Abstract
Type 2 diabetes mellitus (T2DM) accounts for 90% of diabetes cases worldwide. The majority of T2DM patients are obese. Dysbiosis in the gut microflora is strongly associated with the pathogenesis of obesity and T2DM; however, the microbiome of obese-T2DM individuals in the Pakistani population remains unexplored. The gut microbiota signature of 60 Pakistani adults was studied using 16S rRNA sequencing targeting V3–V4 hypervariable regions. The sequence analysis revealed that bacteria from Firmicutes were predominant along with those from Clostridia and Negativicutes, whereas bacteria from Verrucomicrobia, Bacteroidetes, Proteobacteria, and Elusimicrobia were less abundant among the obese T2DM patients. These data distinctively vary from those in reports on the Indian population. The difference in gut microbiota could presumably be related to the distinct lifestyle and eastern dietary habits (high carbohydrate and fat intake, low fiber intake) and unregulated antibiotic consumption. This is the first study carried out to understand the gut microbiome and its correlation with individual life style of obese T2DM patients in the Pakistani population.
Introduction
The prevalence of diabetes mellitus is increasing at an alarming rate worldwide with more than 422 million people diagnosed with the disease, 90% of which have type 2 diabetes mellitus (T2DM) [1]. Obesity-induced insulin resistance is one of the primary causes of this metabolic disorder [2], with over 90% patients being overweight [3]. In addition to being governed by genetic and environmental factors, T2DM is also influenced by the patient’s gut microbiota specifically, their abundance and diversity [4–6].
The role of gut microbes in the synthesis of essential vitamins, amino acids, and degradation of toxins has been studied [7]. However, during the past decade, evidence has suggested that the gut microbiome plays a crucial role in diseases and dysbiosis and is significantly associated with T2DM [3]. Interestingly, large-scale 16S rRNA sequencing studies revealed a strong association between the gut microbiota composition and metabolic disorders such as obesity and T2DM [8,9]. More than 1000 species of bacteria belonging to different phyla reside in the human gut, the dominant ones being Firmicutes and Bacteroidetes together with Actinobacteria and Proteobacteria [10,11]. The aforementioned phyla enhance the uptake of monosaccharides, thus elevating the production of hepatic triglycerides, which in turn trigger insulin resistance [12]. Additionally, Bacteroides have strong positive correlation with intake of fat-rich diet [13–15]. The disturbance in relative bacterial abundance and diversity eventually results in metabolic disorders such as inflammatory bowel disease, diabetes mellitus, Crohn’s disease, and multiple sclerosis [16]. Previous studies have also reported that the gut microbiome has a strong association with T2DM [6,7,10]. The gut microbiota of T2DM patients was characterized by reduced diversity with increased proportion of Firmicutes relative to Bacteroidetes, in reports from the populations of eastern countries (e.g., Japan and Saudi Arabia) [17–19] compared to that in the Western population where this trend was not consistently observed in different studies [8,20–22].
The dysbiosis may lead to metabolic disorders including T2DM by hampering the production of carbohydrate metabolism end products specifically associated with decreased number of butyrate-producing microbes [23]. Acetate and butyrate are short chain fatty acids (SCFAs); butyrate can modulate immune response and cause low-grade inflammation and could be one of the causes of T2DM. Therefore, differences in the gut microbiota could be influenced by ethnic, environmental, dietary, and socioeconomic factors. The majority of the aforementioned factors can vary greatly among different ethnic groups worldwide [24]. Furthermore, gut microbial communities (Clostridium hystoliticum, Eubacterium rectale, and Clostridium coccoides) can affect neurotransmitters involved in gut–brain signaling pathways, and thus, regulate food intake and body weight. These neurotransmitters directly control the intestinal transit time, serotonin levels, physical activity, and intestinal permeability [4,25,26].
There is a paucity of information available on gut microbiota composition of Muslim populations with low socioeconomic status and consumption of a typical eastern diet. In a preliminary study in 2018, Batool et al. examined the gut microbiome structure of healthy individuals in the Pakistani population [27]. The present study is the first to report the gut microbiota of obese Pakistani individuals with T2DM.
Material and methods
Sample collection
Blood samples were collected during the first quarter of 2017 from the diabetic clinic of the Social Security Hospital, Islamabad. All volunteers were interviewed and their age, gender, weight, ethnicity, diet, medical/drug history, fasting glucose level, lipid profile, smoking, physical activities, and alcohol intake was documented. Fasting blood glucose and lipid profile (triglycerides, total cholesterol, HDL, and LDL) were measured using an automated enzymatic analyzer (Cobas Integra 700; Hoffman-La Roche, Basel, Switzerland).
In general, adults aged 25–55 years, belonging to ethnic Punjabi (inhabitants of largest province) population of Pakistan, were included in the present study. Participants suffering from any symptoms of constipation, bloody stool, diarrhea, or any other gastrointestinal disease and those who were administered antibiotics (oral or injectable) in past three months, were excluded. All stool samples were collected within 24 h after compiling the questionnaire. Samples were collected in sterile containers provided to the volunteers and were stored at −80 °C. Sampling was performed using all standard protocols and regulation.
Data collection, 16S rRNA sequencing, and quality control
Microbial DNA was isolated from 200 mg of each homogenized fecal sample, following the protocols of TIAGEN® DNA Stool kit (Tiagen, China). It was then quantified using Nanodrop spectrophotometer (Nanodrop Technologies, USA). Isolated DNA was stored at −20 °C for further study. The V3–V4 region of 16S rRNA gene was amplified in each sample using universal primers 343 F (5′-TACGGRAGGCAGCAG-3′) and 798 R (5′-AGGGTATCTAATCCT-3′) [28]. The resulting amplicons (456 bp) were subsequently subjected to high-throughput sequencing using the Illumina MiSeq platform. All low quality (<Q20) terminal bases were removed using Trimmomatic (v0.35), thus retaining the high quality reads only. All sequencing results after quality control, which are used in the project along with the metadata, have been submitted to Sequence Read Archive (SRA) NCBI database (accession number PRJNA554535).
