Abstract
Metagenomics has altered our understanding of microbial diversity and ecology. This includes its applications to viruses in marine environments that have demonstrated their enormous diversity. Within these are RNA viruses, many of which share genetic features with members of the order Picornavirales; yet, very few of these have been taxonomically classified. The only recognized family of marine RNA viruses is the Marnaviridae, which was founded based on discovery and characterization of the species Heterosigma akashiwo RNA virus. Two additional genera of marine RNA viruses, Labyrnavirus (one species) and Bacillarnavirus (three species), were subsequently defined within the order Picornavirales but not assigned to a family. We have defined a sequence-based framework for taxonomic classification of twenty marine RNA viruses into the family Marnaviridae. Using RNA-dependent RNA polymerase (RdRp) phylogeny and distance-based analyses, we assigned the genera Labyrnavirus and Bacillarnavirus to the family Marnaviridae and created four additional genera in the family: Locarnavirus (four species), Kusarnavirus (one species), Salisharnavirus (four species) and Sogarnavirus (six species). We used pairwise capsid protein comparisons to delineate species within families, with 75 per cent identity as the species demarcation threshold. The family displays high sequence diversities and Jukes–Cantor distances for both the RdRp and capsid genes, suggesting that the classified viruses are not representative of all of the virus diversity within the family and that there are many more extant taxa. Our proposed taxonomic framework provides a sound classification system for this group of viruses that will have broadly applicable principles for other viral groups. It is based on sequence data alone and provides a robust taxonomic framework to include viruses discovered via metagenomic studies, thereby greatly expanding the realm of viruses subject to taxonomic classification.
Keywords: Picornavirales, Marnaviridae, taxonomy, metagenomics, RNA viruses, classification
1. Introduction
The isolation and subsequent genomic analysis of an RNA virus infecting the single-celled eukaryotic marine alga, Heterosigma akashiwo, altered our view of marine viruses due to its clear evolutionary relationship to viruses infecting higher plants and animals (Tai et al.2003; Lang, Culley, and Suttle 2004). Moreover, there is evidence that RNA viruses constitute approximately 50 per cent of marine virus assemblages (Steward et al. 2013), and play an important ecological role through their infection of marine eukaryotic phytoplankton (Brussaard 2004; Lawrence and Suttle 2004; Nagasaki et al. 2004; Tomaru et al. 2009, 2012, 2013; Gustavsen et al. 2014; Miranda et al. 2016). To date, there are ten described RNA virus species represented by isolates that infect marine single-celled eukaryotes (Tai et al. 2003; Lang, Culley, and Suttle 2004; Brussaard et al. 2004; Nagasaki et al. 2004; Tomaru et al. 2004; Shirai et al. 2008; Tomaru et al. 2009, 2012, 2013; Kimura and Tomaru 2015). Eight of these (Heterosigma akashiwo RNA virus [HaRNAV], Aurantiochytrium single-stranded RNA virus, Astarnavirus, Chaetoceros tenuissiumus RNA virus 01, Rhizosolenia setigera RNA virus [RsRNAV], Chaetoceros socialis f. radians RNA virus 01 [CsfrRNAV], Chaetenuissarnavirus II, and Chaetarnavirus 2) have clear affiliations with the order Picornavirales, while two (Heterocapsa circularisquama RNA virus, family Alvernaviridae, and Micromonas pusilla reovirus, family Reoviridae) are divergent and fall outside the Picornavirales.
Culture independent studies indicated that RNA viruses are abundant and genetically diverse in the ocean (Culley, Lang, and Suttle 2003; Culley and Steward 2007; Culley et al. 2014; Gustavsen et al. 2014; Miranda et al. 2016), and metagenomics revealed the existence of multiple picorna-like viruses that are more similar to the above-mentioned marine RNA virus isolates than to other known viruses (Culley, Lang, and Suttle 2007; Culley et al. 2014; Miranda et al. 2016; Shi et al. 2016). Therefore, RNA viruses are now considered to be important members of marine virus assemblages.
The order Picornavirales is comprised of positive-sense single-stranded RNA (+ssRNA) viruses in the families Picornaviridae, Dicistroviridae, Iflaviridae, Secoviridae, Marnaviridae (Sanfaçon et al. 2009, 2011a), and the newly defined Polycipiviridae (King et al. 2018; Olendraite et al. 2018). Marnaviridae existed for many years with Marnavirus as its only genus and HaRNAV its only species. Two additional genera of marine RNA viruses (Labyrnavirus and Bacillarnavirus) were also classified within the order Picornavirales but were unassigned to a family.
All known genomes of Picornavirales members encode proteins with helicase, 3C-like protease, and RNA-dependent RNA polymerase (RdRp) domains, as well as capsid proteins with related structures, although the genome organizations can differ among viruses (Le Gall et al. 2008). The viral proteins are expressed as one or more polyprotein(s) that are cleaved into individual functional proteins and the viruses can vary in their number of genome segments and open reading frames (ORFs) (Le Gall et al. 2008). HaRNAV has an 8.6-kb polyadenylated genome with a single 7.7-kb ORF encoding one polyprotein (Lang, Culley, and Suttle 2004). Members of the genera Labyrnavirus and Bacillarnavirus share sequence similarities with HaRNAV, although their genomes have a dicistronic organization (Nagasaki et al. 2004; Takao et al. 2006; Shirai et al. 2008; Tomaru et al. 2009) similar to viruses belonging to the Dicistroviridae (Chen et al. 2011a).
Virus classification by the International Committee on Taxonomy of Viruses (ICTV) is polythetic, and has traditionally relied on biological information such as host range, replication cycle, virus particle structure and properties, and serology, as well as sequence similarity (Simmonds et al. 2017). Although biological information is absent for viruses identified through metagenomic studies, given their vast numbers and diversity, it is important to also incorporate these viruses within the established taxonomic framework. This argument was recently formalized (Simmonds et al. 2017) and the Genomoviridae, a family of single-stranded DNA (ssDNA) viruses, was the first to include viruses discovered using metagenomics into a taxonomic framework (Varsani and Krupovic 2017). This approach can be applied to other viral groups.
