Abstract
The DNA genome of eukaryotic cells is compacted by histone proteins within the nucleus to form chromatin. Nuclear-replicating viruses such as adenovirus have evolved mechanisms of chromatin manipulation to promote infection and subvert host defenses. Epigenetic factors may also regulate persistent adenovirus infection and reactivation in lymphoid tissues. In this review, we discuss the viral proteins E1A and protein VII that interact with and alter host chromatin, as well as E4orf3, which separates host chromatin from sites of viral replication. We also highlight recent advances in chromatin technologies that offer new insights into virus-directed chromatin manipulation. Beyond the role of chromatin in the viral replication cycle, we discuss the nature of persistent viral genomes in lymphoid tissue and cell lines, and the potential contribution of epigenetic signals in maintaining adenovirus in a quiescent state. By understanding the mechanisms through which adenovirus manipulates host chromatin, we will understand new aspects of this ubiquitous virus and shed light on previously unknown aspects of chromatin biology.
Keywords: adenovirus, chromatin, E1A, epigenetics, histones, host-pathogen interactions, persistence, post-translational modifications, protein VII
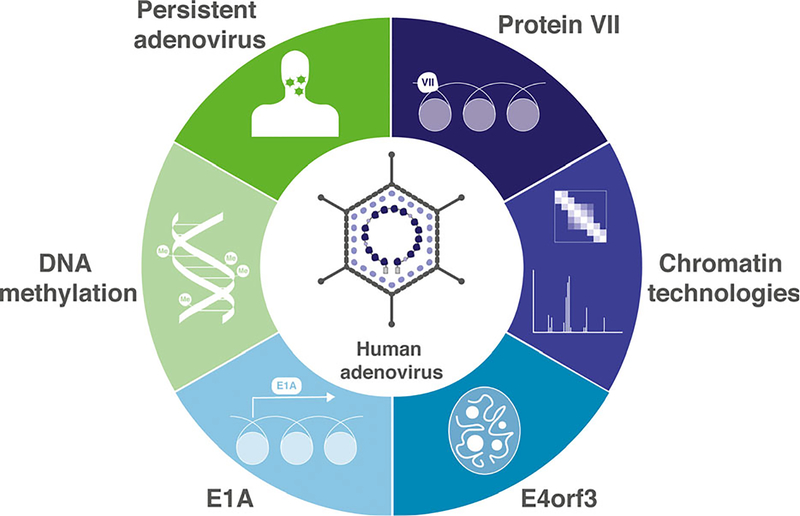
Adenovirus is a human pathogen of growing concern [1]. As the number of successful solid organ and hematopoietic stem cell transplants increases, the proportion of the population on immunosuppressive drug regimens also increases [2]. This immunocompromised population is more susceptible to adenovirus infections, and therefore, the incidence of infection continues to rise [2–5]. Recent adenovirus outbreaks in the United States illustrate that both healthy and immunocompromised individuals are vulnerable [6–8]. Efforts to limit the increased morbidity and mortality caused by adenovirus infections in transplant patients are complicated by the poorly understood phenomenon of persistent adenovirus, in which extremely low levels of viral replication are maintained [9]. It is thought that, in the case of transplant patients, acquired immunodeficiency allows for reactivation of persistent adenovirus resulting in acute lytic infection. Evidence suggests that persistent adenovirus may be tied to chromatin state and epigenetic changes in the host cell [10,11]. Here, we provide a brief primer on chromatin structure and examine adenovirus infection through the lens of chromatin biology and epigenetics (Fig. 1).
Fig. 1.
Schematic overview of the content of this review. Host chromatin and epigenetics are central to the adenovirus replication cycle. We focus on core protein VII because it interacts both with the viral genome and with host chromatin. We also address the effects of early viral proteins E1A and E4orf3 on host chromatin and nuclear organization. Although the nature of persistent adenovirus infection is not well-characterized, epigenetic state and chromatin may be important for maintaining adenovirus in lymphoid tissues. Adenovirus infection also impacts host DNA methylation, an important inherited DNA modification. Historically, adenoviruses have provided key insights into chromatin biology, and developing chromatin technologies will reveal new facets of the intersection of adenoviruses and chromatin.
Beginning with incoming viral genomes, we discuss the core protein VII and its role on incoming viral chromatin and in initiation of viral transcription, as well as its emerging effects on host chromatin. The early viral gene product E1A is essential to virus infection and causes global changes to host chromatin resulting in cellular reprogramming. We examine the state of the field of research on E1A and specifically its impact on host chromatin. Adenovirus infection also results in structural changes to the host nucleus to facilitate viral replication, many of which are induced by the early protein E4orf3. We dissect the current understanding of E4orf3 with respect to chromatin biology. Finally, we address the possible role of epigenetics in the understudied field of persistent adenovirus. We explore epigenetic signals, including cellular DNA methylation, that may regulate adenovirus persistence in lymphoid organs. Finally, we discuss pathologies that may be associated with persistent adenovirus infection.
Architecture and function of host chromatin
The repeating unit of chromatin, the nucleosome, consists of approximately 147 base pairs of DNA wrapped around an octamer of histone proteins. The octamer is primarily comprised of a heterotetramer of H3 and H4, and two H2A/H2B heterodimers [12]. The N-terminal tails of histones that protrude from core nucleosomes are modified post-translationally [13]. Modifications include phosphorylation, methylation, acetylation, and ubiquitination, which regulate access to surrounding DNA through binding partners and compaction [14]. Chromatin has been classified into many different sub-types: the two main groups being ‘active’ euchromatin and ‘repressed’ heterochromatin. Active chromatin is associated with higher transcriptional activity and specific histone marks, for instance, histone H3 lysine 4 trimethylation (H3K4me3); histone acetylation (ac), such as H3K18ac; and histone H2B ubiquitination, such as H2BK120Ub. Repressed chromatin varies in definition by species but is generally more transcriptionally silent. For example, in Schizosaccharomyces pombe, there are two main states of heterochromatin: facultative and constitutive [15]. As the names imply, one state is generally not reversible, whereas the other can be altered. In Drosophila melanogaster, a colored naming scheme distinguishes between multiple types of active and repressed chromatin, with two types of active and three of repressed [16]. Although these categories do not to translate simply to mammalian systems, types of heterochromatin are differentiated by description, such as H3K27me3- or H3K9me3-enriched. It remains to be determined whether these categories form a discernible ‘code’ that can easily be interpreted. The combinatorial nature of histone marks complicates their functional output. For example, H3K9me3 may result in an entirely different outcome on transcription when accompanied by a nearby mark such as H3S10ph [17,18]. In addition to histone modifications, DNA itself can be modified by methylation of cytosine within CG sequences or CpG sites [19,20]. DNA methylation is an important component of gene silencing, especially during development and X inactivation in mammals [21]. Despite its importance in mammalian development, DNA methylation does not occur in many organisms including yeasts, Caenorhabditis elegans, and Drosophila melanogaster [22]. DNA methylation, together with H3K9 trimethylation, contributes to heterochromatin formation [23,24], although DNA methylation is also found in activating contexts [25,26]. Like many epigenetic marks, the genomic context and neighboring histone marks are key factors in determining active or repressed transcriptional outcomes.
Incoming viral genomes are coated with protein VII
Viral genomes in the virion
Adenoviruses encode a unique histone-like protein called protein VII [27–29] that is packaged with the viral genome in what has been described as a ‘beads-on-a-string’ structure [29]. Protein VII is the most abundant core protein [30,31]. It is suggested that approximately 2–6 protein VII molecules form a multimer, around which 90–150 base pairs of DNA may be wrapped within virus particles [32,33]. The interaction of proteins within the adenovirus core was studied in the 1970s and 1980s [27–29,32–36], establishing that protein VII directly interacts with viral DNA. Through interactions with protein V, protein VII also stabilizes the genome within the capsid. Recent structural studies propose that protein VII is integral to the virion assembly process [37]; however, the formation of adenovirus particles in the complete absence of protein VII [38] implies that the viral genome conformation may have multiple states that are amenable to virion formation. It has been suggested recently that one conformation may resemble a more ancient structure similar to archaeal chromatin [39,40], although without further structural insight into protein VII–DNA complexes, the conformation of viral genomes within virions remains unclear. Virus particles lacking protein VII are noninfectious [38], suggesting protein VII has a role in uncoating of the viral genome from the capsid and may have further functions outside of viral packaging.
DNA entry into the nucleus and the activation of viral transcription
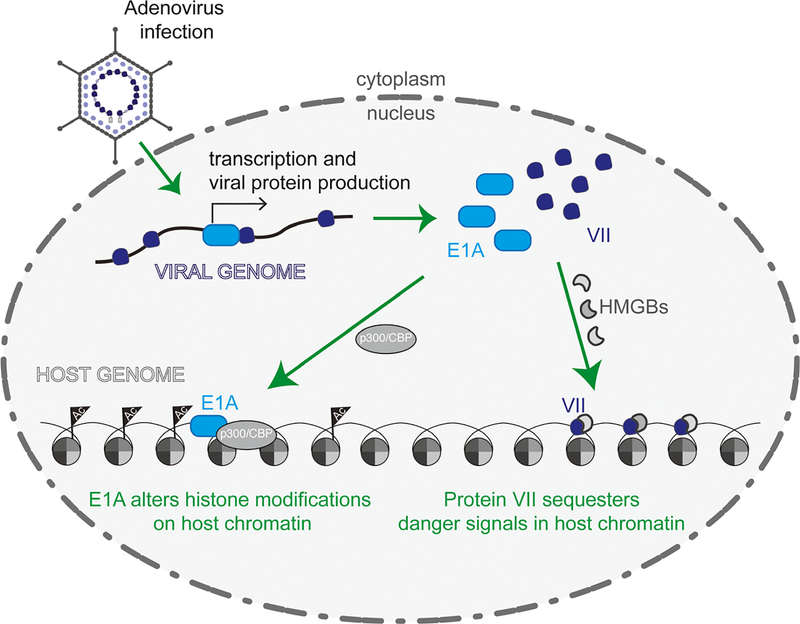
Upon nuclear entry, it is thought that protein VII must be removed from incoming viral DNA to allow transcription initiation [41] (Fig. 2) although the evidence for transcriptional repression by protein VII is not consistent across experimental systems. When Xenopus laevis eggs are injected with mRNA to synthesize protein VII, protein VII binds and condenses Xenopus chromatin as well as represses the formation of R-loops, a marker of active transcription [42]. These data, together with the observation that virion cores are highly compact, have led to the description of protein VII as a ‘repressor’ although it does not fit the canonical definition of a transcriptional repressor with affinity to a specific DNA sequence [43]. It is likely that protein VII bound to DNA alone, without histones, prevents transcription due to its extremely high affinity for DNA. Conversely, precoating plasmids with protein VII prior to transfection into HeLa cells results in greater transgene expression compared to transfection of plasmid alone [44]. Likewise, the transfection of adenovirus cores generated by pre-incubating viral genomes with protein VII results in greater association of histones with viral DNA and increased transcription compared to cores transfected without protein VII bound [44]. These observations suggest that the inherent differences between human and frog cells are important in the function of protein VII on DNA. Given that virus particles lacking protein VII are unable to initiate transcription [38], it is likely that protein VII functions to activate, rather than repress, initiation of viral transcription in human cells. It is also thought that protein VII functions cooperatively with the early-transcribed E1A gene to promote transcription from the viral genome [42] (see the section titled E1A: The first viral protein produced attacks host chromatin below). Furthermore, protein VII may function differently on viral and host genomes, where the chromatin landscape is vastly different. The presence of histones on viral genomes also depends on histone chaperones such as HIRA and SET [45,46] (see Weitzman review [47]) where the interaction of SET and protein VII may also facilitate transcription from the incoming viral genomes [44,45].
Fig. 2.
