Abstract
Confocal and scanning electron microscopic observations have previously shown the strong bacterial association of Microcystis aeruginosa cells on their surfaces. DNA-based analyses of the associated bacterial communities were carried out using two M. aeruginosa strains grown in the laboratory and eight newly collected cyanobacterial bloom samples. M. aeruginosa was the most predominant species (66–100%) within the phylum Cyanobacteria. Rhizobium, Hydrogenophaga and Brevundimonas species were commonly found, and Flavobacterium species were present in all the cyanobacterial bloom samples. In total, 396 colonies from various samples were screened, revealing that most culturable bacteria belonged to the class Alphaproteobacteria (19%) including Rhizobium, Brevundimonas, and Porphyrobacter species. The genetic variation among the M. aeruginosa strains and different habitat conditions may have led to the presence of distinct bacterial populations among the tested samples. Among all the tested seven culturable isolates, Rhizobium sp. MK23 showed the best growth-promotion effect on the axenic M. aeruginosa strains. H2O2 was observed to be produced during the growth of M. aeruginosa PCC7806 under light conditions, this strain was more resistant to H2O2 when associated with Rhizobium sp. MK23. Our data suggested that Rhizobium species along with other associated bacteria might help the growth of M. aeruginosa by decomposing H2O2 under the aerobic growing conditions.
Subject terms: Water microbiology, Microbial ecology
Introduction
Cyanobacterial blooms are common phenomena in several freshwater environments, including drinking water sources, around the world. These blooms are often associated with production of cyanotoxins, oxygen depletion, unpleasant odor, and ecosystem health issues1. The bloom-forming cyanobacteria such as Microcystis, Anabaena, and Oscillatoria species decrease the esthetic and recreational values of the water bodies and cause a serious threat to the ecosystems2. Cyanobacterial blooms are caused by high nutrient loading, elevated water temperature and radiation levels, global warming due to high CO2 concentration, drought, and increased water salinity2,3. The factors that influence cyanobacterial growth are water temperature above 25 °C, weak water current2, sufficient phosphorus supply, and low nitrogen/phosphorous (N/P) ratio3. The appearance of floating cyanobacterial blooms has been one of the most unpleasant symptoms of eutrophication4. Previous studies have emphasized on phosphorous as the main nutrient source affecting phytoplankton mass and eutrophication in the majority of lakes5. Representative bloom-forming cyanobacterial genera include Microcystis, Anabaena, and Cylindrospermopsis from freshwater sources, and Nodularia and Aphanizomenon from estuaries6. The Microcystis species, such as M. aeruginosa, M. flosaquae, M. ichthyoblabe, M. wesenbergii, and M. pherta are responsible for causing almost 90% of the cyanobacterial blooms in freshwater bodies. Of these, M. aeruginosa is the most commonly observed to cause cyanobacterial blooms2,7,8.
The colonies of Microcystis genus are generally about 4–5 µm in size; however, coagulated colonies range from 52–200 µm in size9, consequently exhibits thick aggregations of cells under natural environmental conditions10. Microcystis species are widely known to produce anatoxin-a, a neurotoxin, and microcystin which are the primary toxins found in freshwaters worldwide11. Microcystin synthetase gene clusters (mcy) comprise genes coding for polyketide synthase, peptide synthase, and mixed polyketide/peptide synthase12. Microcystin concentrations have been directly or indirectly determined using protein phosphatase inhibition assays, enzyme-linked immunosorbent assay (ELISA), chemical derivatization with gas chromatography– mass spectrometry analysis, and high-performance liquid chromatography coupled to either ultra-violet, photodiode array detection or mass spectrometry detection13. PCR amplification of the mcy genes has proven to be effective in distinguishing between the microcystin-producing hepatotoxic and non-toxic Microcystis strains14. For identifying Microcystis species in environmental samples, the most common PCR targets employed are the microcystin synthetase gene operon and Microcystis-specific 16S rRNA and phycocyanin (cpcBA) genes15,16, in addition to the genes related to nutrient transport and metabolism17. The size of Microcystis genomes range from 4.26 Mbp (M. aeruginosa PCC 9806) to 5.84 Mbp (M. aeruginosa NIES 843) in size; however, only 27 draft or closed genomes are available, and ≥12,000 predicted genes remain uncharacterized18. Moreover, since there is a large genetic variation among Microcystis species, the lack of species-specific genetic information complicated PCR-based differentiation of individual species17.
Microcystis species grow into large mucilaginous aggregates, which comprise a microscale mucus region called the phycosphere, and this region is generally colonized by associated bacteria19. Fluctuations in the bacterial diversity associated with Microcystis-blooms are often observed in many environmental samples20. Microcystis-associated bacteria can directly adhere to the cells on the surface of a Microcystis colony21 or colonize within the enclosed region22. Many associated bacteria can enhance or suppress the growth of cyanobacteria23, or even kill them24. Difficulty in obtaining the axenic cultures of Microcystis species is probably due to the lack of knowledge about the roles of associated bacteria in xenic cultures25. The photosynthetic performances and growth rates of Microcystis species in xenic culture are higher than those in the axenic cultures26. Nevertheless, the effects of bacterial association on Microcystis species and their underlying mechanism remain unclear. Interactions between algae and bacteria have been intensively studied (but not within phycosphere); for example, the Emiliania huxleyi-Roseobacter interaction in the marine ecosystem27. Roseobacter species promote algal growth for a short period by supplying the algae with vitamin B12 and phytohormones, such as abscisic acid, auxin, and gibberellins and by providing antimicrobial protection against the other bacteria27. In addition, the ammonium-excreting bacterium Azotobacter vinelandii has been found to promote microalgal growth28. Mutualism has been observed between Chlorella vulgaris and Pseudomonas species under photoautotrophic condition29.
This study examined the bacterial species that are associated with the phycospheres of 10 bloom-forming ten Microcystis aeruginosa (two strains were laboratory-grown and eight srains were recently collected from various cyanobacterial-bloom-forming areas in August, 2018). Culture-independent and -dependent studies were conducted in order to explore the diversity of Microsytis-associated bacteria. Our data here demonstrated that many Alphaproteobacteria including Rhizobium species, are the dominant bacteria in the phycosphere of M. aeruginosa. Rhizobium species along with other associated bacteria likely protect M. aeruginosa against oxidative stress under aerobic growth conditions and M. aeruginosa produces acetate for associated bacteria.
Results
Cyanobacterial bloom related to variation in the water characteristics
Water quality at each sampling site was assessed in order to understand the cyanobacterial bloom conditions in the environment (Fig. S1). The temperature ranged from 31.0 °C [at Daecheong lake (DC) sample] to 33.0 °C [at Hapcheon Changnyeong barrage (HC) sample] among the sample locations (Table 1, Fig. S1). The pH value ranged from 7.0 to 9.4, and the most alkaline condition was found in the DC sample. Higher alkaline conditions, such as pH 9.5, have been shown to promote the growth of cyanobacteria30. The dissolved oxygen (DO) levels in the DC sample was low (2.0 ± 0.22 mg/L); similarly, the other samples also presented with low DO levels (<1 mg/L). It was difficult to judge the correlation between the chlorophyll-a (chl-a) concentration and organic contents because the chemical oxygen demand (COD) values of our environmental samples were slightly lower (9.6–9.82 mg/L) than the previously reported the COD values (10.5–17.5 mg/L) of bloom-containing water samples31. The concentration of total nitrogen (TN) in the HC sample was 13.84 mg/L, and this level was significantly higher than those in the other sites. Additionally, HC sample also had the highest total phosphorus (TP) concentration (0.38 mg/L). The concentration of chl-a increased with TN and TP values, except for the Juksan barrage (JS) sample differed (Fig. S2), and indicated the severity of the cyanobacterial bloom at the corresponding sampling site (Fig. S3). With 1111.04 mg/m3, HC sample had the maximum chl-a concentration. The cyanobacteria in samples collected from Murwang reservoir (MW), Wangsong reservoir (WS), Gangjeong-Goryeong barrage (GJ), and Bohyun mountain Dam (BH) had formed small coagulates although these sites had cyanobacterial blooms. Hence, the amount of chl-a per volume was lower (below 10 mg/m3) than expected because of variation of chl-a concentration at different sampling points. Although the main cause of a cyanobacterial bloom could not be identified from our bloom-water characteristics, the cyanobacterial blooms and high concentrations of chl-a and cyanobacterial blooms in JS and HC samples were correlated with elevated TN and TP concentrations. However, regarding DC sample, the main cyanobacterial bloom was likely correlated with the high pH rather than TN or TP concentration.
