Abstract
Computed tomographic (CT) is a fundamental imaging modality to generate cross-sectional views of internal anatomy in a living subject or interrogate material composition of an object, and it has been routinely used in clinical applications and nondestructive testing. In a standard CT image, pixels having the same Hounsfield Units (HU) can correspond to different materials, and it is therefore challenging to differentiate and quantify materials. Dual-energy CT (DECT) is desirable to differentiate multiple materials, but the costly DECT scanners are not widely available as single-energy CT (SECT) scanners. Recent advancement in deep learning provides an enabling tool to map images between different modalities with incorporated prior knowledge. Here we develop a deep learning approach to perform DECT imaging by using the standard SECT data. The end point of the approach is a model capable of providing the high-energy CT image for a given input low-energy CT image. The feasibility of the deep learning-based DECT imaging method using a SECT data is demonstrated using contrast-enhanced DECT images and evaluated using clinical relevant indexes. This work opens new opportunities for numerous DECT clinical applications with a standard SECT data and may enable significantly simplified hardware design, scanning dose and image cost reduction for future DECT systems.
Keywords: Dual-energy computed tomography, Single-energy computed tomography, Deep learning, Convolutional neural network, Material decomposition, Virtual non-contrast, Iodine quantification
1. Introduction
Pixel number in a standard single-energy computed tomography (SECT) image represents effective linear attenuation coefficient and it is an averaged contribution of all materials or chemical elements in the pixel. It therefore does not give a unique description for any given material, and pixels having the same CT numbers can represent materials with different elemental compositions, making the differentiation and quantification of materials extremely challenging. Dual-energy CT (DECT) scans the object using two different energy spectra and is able to take advantage of the energy dependence of the linear attenuation coefficients to yield material-specific images.1–20 This enables DECT to be applied in several emerging clinical applications, including virtual monoenergetic imaging, automated bone removal in CT angiography, perfused blood volume, virtual noncontrast-enhanced images, urinary stone characterization and so on.21
In practice, leading industrial CT vendors have used different techniques to acquire dual-energy data. For examples, GE Healthcare scanners use rapid switching of x-ray tube potential to acquire alternate projection measurements at low and high-energy spectra.11,22 This technique requires the transition time from low to high tube potential between consecutive views to be less than a millisecond while separating the spectra as much as possible, which is very technically challenging. Siemens Healthineers scanners use dual x-ray sources and two data acquisition systems, both of which are mounted on the same gantry.7 Thus, the dual-source scanner cost is much higher than the standard SECT scanner. Philips Healthcare scanner acquires DECT projection data using a layered detector, with which the low-energy data and the high-energy data are collected by the front detector layer and the back-detector layer, respectively.6,23 All the DECT data acquisition techniques have posed a significant burden on CT system hardware. Hence, DECT scanners are not widely available as SECT scanners, especially for less-developed regions. In addition to the increased complexity of the imaging system and cost, DECT may also increase the radiation dose to patients due to the additional CT scan. In this work, we demonstrate a standard DECT imaging using images acquired by a SECT scanner is feasible by leveraging from the state-of-the-art deep learning technique and seamless integration of prior DECT knowledge in the deep learning model training process. It is intriguing to note that the proposed method enables DECT clinical applications (such as iodine quantification, virtual noncontrast-enhanced imaging) to be performed using a SECT data, which has potential to provide a fundamental paradigm for DECT imaging applications for less developed regions where only SECT scanners are available and high-end DECT scanners are not affordable.
Deep neural network has recently attracted much attention for its unprecedented ability to learn complex relationships and incorporate existing knowledge into the inference model through feature extraction and representation learning.24 The method has found widespread applications in biomedicine.25–32 Here we introduce a hierarchical neural network for DECT imaging with a SECE data and demonstrate the superior performance of the deep learning-based DECT method using a popular DECT clinical application. The essences of our approach are the extraction of the intrinsic features between the differences of the high- and low-energy CT images by exclusion of the image noise, and the construction of a robust encoder/decoder network architecture to map the differences. By using the network architecture, we incorporate the intrinsic differences learned from paired DECT images acquired from commercial scanners into a deep learning model for the high-energy CT image prediction in the subsequent new low-energy CT image input.
