SUMMARY
Transcripts of noxious stimulus-detecting TrpA1 channels are alternatively spliced. Despite the importance of nociception for survival, the in vivo significance of expressing different TrpA1 isoforms is largely unknown. Here we develop a novel genetic approach to generate Drosophila knock-in strains expressing single TrpA1 isoforms. Drosophila TrpA1 mediates heat and UVC-triggered nociception. We show TrpA1-C and TrpA1-D, two alternative isoforms, are co-expressed in nociceptors. When examined in heterologous cells, both TrpA1-C and TrpA1-D are activated by heat and UVC. By contrast, analysis of knock-in flies reveals the striking functional specificity; TrpA1-C mediates UVC-nociception, whereas TrpA1-D mediates heat-nociception. Therefore, in vivo functions of TrpA1-C and TrpA1-D are different from each other, and are different from their in vitro properties. Our results indicate that a given sensory stimulus preferentially activates a single TrpA1 isoform in vivo, and that polymodal nociception requires co-expression of TrpA1 isoforms, providing novel insights of how alternative splicing regulates nociception.
Keywords: Alternative splicing, nociception, polymodal nociceptors, Drosophila, transient receptor potential (Trp), genome engineering, translational reporters
In Brief
Gu et al. demonstrate that heat and UVC light preferentially activate distinct Drosophila TrpA1 isoforms in vivo, and polymodal nociception requires co-expression of TrpA1-C and TrpA1-D.
INTRODUCTION
Alternative splicing is highly enriched in the nervous system [1, 2], suggesting that alternative protein isoforms contribute to the structural and functional complexity of the nervous system [3-5]. Supporting this idea, extensive alternative splicing of genes encoding cell surface recognition molecules, such as Down syndrome cell adhesion molecule (Dscam) [6-8], neurexin [9], and protocadherin [4], has been shown to regulate neuronal morphogenesis and circuit assembly. Genes encoding ion channels and neurotransmitter receptors are often alternative spliced. Electrophysiological analysis, mostly in heterologous cells, has suggested that alternative splicing fine-tunes ion channel properties [10-12].
Very few alternative isoforms have been characterized in vivo. How do alternative splicing of ion channels regulate neuronal physiology and animal behavior? Are functions of alternative isoforms in vivo corresponding to functions obtained in heterologous cells? Addressing these questions requires genetic manipulation of single isoforms in vivo.
Nociception is an ancient mechanism of the nervous system enabling animals to detect and avoid danger. Harmful stimuli activate nociceptors to trigger appropriate avoidance behavior and produce pain. The majority of nociceptors are polymodal [13, 14]. One good example is the Drosophila larval nociceptor, which extends dendrites to cover the entire body wall and detects noxious heat, irritant chemicals, reactive oxygen species (ROS), and tissue-damaging UVC light [15-18].
Transient receptor potential (Trp) ion channels are the principal detectors of noxious stimulus [14]. Widespread alternative splicing for trp transcripts has been found in species from Drosophila to human [19]. Drosophila TrpA1, which shares conserved sensory functions with its mammalian ortholog and is activated by heat, ROS, UV light, and irritant chemicals [15, 17, 20-24], is alternatively spliced to produce five isoforms. In vitro heterologous studies and in vivo overexpression studies have suggested that Drosophila TrpA1 isoforms are functionally different [17, 25], however, the in vivo expression and functions of endogenous TrpA1 isoforms are unknown.
In this study, we design a novel and versatile genetic method for targeting alternative splicing. Applying this method, we modify the Drosophila TrpA1 locus, to generate a large cohort of mutants, including knock-in flies each expressing a single TrpA1 isoform and knockout flies lacking select TrpA1 isoforms. Examining these novel alleles, we determine the expression of TrpA1 at single-cell and single-isoform resolutions, and nociceptive functions of TrpA1 isoforms by behavioral analysis and Ca2+ imaging.
RESULTS
Generation of TrpA1 Isoform-specific Knock-in and Knockout Alleles by isoEXPRESS
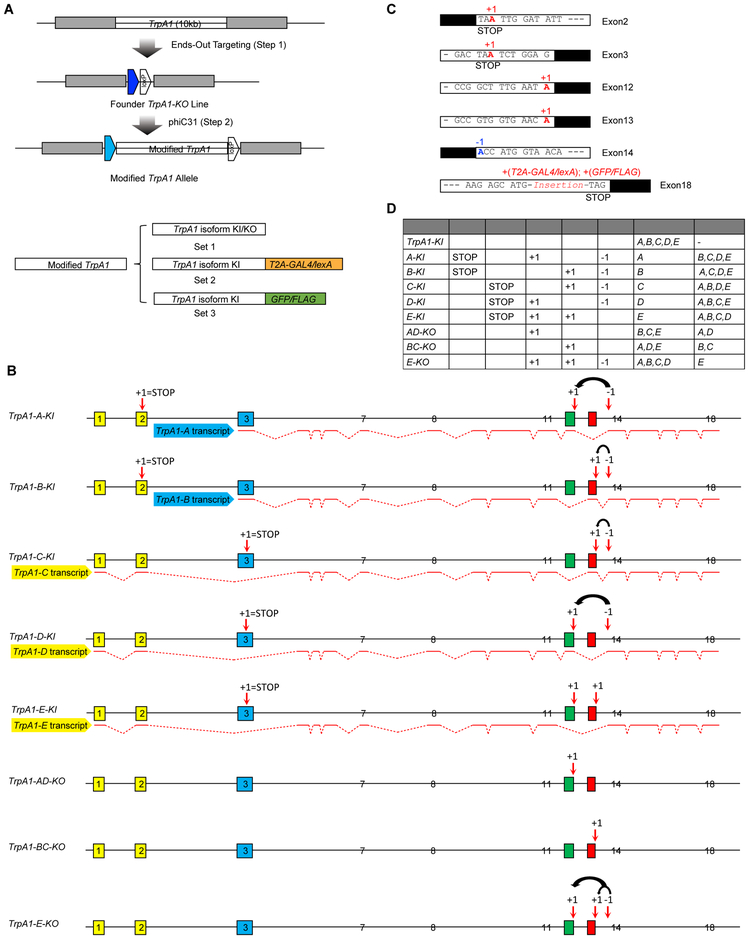
Three alternative splicing types—alternative first exon, mutually exclusive exon and cassette exon, together produce five alternative transcripts of Drosophila TrpA1 (Figure S1A). Here, we assign alphabetic orders to each TrpA1 isoforms (i.e. TrpA1-A, B, C, D and E, see Figure S1E for nomenclature comparison) [17, 26]. The five TrpA1 protein isoforms have identical ankyrin repeats and transmembrane domains, but differ either at the N-terminal domain preceding the ankyrin repeats (encoded by exons 1-2 or 3), or at the linker domain (encoded by exon 12 or 13) connecting the ankyrin repeats to the transmembrane domains (Figure S1A). To create TrpA1 isoform-specific alleles, we employed the two-step genomic engineering strategy (Figure 1A, S1B-C) [27, 28]. In the first step, homologous recombination-mediated gene targeting removed the entire TrpA1 gene, followed by Cre-mediated recombination to remove the construct backbone, to create a TrpA1-KO founder carrying an attP at the native TrpA1 locus (Figure 1A, S1B, D). In the second step, we reintroduced the full length TrpA1 genomic sequence containing various modifications, into the TrpA1-KO line by attP-mediated site-specific recombination (Figure 1A, D, S1C).
Figure 1. isoEXPRESS Design to Target Drosophila TrpA1 Isoforms.
(A) Schematics of the two-step genomic engineering. Three sets of TrpA1 isoform-specific alleles were generated.
(B) Detailed genomic modifications for each TrpA1 isoform-specific knock-in and knockout allele. isoEXPRESS modification, which is indicated by the black arrow, involved moving the first nucleotide of exon 14 (shown as ‘−1’) to the end of either exon 12 or 13 (shown as ‘+1’). Stop codons were introduced at exons 2 or 3 by adding a nucleotide. Alternative exons are shown in colors. Splicing patterns for each TrpA1 Transcript are denoted by red lines.
(C) Detailed modifications made at TrpA1 exons. Sequences of exons and introns/UTR are represented in white and black boxes, respectively.
(D) Summary of TrpA1 isoform-specific alleles, their associated genomic modifications, and effects of these modifications on each of the five TrpA1 alternative isoforms.
See also Figures S1, S2.
Alternative splicing of exons 12 and 13, by mutually exclusive exon and cassette exon skipping, produces three TrpA1 isoform groups (i.e. A/D, B/C and E, Figure 1B, S1A). Here, we designed the genomic engineering framework isoform expression by relocating a nucleotide at splice sites (isoEXPRESS), to selectively mutate any two groups of TrpA1 isoforms while leaving one group intact. The essence of isoEXPRESS was to relocate a single nucleotide between the boundaries of two exons. For example, relocation of adenine—the first nucleotide of exon 14, to the end of exon 12, resulted in intact TrpA1-A and TrpA1-D isoforms which connect exon 12 to exon 14. By contrast, TrpA1-B, TrpA1-C, and TrpA1-E had shifted reading frames and encountered early stop codons due to the exon 14 mutation (Figure 1B, C). Therefore, by simply repositioning of a single nucleotide from exon 14 to exon 12, we mutated TrpA1-B, TrpA1-C and TrpA1-E while leaving TrpA1-A and TrpA1-D intact. Further introduction of a single point mutation at either exon 2 or exon 3 will allow the expression of only intact TrpA1-A or TrpA1-D (Figure 1B-D). Thus, by reintroducing into TrpA1-KO the TrpA1 genomic sequence bearing these isoEXPRESS modifications, we created alleles that expressed a single isoform and named them isoform-specific knock-in alleles (i.e. TrpA1-A-KI or TrpA1-D-KI, Figure 1B-D). Employing the isoEXPRESS strategy, we generated TrpA1-B-KI and TrpA1-C-KI (Figure 1B-D). TrpA1-E-KI was created by introducing point mutations in exons 3, 12 and 13 (Figure 1B-D). Sequencing of the cDNAs from these isoform-specific knock-in alleles confirmed that isoEXPRESS did not alter the TrpA1 alternative splicing patterns (Figure S1F). We also generated TrpA1 isoform-specific knockout alleles, namely TrpA1-AD-KO, TrpA1-BC-KO and TrpA1-E-KO, using the similar startegy (Figure 1B-D). These knock-in and knockout alleles, collectively named set 1, were used to determine in vivo functions of single TrpA1 isoforms.
