Abstract
The numerous quality control pathways that target defective ribonucleic acids (RNAs) for degradation play key roles in shaping mammalian transcriptomes and preventing disease. These pathways monitor most steps in the biogenesis of both non-coding RNAs (ncRNAs) and protein-coding messenger RNAs (mRNAs), degrading ncRNAs that fail to form functional complexes with one or more proteins and eliminating mRNAs that encode abnormal, potentially toxic proteins. Mutations in components of diverse RNA surveillance pathways manifest as disease. Some mutations are characterized by elevated interferon production, suggesting a major role of these pathways is to prevent aberrant cellular RNAs from being recognized as “non-self”. Other mutations are common in cancer, or result in developmental defects, revealing the importance of RNA surveillance to cell and organismal function.
RNA surveillance pathways sculpt cellular transcriptomes by degrading aberrant, potentially harmful RNAs. Most, if not all, RNAs are subject to surveillance. By far, the most abundant RNAs are long-studied “classical” ncRNAs such as the ribosomal RNAs (rRNAs) and transfer RNAs (tRNAs) that function in protein synthesis and the small nuclear RNAs (snRNAs) that carry out pre-mRNA splicing. These and many other ncRNAs, including more recently described ncRNAs such as microRNAs (miRNAs), are synthesized as precursors that fold into intricate three-dimensional structures, assemble with proteins and undergo one or more processing events to achieve their functional forms. As ncRNA precursors can misfold, fail to assemble into larger complexes or be incorrectly processed, cells must distinguish correct ncRNAs from RNAs that should be degraded. All cells also contain many less abundant ncRNAs, some of which derive from bidirectional transcription at the promoters of protein-coding genes. Many of these ncRNAs are surveillance targets, as they increase in amount when specific ribonucleases are inactivated. Failure to degrade some of these ncRNAs can alter transcription of adjacent protein-coding genes and can result in formation of RNA−DNA hybrids (R-loops) that render cells prone to DNA damage and chromosomal rearrangements (1). Although mRNAs constitute only a small fraction of cellular RNA, incompletely processed mRNAs can form RNA−DNA hybrids with their DNA templates, and mutation-containing mRNAs can encode abnormal, e.g. truncated and toxic proteins (2). Thus, all cells contain extensive surveillance pathways that recognize and target aberrant ncRNAs and mRNAs for degradation.
A feature of surveillance pathways for both ncRNAs and protein-coding RNAs is that they target newly synthesized RNAs in competition with their normal processing pathways. For ncRNAs, surveillance occurs in kinetic competition with (i) assembly of nascent ncRNAs with their core proteins to form ribonucleoproteins (RNPs), (ii) enzymes that trim or modify nascent ncRNA 5′ or 3′ ends and (iii) binding of proteins required for nuclear export of the assembled and matured RNPs (1). Consistent with competition, ncRNAs that fail to undergo RNP assembly and correct maturation are primarily targeted for degradation. Similarly, for mRNAs and long noncoding RNAs (lncRNAs, which resemble mRNAs in that they are synthesized by RNA polymerase II but are computationally defined as not encoding protein), transcription elongation, capping of the newly synthesized 5′ end, splicing, 3′ end formation and nuclear export compete with targeting for decay. Since surveillance takes place at multiple steps in RNA biogenesis, aberrant RNAs that escape one surveillance pathway are often subsequently targeted by another pathway.