Analysis of bacterial composition
The filtered high-quality reads were stitched by using FLASH (v1.2.11). Script multiple_join_paired_ends.py of QIIME (1.8.0) was used to assemble the 16S rRNA amplicons. Moreover, mismatched barcodes and sequences with length less than the threshold (200 bases), were removed. Chimeric sequences were removed using UCHIME (v4.2) providing valid tags (a high-quality sequence) for downstream microbial diversity analysis. These quality-filtered reads were clustered into operational taxonomic units (OTUs; 97% identity) using de novo OTU picking and taxonomic assignment by implementing VSEARCH (v2.4.2) against Silva (v123), and Greengenes (gg13.8) reference databases utilizing QIIME script (pick_de_novo_otus.py). The representative sequences were classified and annotated by the Naive Bayesian classification algorithm of RDP classifier (v2.2).
Diversity index calculation statistics
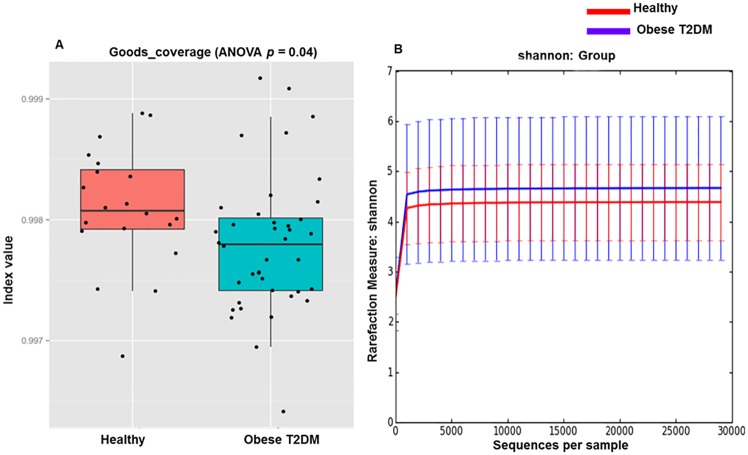
QIIME was used to perform diversity analysis, using the default parameters. Filtration and closed-reference OTU clustering ended with a variable count, possessing a wide range of sequences per sample. To address this randomness, the sequence data was rarefied to 829 OTUs per sample for alpha and beta diversity analyses. Furthermore, the diversity boxplot diagram and ANOVA was used to find the diversity and Shannon index in order to enumerate the species count in obese-T2DM and healthy individuals (Fig 1). Additionally, species accumulation curve was used to determine the sampling depth (S1A Fig), and the sample diversity and degree of uniformity were measured via rank abundance curve method (S1B Fig).
Fig 1. Alpha diversity between obese-T2DM patients.
(A) Comparison of Boxplots depicting OTU/Diversity between obese-T2DM patients (n = 40) and healthy participants (n = 20). (B) The Shannon Wiener index representing the number of species and uniformity of individual distribution between obese-T2DM patients (n = 40) and healthy participants (n = 20).
α-diversity and β-diversity analysis
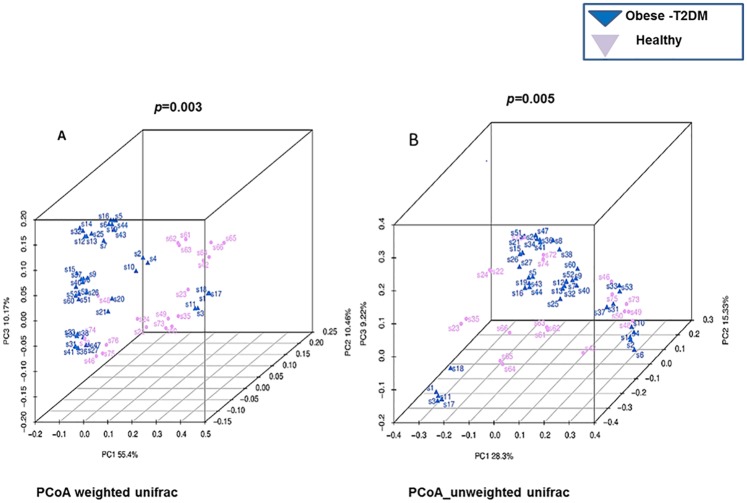
α-Diversity (diversity within samples) was analyzed based on rarefied data (using minimum number of sequences among samples), using the PD whole-tree box plot, observed species box plot and Chao1 (frequency-based richness estimator) (S2A–S2C Fig). β-diversity (diversity between samples) was assessed by weighted UniFrac, unweighted UniFrac, and Bray Curtis distances. Adonis analysis was used to evaluate the significance between groups (S1 Table). A Principal Coordinate Analysis (PCoA) plot, based on weighted-UniFrac and unweighted-UniFrac and Bray-Curtis distance matrices was constructed to demonstrate the overall dissimilarity of bacterial communities in the two groups of individuals from the Pakistani population (Fig 2).
Fig 2. Beta-diversity of the gut microbial communities in obese-T2DM patients and healthy participants.
Principal Coordinates Analysis (PCoA) plot based on weighted and unweighted UniFrac distance. Each dot represents one sample from each group.
Statistical analysis
Welch t-test was used to calculate significant differences in age, BMI, and lipid profile of obese-T2DM and healthy individuals (Table 1). Moreover, Kruskal–Wallis rank-sum test was used to compare the relative abundance between obese-T2DM patients and healthy individuals, and false discovery rate (FDR) using the Benjamini-Hochberg method was applied to correct the significant p-values (Table 2).
