Abstract
Crassulacean acid metabolism (CAM) photosynthesis is an innovation of carbon concentrating mechanism that is characterized by nocturnal CO2 fixation. Recent progresses in genomics, transcriptomics, proteomics, and metabolomics of CAM species yielded new knowledge and abundant genomic resources. In this review, we will discuss the pattern of cis-elements in stomata movement-related genes and CAM CO2 fixation genes, and analyze the expression dynamic of CAM related genes in green leaf tissues. We propose that CAM photosynthesis evolved through the re-organization of existing enzymes and associated membrane transporters in central metabolism and stomatal movement-related genes, at least in part by selection of existing circadian clock cis-regulatory elements in their promoter regions. Better understanding of CAM evolution will help us to design crops that can thrive in arid or semi-arid regions, which are likely to expand due to global climate change.
Subject terms: Evolutionary biology, Plant evolution
Introduction
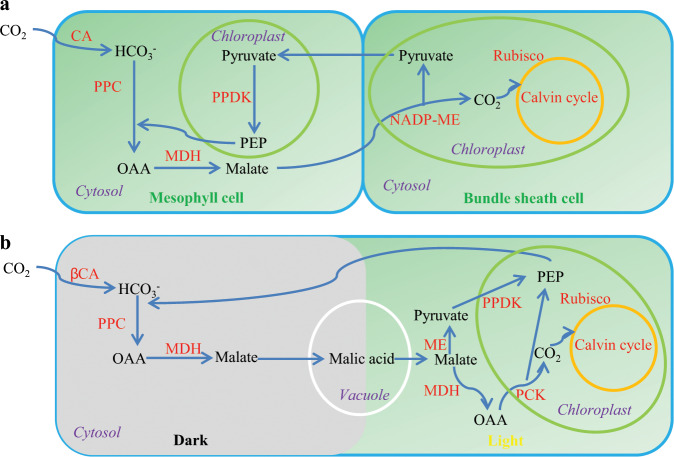
Photosynthesis is the process that harvests solar energy to synthesize organic compounds that can ultimately be utilized to drive cellular processes by all forms of life. Photosynthesis is known from cyanobacteria to their descendants including algae and vascular plants1. There are three different photosynthetic pathways in terrestrial plants for fixation of carbon dioxide (CO2): C3, C4, and CAM. C3 photosynthesis is employed by most vascular plants. C4 plants represent about 3% of vascular plants2, while CAM plants represent about 6%3. Both C4 and CAM are add-ons to the C3 pathway. C4 and CAM metabolisms are similar in biochemistry but CO2 concentration steps are spatially separated in C4 rather than temporally as in CAM. C4 minimizes photorespiration by concentrating CO2 in bundle sheath cells, which relies in part on the unique cellular structure (Fig. 1a). Many C4 plants are agronomically important species, such as maize and sugarcane4. CAM plants have high water-use efficiency (WUE, expressed as mmol CO2 mol−1 H2O), which is a direct consequence of the fact that they open their stomata at night and keep them closed during the daytime5. WUE for carbon assimilation in CAM plants is much higher than in C3 or C4 plants. It will be 2–10 times higher than that of C4 plants and 2.6–20 times higher than that of C3 plants6. One of the major differences between C4 and CAM photosynthesis centers on the temporal regulation of CO2 absorption and fixation (Fig. 1).
Fig. 1. Photosynthetic reactions in C4 and CAM plants.
a NADP-malic enzyme type of C4 pathway. b Carbon fixation in CAM plants.
CAM is found in over 400 genera across 36 families of vascular plants7 and has evolved multiple times independently from diverse ancestral C3 plants3. While we know ecological factors such as drought condition and CO2 concentration drive the evolution of CAM8–10, far less is known about genetics. Gene family duplication was previously proposed as the driver of CAM metabolism evolution through neofunctionalization of newly duplicated paralogous genes3, while others proposed that C4 and CAM photosynthesis may have arisen through the re-organization of metabolic processes already present in C3 plants11–13.
The past few years have seen the rapid progresses in genomics, transcriptomics, proteomics, and metabolomics for an increasing number of plant species including CAM species. The orchid Phalaenopsis equestris was the first CAM species for which the genome was assembled in 201514. In the same year, the genome of fruit pineapple (Ananas comosus var. comosus) cultivar ‘F153’, which has been cultivated by Del Monte for 50 years, was sequenced, and the evolution of CAM photosynthesis was investigated15. In a following work, temporal and spatial transcriptomic profiles of CAM-performing mature leaves have also been studied in fruit pineapple16. Recently, we sequenced the bracteatus pineapple (Ananas comosus var. bracteatus) accession CB5 genome, and assembled to chromosomal level17. The genome of Kalanchoë fedtschenkoi, a eudicot CAM species was also available in 201718. Furthermore, multi-dimensional omics data were available for CAM species, such as Agave americana19,20, ochids21,22, and Talinum triangulare23. These progresses for species that evolved CAM independently provide an excellent resource for comparative analyses, which will help us have a better understanding on the evolution of CAM photosynthesis.
Cis-elements of stomatal movement-related genes
Stomata were first described as ‘pore-like’ structures on the surface of leaves over three centuries ago24. Since the earliest examples of stomata were discovered in the leaf fossil record, plants have been evolving in terms of size and density of stomata to maintain the maximum leaf conductance as the atmosphere CO2 changed25. Stomata play an essential role in controlling of transpiration rate and water homeostasis in plants26. Stomatal movement can be stimulated by different environmental factors, such as light, abscisic acid (ABA), pathogens, CO2 and air humidity27. Among them, air humidity and ABA are directly related to water status in plants28. In CAM plants, the diel rhythms of stomatal conductance and transpiration are closely linked to the net CO2-uptake rhythm5.