Here, we present a sequence-based taxonomic framework for the family Marnaviridae that has been successfully applied to restructure this family (Walker et al. 2019). We analyzed the amino acid sequences of the capsid proteins and the RdRp domains of eight isolates and twelve metagenomically assembled genomes (Table 1) of +ssRNA viruses affiliated with the order Picornavirales. Phylogeny, pairwise identity and domain analyses yielded a framework for classifying these viruses in seven genera within the Marnaviridae; this taxonomic framework was further validated by classifying 187 related metagenomic +ssRNA viruses and therefore can be applied to novel members of the family discovered in the future. This has revealed the enormous diversity of viruses within the Marnaviridae and enables their appropriate classification, which is important to further define their roles in marine environments.
Table 1.
Summary of genome and isolation information for twenty viruses classified within the family Marnaviridae.
| Genus | Species | Virus name | Abbreviation | Accession |
Genome |
Source | Country | References | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino acid | Size (nt) | No. of ORFs | |||||||
| Marnavirus | Heterosigma akashiwo RNA virus a | Heterosigma akashiwo RNA virus | HaRNAV | AY337486 | AAP97137 | 8587 | One | Heterosigma akashiwo | Canada | Lang et al. (2004) and Tai et al. (2003) |
| Labyrnavirus | Aurantiochytrium single-stranded RNA virus a | Aurantiochytrium single-stranded RNA virus | AuRNAV | AB193726 | 9035 | Threeb | Aurantiochytrium sp. | Japan | Takao et al. (2006) | |
| Locarnavirus | Jericarnavirus B c | Marine RNA virus JP-B | JP-B | EF198242 | 8926 | Two | Coastal marine | Canada | Culley, Lang, and Suttle (2007) | |
| Sanfarnavirus 2 | Marine RNA virus SF-2 | SF-2 | KF412901 | 9321 | Two | Coastal wastewater | USA | Greninger and DeRisi (2015) | ||
| Sanfarnavirus 1 | Marine RNA virus SF-1 | SF-1 | JN661160 | 8970 | Two | Coastal wastewater | USA | Greninger and DeRisi (2015) | ||
| Sanfarnavirus 3 | Marine RNA virus SF-3 | SF-3 | KF478836 | AHA44480 | 8648 | One | Coastal wastewater | USA | Greninger and DeRisi (2015) | |
| Kusarnavirus | Astarnavirus c | Asterionellopsis glacialis RNA virus | AglaRNAV | AB973945 | 8842 | Two | Asterionellopsis glacialis | Japan | Tomaru et al. (2012) | |
| Bacillarnavirus | Chaetoceros tenuissimus RNA virus 01 | Chaetoceros tenuissimus RNA virus 01 | CtenRNAV01 | AB375474 | 9431 | Two | Chaetoceros tenuissimus | Japan | Shirai et al. (2008 | |
| Rhizosolenia setigera RNA virus a | Rhizosolenia setigera RNA virusa | RsRNAV | AB243297 | 8877 | Two | Rhizosolenia setigera | Japan | Nagasaki et al. (2004 | ||
| Chaetoceros socialis f. radians RNA virus 01 | Chaetoceros socialis f. radians RNA virus 01 | CsfrRNAV | AB469874 | 9467 | Two | Chaetoceros socialis f. radians | Japan | Tomaru et al. (2009 | ||
| Salisharnavirus | Britarnavirus 4 c | Marine RNA virus BC-4 | BC-4 | MH171300 | 8593 | Two | Coastal/oceanic marine | Canada | Vlok et al. (unpublished) | |
| Palmarnavirus 473 | Marine RNA virus PAL473 | PAL473 | KT727026 | 6360 | Two | Coastal marine | USA | Miranda et al. (2016) | ||
| Britarnavirus 1 | Marine RNA virus BC-1 | BC-1 | MG584187 | 8638 | Two | Coastal marine | Canada | Vlok et al. (2019) | ||
| Palmarnavirus 128 | Marine RNA virus PAL128 | PAL128 | KT727023 | 8660 | Two | Coastal marine | USA | Miranda et al. (2016 | ||
| Sogarnavirus | Britarnavirus 2 | Marine RNA virus BC-2 | BC-2 | MG584188 | 8843 | Two | Coastal marine | Canada | Vlok et al. (2019) | |
| Palmarnavirus 156 | Marine RNA virus PAL156 | PAL156 | KT727024 | 7897 | Two | Coastal marine | USA | Miranda et al. (2016) | ||
| Britarnavirus 3 | Marine RNA virus BC-3 | BC-3 | MG584189 | 8496 | Two | Coastal marine | Canada | Vlok et al. (2019) | ||
| Jericarnavirus A | Marine RNA virus JP-A | JP-A | EF198241 | 9236 | Two | Coastal marine | Canada | Culley, Lang, and Suttle (2007) | ||
| Chaetenuissarnavirus II c | Chaetoceros tenuissimus RNA virus type IIc | CtenRNAVII | AB971661 | 9562 | Two | Chaetoceros tenuissimus | Japan | Kimura and Tomaru (2015) | ||
| Chaetarnavirus 2 | Chaetoceros species RNA virus 02 | CspRNAV2 | AB639040 | 9417 | Two | Chaetoceros sp. | Japan | Tomaru et al. (2013) | ||
Assigned type species in existing genera.
AuRNAV has a third small overlapping ORF that produces a sub-genomic RNA. While we cannot exclude the existence of similar ORFs in other genomes, none has been experimentally demonstrated to encode protein.
Type species in new genera.
2. Materials and Methods
2.1 Origin of metagenomic and isolate sequences
Genome sequences were downloaded from the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) using the accession numbers in Table 1. Methods pertaining to the isolation and sequencing of both metagenome-assembled and isolate genomes are described in the original papers (Table 1). Marine RNA virus BC-4 is the only unpublished genome (Vlok, Short. and Suttle, unpublished), but it was obtained using the methods described in Vlok, Lang, and Suttle (2019). One hundred and eighty-seven additional new viral genomes assembled from aquatic metagenomic and meta-transcriptomic datasets (López-Bueno et al. 2015; Shi et al. 2016; Lachnit, Thomas, and Steinberg 2016; Moniruzzaman et al. 2017) were downloaded from GenBank using the accession numbers in Supplementary Tables S1 and S2.
