Abstract
Tea is among the world’s most widely consumed non-alcoholic beverages and possesses enormous economic, health, and cultural values. It is produced from the cured leaves of tea plants, which are important evergreen crops globally cultivated in over 50 countries. Along with recent innovations and advances in biotechnologies, great progress in tea plant genomics and genetics has been achieved, which has facilitated our understanding of the molecular mechanisms of tea quality and the evolution of the tea plant genome. In this review, we briefly summarize the achievements of the past two decades, which primarily include diverse genome and transcriptome sequencing projects, gene discovery and regulation studies, investigation of the epigenetics and noncoding RNAs, origin and domestication, phylogenetics and germplasm utilization of tea plant as well as newly developed tools/platforms. We also present perspectives and possible challenges for future functional genomic studies that will contribute to the acceleration of breeding programs in tea plants.
Subject terms: Genomics, Plant genetics
Introduction
The tea plant (Camellia sinensis) is an economic crop with significant importance1. Its leaves are generally used to produce tea—the world’s most popular non-alcoholic beverage. Tea comprises abundant characteristic compounds, such as tea polyphenols, theanine, and caffeine, which not only determine the quality of tea but also confer it with tremendous health benefits2. However, compared to other crops, such as rice3, research on the functional genomics of tea plants is lagging behind, which has hampered the utilization of genetic resources for modern molecular breeding, which is urgently needed. The rapid development and innovation of biotechnologies, particularly next-generation sequencing (NGS), has resulted in significant progress in understanding the genomics and genetics of tea plants in the past two decades, which can be briefly summarized according to five aspects: (1) the deciphering of the genome and transcriptome of tea plants using diverse NGS technologies; (2) the identification and functional analysis of tea quality-related genes and their regulatory networks; (3) the investigation of the epigenetics and noncoding RNAs (ncRNAs) involved in stress responses and other biological processes; (4) the exploration of the diversification and domestication of tea plants; and (5) the development of innovative tools and techniques for the characterization of novel genes and alleles. In this review, we briefly summarize the main achievements of the last two decades and present potential challenges and perspectives for future functional genomic studies that would contribute to the acceleration of breeding programs in tea plants.
Whole-genome sequencing of tea plants
Cytogenetics of tea plants
Tea plants are an economically important crop. Extensive investigations of tea plant karyotypes have facilitated our fundamental understanding of their chromosomal biology4–7. The first report of the chromosome karyotype analysis of tea plants can be traced back to the last century. Morinaga and colleagues observed that 15 chromosomes (n = 15) were present in the gametes of C. sinensis tea plants and that 30 chromosomes (2n = 30) occurred in the zygotes of C. japonica, a closely related species of tea plant8,9. These findings suggested that the diploid tea plant has 30 chromosomes and that the chromosome number might be conserved in genus Camellia. New innovations in biotechnologies gradually confirmed these conclusions among globally collected tea clones5,7,10–12. In particular, the investigation of the chromosome numbers and ploidy levels of 139 tea individuals representing almost all sections of genus Camellia revealed the diploid nature of the majority (90.65%) of Camellia species, with a chromosome number of 2n = 306. Only a few tea plant species are polyploid, which exhibit 45–120 chromosomes. These species predominantly come from section Camellia (C. chekiangoleosa, C. mairei, and C. reticulata), section Oleifera (C. oleifera and C. sasanqua), and section Archecamellia (C. granthamiana).
Genome size refers to the DNA content within the haploid genome of an organism. The estimation of genome size is not only of great significance for cytogenetic studies but also provides indispensable basic data for genome sequencing, comparative genomics, and evolutionary analyses. The genome size of tea plants was initially estimated to be 3.8–4.0 Gb13,14, which was further demonstrated by the investigation of 36 Indian tea accessions, where all the investigated accessions exhibited diploid genomes with sizes ranging from 3.5 to 3.8 Gb15. The recent genome sequencing of two representative elite tea plant cultivars, C. sinensis var. sinensis cv. shuchazao and C. sinensis var. assamica cv. yunkang#10, showed an ~3.0 Gb genome size, which is similar to that of maize but much larger than those of coffee and cocoa1,2. While genome size is likely to be conserved in cultivated tea plants, intraspecific and interspecific variations are also observed in genus Camellia, possibly due to the frequent hybridization and polyploidization events that occur after speciation6.
Genetic linkage maps of tea plants
The construction of genetic linkage maps is the basis of molecular biology and is essential for a wide range of genetics and genomic studies, such as quantitative trait mapping, molecular marker-assisted breeding and comparative genomic studies16. Unlike other crops, tea plants are self-incompatible species with relatively low cross-fertility rates, resulting in a lack of a high generation segregation populations and sufficient offspring for genetic map construction17. More than 10 genetic linkage maps have been generated for tea plants, most of which are produced based on an F1 population consisting of <150 offspring18–22 (Table 1). The dominant molecular markers used for genetic map construction include random amplification of polymorphic DNA (RAPD) markers18,20,23–26, amplified fragment length polymorphism (AFLPs)18–20, inter-simple sequence repeats (ISSRs)24, simple sequence repeats (SSRs)20,25–28, and single nucleotide polymorphisms (SNPs)21,29. These reported genetic maps vary in length and density, with the total map length ranging from 1180.9 to 3314.3 cM and the average distance between adjacent markers from 0.4 to 20.1 cM. Such differences are principally affected by the total number of individuals and types of molecular markers applied for map construction. The highest-density genetic map of tea plants obtained to date was constructed using a total of 4217 polymorphic SNP markers developed from 327 individuals of the F1 segregating population (LJ43 × BHZ) via 2b-RAD sequencing29. The resultant genetic maps consisted of 15 linkage groups ranging from 87.46 to 146.93 cM. The total map length was 1679 cM, with an average interval of 0.4 cM between 4217 adjacent markers. LG01 and LG03 were the two largest linkage groups, with genetic distances of 146.93 cM (374 markers) and 146.61 cM (374 markers), respectively. Despite the numerous genetic maps currently available, the density and quality of these maps need to be further improved by investigating larger populations and applying more advanced biotechnologies. The resultant data will be important for future investigations of tea plant biology that will benefit the whole tea industry.
Table 1.
Summary of the representative linkage maps of tea plants.
| Mapping population | Sizea | Marker type | No. of markers | Total length (cM) | Marker density (cM) | No. of linkage groups | Reference |
|---|---|---|---|---|---|---|---|
| Sayamakaori × Kana-CK17 | 54 | RAPD | 140 | 1640 | 11.7 | 17 | Ota et al.23 |
| SFS150 × TN14/3 | 90 |
RAPD AFLP |
126 | 1350 | 11.7 | 15 | Hackett et al.18 |
| Keemun #4 × Chaoandawuye | 69 | AFLP | 208 | 2458 | 11.9 | 17 | Huang et al.19 |
| Fudingdabaicha × Zhenong #12 | 94 |
ISSR RAPD |
62 | 1181 | 20.1 | 7 | Huang et al.24 |
| SFS150 × S15/10 | 42 |
SSR RAPD AFLP |
100 | 1412 | 14.1 | 30 | Kamunya et al.20 |
| Sayamakaori × Kana-CK17 | 54 |
SSR STS CAPS RAPD |
279 | 1218 | 4.35 | 15 | Taniguchi et al.25 |
| Yingshuang × Beiyuedanzhu | 183 | SSR | 406 | 1144 | 2.9 | 15 | Ma et al.27 |
| Yingshuang × Beiyuedanzhu | 148 |
SSR SNP |
6448 | 3965 | 1.0 | 15 | Ma et al.21 |
| Long #43 × Baihaozao | 170 | SSR | 483 | 1226 | 2.5 | 15 | Tan et al.28 |
| Fushun × Kemsull | 79 |
SSR RAPD AFLP |
678 | 1442 | 4.7 | 15 | Chang et al.26 |
| Long #43 × Baihaozao | 327 | SNP | 4217 | 1679 | 0.4 | 15 | Xu et al.29 |
aPopulation size
Current progress in tea plant genome sequencing
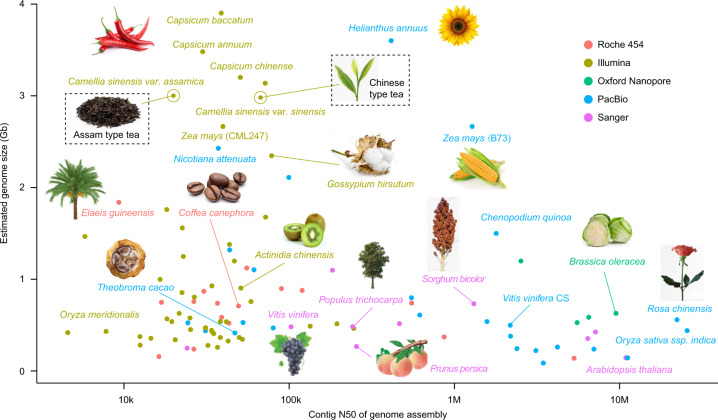
With the revolution of sequencing technology, an increasing number of labs around the world have successfully released more than 236 plant genomes, and the tea plant genome is probably among the most complex of these30 (Fig. 1). This is chiefly because tea plants have an extremely large, highly heterozygous, repeat-rich nuclear genome, presenting major challenges in genome assembly. The tea community has put forth its best effort to assemble and release the genomic sequences of two primary tea plant varieties: C. sinensis var. sinensis (CSS; Chinese type tea) and C. sinensis var. assamica (CSA; Assam type tea), using NGS technologies1,2. Both of the assemblies are composed of ~3 Gb of genomic sequences with an average scaffold N50 size of 920 Kb, which is much larger than those from other plants with complex genomes, such as the orchid (scaffold N50 = 359 Kb)31 and moso bamboo (scaffold N50 = 329 Kb)32. Nevertheless, compared to other model species, such as rice33, the scaffold N50 sizes of the current assemblies are still somewhat disappointing, due largely to the limitation imposed by the short read length generated from NGS technology (Fig. 1). Unassembled regions account for ~5% of the current assemblies. In the future, additional efforts will be needed to further improve the quality and completeness of the genome assemblies of tea plants, particularly with the upcoming advances in sequencing technologies (e.g., 3rd-generation sequencing) and new algorithm-derived assemblers.
Fig. 1. Current genome sequencing progress in tea and other plants.
The x-axis represents the contig N50 of the genome assembly, while the y-axis shows the estimated genome size of each plant. The sequencing platforms are indicated in red (Roche 454), brown (Illumina), green (Oxford nanopore), blue (PacBio SMRT), and pink (Sanger). Tea plants are highlighted with a rectangular box.
There are 33,932 and 36,951 protein-coding genes in the CSS and CSA tea plant genomes, respectively. Transposable elements (TEs) account for 64% and 80% of the CSS and CSA genomes, respectively. LTR retrotransposons are among the most dominant TEs, independently representing 58% and 67% of the CSS and CSA genomes, respectively. The rapid propagation of TEs, particularly the proliferation and persistence of a single Ty3/gypsy retrotransposon family (TL001) for over 50 million years, has been proven to have led to a considerable increase in genome size in tea plants1. CSA and CSS diverged from their common ancestor ~0.38–1.54 million years ago (mya). However, this divergence time might have been underestimated because of the draft nature of the two current genome assemblies, which generated only a small proportion of collinear genes for dating. Similar to other plants, tea plants underwent two whole-genome duplication (WGD) events, including recent and ancient events occurring 30–40 and 90–100 mya, respectively2.
Tea plants contain rich and diverse secondary metabolites that not only confer tea quality characteristics, such as color, aroma and taste, but also play critical roles in the responses to biotic and abiotic stresses34. Several genes encoding enzymes associated with the biosynthesis of secondary metabolites were significantly amplified in the tea plant genome. In particular, the striking expansion of serine carboxypeptidase-like (SCPL) acyltransferase-encoding genes might directly contribute to the high accumulation of galloylated catechins that determine tea palatability2. Compared to kiwifruit, tomato, potato, cacao, and Arabidopsis thaliana, tea plants show expansion of disease resistance genes, including nucleotide-binding sites with leucine-rich repeats (NBS-LRRs) and pattern-recognition receptors (RLK-LRRs)1. This contributes to the enhancement of the immune system of tea plants and, thus, their adaptation to diverse global environments. WGD events and subsequent tandem duplications are the two major evolutionary forces that drive such gene expansions.
Tea-processing suitability refers to the characteristics of tea varieties that make them suitable for the manufacture of certain types of tea to achieve the best quality. It is accepted that different varieties of tea plants differ in terms of flavors and tastes. The sequencing of the tea plant genome together with the transcriptomes and metabolomes of 25 Camellia species representing almost all the sections from genus Camellia showed that, despite the conservation of three metabolic pathways (catechins, theanine, and caffeine) among Camellia species, most of the flavonoid and caffeine, but not the theanine-related genes, exhibited higher expression levels in species from section Thea than in non-Thea species1. This elucidates why the leaves of tea plants from non-Thea sections, such as well-known ornamental camellias (e.g., C. japonica) and the traditional oil tea plant (C. oleifera), accumulate such low contents of catechins and caffeine but not theanine, making them unsuitable for tea production. Further investigation of the roles of natural selection and artificial selection in shaping the genes involved in the three metabolic pathways will contribute to a deep understanding of the processing suitability and quality of tea. In addition, caffeine (1,3,7-trimethylxanthine) is among the most well-known purine alkaloids in plants, and its biosynthesis is mainly catalyzed by the N-methyltransferases (NMTs)35. The NMT genes have undergone recent rapid and independent evolution in tea plants relative to cocoa and coffee1.