Table 1.
Measurement of the Temperature (°C), pH, DO (mg/L), Salinity (ppt), COD (mg/L), Total N (mg/L), and Total P (mg/L) of water quality of the environmental samples for determining the water quality; MW (Murwang reservoir), DC (Daecheong lake), BJ (Baekje barrage), HC (Hapcheon-Changnyeong barrage), WS (Wangsong reservoir), JS (Juksan barrage), and GJ (Gangjeong-Goryeong barrage).
| Location | Temperature | pH | DO | Salinity | COD | Total N | Total P |
|---|---|---|---|---|---|---|---|
| MW | 32.35 ± 1.04 | 7.04 ± 0.13 | 0.43 ± 0.06 | 0.13 ± 0.005 | 9.631 | ND | 0.122 |
| WS | 31.90 ± 0.67 | 8.40 ± 0.36 | 1.01 ± 0.18 | 0.21 ± 0.000 | 9.783 | 1.62 | 0.121 |
| DC | 31.00 ± 0.14 | 9.44 ± 0.14 | 1.95 ± 0.22 | 0.08 ± 0.000 | 9.774 | 1.97 | 0.082 |
| BJ | 32.67 ± 0.23 | 7.83 ± 0.17 | 0.89 ± 0.50 | 0.18 ± 0.000 | 9.599 | 1.83 | 0.078 |
| JS | 32.28 ± 0.08 | 7.47 ± 0.18 | 0.56 ± 0.19 | 0.15 ± 0.000 | 9.659 | 2.13 | 0.239 |
| HC | 32.97 ± 0.14 | 7.89 ± 0.11 | 0.51 ± 0.20 | 0.14 ± 0.000 | 9.653 | 13.84 | 0.38 |
| GJ | 31.22 ± 0.46 | 7.52 ± 0.26 | 0.74 ± 0.59 | 0.36 ± 0.005 | 9.820 | ND | 0.024 |
ND: not detected.
Cyanobacterial coexistence with bacteria
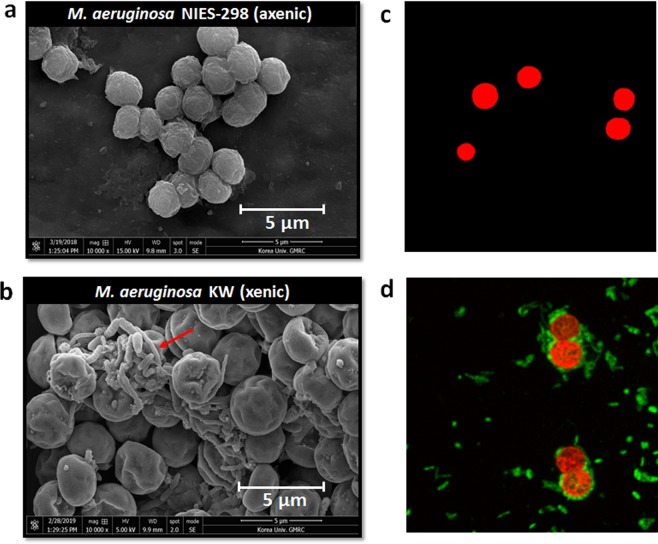
To test whether M. aeruginosa-associated bacteria were attached to M. aeruginosa cells, an M. aeruginosa KW sample was examined by field emission scanning electron microscopy (FE-SEM) and confocal laser scanning microscopy (CLSM). The cells of M. aeruginosa KW coexisted with various bacterium species unlike compared to the axenic M. aeruginosa NIES-298 (Fig. 1a,b). Some bacterium cells were found attached to the surface of M. aeruginosa KW. However, the bacteria were not attached to all the cells of M.aeruginosa, but rather clustered intensively. CLSM following SYTO 9 staining also confirmed the presence of epiphytic bacteria on the surface of M. aeruginosa KW cells (Fig. 1c,d). Notably, the bacteria were observed to be surrounding the M. aeruginosa KW cells.
Figure 1.
Microcystis-bacteria co-existence. Scanning electron microscopy (SEM) image of the (a) axenic M. aeruginosa NIES-298 culture, and (b) xenic M. aeruginosa KW. SYTOTM 9 staining analysis of the (c) axenic M. aeruginosa NIES-298 and (d) xenic M. aeruginosa KW. The red and green represents M. aeruginosa and bacteria, respectively. The bacteria associated with the M. aeruginosa surface.
Bacterial communities in cyanobacterial phycospheres
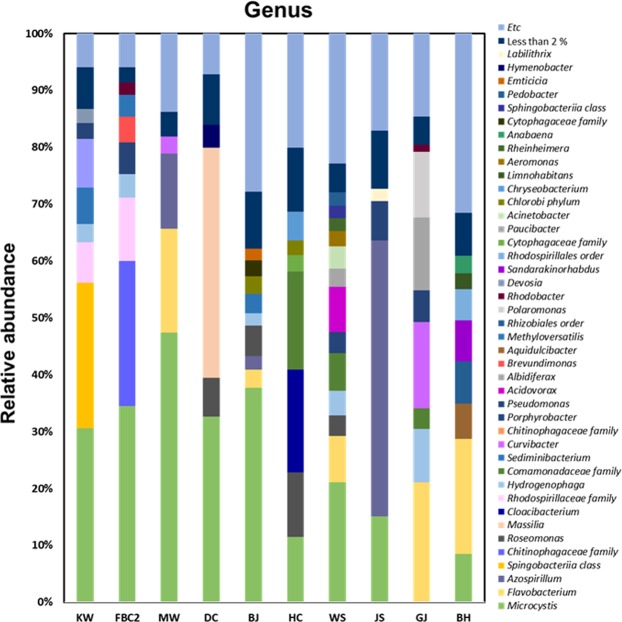
Bacterial communities of two laboratory grown samples (KW and FBC2) and eight environmental samples were analyzed by microbiome taxonomic profiling of the epiphytic bacteria isolated using 3-μm pore filters from the cyanobacterium samples. Differential interference contrast microscopy results showed that Microcystis species and various phytoplanktons were present together in the environmental samples (Fig. S4). M. aeruginosa was the most predominant species (66–100%) within the phylum cyanobacteria (Fig. 2). At genus level, Flavobacterium species were represented in all the environmental samples and were a dominant species in the MW, WS, GJ, and BH samples (10–35%; Fig. 2); however, it was absent from the laboratory samples. Approximately 22 species (except Flavobacterium species) were exclusively present in the environmental samples but not abundantly (<8%), and 11 species, including Rhizobium, Brevundimonas, Porphyrobacter, Hydrogenophaga, and Sediminibacterium were present in all the samples (Fig. S5). Two laboratory M. aeruginosa strains were dominated by different unclassified bacteria of the class Sphingobacteriia (KW: 36.86% and FBC2: 38.9%). Moreover, the predominant species in KW sample also existed in the JS sample, and the primary species of FBC2 was present in all the environmental samples, except for DC and MW samples. Unclassified Chitinophagaeceae species, the second dominant species in KW sample (12.37%), were present in all the samples, except for DC sample. Interestingly, at genus level, the bacterial community of the DC sample revealed that genus Massilia (40.45%) was more abundant than M. aeruginosa (32.66%) at the genus level. M. aeruginosa KW and M. aeruginosa FBC2 were similar in bacterial community to each other and simpler than the environmental samples, except for the five bacteria at the genus level (<1%). The environmental samples presented a wider variety of phyla (>15 phyla) than the laboratory samples (8 and 10 phyla in KW and FBC2, respectively). The OTUs of the environmental samples were significantly higher than those of the laboratory strains; among the environmental samples, the highest OTU value was observed in the BH sample (1380), and the lowest in the DC sample (384) (Table 2). ETC is a group of strains that contributed <1% of the total abundance of genus. The presence of ETC also revealed that the environmental samples were more heterogeneous than the laboratory samples.
Figure 2.