2.2 Phylogenetic analysis
All sequences were either aligned or added to existing alignments using MUSCLE v3.8.425 with the default parameters (Edgar 2004), and then manually refined with Aliview version 1.17.1 (Larsson 2014). To corroborate amino acid model selection, both the Smart Model Selection in PhyML (Lefort, Longueville, and Gascuel 2017) and ProtTest 3.2 (Darriba et al. 2011) as part of the Phylemon2 package (Sánchez et al. 2011) were used. Maximum-likelihood trees were constructed with PhyML 3.0 (Guindon et al. 2010) and the LG+I + G+F amino acid model, and branch support was evaluated with the Shimodaira–Hasegawa approximate-likelihood ratio test. The resultant trees were edited in iTOL v3 (Letunic and Bork 2016).
RdRp protein sequences of Picornavirales type members, spanning the eight conserved domains (Koonin, Dolja, and Morris 1993), were aligned as described earlier. Capsid protein sequences were aligned using domains as identified by HMMER 1.9 (https://www.ebi.ac.uk/Tools/hmmer/) using the Pfam database. N- and C-termini were trimmed as they have lower similarity and do not align well. GenBank accession numbers of relevant sequences are provided in Table 1 and Supplementary Tables S1 and S2.
2.3 Pairwise similarities and diversity metrics
Relevant alignments were analyzed for Jukes–Cantor distances and sequence identities using the CLC genomics workbench v7.5 (CLCBio). Mean distances within and among genera, as well as the number of amino acid differences per site from mean diversity calculations (amino acid sequence diversity) for each population or genus, were calculated for the capsid and RdRp sequences using the p-distance method in MEGA7 (Kumar, Stecher, and Tamura 2016).
2.4 Domain analysis
The RdRp-conserved domains were identified as per Koonin, Dolja, and Morris (1993) and illustrated using WebLogo3 (Crooks et al. 2004).
3. Results
3.1 Family and genera classifications
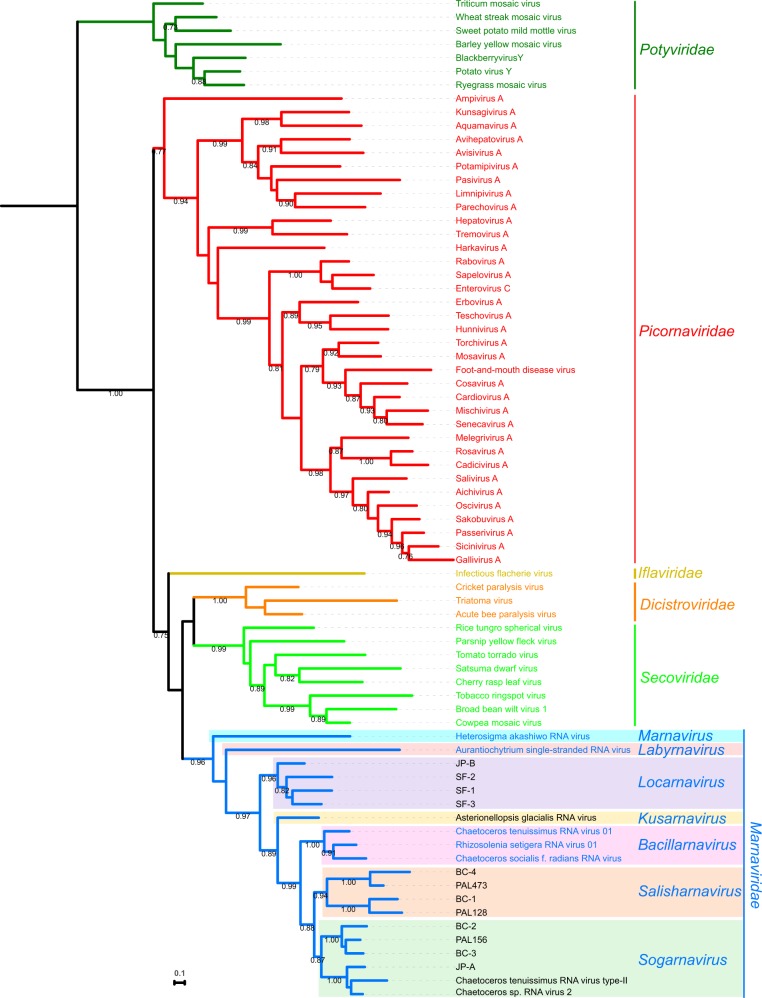
Maximum-likelihood phylogenetic analysis of the RdRp domain sequences placed the 20 marine RNA virus sequences in a strongly supported monophyletic group relative to other Picornavirales sequences (Fig. 1). HaRNAV and the Aurantiochytrium viruses were most distant (Supplementary Figs S1–S3) and basal within the clade (Fig. 1). Within this Marnaviridae grouping, mean RdRp amino acid sequence diversity was 59 per cent.
Figure 1.
Maximum-likelihood phylogeny of the Picornavirales RdRp amino acid sequences. The tree was rooted with sequences from the Potyviridae. Branches are coloured based on virus families: Potyviridae (dark green), Picornaviridae (red), Iflaviridae (yellow), Dicistroviridae (orange), Secoviridae (lime green), and Marnaviridae (blue). Genera within the Marnaviridae are indicated by coloured boxes. SH-like branch support values are indicated at the nodes when >0.70 and the maximum-likelihood scale bar indicates average residue substitution per site.
The analyses placed the twenty viruses into seven clades that we defined as genera within the Marnaviridae, including the previously classified genus Marnavirus and unassigned genera Bacillarnavirus and Labyrnavirus. The new genera were named Kusarnavirus, Locarnavirus, Salisharnavirus, and Sogarnavirus; as elaborated in Section 4, the names comprise prefixes to ‘rnavirus’ representing the geographical areas where the ‘type’ viruses were found. Mean distances between RdRp sequences ranged from 53 per cent to 54 per cent for Locarnavirus and Kusarnavirus, and Bacillarnavirus and Sogarnavirus, respectively, to 75 per cent for Marnavirus and Labyrnavirus (Supplementary Table S3).
Capsid amino acid sequence analysis corroborated the findings based on the RdRP sequences. Within the Marnaviridae, the capsid sequence diversity was 69 per cent with mean distances ranging from 58 per cent for Kusarnavirus and Sogarnavirus to 76 per cent between Marnaviridae, Labyrnavirus, and Locarnavirus. The seven genera were also supported by the capsid phylogeny with the exception of Sogarnavirus, which was split into two distinct groups (Fig. 2 and Supplementary Fig. S4).
Figure 2.