Organelle genome sequencing
Plants generally harbor two independent organelle genomes (chloroplast and mitochondria), which provide invaluable resources for a range of functional, evolutionary and comparative genomic studies36. The cultivated tea plant harbors a circular chloroplast (cp) genome of 157,096 bp in length with an overall GC content of 37.3%37. It exhibits the typical plant cp genome structure, including a pair of inverted repeat regions (IRs, 26,080 bp) separated by a large single-copy region (LSC, 86,653 bp) and a small single-copy region (SSC, 18,283 bp). The whole-genome sequence encodes a total of 133 cp genes, 86 of which are protein-coding genes, while 8 are rRNA genes, and 39 are tRNA genes. The cp genomes have been assembled for diverse well-known tea cultivars, such as “Longjing #43”37, “Yunkang #10”38, and “Shuchazao”2. In addition to cultivated tea plants, an increasing number of cp genomes from species of genus Camellia have also been released, including those of C. japonica (NC_036830), C. mairei (NC_035688), C. szechuanensis (NC_035651), C. elongata (NC_035652), C. azalea (NC_035574), C. pubicosta (NC_024662), C. petelotii (NC_024661), C. reticulata (NC_024663), C. grandibracteata (NC_024659), C. leptophylla (NC_024660), C. crapnelliana (NC_024541), C. oleifera (NC_023084), C. danzaiensis (NC_022460), C. impressinervis (NC_022461), C. yunnanensis (NC_022463), C. cuspidata (NC_022459), C. pitardii (NC_022462), and C. taliensis (NC_022264)39–45. The Camellia cp genomes possess quite similar genome sizes, total numbers of genes, and sequence homology, suggesting extreme genome conservation during evolution.
In sharp contrast to the chloroplast genome, only a partially assembled mitochondrial (mt) genome is available for cultivated tea plants38. The current release of the tea plant mt genome consists of two circular scaffolds with total lengths of 702,253 bp (45.63% GC) and 178,082 bp (45.81% GC). It encodes 71 mt genes, including 44 protein-coding genes, 24 tRNA genes, and 3 rRNA genes. Compared to the cp genome, the tea plant mt genome shows a repeat-rich nature, including a total of 38,027 bp of long repeat sequences38.
Transcriptome sequencing and gene discovery in tea plants
Transcriptome sequencing of tea plants
Transcriptome sequencing has revolutionized genetic and functional genomic studies of organisms, particularly for nonmodel species without available sequenced genomes46. In the last decade, innovations overcoming challenges in tea plant genome sequencing have greatly accelerated the transcriptomic investigation of this economically important crop. Shi and colleagues examined the first transcriptome of tea plants (cultivar shuchazao) using Illumina sequencing technology and obtained a total of 127,094 unigenes, which were applied for the in-depth exploration of candidate genes involved in the biosynthesis of characteristic compounds of tea plants that determine tea quality47. Subsequently, growing transcriptome-sequencing projects were launched to further investigate the gene expression dynamics of tea plants under cold acclimation48–51, drought stress52,53, and hormone responses54 and the mechanisms underlying self-incompatibility17,55, nitrogen utilization56,57, trichome formation58, and tea quality59–62. These results greatly broadened our understanding of tea plant biology. With the advent of single-molecule sequencing technology, a recent study produced a more accurate full-length transcriptome of tea plants63. In addition to the cultivated tea plants, several transcriptomes from closely related species from genus Camellia have also been reported, which have provided indispensable resources for comparative transcriptomic studies64–66 (Supplementary Table 1). The sequencing of the transcriptome of oil tea plants (C. oleifera) identified 3022 orthologous gene pairs between cultivated tea plants and oil tea plants, among which 211 exhibited evidence of positive selection64. Compared to the cultivated tea plants, C. taliensis showed extraordinary amplification of cold tolerance-related genes65. Most of the genes associated with triacylglycerol biosynthesis in C. reticulata and C. sinensis are multiple-copy genes, suggesting the potential occurrence of WGD events in the common ancestor of genus Camellia66. An increasing number of transcriptome-sequencing projects have also been carried out in other Camellia species, such as C. sasanqua67, C. chekiangoleosa68, and C. nitidissima69. These datasets expanded the gene pool of tea plants and will be of particular importance for future tea plant breeding, as well as investigations of functional genomics, phylogenomics, and comparative transcriptomics. It should be noted that the current reported transcriptomes from both tea plants and other Camellia species vary in the total number of assembled transcripts and N50 sizes, primarily due to the different sequencing depths and assemblers adopted. In the future, more efforts should be made to evaluate the optimized-sequencing depth and assembler to better construct the transcriptomes of tea plants.
Functional gene sets and their regulatory networks
One fundamental mission of the molecular biology research on tea plants is to understand the functions and regulatory mechanisms of genes encoded by the genome. During recent decades, rapid advances in biotechnologies have facilitated the cloning and functional characterization of an increasing number of genes in tea plants70–72. Most of these genes have been identified from CSS and CSA, in which the numbers of cloned genes are much greater than those from other closely related tea plant species. Their functions can generally be classified into three major categories, including secondary metabolite biosynthesis73–80, abiotic and biotic stress responses81–86, and aroma formation70,71,81,87–91 (Table 2). Among these genes, those associated with secondary metabolite biosynthesis and aroma formation are the most studied because they directly determine the quality of tea. Supplementary Table 2 summarizes the details of the currently available cloned genes in tea plants. We here selectively highlight some representatives that would be preferred candidates for the future genetic improvement of tea plants.
Table 2.
List of representative functional genes in tea plants.
| Gene symbol | NCBI accession | Length (bp) | Function description | Ref. |
|---|---|---|---|---|
| Polyphenol biosynthesis | ||||
| CsANR1 | GU992402 | 1044 | Encoding anthocyanidin reductase | 77 |
| CsANR2 | GU992400 | 1014 | Encoding anthocyanidin reductase | 77 |
| CsLAR | GU992401 | 1282 | Involved in flavan-3-ol biosynthesis | 77 |
| CsF3'H | KT180309 | 1706 | Encoding flavonoid 3'-hydroxylase | 92 |
| CsC4H | KY615675 | 1518 | Encoding cinnamate 4-hydroxylase in synthesis of flavonoids and lignins | 93 |
| CsF3H | KY615688 | 1107 | Encoding flavanone-3-hydroxylase | 94 |
| CsMYB4a | KY774676 | 735 | Negatively regulates phenylpropanoid and shikimate pathway | 78 |
| CsMYB5 | KY827396 | 930 | Regulate anthocyanin and proanthocyanidin biosynthesis | 79 |
| CsMYB75 | GGTM01017033 | 765 | Regulate anthocyanin hyperaccumulation | 76 |
| CsGST | MK431867 | 642 | Encoding glutathione-S-transferase | 95 |
| Caffeine biosynthesis | ||||
| CsTCS | AB031280 | 1438 | Caffeine synthase gene | 73 |
| Theanine biosynthesis | ||||
| CsTSI | TEA015198a | 2544 | Theanine synthetase gene | 2 |
| CsAAPs | TEA031577a | 2339 | Encoding amino acid permease involved in theanine transportation | 96 |
| Aroma formation | ||||
| Csβ-PD | AB088027 | 1729 | Encoding beta-primeverosidase in floral aroma formation | 88 |
| CsGT1 | AB847092 | 1458 | Converting volatile organic compounds into β-primeverosides for aroma biosynthesis | 90 |
| CsSAMT | MG459470 | 1104 | Encoding the salicylic acid carboxyl methyltransferase for methyl salicylate formation | 97 |
| CsLIS/NES | KF006849 | 2050 | Producing (E)-nerolidol and linalool in vitro | 87 |
| CsUGT85A53 | NA | 1455 | Formation of (Z)‐3‐hexenyl glucoside, novel insect pest control | 81 |
| Abiotic and biotic stress | ||||
| CsCOR1 | EU563236 | 755 | Involved in cold, salinity and dehydration tolerance | 82 |
| CsHSP17.2 | KU244518 | 734 | Mediates tea plant thermo tolerance | 98 |
| CsICE1 | GQ229032 | 1964 | Inducer of CBF expression | 84 |
| CsCBF1 | EU563238 | 1210 | C-repeat-binding factor | 84 |
| CsTLP | DQ444296 | 681 | Increases tolerance to fungal pathogens overexpressed in potato | 83 |
| CsGolS2 | KP757767 | 1574 | Response to Ectropic oblique attack | 86 |
aCSS locus ID (from TPIA: http://tpia.teaplant.org)
Caffeine is the most abundant purine alkaloid in the majority of tea plants99. It is a crucial component of tea quality and is significantly related to the bitterness of tea. Nevertheless, excessive intake of caffeine has been reported to have some side effects for human health, such as increasing the risk of cardiovascular disease, palpitations, and insomnia100,101. The documentation of the genes controlling caffeine biosynthesis is therefore essential for the future of breeding new varieties with a low caffeine content. Kato et al.73 cloned the gene coding caffeine synthase (TCS) from the young leaves of tea plants using the rapid amplification of complementary DNA ends (RACE) technique73. This gene is 1438 bp in size and encodes 369 amino acids. The expression of the TCS gene in Escherichia coli enables high production of caffeine after feeding on xanthine and S-adenosylmethionine as substrates, confirming the caffeine synthase activity of TCS. Theanine is another characteristic amino acid of tea plants and is related to the fresh tastes of tea. The gene encoding enzyme associated with theanine biosynthesis (CsTSI) was identified through a combination of genomic and transcriptomic analyses2. This gene shares high homology with the glutamine synthetase (PtGS) gene of Pseudomonas taetrolens, indicating its potential bacterial origin102. The overexpression of CsTSI in A. thaliana significantly increases the accumulation of theanine after ethylamine feeding. Tea polyphenols are the major secondary metabolites of tea plants103 and are closely related to the astringent and bitter taste of tea. Two enzymes, UDPglucose:galloyl-1-O-β-d-glucosyltransferase (UGGT) and epicatechin:1-O-galloyl-β-d-glucose O-galloyltransferase (ECGT), were purified and shown to be involved in the biosynthesis of galloylated catechins74. Three UDP-glycosyltransferase (UGT) genes, CsUGT84A22, CsUGT78A14, and CsUGT78A15, were found to be involved in the biosynthesis of β-glucogallin and glycosylated flavonols75. Anthocyanin is another type of polyphenol that typically accumulates in purple tea varieties. It has considerable health benefits and has been used to develop anthocyanin-enriched beverages. The expression of a gene encoding leucoanthocyanidin reductase (CsLAR) in E. coli results in the production of 2R,3S-trans-flavan-ol (+)-catechin after the feeding of leucocyanidin as a substrate77. Similarly, the expression of two anthocyanidin reductase (CsANR1 and CsANR2)-encoding genes in E. coli enables the conversion of cyanidin to a mixture of (+)-epicatechin and (−)-catechin77. More recently, CsMYB75 and CsGSTF1 were found to be involved in anthocyanin accumulation in purple tea76. CsMYB75 can promote the expression of CsGSTF1 in transgenic tobacco plants. CsGSTF1 enables the transfer of anthocyanin from the endoplasmic reticulum (ER) to vacuoles76. In tea plants, the regulation of the catechin metabolic pathway is complex, and several transcription factors (e.g., MBW complexes) are involved76,78–80,104.
Aroma is vital for tea quality and for attracting global interest. Since the advent of new chemical analytical techniques, such as mass spectrometry, considerable efforts have been made to identify volatile constituents of different types of teas and to assess volatile odor activities and their contribution to tea aroma70,71,88,89,105–109. Significant studies on tea aroma biology have revealed that hundreds of volatile terpenoids, in addition to some other volatiles, such as cis-3-Hexen-1-ol, are present in tea as glycosides110–112, which can be released during the tea manufacturing process113. A tea beta-primeverosidase gene was isolated and expressed in E. coli. The prokaryotically expressed mature protein was found to be able to hydrolyze beta-primeverosides, liberating a primeverose unit and aglycons and playing roles in tea plant defense and floral aroma formation during the tea manufacturing process88. Two UGTs from C. sinensis, UGT85K11 (CsGT1) and UGT94P1 (CsGT2), were found to convert volatiles into β-primeverosides via sequential glucosylation and xylosylation, respectively90. More recently, a UGT gene (CsUGT85A53) that was functionally validated was shown to enable the conversion of exogenous (Z)-3-hexenol from damaged adjacent tea leaves to its glucoside form, enhancing the ability of tea plants to defend against pests81. Studies on tea volatile biosynthesis have also revealed an interesting bifunctional linalool/nerolidol synthase gene (CsLIS/NES) in tea plants87. This gene transcribes two transcript isoforms, CsLIS/NES-1 and CsLIS/NES-2, which possess distinct subcellular localizations and molecular functions, with CsLIS/NES-1 localizing to chloroplasts and functioning as linalool synthase, while CsLIS/NES-2 localizes to the cytosol and potentially acts as a nerolidol synthase. In addition, the biosynthesis of the tea volatiles indole, linalool, and nerolidol was found to be controlled by the corresponding genes in tea89,91,114. Collectively, the aforementioned genes that were cloned and functionally validated in tea plants have broadened our understanding of the genetic basis of tea quality and will be particularly useful for the future genetic improvement of tea plants.
DNA methylation of tea plants
DNA methylation is among the most essential and ubiquitous epigenetic modifications in plants115. It plays crucial roles in gene expression regulation, cell differentiation, and transposon element (TE) silencing116. DNA methylation are mostly investigated in model plants, such as Arabidopsis and rice, due to their small genome sizes and low genome complexity117,118. Tea plants are nonmodel woody plants with complex nuclear genomes. The sequencing of the tea plant genome has revealed an extraordinary amplification of genome size driven primarily by a burst of TEs1,2. DNA methylation was confirmed to be closely linked with TE burst in tea plants119. Compared to asterids such as potato, tea plants exhibit higher CHG methylation levels, similar to those found in maize and Norway spruce, two plant species with a comparable genome size and repeat content to tea plants120,121. Remarkably, the DNA methylation levels of TEs vary with their evolutionary distance in tea plants, with the methylation level increasing in recent, active TEs and decreasing in ancient TEs. It is widely accepted that DNA methylation can spread from TE boundaries to close genomic regions, resulting in increased methylation levels of adjacent genes122. High TE contents in tea plants might therefore contribute to the high level of DNA methylation. Several genes involved in the biosynthesis of secondary metabolites (catechins, theanine, and caffeine) were found to be TE-related in tea plants. This suggests that TE-mediated DNA methylation may have some effects on the formation of tea quality.