Table 1. Anthropometric and biochemical parameters of participants.
| Parameters | Obese-T2DM | Healthy | Welch t-test p-value |
|---|---|---|---|
| N = 60 | 40 | 20 | - |
| Age (years) | 38.1± 12.1 | 37.7 ± 12.1 | > 0.06 |
| BMI (Kg/ m2) | 32.4 ± 3.6* | 22.08 ± 3.1* | < 2.2E-16 |
| Glucose (mg/dL) | 221.5 ± 61.5* | 84.8± 30.4* | < 2.2E-16 |
| Total cholesterol (mg/dL) | 202.2 ± 25.2* | 124.3 ± 31.6* | < 2.2E-16 |
| Triglycerides (mg/dL) | 164.9 ± 26.4* | 85.7± 26.4* | < 2.2E-16 |
| HDL(mg/dL) | 46.2 ± 10.4* | 36.9 ± 8.5* | = 4.326E-05 |
| LDL(mg/dL) | 131.2 ± 33.4* | 49.2 ± 27.7* | < 2.2E-16 |
Results of Welch two sample t-test (obese-T2DM, healthy age, BMI, glucose, cholesterol, triglycerides, HDL, LDL, mu = 0, alt = “two. sided.”, conf. level = 0.95)
* Indicates p < 0.05
Table 2. The relative abundance of gut microbiota at phylum, class and genus level between obese-T2DM patients (n = 40) and healthy participants (n = 20) after FDR adjustment using Kruskal-Wallis rank-sum tests.
| Bacterial Taxonomy | Mean relative Abundance | p-Value | ||||
|---|---|---|---|---|---|---|
| ObeseT2DM | Healthy | Test-Statistics | p-value | FDR | ||
| Verrucomicrobia | Phylum | 0.0% | 0.1% | 11.90 | 0.00002 | 0.0003 |
| Verrucomicrobiae | Class | 0.0% | 0.1% | 18.03 | 0.00002 | 0.0006 |
| Firmicutes | Phylum | 55.7% | 36.9% | 11.90 | 0.0005 | 0.004 |
| Bacilli | Class | 5.7% | 6.5% | 4.61 | 0.03 | 0.1 |
| Bacillus | Genus | 0.008% | 0.04% | 6.30 | 0.01 | 0.06 |
| Lactobacillus | Genus | 2.9% | 1.4% | 6.29 | 0.01 | 0.06 |
| Clostridia | Class | 36.2% | 22.5% | 10.84 | 0.0009 | 0.01 |
| Eubacterium coprostanoligenes group | Genus | 7.3% | 1.9% | 13.23 | 0.0002 | 0.006 |
| Subdoligranulum | Genus | 5.3% | 4.1% | 5.60 | 0.01 | 0.07 |
| Christensenellaceae_R_7_group | Genus | 1.2% | 0.5% | 4.38 | 0.03 | 0.1 |
| Ruminococcus_2 | Genus | 0.09% | 0.1% | 9.02 | 0.002 | 0.02 |
| Negativicutes | Class | 9.6% | 2.2% | 13.00 | 0.0003 | 0.004 |
| Dialister | Genus | 8.8% | 1.7% | 12.44 | 0.0004 | 0.007 |
| Allisonella | Genus | 0.1% | 0.002% | 28.20 | 1.09e07 | 0.00002 |
| Bacteroidetes | Phylum | 19.5% | 32.1% | 9.73 | 0.001 | 0.009 |
| Bacteroidia | Class | 19.5% | 32.1% | 9.73 | 0.001 | 0.01 |
| Prevotella_9 | Genus | 15.4% | 23.0% | 12.89 | 0.0003 | 0.006 |
| Proteobacteria | Phylum | 8.9% | 15.8% | 9.06 | 0.002 | 0.01 |
| Gamaproteobacteria | Class | 8.6% | 15.0% | 9.15 | 0.002 | 0.01 |
| Escherichia_Shigella | Genus | 7.0% | 11.9% | 6.53 | 0.01 | 0.05 |
| Deltaproteobacteria | Class | 0.02% | 0.2% | 5.83 | 0.01 | 0.06 |
| Elusimicrobia | Phylum | 0.0% | 0.00001% | 6.20 | 0.01 | 0.04 |
| Elusimicrobia | Class | 0.0% | 0.001% | 6.20 | 0.01 | 0.05 |
| Coriobacteriia | Class | 8.9% | 5.3% | 7.53 | 0.006 | 0.03 |
Ethical approval and participants’ consent
Our analysis was conducted on total 60 fecal samples subdivided as obese-T2DM (40 samples) and lean, non-T2DM, and healthy (20 samples), isolated from Punjabi individuals in Pakistan. Written consent was obtained from all the participants enrolled in the study. The study is approved by the Ethical Research Review Committees of COMSATS University, Islamabad, Pakistan. The experiments were performed according to the standard procedures and regulations.
Results
Sequencing output and data preprocessing
The process of filtering and adapter trimming resulted in 2,402,668 high-quality paired-end reads (median read length of 428 bp per sample).
Anthropometric and biochemical parameters of participants
The age and sex distribution were similar for the obese-T2DM (mean age 38.1± 12.1 years) and healthy individuals (mean age 37.7 ± 12.1 years). The BMI, and fasting blood sugar, total cholesterol, HDL, LDL, and triglyceride levels of obese-T2DM patients were significantly higher as compared to those of the healthy participants (Table 1).
Association of α-diversity with obese-T2DM status
Alpha diversity was quantified using the Shannon diversity index, which relates both OTU richness and evenness, and by the total number of observed species. Fig 1 depicts the alpha diversity measurements for obese T2DM patients versus controls. Statistical testing presented no difference for the PD_whole tree (pPD_whole = 0.91; S2B Fig), observed species (pObserved = 0.23; S2B Fig), and Chao1 richness estimator (pChao1 = 0.15; S2C Fig), while the Good’s coverage (diversity index) that reflects that the depth of sequencing was significantly decreased in obese-T2DM patients compared to that in the controls (pGoods = 0.04).
