Abstract
Understanding the mechanisms triggering variation of cell wall degradability is a prerequisite to improving the energy value of lignocellulosic biomass for animal feed or biorefinery. Here, we implemented a multiscale systems approach to shed light on the genetic basis of cell wall degradability in maize. We demonstrated that allele replacement in two pairs of near-isogenic lines at a region encompassing a major quantitative trait locus (QTL) for cell wall degradability led to phenotypic variation of a similar magnitude and sign to that expected from a QTL analysis of cell wall degradability in the F271 × F288 recombinant inbred line progeny. Using DNA sequences within the QTL interval of both F271 and F288 inbred lines and Illumina RNA sequencing datasets from internodes of the selected near-isogenic lines, we annotated the genes present in the QTL interval and provided evidence that allelic variation at the introgressed QTL region gives rise to coordinated changes in gene expression. The identification of a gene co-expression network associated with cell wall-related trait variation revealed that the favorable F288 alleles exploit biological processes related to oxidation-reduction, regulation of hydrogen peroxide metabolism, protein folding and hormone responses. Nested in modules of co-expressed genes, potential new cell-wall regulators were identified, including two transcription factors of the group VII ethylene response factor family, that could be exploited to fine-tune cell wall degradability. Overall, these findings provide new insights into the regulatory mechanisms by which a major locus influences cell wall degradability, paving the way for its map-based cloning in maize.
Introduction
Maize (Zea mays) is one of the most important staple crops, providing carbohydrates, proteins, lipids and vitamins for billions of people. It also serves as an important energy resource for ruminant animal. Indeed, maize silage is largely used for cattle feeding during winter and summer seasons, as well as a complementary resource with high-energy content in cow diets during the year [1]. Thus, improved feeding value is a key target of silage maize breeding.
Investigations with sheep in digestibility crates have shown that silage maize energy value is first related to cell wall degradability [2], which refers to the complex characteristics of cell wall to protect its carbohydrates from degradation by enzymes [3–6]. Importantly, in vivo cell wall degradability ranges from 36 to 60% in silage maize [2]. Correlatively, a similar two-fold variation was observed for in vitro cell wall degradability, which varies from 25 to 50% when determined as neutral detergent fiber (NDF) digestibility (IVNDFD) [7,8]. Consequently, numerous studies have been successfully carried out in silage maize to identify genomic regions (quantitative trait loci; QTL) involved in the variation of cell wall degradability [9–25]. Overall, they demonstrated that cell wall degradability is a highly multifactorial trait, primarily related to lignin content, lignin structure, hemicellulose content, and p-hydroxycinnamic acid cross-linkages [7,23,26–28].
In the majority of cases, QTL with ‘minor’ R2 values (R2 up to 15%) have been identified for cell wall component and degradability traits [23,29]. However, in the QTL studies of the maize recombinant inbred line (RIL) population derived from the cross between early dent lines F288 and F271, a cluster of 10 stable QTL with ‘major’ effects (R2 values up to 42.5% for cell wall components and accounting for 33.1% of the observed variation for IVNDFD) was mapped in bin 6.05 of the maize chromosome 6 [22]. The identified QTL colocalized in the 124–134 cM interval (referred to as QTL6.05), corresponding to the physical position 150.4–153.8 Mp in the B73 reference genome (AGPv3 sequence release). It is noteworthy that cell wall degradability of F271 was reported to be lower than that of F288 and, according to QTL detection [22], alleles of F288 at QTL6.05 increased cell wall degradability in the RIL population derived from F288 x F271.
The dissection of highly complex traits into their underlying components offers practical, technical, and commercial advantages. Specifically, trait dissection leads to the identification of phenotypic attributes which can be measured with greater precision and at a lower cost, but most importantly, these component traits typically display simpler inheritance patterns and provide additional information as to how genes and their variants act together in biological pathways to influence trait variation [30]. However, in complex genomes such as maize which exhibits extensive genome content variation among lines [31–37], including massive expansion of gene families, fine mapping remains a non-trivial process. Despite several attempts [29,38], the mechanisms and genes underlying the QTL6.05 effects remain largely unresolved. Selection of near-isogenic lines (NILs) have been performed to validate and fine map QTL in several plant species including maize [39]. Additionally, coupling genetic information with molecular phenotypes (e.g. transcript levels) can contribute to a better mechanistic understanding of trait variation [35,40–43].
Here, we implemented a multiscale systems approach to shed light on the mechanisms by which the QTL6.05 locus influences cell wall degradability. We developed and selected NILs in which the favorable F288 alleles at an 18–20 Mb region encompassing the QTL6.05 (referred to as the introgressed QTL6.05 region or QTL6.05i) had been introgressed into the background of the recurrent parent F271. We demonstrated that substitution of F271 alleles with that of F288 at QTL6.05i led to variation in cell wall degradability. Using Illumina RNA sequencing (RNA-seq) datasets from NIL internodes, we then captured the QTL6.05i allelic variation consequences at the transcript level relative to a maize reference genome that included F271- and F288-specific genomic sequences at the targeted QTL6.05 locus. Finally, we explored how the variation in gene expression contribute to the variation in cell wall-related traits observed between the selected NILs. Genes associated with the cell wall-related traits were identified, paving the way for pinpointing the causal genes at the QTL6.05 locus in future follow-up studies.
Materials and methods
Plant materials
The RIL122 from the F288 x F271 RIL progeny was crossed with F271 to produce F1 seeds. Then, 31 BC1 lines from the F1 plants x F271 progeny were backcrossed with F271. Genotyping of 31 BC2 lines (25 plants sown per BC1 progeny) was performed using a 50K single-nucleotide polymorphism chip from TraitGenetics. After quality controls based on missing data, heterozygosity, and minor allele frequency, 12,758 markers were retained for genotyping and two BC2 lines (30–16 and 31–04) were selected (S1 Table). Genotyping of BC2S1 lines (30–16 progeny referred to as 1F271 and 1F288; 31–04 progeny referred to as 2F271 and 2F288) was confirmed by High Resolution Melting (S2 Table). The selected BC2S2 lines were then grown in the field at Mauguio (Hérault, France) in 2015 with two row replicates. Irrigation was applied during summer to prevent water stress. For RNA-seq analysis, the bottom third of upstream-ear internodes (without nodes) of three representative plants in each of the two replicates were harvested two days before silking at tasseling stage. All samples were immediately frozen in liquid nitrogen and stored at -80°C. Because of the presence of borer on the stem in two plants, one of the two 1F288 samples was removed from the downstream analysis. For cell wall investigations, ears were removed by hand from plants at silage stage the day before and/or the day of harvest. Representative samples of 1 kg chopped material per plot were collected, dried in a ventilated oven 72 h at 55°C and ground with a hammer mill gondard to pass through a 1-mm screen. Genotyping of BC2S2 lines was further assessed by PCR (S2 Table and S1 Fig).
Cell wall composition and degradability assessment
Dried samples from plants without ears were dried at 50°C for at least 14 h to exclude any moisture, before scanning through the Antaris II near-infrared reflectance analyzer (ThermoFisher). Briefly, for each sample, the measurement consisted of an average of 16 scans done while rotating the cups over the 10,000 cm-1–4,000 cm-1 range with a resolution of 8 cm-1, resulting in absorbance data every 4 cm-1. The resulting spectra were loaded into the TQ analyst software (v.9.4.45; ThermoFisher) and smoothed with a running average in a window size of 9 data points. Their first derivate was computed, normalized and used to estimate the cell wall-related traits of each sample using a formula that was previously calibrated for plants without ears [16]. A two-way ANOVA analysis was then conducted using R software (v.3.4.1; http://cran.at.r-project.org) as described [44].
Pangenome construction
Oriented reads (sizing 150) of both F288 and F271 1.5 Mb-genomic sequences which included the QTL6.05 peak were first simulated using wgsim (v.0.3.0, depth = 20). Mapping of these simulated reads and AGPv3.22 real reads to one parental DNA sequence was then performed using Bowtie2 (v2.2.3, sensitive parameter). Then 2,322 representative transcript assemblies (RTAs) [34] that were lower than 45% coverage and 80% identity to the AGPv3.22 or absent in the AGPv3.22 sequence were mapped to both genomic sequences using gmap (v.2011-08-15, default parameter). Genomic sequences aligned with at least one of the previous sequences were then hard masked with N using maskfasta (v.2.25.0) with default parameters (S2 Fig).
RNA-seq datasets
The internodes were crushed in liquid nitrogen using an Ika Mill crusher (IKA, Staufen-Im-Breisgau, Germany) prior RNA extraction. Total RNA was isolated using the TRIzol Reagent (Invitrogen) and purified with the Qiagen RNeasy Plant Mini kit and RNase-free DNase set according to the suppliers’ instructions. Total RNA integrity was evaluated using the Agilent 2100 bioanalyzer according to the Agilent technologies (Waldbroon, Germany). Individual Illumina TruSeq stranded RNA-seq libraries were prepared for each of the genotypes according to the supplier’s instructions. Libraries were sequenced in four lanes on the Illumina HiSeq 2000 platform at the Centre National de Séquençage (CNS, Evry, France) to generate paired-end stranded reads (sizing 260). For each library, RNA-Seq pre-processing was performed including adapters removing and quality control assessment with fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). The fastq datasets were trimmed for Phred Quality Score (Qscore >20, read length >30 bases), and free of ribosome sequences with tool sortMeRNA [45]. All steps of the experiment, from growth conditions to bioinformatic analyses, were managed in CATdb database [46] according to the international standard MINSEQE ‘minimum information about a high-throughput sequencing experiment’.
RNA-seq reads (35 million to 55 million quality-filtered RNA-seq reads per RNA-seq library) were then mapped to panB using TopHat2 (v. 2.0.14) without gene annotation file with minimum intron length set to 5 bp and maximum intron length to 60 kb, with default settings for other parameters. Next, transcript prediction was performed using Cufflinks (v.2.2.1, default parameters). Using bedtools getfasta (v.2.25.0, default parameters), coding sequences were then extracted leading to the CuffpanB dataset. To quantify expression levels, RNA-seq reads were mapped to panB using TopHat2 (v. 2.0.14) with a gene annotation file and the parameters described above. Expression quantification was determined using HTseq-count (v.0.6.0) with stranded orientation and ‘union’ option. The RNA-Seq project was submitted into the international repository NCBI-Gene Expression Omnibus [47] project ID GSE115157.
Gene annotation
Genes were predicted using an improved and maize dedicated version of the TriAnnot pipeline (v.5.2p04) [48,49] as described below.
Step 1: Similarity search against mitochondria, chloroplast, non-coding RNAs and transposable elements, and masking
Mitochondrial and chloroplast similarities were identified using BLAST [50] against EMBL databanks release 129; non-coding RNAs were identified using tRNAscan [51], RNAmmer [52] and Infernal (http://infernal.janelia.org - infernal-1.1) based on RFAM database (EMBL RFAMEcm_v11). Transposable elements were identified using the maize transposable element database (http://maizetedb.org/~maize/). They were then all masked using RepeatMasker (cross_match engine, cutoff 250; http://repeatmasker.org).
Step 2: Similarity search against transcripts and related proteomes
This was performed using BLAST on the transposable element-masked sequence. Spliced alignments of BLAST hits were then generated using EXONERATE [53]. The following datasets were used: the CuffpanB dataset; all available Zea ESTs and full-length cDNAs (EMBL release 129), non-redundant proteomes of barley (UniProt Release 2016_03 and High confidence proteins IBGSC 2012), Brachypodium (UniProt Release 2016_10 and phytozome v11), rice (UniProt Release 2016_10, IRGSP 2005 and phytozome v11), Sorghum (UniProt Release 2016_10 and phytozome v11), Triticeae (UniProt Release 2016_10 and phytozome v11) and maize (UniProt Release 2016_10).
Step 3: Gene modeling
Two ab initio gene finders trained with a monocot and maize-specific matrix were used: FGeneSH (SOFTBERRY, http://linux1.softberry.com/berry.phtml) and AUGUSTUS [54] respectively. Evidence-driven gene prediction was also computed through the use of two different modules implemented in TriAnnot. The first module is based on BLASTX-EXONERATE spliced alignments of protein sequences from rice (phytozome v11), Brachypodium (phytozome v11), Sorghum (phytozome v11) and barley (High confidence proteins IBGSC 2012) which have been checked for Methionine start. In case of missing start and/or stop codons in the derived coding sequence model, iterative extension was applied in a range of 200 codons until an in frame start and stop codon could be found. If no start and/or stop codon could be find, the model was flagged as pseudogene. The second module (named SIMsearch, derived from FPGP pipeline [55] focused on similarity with maize transcripts: full-length cDNAs (EMBL release 129), CuffpanB and coding sequence-derived gene models from rice (phytozome v11 and IRGSP 2005), Brachypodium (phytozome v11), Sorghum (phytozome v11) and barley (High confidence proteins IBGSC 2012). For every locus showing similarity with transcripts, SIMSearch predicts the coding sequence borders by considering similarity with known proteins from related Poaceae.
Step 4: Selection and functional annotation of the best gene model at every locus
TriAnnot delivered six outputs with gene models: two ab initio gene finders, BLASTX-EXONERATE, and three SIMsearch-derived models. Therefore, a final step was required to select the best model at each locus. This was made according to a scoring system that considers the metrics of the alignments of each gene model against known proteins (taking into account the percentage of identity and coverage). Finally, if the retained gene model had a canonical structure but shared similarity over less than 70% of the length of its best BLAST hit, it was classified as pseudogene. Putative function for the best gene model was then assigned via a combination of similarity search (BLASTP) against several protein databanks, including Arabidopsis, Brachypodium, Hordeum, Glycine, Oryza, Prunus, Populus, Saccharum, Sorghum, Setaria and Vitis families, as well as the Pfam [56,57] protein domain collection using HMMER 3.0 (http://hmmer.janelia.org/software). The alignment with the best hit was parsed in order to check for the presence of gaps (>9 amino acids). TriAnnot follows a nomenclature based on the guideline established in 2006 by the IWGSC annotation working group and provides gene ontology (GO) terms for each gene model and protein domain predictions based on InterProScan [58] search against Pfam, Prosite [59], and SMART [60]. Final GFF output files were post-treated to be used with the graphical editor GenomeView [61]. Several round of manual expertise, made by different curators, were intensively followed to obtain a final structural and functional annotation of F271 and F288 DNA sequences at the targeted QTL6.05 locus (S3 Table).
