Abstract
The bacterial and fungal communities from the olive (Olea europaea L.) root systems have not yet been simultaneously studied. We show in this work that microbial communities from the olive root endosphere are less diverse than those from the rhizosphere. But more relevant was to unveil that olive belowground communities are mainly shaped by the genotype of the cultivar when growing under the same environmental, pedological and agronomic conditions. Furthermore, Actinophytocola, Streptomyces and Pseudonocardia are the most abundant bacterial genera in the olive root endosphere, Actinophytocola being the most prevalent genus by far. In contrast, Gp6, Gp4, Rhizobium and Sphingomonas are the main genera in the olive rhizosphere. Canalisporium, Aspergillus, Minimelanolocus and Macrophomina are the main fungal genera present in the olive root system. Interestingly enough, a large number of as yet unclassified fungal sequences (class level) were detected in the rhizosphere. From the belowground microbial profiles here reported, it can be concluded that the genus Actinophytocola may play an important role in olive adaptation to environmental stresses. Moreover, the huge unknown fungal diversity here uncovered suggests that fungi with important ecological function and biotechnological potential are yet to be identified.
Subject terms: Microbiome, Agroecology
Introduction
The cultivated olive (Olea europaea L. subsp. europaea var. europaea) is not only one of the oldest domesticated trees1, but also constitutes one of the most important and outstanding agro-ecosystems in the Mediterranean Basin, shaped along millennia2. In this area, there is an olive belt with more than 10 million ha in countries of the coastal regions, accounting for nearly 80% of the olive cultivation area worldwide3. In some of these countries such as Spain, the world’s largest olive oil and table olive producer, this woody crop has undisputable social, economic and agro-ecological relevance4. In addition to its ecological and social importance, the main product obtained from this iconic tree (the virgin olive oil), has a number of health and nutritional benefits so that its consumption is increasing worldwide5.
Olive cultivation is threatened by several abiotic (for example soil erosion) and biotic (attacks from insects, nematodes and pathogenic microbes) constraints. Among relevant phytopathogens present in the soil microbiota affecting olive health, representatives of the Oomycota class (for example Phytophthora spp.) as well as higher fungi (for instance Verticillium dahliae Kleb.) must be highlighted2,6–8. In addition to the traditional and well-known microbiological menaces affecting olive crop (for example, anthracnose [caused by Colletotrichum spp.], Verticillium wilt [VWO, V. dahliae], peacock spot [Spilocea oleagina (Cast.) Hughes.], or knot disease [Pseudomonas savastanoi pv. savastanoi Smith.])9–12, emerging diseases like the olive quick decline syndrome caused by Xylella fastidiosa Wells. ssp. pauca must be considered13. In addition to new threats, some reports warn on the increase in pathogen and arthropod attacks as a consequence of changing from traditional olive cropping systems to high-density tree orchards. However, the impact of high-density olive groves on, for instance, soil-borne diseases has not been yet studied14. Another important menace to take into account is climate change, which is expected to affect the incidence and severity of olive diseases6. Finally, the reduction in the number of olive cultivars due to either commercial (for example improved yield, etc.) or phytopathological (as tolerance to diseases) reasons, a trend observed in many areas, will eventually lessen olive genetic diversity. All these factors may have a profound, yet not evaluated impact on the composition, structure and functioning of belowground microbial communities8.
A comprehensive knowledge of microbial communities associated to the olive root system, including the root endosphere and the rhizosphere soil, is therefore instrumental to better understand their influence on the development, health and fitness of this tree. A priori, the vast majority of the olive-associated microbiota must be composed of microorganisms providing either neutral or positive effects to the host. Indeed, recent literature provides solid evidence that olive roots are a good reservoir of beneficial microorganisms, including effective biocontrol agents (BCA)15–18. Among the beneficial components of the plant-associated microbiota, endophytic bacteria and fungi are of particular interest to develop novel biotechnological tools aiming to enhance plant growth promotion and/or control of plant diseases. Moreover, microorganisms able to colonize and endure within the plant tissue pose the additional advantage to be adapted to the specific microhabitat/niche where they can provide their beneficial effects19. Besides endophytes, beneficial components of tree root-associated microbiota colonizing the rhizoplane and/or the rhizosphere soil can also directly promote plant growth (as bio-fertilization, phyto-stimulation) or alleviate stress caused by either abiotic (for example environmental pollutants, drought, salinity resistance) or biotic (see above) constraints8.
Our knowledge on olive-associated microbiota is still very scarce and fragmentary. So far, bacterial communities associated with wild olive (Olea europaea L. subsp. europaea var. sylvestris) roots (endo- and rhizosphere) have been studied using fluorescent terminal restriction fragment length polymorphism (FT-RFLP) as well as by bacteria isolation in culturing media15. Endophytic fungi from the phyllosphere and roots of the olive cultivar (cv.) Cobrançosa have been also compared using a culture-dependent approach20. Microbial communities of the olive phyllosphere and carposphere have been analyzed using denaturing gradient gel electrophoresis (DGGE)21, isolation of fungi in culturing media22 and high-throughput sequencing of both fungal23 and prokaryotic24 communities.
In this study we aim, for the first time, to unravel the composition and structure of belowground prokaryotic and fungal communities of cultivated olive by high-throughput sequencing. A core collection of olive cultivars (36 originating from 9 different countries, Table 1) present at the World Olive Germplasm Collection (WOGC; Córdoba, Spain) and representative of enough genetic diversity within the Mediterranean Basin have been analyzed when grown under the same climatic, pedological and agronomic conditions. The following objectives were pursued: (a) to perform an in-depth study of the belowground microbial communities (root endosphere and rhizosphere) in a wide range of olive genotypes; (b) to assess what is/are the determinant factor(s) contributing to build up such communities; (c) to establish the core and accessory microbiota of the olive rhizosphere and root endosphere. The hypothesis to-be-tested is that under specific agro-climatic and edaphic conditions the olive genotype is the key factor for building up belowground microbial communities. Moreover, we also questioned whether this factor can play a more crucial role in the root endosphere than in the rhizosphere.
Table 1.
The 36 olive cultivars sampled in the World Olive Germplasm Collection (WOGC).