β-diversity of obese-T2DM patients and healthy individuals in Pakistani population
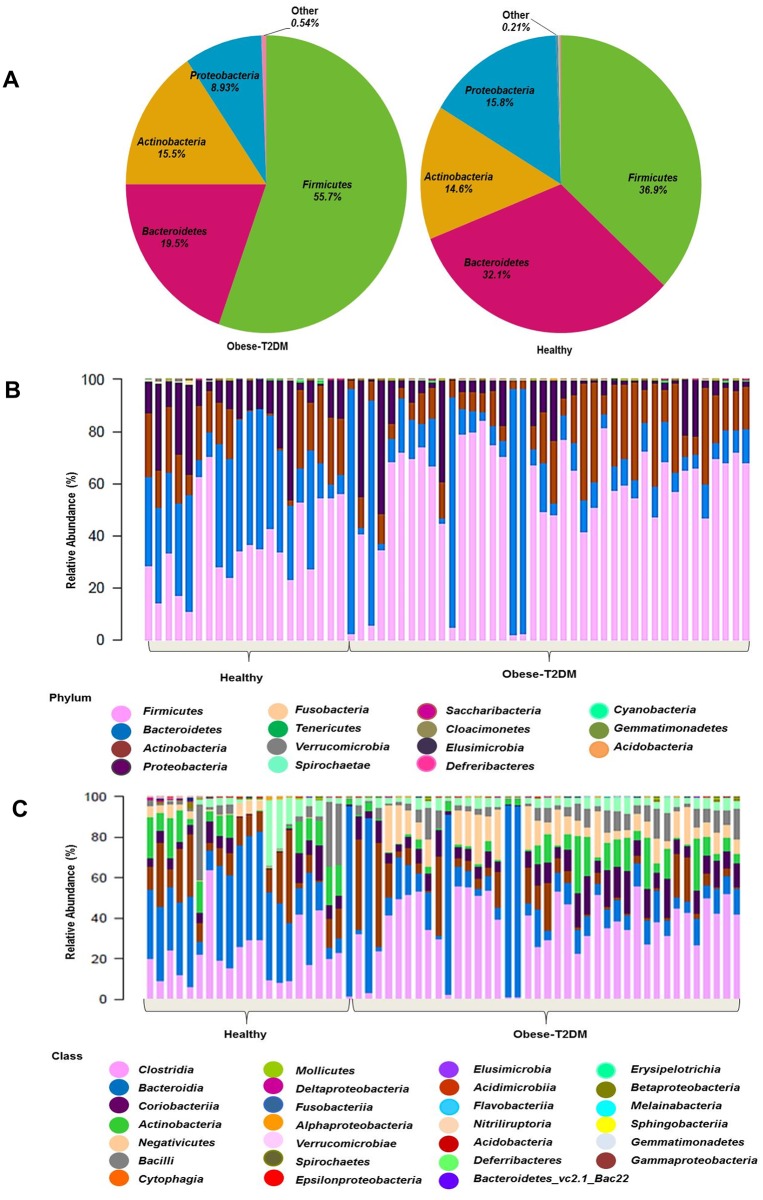
In majority of the obese-T2DM samples, the Firmicutes phyla constituted the highest proportion of the bacterial population compared to that in the healthy individuals, with relative abundance of 55.7% and 36.9%, respectively (Fig 3A–3C). Moreover, the relative abundance of Verrucomicrobia, Bacteroidetes, Proteobacteria, and Elusimicrobia was less or even completely absent in obese-T2DM patients (Table 2).
Fig 3. The relative abundance of gut bacteria in obese-T2DM patients and healthy participants using Kruskal-Wallis rank-sum tests.
(A) Relative percentage of most abundant phyla between obese-T2DM patients (n = 40) and healthy individuals (n = 20). (B) Relative percentage of most abundant phyla in each sample between obese-T2DM patients (n = 40) and healthy individuals. (C) Relative abundance of bacteria at class level in obese-T2DM patients (n = 40) and healthy participants (n = 20).
Relative abundance of Clostridia, Negativicutes, and Coriobacteriia was higher in obese-T2DM patients. At the genus level, the Eubacterium coprostanoligenes group, Dialister, and Allisonella presented increased relative abundance in obese-T2DM individuals (Table 2).
Microbial configuration in T2DM presents differential clustering
Microbial community distribution between obese-T2DM and healthy individuals was assessed by Adonis analysis based on permutation pMANOVA to analyze the variance. The weighted UniFrac patterns indicated that the obese-T2DM and healthy individuals clustered separately, thus presenting 55.4% and 10.17% of the total variance on the x-axis and y-axis, respectively. In contrast, the unweighted UniFrac clustering pattern reflects 28.3% and 9.22% of the total variance along the x-axis and y-axis, respectively (Fig 2), and UPGMA tree analysis presented a noticeable difference in the gut microbial compositions between obese-T2DM and healthy groups (S3 Fig).
Distinct gut microbiota in obese-T2DM individuals
As indicated in the Shannon index (Fig 1), a non-significant correlation of the microbial community richness exists between obese-T2DM and healthy individuals. An evident difference in the gut microbiota, at all taxonomic levels, was observed between these two groups (Table 3). The presence of Verrucomicrobia and Elusimicrobia exclusively, in the healthy Pakistani population revealed positive correlation with the healthy controls (Table 3). Similarly, the exclusive presence of Acidobacteria, Deferribacteres, and Gemmatimonadetes in the obese-T2DM individuals demonstrated their positive association with the diabetes state (Table 3). Furthermore, numerous bacterial species namely, Prevotella P4_76, Clostridiales bacterium canine oral taxon 100, Porphyromonadaceae bacterium DJF B175, rumen bacterium RF9, Candidatus Alistipes marseilloanorexic AP11, bacterium ASC802, Bacillus sporothermodurans, Staphylococcus SV3, and Iamia, were present in the obese-T2DM group of individuals only; whereas, methanogenic archaeon was absent in the diseased individuals and was observed only in healthy individuals (Table 3).
Table 3. Gut microbiota present exclusively in either obese-T2DM patients (n = 40) or healthy participants (n = 20).