CAM plants present a reverse stomatal conductance pattern by assimilating CO2 during the night when the temperature is low resulting in lower evapotranspiration rate compared to C3 and C4 plants29,30. This unique pattern of stomatal movement leads to the higher WUE in CAM plants31. The reverse stomatal rhythm has aroused curiosity and investigation for centuries32. Understanding the regulation of stomatal movement-related genes in CAM species may provide promising opportunities for engineering crops with higher WUE32.
We identified 118 stomatal movement-related genes in A. comosus var. comosus, 95 in A. comosus var. bracteatus, 121 in P. equestris, 140 in Arabidopsis, 123 in rice, and 121 in sorghum (Supplementary Table S1). Based on the GO annotation, the stomatal movement-related genes were divided into three categories, including genes involved in stomatal opening, stomatal closure, and regulation of stomatal movement. For genes involved in stomatal movement, the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1)-binding site (CBS; AAAAATCT) and G-box binding site (CACGTG) showed more than 10% or higher frequency than the expected frequencies based on random chance in A. comosus var. comosus (Table 1). The G-box element was enriched in genes involved in all three categories in A. comosus var. comosus (Table 1). The evening element (EE; AAAATATC) and CBS were enriched in 123 stomatal movement-related genes in rice, whereas the morning element (MOE; CCACAC) was only enriched in stomatal opening category in rice (Table 1). When comparing with non-CAM species, Motif ERF73, ERF7, and ABR1 were enriched in CAM species (Supplementary Table S2). Based on these in silico findings, we propose the hypothesis that these different sets of cis elements regulate stomatal opening during the day and closure during the night for these three C3 and C4 species. Interestingly, stomatal movement-related genes in A. comosus var. comosus have higher frequencies of circadian clock cis-regulatory elements than A. comosus var. bracteatus (Table 1).
Table 1.
Frequency of circadian clock-associated motifs (per kb) in 2 kb promoter regions of genes involved in stomatal movementa.
| Circadian Motif | A. comosus var. comosus | A. comosus var. bracteatus | P. equestris | Arabidopsis | rice | sorghum |
|---|---|---|---|---|---|---|
| Genes involved in stomatal movementb | ||||||
| MOE | 0.212 (0.250) | 0.170 (0.269) | 0.132 (0.152) | 0.164 (0.215) | 0.333 (0.367) | 0.281 (0.327) |
| EE | 0.110 (0.105) | 0.101 (0.096) | 0.107 (0.168) | 0.125 (0.124) | 0.106 (0.059) | 0.074 (0.072) |
| CBS | 0.148 (0.105) | 0.160 (0.096) | 0.087 (0.168) | 0.118 (0.124) | 0.069 (0.059) | 0.058 (0.072) |
| G-box | 0.288 (0.250) | 0.250 (0.269) | 0.120 (0.152) | 0.232 (0.215) | 0.317 (0.367) | 0.248 (0.327) |
| Regulation of stomatal movement | ||||||
| MOE | 0.209 | 0.180 | 0.156 | 0.174 | 0.276 | 0.256 |
| EE | 0.115 | 0.090 | 0.065 | 0.107 | 0.141 | 0.103 |
| CBS | 0.203 | 0.172 | 0.097 | 0.112 | 0.090 | 0.064 |
| G-box | 0.284 | 0.270 | 0.130 | 0.247 | 0.359 | 0.231 |
| Stomatal closure | ||||||
| MOE | 0.185 | 0.188 | 0.089 | 0.121 | 0.397 | 0.296 |
| EE | 0.093 | 0.094 | 0.214 | 0.136 | 0.069 | 0.037 |
| CBS | 0.074 | 0.188 | 0.071 | 0.152 | 0.034 | 0.074 |
| G-box | 0.296 | 0.000 | 0.089 | 0.152 | 0.259 | 0.352 |
| Stomatal opening | ||||||
| MOE | 0.250 | 0.206 | 0.088 | 0.184 | 0.471 | 0.353 |
| EE | 0.111 | 0.176 | 0.118 | 0.184 | 0.000 | 0.000 |
| CBS | 0.028 | 0.088 | 0.059 | 0.079 | 0.029 | 0.000 |
| G-box | 0.278 | 0.235 | 0.118 | 0.289 | 0.206 | 0.147 |
aClock-associated motifs with ≥10% frequency higher than expected frequency appearance in stomatal movement-related genes than expected genome wide frequency is highlighted in bold
bSpecies-specific expected frequencies are indicated in brackets. For example, in A. comosus var. comosus, since the genome GC content is 38%, G/C and A/T occurrence probabilities are 0.19 and 0.31. Both forward and reversed strands were included to calculate the expected frequency of the motif occurrence per kb (For MOE (CCACAC): 0.19 × 0.19 × 0.31 × 0.19 × 0.31 × 0.19 × 2 × 1,000 = 0.250)
In A. americana, the temporal re-programming of particular genes, including CO2 and ABA signaling and turgor pressure regulating genes are essential to regulate stomatal movement19. Comparative transcriptomic analyses between the C3 and CAM Erycina species also showed that genes involved in light and ABA signaling are altered22. The numbers of genes that contain cis-elements involved in several key stomatal movement pathways, such as light, ABA, and stress, are summarized (Table 2). ABA responsiveness-related motif (ABRE) appeared most frequently compared to other signaling pathways in the three species (Arabidopsis, P. equestris, and sorghum) with different photosynthetic pathways. From previous studies, exogenous ABA can induce stomatal closure and the expression and activity of CAM33,34. Moreover, stress-related motif (STRE) was the most frequent in the stomatal related genes of rice and pineapple (Table 2). The stress-induced stomatal movement signaling pathway is closely related to the water status of the plant35. Further genomic and molecular analysis of potential stomatal movement genes will enable us to have a comprehensive understanding of stomatal biology of CAM plants, and might provide candidate genes for engineering crop plants with higher sustainable production32,36.