Surveillance of aberrant ncRNAs
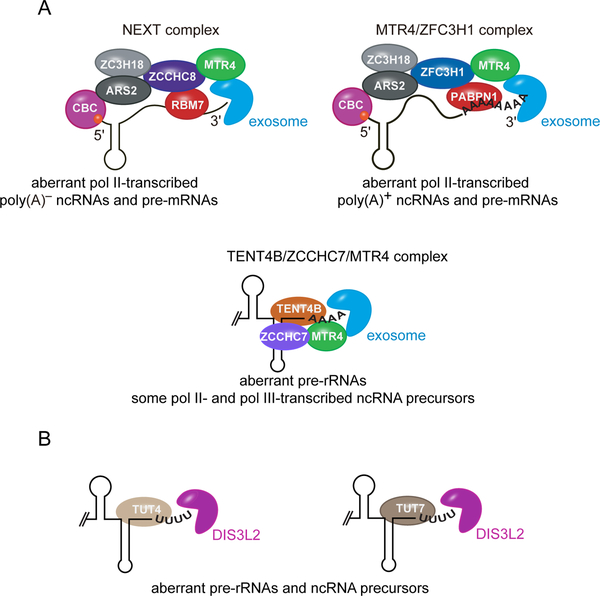
Although the ribonucleases that degrade defective RNAs are the major effectors of RNA surveillance pathways, they do not operate in isolation. Instead, most ribonucleases rely on protein cofactors, or adaptors, that recognize defective RNAs and recruit the nucleases. For example, a critical ncRNA surveillance pathway in eukaryotic nuclei involves a multiprotein nuclease complex called the RNA exosome, which manifests both 3′-to-5′ exoribonucleolytic and endoribonucleolytic activities. In all studied eukaryotic species, adaptor complexes containing the RNA helicase MTR4 and one or more RNA-binding proteins recognize defective ncRNAs and recruit the exosome (3). In these complexes, the RNA-binding moieties recognize the defective RNAs, while MTR4 binds a conserved surface on the exosome. Upon association of MTR4 with the exosome, the unwinding activity of MTR4 is enhanced, allowing MTR4 to deliver single-stranded RNA to the exosome for degradation (4). A major adaptor complex in human nuclei, called the nuclear exosome targeting complex (NEXT), is comprised of MTR4, the zinc finger protein ZCCHC8 and the RNA-binding protein RBM7 (5) (Fig. 1A). RBM7 binds single-stranded U-rich sequences near the ends of many aberrant nascent RNA polymerase II-transcribed ncRNAs, while ZCCHC8 scaffolds the interaction of RBM7 with MTR4 and may also contribute to RNA binding (6). A second complex, consisting of MTR4 and the zinc finger protein ZFC3H1, targets polyadenylated RNA polymerase II transcripts that are prematurely terminated or are generated by bidirectional promoter activity for degradation, most likely through interactions with the nuclear poly(A)-binding protein, PABPN1 (7, 8) (Fig. 1A).
Fig. 1. Protein adaptors target aberrant RNAs for degradation.
(A) In the nucleus, NEXT (top left) targets newly synthesized RNA polymerase II-transcribed poly(A)− ncRNAs and pre-mRNAs for degradation by the RNA exosome, while MTR4-ZFC3H1 (top right) targets nascent RNA polymerase II-transcribed poly(A)+ ncRNAs and poly(A)+ pre-mRNAs for exosome degradation. Prematurely terminated pre-rRNA precursors, as well as some RNA polymerase II- and polymerase III-transcribed ncRNA precursors, are adenylated by the TENT4B-ZCCHC7-MTR4 complex (bottom), facilitating degradation by the RNA exosome.
(B) In the cytoplasm, TUT4 (left) and TUT7 (right) add a short U tail to defective ncRNA precursors, stimulating degradation by the DIS3L2 exoribonuclease.
Another major category of ribonuclease adaptors includes or consists of a terminal nucleotidyl transferase (TENT) that adds short A or U tails to defective ncRNAs. In human nucleoli, a complex consisting of TENT4B, the RNA-binding protein ZCCHC7 and MTR4 adds a short A tail to the 3′ ends of prematurely terminated pre-rRNAs (Fig. 1A) (5). These tails, which are too short to be bound by PABPN1, provide a single-stranded end for MTR4 binding and degradation by exosome exoribonucleases. In the cytoplasm, terminal uridyl transferases TUT4 and TUT7 (also called TENT3A and TENT3B) add short U tails to the ends of many defective ncRNAs, stimulating degradation by the exoribonuclease DIS3L2 (Fig. 1B) (9, 10).
How are ncRNAs recognized as aberrant? For those adaptors that have been studied, specificity is conferred through some combination of subcellular location, association with the RNA synthesis machinery, affinity for specific RNA sequences and the presence of an accessible 3′ end. For example, the NEXT and MTR4-ZFC3H1 complexes are located in the nucleoplasm (5, 7, 8) while TENT4B-ZCCHC7-MTR4 is primarily nucleolar (5). NEXT and MTR4-ZFC3H1 are also in proximity to their targets because they associate, through their respective ZCCHC8 and ZFC3H1 subunits, with the cap-binding complex (CBC, consisting of cap-binding proteins CBP80 and CBP20) that binds the 5′ cap of nascent RNA polymerase II-transcribed RNAs (Fig. 1A) (11). Since TENT proteins add non-templated tails, they recognize RNAs with an accessible single-stranded 3′ end.