| Bacterial Taxonomy | Annotation | Obese-T2DM | Healthy | Test- statistics | p-value | FDR |
|---|---|---|---|---|---|---|
| Verrucomicrobia | Phylum | 0 | 0.001 | 18.03 | 0.00002 | 0.0003 |
| Elusimicrobia | Phylum | 0 | 0.00001 | 6.205 | 0.01 | 0.04 |
| Acidobacteria | Phylum | 2.93E-06 | 0 | 0.5 | 0.4 | 0.6 |
| Deferribacteres | Phylum | 2.26E-06 | 0 | 0.5 | 0.4 | 0.6 |
| Gemmatimonadetes | Phylum | 1.61E-06 | 0 | 0.5 | 0.4 | 0.6 |
| Prevotella_sp._P4_76 | Species | 0.000136 | 0 | 8.00 | 0.004 | 0.05 |
| uncultured_methanogenic_archaeon | species | 0 | 0.00005 | 6.20 | 0.01 | 0.08 |
| Clostridiales_bacterium_canine_oral_taxon_100 | species | 8.49E-06 | 0 | 4.50 | 0.03 | 0.1 |
| Porphyromonadaceae_bacterium_DJF_B175 | species | 1.19E-05 | 0 | 3.87 | 0.04 | 0.1 |
| unidentified_rumen_bacterium_RF9 | species | 0.000349 | 0 | 3.26 | 0.07 | 0.1 |
| bacterium_ASC802 | species | 1.51E-06 | 0 | 1.01 | 0.3 | 0.4 |
| Candidatus_Alistipes_marseilloanorexicus_AP11 | species | 6.00E-06 | 0 | 1.55 | 0.2 | 0.3 |
| Bacillus_sporothermodurans | species | 1.53E-06 | 0 | 0.5 | 0.4 | 0.6 |
| Staphylococcus_sp._SV3 | species | 1.61E-06 | 0 | 0.5 | 0.4 | 0.6 |
| uncultured_Iamia_sp. | species | 2.27E-06 | 0 | 0.5 | 0.4 | 0.6 |
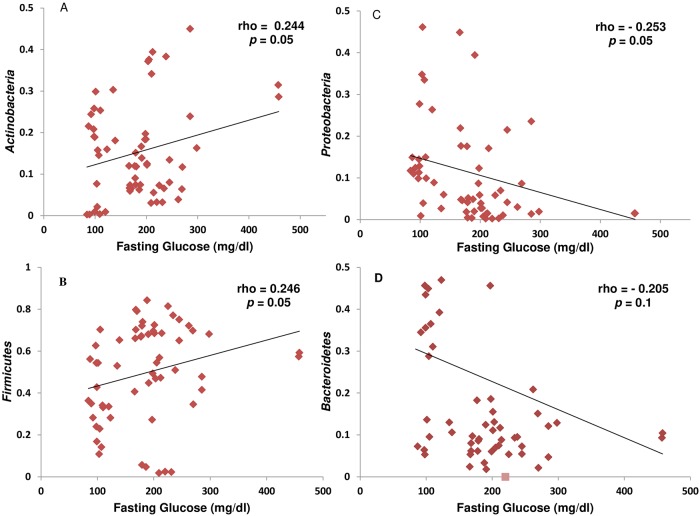
Correlations between gut microbiota in fecal samples and fasting glucose levels
Spearman’s rank correlation is a univariate analysis that was carried out with adjusted weight to evaluate the possible effect of glucose metabolism on gut microbial abundances (Fig 4). It was revealed that the relative abundance of Actinobacteria and Firmicutes was positively correlated with fasting glucose levels (Fig 4A and 4B), whereas that of Proteobacteria and Bacteroidetes demonstrated a negative correlation (Fig 4C and 4D).
Fig 4. Correlations between fasting glucose and gut microbiota in fecal samples of participants.
Spearman’s rank correlation coefficient was used to analyze the correlation between obese-T2DM (n = 40) and healthy (n = 20). (A) Correlation between fasting glucose and phylum Actinobacteria. (B) Correlation between fasting glucose and phylum Firmicutes (ρ = 0.246, p = 0.05). (C) Correlation between fasting glucose and phylum Proteobacteria (ρ = -0.253, p = 0.05). (D) Correlation between fasting glucose and Bacteroidetes (ρ = -0.205, p = 0.11).
Discussion
This is the first study to characterize the gut microbiome of obese-T2DM and healthy individuals in the Pakistani population. A total of 60 participants (40 obese-T2DM and 20 healthy) were recruited in the study, from January to March 2017. We evaluated the diversity, richness, and compositional variation of the gut microbiome among the healthy controls and obese-T2DM groups from the Punjabi Pakistani population. In contrast to those among the healthy individuals, the baseline clinical characteristics such as fasting glucose concentration, total cholesterol, triglycerides, HDL, LDL, and BMI were significantly high among individuals with obese-T2DM. Elevated levels of HDL and triglycerides are the most common factors associated with metabolic disorders in 85.5% of the Pakistani obese and T2DM individuals [29]. This is presumably due to frequent intake of eastern food comprising carbohydrate-rich compounds and edible oils, which are an essential part of the Pakistani diet [27].
We observed an altered microbial composition in obese-T2DM individuals, thus suggesting a distinct signature of microbiota associated with T2DM in obese individuals from the Pakistani population. Previous studies on healthy individuals in the Pakistani population reported that the microbiome of Pakistani individuals is more similar to that of people from Western countries as compared to that of the Indian population [27,30]. Moreover, the microbiomes of obese-T2DM individuals in the Pakistani population revealed an altered composition with an increased abundance of Firmicutes (55.7%), whereas Proteobacteria (58.8%) is reported to dominate the gut microbial diversity of obese-T2DM individuals from India [31]. Furthermore, at the generic level, Prevotella_9 and Eubacterium coprostanoligenes demonstrated a declining trend in the obese-T2DM Pakistani individuals compared to those from India [31]. This study sets a benchmark for the comparative analysis of obese-T2DM and healthy metagenomic samples in the Pakistani population.
We discovered a high percentage of Firmicutes and a reduced abundance of Bacteroidetes in the obese-T2DM samples similar to that in the earlier studies [8,9,12,32–34]. Furthermore, Firmicutes and Actinobacteria demonstrated a positive correlation with the fasting glucose levels, whereas the abundance of Bacteroidetes and Proteobacteria presented a negative relationship [35]. The Firmicutes are well known for fat digestion, and their increased abundance is also known to be associated with obesity. Similarly, Bacteroidetes play a crucial role in producing short-chain fatty acids (SCFAs), and Actinobacteria execute a key role in the biodegradation of resistant starch. It is also suggested that Firmicutes and Bacteroidetes enhance the monosaccharide uptake from the host gut, which elevates the production level of hepatic triglycerides, thereby resulting in insulin resistance [9,20,33]. This change may contribute positively to low-grade inflammation mainly achieved via activation of SCFA-linked G-protein-coupled receptors (GPCR), thus leading to metabolic disorders [12,36,37]. Additionally, resistance to resistin due to prolonged gut dysbiosis makes an individual susceptible to insulin resistance [38]. Factors such as ethnicity or genetic makeup may contribute to an increased susceptibility of Pakistani individuals to insulin resistance. In a comparative study between Pakistani immigrants and Norwegian individuals with T2DM in Norway, despite the fact that Pakistani immigrants had lower BMI and body fat mass (BFM) than Norwegians, they were more susceptible to insulin resistance and inflammation [39].
