Abstract
Introduction:
Some models of therapy for neurodegenerative diseases envision starting treatment before symptoms develop. Demonstrating that such treatments are effective requires accurate knowledge of when symptoms would have started without treatment. Familial frontotemporal lobar degeneration offers a unique opportunity to develop predictors of symptom onset.
Methods:
We created dementia risk scores in 268 familial frontotemporal lobar degeneration family members by entering covariate-adjusted standardized estimates of brain atrophy into a logistic regression to classify asymptomatic versus demented participants. The score’s predictive value was tested in a separate group who were followed up longitudinally (stable vs. converted to dementia) using Cox proportional regressions with dementia risk score as the predictor.
Results:
Cross-validated logistic regression achieved good separation of asymptomatic versus demented (accuracy = 90%, SE = 0.06). Atrophy scores predicted conversion from asymptomatic or mildly/questionably symptomatic to dementia (HR = 1.51, 95% CI: [1.16,1.98]).
Discussion:
Individualized quantification of baseline brain atrophy is a promising predictor of progression in asymptomatic familial frontotemporal lobar degeneration mutation carriers.
Keywords: Frontotemporal dementia, Genetics, Tau, TDP-43, Magnetic resonance imaging (MRI)
1. Introduction
Neurodegenerative diseases such as Alzheimer’s disease (AD) and frontotemporal lobar degeneration (FTLD) dementia are devastating conditions caused by the accumulation of toxic proteins in the brain. Drugs designed to reduce the levels of these proteins and modify the course of disease are being aggressively developed [1,2]. Advances in biomarker research have demonstrated that the causative proteins in neurodegenerative disease begin to accumulate years before the onset of symptoms [3,4], which suggests that very early treatment in this prodromal stage might prevent or delay the development of symptoms [5,6]. Demonstrating that a treatment has delayed onset of symptoms requires accurate knowledge of when symptoms would have started had the treatment not been initiated. In the general population, this is very difficult to predict. These considerations have spurred the initiation of studies of individuals at high risk of neurodegenerative disease to identify markers that predict that symptoms are likely to develop within a predictable amount of time [3,7,8].
FTLD is a neurodegenerative disorder characterized by relatively early age of onset [9] and, in many cases, rapid progression to disability and death once symptoms manifest [10]. Thirty percent to 40% of FTLD is considered familial (f-FTLD), and several causative mutations have been identified, the three most common being mutations in the MAPT, GRN, and C9orf72 genes [11]. Although all mutations are highly penetrant, such that the likelihood of symptoms is close to 100%, the age of onset can vary dramatically, even within a family (e.g., onset in the 30s vs. the 70s in the same family) [12]. Unlike other familial disorders such as familial AD [13] and Huntington’s disease [14], there are no accurate predictors of when symptoms will develop in an individual at risk for FTLD. Two projects, Longitudinal Evaluation of Familial Frontotemporal Dementia Subjects (LEFFTDS) and Advancing Research and Treatment in Frontotemporal Lobar Degeneration (ARTFL), were recently initiated to identify better markers of disease in f-FTLD.
Several studies of f-FTLD have suggested that brain volume is reduced in mutation carriers, even when still asymptomatic, making brain volume attractive as a potential biomarker [15,16]. Early tracking of imaging abnormalities in f-FTLD is difficult, however, because of the broad spectrum of presenting symptoms, which range from motor neuron disease and parkinsonism to socioemotional, language, and speech disorders that can initially occur alone or in various combinations; some syndromes even mimic AD [17]. Each of these syndromes is associated with a specific pattern of brain atrophy [18], and thus the earliest imaging changes vary across individuals. Because there are currently no known indicators that predict which neural systems will be affected first, previous work in f-FTLD has focused on tracking whole brain changes or regional changes that might be specific to the type of mutation [19], but this type of approach does not permit optimal tracking because the measures are not tailored for each individual.
The analysis presented here examines individualized patterns of brain atrophy as a biomarker in ARTFL/LEFFTDS participants. We reasoned that symptoms in each individual might predictably begin when brain atrophy reaches a specific level of severity. In some individuals, this may represent severe atrophy in a limited number of brain regions, whereas in others, this may be caused by milder but more extensive changes. This would suggest that quantifying the specific pattern of brain atrophy in each individual, accounting for both severity and extent but ignoring portions of the brain that are less affected and presumably healthy, could produce a valuable marker for predicting symptom onset. The analysis presented here examines the utility of using individualized maps of brain atrophy, which account for variability in age, to predict symptom onset.