| Cultivar | Country | Sample |
|---|---|---|
| Klon-14-1812 | Albania | 7 |
| Chemlal de Kabylie | Algeria | 8 |
| Kalamon | Greece | 15 |
| Koroneiki | Greece | 16 |
| Mastoidis | Greece | 23 |
| Mavreya | Greece | 24 |
| Megaritiki | Greece | 27 |
| Myrtolia | Greece | 30 |
| Shengeh | Iran | 9 |
| Mari | Iran | 22 |
| Barnea | Israel | 5 |
| Frantoio | Italy | 12 |
| Grappolo | Italy | 13 |
| Leccino | Italy | 17 |
| Arbequina | Spain | 4 |
| Forastera de Tortosa | Spain | 11 |
| Llumeta | Spain | 18 |
| Manzanilla de Sevilla | Spain | 20 |
| Manzanillera de Huercal Overa | Spain | 21 |
| Menya | Spain | 28 |
| Morrut | Spain | 29 |
| Picual | Spain | 31 |
| Picudo | Spain | 32 |
| Piñonera | Spain | 33 |
| Temprano | Spain | 34 |
| Verdial de Vélez-Málaga-1 | Spain | 36 |
| Abbadi Abou Gabra-842 | Syria | 1 |
| Abou Satl Mohazam | Syria | 2 |
| Abou Kanani | Syria | 3 |
| Barri | Syria | 6 |
| Jabali | Syria | 14 |
| Maarri | Syria | 19 |
| Majhol-1013 | Syria | 25 |
| Majhol-152 | Syria | 26 |
| Dokkar | Turkey | 10 |
| Uslu | Turkey | 35 |
Results
Microbial communities clustered by compartments (endosphere and rhizosphere), and by olive cultivar in each compartment
From about 37 million raw reads, 1,404,769 (prokaryotic) and 1,005,148 (fungal), and 5,330,385 (prokaryotic) and 912,302 (fungal) good quality reads from the root endosphere and the rhizosphere, respectively, were eventually retained from the 36 olive cultivars here analyzed (Table 1). The smallest samples had 2,061 prokaryotic and 442 fungal sequences (originating from the root endosphere), and the largest ones reached 78,913 prokaryotic and 55,072 fungal sequences (from the rhizosphere in this case) (Tables S1 and S2). After rarefying to the smallest sample, alpha diversity indices showed statistically significant differences between the two compartments (that is to say the endosphere and the rhizosphere), showing the rhizosphere samples the highest richness and diversity values (Fig. S1). Subsequently, both compartments were split and rarefied independently for further alpha diversity analyses to 2,061 (442 in fungi) and 15,565 (665 in fungi) sequences from endosphere and rhizosphere, respectively.
With regard to prokaryotic communities, richness showed significant differences when comparing the root endosphere of olive cultivars, showing just marginal differences in diversity. Considering the rhizosphere, only the diversity showed statistically significant differences among cultivars (Table 2, Fig. S2a,b). Concerning the fungal communities, both richness and diversity indices showed statistically significant differences when comparing olive cultivars for each compartment (Table 2, Fig. S2c,d).
Table 2.
Comparisons of alpha diversity indices in the different microbial communities.
| Prokaryotes | Cultivar | Endosphere vs rhizosphere | |
|---|---|---|---|
| Index | Root endosphere | Rhizosphere | Whole community |
| Sobs | 0.0178 (36.8)* | 0.0500 (49.9) | <2.2e−16 (122.2)* |
| Chao1 | 0.0357 (34.1)* | 0.2117 (41.4) | <2.2e−16 (122.2)* |
| Shannon | 0.0774 (30.8) | 4.6e−05 (77.6)* | <2.2e−16 (122.2)* |
| InvSimpson | 0.0602 (31.9) | 8.5e−05 (83.2)* | <2.2e−16 (122.2)* |
| df | 21 | 35 | 1 |
| Fungi | Cultivar | Endosphere vs rhizosphere | |
| Index | Root endosphere | Rhizosphere | Whole community |
| Sobs | 0.0018 (60.4)* | 0.0096 (57.5)* | <2.2e−16 (147.1)* |
| Chao1 | 0.0133 (52.3)* | 0.0119 (56.6)* | <2.2e−16 (142.5)* |
| Shannon | 0.0014 (61.3)* | 0.0276 (52.8)* | <2.2e−16 (110.9)* |
| InvSimpson | 0.0127 (52.5)* | 0.0593 (48.9) | <2.2e−16 (82.8)* |
| df | 32 | 35 | 1 |
Sobs: Observed richness.
df: degree of freedom.
Asterisk means significant p-values considering a confidence interval of 95%.
In brackets: chi-squared values.
We compared the distribution of the samples from rhizosphere and root endosphere compartments. Results showed significantly different prokaryotic (PERMANOVA R2 0.43; p-value < 0.0001) and fungal (PERMANOVA R2 0.06; p-value < 0.0001) communities (Fig. 1a,b). Regarding to bacterial communities of the root endosphere, the olive cultivar explained 42% of the variation (PERMANOVA R2 0.42; p-value < 0.0001) (Fig. 2). In the rhizosphere, the olive cultivar explained more than 53% of the distribution (PERMANOVA R2 0.53; p-value < 0.0001) (Fig. 3).
Figure 1.
NMDS (Nonmetric MultiDimensional Scaling) of bacterial (a) and fungal (b) communities by compartment. The letters A, B and C after the numbers were used to distinguish the 3 replicates of each cultivar. The different colors indicate the country of origin of the cultivars.
Figure 2.
NMDS (Nonmetric MultiDimensional Scaling) of bacterial communities from rhizosphere. The letters A, B and C after the numbers were used to distinguish the 3 replicates of each cultivar. The different colors indicate the country of origin of the cultivars.
Figure 3.
NMDS (Nonmetric MultiDimensional Scaling) of bacterial communities from root endosphere. The letters A, B and C after the numbers were used to distinguish the 3 replicates of each cultivar. The different colors indicate the country of origin of the cultivars.
Concerning fungal communities, the cultivar explained 39% of the variation in the root endosphere (PERMANOVA R2 0.39; p-value < 0.0001). In the rhizosphere, this factor explained 44% of the variation (PERMANOVA R2 0.44; p-value < 0.0001). Data were not plotted because of the high NMDS stress value (0.22 with 3 dimensions). In both cases, prokaryotes and fungi, the soil physicochemical properties (Table S3) were not relevant for composition or structure of the microbial communities (data not shown).
The olive root endosphere and soil rhizosphere show different prokaryotic taxonomic profiles
Completely different taxonomic profiles at phylum level (class level for Proteobacteria) were obtained when comparing the prokaryotic communities inhabiting the olive root endosphere with those ones present on the rhizosphere (Fig. 4). Despite the fact that universal primers for both prokaryotic kingdoms were used (see Materials and Methods section), no OTU was classified as Archaea. On the one hand, predominant phyla (or class) in the endophytic communities of the 22 olive cultivars examined (see Methods for exclusion criteria) were Actinobacteria, Alphaproteobacteria, Gammaproteobacteria, Bacteroidetes and Deltaproteobacteria, accounting for more than 90% of the sequences. Remarkably, Actinobacteria exceeded 50% in all of them, highlighting cultivars Chemlal de Kabylie, Llumeta and Mavreya (from Algeria, Spain and Greece, respectively) that represented more than 80% of the total number of sequences (Fig. 4a). On the other hand, rhizosphere communities showed more uniform profiles with the phylum Acidobacteria accounting for an average of 27.5% of the sequences in the 36 olive cultivars examined. Acidobacteria was followed by Alphaproteobacteria (18.8%), Actinobacteria (9.8%), Gemmatimonadetes (5.2%) and Betaproteobacteria (4.5%). Overall, the average sum of all of them represented nearly 70% of the total number of sequences (Fig. 4b). In contrast, Gemmatimonadetes and Betaproteobacteria were minor phyla in the olive root endosphere (0.06% and 0.8%, respectively).
Figure 4.
Bacterial phyla (class for Proteobacteria) in the root endosphere (a) and rhizosphere (b). The horizontal colored lines indicate the country of origin of the cultivars.