Differential expression analysis
RNA-seq data were normalized using TMM implemented in the edgeR package (v 3.18.1) [62,63] in R. Gene-wise RNA-seq counts were then analyzed using a negative binomial generalized linear model with a stepwise procedure for increasing the power to detect QTL6.05i-specific differentially expressed (DE) genes between NILsF288 (i.e. both 1F288 and 2F88) and NILsF271 (i.e. both 1F271 and 2F271) as follows:
In step I, the differential expression was assessed for the two pairs of NILs using a negative binomial generalized linear model with allele and NIL main effects, and an allele × NIL interaction. A Benjamini-Hochberg adjusted P-value of 0.05 was used as the cut-off criterion to identify genes with significant allele × NIL interactions.
In step II, the differential expression was assessed for each pair of NILs (e.g. 1F288 vs 1F271) using a negative binomial generalized linear model with allele main effect for each gene. A Benjamini-Hochberg adjusted P-value of 0.05 was used as the cut-off criterion to identify genes with significant allele effect for both NILs 1 and 2.
In step III, to identify DE genes that were not a result of NIL effects but were due to the QTL6.05i region only, step II DE genes were further dissected by making the intersection between the step I genes with no significant differential expression for NIL effects (i.e. no significant differential expression for both NIL and allele × NIL interaction) and the step II DE genes for both NILs 1 and 2.
In the final step, the intersection between the step III DE genes for NILs 1 and the step III DE genes for NILs 2 delivered DE genes for which the differential expression between the NILsF288 and the NILsF271 was declared QTL6.05i-specific.
Hierarchical clustering of gene expression profile
Hierarchical clustering analyses of the set of common QTL6.05i DE genes were performed using Euclidean distances and unweighted pair group averages as the aggregation method with the R function hclust.
Functional annotation of common QTL6.05i genes
MapMan [64] was first used to identify homologous genes in Arabidopsis. GO term enrichment analysis based on the GO classification was then performed by comparing the relative occurrence of a GO term into each DE list (downregulated or upregulated gene list) to its relative occurrence in the genome (reference list) by a hypergeometric test with the R function phyper. A Benjamini-Hochberg adjusted P-value of 0.05 was used as the cut-off criterion.
Correlation analyses
For correlation analysis between gene expression levels and cell wall-related traits, Pearson correlation coefficients (PCCs) were calculated between each DE gene and each trait over the two pairs of NILs using the R cor function. The Student’s t-distribution was used to test the significance of the PCC as follows: t = r/√[(1-r2)/(n-2)] with a level of significance set at 0.05 and n = 4, absolute PCCs higher than 0.95 were significant. The expression pattern of the set of DE genes correlating positively or negatively with at least one cell wall-related trait in both pairs of NILs were visualized using the R cim function from MixOmics (v. 6.3.1) [65].
Gene co-expression network analysis was performed to group DE genes into connected components based on absolute PCCs higher than 0.995 (level of significance of 0.01) and visualized using Cytoscape (v. 3.3.0) [66]. A node corresponded to a DE gene; an edge was determined by the similarity between expression profiles of paired genes calculated with PCCs. Connected modules were identified using NetworkAnalyzer (v. 2.7) [67].
Promoter motif analysis
The presence of conserved motifs in the 5’ regions of the co-expressed genes was determined using the preferentially located motif (PLM) method [68], with minor modifications. Briefly, the dataset was set up using 1,000 bases upstream the transcription start site and 5’ untranslated region (UTR) from the maize AGPv3 plus [69] and the final GFF of the 1.5 Mb-genomic sequences from F271 and F288. Promoters with a 5’ UTR smaller than 10 bases were excluded in order to avoid false promoter regions. The final promoter dataset consisted of 119 promoter sequences, which were divided into two regions. First, the [−1,000, −300] region was used to learn the distribution model using a simple linear regression and determine a 95% confidence interval. Second, non-evenly distributed motifs (i.e. those exhibiting a peak above the confidence interval) were searched within the [−300, UTR] region, leading to the identification of 33 PLMs among 419 motifs tested coming from PLACE [70] and AGRIS [71].
Quantitative RT-PCR analysis of candidate gene expression
Primer pairs were designed from the F271 and F288 1.5 Mb-genomic sequences at the QTL6.05 interval (S2 Table) using Primer-BLAST [72]. Quantitative RT (qRT)-PCR reactions were performed using the Bio-Rad CFX384 touch (Bio-Rad, France) and the SYBR Premix Ex Taq (Tli RNAseH Plus) (Takara, Ozyme, France) following the supplier’s instructions. A dilution series of the pooled cDNAs and a melting analysis were used to validate the primer pairs. Each sample was performed in duplicate. Gene expression was normalized with the mean of the two reference genes ZmGRP2 (GRMZM2G080603) and ZmUBC30 (GRMZM2G102471) coding for Glycine-rich RNA-binding protein 2 and Ubiquitin-conjugating enzyme E2, respectively. Differential expression for each gene was tested with a two-way ANOVA as described [44].
Results
Allelic variation at QTL6.05i leads to higher cell wall degradability
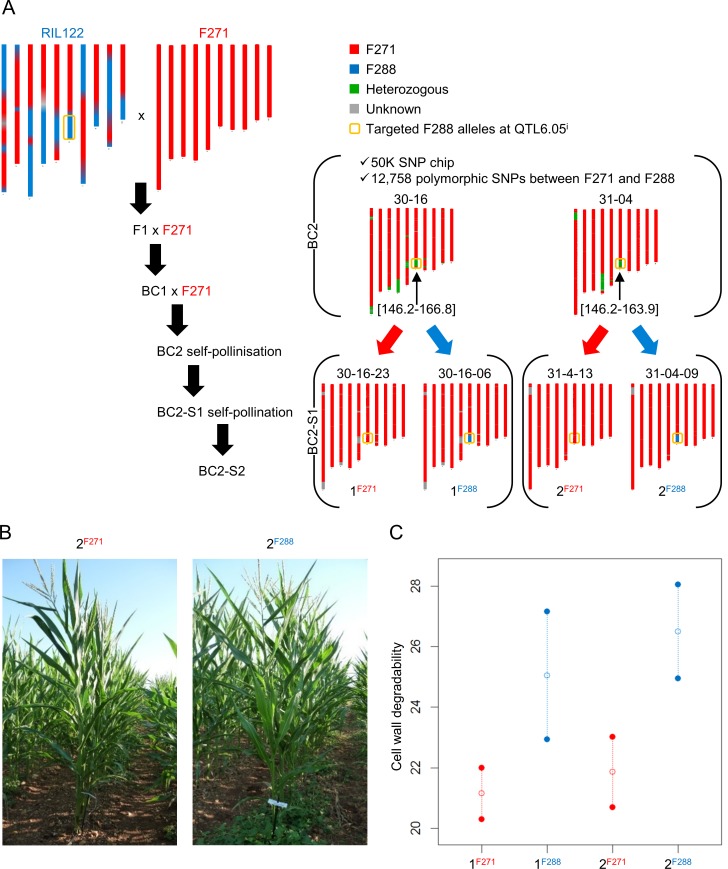
The RIL122 from the F288 x F271 RIL progeny carried F288 alleles at QTL6.05, although F271 alleles were already fixed at roughly more than half of the genome (Fig 1A) [21]. Therefore, this RIL was backcrossed with the parental inbred line F271 for three generations to fasten the introgression of F288 alleles at QTL6.05 into the background of F271. With the help of deep DNA genotyping, two BC2 lines (30–16 and 31–04) were selected that (1) showed a tremendous increase of F271 alleles fixed throughout the genome (93% and 94.3% for 30–16 and 31–04, respectively) compared to the RIL122 (53.8%) and (2) remained heterozygous at QTL6.05i with a relatively similar introgressed genomic region size, ranging from 18 to 20 Mb (Fig 1A and S1 Table). According to QTL detection [22], the QTL6.05i region included the QTL6.05 as well as a cluster of four QTL for cell wall component and degradability (referred to as QTL6.07) with ‘minor’ effects relative to QTL6.05 (R2 values ranging between 8 and 12% whatever the trait; Fig 2A). The two selected BC2 lines were then selfed twice to fix the introgressed genomic region at homozygous state and to multiply the progenies of 30–16 (1F271 and 1F288) and 31–04 (2F271 and 2F288) lines.
Fig 1. Selection and characterization of NILs with an introgressed genomic region at QTL positions for cell wall degradability.
(A) Experimental workflow used for introgression of F288 alleles at an 18–20 Mb region encompassing the QTL6.05 (referred to as the introgressed QTL6.05 region or QTL6.05i) into the background of F271. SNP: single nucleotide polymorphism. (B) The selected BC2S2 NILs 1 (referred to as 1F271 and 1F288) and NILs 2 (referred to as 2F271 and 2F288) were grown in the field under well-watered conditions for DNA and mRNA sampling and phenotyping. (C) Comparison of the cell wall degradability IVNDFD performance of NILs introgressed with F288 alleles at QTL6.05i (blue) and their respective recipient lines with F271 alleles (red). Open circles represent the means of two biological replicates.
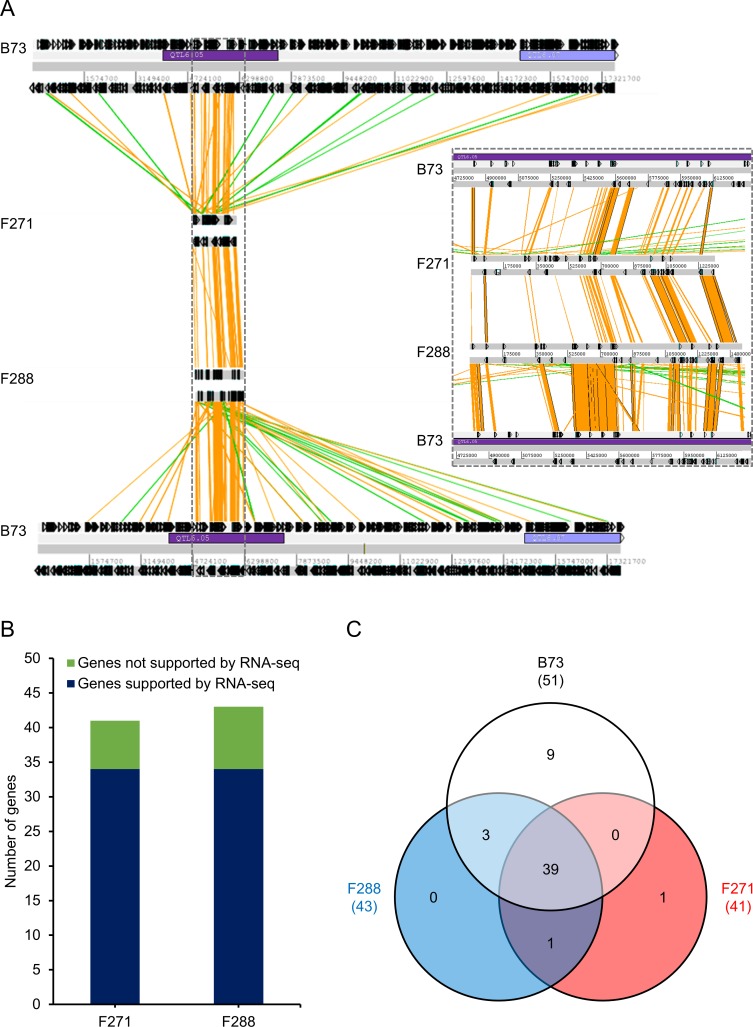
Fig 2. Structure of the introgressed genomic QTL6.05i region.
(A) Large structural variation between B73, F271 and F288 in the introgressed genomic QTL6.05i region. Dark and light purple boxes represent clusters of QTL in bins 6.05 and 6.07, respectively, as previously defined [22]. The grey dotted line insertion represents a zoom of the genomic region that included the 1.5 Mb-genomic sequences from F271 and F288. Orange and green lines indicate sense and antisense DNA strands, respectively, of at least 910 bp and sharing at least 98% of identity; arrowed boxes represent genes and pseudogenes. (B) Number of annotated genes supported by RNA-seq predicted genes in the targeted QTL6.05 locus. (C) Gene content distribution in B73, F288 and F271 in the targeted QTL6.05 locus.
To assess the cell wall properties of the two pairs of NILs grown in the field (Fig 1B), near-infrared spectroscopy data were then collected as was done in the past for QTL6.05 detection [12,22]. The introgressed F288 alleles at QTL6.05i conferred on the NILs 1F288 and 2F288 (NILsF288) a higher cell wall degradability (as estimated by IVNDFD) than the F271 alleles on the NILs 1F271 and 2F271 (NILsF271) (Fig 1B and 1C). Although the two pairs of NILs had similar lignin content, the NILsF288 exhibited lower cellulose content in the cell wall as compared to the NILsF271 (S4 Table). The NILsF288 also tended to have higher hemicellulose content in the cell wall as compared to the NILsF271 (S4 Table). Overall, these results evidenced that substitution of F271 alleles with those from F288 at the QTL6.05i region led to cell wall composition variation and confirmed the presence of the detected QTL.