Impact of E1A and protein VII on host chromatin. Adenovirus E1A and protein VII significantly affect both viral genomes and host chromatin. Viral DNA is bound by the histone-like protein VII as it enters the nucleus. E1A is the first protein produced during infection and has been implicated in initiating transcription and displacing protein VII from incoming viral genomes. E1A binds the host acetyltransferase proteins p300/CBP to disrupt acetylation of histones and alter host transcription to promote viral replication. Late-expressed protein VII binds host chromatin and sequesters the danger signaling HMGB proteins in chromatin. For more details on viral entry, see reference [39].
SET, also known as template activating factor 1 (TAF-1β), was first identified as an oncoprotein [48]. SET is implicated in diverse cellular pathways, including membrane protein localization [49], inhibition of histone acetylation [50], DNA damage responses (DDRs) [51], regulation of p53 acetylation [52], and histone chaperone activity [53,54]. In the context of adenovirus infection, the histone chaperone activity of SET is its best characterized function. SET binds to protein VII on the incoming viral genomes [55–57], which protects the viral genome from the cellular DDR [57,58] and facilitates the deposition of histones on viral DNA, potentially to replace protein VII. This histone deposition then promotes the initiation of transcription from the early viral gene promoters, before the transcriptional activator E1A is expressed. siRNA-mediated knockdown of SET results in decreased initial viral transcription [44,45], which is thought to depend on the deposition of histones on incoming viral genomes. Developing technologies to visualize viral genomes immediately after nuclear entry has been challenging, but detection of protein VII together with SET by immunofluorescence microscopy allows for tracking incoming viral DNA [56]. Live-imaging of incoming adenoviral genomes is also possible using eGFP-labeled SET [56] or direct labeling of incoming viral DNA [59]. In these direct labeling experiments, known as ANCHOR3, the deletion of E1A and E3 allows infected cells to enter mitosis during which viral genomes associate tightly with the condensed host chromosomes [59]. This observation suggests that incoming viral genomes are physically linked to host chromatin and may have implications in the context of persistent virus.
It is not clear how protein VII is removed from incoming viral DNA. Sequence analysis of protein VII reveals stretches of arginine residues that are similar to protamine arginine domains [60,61], suggesting properties similar to protamines that compact sperm DNA [62]. It is currently unclear whether the removal of protein VII occurs via a similar mechanism to protamine removal whereby post-translational modifications alter the protamine–DNA interaction and allow for histone replacement of protamines [46,62–65]. Within virus particles, protein VII is phosphorylated at three sites, while two acetylation sites are present only during infection [66,67], suggesting these moieties are relevant for protein VII removal from incoming viral genomes. Interestingly, replacing the protein VII acetylation site on lysine 2 or lysine 3 with alanine (K2A or K3A) is sufficient to alter protein VII nuclear localization from dispersed throughout chromatin to primarily localizing to the nucleolus [66]. This modification-dependent change in localization suggests that post-translational modifications of VII have important but uncharacterized functions in protein VII’s interaction with viral genomes both during initial infection and during packaging. However, K2A or K3A substitutions of the precursor protein VII, which has an additional 24 amino acids at the N terminus, result in the opposite effect on nuclear localization, with precursor VII unable to localize to the nucleolus and found throughout the nucleus [68]. This observation suggests that the regulation of protein VII localization through post-translational modification is complex and not yet fully understood.
Protein VII expressed late during infection localizes to host chromatin
Protein VII also associates with host chromatin during infection and is necessary and sufficient to alter chromatin composition [66]. Purified recombinant protein VII binds DNA and cellular nucleosomes. Protein VII can also limit DNA accessibility in vitro, suggesting a novel mechanism for host chromatin disruption [66]. Mass spectrometry analysis of fractionated chromatin from cells ectopically expressing protein VII [69] demonstrates that several proteins are significantly enriched in the chromatin fraction upon expression of protein VII, including the previously identified interacting partner SET [45,53,70] and the family of high-mobility group box (HMGB) proteins [71,72]. The functional consequence of VII-induced enrichment of SET on host chromatin is not known, but retention of HMGB may benefit viral infection by altering inflammatory responses. HMGB1 is a prototypic danger signal involved in orchestrating inflammatory responses and acts both within and outside the cell [73,74]. Extracellular HMGB1 binds to key receptors to amplify and sustain inflammation, making HMGB1 the focus of many anti-inflammatory therapeutic efforts [75]. HMGB1 also functions in the nucleus where it is thought to facilitate transcription factor binding [76,77]. In vitro purified protein VII and HMGB1 interact directly, and this interaction is sufficient to downregulate the cell’s ability to mount HMGB1-dependent immune responses in cells and in vivo [66] (Fig. 2). To date, it is unclear how these diverse binding partners contribute to the multiple functions of protein VII both on viral and host genomes. It is possible that post-translational modifications of protein VII determine its binding partners during different phases of the infectious cycle. On the host genome, protein VII localization also downregulates the DDR, likely due to limited DNA access by repair proteins [57], which is similar to protein VII’s protection of the viral genome from the DDR early during infection [58]. The protein VII amino acid sequence is strongly conserved across human adenovirus serotypes [78]. Protein VII is among the 16 genes common to all adenoviruses [79], both human and nonhuman serotypes, which suggests that its function in viral genome compaction is conserved. However, the amino acid sequence of protein VII has diverged significantly depending on the host species [79], which implies that protein VII is under positive evolutionary selection, likely reflecting the divergence in host immune responses. These observations taken together show that protein VII is typical of other multifunctional adenoviral proteins in that it compacts viral genomes, manipulates host chromatin composition, and modulates host immune responses.
E1A: The first viral protein produced attacks host chromatin
Protein VII recruits E1A to viral genomes to initiate viral transcription
The first viral protein produced during adenovirus infection, E1A [80,81], consists of two main isoforms during early infection via splicing – small and large E1A [82]. E1A, while not itself a DNA-binding protein, interacts with myriad chromatin-associated proteins such as transcriptional regulators to initiate viral transcription and overhaul host processes (see review [83]). Small E1A is considered a viral oncogene that can contribute to cellular transformation (see reviews [84,85]). Upon nuclear entry, the adenovirus genome is bound by the small, highly charged histone-like protein VII [27–33]. Upon co-expression of large E1A with protein VII in the X. laevis oocyte system, protein VII recruits large E1A to chromosomal DNA [42] and presumably facilitates the same interaction on viral genomes (Fig. 2). In support of this claim, the purified N terminus of large E1A and purified protein VII interact directly in vitro [42]. Additional studies show that expression of E1A along with transcription from the viral genome allows for release of protein VII from adenovirus DNA early in infection, although E1A-independent transcription from viral DNA is also sufficient for protein VII release [86]. Taken together, these observations suggest that protein VII directly interacts with and recruits E1A to viral genomes in order to facilitate viral transcription and release a subpopulation of protein VII from viral DNA [55,86]. Since this model is derived from population-scope experiments in which asynchrony of infection cycles is inherent [55,86], it does not explain how E1A itself is first expressed from the viral genome. How the earliest transcriptional activity from the viral genome is coordinated is unknown, although the recent advent of single-cell genomics [87,88], in which RNA-seq and ChIP-seq are performed on a single cell, may soon offer insights into the earliest stages of adenovirus infection.
E1A localizes to host chromatin, alters host histone modifications, and overhauls transcription
Small E1A has profound effects on host chromatin that are enacted through redistribution of histone post-translational modifications (Fig. 2). These effects were first identified by genome-wide assays of small E1A’s binding of host chromatin in diploid human lung fibroblast (IMR-90) cells [89] infected with adenovirus that expresses small E1A but does not result in a complete viral infection cycle [90]. Use of the replication-incompetent virus allows characterization of the effects of E1A expression for a much longer timeframe (6, 12, and 24 hpi) than E1A is active during wild-type infection [89]. Over time, E1A localizes to three different sets of host gene promoters, defined by similarities in E1A binding pattern. The first cluster, which is enriched for genes related to host response to pathogen infection and the inflammatory response, has high E1A occupancy at 6 hpi that does not persist into the later timepoints, and transcription of these genes is rapidly downregulated. E1A occupancy is also highly enriched at the promoters of a second cluster, which mainly consists of cell cycle-regulated genes that are quickly upregulated. The association of E1A with the second cluster is observed immediately after initial E1A expression and persists up to 24 hpi. The third cluster of E1A-bound genes are implicated in differentiation and development and are bound by E1A between 12 and 24 hpi correlating with reduced gene expression throughout. These changes in transcription imply dramatic changes to host chromatin that are converted to large-scale alterations in transcriptional regulation.
It is notable that E1A binding can either upregulate or downregulate gene expression depending on the genomic context. Twenty-four hours of E1A expression results in a global reduction in histone acetylation [91], a potent regulator of transcription [92]. Acetylation of the N-terminal H3 histone tail generates a more open, relaxed chromatin state that is permissive to transcription, while deacetylated H3 chromatin is more compact and therefore refractory to transcription [93]. Though H3 can be post-translationally modified on a number of different amino acid residues, acetylation marks on K9, K18, or K27 are enriched in the regulatory regions such as enhancers and promoters of active genes [94]. In response to E1A expression, loss of H3K18 acetylation is the most significant change in histone post-translational modifications, as determined by mass spectrometry [91]. E1A interacts directly with two closely related lysine acetyltransferases, CBP and p300 [95–99], both of which are required to establish H3K18ac in uninfected cells [91]. E1A mutants unable to bind p300/CBP do not localize to the promoters of gene clusters 1, 2, and 3, nor are H3K18ac marks reduced at those loci [89]. The implication of this observation is that E1A binds CBP/p300 in order to locate E1A-targeted genes and alter their regulation through aberrant histone acetylation.
While E1A expression causes an overall loss of H3 acetylation on the host genome, mostly of H3K18ac [89,91], genome-wide chromatin profiling of both H3K9ac and H3K18ac demonstrates that these histone marks are depleted from specific loci [100,101]. While most sites of H3K18ac are lost, a new set of H3K18ac peaks form, primarily in the regulatory regions of genes that increase in expression such as cell cycle genes [100,101]. These changes in H3K18ac distribution are largely abrogated if E1A is unable to interact with its lysine acetyltransferase partners, p300 and CBP, suggesting that p300/CBP is the main effector of E1A-induced changes to histone acetylation [101]. Intriguingly, E1A can inhibit H3 acetylation via its interaction with p300/CBP at some genomic loci, while in other contexts, E1A stimulates acetylation via p300/CBP. How E1A differentiates between these different regions remains mysterious.
Given that p300 is an important transcriptional coactivator [102], it is not surprising that the interaction of E1A with p300 is required to alter transcription [101]. The E1A-p300-repressed genes are enriched for secreted extracellular matrix proteins, which are normally highly expressed in fibroblast cells. This down-regulation of highly expressed genes may benefit viral infection both by liberating transcriptional resources that are redirected to viral transcription and by facilitating the cellular dedifferentiation orchestrated by E1A that will be discussed in greater detail below (see next section E1A-induced epigenetic reprogramming promotes cellular transformation).
E1A epigenetic reprogramming also subverts the transcriptional regulation of interferon-directed innate immune response to adenovirus [103]. E1A blocks the formation of a functional hBRe1 ubiquitin ligase complex that is required to upregulate interferon-stimulated genes (ISGs) in response to adenovirus infection [103]. Without E1A, the hBre1 complex monoubiquitinates histone H2B at ISGs, which allows for efficient upregulation of transcription. The interaction of E1A and hBre1 also regulates transcription from the viral genome albeit through a different mechanism [104]. On the viral genome, the interaction of hBre1 and its complex partner RNF20 recruits the human RNA polymerase II-associated factor 1 (hPaf1) complex to the viral DNA. Since the hPaf1 complex promotes transcription elongation and histone modifications associated with active transcription [105], E1A recruitment of the complex allows for efficient transcription of viral genes [104,106]. The targeting of hBre by E1A, which both downregulates the host viral defense mechanism and upregulates viral transcription, illustrates the economy of E1A’s epigenetic dysregulation via host factors.