2. Methods
In this study, we aim to train a deep learning model to transform low-energy CT image IL to high-energy CT image IH using routinely available paired DECT images. To this end, we first use a fully convolutional network derived from ResNet to significantly reduce the image noise.33 This procedure is performed in an ene-to-end fashion to provide low noise DECT images IHD and ILD. With these images, instead of directly mapping the high-energy CT image from the low-energy CT image, an independent mapping convolutional neural network (CNN), which we call DECT-CNN, is trained to learn an difference Idiff image between IHD and ILD for the given input ILD. Then a predicted high-energy CT image Ipred is calculated as the summation of the origianl low-energy CT image IL and the predicted difference image Idiff during inference procedure. The DECT-CNN model is based on an U-Net-type architecture and the mean-squared-error is used as the loss function. During training, the loss function is minimized using the adaptive moment estimation (ADAM) algorithm. Weights in the convolutional kernels are updated using back-propagation method.
The proposed deep learning-based DECT imaging approach was evaluated using a popular DE application: virtual non-contrast (VNC) imaging and iodine contrast agent quantification. The VNC images and the iodine maps were decomposed using a statistically optimal image-domain material decomposition algorithm.34 To assess the accuracy of the approach, we retrospectively acquired DECT images from 22 patients who received contrast-enhanced abdomen CT scan. Quantitative comparisons between the original high-energy CT images and the predicted high-energy CT images were performed using clinically relevant HU values for different types of tissues. Material-specific images (VNC images and iodine maps) quantification obtained from the original DECT images and the deep learning-predicted DECT images were also compared and quantitatively evaluated using HU value and noise level in region-of-interests (ROIs).
3. Results
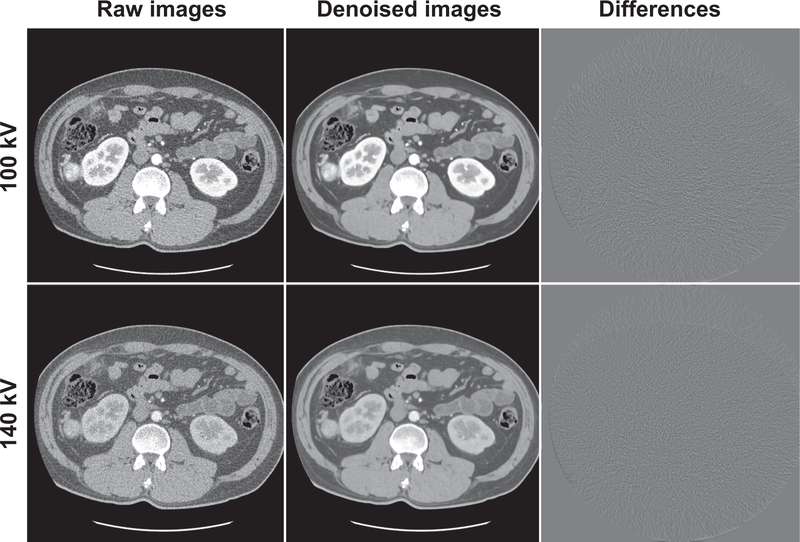
We found that the mapping CNN yields inferior high-energy CT images with original noisy low-energy CT images as input, indicating the CNN wastes its ability of expression on mapping the image difference between high- and low-energy levels with the presence of noise. Fig. 1 shows an example of the DECT images with and without noise reduction. As can be seen, the denoised images provide significantly improved quality compared to the raw CT images. There are no anatomical structure information in the difference images, suggesting the spatial resolution of the denoised CT images is well preserved after noise reduction. Quantitative assessments of the images with and without noise reduction using ROIs on different types of tissues are shown in Table. 1. The HU accuracy of the images is well preserved after noise reduction. The HU differences between the original DECT images and noise reduced DECT images are smaller than 3 HU, while the standard deviations of the ROIs suggest the noise has significantly reduced.
Fig. 1.
Low- and high-energy CT images with and without noise reduction. The difference images in column three are obtained by subtracting the denoised images from the raw images. All images are displayed in (C=0HU and W=500HU).
Table 1.