Although numerous bacterial species were particularly present or absent in the obese-T2DM samples, due to low sequencing depth and few number of samples, we could not effectively compare the species-level composition. Nevertheless, our preliminary findings at the species level provide interesting evidence warranting further investigations. For instance, Prevotella P4 76 was the only bacterial species observed particularly in the obese-T2DM individuals. The species is involved in elevating levels of proinflammatory cytokines, onset of low-grade inflammation, and insulin resistance [40–43].
Another important observation was the absence of Verrucomicrobia from obese-T2DM samples, and this phyla is considered as an efficient agent in maintaining the anti-inflammatory state of gut and improved insulin sensitivity [44]. Moreover, Verrucomicrobia is known to improve glucose metabolism in animals [45]. The depletion of Verrucomicrobia might have resulted in growth of rogue Protobacteria, such as Escherichia and Shigella. This notable change might contribute toward the development of T2DM [12].
The comparison of obese-T2DM and healthy microbiome samples during the present study suggested significantly reduced abundance of Gammaproteobacteria in obese-T2DM. Additionally, we observed that the abundance of gram-negative bacteria (Dialister and Allisonella) increased drastically in the obese-T2DM samples from the Pakistani population. These bacteria are extremely important for their contribution toward the development of disease as their increased abundance might have elevated the lipopolysaccharide (LPS) level in obese-T2DM individual. The circulating LPS binds with CD14 and mediates an inflammatory response, thus contributing positively to the development of obesity and insulin resistance [46–49]. We also detected an abundance of class Fusobacteria in the obese-T2DM samples. This particular class plays a pivotal role in energy generation, mounting adhesiveness to host epithelial cells, and inflammatory responses [50,51]. Studies suggest that high abundance of Fusobacteria is contributing significantly in T2DM along with other conditions [52].
The recruitment of healthy individuals from the Pakistani population was difficult as >85% of the individuals interviewed did not fall in the normal BMI range. This was due to several factors including physical inactivity, poverty (malnutrition), dietary habits, and overeating. A recent survey also revealed that around 60% of the Pakistani population are inactive [53]. Frequent unregulated use of antibiotics was another reason for the exclusion of several healthy volunteers from the present study.
Although, this is a preliminary study to explore the obese-T2DM gut microbiome in comparison to the healthy gut microbiome of Pakistani population, it includes certain limitation such as low sample number as well as sequencing depth. The majority of the participants were sampled only once, a time-series monitoring of multiple samples at various time points and of a higher number of participants would provide more insights that might have been missed due to low sequencing depth.
Conclusion
This preliminary study based on a small sample size depicts the obese-T2DM microbiome in the Pakistani population. Our study revealed high variability in the microbiome of healthy and obese-T2DM individuals within this population. The variation between normal and obese-T2DM fecal microbiome trends have been reported in other studies as well. Nonetheless, the significance of the present study cannot be undervalued as it provides the scientific community with a first glimpse of the signature microbiota associated with Pakistani obese-T2DM individuals. Furthermore, it also provides the road map for future studies. We believe that this signature of obese-T2DM individuals in the Pakistani population will aid future studies.
Supporting information
(A) Alpha Diversity: Species Accumulation Curve for each sample, the horizontal axis represents the sample size, and the vertical axis represents the number of OTUs detected in the sample. Cross symbols represent the possibility of new species (OTU) addition, with the addition of new samples. (B) Rank Abundance curve was used to explain sample diversity and abundance of species in each sample. The abundance of species is reflected by the length of the curve on the horizontal axis while uniformity of species composition is reflected by the shape of the curve.
(TIF)
(A) PD_whole_tree measures species richness or diversity (B) Observed Species indicates the number of actually observed species (C) Chao1 estimates observed species frequency.
(TIF)
(TIF)
R2: indicates the degree of interpretation of the difference between the different groups, that is, the ratio of the variance of the group to the total variance.
(DOCX)
Acknowledgments
Authors appreciate the technical support of Oebiotech Co., Ltd. (Shanghai, China) and would like to express their thanks and gratitude. The authors acknowledge the patient recruitment support from doctors of PESSI Hospital Islamabad and sincerely thank the patients for their participation in this study.
Data Availability
All sequence files are available from the SRA NCBI database (accession number(s) PRJNA554535. All relevant meta data are within the manuscript and its Supporting Information files.
Funding Statement
All sequence files and meta data information can be accessed using the following information: Bioproject; Accession: PRJNA55453, URL: (https://www.ncbi.nlm.nih.gov/bioproject/?term=prjna554535). URL for all samples: (https://www.ncbi.nlm.nih.gov/biosample?LinkName=bioproject_biosample_all&from_uid=554535).