Aligning RNA-seq reads to a pan-genomic reference genome: a tool to assess the number and type of genes predicted at the targeted QTL6.05 locus
RNA-seq datasets from internodes of the selected NILs were produced at tasseling stage in which transcript patterns of secondary cell wall cellulose synthase genes and lignin-related genes were shown to be highly abundant [73,74]. To take into account any genetic polymorphism, particularly putative present/absent genes, between F271 and F288 at the targeted QTL6.05 locus, a pan-genomic sequence (panB) was assembled through an in lab pipeline and used as a reference sequence for RNA-seq read mapping (Fig 2A and S2 Fig). This pan-genomic reference sequence included (1) the B73 transcript reference sequences (AGPv3), (2) 2,322 RTAs absent in B73 [34] and (3) genomic sequences specific to either F271 or F288 at the QTL6.05 locus which were kindly provided by the French Plant Genomic Resources Center (CNRGV) (Fig 2A and S2 Fig). The pipeline evidenced total lengths of 40.7 kb and 13.1 kb that were specific to F271 (3.1% of the F271 genomic region) and F288 (0.9% of the F288 genomic region), respectively.
To achieve better mapping of the RNA-seq reads to the panB reference sequence, gene prediction at the targeted QTL6.05 locus was then performed using (1) the TriAnnot pipeline [48] through de novo and similarity-based methods and (2) the transcript prediction based on the mapping of the RNA-seq dataset. About 80% of the predicted genes through the TriAnnot pipeline were supported by the mapped RNA-seq dataset (Fig 2B), highlighting the quality of the genomic sequences of each two lines at the QTL6.05 locus and the annotation. Of the 51 genes present in B73 at the targeted QTL6.05 locus, 39 (76%) were found in both F271 and F288 (Fig 2C and S3 Table). Three genes (genes 23, 41 and 42) were found only in F288 and B73; one gene (gene 43a) provided by the RNA-seq dataset was found only in F288 and F271; one gene (gene 12) was found only in F271. Overall, four genes (genes 12, 23, 41 and 42) were present in only one of the two F271 and F288 parental inbred lines at the targeted QTL6.05 locus.
Mapping of the RNA-seq dataset to the panB reference sequence yielded 32,446 genes, including 1,198 RTAs. It is noteworthy that RNA-seq counts from the F271- and F288-specific genomic sequences present in the panB reference sequence were identified (including counts for genes 10, 11, 12, 13 and 21), highlighting the relevancy of the pan-genomic approach. Two (genes 23 and 41) of the four genes present in only one of the two F271 and F288 parental inbred lines at the QTL6.05 locus did not exhibited measurable expression in all the experimental samples. In contrast, the gene 12 was expressed in all the experimental samples, although it was found only in F271 at the QTL6.05 locus (S3 Table). The gene 42 was consistently expressed at a very low level in the NILs 1F288 and 2F288, but not measurable in the NILs 1F271 and 2F271. Hence, this gene represents a case of single pattern expression (SPE) [75]. We designated an observed pattern as SPE_F288 if genes were expressed in the NILsF288 (i.e. in both 1F288 and 2F288), but not in the NILsF271 (i.e. in both 1F271 and 2F271). In contrast, we referred to genes that were expressed in the NILsF271, but not in the NILsF288 as SPE_F271.
Allelic variation at QTL6.05i leads to transcriptional variation
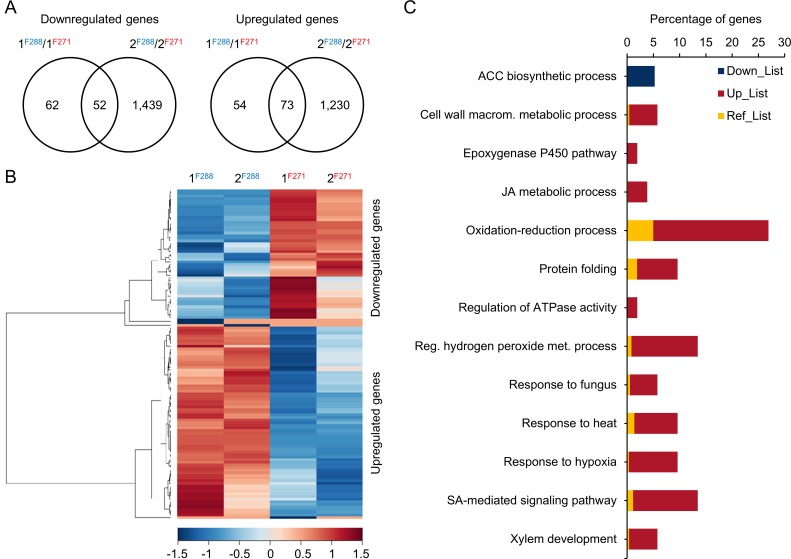
To test the hypothesis that the substitution of F271 alleles with those of F288 at QTL6.05i led to transcriptional variation, RNA-seq counts were analyzed using a stepwise procedure to detect DE genes upon the NILsF288 relative to the NILsF271 that depend only on the QTL6.05i region. As shown in Fig 3A, this stepwise statistical analysis identified 241 and 2,794 DE genes for the NILs 1 and the NILs 2, respectively. Since our goal was to identify DE genes that were due to the introgressed QTL region, we ensured the robustness of the DE gene dataset by keeping only the DE genes that were common to both NILs. This selective approach delivered 125 DE genes, which are referred to as common QTL6.05i DE genes (Fig 3A and S5 Table). As shown in Table 1, these included one RTA (cov45_id80_joint_Locus_58585), which highlighted once again the relevancy of the pan-genomic approach, and 18 genes located in QTL6.05i, of which five were located in the targeted QTL6.05 locus: (1) a δ-1-pyrroline-5-carboxylate synthase (gene 03; ZmP5CS1), (2) an aldo-keto reductase (AKR) (gene 06a; hereafter named ZmAKR4C9 according to [76]), (3) a FK506-binding protein (gene 10; hereafter named ZmFKB20-1 according to [77]), (4) one gene (gene 14a) with homology to Arabidopsis ESMERALDA1 (ESMD1), a putative O-fucosyltransferase [78], hereafter named ZmESMD1, and (5) a 40 S ribosomal protein S23 (gene 36; ZmRPS23).
Fig 3. Transcriptomic adjustments in internodes of NILs introgressed at QTL6.05i.
(A) Venn diagram of upregulated or downregulated genes in the NILsF288 relative to the NILsF271. (B) Hierarchical clustering of DE genes in the NILsF288 relative to the NILsF271. The indicated scale is the natural log value of the normalized level of gene expression. (C) GO Biological Process analysis of DE genes in the NILsF288 relative to the NILsF271. Only Biological Processes with a Benjamini and Hochberg adjusted P-value below 0.05 are represented. Down_List: analyzed downregulated gene list; Up_List: analyzed upregulated gene list; Ref_List: reference list; ACC biosynthetic process: 1-aminocyclopropane-1-carboxylate biosynthetic process; Cell wall macrom. metabolic process: Cell wall macromolecule metabolic process; JA metabolic process: jasmonic acid metabolic process; Reg. hydrogen peroxide met. Process: Regulation of hydrogen peroxide metabolic process; SA-mediated signaling pathway: salicylic acid-mediated signaling pathway.
Table 1. Comprehensive list of the 125 common QTL6.05i DE genes.
| Correlated cell wall-related traits | |||||||
|---|---|---|---|---|---|---|---|
| AGPv3 gene ID and/or gene name | Trend in NILF288 | Cell wall degradabilty | Hemicellulose content | Cellulose content | Lignin content | Connected modules in the gene co-expression network | MapMan annotation |
| GRMZM2G051683 | Down | -0.9968 | -0.9825 | 0.9570 | 0.9668 | M1 | flavonoids.dihydroflavonols |
|
AC203754.4_FG008 ZmP5CS1 (gene 03) |
Down | -0.9955 | -0.9853 | 0.9572 | 0.9769 | M1 | glutamate family.proline |
| GRMZM2G094666 | Down | -0.9978 | -0.9849 | 0.9597 | 0.9678 | M1 | myo-inositol.InsP-Kinases |
| GRMZM2G119766 | Down | -0.9965 | -0.9924 | 0.9693 | 0.9682 | M1 | not assigned |
| GRMZM2G162127 | Down | -0.9825 | -0.9810 | 0.9561 | 0.9633 | M1 | not assigned |
| Locus_58585SPE_F271 | Down | -0.9918 | -0.9978 | 0.9837 | 0.9469 | M1 | NA |
| GRMZM2G050917SPE_F271 | Down | -0.9918 | -0.9978 | 0.9837 | 0.9469 | M1 | not assigned |
| GRMZM2G088349 | Down | -0.9724 | -0.9936 | 0.9852 | 0.9264 | M1 | not assigned |
| GRMZM5G894619 | Down | -0.9921 | -0.9606 | 0.9187 | 0.9951 | M1 | ACC synthase* |
| GRMZM2G368838 | Down | -0.9892 | -0.9718 | 0.9374 | 0.9832 | M1 | AP2/EREBP, APETALA2 |
| GRMZM2G066153 | Down | -0.9673 | -0.9606 | 0.9302 | 0.9612 | M1 | aromatic aa.tryptophan |
| GRMZM2G342509 | Down | -0.9958 | -0.9703 | 0.9326 | 0.9912 | M1 | cell wall proteins.LRR |
| GRMZM2G353276 | Down | -0.9848 | -0.9591 | 0.9188 | 0.9890 | M1 | cellulose synthesis.COBRA |
| GRMZM2G106190 | Down | -0.9771 | -0.9543 | 0.9146 | 0.9827 | M1 | ferredoxin |
| GRMZM2G465835 | Down | -0.9986 | -0.9743 | 0.9410 | 0.9821 | M1 | NAC domain transcription factor family |
| GRMZM2G467184 | Down | -0.9954 | -0.9693 | 0.9312 | 0.9917 | M1 | not assigned |
| GRMZM2G125649 | Down | -0.9934 | -0.9749 | 0.9478 | 0.9643 | M1 | not assigned |
| GRMZM2G463493 | Down | -0.9916 | -0.9614 | 0.9199 | 0.9944 | M1 | receptor kinases.leucine rich repeat XI |
| GRMZM2G170602 | Down | -0.9850 | -0.9396 | 0.8897 | 0.9996 | M1 | C1-metabolism |
| GRMZM2G040965 | Down | -0.9735 | -0.9235 | 0.8687 | 0.9990 | M1 | not assigned |
| GRMZM2G069335 | Down | -0.9541 | -0.9389 | 0.9016 | 0.9615 | M1 | not assigned |
| GRMZM2G430942 | Down | -0.9874 | -0.9485 | 0.9081 | 0.9794 | M1 | stress.biotic |
| GRMZM2G169382 ZmERF71 |
Up | 0.9967 | 0.9937 | -0.9720 | -0.9657 | M1 | AP2/EREBP, APETALA2 |
| GRMZM2G147399 | Up | 0.9986 | 0.9899 | -0.9662 | -0.9678 | M1 | development.unspecified |
| GRMZM2G108997 | Up | 0.9993 | 0.9890 | -0.9637 | -0.9720 | M1 | not assigned |
| GRMZM2G084794SPE_F288 | Up | 0.9866 | 0.9945 | -0.9779 | -0.9513 | M1 | protein.postranslational modification |
|
GRMZM2G140609 ZmRPS23 (gene 36) |
Up | 0.9923 | 0.9977 | -0.9816 | -0.9529 | M1 | ribosomal protein.eukaryotic.40S subunit.S23 |
| GRMZM2G085964 ZmERF72 |
Up | 0.9817 | 0.9962 | -0.9895 | -0.9236 | M1 | AP2/EREBP, APETALA2 |
| GRMZM2G142802 | Up | 0.9737 | 0.9982 | -0.9968 | -0.9102 | M1 | auxin.induced-regulated-responsive-activated* |
| GRMZM2G175805SPE_F288 | Up | 0.9918 | 0.9978 | -0.9837 | -0.9469 | M1 | development.late embryogenesis abundant |
| GRMZM2G098875 | Up | 0.9919 | 0.9941 | -0.9790 | -0.9465 | M1 | glutamate decarboxylase |
|
GRMZM2G059285 ZmAKR4C9 (gene 06a) |
Up | 0.9856 | 0.9919 | -0.9809 | -0.9328 | M1 | minor CHO metabolism.others* |
| GRMZM2G129234 | Up | 0.9570 | 0.9914 | -0.9923 | -0.8969 | M1 | not assigned |
| AC214451.3_FG005SPE_F288 | Up | 0.9918 | 0.9978 | -0.9837 | -0.9469 | M1 | not assigned |
| GRMZM2G005344SPE_F288 | Up | 0.9918 | 0.9978 | -0.9837 | -0.9469 | M1 | not assigned |
| GRMZM2G554314SPE_F288 | Up | 0.9918 | 0.9978 | -0.9837 | -0.9469 | M1 | not assigned |
| AC203989.4_FG001 | Up | 0.9674 | 0.9781 | -0.9601 | -0.9405 | M1 | serine-glycine-cysteine group* |