Table 2.
The number of genes and their percentages to the total genes of the genomes that contain cis-elements involved in partial key stomatal movement pathways annotated at promoter regions of orthologs in A. comosus var. comosus, A. comosus var. bracteatus, P. equestris, Arabidopsis, rice, and sorghum.
| Pathways | A. comosus var. comosus | A. comosus var. bracteatus | P. equestris | Arabidopsis | rice | sorghum | |
|---|---|---|---|---|---|---|---|
| Light responsiveness | Box 4a | 107 (0.40%) | 69 (0.23%) | 30 (0.10%) | 116 (0.24%) | 89 (0.17%) | 76 (0.22%) |
| GT1-motifb | 73 (0.27%) | 42 (0.14%) | 38 (0.13%) | 101 (0.21%) | 80 (0.15%) | 61 (0.18%) | |
| GA-motifc | 25 (0.09%) | 11 (0.04%) | 19 (0.06%) | 29 (0.06%) | 17 (0.03%) | 33 (0.10%) | |
| G-Boxd | 40 (0.15%) | 18 (0.06%) | 36 (0.12%) | 40 (0.08%) | 50 (0.10%) | 105 (0.31%) | |
| TCT-motife | 75 (0.28%) | 46 (0.16%) | 48 (0.16%) | 112 (0.23%) | 55 (0.10%) | 56 (0.16) | |
| ABA responsiveness | ABREf | 101 (0.37%) | 65 (0.22%) | 58 (0.20%) | 123 (0.25%) | 115 (0.22%) | 112 (0.33%) |
| Stress | WUN-motifg | 29 (0.11%) | 27 (0.09%) | 42 (0.14%) | 62 (0.13%) | 47 (0.09%) | 43 (0.13%) |
| STREh | 116 (0.43%) | 89 (0 .30%) | 35 (0.12%) | 82 (0.17%) | 116 (0.22%) | 104 (0.30%) |
a Box 4-motif (ATTAAT)
bGT1-motif (GGTTAA)
cGA-motif (ATAGATAA)
dG-Box (CACGTG)
eTCT-motif (TCTTAC)
fABRE-motif (ACGTG)
gWUN-motif (AAATTACT)
hSTRE motif (AGGGG)
Diurnal transcript abundance patterns of CAM pathway genes: pineapple as an example
The pineapple genome assembly also allowed the identification of full- and partial-length predicted amino-acid sequences of the key metabolic enzymes comprising the core carboxylation module of CAM responsible for nocturnal fixation of CO215,31,37. Carbonic anhydrase (CA), catalyzing the conversion of CO2 into HCO3–, is responsible for the first step in CO2 assimilation both in C4 and CAM plants. All three CA subfamily (α, β, and γ) enzymes were identified in pineapple genome (Supplementary Table S3). Only βCA genes (AccβCA2–1 and AccβCA2–2) implicated in CAM-specific roles due to their mRNA abundance in green leaf tissue15, indicating that βCA may acts as the enzyme in the initiation of CO2 fixation.
Three genes encoding the key enzyme PPC responsible for nocturnal CO2 fixation were identified in the genome assembly, all of which are predicted to be localized to the cytosol as expected15. Three PPC genes were identified in comosus pineapple genome (Supplementary Table S3, Supplementary Fig. S1). Among these three PPC genes, AccPPC1 is the most abundant transcript (>3000 FPKM, fragments per kilobase of exon per million fragments mapped) and displayed highest abundance at 6 pm (>5500 FPKM) in CAM-performing leaf tissues. In T. triangulare, a facultative CAM species, PPC was upregulated 25-fold (to 15,510 rpm, reads per million) at midnight on day 9 and 12 of water limitation when indicative of CAM was observed23. Comparative transcriptomic analyses between the C3 and CAM Erycina species also demonstrated that PPC gene in CAM Erycina displayed higher abundance than in C3 Erycina22. These results suggest that high levels of PPC transcripts are important for CAM.
PPC undergoes reversible N-terminal phosphorylation by a circadian clock-controlled PPC kinase (PPCK), which reduces the sensitivity of the enzyme to allosteric inhibition by L-malate and increases its affinity for its substrate phosphoenolpyruvate (PEP)38,39. In A. americana, which is an obligate CAM plant, PPCK1 gene displayed diel transcripts abundance pattern, suggesting its important role in temporal re-programming of CAM20. In K. fedtschenkoi, PPCK1 is also essential for nocturnal CO2 fixation; moreover, knock-down of oscillations in the transcript abundance of PPCK1 will lead to the altered accumulation and periodicity of core circadian clock-related transcripts40. In pineapple, AccPPCK2 was found to exhibit greater mRNA abundance than AccPPCK1, and AccPPCK2 also displayed diel mRNA abundance with high levels at night, suggesting that it functions in CAM15.