Comparative RdRp and capsid maximum-likelihood phylogenies of representative members of the family Marnaviridae. Branches are coloured based on virus genera: Marnavirus (blue), Labyrnavirus (red), Locarnavirus (purple), Kusarnavirus (yellow), Bacillarnavirus (pink), Sogarnavirus (green), and Salisharnavirus (orange). SH-like branch support values are shown at the nodes when >0.70 and the maximum-likelihood scale bars indicate average residue substitutions per site.
Amino acid diversity was not directly related to the number of genomes analyzed (Fig. 3 and Supplementary Fig. S5). The mean diversity for the capsid ORFs for each genus was higher than for the RdRp but was not proportional to the RdRp diversity. For example, the genus Bacillarnavirus had the highest capsid (66%) and lowest RdRp (37%) diversities, while the genus Salisharnavirus had the highest RdRp diversity (51%) with the second-lowest capsid diversity (60%). On average, the amino acid diversities, or genetic variation, of the Marnaviridae sit in the middle of the scale of 0–1.
Figure 3.
Summary of within-genus details for genera within the Marnaviridae that contain multiple viruses. The numbers of viruses are depicted as black circles (●) while diversities are represented as bars: capsid (light grey) and RdRp (dark grey).
3.1.1 Marnavirus
The only genus originally classified within the Marnaviridae was Marnavirus, which consisted of one species and representative virus, HaRNAV (Table 1). In our analyses, it was the most divergent taxon in the family, sharing 22.4–30.3 per cent and 18.5–26.4 per cent amino acid sequence identity for the RdRp and capsid sequences, respectively, with viruses in the other genera (Supplementary Figs. S1 and S2). It was the most deeply branching taxon in the family in both phylogenetic analyses.
3.1.2 Labyrnavirus
This previously unassigned genus contains one representative, AuRNAV (Table 1). At the amino acid level, it shared 22.4–33.6 per cent and 17.1–22.5 per cent identities with RdRp and capsid sequences, respectively, from other genera (Supplementary Figs. S1 and S2). It was a deep-branching taxon in both phylogenetic analyses.
3.1.3 Locarnavirus
Four viruses, JP-B, SF-1, SF-2, and SF-3, originating from metagenomic data (Table 1), grouped within the genus Locarnavirus. The RdRp and capsid diversities among these four viruses were 44 per cent and 63 per cent, respectively (Fig. 3). The members of this genus shared 52.4–59.1 per cent and 32.2–38.3 per cent identities for the RdRp and capsid amino acid sequences, respectively, with one another. Members of this genus shared 24.0–48.2 per cent and 17.2–27.4 per cent identities for the RdRp and capsid amino acid sequences, respectively, with other members of the family (Supplementary Figs. S1 and S2). The type species is defined by Marine RNA virus JP-B (species name Jericarnavirus B).
3.1.4 Kusarnavirus
The previously unclassified virus isolate AglaRNAV, which infects the pennate diatom Asterionellopsis glacialis, is the only member of this genus and therefore defines the type species (Table 1; species name Astarnavirus). It shared 25.4–48.2 per cent and 21.7–47.9 per cent identities for the RdRp and capsid amino acid sequences, respectively, with viruses from other Marnaviridae genera. The capsid sequence shared the highest amino acid identity (44.3–47.9%) with members of the genus Sogarnavirus (Supplementary Figs. S1 and S2).
3.1.5 Bacillarnavirus
This previously unassigned genus contains three isolates, Chaetoceros tenuissimus RNA virus 01 (CtenRNAV01), RsRNAV, and CsfrRNAV (Table 1), all of which infect centric diatoms. The RdRp and capsid diversities among these three viruses were 38 per cent and 66 per cent, respectively (Fig. 3). The members of this genus shared 57.8–64.4 per cent and 29.1–33.4 per cent identities for the RdRp and capsid amino acid sequences, respectively. Members of this genus shared 23.8–47.8 per cent and 18.3–29.4 per cent identities for the RdRp and capsid amino acid sequences, respectively, with other members of the family (Supplementary Figs. S1 and S2). The type species is defined by RsRNAV (species name Rhizosolenia setigera RNA virus).
3.1.6 Salisharnavirus
This genus includes four viruses, BC-4, PAL473, BC-3, and PAL128 (Table 1), which were all discovered through metagenomics. The RdRp and capsid diversities among these three viruses were 51 per cent and 60 per cent, respectively (Fig. 3). Within this genus, members shared 35.6–63.6 per cent and 24.9–34.4 per cent identities for the RdRp and capsid amino acid sequences, respectively. Amino acid identities between members of this genus and other members of the Marnaviridae ranged from 24.3 to 47.2 per cent for the RdRp and 18.2 to 31.5 per cent for the capsid (Supplementary Figs. S1 and S2). The type species is defined by Marine RNA virus BC-4 (species name Britarnavirus 4).
3.1.7 Sogarnavirus
This was the largest genus and includes six new species, with four derived from metagenomic studies and two isolates (Table 1). The RdRp and capsid diversities among these viruses were 38 per cent and 56 per cent, respectively (Fig. 3). The members of this genus shared 48.8–77.0 per cent and 27.6–59.0 per cent identities for the RdRp and capsid amino acid sequences, respectively, with one another, and 24.3–47.2 per cent and 17.1–47.9 per cent identities for the RdRp and capsid amino acid sequences, respectively, with other members of the family (Supplementary Figs. S1 and S2). The type species is defined by CtenRNAV type-II (species name Chaetenuissarnavirus II).
3.2 Species demarcations
Pairwise comparisons of the RdRp and capsid amino acid sequences revealed a large range of identity values (Fig. 4 and Supplementary Figs. S1 and S2). Most of the capsid sequences analyzed shared 22–30 per cent pairwise amino acid identities compared to 26–46 per cent for the RdRp sequences. The highest pairwise identity scores were 59 per cent and 77 per cent for the capsid and RdRp, respectively. Based on the pairwise scores (Fig. 4) we suggest conservative species demarcation cut-offs of 75 per cent for the capsid and 90 per cent for the RdRp.
Figure 4.
Distribution of pairwise amino acid sequence identities for Marnaviridae members. Identities calculated for the entire capsid polyprotein region are in grey (A) and for the RdRp domain in black (B) for the twenty viruses used to develop the classification system. Identities for the additional viruses used to test the system are in the bottom row with the capsid polyprotein region in grey (C) and RdRp in black (D). Dotted lines represent species demarcation cut-offs.