Chilling damage caused by low temperature occurs commonly in tea plants and has severely affected the sustainable development of the global tea industry. Approximately 49–51% of cytosine residues are methylated during the cold acclimation of tea plants123. Compared to preacclimation, the DNA methylation level increases significantly during cold acclimation and is maintained at a higher level after deacclimation. The gene encoding DNA methyltransferase, CsDRM2, was cloned from an elite cultivar of tea plant (Longjing #43) and found to exhibit high expression levels under cold acclimation124. In the future, additional DNA methylation projects need to be performed in relation to other aspects of tea plants to elucidate the roles of these modifications in plant development, environmental adaptation, stress responses, and even genome structure variation and stabilization.
ncRNAs of tea plants
MicroRNAs (miRNAs), a class of endogenous small ncRNAs, have been recognized as a critical genetic regulator primarily engaged in posttranscriptional regulation. In tea plants, a series of studies have been undertaken to examine the posttranscriptional regulation of miRNAs in development125,126, metabolism126–128, and responses to biotic/abiotic stresses129–133. Tea plant miRNAs can negatively regulate catechins and terpenoid biosynthesis by down-regulating target genes related to their biosynthesis at both the transcriptional and posttranscriptional levels126,128. For instance, Cs-miR156 regulates catechin accumulation in tea plants by suppressing the expression of the target gene SPL in the presence of different nitrogen forms128. The characterization of tea plant miRNA regulatory networks in response to Colletotrichum gloeosporioides, which is considered one of the dominant endophytic taxa in tea plants, indicated that miRNAs may be involved in the response to C. gloeosporioides attack134. miRNA characterization in tea plants under cold and drought stresses suggested the potential existence of an miRNA-mediated regulatory mechanism under abiotic stresses showing coherent or incoherent relationships with target genes to prevent tea plants from being injured129–132. Csn-miR398a-3p-1 of tea plants was experimentally proven to directly cleave CsCSD4, a superoxide dismutase (SOD) gene related to the removal of reactive oxygen species (ROS), and the expression pattern of csn-miR398-3p-1 is opposite that of CsCSD4 under cold treatment135. Roles of miRNAs have also been found in the bud dormancy of tea plants. Cs-miR139c regulates the release of tea plants from bud dormancy by suppressing the expression of its target gene, CsnTCP2 (Teosinte branched/Cycloidea/Proliferating cell factor 2)136. There is no doubt that the investigation of miRNAs provides a broad understanding of the posttranscriptional regulatory networks between protein-coding and ncRNAs; however, most current progress is limited to the identification, but not the functional verification, of miRNAs, which will require further investigation in tea plants.
In addition to miRNAs, long noncoding RNAs (lncRNAs) have been also identified in tea plants using 170 RNA-seq data (~7157 million reads) from 11 tissues, generating more than 33,000 putative novel lncRNAs137. The tea plant lncRNAs showed tissue-specific expression patterns and were suggested to be involved in major aroma formation pathways of black tea137. circRNAs (circular RNAs) were recently discovered as a new class of ncRNAs that are covalently closed, single-stranded and generated via back-splicing events138,139. Since their first documentation in A. thaliana138, circRNAs have been detected in several other plants138,140–142. A total of 3174 circRNAs have been characterized in tea plants141. As similarly observed in most other plants139, tea plant circRNAs exhibit tissue-specific expression patterns, and their expression shows a positive relationship with that of their parental genes141. Tea plant circRNAs were found to exhibit potential miRNA-binding sites and to display crucial functions in the photosynthetic machinery and metabolite biosynthesis during leaf development141. In other plants, circRNAs can act as miRNA sponges under tomato chilling injury142, dehydration stress in wheat143, and infection with the bacterial canker pathogen in kiwifruit144. These studies have increasingly complicated our understanding of ncRNAs and their regulatory mechanisms in tea plants. Additionally, none of these identified ncRNAs (miRNAs, lncRNAs, and circRNAs) have been functionally well-verified regarding their roles in regulating posttranscriptional processes in tea plants. Further efforts aimed at accurate lncRNA characterization and the functional examination of their interaction mechanisms in competing endogenous RNA (ceRNA) networks should receive more attention to elucidate their roles in response to stress and the regulation of tea plant secondary metabolite formation.
Genetic diversity and population structure of tea plants
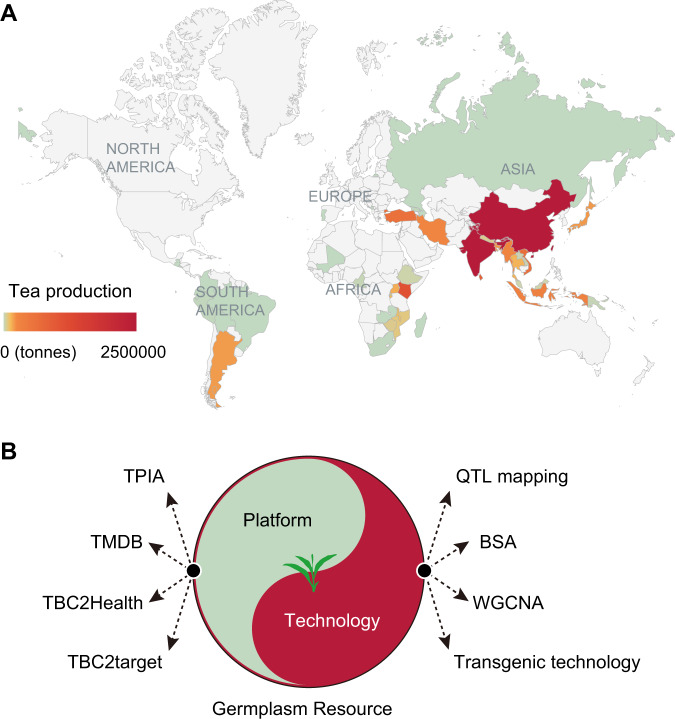
In the thousands of years since the tea plant was first discovered, recorded, and cultivated to produce tea in China, it has spread to more than 50 countries around the world in tropical and subtropical regions1 (Fig. 2a). The wide range of growing areas, long history of cultivation, self-incompatibility and allogamy of tea plants make these cultivars highly heterogeneous, resulting in high diversity in both their genetics and morphology145,146. As the key component of crop genetic improvement, the available germplasms and the genetic diversity within a gene pool determine the success of breeding programs. The development of molecular markers, such as RAPD, AFLP, SSR, and SNP markers, has allowed the successful utilization of these markers to estimate the genetic diversity and determine the phylogenetic relationships of different tea germplasms147–153. Early in 1995, the systematic assessment of the genetic variability of three major types (CSA, CSS, and Cambod type) of tea plants from Kenya and the UK suggested that the Cambod population possessed the lowest diversity153. The further investigation of genetic and morphological traits revealed consistent findings in populations from other major tea plantation countries145,147,154–158. Germplasm from the Korean population shows higher diversity than that from Japan due to more extensive plantations in Korea and longer, intensive tea selection programs in Japan157. The genetic diversity of the Chinese, Indian, and Sri Lankan populations is higher than those from other countries151. Most Italian tea plants exhibit a close relationship with cultivars from Zhejiang Province159. Kenyan tea plants show the highest diversity among all African germplasms, while the lowest is found in South Africa160,161. CSA was indicated to have donated the largest amount of genetic material in African tea breeding161. Narrow genetic diversity is detected in the Sri Lankan population, which is dominated by Assam-type tea162. The estimation of worldwide tea plant genetic diversity has increased our understanding of the population structures and origins of tea plants, which will benefit the global tea industry.
Fig. 2. Global tea production and platforms/techniques for scientific studies of tea.
a Global tea production across the majority of tea-producing countries. The data were collected from 2017 FAO statistics (http://www.fao.org/). b Resource-centered research platforms and technologies for tea molecular biology.
China hosts abundant tea plant germplasms. A wide range of genetic diversity and population studies have been undertaken, particularly for Chinese germplasms148,149,152,163–167. Most of the Chinese tea germplasms can be clustered based on their geographical origin and genetic background149. Higher levels of genetic diversity are observed in Guangxi, Yunnan, and Guizhou Provinces148,167. Compared to the wild tea plants, such as C. taliensis, the cultivated tea plants display higher heterozygosity152.
Molecular markers are useful and are commonly employed in crop breeding. Traditional experimental screening of marker polymorphism is laborious and time-consuming, especially for the broadly used SSR markers. The development of the CandiSSR pipeline has facilitated the efficient identification of polymorphic SSRs (PolySSRs) based on high-throughput sequencing data, and this pipeline has successfully identified 450 PolySSRs in the transcriptomes of four Camellia species168. Despite the diverse molecular and morphological markers developed, more efforts are still needed to develop extra-robust markers (e.g., SNPs) to better facilitate the conservation and utilization of global tea resources.
Platforms and tools for tea plant functional genomics
Databases
Biological databases provide scientists with an opportunity to centrally access a variety of biological data. Various tea plant databases have been established to help the tea-related research community to better understand the metabolome, health benefits, and genomics of tea169–171 (Fig. 2b). Yue and colleagues constructed the first comprehensive manually curated Tea Metabolome Database (TMDB)171, which contains over 1400 metabolites and 600 NMR datasets collected from 364 publications. Retrieval from the TMDB can thoroughly access a total of 24 basic attributes of tea compounds of interest, such as compound structure, molecular weight, and compound uses. The beneficial effects of tea are principally derived from diverse bioactive compounds. The establishment of the searchable TBC2health database has connected the relationships between tea compounds and their health beneficial effects well169. The current release of TBC2health compromises 497 bioactive tea compounds and their associated health effects on 206 diseases. The user-friendly response interface contains multiple easily visualized results, aiding in the efficient discovery of potential mechanisms of the health benefits of tea. Increasingly, studies have confirmed that the health benefits of tea are primarily achieved through the regulation of target gene expression or protein activities172,173. More recently, the TBC2target database was developed to provide candidate target genes for a total of 240 bioactive tea compounds170. The target genes were predicted using PharmMapper via a pharmacophore-mapping strategy174. With TBC2target in hand, scientists can explore the potential genes targeted by tea compounds and then reveal the possible health-promoting mechanisms of bioactive tea compounds.
Tea plants are rich in secondary metabolites that essentially determine tea quality2. Hundreds of studies on tea quality and physiology have been reported, which have generated a wide variety of specific biological data1,2. The recently developed Tea Plant Information Archive (TPIA) database employs the published tea plant genome as a basic framework and incorporates a wide range of publicly available genomic, transcriptomic, and metabolomic data of tea plants together with globally collected tea germplasms175. It also comprises over 70 transcriptomes widely collected from 21 Camellia species, plentiful gene expression data from various conditions (across species, stresses, and hormone treatments), orthologs, transcription factors, SSRs, metabolites (mainly catechins, theanine, and caffeine), correlations, and manually curated functional genes. Through long-term maintenance and timely updating with novel datasets, the TPIA is gradually becoming the central gateway for tea communities to investigate the biology of tea plants in more detail, thus benefitting the sustainable development of the global tea industry.
Quantitative trait loci (QTL) mapping and bulked-segregant analysis (BSA)
QTL mapping is an efficient tool for revealing the molecular basis of complex traits of organisms176. Despite the fact that QTL mapping is widely used in several crops177,178, the application of this method to tea plants is still challenged, mainly due to the self-incompatible nature of tea plants, which exhibit a low seed yield, leading to the propagation of insufficient populations for QTL mapping. Nevertheless, considering the importance of tea plants in global tea consumption, increasing efforts are being focused on the QTL mapping of several agronomic traits of tea plants, particularly tea yield20 and secondary metabolites22,27–29 (Fig. 2b). A total of 23 putative QTLs associated with tea yield were detected using 5-year yield data from 42 F1 individuals collected across two sites in Kenya20. These efforts led to the first QTL-mapping investigation in tea plants, but there was no shared QTLs discovered at the two sites, indicating an overestimated QTL effect due to insufficient mapping populations. In contrast to yield traits, QTLs related to tea quality have been widely explored. Using 2-year catechins data from 183 individuals crossbred from two tea plant varieties with distinct catechins composition, a total of 25 catechin-related QTLs were identified27. Nine of these QTLs were validated across years and found to cluster together in the chromosomal regions of LG03 and LG11, suggesting a potential tandem duplication origin of tea plant catechin genes. Similarly, 10 QTLs were found to be involved in the determination of theobromine and caffeine contents using 148 individuals from a pseudo-testcross population22. With the exception of one QTL related to caffeine content, which was validated across years and mapped on LG01, the other QTLs were validated from only 1 or 2 years of data, possibly due to the relatively small size of the mapping populations. With the expansion of mapping populations, a recent QTL mapping study detected several novel QTLs related to flavonoids29. Among the 27 identified QTLs associated with flavonoids, 7 were newly revealed to be involved in the determination of anthocyanin content and young shoot color. Interestingly, quite different from other recent reports22, QTLs controlling caffeine content were identified on LG11 in this study29. This implies that the caffeine content of tea plants is probably controlled by multiple genes located on different chromosomes or that the QTL effects have been overestimated because of the limitations of the mapping populations. In addition to tea quality and yield traits, QTL mapping has been applied to agronomic traits such as spring bud flush and leaf size (e.g., mature leaf length, width, and shape indexes)28. In the future, efforts will be needed to further increase the mapping populations and improve the density of genetic linkage maps to precisely identify QTLs in tea plants.
Although over 80 QTLs have been detected for diverse agronomic traits of tea plants, it is still difficult to explore the candidate genes and/or alleles related to QTLs further owing to the insufficient genetic markers and genomic information available. The recently developed BSA method selects and pools samples from individuals with extreme phenotypes from biparental segregation populations and enables the efficient identification of genes and alleles governing complex traits through statistical genomic and phenotypic analyses. In tea plants, the utilization of the BSA method coupled with RNA sequencing has facilitated the discovery of a flavonoid 3′,5′-hydroxylase (F3′5′H) gene that is significantly associated with catechin content based on individuals from the two tails of an F1 population that segregates with catechin content179.
