiable amino acid targets of various Inc proteins
| Inc protein Serovar D naming convention |
Targets (#a) |
Length (AAb) |
modifiable residuesc (%) |
|---|---|---|---|
| CT101d | 18 | 153 | 11.76 |
| CT249 | 9 | 116 | 7.76 |
| CT058 | 25 | 367 | 6.81 |
| CT222d | 8 | 128 | 6.25 |
| IncE | 7 | 132 | 5.3 |
| CT223d | 14 | 268 | 5.22 |
| IncBd | 6 | 115 | 5.22 |
| CT850d | 21 | 405 | 5.19 |
| CT813 | 13 | 264 | 4.92 |
| CT005 | 16 | 363 | 4.41 |
| IncD | 6 | 146 | 4.11 |
| IncA | 11 | 273 | 4.03 |
| IncF | 4 | 104 | 3.85 |
| IncG | 6 | 167 | 3.59 |
| CT226 | 6 | 176 | 3.41 |
| CT228 | 6 | 196 | 3.06 |
number of amino acid targets for APEX2
amino acid (AA)
number of AA targets divided by Inc length
endogenous protein localizes in microdomains
In contrast, Inc proteins with fewer than five modifiable residues were less frequently detected by mass spectrometry and were not statistically significant. For example, IncA (serovar L2) has 11 modifiable residues (not in the transmembrane domain region), where CT226 has only six residues (not in the transmembrane domain region) with three of those biotin-modifiable residues in the N-terminal type three secretion signal region. In contrast, IncA contains only one residue in the N-terminal T3SS region of the 11 biotin-modifiable residues. IncA was statistically significant for IncA-APEX2 and IncATM-APEX2 and was detected by western blot in the eluates from the streptavidin affinity purification [12]. Another statistically significant Inc detected in our dataset, CT223, has 17 modifiable residues [12]. One important note is that because the organization of Incs in the inclusion is not understood [13], we cannot exclude the impact of the proximity of Incs (Inc-APEX2 constructs) to other Incs in the inclusion membrane on the labeling efficiency. We also identified chlamydial outer membrane proteins, OmcB and MOMP, as significant for our Chlamydia dataset [12]. Both proteins have numerous APEX2 targets with OmcB containing 49 APEX2 biotin-modifiable residues and MOMP containing 29 APEX2 modifiable residues, respectively. It is possible that during the short biotinylation reaction step the small biotin-phenoxyl radicals diffuse across the inclusion membrane and label the outer membranes of chlamydial developmental forms [23, 25].
CONCLUSION
With the recent advances in genetic tools for the manipulation of C. trachomatis L2, there has also been an expansion in the acquisition of large-scale AP-MS data to determine protein-protein interactions at the inclusion membrane. Four large-scale AP-MS experiments have been published in the last five years, each of which each aims to either identify eukaryotic proteins recruited to the chlamydial inclusion [19] or to understand the role of Incs in the inclusion, beginning with the identification of protein-protein interaction partners [12, 18, 20]. Each experiment was approached in a different fashion (e.g., Inc fused to a Strep-tag, inclusion purification, or proximity labeling), yielding over approximately 200 statistically significant eukaryotic proteins at the inclusion membrane. Large-scale proteomics studies, regardless of the software used in the analysis, frequently generate lists of hundreds of proteins detected in the sample. It is necessary to validate the localization and interaction by independent means to discern true interactions from these protein lists. This is highlighted in this meta-analysis as only seven of over 1,000 proteins were commonly identified from the four large proteomics experiments [12, 18-20]. We compared the statistically significant proteins from each study to highlight commonly identified proteins, which likely reflect high confidence interacting proteins at the inclusion (Fig. 1, Table 8, Table S1, Table S9). In support, three of the four high-confidence hits: LRRF1 [12], MYPT1 [30, 31], and 14-3-3β [14] have been previously validated at the inclusion.
Table 8.