3.3 Applying the taxonomic framework to additional new viruses
An RdRp maximum-likelihood analysis with 187 new viral genomes, in addition to the 20 viruses used to define the seven genera within the Marnaviridae, revealed that the proposed genera encompassed the majority of this viral diversity (Fig. 5 and Supplementary Table S1). In addition to the seven proposed genera, two new supported clades were observed as well as nine viruses that did not have strong support within the tree. However, in this larger analysis, less congruence was observed for the RdRp and capsid phylogenies (Fig. 5 and Supplementary Fig. S6).
Figure 5.
Phylogenetic analysis of aquatic RNA virus RdRp sequences. The revised seven-genus Marnaviridae taxonomy encompasses most of the viral diversity that would potentially be assigned to the Marnaviridae based on RdRp sequence similarities. Colours denote genera within the Marnaviridae: Marnavirus (blue), Labyrnavirus (red), Locarnavirus (purple), Kusarnavirus (yellow), Bacillarnavirus (pink), Salisharnavirus (orange). and Sogarnavirus (green). The twenty viruses used to develop the classification system are in bold. The SH-like branch support values shown are from the Picornavirales RdRp maximum-likelihood phylogeny (Fig. 1) and the scale bar indicates average residue substitutions per site.
Frequency profiles of pairwise amino acid identities of the expanded dataset (Fig. 4C and D) were similar to those observed for the 20-virus dataset. The majority of the viruses analyzed were within the species demarcation cut-offs. Some genomes which were denoted as strains in one study (Shi et al. 2016) were also indicated as such by this taxonomic framework. Exceptions were strains of the viruses Wenzhou picorna-like virus 2 and Hubei picorna-like virus 7, which each had 100 per cent RdRp sequence identities but 69 per cent and 48 per cent capsid sequence identities, respectively. This is likely a result of recombination. For seven other virus pairs, identities of one of the two protein regions fell outside the species demarcation criteria (Supplementary Table S4).
Analysis of the conserved RdRp domains (I–VIII; Koonin, Dolja, and Morris 1993) showed high conservation of specific amino acids in the Marnaviridae, although genera were distinguished by unique amino acid motifs (Supplementary Fig. S7). Domains I, IV, VI, and VII showed the highest degree of conservation within genera and at the family level (Supplementary Figs. S7 and S8).
4. Discussion
Sequence analysis of previously unclassified marine virus isolates and twelve viruses described from metagenomic studies, as well as members of the previously unassigned genera Labyrnavirus and Bacillarnavirus, and the genus Marnavirus in the family Marnaviridae, allow us to greatly expand classification of viruses within this family. Arguments supporting this expansion are elaborated in the following sections.
4.1 Expanding the family Marnaviridae to encompass seven genera
The RdRp phylogeny for the viruses analyzed in this study shows the 20 marine viruses form a well-supported monophyletic group (Fig. 1). These viruses are more similar to each other than to other members of the order Picornavirales (Supplementary Figs. S3 and S8). Because of this relationship, and the basal location of the originally classified representative, Marnavirus, we argued that the 20 analyzed viruses (Table 1) belong within the family Marnaviridae, and this has been ratified by the ICTV (Walker et al. 2019).
The RdRp phylogeny and measurements of sequence identities define seven genera in the family (Fig. 1). Polymerases are sufficiently conserved at the amino acid level that similarities can be used to establish viral classification at the genus level, as has been demonstrated for multiple virus families (Baker and Schroeder 2008; Nibert et al. 2014; Varsani and Krupovic 2017). The genera newly assigned to the Marnaviridae were selected based primarily on robust phylogenetic relationships. Expanding the analysis to include an additional 187 viruses validate the seven genera as representative of most of the currently known Marnaviridae diversity (Fig. 5 and Supplementary Table S1). While all genera share some conserved RdRp amino acid motifs (Supplementary Fig. S7), such as the catalytic GDD (Wang and Gillam 2001; Kok and McMinn 2009), genus-specific motifs are also present. Amino acid substitutions that are chemically different suggest that these positions may not be required for the catalytic function of the protein but are useful for this level of sequence classification.
Genome organization is useful for comparing virus groups but is not a sufficient marker for either family- or genus-level demarcations. While the majority of genomes analyzed here have a dicistronic organization (Table 1), HaRNAV and SF-3 have a single predicted polyprotein encoded by their genomes. HaRNAV is the only representative of the genus Marnavirus, but SF-3 is one of four viruses placed within the genus Locarnavirus. While the remaining isolates and most of the viruses identified in metagenomes considered here (Table 1) have dicistronic genomes, other monocistronic genomes that fall within the Marnaviridae as defined here are known for viruses from the coastal waters of Hawaii (Culley et al. 2014). Sequences for these viruses were unavailable and are not included in our analysis. Furthermore, the Secoviridae, another family within the order Picornavirales, contains members with both mono- and bipartite genomes. Similar to our Marnaviridae findings, the Secoviridae form a monophyletic clade based on RdRp sequences; thus, genome organization is not a robust criterion for family demarcation within the Picornavirales. We also note that AuRNAV, genus Labyrnavirus, contains a third annotated ORF that is transcribed as a sub-genomic RNA (Takao et al. 2006).
With the exception of the sogarnaviruses, all multi-member genera within the Marnaviridae form independent clades based on capsid phylogeny (Fig. 2 and Supplementary Fig. S4); however, there are incongruences between the capsid and RdRp phylogenies (Fig. 2). Some of this may be due to recombination within the family. While it is unlikely that recombination occurs within the polymerase domain (Greenspan et al. 2004), recombination has been observed in the genomes of other members of the Picornavirales (Simmonds 2006; Moore et al. 2011; Elbeaino et al. 2012). Recombination would also explain the similarity between the capsid proteins of AglaRNAV and BC-2, as well as BC-3 and Pal156, but not the rest of the genus Sogarnavirus (Supplementary Figs. S1 and S4). The structural and non-structural proteins could also have different evolutionary trajectories that are not evident from the phylogenies. Regardless, the presence of two groups of sogarnaviruses suggests a sub-genus classification may be warranted, which may become clearer with the discovery and addition of new species.