To map the in vivo expression of single TrpA1 isoforms, we generated set 2 and set 3 of isoform-specific knock-in alleles harboring additional modifications at the C-terminus (Figure 1A). In set 2, we inserted a T2A-GAL4 or T2A-lexA immediately before the stop codon (Figure 1A, C). During protein translation, the T2A peptide should mediate self-cleavage to produce the transcriptional activator GAL4 or lexA, which can drive transgene expression [29]. We referred to set 2 as isoform-specific translational driver lines (e.g. TrpA1-isoform-KI-T2A-GAL4 or TrpA1-isoform-KI-T2A-lexA, Figure 1A, C), to distinguish them from transcriptional driver lines. In set 3, we inserted a GFP or a FLAG tag immediately before the stop codon (Figure 1A, C).
In summary, we created three sets, for a total of 35 novel alleles carrying TrpA1 isoform-specific modifications (Figure 1D). In addition to the three alternative splicing types present in TrpA1, isoEXPRESS should also be able to target other alternative splicing types such as alternative 5’ splice site and alternative 3’ splice site (Figure S2). In all cases, isoEXPRESS targets a single isoform by introducing minimal modifications to the genome.
Translational Driver Lines Reveal Broad TrpA1 Expression
By crossing the TrpA1-KI-T2A-GAL4 translational driver to UAS reporters, we detected TrpA1 expression in larval midgut, corpus cardiacum (CC) cells and the nervous system. In the nervous system, TrpA1 expression was detected in the central nervous system (CNS) including the brain and ventral nerve cord (VNC), and in the peripheral nervous system (PNS) in class 4 dendritic arborization (C4da) neurons—the larval nociceptors (Figure S3A, B) [15, 30, 31]. Most of TrpA1-positive cells in larval CNS were labelled by the neuronal driver elav-GAL4 but not the glial driver repo-GAL4, indicating their neuronal identities (Figure S3B). To validate that the translational driver line reports TrpA1 expression, we performed immunostaining in TrpA1-KI-T2A-GAL4 larvae using a previously reported anti-TrpA1 serum [32]. TrpA1 antibody labelled larval CC cells and a few cells in the brain, but not C4da nociceptors or VNC (Figure S3D) [32]. Intriguingly, we noticed that brain cells labelled by the antibody belonged to a subgroup of TrpA1-KI-T2A-GAL4-positive cells (Figure S3D). These results suggest that TrpA1 translational driver lines report bona fide cellular expression patterns of TrpA1. Supporting this notion, we detected a significant overlap between TrpA1-KI-T2A-GAL4-positive cells and cells labeled by TrpA1-ABlexA and R21G01-lexA, two other TrpA1 driver lines (Figure S3C) [33, 34]. The high sensitivity of TrpA1-KI-T2A-GAL4 thus reveal widespread TrpA1 expression, including cells that expressed TrpA1 at a level below the antibody detection threshold.
Alternative Splicing Specifies TrpA1 Isoform Expression Patterns
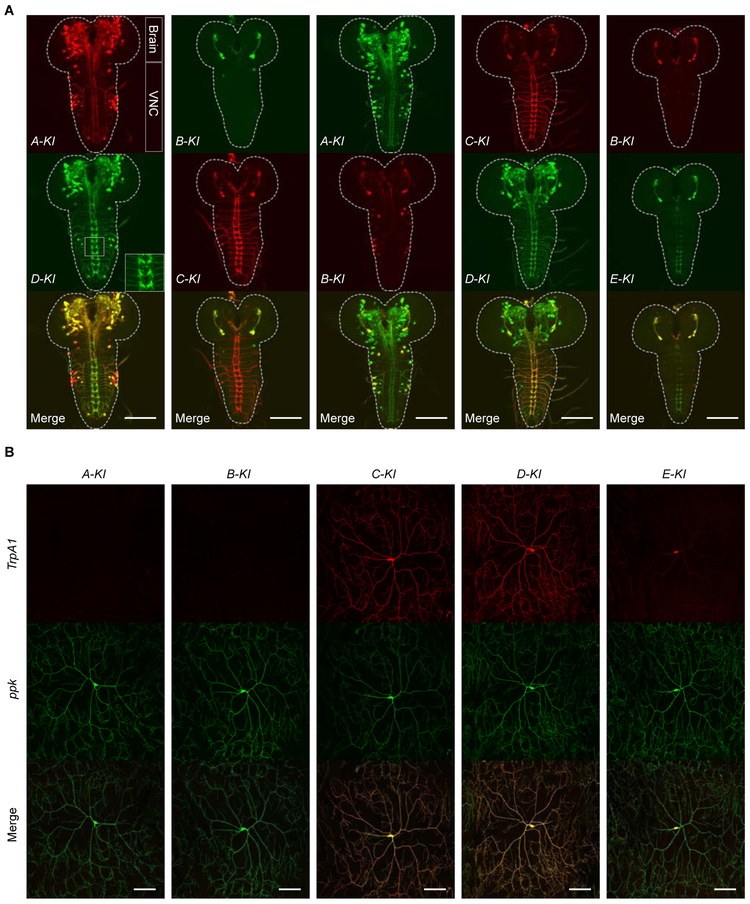
To assess the expression patterns of individual TrpA1 isoforms, we crossed TrpA1-isoform-KI-T2A-GAL4 and TrpA1-isoform-KI-T2A-lexA alleles to UAS or lexAop reporter lines. We detected the expression of all five TrpA1 isoforms in larval CNS (Figure 2A). In C4da nociceptors, we detected the expression of C, D and E, but not A or B isoforms (Figure 2B). Notably, GAL4 activities associated with TrpA1-E were consistently lower than other isoforms (Figure 2A, B), suggesting that the TrpA1-E expression level is lower. Next, we determined the relative expression patterns between pairs of TrpA1 isoforms. We found most TrpA1-positive neurons expressed more than one isoforms (Figure 2A). Remarkably, no two TrpA1 isoforms showed identical expression patterns (Figure 2A). Expression between TrpA1-A and TrpA1-B, or between TrpA1-C and TrpA1-D, was significantly different from each other and only overlapped in a few cells (Figure 2A). On the other hand, a significant overlap was detected between TrpA1-A and TrpA1-D, and between TrpA1-B and TrpA1-C (Figure 2A). Intriguingly, TrpA1-B/C was expressed in a small subgroup of cells that expressed TrpA1-A/D (Figure 2A). These results indicate that each TrpA1 isoform has its unique expression pattern, and multiple isoforms are often co-expressed in the same cells. Although alternative first exon splicing specifies whether an isoform is expressed in the PNS, possibly related to different promoter activities, alternative splicing of exons 12 and 13 critically regulate the TrpA1 expression broadness in the CNS.
Figure 2. Alternative Splicing Specifies TrpA1 Isoform-specific Expression Patterns in Drosophila Larval Nervous System.
(A) Representative images showing TrpA1-positive cells in larval brain lobes and VNC, reported by translational driver lines for each of the five TrpA1 isoforms. Each vertical panel shows expression of a pair of TrpA1 isoforms and their overlap, in the transheterozygotes carrying TrpA1-X-KI-T2A-GAL4>UAS-6XGFP and TrpA1-Y-KI-T2A-lexA>lexAop2-6XmCherry. X and Y referred to different TrpA1 isoforms. The inset in the D-KI panel showed the axon projection patterns of C4da nociceptors in the VNC. Dashed lines outline the CNS boundary. Scale bar; 100 μm.
(B) Representative images showing expression of translational driver lines in larval C4da nociceptors, in transheterozygotes bearing TrpA1-isoform-KI-T2A-GAL4>UAS-CD4TdTamoto and C4da neuronal marker ppk-CD4TdGFP. Scale bar, 100 μm.
See also Figures S3, S4, S5.
Previous studies have suggested that TrpA1-C and TrpA1-D are not expressed in the larval brain [17, 33, 35]. To further assess their expression, we performed GFP staining in TrpA1-C-KI-GFP and TrpA1-D-KI-GFP larvae. Indeed, we confirmed that both TrpA1-C and TrpA1-D were expressed in the brain (Figure S3E).
GFP staining in TrpA1-KI-GFP larvae revealed TrpA1 expression in C4da nociceptors (Figure S4A). Similar results were obtained by FLAG staining in TrpA1-KI-FLAG larvae (Figure S4C). By GFP staining in TrpA1-C-KI-GFP, TrpA1-D-KI-GFP and TrpA1-E-KI-GFP larvae, we found that TrpA1-C was expressed in C4da nociceptors with a level comparable to TrpA1-D but much higher than TrpA1-E (Figure S4A, B). These results provide direct evidence that three isoforms—TrpA1-C, TrpA1-D and TrpA1-E, are co-expressed in larval C4da nociceptors, prompting us to determine their respective nociceptive functions.
We next determined the TrpA1 isoform expression in adult flies. We found that each TrpA1 isoform had its unique expression pattern in the brain, taste and olfactory neurons, and TrpA1-A and TrpA1-D were widely expressed in the brain when comparing to TrpA1-B, TrpA1-C, or TrpA1-E (Figure S5A, B). Therefore, similar to larvae, alternative splicing of exons 12 and 13 govern the expression broadness in adult flies.
Heat Nociception Is Mediated by TrpA1 Activities in C4da Nociceptors
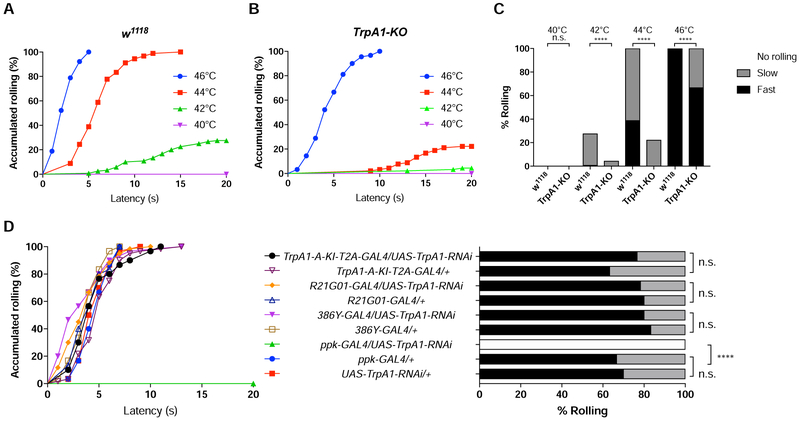
Using a previously described heat avoidance assay wherein heat stimulation was delivered locally to a 3rd instar larva by a heat probe clamped at a preset temperature [31, 36], we found that w1118 control larvae exhibited stereotyped corkscrew-like rolling behavior only when the probe was set to or above 42°C, with higher temperature increasing the response ratio and triggering faster rolling (Figure 3A, C, Video S1).