Bacterial community analysis of all the following M. aeruginosa samples: M. aeruginosa KW (KW), M. aeruginosa FBC000002 (FBC2), Murwang (MW), Daecheong (DC), Baekje (BJ), Hapcheon (HC), Wangsong (WS), Juksan (JS), Gangjeong (GJ), and Bohyun (BH). The genera with <1% abundance were included as well. The laboratory-cultured samples consisted of simple bacterial communities, whereas the environmental samples had various bacterial populations.
Table 2.
Results of the bar-coded pyrosequencing and diversity indices of two laboratory strains and eight environmental samples.
| Sample | Reads per sample | Average length (bp) | OTUs | Number of species found | Chao1 | Shannon |
|---|---|---|---|---|---|---|
| KW | 138,693 | 412.6 | 350 | 99 | 350.55 | 2.44 |
| FBC2 | 135,777 | 410.1 | 342 | 110 | 343.70 | 2.23 |
| MW | 83,642 | 408.8 | 655 | 373 | 670.05 | 2.20 |
| DC | 38,436 | 414.9 | 384 | 184 | 396.66 | 2.29 |
| BJ | 64,915 | 410.7 | 1306 | 570 | 1320.99 | 3.83 |
| HC | 63,341 | 415.8 | 815 | 324 | 822.97 | 3.51 |
| WS | 80,158 | 418.0 | 949 | 674 | 974.88 | 4.28 |
| JS | 74,589 | 409.3 | 1060 | 445 | 1068.14 | 2.84 |
| GJ | 59,841 | 422.3 | 1174 | 613 | 1194.81 | 4.21 |
| BH | 72,252 | 411.0 | 1380 | 629 | 1394.12 | 4.43 |
Different genotypes in Microcystis aeruginosa strains
In our growth experiments, which were performed using a PhotoBiobox, the optimum light and temperature conditions were different for four cyanobacterial bloom samples (Fig. S6). Interestingly the growth of all the strains was affected by temperature rather than light intensity, and optimal growth was observed at <25 °C rather than 30 °C. The 16S rRNA gene sequencing revealed a high similarity of >99% among the Microcystis species in all the samples, and this observation suggested that the 16S rRNA analysis alone was not enough to distinguish the genotypes of M. aeruginosa strains. Nevertheless, a few sequence differences were observed among the sequences. The 16S rRNA gene sequencing revealed the difference of T vs. C at nucleotide 223, and T vs. G at nucleotides 235 and 236 (Fig. S7). The result of PCR analysis, targeting the microcystin gene cluster (mcyA, mcyB, and mcyC), confirmed the presence of toxic Microcystis species in our samples. An electropherogram of the PCR products for mcyA revealed that MW, DC, BJ, JS, and BH samples contained the mcyA gene (Fig. S8a). HC and GJ samples did not contain the mcyB gene, whereas the mcyC gene was detected in all the environmental samples. The 16S rRNA gene sequencing indicated that GJ and BH samples contained the same species, but the two species differed in mcy-gene-targeting PCR analysis (Fig. S8). Interestingly, the species in the MW and BJ samples have the mcyA, mcyB, and mcyC genes, but their microcystin-LR concentrations were <0.1 µg/L (Fig. S8b). This data suggested that these samples had M. aeruginosa strains producing structurally different microcystins that could not be detected with the kits used in this study. HC sample did not generate a product with the conventional microcytin PCR primer set32–34, but produced microcystin-LR, indicating the complexity of the genotypes and evolution of M. aeruginosa strains. Our data suggested that genotypic differences among the M. aeruginosa strains may contribute to the variations in associated bacterial community structures.
Diversity structure and phylogenetic analyses of the culturable bacteria
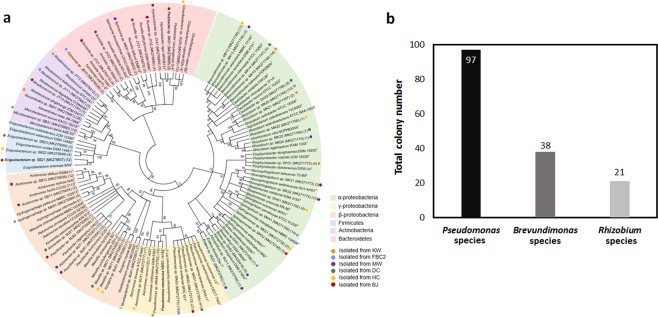
To characterize the culturable bacteria, 396 colonies were isolated from six samples including the laboratory samples (KW, FBC2, MW, DC, BJ, and HC), indicating the presence of high percentage of Microcystis species in the bacterial community. Although the chl-a concentration in the MW sample was low (6.29 mg/m3), this sample was selected for culture-dependent analysis because it had 100% M. aeruginosa presence as the phylum cyanobacteria. Three types of culture media used for bacterial growth did not reveal any significant difference in colony formation. The strains with identical BOX-PCR patterns were used to perform the amplification of 16S rRNA genes (Fig. S9). A phylogenetic analysis was performed using the identified gene sequences and closely related type strains from the EzTaxon Database (Fig. 3a). Rhizobium species were commonly found in five samples, except for the BJ sample (Fig. 3b); however, it was present in all the samples from the culture-independent analysis. Pseudomonas species were abundantly found in KW, FBC2, DC, and BJ samples, and Aeromonas species were dominant in HC sample. With respect to culturable bacteria in MW sample, Acidovorax, Pelomonas, and Paucibacter species were found in similar proportions. Various bacterial species were isolated (12 genera) from the environmental MW sample, however, only six bacterial species were identified in the laboratory-grown KW sample. Interestingly, one-third of all the culturable genera belonged to the Alphaproteobacteria class. These results indicate that culture analysis and DNA-based bacterial community analysis resulted in different diversity structures probably because of different bacterial growth properties.
Figure 3.
Culture-dependent analysis of the 16S rRNA gene sequences. (a) Neighbor‐joining phylogenetic tree of the isolated bacteria. Each color represents a phylum level of the bacterial community. The associated bacteria were isolated from KW, FBC2, MW, DC, BJ and HC samples. The number in parentheses indicates the number of isolated associated bacteria. Most bacteria belonged to the class Alpha-proteobacteria. GenBank accession numbers are provided in parentheses. (b) Pseudomonas species were the most frequently isolated species from the culturable bacteria.