4.2 Derivation of genus names
The names of the three established genera are acronyms based on a prefix reflecting the host of the ‘type’ virus (e.g. Labyrnavirus and Bacillarnavirus) or its origin (e.g. Marnavirus), added to ‘rnavirus’ to reflect the genome type. Labyrnavirus and Bacillarnavirus refer to the protist classes Labyrinthulomycetes and Bacillariophyceae (diatoms), which are hosts for the type viruses, while Marnavirus is derived from the Latin for sea, Mare. We kept to this general scheme of including ‘rnavirus’ for consistency in naming for the new genera. Locarnavirus is based on Locarno Beach, where Jericho Pier is located and the first marine RNA virus metagenome and type virus (JP-B) for the genus originated (Culley, Lang, and Suttle 2003); JP-B is also basal to the clade. Salisharnavirus is derived from ‘Salish Sea’, the water mass around coastal southern British Columbia from which the first marine RNA virus metagenomes were assembled, including the type virus BC-1. ‘Sog in Sogarnavirus is an acronym for the Strait of Georgia, a major water body within the Salish Sea that includes the area from which the first marine RNA viromes were assembled (Culley, Lang, and Suttle 2003, 2007). Chaetoceros tenuissimus RNA virus type-II (CtenRNAVII) was selected as the type Sogarnavirus, because it is the better studied of the two isolates in the genus. Kusarnavirus is derived from the Afrikaans word for coast, kus, and the only current member of the genus, Asterionellopsis glacialis RNA virus (AglaRNAV) that infects a coastal diatom host, by default defines the type species. Rhizosolenia setigera RNA virus 01 (CtenRNAV01) is already established as the type species of the genus Bacillarnavirus.
4.3 Classifying new species
The amino acid divergence of the capsid sequences compared to that for RdRp makes capsid sequence useful for species demarcation (Fig. 4 and Supplementary Figs. S1 and S2). Functional differences likely cause the differences between the two regions, as capsid proteins are involved in host recognition and co-evolve with cellular receptors (Nagasaki et al. 2005; Tully and Fares 2006). This may partly explain the lack of perfect congruence between the two phylogenies (Fig. 2). Thus, the percent amino acid identities for both the RdRp and capsid proteins were considered for species demarcation.
For the capsid polyprotein, we suggest a conservative cut-off of 75 per cent pairwise amino acid identity for species demarcation. This is 15 per cent higher than the lowest observed inter-species identity (59–60% bin) (Fig. 4). The large genetic distances among the proposed genera and the high estimates of diversity suggest that other genera remain to be discovered within the Marnaviridae, and this is also supported by our expanded analysis of additional viruses (Fig. 5), which need revisited in the future for formal taxonomic assignments. Furthermore, a 75-per cent capsid protein sequence identity for species demarcation is in accordance with the parameters required for species classification in the Secoviridae (Sanfaçon et al. 2009, 2011b), although less stringent than the 90 per cent cut-off for species in the families Iflaviridae and Dicistroviridae (Chen et al. 2011a,b).
The second parameter we considered in species demarcation is the RdRp amino acid identity. The employed cut-off for RdRp amino acid identity is 90 per cent, which is higher than the 77–78 per cent bin that functionally defined the species in our analyses. The family Secoviridae employs an 80-per cent identity cut-off for the protease-polymerase region (Sanfaçon et al. 2009, 2011b). We were not able to employ the protease region for the Marnaviridae as it is too divergent and is difficult to identify with confidence. This might reflect greater evolutionary distance among Marnaviridae hosts compared with plants.
4.4 Testing the classification system on other metagenomic viruses
A subset of the available potential Marnaviridae genome sequences was used to establish this taxonomic classification. The genomes used to test our taxonomic definitions had the potential to be very different, because they were from freshwater (López-Bueno et al. 2015), holobiont (Lachnit, Thomas, and Steinberg 2016) and (meta-)transcriptome (Shi et al. 2016; Moniruzzaman et al. 2017) studies. An important issue with transcriptome data from metazoans and phagotrophic protists is that for RNA virus genomes, particularly those with poly-A tails that get captured during poly-A selection, it is not possible to determine whether the viruses are actively replicating within the study organism or a microbial symbiont, are present inside food, or are from the surrounding environment. To avoid these possible complications, we limited our initial analysis to viruses from culture systems or identified by metagenomics with free virus particles in aquatic environments.
Most of the additional genomes analyzed could be placed within the proposed genera based on RdRp amino acid sequences. The potential expansion of the genera this revealed may require the subsequent definition of subgenera, especially for the salisha- and sogarnaviruses, where distinct, deeper-branching clades are present (Fig. 5).
Recombination can complicate species demarcation. Because of this, it is important to take both the capsid and RdRp into account when classifying new species. This is evident from the expanded dataset results, where seven virus pairs shared high percent identities for one of the two proteins while the other shared much lower identities (Supplementary Table S4). Such viruses would have to be flagged as potential recombinants and require special consideration.
5. Conclusions
In this study, a sequence-based approach was used to expand the family Marnaviridae to include both viruses discovered through culturing and metagenomic studies. This places two previously unclassified genera, Labyrnavirus and Bacillarnavirus, as well as four new genera within the Marnaviridae. Because of differences in the levels of sequence conservation for the RdRp and capsid proteins, we propose the RdRp be used for genus-level classification, while the capsid and RdRp be used for species demarcation. A previous analysis of a ssDNA virus family employed an approach that deviates from classic taxonomic methods (Varsani and Krupovic 2017), and we have now employed a similar non-classical method to a group of +ssRNA viruses. Although this approach does not take into account any biological properties such as pathology or host range, it does provide a means for creating structure and evaluating relationships within a taxonomic framework for the large pool of new viruses that are being discovered through metagenomic analyses.
Supplementary Material
Acknowledgements
We thank Hélène Sanfaçon (University of British Columbia and Agriculture and Agri-Food Canada) and Valerian Dolja (Oregon State University), who provided feedback on the manuscript, Amy M. Chan, Emma Shelford, Jan Finke, and Julia Gustavsen, for enabling the work through advice, logistics and sample collection, and the crews of the CCGS Vector.
Funding
This project was supported by funding to C.A.S. from the Natural Science and Engineering Research Council of Canada Discovery (RGPIN-2015-05896) and Ship-time Grants, the Canada Foundation for Innovation, and the British Columbia Knowledge Development Fund. A.S.L. was supported by a salary-based research grant from Memorial University of Newfoundland.
Conflict of interest: None declared.