There are important caveats to these studies of E1A. First, in using replication-incompetent adenovirus to study E1A-induced epigenetic alterations, the timeframe of E1A expression is prolonged compared to wild-type infection. As a result, many of the changes to histone acetylation observed at 24 hpi [89,91] may not occur during wild-type infection. Mass spectrometry of all histone modifications during adenovirus infection of the same cell type, IMR-90, demonstrates that most alterations to histone modifications are not statistically significant [107]. In fact, by mass spectrometry, H3K18ac is increased 24 hpi during wild-type infection [107] in contrast to the decrease in H3K18ac induced by 24 h of E1A expression [89,91]. Exactly how the viral replication cycle and expression of other viral proteins account for the difference in these observations remains to be uncovered. A second consideration is that when expressed in host cells, E1A establishes a complex network of interactions with the host proteome; E1A is known to directly bind and modulate the function of more than 30 host proteins and therefore may have secondary impact on the function of up to 2000 host proteins [83]. This high degree of E1A linkage is thought to be coordinated by E1A’s preponderance of short linear interaction motifs (SLiMs), which are short peptide sequences that facilitate protein–protein interactions [83]. Since the mutant versions of E1A employed to study the impact of individual E1A binding partners [89,101,103] may change the affinity of E1A for other proteins besides the protein of interest, it is challenging to differentiate between direct and indirect effects of E1A expression as well as how other viral proteins may contribute to these effects.
E1A-induced epigenetic reprogramming promotes cellular transformation
Oncogenes are dysregulated cellular genes that allow for cancerous proliferation of cells [108]. Viral oncogenes are virally delivered genes that can also lead to oncogenesis [109]. Adenovirus E1A, when expressed alone in human cells does not initiate transformation, but in conjunction with other processes can contribute to cellular transformation [84]. In fact, the commonly used transformed cell line HEK293 was created through the integration and expression of adenovirus E1A, along with E1B, in aneuploid cells isolated from human embryonic kidney [110]. The transformation enacted by E1A occurs through global epigenetic alterations, primarily of histone acetylation resulting in altered cell cycle gene expression (detailed above). E1A may also effect changes to host chromatin by increasing chromatin condensation [101], although the evidence for this observation is derived from a fluorescent reporter system in Chinese hamster ovary cells, and the same may not occur in human cells. Cell cycle reprogramming by E1A also requires the interaction of E1A with Rb proteins, which prompts infected cells to initiate a program of gene expression typical of S phase [111]. Rb proteins bind and inhibit E2F transcription factors at the promoters of cell cycle-regulated genes [112,113]. When Rb suppression is lifted, transcription from E2F-responsive promoters increases, leading to S phase gene expression [112,113]. By binding to and inactivating Rb, E1A can promote the aberrant initiation of the S phase gene expression program [114] and contribute to cellular transformation [110]. This upregulation of S phase-specific genes is thought to enhance viral infection since the same resources required to support cellular DNA replication can be redirected to support viral replication.
Remarkably, cell cycle perturbations and redistribution of histone acetylation at gene regulatory regions can be reversed upon E1A inactivation [115]. siRNA-mediated knockdown of E1A in HEK293 cells increases H3K18ac and H3K27ac especially at enhancers. Loss of E1A causes shifts in gene expression with cell cycle and mitosis genes among the most highly repressed and differentiation-associated genes among the most highly upregulated. These shifts in expression are essentially a reversal of the changes in gene expression caused by E1A [89,100]. Unexpectedly, the reversal of the effects of E1A suggests that viral chromatin-binding proteins can act as exogenous switches to manipulate cell fate. The E1A knockdown approach also illuminated the role of the Hippo pathway, an important regulator of development, in the E1A transformation process. The Hippo pathway, in addition to being essential for the earliest stages of development, is dysregulated in a number of cancers [116]. In response to extracellular cues, inactivation of the Hippo pathway stabilizes the effector proteins YAP and TAZ resulting in their accumulation in the nucleus [116]. In the nucleus, YAP and TAZ function as transcriptional coactivators binding the TEAD (TEA domain) family transcription factors that regulate apoptotic and cell proliferation genes [117]. Knockdown of E1A in HEK293 cells results in nuclear relocalization of YAP and TAZ and increased YAP occupancy at new sites of open, highly acetylated chromatin that correlates with increased TEAD transcription factor binding and increased transcription from those loci [115]. YAP and TAZ relocalization to the nucleus is required in order for E1A knockdown-induced changes in H3 acetylation and upregulation of gene expression [115]. Further studies will be needed to determine whether relocalization of YAP and TAZ is required for E1A-induced epigenetic reprogramming and whether Hippo pathway signaling is altered during adenovirus infection.
Current approaches to understand chromatin changes during adenovirus infection
Adenovirus E1A reprograms infected cells and induces shifts in their epigenetic identity that are not unlike the changes that contribute to oncogenesis. Historically, DNA viruses have provided valuable insights into cancer biology, such as the discovery of the tumor suppressor p53 via its interaction with SV40 T antigen [118]. Emergent technologies such as next-generation sequencing, chromosome conformation capture, high-throughput chromatin profiling methods, and highly sensitive mass spectrometry will allow for further exploration of fundamental nuclear processes through the lens of adenovirus infection. To date, global histone mass spectrometry has identified gross changes to histone modifications in a more unbiased way than traditional western blot approaches [119]; however, further work is needed to determine the genomic localization of these histone modifications in addition to their contribution to gene expression and the progression of infection. Furthermore, as chromosome capture technologies increase in their precision [120] and other approaches such as CUT&RUN [121] allow for smaller and smaller populations of cells to be used per assay, it is conceivable that in the near future, more precise detail of the dynamics of chromatin changes during adenovirus infection will be attainable.
E4orf3 separates host chromatin from viral replication centers
In addition to adenoviral proteins that interact directly with host chromatin, viral proteins that interact with host nuclear factors, such as the E4 proteins, also impact chromatin. The E4 transcriptional unit produces eight proteins [122–125] and deletion of the entire unit is deleterious to viral replication and late gene expression [124]. Paradoxically, disrupting single E4 ORFs by point mutation has moderate-to-no effect on viral fitness [124], suggesting that there is functional redundancy among E4 proteins. Disrupting both E4orf3 and E4orf6 significantly compromises viral replication and progeny production to nearly the same degree as complete deletion of E4 [125]. Loss of both E4orf3 and E4orf6 function leads to concatemerization of postreplicative viral genomes [126] that are presumably too large to be packaged and explains the defects in progeny production observed [125]. While these genetic analyses suggest significant functional overlap between E4orf3 and E4orf6, the two proteins promote the viral replication cycle through distinct mechanisms. For example, E4orf6, a highly multifunctional protein (reviewed more extensively by [127,128]), forms a ubiquitin ligase complex with the early adenovirus E1B-55K protein, which then binds and degrades multiple proteins including the tumor suppressor and cell cycle regulator p53 [129–131]. The degradation of p53 by E4orf6/E1B-55K allows for adenoviral reprogramming of the cell cycle and more productive viral infection (see E1A section for more details). In contrast, E4orf3 seems to primarily disrupt nuclear structure to benefit viral replication.
E4orf3 forms polymers and sequesters host proteins during infection
Like all nuclear-replicating viruses, adenovirus disrupts host nuclear structure, enabling formation of replication centers (see Gonzalez review [132]) and progeny production. The role of E4orf3 in the adenovirus replication program was initially identified through one class of nuclear body, promyelocytic leukemia (PML) bodies. PML bodies are an aggregate of nuclear proteins surrounded by a shell of PML protein (reviewed by [133]). PML bodies were first identified because they are disrupted in acute PML although their exact function remains enigmatic [133]. Two independent studies of PML body structure during adenovirus infection found that infection causes a striking conversion of PML protein from a circular conformation at several PML bodies per nucleus into numerous ‘tracks’ of PML distributed throughout the nucleus [134,135] (see Weitzman review [47]). Deletion of E4orf3 prevents the formation of PML tracks because E4orf3 itself polymerizes to form a twisting ribbon-like ultrastructure with which PML and SP100, another PML body component, colocalize forming distinctive tracks throughout the nucleus [134,135]. Transfection of E4orf3 into cells is sufficient for PML body disruption and for E4orf3 tracks to form [134,135]. Intriguingly, E4orf3 tracks are able to form outside the nucleus when the protein is expressed in plant cells, implying that the polymers assemble without the need for human-specific proteins [136]. The functional implications of E4orf3-based protein targeting extend beyond PML bodies.
E4orf3 tracks also target host DNA repair machinery [137]. The concatemerization of viral genomes observed during E4orf3Δ/E4orf6Δ virus infection relies on the host DNA repair proteins Mre11 and NBS1 [137]. Mre11, Rad50, and NBS1 together form the MRN complex that senses DNA double-stranded breaks, activates the cell cycle checkpoint, and initiates repair (reviewed by [138]). All three complex proteins are relocalized from being dispersed throughout the nucleus to forming small foci that are excluded from sites of viral replication as determined by immunofluorescent microscopy during wild-type infection [137]. The abundance of all three proteins also decreases in a proteasome-dependent manner during infection [137]. Adenoviruses lacking both E4orf3 and E4orf6 are unable to relocalize or degrade MRN complex proteins. Ectopic expression of E4orf3 alone is sufficient to relocalize MRN proteins [137], much as it is sufficient to relocalize PML protein [134,135]. Degradation of MRN complex proteins, however, is dependent upon the interaction of E4orf6 and E1B-55K [137]. These observations, coupled with the fact that both E4orf3 and E4orf6 must be inactivated to reduce viral fitness and observe genome concatemers, demonstrate that adenovirus type 5 has evolved redundant strategies to inhibit host DNA repair – one based on sequestration of host factors and the other on active degradation of host proteins. This dual targeting of MRN proteins is not universally conserved across other human adenovirus serotypes [139,140]. A comparison of MRN complex degradation among Ad12, Ad35, Ad2, Ad5, Ad9, and Ad4 human adenovirus serotypes demonstrates that degradation of some or all MRN proteins is a conserved effect, except for the case of Ad9 [139,140]. The relocalization of MRN complex proteins is somewhat less conserved with Ad12 and Ad35 infection failing to produce characteristic Mre11 puncta [140], while expression of E4orf3 from Ad4 fails to mislocalize NBS1 [139]. As a consequence, Ad9 and Ad12 viral DNA replication is restricted by the MRN complex [140]. The Ad4- and Ad12-E4orf3 protein tracks reorganize PML protein into tracks [139] reinforcing the notion that E4orf3 has serotype specificity for its target proteins.
Despite targeting PML protein, E4orf3 does not target all PML body-associated proteins. While E4orf3 reorganizes PML protein, SP100, SUMO-1, and DAXX into nuclear tracks, it fails to do so in the case of BLM and TopBP1, which are DNA repair-associated PML body proteins [134,135,139]. An important function of E4orf3’s specific interaction with host factors seems to be selective sequestration of some host factors away from sites of viral replication or viral replication centers (VRCs). In the case of E4orf3-driven sequestration of the MRN complex, repair complex proteins are directed away from VRCs to prevent the host DSB repair machinery from ligating linear viral genomes. Mass spectrometry analysis of proteins associated with nascent viral DNA during adenovirus replication has provided a comprehensive view of which host factors are selectively excluded or enriched at VRCs [141]. E4orf3 sequesters other host factors besides PML body proteins and DNA repair factors. For example, the transcriptional regulator TFII-I is underrepresented on replicated viral genomes compared to host DNA and forms foci that are similar to MRN complex foci formed during adenovirus infection. TFII-I relocalization and its degradation by the proteasome are dependent upon E4orf3 expression. Likewise, ectopic E4orf3 expression is sufficient to reorganize TFII-I into nuclear tracks. Exclusion of TFII-I from replicated viral genomes may be significant during the viral replication cycle as a regulator of viral transcription, although the connection between TFII-I and viral transcription has not been well-established. These studies indicate that E4orf3 acts directly on specific host factors to promote viral fitness (see Weitzman review [47]).