High-confidence proteins commonly identified in all four AP-MS studies at the inclusion membrane
| Venny comparison using the reported significant proteins |
Venny comparison of significant proteins post-SAINT analysis of Dickinson et al. 24 hpi datasets |
|---|---|
| LRRF1_HUMAN | LRRF1_HUMAN |
| MYPT1_HUMAN | MYPT1_HUMAN |
| TMOD3_HUMAN | TMOD3_HUMAN |
| 1433B_HUMAN | 1433B_HUMAN |
| 1433F_HUMAN | |
| RTN4_HUMAN | |
| SNX1_HUMAN |
We also used this opportunity to compare the limitations of each experimental system. The two APEX2 proximity labeling experiments were directly compared using the same statistical analysis tool, which revealed that different statistical analysis tools can greatly impact the outcome of an individual experimental dataset (Table 3-4 and Table S2-S3). We did not apply the G- and t-test to our datasets as the exact methods implemented for these analyses were poorly described. However, we did analyze our datasets using increased peptide threshold minimums (Fig.5., Table 6, TableS7 and Table S8). These data indicated that a two-peptide minimum threshold might decrease the overall false positives for our eukaryotic protein dataset (Fig. 5, Table S8) but did not impact the chlamydial protein dataset (Table 6, Table S8). Overall, as more molecular tools are developed and adapted to understand the complex interactions at the chlamydial inclusion membrane and to understand host-pathogen interactions at the bacterial-containing vacuole of other intracellular bacteria, it is important to understand both the limitations and advantages of these different tools. Finally, as a field, it will be important to use statistical analysis tools that allow for efficient and meaningful interpretation of AP-MS data.
Supplementary Material
SFig. 1. Analysis of APEX2 modifiable target residues of Inc proteins.
Table S1. Venny comparison of reported significant proteins from AP-MS studies at the C. trachomatis inclusion membrane.
Table S2. SAINT analysis of Dickinson et al. datasets.
Table S3. Venny comparison of Dickinson et al. SAINT analyzed datasets with Dickinson et al. reported G- and t-test analyzed datasets.
Table S4. Comparison of SAINT analyzed Dickinson et al. datasets with Dickinson et al. reported RNAi experiments.
Table S5. CRAPome analysis of Dickinson et al. and Olson et al. significant eukaryotic proteins.
Table S6. Contaminant proteins identified from streptavidin AP-MS of uninfected HeLa cell lysates.
Table S7. SAINT analysis of Olson et al. eukaryotic proteins with a two-peptide minimum threshold.
Table S8. SAINT analysis of Olson et al. C. trachomatis L2 proteins with a two-peptide minimum threshold.
SIGNIFICANCE.
Chlamydia trachomatis, an obligate intracellular pathogen, grows within a membrane-bound vacuole termed the inclusion. The inclusion is studded with bacterial membrane proteins that likely orchestrate numerous interactions with the host cell. Although maintenance of the intracellular niche is vital, an understanding of the host-pathogen interactions that occur at the inclusion membrane is limited by the difficulty in purifying membrane protein fractions from infected host cells. The experimental procedures necessary to solubilize hydrophobic proteins fail to maintain transient protein-protein interactions. Advances in C. trachomatis genetics has allowed us and others to use various experimental approaches in combination with affinity purification mass spectrometry (AP-MS) to study the interactions that occur at the chlamydial vacuolar, or inclusion, membrane. For the first time, two groups have published AP-MS studies using the same tool, the ascorbate peroxidase proximity labeling system (APEX2), which overcomes past experimental limitations because membrane protein interactions are labeled in vivo in the context of infection. The utility of this system is highlighted by its ability to study chlamydial type III secreted inclusion membrane protein (Inc) interactions. Incs act as the mediators of host-pathogen interactions at the inclusion during C. trachomatis infection. When carefully controlled and analyzed, the data obtained can yield copious amounts of useful information. Here, we critically analyzed four previously published studies, including statistical analysis of AP-MS datasets related to Chlamydia-host interactions, to contextualize the data and to identify the best practices in interpreting these types of complex outputs.
Highlights.
Direct critical and statistical comparison of AP-MS proteomes of Chlamydia trachomatis inclusion (bacteria-containing vacuole)-host interactions
For the first time, we were able to compare datasets from two APEX2 proximity labeling experiments to contextualize our understanding of the inclusion proteome
Our direct comparison of statistical tools affirmed that SAINT statistical analysis is more rigorous than a G- and t-test.