References
- Baker A. C., Schroeder D. C. (2008) ‘The Use of RNA-Dependent RNA Polymerase for the Taxonomic Assignment of Picorna-Like Viruses (Order Picornavirales) Infecting Apis mellifera L. populations’, Virology Journal, 5: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussaard C. (2004) ‘Viral Control of Phytoplankton Populatons - A Review’, The Journal of Eukaryotic Microbiology, 51: 125–38. [DOI] [PubMed] [Google Scholar]
- Brussaard C. P. D. et al. (2004) ‘Discovery of a dsRNA Virus Infecting the Marine Photosynthetic Protist Micromonas pusilla’, Virology, 319: 280–91. [DOI] [PubMed] [Google Scholar]
- Chen Y. P. et al. (2011a) ‘Dicistroviridae’, in King A. M. Q.et al. (eds.) Virus Taxonomy: 9th Report of the International Committee on the Taxonomy of Viruses, pp. 840–5. Elsevier Academic Press, San Diego, USA. [Google Scholar]
- Chen, Y. P. et al. (2011b) ‘Iflaviridae’, in King, A. M. Q. et al. (eds.) Virus taxonomy: 9th Report of the International Committee on the Taxonomy of Viruses, pp. 846–9. San Diego, USA: Elsevier Academic Press. [Google Scholar]
- Crooks G. E. et al. (2004) ‘WebLogo: A Sequence Logo Generator’, Genome Research, 14: 1188–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley A. I., Lang A. S., Suttle C. A. (2003) ‘High Diversity of Unknown Picorna-Like Viruses in the Sea’, Nature, 424: 1054–7. [DOI] [PubMed] [Google Scholar]
- Culley A. I., Lang A. S., Suttle C. A. (2007) ‘The Complete Genomes of Three Viruses Assembled from Shotgun Libraries of Marine RNA Virus Communities’, Virology Journal, 4: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley A. I. et al. (2014) ‘The Characterization of RNA Viruses in Tropical Seawater Using Targeted PCR and Metagenomics’, MBio, 5: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culley A. I., Steward G. F. (2007) ‘New Genera of RNA Viruses in Subtropical Seawater, Inferred from Polymerase Gene Sequences’, Applied and Environmental Microbiology, 73: 5937–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba D. et al. (2011) ‘ProtTest 3: fast Selection of Best-Fit Models of Protein Evolution’, Bioinformatics, 27: 1164–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. (2004) ‘MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput’, Nucleic Acids Research, 32: 1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbeaino T. et al. (2012) ‘Grapevine Deformation Virus: Completion of the Sequence and Evidence on Its Origin from Recombination Events between Grapevine fanleaf Virus and Arabis mosaic Virus’, Virus Research, 166: 136–40. [DOI] [PubMed] [Google Scholar]
- Le Gall O. et al. (2008) ‘Picornavirales, a Proposed Order of Positive-Sense Single-Stranded RNA Viruses with a pseudo-T = 3 Virion Architecture’, Archives of Virology, 153: 715–27. [DOI] [PubMed] [Google Scholar]
- Greenspan G. et al. (2004) ‘Recombination Does Not Occur in Newly Identified Diverged Oceanic Picornaviruses’, Journal of Molecular Evolution, 58: 359–60. [DOI] [PubMed] [Google Scholar]
- Greninger A. L., DeRisi J. L. (2015) ‘Draft Genome Sequences of Marine RNA Viruses SF-1, SF-2 and SF-3 Recovered from San Francisco Wastewater', Genome Announcements, 3: e00653–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S. et al. (2010) ‘New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0’, Systematic Biology, 59: 307–21. [DOI] [PubMed] [Google Scholar]
- Gustavsen J. A. et al. (2014) ‘High Temporal and Spatial Diversity in Marine RNA Viruses Implies That They Have an Important Role in Mortality and Structuring Plankton Communities’, Front Microbiol, 5: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Tomaru Y. (2015) ‘Discovery of Two Novel Viruses Expands the Diversity of Single-Stranded DNA and Single-Stranded RNA Viruses Infecting a Cosmopolitan Marine Diatom’, Applied and Environmental Microbiology, 81: 1120–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A. M. Q. et al. (2018) ‘Changes to Taxonomy and the International Code of Virus Classification and Nomenclature Ratified by the International Committee on Taxonomy of Viruses (2018)’, Archives of Virology, 163: 2601–31. [DOI] [PubMed] [Google Scholar]
- Kok C. C., McMinn P. C. (2009) ‘Picornavirus RNA-Dependent RNA Polymerase’, The International Journal of Biochemistry and Cell Biology, 41: 498–502. [DOI] [PubMed] [Google Scholar]
- Koonin E. V., Dolja V. V., Morris T. J. (1993) ‘Evolution and Taxonomy of Positive-Strand RNA Viruses: Implications of Comparative Analysis of Amino Acid Sequences’, Critical Reviews in Biochemistry and Molecular Biology, 28: 375–430. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Tamura K. (2016) ‘MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets’, Molecular Biology and Evolution, 33: 1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachnit T., Thomas T., Steinberg P. (2016) ‘Expanding Our Understanding of the Seaweed Holobiont: RNA Viruses of the Red Alga Delisea pulchra’, Frontiers in Microbiology, 6: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang A. S., Culley A. I., Suttle C. A. (2004) ‘Genome Sequence and Characterization of a Virus (HaRNAV) Related to Picorna-Like Viruses That Infects the Marine Toxic Bloom-Forming Alga Heterosigma akashiwo’, Virology, 320: 206–17. [DOI] [PubMed] [Google Scholar]
- Larsson A. (2014) ‘AliView: A Fast and Lightweight Alignment Viewer and Editor for Large Datasets’, Bioinformatics, 30: 3276–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J. E., Suttle C. A. (2004) ‘Effect of Viral Infection on Sinking Rates of Heterosigma akashiwo and Its Implications for Bloom Termination’, Aquatic Microbial Ecology, 37: 1–7. [Google Scholar]
- Lefort V., Longueville J.-E., Gascuel O. (2017) ‘SMS: Smart Model Selection in PhyML’, Molecular Biology and Evolution, 34: 2422–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I., Bork P. (2016) ‘Interactive Tree of Life (iTOL) v3: An Online Tool for the Display and Annotation of Phylogenetic and Other Trees’, Nucleic Acids Research, 44: W242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Bueno A. et al. (2015) ‘Ecological Connectivity Shapes Quasispecies Structure of RNA Viruses in an Antarctic Lake’, Molecular Ecology, 24: 4812–25. [DOI] [PubMed] [Google Scholar]