Quantitative analysis of the DECT images with and without noise reduction. Region-of-interests (ROIs) assessments on different tissues (arota, liver, spine, and stomach) show the HU accuracy of the CT images is well preserved, while the noise is significantly reduced after noise reduction.
| ROIs | HUraw | HUdenoised | ΔHU | stdraw | stddenoised | |
|---|---|---|---|---|---|---|
| Arota | 100 kV | 314.5 | 312.7 | 1.8 | 35.4 | 6.8 |
| 140 kV | 166.7 | 164.8 | 1.9 | 33.0 | 7.6 | |
| Liver | 100 kV | 71.8 | 69.5 | 2.3 | 31.6 | 6.8 |
| 140 kV | 60.5 | 63.4 | −2.9 | 24.8 | 5.4 | |
| Spine | 100 kV | 214.6 | 214.7 | −0.1 | 39.7 | 20.7 |
| 140 kV | 149.5 | 149.0 | 0.5 | 32.2 | 13.6 | |
| Stomach | 100 kV | 2.2 | 2.6 | −0.4 | 36.7 | 8.1 |
| 140 kV | −1 | −1 | 0 | 38.8 | 7.7 | |
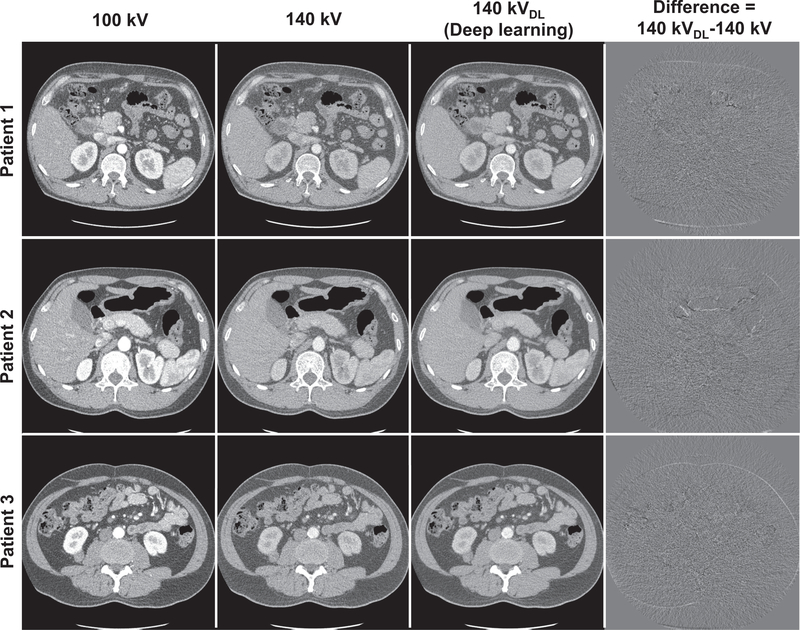
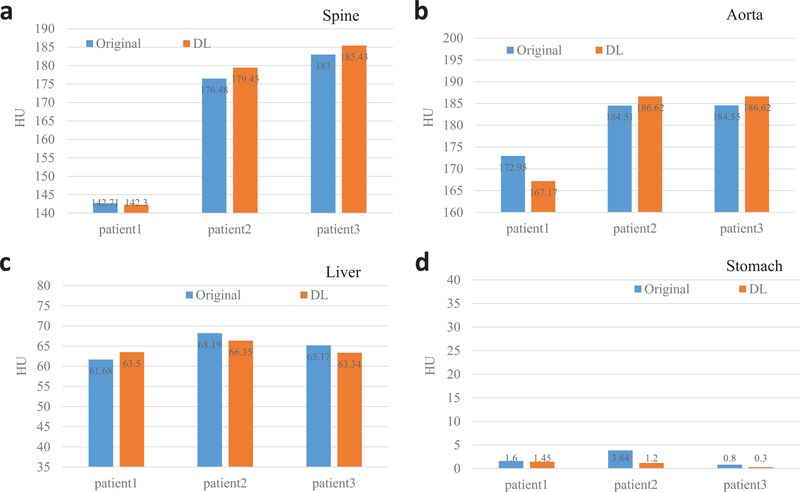
With noise significantly reduced DECT images, the CNN can appreciate the ingenuous CT image difference at different energy levels and ultimately yield superior high-energy CT image. Fig. 2 shows original DECT images, deep learning predicted high-energy CT images as well as the differences images between the predicted images and their corresponding original images for three patients. As can be seen, the predicted 140kV high-energy CT images are highly consistent with original 140 kV images. The difference images show marginal anatomical structures and suggest the spatial resolution is greatly preserved in the deep learning predicted 140 kV images. Quantitative evaluation using clinical relevant metrics shows the HU values of the original images and the deep learning predicted images are very close to each other. The HU difference between the predicted and original high-energy CT images are 1.94 HU, 3.32 HU, 1.83 HU and 1.10 HU for ROIs on spine, aorta, liver and stomach, respectively (Fig. 3).
