Abstract
Dense-core vesicles (DCVs) are secretory vesicles found in neurons and endocrine cells. DCVs package and release cargoes including neuropeptides, biogenic amines, and peptide hormones. We recently identified the endosome-associated recycling protein (EARP) complex and the EARP-interacting-protein EIPR-1 as proteins important for controlling levels of DCV cargoes in Caenorhabditis elegans neurons. Here we determine the role of mammalian EIPR1 in insulinoma cells. We find that in Eipr1 KO cells, there is reduced insulin secretion, and mature DCV cargoes such as insulin and carboxypeptidase E (CPE) accumulate near the trans-Golgi network and are not retained in mature DCVs in the cell periphery. In addition, we find that EIPR1 is required for the stability of the EARP complex subunits and for the localization of EARP and its association with membranes, but EIPR1 does not affect localization or function of the related Golgi-associated retrograde protein (GARP) complex. EARP is localized to two distinct compartments related to its function: an endosomal compartment and a DCV biogenesis-related compartment. We propose that EIPR1 functions with EARP to control both endocytic recycling and DCV maturation.
INTRODUCTION
Dense-core vesicles (DCVs) are regulated secretory vesicles found in neurons and endocrine cells, where they are also called secretory granules. DCVs package several types of cargo, including neuropeptides and peptide hormones, for release at the cell membrane (Gondré-Lewis et al., 2012). The secreted cargoes modulate a variety of processes including development, growth, glucose metabolism, and mental state. DCVs are generated at the trans-Golgi network (TGN) in a process that includes correct sorting of cargo and acquisition of proper compartmental identity. Because DCVs are not regenerated locally at release sites, the DCV pool needs to be continuously supplied by the TGN.
Genetic studies in the nematode Caenorhabditis elegans have identified several molecules that function in DCV biogenesis, including the endosome-associated recycling protein (EARP) complex and the EARP-interacting-protein EIPR-1, a WD40 domain protein that interacts with EARP (Edwards et al., 2009; Sumakovic et al., 2009; Mesa et al., 2011; Yu, Wang, Jiu, et al., 2011; Hannemann et al., 2012; Ailion et al., 2014; Topalidou et al., 2016). The EARP complex is structurally similar to the Golgi-associated retrograde protein (GARP) complex. EARP shares the VPS51, VPS52, and VPS53 subunits with the GARP complex, but uses VPS50 instead of VPS54 as the fourth subunit (Gillingham et al., 2014; Schindler, Chen, et al., 2015). Whereas GARP functions in retrograde trafficking from endosomes to the TGN (Conibear and Stevens, 2000; Conibear et al., 2003; Pérez-Victoria et al., 2008, 2010; Pérez-Victoria and Bonifacino, 2009), EARP was shown to act in recycling cargoes from endosomes back to the plasma membrane (Schindler, Chen, et al., 2015). In C. elegans, the EARP complex and EIPR-1 were shown to be required for controlling levels of DCV cargoes (Topalidou et al., 2016). The VPS50 subunit of the EARP complex was also shown to be required for the maturation of DCV cargoes and DCV acidification (Paquin et al., 2016).
EIPR1 physically interacts with the EARP complex in rat insulinoma cells (Topalidou et al., 2016). EIPR1 (also named TSSC1) was also independently identified as a physical interactor and functional partner of both the GARP and EARP complexes in human cell lines (Gershlick et al., 2016). Moreover, two mass spectrometry interactome data sets identified EIPR1 as an interactor of EARP subunits in human HEK293T and HeLa cells (Hein, Hubner, et al., 2015; Huttlin et al., 2015). WD40 domain proteins like EIPR1 often act as scaffolds for the assembly of protein complexes (Stirnimann, Petsalaki, et al., 2010). Though EIPR1 interacts with EARP, it has not been determined whether EIPR1 is required for the localization or stability of the EARP complex. Fluorescence recovery after photobleaching in Eipr1 knockdown cells showed that EIPR1 is required for efficient recruitment of GARP to the TGN (Gershlick et al., 2016).
Here we investigated the role of mammalian EIPR1 in DCV function and EARP complex formation using insulin-secreting insulinoma cells. Specifically, we used Eipr1 knockout and rescue experiments to demonstrate that EIPR1 controls proper insulin distribution and secretion, and retention of cargo in mature DCVs. We also found that EIPR1 is required for the stability of the EARP complex subunits and for proper localization and association of EARP with membranes. Finally, we found that EARP localizes to two distinct compartments relevant to its functions in endocytic recycling and DCV maturation.
RESULTS
EIPR1 is required for insulin secretion
The C. elegans WD40 domain protein EIPR-1 is needed for DCV cargo trafficking in C. elegans neurons (Topalidou et al., 2016). To investigate the role of EIPR1 in the trafficking of mammalian DCV cargo in endocrine cells, we generated Eipr1 knockout (KO) insulinoma 832/13 cells using the CRISPR technology by inserting a puromycin cassette in the first exon of Eipr1 (Figure 1A and Supplemental Figure S1, A and B). We identified positive clones by PCR (Supplemental Figure S1C). To confirm that EIPR1 is lost in the Eipr1 KO line, we analyzed the cells for EIPR1 expression by Western blot. Wild-type (WT) cells displayed a band at around 45 kD, the approximate molecular weight of EIPR1, which was missing from Eipr1 KO cells (Figure 1B).
FIGURE 1:
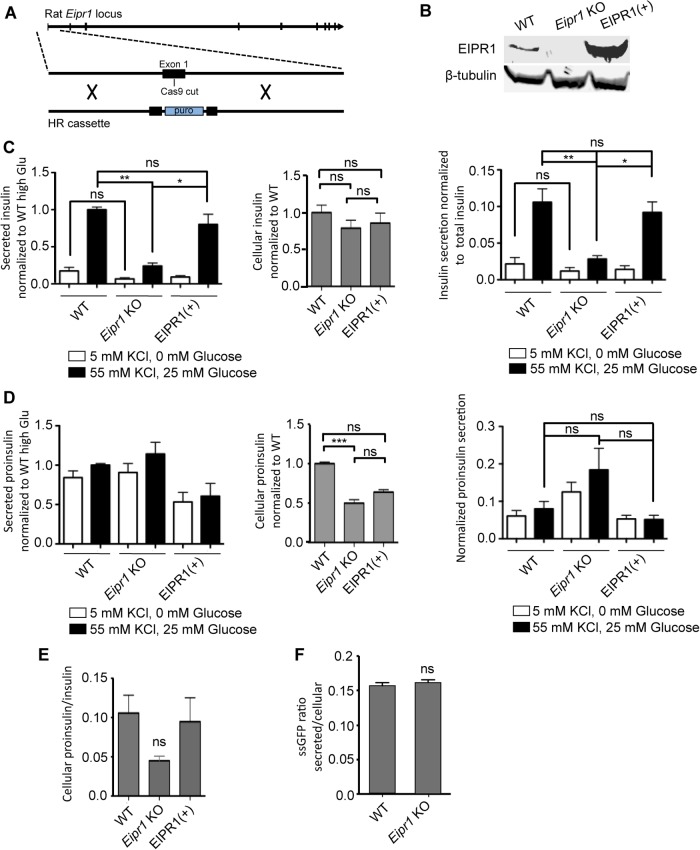
Insulin secretion is reduced in Eipr1 KO cells. (A) Strategy used to create the Eipr1 KO 832/13 cell line. Cas9 was targeted to cut in the first exon of the rat Eipr1 locus and homologous recombination (HR) was used to insert a puromycin cassette. (B) Eipr1 KO cells do not express wild-type (WT) EIPR1. Protein extracts from 832/13 (WT), Eipr1 KO 832/13 (Eipr1 KO), and Eipr1 KO 832/13 cells expressing a WT Eipr1 cDNA (EIPR1[+]) were blotted with an EIPR1 antibody. β-Tubulin served as a loading control. (C) Left panel, insulin secretion under resting (5 mM KCl, 0 mM glucose) and stimulating conditions (55 mM KCl, 25 mM glucose) from 832/13 cells (WT), Eipr1 KO 832/13 cells (Eipr1 KO), and an Eipr1 KO stable line expressing WT Eipr1 (EIPR1[+]). All values were normalized to the value of the WT under stimulating conditions. n = 7; *, p < 0.05; **, p < 0.01; ns, p > 0.05; error bars = SEM. Middle panel, total insulin content in WT, Eipr1 KO, and EIPR1(+) cells. All values were normalized to the WT. n = 5–7; ns, p > 0.05; error bars = SEM. Right panel, insulin secretion normalized to insulin content under resting (5 mM KCl, 0 mM glucose) and stimulating conditions (55 mM KCl, 25 mM glucose) from WT, Eipr1 KO, and EIPR1(+) cells. n = 5–7; *, p < 0.05; **, p < 0.01; ns, p > 0.05; error bars = SEM. We performed three biological replicates. For each replicate, the same cells were used to determine the amount of insulin secreted under resting conditions, stimulating conditions, and the amount of total cellular insulin. (D) Left panel, proinsulin secretion under resting (5 mM KCl, 0 mM glucose) and stimulating conditions (55 mM KCl, 25 mM glucose) from WT, Eipr1 KO, and EIPR1(+) cells. All values were normalized to the value of the WT under stimulating conditions. n = 6; error bars = SEM. Middle panel, total proinsulin content in WT, Eipr1 KO, and EIPR1(+) cells. All values were normalized to the WT. n = 6; ***, p < 0.001; ns, p > 0.05; error bars = SEM. Right panel, proinsulin secretion normalized to proinsulin content under resting (5 mM KCl, 0 mM glucose) and stimulating conditions (55 mM KCl, 25 mM glucose) from WT, Eipr1 KO, and EIPR1(+) cells. All values were normalized to the WT under stimulating conditions. n = 6; ns, p > 0.05; error bars = SEM. We performed three biological replicates. (E) Ratio of total cellular proinsulin to total insulin. n = 4–6; ns, p > 0.05; error bars = SEM. (F) The absence of EIPR1 does not affect the constitutive secretory pathway. Eipr1 KO 832/13 cells secrete normal levels of ssGFP (GFP fused to a signal peptide at its N-terminus). Values of secreted GFP were normalized to total. n = 6; error bars = SEM. The data shown were combined from two independent experiments with similar results. The data shown for the WT are the same as shown in Figure 1D of Cattin-Ortolá, Topalidou, et al. (2019) because these experiments were run in parallel with the same WT control.
To examine whether EIPR1 is needed for DCV cargo trafficking in insulinoma cells, we measured insulin secretion of WT and Eipr1 KO cells under resting (5 mM KCl, 0 mM glucose) and stimulating (55 mM KCl, 25 mM glucose) conditions. Insulin secretion under stimulating conditions was lower in Eipr1 KO cells than in WT cells (stimulated secretion was reduced to ∼25% of WT; Figure 1C, left panel). Insulin secretion under resting conditions was also lower, but the difference was not statistically significant (Figure 1C, left panel). To verify that the effects were due to loss of EIPR1, we introduced a WT Eipr1 cDNA into the Eipr1 KO cells (Figure 1B). Expression of WT EIPR1 in Eipr1 KO cells rescued the stimulated insulin secretion defect of the Eipr1 KO line, confirming that this defect is due to loss of EIPR1 (Figure 1C, left panel, EIPR1[+]).
Defective insulin secretion can be due to reduced insulin content. The total insulin content of Eipr1 KO was slightly reduced (to ∼80% of WT; Figure 1C, middle panel), but the difference was not statistically significant. After normalizing insulin secretion to total insulin content, we found that insulin secretion under stimulating conditions was still significantly reduced in the Eipr1 KO line (∼30% of WT; Figure 1C, right panel).
The observed decrease in insulin secretion in the Eipr1 KO could be due to a defect in the processing of proinsulin to insulin. Thus, we measured the total and secreted levels of proinsulin. Although secretion of proinsulin was not altered in Eipr1 KO (Figure 1D, left panel), total proinsulin content was reduced (∼50% of WT; Figure 1D, middle panel). To examine whether the reduced level of total proinsulin is due to a transcription defect, we performed quantitative reverse transcription-PCR (RT-PCR). No difference was observed in the level of proinsulin mRNA in Eipr1 KO cells (Supplemental Figure S2A). The reduction in total proinsulin was not significantly rescued in EIPR1(+) cells (Figure 1D, middle), so it may be due to an EIPR1-independent defect of the Eipr1 KO cell line. The ratio of total cellular proinsulin/insulin was also reduced in the Eipr1 KO, but the effect was not statistically significant (Figure 1E). These results suggest that proinsulin processing and secretion are not strongly affected by the absence of EIPR1.
One possible explanation for the reduced insulin secretion in Eipr1 KO cells is that exocytosis is impaired. We counted exocytotic events in WT and Eipr1 KO cells expressing the neuropeptide NPY tagged to pH-sensitive GFP (NPY::pHluorin). Exocytotic events were reduced in Eipr1 KO cells under stimulating conditions (Supplemental Figure S3), suggesting that Eipr1 KO cells have an exocytosis defect. However, a caveat to this experiment is that this assay may have underestimated the true exocytosis rate if it was not possible to detect DCVs carrying reduced amounts of NPY::pHluorin. As Eipr1 KO cells appear to carry reduced amounts of cargoes in mature DCVs (see below), it is possible that DCV exocytotic events in this mutant line were more difficult to detect.
To investigate whether EIPR1 is needed for constitutive secretion, we measured the secretion of GFP fused to a signal peptide (ssGFP) under resting conditions (Hummer et al., 2017). We found that secretion of ssGFP in Eipr1 KO cells is similar to WT, suggesting that EIPR1 is not needed for constitutive secretion (Figure 1F).