E4orf3 polymers divide replication compartments from host chromatin
A comparison of E4orf3-PML tracks with sites of viral replication using BrdU incorporation and sites of active viral transcription, visualized by costaining for a splicing-related snRNP, suggests that E4orf3 tracks physically separate sites of viral replication and transcription from the rest of the nucleus [134]. A more thorough structural and 3D-imaging study of E4orf3 polymers in a nucleus infected with E1B-55KΔ Ad5 supports this model [136]. The integration of a photo-oxidative domain between two E4orf3 β-sheets is necessary to increase the electron density of E4orf3 polymers in order to detect E4orf3 by electron microscopy. This modified protein forms tracks throughout the nucleus and allows for imaging sections of the nucleus to generate 3D nuclear structures. Consistent with earlier observations [134], E4orf3 tracks wind throughout the nucleus but do not colocalize with VRCs or nucleoli [136]. In addition, the E4orf3 polymerization mutant (N82A), which cannot form nuclear tracks, is unable to disrupt PML bodies or sequester the MRN complex member NBS1 [136]. This observation suggests that the assembly of the E4orf3 ultrastructure is required for PML dysregulation, inhibition of DNA repair pathways, and therefore productive infection. Together, these observations indicate that E4orf3 facilitates compartmentalization of the host nucleus to favor efficient viral replication. However, it remains to be determined how the assembled E4orf3 ultrastructure is able to wind throughout the nucleus while avoiding and excluding VRCs as well as the nuclear periphery (see also Gonzalez review [132]). This elegant and striking mechanism thus far appears specific to adenovirus; however, other viruses may have evolved similar compartmentalization mechanisms that have yet to be uncovered.
Impact of E4orf3 on host chromatin and chromatinization of viral genomes
E4orf3 may regulate gene expression both on the host and viral genomes. On the host genome, E4orf3 causes broad changes to host chromatin that alters gene expression, including p53 targets [142]. In a study of the dynamics of p53 and its downstream effectors during infection with an E1B-55KΔ virus, in which p53 is not degraded, unexpectedly downstream DNA damage genes cannot be upregulated. Upon additional deletion of E4orf3, the p53 target genes are induced in response to viral infection. Since a p53-regulated reporter gene can still be activated on a plasmid in this context, the authors conclude that changes occur on host chromatin that does not take place on the plasmid. ChIP-PCR demonstrates that there is less p53 occupancy at select p53-regulated gene promoters on the host genome. While the amount of H3K9me3 is not changed overall, there is more H3K9me3 at p53-regulated promoters compared to infection lacking E4orf3. As in many chromatin studies, it can be difficult to determine whether a change to a histone modification level is a cause or an effect of the phenomenon under scrutiny. The case of increased H3K9me3 at specific p53-bound promoters in the absence of E4orf3 is no exception. It is unclear whether the decrease in transcription is a result of the change in histone modification, or the histone modification is deposited as a consequence of decreased transcription. Additionally, the mutant viruses used in this study lack expression of late viral proteins, which excludes the possibility that chromatin-bound late proteins, such as protein VII, may also impact the outcome of transcription at p53 target genes. Future work including ectopic expression of E4orf3 alone will clarify this point.
A recent study suggests that E4orf3 may have a similarly repressive effect on transcription from viral genomes that is mediated through a heretofore unreported interaction between E4orf3 and E1A [143]. E1A co-immunoprecipitates with E4orf3 from cells that ectopically express an epitope-tagged copy of E4orf3 and are infected with E3Δ Ad5 demonstrating that E1A and E4orf3 interact during infection. The interaction between E4orf3 and E1A is likely direct since purified E1A co-immunoprecipitates with GST-tagged and purified E4orf3 in vitro. Deletion analysis of E1A demonstrates that the N-terminal amino acid residues 11–13 are responsible for E1A’s interaction with E4orf3. During E1AΔ11–13 virus infection, histone occupancy is higher, E1A occupancy reduced, and transcription is lower at specific early-transcribed adenovirus genes as well as at the late gene promoter, suggesting that E4orf3 represses transcription from the viral genome. Since E1A is known to interact with and modulate the activity of many host factors (see a comprehensive review by [83]), it is possible that the deletion of E1A residues 11–13 impacts more than its interaction with E4orf3. On viral genomes, transcription is reduced, histone occupancy increases, and E1A occupancy decreases only when E4orf3 is expressed in the presence of E1A compared to a control vector [143]. Based on these observations, E4orf3 seems to facilitate the association of host histone proteins with viral genomes and therefore modulate transcription. Given that both E1A and E4orf3 have previously been associated with regulators of host genome transcription (see E1A section of this review and [83,141]), it is plausible that the interaction of E1A and E4orf3 may impact transcription from viral DNA. While many mechanistic details of this process remain unknown, these recent studies of E4orf3 make it clear that there is still much to be learned about E4orf3 and its interaction with host factors.
In addition to viral proteins interacting with host chromatin, host factors also interact with incoming viral genomes as a defense mechanism. Recent evidence demonstrates that the host cell broadly restricts chromatinization of the incoming viral genome during the earliest stages of infection. It is proposed that this restriction occurs via PML-associated proteins Daxx, ATRX, and Sp100 [144–146] as well as the chromatin regulator SPOC1 [147]. Outside of viral infection, the Daxx/ATRX histone chaperone complex is thought to promote chromatin silencing via deposition of the H3.3 histone variant. This deposition is proposed to maintain the silencing marker H3K9me3 at specific heterochromatic loci [148]. To combat this restriction, adenovirus targets Daxx/ATRX for proteasomal degradation via the E1B-55K and E4orf6 ubiquitin ligase complex [144,145]. In the absence of Daxx/ATRX degradation, adenovirus early gene transcription is reduced and fewer viral progeny are produced [145]. Daxx may also mediate latency of other viruses, suggesting that histone incorporation onto viral genomes is a common host defense strategy [149]. Furthermore, it is possible that Daxx-mediated regulation of adenovirus transcription and subsequent reduction of viral progeny may play a role in persistent adenovirus infection.
Epigenetic alterations in persistently infected cells
Although rarely detected in peripheral blood lymphocytes [150–152], adenovirus is frequently found in lymphocytes residing in lymphoid organs of the oropharynx [153–155] and intestine [156–158]. Adenovirus may persist as a smoldering infection in the intestine from which it can be shed in the feces of asymptomatic individuals [150,159]. The smoldering nature of the infection in the gut may reflect a balance between viral replication and an active antiviral response because immune suppressive treatments such as those required for allogeneic hematopoietic stem cell transplant often lead to an explosive degree of adenovirus replication with an increased risk of disease and death for the stem cell recipient [6,157,160]. Given that adenovirus infection can cause significant morbidity and mortality in immunocompromised transplant patients, it is important to better characterize persistent adenovirus infection. The studies of persistent infection summarized here suggest that epigenetic regulation plays a role in establishment and maintenance of persistent adenovirus.
Viral and cellular determinants of persistence
Although the determinants of adenovirus persistence are poorly characterized, some evidence suggests that both host and viral factors contribute to persistence. For example, in the case of latent infection of mucosal T cells from the adenoids and tonsils [153,154], when purified lymphocytes containing viral DNA but not replication-competent virions were cultured in the presence of signals that activate T cells, nearly 90% (13 of 15) of the isolates released infectious species C viruses [154]. When passaged, these viruses replicate at a slower rate than laboratory-adapted strains, suggesting that new viruses produced by reactivation of a persistent infection may be subject to heritable changes in viral gene expression. Furthermore, studies performed with HAdV-A12 in nonpermissive baby hamster kidney (BHK) cells point to DNA methylation as a consequence of viral infection and a regulator of viral gene expression (reviewed by [161]). Although the limited studies performed in a nonpermissive cell line may not reflect the state in humans, together these studies suggest that epigenetic factors such as DNA methylation may influence persistent infection.
One challenge to understanding persistent infection has been the dearth of model systems although human lymphocytic cell lines that sustain noncytolytic adenovirus infection now permit the study of persistence. In this model, T- and B-cell lines grow at the same rate as noninfected cells over multiple months of culture when persistently infected with HAdV-C5 even though viral DNA levels exceed 50 000 genomes per cell [10]. The model provides some insights into the potential role of the viral factor adenovirus death protein (ADP) in persistent infection. ADP expression is repressed in cells that sustain a persistent infection but is not repressed in Jurkat T cells, which support a transient and lytic infection [162]. Jurkat cells infected with a virus deleted for the ADP gene retain viral DNA and survive for over a month. A separate study of the same persistently infected B and T cells shows that virus-associated RNAs I and II are expressed at a high level in persistently infected BJAB cells [163], which suggests that viral miRNAs play a role in facilitating a persistent infection. Together, these studies suggest that a program of repression is required to maintain persistent infection although further work is undoubtedly needed to elucidate the details.
Adenovirus can elicit persistent changes in chromatin and DNA methylation
Persistent infection may also alter host gene expression possibly through epigenetic mechanisms. In the B- and T-cell lines used to model adenovirus persistence, cellular gene expression is modified during persistent infection [10]. Following infection, expression of the adenovirus receptor CAR is transcriptionally repressed and repression persists in cells that no longer retain the viral genome. CAR expression is restored by treating cells with the histone deacetylase inhibitor trichostatin A and DNA methylase inhibitor 5′azacytidine, suggesting that CAR repression is epigenetic. Remarkably, CAR is one of only six host genes identified in chronically infected B cells and T cells as being repressed using microarray analysis of gene expression [164]. The repression of these genes is released by the inhibition of DNA methyltransferases with 5′azacytidine. Furthermore, it is thought that transient expression of viral factors can induce long-lasting heritable changes to host gene expression based on the observation that expression of the E1A gene and E4 oncogenes [165–167] transforms primary baby rat kidney cells and elicits mutations at the hypoxanthine phosphoribosyltransferase locus [168]. Because the majority of transformed cells do not retain E1A or E4 DNA sequences, this mechanism has been termed ‘hit-and-run’ transformation [168]. The capacity of these proteins to suppress the sensing and repair of damaged DNA is postulated to be the basis for the mutagenic nature of these viral oncoproteins. However, adenovirus may also elicit heritable and transforming changes in the epigenetic landscape following transient expression of viral genes during infection [169]. Further work is required to determine the epigenetic mechanisms behind these changes in host and viral gene expression.
One possible mechanism by which host and viral gene expression could be modulated is DNA methylation. The incorporation of foreign DNA into a mammalian cell, whether through mechanical delivery or viral infection, alters the pattern of CpG methylation on cellular DNA to a greater extent than on viral DNA. For example, there is 50- to 100-fold more methylated cytosine in host DNA than in HAdV-C2 or HAdV-A12 DNA [170]. Additional studies have shown that integration of HAdV-A12 DNA into the genome of BHK cells leads to extensive changes in DNA methylation [171]. Notably, high levels of DNA methylation persist in cell lines that ostensibly have lost all of the detectable HAdV-A12 genome [171,172], reminiscent of B lymphocytes infected with Epstein–Barr virus that shows widespread and increased levels of DNA methylation [173,174]. These observations suggest that the introduction of foreign DNA may have consequences on gene expression through epigenetic means [175].