Figure 3. TrpA1 Activities in Larval C4da Nociceptors Mediates Heat Nociception.
(A-C) Rolling behavior responses of w1118 control and TrpA1-KO larvae to heat stimulation. Results were displayed in both “non-categorial” accumulated curve and “categorial” bar graphs.
(A-B) Accumulated curve showing percentage of larvae exhibiting rolling behavior (C) Rolling behavioral responses were divided into 3 categories, Fast (≤5 seconds), Slow (>5 but ≤20 seconds) and No rolling (>20 seconds). At least 90 larvae were tested for each group.
(D) Rolling behavior responses of larvae bearing UAS-TrpA1 RNAi under the control of different GAL4 drivers, to a 44°C heat probe. 30 larvae were tested except for R21G01-GAL4/UAS-TrpA1-RNAi and TrpA1-A-T2A-Gal4/+ groups in which 60 larvae were tested. Log-rank test was performed to compare dataset displayed in accumulated response curves. For clarity, results of statistical analysis are instead marked in the accompanying bar graph. n.s. no significant difference. **** p< 0.0001.
See also Video S1.
Drosophila TrpA1 is gated by heat [21, 37, 38]. Consistent with previous studies [17, 20], we found that rolling of TrpA1-KO larvae was significantly reduced when examined at 42°C and 44°C (Figure 3A-C). Surprisingly, despite an increased latency, all TrpA1-KO larvae rolled in response to 46°C or higher (Figure 3B, C). Our results suggest that TrpA1 is the predominant heat sensor for a narrow temperature range of 42-44°C. However, there exist TrpA1-independent mechanisms for detection of 46°C or higher. In the following studies, we chose 44°C to study heat nociception unless specified.
We next knocked down TrpA1 expression in a tissue-specific manner by RNAi, to determine which cells possess the TrpA1 activities necessary for rolling behavior. We found that knocking down TrpA1 with TrpA1-A-KI-T2A-GAL4, which was expressed in almost all TrpA1-positive cells except for C4da nociceptors (Figure 2A-B), caused no behavioral defects (Figure 3D). Similarly, knocking down TrpA1 by 386Y-GAL4 or R21G01-GAL4, two driver lines expressed in most TrpA1 positive cells in larval CNS but not in C4da nociceptors (Figure S3C) [33], caused no behavioral defects (Figure 3D). By contrast, knocking down TrpA1 by ppk-GAL4, a driver line specifically expressed in larval C4da nociceptors [15], abolished rolling behavior (Figure 3D). Together, these results suggest that TrpA1 activities in C4da nociceptors, but not in the CNS, are necessary and sufficient for heat nociception.
Noxious Heat Preferentially Activates A Single Isoform TrpA1-D
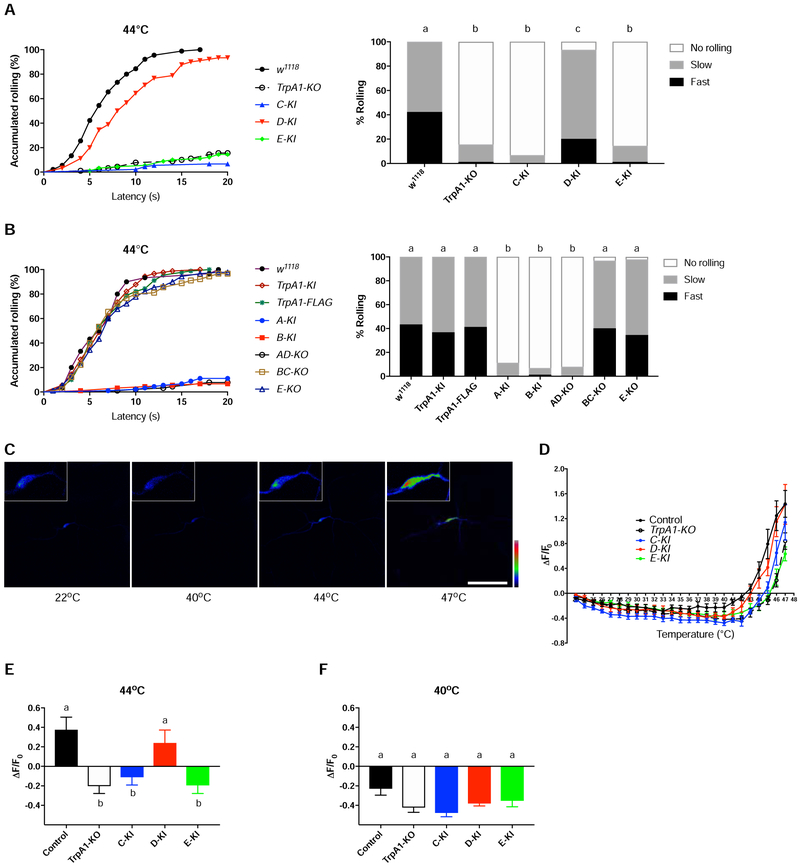
To investigate which isoforms underlie heat nociception, we examined rolling of TrpA1-isoform-KI larvae. Strikingly, we found that more than 90% of TrpA1-D-KI larvae rolled in response to a 44°C probe, whereas responses of TrpA1-C-KI or TrpA1-E-KI larvae were indistinguishable from TrpA1-KO (Figure 4A). Diminished rolling was also observed in TrpA1-A-KI and TrpA1-B-KI larvae (Figure 4B), consistent with the lack of TrpA1-A and TrpA1-B expression in C4da nociceptors (Figure 2B). These results indicate that heat nociception is largely restored to the wild-type level if and only if a single isoform, TrpA1-D, is expressed. Next, we examined necessity. We found significantly decreased rolling in TrpA1-AD-KO larvae, whereas TrpA1-BC-KO or TrpA1-E-KO larvae showed normal responses indistinguishable from w1118 control (Figure 4B). Because TrpA1 activities in C4da nociceptors mediated rolling (Figure 3D), and C4da nociceptors expressed TrpA1-C, D and E isoforms (Figure 2B, S4B), behavioral defects observed specifically in TrpA1-AD-KO larvae were best explained if TrpA1-D, but not TrpA1-C or TrpA1-E, was required for heat nociception. We also found that a 46°C probe triggered robust rolling behavior in all TrpA1 isoform alleles examined (Figure S6), supporting the notion that detection of higher temperature involves mechanisms in addition to TrpA1 (Figure 3C).
Figure 4. A Single Isoform TrpA1-D Is the Primary Heat Sensor.
(A-B) Rolling behavior responses of control w1118 larvae and larvae bearing TrpA1 alleles to a 44°C heat probe. For clarity, data were split and displayed in two separate panels. 90 larvae were tested for each genotype except for w1118 group in panel B in which 30 larvae were tested. Log-rank test was performed, corrected by total comparing group numbers. Columns with different superscript (a, b and c) are significantly different from each other (p<0.05).
(C) GCaMP6s responses of a single C4da nociceptor in a w1118 larva to heat stimulation. Insets show signals of the somatic area. Fluorescence intensities are represented in pseudocolor with the scale from zero (black) to the maximal intensity (red). Scale bar, 100 μm.
(D) Summary results of heat responses of C4da nociceptors in w1118 control and TrpA1 alleles carrying ppk-GAL4>UAS-GCaMP6s. GCaMP6s fluorescence intensities, measured from the soma of C4da nociceptors, are shown in Y axis versus temperature in X axis. n≥8 for each genotype. Data are represented as mean ± SEM.
(E-F) Heat responses of C4da nociceptors at 44°C (E) and 40°C (F). Data were extracted from (D), and are represented as mean ± SEM. n≥8 per group. One-way ANOVA with Tukey’s post comparison test. Columns with different superscript (a and b) are significantly different from each other (p<0.05).
See also Figure S6.
We next assessed heat responses of C4da nociceptors with GCaMP6s imaging [39]. We found that C4da nociceptors in wild-type larvae were activated by heat with a threshold of ~43°C, and heat threshold was increased to ~46°C in TrpA1-KO larvae (Figure 4C, D). These heat thresholds are in agreement with the behavioral analysis. Examining knock-in flies, we found that C4da nociceptors in TrpA1-D-KI larvae had a lower heat threshold than TrpA1-C-KI or TrpA1-E-KI (Figure 4D). When examined at 44°C, a behaviorally relevant temperature, C4da nociceptors in TrpA1-D-KI larvae were activated with the response amplitude comparable to w1118 control (Figure 4E). By contrast, responses in TrpA1-C-KI and TrpA1-E-KI larvae were lower and indistinguishable from TrpA1-KO (Figure 4E). We detected no difference in C4da neuronal responses to subthreshold heat of 40°C (Figure 4F).
These behavioral and Ca2+ imaging analysis reveal that noxious heat preferentially activates TrpA1-D to excite C4da nociceptors and elicit rolling behavioe. By contrast, TrpA1-C and TrpA1-E contribute little to heat sensation. Surprisingly, TrpA1-C and TrpA1-D were acrivated by heat when examined in heterologous cells (see the last result section, Figure 7), indicating both isoforms are intrinsically heat-sensitive. Thus, in vivo heat sensitivities of TrpA1-C and TrpA1-D are different from each other, and cannot be predicted from their in vitro properties.
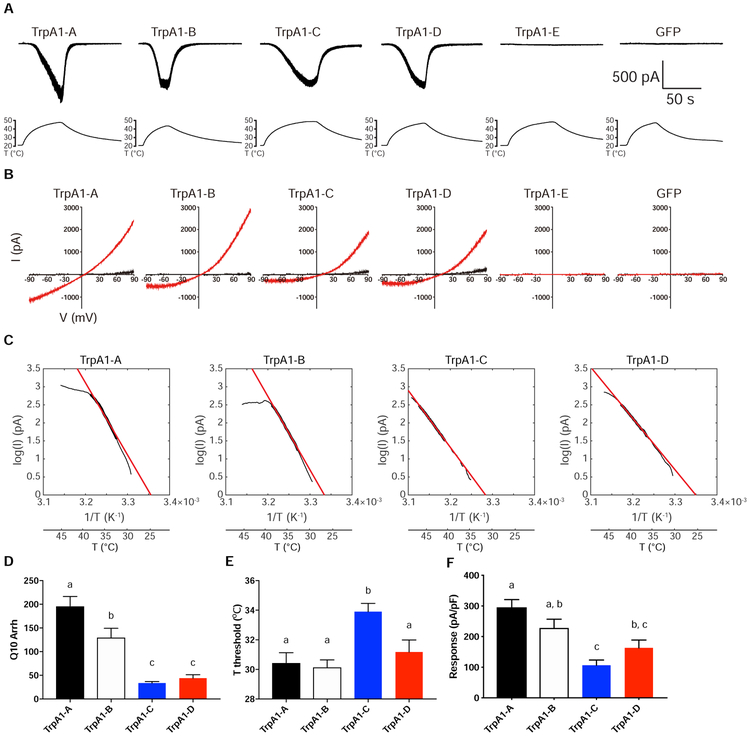
Figure 7. Heat Sensitivity of TrpA1 Isoforms in Drosophila S2 Cells.