In the endosphere, and at the genus level, only two genera were significantly differentially represented among the 22 cultivars eventually analyzed: Flavitalea (Bacteroidetes) and Actinophytocola (Actinobacteria). Indeed, Flavitalea was most abundantly represented in cv. Myrtolia but absent in cv. Uslu (Fig. S3a). Conversely, Actinophytocola was highly prevalent in 8 cultivars including Uslu (the highest) and Myrtolia (Fig. S3b). Furthermore, Actinophytocola was the most abundant genus inhabiting the olive root endosphere accounting for an average of 22.1 ± 15.0% of the sequences, followed by Streptomyces (13.2 ± 8.2%), Pseudonocardia (9.4 ± 3.8%), Bradyrhizobium (2.6 ± 1.4%), Ensifer (2.6 ± 6.6%) and Rhizobium (2.0 ± 2.8%). The sum of relative abundances of these six main endophytic genera ranged from 33.3% in cv. Barri (Syria) to 73.1% in cv. Uslu (Turkey) (Fig. S3c).
With regard to rhizosphere soil samples, our results showed that 63 genera were significantly more abundant in the cultivars examined. Moreover, eight out of the eleven (accounting for relative abundances ranging from 49.4% in the Spanish cultivar Temprano to 64% in the Israeli cultivar Barnea; Fig. S4) main rhizosphere genera with relative abundance > 1%, showed statistically significant differences among cultivars (Fig. S4). Three of the most prevalent genera, namely Gp6, Gp4 and Gp7, belong to the main rhizosphere phylum Acidobacteria, but only Gp6 and Gp4 showed significant differences. Belonging to the second most abundant phylum (Proteobacteria), the α-Proteobacteria Rhizobium and Sphingomonas were also relatively highly abundant, both genera showing significant differences among cultivars.
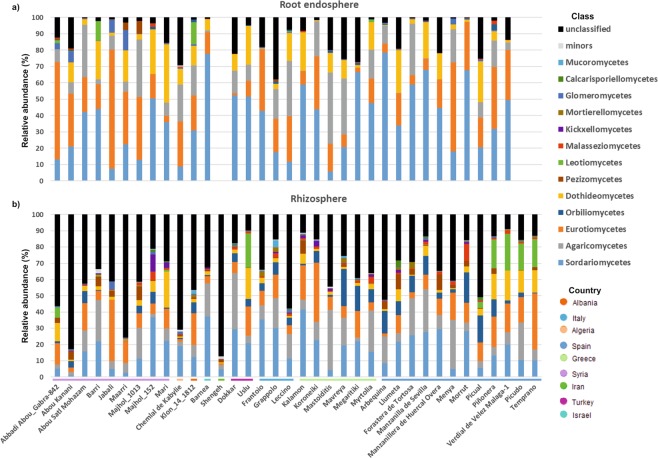
Fungal taxonomic profiles only showed minor differences between the olive root endosphere and the soil rhizosphere
In contrast to prokaryotic communities, fungal communities showed more similar taxonomic profiles at the class level. The main difference between the two compartments was the percentage of sequences that remained unclassified (10.7% in the root endosphere versus 35.4% in the rhizosphere) (Fig. 5). This proportion was very heterogeneous among olive cultivars, Grappolo (Italy) and Chemlal de Kabylie (Algeria) being the two cultivars that harbored more unclassified sequences in the root endosphere (37.8 and 29.4%, respectively), and cultivars Shengeh (Iran) and Abou Kanani (Syria) in the rhizosphere (87.3 and 82.8%, respectively). The prevalent classes present in the olive root endosphere were Sordariomycetes (38.1%), Eurotiomycetes (23%), Agaricomycetes (13.2%) and Dothideomycetes (11.5%), accounting for more than 85% of the sequences obtained from the 33 olive cultivars eventually assessed (see Methods for exclusion criteria) (Fig. 5a). The remaining classes were clearly less relatively abundant, Glomeromycetes being the only one reaching 1%, on average, in all cultivars. Nevertheless, due to the heterogeneity found among the cultivars, this class represented more than 12 and 8% of relative abundance in the Syrian cultivars Maarri and Jabali, respectively.
Figure 5.
Fungal class in the root endosphere (a) and rhizosphere (b). The horizontal colored lines indicate the country of origin of the cultivars.
Regarding to rhizosphere communities, a smaller difference between the prevalent classes and the remaining ones was found in comparison to those found in the root endosphere. Similar to endophytic communities, Sordariomycetes was the predominant class in the rhizosphere (19%), followed by Agaricomycetes (12.9%), Eurotiomycetes (12.2%), Orbiliomycetes (6.5%) and Dothideomycetes (4.9%). While in general Pezizomycetes (2.4%) was more abundant than Leotiomycetes (2.3%), the latter class was exceptionally more abundant in the Spanish cultivars Piñonera, Picudo, Verdial de Velez Málaga-1 and Temprano (relative abundance ranging from 16.5 to 22.6%), and in the Turkish cv. Uslu (21.1%) (Fig. 5b).
Concerning the genus level only five fungal genera with statistically significant differences in relative abundance, namely Scutellinia (Pezizomycetes), Acaulium, Purpureocillium (Sordariomycetes), Entoloma (Agaricomycetes) and Minimelanolocus (Eurotiomycetes), were found in the root endosphere of the 33 olive cultivars examined (Fig. S5a). Ten fungal endophytic genera were found with relative abundance > 1%, accounting for an average proportion of sequences ranging from 24.7% in cv. Grappolo (Italy) to 97.4% in cv. Forastera de Tortosa (Spain) (Fig. S5c). However, due to the high heterogeneity of relative abundances observed for these genera, Minimelanolocus was the only genus showing statistically significant differences.
Seven fungal genera with statistically significant differences in relative abundance, Macrophomina, Polyschema (Dothideomycetes), Minimelanolocus, Spiromastix (Eurotiomycetes), Cunninghamella (Mucoromycetes), Chlorophyllum (Agaricomycetes) and Dichotomopilus (Sordariomycetes), were found in the rhizosphere of the 36 cultivars examined (Fig. S5b). Only Macrophomina and Minimelanolocus showed enough relative abundance to be considered as part of the main fungal rhizosphere genera. On average, Macrophomina was the third most abundant genus in the olive cultivars core collection highlighting the Spanish cultivars Picual, Piñonera, Verdial de Velez Málaga-1, Picudo and Temprano, and cultivar Uslu from Turkey (Fig. S5d).
Defining the belowground core microbiota of olive trees
Regarding bacterial communities, 46 (root endosphere) and 109 (rhizosphere) genera were found in all examined cultivars of each compartment. Furthermore, 40 genera were found in all cultivars and in both compartments. Interestingly, 26 genera had a relative abundance higher than 1% in at least one compartment. The top 10 genera in the core olive root bacteriota were Actinophytocola, Streptomyces, Gp6, Gp4, Pseudonocardia, Rhizobium, Sphingomonas, Gemmatimonas, candidate division WPS-1 and Gp7, accounting for almost 50% of the sequences in each compartment (Tables 3, S4). Finally, all the main bacterial genera found in both compartments (Figs. S3c and S4) were part of the core olive belowground bacteriota.
Table 3.