EIPR1 is required for the normal cellular distribution of insulin
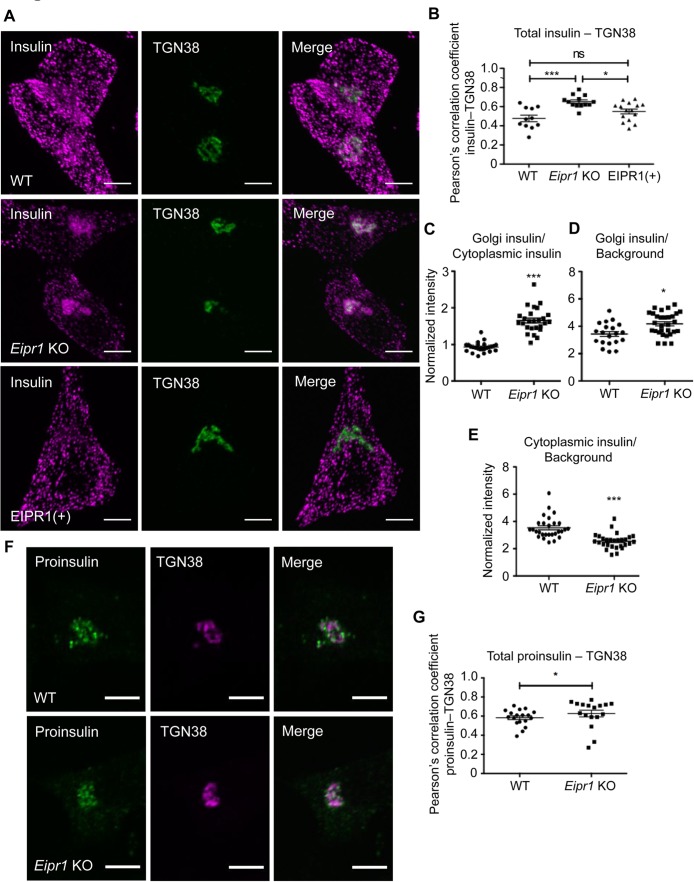
To further investigate the role of EIPR1 in DCV cargo trafficking, we examined the subcellular localization of insulin by immunostaining. In WT cells, insulin is detected as puncta spread throughout the cytoplasm (Figure 2A). By contrast, in Eipr1 KO cells, insulin is localized primarily to a perinuclear region that partially overlaps with the trans-Golgi marker TGN38 (Figure 2, A and B). This phenotype was rescued in Eipr1 KO cells that stably express WT EIPR1 (Figure 2, A and B). The ratio of TGN38-localized insulin to the level of insulin in the cytoplasm was increased in Eipr1 KO cells (Figure 2C), suggesting accumulation of insulin near the TGN, decreased insulin in the periphery of the cell, or both. To distinguish between these possibilities, we quantified the levels of insulin localized near the TGN and in the cytoplasm. We found that Eipr1 KO cells have a small increase in insulin near the TGN (Figure 2D) and a decrease in insulin in the cell periphery (Figure 2E). The reduction in insulin in the cell periphery might contribute to the reduction in insulin secretion in Eipr1 KO cells under stimulating conditions and reduced frequency of detectable exocytotic events (Figure 1C and Supplemental Figure S3).
FIGURE 2:
Insulin localization is disrupted in Eipr1 KO cells. (A) Representative images of 832/13 (WT), Eipr1 KO 832/13 (Eipr1 KO), and Eipr1 KO 832/13 cells expressing WT Eipr1 (EIPR1[+]) costained for endogenous insulin and TGN38. In WT and EIPR1(+) cells, insulin is spread throughout the cytoplasm, but in Eipr1 KO cells insulin accumulates in a perinuclear region that partially overlaps with TGN38. The experiment was repeated three times, and the experimenter was blinded to the genotypes of the stained cells. Scale bars = 5 μm. (B) Pearson’s correlation coefficient was measured to quantify the localization between insulin and the TGN marker TGN38. n = 11 for WT, n = 13 for Eipr1 KO, and n = 15 for EIPR1(+); ***, p < 0.001; *, p < 0.05; ns, p > 0.05; error bars = SEM. The experiment was repeated three times. (C) Eipr1 KO cells have an increased Golgi/cytoplasmic ratio of insulin relative to WT cells. Fluorescence of a region of interest that includes the TGN divided by the fluorescence of a region of the same size in the cytoplasm, in WT and Eipr1 KO. (n = 25 for WT and Eipr1 KO; error bars = SEM; ***, p < 0.001). The data shown for the WT are the same as shown in Figure 2C of Cattin-Ortolá, Topalidou, et al. (2019) because these experiments were run in parallel with the same WT control. (D) Eipr1 KO cells have a slightly increased amount of insulin localized at or near the TGN. Fluorescence of a region of interest that includes the TGN divided by the fluorescence of a region of the same size in the background, in WT and Eipr1 KO. (n = 20 for WT and n = 30 for Eipr1 KO; error bars = SEM; *, p < 0.05). (E) Eipr1 KO cells have a decreased amount of insulin localized to the cytoplasm. Fluorescence of a region of interest in the cytoplasm divided by the fluorescence of a region of the same size in the background (n = 28 for WT and Eipr1 KO; error bars = SEM;***, p < 0.001). (F) Representative images of 832/13 (WT) and Eipr1 KO 832/13 (Eipr1 KO) cells costained for endogenous proinsulin and TGN38. In both WT and Eipr1 KO cells, proinsulin is localized in a perinuclear region that partially overlaps with TGN38. Scale bars = 5 μm. (G) Pearson’s correlation coefficient was measured to quantify the localization between proinsulin and the TGN marker TGN38. n = 18 for WT and n = 17 for Eipr1 KO; *, p < 0.05; error bars = SEM. The experiment was repeated three times.
We also examined the localization of proinsulin, a cargo of immature DCVs. Proinsulin is localized in a similar perinuclear region in both WT and Eipr1 KO cells, with perhaps a very slight shift toward tighter colocalization with TGN38 in the Eipr1 KO (Figure 2, F and G). Because insulin seems to be shifted to the TGN in Eipr1 KO cells (Figure 2, A–D), we wondered whether there might be increased acidification of the Golgi in these mutant cells that would allow processing of proinsulin into insulin in this compartment. To assay the pH of the Golgi, we measured the fluorescence of the pH-sensitive GFP variant pHluorin fused to the Golgi-localization domain of sialyltransferase (St6Gal1), a late-Golgi enzyme (Wong et al., 1992; Hummer et al., 2017). The pH of the Golgi is similar in Eipr1 KO and WT cells (Supplemental Figure S4), suggesting that the accumulation of insulin near the TGN in Eipr1 KO cells is not due to proinsulin processing in the Golgi.
EIPR1 is needed for the normal levels and distribution of mature DCV cargo
We next examined the levels of other known DCV cargoes, such as the proprotein convertase 1/3 (PC1/3), proprotein convertase 2 (PC2), and carboxypetidase E (CPE), that mediate the processing of proinsulin to insulin (Smeekens et al., 1992; Naggert et al., 1995). These enzymes are themselves processed from their inactive pro forms to their active mature forms (Vindrola and Lindberg, 1992; Zhou and Lindberg, 1993; Song and Fricker, 1995; Muller and Lindberg, 1999).
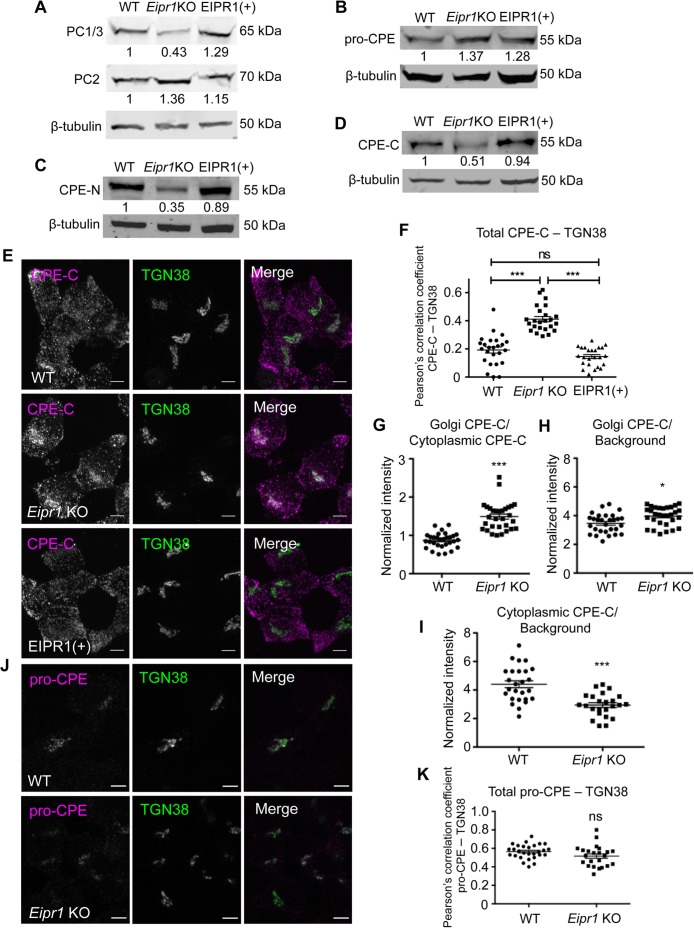
The Eipr1 KO cells had reduced levels of the processed form of PC1/3 but normal levels of PC2 (Figure 3A). Expression of WT EIPR1 in Eipr1 KO cells rescued the PC1/3 defect (Figure 3A). To examine whether the reduced levels of total PC1/3 are due to a transcription defect, we performed quantitative RT-PCR but found no difference between WT and Eipr1 KO cells (Supplemental Figure S2A).
FIGURE 3:
EIPR1 is required for normal levels and distribution of mature DCV cargoes. (A) Eipr1 KO cells have reduced levels of the processed form of the proprotein convertase PC1/3 but normal levels of PC2. Lysates from WT, Eipr1 KO, and EIPR1(+) 832/13 cells were blotted with an antibody to PC1/3 or PC2. β-Tubulin was used as a loading control. Shown are representative blots of three independent experiments with similar results. (B) The levels of the unprocessed form of CPE (pro-CPE) are not affected by the loss of EIPR1. Lysates from the indicated cell lines were blotted with an antibody to pro-CPE. The experiment was repeated three times with similar results. The data shown for the WT are the same as shown in Figure 3B of Cattin-Ortolá, Topalidou, et al. (2019) because these experiments were run in parallel with the same WT control. (C, D) Eipr1 KO cells have reduced levels of the processed form of CPE. Detergent lysates from WT, Eipr1 KO, and EIPR1(+) 832/13 cells were blotted with an antibody to the N-terminus of CPE (CPE-N) or the C-terminus of CPE (CPE-C). β-Tubulin was used as a loading control. The experiment was repeated three times. The data shown for the WT in Figure 3C are the same as shown in Figure 3C of Cattin-Ortolá, Topalidou, et al. (2019) because these experiments were run in parallel with the same WT control. (E) The mature processed form of CPE is localized at or near the TGN in Eipr1 KO cells. Representative confocal images of WT, Eipr1 KO, and EIPR1(+) cells costained with the CPE C-terminal antibody (CPE-C) and TGN38. Maximum intensity projections. Scale bars = 5 μm. The experiment was repeated three times with similar results. (F) Quantification of the colocalization between the mature form of CPE (CPE-C) and the TGN marker TGN38. Maximum intensity projection images were obtained and Pearson’s correlation coefficients were determined by drawing a line around each cell. (n = 25 for WT; n = 24 for Eipr1 KO; n = 24 for EIPR1(+); error bars = SEM; ***, p < 0.001; ns, p > 0.05). (G) Eipr1 KO cells have an increased Golgi/cytoplasmic ratio of CPE relative to WT cells. Fluorescence of a region of interest that includes the TGN divided by the fluorescence of a region of the same size in the cytoplasm, in WT and Eipr1 KO. (n = 30 for WT and Eipr1 KO; error bars = SEM; ***, p < 0.001). The data shown for the WT are the same as shown in Figure 3G of Cattin-Ortolá, Topalidou, et al. (2019) because these experiments were run in parallel with the same WT control. (H) Eipr1 KO cells have an increased amount of mature CPE localized at the Golgi. Fluorescence of a region of interest at the Golgi divided by the fluorescence of a region of the same size in the background, in WT and Eipr1 KO. (n = 29 for WT and n = 31 for Eipr1 KO; error bars = SEM; *, p < 0.05). (I) Eipr1 KO cells have a decreased amount of mature CPE localized to the cytoplasm. Fluorescence of a region of interest in the cytoplasm divided by the fluorescence of a region of the same size in the background (n = 26 for WT and n = 24 for Eipr1 KO; error bars = SEM; ***, p < 0.001). (J) The unprocessed form of CPE (pro-CPE) is localized at or near the TGN in both WT and Eipr1 KO cells. Representative confocal images of WT and Eipr1 KO cells costained for pro-CPE antibody and TGN38. Single slices. Scale bars = 5 μm. The experiment was repeated twice. (K) Quantification of the colocalization between the unprocessed form of CPE (pro-CPE) and the TGN marker TGN38. Maximum intensity projection images were obtained and Pearson’s correlation coefficients were determined by drawing a line around each cell. (n = 28 for WT; n = 23 for Eipr1 KO; error bars = SEM; ns, p > 0.05).
We measured the levels of CPE with antibodies that recognize the unprocessed CPE form that resides in the Golgi and immature DCVs (pro-CPE antibody), or the processed forms localized to mature DCVs (CPE-N and CPE-C antibodies; Fricker et al., 1990, 1996). The Eipr1 KO had normal levels of the unprocessed form of CPE (Figure 3B), but reduced levels of the processed form (Figure 3, C and D). Expression of WT EIPR1 in Eipr1 KO cells rescued the mature CPE defects (Figure 3, C and D). These results suggest that EIPR1 acts in a postprocessing step and is required for the normal levels of mature DCV cargo.
We next examined the subcellular localization of CPE by immunostaining. In WT cells, mature CPE was detected as puncta spread throughout the cytoplasm that almost fully colocalize with insulin (Figure 3, E and F, and Supplemental Figure S5). By contrast, in Eipr1 KO cells, mature CPE was reduced in the cell periphery and increased in the TGN region (Figure 3, E and F). This phenotype was rescued in Eipr1 KO cells that stably expressed WT EIPR1 (Figure 3, E and F). As with insulin, the ratio of TGN38-localized mature CPE to mature CPE in the cytoplasm was increased in Eipr1 KO cells (Figure 3G). Eipr1 KO cells have slightly more mature CPE near the TGN (Figure 3H) and significantly less mature CPE in the cell periphery (Figure 3I). This loss of mature CPE from the cell periphery supports our finding that there are reduced levels of mature CPE in the Eipr1 KO (Figure 3, C and D). Pro-CPE was localized in a perinuclear region in both WT and Eipr1 KO cells (Figure 3, J and K), similar to proinsulin (Figure 2, F and G), consistent with EIPR1 acting in a postprocessing step.