Fig. 2.
Original low- (1st column) and high-energy (2nd column) DECT images and predicted high-energy CT images (3rd column), and difference images (4th column) between the predicted and original 140 kV images. All images are displayed in (C=0HU and W=500HU).
Fig. 3.
Quantitative measurement of the 140 kV images using ROIs on spine (a), aorta (b), liver (c) and stomach (d) for different patients. The deep learning (DL) predicted images are highly consistent with the original CT images.
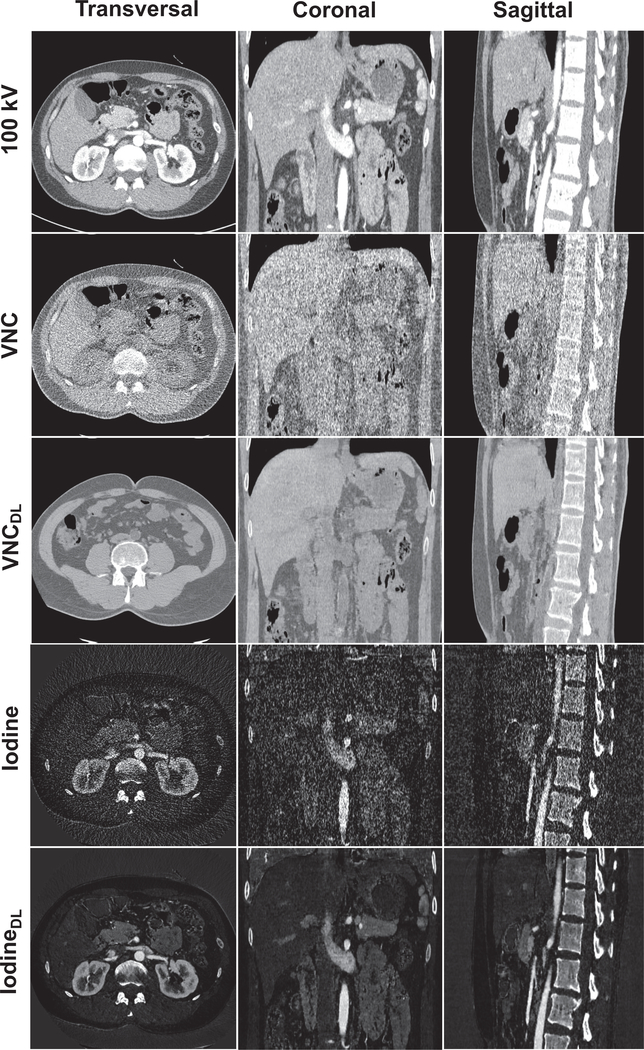
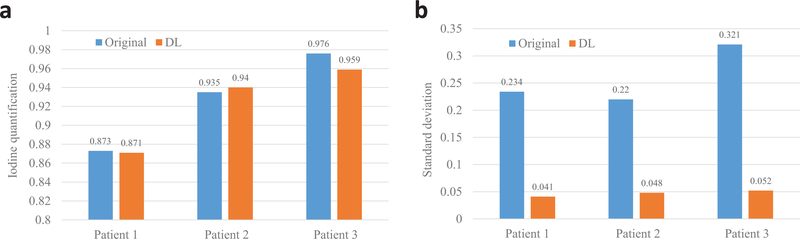
Fig. 4 shows the three-dimensional VNC images and iodine maps obtained from the original 100 kV/140 kV DECT images and the deep learning-based DECT images. As can be seen, the deep learning-based DECT approach provides high quality VNC and iodine maps. Since material decomposition uses matrix inversion and yields amplified image noise, the noise levels in VNC images and iodine maps obtained from the original DECT images are much higher than the 100 kV images. Due to the noise correlation between the predicted high-energy CT images and the original low-energy CT images, deep learning-based DECT imaging provides noise significantly reduced VNC images and iodine maps. The HU differences between VNC images obtained from original DECT and deep learning DECT are 4.10 HU, 3.75 HU, 2.33 HU and 2.92 HU for ROIs on spine, aorta, liver and stomach, respectively. The aorta iodine quantification differences between iodine maps obtained from original DECT and deep learning DECT images are 0.2%, 0.5%, and 1.7% for the three patients, respectively, suggesting high consistency between the predicted and the original high-energy CT images. More importantly, the noise of the iodine maps obtained from the deep leaning predicted DECT images are about 6-fold smaller than that obtained from the original DECT images.