Main (relative abundance ≥1%) core bacterial and fungal genera.
| Bacterial core | ||
|---|---|---|
| Genus | Root endosphere (%)a | Rhizosphere (%)b |
| Actinophytocola | 22.07 | 0.07 |
| Streptomyces | 13.17 | 0.31 |
| Gp6 | 0.58 | 11.08 |
| Gp4 | 0.26 | 9.31 |
| Pseudonocardia | 9.37 | 0.14 |
| Rhizobium | 2.00 | 7.71 |
| Sphingomonas | 0.77 | 5.92 |
| Gemmatimonas | 0.06 | 5.24 |
| candidate_division_WPS-1d | 0.08 | 3.92 |
| Gp7 | 0.04 | 4.08 |
| Bacillus | 0.68 | 2.31 |
| Bradyrhizobium | 2.57 | 0.20 |
| Ensifer | 2.56 | 0.15 |
| Rubrobacter | 0.05 | 2.48 |
| Subdivision3d | 0.02 | 2.35 |
| Steroidobacter | 1.78 | 0.34 |
| Candidatus_Saccharibacteriad | 1.03 | 0.40 |
| Saccharothrix | 1.18 | 0.07 |
| Ohtaekwangia | 0.21 | 1.03 |
| Mycobacterium | 0.98 | 0.09 |
| Nonomuraea | 1.04 | 0.04 |
| Fungal core | ||
| Genus | Root endosphere (%)c | Rhizosphere (%)b |
| Canalisporium | 29.53 | 6.05 |
| Macrophomina | 10.93 | 2.44 |
| Aspergillus | 1.66 | 3.84 |
| Malassezia | 0.28 | 1.37 |
aRelative abundance average of 22 cultivars.
bRelative abundance average of 36 cultivars.
cRelative abundance average of 33 cultivars.
dName of phylum/class to which this incertae sedis genus belongs.
Regarding fungal communities, only four (root endosphere) and eight (rhizosphere) genera were found in all examined cultivars. Interestingly, the four core endophytic fungal genera were also part of the rhizosphere fungal core. Only five genera had a relative abundance >1% in at least one compartment. The four fungal genera constituting the olive belowground fungal core were Canalisporium, Macrophomina, Aspergillus and Malassezia. They represented more than 40% of the endophytic sequences, but only 13.08% of the rhizosphere sequences. Furthermore, the eight core rhizosphere fungal genera represented only 15.88% of the sequences (Tables 3, S5).
Discussion
Besides the higher alpha diversity (richness and evenness) found in the olive rhizosphere microbiota compared to that in the root endosphere, and the finding that quite different communities were found in each compartment, a common scenario described in several studies25,26, the following major results must be highlighted from our work. Concerning the endosphere, cultivars originating from Syria showed the highest diversity level in contrast to the Turkish cultivars that showed the lowest one. With regard to the rhizosphere, fungal communities of cultivars from Albania and Syria appeared as the most diverse, while the Iranian and Israeli cultivars harbored the least diverse communities. Rhizosphere bacterial communities were not different in richness but showed dissimilar evenness. As observed for fungal communities, cultivars from Iran and Israel were also the least diverse in their rhizosphere bacterial assemblages.
Results here presented are in overall agreement with the major conclusion reported by Müller et al., even considering that these authors focused on aerial organs. Indeed, they concluded that the structure of endophytic prokaryotic communities residing in aboveground tissues was mainly driven by the geographical origin of the olive cultivars evaluated (Eastern: Greece, Syria; Central: France, Italy, Tunisia; and Western Mediterranean: Portugal, Spain, Morocco). However, the main general conclusion from our study, based on a larger number of cultivars, is slightly different. Indeed, the main factor in our study was the genotype (cultivar) rather than the geographical origin. While the geographical origin was a statistically significant factor too, its variation was nested within cultivar variation (data from PERMANOVA test). In addition, more detailed information was obtained in our study. Thus, communities harbored by olive cultivars originating from Greece (in olive green color; see colors and distribution in Figs. 2 and 3) and Spain (in blue) showed more similarities among them than to those from Syrian (in purple) cultivars. Moreover, the Italian (in light blue) cultivar was intermingled between these two clusters and the unique Turkish (in pink) representative tested in our work appeared as distantly related to the Syrian genotypes. Although a distinction among different geographical origins was observed, these clusters did not correspond to a longitudinal gradient (eastern, central, western Mediterranean countries), as reported by Müller et al. Our results indicate that the endophytic and rhizosphere microbial (bacteria and fungi) communities are mainly shaped by the olive genotype. We therefore conclude that the genotype is the main factor shaping olive belowground microbial communities, this factor being more determinant for the rhizosphere than for the endosphere, and more crucial for the bacteriota than for the mycobiota (see PERMANOVA R2 in results).
Proteobacteria has been described as the predominant prokaryotic phylum (about 90% of the relative abundance) present in root endophytic communities26,27. The same was observed for prokaryotic communities of the olive phyllosphere24. However, in our study, Proteobacteria (26% average relative abundance) was clearly overcome by Actinobacteria (64% average relative abundance) in the root endosphere. A similar finding has also been reported in Agave spp., particularly during the dry season25. Interestingly, no sequences belonging to the kingdom Archaea were detected in the root endosphere in our study, in contrast to the results by Müller et al.24 who reported that Archaea was a major group in the olive phyllosphere. In this latter study as well as in ours, the reverse primer used was the same. However, the forward primer used in our study has 94.6% archaeal amplification efficiency28. Archaea representatives were not found in the olive rhizosphere indicating that, without excluding the potential bias introduced by the primer pair here used, this kingdom is poorly represented in the olive belowground microbiota at least at the sampling time and under environmental conditions in which olive trees are cultivated in the WOGC.
The olive-associated microbiota harbors an important reservoir of beneficial microorganisms that can be used as plant growth promotion and/or biocontrol tools15,24. Moreover, bacterial antagonists of olive pathogens isolated from the olive root endosphere or the rhizosphere have the advantage to be adapted to the ecological niche where they can potentially exert their beneficial effect18. For instance, Proteobacteria and Firmicutes representatives, usually found as natural inhabitants from the olive rhizosphere, are thus good examples of effective antagonists against V. dahliae16–18,29. Besides, representatives of these phyla such as the genera Pseudomonas and Bacillus are easy to isolate, manipulate, propagate and formulate as BCAs. In addition to these well-known genera, species of the actinobacterial genus Strepytomyces have also been demonstrated as excellent BCAs in different pathosystems30,31. Moreover, the potential biocontrol of non-streptomycete Actinobacteria genera has been reported as well30,32–34. Taking into account that the prevalence of Actinobacteria found in our study (the genera Actinophytocola, Streptomyces and Pseudonocardia ranged from 30 to 60% of the bacterial olive root endophytic community), the isolation and in-depth characterization of culturable representatives of these genera will be of interest for their assessment as potential PGPR (Plant Growth Promoting Rhizobacteria) and/or BCA against olive tree pathogens. The genus Actinophytocola, described for the first time in 201035 as a root endophytic actinobacteria (Pseudonocardiaceae family), has been isolated from Saharan non-rhizosphere soils in the south of Algeria34. Interestingly enough, these authors demonstrated its antimicrobial ability against some bacteria and fungi. Actinophytocola sp., in addition to other actinomycetes, has also been demonstrated to inhibit the growth of well-known human pathogens (B. subtilis and S. aureus)36. Finally, Actinophytocola gilvus was recently isolated from extremely dry conditions, from a soil crusts sample collected in the Tengger Desert in China37. Considering that this genus was ubiquitously and abundantly found in our study, Actinophytocola spp. inhabiting olive roots can be relevant for olive fitness and health (that is to say drought tolerance, broad antimicrobial activity range, etc.), what grants further research efforts aiming to isolate and characterize members of this relevant component of the olive belowground microbiome.