Nuclear surveillance of protein-coding transcripts
Many of the mechanisms that surveil aberrant RNA polymerase II-transcribed ncRNAs also recognize and degrade aberrant pre-mRNAs. Pre-mRNA processing, including capping of the newly made 5′ end, splicing-mediated removal of introns and formation of the 3′ end, usually occurs while the pre-mRNA is being synthesized. Surveillance mechanisms often sense the absence of one or more RNA-binding proteins during a critical window of time, reducing transcript half-life concurrently with or subsequent to the affected processing step.
A critical step for both mRNA biogenesis and mRNA surveillance is binding of the CBC to the capped 5′ end. Transcripts that fail to bind the CBC as the 5′ end emerges from elongating RNA polymerase II undergo decapping by the DXO enzyme and degradation by the 5′-to-3′ exoribonuclease XRN2 (12). Capping occurs concomitantly with release of the polymerase from promoter-proximal pausing (13). Those transcripts that prematurely terminate (instead of undergoing productive elongation) are recognized by NEXT and degraded by the exosome. During elongation, the CBC associates with the ARS2 protein, which serves as an interaction hub for the formation of mutually exclusive complexes that either promote pre-mRNA splicing, 3′-end formation and nuclear export or target incomplete and unprocessed transcripts for degradation by NEXT and MTR4-ZFC3H1 (14). In general, NEXT targets transcripts that lack poly(A) tails, while MTR4-ZFC3H1, together with PABPN1, targets unspliced polyadenylated mRNAs for degradation (Fig. 1A) (7, 8).
The exon-junction complex (EJC) is another central player that links mRNA biogenesis to quality control (15). Deposition of this multiprotein complex upstream of a splicing-generated exon−exon junction marks an mRNA as having undergone splicing (Fig. 2). The EJC, by interacting with additional proteins, enhances subsequent steps of mRNA transport to the cytoplasm, protein synthesis and, if appropriate, nonsense-mediated mRNA decay (NMD, see below). The human genome also contains a sea of cryptic splice sites that are similar in sequence to the canonical sites whose regulated use results in mRNAs encoding functional proteins. The EJC, through association with the splicing factor RNPS1, suppresses the inappropriate recognition of nearby cryptic splice sites, thus protecting transcriptome integrity (16).
Fig. 2. Nuclear synthesis and surveillance of protein-coding transcripts.
Following transcription, pre-mRNA splicing, EJC deposition and 3′-end formation, mRNAs are subject to quality control at the nuclear pore. Upon export, ribosomes engaged in the pioneer round of translation remove EJCs in their path, the CBC is replaced by eIF4E and PABPN1 is replaced by the cytoplasmic poly(A) binding protein PABPC1.
Termination of transcription and formation of pre-mRNA 3′-ends are also subject to surveillance. Pre-mRNAs contain numerous cryptic polyadenylation sites that, if utilized, can result in synthesis of truncated, nonfunctional proteins. Use of these sites is suppressed by binding of the U1 snRNP, a component of the spliceosome, to upstream 5′-splice sites (17). At least some pre-mRNAs that undergo premature termination and polyadenylation are degraded by the exosome, most likely through binding of PABPN1 to the polyadenylated mRNA and recruitment of MTR4-ZFC3H1 (Fig. 1A) (18).
Nuclear export is yet another step at which nonfunctional and incompletely processed mRNAs are subject to quality control. The nuclear export receptor NXF1 is preferentially loaded on spliced mRNAs through connections with the CBC, splicing factors, the EJC and the 3′-end formation machinery (19, 20). In addition, the nuclear basket protein Translocated Promoter Region (TPR) is part of a nuclear pore-based surveillance mechanism that prevents export of incompletely spliced mRNAs (Fig. 2) (21). Together, these mechanisms help ensure that only properly spliced and processed mRNAs are exported. These surveillance mechanisms can be subverted, as some mRNAs with retained introns reach the cytoplasm (22). Some cytoplasmic intron-containing mRNAs are rapidly degraded through surveillance mechanisms described below, particularly NMD, while others remain stable.