Effect of associated bacteria on the growth of M. aeruginosa cells
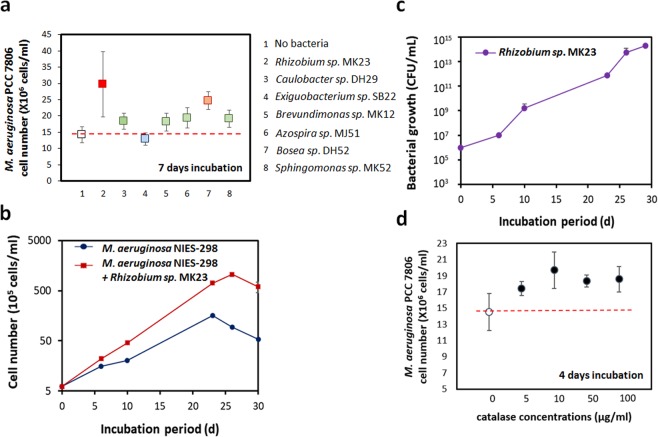
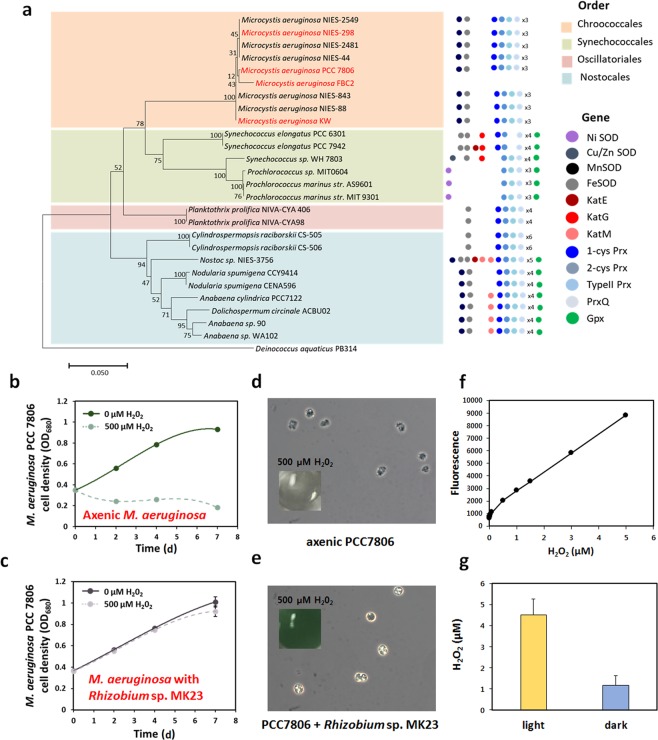
Seven cultured genera were randomly selected, and co-culture experiments were performed with axenic M. aeruginosa PCC7806. All the tested bacteria except Exiguobacterium species had positive effects on the growth of M. aeruginosa PCC7806 compared with the growth of axenic PCC7806 (Fig. 4a). Among the seven genera we tested, Rhizobium and Bosea species appeared to be the best M. aeruginosa’ growth-promoting bacteria for M. aeruginosa (Fig. 4a). Rhizobium species were present with different degrees in all the samples, and Bosea species were present in all the samples, except for WS sample. Rhizobium sp. MK23 also promoted the growth of the axenic M. aeruginosa NIES-298 strain (Fig. 4b). The maximum density of axenic M. aeruginosa was 1.6 × 107 cells/ml on day 23; however, that of the xenic culture (incubated with Rhizobium sp. MK23) was 1.1 × 108 cells/ml on day 26. Interestingly, the cell number of M. aeruginosa NIES-298 started to decreases on the 23rd day, whereas the xenic culture began to decreases on the 26th day. Furthermore, Rhizobium sp. MK23 continued to grow, with cell population increasing from 1 × 106 cells/ml to 2 × 1014 cells/ ml within 30 days (Fig. 4c). The addition of catalase resulted in the enhanced growth of the axenic M. aeruginosa PCC7806 (Fig. 4d). Our genome analysis indicated that M. aeruginosa strain does not have a gene for producing catalase (Fig. 5a). The sensitivity of M. aeruginosa PCC7806 with Rhizobium sp. MK23 to H2O2 (500 µM) was much lesser than that of the axenic M. aeruginosa PCC7806 (Fig. 5b,c). The axenic PCC7806 treated with H2O2 observed an unusual cell shape. However, PCC7806 co-cultured with Rhizobium sp. MK23 had still a normal, round shape (Fig. 5d,e). Under the light condition, the concentration of H2O2 was 4.5 µM in the axenic culture of axenic M. aeruginosa PCC7806, and the concentration of H2O2 was decreased under the dark condition (Fig. 5f,g). Our data strongly suggested that associated bacteria protect M. aeruginosa cells under oxidative stress, leading to better growth of the xenic culture.
Figure 4.
(a) Co-culture experiments of M. aeruginosa PCC7806 with seven genera, Rhizobium sp. MK23 apparently promoted the growth of M. aeruginosa PCC7806 for 7 days. **p < 0.05. (b) M. aeruginosa NIES-298 grew faster with Rhizobium sp. MK23. The M. aeruginosa NIES-298 culture and co-culture started degrading on 23 and 26, repectively. (c) Rhizobium sp. MK23 in co-culture grew at a density of 1014 cells/ml in 30 days. (d) Addition of catalase promoted the growth of M. aeruginosa PCC7806 for 4 days.
Figure 5.
M. aeruginosa cells are sensitive to oxidative stress. (a) The listed cyanobacterial antioxidant genes are SOD, superoxide dismutase; Kat, catalase; Prx, peroxidase; and Gpx, glutathione peroxidase. M. aeruginosa does not have a gene for producing a catalase. (b) Axenic of M. aeruginosa PCC7806 and M. aeruginosa PCC7806 with Rhizobium sp. MK23 were treated with 500 µM concentration of H2O2. After 7 days of treatment, the growth of M. aeruginosa PCC7806 was retarded under H2O2. (c) The growth of axenic M. aeruginosa PCC7806 with Rhizobium sp. MK23 did not change significantly. (d) The axenic PCC7806 treated with H2O2 was transparent in color and had an unusual cell shape. (e) PCC7806 co-cultured with Rhizobium sp. MK23 were still green and had a normal, round shape. (f) The standard curve for H202 concentrations in H2O2 assay. (g) In the H2O2 assay, the concentrations of H2O2 were much higher in the light condition than in dark.
Discussion
Cyanobacterial blooms of Microcystis species are characterized by the formation of large colonies through collision after cell division and proliferation35. Flocculated colonies float with buoyancy and form a layer on the surface of the water, gradually covering the entire water surface36. At the sampling sites, cyanobacterial blooms were found as dense layers of cells on the water surfaces or as sparsely floating coagulates. Owing to the differences in the timing of sampling, the concentrations of chl-a widely differed among the sites. Previous studies have reported significant differences among the communities of free-living and cyanobacteria-associated bacteria as a result of several weeks of blooming36. Therefore, bacterial communities and nutrient qualities may change depending on the cyanobacterial bloom periods.
The major factors that promote the growth of cyanobacterial biomass include high concentration of nutrients such as phosphorus and nitrogen37. Bacterial communities, as well as the levels of water nutrients, are affected by the surrounding area was near a farmland or livestock farms, the concentrations of TN and TP were relatively higher, suggesting that nutrients present in water are related to the surrounding environment. Globally, approximately 50% of the N fertilizer applied to cultivation systems is not absorbed by plants and is released into the environment as ammonia (NH3), nitrate (NO3−), and nitrous oxide (N2O)38. Many of our sampling areas were surrounded by farmlands except the DC sample, which was collected from a drinking water reservoir. Therefore, the number of OTUs in DC sample was not significantly lower than those in other environmental samples (Table 2). The Rhizobium and Azospirillum species were the predominant bacteria in the JS sample. These species have the nitrogen-fixing ability, which might help the growth of M. aeruginosa39,40. WS sample had the lowest concentration of total nitrogen, which may be related to the presence of a second dominant bacterium, a nitrate-reducing Acidovorax species41. The growth of Microcystis species in natural environments can be affected by not only abiotic factors but also biotic facters involved in the N and P cycles of associated bacteria.
The culture-independent analysis results revealed that the two laboratory strains had lower levels of ETC (<1%) than the environmental samples, indicating that there is less species richness in domesticated samples (Fig. 2). Our data suggested that only important bacteria interacting with Microcystis species remain as cyanobacterial bloom samples continued growing in the laboratory. The culture-independent analysis results revealed that some bacteria were common to all the samples, but the dominant species were different. Different optimal temperature and light conditions suggested that four samples had genotypically different M. aeruginosa species (Fig. S6). An electropherogram of PCR products for the microcystin gene was likely to miss the presence of genes because of primer mis-matches. It is difficult to distinguish the diversity of Microcystis species only by microcystin gene PCR or 16S rRNA gene sequencing because of the significant genetic variations among Microcystis strains17. A previous study has shown that seventeen various strains of M. aeruginosa different from M. aeruginosa NIES-843 had >99% 16S rRNA sequence identity10. Therefore, some associated bacteria may be specifically to certain Microcystis species.
The genus Polynucleobacter, which is the most well-known genus predominantly found in freshwater habitats, can be regarded as a good indicator of water quality. Polynucleobacter species accounted for <1% of all the species, indicating poor water qualities of our environmental samples. Actinobacteria are also known to be abundant in freshwater, but their number is decreased in nutrient-rich ecosystems and is inversely proportional to cyanobacteria42–44. The culture-independent analysis results showed that the proportion of Actinobacteria was also <1%. Flavobacterium species was predominant in four samples and abundantly present within the cyanobacterial aggregates45. This species may be crucial for degrading the cyanobacterial hepatotoxin46. The bacterial community structure determined by the culture-dependent analysis was different from that of the DNA-based bacterial community analysis. According to the culture-dependent analysis results, Pseudomonas species was the most prevalent in the DC sample but accounted for only 0.14% according to the results of the DNA-based bacterial community analysis. Pseudomonas, Brevundimonas and Rhizobium species were the most abundant according to the results of the culture-dependent analysis, by which 396 colonies were screened from six samples (Fig. 3). However, Brevundimonas species, but not Pseudomonas strains, were estimated to be more abundant in many samples by DNA-based analysis.