Mature cargo is able to exit the TGN in Eipr1 KO cells
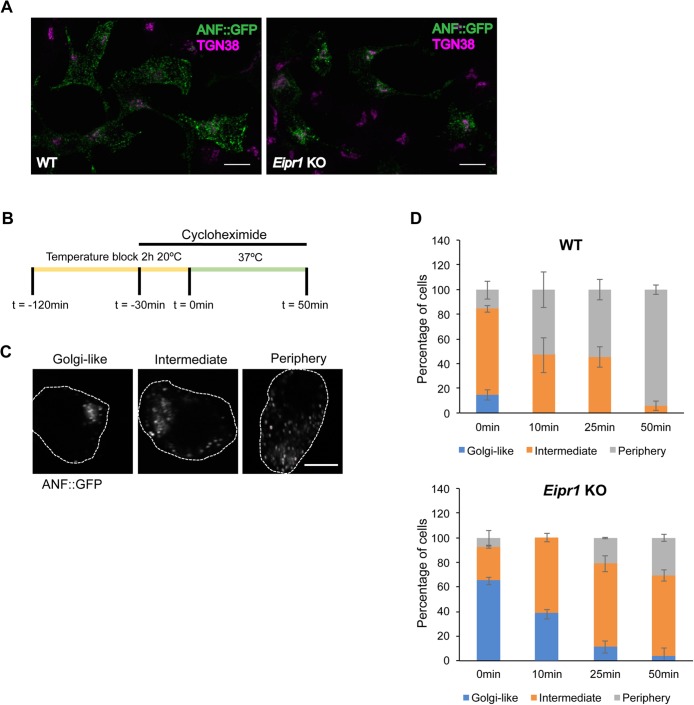
To investigate whether the increased insulin and mature CPE at the TGN in Eipr1 KO cells is due to mature DCV cargo being stuck at the TGN, we used a pulse-chase method to monitor cargo exit from the TGN. We transiently transfected the DCV cargo ANF::GFP into WT and Eipr1 KO 832/13 cells and blocked cargo exit from the Golgi by incubating for 2 h at 20°C (Kögel et al., 2013). At steady state (before the temperature block), ANF::GFP is localized more in a perinuclear region in Eipr1 KO cells (Figure 4A), similar to insulin and mature CPE. After the temperature block (pulse), cells were returned to 37°C and incubated for different times (chase; Figure 4B). They were then processed for GFP immunostaining and imaging. Cells were scored based on whether ANF::GFP was: 1) predominantly at the TGN region (“Golgi-like” in Figure 4C), 2) found both at the TGN region and at the cell periphery (“Intermediate” in Figure 4C), or 3) excluded from the Golgi (“Periphery” in Figure 4C). We observed that the localization of ANF::GFP at the TGN was different between WT and Eipr1 KO cells at all time points (Figure 4D), confirming that EIPR1 affects the distribution of DCV cargo. The distribution of ANF::GFP changed in Eipr1 KO cells during the chase period, with most of the cells having a Golgi-like distribution at t = 0, and most of the cells having an Intermediate distribution at t = 50 min (Figure 4D). This indicates that the DCV cargo is not permanently stuck at the TGN region in Eipr1 KO cells but is able to reach the cell periphery, although probably at a slower rate than in WT cells.
FIGURE 4:
DCV cargoes exit the TGN in Eipr1 KO cells. (A) Representative images of WT and Eipr1 KO 832/13 cells transfected with ANF::GFP and costained with anti-GFP and anti-TGN38 antibodies. In WT cells, ANF::GFP is distributed in cytoplasmic puncta, but in Eipr1 KO cells, ANF::GFP is restricted to perinuclear puncta. Not all cells in this field of view are transfected with ANF::GFP. Scale bars = 10 μm. (B) Schematic of the pulse-chase experiment. Cells transiently transfected with ANF::GFP were incubated at 20°C for 2 h to cause the accumulation of DCV cargoes at the TGN (pulse). Before the end of the temperature block (30 min), cyclohexamide was added to block protein translation. At the end of the temperature block, cells were returned to 37°C and incubated for various times (chase) before fixation and immunostaining. (C) Representative images of the cell categories used for qualitative assessment of TGN exit: 1) ANF::GFP concentrated at the TGN region (Golgi-like), 2) ANF::GFP distributed both at the TGN and at the cell periphery (Intermediate), or 3) ANF::GFP excluded from the TGN (Periphery). Scale bar = 5 μm. (D) Percentage of WT and Eipr1 KO cells with the indicated ANF::GFP distribution at the indicated time points. For each data point and genotype, 50–100 cells were counted blindly. The experiment was repeated three times with similar results. The data shown for the WT are the same as shown in Supplemental Figure S5D of Cattin-Ortolá, Topalidou, et al. (2019) because these experiments were run in parallel with the same WT control.
One possible explanation for the reduced cargo in mature DCVs in the Eipr1 KO could be that cargo is lost through the endolysosomal system. C. elegans EIPR-1 acts in the same genetic pathway as RAB-2 (Topalidou et al., 2016) and RAB-2 has been proposed to act by preventing DCV cargo from getting lost through the endolysosomal system (Edwards et al., 2009; Sumakovic et al., 2009). To test this possibility, we expressed a constitutively active form of RAB-5 (RAB-5[QL]) in WT and eipr-1 mutants in C. elegans to inhibit trafficking to early endosomes (Sumakovic et al., 2009). We found that this construct partially rescued the reduced DCV cargo levels in eipr-1 mutant neuronal axons, similar to rab-2 (Supplemental Figure S6, A and B). However, expression of rat RAB5A(QL) in Eipr1 KO insulinoma cells did not rescue the CPE-C localization defect (Supplemental Figure S6, C and D). It is unclear why C. elegans neurons and insulinoma cells showed different effects of expressing activated RAB5.
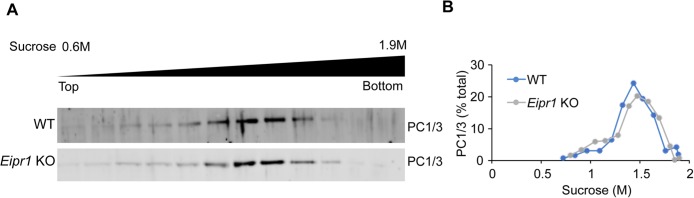
Loss of some proteins required for DCV biogenesis (e.g., AP-3, VPS41, HID-1) leads to both loss of cargoes and alterations in DCV biophysical properties such as density (Asensio et al., 2010, 2013; Hummer et al., 2017). However, we found no difference in DCV density in Eipr1 KO cells as measured by the distribution of mature PC1/3 in membranes separated by equilibrium sedimentation through a sucrose gradient (Figure 5, A and B). Thus, though levels of mature PC1/3 are reduced in Eipr1 KO cells, the remaining PC1/3 appears to still be found in DCVs of normal density.
FIGURE 5:
The density of DCVs is not affected in Eipr1 KO cells. (A) Postnuclear supernatants from WT and Eipr1 KO cells were separated by equilibrium sedimentation through 0.6–1.9 M sucrose. Fractions were blotted with an antibody against PC1/3 (mature DCV marker). The data shown are from one representative experiment of three independent experiments with similar results. (B) Band intensity for each fraction was quantified using Fiji, presented as a percentage of total band intensity, and plotted against the sucrose concentration of the fraction.
EIPR1 is needed for the localization of EARP subunits and their association with membranes
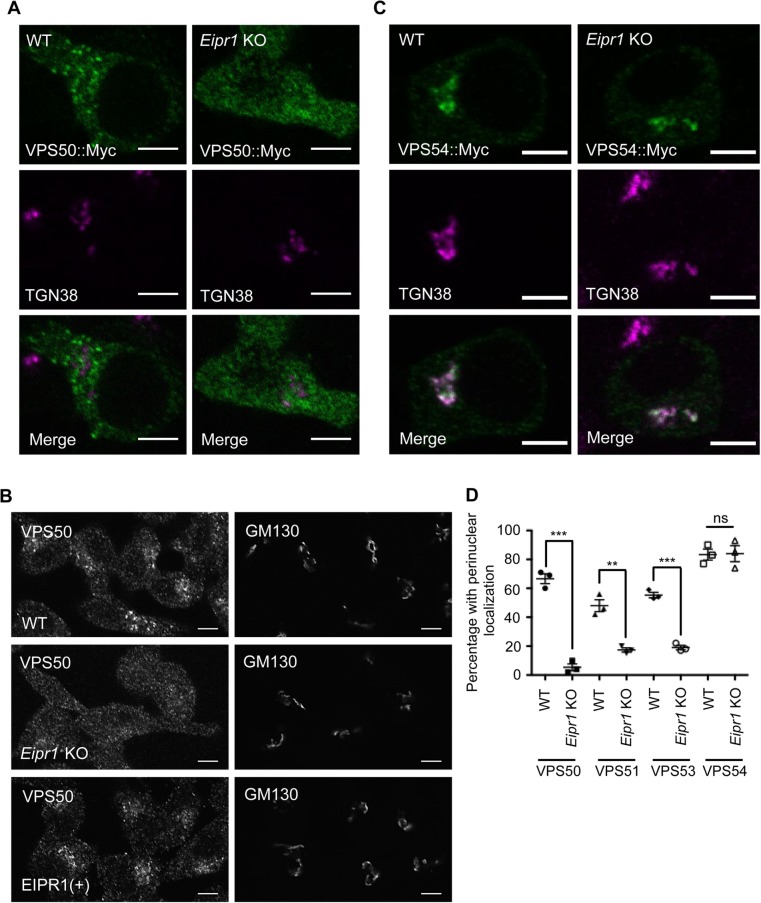
It was recently shown that mammalian EIPR1 interacts with the EARP and GARP complex subunits and functions with EARP in endosomal recycling and with GARP in endosome-Golgi retrograde trafficking (Gershlick et al., 2016; Topalidou et al., 2016). However, eipr-1 mutants in C. elegans have behavioral and cellular phenotypes similar to EARP-specific mutants, but not GARP-specific mutants (Topalidou et al., 2016). EIPR1 is a WD40 domain protein, and WD40 domains often act as scaffolds for mediating protein interactions and multiprotein complex assembly (Stirnimann, Petsalaki, et al., 2010). To investigate whether EIPR1 is required for the localization of EARP or GARP complex subunits in insulin-secreting cells, we examined the localization of transiently transfected Myc-tagged VPS50 (the sole EARP-specific subunit), GFP and Myc-tagged VPS54 (the sole GARP-specific subunit), and Myc-tagged VPS51 and VPS53 (subunits present in both the GARP and EARP complexes). We also examined the localization of endogenous VPS50 using a commercial antibody, the only antibody we have for EARP or GARP subunits that is suitable for immunofluorescence. Unfortunately, we were unable to determine the localization of EIPR1 as we do not have an antibody that works for immunofluorescence, and tagging the C. elegans or rat EIPR1 proteins at either the N- or C-terminus (with either GFP or Myc) leads to a diffuse cytoplasmic localization that is probably nonphysiological. A similar issue with tagging EIPR1 was reported elsewhere (Gershlick et al., 2016).
In WT cells, VPS50, VPS51, VPS53, and VPS54 all showed a punctate pattern of localization. Interestingly, the punctate localization of the EARP-specific subunit VPS50 and the EARP/GARP common subunits VPS51 and VPS53 was disrupted in Eipr1 KO cells, with diffuse fluorescence throughout the cytoplasm (Figure 6, A, B, and D, and Supplemental Figure S7, A and B). By contrast, the GARP-specific subunit VPS54 was still punctate and localized near the TGN in Eipr1 KO cells (Figure 6, C and D, and Supplemental Figure S7C). We conclude that EIPR1 is needed for the localization of the EARP complex subunit VPS50 and the EARP/GARP common subunits, but not the GARP complex–specific subunit VPS54. This result suggests that EIPR1 is preferentially required for the localization of the EARP complex.
FIGURE 6:
EIPR1 is required for the localization of EARP in insulin-secreting cells. (A) Representative images of 832/13 WT and Eipr1 KO cells transfected with VPS50::13Myc (VPS50::Myc) and costained with anti-Myc and anti-TGN38 antibodies. In WT cells, VPS50::Myc is localized to puncta but in Eipr1 KO cells, fluorescence is diffuse throughout the cytoplasm. The punctate pattern of localization of VPS50::Myc overlaps only partially with TGN38. Scale bars = 5 μm. (B) Representative images of 832/13 (WT), Eipr1 KO, and EIPR1(+) cells stained with anti-VPS50 antibody. Endogenous VPS50 is punctate in WT cells, but diffuse throughout the cytoplasm in Eipr1 KO cells. Scale bars = 5 μm. (C) Representative images of 832/13 cells (WT) and Eipr1 KO 832/13 (Eipr1 KO) cells transfected with VPS54::13Myc (VPS54::Myc) and costained with anti-Myc and anti-TGN38 antibodies. In both WT and Eipr1 KO cells, VPS54::Myc is localized to perinuclear puncta that largely overlap with TGN38. Scale bars = 5 μm. (D) Localization of Myc-tagged VPS50, VPS51, and VPS53, but not VPS54, is disrupted in Eipr1 KO cells. Shown is the percentage of cells with perinuclear and punctate staining of each indicated subunit. The experiment was repeated three times and ∼100 cells were imaged per experiment. Cells with very high expression level were not included in the counting because overexpression of the individual subunits leads to their mislocalization to the cytoplasm in WT cells. ***, p < 0.001; **, p < 0.01; ns, p > 0.05.