Cytoplasmic surveillance of protein-coding transcripts
A unique feature of protein-coding RNAs is the ability to use translation to monitor RNA quality. In the best-characterized of these pathways, NMD, mRNAs containing premature termination codons (PTCs; also called nonsense codons) are identified and targeted for decay during the first round of translation, before the CBC is replaced with the cap-binding eukaryotic translation initiation factor 4E (eIF4E). During this pioneer round of translation, which is defined as the translation of CBC-bound mRNA, ribosomes displace EJCs on those mRNAs that have undergone splicing (Fig. 2). In this way, the NMD pathway “inspects” mRNAs for genetic or processing defects that manifest in the premature termination of translation (2). As a rule, if the pioneer round of translation terminates more than ~50−55-nucleotides upstream of an EJC-bound exon−exon junction, the terminating ribosome will fail to physically displace that EJC (and any remaining downstream EJCs) and target the mRNA for NMD (Fig. 3A).
Fig. 3. Cytoplasmic surveillance of protein-coding transcripts.

(A) If the ribosome terminates before reaching the final EJC, the NMD machinery interacts with the EJC and targets the mRNA for degradation.
(B) In no-go and possibly nonstop decay, a ribosome stalls during translation elongation. As a consequence, ribosomes collide, providing a surface for ubiquitination by ZNF598. Afterwards, the N4BP2/Cue2 endonuclease cleaves at the 5′ side of the stalled ribosome, allowing rescue and recycling by PELO and HBS1.
A translation-dependent quality control mechanism that depends on a PTC and also feeds back to the nucleus is transcriptional adaptation (23, 24). Transcriptional adaptation, which occurs when transcription of paralogous genes increases to compensate for a defective gene, explains the observation that inactivating a gene can be less deleterious than depleting the mRNA with siRNAs or shRNAs. Two recent studies showed that those mutant genes that result in increased transcription of paralogs produce mRNAs containing PTCs (23, 24). Degradation of the mRNA via the NMD machinery triggers transcriptional adaptation through a mechanism that involves the UPF3 component of the NMD machinery (also called UPF3A). UPF3 interacts with the COMPASS complex to recruit chromatin modifiers that upregulate genes with sequence complementarity to the PTC-containing mRNA (23, 24). How the COMPASS complex is guided to paralogous gene promoters remains unclear but could involve fragments of the degraded mRNAs. Future therapeutic strategies targeting defective genes will undoubtedly take note of those genes that, when engineered to harbor a PTC, would allow one or more paralogs to functionally substitute for the deficiency.
A third translation-dependent pathway is triggered when ribosomes move pathologically slowly or stall during translation elongation. Causes of so-called “no-go” mRNA decay include difficulties forming peptide bonds between particular amino acids in the growing polypeptide chain; rare or suboptimal codons due to inadequate amounts of the corresponding tRNA(s) and mRNA structures that impede ribosome progression (25). A collided di-ribosome constitutes the decisive indicator that triggers both ribosome and mRNA quality control by forming a surface recognized by the ubiquitin ligase ZNF598, a prerequisite for triggering endonucleolytic cleavage of the mRNA (26, 27) (Fig. 3B). The responsible endonuclease in yeast no-go mRNA decay was identified recently as the Cue2 protein, the putative human ortholog of which is called N4BP2 (28). Following cleavage, ribosome rescue factors PELO and HBS1 bind and initiate ribosome subunit dissociation and recycling for further use. A related pathway, called “nonstop” mRNA decay, occurs when mRNAs lack a stop codon, either because they are truncated or have undergone premature 3′-end cleavage and polyadenylation in the nucleus. In either case, the ribosome stalls after translating to the end of the mRNA. As in no-go decay, removal of the arrested ribosome involves endonucleolytic cleavage of the mRNA and ribosome rescue by PELO and HBS1 (25), and a preliminary report has identified Cue2/N4BP2 as the required endonuclease (29).