One example of DNA methylation-based modulation of viral gene expression is the observation that methylation of one of three CCGG sites in the HAdV-C2 E2A gene promoter, one of three viral genes required for viral DNA replication, prevents its transcription in a X. laevis oocyte [176]. In contrast, methylation at eleven sites in the remainder of the E2A gene has little effect on expression. Methylation of the HAdV-A12 E1A promoter also prevents transcription in this model system [177]. These findings suggest potential points of control for viral gene expression during latency and reactivation from latency. Methylation at both E1A and E2A promoters could silence all viral gene expression that depends on E1A expression and viral DNA replication. By contrast, methylation at only the E2A promoter would enable expression of E1A-dependent early genes with no viral DNA replication and consequently no late gene expression. Such control would permit adenovirus to remain latent and emerge if an appropriate signal impinges on the relevant promoters. As noted above, lymphoid cells from individuals harboring a latent adenovirus infection have no measurable levels of viral mRNA, consistent with the complete silencing of RNA Pol II-dependent promoters [153,154]. However, viral gene expression and viral replication are restored following exposure to T-cell-activating stimuli [154]. In a transgenic mouse model, T-cell-activating stimuli lead to rapid demethylation of the interleukin 2 gene promoter with a concomitant increase in transcription [178]. In the immune-competent organism, successful emergence from latency may require not only activation of replicative viral genes but activation of viral genes that defeat immune surveillance. The E3 region encodes many of the activities that defeat immune surveillance [179,180]. Remarkably, the E3A promoter is nearly 20-fold more responsive to T-cell-activating signals than to E1A expression [181]. The E3 promoter may be exquisitely configured to respond to T-cell stimulation in order to avoid immune surveillance during a persistent infection, and to facilitate successful viral replication in the face of an activated immune response. To reiterate, the molecular mechanisms underlying adenovirus persistence and reactivation of infection are largely unknown, but based on the studies discussed here, it is thought that epigenetic regulation plays a role.
Pathologies associated with adenovirus reactivation
Tissue harboring cells that are latently or persistently infected with adenovirus may contribute to disease in a variety of settings. Persistently infected lymphocytes in the gastrointestinal tract are the primary source of disseminated adenoviral infections after pediatric allogeneic stem cell transplants (see Lion review [182]). The systemic immunosuppression required to prevent graft-versus-host disease in this setting most likely contributes to the emergence of actively replicating viruses. A greater understanding of the signals that govern latency and persistence would aid in the prevention of the emergence of this life-threatening infection when immunosuppression cannot be avoided. Intussusception, in which a segment of intestine ‘telescopes’ into another segment causing intestinal blockage, is the most common abdominal crisis in young children [183]. Adenovirus DNA was associated with nearly one-third of the cases of intussusception lacking anatomical irregularities associated with this disease [184]. Both molecular and epidemiological analyses identified nonenteric adenoviruses as a risk factor for intussusception [185]. Another pathology associated with adenovirus infection is otitis media, an inflammatory disease associated with the ascension of bacteria from the nasopharynx to the middle ear. An experimental animal model demonstrated that adenovirus infection can promote the ascension of Streptococcus pneumoniae [186] and nontypeable Haemophilus influenzae into the middle ear [187,188]. Adenovirus was detected more frequently in the adenoids of children that developed otitis media with effusion [189], and children infected with adenovirus are more likely to develop acute otitis media [190]. Finally, adenovirus is among the most frequently detected viruses in amniotic fluid (reviewed in [191]) and is found in the cord blood lymphocytes of approximately 4% of all newborns [191]. Although the nature of the viral infection in neonatal cord blood is not known, the capacity of adenovirus to elicit epigenetic and even genetic modifications in this population of cells enriched in stem and progenitor cells warrants further study. Broadly, new work is needed to identify, understand, and combat complications associated with the persistent form of adenovirus infection.
Conclusions and perspectives
Understanding how virus replication compartments develop to recruit selectively or exclude chromatin modifiers will provide insights into chromatin regulation and nuclear architecture. Recent work in chromatin biology has focused on phase separation as a means to segregate non-organelle-bound regions within the nuclear compartment [192]. For example, heterochromatin may be separated from actively transcribing genes through a liquid–liquid separation akin to oil and water [193,194]. This separation is an attractive idea to explain how viral compartments are separated from the host chromatin during infection and future work will determine how biophysical properties define viral genome segregation. As chromatin technologies continue to advance, the mechanisms by which viruses take advantage of every aspect of genome organization to benefit viral infection will continue to be elucidated. By understanding viral manipulation at an epigenetic level, future work will identify mechanisms of perturbation of genome architecture.
The three-dimensional (3D) context of the genome within the nucleus must also be considered when examining the outcome of infection. Recent advances in chromatin technologies have resulted in a leap from one-dimensional sequencing approaches to 3D chromosome capture [195,196]. In parallel, microscopy techniques have also developed allowing visualization down to individual nucleosomes [197]. These advances represent significant progress in understanding chromatin in the context of the 3D nucleus. A few groups have begun to apply these cutting-edge techniques to the study of virus infection. For example, studies examining how the architectural CTCF protein, which generates chromatin loops to maintain the 3D structure of the genome [198], is co-opted during infection [199] have demonstrated that CTCF controls viral gene expression among many DNA viruses, including adenovirus [200]. We are only beginning to understand how viruses evolved to take advantage of all features of genome architecture. Reconciling complex datasets with biological outcomes remains a challenge with these emerging technologies [120]. It will be interesting to follow how virus infection both informs and directs the evolution of technologies to probe chromatin structure.
Probing the interactions between adenovirus and chromatin architecture will also provide new targets that can be used for development of antiviral therapies. Expanding the repertoire of adenovirus antiviral therapies will be extremely important for improving transplant outcomes. Currently, the treatment for acute adenovirus infection, the nucleoside analogue cidofovir, is neither specific for adenovirus nor efficacious and poses the risk of kidney damage for patients [2,6]. Prophylactic antiviral therapy has been of limited success in transplant patients [6]. Drugs capable of targeting chromatin modifiers exploited by reactivated viruses may prove to be useful in preventing viral spread especially in the immunocompromised. Following the battle between epigenomes during viral infection may also reveal key vulnerabilities in chromatin architecture that can be exploited for therapies of other diseases such as cancer and autoimmune conditions. The examples of intussusception and otitis media illustrate that persistent adenovirus likely has an underappreciated role in myriad human diseases and that more work is needed to fully comprehend adenovirus as an etiologic agent.
Adenoviruses, while a threat to human health, have also unlocked key biological insights into the inner workings of the cell. Chromatin biology is a burgeoning field that is still in its infancy by some measures. While recent efforts such as the ENCODE project have painted a highly descriptive picture of the human genome, how the vast chromatin landscape functions dynamically and in concert largely remains unknown. By tracing how adenoviruses co-opt and undermine host chromatin processes, we will gain valuable insights into chromosome biology and expand the currently limited options for adenovirus treatment.
Acknowledgements
We thank members of the Avgousti, Ornelles, and Garnett-Benson laboratories for critical reading of the manuscript and helpful discussions. We apologize to those whose primary research papers could not be cited due to space constraints. Research in the Avgousti laboratory is supported by start-up funds from Fred Hutchinson Cancer Research Center and grants from the National Institutes of Health to DCA (GM133441), to KLL (CA9657), to DAO (CA127621) and to CG-B (CA162235).
Abbreviations
- ADP
adenovirus death protein
- Ad5 or Ad 12
human adenovirus type 5 or 12, or other type referenced
- BHK
baby hamster kidney
- BrdU
bromodeoxyuridine
- CTCF
CCCTC binding factor
- DBS
double stranded break
- DDR
DNA damage response
- E1A
early 1 A
- E4orf3 or E4orf6
early 4 open reading frame 3 or 6
- HAdV-A12
human adenovirus subgroup A type 12, or different subgroup or type referenced
- HMGB
high-mobility group box
- hPaf1
human RNA polymerase II-associated factor 1
- ISGs
interferon-stimulated genes
- PML
promyelocytic leukemia
- snRNP
small nuclear ribonuclear protein
- SET or TAF-1
SE translocation or template activating factor
- VRCs
viral replication centers
References
- 1.Ghebremedhin B (2014) Human adenovirus: viral pathogen with increasing importance. Eur J Microbiol Immunol 4, 26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Echavarría M (2008) Adenoviruses in immunocompromised hosts. Clin Microbiol Rev 21, 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esquer Garrigos Z, Barth D, Hamdi AM, Abu Saleh OM and Sohail MR (2018) Nitazoxanide is a therapeutic option for adenovirus-related enteritis in immunocompromised adults. Antimicrob Agents Chemother 62, e01937–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majorant D, Qiu F, Kalil AC, Wilson N and Florescu DF (2018) Adenovirus – a deadly disease in the solid organ transplant population: risk factors and outcomes. Transpl Proc 50, 3769–3774. [DOI] [PubMed] [Google Scholar]
- 5.Pochon C and Voigt S (2018) Respiratory virus infections in hematopoietic cell transplant recipients. Front Microbiol 9, 3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lion T (2014) Adenovirus infections in immunocompetent and immunocompromised patients. Clin Microbiol Rev 27, 441–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nedelman M (2018) 11th child dies in adenovirus outbreak in New Jersey. CNN.com https://www.cnn.com/2018/11/16/health/wanaque-adenovirus-deaths-new-jersey-bn/index.html [Google Scholar]
- 8.Abelson J, Brittain A and Larimer S (2019) A dangerous delay. The Washington Post. https://www.washingtonpost.com/news/national/wp/2019/05/16/feature/university-of-maryland-moldadenovirus/ [Google Scholar]
- 9.King CR, Zhang A and Mymryk JS (2016) The persistent mystery of adenovirus persistence. Trends Microbiol 24, 323–324. [DOI] [PubMed] [Google Scholar]
- 10.Zhang Y, Huang W, Ornelles DA and Gooding LR (2010) Modeling adenovirus latency in human lymphocyte cell lines. J Virol 84, 8799–8810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng Y, Stamminger T and Hearing P (2016) E2F/Rb family proteins mediate interferon induced repression of adenovirus immediate early transcription to promote persistent viral infection. PLoS Pathog 12, e1005415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luger K, Mäder AW, Richmond RK, Sargent DF and Richmond TJ (1997) Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- 13.Kouzarides T (2007) Chromatin modifications and their function. Cell 128, 693–705. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y and Garcia BA (2015) Comprehensive catalog of currently documented histone modifications. Cold Spring Harb Perspect Biol 7, a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizuguchi T, Barrowman J and Grewal SIS (2015) Chromosome domain architecture and dynamic organization of the fission yeast genome. FEBS Lett 589, 2975–2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ et al. (2010) Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143, 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H and Allis CD (2005) Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature 438, 1116–1122. [DOI] [PubMed] [Google Scholar]