2. Materials and methods
2.1. Participants
The study included 268 members of families affected by f-FTLD, most of whom were enrolled in the ARTFL and/or LEFFTDS studies, which are conducted through a consortium of 18 centers across the United States and Canada (https://www.rarediseasesnetwork.org/cms/artfl/). These studies enroll individuals based on a family history suggestive of f-FTLD, usually identified through interview of a symptomatic proband. Once a family member with a mutation is identified, other members of the family are contacted and invited to participate. Enrollees do not need to know their mutation status. For LEFFTDS, at least one individual in the family must have a mutation in the MAPT, GRN, or C9orf72 genes. For ARTFL, families with any f-FTLD mutation or without a known mutation can enroll, but only patients with MAPT, GRN, or C9orf72 mutations were included in the analysis reported here. The ARTFL and LEFFTDS protocols include annual follow-up with clinical reassessment. Additional participants with f-FTLD were included who had been enrolled in a longstanding study of FTLD at the University of California, San Francisco, and who had undergone a similar brain imaging protocol (AG032306-05).
Reference images for creation of atrophy maps in each of the f-FTLD family members were obtained from 383 control participants chosen to cover the age range of the mutation carriers, including 28 who enrolled a previous study of neuroimaging in FTLD at the University of California, San Francisco (AG032306), 143 nonmutation carriers from the Dominantly Inherited Alzheimer’s Network () [20], 31 who participated in the Parkinson’s Progression Markers Initiative () [21], and 181 from the Alzheimer’s Disease Neuroimaging Initiative ().
2.2. Clinical and genetic assessment
The uniform multidisciplinary assessment includes neurological history and examination, informant interview, and neuropsychological assessment. Cognitive testing was accomplished using the third version of the NIA Alzheimer’s Disease Centers’ Uniform Dataset neuropsychological battery (UDS-3) [22], supplemented with a UDS module for assessment of FTLD (https://www.alz.washington.edu/WEB/forms_uds.html). Functional status was quantified using the Clinical Dementia Rating Scale (CDR®) plus Behavioral and Language Domains from the National Alzheimer’s Coordinating Center (NACC) FTLD module (CDR® plus NACC FTLD) [23]. The traditional CDR can be used to generate a total score that represents a weighted average of six functional domain scores to categorize each patient as having questionable or mild symptoms of neurodegenerative disease (CDR® = 0.5) or clear symptoms of dementia (CDR® 1, 2, or 3). This system is biased toward memory complaints, which are not the presenting symptom in many patients with FTLD. The scoring algorithm is described in greater detail elsewhere (Olney et al., this issue). Brain imaging was not used for patient diagnosis or severity rating.
All participants had genetic testing at the same laboratory at UCLA using previously published methods [24].
2.3. Image acquisition
Participants were scanned at 3 Tesla on MRI scanners from one of three vendors: Philips Medical Systems, Siemens, or General Electric Medical Systems. A standard imaging protocol was used at all centers, managed, and reviewed for quality by a core group at the Mayo Clinic, Rochester. The current analysis used the T1-weighted images, which were acquired as Magnetization Prepared Rapid Gradient Echo images using the following parameters: 240×256×256 matrix; about 170 slices; voxel size = 1.05×1.05 × 1.25 mm3; flip angle, TE and TR varied by vendor.
2.4. Image processing and creation of atrophy scores
Before processing, all T1-weighted images were visually inspected for quality. Images with excessive motion or image artifact were excluded. Magnetic field bias was corrected using the N3 algorithm [25]. Tissue segmentation was performed using the unified segmentation procedure in Statistical Parameter Mapping 12. For the cross-sectional analysis, each participant’s T1-weighted image was warped to create a study-specific template using Diffeomorphic Anatomical Registration using Exponentiated Lie algebra [26]; subsequently, the images were normalized and modulated in the study-specific template space using nonlinear and rigid-body registration. For longitudinal analysis, an intraparticipant template was created using nonlinear diffeomorphic and rigid-body registration implemented with Statistical Parameter Mapping [27]. The intraparticipant template was segmented and a group template was generated from the within-participant averaged gray matter, white matter, and CSF tissue maps by nonlinear and rigid-body registration. Images were finally warped into the Montreal Neurological Institute standard space, and smoothed using a 4 mm isotropic Gaussian kernel. Total intracranial volume was estimated for each participant in Montreal Neurological Institute space.