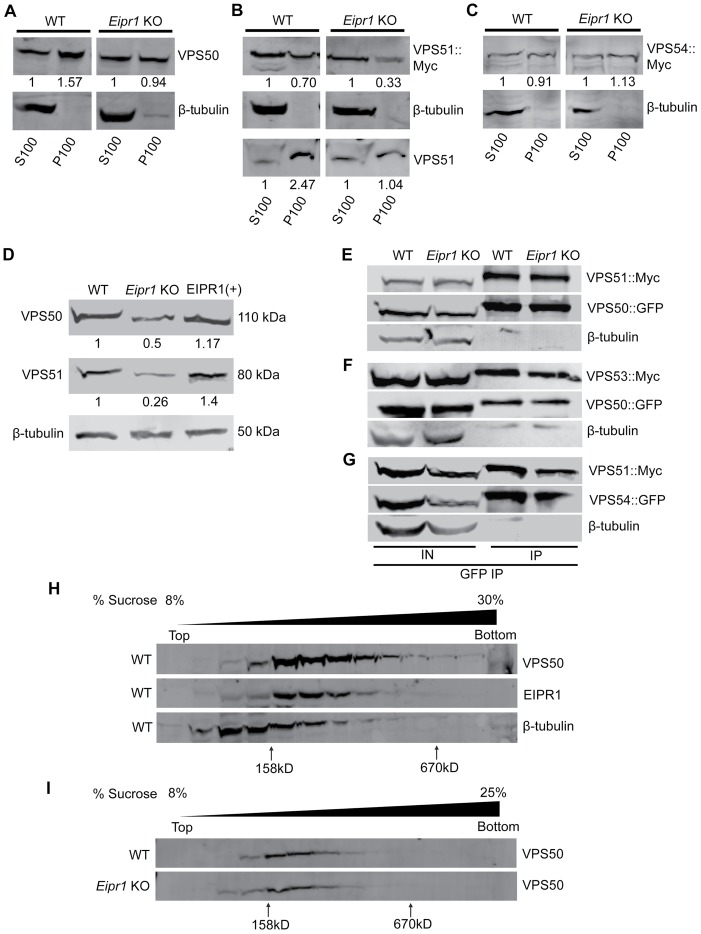
To test whether the diffuse localization of EARP subunits in the Eipr1 KO is due to a reduced association of EARP with membranes, we fractionated 832/13 cell lysates and probed for VPS50. VPS50 was found primarily in the membrane fraction (P100) in WT cells, but the association of VPS50 with membranes was partially lost in Eipr1 KO cells (Figure 7A). Similarly, VPS51 was found in the membrane fraction in WT cells and this association was reduced in the Eipr1 KO (Figure 7B). By contrast, the association of the GARP-specific subunit VPS54 with membranes was not altered in Eipr1 KO cells (Figure 7C). We conclude that EIPR1 is partially required for the proper association of EARP with membranes, but is not required for membrane association of GARP.
FIGURE 7:
Membrane association and stability of EARP is disrupted in Eipr1 KO insulin-secreting cells. (A) VPS50 associates with membranes in an EIPR1-dependent manner. In WT cell fractions, endogenous VPS50 was found primarily in the postnuclear P100 membrane fraction. In Eipr1 KO 832/13 cells, VPS50 was equally distributed between the P100 membrane fraction and the S100 cytosolic fraction. β-Tubulin served as a control soluble protein. S100, P100: supernatant and pellet fractions obtained by a 100,000 × g spin of the cell lysate, containing cytosolic and membrane-associated proteins, respectively. (B) VPS51 associates with membranes in an EIPR1-dependent manner. In WT cell fractions, VPS51::Myc was roughly equally distributed between the postnuclear P100 membrane fraction and the S100 cytosolic fraction, and endogenous VPS51 was found primarily in the P100 membrane fraction. In Eipr1 KO 832/13 cells (Eipr1 KO), VPS51::Myc was found mostly in the soluble fraction and endogenous VPS51 was equally distributed between the membrane fraction and the cytosolic fraction. (C) VPS54 associates with membranes in an EIPR1-independent manner. In both WT and Eipr1 KO 832/13 cell fractions, VPS54::Myc was equally distributed between the postnuclear P100 membrane fraction and the S100 cytosolic fraction. (D) VPS50 and VPS51 levels are reduced in Eipr1 KO cells. Protein extracts from 832/13 (WT), Eipr1 KO 832/13 (Eipr1 KO), and Eipr1 KO 832/13 cells expressing WT Eipr1 (EIPR1[+]) were blotted with antibodies against VPS50 and VPS51. β-Tubulin served as a loading control. (E) VPS50 interacts with VPS51 in an EIPR1-independent way. EGFP-tagged VPS50 was coexpressed with 13Myc-tagged VPS51 in 832/13 (WT) and Eipr1 KO cells. Immunoprecipitation of VPS50::GFP pulled down VPS51::Myc independently of EIPR1, but did not pull down β-tubulin. IN, input; IP, immunoprecipitated. (F) VPS50 interacts with VPS53 in an EIPR1-independent way. EGFP-tagged VPS50 was coexpressed with 13Myc-tagged VPS53 in 832/13 (WT) and Eipr1 KO cells. Immunoprecipitation of VPS50::GFP pulled down VPS53::Myc independently of EIPR1, but did not pull down β-tubulin. IN, input; IP, immunoprecipitated. (G) VPS51 interacts with VPS54 in an EIPR1-independent way. EGFP-tagged VPS54 was coexpressed with 13Myc-tagged VPS51 in 832/13 (WT) and Eipr1 KO cells. Immunoprecipitation of VPS54::GFP pulled down VPS51::Myc independently of EIPR1, but did not pull down β-tubulin. IN, input; IP, immunoprecipitated. (H) VPS50 and EIPR1 cofractionate on a linear 8–30% sucrose velocity gradient. Fractions from 832/13 (WT) cell lysate were blotted with antibodies against VPS50 and EIPR1. β-Tubulin served as a control soluble protein. (I) VPS50 fractionates similarly from cell lysates of 832/13 (WT) and Eipr1 KO cells on a linear 8–25% sucrose velocity gradient. Fractions from WT and Eipr1 KO cell lysates were blotted with antibodies against VPS50.
EIPR1 is not required for the physical interactions between EARP complex subunits
Because EIPR1 is needed for the proper localization of EARP complex subunits and WD40 domain proteins often serve as scaffolds for complex assembly, we examined whether EIPR1 is needed for the formation of the EARP complex. We first compared the levels of endogenous VPS50 and VPS51 in WT and Eipr1 KO 832/13 cells. Eipr1 KO cells had reduced levels of VPS50 and VPS51, and this defect was rescued by expression of WT EIPR1 (Figure 7D). Thus, EIPR1 is required for expression or stability of the individual EARP subunits. To test whether these reduced protein levels were due to reduced transcription, we examined the levels of the VPS50 and VPS51 mRNAs in WT and Eipr1 KO cells. Quantitative PCR using total cDNA from WT and Eipr1 KO cells showed no difference in the mRNA levels of these EARP complex subunits (Supplemental Figure S2B). Thus, the EARP protein subunits are less stable in the absence of EIPR1.
To determine whether EIPR1 is required for physical interactions between the individual EARP subunits, we expressed GFP-tagged VPS50 with Myc-tagged VPS51 or Myc-tagged VPS53 in WT and Eipr1 KO 832/13 cells and performed coimmunoprecipitation experiments. Unlike with endogenous VPS50 and VPS51, the levels of VPS50::GFP and VPS51::Myc were not reduced in Eipr1 KO cells, probably because of overexpression (Figure 7, E–G; IN: input). GFP-tagged VPS50 coimmunoprecipitated with either Myc-tagged VPS51 or Myc-tagged VPS53, and these interactions were not disrupted by loss of EIPR1 (Figure 7, E and F). To test whether EIPR1 is required for the interactions between GARP complex subunits, we expressed GFP-tagged VPS54 with Myc-tagged VPS51 in WT and Eipr1 KO 832/13 cells. GFP-tagged VPS54 coimmunoprecipitated with Myc-tagged VPS51 as expected, and this interaction was not changed in Eipr1 KO cells (Figure 7G). These data indicate that EIPR1 is not required for interactions between the individual subunits of the EARP or GARP complexes, at least under these conditions where some subunits are overexpressed.
Finally, to further examine whether the EARP complex is disrupted in the absence of EIPR1, we subjected cell lysates from WT and Eipr1 KO 832/13 cells to velocity sedimentation through an 8–30% linear sucrose gradient and blotted for VPS50. VPS50 sedimented in a broad peak between the 158 and 670 kDa standards, indicating that the protein is part of a complex (Figure 7H). EIPR1 sedimentation from the same cell lysate showed a peak similar to that of VPS50, suggesting that EIPR1 and VPS50 might be part of the same complex (Figure 7H). Of note, sedimentation of VPS50 was not affected by loss of EIPR1 (Figure 7I), suggesting that the EARP complex still forms in the absence of EIPR1.
EIPR1 is required for EARP function but not GARP function
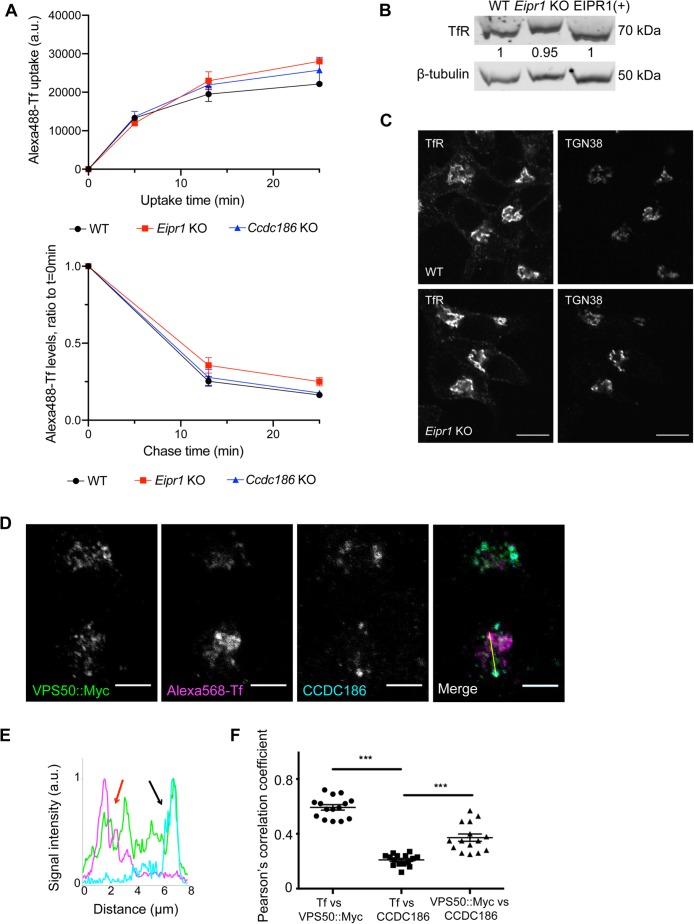
EARP is required for the recycling of cargoes from recycling endosomes back to the plasma membrane (Schindler, Chen, et al., 2015). EIPR1 was also shown to be required for the endocytic recycling of transferrin in HAP1 cells (Gershlick et al., 2016). We examined whether EIPR1 is required for the recycling of transferrin in 832/13 cells and found that Eipr1 KO 832/13 cells take up and retain slightly more transferrin than WT cells (Figure 8A), similar to results seen in HAP1 cells (Gershlick et al., 2016). The steady-state level and distribution of the transferrin receptor were not obviously altered in Eipr1 KO cells (Figure 8, B and C).
FIGURE 8:
EIPR1 has a slight defect in transferrin recycling in insulinoma cells and EARP localizes to two distinct compartments. (A) EIPR1, but not CCDC186, has a slight defect in transferrin (Tf) recycling. FACS analysis of Alexa 488–labeled transferrin in WT, Eipr1 KO, and Ccdc186 KO cells after transferrin uptake for the indicated times (top) and at different chase times following 25-min transferrin uptake (bottom). The top plot represents the median of Alexa 488–Tf intensity of the population of cells as a function of time. The bottom plot represents the median of Alexa 488–Tf intensity of the population of cells at each time point normalized to the median intensity at t = 0 as a function of time. The experiment was repeated three times. For Tf uptake at 25 min, p = 0.16, WT vs. Eipr1 KO. For Tf recycling at 25 min, p = 0.11, WT vs. Eipr1 KO, one-way ANOVA with Bonferroni correction for multiple comparisons. (B) Levels of expression of the transferrin receptor (TfR) are not affected in Eipr1 KO cells. Protein extracts from 832/13 (WT), Eipr1 KO 832/13 (Eipr1 KO), and Eipr1 KO 832/13 cells expressing WT Eipr1 (EIPR1[+]) were blotted with an antibody against (TfR). β-Tubulin served as a loading control. (C) Localization of the TfR is not affected in Eipr1 KO cells. Representative images of 832/13 (WT) and Eipr1 KO (Eipr1 KO) 832/13 cells costained with antibodies for endogenous TfR and TGN38. Scale bars = 5 μm. (D–F) EARP localizes to two distinct compartments: a CCDC186-positive compartment near the TGN and immature DCVs and a transferrin-positive compartment that is associated with endosomes. 832/13 cells were transiently transfected with Myc-tagged VPS50, incubated with Alexa 568–labeled transferrin (Tf), and immunostained for Myc and CCDC186. (D) Representative confocal images of cells costained for endogenous CCDC186, Alexa 568–Tf, and VPS50::Myc. Scale bars = 5 μm. (E) The graph shows a representative intensity plot of normalized signal intensity vs. distance across the yellow line shown in D. VPS50::Myc is shown in green, Alexa 568–Tf is in magenta, and CCDC186 in cyan. Red arrow: overlap between VPS50 and Tf. Black arrow: overlap between VPS50 and CCDC186. (F) Pearson’s correlation coefficient was measured to quantify the localization between transferrin and VPS50::Myc, transferrin and CCDC186, and CCDC186 and VPS50::Myc. n = 15; ***, p < 0.001; error bars = SEM. The experiment was repeated twice.
Depletion of GARP leads to redistribution of TGN38 to cytoplasmic vesicles thought to correspond to retrograde transport intermediates (Pérez-Victoria et al., 2008). We too found that siRNA knockdown of the VPS51 subunit of the GARP complex causes partial redistribution of TGN38 to cytoplasmic puncta, but the Eipr1 KO had no obvious change in the distribution of TGN38 (Supplemental Figure S8). Thus, EIPR1 is required for both the localization and function of EARP, but is not required for the localization of GARP or the function of GARP in the retrograde trafficking of TGN38.