(A) Sample traces of heat-induced current in S2 cells expressing TrpA1 isoforms. Temperature ramps were shown in the bottom accompanying each recoding trace.
(B) Sample I-V curves of heat-induced responses. Black I-V traces were obtained at room-temperature. Red I-V traces were acquired at the time point when temperature was at the peak and heat responses reached steady state.
(C) Sample Arrhenius plots (black trace) of heat-induced current in log unit over the inverse of absolute temperature. The portion of data used for linear fitting (red line) is indicated by the thickened black trace.
(D) Summary of Q10 values of TrpA1-A, B, C and D derived from Arrhenius plot.
(E) Summary of heat activation threshold of TrpA1-A, B, C and D.
(F) Summary of maximal heat responses of TrpA1-A, B, C and D.
n≥7 for each isoform. Data in D-F are represented as mean ± SEM. One-way ANOVA with Tukey’s post comparison test. Bars with different superscript (a, b and c) in D-F are significantly different from each other (p<0.05).
UVC/ROS and AITC Preferentially Activate TrpA1-C and TrpA1-D Respectively
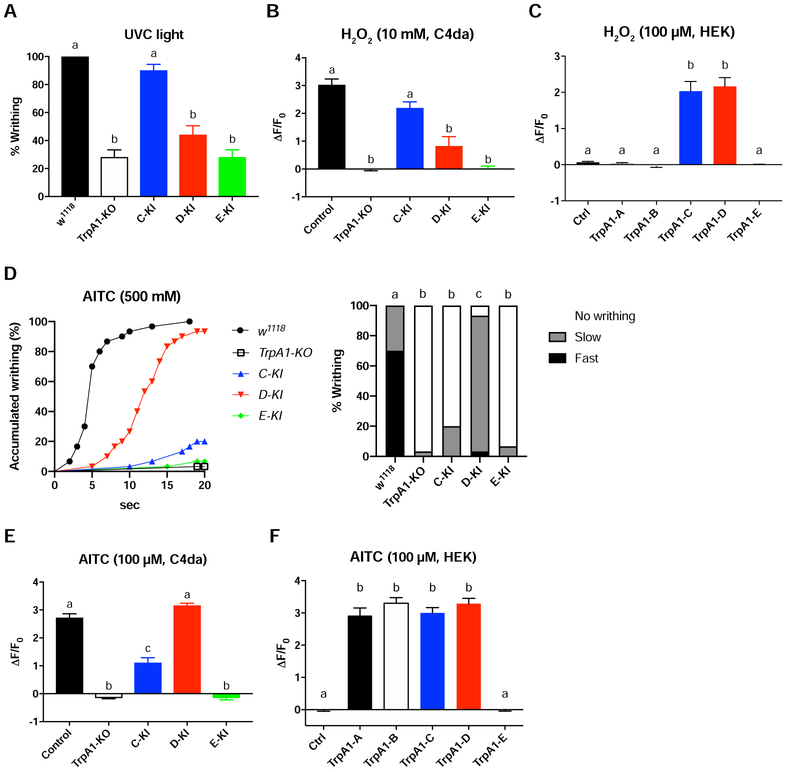
In addition to heat, C4da nociceptors mediate UVC light-induced writhing behavior by responding to photochemically produced H2O2 [16]. Here, we group UVC and H2O2 into one noxious modality (UVC/H2O2). We confirmed that acute exposure of larvae to 254 nm UVC triggers robust writhing behavior characterized by body twisting (Video S2) [16, 40]. This writhing behavior was significantly diminished in TrpA1-KO larvae (Figure 5A). Surprisingly, TrpA1-C-KI larvae restored writhing behavior to wild-type levels (Figure 5A). In contrast, responses of TrpA1-D-KI and TrpA1-E-KI larvae were significantly reduced and indistinguishable from TrpA1-KO (Figure 5A). GCaMP6s imaging revealed that C4da nociceptors were activated by H2O2 in a TrpA1-depdendent manner, and that H2O2 responses of TrpA1-C-KI larvae were restored to wild-type levels, but responses of TrpA1-D-KI or TrpA1-E-KI larvae were dramatically reduced and indistinguishable from TrpA1-KO (Figure 5B).
Figure 5. UVC/ROS and AITC Preferentially Activate TrpA1-C and TrpA1-D Respectively.
(A) Larval writhing behavior in responses to 254 nm UVC (5 mW/cm2) irradiation. 10 replicates for each genotype.
(B) Summary of GCaMP6s responses of C4da nociceptors to 10 mM H2O2 in control and TrpA1 isoform alleles carrying ppk-GAL4>UAS-GCaMP6s. n≥7 for each genotype.
(C) Summary of GCaMP6s responses to 100 μM H2O2 in HEK293 cells that expressed different TrpA1 isoforms. n≥7 for each group.
(D) Larval writhing behavior in response to 500 mM AITC. Accumulated response curves are displayed in left panel. Statistical analysis is marked in right bar graph. Writhing behavioral responses were divided into 3 categories, Fast (≤5 seconds), Slow (>5 but ≤20 seconds) and No rolling (>20 seconds) . 30 larvae were tested for each group. Log-rank test, corrected by total comparing group numbers. Columns with different superscript (a, b and c) are significantly different from each other (p<0.05).
(E) Summary of GCaMP6s responses of C4da nociceptors to 100 μM AITC in control and TrpA1 isoform alleles carrying ppk-GAL4>UAS-GCaMP6s. n≥7 for each genotype.
(F) Summary of GCaMP6s responses to 100 μM AITC in HEK293 cells that expressed different TrpA1 isoforms. n≥7 for each group.
Data in A-C and E-F are represented as mean ± SEM. One-way ANOVA with Tukey’s post comparison test. Columns with different superscript (a, b and c) are significantly different from each other (p<0.05).
See also Video S2.
These behavioral and Ca2+ imaging analysis indicate that UVC/H2O2 preferentially activates TrpA1-C to excite C4da nociceptors, by contrast, TrpA1-D or TrpA1-E contribute little to UVC/H2O2 detection. Surprisingly, both TrpA1-C and TrpA1-D were activated by H2O2 when examined in HEK293 cells (Figure 5C), indicating that TrpA1-C and TrpA1-D are intrinsically H2O2-sensitive [24]. Thus, in vivo UVC/H2O2 sensitivities of TrpA1-C and TrpA1-D are different from each other, and are different from their in vitro properties.
Irritant chemicals such as allyl isothiocyanate (AITC) activate both mammalian and Drosophila TrpA1 [22, 41, 42]. We found that AITC triggered writhing behavior in larvae in a TrpA1-depedent manner, and writhing behavior was stronger in TrpA1-D-KI larvae when comparing to TrpA1-C-KI and TrpA1-E-KI larvae (Figure 5D). Ca2+ imaging revealed that AITC strongly activated C4da nociceptors, and responses of TrpA1-D-KI larvae were stronger than TrpA1-C-KI and TrpA1-E-KI larvae (Figure 5E). These behavioral and Ca2+ imaging analysis indicate that AITC preferentially activates TrpA1-D to excite C4da nociceptors. However, when examined in HEK293 cells, both TrpA1-C and TrpA1-D were strongly activated by AITC and their responses were comparable (Figure 5F).
Expression Level Regulates TrpA1 Functionality
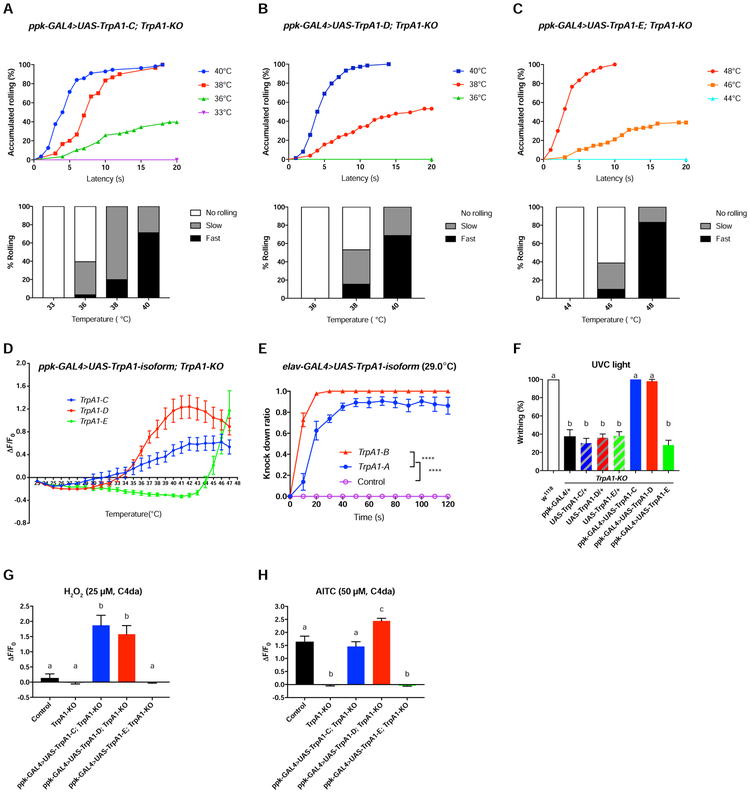
TrpA1 was expressed at a very low level in C4da nociceptors (Figure S4), however, its expression level should be drastically higher in heterologous cells. To investigate whether expression levels could regulate TrpA1 functions, we drastically increased TrpA1 levels in C4da nociceptors. To ensure expression levels of different transgenes are comparable, we generated UAS-TrpA1 transgenic flies for each of the five TrpA1 isoforms by inserting constructs at the same genomic locus [43]. We found that TrpA1-KO larvae overexpressing in C4da nociceptors either TrpA1-C or TrpA1-D caused heat allodynia, with rolling behavioral thresholds being lowered to ~36°C and ~38°C, respectively (Figure 6A, B), from 42°C in w1118 control (Figure 3A). Consistently, Ca2+ imaging of C4da nociceptors revealed significantly lowered heat threshold when TrpA1-C or TrpA1-D was overexpressed (32°C for UAS-TrpA1-C and 34°C for UAS-TrpA1-D, Figure 6D), comparing to 43°C in control larvae (Figure 4D).