- 18.Hirota T, Lipp JJ, Toh B-H and Peters J-M (2005) Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438, 1176–1180. [DOI] [PubMed] [Google Scholar]
- 19.Luo C, Hajkova P and Ecker JR (2018) Dynamic DNA methylation: in the right place at the right time. Science 361, 1336–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg MVC and Bourc’his D (2019) The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 20, 590–607. [DOI] [PubMed] [Google Scholar]
- 21.Li E, Beard C and Jaenisch R (1993) Role for DNA methylation in genomic imprinting. Nature 366, 362–365. [DOI] [PubMed] [Google Scholar]
- 22.Zemach A and Zilberman D (2010) Evolution of eukaryotic DNA methylation and the pursuit of safer sex. Curr Biol 20, R780–R785. [DOI] [PubMed] [Google Scholar]
- 23.Rose NR and Klose RJ (2014) Understanding the relationship between DNA methylation and histone lysine methylation. Bioch Biophys Acta Gene Regul Mech 1839, 1362–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao Q, Zhang J, Chen R, Wang L, Li B, Cheng H, Duan X, Zhu H, Wei W, Li J et al. (2016) Dissecting the precise role of H3K9 methylation in crosstalk with DNA maintenance methylation in mammals. Nat Commun 7, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo Q-M et al. (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462, 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zemach A, McDaniel IE, Silva P and Zilberman D (2010) Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919. [DOI] [PubMed] [Google Scholar]
- 27.Corden J, Engelking HM and Pearson GD (1976) Chromatin-like organization of the adenovirus chromosome. Proc Natl Acad Sci USA 73, 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lischwe MA and Sung MT (1977) A histone-like protein from adenovirus chromatin. Nature 267, 552–554. [DOI] [PubMed] [Google Scholar]
- 29.Mirza MA and Weber J (1982) Structure of adenovirus chromatin. Biochim Biophys Acta 696, 76–86. [DOI] [PubMed] [Google Scholar]
- 30.Chelius D, Hühmer AFR, Shieh CH, Lehmberg E, Traina JA, Slattery TK and Pungor E (2002) Analysis of the adenovirus type 5 proteome by liquid chromatography and tandem mass spectrometry methods. J Proteome Res 1, 501–513. [DOI] [PubMed] [Google Scholar]
- 31.Alqahtani A, Heesom K, Bramson JL, Curiel D, Ugai H and Matthews DA (2014) Analysis of purified Wild type and mutant adenovirus particles by SILAC based quantitative proteomics. J Gen Virol 95, 2504–2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vayda ME, Rogers AE and Flint SJ (1983) The structure of nucleoprotein cores released from adenovirions. Nucleic Acids Res 11, 441–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vayda ME and Flint SJ (1987) Isolation and characterization of adenovirus core nucleoprotein subunits. J Virol 61, 3335–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatterjee PK, Vayda ME and Flint SJ (1985) Interactions among the three adenovirus core proteins. J Virol 55, 379–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chatterjee PK, Vayda ME and Flint SJ (1986) Adenoviral protein VII packages intracellular viral DNA throughout the early phase of infection. EMBO J 5, 1633–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatterjee PK, Vayda ME and Flint SJ (1986) Identification of proteins and protein domains that contact DNA within adenovirus nucleoprotein cores by ultraviolet light crosslinking of oligonucleotides 32P-labelled in vivo. J Mol Biol 188, 23–37. [DOI] [PubMed] [Google Scholar]
- 37.Dai X, Wu L, Sun R and Zhou ZH (2017) Atomic structures of minor proteins VI and VII in the human adenovirus. J Virol 91, JVI00850–17. 10.1128/JVI.00850-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostapchuk P, Suomalainen M, Zheng Y, Boucke K, Greber UF and Hearing P (2017) The adenovirus major core protein VII is dispensable for virion assembly but is essential for lytic infection. PLoS Pathog 13, e1006455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Greber UF and Flatt JW (2019) Adenovirus entry: from infection to immunity. Ann Rev Virol 6, 177–197. [DOI] [PubMed] [Google Scholar]
- 40.Mattiroli F, Bhattacharyya S, Dyer PN, White AE, Sandman K, Burkhart BW, Byrne KR, Lee T, Ahn NG, Santangelo TJ et al. (2017) Structure of histone-based chromatin in Archaea. Science 357, 609–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong CM, McFall ER, Burns JK and Parks RJ (2013) The role of chromatin in adenoviral vector function. Viruses 5, 1500–1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson JS, Osheim YN, Xue Y, Emanuel MR, Lewis PW, Bankovich A, Beyer AL and Engel DA (2004) Adenovirus protein VII condenses DNA, represses transcription, and associates with transcriptional activator E1A. J Virol 78, 6459–6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reynolds N, O’Shaughnessy A and Hendrich B (2013) Transcriptional repressors: multifaceted regulators of gene expression. Development 140, 505–512. [DOI] [PubMed] [Google Scholar]
- 44.Komatsu T, Haruki H and Nagata K (2011) Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res 39, 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haruki H, Okuwaki M, Miyagishi M, Taira K and Nagata K (2006) Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J Virol 80, 794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Giberson AN, Davidson AR and Parks RJ (2012) Chromatin structure of adenovirus DNA throughout infection. Nucleic Acids Res 40, 2369–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Charman M, Herrmann C and Weitzman MD (2019) Viral and cellular interactions during viral DNA replication. FEBS Lett. 10.1002/1873-3468.13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.von Lindern M, van Baal S, Wiegant J, Raap A, Hagemeijer A and Grosveld G (1992) Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3’ half to different genes: characterization of the set gene. Mol Cell Biol 12, 3346–3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Berkovits BD and Mayr C (2015) Alternative 3’ UTRs act as scaffolds to regulate membrane protein localization. Nature 522, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Seo SB, McNamara P, Heo S, Turner A, Lane WS and Chakravarti D (2001) Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104, 119–130. [DOI] [PubMed] [Google Scholar]
- 51.Kalousi A, Hoffbeck A-S, Selemenakis PN, Pinder J, Savage KI, Khanna KK, Brino L, Dellaire G, Gorgoulis VG and Soutoglou E (2015) The nuclear oncogene SET controls DNA repair by KAP1 and HP1 retention to chromatin. Cell Rep 11, 149–163. [DOI] [PubMed] [Google Scholar]
- 52.Wang D, Kon N, Lasso G, Jiang L, Leng W, Zhu W-G, Qin J, Honig B and Gu W (2016) Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature 538, 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haruki H, Gyurcsik B, Okuwaki M and Nagata K (2003) Ternary complex formation between DNA-adenovirus core protein VII and TAF-Ibeta/SET, an acidic molecular chaperone. FEBS Lett 555, 521–527. [DOI] [PubMed] [Google Scholar]
- 54.Muto S, Senda M, Akai Y, Sato L, Suzuki T, Nagai R, Senda T and Horikoshi M (2007) Relationship between the structure of SET/TAF-Ibeta/INHAT and its histone chaperone activity. Proc Natl Acad Sci USA 104, 4285–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xue Y, Johnson JS, Ornelles DA, Lieberman J and Engel DA (2005) Adenovirus protein VII functions throughout early phase and interacts with cellular proteins SET and pp32. J Virol 79, 2474–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Komatsu T, Dacheux D, Kreppel F, Nagata K and Wodrich H (2015) A method for visualization of incoming adenovirus chromatin complexes in fixed and living cells. PLoS ONE 10, e0137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Avgousti DC, Della Fera AN, Otter CJ, Herrmann C, Pancholi NJ and Weitzman MD (2017) Adenovirus core protein VII downregulates the DNA damage response on the host genome. J. Virol 91, e01089–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karen KA and Hearing P (2011) Adenovirus core protein VII protects the viral genome from a DNA damage response at early times after infection. J Virol 85, 4135–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Komatsu T, Quentin-Froignant C, Carlon-Andres I, Lagadec F, Rayne F, Ragues J, Kehlenbach RH, Zhang W, Ehrhardt A, Bystricky K et al. (2018) In vivo labelling of adenovirus DNA identifies chromatin anchoring and biphasic genome replication. J Virol 92, e00795–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cao TM and Sung MT (1982) A protamine-like domain in basic adenovirus core protein. Biochem Biophys Res Comm 108, 1061–1066. [DOI] [PubMed] [Google Scholar]
- 61.Sung MT, Cao TM, Coleman RT and Budelier KA (1983) Gene and protein sequences of adenovirus protein VII, a hybrid basic chromosomal protein. Proc Natl Acad Sci USA 80, 2902–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wykes SM and Krawetz SA (2003) The structural organization of sperm chromatin. J Biol Chem 278, 29471–29477. [DOI] [PubMed] [Google Scholar]
- 63.Wykes SM, Nelson JE, Visscher DW, Djakiew D and Krawetz SA (1995) Coordinate expression of the PRM1, PRM2, and TNP2 multigene locus in human testis. DNA Cell Biol 14, 155–161. [DOI] [PubMed] [Google Scholar]
- 64.Biegeleisen K (2006) The probable structure of the protamine-DNA complex. J Theor Biol 241, 533–540. [DOI] [PubMed] [Google Scholar]
- 65.Montellier E, Boussouar F, Rousseaux S, Zhang K, Buchou T, Fenaille F, Shiota H, Debernardi A, Héry P, Curtet S et al. (2013) Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev 27, 1680–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Avgousti DC, Herrmann C, Kulej K, Pancholi NJ, Sekulic N, Petrescu J, Molden RC, Blumenthal D, Paris AJ, Reyes ED et al. (2016) A core viral protein binds host nucleosomes to sequester immune danger signals. Nature 535, 173–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fedor MJ and Daniell E (1980) Acetylation of histone-like proteins of adenovirus type 5. J Virol 35, 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Inturi R, Thaduri S and Punga T (2013) Adenovirus precursor pVII protein stability is regulated by its propeptide sequence. PLoS ONE 8, e80617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Herrmann C, Avgousti DC and Weitzman MD (2017) Differential salt fractionation of nuclei to analyze chromatin-associated proteins from cultured mammalian cells. Bio Protoc 7, e2175 https://www.ncbi.nlm.nih.gov/pubmed/28845440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gyurcsik B, Haruki H, Takahashi T, Mihara H and Nagata K (2006) Binding modes of the precursor of adenovirus major core protein VII to DNA and template activating factor I: implication for the mechanism of remodeling of the adenovirus chromatin. Biochemistry 45, 303–313. [DOI] [PubMed] [Google Scholar]
- 71.Stros M (2010) HMGB proteins: interactions with DNA and chromatin. Biochem Biophys Acta 1799, 101–113. [DOI] [PubMed] [Google Scholar]
- 72.Kang R, Chen R, Zhang Q, Hou W, Wu S, Cao L, Huang J, Yu Y, Fan X-G, Yan Z et al. (2014) HMGB1 in health and disease. Mol Aspects Med 40, 1–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang H, Antoine DJ, Andersson U and Tracey KJ (2013) The many faces of HMGB1: molecular structure-functional activity in inflammation, apoptosis, and chemotaxis. J Leukoc Biol 93, 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang H, Wang H, Chavan SS and Andersson U (2015) High Mobility Group Box Protein 1 (HMGB1): The Prototypical Endogenous Danger Molecule. Mol Med 21 (Suppl 1), S6–S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yang H, Wang H, Czura CJ and Tracey KJ (2002) HMGB1 as a cytokine and therapeutic target. J Endotoxin Res 8, 469–472. [DOI] [PubMed] [Google Scholar]
- 76.Agresti A, Lupo R, Bianchi ME and Müller S (2003) HMGB1 interacts differentially with members of the Rel family of transcription factors. Biochem Biophys Res Comm 302, 421–426. [DOI] [PubMed] [Google Scholar]
- 77.Rowell JP, Simpson KL, Stott K, Watson M and Thomas JO. (2012) HMGB1-facilitated p53 DNA binding occurs via HMG-Box/p53 transactivation domain interaction, regulated by the acidic tail. Structure 20, 2014–2024. [DOI] [PubMed] [Google Scholar]
- 78.Robinson CM, Singh G, Lee JY, Dehghan S, Rajaiya J, Liu EB, Yousuf MA, Betensky RA, Jones MS, Dyer DW et al. (2013) Molecular evolution of human adenoviruses. Sci Rep 3, 1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Davison AJ (2003) Genetic content and evolution of adenoviruses. J Gen Virol 84, 2895–2908. [DOI] [PubMed] [Google Scholar]