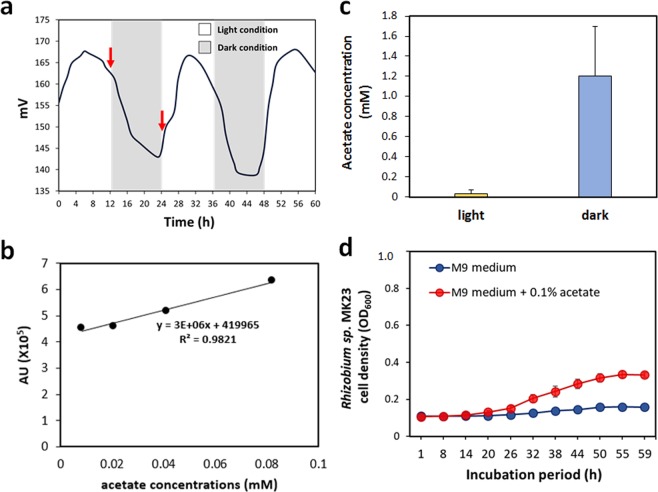
The growth of axenic M. aeruginosa PCC7806 with several isolates including Rhizobium species was boosted for 7 days-culture experimement (Fig. 4a). The Rhizobium species can do nitrogen fixation and produce catalases47. However, Microcystis species lack the genes for catalase48 and nitrogenase. Our catalase test showed that Rhizobium sp. MK23 has catalase activity (Fig. S10a). The cell number of the axenic PCC7806 in BG11 without nitrogen source was more decreased than that of the PCC7806 with Rhizobium sp. MK23 (Fig. S10b). We speculated that Rhizobium species help the growth of M. aeruginosa by providing catalase function and fixed nitrogen resources. Indeed, the addition of catalase improved the axenic M. aeruginosa culture growth, which partly supporting our speculation (Fig. 4d). Under the light conditions, the concentration of H2O2 in M. aeruginosa PCC7806 culture was 4.5 µM, and the M. aeruginosa PCC7806 with Rhizobium sp. MK23 was less sensitive to H2O2 than axenic PCC7806 (Fig. 5). DO changes in axenic M. aeruginosa PCC7806 culture under the light and dark conditions were measured for 60 h (Fig. 6a). Under the light condition, O2 was produced during photosynthesis (Fig. 6a). M. aeruginosa PCC7806 is known to ferment endogenously stored glycogen to ethanol, acetate, CO2, and H2 under the dark and anaerobic conditions49. The acetate level in axenic M. aeruginosa PCC7806 culture increased under the dark conditions (Fig. 6b,c). Rhizobium sp. MK23 could use acetate as a carbon source (Fig. 6d). Hydrogen-oxidizing Hydrogenophaga species was present in all the samples although it occupied a minor proportion. Our M. aeruginosa species might produce H2 gas as a fermentation product, which can be a good substrate for Hydrogenophaga species. More experiments seem necessary to prove this hypothesis. Taken together, our data suggested that different associated bacteria were identified in the phycospheres of M. aeruginosa strains probably due to genetic and physiological heterogeneities among these strains. Although further analysis is needed to have a full understanding of their mutual interaction, we demonstrated that metabolites, including acetate, and catalase activities could be shared by two different microorganisms.
Figure 6.
(a) Dissolved oxygen (DO) changes during the growth of M. aeruginosa PCC7806 under the light and dark conditions for 60 h. In M. aeruginosa PCC7806 cultures, DO increased in the light condition and decreased in the dark condition. Cell supernatant was extracted at 12 h and 24 h (red arrows). (b) The standard curve for the acetate concentrations in HPLC analysis. (c) The HPLC analysis results showed that the concentration of acetate was much higher in the dark condition than in the light. (d) Growth curve of Rhizobium sp. MK23 under 0.1% acetate.
Methods
Cyanobacterial strains and culture conditions
Experiments were performed in vitro using M. aeruginosa KW, M. aeruginosa FBC000002 (M. aeruginosa FBC2), and eight collected cyanobloom samples. M. aeruginosa KW was isolated from Wangsong reservoir, Republic of Korea by researchers from the Korea Research Institute of Bioscience & Biotechnology. M. aeruginosa FBC2 was isolated from the Nakdong River, Republic of Korea and cultured by researchers from Freshwater Bioresources Culture Collection. Two axenic strains were used in the experiments to prevent strain-specific bias. The axenic culture of M. aeruginosa NIES-298 was established by the National Institute for Environmental Studies, Japan, and the axenic culture of M. aeruginosa PCC7806 was established by the Pasteur Culture collection of Cyanobacteria, France. These strains were cultured in BG11 medium and at 25 °C and a light intensity of 50 µmol m−2s−1 with a photoperiod of 12 h light/12 h dark under controlled conditions in a growth chamber
Experimental sites and water sampling
Cyanobacteria samples were collected from the following eight sites of cyanobacterial blooms in the Republic of Korea: Murwang reservoir (MW; 37°22′46.5′′N, 126°49′59.5′′E), Wangsong reservoir (WS; 37°18′47.0′′N, 126°56′53.8′′E), Daecheong lake (DC; 36°21′21.8′′N, 127°33′30.4′′E), Baekje barrage (BJ; 36°16′30.5′′N, 126°53′27.9′′E), Juksan barrage (JS; 34°58′29.9′′N 126°36′37.0′′E), Hapcheon Changnyeong barrage (HC; 35°35′32.5′′N, 128°21′25.4′′E), Gangjeong Goryeong barrage (GJ; 35°48′43.0′′N 128°28′38.9′′E), and Bohyun mountain dam (BH; 36°07′25.4′′N 128°56′38.0′′E) in August of 2018 (Fig. S1). The samples were collected from surface water and the water source was profiled in situ at 1 m depth for temperature, pH, DO, and salinity using a multiparameter water quality meter (Table 1) (YSI 556, Yellow Springs Instruments, USA). Regarding BH sample, it was impossible to measure the water quality. In addition, the samples were analyzed for chemical oxygen demand (CODMn) and total nitrogen (TN), and total phosphorus (TP) concentrations using HS-CODMn-L, HS-TN-L (CA), and HS-TP-L kits (Humas, Republic of Korea). Chl-a was extracted with 90% acetone using a spectrophotometer (Spark, Germany). Each water sample was mixed with 1 L of water was added in a plastic bottle and preserved at 4 °C while transporting to the laboratory.
Confocal and scanning electron microscopy
M. aeruginosa and epiphytic bacterial co-cultures were examined via confocal and scanning electron microscopes (SEM). M. aeruginosa KW was cultured in BG11; 1 ml of suspension was added to 5.9 µl of SYTO 9 green-fluorescent nucleic acid stain and vortexed for 30 s. M. aeruginosa cells were stained, for 5 min in the dark. SEM observation was also performed to confirm that various bacteria were attached to the cells of M. aeruginosa. To visualize the coexisting condition, 1 ml of cells was harvested via centrifugation (1 min at 11,500 rcf) and gently washed twice with PBS. The cells were primarily fixed at 4 °C for 4 h (Karnovsky’s fixation method). Thereafter, they were washed thrice at 4 °C with 0.05 M potassium phosphate buffer for 10 min each at 4 °C. The cells were additionally fixed with a mixture of 0.1 M potassium phosphate buffer and 2% osmium tetroxide at 4 °C for 2 h. Next, they were washed twice with distilled water at room temperature. The cells were dehydrated using increasing ethanol concentrations (30, 50, 70, 80, and 90%, and then treated thrice with 100% ethanol, for 10 min each) at room temperature. The samples were coated with platinum prior to examination by FE-SEM (FEI, Japan).