There are also examples of mRNA surveillance in which the precipitating event is the failure of the nascent polypeptide to bind a partner as it emerges from the ribosome tunnel. One example involves mRNAs encoding secretory proteins. Although the synthesis of proteins destined for the endoplasmic reticulum (ER) initiates in the cytosol, the translating ribosomes are targeted to the ER through binding of the signal recognition particle (SRP) to the signal peptide as it emerges from the elongating ribosome. Failure of the signal peptide to engage SRP, due to a mutant signal peptide or SRP depletion, results in degradation of the mRNA (30). Although the mechanisms that connect the nascent polypeptide to co-translational mRNA decay are not understood, this and other examples where coding region sequences are coupled to mRNA decay support the idea of crosstalk between mRNA and protein quality control pathways.
Diseases Linked to Defective RNA Surveillance
Because the nucleases that degrade defective RNAs have myriad targets, and have roles outside surveillance, it is difficult to attribute mutant phenotypes to a specific role in quality control, or even to quality control at all. Nonetheless, missense mutations that partly inactivate RNA exosome function cause multiple developmental disorders. Mutations in exosome core subunits EXOSC3 and EXOSC8 cause pontocerebellar hypoplasia (PCH) 1B and 1C, respectively, which are autosomal recessive syndromes presenting early in childhood characterized by cerebellar atrophy and spinal motor neuron degeneration (31). Although only one patient with a homozygous mutation in the RBM7 component of NEXT has been described, this patient exhibited spinal motor neuron degeneration similar to PCH1 patients, suggesting PCH1 could be due to altered RNA surveillance (31). Several other PCH subtypes (PCH2–7 and PCH10) are caused by mutations that specifically disrupt processing of tRNAs or snRNAs (32), suggesting that accumulation of unprocessed and defective ncRNAs could contribute to PCH.
Other mutations in RNA surveillance pathway components are also associated with childhood disease. Mutations is DIS3L2 cause Perlman syndrome, an autosomal recessive overgrowth syndrome associated with increased frequency of Wilms tumor, the most common kidney tumor in children (33). Although the mechanisms by which DIS3L2 mutations cause Perlman syndrome have not been fully elucidated, kidney progenitor cells lacking DIS3L2 upregulate IGF2 (insulin-like growth factor 2), which is strongly associated with Wilms tumorigenesis (34). Disease may reflect the toxic accumulation of one or more of the many abnormal RNAs that are routinely eliminated by DIS3L2 (1). Mutations in genes encoding the NMD factors UPF2 or UPF3X (also called UPF3B) are associated with neurodevelopmental disorders, including autism spectrum disorder and schizophrenia (35).
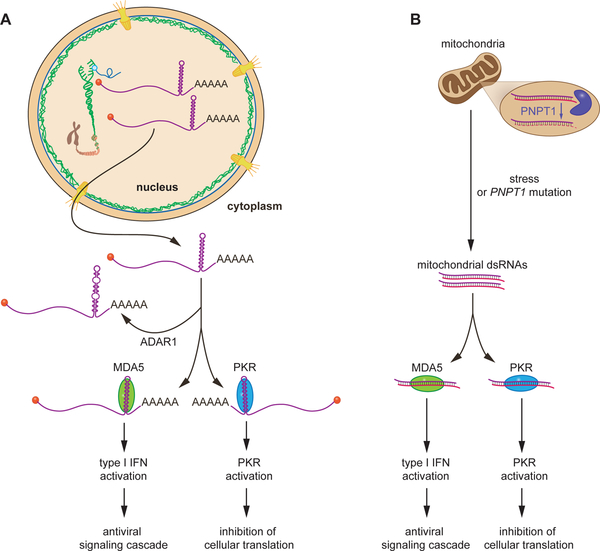
In other cases, failure to degrade aberrant RNAs is clearly linked to disease. For example, failure of translation-based surveillance mechanisms, such as NMD, affect disease through the synthesis of an aberrant, often truncated, and harmful protein that functions in dominant-negative ways. Failure to degrade excess RNAs results in activation of cytoplasmic innate immune sensors whose normal role is to recognize viral RNAs and trigger an antiviral response mediated by signaling molecules called interferons. These sensors, which recognize double-stranded RNA (dsRNA) generated during virus infection, include retinoic acid inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA5), which activate an antiviral signaling cascade, and protein kinase R (PKR), which phosphorylates the eIF2α translation initiation factor to inhibit protein synthesis (36). Mutations that inactivate proteins important for eliminating cellular dsRNAs, such as the adenosine deaminase acting on RNA 1 (ADAR1), which disrupts dsRNA by deaminating adenosines in double-stranded regions to inosines (37), or polynucleotide phosphorylase (PNPT1), a 3′-to-5′ exoribonuclease that degrades mitochondrial dsRNAs (38), result in neurologic diseases characterized by elevated interferon (39). Although the full catalog of aberrant cellular RNAs that can activate RIG-I and MDA5 is not known, loss of ADAR1 results in increased amounts of dsRNA formed from members of the Alu family of repetitive elements. These dsRNAs, many of which derive from Alu−Alu inverted repeats in mRNA untranslated regions, activate MDA5 to trigger interferon production and PKR to shut down translation (40, 41) (Fig. 4A). Mutations in PNPT1 result in accumulation of mitochondrial dsRNAs that enter the cytoplasm and activate MDA5 (38) and PKR (42) (Fig. 4B).