| AC209215.4_FG004SPE_F288 | Up | 0.9827 | 1.0000 | -0.9922 | -0.9304 | M1 | transport.sugars |
| GRMZM2G136344 | Up | 0.9676 | 0.9957 | -0.9932 | -0.9111 | M1 | zeaxanthin epoxidase* |
| GRMZM5G859099 | Up | 0.9926 | 0.9643 | -0.9307 | -0.9740 | M1 | auxin.signal transduction |
| GRMZM2G131421 | Up | 0.9745 | 0.9603 | -0.9256 | -0.9732 | M1 | development.unspecified |
| GRMZM2G018820 | Up | 0.9975 | 0.9677 | -0.9300 | -0.9891 | M1 | glycerophosphodiester phosphodiesterase |
| GRMZM2G123480 | Up | 0.9875 | 0.9557 | -0.9125 | -0.9945 | M1 | not assigned |
| GRMZM2G050384 | Up | 0.9700 | 0.9598 | -0.9274 | -0.9665 | M1 | not assigned |
| GRMZM2G010673 | Up | 0.9994 | 0.9803 | -0.9481 | -0.9842 | M1 | not assigned |
| GRMZM2G387341 | Up | 0.9904 | 0.9671 | -0.9292 | -0.9889 | M1 | not assigned |
| GRMZM2G399136 | Up | 0.9924 | 0.9533 | -0.9100 | -0.9924 | M1 | stress.abiotic.heat |
| GRMZM2G009719 | Up | 0.9971 | 0.9667 | -0.9277 | -0.9919 | M1 | stress.abiotic.unspecified |
| GRMZM2G026523 | Up | 0.9830 | 0.9666 | -0.9431 | -0.9460 | M1 | transport.peptides and oligopeptides |
| GRMZM2G376661 | Up | 0.9847 | 0.9401 | -0.8903 | -0.9996 | M1 | allene oxidase synthase* |
| GRMZM2G004377 | Up | 0.9618 | 0.8989 | -0.8373 | -0.9969 | M1 | not assigned |
| GRMZM2G371462 | Up | 0.9628 | 0.9189 | -0.8656 | -0.9907 | M1 | not assigned |
| GRMZM2G012631 | Up | 0.9564 | 0.9184 | -0.8676 | -0.9828 | M1 | stress.abiotic.heat* |
| GRMZM2G069651 | Up | 0.9674 | 0.9159 | -0.8594 | -0.9971 | M1 | stress.abiotic.heat* |
| GRMZM2G316362 | Up | 0.9497 | 0.9391 | -0.9044 | -0.9539 | M1 | ACP desaturase* |
| GRMZM2G083716 | Up | 0.9364 | 0.9030 | -0.8534 | -0.9655 | M1 | protein.folding* |
| GRMZM2G024668 | Up | 0.9409 | 0.8949 | -0.8384 | -0.9783 | M1 | stress.abiotic.heat* |
| GRMZM2G028218 | Up | 0.9375 | 0.8816 | -0.8196 | -0.9823 | M1 | stress.abiotic.heat* |
| GRMZM2G164405 | Down | -0.8584 | -0.8310 | 0.7812 | 0.9004 | M1 | ACC synthase* |
| GRMZM2G092432 | Down | -0.9418 | -0.9350 | 0.9024 | 0.9445 | M1 | amino acid metabolism.misc |
| GRMZM2G302245 | Up | 0.8952 | 0.8675 | -0.8190 | -0.9298 | M1 | cell.division |
| GRMZM2G179685 | Down | -0.8536 | -0.8496 | 0.8145 | 0.8740 | M1 | flavonoids.dihydroflavonols |
| GRMZM5G881369 | Down | -0.9043 | -0.8614 | 0.8042 | 0.9496 | M1 | N-metabolism.misc |
| GRMZM2G140763 | Down | -0.8631 | -0.8682 | 0.8395 | 0.8721 | M1 | not assigned |
| GRMZM2G034623 | Up | 0.9224 | 0.9251 | -0.8976 | -0.9205 | M1 | not assigned |
| GRMZM2G177458 | Up | 0.9351 | 0.9353 | -0.9069 | -0.9323 | M1 | not assigned |
| GRMZM2G041714 | Up | 0.8949 | 0.8619 | -0.8101 | -0.9342 | M1 | not assigned |
| GRMZM2G113355 | Up | 0.9106 | 0.8732 | -0.8196 | -0.9500 | M1 | not assigned |
| GRMZM2G316721 | Up | 0.9060 | 0.9093 | -0.8814 | -0.9074 | M1 | not assigned |
| GRMZM2G440313 | Up | 0.8613 | 0.8396 | -0.7934 | -0.8975 | M1 | not assigned |
| GRMZM5G822947 | Up | 0.9161 | 0.8959 | -0.8531 | -0.9390 | M1 | not assigned |
| GRMZM2G056252 | Up | 0.8884 | 0.8765 | -0.8377 | -0.9097 | M1 | omega 6 desaturase* |
| GRMZM2G118610 | Up | 0.8853 | 0.9017 | -0.8818 | -0.8768 | M1 | phenylpropanoids.lignin biosynthesis.CAD* |
| GRMZM2G149422 | Down | -0.8683 | -0.8354 | 0.7826 | 0.9132 | M1 | signalling.in sugar and nutrient physiology |
| GRMZM2G128179 | Down | -0.8939 | -0.9152 | 0.8993 | 0.8775 | M1 | stress.abiotic.drought/salt |
| GRMZM2G300965 | Up | 0.8939 | 0.9187 | -0.9053 | -0.8733 | M1 | stress.biotic.respiratory burst* |
| GRMZM2G158328 | Down | -0.9097 | -0.8873 | 0.8427 | 0.9362 | M1 | WRKY domain transcription factor family |
| GRMZM2G173192 | Up | 0.9578 | 0.9018 | -0.8537 | -0.9596 | M2 | fermentation.LDH* |
| GRMZM2G035890 | Up | 0.9671 | 0.9278 | -0.8913 | -0.9491 | M2 | not assigned |
| GRMZM2G411216 | Up | 0.9528 | 0.8993 | -0.8541 | -0.9489 | M2 | not assigned |
| GRMZM2G165530 | Down | -0.9452 | -0.8774 | 0.8212 | 0.9613 | M2 | not assigned |
| GRMZM2G179827 | Down | -0.9176 | -0.9142 | 0.9069 | 0.8510 | M2 | ARR |
| GRMZM2G313272 | Up | 0.8990 | 0.8926 | -0.8851 | -0.8320 | M2 | aspartate family.asparagine |
| GRMZM2G124921 | Up | 0.9057 | 0.8377 | -0.7870 | -0.9095 | M2 | development.storage proteins |
| GRMZM2G126732 | Up | 0.8616 | 0.8439 | -0.8321 | -0.8003 | M2 | ethylene.synthesis-degradation* |
| GRMZM2G087186 | Up | 0.8691 | 0.8802 | -0.8858 | -0.7825 | M2 | fermentation.PDC* |
| GRMZM2G170958 | Up | 0.9004 | 0.9130 | -0.9167 | -0.8176 | M2 | HB,Homeobox transcription factor family |
| GRMZM2G178546 | Down | -0.8441 | -0.8457 | 0.8473 | 0.7627 | M2 | minor CHO metabolism.trehalose.TPP |
| GRMZM2G100158 | Up | 0.8619 | 0.8353 | -0.8177 | -0.8096 | M2 | misc.cytochrome P450* |
| GRMZM2G133407 | Up | 0.8471 | 0.8207 | -0.8044 | -0.7928 | M2 | misc.misc2 |
| GRMZM2G021388 | Up | 0.9020 | 0.8502 | -0.8120 | -0.8838 | M2 | N misc.alkaloid-like |
| GRMZM2G008972 | Down | -0.9132 | -0.8350 | 0.7747 | 0.9349 | M2 | not assigned |
| GRMZM2G050556 | Down | -0.9090 | -0.8697 | 0.8399 | 0.8772 | M2 | not assigned |
| GRMZM2G552956 | Down | -0.9096 | -0.8857 | 0.8662 | 0.8612 | M2 | not assigned |
| GRMZM2G055802 | Up | 0.8067 | 0.8213 | -0.8332 | -0.7103 | M2 | not assigned |
| GRMZM2G119705 | Up | 0.8766 | 0.8460 | -0.8247 | -0.8304 | M2 | not assigned |
| GRMZM2G122543 | Up | 0.8544 | 0.8188 | -0.7957 | -0.8108 | M2 | not assigned |
| GRMZM2G127418 | Up | 0.8944 | 0.9171 | -0.9271 | -0.8029 | M2 | phenylpropanoids |
| GRMZM2G039993 | Up | 0.9138 | 0.8562 | -0.8126 | -0.9053 | M2 | salicylic acid.synthesis-degradation |
| GRMZM2G123973 | Down | -0.9190 | -0.8483 | 0.7937 | 0.9301 | M2 | transport.misc |
| GRMZM2G019183 | Up | 0.8756 | 0.8887 | -0.8949 | -0.7884 | M2 | trehalose.potential TPS/TPP |
| GRMZM2G101000 | Up | 0.7969 | 0.7826 | -0.7393 | -0.8373 | M3 | DNA.synthesis/chromatin structure |
| GRMZM2G311961 | Down | -0.7418 | -0.7117 | 0.6575 | 0.8051 | M3 | G-proteins |
| GRMZM2G381404 | Down | -0.7918 | -0.7719 | 0.7250 | 0.8383 | M3 | not assigned |
| GRMZM2G176774 | Down | -0.7895 | -0.7898 | 0.7559 | 0.8165 | M3 | protein.glycosylation |
| GRMZM2G015333 | Down | -0.7922 | -0.7849 | 0.7460 | 0.8265 | M3 | stress.biotic |
|
AC213621.5_FG002 ZmESMD1 (gene 14a) |
Up | 0.9530 | 0.9601 | -0.9552 | -0.8857 | M4 | auxin.induced-regulated-responsive-activated |
| GRMZM2G023982 | Up | 0.9560 | 0.9694 | -0.9676 | -0.8852 | M4 | not assigned |
|
GRMZM2G035922 ZmFKBP20-1 (gene 10) |
Down | -0.9458 | -0.9710 | 0.9766 | 0.8649 | M4 | protein.folding |
|
GRMZM5G869403 ZmEXO70B1 |
Up | 0.9289 | 0.9657 | -0.9790 | -0.8375 | M4 | RNA.regulation of transcription.unclassified |
| GRMZM2G464572 | Down | -0.9551 | -0.8847 | 0.8220 | 0.9870 | M5 | protein.degradation.ubiquitin.E2 |
| GRMZM2G428391 | Up | 0.9290 | 0.8454 | -0.7750 | -0.9742 | M5 | stress.abiotic.heat* |
| GRMZM2G389416 | Down | -0.6438 | -0.5225 | 0.4477 | 0.6940 | M6 | glutamate family.proline |
| GRMZM2G086590 | Down | -0.6438 | -0.5225 | 0.4477 | 0.6940 | M6 | isoprenoids.terpenoids |
| GRMZM2G006206 | Down | -0.9286 | -0.9805 | 0.9947 | 0.8479 | M7 | G-proteins |
| GRMZM2G159393 | Down | -0.9227 | -0.9776 | 0.9960 | 0.8331 | M7 | misc.oxidases |
| GRMZM2G077662 | Down | -0.9182 | -0.9667 | 0.9744 | 0.8547 | NA | G-proteins |
| GRMZM2G067122 | Up | 0.9408 | 0.9827 | -0.9955 | -0.8542 | NA | not assigned |
| GRMZM2G431039 | Down | -0.7376 | -0.7720 | 0.7604 | 0.7348 | NA | misc.beta 1,3 glucan hydrolases |
| GRMZM2G164074 | Up | 0.7721 | 0.7447 | -0.7324 | -0.7121 | NA | misc.cytochrome P450* |
| GRMZM2G093246 | Down | -0.5015 | -0.6296 | 0.6882 | 0.3994 | NA | misc.myrosinases-lectin-jacalin |
| GRMZM2G087824 | Down | -0.8454 | -0.7783 | 0.7336 | 0.8378 | NA | not assigned |
| GRMZM2G071846 | Up | 0.8348 | 0.7815 | -0.7156 | -0.9027 | NA | phosphoribosyltransferases.aprt |
| GRMZM2G054448SPE_F288 | Up | 0.6488 | 0.5684 | -0.4839 | -0.7639 | NA | protein.degradation |
| GRMZM2G105348 | Down | -0.6933 | -0.6953 | 0.6608 | 0.7321 | NA | stress.abiotic.heat |
DE genes located at QTL6.05i and/or displaying SPE patterns are in bold and/or underlined, respectively. Dark gray indicates DE genes with expression levels negatively and significantly correlating with cell wall-related traits. Light gray indicates DE genes with expression levels positively and significantly correlating with cell wall-related traits. Co-expression modules as determined by NetworkAnalyzer are designated M1-M7. Cell wall degradability, sugar content and lignin content were estimated by IVNDFD, hemicellulose content in % NDF, cellulose content in % NDF and acid detergent lignin in % NDF. Locus_58585: cov45_id80_joint_Locus_58585; NA: not appropriate.
* DE genes with significant enrichment in Biological processes.
This prompted us to compare our common QTL6.05i DE gene dataset to the deregulated transcriptome of four RILs derived from the cross of F288 x F271 [38]. Interestingly, five of the 125 common QTL6.05i DE genes exhibited DE patterns between F271 and the set of four RILs (Table 1): (1) ZmFKB20-1 described above, (2) an universal stress protein (USP) (GRMZM2G009719), (3) the maize J3 ortholog (GRMZM2G028218; ZmHSP40) encoding an HSP40 known to function in meristem size control in Arabidopsis [79], (4) one gene (GRMZM2G041714) with homology to Arabidopsis hypoxia-induced gene domain 3, and (5) GRMZM2G169382, an ortholog of Arabidopsis group VII ethylene response factor (ERF-VII) ERF71/HYPOXIA RESPONSIVE ERF2 (ZmERF71/HRE2) that may supervise plant intracellular reactive oxygen species (ROS) homeostasis [80]. It is noteworthy that the gene 42 with unknown function did not belong to the common QTL6.05i DE gene dataset, which is not in favor of a direct role of this gene in cell wall-related trait variation. Importantly, none of the common QTL6.05i DE genes were located in the QTL6.07 confidence interval.