In the final metabolic step of phase I, the OAA formed as a result of PEP carboxylation is reduced to malate by NAD(P)-dependent malate dehydrogenase (MDH). Fourteen genes in pineapple encode MDH: three genes (AccMDH4, AccMDH5, and AccMDH8) are predicted to be cytosolic-localized and strongly expressed in leaves, suggesting their potential to perform functional roles in CAM; four genes (AccMDH10, AccMDH11, AccMDH12, and AccMDH13) are tandemly duplicated and lowly expressed except AccMDH1315.
In Arabidopsis, the malate is transported into the vacuole by an inward-rectifying anion-selective ion channel belonging to the aluminium-activated malate transporter (ALMT) family41. In K. fedtschenkoi, a putative ALMT6 gene (Kaladp0062s0038) displays diel mRNA abundance in leaves18. There are eight candidate ALMT family genes in pineapple, including three ALMT9 genes (AccALMT9-1–3) and five ALMT1 genes (AccALMT1-1–5). Only two ALMT9 genes (AccALMT9-1 and AccALMT9-3) showed high abundant transcript levels in photosynthetic leaf tissues. ALMT1 only has higher steady-state transcript levels at the midday on day 9 of water limitation in T. triangulare23. The malate then undergoes protonation, with protons supplied by the tonoplast H+-ATPase and H+-PPiase, and is stored as malic acid. In the daytime, malic acid is effluxed out of the vacuole possibly through a putative tonoplast dicarboxylate transporter (tDT)42. There are five DT genes (AccDT1–5) in the pineapple genome, and AccDT2 and AccDT3 display specifically high abundant transcripts in daytime in photosynthetic leaf tissues, indicating that they may play a role in malic acid efflux in CAM. Decarboxylation of the malate during phase III of the CAM cycle occurs in pineapple primarily via PEP carboxykinase (PCK)30,43, which, following oxidation of malate to OAA by NAD(P)-dependent MDH, decarboxylates OAA to PEP. A single PCK gene (AccPCK1) is present in the pineapple genome and it is predicted to encode a cytosolic enzyme15. It is an ortholog of AtPCK1 (AT4G37870.1), one of two PCK genes in Arabidopsis, which is expressed in guard cells and is implicated in stomatal closure44. Despite the fact that extractable PCK activity from pineapple leaves is over 15 times higher than that of the malic enzymes (MEs)45, and it remains possible that malate may also be decarboxylated, in part, by ME in pineapple46. The comosus pineapple genome contains five ME genes encoding both NAD- and NADP-ME (Supplementary Table S3): two NADP-ME genes (AccNADP-ME1 and AccNADP-ME3) exhibit higher mRNA levels during the daytime in photosynthetic leaf tissues and one additional NADP-ME gene (AccNADP-ME2) shows none mRNA transcript in leaves; two NAD-ME genes (AccNAD-ME1 and AccNAD-ME2) encoding isoforms predicted to be localized to the mitochondria exhibit moderate abundant mRNA expression and AccNAD-ME2 also displayed higher mRNA level during the daytime15.
The abundant transcript level for ME genes in pineapple suggests that malate decarboxylation also results in the formation of pyruvate, which must then be phosphorylated to PEP by pyruvate phosphate dikinase (PPDK). Consistent with this supposition, a single candidate PPDK1 gene (AccPPDK1) was identified in the pineapple genome15, providing the metabolic flexibility to allow gluconeogenesis via both the PCK and ME/PPDK routes47. AccPPDK1 displayed higher transcript abundance during the daytime. The AtPPDK1 gene encodes an enzyme predicted to be localized to the cytosol, but this enzyme might be localized to either the chloroplast or the cytosol depending upon the production of alternative transcripts arising from two different promoters48. More detailed examination of this locus in pineapple is needed to verify this possibility. Overall, the enzymes making up the carboxylation and decarboxylation pathways in the CAM cycle in pineapple are encoded by gene families that are generally smaller than those encoded by the A. thaliana genome, because pineapple has one fewer whole-genome duplications than that have been reported for Arabidopsis and the grass family49.
Circadian clock-associated cis-elements in CAM genes
In most living organisms, internally synchronized circadian clocks make it possible for them to coordinate behavior and physiology corresponding with the 24 h light-dark cycle. CCA1 and LATE ELONGATED HYPOCOTYL (LHY), two single-MYB domain transcription factors, are central to the circadian oscillator of angiosperms50,51. CCA1 and LHY are morning expressed genes. They act to suppress the expression of the DNA sequence they bind to. CCA1 and LHY are partially redundant, and they can directly bind to the TIMING OF CAB EXPRESSION 1 (TOC1) also known as PRR1 (PSEUDO-RESPONSE REGULATOR 1) promoter to negatively regulate its expression52.