We also performed an intraexperimental analysis to understand how APEX2 targets chlamydial and eukaryotic proteins.
This study provides important guidelines to help map the chlamydial inclusion proteome using additional Inc-APEX2 constructs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest Statement
The authors declare no conflict of interest.
REFERENCES:
- [1].CDC, Sexually Transmitted Disease Surveillance 2017, (2017). [Google Scholar]
- [2].Kagan JC, Roy CR, Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites, Nature cell biology 4(12) (2002) 945–54. [DOI] [PubMed] [Google Scholar]
- [3].Derré I, Isberg RR, Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system, Infection and immunity 72(5) (2004) 3048–3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Creasey EA, Isberg RR, Maintenance of vacuole integrity by bacterial pathogens, Current opinion in microbiology 17 (2014) 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Rockey DD, Chlamydia psittaci IncA is phosphorylated by the host cell and is exposed on the cytoplasmic face of the developing inclusion, Molecular microbiology 24(1) (1997) 217–228. [DOI] [PubMed] [Google Scholar]
- [6].Bannantine JP, Stamm WE, Suchland RJ, Rockey DD, Chlamydia trachomatis IncA Is Localized to the Inclusion Membrane and Is Recognized by Antisera from Infected Humans and Primates, Infection and Immunity 66(12) (1998) 6017–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Scidmore-Carlson MA, E.I. Shaw, C.A. Dooley, E.R. Fischer, T. Hackstadt, Identification and characterization of a Chlamydia trachomatis early operon encoding four novel inclusion membrane proteins, Molecular microbiology 33(4) (1999) 753–65. [DOI] [PubMed] [Google Scholar]
- [8].Bannantine JP, Griffiths RS, Viratyosin W, Brown WJ, Rockey DD, A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane, Cellular microbiology 2(1) (2000) 35–47. [DOI] [PubMed] [Google Scholar]
- [9].Bauler LD, Hackstadt T, Expression and Targeting of Secreted Proteins from Chlamydia trachomatis, Journal of bacteriology 196(7) (2014) 1325–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Weber MM, Bauler LD, Lam J, Hackstadt T, Expression and localization of predicted inclusion membrane proteins in Chlamydia trachomatis, Infect Immun 83(12) (2015) 4710–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rucks EA, Olson MG, Jorgenson LM, Srinivasan RR, Ouellette SP, Development of a Proximity Labeling System to Map the Chlamydia trachomatis Inclusion Membrane, Frontiers in Cellular and Infection Microbiology 7 (2017) 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Olson MG, Widner RE, Jorgenson LM, Lawrence A, Lagundzin D, Woods NT, Ouellette SP, Rucks EA, Proximity Labeling to Map Host-Pathogen Interactions at the Membrane of a Bacteria Containing Vacuole in Chlamydia trachomatis Infected Human Cells, Infect Immun (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Moore ER, Ouellette SP, Reconceptualizing the chlamydial inclusion as a pathogen-specified parasitic organelle: an expanded role for Inc proteins, Front Cell Infect Microbiol 4 (2014) 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Scidmore MA, Hackstadt T, Mammalian 14-3-3β associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG, Molecular microbiology 39(6) (2001) 1638–1650. [DOI] [PubMed] [Google Scholar]