Figure 6. Both TrpA1-C and TrpA1-D Rescue Heat and UVC/ROS Defects of TrpA1-KO.
(A-C) Summary of heat-induced rolling behavioral responses of TrpA1-KO larvae expressing UAS-TrpA1-C, UAS-TrpA1-D, or UAS-TrpA1-E under the control of ppk-GAL4. For each genotype, at least 30 larvae were tested at each temperature point.
(D) Summary of heat-induced GCaMP6s responses of C4da nociceptors in TrpA1-KO larvae expressing UAS-GCaMP6s as well as UAS-TrpA1-C, UAS-TrpA1-D, or UAS-TrpA1-E, under the control of ppk-GAL4. Data are represented as mean ± SEM. n=8 for each genotype.
(E) Over-expression of UAS-TrpA1-A or UAS-TrpA1-B by elav-GAL4 caused adult incapacitation when exposed to 29°C, with UAS-TrpA1-B showing a stronger effect. Control group: elav-GAL4/+. 10 flies were tested in a group to calculate the knockdown ratio at different time points, and at least 7 replicates were performed. Data are represented as mean ± SEM. Two-way ANOVA with Tukey’s post comparison test, ****p<0.0001.
(F) Summary of writhing responses to 254 nm UVC (5 mW/cm2) of TrpA1-KO larvae expressing UAS-TrpA1-C, UAS-TrpA1-D, or UAS-TrpA1-E, under the control of ppk-GAL4. n=10 for each genotype.
(G) Summary of 25 μM H2O2-induced GCaMP6s responses of C4da nociceptors in TrpA1-KO larvae expressing UAS-GCaMP6s as well as UAS-TrpA1-C, UAS-TrpA1-D, or UAS-TrpA1-E, under the control of ppk-GAL4. w1118 larvae served as control. n=7 for each genotype.
(H) Summary of 50 μM AITC-induced GCaMP6s responses of C4da nociceptors in TrpA1-KO larvae expressing UAS-GCaMP6s as well as UAS-TrpA1-C, UAS-TrpA1-D, or UAS-TrpA1-E, under the control of ppk-GAL4. w1118 larvae served as control. n≥7 for each genotype.
Data in F-H are represented as mean ± SEM. One-way ANOVA with Tukey’s post comparison test. Columns with different superscript (a, b and c) are significantly different from each other (p<0.05).
Next, we examined the overexpression effect on UVC/H2O2 nociception. We found that TrpA1-KO larvae overexpressing in their C4da nociceptors either TrpA1-C or TrpA1-D restored writhing behavior to the level of w1118 control larvae (Figure 6F). Ca2+ imaging studies showed that C4da nociceptors overexpressing either TrpA1-C or TrpA1-D were strongly activated by 25 μM H2O2, a sub-threshold concentration that was ineffective in activating C4da neurons in control larvae (Figure 6G). Finally, we examined genetic rescue effects on AITC responses. We found that overexpression in C4da nociceptors of either TrpA1-C or TrpA1-D restored AITC responses to the magnitude equal or higher than that in control larvae (Figure 6H).
Next, we compared responses between TrpA1-C-KI and rescue flies overexpressing TrpA1-C, or between TrpA1-D-KI and rescue flies overexpressing TrpA1-D. In both conditions C4da nociceptors expressed a single TrpA1 isoform but at drastically different levels. We found that heat, UVC/H2O2 and AITC preferentially activated a single TrpA1 isoform at the endogenous expression levels (Figure 4A, E, 5A-B, D-E). However, this stimulus-specificity was lost when TrpA1-D and TrpA1-C were overexpressed (Figure 6A-B, D, F-H). Notably, overexpressed TrpA1-C and TrpA1-D in C4da nociceptors shared similar properties—being activable by heat, H2O2 and AITC (Figure 6D, G-H), just as they did in heterologous cells (Figure 5C, F and Figure 7). These findings demonstrate that properties of nociceptors are determined not only by the identity of transducer proteins, but also by their expression levels.
TrpA1-B Is a Thermogenetic Tool Activatable by Moderate Heat
We next overexpressed UAS-TrpA1 for each isoform in C4da nociceptors, this time in wild-type background, to investigate heat-induced rolling behavior. We found that overexpression of any single TrpA1 isoform except UAS-TrpA1-E triggered heat allodynia (i.e. lowered thresholds) (Figure S7). Intriguingly, UAS-TrpA1-B expression caused the strongest sensitization, with the effect stonger than the widely-used thermogenetic TrpA1-A (Figure S7) [37]. To further compare the heat sensitivity of TrpA1-B with TrpA1-A, we explored the heat-induced incapacitation assay in adult flies that expressed UAS-TrpA1-B or UAS-TrpA1-A pan-neuronally. We found that heating to 29°C triggered seizure-like immobilization in 100% flies expressing UAS-TrpA1-B within 30s, by contrast, immobilization was not complete even after 120s in flies expressing UAS-TrpA1-A (Figure 6E, Video S3). These results suggest that TrpA1-B is an efficient thermogenetic tool whenever response to moderate heating is desired.
TrpA1-A, B, C and D Isoforms Are Heat Sensors
Previous studies in S2 cells suggest that TrpA1-A and TrpA1-D, but not TrpA1-B or TrpA1-C, are heat-sensitive [17]. To further investigate thermal properties of TrpA1, we performed whole-cell recordings in S2 cells expressing TrpA1 isoforms. We found that all TrpA1 isoforms except TrpA1-E were activated by heat (Figure 7A). I-V plot showed that TrpA1 currents had the reversal potential close to 0 mV (Figure 7B), consistent with the notion that TrpA1 is a non-selective cation channel [21]. Analysis of temperature coefficient Q10 revealed that TrpA1-A and TrpA1-B had higher heat sensitivities than TrpA1-C and TrpA1-D (Figure 7C, D), as also indicated by our behavioral analysis (Figure 6E, S7). Therefore, alternative first exon choices, possibly related to distinct promoter activities, are the primary determinant of heat sensitivities of TrpA1, while mutually exclusive splicing of the linker domain-encoding exons 12 and 13 has less impact. On the other hand, the lack of heat sensitivities of TrpA1-E demonstrates that the linker domain is indispensable for heat activation.
Of all sensory modalities we examined in vivo or in vitro, TrpA1-E is non-functional regardless of expression levels. It is possible that TrpA1-E is a byproduct of alternative splicing, alternatively, TrpA1-E might be activated by yet unknown stimuli.
DISCUSSION
In the present study, we develop a new and versatile genetic method to target alternative splicing, to enable the systematic mapping of expression and functions of single isoforms. Our studies demonstrate that alternative splicing critically regulates TrpA1 expression, and reveal an unexpected functional specificity of Drosophila TrpA1 isoforms—a given noxious stimulus is transduced predominantly by a single TrpA1 isoform. Therefore, detection of heat and UV light, two ancient modalities, is molecularly segregated through alternative splicing of TrpA1. Together, our study provides a new molecular framework to understand nociception at alternative splicing levels.
Polymodality of Drosophila Nociceptors Requires TrpA1 Alternative Splicing
Polymodality, the hallmark of nociceptors, is generally attributed to co-expression of different transducers, or to the intrinsic polymodality of a single transducer [14, 44]. What appears unique to our findings is that although TrpA1-C and TrpA1-D are polymodal transducers by nature as suggested by their in vitro properties, their in vivo functions are modulated to confer stimulus-specificity. As a result, polymodal nociception in C4da nociceptors requires TrpA1 alternative splicing, and co-expression of TrpA1-C and TrpA1-D.
To explain the stimulus specificity of TrpA1-C and TrpA1-D observed in vivo, we propose that heat-sensitivity of TrpA1-C, and UVC/H2O2-sensitivity of TrpA1-D, are suppressed in C4da nociceptors. One possible mechanism underlying this modulation could be that TrpA1-C and TrpA1-D interact with different protein or lipid partners to regulate their in vivo functions. Based on the structure of human TrpA1 [45], it is predicted that the linker domain, which represents the only difference between TrpA1-C and TrpA1-D, is exposed thus allowing interaction with other partners. Indeed, recent large-scaled proteomic studies suggest that alternatively protein isoforms tend to form completely different interactome, just as different genes will do [46]. Future studies, such as interactome analysis by mass spectrometry, will help elucidate the biochemical mechanisms underlying functional modulation of TrpA1-C and TrpA1-D. Although detailed mechanisms are yet unknown, genetic rescue experiments suggest that the functional modulation has limited capacity, as overexpressed TrpA1-C and TrpA1-D in C4da nociceptors escape the control to exhibit their intrinsic sensitivities towards both heat and UVC/H2O2.
Expression Level Regulates TrpA1 Heat Sensitivities
Noxious heat is primarily transduced by TrpA1-D, which has a threshold of ~43°C (Figure 4A, D), in agreement with previous electrophysiological studies [15, 18]. However, TrpA1-D reconstituted in S2 cells shows a much lower heat threshold of ~31°C (Figure 7E) [17]. In this study, we tested the idea that expression levels regulate TrpA1 heat sensitivity, by comparing GCaMP6s responses in the TrpA1-D-KI allele versus in the genetic rescue flies overexpressing TrpA1-D. We found that TrpA1-D heat sensitivity differs significantly in these two in vivo conditions, with threshold being 43°C in TrpA1-D-KI larvae and 34°C in the rescue strain (Figure 4D, 6D). As the only difference in C4da nociceptors in these two conditions is the TrpA1-D expression level, these findings indicate that overexpression by the GAL4-UAS system in C4da nociceptors significantly enhances TrpA1-D heat sensitivity. Therefore, the heat sensitivity of C4da nociceptors is determined by the biophysical property and expression levels of TrpA1-D.