Circadian control of CAM has been implicated as a core component in diel re-programming of metabolism in CAM plants20,53. A comprehensive spatial and temporal survey of gene co-expression clusters in pineapple leaf tissues reveals CAM pathway genes are enriched with clock-associated cis-elements, suggesting circadian regulation of CAM15,16. At dawn, CCA1 and LHY repress evening-phased genes by binding to CBS and EE49. In addition to CBS and EE, the G-box is also enriched in the CCA1 binding regions54,55. TOC1 can bind to MOE as a negative regulator56. In pineapple, all of the three βCA genes contain CBS in their promoter regions (Table 3), suggesting they may have function in βCA genes’ nighttime and early-morning transcripts abundance pattern in photosynthetic leaf tissues. All three copies of PPC genes also contain CBS in their promoter regions, along with MOE or G-box (Table 3). Interestingly, CAM pathway genes in A. comosus var. comosus, contain more circadian clock cis-regulatory elements than A. comosus var. bracteatus (Table 3). Besides the core CAM genes, more than 40% of transcription factors and transcription co-regulators displayed diel rhythmic expression in pineapple, suggesting it is a global adaptation57. In a recent work by Heyduk and colleagues (2018), they demonstrated that some canonical CAM genes were unaltered by comparative transcriptomic analyses between the C3 and CAM Erycina species. However, 149 gene families, including genes involved in light and ABA signaling, had significant differences in network connectivity, indicating that transcriptional cascades changes are critical for the transition from C3 to CAM in Erycina22.
Table 3.
Cis-elements annotated at promoter regions of selected CAM photosynthetic genes in pineapple.
| CAM enzyme | A. comosus var. comosus | A. comosus var. bracteatus | ||
|---|---|---|---|---|
| Gene ID | TF binding motif | Gene ID | TF binding motif | |
| βCA | Aco002732 | CBS (2) | CB5.v30091940 | CBS (2), G-Box (3) |
| Aco005402 | CBS (1) | CB5.v30297520 | CBS (2) | |
| Aco006181 | CBS (2) | CB5.v30069370 | – | |
| PPC | Aco010025 | EE (1), MOE (2), CBS (2) | CB5.v30072230 | – |
| Aco018093 | MOE (2), CBS (1) | CB5.v30160110 | EE (1), MOE (1) | |
| Aco016429 | G-box (1), CBS (1) | CB5.v30185780 | – | |
| CB5.v30098990 | G-box (1) | |||
| PPCK | Aco010095 | G-box (1) | CB5.v30291670 | – |
| Aco013938 | G-box (1) | CB5.v30308180 | G-box (3) | |
| MDH | Aco006122 | CBS (1) | CB5.v30030450 | – |
| Aco007734 | CBS (2) | CB5.v30063990 | – | |
| Aco013935 | MOE (2) | CB5.v30119070 | – | |
| Aco002885 | MOE (2), G-box (1) | CB5.v30217860 | – | |
| Aco004349 | MOE (2), G-box (1) | CB5.v30283140 | CBS (2), G-box (2) | |
| Aco014690 | G-box (2) | CB5.v30016950 | CBS (1) | |
| Aco017525 | CBS (3) | CB5.v30081100 | MOE (1) | |
| Aco017526 | – | CB5.v30308160 | MOE (2) | |
| Aco017527 | CBS (1) | CB5.v30167990 | – | |
| Aco017528 | CBS (2) | CB5.v30057750 | EE (1), CBS (1) | |
| Aco019631 | CBS (1) | CB5.v30057770 | – | |
| Aco010232 | MOE (1), G-box (1) | CB5.v30175150 | CBS (1) | |
| Aco004996 | MOE (3) | CB5.v30175160 | CBS (2) | |
| Aco008626 | – | CB5.v30057740 | CBS (1) | |
| CB5.v30057760 | G-box (1) | |||
| NAD_ME | Aco016569 | – | CB5.v30106390 | – |
| Aco007622 | CBS (1), G-box (1) | CB5.v30132860 | CBS (1), G-box (2) | |
| NADP_ME | Aco009967 | G-box (4), MOE (1) | CB5.v30038850 | G-box (2) |
| Aco005631 | CBS (1) | CB5.v30285200 | CBS (1) | |
| Aco005989 | G-box (2) | CB5.v30300470 | G-box (3), ME (1) | |
| PCK | Aco017762 | MOE (2), G-box (1), CBS (1) | CB5.v30124740 | MOE (1), G-box (1), CBS (1) |
| PPDK | Aco024818 | EE (2), G-box (1), CBS (1) | CB5.v30137360 | EE (1), G-box (1), CBS (1) |
Evolution of CAM photosynthesis
C4 and CAM photosynthesis are innovations that evolved in response to decreasing atmospheric levels of CO2 and water-limiting environments2,9. CAM has a higher incidence3, and mutation of CAM genes in CAM species is not lethal40,58. Both C4 and CAM have evolved independently multiple times, even within individual families, or even genera during angiosperm evolution59–61. For example, in the Neotropical family Bromeliaceae, to which pineapple belongs, CAM photosynthesis evolved independently at least four, and probably five times59.
Recruitment of pre-existing mechanisms underlying C3 photosynthesis is adopted in Gynandropsis gynandra (referred to previously as Cleome gynandra), a C4 plant which is relatively closely related to Arabidopsis62. Furthermore, gene duplication also plays a profound role in the evolution of C4. For example, βCA genes are tandemly duplicated in sorghum63. After duplication, some C4 genes, such as C4 PPC genes, NADP-MDH genes, and PPDK genes, underwent adaptive evolution63.
Comparative analyses demonstrated signatures of convergence in protein sequence and re-scheduling of diel transcript abundance of genes involved in nocturnal CO2 fixation, stomatal movement, heat tolerance, the circadian clock, and carbohydrate metabolism14,18,21. Firstly, convergent evolution has been detected in terms of diel cycles of gene transcript abundance18. PPCK is a key regulator of PPC, which can activate PPC by phosphorylating it. Both AccPPCK2 and KfPPCK2 showed diel expression patterns18. Secondly, a convergent amino-acid change in PPC2 was discovered to be shared by K. fedtschenkoi and P. equestris and the PPC2 gene in K. fedtschenkoi is a much lower abundance transcript relative to the CAM-associated PPC1 gene, so the function of PPC2 has yet to be linked to CAM directly in either K. fedtschenkoi or P. equestris18.