- [15].Derre I, Swiss R, Agaisse H, The lipid transfer protein CERT interacts with the Chlamydia inclusion protein IncD and participates to ER-Chlamydia inclusion membrane contact sites, PLoS Pathog 7(6) (2011) e1002092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Elwell CA, Jiang S, Kim JH, Lee A, Wittmann T, Hanada K, Melancon P, Engel JN, Chlamydia trachomatis co-opts GBF1 and CERT to acquire host sphingomyelin for distinct roles during intracellular development, PLoS Pathog 7(9) (2011) e1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Wang Y, Kahane S, Cutcliffe LT, Skilton RJ, Lambden PR, Clarke IN, Development of a transformation system for Chlamydia trachomatis: restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector, PLoS Pathog 7(9) (2011) e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mirrashidi KM, Elwell CA, Verschueren E, Johnson JR, Frando A, Von Dollen J, Rosenberg O, Gulbahce N, Jang G, Johnson T, Jager S, A.M. Gopalakrishnan, J. Sherry, J.D. Dunn, A. Olive, B. Penn, M. Shales, M.N. Starnbach, I. Derre, R. Valdivia, N.J. Krogan, J. Engel, Global Mapping of the Inc-Human Interactome Reveals that Retromer Restricts Chlamydia Infection, Cell host & microbe 18(1) (2015) 109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Aeberhard L, Banhart S, Fischer M, Jehmlich N, Rose L, Koch S, Laue M, Renard BY, Schmidt F, Heuer D, The Proteome of the Isolated Chlamydia trachomatis Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components, PLOS Pathogens 11(6) (2015) e1004883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dickinson MS, Anderson LN, Webb-Robertson B-JM, Hansen JR, Smith RD, Wright AT, Hybiske K, Proximity-dependent proteomics of the Chlamydia trachomatis inclusion membrane reveals functional interactions with endoplasmic reticulum exit sites, PLOS Pathogens 15(4) (2019) e1007698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Paul B, Kim HS, Kerr MC, Huston WM, Teasdale RD, Collins BM, Structural basis for the hijacking of endosomal sorting nexin proteins by Chlamydia trachomatis, eLife 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mital J, Miller NJ, Dorward DW, Dooley CA, Hackstadt T, Role for Chlamydial Inclusion Membrane Proteins in Inclusion Membrane Structure and Biogenesis, PLOS ONE 8(5) (2013) e63426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Martell JD, Deerinck TJ, Sancak Y, Poulos TL, Mootha VK, Sosinsky GE, Ellisman MH, Ting AY, Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy, Nature biotechnology 30(11) (2012) 1143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Rhee HW, Zou P, Udeshi ND, Martell JD, Mootha VK, Carr SA, Ting AY, Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging, Science (New York, N.Y.) 339(6125) (2013) 1328–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lam SS, Martell JD, Kamer KJ, Deerinck TJ, Ellisman MH, Mootha VK, Ting AY, Directed evolution of APEX2 for electron microscopy and proximity labeling, Nat Meth 12(1) (2015) 51–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Choi H, Larsen B, Lin Z-Y, Breitkreutz A, Mellacheruvu D, Fermin D, Qin ZS, Tyers M, Gingras A-C, Nesvizhskii AI, SAINT: Probabilistic Scoring of Affinity Purification - Mass Spectrometry Data, Nature methods 8(1) (2011) 70–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Oliveros JC, Venny. An interactive tool for comparing lists with Venn's diagrams., 2007-2015. https://bioinfogp.cnb.csic.es/tools/venny/index.html.