This is the first study in which a high-throughput sequencing approach has been implemented to unravel the olive belowground fungal communities. Sordariomycetes (38%) and Eurotiomycetes (23%), both belonging to Ascomycota, were found as the most abundant fungal classes in the root endosphere of olive. Sordariomycetes was previously found as the main endophytic fungal class in olive roots using a culture-dependent approach20. The endophytic fungal communities earlier found in aboveground olive tree compartments (phyllosphere and carposphere) by high-throughput sequencing23 or a culture-dependent approach20, differed from belowground communities reported in our study. This outcome reinforces previous reports showing important differences between above- and belowground olive fungal communities, irrespective the methodological approach implemented20,22,23. Interestingly enough, Sordariomycetes was also the most abundant class found in olive fruits regardless the presence or not of anthracnose symptoms23, pointing to the fact that this fungal class seems to be ubiquitously colonizing the interior of olive tissues. In the rhizosphere, Agaricomycetes (12.7%), belonging to Basidiomycota, and Eurotiomycetes (12.6%) were the most abundant classes. At this taxonomic level, the main difference between the two compartments was the percentage of unclassified sequences (12.4% in the root endosphere and 35.6% in the rhizosphere). Furthermore, in the particular case of cvs. Abou Kanani and Shengeh, unclassified sequences represented more than 80% of the good quality sequences found in the rhizosphere. The high percentage of unclassified sequences in this compartment seems to be a common finding when using the same fungal database38, less pronounced in annual plants though39. According to data here obtained, the olive rhizosphere carries a huge fungal diversity yet to be discovered. It is worth mentioning that many of the unclassified sequences here reported may likely correspond to inaccurate identification due to limitations in the method and/or errors in the currently-available fungal database rather than to the actual presence of unidentified fungi. In this sense, we cannot discard that some of these sequences can belong to Glomeromycetes, besides the identified in this work since it is known that olive trees are colonized by AM fungi. Notwithstanding, this may have important ecological implications for the tree, and pose novel agro-biotechnological avenues to be explored.
At the genus level, the structure and composition of olive belowground fungal communities also showed important differences compared to previous reports. For instance, in the particular case of phytopathogenic fungi, Phomopsis columnaris (fungus species causing twig dieback of Vaccinium vitis-idaea [lingonberry])40 and Fusarium oxyporum were found by Martins et al. as the most relative abundant species, although sampled trees did not show visible symptoms. In our study, however, the above-mentioned species/genera were absent. However, the pathogenic fungi Macrophomina phaseolina showed relevant relative abundance in several cultivars and for both compartments. Macrophina phaseolina is a well-known pathogen causing charcoal rot in important crops including olive16,41–44, and it has also been shown that olive leaves produce compounds able to reduce its pathogenic activity45. This finding raises the possibility that M. phaseolina could be a common component of the olive-associated microbiota, but may reside within olive tissues without causing visible symptoms until external factors and/or microbiota alterations (dysbiosis) trigger a pathogenic stage. With regard to relevant soil-borne olive pathogens, it is worth mentioning that neither sequences corresponding to Verticillium spp. and Fusarium spp. nor to the oomycetes Phytophthora spp. and Phytium spp. were found in our study, confirming the good phytosanitary status in the WOGC soil. Finally, and regarding beneficial fungi, representatives of the genus Trichoderma were found in the rhizosphere of all cultivars but Chemlal de Kabylie and Llumeta. Species of this genus have been used as BCA against VW of olive46,47.
In the olive belowground (endophytic and rhizosphere) core bacteriota here reported, genera from which some species have been well characterized and described as BCA werepresent. For instance, Streptomyces was the second most abundant genus in the endosphere whereas Bacillus was the tenth more abundant in the rhizosphere. While Pseudomonas was part of the rhizosphere core bacteriota, it was not considered as constituent of the endophytic core because it was absent in the root endosphere of cv. Mavreya. Nevertheless, Pseudomonas was relatively much more abundant inside olive root tissues than in the rhizosphere. Regarding the core mycobiota, and as mentioned above, the most noticeable presence of a pathogenic fungus was that of Macrophomina, and to a lesser extent Colletotrichum. The reported core microbiota indicates that, under the conditions found in the WOGC, olive trees harbor an important reservoir of beneficial/neutral microbes, and that the presence of deleterious microorganisms is nearly anecdotal. This correlates with the good development and appearance of the trees in the examined orchard, showing no visible symptoms of biotic stresses. The role of native microbiota in protecting plants from soil-borne pathogens has been highlighted in previous studies48. Nonetheless, further studies must be conducted in the presence of soil-borne pathogens, such as Verticillium dahliae, to study the community alterations and confirm the protective role of some of the core microorganisms described in the present study.
Materials and Methods
Sample collection
Soil and root samples were collected from the World Olive Germplasm Collection (WOGC) (37°51′38.11″N; 4°48′28.61″W; 102 m.a.s.l.) located at the Instituto de Investigación y Formación Agraria y Pesquera (IFAPA, Córdoba, Spain) in the spring of 2017, when the trees were in full bloom. The selected 36 olive cultivars (Table 1) surveyed are grown in the same orchard to avoid differences related to the physicochemical characteristics of the soil, water availability, agricultural management, weather conditions or any other influencing factor. The cultivars selected represent the subset of the working olive core collection from the WOGC49. Geographical origin and commercial interest of varieties were the main criteria to choose these cultivars for downstream studies. The upper layer (first 5 cm) of soil was removed and rhizosphere soil samples were collected (5 to 20-cm depth) following the main roots of each plant until finding non-suberified roots, where we took manually the soil firmly attached to the roots. These same root samples were also collected to assess the root endophytic communities. Three rhizosphere soil and three root samples from different trees of each cultivar were collected (n = 108). Furthermore, 10 bulk soil samples (1 kg) were collected at 1–1.5 m trunk distance of randomly selected trees (among the ones chosen for soil/root sampling) to analyze a number of physicochemical parameters of the WOGC soil (Table S5). These spots were randomly scattered along the orchard. Bulk soil samples in plastic bags were then transferred to the Agri-Food Laboratory of the Andalusian Regional Government at Córdoba (Spain), where physiochemical analyses were performed using standardized procedures.