The EARP complex localizes to two distinct compartments
To determine whether EARP functions at distinct cellular sites to mediate endocytic recycling and DCV cargo sorting, we examined colocalization of the EARP subunit VPS50 with CCDC186 and transferrin. Transferrin is found at early and recycling endosomes and VPS50 localizes to RAB4-positive endosomes involved in transferrin receptor recycling (Schindler, Chen, et al., 2015). CCDC186/CCCP-1 is a coiled-coil protein that functions in DCV biogenesis in the same genetic pathway as EIPR1 and EARP (Cattin-Ortolá, Topalidou, et al., 2019), but is not required for transferrin recycling (Figure 8A). CCDC186 is localized near immature DCVs and the TGN (Ailion et al., 2014; Cattin-Ortolá et al., 2017; Cattin-Ortolá, Topalidou, et al., 2019), but is distinct from recycling endosomes marked by transferrin (Supplemental Figure S9A). Notably, localization of CCDC186 does not require EIPR1 (Supplemental Figure S9B). We observed that VPS50 is found at two distinct sites. At one site, VPS50 colocalizes with CCDC186 and at the other VPS50 colocalizes with transferrin (Figure 8, D–F, and Supplemental Figure S9A), consistent with EARP acting both at recycling endosomes and at the site of DCV maturation. VPS50 is found in puncta both in the perinuclear region and more peripherally and has a localization pattern that shows little overlap with mature insulin granules in the cell periphery or the immature DCV markers syntaxin 6 and proinsulin (Supplemental Figure S9, C–E). By contrast, the GARP-specific VPS54 subunit was specifically localized to a perinuclear region largely overlapping TGN38 and proinsulin (Figure 6C and Supplemental Figure S9E), consistent with its role in retrograde trafficking to the Golgi as a component of the GARP complex.
DISCUSSION
In this study, we demonstrate that the EARP complex interacting-protein EIPR1 acts in a postprocessing maturation step to regulate proper insulin secretion and distribution of mature DCV cargo. Our data indicate that cells lacking EIPR1 are capable of stimulated insulin secretion but secrete less insulin, and have less insulin and other mature DCV cargoes in DCVs, supporting a role for EIPR1 in the maturation of DCVs. We further show that EIPR1 is required for localization of EARP, but not GARP, and that EIPR1 functions to control endocytic recycling and DCV cargo sorting. Consistent with its dual function, we find that the EARP complex localizes to two distinct compartments: a DCV biogenesis-related compartment and an endocytic-recycling compartment.
EIPR1 regulates DCV cargo levels and distribution
Our studies in C. elegans support a role for EIPR1 and EARP in controlling DCV cargo levels. C. elegans mutants in eipr-1 and the EARP complex subunits have reduced levels of cargo in mature DCVs and secrete less cargo (Paquin et al., 2016; Topalidou et al., 2016). Because these studies were based on the overexpression of exogenous DCV cargoes, here we investigated the role of EIPR1 in the distribution and secretion of endogenous DCV cargo using the insulin-secreting 832/13 cell line. We found that cells lacking EIPR1 remain responsive to stimulated secretion, but secrete less insulin and contain less insulin and mature cargo in DCVs. These phenotypes are similar to those observed in neurons in eipr-1 mutants in C. elegans, suggesting that neurons and endocrine cells share a conserved pathway for DCV biogenesis.
Our analysis of the levels and distribution of immature and mature DCV cargo in Eipr1 KO cells suggests that EIPR1 acts in a post-Golgi and postprocessing step to regulate DCV biogenesis. The total level of processed proprotein convertase 1/3 (PC1/3) is reduced in the absence of Eipr1. Also, processed mature CPE is both reduced and misdistributed in Eipr1 KO cells, but the level and distribution of the unprocessed form of CPE are not affected. If the reduction of PC1/3 and CPE led to a defect in the processing of proinsulin, we would expect an increase in proinsulin content in Eipr1 KO. However, total proinsulin levels in the Eipr1 KO were not increased, again suggesting that EIPR1 does not affect the processing of DCV cargoes. Additionally, Golgi pH is unaffected in Eipr1 KO cells. Together, these results suggest that EIPR1 is not required for early steps in the sorting and processing of DCV cargo.
Interestingly, Eipr1 KO cells have similar defects as Ccdc186 KO cells (Cattin-Ortolá, Topalidou, et al., 2019), suggesting that they both act at a post-Golgi step during DCV maturation. This is in contrast to other known regulators of DCV biogenesis such as PICK1, ICA69, and HID-1 that are needed to control the budding of immature DCVs from the TGN and whose loss leads to a defect in the processing of proinsulin to insulin (Cao, Mao, et al., 2013; Holst, Madsen, et al., 2013; Du, Zhou, Zhao, Cheng, et al., 2016; Hummer et al., 2017). Our findings suggest that EIPR1 and CCDC186 act at a later step in DCV biogenesis than PICK1, ICA69, and HID-1.
EIPR1 regulates the localization of the EARP complex and its association with membranes
The WD40 domain protein EIPR1 was identified as an interactor of the EARP and GARP complexes in rat insulin-secreting cells and human neuroglioma cells (Gershlick et al., 2016; Topalidou et al., 2016). Additionally, two mass spectrometry interactome data sets found that EIPR1 interacts with EARP subunits in human HEK293T or HeLa cells (Hein, Hubner, et al., 2015; Huttlin et al., 2015). VPS50 was shown to pull down VPS51, VPS52, VPS53, and EIPR1 as a stoichiometric complex (Hein, Hubner, et al., 2015), indicating that EIPR1 may form a stable complex with EARP. Although WD40 domain proteins often act as scaffolds for the assembly of large protein complexes (Stirnimann, Petsalaki, et al., 2010), our data suggest that EIPR1 is not needed for the formation of the EARP or GARP complex. First, EIPR1 is not required for interactions between individual subunits of the EARP and GARP complexes, as shown by coimmunoprecipitation experiments. Second, sedimentation of VPS50 was not affected by loss of EIPR1, suggesting that the EARP complex still forms in the absence of EIPR1. By contrast, we find that EIPR1 is needed for the stability of the individual EARP subunits, the localization of the EARP complex subunits, and association of EARP with membranes, supporting the model that EIPR1 recruits the EARP complex to its site of action and stabilizes it there.
Although localization of the EARP subunits is disrupted in the absence of EIPR1, localization of the GARP-specific VPS54 subunit is not affected. A recent study also showed that VPS54::GFP stably expressed in H4 cells is localized at the TGN region in both WT and Eipr1 knockdown (KD) cells, but fluorescence recovery after photobleaching analysis showed that EIPR1 contributes to GARP recruitment to the TGN (Gershlick et al., 2016), suggesting that EIPR1 is not needed for localization of GARP to the TGN per se, but is required to recruit GARP efficiently.
EIPR1 participates in EARP-specific functions
EARP and EIPR1 were recently shown to participate in the endocytic recycling of transferrin (Schindler, Chen, et al., 2015; Gershlick et al., 2016). We also found that EIPR1 is needed for normal endocytic recycling of transferrin in insulinoma cells. To test whether EIPR1 might also function with GARP, we examined the distribution of TGN38 in Eipr1 KO and Vps51 KD cells. In the absence of GARP, TGN38 is partially redistributed to cytoplasmic vesicles (Pérez-Victoria et al., 2008), but loss of EIPR1 had no obvious effect on the distribution of TGN38. Additionally, C. elegans GARP mutants were shown to have enlarged lysosomes in coelomocytes, but eipr-1 and vps-50 mutants do not have enlarged lysosomes (Topalidou et al., 2016). These results suggest that EIPR1 does not participate in GARP-specific functions, which agrees with the model that EIPR1 is required for the localization of EARP but not GARP. However, EIPR1 was reported to be required for the retrograde traffic of the Shiga toxin B subunit (STxB), whose trafficking also depends on GARP (Gershlick et al., 2016). We were unable to examine the trafficking of STxB in insulinoma 832/13 cells because we found that these cells do not take up STxB. These data together suggest that if EIPR1 functions with GARP, it is required for only a subset of GARP functions.
EARP localizes to two distinct compartments
The dual functionality of EIPR1 and EARP in endocytic recycling and DCV cargo retention prompted us to examine the cellular distribution of EARP. We found that EARP localizes to two distinct cellular sites, one that is associated with endosomes and one associated with CCDC186 near the TGN and immature DCVs. The endosomal recycling and DCV cargo sorting functions of EARP and EIPR1 may reflect two independent functions or may be interconnected. It is possible that the EARP complex moves from endosomes to the Golgi to participate in the retrieval of DCV cargo or sorting factors from an endosomal compartment. Alternatively, a pool of EARP localized near the TGN and immature DCVs may function in DCV biogenesis independently of a second pool of EARP that acts to traffic cargo out of endosomes.
A connection between endosomal trafficking and DCV biogenesis?
We have demonstrated here and elsewhere that EARP, an endosomal-recycling complex, and the EARP-interacting-protein EIPR1 are involved in DCV cargo retention (Topalidou et al., 2016), raising the possibility that DCV maturation at or near the TGN may require input from endosomal compartments. Several other studies have also suggested that there may be a role of endosomes in DCV biogenesis or maturation (Klumperman et al., 1998; Vo et al., 2004; Edwards et al., 2009; Sumakovic et al., 2009; Bäck et al., 2010; Topalidou et al., 2016; Zhang et al., 2017).
There are several possible ways that DCV maturation may be connected to endosomes. Studies in C. elegans suggested that RAB-2 acts in DCV biogenesis by ensuring that cargo is retained in mature DCVs and is not lost to the endolysosomal system (Edwards et al., 2009; Sumakovic et al., 2009). EIPR-1 and RAB-2 act in the same genetic pathway (Topalidou et al., 2016), suggesting that EIPR1 might also act to retain cargoes in mature DCVs and prevent their loss to the endolysosomal system. In agreement with this model, we found that disrupting trafficking to the endolysosomal system by overexpressing a constitutively active form of RAB-5 partially restored levels of DCV cargo in C. elegans eipr-1 mutants. However, expression of an analogous activated RAB5A construct did not restore the CPE localization defect in Eipr1 KO insulinoma cells, so the precise relationship between DCV maturation and endolysosomal trafficking remains unclear.
A second possible connection of endosomes to DCV maturation is the removal of cargoes from immature DCVs. The AP-1 adaptor that is involved in trafficking between the trans-Golgi and endosomes has been shown to associate with immature DCVs and to mediate the removal of syntaxin 6 and mannose 6-phosphate receptors from immature DCVs (Dittie et al., 1996; Klumperman et al., 1998). Once removed, such proteins possibly follow the endosomal route (Feng and Arvan, 2003; Arvan and Halban, 2004). In support of this idea, we found that EIPR1 is needed for the removal of carboxypeptidase D (CPD) from immature DCVs (Cattin-Ortolá, Topalidou, et al., 2019). CPD is localized to the TGN and immature DCVs, but is mostly absent from mature DCVs (Varlamov et al., 1999). In the absence of EIPR1, CPD remains in mature DCVs (Cattin-Ortolá, Topalidou, et al., 2019), suggesting that EIPR1 ensures the removal of CPD from immature DCVs. The RAB-2 effector CCDC186 was also shown to act in a similar way (Cattin-Ortolá, Topalidou, et al., 2019). We propose that EIPR1 and CCDC186 play a role in ensuring that the proper amount and type of cargo remains in mature DCVs.
Another possible connection of endosomes to DCV maturation is the recycling and retrieval of DCV cargoes from the plasma membrane. Following DCV exocytosis, transmembrane DCV cargoes may be recycled back to nascent DCVs via an endosomal pathway (Vo et al., 2004; Bäck et al., 2010). Additionally, a recent study identified a possible role for the secretory cell-specific Munc13-4 paralogue BAIAP3 in this DCV recycling pathway (Zhang et al., 2017). BAIAP3 was shown to localize to late and recycling endosomes and to be needed for DCV maturation and for more general endosome recycling to the TGN (Zhang et al., 2017). Because EARP and EIPR1 act in recycling plasma membrane proteins like transferrin out of endosomes, perhaps EARP and EIPR1 are also needed for the trafficking of recycled DCV membrane cargo out of endosomes to the cellular compartments where DCVs are formed and mature. It will be interesting to determine whether DCV membrane cargoes are recycled from the plasma membrane through endosomes in an EIPR1 and EARP-dependent manner.
MATERIALS AND METHODS
Cell culture
The 832/13 cell line is an INS-1–derived clone isolated by the lab of Christopher Newgard (Duke University School of Medicine; Hohmeier, Mulder, et al., 2000) and obtained by Duk-Su Koh via Ian Sweet (University of Washington). Cell lines were grown in RPMI 1640-GlutaMAX (GIBCO) medium supplemented with 10% fetal bovine serum (FBS; RMBIO), 1 mM sodium pyruvate (GIBCO), 10 mM HEPES (GIBCO), 1× Pen/Strep (GIBCO), and 0.0005% 2-β-mercaptoethanol at 5% CO2 and 37°C. Cells were transfected using Lipofectamine 2000 (Thermo Fisher) according to the manufacturer’s instructions.
Constructs
The plasmids VPS50::13Myc, VPS51::13Myc, VPS53::13Myc, VPS54::13Myc, VPS54::GFP, and VPS50::GFP were a gift from Juan Bonifacino, National Institutes of Health, Bethesda, MD (Pérez-Victoria et al., 2008; Pérez-Victoria and Bonifacino, 2009; Schindler, Chen, et al., 2015). The ANF::GFP plasmid (rat atrial natriuretic factor fused to GFP) was described (Hummer et al., 2017). The EIPR1_pBabe-hygro construct (pET222) used for making EIPR1(+) stable lines was constructed by amplifying rat EIPR1 cDNA from an 832/13 cDNA library using primers
oET513: 5′-ccatggatccatggaagacgacgccccg-3′
oET514: 5′-ctgagaattctcagagcagtatgtggtacttcagtgc-3′
The PCR product was digested by EcoRI/BamHI and cloned into EcoRI/BamHI-digested pBabe-hygro (a gift from Suzanne Hoppins, University of Washington, Seattle, WA).