Of the common QTL6.05i DE genes, 52 and 73 were found to be downregulated and upregulated in the NILsF288 relative to the NILsF271, respectively (Fig 3A and 3B, and Table 1). Interestingly, two of the 52 downregulated genes showed SPE_F271 patterns, while seven of the 73 upregulated genes showed SPE_F288 patterns (Table 1). Additionally, GO analysis revealed that the downregulated genes had significant enrichment in the “1-aminocyclopropane-1-carboxylate (ACC) biosynthetic process”, which is the first committed and in most instances the rate-limiting step in ethylene biosynthesis (Fig 3C and S5 Table). For the upregulated genes, enriched GO terms associated with biological processes were related to oxidation-reduction, regulation of hydrogen peroxide metabolism, response to hypoxia, jasmonic acid (JA) metabolism, salicylic acid (SA)-mediated signaling pathway, protein folding, response to heat, cell wall macromolecule metabolism and xylem development (Fig 3C and S5 Table).
Almost two-thirds of the common QTL6.05i DE genes are associated with the cell wall-related traits
To test the ability of the 125 common QTL6.05i DE genes to be associated with the cell wall-related traits, linear correlation between gene expression levels and traits was determined by calculating PCCs between the expression level of each of the common QTL6.05i DE genes and each cell wall-related trait. We found 79 DE genes significantly associated with at least one of the traits, referred to below as the correlating DE gene set (Table 1 and S6 Table). This included the cov45_id80_joint_Locus_58585 and the five DE genes (ZmP5CS1, ZmAKR4C9, ZmFKB20-1, ZmESMD1 and ZmRPS23) located in the QTL6.05 locus (Table 1).
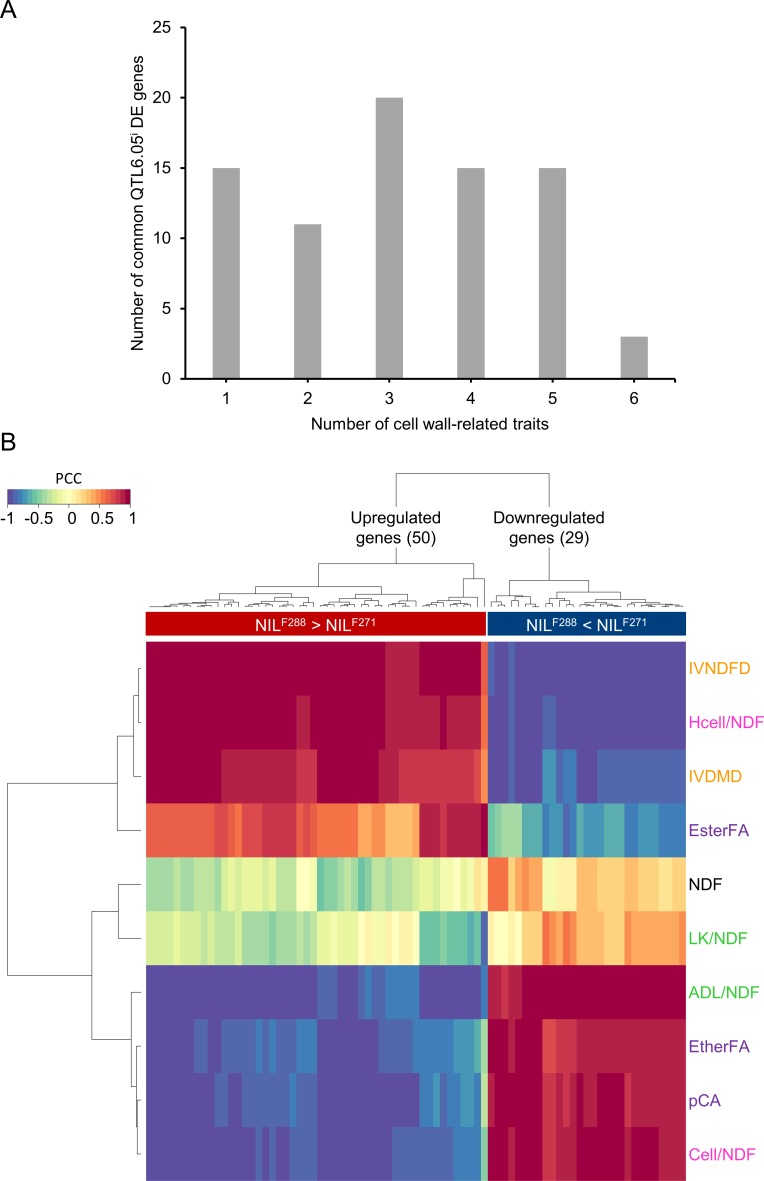
The number of correlating DE genes ranged from 15 for one trait up to 20 for three traits (Fig 4A and S6 Table). A subset of three upregulated genes (AC209215.4_FG004, GRMZM2G085964 and GRMZM2G142802) were associated with six traits (S6 Table). Among them, one gene (AC209215.4_FG004) with homology to Arabidopsis voltage dependent anion channel 1 showed SPE_F288 pattern (Table 1). Interestingly, the subset of three upregulated genes also included a transcription factor (TF) with homology with Arabidopsis ERF72/RELATED TO APETALA2.3 (RAP2.3) (GRMZM2G085964; hereafter named ZmERF72/RAP2.3) linked to the hypoxia response [81]. The third upregulated gene (GRMZM2G142802) was annotated as a stem-specific protein TSJT1 and related to enriched GO terms “SA-mediated signaling pathway”, “regulation of hydrogen peroxide metabolic process” and “hypoxia” (Table 1 and S5 Table).
Fig 4. Identification of DE genes associated with cell wall-related traits.
(A) Number of common QTL6.05i DE genes correlating with at least one of the cell wall-related traits. (B) Gene expression levels of the 79 correlating DE genes in NILs introgressed into QTL6.05i. Columns represent gene expression profiles, and rows represent cell wall-related traits as follows: cell wall content (black), degradability (orange), sugar content (pink), lignin content (green) and p-hydroxycinnamic acids content (purple). IVNDFD: in vitro neutral detergent fiber digestibility; Hcell/NDF: hemicellulose content in % NDF; IVDMD: in vitro dry matter digestibility; EsterFA: esterified ferulic acid content in mg g-1 NDF; LK/NDF: lignin klason in % NDF; NDF: neutral detergent fiber; ADL/NDF: acid detergent lignin in % NDF; Cell/NDF: cellulose content in % NDF; pCA: p-coumaric acid content mg g-1 NDF; EtherFA: etherified ferulic acid content in mg g-1 NDF.
The expression levels of the 79 correlating DE genes and their correlation with the cell wall-traits were visualized in a clustered map, revealing opposite correlation of individual DE genes with multiple traits (Fig 4B). It is worth noting that upregulated genes (including ZmAKR4C9, ZmESMD1 and ZmRPS23 located in the QTL6.05 locus) showed positive correlations with cell wall degradability and hemicellulose content, and negative correlations with cellulose content (Fig 4B). In contrast, downregulated genes (including ZmP5CS1 and ZmFKB20-1 located in the QTL6.05 locus) showed negative correlations with cell wall degradability and hemicellulose content, while exhibiting positive correlations with cellulose content (Fig 4B).
Toward a co-expression network of genes associated with QTL6.05i
To get more insights into the putative coregulation of the 125 common QTL6.05i DE genes, a network analysis of co-expression relationships among the common QTL6.05i DE genes was performed using PCCs between the gene expression levels. The resulting gene co-expression network incorporated 116 DE genes and 329 edges (Fig 5 and S7 Table). Seven connected modules (designated M1-M7) were determined and significant correlations with cell wall degradability and sugar content were visualized in the gene co-expression network for each DE gene (Table 1). As shown in Fig 5, modules M1 (77 DE genes and 283 edges), M4 (4 DE genes and 3 edges) and M7 (2 DE genes and 1 edge) brought together the majority of the correlating DE genes since 58% (45/77), 100% (4/4) and 100% (2/2) of their nodes, respectively, were correlated with at least two of the three cell wall-related traits. It is also worth noting that 70% (54/77) of the genes in module M1 correlated with cell wall degradability (Table 1). In contrast, DE genes within modules M2 (24 DE genes and 34 edges), M3 (5 DE genes and 6 edges), M5 (2 DE genes and 1 edge) and M6 (2 DE genes and 1 edge) tended to be almost not correlated with cell wall-related traits (Table 1 and Fig 5). Interestingly, up to two-thirds of the genes in module M1 that were associated with at least one of the three cell wall-related traits were related to the enriched biological processes we identified. One noteworthy gene of module M1 that negatively correlated with cell wall degradability and hemicellulose content was an ACC synthase (ACS) gene (GRMZM5G894619; hereafter named ZmACS7) involved in the ethylene biosynthetic pathway (Fig 5). Also related to hormone responses were the two ERF-VIIs (ZmERF71/HRE2 and ZmERF72/RAP2.3) that showed positive correlation with the three cell wall-related traits (Fig 5). Another class of genes correlated with cell wall degradability are heat shock proteins (HSPs) known to affect protein folding and response to heat (Fig 5). Two positively correlating genes (GRMZM2G012631 and GRMZM2G069651) with homology to Arabidopsis HSP90 were connected to a limited set of other genes, including another maize HSP90 ortholog (GRMZM2G024668) and ZmHSP40.
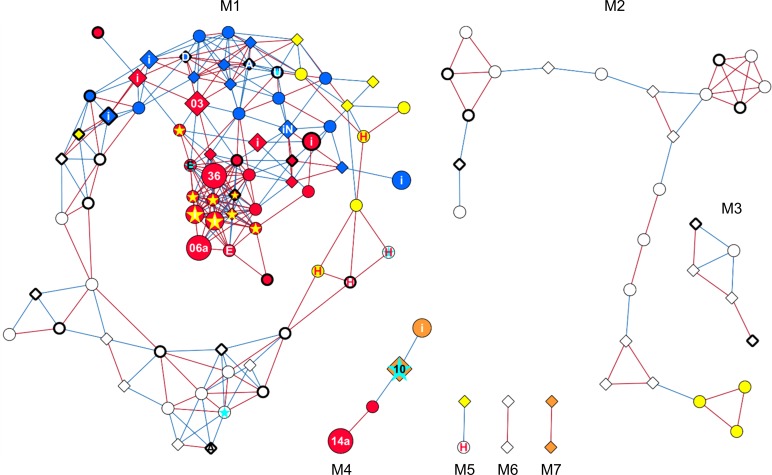
Fig 5. Co-expression network of common QTL6.05i DE genes.
Downregulated genes in the NILsF288 relative to the NILsF271 are represented by diamonds, and upregulated genes by circles. Colored diamonds indicate genes with expression levels negatively correlating with cell wall degradability (IVNDFD) and/or hemicellulose content (Hcell/NDF), and/or positively correlating with cellulose content (Cell/NDF). Colored circles indicate genes with expression levels positively correlating with IVNDFD and/or Hcell/NDF, and/or negatively correlating with Cell/NDF. Node colors represent correlation with IVNDFD (yellow), IVNDFD and Hcell/NDF (blue); IVNDFD, Hcell/NDF and Cell/NDF (red); Hcell/NDF and Cell/NDF (orange). The “i” present in the nodes indicates DE genes located in the QTL6.05i region; numbered and larger nodes indicate genes located in the QTL6.05 locus (gene 03: ZmP5CS1; gene 06: ZmAKR4C9; gene 10: ZmFKB20-1; gene 14a: ZmESMD1; gene 36: ZmRPS23). The letters “A”, “D”, “E”, “H”, “N” and “U” present in the nodes indicate genes encoding ACC synthase, DREB TF, ERF-VII TF, HSP, NAC TF and universal stress protein, respectively. Blue stars indicate DE genes previously identified [38]; yellow stars indicate DE genes with SPE patterns co-expressed with ZmRPS23 (gene 36). Thick borders indicate gene with the GCCGCC motif in their promoter. Edges colored blue connect co-expressed genes with a PCC less than -0.995, red edges represent a PCC bigger than 0.995. Modules as determined by NetworkAnalyzer are designated M1-M7.
Of the 116 DE genes incorporated in the gene co-expression network, 16 (almost 14% of the DE genes incorporated in the gene co-expression network) located in QTL6.05i, including the fives genes (ZmP5CS1, ZmAKR4C9, ZmFKB20-1, ZmESMD1 and ZmRPS23) located in the QTL6.05 locus (Table 1 and Fig 5). Furthermore, we observed that 13 of these 16 DE genes were in module M1 (including ZmP5CS1, ZmAKR4C9 and ZmRPS23), while three were in module M4 (including ZmFKB20-1 and ZmESMD1). All these 16 DE genes were correlating with at least two of the three cell wall-related traits. Of note, ZmRPS23 in module M1 had high connectivity with 13 DE genes (including ZmERF71/HRE2 and eight of the nine DE genes displaying SPE_patterns) that were all correlated with cell wall degradability and sugar content (Fig 5). Moreover, seven neighbors of ZmRPS23 were connected with ZmAKR4C9, while four neighbors of ZmRPS23 were connected with ZmP5CS1. It is also worth noting that ZmAKR4C9 had high connectivity with eight DE genes (including ZmERF2/RAP2.3) that were all correlated with cell wall degradability and sugar content (Fig 5). Additionally, three of the four DE genes of module M4 were located in QTL6.05i, namely ZmFKB20-1, ZmESMD1 and one gene (GRMZM5G869403) annotated as exocyst subunit EXO70 family protein B1 (ZmEXO70B1).