References
- 1.World Health Organization. World health statistics 2016: monitoring health for the SDGs, sustainable development goals. WHO Library Cataloguing-in-Publication Data World; 2016. [Google Scholar]
- 2.Kahn BB, Flier JS. On diabetes : insulin resistance Obesity and insulin resistance. The Journal of Clinical Investigation. 2000; 10(4):473–481. 10.1172/JCI10842.on [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okazaki F, Zang L, Nakayama H, Chen Z, Gao ZJ, Chiba H, et al. Microbiome Alteration in Type 2 Diabetes Mellitus Model of Zebrafish. Scientific Reports. 2019; 29;9(1):87 10.1038/s41598-018-36442-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koopman KE, Booij J, Fliers E, Serlie MJ, la Fleur SE. Diet-induced changes in the lean brain: Hypercaloric high-fat-high-sugar snacking decreases serotonin transporters in the human hypothalamic region. Molecular Metabolism. 2013;2(4):417–422. 10.1016/j.molmet.2013.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukumo DM, Carvalho BM, Carvalho-Filho MA, Saad MJA. Translational research into gut microbiota: new horizons in obesity treatment Pesquisa translacional em microbiota intestinal: novos horizontes no tratamento da obesidade. Arq Bras Endocrinol Metab Arq Bras Endocrinol Metab. 2009;53(2):139–144. [DOI] [PubMed] [Google Scholar]
- 6.Harsch I, Konturek P. The Role of Gut Microbiota in Obesity and Type 2 and Type 1 Diabetes Mellitus: New Insights into “Old” Diseases. Medical Sciences. 2018;6(2):32 10.3390/medsci6020032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komaroff AL. The microbiome and risk for obesity and diabetes. JAMA—Journal of the American Medical Association. 2017;317(4):355–356. 10.1001/jama.2016.20099 [DOI] [PubMed] [Google Scholar]
- 8.Larsen N, Vogensen FK, Van Den Berg FWJ, Nielsen DS, Andreasen AS, Pedersen BK, et al. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE. 2010;136(5):1476–1483. 10.1371/journal.pone.0009085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang X, Shen D, Fang Z, Jie Z, Qiu X, Zhang C, et al. Human Gut Microbiota Changes Reveal the Progression of Glucose Intolerance. PLoS ONE. 2013;8(8):e71108 10.1371/journal.pone.0071108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naseer M, Bibi F, Alqahtani M, Chaudhary A, Azhar E, Kamal M, et al. Role of Gut Microbiota in Obesity, Type 2 Diabetes and Alzheimer’s Disease. CNS & Neurological Disorders—Drug Targets. 2014; 13(2):305–311. 10.2174/18715273113126660147 [DOI] [PubMed] [Google Scholar]
- 11.Dicksved J, Flöistrup H, Bergström A, Rosenquist M, Pershagen G, Scheynius A, et al. Molecular fingerprinting of the fecal microbiota of children raised according to different lifestyles. Applied and Environmental Microbiology. 2007;73(7):2284–2289. 10.1128/AEM.02223-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- 13.Clemente JC, Ursell LK, Parfrey LW, Knight R. The impact of the gut microbiota on human health: An integrative view. Cell. 2012;148(6): 1258–1270. 10.1016/j.cell.2012.01.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barengolts E. Gut microbiota, prebiotics, probiotics, and synbiotics in management of obesity and prediabetes: review of randomized controlled trials. Endocrine Practice. 2016;22(10):1224–1234. 10.4158/EP151157.RA [DOI] [PubMed] [Google Scholar]
- 15.Valdes AM, Walter J, Segal E, Spector TD. Role of the gut microbiota in nutrition and health. BMJ. 2018; 361:2179 10.1136/bmj.k2179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin NR, Whon TW, Bae JW. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends in Biotechnology. 2015; 33(9):496–503. 10.1016/j.tibtech.2015.06.011 [DOI] [PubMed] [Google Scholar]
- 17.Kasai C, Sugimoto K, Moritani I, Tanaka J, Oya Y, Inoue H, et al. Comparison of the gut microbiota composition between obese and non-obese individuals in a Japanese population, as analyzed by terminal restriction fragment length polymorphism and next-generation sequencing. BMC Gastroenterology. 2015; 15(1):100 10.1186/s12876-015-0330-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasir M, Angelakis E, Bibi F, Azhar EI, Bachar D, Lagier JC, et al. Comparison of the gut microbiota of people in France and Saudi Arabia. Nutrition and Diabetes. 2015; 5(4):e153 10.1038/nutd.2015.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sedighi M, Razavi S, Navab-Moghadam F, Khamseh ME, Alaei-Shahmiri F, Mehrtash A, et al. Comparison of gut microbiota in adult patients with type 2 diabetes and healthy individuals. Microbial Pathogenesis. 2017; 111:362–369. 10.1016/j.micpath.2017.08.038 [DOI] [PubMed] [Google Scholar]
- 20.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009; 457(7228):480 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tilg H, Moschen AR, Kaser A. Obesity and the Microbiota. Gastroenterology. 2009; 136(5):1476–1483. 10.1053/j.gastro.2009.03.030 [DOI] [PubMed] [Google Scholar]
- 22.Verdam FJ, Fuentes S, De Jonge C, Zoetendal EG, Erbil R, Greve JW, et al. Human intestinal microbiota composition is associated with local and systemic inflammation in obesity. Obesity. 2013; 21(12):e607–615. 10.1002/oby.20466 [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Qin J, Li Y, Cai Z, Li S, Zhu J, et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature. 2012; 490(7418):55 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- 24.Brooks AW, Priya S, Blekhman R, Bordenstein SR. Gut microbiota diversity across ethnicities in the United States. PLoS Biology. 2018; 16(12):e200842 10.1371/journal.pbio.2006842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gill RK, Kumar A, Malhotra P, Maher D, Singh V, Dudeja PK, et al. Regulation of intestinal serotonin transporter expression via epigenetic mechanisms: Role of HDAC2. American Journal of Physiology—Cell Physiology. 2013; 304(4):c334–341. 10.1152/ajpcell.00361.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Won Y-J, Lu VB, Puhl HL, Ikeda SR. -Hydroxybutyrate Modulates N-Type Calcium Channels in Rat Sympathetic Neurons by Acting as an Agonist for the G-Protein-Coupled Receptor FFA3. Journal of Neuroscience. 2013; 33(49):19314–19325. 10.1523/JNEUROSCI.3102-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Batool M, Ali SB, Jaan A, Khalid K, Ali SA, Kamal K, et al. Initial Sequencing and Characterization of Gastrointestinal and Oral Microbiota in Urban Pakistani Adults Reveals Abnormally High Levels of Potentially Starch Metabolizing Bacteria in the General Population. bioRxiv. 2018; 1:419598 10.1101/419598 [DOI] [Google Scholar]
- 28.Nossa CW, Oberdorf WE, Yang L, Aas JA, Paster BJ, de Santis TZ, et al. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World Journal of Gastroenterology. 2010; 16(33):4135 10.3748/wjg.v16.i33.4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mohsin A, Zafar J, Nisar Y Bin, Imran SM, Zaheer K, Khizar B, et al. Frequency of the metabolic syndrome in adult type2 diabetics presenting to Pakistan Institute of Medical Sciences. Journal of the Pakistan Medical Association. 2007; 57(5):235 [PubMed] [Google Scholar]