Analysis of bacterial communities in the M. aeruginosa phycospheres
Each sample was filtered through a 3-μm (47 mm, Macherey-Nagel, Germany) pore size polytetrafluoroethylene filter paper and washed twice to remove the free-living bacteria. Metagenomic DNA was extracted from microorganisms in the water using a FastDNA Spin Kit (MP Biomedicals, USA) according to the manufacturer’s instructions, and the DNA yield was quantified using a NanoDrop spectrophotometer (BioTek, USA). The V3-V4 hypervariable region of 16S rRNA gene from the genomic DNA was amplified using primers 341 F (5′-TCGTCGGCAGCGTC-AGATGTGTATAAGAGACAG-CCTACGGGNGGCWGCAG-3′) and 805 R (5′-GTCTCGTGGGCTCGG-AGATGTGTATAAGAGACAG-GACTACHVGGGTATCTAATCC-3′). The amplified products were confirmed by agarose gel electrophoresis. The amplicons were purified by Agencourt AMPure XP (Beckman Coulter, Republic of Korea) and quantified using a Quanti-iT Picogreen dsDNA Assay kit. Equimolar concentrations of each amplicon from the different samples were pooled and purified using Agencourt AMPure XP (Beckman Coulter, Republic of Korea). All sequencing procedures were conducted by ChunLab (Republic of Korea). The sequences obtained were compared and classified using the EzTaxon Database (http://www.ezbiocloud.net). The operational taxonomic units (OTUs) among the samples were obtained with a taxonomic composition using the CLcommunity program (ChunLab, Republic of Korea). The raw sequences obtained from each sample were deposited in the GenBank SRA archive and are available with the following accession numbers: BioProject accession number PRJNA528487; Samples accession numbers: SRX5561067~SRX5561076.
Comparative analysis of the Microcystis strains
The similarity in 16S rRNA sequence among the strains was analyzed using MegAlign software. Percentage identity showed the difference in M. aeruginosa sequence among the environmental samples, based on the KW strains. To compare the optimal culture conditions in BG11 medium for each strain, screening of cyanobacteria was performed with a PhotoBiobox50 at a temperature of 15–40 °C and light intensity of 20–160 µmol m−2 s−1 with a photoperiod of 10 h light/10 h dark. All the strains were incubated in 96-well plates with a working volume of 200 µL. After cultivation for 3 days, fluorescence was measured with the excitation at 420 nm and emission detection at 680 nm using a spectrophotometer (Spark, Germany). Three pairs of primers targeting the mcyA, mcyB, and mcyC genes were used for the amplification (Table 3)32–34. After separating the aggregated colonies by a pipette, the cells were sonicated. A total volume of 20 µL containing 10 µl of cells, 2 µl of each primer, 1.6 µl of dNTPs, 2 µL of 10 × buffer, 0.2 µL of Han-taq (Genenmed, Republic of Korea) polymerase, and 10 µL ddH2O was used for each PCR amplification. The following PCR protocol was used: 94 °C for 5 min (1 cycle); 94 °C for 1 min, 60 °C for 30 sec (mcyA) or 52 °C for 30 sec (mcyB and mcyC), and 72 °C for 1 min (34 cycles), and 72 °C for 10 min (1 cycle). The amplificons were analyzed by agarose (0.8%) gel electrophoresis. The microcystin-LR concentrations in the environmental sample were measured using an ELISA kit (Abnova, Taiwan),.
Table 3.
Primers used in the study.
| Gene region and primer | Sequence (5′-3′) |
|---|---|
| Microcystis 16S rRNA | |
| 209F | ATGTGCCGCGAGGTAAACCTAAT |
| 409R | TTACAATCCAAAGACCTTCCTCCC |
| mcyA | |
| MSF | ATCCAGCAGTTGAGCAAGC |
| MSR | TGCAGATAACTCCGCAGTTG |
| mcyB | |
| 2156F | ATCACTTCAATCTAACGACT |
| 3111R | AGTTGCTGCTGTAAGAAA |
| mcyC | |
| PSCF1 | GCAACATCCCAAGAGCAAAG |
| PSCR1 | CCGACAACATCACAAAGGC |
Isolation of culturable bacteria from M. aeruginosa
Experiments were conducted after filtering the samples using a 3-μm (47 mm, Macherey-Nagel, Germany) pore size polytetrafluoroethylene filter paper, followed by washing. To obtain various bacteria, the culturabilities of the bacteria were investigated by using R2A medium, R2A medium at 1/10 of its normal concentrations, or nutrient agar medium at 1/100 of its normal concentrations as the growth medium51. The colonies were isolated after 4 days of incubation at 30 °C, and at the end of microcosm incubation, 66 colonies per sample were streaked on a fresh medium. Approximately 34 colonies were selected in R2A medium, with 16 colonies in 1/10 R2A, and 1/100 NB mediums each. Prior to bacterial identification, BOX PCR was performed to determine the presence of the same bacteria. The BOX primer52 was inserted in a 20 μl reaction volume that contained 10 μl of cells, 2 μl of 10× taq buffer, 4 μl of primer (5 pmol; BOXA1R, 5′-CTA CGG CAA GGC GAC GCT GAC G-3′), 1.6 μl of dNTPs (10 mM of each), and 0.2 μl of Han-taq (Genenmed, Republic of Korea) polymerase. The reaction mixture was amplified using a Mastercycler nexus X2 (Eppendorf, Germany) with the PCR conditions as follows: an initial denaturation step at 95 °C for 2 min (1 cycle); 94 °C for 3 s, 92 °C for 30 s, 50 °C for 1 min, and 65 °C for 8 min (34 cycle), with a final extension at 65 °C for 8 min (1 cycle). The PCR amplification products (10 μl aliquots) were analyzed by 0.8% agarose gels electrophoresis in Tris-acetate EDTA (TAE) buffer (Bioneer, Republic of Korea). Subsequently, the gels were stained with ethidium bromide (EtBr, 5 μl), and were visualized under UV light.
Phylogenetic analysis of isolated bacteria
In this study, culturable bacteria were identified by partial sequencing of 16S rRNA genes. PCR amplification and sequencing of the 16S rRNA gene from the isolates were performed using universal primers (27 F, 5′-AGAGTTTGATCMTGGCTCAG-3′ and 1492 R, 5′-GGTTACCTTGTTACGACTT-3′). The PCR was performed with 2 µl of each primer, 1.6 µl of dNTPs, 2 µL of 10 × buffer, and 0.2 µL of Han-taq (Genenmed, Republic of Korea) polymerase. The following PCR protocol was used: 94 °C for 90 s (1 cycle); 94 °C for 45 s, 50 °C for 45 s, and 72 °C for 45 s (40 cycles) and 72 °C for 5 min (1 cycle). Pyrosequencing of the PCR product was performed by Macrogen (Republic of Korea). The obtained sequences were compared and classified using the EzTaxon Database (http://www.ezbiocloud.net). Similar sequences were grouped into OTUs based on the manual comparison. The 16S rRNA gene sequences of each sample of the culturable bacteria were aligned using MEGA 7.0 software. After trimming the unaligned regions, a neighbor-joining phylogenetic tree was constructed using MEGA software. The 16S rRNA sequences of the culturable bacteria have been deposited in the National Center for Biotechnology Information (NCBI) GenBank.
Co-culture experiment of M. aeruginosa with culturable bacteria
The bacteria used in the experiment were selected from those that were commonly detected or occupied a large proportion in all the samples and those that were present in only 1–2 samples. M. aeruginosa PCC7806 culture in BG11 media was inoculated with 106 cells/ml bacteria, and one of the M. aeruginosa PCC7806 cultures was left uninoculated. The cultures were grown with 12 h light/12 h dark cycle for 7 days at 25 °C. The growth of M. aeruginosa PCC7806 cells was measured by a hemocytometer. Becasue the Rhizobium species was the best growth- stimulating bacteria for M. aeruginosa PCC7806, co-culture experiments with M. aeruginosa NIES-298 and Rhizobium species were intensively conducted for 30 days. The bovine liver catalase (20,658 units/ml enzyme; Sigma-Aldrich, USA) was added to axenic M. aeruginosa PCC7806 culture for 4 days, and the culture growth was monitored by a hemocytometer. Catalase activity depends on the conversion of the oxidation state cobalt (II) to cobalt (III) by H2O2 in the presence of biocarbonate solution as previously reported53. The concentration of H2O2 in the culture of M. aeruginosa PCC7806 was measured using the Amplex red hydrogen peroxide/peroxidase assay kit (Thermo Fisher Scientific, USA). M. aeruginosa PCC7806 culture in BG11 media without nitrogen source was inoculated with 107 bacterial cells/ml and negative control has no Rhizobium sp. MK23 cells. Because the growth of Rhizobium sp. MK23 depends on carbon sources from PCC7806, the 1% acetate was added to BG11 without nitogen source. The DO levels in the M. aeruginosa PCC7806 culture under the light and dark conditions were measured using a Unisense O2 sensor (Aarhus, Denmark) for 60 h. The same amount of cell supernatant was extracted after 12 h in the light and after 12 h in the dark. Samples were filtered with a 0.45-µm filter and stored at −20 °C until measurement. The concentrations of acetate in the supernatants of M. aeruginosa PCC7806 growth media were measured by HPLC (Waters Co. USA) equipped with refractive index and photodiode array detector. The chromatography was performed on a Bio-Rad Aminex HPX-87H ion-exchange column (7.8 × 300 mm; Bio-Rad Laboratories, USA) at room temperature using 8 mM H2SO4 as the mobile phase at a flow rate of 0.1 mL/min. The injection volume was set to 15 µL, with the detection at 210 nm. The growth of Rizobium sp. MK23 was evaluating the OD600 after adding 0.1% acetate to the M9 media.