Transcription factors associated with QTL6.05i
Genes having the same TF binding sites are assumed to be functional partners since they are regulated by the same TFs. Therefore, a PLM study [68] was carried out to determine whether there were any over-represented TF binding sites in the promoter of the common QTL6.05i DE gene dataset. Among the promoters of the common QTL6.05i DE genes, we identified the CATGTG motif (S8 Table) which is bound by NAC TFs [82]. Interestingly, there was one gene (GRMZM2G465835) in module M1 (Fig 5) located in QTL6.05i and showing homology to Arabidopsis NAC090, which is a negative regulator of SA-mediated leaf senescence [83]. Also present in the promoters of the common QTL6.05i DE genes were (1) the TGACG motif which is involved in transcriptional activation of several genes by auxin and/or SA [84] and (2) the GCCGCC motif which is the core site of GCC-box required for the binding of AP2/ERF TF family and for the regulation of JA-responsive gene expression [85,86]. It is noteworthy that in module M1 the two ERF-VIIs (ZmERF71/HRE2 and ZmERF72/RAP2.3) were central in the sense that together they covered 73% (19/26) of the DE genes (including ZmAKR4C9 and ZmRPS23 located in the QTL6.05 locus) that were correlated with cell wall degradability and sugar content. Additionally, a member (GRMZM2G368838) of the DREB (dehydration-responsive-element-binding) subfamily A-4 of AP2/ERF TF family was also present in module M1 (Fig 5). One noteworthy neighbors of GRMZM2G368838 was ZmACS7 (Fig 5). Finally, several F271 or F288-specific PLMs were identified in the promoter of ZmAKR4C9, ZmFKBP20-1, ZmESD1 and ZmRPS23 (S3 Fig). Although, the biological importance of these genotype-specific motifs will require further investigation, these results, together with those of Fig 5, support ZmAKR4C9, ZmFKBP20-1, ZmESD1 and ZmRPS23 as candidate genes for the targeted QTL6.05 locus. Therefore, to confirm the differential expression, qRT-PCR experiments were performed based on the same samples that had been used for the initial RNA-seq analysis. Transcript levels of ZmAKR4C9 were not quantified because it was not possible to design gene-specific primers of sufficient quality for qRT-PCR. The other three candidate genes were significantly affected by the substitution of F271 alleles with that of F288 at QTL6.05i with opposite effects: (1) downregulation of ZmFKBP20-1 in the NILsF288 relative to the NILsF271; (2) upregulation of ZmESD1 and ZmRPS23 in the NILsF288 relative to the NILsF271 (S9 Table). These results were in agreement with those obtained by RNA-seq (Table 1 and S5 Table), thus validating our differential expression analysis.
Discussion
Expanding maize genetic and genomic resources to shed light on the mechanisms whereby the QTL6.05 locus influences cell wall degradability
We implemented a systems biology approach that combined genetic, genomic, transcriptomic and phenotypic data to begin answering the mechanisms whereby the major QTL6.05 locus influences cell wall degradability. The use of the NILs that we developed using the F271 inbred line as the recurrent parent and the RIL122 carrying F288 alleles at QTL6.05 as donor enabled us to show that substitution of F271 alleles with those of F288 at QTL6.05i led to an increase of four percentage points of IVNDFD, as previously reported for QTL6.05 in QTL mapping experiments [21,22]. Consequently, the results obtained with the selected NILs most likely reflected the QTL6.05 rather the QTL6.07, which had ‘minor’ effects [21,22]. Consistently, none of the 125 common QTL6.05i DE genes were located in the QTL6.07 support interval. Additionally, similar to what we observed for cell wall degradability, we found that hemicellulose content tended to be higher in the NILsF288 compared to the NILsF271. In contrast, cellulose content in the NILsF288 was significantly lower relative to the NILsF271. These data are comparable to the previous characterizations of the F288 x F271 RIL progeny [12], as well as those of a forage maize doubled haploid population [23], an association panel [87] and six mapping populations derived from European elite maize germplasm [25]. Therefore, these results, together with those of cell wall degradability, showed that the NILs developed in this study provided the suitable biological tools to understand the underlying mechanisms and genes that control cell wall-related trait variation at QTL6.05.
Gene prediction at the QTL6.05 locus from both F271 and F288 inbred lines highlighted the three genes 23, 41 and 42 that are present in F288 but not in F271, and the gene 12 that is present in F271 and not in F288. It is noteworthy that the gene 12 was expressed in the two pairs of NILs, which might reflect relocated single-copy gene in F288. In contrast, we evidenced 10 genes displaying SPE patterns, including the gene 42. Hence, such SPE pattern was consistent with the absence of the gene 42 in F271 at the QTL6.05 locus interval and suggests that this gene is completely absent from the genome of F271. It was not possible to determine the fraction of genes displaying presence/absence variation among the nine other identified SPE patterns because of the lack of genomic data from both F271 and F288. Considering that low proportions of presence/absence variations had been observed among genes displaying SPE patterns [37,75,88,89], we anticipate that these nine genes (two-thirds of which have unknown function) represent cases of genotype-dependent differential gene expression. Alleles from the unfavorable NILs for cell wall degradability were activated in the favorable NILs for cell wall degradability presumably via interactions with QTL6.05i-specific regulatory factors from the favorable NILs. Consistently, the group of co-expressed genes that were located in QTL6.05i included the maize NAC090 ortholog involved in SA-mediated signaling pathway [83], which is one of the enriched biological processes observed here. Moreover, the nine SPE patterns were correlated with cell wall-related traits. This observation confirms the expectation that SPE can affect the phenotype variation directly [35,37] and suggests a biological role of these SPE patterns in the variation of cell wall degradability controlled by the QTL6.05 locus.
Identification of hub genes associated with cell wall-related trait variation at QTL6.05i
Additionally, the systems biology approach employed in this study revealed biological processes occurring in response to allelic variation at QTL6.05i. First, a set of processes related to oxidation-reduction and regulation of hydrogen peroxide metabolism was included in the gene co-expression network. Two noteworthy genes are a ferulate 5-hydroxylase (GRMZM2G100158) and a cinnamyl alcohol dehydrogenase (GRMZM2G118610) involved in lignin biosynthesis [90]. In addition, two plant cysteine oxidases (AC203989.4_FG001 and GRMZM2G113355; PCOs), one ACC oxidase (GRMZM2G126732) involved in the ethylene biosynthetic pathway, one respiratory burst oxidase homolog gene (GRMZM2G300965 or ZmRbohB) encoding a NADPH-oxidase and ZmAKR4C9 were also observed in the gene co-expression network. These results, together with those described previously [38], support the idea that a tight and complex regulation, a long way upstream of the monolignol biosynthetic pathway, is driven by the F288 alleles at QTL6.05i.
Second, genes involved in hormone responses are co-expressed in the network. Notably, ZmACS7, the maize PCO ortholog AC203989.4_FG001 and the two ERF-VIIs, ZmERF71/HRE2 and ZmERF72/RAP2.3, were co-expressed in module M1 and associated with cell wall-related trait variation. PCOs were shown to oxidize the penultimate cysteine of ERF-VIIs by using oxygen as co-substrate, thereby controlling the lifetime of these proteins in Arabidopsis [91,92]. A perspective is emerging in which a diversified set of mechanisms can influence ERF-VII expression in order to specify their functions in a wider network of physiological pathways, in particular those activated by hormones [81,93]. In that respect, a target of ERF-VIIs, namely hypoxia responsive universal stress protein 1, has been shown to coordinate oxygen sensing by PCO/ERF74 (also known as RAP2.12) with H2O2 production by NADPH oxidases in Arabidopsis [94]. Consistently, module M1 also included one USP ortholog already described [38] and one maize NADPH oxidase ortholog ZmRbohB. Co-expression of these genes in module M1, together with the identification of the TGACG and GCCGCC motifs (included in the maize PCO ortholog AC203989.4_FG001 and the USP gene) and the recent observation that methyl jasmonate and SA are able to modify the cell wall structure in Brachypodium [95], thus opens the exciting possibility that ERF-VIIs might play a role in the mechanisms whereby the QTL6.05 influences cell wall degradability.
Third, genes involved in protein folding and response to heat are co-expressed in the network, including ZmHSP40 already described [38] and three maize HSP90 orthologs. HSP40 proteins can initiate a conserved chaperone assembly line that mediates conformational changes required for the activity of many native proteins, including HSP70 and HSP90. In plants, HSP90 chaperone pathway plays crucial role in development, and known clients include auxin and JA receptors [96,97]. Furthermore, transcripts of the Arabidopsis HSP90 orthologs of GRMZM2G024668 and GRMZM2G069651 were reported to move in root-to-shoot direction [98], which may indicate that they function in shoot. Overall, these results also implicate protein folding in the mechanisms underlying the QTL6.05 effects.
Identification of four candidate genes for the targeted QTL6.05 locus
Finally, combining meta-analysis of gene co-expression network, phenotypic NIL data and specific TF binding sites within promoters revealed ZmFKB20-1, ZmESMD1, ZmAKR4C9 and ZmRPS23 as promising candidate genes for the targeted QTL6.05 locus. The first two genes belonged to module M4. ZmFKB20-1 which was one of only two DE genes already identified in the targeted QTL6.05 support interval [38] belongs to the FK506-binding protein family, which contains up to 30 paralogous genes in the maize genome [77,99]. Although historically linked to immunosuppression and proline bond rotation, the physiological importance of FK506-binding proteins extends general protein folding to diverse biological processes, including hormone signaling, DNA transcription and protein trafficking. In module M4, ZmFKBP20-1 was co-expressed with a limited set of other genes, including ZmEXO70B1 involved in cell polarity and morphogenesis [100] and ZmESMD1 involved in cell adhesion through as yet unidentified cell wall modifications in Arabidopsis [78]. Together, these results indicate that ZmFKBP20-1 may function in cell wall-related trait variation and highlight the potential importance of cell adhesion in the mechanisms underlying the QTL6.05 effects.
The two other candidate genes ZmAKR4C9 and ZmRPS23 belonged to module M1. They both had high connectivity with other DE genes in the gene co-expression network, including the two ERF-VIIs and eight of the nine DE genes displaying SPE_patterns, which were found to be associated with cell wall-related traits. Comparison of multiple sequence alignments suggests that ZmAKR4C9 may be involved in detoxification mechanisms similar to those of AtAKR4C9 [101]. Consistently, gene expression of tomato AKR4C9 was shown to be induced by hydrogen peroxide and plant hormones such as SA and JA [102], which coincide with the enriched biological processes we observed here. In contrast to ZmAKR4C9, ZmRPS23 belongs to the ribosomal protein S12/S23 family that functions in translation fidelity [103]. Interestingly, several studies identified RPS23 as an enzyme substrate for the oxygenase OFD1 in fission yeast [104]. When little oxygen is around, OFD1 cannot hydroxylate RPS23. This traps OFD1 away from a TF called SRE1. As a result, unassembled RPS23 regulates SRE1 signaling by sequestering OFD1 in an oxygen-dependent manner, thereby coupling hypoxic gene expression to rates of ribosomal synthesis. Therefore, our findings raise the intriguing possibility that unassembled ZmRPS23 regulates an as yet undetermined TF-mediated signaling pathway in which ERF-VIIs may be involved.
Conclusion
We expanded maize genetic and genomic resources to shed light on the mechanisms whereby the QTL6.05 locus influences cell wall degradability. Our data showed that the F288 alleles exploit processes related to oxidation-reduction, regulation of hydrogen peroxide metabolism and protein folding to modify cell wall degradability. Involvement of the SA, JA and ethylene pathways in the mechanisms underlying the QTL6.05 effects is also supported by expression of several related genes, identification of the TGACG and GCCGCC regulatory motifs, and co-expression of two ERF-VIIs that function as central nodes in the gene co-expression network. Overall, our work led to the identification of a subset of potential new cell wall regulatory genes that could be exploited to fine-tune cell wall degradability.
Supporting information
The two selected pairs of NILs (BC2-S2) were field grown under well-watered conditions for DNA and mRNA sampling and phenotyping. DNA samples were used for genotyping by PCR using three markers located in the introgressed genomic region (bngl1702, bnlg1732 and bnlg345) and one outside (umc1127) as indicated in the S2 Table. L: 50 bp DNA ladder.
(PDF)
(i) Simulation of reads from the F288 genomic sequence using wgsim v.0.3.0. (i-ii) Alignment of F288 simulated reads and B73 (AGPv3) real reads to the F271 genomic sequence using Bowtie2 v2.2.3. (iii) Alignment of 2,322 representative transcript assemblies (RTAs) [34] to the F271 genomic sequence using gmap v.2011-08-15. (i-iii) Hard masking (with N) of common F271 genomic sequence. The same pipeline was followed to identify F288-specific sequences in the F288 genomic sequence. Together, the identified F271- and F288-specific sequences were added to the maize B73 reference genome (AGPv3) to which 2,322 RTAs were also added.
(PDF)
PLMs colored red and blue indicate PLMs specifically present in the promoter of the considered DE gene in F271 and F288, respectively. 2: ASF1MOTIFCAMV; 5: MYBCOREATCYCB1; 6: TATCCAOSAMY; 7: ARR1AT; 8: MYCATERD1; 9: WBOXNTCHN48; 11: SITEIIATCYTC; 12: SORLIP2AT; 13: DPBFCOREDCDC3; 14: MARTBOX; 15: TATA-box; 18: part of VOZ-binding sequence; 19: TATABOX4; 21: VIP1 response elements. For detailed explanation, see ‘Materials and Methods‘ and S8 Table.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Acknowledgments
We are grateful to Audrey Courtial (INRA CNRGV, Castanet-Tolosan, France), Yves Barrière (UGAPF, INRA Lusignan, France) and the CNRGV for sharing the F271 and F288 genomic sequences at the QTL6.05 interval. We also thank Audrey Courtial and Yves Barrière for their contribution to sequence annotation and comments that improved the manuscript, Sandrine Balzergue (IRHS, Beaucouzé, France) for preliminary RNA-seq discussion, and Cyril Bauland (GQE-Le Moulon, Gif-sur-Yvette, France), Serge Malavieille and Pascal Sartre (INRA Domaine de Melgueil, Mauguio, France) for their precious contribution to field experiments.