- 30.Tandon D, Haque MM, Saravanan R, Shaikh S, Sriram P, Dubey AK, et al. A snapshot of gut microbiota of an adult urban population from Western region of India. PLoS ONE. 2018; 13(4):e0195643 10.1371/journal.pone.0195643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pushpanathan P, Srikanth P, Seshadri K, Selvarajan S, Pitani R, Kumar T, et al. Gut Microbiota in Type 2 Diabetes Individuals and Correlation with Monocyte Chemoattractant Protein1 and Interferon Gamma from Patients Attending a Tertiary Care Centre in Chennai, India. Indian Journal of Endocrinology and Metabolism. 2016; 20(4):523 10.4103/2230-8210.183474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: Human gut microbes associated with obesity. Nature. 2006; 444(7122):1022 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- 33.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, et al. Human gut microbiota in obesity and after gastric bypass. Proceedings of the National Academy of Sciences. 2009; 106(7):2365–2370. 10.1073/pnas.0812600106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson J, Garges S, Giovanni M, McInnes P, Wang L, Schloss JA, et al. The NIH Human Microbiome Project. Genome Research. 2009; 19(12):2317–2323. 10.1101/gr.096651.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sepp E, Kolk H, Lõivukene K, Mikelsaar M. Higher blood glucose level associated with body mass index and gut microbiota in elderly people. Microbial Ecology in Health & Disease. 2014; 25(1):22857 10.3402/mehd.v25.22857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L, Yu Z, Ye X, Zou S, Li H, Yu D, et al. A marker of endotoxemia is associated with obesity and related metabolic disorders in apparently healthy Chinese. Diabetes Care. 2010; 33(9):1925–1932. 10.2337/dc10-0340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Erejuwa OO, Sulaiman SA, Ab Wahab MS. Modulation of gut microbiota in the management of metabolic disorders: The prospects and challenges. International Journal of Molecular Sciences. 2014; 15(3):4158–4188. 10.3390/ijms15034158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steppan CM, Bailey ST, Bhat S, Brown EJ, Banerjee RR, Wright CM, et al. The hormone resistin links obesity to diabetes. Nature. 2001; 409(6818):307 10.1038/35053000 [DOI] [PubMed] [Google Scholar]
- 39.Wium C, Eggesbø HB, Ueland T, Michelsen AE, Torjesen PA, Aukrust P, et al. Adipose tissue distribution in relation to insulin sensitivity and inflammation in Pakistani and Norwegian subjects with type 2 diabetes. Scandinavian Journal of Clinical and Laboratory Investigation. 2014; 74(8):700–707. 10.3109/00365513.2014.953571 [DOI] [PubMed] [Google Scholar]
- 40.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012; 3(4):289–306. 10.4161/gmic.19897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nature Reviews Microbiology. 2013; 11(7): 497 10.1038/nrmicro3050 [DOI] [PubMed] [Google Scholar]
- 42.Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nature Medicine. 2013; 19(5):576 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leite AZ, de Rodrigues NC, Gonzaga MI, Paiolo JCC, de Souza CA, Stefanutto NAV, et al. Detection of increased plasma interleukin-6 levels and prevalence of Prevotella copri and Bacteroides vulgatus in the feces of type 2 diabetes patients. Frontiers in Immunology. 2017; 8:1107 10.3389/fimmu.2017.01107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujio-Vejar S, Vasquez Y, Morales P, Magne F, Vera-Wolf P, Ugalde JA, et al. The gut microbiota of healthy Chilean subjects reveals a high abundance of the phylum Verrucomicrobia. Frontiers in Microbiology. 2017; 8:11221 10.3389/fmicb.2017.01221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014; 63(5):727–735. 10.1136/gutjnl-2012-303839 [DOI] [PubMed] [Google Scholar]
- 46.Shen J, Obin MS, Zhao L. The gut microbiota, obesity and insulin resistance. Molecular Aspects of Medicine. 2013. 34(1):39–58. 10.1016/j.mam.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 47.Cani PD, Osto M, Geurts L, Everard A. Involvement of gut microbiota in the development of low-grade inflammation and type 2 diabetes associated with obesity. Gut Microbes. 2012; 3(4):279–288. 10.4161/gmic.19625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vrieze A, Holleman F, Zoetendal EG, De Vos WM, Hoekstra JBL, Nieuwdorp M. The environment within: How gut microbiota may influence metabolism and body composition. Diabetologia. 2010; 53(4):606–613. 10.1007/s00125-010-1662-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proceedings of the National Academy of Sciences of the United States of America. 2007; 104(3):979–984. 10.1073/pnas.0605374104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cani PD, Possemiers S, Van De Wiele T, Guiot Y, Everard A, Rottier O, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009; 58(8):1091–1103. 10.1136/gut.2008.165886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pflughoeft KJ, Versalovic J. Human Microbiome in Health and Disease. Annual Review of Pathology: Mechanisms of Disease. 2012; 7:99–122. 10.1146/annurev-pathol-011811-132421 [DOI] [PubMed] [Google Scholar]
- 52.Afra K, Laupland K, Leal J, Lloyd T, Gregson D. Incidence, risk factors, and outcomes of Fusobacterium species bacteremia. BMC Infectious Diseases. 2013; 13(1):24 10.1186/1471-2334-13-264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Karlsson FH, Tremaroli V, Nookaew I, Bergström G, Behre CJ, Fagerberg B, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013; 498(7452):99 10.1038/nature12198 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Alpha Diversity: Species Accumulation Curve for each sample, the horizontal axis represents the sample size, and the vertical axis represents the number of OTUs detected in the sample. Cross symbols represent the possibility of new species (OTU) addition, with the addition of new samples. (B) Rank Abundance curve was used to explain sample diversity and abundance of species in each sample. The abundance of species is reflected by the length of the curve on the horizontal axis while uniformity of species composition is reflected by the shape of the curve.
(TIF)
(A) PD_whole_tree measures species richness or diversity (B) Observed Species indicates the number of actually observed species (C) Chao1 estimates observed species frequency.
(TIF)
(TIF)
R2: indicates the degree of interpretation of the difference between the different groups, that is, the ratio of the variance of the group to the total variance.
(DOCX)
Data Availability Statement
All sequence files are available from the SRA NCBI database (accession number(s) PRJNA554535. All relevant meta data are within the manuscript and its Supporting Information files.