DNA extraction and Illumina sequencing
The soil DNA from each individual sample (n = 108) was obtained using the Power Soil DNA Isolation kit (MoBio, Laboratories Inc., CA), following the manufacturer’s recommendations within 24 hours of samples collection. The root DNA (n = 108) was obtained, after root surface sterilization and grinding, using’Illustra DNA extraction kit Phytopure’ (GE Healthcare, Little Chalfont, UK). To ensure that DNA originated from endophytic microorganisms, and that microorganisms attached to the rhizoplane were eliminated, a thorough root surface sterilization protocol was implemented. Firstly, 20 ml of NaCl 0.8% were added to 50 ml screw cap polypropylene tubes containing each root sample. Tubes were then vigorously shaken in order to remove adhering soil particles. After discarding the supernatant, roots were washed five times with distilled water. Secondly, the following root surface sterilization protocol was implemented: 70% ethanol for 5 min, NaClO (3.7%) containing Tween 20 0.01% for 3 min, and finally 3 rinses in sterile and distilled water. To confirm that the disinfection protocol was successful, aliquots (100 µl) of water from the final rinse were plated in NA (Nutrient Agar) and PDA (Potato Dextrose Agar) plates that were incubated at 28 °C for 7 days. Then, plates were examined to confirm the absence of microbial growth. DNA yields and quality were checked both by electrophoresis in 0.8% (w/v) agarose gels stained with GelRed and visualized under UV light, and using a Qubit 3.0 fluorometer (Life Technologies, Grand Island, NY). The DNA was sequenced with Illumina MiSeq platform in a commercial sequencing service (The Institute of Parasitology and Biomedicine “López Neyra”, CSIC, Granada, Spain). In the first run, a prokaryotic library was constructed amplifying the hyper-variable regions V3-V4 of the 16S rRNA gene using the primer pair Pro341F and Pro805R according to Takahashi et al. In the second run, a eukaryotic library was constructed amplifying the ITS1 region using the primer pair ITS1FI2 and ITS2 according to Schmidt et al.50 and developed by White et al.51. Each library was prepared by amplifying each DNA sample (three biological replicas per olive cultivar; see above) in three independent PCR reactions (three technical replicas per biological replica). Then, PCR products of each biological replica were pooled and, finally, the 216 PCR products (108 from rhizosphere samples and 108 from root endosphere samples) were equimolarly mixed and used for sequencing. Both runs were sequenced using a paired-end 2 × 300-bp (PE 300) strategy. These sequence data have been submitted to the NCBI Sequence Read Archive (SRA) under the BioProject number PRJNA498945.
Data quality screening and overlapping
Demultiplexed and Phi-X174-free reads were quality checked with FastQC v.0.11.552 and end-trimmed with FASTX-Toolkit v.0.01453. All the 3′ end nucleotides were removed until the first position which reached an average quality value bigger than Q25. The paired reads were overlapped with fastq-join v.1.3.154 requesting a minimum overlap of 40 bp and a maximum of 15% of difference in the overlapping region. Both libraries were processed with the same bioinformatics tools but following different pathways detailed below.
Prokaryotic data processing
The overlapped reads from the prokaryotic (Bacteria and Archaea) library were initially classified with an 80% bootstrap cutoff to the Ribosomal Database Project (RDP-II) 16S rRNA reference database, training set v.16 MOTHUR-formatted55, with MOTHUR v.1.39.556. This initial step was performed to remove reads belonging to mitochondria, chloroplast and not identified at kingdom level (unknown). Then, using the software SEED2 v.2.1.0557 the prokaryotic sequences were trimmed and clustered. Firstly, by trimming the specific primers; then, by removing sequences with ambiguities and shorter than 400 bp as well as reads with an average read quality lower than Q30. Secondly, chimeric reads were removed by VSEARCH “De Novo” v.2.4.358 implemented in SEED2 and OTUs were clustered with the same tool at 97% similarity. Finally, the OTU table was saved and OTUs accounting for less than 0.005% of the total sequences were removed according to Bokulich et al.59 for further analyses. The most abundant OTU sequences were retrieved in SEED2 and classified as mentioned above. This classification was considered as the taxonomic information of each OTU.
Eukaryotic data processing
The eukaryotic library was directly quality-trimmed in SEED2 by the removal of sequences with ambiguities and an average read quality lower than Q30. There was not size exclusion and the primers were initially kept for the next step. Subsequently, ITSx v.1.0.1160 was performed but the result was discarded because of it was unable to properly recognize and remove the forward primer (ITS1FI2). Then, to accurately extract the ITS1 region, the high quality reads were aligned against the ribosomal operons of Saccharomyces cerevisiae S288c using Geneious R1161. As expected, the forward primer plus 4 nt matched the end of the 18S rRNA gene, and the reverse primer plus 30 nt matched the beginning of the 5.8S rRNA gene. Both intragenic ends were removed using SEED2 and chimeric sequences identified and discarded with VSEARCH “De Novo” implemented in SEED2. Then, the good quality sequences were distance-based greedy clustered at 97% similarity with VSEARCH algorithm implemented in MOTHUR. The most abundant OTU sequences were classified using the UNITE v.7.2 dynamic database62 with MOTHUR following the parameters recommended in the website and used by Findley et al.63. Finally, only OTUs with more than 0.005% of the sequences and assigned to kingdom Fungi were kept for further analyses. Furthermore, OTUs assigned to the phylum Oomycota were manually checked to examine the (possible) presence of the phytopathogenic genera.
Statistical analyses
Alpha diversity indices (observed and Chao1 richness; Shannon and inverse of Simpson diversity) were compared with Kruskal-Wallis test and p-values were FDR corrected by the Benjamini-Hochberg method using the R package agricolae64. Concerning the beta diversity, a normalization of the filtered OTU sequence counts was performed using the “trimmed means of M” (TMM) method with the BioConductor package edgeR65. The normalized data were considered to perform Nonmetric MultiDimensional Scaling (NMDS) on Bray-Curtis dissimilarities to ordinate in two dimensions the variance of beta diversity between compartments (root endosphere and rhizosphere) and among cultivars in each compartment, in both kingdoms. Ordination analyses were performed using the R package phyloseq66. We analyzed compartment and olive cultivar effects on community dissimilarities with permutational analysis of variance (PERMANOVA) and permutational analysis of multivariate dispersions (BETADISPER) using the functions adonis and betadisper in the vegan package with 9,999 permutations67. Significant prokaryotic or fungal genera in olive cultivar were obtained with the following protocol: (i) we tested for differential genus abundance using likelihood ratio tests (LRT) in the normalized data with the R package edgeR; (ii) we tested for differential genus abundance using proportions in non-normalized counts with the STAMP v.2.1.3 software68, selecting default statistical comparisons for multiple groups and firstly considering both Benjamini-Hochberg FDR for multiple test correction and without FDR correction; (iii) those genera significantly different in the two methods previously described were plotted and manually checked to generate the final selection. Most of the steps performed on R were carried out following the R script publicly donated by Hartman et al.69. In every case, statistically significant differences were considered when obtaining an adjusted p-value FDR corrected by Benjamini-Hochberg lower than 0.05.
Supplementary information
Acknowledgements
This work was supported by grant AGL2016-75729-C2-1-R from the Spanish Ministerio de Economía, Industria y Competitividad/Agencia Estatal de Investigación, and co-financed by the European Regional Development Fund (ERDF). We thank Dr. José F. Cobo-Díaz from the Université de Brest, Laboratoire Universitaire de Biodiversité et Ecologie Microbieene IBSAM, ESIAB, Plouzané, France for technical support in eukaryotic data analysis; and to PhD student Ana V. Lasa from Departamento de Microbiología del Suelo y Sistemas Simbióticos, Estación Experimental del Zaidín – CSIC for technical support in R scripts.
Author contributions
J.M.B. and M.F.L. conceived the study and performed its design. A.J.F.G., J.M.B. and M.F.L. wrote the manuscript. A.B. maintains the crop system and helped with sampling. P.J.V., C.G.L.C. and A.V.C. conducted the sampling and DNA extractions. A.J.F.G. performed the bioinformatics analysis and analyzed the data. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-56977-9.