Data Availability
The sequence data underlying the results presented in this study have been deposited to the NCBI Gene Expression Omnibus and may be accessed via the accession number GSE115157.
Funding Statement
This research was supported by grants from ProMaïs (ZeaWall II project to MR) and the French Government (LabEx Saclay Plant Sciences-SPS, ref. ANR-10-LABX-0040-SPS to SC, and ANR-11-BTBR-0006 BIOMASS FOR THE FUTURE to SC), managed by the French National Research Agency under an Investment for the Future program (ref. ANR-11-IDEX-0003-02 to SC).
References
- 1.Barrière Y. Brown-midrib genes in maize and their efficiency in dairy cow feeding. Perspectives for breeding improved silage maize targeting gene modifications in the monolignol and p-hydroxycinnamate pathways. Maydica. 2017;62: 1–19. [Google Scholar]
- 2.Barrière Y, Emile JC, Traineau R, Surault F, Briand M, Gallais A. Genetic variation for organic matter and cell wall digestibility in silage maize. Lessons from a 34-year long experiment with sheep in digestibility crates. Maydica. 2004;49: 115–126. [Google Scholar]
- 3.Grabber JH, Ralph J, Lapierre C, Barrière Y. Genetic and molecular basis of grass cell-wall degradability. I. Lignin-cell wall matrix interactions. Comptes Rendus Biologies. 2004;327: 455–465. 10.1016/j.crvi.2004.02.009 [DOI] [PubMed] [Google Scholar]
- 4.Pauly M, Keegstra K. Plant cell wall polymers as precursors for biofuels. Curr Opin Plant Biol. 2010;13: 305–312. 10.1016/j.pbi.2009.12.009 [DOI] [PubMed] [Google Scholar]
- 5.Zhao X, Zhang L, Liu D. Biomass recalcitrance. Part I: The chemical compositions and physical structures affecting the enzymatic hydrolysis of lignocellulose. Biofuels Bioproducts and Biorefining. 2012;6: 465–482. [Google Scholar]
- 6.Meng X, Ragauskas AJ. Recent advances in understanding the role of cellulose accessibility in enzymatic hydrolysis of lignocellulosic substrates. Curr Opin Biotechnol. 2014;27: 150–158. 10.1016/j.copbio.2014.01.014 [DOI] [PubMed] [Google Scholar]
- 7.Méchin V, Argillier O, Rocher F, Hébert Y, Mila I, Pollet B, et al. In Search of a Maize Ideotype for Cell Wall Enzymatic Degradability Using Histological and Biochemical Lignin Characterization. J Agric Food Chem. 2005;53: 5872–5881. 10.1021/jf050722f [DOI] [PubMed] [Google Scholar]
- 8.Barrière Y, Méchin V, Riboulet C, Guillaumie S, Thomas J, Bosio M, et al. Genetic and genomic approaches for improving biofuel production from maize. Euphytica. 2009;170: 183–202. [Google Scholar]
- 9.Lübberstedt T, Melchinger AE, Klein D, Degenhardt H, Paul C. QTL mapping in testcrosses of European flint lines of maize: II. Comparison of different testers for forage quality traits. Crop science. 37: 1913–1922. [Google Scholar]
- 10.Bohn M, Schulz B, Kreps R, Klein D, Melchinger AE. QTL mapping for resistance against the European corn borer (Ostrinia nubilalis H.) in early maturing European dent germplasm. Theoretical and Applied Genetics. 101: 907–917. [DOI] [PubMed] [Google Scholar]
- 11.Méchin V, Argillier O, Hébert Y, Guingo E, Moreau L, Charcosset A, et al. Genetic Analysis and QTL Mapping of Cell Wall Digestibility and Lignification in Silage Maize. Crop Science. 2001;41: 690 10.2135/cropsci2001.413690x [DOI] [Google Scholar]
- 12.Roussel V, Gibelin C, Fontaine A, Barrière Y. Genetic analysis in recombinant inbred lines of early dent forage maize. 2002;47: 9–20. [Google Scholar]
- 13.Cardinal AJ, Lee M, Moore KJ. Genetic mapping and analysis of quantitative trait loci affecting fiber and lignin content in maize. Theor Appl Genet. 2003;106: 866–874. 10.1007/s00122-002-1136-5 [DOI] [PubMed] [Google Scholar]
- 14.Fontaine A-S, Briand M, Barrière Y. Genetic variation and QTL mapping of para-coumaric and ferulic acid contents in maize stover at silage harvest. Maydica. 2003;48: 75–84. [Google Scholar]
- 15.Krakowsky M, Lee M, Beeghly H, Coors J. Characterization of quantitative trait loci affecting fiber and lignin in maize (Zea mays L.). Maydica. 2003;48: 283–292. [Google Scholar]
- 16.Riboulet C, Fabre F, Denoue D, Martinant JP, Lefevre B, Barriere Y. QTL mapping and candidate gene research from lignin content and cell wall digestibility in a top-cross of a flint maize recombinant inbred line progeny harvested at silage stage. Maydica. 2008;53: 1–9. [Google Scholar]
- 17.Barrière Y, Thomas J, Denoue D. QTL mapping for lignin content, lignin monomeric composition, p-hydroxycinnamate content, and cell wall digestibility in the maize recombinant inbred line progeny F838 x F286. Plant Science. 2008;175: 585–595. [Google Scholar]
- 18.Barrière Y, Méchin V, Denoue D, Bauland C, Laborde J. QTL for Yield, Earliness, and Cell Wall Quality Traits in Topcross Experiments of the F838 × F286 Early Maize RIL Progeny. Crop Science. 2010;50: 1761 10.2135/cropsci2009.11.0671 [DOI] [Google Scholar]
- 19.Barrière Y, Méchin V, Lefevre B, Maltese S. QTLs for agronomic and cell wall traits in a maize RIL progeny derived from a cross between an old Minnesota13 line and a modern Iodent line. Theor Appl Genet. 2012;125: 531–549. 10.1007/s00122-012-1851-5 [DOI] [PubMed] [Google Scholar]
- 20.Thomas J, Guillaumie S, Verdu C, Denoue D, Pichon M, Barrière Y. Cell wall phenylpropanoid-related gene expression in early maize recombinant inbred lines differing in parental alleles at a major lignin QTL position. Mol Breeding. 2010;25: 105–124. 10.1007/s11032-009-9311-x [DOI] [Google Scholar]
- 21.Courtial A, Thomas J, Reymond M, Méchin V, Grima-Pettenati J, Barrière Y. Targeted linkage map densification to improve cell wall related QTL detection and interpretation in maize. Theor Appl Genet. 2013;126: 1151–1165. 10.1007/s00122-013-2043-7 [DOI] [PubMed] [Google Scholar]
- 22.Courtial A, Méchin V, Reymond M, Grima-Pettenati J, Barrière Y. Colocalizations Between Several QTLs for Cell Wall Degradability and Composition in the F288 × F271 Early Maize RIL Progeny Raise the Question of the Nature of the Possible Underlying Determinants and Breeding Targets for Biofuel Capacity. Bioenerg Res. 2014;7: 142–156. 10.1007/s12155-013-9358-8 [DOI] [Google Scholar]
- 23.Torres AF, Noordam-Boot CMM, Dolstra O, van der Weijde T, Combes E, Dufour P, et al. Cell Wall Diversity in Forage Maize: Genetic Complexity and Bioenergy Potential. Bioenerg Res. 2015;8: 187–202. 10.1007/s12155-014-9507-8 [DOI] [Google Scholar]
- 24.Li K, Wang H, Hu X, Ma F, Wu Y, Wang Q, et al. Genetic and Quantitative Trait Locus Analysis of Cell Wall Components and Forage Digestibility in the Zheng58 × HD568 Maize RIL Population at Anthesis Stage. Front Plant Sci. 2017;8: 1472 10.3389/fpls.2017.01472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leng P, Ouzunova M, Landbeck M, Wenzel G, Eder J, Darnhofer B, et al. Quantitative trait loci mapping of forage stover quality traits in six mapping populations derived from European elite maize germplasm. Leon J, editor. Plant Breed. 2018;137: 139–147. 10.1111/pbr.12572 [DOI] [Google Scholar]
- 26.Ralph J, Guillaumie S, Grabber JH, Lapierre C, Barrière Y. Genetic and molecular basis of grass cell-wall biosynthesis and degradability. III. Towards a forage grass ideotype. Comptes Rendus Biologies. 2004;327: 467–479. 10.1016/j.crvi.2004.03.004 [DOI] [PubMed] [Google Scholar]
- 27.Truntzler M, Barrière Y, Sawkins MC, Lespinasse D, Betran J, Charcosset A, et al. Meta-analysis of QTL involved in silage quality of maize and comparison with the position of candidate genes. Theor Appl Genet. 2010;121: 1465–1482. 10.1007/s00122-010-1402-x [DOI] [PubMed] [Google Scholar]
- 28.Penning BW, Sykes RW, Babcock NC, Dugard CK, Held MA, Klimek JF, et al. Genetic Determinants for Enzymatic Digestion of Lignocellulosic Biomass Are Independent of Those for Lignin Abundance in a Maize Recombinant Inbred Population. Plant Physiol. 2014;165: 1475–1487. 10.1104/pp.114.242446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrière Y, Courtial A, Chateigner-Boutin A-L, Denoue D, Grima-Pettenati J. Breeding maize for silage and biofuel production, an illustration of a step forward with the genome sequence. Plant Science. 2016;242: 310–329. 10.1016/j.plantsci.2015.08.007 [DOI] [PubMed] [Google Scholar]
- 30.Yin X, Struik PC, Kropff MJ. Role of crop physiology in predicting gene-to-phenotype relationships. Trends Plant Sci. 2004;9: 426–432. 10.1016/j.tplants.2004.07.007 [DOI] [PubMed] [Google Scholar]
- 31.Rafalski A, Morgante M. Corn and humans: recombination and linkage disequilibrium in two genomes of similar size. Trends Genet. 2004;20: 103–111. 10.1016/j.tig.2003.12.002 [DOI] [PubMed] [Google Scholar]
- 32.Springer NM, Ying K, Fu Y, Ji T, Yeh C-T, Jia Y, et al. Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet. 2009;5: e1000734 10.1371/journal.pgen.1000734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swanson-Wagner RA, Eichten SR, Kumari S, Tiffin P, Stein JC, Ware D, et al. Pervasive gene content variation and copy number variation in maize and its undomesticated progenitor. Genome Res. 2010;20: 1689–1699. 10.1101/gr.109165.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, et al. Insights into the Maize Pan-Genome and Pan-Transcriptome. Plant Cell. 2014;26: 121–135. 10.1105/tpc.113.119982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin M, Liu H, He C, Fu J, Xiao Y, Wang Y, et al. Maize pan-transcriptome provides novel insights into genome complexity and quantitative trait variation. Sci Rep. 2016;6: 18936 10.1038/srep18936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Darracq A, Vitte C, Nicolas S, Duarte J, Pichon J-P, Mary-Huard T, et al. Sequence analysis of European maize inbred line F2 provides new insights into molecular and chromosomal characteristics of presence/absence variants. BMC Genomics. 2018;19: 119 10.1186/s12864-018-4490-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Baldauf JA, Marcon C, Lithio A, Vedder L, Altrogge L, Piepho H-P, et al. Single-Parent Expression Is a General Mechanism Driving Extensive Complementation of Non-syntenic Genes in Maize Hybrids. Current Biology. 2018;28: 431–437.e4. 10.1016/j.cub.2017.12.027 [DOI] [PubMed] [Google Scholar]
- 38.Courtial A, Jourda C, Arribat S, Balzergue S. Comparative expression of cell wall related genes in four maize RILs and one parental line of variable lignin content and cell wall degradability. Maydica. 2012; 19. [Google Scholar]
- 39.Szalma SJ, Hostert BM, Ledeaux JR, Stuber CW, Holland JB. QTL mapping with near-isogenic lines in maize. Theor Appl Genet. 2007;114: 1211–1228. 10.1007/s00122-007-0512-6 [DOI] [PubMed] [Google Scholar]
- 40.Feltus FA. Systems genetics: a paradigm to improve discovery of candidate genes and mechanisms underlying complex traits. Plant Sci. 2014;223: 45–48. 10.1016/j.plantsci.2014.03.003 [DOI] [PubMed] [Google Scholar]
- 41.Baute J, Herman D, Coppens F, De Block J, Slabbinck B, Dell’Acqua M, et al. Combined Large-Scale Phenotyping and Transcriptomics in Maize Reveals a Robust Growth Regulatory Network. Plant Physiol. 2016;170: 1848–1867. 10.1104/pp.15.01883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wen W, Liu H, Zhou Y, Jin M, Yang N, Li D, et al. Combining Quantitative Genetics Approaches with Regulatory Network Analysis to Dissect the Complex Metabolism of the Maize Kernel. Plant Physiol. 2016;170: 136–146. 10.1104/pp.15.01444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu Y-B, Yang M-H, Zeng F, Biligetu B. Searching for an Accurate Marker-Based Prediction of an Individual Quantitative Trait in Molecular Plant Breeding. Front Plant Sci. 2017;8: 1182 10.3389/fpls.2017.01182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Virlouvet L, Jacquemot M-P, Gerentes D, Corti H, Bouton S, Gilard F, et al. The ZmASR1 protein influences branched-chain amino acid biosynthesis and maintains kernel yield in maize under water-limited conditions. Plant Physiol. 2011;157: 917–936. 10.1104/pp.111.176818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kopylova E, Noé L, Touzet H. SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28: 3211–3217. 10.1093/bioinformatics/bts611 [DOI] [PubMed] [Google Scholar]
- 46.Gagnot S, Tamby J-P, Martin-Magniette M-L, Bitton F, Taconnat L, Balzergue S, et al. CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 2008;36: D986–990. 10.1093/nar/gkm757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30: 207–210. 10.1093/nar/30.1.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Leroy P, Guilhot N, Sakai H, Bernard A, Choulet F, Theil S, et al. TriAnnot: A Versatile and High Performance Pipeline for the Automated Annotation of Plant Genomes. Front Plant Sci. 2012;3: 5 10.3389/fpls.2012.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Choulet F, Alberti A, Theil S, Glover N, Barbe V, Daron J, et al. Structural and functional partitioning of bread wheat chromosome 3B. Science. 2014;345: 1249721 10.1126/science.1249721 [DOI] [PubMed] [Google Scholar]