These findings are consistent with the hypothesis that the CAM photosynthesis evolved as a result of a re-organization of pre-existing metabolic pathways11,15. These different features were later coordinated to form the functional CAM photosynthesis.
Concluding remarks
Genomic studies have led to a renaissance in CAM research. Recent genomic and transcriptomic information from CAM species has improved our understanding of the evolution of CAM photosynthesis14–21. The identified candidate genes provide initial targets for detailed functional studies of how the CAM genes have evolved through regulation of gene expression to gain the observed spatial and temporal expression patterns, and loss of repressors is certainly involved. It may be possible for us to apply genome editing to verify functions of candidate CAM genes. CRISPR/Cas9 technology will be a powerful tool to get higher order mutants of tandemly duplicated genes in the same chromosome, which is impossible to generate by traditional mutagenesis methods.
Water loss from stomata for C3 plants can be very substantial under hot and dry condition. Adjusting the temporal pattern of stomatal movement genes may be a key evolutionary step for switching stomatal opening from the light period to dark32. Enrichment of different sets of circadian clock regulatory cis elements may have played a role in this dramatic shift in gene regulation in pineapple and P. equestris. CAM photosynthesis and its associated high WUE are key evolutionary innovations that adapted to arid environments and/or low CO2 environment and this valuable trait is a direct consequence of stomatal closure throughout hottest and driest part for the 24 h cycle, and leaf succulence.
Supplementary information
Acknowledgements
This work was supported by the grant 2016NZ0001-1 from the Department of Science and Technology of Fujian Province, National Natural Science Foundation of China (31701052), Distinguished Young Scholars Fund in Fujian Agriculture and Forestry University (xjq201609), Natural Science Foundation of Fujian Province (KJd18032A), and Program for Excellent Youth Talents in Fujian Province University.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-019-0229-0).
References
- 1.Margulis, L. Symbiosis in Cell Evolution. (WH Freeman, 1992).
- 2.Sage RF. The evolution of C4 photosynthesis. New Phytol. 2004;161:341–370. doi: 10.1111/j.1469-8137.2004.00974.x. [DOI] [PubMed] [Google Scholar]
- 3.Silvera, K. et al. Evolution along the crassulacean acid metabolism continuum. Funct. Plant Biol. 37, 995–1010 (2010).
- 4.Hibberd JM, Sheehy JE, Langdale JA. Using C4 photosynthesis to increase the yield of rice—rationale and feasibility. Curr. Opin. Plant Biol. 2008;11:228–231. doi: 10.1016/j.pbi.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Borland AM, et al. Engineering crassulacean acid metabolism to improve water-use efficiency. Trends Plant Sci. 2014;19:327–338. doi: 10.1016/j.tplants.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borland AM, Griffiths H, Hartwell J, Smith JAC. Exploiting the potential of plants with crassulacean acid metabolism for bioenergy production on marginal lands. J. Exp. Bot. 2009;60:2879–2896. doi: 10.1093/jxb/erp118. [DOI] [PubMed] [Google Scholar]
- 7.Smith, J. A. C. & Winter, K. in Crassulacean Acid Metabolism: Biochemistry, Ecophysiologyand Evolution (eds K. Winter & J. A. C. Smith) 1–13 (Springer, 1996).
- 8.Borland AM, et al. Climate-resilient agroforestry: physiological responses to climate change and engineering of crassulacean acid metabolism (CAM) as a mitigation strategy. Plant Cell Environ. 2015;38:1833–1849. doi: 10.1111/pce.12479. [DOI] [PubMed] [Google Scholar]
- 9.Cushman JC. Crassulacean acid metabolism. A plastic photosynthetic adaptation to arid environments. Plant Physiol. 2001;127:1439–1448. doi: 10.1104/pp.010818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffiths H, Cousins AB, Badger MR, von Caemmerer S. Discrimination in the dark. Resolving the interplay between metabolic and physical constraints to phosphoenolpyruvate carboxylase activity during the crassulacean acid metabolism cycle. Plant Physiol. 2007;143:1055–1067. doi: 10.1104/pp.106.088302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.West-Eberhard MJ, Smith JAC, Winter K. Photosynthesis, reorganized. Science. 2011;332:311–312. doi: 10.1126/science.1205336. [DOI] [PubMed] [Google Scholar]
- 12.Brautigam A, Schluter U, Eisenhut M, Gowik U. On the evolutionary origin of CAM photosynthesis. Plant Physiol. 2017;174:473–477. doi: 10.1104/pp.17.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyduk K, Moreno-Villena JJ, Gilman IS, Christin PA, Edwards EJ. The genetics of convergent evolution: insights from plant photosynthesis. Nat. Rev. Genet. 2019;20:485–493. doi: 10.1038/s41576-019-0107-5. [DOI] [PubMed] [Google Scholar]
- 14.Cai J, et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015;47:65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- 15.Ming R, et al. The pineapple genome and the evolution of CAM photosynthesis. Nat. Genet. 2015;47:1435–1442. doi: 10.1038/ng.3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wai CM, et al. Temporal and spatial transcriptomic and microRNA dynamics of CAM photosynthesis in pineapple. Plant J. 2017;92:19–30. doi: 10.1111/tpj.13630. [DOI] [PubMed] [Google Scholar]