Weighted correlation network analysis (WGCNA)
WGCNA is of particular importance for tea plants to establish the relationships between gene expression and secondary metabolites that determine tea quality180,181 (Fig. 2b). It is also helpful for identifying clusters of highly correlated genes and revealing their regulatory networks in tea plant development182,183. The investigation of the expression profiles of tea plants at 10 developmental stages identified a total of 35 coexpression modules using the WGCNA method with differentially expressed genes (DEGs)182. Among these, 20 modules were found to be related to the biosynthesis of catechins, theanine, and caffeine, which were biologically coregulated by several hub genes, suggesting a potential network of coordinated regulation in three characteristic secondary metabolic pathways of tea plants. Building the association between tea aroma accumulation and gene expression using the WGCNA method remarkably revealed 13 transcription factor genes that are probably involved in terpene metabolism181. Among these genes, CsOCS2 was validated in vitro to function in terpenoid biosynthesis, indicating a robustness and reliability of WGCNA for trait-associated gene discovery. Similarly, the application of WGCNA identified a gene module consisting of eight genes significantly associated with the biosynthesis of cyanidin 3-O-glucoside and petunidin 3-O-glucoside, helping to elucidate the mechanism of pigmentation in the anthocyanin-rich tea plants180. These findings suggested that, compared to several previous correlation studies1,2,60, WGCNA is much more powerful; however, the sample sizes and/or traits used in most of the current studies are still limited, which decreases the accuracy and scope of the acquired candidates. The integration of more samples and traits from diverse cultivars and conditions in the future will improve the capability of WGCNA and thus enhance the rapid discovery of candidate genes associated with important agronomic traits in tea plants.
Future challenges and perspectives
Completion of the tea plant genome with advanced technologies
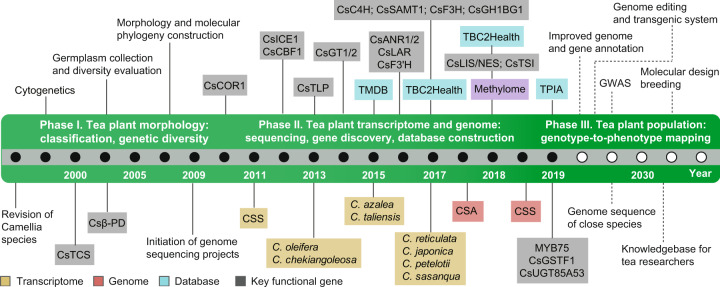
Tea originated from China and has spread to over 160 countries around the world. According to statistics from 2017 from the Food and Agriculture Organization of the United Nations (FAO; http://www.fao.org/), the global tea plantation area has exceeded 4.08 million hectares, and global total tea production has reached 6.10 million tons, with an average increase of 0.33 million hectares and 0.58 million tons over the past 5 years (Fig. 2a). In stark contrast to the flourishing development of the global tea industry, research on the major fundamental biological problems encountered in tea plants is still lagging behind, resulting in a low breeding efficiency and few excellent varieties. The rapid development and application of genomics approaches have effectively promoted the breeding programs of crops; however, genomic investigations of tea plants are still limited and challenging. Although the genome sequences of two major varieties (CSS and CSA) of tea plants were generated recently, their quality and completeness still need to be improved1,2. Given the limitations of current NGS-based approaches related to highly repetitive and heterozygous genomes, advanced single-molecule-sequencing technologies (e.g., PacBio SMRT and Oxford Nanopore) will help resolve the highly repetitive regions of the tea plant genome. With the further inclusion of linkage map and Hi-C technology to interactively address heterozygous genomic regions, the eventually produced chromosome-level reference genome of tea plants will facilitate comparative genomics, evolutionary and population genetics studies and, thus, promote breeding programs in the future (Fig. 3).
Fig. 3. Timeline of research on tea plant genetics and genomics.
The solid black circles indicate past events in tea plant genomics, including Phase I of tea plant morphology and Phase II of tea plant transcriptome and genome studies. Events are highlighted with colored rectangular boxes: yellow (transcriptome sequencing), orange (genome sequencing), cyan (database development), and gray (gene cloning). The solid white circles represent Phase III of tea plant comparative genomics and population genetics studies in the future.
Towards tea plant pan-genomics and the phylogeny of genus Camellia
Tea plants are widely distributed in tropical and subtropical regions around the world184. They contain rich and unique characteristic secondary metabolites, such as catechins, theanine, and caffeine, which are essential to the formation of tea quality. However, the contents of these secondary metabolites vary greatly among different varieties and Camellia species, leading to large differences in tea processing suitability1,185. They also differ significantly in several morphological traits (e.g., leaf size) and stress resistance characteristics (e.g., cold tolerance), showing a divergent genetic makeup50,186. Therefore, the genome sequence of a single individual of a tea plant variety (e.g., shuchazao or yunkang #10) cannot represent the entire gene pool of tea plants. Pan-genome sequencing is a newly developed strategy for investigating the genetic variations among different subspecies and/or individuals that has been widely used in several crops and model species, such as rice187, soybean188, maize189, and Arabidopsis190. With the completion of the tea plant genome, more efforts are still needed to further investigate the pan-genomics of tea plants from different varieties and/or ecotypes to expand the tea gene pool. However, the phylogeny of genus Camellia and the relationships among different tea populations remain largely unclear, which has seriously hindered the in-depth exploration of pan-genomics in tea plants.
Although several studies have attempted to resolve the phylogeny of genus Camellia using chloroplast and/or mitochondrial DNA sequences, the generated phylogenetic trees are somewhat poorly supported, primarily due to the high frequency of hybridization and polyploidization, as well as the relative conservation of organelle DNA sequences among Camellia species39,45,191–193. Compared to organelle fragments, massive nuclear gene sequences rapidly obtained from genome and transcriptome sequencing provide abundant candidate low-copy-number nuclear genes and informative sites for phylogenetic investigation and have been increasingly applied to clarify the phylogenies of some complex plant taxa194–198. Therefore, the sequencing of additional genomes and/or transcriptomes of Camellia species, as well as elite tea cultivars in the foreseeable future will not only help to reveal their complex phylogenetic relationships but will in turn promote pan-genomic studies of tea plants to expand the gene pool.
Genome-wide association study (GWAS) of tea plants
GWAS is a widely accepted strategy to identify genes/alleles associated with complex traits in crops with the rapid innovation of NGS technologies199–202. Genotypes (typically based on SNPs) and phenotypes (usually those of different traits) are the two fundamental requirements in GWAS, in which a higher SNP density and more diverse phenotypes of a population will theoretically produce more reliable and significant associations. However, conventional GWAS design and methods have resulted in challenges in the identification of multiple functional alleles within a single gene as well as rare alleles in the population203,204. GWAS of tea plants has also been confronted with major difficulties in population construction and phenotyping related to germplasm collection and the ultraslow growth of tea plants when transplanted to nursery gardens. Nevertheless, considering the high level of genetic and morphological diversity in tea plants, GWAS is still the most efficient way to detect candidate loci of complex traits undergoing improvement. To apply GWAS to study the genetics underlying tea traits of interest, the following goals should be considered: (1) collection and construction of core germplasms with adequate genetic diversity; (2) transplantation of the germplasms to one or more local gardens for accurate multiple-year phenotyping; (3) further improvement of the quality and annotation accuracy of the current genome assembly; (4) integration of GWAS data with other omics datasets (e.g., transcriptomic and metabolomics data); and (5) construction of an efficient transgenic system to functionally validate the candidates. In this way, GWAS can be effectively applied to understand the genetic basis of agronomic traits associated with tea quality, thus enabling the molecular design of new cultivars in the near future.
Origin and domestication of tea plants
China is documented as the first country to cultivate and utilize tea plants in the world205. It harbors abundant tea germplasms and has long been considered the origin of tea plants206. However, since no wild ancestor of tea plants has been discovered in China, the origin and domestication of tea plants are still a mystery and attract worldwide attention. Although recent progress has been made in understanding the origin and domestication of tea plants based on molecular markers (primarily SSRs), the resultant conclusions are unilateral and even controversial due to the poor representativeness of sample collections and the lack of sufficient molecular markers for diversity evaluation160,161,207–211. The recent release of tea plant genome sequences has laid a solid foundation for solving this problem1,2; however, further efforts focusing on the following issues are still needed: (1) collection of globally representative tea plant samples; (2) investigation of the population structure of tea plants and identification of their putative wild ancestor; (3) estimation of population diversity using genome-wide SNP markers; (4) scanning of genomic regions with significantly lower diversity in cultivated but not wild tea plants and identification of the candidate genomic regions selected during domestication; and (5) functional investigation of the characteristics of genes within regions of domestication. It should be noted that most of the existing tea varieties are bred and propagated directly from natural populations. Artificial domestication may have had little impact on the variation in genome sequences. Therefore, tea plants may actually undergo adaptive evolution compared to other crops (e.g., rice) subjected to strong domestication pressure212.
Establishment of high-efficient transgenic system
Agrobacterium-mediated genetic transformation has been extensively applied to verify the function of genes with the aim of improving the quality and yield of crops. In tea plants, the first transgenic plant was generated using Agrobacterium-mediated transformation in somatic embryos213. However, at least 45 weeks were possibly needed for the transformation procedure alone, and additional years were required for further transplantation213. It also required several years of experimental work to establish stability and germline transmission in these plants. Although several efforts have been made to optimize the transformation system of tea plants using different strategies214–223, transformation efficiency still represents a challenge under the available methods215,224. In the face of these challenges, some key points related to transgenic technology need to be further detailed: (1) optimization of the transformation and regeneration system; (2) clarification of the transformation mechanism of tea plants, particularly to elucidate the effects of tea polyphenols on Agrobacterium infection; and (3) identification of more suitable tea varieties for better transformation and regeneration. The most recent CRISPR system has enabled efficient genome editing in crops and shows great potential for accelerating precise crop improvement225. Considering the difficulties encountered transgenic studies of tea plant, the establishment of the CRISPR system will promote the transgenic progress in tea plants221. More recently, scientists demonstrated an efficient approach to overcome self-incompatibility in diploid potato using the CRISPR/Cas9 system through S-RNase locus knockout, which provided a useful example for the further application of transgenic technology and breeding in tea plants, considering their similar self-incompatibility features226.
Collection and conservation of tea plant germplasms
Resources are clearly the key to crop genetic improvement. The success of plant breeding and conservation is largely dependent on the amount and distribution of genetic variations present in collections. Currently, tea germplasms are becoming the most valuable fundamental material for tea breeding and biotechnology studies, presenting great potential in the whole tea industry227. The remarkable achievements made in worldwide tea plant germplasm surveys, collection, and conservation have preserved more than 15,000 tea plant accessions according to incomplete statistics from the China Tea Science Society (http://www.chinatss.cn/). These invaluable germplasms have driven the extraordinarily rapid development of tea plant genomics, genetics, and breeding151,152,175,207,228,229. However, most of the current accessions that have been collected and used are varieties, and few “wild” or close relatives have been integrated. Another major problem that is currently faced by the field is the imbalanced utilization and protection of cultivars versus “wild” germplasms. The survival environment of “wild” tea resources has been constantly destroyed with rapid economic development and urbanization. In addition, local germplasms (landraces) with specific characteristics are on the edge of being lost due to the popularization of elite varieties in the past few years. Above all, the surveying, collection, and conservation of tea plant germplasms should be urgently emphasized. Otherwise, no diverse tea plant resources or only a few mono varieties will survive and be available for utilization for the genetic improvement of tea in the future. We therefore propose that a tea plant conservation alliance uniting the global tea-planting countries needs to be established to accelerate the preservation and utilization of global tea plant resources.
In-depth understanding of the complex secondary metabolism network
The overall importance of tea plants as an economic crop is due to the health-promoting functions of teas as non-alcoholic drinks that are popular worldwide230. Numerous studies have documented the health benefits of various tea drinks in humans, and studies continue to produce more fundamental discoveries about the effects of functional tea components on the improvement of human heath231,232. It is not only the three major types of tea plant secondary metabolites (catechins, caffeine, and theanine) but also the volatile terpenoids, saponins, polysaccharides, and other phenolic conjugates found in tea that contribute to the beneficial health effects and the enjoyable flavors of various teas70,71,233. As mentioned above, our understanding of these complicated secondary metabolites in tea plants in terms of their biosynthesis, transport, and storage as well as their regulation is very limited. This is one of the major obstacles to the breeding of high-quality tea plant varieties with biofortified nutrients or functional components or other special properties. Future work could focus on basic questions about tea secondary metabolism, which will facilitate the breeding of tea plants with desirable qualities and benefit human health improvement projects.
In conclusion, tea plants are perennial woody plants in nature with a long growth cycle, which severely limits their genetic breeding. Traditional cross-breeding is extremely difficult and time-consuming for tea plants because most of the existing tea varieties are bred and propagated directly from natural populations. Modern transgenic breeding technology has provided us a new solution for the molecular design of breeding strategies, which basically relies on the development of functional genomics and molecular biology. Although great progress has been made in the last two decades, the genomics and molecular biology of tea plants are still not fully understood, mainly due to difficulties in resource collection and identification, the generation and identification of mutant plants, and population construction. In particular, the limited knowledge of the functional genomics and developmental biology of tea plants has narrowed our understanding of their basic biological characteristics. Therefore, compared to other crops such as rice, there is still a long way to go in tea plant genomics and molecular biology research. In the near future, scientists should focus more on industry-oriented major basic biological research based on germplasm collection and utilization, particularly by deciphering the tea plant genome, as an opportunity for enhancing studies on the mechanisms of the biosynthesis and regulation of secondary metabolites, the genetic basis of important agronomic traits, the molecular mechanisms of stress and disease resistance, and the developmental biology of tea plants, to accelerate the molecular design breeding program and, thus, promote industrial development.
Supplementary information
Acknowledgements
We would like to thank Dr. Penghui Li, Dr. Zhaoliang Zhang, and Dr. Chuankui Song for his discussion and comments on the manuscript. We apologize for not being able to cite many of the excellent publications on tea plants due to the limit of paper length. The authors acknowledge support from the National Natural Science Foundation of China (31800180), the Natural Science Foundation of Anhui Province (1908085MC75), the National Key Research and Development Program of China (2018YFD1000601), the Science and Technology Project of Anhui Province (13Z03012), the Special Innovative Province Construction in Anhui Province (15czs08032), the Changjiang Scholars and Innovative Research Team in University (IRT1101), the China Postdoctoral Science Foundation (No. 2017M621992), and the Postdoctoral Science Foundation of Anhui Province, China (No. 2017B189).
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: En-Hua Xia, Wei Tong
Contributor Information
Chao-Ling Wei, Email: weichl@ahau.edu.cn.