Fig. 4.
Illustration of the contrast-enhanced 100 kV CT images (C=0HU and W=500HU) and VNC images (C=0HU and W=500HU) and iodine maps (C=0.6 and W=1.2) obtained using original DECT images and deep learning (DL)-based DECT images in transversal, coronal and sagittal views.
4. Discussion and Conclusion
Since we have used an image domain material decomposition method, artifacts in the C-T images, such as beam hardening artifacts, scatter artifacts, may reduce the accuracy of the material-specific images. This can impact material decomposition using both the original DECT images and the deep learning DECT images. Hence, in the cases where artifacts significantly reduce the HU accuracy of the CT images, beam hardening correction20 and scatter correction35–37 methods can be employed to reduce the CT artifacts and further to enhance the material decomposition accuracy.
Classical U-Net uses cross entropy loss and soft-max for classification and segmentation problem (i.e., 0–1 labelling). Here we are using mean-squared-error because the dual-energy residual mapping is not a classification or segmentation problem. Instead, the encoder-decoder architecture is employed to incorporate the dual-energy residual into multi-resolution multi-scale feature representation. The output of the network is a 1 × 1 convolutional layer which yields a final feature that has the spatial shape as the DECT residual image. The decoded final feature was then compared to the training label (ground truth) using the mean-squared-error. The parameters of the network are updated using backpropagation during training procedure.
The difference of dual-energy attenuation properties of different types of tissues can be characterized by the residual images of the DECT images. By using the residual images and the corresponding low-energy CT images (with consistent anatomical information) to train the deep learning model, the model can learn where the organs are in a normal body and what is the dual-energy attenuation properties. The trained model can then adjust the pixel value of the low-energy CT (100kV) according its location to reflect the attenuation property at the high-energy (140 kV). In this study, all patients were performed contrast-enhanced CT scans, so one of the distinct features of the model learned is the attenuation property of the iodine contrast agent at the dual-energy scenarios, which can be used to map the dual-energy residual during the inference process.
This study demonstrates that highly accurate DECT imaging with single low-energy data is achievable by using a deep learning approach. The proposed algorithm shows superior and reliable performance on the clinical datasets, and provides clinically valuable high quality VNC and iodine maps. Compared to the current standard DECT techniques, the proposed method can significantly simplify the DECT system design, reduce the scanning dose by using only a single kV data acquisition, and reduce the noise level of material decomposition by taking advantage of the noise correlation of the deep learning derived DECT images. The strategy reduces the DECT imaging cost and may find widespread clinical applications, including cardiac imaging, angiography, perfusion imaging and urinary stone characterization and so on.
Fig. 5.
Iodine quantification obtained using both the original DECT images and the DL-based DECT images. (a) Iodine quantifications, (b) standard deviations of the measured ROIs.
5. Acknowledge
This work was partially supported by NIH/NCI (1R01CA176553, 1R01CA223667 and 1R01CA227713) and a Faculty Research Award from Google Inc.
Contributor Information
Wei Zhao, Department of Radiation Oncology, Stanford University, Palo Alto, CA 94306, USA.