- 17.Chen, L. Y., et al. The bracteatus pineapple genome and domestication of clonally propagated crops. Nat. Genet. 51, 1549–1558 (2019). [DOI] [PubMed]
- 18.Yang XH, et al. The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat. Commun. 2017;8:1899. doi: 10.1038/s41467-017-01491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abraham PE, et al. Transcript, protein and metabolite temporal dynamics in the CAM plant Agave. Nat. Plants. 2016;2:16178. doi: 10.1038/nplants.2016.178. [DOI] [PubMed] [Google Scholar]
- 20.Yin HF, et al. Diel rewiring and positive selection of ancient plant proteins enabled evolution of CAM photosynthesis in Agave. BMC Genom. 2018;19:588. doi: 10.1186/s12864-018-4964-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang LS, et al. Origin and mechanism of crassulacean acid metabolism in orchids as implied by comparative transcriptomics and genomics of the carbon fixation pathway. Plant J. 2016;86:545–545. doi: 10.1111/tpj.13159. [DOI] [PubMed] [Google Scholar]
- 22.Heyduk K, et al. Altered gene regulatory networks are associated with the transition from C3 to crassulacean acid metabolism in Erycina (Oncidiinae: Orchidaceae) Front. Plant Sci. 2018;9:2000. doi: 10.3389/fpls.2018.02000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brilhaus D, Brautigam A, Mettler-Altmann T, Winter K, Weber AP. Reversible burst of transcriptional changes during induction of crassulacean acid metabolism in Talinum triangulare. Plant Physiol. 2016;170:102–122. doi: 10.1104/pp.15.01076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woolfenden HC, et al. Models and mechanisms of stomatal mechanics. Trends Plant Sci. 2018;23:822–832. doi: 10.1016/j.tplants.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Beerling DJ, Franks PJ. Evolution of stomatal function in ‘lower’ land plants. New Phytol. 2009;183:921–925. doi: 10.1111/j.1469-8137.2009.02973.x. [DOI] [PubMed] [Google Scholar]
- 26.Grondin A, et al. Aquaporins contribute to ABA-triggered stomatal closure through OST1-mediated phosphorylation. Plant Cell. 2015;27:1945–1954. doi: 10.1105/tpc.15.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kollist H, Nuhkat M, Roelfsema MRG. Closing gaps: linking elements that control stomatal movement. New Phytol. 2014;203:44–62. doi: 10.1111/nph.12832. [DOI] [PubMed] [Google Scholar]
- 28.Taybi T, Cushman JC. Signaling events leading to crassulacean acid metabolism induction in the common ice plant. Plant Physiol. 1999;121:545. doi: 10.1104/pp.121.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.And RAF, Turner NC. Plant productivity in the arid and semiarid zones. Annu. Rev. Plant Physiol. 1978;29:277–317. doi: 10.1146/annurev.pp.29.060178.001425. [DOI] [Google Scholar]
- 30.Osmond CB. Crassulacean acid metabolism: a curiosity in context. Annu. Rev. Plant Biol. 1978;29:379–414. doi: 10.1146/annurev.pp.29.060178.002115. [DOI] [Google Scholar]
- 31.Ming R, Wai CM, Guyot R. Pineapple genome: a reference for monocots and CAM photosynthesis. Trends Genet. 2016;32:690–696. doi: 10.1016/j.tig.2016.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Males J, Griffiths H. Stomatal biology of CAM plants. Plant Physiol. 2017;174:550. doi: 10.1104/pp.17.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taybi T, et al. Differential effects of abscisic acid on phosphoenolpyruvate carboxylase and CAM operation in Kalanchoë blossfeldiana. Plant Biol. 2015;108:240–246. [Google Scholar]
- 34.Chu C, Edwards GE. Induction of crassulacean acid metabolism in the facultative halophyte Mesembryanthemum crystallinum by abscisic acid. Plant Physiol. 1990;93:1253–1260. doi: 10.1104/pp.93.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.El-Sharkaway MA, Cock JH. Water use efficiency of cassava. I. Effects of air humidity and water stress on stomatal conductance and gas exchange. Crop Sci. 1984;24:497–502. doi: 10.2135/cropsci1984.0011183X002400030017x. [DOI] [Google Scholar]
- 36.Lee JS. Stomatal opening mechanism of CAM plants. J. Plant Biol. 2010;53:19–23. doi: 10.1007/s12374-010-9097-8. [DOI] [Google Scholar]
- 37.Xu HM, et al. PGD: Pineapple genomics database. Hortic. Res. 2018;5:66. doi: 10.1038/s41438-018-0078-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nimmo GA, Nimmo HG, Fewson CA, Wilkins MB. Diurnal changes in the properties of phosphoenolpyruvate carboxylase in Bryophyllum leaves: a possible covalent modification. FEBS Lett. 1984;178:199–203. doi: 10.1016/0014-5793(84)80600-6. [DOI] [Google Scholar]
- 39.Carter PJ, Nimmo HG, Fewson CA, Wilkins MB. Circadian rhythms in the activity of a plant protein kinase. EMBO J. 1991;10:2063–2068. doi: 10.1002/j.1460-2075.1991.tb07737.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boxall SF, Dever LV, Knerova J, Gould PD, Hartwell J. Phosphorylation of phosphoenolpyruvate carboxylase is essential for maximal and sustained dark CO2 fixation and core circadian clock operation in the obligate crassulacean acid metabolism species Kalanchoë fedtschenkoi. Plant Cell. 2017;29:2519–2536. doi: 10.1105/tpc.17.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovermann P, et al. The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 2007;52:1169–1180. doi: 10.1111/j.1365-313X.2007.03367.x. [DOI] [PubMed] [Google Scholar]