Xiao-Chun Wan, Email: xcwan@ahau.edu.cn.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41438-019-0225-4).
References
- 1.Xia E-H, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant. 2017;10:866–877. doi: 10.1016/j.molp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 2.Wei C, et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl Acad. Sci. USA. 2018;115:E4151–E4158. doi: 10.1073/pnas.1719622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y, et al. Rice functional genomics research: past decade and future. Mol. Plant. 2018;11:359–380. doi: 10.1016/j.molp.2018.01.007. [DOI] [PubMed] [Google Scholar]
- 4.Li B, Chen X, Chen G, Wang J. The analysis of karyotype in tea plant. J. Tea Sci. 1986;6:7–14. [Google Scholar]
- 5.Sheidai M, Jahanbakht H, Sofi-Siyavash P. Cytogenetic study of various types of tea (Camellia sinensis) cultivars in Iran. Iran. J. Sci. Technol. 2004;28:33–42. [Google Scholar]
- 6.Huang H, Tong Y, Zhang QJ, Gao LZ. Genome size variation among and within Camellia species by using flow cytometric analysis. PLoS ONE. 2013;8:e64981. doi: 10.1371/journal.pone.0064981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furukawa K, Sugiyama S, Ohta T, Ohmido N. Chromosome analysis of tea plant (Camellia sinensis) and ornamental camellia (Camellia japonica) Chromosome Sci. 2017;20:9–15. [Google Scholar]
- 8.Morinaga T, Fukushima E, Kano T, Maruyama Y, Yamasaki Y. Chromosome numbers of cultivated plants II. Bot. Mag. 1929;43:589–594. doi: 10.15281/jplantres1887.43.589. [DOI] [Google Scholar]
- 9.Morinaga T, Fukushima E. Chromosome numbers of cultivated plants III. Bot. Mag. 1931;45:140–145. doi: 10.15281/jplantres1887.45.140. [DOI] [Google Scholar]
- 10.Kondo K. Chromosome numbers in the genus Camellia. Biotropica. 1977;9:86–94. doi: 10.2307/2387663. [DOI] [Google Scholar]
- 11.Sharma S, Raina N. Chromosome constitution of some Indian tea clones. Int. J. Tea Sci. 2006;5:21–28. [Google Scholar]
- 12.Rahman H, et al. Cytomorphological characterization of tea cultivars. Pak. J. Bot. 2010;42:485–495. [Google Scholar]
- 13.Hanson L, Mahon K, Johnson M, Bennett M. First nuclear DNA C-values for another 25 angiosperm families. Ann. Bot. 2001;88:851–858. doi: 10.1006/anbo.2001.1528. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka J, Taniguchi F, Hirai N, Yamaguchi S. Estimation of the genome size of tea (Camellia sinensis), Camellia (C. japonica), and their interspecific hybrids by flow cytometry. Tea Res. J. 2006;1:1–7. doi: 10.5979/cha.2006.1. [DOI] [Google Scholar]
- 15.Sharma S, Kaushik S, Raina S. Estimation of nuclear DNA content and its variation among Indian Tea accessions by flow cytometry. Physiol. Mol. Biol. Plants. 2018;1:1–8. doi: 10.1007/s12298-018-0587-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ott J, Wang J, Leal SM. Genetic linkage analysis in the age of whole-genome sequencing. Nat. Rev. Genet. 2015;16:275–284. doi: 10.1038/nrg3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang CC, et al. Transcriptome analysis reveals self-incompatibility in the tea plant (Camellia sinensis) might be under gametophytic control. BMC Genom. 2016;17:359. doi: 10.1186/s12864-016-2703-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hackett C, Wachira F, Paul S, Powell W, Waugh R. Construction of a genetic linkage map for Camellia sinensis (tea) Heredity. 2000;85:346–355. doi: 10.1046/j.1365-2540.2000.00769.x. [DOI] [PubMed] [Google Scholar]
- 19.Huang J, et al. Construction of AFLP molecular markers linkage map in tea plant. J. Tea Sci. 2005;25:7–15. [Google Scholar]
- 20.Kamunya SM, et al. Genomic mapping and testing for quantitative trait loci in tea (Camellia sinensis (L.) O. Kuntze) Tree Genet. Genomes. 2010;6:915–929. doi: 10.1007/s11295-010-0301-2. [DOI] [Google Scholar]
- 21.Ma JQ, et al. Large-scale SNP discovery and genotyping for constructing a high-density genetic map of tea plant using specific-locus amplified fragment sequencing (SLAF-seq) PLoS ONE. 2015;10:e0128798. doi: 10.1371/journal.pone.0128798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ma JQ, et al. Quantitative trait loci mapping for theobromine and caffeine contents in tea plant (Camellia sinensis) J. Agric. Food Chem. 2018;66:13321–13327. doi: 10.1021/acs.jafc.8b05355. [DOI] [PubMed] [Google Scholar]
- 23.Ota S. RAPD-based linkage mapping using F1 segregating populations derived from crossings between tea cultivar ‘Sayamakaori' and strain ‘Kana-Ck17’. Breed. Res. 1999;1:16. [Google Scholar]
- 24.Huang F, Liang Y, Lu J, Chen R. Genetic mapping of first generation of backcross in tea by RAPD and ISSR markers. J. Tea Sci. 2006;26:171–176. [Google Scholar]
- 25.Taniguchi F, et al. Construction of a high-density reference linkage map of tea (Camellia sinensis) Breed. Sci. 2012;62:263–273. doi: 10.1270/jsbbs.62.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang, Y. et al. Construction of a genetic linkage map based on RAPD, AFLP, and SSR markers for tea plant (Camellia sinensis). Euphytica213, 10.1007/s10681-017-1979-0 (2017).
- 27.Ma JQ, et al. Construction of a SSR-based genetic map and identification of QTLs for catechins content in tea plant (Camellia sinensis) PLoS ONE. 2014;9:e93131. doi: 10.1371/journal.pone.0093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan, L.-Q. et al. SSR-based genetic mapping and QTL analysis for timing of spring bud flush, young shoot color, and mature leaf size in tea plant (Camellia sinensis). Tree Genet. Genomes12, 10.1007/s11295-016-1008-9 (2016).
- 29.Xu LY, et al. High-density SNP linkage map construction and QTL mapping for flavonoid-related traits in a tea plant (Camellia sinensis) using 2b-RAD sequencing. BMC Genom. 2018;19:955. doi: 10.1186/s12864-018-5291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen F, et al. The sequenced angiosperm genomes and genome databases. Front. Plant Sci. 2018;9:418. doi: 10.3389/fpls.2018.00418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cai J, et al. The genome sequence of the orchid Phalaenopsis equestris. Nat. Genet. 2015;47:65–72. doi: 10.1038/ng.3149. [DOI] [PubMed] [Google Scholar]
- 32.Peng Z, et al. The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla) Nat. Genet. 2013;45:456–461. doi: 10.1038/ng.2569. [DOI] [PubMed] [Google Scholar]
- 33.Goff SA, et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 34.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 35.Ashihara H, Suzuki T. Distribution and biosynthesis of caffeine in plants. Front. Biosci. 2004;9:1864–1876. doi: 10.2741/1367. [DOI] [PubMed] [Google Scholar]
- 36.Petit, R. J. & Vendramin, G. G. in Phylogeography of Southern European Refugia: Evolutionary perspectives on the origins and conservation of European biodiversity (eds Steven Weiss & Nuno Ferrand) 23–97 (Springer, Netherlands, 2007).
- 37.Chen C, Ma C, Ma J, Liu S, Chen L. Sequencing of chloroplast genome of Camellia sinensis and genetic relationship for Camellia plants based on chloroplast DNA sequences. J. Tea Sci. 2014;34:371–380. [Google Scholar]
- 38.Zhang, F., Li, W., Gao, C. & Gao, L. Deciphering tea tree chloroplast and mitochondrial genomes of Camellia sinensis var. assamica. Scientific Data 6, 209 (2019). [DOI] [PMC free article] [PubMed]
- 39.Yang JB, Yang SX, Li HT, Yang J, Li DZ. Comparative chloroplast genomes of Camellia species. PLoS ONE. 2013;8:e73053. doi: 10.1371/journal.pone.0073053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi C, et al. Contradiction between plastid gene transcription and function due to complex posttranscriptional splicing: an exemplary study of ycf15 function and evolution in angiosperms. PLoS ONE. 2013;8:e59620. doi: 10.1371/journal.pone.0059620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong M, et al. The complete chloroplast genome of an economic plant, Camellia sinensis cultivar Anhua, China. Mitochondrial DNA Part B. 2018;3:558–559. doi: 10.1080/23802359.2018.1462124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu Y, Han Y. The complete chloroplast genome sequence of Camellias (Camellia fangchengensis) Mitochondrial DNA Part B. 2017;3:34–35. doi: 10.1080/23802359.2017.1419086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Y, Han Y. The complete chloroplast genome sequence of endangered camellias (Camellia pubifurfuracea) Conserv. Genet. Resour. 2017;10:843–845. doi: 10.1007/s12686-017-0944-5. [DOI] [Google Scholar]
- 44.Wang G, Luo Y, Hou N, Deng L-X. The complete chloroplast genomes of three rare and endangered camellias (Camellia huana, C. liberofilamenta and C. luteoflora) endemic to Southwest China. Conserv. Genet. Resour. 2017;9:583–585. doi: 10.1007/s12686-017-0727-z. [DOI] [Google Scholar]
- 45.Huang H, Shi C, Liu Y, Mao S, Gao L. Thirteen Camellia chloroplast genome sequences determined by high-throughput sequencing genome structure and phylogenetic relationships. BMC Evolut. Biol. 2014;14:151. doi: 10.1186/1471-2148-14-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat. Rev. Genet. 2011;12:87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi C, et al. Deep sequencing of the Camellia sinensis transcriptome revealed candidate genes for major metabolic pathways of tea-specific compounds. BMC genomics. 2011;12:131. doi: 10.1186/1471-2164-12-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, et al. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genome. 2013;14:415. doi: 10.1186/1471-2164-14-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Q, et al. RNA-seq based transcriptomic analysis uncovers alpha-linolenic acid and jasmonic acid biosynthesis pathways respond to cold acclimation in Camellia japonica. Sci. Rep. 2016;6:36463. doi: 10.1038/srep36463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ban Q, et al. Comparative analysis of the response and gene regulation in cold resistant and susceptible tea plants. PLoS ONE. 2017;12:e0188514. doi: 10.1371/journal.pone.0188514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hao X, et al. Comprehensive transcriptome analysis reveals common and specific genes and pathways involved in cold acclimation and cold stress in tea plant leaves. Sci. Hortic. 2018;240:354–368. doi: 10.1016/j.scienta.2018.06.008. [DOI] [Google Scholar]
- 52.Liu SC, et al. Transcriptomic analysis of tea plant responding to drought stress and recovery. PLoS ONE. 2016;11:e0147306. doi: 10.1371/journal.pone.0147306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang Q, et al. Transcriptome dynamics of Camellia sinensis in response to continuous salinity and drought stress. Tree Genet. Genomes. 2017;13:78. doi: 10.1007/s11295-017-1161-9. [DOI] [Google Scholar]
- 54.Shi J, et al. Transcriptional responses and flavor volatiles biosynthesis in methyl jasmonate-treated tea leaves. BMC Plant Biol. 2015;15:233. doi: 10.1186/s12870-015-0609-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma Q, et al. Transcriptomic analysis between self- and cross-pollinated pistils of tea plants (Camellia sinensis) BMC Genom. 2018;19:289. doi: 10.1186/s12864-018-4674-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Huang H, Yao Q, Xia E, Gao L. Metabolomics and transcriptomics analyses reveal nitrogen influences on the accumulation of flavonoids and amino acids in young shoots of tea plant (Camellia sinensis L.) associated with tea flavor. J. Agric. Food Chem. 2018;66:9828–9838. doi: 10.1021/acs.jafc.8b01995. [DOI] [PubMed] [Google Scholar]
- 57.Li W, et al. Transcriptome and metabolite analysis identifies nitrogen utilization genes in tea plant (Camellia sinensis) Sci. Rep. 2017;7:1693. doi: 10.1038/s41598-017-01949-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yue C, et al. Comparative transcriptome study of hairy and hairless tea plant (Camellia sinensis) shoots. J. Plant Physiol. 2018;229:41–52. doi: 10.1016/j.jplph.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Guo F, Guo Y, Wang P, Wang Y, Ni D. Transcriptional profiling of catechins biosynthesis genes during tea plant leaf development. Planta. 2017;246:1139–1152. doi: 10.1007/s00425-017-2760-2. [DOI] [PubMed] [Google Scholar]