- [28].Skarra DV, Goudreault M, Choi H, Mullin M, Nesvizhskii AI, Gingras A-C, Honkanen RE, Label-free quantitative proteomics and SAINT analysis enable interactome mapping for the human Ser/Thr protein phosphatase 5, Proteomics 11(8) (2011) 1508–1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mellacheruvu D, Wright Z, Couzens AL, Lambert JP, St-Denis NA, Li T, Miteva YV, Hauri S, Sardiu ME, Low TY, Halim VA, Bagshaw RD, Hubner NC, Al-Hakim A, Bouchard A, Faubert D, Fermin D, Dunham WH, Goudreault M, Lin ZY, Badillo BG, Pawson T, Durocher D, Coulombe B, Aebersold R, Superti-Furga G, Colinge J, Heck AJ, Choi H, Gstaiger M, Mohammed S, Cristea IM, Bennett KL, Washburn MP, Raught B, Ewing RM, Gingras AC, Nesvizhskii AI, The CRAPome: a contaminant repository for affinity purification-mass spectrometry data, Nature methods 10(8) (2013) 730–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Lutter EI, Barger AC, Nair V, Hackstadt T, Chlamydia trachomatis inclusion membrane protein CT228 recruits elements of the myosin phosphatase pathway to regulate release mechanisms, Cell reports 3(6) (2013) 1921–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shaw JH, Key CE, Snider TA, Sah P, Shaw EI, Fisher DJ, Lutter EI, Genetic Inactivation of Chlamydia trachomatis Inclusion Membrane Protein CT228 Alters MYPT1 Recruitment, Extrusion Production, and Longevity of Infection, Frontiers in Cellular and Infection Microbiology 8(415) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kumar Y, Valdivia RH, Actin and intermediate filaments stabilize the Chlamydia trachomatis vacuole by forming dynamic structural scaffolds, Cell host & microbe 4(2) (2008) 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Wang CLA, Coluccio LM, New insights into the regulation of the actin cytoskeleton by tropomyosin, Int Rev Cell Mol Biol 281 (2010) 91–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Pathan-Chhatbar S, Taft MH, Reindl T, Hundt N, Latham SL, Manstein DJ, Three mammalian tropomyosin isoforms have different regulatory effects on nonmuscle myosin-2B and filamentous β-actin in vitro, The Journal of biological chemistry 293(3) (2018) 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mital J, Miller NJ, Fischer ER, Hackstadt T, Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network, Cellular microbiology 12(9) (2010) 1235–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Fawaz FS, van Ooij C, Homola E, Mutka SC, Engel JN, Infection with Chlamydia trachomatis alters the tyrosine phosphorylation and/or localization of several host cell proteins including cortactin, Infect Immun 65(12) (1997) 5301–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Grieshaber SS, Grieshaber NA, Hackstadt T, Chlamydia trachomatis uses host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process, Journal of cell science 116(18) (2003) 3793–3802. [DOI] [PubMed] [Google Scholar]
- [38].E.I. Shaw, C.A. Dooley, E.R. Fischer, M.A Scidmore, K.A. Fields, T. Hackstadt, Three temporal classes of gene expression during the Chlamydia trachomatis developmental cycle, Molecular microbiology 37(4) (2000) 913–925. [DOI] [PubMed] [Google Scholar]
- [39].Gupta N, Pevzner PA, False discovery rates of protein identifications: a strike against the two-peptide rule, Journal of proteome research 8(9) (2009) 4173–4181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bagag A, Jault J-M, Sidahmed-Adrar N, Réfrégiers M, Giuliani A, Le Naour F, Characterization of Hydrophobic Peptides in the Presence of Detergent by Photoionization Mass Spectrometry, PLOS ONE 8(11) (2013) e79033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Swaney DL, Wenger CD, Coon JJ, Value of using multiple proteases for large-scale mass spectrometry-based proteomics, Journal of proteome research 9(3) (2010) 1323–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Gauliard E, Ouellette SP, Rueden KJ, Ladant D, Characterization of interactions between inclusion membrane proteins from Chlamydia trachomatis, Front Cell Infect Microbiol 5 (2015) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Martell JD, Deerinck TJ, Lam SS, Ellisman MH, A.Y. Ting, Electron microscopy using the genetically encoded APEX2 tag in cultured mammalian cells, Nature Protocols 12 (2017) 1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SFig. 1. Analysis of APEX2 modifiable target residues of Inc proteins.
Table S1. Venny comparison of reported significant proteins from AP-MS studies at the C. trachomatis inclusion membrane.
Table S2. SAINT analysis of Dickinson et al. datasets.
Table S3. Venny comparison of Dickinson et al. SAINT analyzed datasets with Dickinson et al. reported G- and t-test analyzed datasets.
Table S4. Comparison of SAINT analyzed Dickinson et al. datasets with Dickinson et al. reported RNAi experiments.
Table S5. CRAPome analysis of Dickinson et al. and Olson et al. significant eukaryotic proteins.
Table S6. Contaminant proteins identified from streptavidin AP-MS of uninfected HeLa cell lysates.
Table S7. SAINT analysis of Olson et al. eukaryotic proteins with a two-peptide minimum threshold.
Table S8. SAINT analysis of Olson et al. C. trachomatis L2 proteins with a two-peptide minimum threshold.