Fig. 4. Surveillance of dsRNA.
(A) In the cytoplasm, ADAR1 deaminates adenines in dsRNA to inosine, disrupting base-pairing. RNAs that fail to undergo editing can activate MDA5 and PKR.
(B) Formation of mitochondrial dsRNA is normally prevented by PNPT1, which degrades the antisense RNA. Under stress, or when PNPT1 is mutated, mitochondrial dsRNAs can enter the cytosol and activate MDA5 and PKR.
Both the upregulation and downregulation of RNA surveillance pathways make critical contributions to cancer pathogenesis. In many tumors, mutations in ADAR1 are associated with increased interferon secretion, growth inhibition and increased sensitivity to radiotherapy and immunotherapy via pathways mediated by PKR and MDA5 (43). While these effects are likely due to dsRNA accumulation, ADAR1 upregulation in chronic myeloid leukemia progenitor cells drives proliferation by editing primary miRNA precursors and mRNA 3′-untranslated regions, reducing the levels of specific miRNAs and their interactions with key targets (44). Upregulation and downregulation of NMD also contributes to cancers, depending on the cell of origin and how the cancer evolved (45). Downregulation of the RNA exosome likely contributes to some cancers, because mutations in the DIS3 catalytic subunit are common in some hematologic malignancies, particularly multiple myeloma. Although the mechanism(s) by which DIS3 mutations affect cancer progression are unknown, most mutations impair exoribonuclease activity, resulting in accumulation of many RNA targets (46).
Neurodegenerative diseases such as amyotrophic lateral sclerosis (ALS) may also be in part due to the failure of RNA quality control. Remarkably, in animal models of ALS, toxicity caused by expressing mutant versions of the splicing factors FUS or TDP43 in neurons or nervous tissue can be partially suppressed by overexpressing the key NMD factor called upframeshift 1 (UPF1) (35). The toxicity associated with the most common mutation in familial ALS and the related frontotemporal dementia (FTD), a repeat expansion in the C9orf72 gene, can similarly be suppressed by overexpressing UPF1 (47). Cell toxicity may derive from an increase in splicing mistakes caused by the mutant proteins, overwhelming the ability of NMD to degrade the ~10% of physiologic mRNAs that are natural NMD targets, the ~33% of newly synthesized mRNAs that normally contain pre-mRNA splicing errors (2) and an additional load of aberrant mRNAs caused by the mutant protein (35). In addition to misprocessed mRNAs, repeat element transcripts and dsRNAs are also elevated in the brains of patients with the C9ORF72 lesion and in mouse models of disease (48), hinting at a failure of multiple RNA surveillance pathways.