The RAB5A[Q79L]-pEGFP-N1 construct (pET259) used for expressing mutant RAB5A in 832/13 cells was made by PCR amplifying rat RAB5A using primers
oJC388: 5′-gatctcgagctcaagcttcgATGGCTAATCGAGGAGCAACAAGACC-3′
oJC389: 5′-ccatggtggcgaccggtggatcGTTACTACAACACTGACTCCTGGCTG-3′
The PCR product was inserted in a pEGFP-N1 vector using Gibson cloning (Gibson et al., 2009) and mutagenized using the Quickchange II site directed mutagenesis kit (Agilent; #200523) and the following primer set:
oET645: 5′-AATATGGGATACAGCTGGCCtAGAACGGTATCATAGCCTA-3′
oJC388: 5′-TAGGCTATGATACCGTTCTaGGCCAGCTGTATCCCATATT-3′
Eipr1 knockout using CRISPR editing
To knock out EIPR1, we performed Cas9-mediated genome editing via homology-directed repair (HDR) in 832/13 cells using the protocol described (Ran, Hsu, et al., 2013).
For designing guide RNAs we used the online CRISPR design tool (Ran, Hsu, et al., 2013) and selected three guide RNAs that recognize sequences in or near the first exon of rat Eipr1:
guide 1: 5′-gacgacgccccggtgatctacggg-3′
guide 2: 5′-gagcccgagtcccgcctcaccagg-3′
guide 3: 5′-gtatcatggaagacgacgccccgg-3′
The guide RNAs were cloned into a pSpCas9(BB)-2A-GFP vector using the indicated protocol (Ran, Hsu, et al., 2013). The efficiency of the cloned guide RNAs was tested using the SURVEYOR nuclease assay (Supplemental Figure S1B) according to the manufacturer’s instructions (Surveyor Mutation Detection kit; Transgenomic). Guide RNA #1 (plasmid pET45) was selected for all subsequent experiments.
To design the homology-directed repair (HDR) template we used the pPUR (Clontech) vector as a backbone and cloned ∼1.5 kb Eipr1 homology arms upstream of and downstream from the puromycin selection cassette (Figure 1A and Supplemental Figure S1A). The HDR template was constructed using a Gibson assembly (plasmid pET69).
To cotransfect the CRISPR plasmid (carrying Cas9 and the guide RNA) and the HDR template, cells were grown in two 10-cm Petri dishes to near confluency. Cells were cοtransfected with 7 μg CRISPR plasmid and 7 μg nonlinearized HDR template using Lipofectamine 3000 according to the manufacturer’s instructions (Thermo Fisher). After 48 h transfection, the media was removed and replaced with new media together with 1 μg/ml puromycin. The puromycin selection was kept until individual clones could be picked, grown in individual dishes, and tested for CRISPR editing.
Individual puromycin-resistant clones were tested for proper CRISPR editing of the Eipr1 gene by extracting genomic DNA and performing PCR (Supplemental Figure S1C). The primers used for the PCR screening of positive clones were the following:
oET236: 5′-gaggtccgttcacccacag-3′ (hybridizes just upstream of the left homology arm)
oET237: 5′-gcctggggactttccacac-3′ (hybridizes in the SV40 promoter that drives the expression of the puromycin resistance gene)
Five out of 16 puromycin-resistant clones showed the band indicative of insertion of the puromycin cassette into Eipr1. To test for homozygosity of the insertion, we performed PCR using primers that amplify the WT Eipr1 locus:
oET236: 5′-gaggtccgttcacccacag-3′ (hybridizes just upstream of the left homology arm)
oET200: 5′-gagcagtatccaacacacacctc-3′ (hybridizes just downstream from the Cas9 cleavage site and in the first Eipr1 intron)
Of the 16 clones tested, three did not show the WT band with primers oET236 and oET200 (Supplemental Figure S1C) and were subsequently tested for EIPR1 expression by Western blot. Clone #3 showed no EIPR1 expression by Western blot (Figure 1B) and was kept as the Eipr1 KO line.
Lentiviral production, infection of cells, and selection of stable lines
Platinum-E (PlatE) retroviral packaging cells (a gift from Suzanne Hoppins) were grown for a couple of generations in DMEM-GlutaMAX (GIBCO) medium supplemented with 10% FBS (RMBIO), 1× Pen/Strep (GIBCO), 1 μg/ml puromycin, and 10 μg/ml blastocidin at 5% CO2 and 37°C. On day 1, ∼3.6 × 105 PlatE cells per well were plated in a six-well dish in DMEM-GlutaMAX medium supplemented with 10% FBS and 1× Pen/Strep. On day 2, a mix of 152 μl Opti-MEM (Thermo Fisher), 3 μg EIPR1_pBabe-hygro DNA, and 9 μl Fugene HD transfection reagent (Promega) was incubated for 10 min at room temperature and transfected into each well. On day 3, the media was removed and replaced with new PlatE media. On day 4, ∼1.5 × 105 Eipr1 KO 832/13 cells per well were plated in a six-well dish in RPMI 1640-GlutaMAX, supplemented with 10% FBS, 1 mM sodium pyruvate, 10 mM HEPES, 1× Pen/Strep, and 0.0005% 2-β-mercaptoethanol. Hexadimethrine bromide (3 μl of 8 mg/ml; Sigma) was added to each well. The supernatant of the PlatE cells (48 h viral supernatant) was collected with a sterile syringe, passed through a 0.45 micron filter, and added to the Eipr1 KO cells. The Eipr1 KO cells were incubated for 5–8 h at 5% CO2 and 37°C, then the media was changed and replaced with new media. The cells were incubated overnight at 5% CO2 and 37°C. On day 5, the supernatant was removed from the Eipr1 KO cells and replaced with the supernatant from the PlatE cells (72 h viral supernatant) after passing through a 0.45 micron filter. Hexadimethrine bromide (3 μl of 8 mg/ml) was added in each well, and the cells were incubated for 5–8 h. The media was replaced with new RPMI 1640-GlutaMAX media. On day 6, the Eipr1 KO cells were collected, transferred to a 10-cm Petri dish, and 200 μg/ml hygromycin was added. The cells were grown under hygromycin selection until individual clones could be picked and tested for EIPR1 expression.
Insulin and proinsulin secretion
Cells were grown in 24-well plates to near confluency, washed twice with phosphate-buffered saline (PBS), and incubated for 1 h in 200 μl per well resting buffer (5 mM KCl, 120 mM NaCl, 24 mM NaHCO3, 1 mM MgCl2, 15 mM HEPES, pH 7.4). The medium was collected, cleared by centrifugation, and stored at −80°C. The cells were incubated for 1 h in 200 μl per well stimulating buffer (55 mM KCl, 25 mM glucose, 70 mM NaCl, 24 mM NaHCO3, 1 mM MgCl2, 2 mM CaCl2, 15 mM HEPES, pH 7.4). After stimulation, the medium was cleared by centrifugation and stored at −80°C. The cells were washed once with PBS, harvested in PBS, and extracted in 100 μl per well acid-ethanol solution (absolute ethanol:H2O:HCl, 150:47:3). The pH of the acid-ethanol solution was neutralized by addition of 20 μl of 1 M Tris base per 100 μl of acid ethanol, and the samples were stored at −80°C.
Samples were assayed for insulin or proinsulin content using enzyme-linked immunosorbent assay (ELISA) according to the manufacturer’s instructions (Rat/Mouse insulin ELISA; Millipore; #EZRMI-13K; Rat/Mouse proinsulin ELISA; Mercodia; #10-1232-01). Secreted insulin and proinsulin levels were normalized against total cellular protein concentration and were presented as fraction of the WT under stimulating conditions (Figure 1, C and D, left panels). These values were then normalized against total cellular insulin or proinsulin levels (Figure 1, C and D, middle panels) to give the secretion data normalized against total insulin or proinsulin content (Figure 1, C and D, right panels).
Quantitative RT-PCR
WT and Eipr1 KO cells were grown in 10 cm plates, harvested in 1 ml of TRIzol (Invitrogen), and frozen at -80°. Total RNA was isolated following the manufacturer’s protocol and cDNA was synthesized from 1 μg RNA using the QuantiTekt Reverse Transcription kit (Qiagen) according to the manufacturer’s instructions. Each 10 μl qPCR contained 1 μl of cDNA and 5 μl of 2 × Sybr Green Master Mix (Kappa Biosystems). Absorbance was measured over 40 cycles using a CFX Connect Real-Time System (Bio-Rad). The cycle quantification value (Cq, cycle at which the amplification curve crosses a prescribed threshold value) for each sample was measured using the provided software and normalized to actin control. The primers used were the following:
Proinsulin 1 F: 5′-atggccctgtggatgc-3′
Proinsulin 1 R: 5′-tcagttgcagtagttctccagttg-3′
PC1/3 F: 5′-atgaagcaaagaggttggactc-3′
PC1/3 R: 5′-ttaattcttctcattcagaatgtcc-3′
Actin F: 5′-atggatgacgatatcgctgc-3′
Actin R: 5′-ctagaagcatttgcggtgc-3′
VPS50 F: 5′-atgcaaaaaatcaaatctcttatgacccgg-3′
VPS50 R: 5′-tcgtttaggtctgtctatatcatctatagc-3′
VPS51 F: 5′-atggcggccgcggcagctgtggggcctggc-3′
VPS51 R: 5′-gccgcgctcgcagatgacctcgacaacact-3′
Exocytosis assay
832/13 cells stably expressing NPY-pHluorin were transfected (FuGene; Promega) with NPY-mCherry. At 1 d after transfection, cells were washed once with PBS, dislodged using media, and transferred onto poly-l-lysine–coated 22-mm glass coverslips. After an additional 2 d, cells were reset for 2 h in low K+ Krebs-Ringer buffer with 1.5 mM glucose, washed once with low K+ Krebs-Ringer buffer with 1.5 mM glucose, and coverslips were transferred to an open imaging chamber (Life Technologies). Cells were imaged close to the coverslips, focusing on the plasma membrane (determined by the presence of NPY-mCherry–positive plasma membrane docked vesicles), using a custom-built Nikon spinning-disk confocal microscope at a resolution of 512 × 512 pixels. Images were collected for 100 ms at 10 Hz at room temperature with a 63× objective (Oil Plan Apo NA 1.49) and an ImageEM X2 EM-CCD camera (Hamamatsu, Japan). Following baseline data collection (15 s), an equal volume of Krebs-Ringer buffer containing 110 mM KCl and 30.4 mM glucose was added to stimulate secretion and cells were imaged for an additional 80 s. At the end of the experiment, cells were incubated with Krebs-Ringer buffer containing 50 mM NH4Cl, pH 7.4, to reveal total fluorescence and to confirm that the imaged cells were indeed transfected. Movies were acquired in MicroManager (UCSF) and exported as tiff files. Movies were analyzed using ImageJ software by counting events and measuring cell area.
Constitutive secretion assay
WT and Eipr1 KO cells were seeded on 12-well microtiter plates. When they reached subconfluency they were transfected using Lipofectamine 2000 with a plasmid expressing GFP fused to a signal peptide at its N-terminus (ssGFP; Hummer et al., 2017). After 48 h transfection, cells were incubated with resting media (114 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.16 mM MgSO4, 20 mM HEPES, 2.5 mM CaCl2, 25.5 mM NaHCO3, 3 mM glucose) for 1 h at 37°C. The secretion media was collected, centrifuged at 20,000 × g for 10 min at 4°C to remove cells and cell debris, and the supernatant was collected. The pelleted cells were lysed on ice in lysis buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, protease inhibitor cocktail without EDTA; Pierce). The cell lysate was centrifuged at 20,000 × g at 4°C for 10 min and the supernatant was collected. Protein concentration was measured using the BCA assay. GFP fluorescence of the media (secreted GFP) and the lysate were measured using a plate reader (Excitation = 485 nm; Emission = 525 nm; cutoff = 515 nm). Background fluorescence was measured from the media and lysate from nontransfected cells.
Immunostaining
Approximately 1–2 × 105 cells per well were plated onto coverslips (Thomas Scientific; #1217N79) placed in 24-well cell culture plates. Cells were transfected with Lipofectamine 2000 according to the manufacturer’s instructions at least 24 h after seeding. After 24–48 h, the cells were rinsed twice with PBS and fixed with 4% paraformaldehyde (PFA; made in PBS) for 20 min at room temperature. The cells were rinsed twice with PBS and permeabilized with 0.5% Triton X-100 in PBS for 5 min at room temperature. The cells were rinsed twice with PBS and placed in 5% milk in PBS for 1 h at room temperature. Cells were stained with primary antibodies in 0.5% milk in PBS at room temperature for 1 h. The following primary antibodies were used: mouse monoclonal anti–c-Myc (1:1000; Santa Cruz; sc-40), rabbit polyclonal anti-CCDC132/VPS50 (1:50; Sigma; #HPA026679), mouse monoclonal anti-GFP (1:200–1:350; Santa Cruz; #sc-9996), mouse monoclonal anti-insulin (1:350; Sigma; #K36AC10), mouse monoclonal anti-proinsulin (1:100; Abcam; #ab8301), rabbit polyclonal anti-TGN38 (1:350; Sigma; #T9826), rabbit polyclonal anti–pro CPE (1:100), rabbit polyclonal anti–CPE-C-terminus (1:100; Fricker et al., 1996), rabbit polyclonal anti–CPE-N-terminus (1:100; Fricker et al., 1990), mouse monoclonal anti–transferrin receptor (1:100; ThermoFisher; #13-6800), mouse monoclonal anti–syntaxin 6 (1:100; Abcam; #ab12370), rabbit polyclonal anti-CCDC186 (1:150; Novus; #NBP1-90440). The cells were then washed with PBS three times for 5 min each, and incubated with rhodamine anti-rabbit secondary antibody (1:1000; Jackson Immunoresearch; #111-025-144), Alexa Fluor 488 anti-rabbit secondary antibody (1:1000; Jackson Immunoresearch; #711-545-152), Alexa Fluor 488 anti-mouse secondary antibody (1:1000; Jackson Immunoresearch; #115-545-146), or Rhodamine Red-X anti-mouse secondary antibody (1:1000; Jackson Immunoresearch; #715-295-150) at room temperature for 1 h. The cells were washed with PBS three times for 5 min each, mounted onto glass slides using Vectashield (Vector Laboratories; H1000) or Prolong Diamond (Life Technologies; P36965) and examined by fluorescence microscopy. Images were obtained using a Nikon 80i wide-field compound microscope with a 60× oil objective (NA = 1.4) or an Olympus FLUOVIEW FV1200 confocal microscope with a 60× UPlanSApo oil objective (NA = 1.35). The acquisition software used for the Nikon was NIS-Elements and for the Olympus it was Fluoview v4.2. Pearson’s correlation coefficients were determined using Fiji and the coloc-2 plug-in by taking maximum intensity projections of z-stacks and drawing a line around each individual cell. For the quantification of the insulin or CPE that is retained at the Golgi, z-stack images were obtained using a Nikon 80i wide-field compound microscope. Maximum intensity projections were quantified using Fiji. The final fluorescence intensity is either the total fluorescence in a region of interest that includes the Golgi divided by either the fluorescence of a region of the same size in the cytoplasm of the cell (Golgi/cytoplasm) or by the background fluorescence of a region of the same size outside the cell (Golgi/background). We were not able to examine the localization of EIPR1 by immunostaining because our antibody does not work in immunofluorescence.