- 50.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25: 3389–3402. 10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25: 955–964. 10.1093/nar/25.5.955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lagesen K, Hallin P, Rødland EA, Staerfeldt H-H, Rognes T, Ussery DW. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35: 3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Slater GSC, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6: 31 10.1186/1471-2105-6-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19 Suppl 2: ii215–225. 10.1093/bioinformatics/btg1080 [DOI] [PubMed] [Google Scholar]
- 55.Amano N, Tanaka T, Numa H, Sakai H, Itoh T. Efficient plant gene identification based on interspecies mapping of full-length cDNAs. DNA Res. 2010;17: 271–279. 10.1093/dnares/dsq017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sammut SJ, Finn RD, Bateman A. Pfam 10 years on: 10,000 families and still growing. Brief Bioinformatics. 2008;9: 210–219. 10.1093/bib/bbn010 [DOI] [PubMed] [Google Scholar]
- 57.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38: D211–222. 10.1093/nar/gkp985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zdobnov EM, Apweiler R. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17: 847–848. 10.1093/bioinformatics/17.9.847 [DOI] [PubMed] [Google Scholar]
- 59.Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38: D161–166. 10.1093/nar/gkp885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37: D229–232. 10.1093/nar/gkn808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abeel T, Van Parys T, Saeys Y, Galagan J, Van de Peer Y. GenomeView: a next-generation genome browser. Nucleic Acids Res. 2012;40: e12 10.1093/nar/gkr995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40: 4288–4297. 10.1093/nar/gks042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37: 914–939. 10.1111/j.1365-313x.2004.02016.x [DOI] [PubMed] [Google Scholar]
- 65.Rohart F, Gautier B, Singh A, Lê Cao K-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13: e1005752 10.1371/journal.pcbi.1005752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13: 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Assenov Y, Ramírez F, Schelhorn S-E, Lengauer T, Albrecht M. Computing topological parameters of biological networks. Bioinformatics. 2008;24: 282–284. 10.1093/bioinformatics/btm554 [DOI] [PubMed] [Google Scholar]
- 68.Bernard V, Lecharny A, Brunaud V. Improved detection of motifs with preferential location in promoters. Genome. 2010;53: 739–752. 10.1139/g10-042 [DOI] [PubMed] [Google Scholar]
- 69.Mejía-Guerra MK, Li W, Galeano NF, Vidal M, Gray J, Doseff AI, et al. Core Promoter Plasticity Between Maize Tissues and Genotypes Contrasts with Predominance of Sharp Transcription Initiation Sites. Plant Cell. 2015;27: 3309–3320. 10.1105/tpc.15.00630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Higo K, Ugawa Y, Iwamoto M, Higo H. PLACE: a database of plant cis-acting regulatory DNA elements. Nucleic Acids Res. 1998;26: 358–359. 10.1093/nar/26.1.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, et al. AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics. 2003;4: 25 10.1186/1471-2105-4-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics. 2012;13: 134 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Riboulet C, Guillaumie S, Méchin V, Bosio M, Pichon M, Goffner D, et al. Kinetics of Phenylpropanoid Gene Expression in Maize Growing Internodes: Relationships with Cell Wall Deposition. Crop Science. 2009;49: 211 10.2135/cropsci2008.03.0130 [DOI] [Google Scholar]
- 74.Zhang Q, Cheetamun R, Dhugga KS, Rafalski J, Tingey SV, Shirley NJ, et al. Spatial gradients in cell wall composition and transcriptional profiles along elongating maize internodes. BMC Plant Biol. 2014;14: 27 10.1186/1471-2229-14-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Paschold A, Jia Y, Marcon C, Lund S, Larson NB, Yeh C-T, et al. Complementation contributes to transcriptome complexity in maize (Zea mays L.) hybrids relative to their inbred parents. Genome Research. 2012;22: 2445–2454. 10.1101/gr.138461.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sengupta D, Naik D, Reddy AR. Plant aldo-keto reductases (AKRs) as multi-tasking soldiers involved in diverse plant metabolic processes and stress defense: A structure-function update. J Plant Physiol. 2015;179: 40–55. 10.1016/j.jplph.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 77.Yu Y, Zhang H, Li W, Mu C, Zhang F, Wang L, et al. Genome-wide analysis and environmental response profiling of the FK506-binding protein gene family in maize (Zea mays L.). Gene. 2012;498: 212–222. 10.1016/j.gene.2012.01.094 [DOI] [PubMed] [Google Scholar]
- 78.Verger S, Chabout S, Gineau E, Mouille G. Cell adhesion in plants is under the control of putative O-fucosyltransferases. Development. 2016;143: 2536–2540. 10.1242/dev.132308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barghetti A, Sjögren L, Floris M, Paredes EB, Wenkel S, Brodersen P. Heat-shock protein 40 is the key farnesylation target in meristem size control, abscisic acid signaling, and drought resistance. Genes Dev. 2018;31: 2282–2295. 10.1101/gad.301242.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yao Y, Chen X, Wu A-M. ERF-VII members exhibit synergistic and separate roles in Arabidopsis. Plant Signaling & Behavior. 2017;12: e1329073 10.1080/15592324.2017.1329073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giuntoli B, Perata P. Group VII Ethylene Response Factors in Arabidopsis: Regulation and Physiological Roles. Plant Physiol. 2018;176: 1143–1155. 10.1104/pp.17.01225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tran L-SP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16: 2481–2498. 10.1105/tpc.104.022699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim HJ, Park J-H, Kim J, Kim JJ, Hong S, Kim J, et al. Time-evolving genetic networks reveal a NAC troika that negatively regulates leaf senescence in Arabidopsis. Proc Natl Acad Sci USA. 2018;115: E4930–E4939. 10.1073/pnas.1721523115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Després C, Chubak C, Rochon A, Clark R, Bethune T, Desveaux D, et al. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell. 2003;15: 2181–2191. 10.1105/tpc.012849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12: 3703–3714. 10.1101/gad.12.23.3703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 2003;132: 1020–1032. 10.1104/pp.102.017814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li K, Wang H, Hu X, Liu Z, Wu Y, Huang C. Genome-Wide Association Study Reveals the Genetic Basis of Stalk Cell Wall Components in Maize. PLoS ONE. 2016;11: e0158906 10.1371/journal.pone.0158906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Paschold A, Larson NB, Marcon C, Schnable JC, Yeh C-T, Lanz C, et al. Nonsyntenic genes drive highly dynamic complementation of gene expression in maize hybrids. Plant Cell. 2014;26: 3939–3948. 10.1105/tpc.114.130948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marcon C, Paschold A, Malik WA, Lithio A, Baldauf JA, Altrogge L, et al. Stability of Single-Parent Gene Expression Complementation in Maize Hybrids upon Water Deficit Stress. Plant Physiol. 2017;173: 1247–1257. 10.1104/pp.16.01045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vélez-Bermúdez I-C, Salazar-Henao JE, Fornalé S, López-Vidriero I, Franco-Zorrilla J-M, Grotewold E, et al. A MYB/ZML Complex Regulates Wound-Induced Lignin Genes in Maize. Plant Cell. 2015;27: 3245–3259. 10.1105/tpc.15.00545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Weits DA, Giuntoli B, Kosmacz M, Parlanti S, Hubberten H-M, Riegler H, et al. Plant cysteine oxidases control the oxygen-dependent branch of the N-end-rule pathway. Nat Commun. 2014;5: 3425 10.1038/ncomms4425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.White MD, Klecker M, Hopkinson RJ, Weits DA, Mueller C, Naumann C, et al. Plant cysteine oxidases are dioxygenases that directly enable arginyl transferase-catalysed arginylation of N-end rule targets. Nat Commun. 2017;8: 14690 10.1038/ncomms14690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mustroph A, Lee SC, Oosumi T, Zanetti ME, Yang H, Ma K, et al. Cross-kingdom comparison of transcriptomic adjustments to low-oxygen stress highlights conserved and plant-specific responses. Plant Physiol. 2010;152: 1484–1500. 10.1104/pp.109.151845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gonzali S, Loreti E, Cardarelli F, Novi G, Parlanti S, Pucciariello C, et al. Universal stress protein HRU1 mediates ROS homeostasis under anoxia. Nature Plants. 2015;1: 15151 10.1038/nplants.2015.151 [DOI] [PubMed] [Google Scholar]
- 95.Napoleão TA, Soares G, Vital CE, Bastos C, Castro R, Loureiro ME, et al. Methyl jasmonate and salicylic acid are able to modify cell wall but only salicylic acid alters biomass digestibility in the model grass Brachypodium distachyon. Plant Science. 2017;263: 46–54. 10.1016/j.plantsci.2017.06.014 [DOI] [PubMed] [Google Scholar]
- 96.Zhang X-C, Millet YA, Cheng Z, Bush J, Ausubel FM. Jasmonate signalling in Arabidopsis involves SGT1b-HSP70-HSP90 chaperone complexes. Nat Plants. 2015;1 10.1038/nplants.2015.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wang R, Zhang Y, Kieffer M, Yu H, Kepinski S, Estelle M. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat Commun. 2016;7: 10269 10.1038/ncomms10269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Thieme CJ, Rojas-Triana M, Stecyk E, Schudoma C, Zhang W, Yang L, et al. Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat Plants. 2015;1: 15025 10.1038/nplants.2015.25 [DOI] [PubMed] [Google Scholar]
- 99.Wang W-W, Ma Q, Xiang Y, Zhu S-W, Cheng B-J. Genome-wide analysis of immunophilin FKBP genes and expression patterns in Zea mays. Genet Mol Res. 2012;11: 1690–1700. 10.4238/2012.June.25.2 [DOI] [PubMed] [Google Scholar]
- 100.Kulich I, Pečenková T, Sekereš J, Smetana O, Fendrych M, Foissner I, et al. Arabidopsis exocyst subcomplex containing subunit EXO70B1 is involved in autophagy-related transport to the vacuole. Traffic. 2013;14: 1155–1165. 10.1111/tra.12101 [DOI] [PubMed] [Google Scholar]
- 101.Simpson PJ, Tantitadapitak C, Reed AM, Mather OC, Bunce CM, White SA, et al. Characterization of two novel aldo-keto reductases from Arabidopsis: expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress. J Mol Biol. 2009;392: 465–480. 10.1016/j.jmb.2009.07.023 [DOI] [PubMed] [Google Scholar]
- 102.Suekawa M, Fujikawa Y, Inada S, Murano A, Esaka M. Gene expression and promoter analysis of a novel tomato aldo-keto reductase in response to environmental stresses. J Plant Physiol. 2016;200: 35–44. 10.1016/j.jplph.2016.05.015 [DOI] [PubMed] [Google Scholar]
- 103.Sharma D, Cukras AR, Rogers EJ, Southworth DR, Green R. Mutational analysis of S12 protein and implications for the accuracy of decoding by the ribosome. J Mol Biol. 2007;374: 1065–1076. 10.1016/j.jmb.2007.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Clasen SJ, Shao W, Gu H, Espenshade PJ. Prolyl dihydroxylation of unassembled uS12/Rps23 regulates fungal hypoxic adaptation. eLife. 2017;6: e28563 10.7554/eLife.28563 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The two selected pairs of NILs (BC2-S2) were field grown under well-watered conditions for DNA and mRNA sampling and phenotyping. DNA samples were used for genotyping by PCR using three markers located in the introgressed genomic region (bngl1702, bnlg1732 and bnlg345) and one outside (umc1127) as indicated in the S2 Table. L: 50 bp DNA ladder.
(PDF)
(i) Simulation of reads from the F288 genomic sequence using wgsim v.0.3.0. (i-ii) Alignment of F288 simulated reads and B73 (AGPv3) real reads to the F271 genomic sequence using Bowtie2 v2.2.3. (iii) Alignment of 2,322 representative transcript assemblies (RTAs) [34] to the F271 genomic sequence using gmap v.2011-08-15. (i-iii) Hard masking (with N) of common F271 genomic sequence. The same pipeline was followed to identify F288-specific sequences in the F288 genomic sequence. Together, the identified F271- and F288-specific sequences were added to the maize B73 reference genome (AGPv3) to which 2,322 RTAs were also added.
(PDF)
PLMs colored red and blue indicate PLMs specifically present in the promoter of the considered DE gene in F271 and F288, respectively. 2: ASF1MOTIFCAMV; 5: MYBCOREATCYCB1; 6: TATCCAOSAMY; 7: ARR1AT; 8: MYCATERD1; 9: WBOXNTCHN48; 11: SITEIIATCYTC; 12: SORLIP2AT; 13: DPBFCOREDCDC3; 14: MARTBOX; 15: TATA-box; 18: part of VOZ-binding sequence; 19: TATABOX4; 21: VIP1 response elements. For detailed explanation, see ‘Materials and Methods‘ and S8 Table.
(PDF)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
(XLSX)
Data Availability Statement
The sequence data underlying the results presented in this study have been deposited to the NCBI Gene Expression Omnibus and may be accessed via the accession number GSE115157.