Perspectives
The examples discussed above illustrate only some of the ways in which surveillance systems monitor cellular RNA quality. Numerous other mechanisms monitor mRNAs and ncRNAs for correct processing, RNP assembly, nuclear export and function. Also not discussed is the role of RNA modifications, which stabilize tRNA structure, and have an emerging role in mRNA stabilization and surveillance (49). Nonetheless, it is clear that gene expression can be viewed as a tightly orchestrated homeostatic continuum of RNA checks and balances – of spatially and temporally connected RNA processing steps that begin in the nucleus, continue in the cytoplasm and, in specialized instances, feedback to the nucleus. Most if not all steps rely on the success of the previous step within a defined period of time or the process will be stopped, usually through RNA destruction, to minimize the ensuing cellular damage. This seemingly straightforward concept behind transcriptomic quality control is complicated by the susceptibility of many steps to genetic or environmental changes and the multitasking abilities of many of the proteins that mediate each step. It follows that disease would result from defects in both the mediators and the sensors of such homeostasis-maintaining systems. Awareness of what defines a transcript that passes quality control is key to the burgeoning use of RNAs as therapeutic targets and tools (50).
Acknowledgements
We apologize to our many colleagues whose work we omitted due to space constraints. We thank Tatsuaki Kurosaki (University of Rochester) and Allen Kane (National Cancer Institute) for help generating figures.
Funding: Research in the Wolin lab is supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research. Research in the Maquat lab is supported by National Institutes of Health grants R37 GM074593 and R01 GM059614 to L.E.M.
Footnotes
Footnote: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Competing interests: The authors declare no competing interests.
References and notes
- 1.Belair C, Sim S, Wolin SL, Noncoding RNA surveillance: The ends justify the means. Chem. Rev 118, 4422–4447 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurosaki T, Popp MW, Maquat LE, Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat Rev Mol Cell Biol 20, 406–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zinder JC, Lima CD, Targeting RNA for processing or destruction by the eukaryotic RNA exosome and its cofactors. Genes Dev 31, 88–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weick EM et al. , Helicase-dependent RNA decay illuminated by a cryo-EM structure of a human nuclear RNA exosome-MTR4 complex. Cell 173, 1663–1677 e1621 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lubas M et al. , Interaction profiling identifies the human nuclear exosome targeting complex. Mol. Cell 43, 624–637 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Puno MR, Lima CD, Structural basis for MTR4-ZCCHC8 interactions that stimulate the MTR4 helicase in the nuclear exosome-targeting complex. Proc. Natl. Acad. Sci. U. S. A 115, E5506–E5515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ogami K et al. , An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 31, 1257–1271 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meola N et al. , Identification of a nuclear exosome decay pathway for processed transcripts. Mol. Cell 64, 520–533 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Eckwahl MJ, Sim S, Smith D, Telesnitsky A, Wolin SL, A retrovirus packages nascent host noncoding RNAs from a novel surveillance pathway. Genes Dev. 29, 646–657 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pirouz M, Du P, Munafo M, Gregory RI, Dis3l2-mediated decay Is a quality control pathway for noncoding RNAs. Cell Rep. 16, 1861–1873 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersen PR et al. , The human cap-binding complex is functionally connected to the nuclear RNA exosome. Nat. Struct. Mol. Biol 20, 1367–1376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiao X, Chang JH, Kilic T, Tong L, Kiledjian M, A mammalian pre-mRNA 5′ end capping quality control mechanism and an unexpected link of capping to pre-mRNA processing. Mol. Cell 50, 104–115 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen FX, Smith ER, Shilatifard A, Born to run: control of transcription elongation by RNA polymerase II. Nat. Rev. Mol. Cell Biol 19, 464–478 (2018). [DOI] [PubMed] [Google Scholar]
- 14.Schulze WM, Stein F, Rettel M, Nanao M, Cusack S, Structural analysis of human ARS2 as a platform for co-transcriptional RNA sorting. Nat. Commun 9, 1701 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Le Hir H, Sauliere J, Wang Z, The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 17, 41–54 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Boehm V et al. , Exon junction complexes suppress spurious splice sites to safeguard transcriptome integrity. Mol. Cell 72, 482–495 e487 (2018). [DOI] [PubMed] [Google Scholar]