Protein extraction
For protein extraction or coimmunoprecipitation, ∼ 4 × 106 832/13 cells were plated onto 10-cm plates. When transfection was required, cells were transfected 24 h later with 15 μg of the relevant plasmid (or 8 μg each if two plasmids were transfected). After 24–48 h the cells were washed with cold PBS twice and harvested in lysis buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP40, and protease inhibitor cocktail (Pierce). Lysates were transferred to microcentrifuge tubes and passed 10 times through a 20-gauge needle followed by incubation for 30 min at 4°. Lysates were centrifuged at 20,000 × g for 15 min at 4°.
Immunoblotting
For the blots shown in Figures 1B, 7, D–G, and 8B, membranes were blocked in 3% milk in TBST (50 mM Tris, pH 7.4, 150 mM NaCl, 0.1% Tween 20) for 1 h at room temperature and stained with the relevant antibodies in 3% milk in TBST overnight, followed by three 5-min washes in TBST. For the blots shown in Figures 3, A–D, 5A, and 7, A–C, H, and I, membranes were blocked with Odyssey Blocking Buffer in PBS (#927-40000). Antibodies were incubated in the same buffer supplemented with 0.2% Tween 20 and washed with PBST (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4, supplemented with 0.1% Tween 20). The following primary antibodies were used: rabbit polyclonal anti-GFP (1:1000; a gift from Alexey Merz, University of Washington, Seattle, WA), mouse monoclonal anti–c-Myc (1:1000; Santa Cruz; sc-40), mouse monoclonal anti–β-tubulin (1:1000; ThermoFisher; BT7R; #MA5-16308), mouse monoclonal anti–β-tubulin (1:1,000; DHSB; E7), rabbit polyclonal anti-PCSK1 (PC1/3; 1:1000; Sigma; #SAB1100415), rabbit polyclonal anti-TSSC1/EIPR1 (1:1000; Thermo Scientific; #PA5-22360), rabbit polyclonal anti-CCDC132/VPS50 (1:1000; Sigma; #HPA026679), rabbit polyclonal anti-VPS51 (1:1000; Atlas antibodies; #HPA061447), rabbit polyclonal PC2 (1:1000; #13/4; a gift from Sharon Tooze, The Francis Crick Institute, London, UK; Dittié and Tooze, 1995), rabbit polyclonal anti–pro-CPE (1:1000; a gift from Lloyd D. Fricker, Albert Einstein College of Medicine, Bronx, NY; antiserum to pro-CPE was generated to the peptide QEPGAPAAGMRRC coupled to maleimide-activated keyhole limpet hemocyanin [KLH]; this peptide corresponds to the 12 residues of mouse/rat/human pro-CPE with an added Cys on the C-terminus [for coupling to the carrier protein, KLH]), rabbit polyclonal anti–CPE-C (1:1000; antiserum was raised against the nine-residue peptide KMMSETLNF corresponding to the C-terminus of mouse CPE; Fricker et al., 1996), rabbit polyclonal anti–CPE-N (1:1000; antiserum was raised to the N-terminal 15 amino acids of bovine CPE; Fricker et al., 1990), mouse monoclonal anti-transferrin receptor (1:1000; ThermoFisher; #13-6800). Membranes were stained with the relevant secondary antibodies either in 3% milk in TBST, followed by three 5-min washes in TBST, or in Odyssey Blocking Buffer (PBS) supplemented with 0.2% Tween 20 and 0.01% SDS, and washed with PBST. The secondary antibodies used were an Alexa Fluor 680–conjugated goat anti-mouse antibody (1:20,000; Jackson Laboratory; #115-625-166), Alexa Fluor 790–conjugated donkey anti-mouse antibody (1:20,000; Jackson Laboratory; #715-655-150), or Alexa Fluor 680–conjugated goat anti-rabbit antibody (1:20,000; Jackson Laboratory; #111-625-144). A LI-COR processor was used to develop images.
pH measurement of the late-Golgi compartment
WT and Eipr1 KO cells stably expressing St6Gal1::pHluorin were generated as follows. HEK293T cells were maintained in DMEM with 10% FBS under 5% CO2 at 37°C. Lentivirus was produced by transfecting HEK293T cells with FUGW, psPAX2, and pVSVG using Fugene HD according to the manufacturer’s instructions. After 2 d transfection, the medium was collected and filtered (0.45 µm). Medium containing the lentivirus was then applied to 832/13 cells in suspension at various ratios. Transduction efficiency was screened using an epifluorescence microscope. The protocol to measure the pH of the late-Golgi compartment (Hummer et al., 2017) was adapted to a plate-reader format. An equal number of WT and Eipr1 KO 832/13 cells stably expressing St6Gal1::pHluorin were plated on clear-bottom black-wall 96-well plates and grown to confluence. Cells were washed once with Tyrode buffer (119 mM NaCl, 2.5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 25 mM HEPES, 30 mM glucose, pH 7.4) and incubated for 10 min either in Tyrode buffer or in KCl-enriched buffer at different pHs (125 mM KCl, 20 mM NaCl, 0.5 mM CaCl2, 0.5 mM MgCl2, and 25 mM MES, at pH 5.5 or 6, or 25 mM HEPES, at pH 6.5, 7, 7.5, 8, or 8.5). The KCl-enriched buffer was supplemented with 5 μM nigericin (Sigma Aldrich) and 5 nM monensin (Sigma Aldrich). The fluorescence of each well was measured using a Varioskan Lux plate reader (Thermo Scientific; Excitation = 485 nm; Emission = 520 nm). For each buffer, cell type, and independent biological replicate, the reading was repeated three times. Calibration curves were generated for each cell type and each independent repetition. The absolute pH values were extrapolated from the calibration curve using the fluorescence of the cells incubated with Tyrode buffer.
ANF-GFP pulse chase
To monitor the exit of DCV cargo from the TGN, we used a protocol similar to the one described (Kögel et al., 2013). WT and Eipr1 KO 832/13 cells were seeded on glass coverslips and grown for 24 h. We then transfected 100 ng of ANF::GFP (Hummer et al., 2017) with Lipofectamine 2000 for 12–16 h at 37°C in complete growth medium. Cells were then incubated at 20°C in PBS for 2 h in a conventional incubator (pulse) to block the formation of DCVs. At 30 min before the end of the low temperature block, 10 μg/ml cyclohexamide was added to the PBS to block the synthesis of new ANF::GFP. The PBS was then exchanged for growth medium and the cells were shifted to 37°C (chase) for the indicated times and then were fixed with 4% PFA, stained, and imaged as described (see Immunostaining). Cells were scored in three categories: those that had most of the ANF::GFP concentrated at the TGN (“Golgi-like”), those that had ANF::GFP both at the TGN region and at the cell periphery (“Intermediate”), and those where the ANF::GFP was excluded from the TGN (“Periphery”). For each genotype, 50–100 cells per time point were counted. The experimenter was blind to the genotypes of the cell lines used and the time point. The experiment was repeated three times with similar results.
Worm strains
Worm strains were cultured using standard methods (Brenner, 1974). The following strains were used in this study:
KG1395 nuIs183[Punc-129::NLP-21-Venus, Pmyo-2::NLS-GFP] III
XZ1026 rab-2(nu415) I; nuIs183[Punc-129::NLP-21-Venus, Pmyo-2::NLS-GFP] III
-
XZ1055 eipr-1(tm4790) I; nuIs183[Punc-129::NLP-21-Venus, Pmyo-2::NLS-GFP] III
XZ1996 yakIs30[Prab-3::mCherry::RAB-5(QL), Pmyo-2::mCherry]
XZ2021 rab-2(nu415) I; nuIs183[Punc-129::NLP-21-Venus, Pmyo-2::NLS-GFP] III; yakIs30[Prab-3::mCherry::RAB-5(QL), Pmyo-2::mCherry]
XZ2022 eipr-1(tm4790) I; nuIs183[Punc-129::NLP-21-Venus, Pmyo-2::NLS-GFP] III; yakIs30[Prab-3::mCherry::RAB-5(QL); Pmyo-2::mCherry]
Worm imaging and image analysis
Young adult worms were washed off plates with M9 and centrifuged for 3 min at 2000 rpm. Five microliters of the worm pellet was placed on 2% agarose pads together with 5 μl of 100 mM sodium azide and the animals were anesthetized for 10 min. The dorsal cord of the animals around the vulva was imaged in all worms using a Nikon 80i wide-field compound microscope. Maximum intensity projections of z-stacks were quantified using ImageJ. The final fluorescence intensity for each animal is the averaged total fluorescence in five regions of interest minus the background fluorescence of a region of the same size next to the dorsal cord.
Equilibrium sedimentation
WT and Eipr1 KO 832/13 cells were grown in 15-cm tissue culture plates until confluent. Cells were washed twice with ice-cold PBS, transferred to 15 ml conical tubes, and centrifuged at 300 × g for 10 min at 4°C. Cells were washed once with 5 ml of SH buffer (10 mM HEPES, pH 7.2, 0.3 M sucrose, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail), resuspended in 1 ml SH buffer, passed once through a 21-gauge needle, and homogenized using a ball bearing device (Isobiotec; 18 μm clearance). The postnuclear lysate was collected after centrifugation at 1000 × g for 8 min. A 0.6–1.9 M continuous sucrose gradient was prepared in 10 mM HEPES, pH 7.2, in ultracentrifuge tubes (Beckman Coulter) that were coated with Sigmacote (Sigma; #SL2). The postnuclear supernatant was loaded onto the gradient and centrifuged at 30,000 rpm in an SW41 rotor for 14–16 h at 4°C. Fractions of 750 μl each were collected from top to bottom. The sucrose concentration for each fraction was determined by measuring the Brix using a Misco palm abbe digital refractometer. Fractions were analyzed by immunoblotting using an antibody to PC1/3 (Sigma; #SAB1100415). Band intensity was quantified using Fiji and plotted against the sucrose concentration of the fraction.
Cell fractionation
We used a method similar to the one described (Cattin-Ortolá et al., 2017). Specifically, WT 832/13 or Eipr1 KO 832/13 cells were seeded on a 24-well cell culture plate and grown until subconfluence. Where indicated, cells were transfected with VPS51::13Myc or VPS54::13Myc for 48 h before fractionation using Lipofectamine 2000, according to the manufacturer’s instructions. Cells were washed twice with ice-cold PBS, transferred into microcentrifuge tubes in PBS, and centrifuged at 500 × g for 10 min at 4°C. Cells were resuspended in lysis buffer (20 mM HEPES, pH 7.4, 250 mM sucrose supplemented with protease inhibitors from Pierce) and disrupted by repeated passage through a 30-gauge needle. Cell lysates were centrifuged twice at 1000 × g for 10 min at 4°C. The postnuclear supernatant was transferred into an ultracentrifuge tube (Beckman Coulter; #343775) and centrifuged at 100,000 × g for 1 h at 4°C in a TLA100 rotor. The supernatant was removed and supplemented with 6× SDS loading dye. The pellet was washed once with lysis buffer, resuspended in an equal volume of lysis buffer, and supplemented with SDS loading dye. Samples were analyzed by immunoblotting as described above.
Coimmunoprecipitation
The anti-GFP nanobody was expressed and purified as described (Topalidou et al., 2016). Approximately 1/10th of the supernatant (see protein extraction) was kept as input control for subsequent immunoblot analysis. The remaining lysate was incubated with 20 μg anti-GFP nanobody bound to magnetic beads for 2–4 h at 4°C. The beads were washed three times with lysis buffer and resuspended in Laemmli loading buffer. The input and immunoprecipitated samples were resolved on 8% SDS–polyacrylamide gels and blotted onto PVDF membranes.
Cofractionation on velocity sucrose gradients
WT or Eipr1 KO 832/13 cells were grown to confluence in 15-cm tissue culture plates. Cells were washed twice with ice-cold PBS, transferred into microcentrifuge tubes in PBS, and centrifuged at 500 × g for 10 min at 4°C. Cells were resuspended in lysis buffer (50 mM Tris-Cl, pH 7.6, 150 mM NaCl, 1% NP-40, 1 mM EDTA, protease inhibitor cocktail from Pierce), kept on ice for 20 min, and centrifuged at 20,000 × g for 10 min at 4°C. Continuous sucrose gradients were prepared by overlaying 8% sucrose on 30% sucrose (or 8–25% as indicated) in 50 mM Tris-Cl, pH 7.6, 150 mM NaCl, 1% NP-40, 1 mM EDTA, in a 3.5-ml ultracentrifuge tube (Beckman Coulter; #349622). The tubes were sealed with parafilm, set horizontally for 2 h at 4°C, and then vertically for 1 h at 4°C. The clarified lysate (or sizing standards) was loaded on top of the gradient. Tubes were centrifuged at 100,000 × g for 16 h at 4°C in a SW50 rotor. Fractions were collected from top to bottom and supplemented with 6× SDS sample loading buffer. EARP complex subunits were analyzed by immunoblotting using antibodies to the endogenous proteins. Sizing standards (Bio-Rad; #151-1901) were analyzed by Coomassie-stained SDS–PAGE gels.
Transferrin uptake and recycling assays and immunostaining assays
For the transferrin uptake and recycling assays, WT, Eipr1 KO, and Ccdc186 KO 832/13 cells were seeded onto 24-well plates and grown in complete media until they reached confluence. For the uptake, cells were placed in warm uptake medium (serum-free RPMI + 1% BSA + 25 mM HEPES + 50 µg/ml Alexa 488 transferrin; Invitrogen; #T13342) for the indicated times (Figure 8A, top). For the recycling, cells were placed in warm uptake medium for a 25-min pulse. Medium was then exchanged for complete RPMI medium and cells were chased for the indicated times.