- 80.Nevins JR, Ginsberg HS, Blanchard JM, Wilson MC and Darnell JE (1979) Regulation of the primary expression of the early adenovirus transcription units. J Virol 32, 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lewis JB and Mathews MB (1980) Control of adenovirus early gene expression: a class of immediate early products. Cell 21, 303–313. [DOI] [PubMed] [Google Scholar]
- 82.Berk AJ and Sharp PA (1978) Structure of the adenovirus 2 early mRNAs. Cell 14, 695–711. [DOI] [PubMed] [Google Scholar]
- 83.King CR, Zhang A, Tessier TM, Gameiro SF and Mymryk JS (2018) Hacking the cell: network intrusion and exploitation by adenovirus E1A. mBio 9, e00390–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Frisch SM and Mymryk JS (2002) Adenovirus-5 E1A: paradox and paradigm. Nat Rev Mol Cell Biol 3, 441–452. [DOI] [PubMed] [Google Scholar]
- 85.Ferrari R, Berk AJ and Kurdistani SK (2009) Viral manipulation of the host epigenome for oncogenic transformation. Nat Rev Genet 10, 290–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen J, Morral N and Engel DA (2007) Transcription releases protein VII from adenovirus chromatin. Virology 369, 411–422. [DOI] [PubMed] [Google Scholar]
- 87.Gawad C, Koh W and Quake SR (2016) Single-cell genome sequencing: current state of the science. Nat Rev Genet 17, 175–188. [DOI] [PubMed] [Google Scholar]
- 88.Hwang B, Lee JH and Bang D (2018) Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 50, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ferrari R, Pellegrini M, Horwitz GA, Xie W, Berk AJ and Kurdistani SK (2008) Epigenetic reprogramming by adenovirus e1a. Science 321, 1086–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Montell C, Courtois G, Eng C and Berk A (1984) Complete transformation by adenovirus 2 requires both E1A proteins. Cell 36, 951–961. [DOI] [PubMed] [Google Scholar]
- 91.Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK and Berk AJ (2008) Adenovirus small e1a alters global patterns of histone modification. Science 321, 1084–1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Struhl K (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev 12, 599–606. [DOI] [PubMed] [Google Scholar]
- 93.Eberharter A and Becker P (2002) Histone acetylation: a switch between repressive and permissive chromatin: second in review series on chromatin dynamics. EMBO Rep 3, 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Calo E and Wysocka J (2013) Modification of enhancer chromatin: what, how, and why? Mol Cell 49, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Eckner R, Ewen ME, Newsome D, Gerdes M, DeCaprio JA, Lawrence JB and Livingston DM (1994) Molecular cloning and functional analysis of the adenovirus E1A-associated 300-kD protein (p300) reveals a protein with properties of a transcriptional adaptor. Genes Dev 8, 869–884. [DOI] [PubMed] [Google Scholar]
- 96.Arany Z, Newsome D, Oldread E, Livingston DM and Eckner R (1995) A family of transcriptional adaptor proteins targeted by the E1A oncoprotein. Nature 374, 81–84. [DOI] [PubMed] [Google Scholar]
- 97.Chakravarti D, Ogryzko V, Kao H-Y, Nash A, Chen H, Nakatani Y and Evans RM (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell 96, 393–403. [DOI] [PubMed] [Google Scholar]
- 98.Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu H-Y, Wang JYJ, Nakatani Y and Kedes L (1999) Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 96, 405–413. [DOI] [PubMed] [Google Scholar]
- 99.O’Connor MJ, Zimmermann H, Nielsen S, Bernard HU and Kouzarides T (1999) Characterization of an E1A-CBP interaction defines a novel transcriptional adapter motif (TRAM) in CBP/p300. J Virol 73, 3574–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ferrari R, Su T, Li B, Bonora G, Oberai A, Chan Y, Sasidharan R, Berk AJ, Pellegrini M and Kurdistani SK (2012) Reorganization of the host epigenome by a viral oncogene. Genome Res 22, 1212–1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ferrari R, Gou D, Jawdekar G, Johnson SA, Nava M, Su T, Yousef AF, Zemke NR, Pellegrini M, Kurdistani SK et al. (2014) Adenovirus small E1A employs the lysine acetylases p300/CBP and tumor suppressor Rb to repress select host genes and promote productive virus infection. Cell Host Microbe 16, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chan HM and La Thangue NB (2001) p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J Cell Sci 114, 2363–73. [DOI] [PubMed] [Google Scholar]
- 103.Fonseca GJ, Thillainadesan G, Yousef AF, Ablack JN, Mossman KL, Torchia J and Mymryk JS (2012) Adenovirus evasion of interferon-mediated innate immunity by direct antagonism of a cellular histone posttranslational modification. Cell Host Microbe 11, 597–606. [DOI] [PubMed] [Google Scholar]
- 104.Fonseca GJ, Cohen MJ, Nichols AC, Barrett JW and Mymryk JS (2013) Viral retasking of hBre1/RNF20 to recruit hPaf1 for transcriptional activation. PLoS Pathog 9, e1003411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van Oss SB, Cucinotta CE and Arndt KM (2017) Emerging insights into the roles of the Paf1 complex in gene regulation. Trends Biochem Sci 42, 788–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Fonseca GJ, Cohen MJ and Mymryk JS (2014) Adenovirus E1A recruits the hPaf1 complex to enhance transcriptional elongation. J Virol 88, 5630–5637. https://www.ncbi.nlm.nih.gov/pubmed/24600005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kulej K, Avgousti DC, Weitzman MD and Garcia BA (2015) Characterization of histone post-translational modifications during virus infection using mass spectrometry-based proteomics. Methods 90, 8–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Croce CM (2008) Oncogenes and cancer. N Engl J Med 358, 502–511. [DOI] [PubMed] [Google Scholar]
- 109.Zheng Z-M (2010) Viral oncogenes, noncoding RNAs, and RNA splicing in human tumor viruses. Int J Biol Sci 6, 730–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Graham FL, Smiley J, Russell WC and Nairn R (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol 36, 59–72. [DOI] [PubMed] [Google Scholar]
- 111.Howe JA, Mymryk JS, Egan C, Branton PE and Bayley ST (1990) Retinoblastoma growth suppressor and a 300-kDa protein appear to regulate cellular DNA synthesis. Proc Natl Acad Sci USA 87, 5883–5887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weinberg RA (1995) The retinoblastoma protein and cell cycle control. Cell 81, 323–330. [DOI] [PubMed] [Google Scholar]
- 113.Giacinti C and Giordano A (2006) RB and cell cycle progression. Oncogene 25, 5220–5227. [DOI] [PubMed] [Google Scholar]
- 114.Whyte P, Buchkovich K, Horowitz J, Friend S, Raybuck M, Weinberg R and Harlow E (1988) Association between an oncogene and an anti-oncogene: the adenovirus E1A proteins bind to the retinoblastoma gene product. Nature 334, 124–129. [DOI] [PubMed] [Google Scholar]
- 115.Zemke NR, Gou D and Berk AJ (2019) Dedifferentiation by adenovirus E1A due to inactivation of Hippo pathway effectors YAP and TAZ. Genes Dev 33, 828–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yu F-X and Guan K-L (2013) The Hippo pathway: regulators and regulations. Genes Dev 27, 355–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang C-Y, Chinnaiyan AM et al. (2008) TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22, 1962–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Levine AJ and Oren M (2009) The first 30 years of p53: growing ever more complex. Nat Rev Cancer 9, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huang H, Lin S, Garcia BA and Zhao Y (2015) Quantitative proteomic analysis of histone modifications. Chem Rev 115, 2376–2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dekker J (2016) Mapping the 3D genome: aiming for consilience. Nat Rev Mol Cell Biol 17, 741–742. [DOI] [PubMed] [Google Scholar]
- 121.Skene PJ and Henikoff S (2017) An efficient targeted nuclease strategy for high-resolution mapping of DNA binding sites. elife 6, e21856 https://www.ncbi.nlm.nih.gov/pubmed/28079019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Freyer GA, Katoh Y and Roberts RJ (1984) Characterization of the major mRNAs from adenovirus 2 early region 4 by cDNA cloning and sequencing. Nucleic Acids Res 12, 3503–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Virtanen A, Gilardi P, N€aslund A, LeMoullec JM, Pettersson U and Perricaudet M (1984) mRNAs from human adenovirus 2 early region 4. J Virol 51, 822–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Halbert DN, Cutt JR and Shenk T (1985) Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol 56, 250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Huang M-M and Hearing P (1989) Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J Virol 63, 2605–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Weiden MD and Ginsberg HS (1994) Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc Natl Acad Sci USA 91, 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Leppard KN (1997) E4 gene function in adenovirus, adenovirus vector and adeno-associated virus infections. J Gen Virol 78, 2131–2138. [DOI] [PubMed] [Google Scholar]
- 128.Täuber B and Dobner T (2001) Molecular regulation and biological function of adenovirus early genes: the E4 ORFs. Gene 278, 1–23. [DOI] [PubMed] [Google Scholar]
- 129.Dobner T, Horikoshi N, Rubenwolf S and Shenk T (1996) Blockage by adenovirus E4orf6 of transcriptional activation by the p53 tumor suppressor. Science 272, 1470–1473. [DOI] [PubMed] [Google Scholar]
- 130.Steegenga WT, Riteco N, Jochemsen AG, Fallaux FJ and Bos JL (1998) The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene 16, 349–357. [DOI] [PubMed] [Google Scholar]
- 131.Cathomen T and Weitzman MD (2000) A functional complex of adenovirus proteins E1B–55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J Virol 74, 11407–11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hidalgo P and Gonzalez RA (2019) Formation of adenovirus DNA replication compartments. FEBS Lett. 10.1002/1873-3468.13672. [DOI] [PubMed] [Google Scholar]
- 133.Lallemand-Breitenbach V and de Thé H (2018) PML nuclear bodies: from architecture to function. Curr Opin Cell Biol 52, 154–161. [DOI] [PubMed] [Google Scholar]
- 134.Carvalho T (1995) Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix- associated PML bodies. J Cell Biol 131, 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Doucas V, Ishov AM, Romo A, Juguilon H, Weitzman MD, Evans RM and Maul GG (1996) Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev 10, 196–207. [DOI] [PubMed] [Google Scholar]
- 136.Ou HD, Kwiatkowski W, Deerinck TJ, Noske A, Blain KY, Land HS, Soria C, Powers CJ, May AP, Shu X et al. (2012) A structural basis for the assembly and functions of a viral polymer that inactivates multiple tumor suppressors. Cell 151, 304–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Stracker TH, Carson CT and Weitzman MD (2002) Adenovirus oncoproteins inactivate the Mre11–Rad50–NBS1 DNA repair complex. Nature 418, 348–352. [DOI] [PubMed] [Google Scholar]
- 138.Lamarche BJ, Orazio NI and Weitzman MD (2010) The MRN complex in double-strand break repair and telomere maintenance. FEBS Lett 584, 3682–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stracker TH, Lee DV, Carson CT, Araujo FD, Ornelles DA and Weitzman MD (2005) Serotype-specific reorganization of the Mre11 complex by adenoviral E4orf3 proteins. J Virol 79, 6664–6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Pancholi NJ and Weitzman MD (2018) Serotype-specific restriction of wild-type adenoviruses by the cellular Mre11-Rad50-Nbs1 complex. Virology 518, 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Reyes ED, Kulej K, Pancholi NJ, Akhtar LN, Avgousti DC, Kim ET, Bricker DK, Spruce LA, Koniski SA, Seeholzer SH et al. (2017) Identifying host factors associated with DNA replicated during virus infection. Mol Cell Proteomics 16, 2079–2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Soria C, Estermann FE, Espantman KC and O’Shea CC (2010) Heterochromatin silencing of p53 target genes by a small viral protein. Nature 466, 1076–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Soriano AM, Crisostomo L, Mendez M, Graves D, Frost JR, Olanubi O, Whyte PF, Hearing P and Pelka P (2019) Adenovirus 5 E1A interacts with E4orf3 to regulate viral chromatin organization. J Virol 93, e00157–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Schreiner S, Wimmer P, Sirma H, Everett RD, Blanchette P, Groitl P and Dobner T (2010) Proteasome-dependent degradation of Daxx by the viral E1B–55K protein in human adenovirus-infected cells. J Virol 84, 7029–7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schreiner S, Bürck C, Glass M, Groitl P, Wimmer P, Kinkley S, Mund A, Everett RD and Dobner T (2013) Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res 41, 3532–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Berscheminski J, Brun J, Speiseder T, Wimmer P, Ip WH, Terzic M, Dobner T and Schreiner S (2016) Sp100A is a tumor suppressor that activates p53-dependent transcription and counteracts E1A/E1B-55K-mediated transformation. Oncogene 35, 3178–3189. [DOI] [PubMed] [Google Scholar]