Most ion channels are functionally characterized in heterologous cells. Moreover, genetic rescue by GAL4-UAS-based overexpression is often considered one key experiment to determine in vivo functions of ion channels in Drosophila. Our findings provide evidence that ion channel properties obtained from in vitro or genetic rescue studies can differ significantly from their endogenous functions, indicating that precise tuning of ion channel expression levels is critical for proper functional analysis in vivo.
STAR METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Yang Xiang (Yang.Xiang@umassmed.edu). Plasmids and TrpA1 mutant flies generated in this study will be available upon request. Complementary DNA of TrpA1-E will be deposited to Addgene.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
All fly stocks were raised at an incubator (I-36VL, Percival Scientific, Inc.) at 25°C and 60% humidity in a 12: 12h light dark cycle. Low yeast brown food recipe can be found on the website of University of Massachusetts Medical School Drosophila Resource Facility. Both male and female early 3rd instar larvae were used unless otherwise noted. All experiments were conducted on age- and size-matched larvae. We generated all TrpA1 isoform-specific knock-in and knockout stocks using the transgenic service provided by Bestgene Inc. (Chino Hills, CA). The following fly stocks are used in this study. w1118 (gift from Patrick Emery lab); GAL477[w−], 6934-hid (BL#25680) (gift from Yang Hong lab) and y1w67c23 P(Crey)1b; ;D*/TM3, Sb (BL#851) were used in generating TrpA1-KO/KI flies; ppk-GAL4 [15],; UAS-CD4TdTomato and ppk-CD4TdGFP [47] (gifts from Chun Han lab); repo-GAL4 (gift from Marc Freeman lab); UAS-6XGFP (BL#52261, BL#52262), lexAop2-6XmCherry (BL#52271, BL#52272), lexAop-GFP::NLS (BL#29954), UAS-mCherry::NLS (BL#38424, BL#38425), 386Y-GAL4 (BL#25410), R21G01-GAL4 (BL#48951), R21G01-lexA (BL#61521), TrpA1-ABlexA (BL#67130) and elav-GAL4 (BL#8765) were used for reporting gene expression patterns; ppk-GAL4, 386Y-GAL4, R21G01-GAL4, UAS-Dcr (BL#24650) and UAS-TrpA1-RNAi (BL#36780) were used for knock down expression of TrpA1 in specific cells; UAS-GCaMP6s (BL#42746, BL#42749) were used for Ca2+ imaging; elav-GAL4 was also used for the assay of adult fly incapacitation.
METHOD DETAILS
Generation of Transgenic Flies
Full length cDNAs of five Drosophila TrpA1 isoforms were cloned from total RNA extracted from w1118 larvae by RT-PCR, using SuperScript III (Invitrogen) and PrimeStar GXL DNA polymerase (Takara). Sequence of cDNAs corresponding to each of the five alternative isoforms was identical to the Flybase annotation. Each of the five TrpA1 cDNAs was then cloned into the transgenic vector pUASTb containing attB. A Kozak consensus sequence (GCCACC) was introduced immediately before ATG translational start site. Each of these five pUASTb-TrpA1-isoform vectors was injected into Drosophila embryos bearing attP16 site by phiC31 integrase.
For cloning of full length cDNAs of TrpA1-A/B and TrpA1-C/D/E, primer sets TrpA1-AF/TrpA1-R and TrpA1-DF/TrpA1-R were used, respectively.
Generation of TrpA1 Isoform-specific Alleles
Drosophila TrpA1 gene knockout procedure was performed as previously described [27, 28, 48]. Briefly, about four kilobases homolog arms of the TrpA1 gene were cloned and inserted into P element-based transforming vector pGX-attP-WN. This targeting vector was inserted into fly genome by standard transgenic procedure to generate transgenic donor fly stocks.
Targeting crosses. A line with transgenic donor DNA on the 2nd chromosome was chosen for targeting cross to delete the entire TrpA1 gene of ~10kb. In the targeting cross, 40 vials of crosses were set up. Each vial contained 20 virgin females of transgenic donor flies mated with 20 6934-hid males (BL#25680). Crosses were maintained at room temperature and flies were transferred to new vials every 24 hours. Eggs in vials were maintained on 0.2 mg/ml G418-containing food at 25°C, and were heat-shocked at both 48 hours and 72 hours after egg-laying. Heat shock was carried out at 38°C for 90 minutes in a water bath.
Screening crosses. Ten virgin females from the targeting crosses were mated with ten GAL477[w−] (w; GAL477[w−]; TM2/TM6b) males in each vial. Flies were transferred to fresh vials every two days at 25°C with a total of five transfers. Preliminary targeting candidates from targeting crosses were screened based on eye color.
Mapping crosses. A single male candidate was crossed with double balancer virgin females (w; sp/CyO, Wee-P; TM2/TM6b). Targeting candidates with white selection marker on the 3rd chromosome were selected for further genotyping. TrpA1 full gene knockout flies were established and confirmed by genotyping and RT-PCR. To remove the white selection marker and vector backbone, these white+ lines were crossed with Cre recombinase line (BL#851). Single male progeny with white eyes was selected, further balanced to remove Cre. Next, we backcrossed these lines to change their X and 2nd chromosomes into w1118 background, to establish the final TrpA1-KO founder line.
The following primers were used in genotyping of TrpAI-KO founder line.
Mcm7-F/R, mfr-F/R, TrpA1(5’)-F/R and TrpA1(3’)-F/R.
The following RT-PCR primers were used in detection of TrpA1 alternative transcripts.
TrpA1-Ex1,2-F(pair with Ex4,5-R for detecting TrpA1-C/D/E transcripts), TrpA1-Ex3,4-F(pair with Ex4,5-R for detecting TrpA1-A/B transcripts), TrpA1-Ex4,5-R and Rp1-F/R.
To generate TrpA1 isoform-specific knock-in or knockout flies, full length of Drosophila TrpA1 genome fragment (~10 kb) was cloned from bacterial artificial chromosome (Pacman BAC Ch322-152I23, BACPAC Resources Center, Children’s Hospital Oakland Research Institute) and was inserted into pGE-attB-GMR vector [27]. Wild-type TrpA1 allele on pGE-attB-GMR vector was modified in vitro. DNA fragments, amplified by PCR with primers containing designated mutations, were merged together by In-Fusion HD cloning kit (Cat#638909, Takara). Vectors with modified TrpA1 genome were injected into TrpA1-KO founder line embryos to insert at the native TrpA1 locus, by phiC31 mediated attB-attP recombination. Integration events were scored by the white selection marker. These lines were then balanced to confirm that modified TrpA1 was inserted on the 3rd chromosome. To remove unnecessary vector backbone sequences and selection markers, single male of these lines was crossed with flies bearing Cre recombinase strain (BL#851). Single male offspring with white eyes was selected, further balanced to remove Cre. Next, we backcrossed these lines to change their X and 2nd chromosomes into w1118 background, to establish final TrpA1-isoform-KI/KO alleles.
Primers TrpA1-Ex10,11-F and TrpA1-Ex14,15R were paired to clone cDNA fragment from exon 11 to 14. Amplified PCR products were sequenced and aligned to wild-type one for analysis of splicing pattern.
Behavioral Assays
Thermal nociception assay was performed as previously described [31]. A custom-made heat probe with a proportional–integral–derivative (PID) control unit was used to deliver heat stimulation. An early 3rd instar larva was rinsed with distilled water and stimulated along with the segment A4-6. One larva was only stimulated once and positive response behavior was defined as at least one 360° roll. The response latency to each stimulus was recorded up to a 20-second cutoff. According to the responding time, behavioral responses were divided into 3 categories, fast rolling (≤5 seconds), slow rolling (>5 but ≤20 seconds) and no rolling (>20 seconds). The assay was conducted at room temperature under a dissection scope (Nikon SMZ800) and a light source (Fostec ACE).
UVC-induced writhing behavioral assay was performed as previously described [16] with minor modification. Early 3rd instar larvae were rinsed with distilled water. After resting in a petri dish for 1 minute, larvae were placed inside a UVC crosslinker (254 nm, Spectorlinker XL-1000) and exposed to UVC radiation. UVC intensity (5 mW/cm2) was measured by a hand-held UV spectrophotometer (AccuMAX XS-254, Spectroline). Immediately after turning on UVC light, all larvae displayed startle responses regardless of genotype. Only continuous writhing behavior within 5 seconds after initial startle responses was defined as a positive response. 5 larvae were tested in one group and the percentage of larvae showing writhing behavior was calculated. 10 trials were tested for each genotype for a total of 50 larvae.
AITC-induced writhing behavioral assay was performed in 3-well Pyrex glass spot plate (34X85 mm). 500 mM AITC was dissolved in DMSO. 25 μl AITC solution was applied to one larva in glass well. The response latency was recorded up to a 20-second cutoff. Similar to rolling behavior, writhing responses were divided into 3 categories, fast writhing (≤5 seconds), slow writhing (>5 but ≤20 seconds) and no writhing (>20 seconds).
In warmth-induced knockdown assay, adult flies were raised at room temperature (about 22°C) to reduce TrpA1 activation. ~10 adults (5-7 days old) were collected in a vial. The vial was immersed in a circulated water bath with a preset temperature of 29.0°C. The whole procedure was video recorded. Numbers of flies being knocked down to the vial bottom at different time points were analyzed according to the video. At least 7 trials were conducted for each genotype.
Confocal Microscopy
To visualize fluorescence signals associated with UAS-6XGFP, UAS-CD4TdTomato and lexAop2-6XmCherry reporter lines, Drosophila larval or adult tissues were dissected in ice-cold phosphate buffered saline (PBS) and then fixed with 4% paraformaldehyde (PFA/PBS) for 1 hour at room temperature. After fixation, samples were incubated with 0.3% Triton X-100/PBS for 15 minutes each for 4 times, followed by PBS wash for 3 times. Samples were mounted in cover glass with mounting media (Vectashield H-1000, Vector Lab, Inc.). Images were acquired with confocal microscope (Zeiss LSM700).
Immunofluorescence
For TrpA1 staining in larval brains, 3rd instar larvae were dissected in ice-cold PBS, fixed in 4% PFA/PBS for 1 hour at room temperature and incubated in blocking buffer (5% normal donkey serum, 0.3% Triton X-100/PBS) for 1 hour. Samples were then incubated with anti-TrpA1 (1:1000, a gift from Paul Garrity lab) for two days at 4°C. After washing with 0.3% Triton X-100/ PBS, samples were incubated with the secondary antibody Alex 647-Donkey-anti-Rat (Jackson IR Lab, 712-605-153, 1:200) for 2 hours at room temperature. After washing, samples were mounted with mounting media (Vectashield H-1000, Vector Lab, Inc.) and images were acquired.