- 42.Emmerlich V, et al. The plant homolog to the human sodium/dicarboxylic cotransporter is the vacuolar malate carrier. Proc. Natl Acad. Sci. USA. 2003;100:11122–11126. doi: 10.1073/pnas.1832002100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Christopher JT, Holtum JA. Patterns of carbon partitioning in leaves of crassulacean acid metabolism species during deacidification. Plant Physiol. 1996;112:393–399. doi: 10.1104/pp.112.1.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Penfield S, et al. Expression and manipulation of PHOSPHOENOLPYRUVATE CARBOXYKINASE 1 identifies a role for malate metabolism in stomatal closure. Plant J. 2012;69:679–688. doi: 10.1111/j.1365-313X.2011.04822.x. [DOI] [PubMed] [Google Scholar]
- 45.Dittrich P, Campbell WH, Black CC. Phosphoenolpyruvate carboxykinase in plants exhibiting crassulacean acid metabolism. Plant Physiol. 1973;52:357–361. doi: 10.1104/pp.52.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong HTK, Nose A, Agarie S. Respiratory properties and malate metabolism in percoll-purified mitochondria isolated from pineapple, Ananas comosus (L.) Merr. cv. smooth cayenne. J. Exp. Bot. 2004;55:2201–2211. doi: 10.1093/jxb/erh241. [DOI] [PubMed] [Google Scholar]
- 47.Eastmond PJ, et al. Arabidopsis uses two gluconeogenic gateways for organic acids to fuel seedling establishment. Nat. Commun. 2015;6:6659. doi: 10.1038/ncomms7659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Parsley K, Hibberd JM. The Arabidopsis PPDK gene is transcribed from two promoters to produce differentially expressed transcripts responsible for cytosolic and plastidic proteins. Plant Mol. Biol. 2006;62:339–349. doi: 10.1007/s11103-006-9023-0. [DOI] [PubMed] [Google Scholar]
- 49.Harmer SL, Kay SA. Positive and negative factors confer phase-specific circadian regulation of transcription in Arabidopsis. Plant Cell. 2005;17:1926–1940. doi: 10.1105/tpc.105.033035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang ZY, Tobin EM. Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell. 1998;93:1207–1217. doi: 10.1016/S0092-8674(00)81464-6. [DOI] [PubMed] [Google Scholar]
- 51.Schaffer R, et al. The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell. 1998;93:1219–1229. doi: 10.1016/S0092-8674(00)81465-8. [DOI] [PubMed] [Google Scholar]
- 52.Alabadi D, et al. Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science. 2001;293:880–883. doi: 10.1126/science.1061320. [DOI] [PubMed] [Google Scholar]
- 53.Wilkins MB. The role of the epidermis in the generation of the circadian rhythm of carbon dioxide fixation in leaves of Bryophyllum fedtschenkoi. Planta. 1991;185:425–431. doi: 10.1007/BF00201067. [DOI] [PubMed] [Google Scholar]
- 54.Nagel DH, et al. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl Acad. Sci. USA. 2015;112:E4802–E4810. doi: 10.1073/pnas.1513609112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamioka M, et al. Direct repression of evening genes by CIRCADIAN CLOCK-ASSOCIATED1 in the Arabidopsis circadian clock. Plant cell. 2016;28:696–711. doi: 10.1105/tpc.15.00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gendron JM, et al. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc. Natl Acad. Sci. USA. 2012;109:3167–3172. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sharma A, Wai CM, Ming R, Yu QY. Diurnal cycling transcription factors of pineapple revealed by genome-wide annotation and global transcriptomic analysis. Genome Biol. Evol. 2017;9:2170–2190. doi: 10.1093/gbe/evx161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dever LV, Boxall SF, Knerova J, Hartwell J. Transgenic perturbation of the decarboxylation phase of crassulacean acid metabolism alters physiology and metabolism but has only a small effect on growth. Plant Physiol. 2015;167:44–59. doi: 10.1104/pp.114.251827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Crayn DM, Winter K, Schulte K, Smith JAC. Photosynthetic pathways in Bromeliaceae: phylogenetic and ecological significance of CAM and C3 based on carbon isotope ratios for 1893 species. Bot. J. Linn. Soc. 2015;178:169–221. doi: 10.1111/boj.12275. [DOI] [Google Scholar]
- 60.Fisher AE, et al. Evolutionary history of Blepharis (Acanthaceae) and the origin of C4 photosynthesis in section. Acanthodium. Int. J. Plant Sci. 2015;176:770–790. doi: 10.1086/683011. [DOI] [Google Scholar]
- 61.Heyduk K, McKain MR, Lalani F, Leebens-Mack J. Evolution of a CAM anatomy predates the origins of crassulacean acid metabolism in the Agavoideae (Asparagaceae) Mol. Phylogenet. Evol. 2016;105:102–113. doi: 10.1016/j.ympev.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 62.Brown NJ, et al. Independent and parallel recruitment of preexisting mechanisms underlying C4 photosynthesis. Science. 2011;331:1436–1439. doi: 10.1126/science.1201248. [DOI] [PubMed] [Google Scholar]
- 63.Wang XY, et al. Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol. 2009;10:R68. doi: 10.1186/gb-2009-10-6-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.