Tianling Lv, Department of Computer Science and Engineering, Southeast University, Nanjing, Jiangsu 210096, China.
Rena Lee, Department of Bioengineering, Ehwa Womens University, Seoul, Korea.
Yang Chen, Department of Computer Science and Engineering, Southeast University, Nanjing, Jiangsu 210096, China.
Lei Xing, Department of Radiation Oncology, Stanford University, Palo Alto, CA 94306, USA.
References
- 1.Alvarez RE and Macovski A, Energy-selective reconstructions in x-ray computerised tomography, Physics in Medicine and Biology 21, p. 733 (1976). [DOI] [PubMed] [Google Scholar]
- 2.Lehmann L, Alvarez R, Macovski A, Brody W, Pelc N, Riederer SJ and Hall A, Generalized image combinations in dual kVp digital radiography, Medical Physics 8, 659 (1981). [DOI] [PubMed] [Google Scholar]
- 3.Brody WR, Cassel DM, Sommer FG, Lehmann L, Macovski A, Alvarez RE, Pelc NJ, Riederer SJ and Hall AL, Dual-energy projection radiography: initial clinical experience, American Journal of Roentgenology 137, 201 (1981). [DOI] [PubMed] [Google Scholar]
- 4.Kalender WA, Perman W, Vetter J and Klotz E, Evaluation of a prototype dual-energy computed tomographic apparatus. I. phantom studies, Medical Physics 13, 334 (1986). [DOI] [PubMed] [Google Scholar]
- 5.Fessler JA, Elbakri IA, Sukovic P and Clinthorne NH, Maximum-likelihood dual-energy tomographic image reconstruction, in Medical Imaging 2002: Image Processing, 2002. [Google Scholar]
- 6.Carmi R, Naveh G and Altman A, Material separation with dual-layer CT, in Nuclear Science Symposium Conference Record, 2005 IEEE, 2005. [Google Scholar]
- 7.Johnson TR, Krauss B, Sedlmair M, Grasruck M, Bruder H, Morhard D, Fink C, Weckbach S, Lenhard M, Schmidt B et al. , Material differentiation by dual energy CT: initial experience, European Radiology 17, 1510 (2007). [DOI] [PubMed] [Google Scholar]
- 8.La Riviere PJ et al. , Penalized-likelihood sinogram decomposition for dual-energy computed tomography, in 2008 IEEE Nuclear Science Symposium Conference Record, 2008. [Google Scholar]
- 9.Mou X, Chen X, Sun L, Yu H, Zhen J and Zhang L, The impact of calibration phantom errors on dual-energy digital mammography, Physics in Medicine and Biology 53, p. 6321 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brendel B, Roessl E, Schlomka J-P and Proksa R, Empirical projection-based basis-component decomposition method, in SPIE Medical Imaging, 2009. [Google Scholar]
- 11.Matsumoto K, Jinzaki M, Tanami Y, Ueno A, Yamada M and Kuribayashi S, Virtual monochromatic spectral imaging with fast kilovoltage switching: improved image quality as compared with that obtained with conventional 120-kVp CT, Radiology 259, 257 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Maaß C, Meyer E and Kachelrieß M, Exact dual energy material decomposition from inconsistent rays (MDIR), Medical Physics 38, 691 (2011). [DOI] [PubMed] [Google Scholar]
- 13.Hao J, Kang K, Zhang L and Chen Z, A novel image optimization method for dual-energy computed tomography, Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 722, 34 (2013). [Google Scholar]
- 14.Niu T, Dong X, Petrongolo M and Zhu L, Iterative image-domain decomposition for dual-energy CT, Medical Physics 41, p. 041901 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Xia T, Alessio AM and Kinahan PE, Dual energy CT for attenuation correction with PET/CT, Medical Physics 41 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao W, Niu T, Xing L, Xie Y, Xiong G, Elmore K, Zhu J, Wang L and Min JK, Using edge-preserving algorithm with non-local mean for significantly improved image-domain material decomposition in dual-energy CT, Physics in Medicine & Biology 61, p. 1332 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Zhao W, Xing L, Zhang Q, Xie Q and Niu T, Segmentation-free x-ray energy spectrum estimation for computed tomography using dual-energy material decomposition, Journal of Medical Imaging 4, p. 023506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forghani R, De Man B and Gupta R, Dual-energy computed tomography: physical principles, approaches to scanning, usage, and implementation: Part 1, Neuroimaging Clinics 27, 371 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Zhang H, Zeng D, Lin J, Zhang H, Bian Z, Huang J, Gao Y, Zhang S, Zhang H, Feng Q et al. , Iterative reconstruction for dual energy CT with an average image-induced nonlocal means regularization, Physics in Medicine & Biology 62, p. 5556 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao W, Vernekohl D, Han F, Han B, Peng H, Yang Y, Xing L and Min JK, A unified material decomposition framework for quantitative dual-and triple-energy CT imaging, Medical physics 45, 2964 (2018). [DOI] [PubMed] [Google Scholar]