References
- 1.Uylaşer V, Yildiz G. The Historical Development and Nutritional Importance of Olive and Olive Oil Constituted an Important Part of the Mediterranean Diet. Crit. Rev. Food Sci. Nutr. 2014;54:1092–1101. doi: 10.1080/10408398.2011.626874. [DOI] [PubMed] [Google Scholar]
- 2.López-Escudero FJ, Mercado-Blanco J. Verticillium wilt of olive: A case study to implement an integrated strategy to control a soil-borne pathogen. Plant Soil. 2011;344:1–50. doi: 10.1007/s11104-010-0629-2. [DOI] [Google Scholar]
- 3.FAO Food and Agriculture of the United Nations. FAOSTAT Available at, http://www.fao.org/faostat/en/#home (2017).
- 4.IOC. Available at, http://www.internationaloliveoil.org/estaticos/view/131-world-olive-oil-figures?lang=en_US.
- 5.Pérez-Jiménez F, Ruano J, Perez-Martinez P, Lopez-Segura F, Lopez-Miranda J. The influence of olive oil on human health: not a question of fat alone. Mol. Nutr. Food Res. 2007;51:1199–1208. doi: 10.1002/mnfr.200790023. [DOI] [PubMed] [Google Scholar]
- 6.Schena, L., Agosteo, G. E., Cacciola, S. O. & Sergeeva, V. Olive diseases and disorders. (Transworld Research Network, 2011).
- 7.Sanzani SM, et al. Abiotic diseases of olive. Offered Review. 2012;94:469–491. [Google Scholar]
- 8.Mercado-Blanco J, et al. Belowground microbiota and the health of tree crops. Front. Microbiol. 2018;9:1–27. doi: 10.3389/fmicb.2018.01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benitez Y, et al. Molecular analysis of the interaction between Olea europaea and the biotrophic fungus Spilocaea oleagina. Mol. Plant Pathol. 2005;6:425–438. doi: 10.1111/j.1364-3703.2005.00290.x. [DOI] [PubMed] [Google Scholar]
- 10.Lahkim LT. Epidemiology and control of Verticillium wilt on olive. Isr. J. Plant Sci. 2011;59:59–69. doi: 10.1560/IJPS.59.1.59. [DOI] [Google Scholar]
- 11.Ramos C, Matas IM, Bardaji L, Aragón IM, Murillo J. Pseudomonas savastanoi pv. savastanoi: some like it knot. Mol. Plant Pathol. 2012;13:998–1009. doi: 10.1111/j.1364-3703.2012.00816.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moral J, et al. Variability in Susceptibility to Anthracnose in the World Collection of Olive Cultivars of Cordoba (Spain) Front. Plant Sci. 2017;8:1–11. doi: 10.3389/fpls.2017.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saponari M, Boscia D, Nigro F, Martelli GP. Identification of DNA sequences related to Xylella fastidiosa in oleander, almond and olive trees exhibiting leaf scorch symptoms (southern Italy) J. Plant Pahol. 2013;95:659–668. [Google Scholar]
- 14.Rallo L, et al. High-Density Olive Plantations. Horticultural Reviews. 2013;41:303–384. doi: 10.1002/9781118707418.ch07. [DOI] [Google Scholar]
- 15.Aranda S, Montes-Borrego M, Jiménez-Díaz RM, Landa BB. Microbial communities associated with the root system of wild olives (Olea europaea L. subsp. europaea var. sylvestris) are good reservoirs of bacteria with antagonistic potential against Verticillium dahliae. Plant Soil. 2011;343:329–345. doi: 10.1007/s11104-011-0721-2. [DOI] [Google Scholar]
- 16.Sanei SJ, Razavi SE. Charcoal rot in nursery of olive in Golestan province of Iran. Soil Sci. 2012;1:211–217. [Google Scholar]
- 17.Gómez-Lama Cabanás C, et al. Indigenous Pseudomonas spp. strains from the olive (Olea europaea L.) rhizosphere as effective biocontrol agents against Verticillium dahliae: from the host roots to the bacterial genomes. Frontiers in Microbiology. 2018;9:277. doi: 10.3389/fmicb.2018.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez-Lama Cabanas C, et al. Bacillales Members from the olive rhizosphere are effective biological control agents against the defoliating pathotype of Verticillium dahliae. Agriculture. 2018;8:1–23. doi: 10.3390/agriculture8070090. [DOI] [Google Scholar]
- 19.Mercado-blanco J, Lugtenberg BJJ. Biotechnological Applications of Bacterial Endophytes. Curr. Biotechnol. 2014;3:60–75. doi: 10.2174/22115501113026660038. [DOI] [Google Scholar]
- 20.Martins F, Pereira JA, Bota P, Bento A, Baptista P. Fungal endophyte communities in above- and belowground olive tree organs and the effect of season and geographic location on their structures. Fungal Ecol. 2016;20:193–201. doi: 10.1016/j.funeco.2016.01.005. [DOI] [Google Scholar]
- 21.Pascazio S, et al. Phyllosphere and carposphere bacterial communities in olive plants subjected to different cultural practices. Int. J. Plant Biol. 2015;6:15–19. doi: 10.4081/pb.2015.6011. [DOI] [Google Scholar]
- 22.Preto G, Martins F, Pereira JA, Baptista P. Fungal community in olive fruits of cultivars with different susceptibilities to anthracnose and selection of isolates to be used as biocontrol agents. Biol. Control. 2017;110:1–9. doi: 10.1016/j.biocontrol.2017.03.011. [DOI] [Google Scholar]
- 23.Abdelfattah A, et al. Metabarcoding analysis of fungal diversity in the phyllosphere and carposphere of olive (Olea europaea) PLoS One. 2015;10:1–19. doi: 10.1371/journal.pone.0131069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Müller H, et al. Plant genotype-specific archaeal and bacterial endophytes but similar Bacillus antagonists colonize Mediterranean olive trees. Front. Microbiol. 2015;6:1–9. doi: 10.3389/fmicb.2015.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman-derr D, et al. Recent Trends in Valuation. New Phytol. 2016;209:798–811. doi: 10.1111/nph.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beckers B, De Beeck MO, Weyens N, Boerjan W, Vangronsveld J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome. 2017;5:1–17. doi: 10.1186/s40168-017-0241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santoyo G, Moreno-Hagelsieb G, del Carmen Orozco-Mosqueda M, Glick BR. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016;183:92–99. doi: 10.1016/j.micres.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. Development of a Prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS One. 2014;9:1–9. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maldonado-González MM, Schilirò E, Prieto P, Mercado-Blanco J. Endophytic colonization and biocontrol performance of Pseudomonas fluorescens PICF7 in olive (Olea europaea L.) are determined neither by pyoverdine production nor swimming motility. Environ. Microbiol. 2015;17:3139–3153. doi: 10.1111/1462-2920.12725. [DOI] [PubMed] [Google Scholar]
- 30.El-Tarabily KA, Sivasithamparam K. Non-streptomycete actinomycetes as biocontrol agents of soil-borne fungal plant pathogens and as plant growth promoters. Soil Biol. Biochem. 2006;38:1505–1520. doi: 10.1016/j.soilbio.2005.12.017. [DOI] [Google Scholar]