Due to the extremely low expression level of TrpA1, conventional staining by anti-TrpA1 antibody did not detect signals in larval C4da nociceptors. Staining against GFP or FLAG in larvae bearing TrpA1-KI-GFP, TrpA1-isoform-KI-GFP, TrpA1-KI-FLAG and TrpA1-isoform-KI-FLAG alleles was performed using tyramide signal amplification (TSA) to boost the fluorescence signals. TSA Plus Cyanine 5 (Cy5) detection kit (PerkinElmer, NEL745001KT) was used in the present study. 3rd instar larvae were dissected and fillets were fixed in 4% PFA/PBS for 0.5 hour at room temperature, then incubated with 3% H2O2/PBS solution to quench endogenous peroxidase. Fillets were blocked in blocking buffer (5% normal donkey serum, 0.3% Triton X-100/ PBS) for 1 hour at room temperature and incubated with anti-GFP (Thermo Fisher, A-11122, 1:1500) or anti-FLAG (Sigma, F3165, 1:500) antibody at 4°C overnight. Next, samples were incubated with HRP-conjugated anti-rabbit (GE Healthcare, NA9340, 1:100) or antimouse (GE Healthcare, NA931, 1:100) antibody for 2 hours at room temperature. Fillets were then incubated with TSA working solution for 3 minutes at room temperature, followed by incubation with Alex 488 Goat-anti-HRP (Jackson IR Lab, 123-545-021, 1:200) for 2 hours at room temperature to label all sensory neurons. Next, samples were mounted with mounting media (Thermo Fisher, P36961) and processed with image acquisition. For each solution changing step, fillets were washed with 0.3% Triton X-100 in PBS for 3 times of 10 minutes each. Fluorescence intensities from the somatic region of 6-8 C4da nociceptors of a single larva were averaged to represent the value for that larva. For each genotype, data represent values from at least 8 larvae.
GCaMP6s Imaging
For chemical stimulation assays, the 3rd instar larval fillets were dissected in fly physiological saline (120 mM NaCl, 4 mM MgCl2, 3 mM KCl, 10 mM NaHCO3, 10 mM Glucose, 10 mM Sucrose, 10 mM Trehalose, 5 mM TES, 10 mM HEPES, 1.5 mM CaCl2, pH7.2). AITC (100 μM or 50 μM), H2O2 (10 mM or 25 μM), dissolved in physiological saline, were perfused through the imaging chamber. ppk-GAL4>UAS-GCaMP6s strain was used to monitor Ca2+ signal changes within C4da nociceptors upon stimulation. Data were collected on a Zeiss LSM700 confocal microscope with a Zeiss 20x/1.0 NA water immersion objective lens (421452-9800). GCaMP6s fluorescence was excited with a 488-nm laser. Images were acquired at 512 × 512 pixels, 12-bit dynamic range in a time-lapse mode. Region of interest (ROI) was selected from the somatic region of C4da nociceptors, although axons and dendrites also responded. To quantify Ca2+ responses, changes in GCaMP6s fluorescence were calculated by ΔF/F0 = (Ft - F0)/F0, where Ft was the fluorescence value of an ROI in a given frame. Averaged value of ROI from 30 s before stimulation was taken as F0, which was adjusted to ~900 A.U. by gain and laser intensity. Max ΔF/F0 was the maximum value during the entire imaging period. To analyze AITC and H2O2 responses of C4da nociceptors, Max ΔF/F0 was plotted on the graph and used for statistical analyses.
Heat responses of C4da nociceptors was measured in a similar way. A 3rd instar larva was dissected in fly physiological saline by removing the head and the internal organs. The remaining body parts, including sensory neurons in their native tissue environment, were mounted on a copper block (WBA-1.62-0.55-CU-01, Custom Thermoelectric) with fly physiological saline and covered by glass coverslip. After resting for 10 minutes at room temperature, pre-heated hot water (70°C) was perfused through the inner cavity of the copper block to apply heat stimulation. The surface temperature of copper block was monitored by a T-type thermal coupler (Omega 5SRTC-TT-T-36-36) connected with a thermometer (Physitemp BAT-10), and recorded by the Clampex software. Region of interest (ROI) was selected from the somatic region of C4da nociceptors, although axons and dendrites also responded to heat. ΔF/F0 was plotted against the corresponding temperature to display the heat responses of C4da nociceptors. Averaged value of ROI from 30 s before heat stimulation was taken as F0. Note, temperature increases caused decreased fluorescence intensity of GCaMP6s. The heat activation thresholds of TrpA1 isoforms were determined by the temperature point when the corresponding fluorescence value returned to zero.
Chemical stimulation assays were also applied on HEK cells. HEK cells were grown on cover glasses in 3.5-cm culture dishes with DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin and transfected by 0.5 μg of TrpA1 plasmids, 0.5 μg of GCaMP6s plasmids and 5 μg of polyethylenimine. The imaging solution (pH 7.2) contained the following ingredients: 130 mM NaCl, 3 mM KCl, 0.6 mM MgCl2, 1.2 mM NaHCO3, 10 mM glucose, 10 mM HEPES, and 1 mM CaCl2. Data were collected by Zeiss LSM700 confocal system. The average peak response was calculated with Zeiss Zen software.
S2 Cell Culture and Electrophysiology
Drosophila S2 cell line was maintained in SFX-insect medium (HyClone SH30728) supplemented with 10% heat-inactivated fetal bovine serum in 25°C incubator. For transfection, S2 cells were cultured on glass coverslips in 3.5 cm culture dishes. pAct5C-GAL4, pUASTb-TrpA1-isoform and pUASTb-GFP (1 μg plasmid each) were co-transfected into cells with 10 μl transfection reagent (Fugene HD, Promega E2311) for electrophysiology experiments.
Whole-cell patch clamp recordings were made on GFP-positive cells 24 hours after transfection. External solution contained 120 mM NaCl, 3 mM KCl, 4 mM MgCl2, 1.5 mM CaCl2, 10 mM NaHCO3, 10 mM Glucose, 10 mM Sucrose, 10 mM Trehalose, 5 mM TES and 10 mM HEPES (pH7.2). Intracellular solution contains 140 mM potassium gluconate, 1 mM KCl, 4 mM Mg-ATP, 0.5 mM Na3-GTP, 1 mM EGTA and 10 mM HEPES (pH7.2). Membrane potential was held at −60 mV. Cells with leak current larger than 10 pA were discarded. Currents were measured by Multiclamp 700B amplifier, digitized by analog-to-digital converter Digidata 1440 and recorded in Clampex 10 software (Molecular Devices), with a sampling rate of 10 kHz and low pass filter of 2 kHz. After whole-cell configurations were established, cells were detached and lifted from glass coverslips, to avoid damages to the recording by heat-induced chamber deformation.
Heat stimulation was conducted in a custom-made chamber. The chamber had two embedded metal tubes. Pre-heated water flowed through the tubes to increase the temperature of bath solution from ~22°C to ~48°C in ~60 seconds. A temperature probe (IT-24P) was positioned adjacent to recorded cells, and temperature was measured by a Physitemp BAT-10 thermometer (Physitemp Instruments). Temperature values were recorded by Clampex simultaneously with whole-cell currents. Cells with < 100 pA maximal heat responses were excluded for further analysis.
Recording data were processed and analyzed in MATLAB (MathWorks). Current values corresponding to the temperature range 23-24°C were set as baseline, and standard deviation of baseline currents was calculated. Heat activation threshold for TrpA1 isoforms was determined as the temperature value when heat-induced current was above 1-fold standard deviation of baseline currents. Arrhenius plot was derived as logarithm of TrpA1 currents versus the inverse of absolute temperature. To quantify heat sensitivities of TrpA1 isoforms, the linear portion of Arrhenius plot was determined with linear fitting (r2>0.99), and Arrhenius Q10 was calculated as 10−10•S/(T1•T2) , where S was the slope and T1/T2 were the temperature limits of the linear portion of Arrhenius plot.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using Graphpad Prism software. Latency of rolling behavior under stimulation were analyzed using the Log-rank (Mantel-Cox) test. Unless specified, 90 larvae were tested for each genotype. For warmth triggered knockdown assay (adult fly incapacitation), Two-way ANOVA with Tukey’s post comparison test was used. GCaMP6s imaging of C4da nociceptors were analyzed using One-way ANOVA with Tukey’s post comparison test. In all figures, n.s. and ***, **** represent p>0.05, p<0.001 and p<0.0001, respectively. Columns with different superscripts (a, b and c) are significantly different from each other (p<0.05). Error bars are standard errors of the mean (SEM).
DATA AND CODE AVAILABILITY
MATLAB code for this study is available upon request from the Lead Contact.
Supplementary Material
Video S1. Heat Probe-induced Rolling Behavior. Related to Figure 3.
Video S2. UVC-induced Writhing Behavior. Related to Figure 5.
Video S3. Warmth triggered knockdown. Related to Figure 6.
HIGHLIGHTS.