- 21.McCollough CH, Leng S, Yu L and Fletcher JG, Dual-and multi-energy CT: principles, technical approaches, and clinical applications, Radiology 276, 637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silva AC, Morse BG, Hara AK, Paden RG, Hongo N and Pavlicek W, Dual-energy (spectral) CT: applications in abdominal imaging, Radiographics 31, 1031 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Hao J, Kang K, Zhang L and Chen Z, A novel image optimization method for dual-energy computed tomography, Nuclear Instruments and Methods in Physics Research Section A: Accelerators, Spectrometers, Detectors and Associated Equipment 722, 34 (2013). [Google Scholar]
- 24.LeCun Y, Bengio Y and Hinton G, Deep learning, Nature 521, p. 436 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Ibragimov B, Xing L, Segmentation of organs-at-risks in head and neck CT images using convolutional neural networks, Medical Physics 44, 547–557 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Jang H, Kijowski R, Bradshaw T and McMillan AB, Deep learning MR imaging-based attenuation correction for PET/MR imaging, Radiology 286, 676 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu F, Zhou Z, Jang H, Samsonov A, Zhao G and Kijowski R, Deep convolutional neural network and 3D deformable approach for tissue segmentation in musculoskeletal magnetic resonance imaging, Magnetic Resonance in Medicine 79, 2379 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing L, Krupinski E, Cai J, Artificial intelligence will soon change the landscape of medical physics research and practice, Medical Physics 45, 1791–1793 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deng J, Xing L, Big Data in Radiation Oncology, Taylor & Francis Books, Inc, San Francisco, CA: (2018). [Google Scholar]
- 30.Zhao W, Han B, Yang Y, Buyyounouski M, Hancock SL, Bagshaw H and Xing L, Incorporating imaging information from deep neural network layers into image guided radiation therapy (IGRT), Radiotherapy and Oncology 140, 167 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao W, Shen L, Han B, Yang Y, Cheng K, Toesca DA, Koong AC, Chang DT and Xing L, Markerless pancreatic tumor target localization enabled by deep learning, International Journal of Radiation Oncology* Biology* Physics (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen L, Zhao W, Xing L, Patient-specific reconstruction of volumetric computed tomography images from a single projection view via deep learning, Nature Biomedical Engineering, In Press (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang W, Zhang H, Yang J, Wu J, Yin X, Chen Y, Shu H, Luo L, Coatrieux G, Gui Z et al. , Improving low-dose CT image using residual convolutional network, IEEE Access 5, 24698 (2017). [Google Scholar]
- 34.Faby S, Kuchenbecker S, Sawall S, Simons D, Schlemmer H-P, Lell M and Kachelrieß M, Performance of today’s dual energy ct and future multi energy CT in virtual non-contrast imaging and in iodine quantification: A simulation study, Medical Physics 42, 4349 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Zhao W, Brunner S, Niu K, Schafer S, Royalty K and Chen G-H, Patient-specific scatter correction for flat-panel detector-based cone-beam CT imaging, Physics in Medicine & Biology 60, p. 1339 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Zhao W, Vernekohl D, Zhu J, Wang L and Xing L, A model-based scatter artifacts correction for cone beam CT, Medical Physics 43, 1736 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi L, Wang A, Wei J and Zhu L, Fast shading correction for cone-beam CT via partitioned tissue classification, Physics in Medicine & Biology 64, p. 065015 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]