Ethical statement
This study did not comprise any experiment or analysis performed on human participants or animals.
Supplementary information
Acknowledgements
This work was supported by a grant from the National Institute of Biological Resources (NIBR), funded by the Ministry of Environment (MOE) of the Republic of Korea (NIBR201920201).
Author contributions
M.K. and W.P. designed this study. M.K., B.S., and J.L. performed the experiments. M.K. analyzed the data and drafted the manuscript. M.K., H.Y.P., and W.P. substantially participated in the discussion and modification of the manuscript. All the authors contributed to and approved the final version of this manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-56882-1.
References
- 1.Rabalais NN, et al. Dynamics and distribution of natural and human-caused hypoxia. Biogeosciences. 2010;7:585–619. doi: 10.5194/bg-7-585-2010. [DOI] [Google Scholar]
- 2.Paerl HW, Otten TG. Harmful cyanobacterial blooms: causes, consequences, and controls. Microb. Ecol. 2013;65:995–1010. doi: 10.1007/s00248-012-0159-y. [DOI] [PubMed] [Google Scholar]
- 3.Schindler DW. Evolution of phosphorus limitation in lakes. Science. 1977;195:260–262. doi: 10.1126/science.195.4275.260. [DOI] [PubMed] [Google Scholar]
- 4.Stone R. China aims to turn tide against toxic lake pollution. Science. 2011;333:1210–1211. doi: 10.1126/science.333.6047.1210. [DOI] [PubMed] [Google Scholar]
- 5.Lewis WM, Wurtsbaugh WA. Control of lacustrine phytoplankton by nutrients: erosion of the phosphorus paradigm. Intern. Rev. Hydrobiol. 2008;93:446–465. doi: 10.1002/iroh.200811065. [DOI] [Google Scholar]
- 6.O’neil JM, Davis TW, Burford MA, Gobler CJ. The rise of harmful cyanobacteria blooms: the potential roles of eutrophication and climate change. Harmful Algae. 2012;14:313–334. doi: 10.1016/j.hal.2011.10.027. [DOI] [Google Scholar]
- 7.Chen Y, Qin B, Teubner K, Dokulil MT. Long-term dynamics of phytoplankton assemblages: Microcystis-domination in Lake Taihu, a large shallow lake in China. J. Plankton Res. 2003;25:445–453. doi: 10.1093/plankt/25.4.445. [DOI] [Google Scholar]
- 8.Preston T, Stewart WDP, Reynolds CS. Bloom-forming cyanobacterium Microcystis aeruginosa overwinters on sediment surface. Nature. 1980;288:365–367. doi: 10.1038/288365a0. [DOI] [Google Scholar]
- 9.Joung SH, et al. Simple method for a cell count of the colonial cyanobacterium, Microcystis sp. J. Microbiol. 2006;44:562–565. [PubMed] [Google Scholar]
- 10.Harke MJ, et al. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae. 2016;54:4–20. doi: 10.1016/j.hal.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 11.Bishop CT, Anet EFLJ, Gorham PR. Isolation and identification of the fast-death factor in Microcystis aeruginosa NRC-1. Can. J. Biochem. Physiol. 1959;37:453–471. doi: 10.1139/y59-047. [DOI] [PubMed] [Google Scholar]
- 12.Nishizawa T, Asayama M, Fujii K, Harada K, Shirai M. Genetic analysis of the peptide synthetase genes for a cyclic heptapeptide microcystin in Microcystis spp. J. Biochem. 1999;126:520–529. doi: 10.1093/oxfordjournals.jbchem.a022481. [DOI] [PubMed] [Google Scholar]
- 13.Sangolkar LN, Maske SS, Chakrabarti T. Methods for determining microcystins (peptide hepatotoxins) and microcystin-producing cyanobacteria. Water Res. 2006;40:3485–3496. doi: 10.1016/j.watres.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Meissner K, Dittmann E, Börner T. Toxic and non-toxic strains of the cyanobacterium Microcystis aeruginosa sequences homologous to peptide synthetase genes. FEMS Microbiol. Lett. 1996;135:295–303. doi: 10.1111/j.1574-6968.1996.tb08004.x. [DOI] [PubMed] [Google Scholar]
- 15.Ouellette AJA, Wilhelm SW. Toxic cyanobacteria: the evolving molecular toolbox. Front. Ecol. Environ. 2003;7:359–366. doi: 10.1890/1540-9295(2003)001[0359:TCTEMT]2.0.CO;2. [DOI] [Google Scholar]
- 16.Otten TG, Crosswell JR, Mackey S, Dreher TW. Application of molecular tools for microbial source tracking and public health risk assessment of a Microcystis bloom traversing 300 km of the Klamath River. Harmful Algae. 2015;46:71–81. doi: 10.1016/j.hal.2015.05.007. [DOI] [Google Scholar]
- 17.Harke MJ, Berry DL, Ammerman JW, Gobler CJ. Molecular response of the bloom-forming cyanobacterium, Microcystis aeruginosa, to phosphorus limitation. Microb. Ecol. 2012;63:188–198. doi: 10.1007/s00248-011-9894-8. [DOI] [PubMed] [Google Scholar]
- 18.Harke M, Davis T, Watson S, Gobler CJ. Nutrient-controlled niche differentiation of western Lake Erie cyanobacterial populations revealed via metatranscriptomic surveys. Env. Sci. Technol. 2015;50:604–615. doi: 10.1021/acs.est.5b03931. [DOI] [PubMed] [Google Scholar]
- 19.Jiang L, et al. Quantitative studies on phosphorus transference occurring between Microcystis aeruginosa and its attached bacterium (Pseudomonas sp.) Hydrobiologia. 2007;581:161–165. doi: 10.1007/s10750-006-0518-0. [DOI] [Google Scholar]
- 20.Xing P, Kong FX, Cao HS, Zhang M, Tan X. Variations of bacterioplankton community composition during Microcystis spp. bloom in a shallow eutrophic lake. J. Freshw. Ecol. 2007;22:61–67. doi: 10.1080/02705060.2007.9664146. [DOI] [Google Scholar]
- 21.Gumbo JR, Colet TE. Light and electron microscope assessment of the lytic activity of Bacillus on Microcystis aeruginosa. Afr. J. Biotchnol. 2011;10:8054–8063. doi: 10.5897/AJB10.1311. [DOI] [Google Scholar]
- 22.Parveen B, et al. Bacterial communities associated with Microcystis colonies differ from free-living communities living in the same ecosystem. Env. Microbiol. Rep. 2013;5:716–724. doi: 10.1111/1758-2229.12071. [DOI] [PubMed] [Google Scholar]
- 23.Salomon PS, Janson S, Graneli E. Molecular identification of bacteria associated with filaments of Nodularia spumigena and their effect on the cyanobacterial growth. Harmful Algae. 2003;2:261–272. doi: 10.1016/S1568-9883(03)00045-3. [DOI] [Google Scholar]
- 24.Manage PM, Kawabata Z, Nakano S. Algicidal effect of the bacterium Alcaligenes denitrificans on Microcystis spp. Aquat. Microb. Ecol. 2000;22:111–117. doi: 10.3354/ame022111. [DOI] [Google Scholar]
- 25.Amin, S.A., Parker, M.S. & Armbrust, E.V. Interactions between diatoms and bacteria. Microbiol Mol Biol Rev., 76, 667–684, doi: 101128/MMBR.00007-12 (2012). [DOI] [PMC free article] [PubMed]
- 26.Paerl, H. W. Interactions with bacteria. In: Carr NG, Whitton BA (ed.) The biology of cyanobacteria, 17rd edn. Oxford, UK, pp 441−461 (1982).