A given stimulus preferentially activates a single TrpA1 isoform in vivo
Polymodal nociception requires co-expression of TrpA1 isoforms in nociceptors
Each TrpA1 isoform has a unique expression profile
isoEXPRESS is a new and versatile framework targeting alternative splicing
ACKNOWLEDGMENTS
We thank Drs. Yang Hong, Juan Huang for reagents and advice for genomic engineering, Drs. Michael Galko and Seol Hee Im for instructions on heat nociceptive behavioral assay, Drs. Paul Garrity, Bing Ye, Patrick Emery, and Chun Han for sharing reagents, and the Bloomington Stock Center for providing fly stocks, Drs. Patrick Emery, David Weaver, Bing Ye, Fan Wang, Rebecca Yang, Yang Hong and Weisheng Chen for critical comments on the manuscript. We thank Peng Ding for technical support. Y.X. is supported by the National Institutes of Health under award number 5R01NS089787 and 1R21NS107924, and Human Frontier Science Program Young Investigator award RGY0090/2014. M.F. is supported by National Institutes of Health under award number 5R37NS053538,
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Dillman AA, Hauser DN, Gibbs JR, Nalls MA, McCoy MK, Rudenko IN, Galter D, and Cookson MR (2013). mRNA expression, splicing and editing in the embryonic and adult mouse cerebral cortex. Nat Neurosci 16, 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang X, Chen MH, Wu X, Kodani A, Fan J, Doan R, Ozawa M, Ma J, Yoshida N, Reiter JF, et al. (2016). Cell-Type-Specific Alternative Splicing Governs Cell Fate in the Developing Cerebral Cortex. Cell 166, 1147–1162 e1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Q, Lee JA, and Black DL (2007). Neuronal regulation of alternative pre-mRNA splicing. Nat Rev Neurosci 8, 819–831. [DOI] [PubMed] [Google Scholar]
- 4.Mountoufaris G, Canzio D, Nwakeze CL, Chen WV, and Maniatis T (2018). Writing, Reading, and Translating the Clustered Protocadherin Cell Surface Recognition Code for Neural Circuit Assembly. Annu Rev Cell Dev Biol 34, 471–493. [DOI] [PubMed] [Google Scholar]
- 5.Raj B, and Blencowe BJ (2015). Alternative Splicing in the Mammalian Nervous System: Recent Insights into Mechanisms and Functional Roles. Neuron 87, 14–27. [DOI] [PubMed] [Google Scholar]
- 6.Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, and Grueber WB (2007). Dendrite self-avoidance is controlled by Dscam. Cell 129, 593–604. [DOI] [PubMed] [Google Scholar]
- 7.Hughes ME, Bortnick R, Tsubouchi A, Baumer P, Kondo M, Uemura T, and Schmucker D (2007). Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron 54, 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, and Jan YN (2007). Drosophila sensory neurons require Dscam for dendritic self-avoidance and proper dendritic field organization. Neuron 54, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudhof TC (2017). Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell 171, 745–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kelemen O, Convertini P, Zhang Z, Wen Y, Shen M, Falaleeva M, and Stamm S (2013). Function of alternative splicing. Gene 514, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bell TJ, Thaler C, Castiglioni AJ, Helton TD, and Lipscombe D (2004). Cell-specific alternative splicing increases calcium channel current density in the pain pathway. Neuron 41, 127–138. [DOI] [PubMed] [Google Scholar]
- 12.Emerick MC, Stein R, Kunze R, McNulty MM, Regan MR, Hanck DA, and Agnew WS (2006). Profiling the array of Ca(v)3.1 variants from the human T-type calcium channel gene CACNA1G: alternative structures, developmental expression, and biophysical variations. Proteins 64, 320–342. [DOI] [PubMed] [Google Scholar]
- 13.Albeg A, Smith CJ, Chatzigeorgiou M, Feitelson DG, Hall DH, Schafer WR, Miller DM 3rd, and Treinin M (2011). C. elegans multi-dendritic sensory neurons: morphology and function. Mol Cell Neurosci 46, 308–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basbaum AI, Bautista DM, Scherrer G, and Julius D (2009). Cellular and molecular mechanisms of pain. Cell 139, 267–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang Y, Yuan Q, Vogt N, Looger LL, Jan LY, and Jan YN (2010). Light-avoidance-mediating photoreceptors tile the Drosophila larval body wall. Nature 468, 921–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim MJ, and Johnson WA (2014). ROS-mediated activation of Drosophila larval nociceptor neurons by UVC irradiation. BMC neuroscience 15, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhong L, Bellemer A, Yan H, Ken H, Jessica R, Hwang RY, Pitt GS, and Tracey WD (2012). Thermosensory and nonthermosensory isoforms of Drosophila melanogaster TRPA1 reveal heat-sensor domains of a thermoTRP Channel. Cell Rep 1, 43–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Terada S, Matsubara D, Onodera K, Matsuzaki M, Uemura T, and Usui T (2016). Neuronal processing of noxious thermal stimuli mediated by dendritic Ca(2+) influx in Drosophila somatosensory neurons. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vazquez E, and Valverde MA (2006). A review of TRP channels splicing. Semin Cell Dev Biol 17, 607–617. [DOI] [PubMed] [Google Scholar]
- 20.Babcock DT, Shi S, Jo J, Shaw M, Gutstein HB, and Galko MJ (2011). Hedgehog signaling regulates nociceptive sensitization. Current biology : CB 21, 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang K, Panzano VC, Chang EC, Ni L, Dainis AM, Jenkins AM, Regna K, Muskavitch MA, and Garrity PA (2011). Modulation of TRPA1 thermal sensitivity enables sensory discrimination in Drosophila. Nature 481, 76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang K, Pulver SR, Panzano VC, Chang EC, Griffith LC, Theobald DL, and Garrity PA (2010). Analysis of Drosophila TRPA1 reveals an ancient origin for human chemical nociception. Nature 464, 597–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arenas OM, Zaharieva EE, Para A, Vasquez-Doorman C, Petersen CP, and Gallio M (2017). Activation of planarian TRPA1 by reactive oxygen species reveals a conserved mechanism for animal nociception. Nat Neurosci 20, 1686–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guntur AR, Gu P, Takle K, Chen J, Xiang Y, and Yang CH (2015). Drosophila TRPA1 isoforms detect UV light via photochemical production of H2O2. Proc Natl Acad Sci U S A 112, E5753–5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang K, Panzano VC, Chang EC, Ni L, Dainis AM, Jenkins AM, Regna K, Muskavitch MA, and Garrity PA (2012). Modulation of TRPA1 thermal sensitivity enables sensory discrimination in Drosophila. Nature 481, 76–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwon Y, Kim SH, Ronderos DS, Lee Y, Akitake B, Woodward OM, Guggino WB, Smith DP, and Montell C (2010). Drosophila TRPA1 channel is required to avoid the naturally occurring insect repellent citronellal. Curr Biol 20, 1672–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang J, Zhou W, Dong W, Watson AM, and Hong Y (2009). From the Cover: Directed, efficient, and versatile modifications of the Drosophila genome by genomic engineering. Proc Natl Acad Sci U S A 106, 8284–8289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang J, Zhou W, Watson AM, Jan YN, and Hong Y (2008). Efficient ends-out gene targeting in Drosophila. Genetics 180, 703–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diao F, and White BH (2012). A novel approach for directing transgene expression in Drosophila: T2A-Gal4 in-frame fusion. Genetics 190, 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang RY, Zhong L, Xu Y, Johnson T, Zhang F, Deisseroth K, and Tracey WD (2007). Nociceptive neurons protect Drosophila larvae from parasitoid wasps. Current biology : CB 17, 2105–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Babcock DT, Landry C, and Galko MJ (2009). Cytokine signaling mediates UV-induced nociceptive sensitization in Drosophila larvae. Curr Biol 19, 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenzweig M, Brennan KM, Tayler TD, Phelps PO, Patapoutian A, and Garrity PA (2005). The Drosophila ortholog of vertebrate TRPA1 regulates thermotaxis. Genes Dev 19, 419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo J, Shen WL, and Montell C (2017). TRPA1 mediates sensation of the rate of temperature change in Drosophila larvae. Nat Neurosci 20, 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li HH, Kroll JR, Lennox SM, Ogundeyi O, Jeter J, Depasquale G, and Truman JW (2014). A GAL4 driver resource for developmental and behavioral studies on the larval CNS of Drosophila. Cell Rep 8, 897–908. [DOI] [PubMed] [Google Scholar]
- 35.Petersen LK, and Stowers RS (2011). A Gateway MultiSite recombination cloning toolkit. PLoS One 6, e24531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tracey WD Jr., Wilson RI, Laurent G, and Benzer S (2003). painless, a Drosophila gene essential for nociception. Cell 113, 261–273. [DOI] [PubMed] [Google Scholar]
- 37.Hamada FN, Rosenzweig M, Kang K, Pulver SR, Ghezzi A, Jegla TJ, and Garrity PA (2008). An internal thermal sensor controlling temperature preference in Drosophila. Nature 454, 217–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viswanath V, Story GM, Peier AM, Petrus MJ, Lee VM, Hwang SW, Patapoutian A, and Jegla T (2003). Opposite thermosensor in fruitfly and mouse. Nature 423, 822–823. [DOI] [PubMed] [Google Scholar]
- 39.Chen TW, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, Schreiter ER, Kerr RA, Orger MB, Jayaraman V, et al. (2013). Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baden HP, Kollias N, Anderson RR, Hopkins T, and Raftery L (1996). Drosophila melanogaster larvae detect low doses of UVC radiation as manifested by a writhing response. Arch Insect Biochem Physiol 32, 187–196. [DOI] [PubMed] [Google Scholar]
- 41.Bautista DM, Jordt SE, Nikai T, Tsuruda PR, Read AJ, Poblete J, Yamoah EN, Basbaum AI, and Julius D (2006). TRPA1 mediates the inflammatory actions of environmental irritants and proalgesic agents. Cell 124, 1269–1282. [DOI] [PubMed] [Google Scholar]
- 42.Macpherson LJ, Dubin AE, Evans MJ, Marr F, Schultz PG, Cravatt BF, and Patapoutian A (2007). Noxious compounds activate TRPA1 ion channels through covalent modification of cysteines. Nature 445, 541–545. [DOI] [PubMed] [Google Scholar]
- 43.Groth AC, Fish M, Nusse R, and Calos MP (2004). Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics 166, 1775–1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tominaga M, Caterina MJ, Malmberg AB, Rosen TA, Gilbert H, Skinner K, Raumann BE, Basbaum AI, and Julius D (1998). The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21, 531–543. [DOI] [PubMed] [Google Scholar]
- 45.Paulsen CE, Armache JP, Gao Y, Cheng Y, and Julius D (2015). Structure of the TRPA1 ion channel suggests regulatory mechanisms. Nature 525, 552. [DOI] [PubMed] [Google Scholar]
- 46.Yang X, Coulombe-Huntington J, Kang S, Sheynkman GM, Hao T, Richardson A, Sun S, Yang F, Shen YA, Murray RR, et al. (2016). Widespread Expansion of Protein Interaction Capabilities by Alternative Splicing. Cell 164, 805–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han C, Jan LY, and Jan YN (2011). Enhancer-driven membrane markers for analysis of nonautonomous mechanisms reveal neuron-glia interactions in Drosophila. Proc Natl Acad Sci U S A 108, 9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou W, Huang J, Watson AM, and Hong Y (2012). W::Neo: a novel dual-selection marker for high efficiency gene targeting in Drosophila. PLoS One 7, e31997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video S1. Heat Probe-induced Rolling Behavior. Related to Figure 3.
Video S2. UVC-induced Writhing Behavior. Related to Figure 5.
Video S3. Warmth triggered knockdown. Related to Figure 6.
Data Availability Statement
MATLAB code for this study is available upon request from the Lead Contact.