- 60.Li CF, et al. Global transcriptome and gene regulat etwork for secondary metabolite biosynthesis of tea plant (Camellia sinensis) BMC Genom. 2015;16:560. doi: 10.1186/s12864-015-1773-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang W, et al. Insight into catechins metabolic pathways of Camellia sinensis based on genome and transcriptome analysis. J. Agric. Food Chem. 2018;66:4281–4293. doi: 10.1021/acs.jafc.8b00946. [DOI] [PubMed] [Google Scholar]
- 62.Li J, et al. Transcriptome analysis reveals the accumulation mechanism of anthocyanins in ‘Zijuan’ tea (Camellia sinensis var. asssamica (Masters) kitamura) leaves. Plant Growth Regul. 2016;81:51–61. doi: 10.1007/s10725-016-0183-x. [DOI] [Google Scholar]
- 63.Xu Q, et al. Transcriptome profiling using single-molecule direct RNA sequencing approach for in-depth understanding of genes in secondary metabolism pathways of Camellia sinensis. Front. Plant Sci. 2017;8:1205. doi: 10.3389/fpls.2017.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xia EH, et al. Transcriptome analysis of the oil-rich tea plant, Camellia oleifera, reveals candidate genes related to lipid metabolism. PLoS ONE. 2014;9:e104150. doi: 10.1371/journal.pone.0104150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang HB, et al. De novo transcriptome assembly of the wild relative of tea tree (Camellia taliensis) and comparative analysis with tea transcriptome identified putative genes associated with tea quality and stress response. BMC Genom. 2015;16:298. doi: 10.1186/s12864-015-1494-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yao QY, Huang H, Tong Y, Xia EH, Gao LZ. Transcriptome analysis identifies candidate genes related to triacylglycerol and pigment biosynthesis and photoperiodic flowering in the ornamental and oil-producing plant, Camellia reticulata (Theaceae) Front. Plant Sci. 2016;7:163. doi: 10.3389/fpls.2016.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang H, Xia EH, Zhang HB, Yao QY, Gao LZ. De novo transcriptome sequencing of Camellia sasanqua and the analysis of major candidate genes related to floral traits. Plant Physiol. Biochem. 2017;120:103–111. doi: 10.1016/j.plaphy.2017.08.028. [DOI] [PubMed] [Google Scholar]
- 68.Wang ZW, et al. Deep sequencing of the Camellia chekiangoleosa transcriptome revealed candidate genes for anthocyanin biosynthesis. Gene. 2014;538:1–7. doi: 10.1016/j.gene.2014.01.035. [DOI] [PubMed] [Google Scholar]
- 69.Zhou X, et al. De novo assembly of the Camellia nitidissima transcriptome reveals key genes of flower pigment biosynthesis. Front. Plant Sci. 2017;8:1545. doi: 10.3389/fpls.2017.01545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zeng L, Watanabe N, Yang Z. Understanding the biosyntheses and stress response mechanisms of aroma compounds in tea (Camellia sinensis) to safely and effectively improve tea aroma. Crit. Rev. Food Sci. Nutr. 2018;1:1–14. doi: 10.1080/10408398.2018.1506907. [DOI] [PubMed] [Google Scholar]
- 71.Song C, Hartl K, McGraphery K, Hoffmann T, Schwab W. Attractive but toxic: emerging roles of glycosidically bound volatiles and glycosyltransferases involved in their formation. Mol. Plant. 2018;11:1225–1236. doi: 10.1016/j.molp.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Z, et al. Advances in research on functional genes of tea plant. Gene. 2019;711:143940. doi: 10.1016/j.gene.2019.143940. [DOI] [PubMed] [Google Scholar]
- 73.Kato M, Mizuno K, Crozier A, Fujimura T, Ashihara H. Caffeine synthase gene from tea leaves. Nature. 2000;406:956–957. doi: 10.1038/35023072. [DOI] [PubMed] [Google Scholar]
- 74.Liu Y, et al. Purification and characterization of a novel galloyltransferase involved in catechin galloylation in the tea plant (Camellia sinensis) J. Biol. Chem. 2012;287:44406–44417. doi: 10.1074/jbc.M112.403071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cui L, et al. Identification of UDP-glycosyltransferases involved in the biosynthesis of astringent taste compounds in tea (Camellia sinensis) J. Exp. Bot. 2016;67:2285–2297. doi: 10.1093/jxb/erw053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wei K, et al. A coupled role for CsMYB75 and CsGSTF1 in anthocyanin hyperaccumulation in purple tea. Plant J. 2019;97:825–840. doi: 10.1111/tpj.14161. [DOI] [PubMed] [Google Scholar]
- 77.Pang Y, et al. Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol. 2013;161:1103–1116. doi: 10.1104/pp.112.212050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li M, et al. Functional characterization of tea (Camellia sinensis) MYB4a transcription factor using an integrative approach. Front. Plant Sci. 2017;8:943. doi: 10.3389/fpls.2017.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jiang X, et al. CsMYB5a and CsMYB5e from Camellia sinensis differentially regulate anthocyanin and proanthocyanidin biosynthesis. Plant Sci. 2018;270:209–220. doi: 10.1016/j.plantsci.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 80.Wang P, et al. A sucrose-induced MYB (SIMYB) transcription factor promoting proanthocyanidin accumulation in the tea plant (Camellia sinensis) J. Agric. Food Chem. 2019;67:1418–1428. doi: 10.1021/acs.jafc.8b06207. [DOI] [PubMed] [Google Scholar]
- 81.Jing T, et al. Glucosylation of (Z)-3-hexenol informs intraspecies interactions in plants: a case study in Camellia sinensis. Plant Cell Environ. 2019;42:1352–1367. doi: 10.1111/pce.13479. [DOI] [PubMed] [Google Scholar]
- 82.Li XW, et al. A novel cold-regulated gene from Camellia sinensis, CsCOR1, enhances salt- and dehydration-tolerance in tobacco. Biochem. Biophys. Res. Commun. 2010;394:354–359. doi: 10.1016/j.bbrc.2010.03.011. [DOI] [PubMed] [Google Scholar]
- 83.Acharya K, et al. Overexpression of Camellia sinensis thaumatin-like protein, CsTLP in potato confers enhanced resistance to Macrophomina phaseolina and Phytophthora infestans infection. Mol. Biotechnol. 2013;54:609–622. doi: 10.1007/s12033-012-9603-y. [DOI] [PubMed] [Google Scholar]
- 84.Wang Y, Jiang CJ, Li YY, Wei CL, Deng WW. CsICE1 and CsCBF1: two transcription factors involved in cold responses in Camellia sinensis. Plant Cell Rep. 2012;31:27–34. doi: 10.1007/s00299-011-1136-5. [DOI] [PubMed] [Google Scholar]
- 85.Deng WW, et al. Molecular cloning, functional analysis of three cinnamyl alcohol dehydrogenase (CAD) genes in the leaves of tea plant, Camellia sinensis. J. Plant Physiol. 2013;170:272–282. doi: 10.1016/j.jplph.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 86.Zhou Y, et al. Molecular cloning and characterization of galactinol synthases in Camellia sinensis with different responses to biotic and abiotic stressors. J. Agric. Food Chem. 2017;65:2751–2759. doi: 10.1021/acs.jafc.7b00377. [DOI] [PubMed] [Google Scholar]
- 87.Liu GF, et al. Implementation of CsLIS/NES in linalool biosynthesis involves transcript splicing regulation in Camellia sinensis. Plant Cell Environ. 2018;41:176–186. doi: 10.1111/pce.13080. [DOI] [PubMed] [Google Scholar]
- 88.Mizutani M, et al. Cloning of beta-primeverosidase from tea leaves, a key enzyme in tea aroma formation. Plant Physiol. 2002;130:2164–2176. doi: 10.1104/pp.102.011023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zeng L, et al. Formation of volatile tea constituent indole during the Oolong tea manufacturing process. J. Agric. Food Chem. 2016;64:5011–5019. doi: 10.1021/acs.jafc.6b01742. [DOI] [PubMed] [Google Scholar]
- 90.Ohgami S, et al. Volatile glycosylation in tea plants: sequential glycosylations for the biosynthesis of aroma beta-Primeverosides are catalyzed by two Camellia sinensis glycosyltransferases. Plant Physiol. 2015;168:464–477. doi: 10.1104/pp.15.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou Y, et al. Formation of (E)-nerolidol in tea (Camellia sinensis) leaves exposed to multiple stresses during tea manufacturing. Food Chem. 2017;231:78–86. doi: 10.1016/j.foodchem.2017.03.122. [DOI] [PubMed] [Google Scholar]
- 92.Zhou TS, et al. Cloning and characterization of a flavonoid 3'-hydroxylase gene from tea plant (Camellia sinensis) Int J. Mol. Sci. 2016;17:261. doi: 10.3390/ijms17020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xia J, et al. Characterization and expression profiling of Camellia sinensis cinnamate 4-hydroxylase genes in phenylpropanoid pathways. Genes. 2017;8:193. doi: 10.3390/genes8080193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Han Y, et al. Functional analysis of two flavanone-3-hydroxylase genes from Camellia sinensis: a critical role in flavonoid accumulation. Genes. 2017;8:300. doi: 10.3390/genes8110300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu Y, et al. Three Camellia sinensis glutathione S-transferases are involved in the storage of anthocyanins, flavonols, and proanthocyanidins. Planta. 2019;250:1163–1175. doi: 10.1007/s00425-019-03206-2. [DOI] [PubMed] [Google Scholar]
- 96.Dong, C. et al. Theanine transporters identified in tea plants (Camellia sinensis L.). Plant J.10.1111/tpj.14517 (2019). [DOI] [PubMed]
- 97.Deng W-W, Wang R, Yang T, Jiang LN, Zhang Z-Z. Functional characterization of salicylic acid carboxyl methyltransferase from Camellia sinensis, providing the aroma compound of methyl salicylate during the withering process of white tea. J. Agric. Food Chem. 2017;65:11036–11045. doi: 10.1021/acs.jafc.7b04575. [DOI] [PubMed] [Google Scholar]
- 98.Wang M, et al. Molecular cloning and expression analysis of low molecular weight heat shock protein gene CsHSP17.2 from Camellia sinensis. J. Nanjing Agric. Univ. 2015;38:389–394. [Google Scholar]
- 99.Mohanpuria P, Kumar V, Yadav SK. Tea caffeine: metabolism, functions, and reduction strategies. Food Sci. Biotechnol. 2010;19:275–287. doi: 10.1007/s10068-010-0041-y. [DOI] [Google Scholar]
- 100.Shirlow M, Mathers C. A study of caffeine consumption and symptoms: indigestion, palpitations, tremor, headache and insomnia. Int. J. Epidemiol. 1985;14:239–248. doi: 10.1093/ije/14.2.239. [DOI] [PubMed] [Google Scholar]
- 101.Schaffer SW, et al. Effect of taurine and potential interactions with caffeine on cardiovascular function. Amino Acids. 2014;46:1147–1157. doi: 10.1007/s00726-014-1708-0. [DOI] [PubMed] [Google Scholar]
- 102.Yamamoto S, Wakayama M, Tachiki T. Cloning and expression of Pseudomonas taetrolens Y-30 gene encoding glutamine synthetase: an enzyme available for theanine production by coupled fermentation with energy transfer. Biosci. Biotechnol. Biochem. 2006;70:500–507. doi: 10.1271/bbb.70.500. [DOI] [PubMed] [Google Scholar]
- 103.Higdon JV, Frei B. Tea catechins and polyphenols: health effects, metabolism, and antioxidant functions. Crit. Rev. Food Sci. Nutr. 2003;43:89–143. doi: 10.1080/10408690390826464. [DOI] [PubMed] [Google Scholar]
- 104.He X, et al. Isolation and characterization of key genes that promote flavonoid accumulation in purple-leaf tea (Camellia sinensis L.) Sci. Rep. 2018;8:130. doi: 10.1038/s41598-017-18133-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gohain B, et al. Understanding Darjeeling tea flavour on a molecular basis. Plant Mol. Biol. 2012;78:577–597. doi: 10.1007/s11103-012-9887-0. [DOI] [PubMed] [Google Scholar]
- 106.Han ZX, et al. Green tea flavour determinants and their changes over manufacturing processes. Food Chem. 2016;212:739–748. doi: 10.1016/j.foodchem.2016.06.049. [DOI] [PubMed] [Google Scholar]
- 107.Lee J, Chambers DH, Chambers Et. A comparison of the flavor of green teas from around the world. J. Sci. Food Agric. 2014;94:1315–1324. doi: 10.1002/jsfa.6413. [DOI] [PubMed] [Google Scholar]
- 108.Hu CJ, et al. Formation mechanism of the oolong tea characteristic aroma during bruising and withering treatment. Food Chem. 2018;269:202–211. doi: 10.1016/j.foodchem.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 109.Schuh C, Schieberle P. Characterization of the key aroma compounds in the beverage prepared from Darjeeling black tea: quantitative differences between tea leaves and infusion. J. Agric. Food Chem. 2006;54:916–924. doi: 10.1021/jf052495n. [DOI] [PubMed] [Google Scholar]
- 110.Guo W, et al. (S)-linalyl, 2-phenylethyl, and benzyl disaccharide glycosides isolated as aroma precursors from oolong tea leaves. Biosci. Biotechnol. Biochem. 1994;58:1532–1534. doi: 10.1271/bbb.58.1532. [DOI] [PubMed] [Google Scholar]