- 31.Law JWF, et al. The potential of streptomyces as biocontrol agents against the rice blast fungus, Magnaporthe oryzae (Pyricularia oryzae) Front. Microbiol. 2017;8:3. doi: 10.3389/fmicb.2017.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.El-Tarabily KA. An endophytic chitinase-producing isolate of Actinoplanes missouriensis, with potential for biological control of root rot of lupin caused by Plectosporium tabacinum. Aust. J. Bot. 2003;51:257–266. doi: 10.1071/BT02107. [DOI] [Google Scholar]
- 33.Maheshwari Dinesh K., editor. Bacteria in Agrobiology: Plant Growth Responses. Berlin, Heidelberg: Springer Berlin Heidelberg; 2011. [Google Scholar]
- 34.Chaouch FC, et al. Planomonospora, Saccharothrix and Actinophytocola genera in Saharan soils of Algeria: Isolation, taxonomic identification and antagonistic properties. J. Microbiol. Biotechnol. Food Sci. 2018;7:505–510. [Google Scholar]
- 35.Indananda C, et al. Actinophytocola oryzae gen. nov., sp. nov., isolated from the roots of Thai glutinous rice plants, a new member of the family. Pseudonocardiaceae. Int. J. Syst. Evol. Microbiol. 2010;60:1141–1146. doi: 10.1099/ijs.0.008417-0. [DOI] [PubMed] [Google Scholar]
- 36.Abdul Malek N, Zainuddin Z, Khan Chowdhury AJ, Zainal Abidin ZA. Diversity and antimicrobial activity of mangrove soil actinomycetes isolated from Tanjung Lumpur, Kuanta. J. Teknol. 2015;25:37–43. [Google Scholar]
- 37.Sun HM, et al. Actinophytocola gilvus sp. nov., isolated from desert soil crusts, and emended description of the genus Actinophytocola Indananda et al. 2010. Int. J. Syst. Evol. Microbiol. 2014;64:3120–3125. doi: 10.1099/ijs.0.061051-0. [DOI] [PubMed] [Google Scholar]
- 38.Sanka Loganathachetti D, Poosakkannu A, Muthuraman S. Fungal community assemblage of different soil compartments in mangrove ecosystem. Sci. Rep. 2017;7:1–9. doi: 10.1038/s41598-017-09281-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z, et al. Fungal communities in rhizosphere soil under conservation tillage shift in response to plant growth. Front. Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.01301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Farr DF, Castlebury LA, Rossman AY, Putnam ML. A new species of Phomopsis causing twig dieback of Vaccinium vitis-idaea (lingonberry) Mycol. Res. 2002;106:745–752. doi: 10.1017/S095375620200583X. [DOI] [Google Scholar]
- 41.Sergeeva V, Tesoriero L, Spooner-Hart R, Nair N. First report of Macrophomina phaseolina on olives (Olea europaea) in Australia. Australas. Plant Pathol. 2005;34:273–274. doi: 10.1071/AP05001. [DOI] [Google Scholar]
- 42.Mahdizadeh V, Safaie N, Goltapeh EM. Diversity of Macrophomina phaseolina based on morphological and genotypic characteristics in Iran. Plant Pathol. J. 2011;27:128–137. doi: 10.5423/PPJ.2011.27.2.128. [DOI] [Google Scholar]
- 43.Kaur S, et al. Emerging phytopathogen Macrophomina phaseolina: Biology, economic importance and current diagnostic trends. Crit. Rev. Microbiol. 2012;38:136–151. doi: 10.3109/1040841X.2011.640977. [DOI] [PubMed] [Google Scholar]
- 44.Chamorro M, et al. Evaluation of biosolarization for the control of charcoal rot disease (Macrophomina phaseolina) in strawberry. Crop Prot. 2015;67:279–286. doi: 10.1016/j.cropro.2014.10.021. [DOI] [Google Scholar]
- 45.Iqbal U, Mukhtar T, Iqbal SM. In vitro and in vivo evaluation of antifungal activities of some antagonistic plants against charcoal rot causing fungus Macrophomina phaseolina. Pakistan J. Agric. Sci. 2014;51:689–694. [Google Scholar]
- 46.Ruano-Rosa D, et al. Fate of Trichoderma harzianum in the olive rhizosphere: time course of the root colonization process and interaction with the fungal pathogen Verticillium dahliae. BioControl. 2016;61:269–282. doi: 10.1007/s10526-015-9706-z. [DOI] [Google Scholar]
- 47.Carrero-Carrón I, et al. Interactions between Trichoderma harzianum and defoliating Verticillium dahliae in resistant and susceptible wild olive clones. Plant Pathol. 2018;67:1758–1767. doi: 10.1111/ppa.12879. [DOI] [Google Scholar]
- 48.Mendes R, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332:1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 49.Belaj A, et al. Tree Genet. Genomes. 2012. Developing a core collection of olive (Olea europaea L.) based on molecular markers (DArTs, SSRs, SNPs) and agronomic traits; pp. 365–378. [Google Scholar]
- 50.Schmidt PA, et al. Illumina metabarcoding of a soil fungal community. Soil Biol. Biochem. 2013;65:128–132. doi: 10.1016/j.soilbio.2013.05.014. [DOI] [Google Scholar]
- 51.White, T., Bruns, T., Lee, S. & Taylor, J. In PCR Protocols: A Guide to Methods and Applications (eds. Innis, M., Gelfand, D., Shinsky, J. & White, T.) 315–322 (Academic Press, 1990).
- 52.FastQC. Available at, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. (Accessed: 14th March 2019).
- 53.FASTX-Toolkit. Available at, http://hannonlab.cshl.edu/fastx_toolkit/index.html. (Accessed: 14th March 2019).
- 54.Aronesty. fastq-join. Available at, https://github.com/ExpressionAnalysis/ea-utils. (Accessed: 14th March 2019) (2011).
- 55.RDP reference files - mothur. Available at, https://mothur.org/wiki/RDP_reference_files. (Accessed: 14th March 2019).
- 56.Schloss PD, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537 LP-7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Větrovský T, Baldrian P, Morais D. SEED 2: a user-friendly platform for amplicon high-throughput sequencing data analyses. Bioinformatics. 2018;34:2292–2294. doi: 10.1093/bioinformatics/bty071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bokulich NA, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods. 2013;10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bengtsson-Palme J, et al. Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol. Evol. 2013;4:914–919. [Google Scholar]
- 61.Kearse M, et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Unite Community 2017. (2017). Available at, https://plutof.ut.ee/#/doi/10.15156/BIO/587478. (Accessed: 14th March 2019).
- 63.Findley K, et al. Topographic diversity of fungal and bacterial communities in human skin. Nature. 2013;498:367–370. doi: 10.1038/nature12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Mendiburu. CRAN - Package agricolae. Available at, https://cran.r-project.org/web/packages/agricolae/index.html. (Accessed: 14th March 2019) (2017).
- 65.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mcmurdie, P. J. & Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. 8 (2013). [DOI] [PMC free article] [PubMed]
- 67.Oksanen, et al. Vegan: community ecology package (2015).
- 68.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics. 2014;30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hartman K, et al. Correction to: Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome. 2018;6:1–14. doi: 10.1186/s40168-017-0383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.