Cells were then transferred to ice, washed twice with ice-cold PBS, and detached on ice with 10 mM EDTA in PBS for 30 min to 1 h with manual agitation and gentle pipetting. Detached single cells were transferred into a microcentrifuge tube and fixed at 4°C with 4% PFA (final concentration ∼3.5%) for 10 min on a Nutator to avoid clumping. Fixed cells were washed with PBS and analyzed by FACS using a LSRII (BD Biosciences). Data were analyzed using FlowJo.
For transferrin immunostaining, cells were seeded onto coverslips in 24-well plates for 24–48 h. Cells were then placed in warm uptake medium for a 25-min pulse (serum-free RPMI + 1% BSA + 25 mM HEPES + 50 µg/ml Alexa 568 transferrin; Invitrogen; #T23365), washed twice with PBS, and fixed with 4% PFA as already described.
VPS51 knockdown by RNAi
VPS51 knockdown was performed using stealth siRNA from Life Technologies (#10620312-353281 D10; 5′-GAUGGACAGUGAGAcGGACAUGGUG-3′). Transfection of oligonucleotides (20 nM) was done using Lipofectamine 2000 as follows: cells were seeded on coverslips placed in 24-well cell culture plates until they reached ∼50% confluence. On days 1 and 3, cells were transfected with 1 μl of oligonucleotides and 1 μl Lipofectamine according to the manufacturer’s instructions. On day 5, cells were stained with the relevant antibodies following the standard immunostaining procedure.
Statistics
Data were tested for normality by a Shapiro-Wilk test. When the data did not pass the normality test, we used the Kruskal-Wallis test followed by Dunn’s test to investigate whether there was statistical significance between groups of three or more. When data passed the normality test, we used a one-way analysis of variance (ANOVA) test with Bonferroni correction when making comparisons among groups of three or more, and an unpaired t test for comparisons between groups of two.
Supplementary Material
ACKNOWLEDGMENTS
We thank Christopher Newgard, Ian Sweet, and Duk-Su Koh for the 832/13 cell line, with the support of the UW DRC Cell Function and Analysis Core; Suzanne Hoppins for the pBabe vector, platE cells, and protocol for lentiviral production and infection; Juan Bonifacino for the EARP and GARP subunit plasmids; Alex Merz and Sharon Tooze for antibodies; and Lloyd Fricker for antibodies and helpful discussions. J.C. was supported in part by a National Institutes of Health (NIH) Institutional Training Grant for Neurobiology (Grant no. T32 GM-007108). This work was supported by American Diabetes Association Grant no. 1-17-JDF-064 and by NIH Grant no. R01 GM-124035 to C.S.A., by a University of Washington Diabetes Research Center Pilot and Feasibility Award (NIH Grant no. P30 DK-017047), an Ellison Medical Foundation New Scholar Award, and by NIH Grant no. R00 MH-082109 and Grant no. R01 GM-121481 to M.A.
Abbreviations used:
- CPD
carboxypeptidase D
- CPE
carboxypeptidase E
- DCV
dense-core vesicle
- EARP
endosome-associated recycling protein
- GARP
Golgi-associated retrograde protein
- PC1/3
proprotein convertase 1/3
- TGN
trans-Golgi network.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E18-07-0469) on November 13, 2019.
REFERENCES
Boldface names denote co–first authors.
- Ailion M, Hannemann M, Dalton S, Pappas A, Watanabe S, Hegermann J, Liu Q, Han H-F, Gu M, Goulding MQ, et al. (2014). Two Rab2 interactors regulate dense-core vesicle maturation. Neuron , 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvan P, Halban PA. (2004). Sorting ourselves out: seeking consensus on trafficking in the beta-cell. Traffic , 53–61. [DOI] [PubMed] [Google Scholar]
- Asensio CS, Sirkis DW, Edwards RH. (2010). RNAi screen identifies a role for adaptor protein AP-3 in sorting to the regulated secretory pathway. J Cell Biol , 1173–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asensio CS, Sirkis DW, Maas JW, Jr, Egami K, To T-L, Brodsky FM, Shu X, Cheng Y, Edwards RH. (2013). Self-assembly of VPS41 promotes sorting required for biogenesis of the regulated secretory pathway. Dev Cell , 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäck N, Rajagopal C, Mains RE, Eipper BA. (2010). Secretory granule membrane protein recycles through multivesicular bodies. Traffic , 972–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. (1974). The genetics of Caenorhabditis elegans. Genetics , 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Mao Z, Kam C, Xiao N, Cao X, Shen C, Cheng KKY, Xu A, Lee K-M, Jiang L, et al. (2013). PICK1 and ICA69 control insulin granule trafficking and their deficiencies lead to impaired glucose tolerance. PLoS Biol , e1001541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin-Ortolá J, Topalidou I, Dosey A, Merz AJ, Ailion M. (2017). The dense-core vesicle maturation protein CCCP-1 binds RAB-2 and membranes through its C-terminal domain. Traffic , 720–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattin-Ortolá J, Topalidou I, Lau H-T, Hummer B, Asensio CS, Ong S-E, Ailion M. (2019). CCDC186 recruits EARP and controls dense-core vesicle cargo sorting by exit. bioRxiv 616458, posted 23 April 2019. 10.1101/616458 [DOI] [Google Scholar]
- Conibear E, Cleck JN, Stevens TH. (2003). Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol Biol Cell , 1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conibear E, Stevens TH. (2000). Vps52p, Vps53p, and Vps54p form a novel multisubunit complex required for protein sorting at the yeast late Golgi. Mol Biol Cell , 305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittie AS, Hajibagheri N, Tooze SA. (1996). The AP-1 adaptor complex binds to immature secretory granules from PC12 cells, and is regulated by ADP-ribosylation factor. J Cell Biol , 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittié AS, Tooze SA. (1995). Characterization of the endopeptidase PC2 activity towards secretogranin II in stably transfected PC12 cells. Biochem J , 777–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du W, Zhou M, Zhao W, Cheng D, Wang L, Lu J, Song E, Feng W, Xue Y, Xu P, et al. (2016). HID-1 is required for homotypic fusion of immature secretory granules during maturation. eLife , e18134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SL, Charlie NK, Richmond JE, Hegermann J, Eimer S, Miller KG. (2009). Impaired dense core vesicle maturation in Caenorhabditis elegans mutants lacking Rab2. J Cell Biol , 881–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Arvan P. (2003). The trafficking of alpha 1-antitrypsin, a post-Golgi secretory pathway marker, in INS-1 pancreatic beta cells. J Biol Chem , 31486–31494. [DOI] [PubMed] [Google Scholar]
- Fricker LD, Berman YL, Leiter EH, Devi LA. (1996). Carboxypeptidase E activity is deficient in mice with the fat mutation. Effect on peptide processing. J Biol Chem , 30619–30624. [DOI] [PubMed] [Google Scholar]
- Fricker LD, Das B, Angeletti RH. (1990). Identification of the pH-dependent membrane anchor of carboxypeptidase E (EC 3.4.17.10). J Biol Chem , 2476–2482. [PubMed] [Google Scholar]
- Gershlick DC, Schindler C, Chen Y, Bonifacino JS. (2016). TSSC1 is novel component of the endosomal retrieval machinery. Mol Biol Cell , 2867–2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods , 343–345. [DOI] [PubMed] [Google Scholar]
- Gillingham AK, Sinka R, Torres IL, Lilley KS, Munro S. (2014). Toward a comprehensive map of the effectors of rab GTPases. Dev Cell , 358–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondré-Lewis MC, Park JJ, Loh YP. (2012). Cellular mechanisms for the biogenesis and transport of synaptic and dense-core vesicles. Int Rev Cell Mol Biol , 27–115. [DOI] [PubMed] [Google Scholar]
- Hannemann M, Sasidharan N, Hegermann J, Kutscher LM, Koenig S, Eimer S. (2012). TBC-8, a putative RAB-2 GAP, regulates dense core vesicle maturation in Caenorhabditis elegans. PLoS Genet , e1002722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. (2015). A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell , 712–723. [DOI] [PubMed] [Google Scholar]
- Hohmeier HE, Mulder H, Chen G, Henkel-Rieger R, Prentki M, Newgard CB. (2000). Isolation of INS-1-derived cell lines with robust ATP-sensitive K+ channel-dependent and -independent glucose-stimulated insulin secretion. Diabetes , 424–430. [DOI] [PubMed] [Google Scholar]
- Holst B, Madsen KL, Jansen AM, Jin C, Rickhag M, Lund VK, Jensen M, Bhatia V, Sørensen G, Madsen AN, et al. (2013). PICK1 deficiency impairs secretory vesicle biogenesis and leads to growth retardation and decreased glucose tolerance. PLoS Biol , e1001542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummer BH, de Leeuw NF, Burns C, Chen L, Joens MS, Hosford B, Fitzpatrick JAJ, Asensio CS. (2017). HID-1 controls formation of large dense core vesicles by influencing cargo sorting and trans-Golgi network acidification. Mol Biol Cell , 3870–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Ting L, Bruckner RJ, Gebreab G, Gygi MP, Szpyt J, Tam S, Zarraga G, Colby G, Baltier K, et al. (2015). The BioPlex network: a systematic exploration of the human interactome. Cell , 425–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumperman J, Kuliawat R, Griffith JM, Geuze HJ, Arvan P. (1998). Mannose 6-phosphate receptors are sorted from immature secretory granules via adaptor protein AP-1, clathrin, and syntaxin 6-positive vesicles. J Cell Biol , 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kögel T, Rudolf R, Hodneland E, Copier J, Regazzi R, Tooze SA, Gerdes H-H. (2013). Rab3D is critical for secretory granule maturation in PC12 cells. PLoS One , e57321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesa R, Luo S, Hoover CM, Miller K, Minniti A, Inestrosa N, Nonet ML. (2011). HID-1, a new component of the peptidergic signaling pathway. Genetics , 467–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller L, Lindberg I. (1999). The cell biology of the prohormone convertases PC1 and PC2. Prog Nucleic Acid Res Mol Biol , 69–108. [DOI] [PubMed] [Google Scholar]
- Naggert JK, Fricker LD, Varlamov O, Nishina PM, Rouille Y, Steiner DF, Carroll RJ, Paigen BJ, Leiter EH. (1995). Hyperproinsulinaemia in obese fat/fat mice associated with a carboxypeptidase E mutation which reduces enzyme activity. Nat Genet , 135–142. [DOI] [PubMed] [Google Scholar]
- Paquin N, Murata Y, Froehlich A, Omura DT, Ailion M, Pender CL, Constantine-Paton M, Horvitz HR. (2016). The conserved VPS-50 protein functions in dense-core vesicle maturation and acidification and controls animal behavior. Curr Biol , 862–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Victoria FJ, Bonifacino JS. (2009). Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-Golgi network. Mol Cell Biol , 5251–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Victoria FJ, Mardones GA, Bonifacino JS. (2008). Requirement of the human GARP complex for mannose 6-phosphate-receptor-dependent sorting of cathepsin D to lysosomes. Mol Biol Cell , 2350–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Victoria FJ, Schindler C, Magadan JG, Mardones GA, Delevoye C, Romao M, Raposo G, Bonifacino JS. (2010). Ang2/fat-free is a conserved subunit of the Golgi-associated retrograde protein complex. Mol Biol Cell , 3386–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc , 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Chen Y, Pu J, Guo X, Bonifacino JS. (2015). EARP is a multisubunit tethering complex involved in endocytic recycling. Nat Cell Biol , 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeekens SP, Montag AG, Thomas G, Albiges-Rizo C, Carroll R, Benig M, Phillips LA, Martin S, Ohagi S, Gardner P. (1992). Proinsulin processing by the subtilisin-related proprotein convertases furin, PC2, and PC3. Proc Natl Acad Sci USA , 8822–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Fricker L. (1995). Processing of procarboxypeptidase E into carboxypeptidase E occurs in secretory vesicles. J Neurochem , 444–453. [DOI] [PubMed] [Google Scholar]
- Stirnimann CU, Petsalaki E, Russell RB, Müller CW. (2010). WD40 proteins propel cellular networks. Trends Biochem Sci , 565–574. [DOI] [PubMed] [Google Scholar]
- Sumakovic M, Hegermann J, Luo L, Husson SJ, Schwarze K, Olendrowitz C, Schoofs L, Richmond J, Eimer S. (2009). UNC-108/RAB-2 and its effector RIC-19 are involved in dense core vesicle maturation in Caenorhabditis elegans. J Cell Biol , 897–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalidou I, Cattin-Ortolá J, Pappas AL, Cooper K, Merrihew GE, MacCoss MJ, Ailion M. (2016). The EARP complex and its interactor EIPR-1 are required for cargo sorting to dense-core vesicles. PLoS Genet , e1006074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlamov O, Eng FJ, Novikova EG, Fricker LD. (1999). Localization of metallocarboxypeptidase D in AtT-20 cells. Potential role in prohormone processing. J Biol Chem , 14759–14767. [DOI] [PubMed] [Google Scholar]
- Vindrola O, Lindberg I. (1992). Biosynthesis of the prohormone convertase mPC1 in AtT-20 cells. Mol Endocrinol , 1088–1094. [DOI] [PubMed] [Google Scholar]
- Vo YP, Hutton JC, Angleson JK. (2004). Recycling of the dense-core vesicle membrane protein phogrin in Min6 beta-cells. Biochem Biophys Res Commun , 1004–1010. [DOI] [PubMed] [Google Scholar]
- Wong SH, Low SH, Hong W. (1992). The 17-residue transmembrane domain of beta-galactoside alpha 2,6-sialyltransferase is sufficient for Golgi retention. J Cell Biol , 245–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Wang L, Jiu Y, Zhan Y, Liu L, Xia Z, Song E, Xu P, Xu T. (2011). HID-1 is a novel player in the regulation of neuropeptide sorting. Biochem J , 383–390. [DOI] [PubMed] [Google Scholar]
- Zhang X, Jiang S, Mitok KA, Li L, Attie AD, Martin TFJ. (2017). BAIAP3, a C2 domain-containing Munc13 protein, controls the fate of dense-core vesicles in neuroendocrine cells. J Cell Biol , 2151–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Lindberg I. (1993). Purification and characterization of the prohormone convertase PC1(PC3). J Biol Chem , 5615–5623. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.