- 111.Wang D, Kubota K, Kobayashi A, Juan I. Analysis of glycosidically bound aroma precursors in tea leaves. 3. Change in the glycoside content of tea leaves during the oolong tea manufacturing process. J. Agric. Food Chem. 2001;49:5391–5396. doi: 10.1021/jf010235+. [DOI] [PubMed] [Google Scholar]
- 112.Wang D, Yoshimura T, Kubota K, Kobayashi A. Analysis of glycosidically bound aroma precursors in tea leaves. 1. Qualitative and quantitative analyses of glycosides with aglycons as aroma compounds. J. Agric. Food Chem. 2000;48:5411–5418. doi: 10.1021/jf000443m. [DOI] [PubMed] [Google Scholar]
- 113.Ho C, Zheng X, Li S. Tea aroma formation. Food Sci. Hum. Wellness. 2015;4:9–27. doi: 10.1016/j.fshw.2015.04.001. [DOI] [Google Scholar]
- 114.Mei X, et al. Formation and emission of linalool in tea (Camellia sinensis) leaves infested by tea green leafhopper (Empoasca (Matsumurasca) onukii Matsuda) Food Chem. 2017;237:356–363. doi: 10.1016/j.foodchem.2017.05.124. [DOI] [PubMed] [Google Scholar]
- 115.Tariq M, Paszkowski J. DNA and histone methylation in plants. Trends Genet. 2004;20:244–251. doi: 10.1016/j.tig.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 116.Zhang H, Lang Z, Zhu JK. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018;19:489–506. doi: 10.1038/s41580-018-0016-z. [DOI] [PubMed] [Google Scholar]
- 117.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li X, et al. High-resolution mapping of epigenetic modifications of the rice genome uncovers interplay between DNA methylation, histone methylation, and gene expression. Plant Cell. 2008;20:259–276. doi: 10.1105/tpc.107.056879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang Lei, Shi Yan, Chang Xiaojun, Jing Shengli, Zhang Qunjie, You Chenjiang, Yuan Hongyu, Wang Haifeng. DNAmethylome analysis provides evidence that the expansion of the tea genome is linked toTEbursts. Plant Biotechnology Journal. 2018;17(4):826–835. doi: 10.1111/pbi.13018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nystedt B, et al. The Norway spruce genome sequence and conifer genome evolution. Nature. 2013;497:579–584. doi: 10.1038/nature12211. [DOI] [PubMed] [Google Scholar]
- 121.Schnable PS, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 122.Gent JI, et al. CHH islands: de novo DNA methylation in near-gene chromatin regulation in maize. Genome Res. 2012;23:628–637. doi: 10.1101/gr.146985.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhou Y, et al. Changes of DNA methylation levels and patterns in tea plant (Camellia sinensis) during cold acclimation. Acta Agron. Sin. 2015;41:1047–1055. doi: 10.3724/SP.J.1006.2015.01047. [DOI] [Google Scholar]
- 124.Zhou Y, et al. Cloning and expression analysis on DNA methyltransferase gene, CsDRM2, of Camellia sinensis. Acta Tea Sin. 2015;56:1–7. [Google Scholar]
- 125.Jeyaraj A, et al. Genome-wide identification of conserved and novel microRNAs in one bud and two tender leaves of tea plant (Camellia sinensis) by small RNA sequencing, microarray-based hybridization and genome survey scaffold sequences. BMC Plant Biol. 2017;17:212. doi: 10.1186/s12870-017-1169-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhao S, et al. Revealing of microRNA involved regulatory gene networks on terpenoid biosynthesis in Camellia sinensis in different growing time points. J. Agric. Food Chem. 2018;66:12604–12616. doi: 10.1021/acs.jafc.8b05345. [DOI] [PubMed] [Google Scholar]
- 127.Sun P, et al. Combined small RNA and degradome sequencing reveals complex microRNA regulation of catechin biosynthesis in tea (Camellia sinensis) PloS ONE. 2017;12:e0171173. doi: 10.1371/journal.pone.0171173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Fan K, Fan D, Ding Z, Su Y, Wang X. Cs-miR156 is involved in the nitrogen form regulation of catechins accumulation in tea plant (Camellia sinensis L.) Plant Physiol. Biochem. 2015;97:350–360. doi: 10.1016/j.plaphy.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 129.Zhang Y, et al. Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol. 2014;14:271. doi: 10.1186/s12870-014-0271-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zheng C, et al. Integrated RNA-Seq and sRNA-Seq analysis identifies chilling and freezing responsive key molecular players and pathways in tea plant (Camellia sinensis) PLoS ONE. 2015;10:e0125031. doi: 10.1371/journal.pone.0125031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu SC, et al. Small RNA and degradome profiling reveals important roles for microRNAs and their targets in tea plant response to drought stress. Physiol. Plant. 2016;158:435–451. doi: 10.1111/ppl.12477. [DOI] [PubMed] [Google Scholar]
- 132.Guo YQ, et al. Identification of drought-responsive miRNAs and physiological characterization of tea plant (Camellia sinensis L.) under drought stress. BMC Plant Biol. 2017;17:211. doi: 10.1186/s12870-017-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Jeyaraj A, et al. Genome-wide identification of microRNAs responsive to Ectropis oblique feeding in tea plant (Camellia sinensis L.) Sci. Rep. 2017;7:13634. doi: 10.1038/s41598-017-13692-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Jeyaraj A, et al. Identification of regulatory networks of microRNAs and their targets in response to Colletotrichum gloeosporioides in tea plant (Camellia sinensis L.) Front. Plant Sci. 2019;10:1096. doi: 10.3389/fpls.2019.01096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhou C, et al. Genome-wide investigation of superoxide dismutase (SOD) gene family and their regulatory miRNAs reveal the involvement in abiotic stress and hormone response in tea plant (Camellia sinensis) PLoS ONE. 2019;14:e0223609. doi: 10.1371/journal.pone.0223609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu S, et al. Integrated analysis of miRNAs and their targets reveals that miR319c/TCP2 regulates apical bud burst in tea plant (Camellia sinensis) Planta. 2019;250:1111–1129. doi: 10.1007/s00425-019-03207-1. [DOI] [PubMed] [Google Scholar]
- 137.Varshney D, et al. Tissue specific long non-coding RNAs are involved in aroma formation of black tea. Ind. Crops Products. 2019;133:79–89. doi: 10.1016/j.indcrop.2019.03.020. [DOI] [Google Scholar]
- 138.Wang, P. L. et al. Circular RNA is expressed across the Eukaryotic tree of life. PLoS ONE9, e90859 (2014). [DOI] [PMC free article] [PubMed]
- 139.Ye CY, Chen L, Liu C, Zhu QH, Fan L. Widespread noncoding circular RNAs in plants. New Phytol. 2015;208:88–95. doi: 10.1111/nph.13585. [DOI] [PubMed] [Google Scholar]
- 140.Lu T, et al. Transcriptome-wide investigation of circular RNAs in rice. RNA. 2015;21:2076–2087. doi: 10.1261/rna.052282.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tong W, et al. Circular RNA architecture and differentiation during leaf bud to young leaf development in tea (Camellia sinensis) Planta. 2018;248:1417–1429. doi: 10.1007/s00425-018-2983-x. [DOI] [PubMed] [Google Scholar]
- 142.Zuo J, Wang Q, Zhu B, Luo Y, Gao L. Deciphering the roles of circRNAs on chilling injury in tomato. Biochem. Biophys. Res. Commun. 2016;479:132–138. doi: 10.1016/j.bbrc.2016.07.032. [DOI] [PubMed] [Google Scholar]
- 143.Wang Y, et al. Identification of circular RNAs and their targets in leaves of Triticum aestivum L. under dehydration stress. Front. Plant Sci. 2016;7:2024. doi: 10.3389/fpls.2016.02024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wang ZP, et al. Identification of circular RNAs in Kiwifruit and their species-specific response to bacterial canker pathogen invasion. Front. Plant Sci. 2017;8:413. doi: 10.3389/fpls.2017.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Chen L, Gao Q-k, Chen D-m, Xu C-j. The use of RAPD markers for detecting genetic diversity, relationship and molecular identification of Chinese elite tea genetic resources [Camellia sinensis (L.) O. Kuntze] preserved in a tea germplasm repository. Biodivers. Conserv. 2005;14:1433–1444. doi: 10.1007/s10531-004-9787-y. [DOI] [Google Scholar]
- 146.Kottawa-Arachchi JD, Gunasekare MTK, Ranatunga MAB. Biochemical diversity of global tea [Camellia sinensis (L.) O. Kuntze] germplasm and its exploitation: a review. Genet. Resour. Crop Evol. 2018;66:259–273. doi: 10.1007/s10722-018-0698-2. [DOI] [Google Scholar]
- 147.Afridi SG, Ahmad H, Alam M, Khan IA, Hassan M. DNA landmarks for genetic diversity assessment in tea genotypes using RAPD markers. Afr. J. Biotechnol. 2011;10:15477–11548. doi: 10.5897/AJB11.801. [DOI] [Google Scholar]
- 148.Fang W, Cheng H, Duan Y, Jiang X, Li X. Genetic diversity and relationship of clonal tea (Camellia sinensis) cultivars in China as revealed by SSR markers. Plant Syst. Evol. 2011;298:469–483. doi: 10.1007/s00606-011-0559-3. [DOI] [Google Scholar]
- 149.Liu S, et al. Genome-wide identification of simple sequence repeats and development of polymorphic SSR markers for genetic studies in tea plant (Camellia sinensis) Mol. Breed. 2018;38:59. doi: 10.1007/s11032-018-0824-z. [DOI] [Google Scholar]
- 150.Ohsako T, Ohgushi T, Motosugi H, Oka K. Microsatellite variability within and among local landrace populations of tea, Camellia sinensis (L.) O. Kuntze, in Kyoto, Japan. Genet. Resour. Crop Evol. 2008;55:1047–1053. doi: 10.1007/s10722-008-9311-4. [DOI] [Google Scholar]
- 151.Taniguchi F, et al. Worldwide core collections of tea (Camellia sinensis) based on SSR markers. Tree Genet. Genomes. 2014;10:1555–1565. doi: 10.1007/s11295-014-0779-0. [DOI] [Google Scholar]
- 152.Yang H, et al. Genetic divergence between Camellia sinensis and its wild relatives revealed via genome-wide SNPs from RAD sequencing. PLoS ONE. 2016;11:e0151424. doi: 10.1371/journal.pone.0151424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wachira FN, Waugh R, Powell W, Hackett C. Detection of genetic diversity in tea (Camellia sinensis) using RAPD markers. Genome. 1995;38:201–210. doi: 10.1139/g95-025. [DOI] [PubMed] [Google Scholar]
- 154.Chen L, Yamaguchi S. Genetic diversity and phylogeny of tea plant (Camellia sinensis) and its related species and varieties in the sectionTheagenusCamellia determined by randomly amplified polymorphic DNA analysis. J. Hortic. Sci. Biotechnol. 2002;77:729–732. doi: 10.1080/14620316.2002.11511564. [DOI] [Google Scholar]
- 155.Wachira FN, Powell W, Waugh R. An assessment of genetic diversity among Camellia sinensis L. (cultivated tea) and its wild relatives based on randomly amplified polymorphic DNA and organelle-specific STS. Heredity. 1997;78:603. doi: 10.1038/hdy.1997.99. [DOI] [Google Scholar]
- 156.Balasaravanan T, Pius PK, Raj Kumar R, Muraleedharan N, Shasany AK. Genetic diversity among south Indian tea germplasm (Camellia sinensis, C. assamica and C. assamica spp. lasiocalyx) using AFLP markers. Plant Sci. 2003;165:365–372. doi: 10.1016/S0168-9452(03)00196-1. [DOI] [Google Scholar]
- 157.Kaundun SS, Zhyvoloup A, Park Y-G. Evaluation of the genetic diversity among elite tea (Camellia sinensis var. sinensis) accessions using RAPD markers. Euphytica. 2000;115:7–16. doi: 10.1023/A:1003939120048. [DOI] [Google Scholar]
- 158.Paul S, Wachira F, Powell W, Waugh R. Diversity and genetic differentiation among populations of Indian and Kenyan tea (Camellia sinensis (L.) O. Kuntze) revealed by AFLP markers. Theor. Appl. Genet. 1997;94:255–263. doi: 10.1007/s001220050408. [DOI] [Google Scholar]
- 159.Ori F, et al. DNA-based diversity of tea plants grown in Italy. Genet. Resour. Crop Evol. 2017;64:1905–1915. doi: 10.1007/s10722-016-0480-2. [DOI] [Google Scholar]
- 160.Wambulwa MC, et al. Nuclear microsatellites reveal the genetic architecture and breeding history of tea germplasm of East Africa. Tree Genet. Genomes. 2016;12:11. doi: 10.1007/s11295-015-0963-x. [DOI] [Google Scholar]
- 161.Wambulwa MC, et al. Insights into the genetic relationships and breeding patterns of the African tea germplasm based on nSSR markers and cpDNA sequences. Front. Plant Sci. 2016;7:1244. doi: 10.3389/fpls.2016.01244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Karunarathna KHT, et al. Understanding the genetic relationships and breeding patterns of Sri Lankan tea cultivars with genomic and EST-SSR markers. Sci. Hortic. 2018;240:72–80. doi: 10.1016/j.scienta.2018.05.051. [DOI] [Google Scholar]
- 163.Chen, Y. et al. DNA fingerprinting of oil camellia cultivars with SSR markers. Tree Genet. Genomes12, 10.1007/s11295-015-0966-7 (2016).