- 17.Venters CC, Oh JM, Di C, So BR, Dreyfuss G, U1 snRNP telescripting: suppression of premature transcription termination in introns as a new layer of gene regulation. Cold Spring Harb. Perspect. Biol 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiu AC et al. , Transcriptional pause sites delineate stable nucleosome-associated premature polyadenylation suppressed by U1 snRNP. Mol. Cell 69, 648–663 e647 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stewart M, Polyadenylation and nuclear export of mRNAs. J. Biol. Chem 294, 2977–2987 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S et al. , The mRNA export receptor NXF1 coordinates transcriptional dynamics, alternative polyadenylation, and mRNA export. Mol. Cell 74, 118–131 e117 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Palazzo AF, Lee ES, Sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front. Genet 9, 440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rekosh D, Hammarskjold ML, Intron retention in viruses and cellular genes: Detention, border controls and passports. Wiley Interdiscip. Rev. RNA 9, e1470 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Brolosy MA et al. , Genetic compensation triggered by mutant mRNA degradation. Nature 568, 193–197 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma Z et al. , PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature 568, 259–263 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Schuller AP, Green R, Roadblocks and resolutions in eukaryotic translation. Nat. Rev. Mol. Cell Biol. 19, 526–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Juszkiewicz S et al. , ZNF598 Is a quality control sensor of collided ribosomes. Mol. Cell 72, 469–481 e467 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeuchi K et al. , Collided ribosomes form a unique structural interface to induce Hel2-driven quality control pathways. EMBO J. 38, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Orazio KN et al. , The endonuclease Cue2 cleaves mRNAs at stalled ribosomes during No Go Decay. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glover ML et al. , NONU-1 encodes a conserved endonuclease required for mRNA translation surveillance. bioRxiv 674358 [Preprint], (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karamyshev AL et al. , Inefficient SRP interaction with a nascent chain triggers a mRNA quality control pathway. Cell 156, 146–157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morton DJ et al. , The RNA exosome and RNA exosome-linked disease. RNA 24, 127–142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Dijk T, Baas F, Barth PG, Poll-The BT, What’s new in pontocerebellar hypoplasia? An update on genes and subtypes. Orphanet J. Rare Dis. 13, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Astuti D et al. , Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet 44, 277–284 (2012). [DOI] [PubMed] [Google Scholar]
- 34.Hunter RW et al. , Loss of Dis3l2 partially phenocopies Perlman syndrome in mice and results in up-regulation of Igf2 in nephron progenitor cells. Genes Dev. 32, 903–908 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jaffrey SR, Wilkinson MF, Nonsense-mediated RNA decay in the brain: emerging modulator of neural development and disease. Nat. Rev. Neurosci 19, 715–728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hur S, Double-stranded RNA sensors and modulators in innate immunity. Annu. Rev. Immunol 37, 349–375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reich DP, Bass BL, Mapping the dsRNA World. Cold Spring Harb. Perspect. Biol 11, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dhir A et al. , Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 560, 238–242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uggenti C, Lepelley A, Crow YJ, Self-awareness: Nucleic acid-driven inflammation and the type I interferonopathies. Annu. Rev. Immunol 37, 247–267 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Ahmad S et al. , Breaching self-tolerance to Alu duplex RNA underlies MDA5-mediated inflammation. Cell 172, 797–810 e713 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chung H et al. , Human ADAR1 prevents endogenous RNA from triggering translational shutdown. Cell 172, 811–824 e814 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim Y et al. , PKR senses nuclear and mitochondrial signals by interacting with endogenous double-stranded RNAs. Mol. Cell 71, 1051–1063 e1056 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Bhate A, Sun T, Li JB, ADAR1: A new target for immuno-oncology therapy. Mol Cell 73, 866–868 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Jiang Q et al. , Hyper-editing of cell-cycle regulatory and tumor suppressor RNA promotes malignant progenitor propagation. Cancer Cell 35, 81–94 e87 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Popp MW, Maquat LE, Nonsense-mediated mRNA decay and cancer. Curr. Opin. Genet. Dev 48, 44–50 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomecki R et al. , Multiple myeloma-associated hDIS3 mutations cause perturbations in cellular RNA metabolism and suggest hDIS3 PIN domain as a potential drug target. Nucleic Acids Res. 42, 1270–1290 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu W et al. , Reactivation of nonsense-mediated mRNA decay protects against C9orf72 dipeptide-repeat neurotoxicity. Brain 142, 1349–1364 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang YJ et al. , Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science 363, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jonkhout N et al. , The RNA modification landscape in human disease. RNA 23, 1754–1769 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pastor F et al. , An RNA toolbox for cancer immunotherapy. Nat. Rev. Drug Discov. 17, 751–767 (2018). [DOI] [PubMed] [Google Scholar]