- 147.Schreiner S, Kinkley S, Bürck C, Mund A, Wimmer P, Schubert T, Groitl P, Will H and Dobner T (2013) SPOC1-mediated antiviral host cell response is antagonized early in human adenovirus type 5 infection. PLoS Pathog 9, e1003775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Voon HPJ and Wong LH (2016) New players in heterochromatin silencing: histone variant H3.3 and the ATRX/DAXX chaperone. Nucleic Acids Res 44, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schreiner S and Wodrich H (2013) Virion factors that target Daxx to overcome intrinsic immunity. J Virol 87, 10412–10422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Durepaire N, Rogez J, Verdier M, Rogez S, Weinbreck P and Denis F (1997) Detection of adenovirus DNA by polymerase chain reaction in peripheral blood lymphocytes from HIV-infected patients and a control group: preliminary results. J Acquir Immune Defic Syndr Hum Retrovirol 1, 189–90. [DOI] [PubMed] [Google Scholar]
- 151.Flomenberg P, Gutierrez E, Piaskowski V and Casper JT (1997) Detection of adenovirus DNA in peripheral blood mononuclear cells by polymerase chain reaction assay. J Med Virol 51, 182–188. [DOI] [PubMed] [Google Scholar]
- 152.Shike H, Shimizu C, Kanegaye J, Foley JL and Burns JC (2005) Quantitation of adenovirus genome during acute infection in normal children. Pediatr Infect Dis J 24, 29–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Garnett CT, Erdman D, Xu W and Gooding LR (2002) Prevalence and quantitation of species C adenovirus DNA in human mucosal lymphocytes. J Virol 76, 10608–10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Garnett CT, Talekar G, Mahr JA, Huang W, Zhang Y, Ornelles DA and Gooding LR (2009) Latent species C adenoviruses in human tonsil tissues. J Virol 83, 2417–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Proenca-Modena JL, Valera FCP, Jacob MG, Buzatto GP, Saturno TH, Lopes L, Souza JM, Paula FE, Silva ML, Carenzi LR et al. (2012) High rates of detection of respiratory viruses in tonsillar tissues from children with chronic adenotonsillar disease. PLoS ONE 7, e42136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Roy S, Calcedo R, Medina-Jaszek A, Keough M, Peng Hand Wilson JM (2011) Adenoviruses in lymphocytes of the human gastro-intestinal tract. PLoS One 6, e24859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kosulin K, Geiger E, Vécsei A, Huber W-D, Rauch M, Brenner E, Wrba F, Hammer K, Innerhofer A, Pötschger U et al. (2016) Persistence and reactivation of human adenoviruses in the gastrointestinal tract. Clin Microbiol Infect 22, 381.e1–e8. [DOI] [PubMed] [Google Scholar]
- 158.Kosulin K (2019) Intestinal HAdV infection: tissue specificity, persistence, and implications for antiviral therapy. Viruses 11, 804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Fox JP, Hall CE and Cooney MK (1977) The Seattle virus watch. VII. Observations of adenovirus infections. Am J Epidemiol 105, 362–386. [DOI] [PubMed] [Google Scholar]
- 160.Kosulin K, Berkowitsch B, Matthes S, Pichler H, Lawitschka A, Pötschger U, Fritsch G and Lion T (2018) Intestinal adenovirus shedding before allogeneic stem cell transplantation is a risk factor for invasive infection post-transplant. EBioMedicine 28, 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Doerfler W (2009) Epigenetic mechanisms in human adenovirus type 12 oncogenesis. Semin Cancer Biol 19, 136–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Murali VK, Ornelles DA, Gooding LR, Wilms HT, Huang W, Tollefson AE, Wold WSM and Garnett-Benson C (2014) Adenovirus death protein (ADP) is required for lytic infection of human lymphocytes. J Virol 88, 903–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Furuse Y, Ornelles DA and Cullen BR (2013) Persistently adenovirus-infected lymphoid cells express microRNAs derived from the viral VAI and especially VAII RNA. Virology 447, 140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ornelles DA, Gooding LR, Dickherber ML, Policard M and Garnett-Benson C (2016) Limited but durable changes to cellular gene expression in a model of latent adenovirus infection are reflected in childhood leukemic cell lines. Virology 494, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Nevels M, Rubenwolf S, Spruss T, Wolf H and Dobner T (1997) The adenovirus E4orf6 protein can promote E1A/E1B-induced focus formation by interfering with p53 tumor suppressor function. Proc Natl Acad Sci USA 94, 1206–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Nevels M, Täuber B, Kremmer E, Spruss T, Wolf H and Dobner T (1999) Transforming potential of the adenovirus type 5 E4orf3 protein. J Virol 73, 1591–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nevels M, Spruss T, Wolf H and Dobner T (1999) The adenovirus E4orf6 protein contributes to malignant transformation by antagonizing E1A-induced accumulation of the tumor suppressor protein p53. Oncogene 18, 9–17. [DOI] [PubMed] [Google Scholar]
- 168.Nevels M, Tauber B, Spruss T, Wolf H and Dobner T (2001) “Hit-and-run” transformation by adenovirus oncogenes. J Virol 75, 3089–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Niller HH, Wolf H and Minarovits J (2011) Viral hit and run-oncogenesis: genetic and epigenetic scenarios. Cancer Lett 305, 200–217. [DOI] [PubMed] [Google Scholar]
- 170.Gunthert U, Schweiger M, Stupp M and Doerfler W (1976) DNA methylation in adenovirus, adenovirus-transformed cells, and host cells. Proc Natl Acad Sci USA 73, 3923–3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pfeffer A, Schubbert R, Orend G, Hilger-Eversheim K and Doerfler W (1999) Integrated viral genomes can be lost from adenovirus type 12-induced hamster tumor cells in a clone-specific, multistep process with retention of the oncogenic phenotype. Virus Res 59, 113–127. [DOI] [PubMed] [Google Scholar]
- 172.Heller H, K€ammer C, Wilgenbus P and Doerfler W (1995) Chromosomal insertion of foreign (adenovirus type 12, plasmid, or bacteriophage lambda) DNA is associated with enhanced methylation of cellular DNA segments. Proc Natl Acad Sci USA 92, 5515–5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Grafodatskaya D, Choufani S, Ferreira JC, Butcher DT, Lou Y, Zhao C, Scherer SW and Weksberg R (2010) EBV transformation and cell culturing destabilizes DNA methylation in human lymphoblastoid cell lines. Genomics 95, 73–83. [DOI] [PubMed] [Google Scholar]
- 174.Birdwell CE, Queen KJ, Kilgore PCSR, Rollyson P, Trutschl M, Cvek U and Scott RS(2014) Genome-wide DNA methylation as an epigenetic consequence of Epstein-Barr virus infection of immortalized keratinocytes. J Virol 88, 11442–11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Doerfler W (2019) Epigenetic consequences of genome manipulations: caveats for human germline therapy and genetically modified organisms. Epigenomics 11, 247–250. [DOI] [PubMed] [Google Scholar]
- 176.Langner KD, Vardimon L, Renz D and Doerfler W (1984) DNA methylation of three 5’ C-C-G-G 3’ sites in the promoter and 5’ region inactivate the E2a gene of adenovirus type 2. Proc Natl Acad Sci USA 81, 2950–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kruczek I and Doerfler W (1983) Expression of the chloramphenicol acetyltransferase gene in mammalian cells under the control of adenovirus type 12 promoters: effect of promoter methylation on gene expression. Proc Natl Acad Sci USA 80, 7586–7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bruniquel D and Schwartz RH (2003) Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol 4, 235–240. [DOI] [PubMed] [Google Scholar]
- 179.Mahr JA and Gooding LR (1999) Immune evasion by adenoviruses. Immunol Rev 168, 121–130. [DOI] [PubMed] [Google Scholar]
- 180.McNees AL, Garnett CT and Gooding LR (2002) The adenovirus E3 RID complex protects some cultured human T and B lymphocytes from Fas-induced apoptosis. J Virol 76, 9716–9723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Mahr JA, Boss JM and Gooding LR (2003) The adenovirus e3 promoter is sensitive to activation signals in human T cells. J Virol 77, 1112–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Lion T (2019) Adenovirus persistence, reactivation, and clinical management. FEBS Lett. 10.1002/1873-3468.13576. [DOI] [PubMed] [Google Scholar]
- 183.Edwards EA, Pigg N, Courtier J, Zapala MA, MacKenzie JD and Phelps AS (2017) Intussusception: past, present and future. Pediatr Radiol 47, 1101–1108. [DOI] [PubMed] [Google Scholar]
- 184.Ukarapol N, Khamrin P, Khorana J, Singhavejsakul J, Damrongmanee A and Maneekarn N (2016) Adenovirus infection: a potential risk for developing intussusception in pediatric patients. J Med Virol 88, 1930–1935. [DOI] [PubMed] [Google Scholar]
- 185.Jang J, Lee YJ, Kim JS, Chung J-Y, Chang S, Lee K, Choe B-H, Hong SJ, Song JS and Park KY (2017) Epidemiological correlation between fecal adenovirus subgroups and pediatric intussusception in Korea. J Korean Med Sci 32, 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Murrah KA, Turner RL, Pang B, Perez AC, Reimche JL, King LB, Wren J, Gandhi U, Swords WE and Ornelles DA (2015) Replication of type 5 adenovirus promotes middle ear infection by Streptococcus pneumoniae in the chinchilla model of otitis media. Pathog Dis 73, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Suzuki K and Bakaletz LO (1994) Synergistic effect of adenovirus type 1 and nontypeable Haemophilus influenzae in a chinchilla model of experimental otitis media. Infect Immun 62, 1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Miyamoto N and Bakaletz LO (1997) Kinetics of the ascension of NTHi from the nasopharynx to the middle ear coincident with adenovirus-induced compromise in the chinchilla. Microb Pathog 23, 119–126. [DOI] [PubMed] [Google Scholar]
- 189.Buzatto GP, Tamashiro E, Proenca-Modena JL, Saturno TH, Prates MC, Gagliardi TB, Carenzi LR, Massuda ET, Hyppolito MA, Valera FCP et al. (2017) The pathogens profile in children with otitis media with effusion and adenoid hypertrophy. PLoS ONE 12, e0171049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Pettigrew MM, Gent JF, Pyles RB, Miller AL, Nokso-Koivisto J and Chonmaitree T (2011) Viral-bacterial interactions and risk of acute otitis media complicating upper respiratory tract infection. J Clin Microbiol 49, 3750–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ornelles DA, Gooding LR and Garnett-Benson C (2015) Neonatal infection with species C adenoviruses confirmed in viable cord blood lymphocytes. PLoS ONE 10, e0119256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Erdel F and Rippe K (2018) Formation of chromatin subcompartments by phase separation. Biophys J 114, 2262–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Larson AG, Elnatan D, Keenen MM, Trnka MJ, Johnston JB, Burlingame AL, Agard DA, Redding S and Narlikar GJ (2017) Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547, 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Strom AR, Emelyanov AV, Mir M, Fyodorov DV, Darzacq X and Karpen GH (2017) Phase separation drives heterochromatin domain formation. Nature 547, 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Buenrostro JD, Giresi PG, Zaba LC, Chang HY and Greenleaf WJ (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, Mirny LA, O’Shea CC, Park PJ, Ren B et al. (2017) The 4D nucleome project. Nature 549, 219–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Ricci MA, Manzo C, García-Parajo MF, Lakadamyali M and Cosma MP (2015) Chromatin fibers are formed by heterogeneous groups of nucleosomes in vivo. Cell 160, 1145–1158. [DOI] [PubMed] [Google Scholar]
- 198.Ong C-T and Corces VG (2014) CTCF: an architectural protein bridging genome topology and function. Nat Rev Genet 15, 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Amelio AL, McAnany PK and Bloom DC (2006) A chromatin insulator-like element in the herpes simplex virus type 1 latency-associated transcript region binds CCCTC-binding factor and displays enhancer-blocking and silencing activities. J Virol 80, 2358–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Komatsu T, Sekiya T and Nagata K (2013) DNA replication-dependent binding of CTCF plays a critical role in adenovirus genome functions. Sci Rep 3, 2187. [DOI] [PMC free article] [PubMed] [Google Scholar]