- 164.Jiang C, et al. A treasure reservoir of genetic resource of tea plant (Camellia sinensis) in Dayao Mountain. Genet. Resour. Crop Evol. 2017;65:217–227. doi: 10.1007/s10722-017-0524-2. [DOI] [Google Scholar]
- 165.Liu S, et al. Construction of fingerprinting for tea plant (Camellia sinensis) accessions using new genomic SSR markers. Mol. Breed. 2017;37:93. doi: 10.1007/s11032-017-0692-y. [DOI] [Google Scholar]
- 166.Zhao D-w, Yang J-b, Yang S-x, Kato K, Luo J-p. Genetic diversity and domestication origin of tea plant Camellia taliensis (Theaceae) as revealed by microsatellite markers. BMC Plant Biol. 2014;14:14. doi: 10.1186/1471-2229-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yao M-Z, Ma C-L, Qiao T-T, Jin J-Q, Chen L. Diversity distribution and population structure of tea germplasms in China revealed by EST-SSR markers. Tree Genet. Genomes. 2011;8:205–220. doi: 10.1007/s11295-011-0433-z. [DOI] [Google Scholar]
- 168.Xia EH, et al. CandiSSR: an efficient pipeline used for identifying candidate polymorphic SSRs based on multiple assembled sequences. Front. Plant Sci. 2015;6:1171. doi: 10.3389/fpls.2015.00490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang S, et al. TBC2health: a database of experimentally validated health-beneficial effects of tea bioactive compounds. Brief Bioinforma. 2017;18:830–836. doi: 10.1093/bib/bbv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang S, et al. TBC2target: a resource of predicted target genes of tea bioactive compounds. Front. Plant Sci. 2018;9:211. doi: 10.3389/fpls.2018.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yue Y, et al. TMDB: a literature-curated database for small molecular compounds found from tea. BMC Plant Biol. 2014;14:243. doi: 10.1186/s12870-014-0243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Klein RD, Fischer SM. Black tea polyphenols inhibit IGF-I-induced signaling through Akt in normal prostate epithelial cells and Du145 prostate carcinoma cells. Carcinogenesis. 2002;23:217–221. doi: 10.1093/carcin/23.1.217. [DOI] [PubMed] [Google Scholar]
- 173.Li M, et al. EGCG induces lung cancer A549 cell apoptosis by regulating Ku70 acetylation. Oncol. Rep. 2016;35:2339–2347. doi: 10.3892/or.2016.4587. [DOI] [PubMed] [Google Scholar]
- 174.Wang X, et al. PharmMapper 2017 update: a web server for potential drug target identification with a comprehensive target pharmacophore database. Nucleic Acids Res. 2017;45:W356–W360. doi: 10.1093/nar/gkx374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Xia En‐Hua, Li Fang‐Dong, Tong Wei, Li Peng‐Hui, Wu Qiong, Zhao Hui‐Juan, Ge Ruo‐Heng, Li Ruo‐Pei, Li Ye‐Yun, Zhang Zheng‐Zhu, Wei Chao‐Ling, Wan Xiao‐Chun. Tea Plant Information Archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnology Journal. 2019;17(10):1938–1953. doi: 10.1111/pbi.13111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Wurschum T, Mapping QTL. for agronomic traits in breeding populations. Theor. Appl. Genet. 2012;125:201–210. doi: 10.1007/s00122-012-1887-6. [DOI] [PubMed] [Google Scholar]
- 177.McCough SR, Doerge RW. QTL mapping in rice. Trends Genet. 1995;11:482–487. doi: 10.1016/S0168-9525(00)89157-X. [DOI] [PubMed] [Google Scholar]
- 178.Szalma SJ, Hostert BM, Ledeaux JR, Stuber CW, Holland JB. QTL mapping with near-isogenic lines in maize. Theor. Appl. Genet. 2007;114:1211–1228. doi: 10.1007/s00122-007-0512-6. [DOI] [PubMed] [Google Scholar]
- 179.Jin JQ, Ma JQ, Yao MZ, Ma CL, Chen L. Functional natural allelic variants of flavonoid 3',5'-hydroxylase gene governing catechin traits in tea plant and its relatives. Planta. 2017;245:523–538. doi: 10.1007/s00425-016-2620-5. [DOI] [PubMed] [Google Scholar]
- 180.Rothenberg D, Yang H, Chen M, Zhang W, Zhang L. Metabolome and transcriptome sequencing analysis reveals anthocyanin metabolism in pink flowers of anthocyanin-rich tea (Camellia sinensis) Molecules. 2019;24:1064. doi: 10.3390/molecules24061064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Xu Q, et al. Unraveling a crosstalk regulatory network of temporal aroma accumulation in tea plant (Camellia sinensis) leaves by integration of metabolomics and transcriptomics. Environ. Exp. Bot. 2018;149:81–94. doi: 10.1016/j.envexpbot.2018.02.005. [DOI] [Google Scholar]
- 182.Tai Y, et al. Gene co-expression network analysis reveals coordinated regulation of three characteristic secondary biosynthetic pathways in tea plant (Camellia sinensis) BMC Genom. 2018;19:616. doi: 10.1186/s12864-018-4999-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zhu J, et al. Global dissection of alternative splicing uncovers transcriptional diversity in tissues and associates with the flavonoid pathway in tea plant (Camellia sinensis) BMC Plant Biol. 2018;18:266. doi: 10.1186/s12870-018-1497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ahmed S, et al. Effects of extreme climate events on tea (Camellia sinensis) functional quality validate indigenous farmer knowledge and sensory preferences in tropical China. PLoS ONE. 2014;9:e109126. doi: 10.1371/journal.pone.0109126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Wei K, et al. Comparison of catechins and purine alkaloids in albino and normal green tea cultivars (Camellia sinensis L.) by HPLC. Food Chem. 2012;130:720–724. doi: 10.1016/j.foodchem.2011.07.092. [DOI] [Google Scholar]
- 186.Erxu PI, et al. Leaf morphology and anatomy of Camellia section Camellia (Theaceae) Bot. J. Linn. Soc. 2009;159:456–476. doi: 10.1111/j.1095-8339.2009.00952.x. [DOI] [Google Scholar]
- 187.Zhao Q, et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 2018;50:278–284. doi: 10.1038/s41588-018-0041-z. [DOI] [PubMed] [Google Scholar]
- 188.Li YH, et al. De novo assembly of soybean wild relatives for pan-genome analysis of diversity and agronomic traits. Nat. Biotechnol. 2014;32:1045–1052. doi: 10.1038/nbt.2979. [DOI] [PubMed] [Google Scholar]
- 189.Hirsch CN, et al. Insights into the maize pan-genome and pan-transcriptome. Plant Cell. 2014;26:121–135. doi: 10.1105/tpc.113.119982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yang J, et al. Phylogenetic relationships of theaceae inferred from mitochondrial matR gene sequence data. Acta Bot. Yunnanica. 2005;28:29–36. [Google Scholar]
- 192.Yang J, Li H, Yang S, Li D, Yang Y. The application of four DNA sequences to studying molecular phylogeny of Camellia (Theaceae) Acta Bot. Yunnanica. 2006;28:108–114. [Google Scholar]
- 193.Fang W, Yang J-B, Yang S-X, Li D-Z. Phylogeny of Camelia sects. Longipedicelata, Chrysantha and Longisima (Theaceae) based on sequence data of four chloroplast DNA loci. Acta Bot. Yunnanica. 2010;32:1–13. doi: 10.3724/SP.J.1143.2010.00001. [DOI] [Google Scholar]
- 194.Zeng L, et al. Resolution of deep angiosperm phylogeny using conserved nuclear genes and estimates of early divergence times. Nat. Commun. 2014;5:4956. doi: 10.1038/ncomms5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wickett NJ, et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl Acad. Sci. USA. 2014;111:E4859–E4868. doi: 10.1073/pnas.1323926111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Zeng L, et al. Resolution of deep eudicot phylogeny and their temporal diversification using nuclear genes from transcriptomic and genomic datasets. New Phytol. 2017;214:1338–1354. doi: 10.1111/nph.14503. [DOI] [PubMed] [Google Scholar]
- 197.Yang Y, et al. Dissecting molecular evolution in the highly diverse plant clade Caryophyllales using transcriptome sequencing. Mol. Biol. Evol. 2015;32:2001–2014. doi: 10.1093/molbev/msv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Deng H, Zhang GQ, Lin M, Wang Y, Liu ZJ. Mining from transcriptomes: 315 single‐copy orthologous genes concatenated for the phylogenetic analyses of Orchidaceae. Ecol. Evol. 2015;5:3800–3807. doi: 10.1002/ece3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Atwell S, et al. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465:627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Li H, et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013;45:43–50. doi: 10.1038/ng.2484. [DOI] [PubMed] [Google Scholar]
- 201.Huang XH, et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010;42:961–976. doi: 10.1038/ng.695. [DOI] [PubMed] [Google Scholar]
- 202.Xiao YJ, Liu HJ, Wu LJ, Warburton M, Yan JB. Genome-wide association studies in maize: praise and stargaze. Mol. plant. 2017;10:359–374. doi: 10.1016/j.molp.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 203.Nordborg M, Weigel D. Next-generation genetics in plants. Nature. 2008;456:720–723. doi: 10.1038/nature07629. [DOI] [PubMed] [Google Scholar]
- 204.Zhou X, Huang X. Genome-wide association studies in rice: how to solve the low power problems? Mol. Plant. 2019;12:10–12. doi: 10.1016/j.molp.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 205.Chen, L., Apostolides, Z. & Chen, Z. M. Global Tea Breeding: Achievements, Challenges and Perspectives (Springer, Berlin, Heidelberg, 2012).
- 206.Hasimoto M. The original of the tea plant. In: Proceedings of 2001 International Conference on O-Cha (tea) Culture and Science (Session II), Shizuoka, Japan, J5–J7 (2001).
- 207.Meegahakumbura MK, et al. Domestication origin and breeding history of the tea plant (Camellia sinensis) in China and India based on nuclear microsatellites and cpDNA sequence data. Front. Plant Sci. 2017;8:2270. doi: 10.3389/fpls.2017.02270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Meegahakumbura MK, et al. Indications for three independent domestication events for the tea plant (Camellia sinensis (L.) O. Kuntze) and new insights into the origin of tea germplasm in China and India revealed by nuclear microsatellites. PLoS ONE. 2016;11:e0155369. doi: 10.1371/journal.pone.0155369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Meegahakumbura M, Wambulwa M, Li D, Gao L. Preliminary investigations on the genetic relationships and origin of domestication of the tea plant (Camellia sinensis (L.)) using genotyping by sequencing. Trop. Agric. Res. 2018;29:229–240. doi: 10.4038/tar.v29i3.8263. [DOI] [Google Scholar]
- 210.Li M, Meegahakumbura M, Yan L, Liu J, Gao L. Genetic involvement of Camellia taliensis in the domestication of Camellia sinensis var. assamica (Assamica Tea) revealed by nuclear microsatellite markers. Plant Divers. Resour. 2015;37:29–37. [Google Scholar]
- 211.Wambulwa MC, et al. Multiple origins and a narrow genepool characterise the African tea germplasm: concordant patterns revealed by nuclear and plastid DNA markers. Sci. Rep. 2017;7:4053. doi: 10.1038/s41598-017-04228-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Xu X, et al. Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 2011;30:105–111. doi: 10.1038/nbt.2050. [DOI] [PubMed] [Google Scholar]
- 213.Mondal TK, Bhattacharya A, Ahuja PS, Chang PK. Transgenic tea [Camellia sinensis (L.) O. Kuntze cv. Kangra Jat] plants obtained by -mediated transformation of somatic embryos. Plant Cell Rep. 2001;20:712–720. doi: 10.1007/s002990100382. [DOI] [Google Scholar]
- 214.Lopez SJ, Kumar RR, Pius PK, Muraleedharan N. tumefaciens-Mediated genetic transformation in tea (Camellia sinensis [L.] O. Kuntze) Plant Mol. Biol. Rep. 2004;22:201–202. doi: 10.1007/BF02772730. [DOI] [Google Scholar]
- 215.Mondal TK, Bhattacharya A, Laxmikumaran M, Ahuja PS. Recent advances of tea (Camellia sinensis) biotechnology. Plant Cell Tiss. Org. 2004;76:195–254. doi: 10.1023/B:TICU.0000009254.87882.71. [DOI] [Google Scholar]
- 216.Sandal I, et al. -mediated genetic transformation of tea leaf explants: effects of counteracting bactericidity of leaf polyphenols without loss of bacterial virulence. Plant Cell Rep. 2007;26:169–176. doi: 10.1007/s00299-006-0211-9. [DOI] [PubMed] [Google Scholar]
- 217.Mohanpuria P, Kumar V, Ahuja PS, Yadav SK. -mediated silencing of caffeine synthesis through root transformation in Camellia sinensis L. Mol. Biotechnol. 2011;48:235–243. doi: 10.1007/s12033-010-9364-4. [DOI] [PubMed] [Google Scholar]
- 218.Saini U, et al. Optimising parameters for biolistic gun-mediated genetic transformation of tea [Camellia sinensis(L.) O. Kuntze] J. Hortic. Sci. Biotechnol. 2012;87:605–612. doi: 10.1080/14620316.2012.11512919. [DOI] [Google Scholar]
- 219.Sandal I, et al. Development of transgenic tea plants from leaf explants by the biolistic gun method and their evaluation. Plant Cell Tissue Organ Cult. 2015;123:245–255. doi: 10.1007/s11240-015-0828-x. [DOI] [Google Scholar]
- 220.Rana Mohammad, Han Zhuo-Xiao, Song Da-Peng, Liu Guo-Feng, Li Da-Xiang, Wan Xiao-Chun, Karthikeyan Alagarsamy, Wei Shu. Effect of Medium Supplements on Agrobacterium rhizogenes Mediated Hairy Root Induction from the Callus Tissues of Camellia sinensis var. sinensis. International Journal of Molecular Sciences. 2016;17(7):1132. doi: 10.3390/ijms17071132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tang Y, et al. Development of a CRISPR/Cas9 constructed for genome editing of caffeine synthase in Camellia sinensis. J. Tea Sci. 2016;36:414–426. [Google Scholar]
- 222.Lv Q, et al. Optimization of tumefaciens -mediated transformation systems in tea plant (Camellia sinensis) Hortic. Plant J. 2017;3:105–109. doi: 10.1016/j.hpj.2017.03.001. [DOI] [Google Scholar]
- 223.Alagarsamy K, Shamala LF, Wei S. Protocol: high-efficiency in-planta -mediated transgenic hairy root induction of Camellia sinensis var. sinensis. Plant Methods. 2018;14:17. doi: 10.1186/s13007-018-0285-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Kumar N, Pandey S, Bhattacharya A, Ahuja PS. Do leaf surface characteristics affect infection in tea [Camellia sinensis (L.) O Kuntze]? J. Biosci. 2004;29:309–317. doi: 10.1007/BF02702613. [DOI] [PubMed] [Google Scholar]
- 225.Chen Kunling, Wang Yanpeng, Zhang Rui, Zhang Huawei, Gao Caixia. CRISPR/Cas Genome Editing and Precision Plant Breeding in Agriculture. Annual Review of Plant Biology. 2019;70(1):667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- 226.Ye M, et al. Generation of self-compatible diploid potato by knockout of S-RNase. Nat. Plants. 2018;4:651–654. doi: 10.1038/s41477-018-0218-6. [DOI] [PubMed] [Google Scholar]
- 227.Kottawa-Arachchi JD, Gunasekare MK, Ranatunga MA. Biochemical diversity of global tea [Camellia sinensis (L.) O. Kuntze] germplasm and its exploitation: a review. Genet. Resour. Crop Evol. 2019;66:259–273. doi: 10.1007/s10722-018-0698-2. [DOI] [Google Scholar]
- 228.Koech RK, et al. Functional annotation of putative QTL associated with black tea quality and drought tolerance traits. Sci. Rep. 2019;9:1465. doi: 10.1038/s41598-018-37688-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Koech RK, et al. Identification of novel QTL for black tea quality traits and drought tolerance in tea plants (Camellia sinensis) Tree Genet. Genomes. 2018;14:9. doi: 10.1007/s11295-017-1219-8. [DOI] [Google Scholar]
- 230.da Silva Pinto M. Tea: a new perspective on health benefits. Food Res. Int. 2013;53:558–567. doi: 10.1016/j.foodres.2013.01.038. [DOI] [Google Scholar]
- 231.Trevisanato SI, Kim YI. Tea and health. Nutr. Rev. 2000;58:1–10. doi: 10.1111/j.1753-4887.2000.tb01818.x. [DOI] [PubMed] [Google Scholar]
- 232.Suzuki Y, Miyoshi N, Isemura M. Health-promoting effects of green tea. Proc. Jpn. Acad. Ser. B. 2012;88:88–101. doi: 10.2183/pjab.88.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Chaturvedula VSP, Prakash I. The aroma, taste, color and bioactive constituents of tea. J. Med. Plants Res. 2011;5:2110–2124. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.