- Miranda J. A. et al. (2016) ‘RNA Viruses as Major Contributors to Antarctic Virioplankton’, Environmental Microbiology, 18: 3714–27. [DOI] [PubMed] [Google Scholar]
- Moniruzzaman M. et al. (2017) ‘Virus-Host Relationships of Marine Single-Celled Eukaryotes Resolved from Metatranscriptomics’, Nature Communications, 8: 16054–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. et al. (2011) ‘Recombinants between Deformed Wing Virus and Varroa Destructor Virus-1 May Prevail in Varroa Destructor-Infested Honeybee Colonies’, Journal of General Virology, 92: 156–61. [DOI] [PubMed] [Google Scholar]
- Nagasaki K. et al. (2004) ‘Isolation and Characterization of a Novel Single-Stranded RNA Virus Infecting the Bloom-Forming Diatom Rhizosolenia setigera’, Applied and Environmental Microbiology, 70: 704–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki K. et al. (2005) ‘Previously Unknown Virus Infects Marine Diatom’, Applied and Environmental Microbiology, 71: 3528–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibert M. L. et al. (2014) ‘Taxonomic Reorganization of Family Partitiviridae and Other Recent Progress in Partitivirus Research’, Virus Research, 188: 128–41. [DOI] [PubMed] [Google Scholar]
- Olendraite I. et al. (2018) ‘Polycipiviridae: A Proposed New Family of Polycistronic Picorna-Like RNA Viruses’, Journal of General Virology, 98: 2368–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez R. et al. (2011) ‘Phylemon 2.0: A Suite of Web-Tools for Molecular Evolution, Phylogenetics, Phylogenomics and Hypotheses Testing’, Nucleic Acids Research, 39: 470–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon H. et al. (2009) ‘Secoviridae: A Proposed Family of Plant Viruses within the Order Picornavirales That Combines the Families Sequiviridae and Comoviridae, the Unassigned Genera Cheravirus and Sadwavirus, and the Proposed Genus Torradoviru’, Archives of Virology, 154: 899–907. [DOI] [PubMed] [Google Scholar]
- Sanfaçon H. et al. (2011a) ‘Order Picornavirales’, in King A.et al. (eds.) Virus Taxonomy: 9th Report of the International Committee on the Taxonomy of Viruses, pp. 835–9. San Diego, USA: Elsevier Academic Press. [Google Scholar]
- Sanfaçon H. et al. (2011b) ‘Secoviridae’, in King A. M. Qet al. (eds.) Virus Taxonomy: 9th Report of the International Committee on the Taxonomy of Viruses, pp. 881–99. San Diego, USA: Elsevier Academic Press. [Google Scholar]
- Shi M. et al. (2016) ‘Redefining the Invertebrate RNA Virosphere’, Nature, 540: 539–43. [DOI] [PubMed] [Google Scholar]
- Shirai Y. et al. (2008) ‘Isolation and Characterization of a Single-Stranded RNA Virus Infecting the Marine Planktonic Diatom Chaetoceros tenuissimus Meunier’, Applied and Environmental Microbiology, 74: 4022–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. (2006) ‘Recombination and Selection in the Evolution of Picornaviruses and Other Mammalian Positive-Stranded RNA Viruses’, Journal of Virology, 80: 11124–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. et al. (2017) ‘Virus Taxonomy in the Age of Metagenomics’, Nature Reviews Microbiology, 15: 161–8. [DOI] [PubMed] [Google Scholar]
- Steward G. F. et al. (2013) ‘Are We Missing Half of the Viruses in the Ocean?’, The Isme Journal, 7: 672–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai V. et al. (2003) ‘Characterization of HaRNAV, a Single-Stranded RNA Virus Causing Lysis of Heterosigma akashiwo (Raphidophyceae)’, Journal of Phycology, 39: 343–52. [Google Scholar]
- Takao Y. et al. (2006) ‘Complete Nucleotide Sequence and Genome Organization of a Single-Stranded RNA Virus Infecting the Marine Fungoid Protist Schizochytrium sp’, Journal of General Virology, 87: 723–33. [DOI] [PubMed] [Google Scholar]
- Tomaru Y. et al. (2004) ‘Isolation and Characterization of Two Distinct Types of HcRNAV, a Single-Stranded RNA Virus Infecting the Bivalve-Killing Microalga Heterocapsa circularisquama’, Aquatic Microbial Ecology, 34: 207–18. [Google Scholar]
- Tomaru Y. et al. (2009) ‘Isolation and Characterization of a Single-Stranded RNA Virus Infecting the Bloom-Forming Diatom Chaetoceros socialis’, Applied and Environmental Microbiology, 75: 2375–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru Y. et al. (2012) ‘First Evidence for the Existence of Pennate Diatom Viruses’, The ISME Journal, 6: 1445–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaru Y. et al. (2013) ‘Isolation and Characterization of a Single-Stranded RNA Virus That Infects the Marine Planktonic Diatom Isolation and Characterization of a Single-Stranded RNA Virus That Infects the Marine Planktonic Diatom Chaetoceros sp. (SS08-C03)’, Phycological Research, 61: 27–36. [Google Scholar]
- Tully D. C., Fares M. A. (2006) ‘Unravelling Selection Shifts among Foot-and-Mouth Disease Virus (FMDV) Serotypes’, Evolutionary Bioinformatics, 2: 117693430600200–225. [PMC free article] [PubMed] [Google Scholar]
- Varsani A., Krupovic M. (2017) ‘Sequence-Based Taxonomic Framework for the Classification of Uncultured Single-Stranded DNA Viruses of the Family Genomoviridae’, Virus Evolution, 3: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlok M., Lang A. S., Suttle C. A. (2019) ‘Marine RNA Virus Quasispecies Are Distributed throughout the Oceans’, mSphere, 4: e00157–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker P. et al. (2019) ‘Changes to Virus Taxonomy and the International Code of Virus Classification and Nomenclature Ratified by the International Committee on Taxonomy of Viruses’, Archives of Virology, 164: 2417–2429. [DOI] [PubMed] [Google Scholar]
- Wang X., Gillam S. (2001) ‘Mutations in the GDD Motif of Rubella Virus Putative RNA-Dependent RNA Polymerase Affect Virus Replication’, Virology, 285: 322–31. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.