- 27.Sharifah EN, Eguchi M. The phytoplankton Nannochloropsis oculata enhances the ability of Roseobacter clade bacteria to inhibit the growth of fish pathogen Vibrio anguillarum. PLoS One. 2011;6:e26756. doi: 10.1371/journal.pone.0026756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz-Marquez JCF, Nascimento MD, Dublan MA, Curatti L. Association with an ammonium-excreting bacterium allows diaztrophic culture of oil-rich eukaryotic bacteria. Appl. Env. Microbiol. 2012;78:2345–2352. doi: 10.1128/AEM.06260-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo Z, Tong YW. The interactions between Chlorella vulgaris and algal symbiotic bacteria under photoautotrophic and photoheterotrophic conditions. J. Appl. Phycol. 2014;26:1483–1492. doi: 10.1007/s10811-013-0186-1. [DOI] [Google Scholar]
- 30.Krausfeldt LE, et al. Urea is both a carbon and nitrogen source for Microcystis aeruginosa: tracking 13C incorporation at bloom pH conditions. Front. Microbiol. 2019;10:1064. doi: 10.3389/fmicb.2019.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao CS, et al. Predicting cyanobacteria bloom occurrence in lakes and reservoirs before blooms occur. Sci. Total. Environ. 2019;20:837–848. doi: 10.1016/j.scitotenv.2019.03.161. [DOI] [PubMed] [Google Scholar]
- 32.Tillett D, Parker DL, Neilan BA. Detection of toxigenity by a probe for the microcystin synthetase A gene (mcyA) of the cyanobacterial genus Microcystis: comparison of toxicities with 16S rRNA and phycocyanin operon (phycocyanin intergenic spacer) phylogenies. Appl. Env. Microbiol. 2001;67:2810–2818. doi: 10.1128/AEM.67.6.2810-2818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mikalsen B, et al. Natural variation in the microcystin synthetase operon mcyABC and impact on microcystin production in Microcystis strains. J. Bacteriol. 2003;185:2774–2785. doi: 10.1128/JB.185.9.2774-2785.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouahid Y, Perez-Silva G, del Campo FF. Identification of potentially toxic environmental Microcystis by individual and multiple PCR amplification of specific microcystin synthetase gene regions. Env. Toxicol. 2005;20:235–242. doi: 10.1002/tox.20103. [DOI] [PubMed] [Google Scholar]
- 35.Qin B, et al. Spatiotemporal changes of cyanobacterial bloom in large shallow eutrophic lake Taihu, China. Front. Microbiol. 2018;9:451. doi: 10.3389/fmicb.2018.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akins LN, Ayayee P, Leff LG. Composition and diversity of cyanobacteria-associated and free-living bacterial communities during cyanobacterial blooms. Ann. Microbiol. 2018;68:493–503. doi: 10.1007/s13213-018-1354-y. [DOI] [Google Scholar]
- 37.Yang X, Wu X, Hao H, He Z. Mechanisms and assessment of water eutrophication. J. Zhejiang Univ. Sci. B. 2008;9:197–209. doi: 10.1631/jzus.B0710626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devrim C, Dev TB, Weiming S, Herbert JK. Nitrogen transformations in modern agriculture and the role of biological nitrification inhibition. Nat. plants. 2017;3:17074. doi: 10.1038/nplants.2017.74. [DOI] [PubMed] [Google Scholar]
- 39.Eckert B, et al. Azospirillum doebereinerae sp. Nov., a nitrogen-fixing bacterium associated with the C4-grass Miscanthus. Int. J. Syst. Evol. Microbiol. 2001;51:17–26. doi: 10.1099/00207713-51-1-17. [DOI] [PubMed] [Google Scholar]
- 40.Xie CH, Yokota A. Azospirillum oryzae sp. Nov., a nitrogen-fixing bacterium isolated from the roots of the rice plant Oryza sativa. Int. J. Syst. Evol. Microbiol. 2005;55:1435–1438. doi: 10.1099/ijs.0.63503-0. [DOI] [PubMed] [Google Scholar]
- 41.Riemann L, Winding A. Community dynamics of free-living and particle-associated bacterial assemblages during freshwater phytoplankton bloom. Microb. Ecol. 2001;42:274–285. doi: 10.1007/s00248-001-0018-8. [DOI] [PubMed] [Google Scholar]
- 42.Debroas D, et al. Metagenomic approach studying the taxonomic and functional diversity of the bacterial community in a mesotrophic lake (Lac du Bourget-France) Env. Microbiol. 2009;11:2412–2424. doi: 10.1111/j.1462-2920.2009.01969.x. [DOI] [PubMed] [Google Scholar]
- 43.Haukka K, et al. Effect of nutrient loading on bacterioplankton community composition in lake mesocosms. Microb. Ecol. 2006;51:137–146. doi: 10.1007/s00248-005-0049-7. [DOI] [PubMed] [Google Scholar]
- 44.Parveen B, Mary I, Vellet A, Ravet V, Debroas D. Temporal dynamics and phylogenetic diversity of free-living and particle-associated Verrucomicrobia communities in relation to environmental variables in a mesotrophic lake. FEMS Microbiol. Ecol. 2013;83:189–201. doi: 10.1111/j.1574-6941.2012.01469.x. [DOI] [PubMed] [Google Scholar]
- 45.Eiler A, Olsson JA, Bertilsson S. Diurnal variation in the auto- and heterotrophic activity of cyanobacterial phycospheres (Gloeotrichia echinulata) and the identity of attached bacteria. Freshw. Biol. 2006;51:298–311. doi: 10.1111/j.1365-2427.2005.01493.x. [DOI] [Google Scholar]
- 46.Berg KA, et al. High diversity of cultivable heterotrophic bacteria in association with cyanobacterial water blooms. ISME J. 2009;3:314–325. doi: 10.1038/ismej.2008.110. [DOI] [PubMed] [Google Scholar]
- 47.Hennecke H, et al. Concurrent evolution of nitrogenase genes and 16S rRNA in Rhizobium species and other nitrogen fixing bacteria. Arch. Microbiol. 1985;142:342–348. doi: 10.1007/BF00491901. [DOI] [Google Scholar]
- 48.Rapala J, Sivonen K, Lyra C, Niemelä SI. Variation of microcystins, cyanobacterial hepatotoxins, in Anabaena spp. as a function of growth stimuli. Appl. Env. Microbiol. 1997;63:2206–2212. doi: 10.1128/aem.63.6.2206-2212.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moezlelaar R, Stal LJ. Fermentation in the unicellular cyanobacterium Microcystis PCC7806. Arch. Microbiol. 1994;162:63–69. doi: 10.1007/BF00264374. [DOI] [Google Scholar]
- 50.Heo J, Cho DH, Ramanan R, Oh HM, Kim HS. PhotoBiobox: A tablet sized, low cost, high throughput photobioreactor for microalgal screening and culture optimization for growth, lipid content and CO2 sequestration. Biochem. Eng. J. 2015;103:193–197. doi: 10.1016/j.bej.2015.07.013. [DOI] [Google Scholar]
- 51.Janssen PH, Yates PS, Grinton BE, Talyor PM, Sait M. Improved culturability of soil bacteria and isolation in pure culture of novel members of the divisions Acidobacteria, Actinobacteria, Proteobacteria, and Verrucomicrobia. Appl. Env. Microbiol. 2002;68:2391–2396. doi: 10.1128/AEM.68.5.2391-2396.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Versalovic J, Schneider M, Bruijin FJ, Lupski JR. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol. Cell Biol. 1994;5:25–40. [Google Scholar]
- 53.Hadwan MN. Simple spectrophotometric assay for measuring catalase activity in biological tissues. BMC Biochem. 2018;19:7. doi: 10.1186/s12858-018-0097-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.