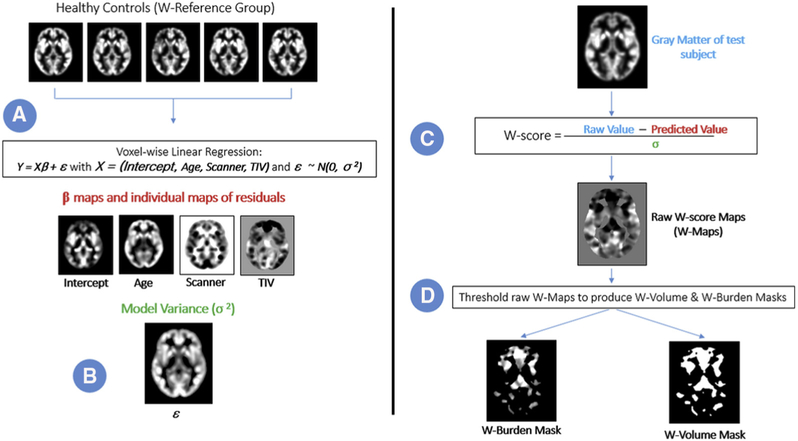
Atrophy was quantified in each family member by creating a W-score representing volume loss at each gray matter voxel. The W-score represents the gray matter content at that voxel as the number of standard deviations away from the expected mean for the reference group after fitting a multiple linear regression accounting for age, total intracranial volume, and scanner platform [28,29] (where the standard deviation is defined as the standard deviation of the residuals for the fitted model). W-scores were integrated within 179 regions of interest (ROIs) as defined by a standard brain atlas [30] to create a measure of atrophy burden for each region of the brain. The process of W-score creation is illustrated in Fig. 1. Fig. 2 shows W-score maps for six symptomatic mutation carriers.
Fig. 1.
Steps to create W-score maps, W-volume masks, and W-burden masks. A linear regression was performed on a healthy control group (1A and 1B). A raw W-score map was then created for each subject (1C). Finally, each patient’s W-map was thresholded to produce the W-burden and W-volume masks (1D).
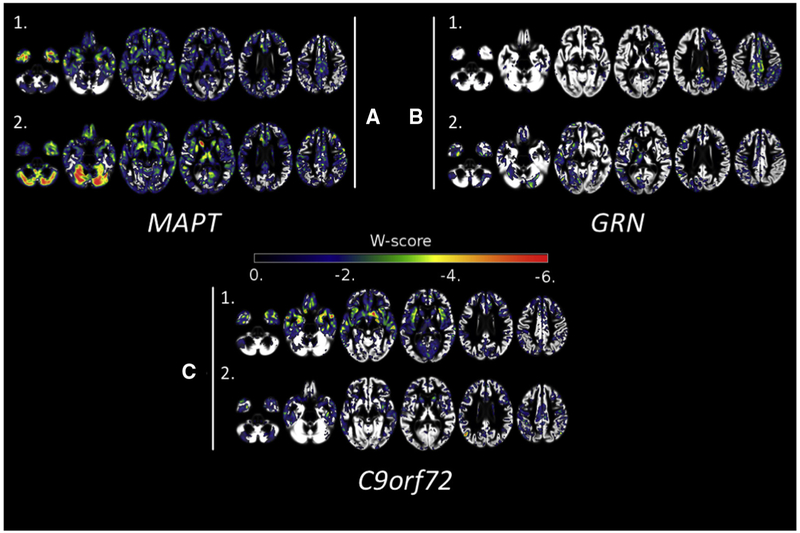
Fig. 2.
Individual W-maps for familial FTLD mutation carriers with CDR® plus NACC FTLD = 1. Axial slices displaying W-scores for 6 f-FTLD mutation carriers with (A) MAPT, (B) GRN, and (C) C9orf72 mutations. Abbreviations: FTLD, Frontotemporal lobar degeneration; CDR, Clinical Dementia Rating Scale; NACC, National Alzheimer’s Coordinating Center.
2.5. Calculation of atrophy-based dementia risk score
We used family members carrying mutations who were clinically normal (CDR® plus NACC FTLD = 0) versus those diagnosed with dementia (CDR® plus NACC FTLD = 1) as a categorical outcome for developing a dementia risk score based on each person’s pattern and degree of atrophy. To retain as large a sample size as possible, participants with the MAPT, GRN, and C9orf72 mutations were all included in the analysis as one group. To be considered normal, an individual had to have been rated as CDR® plus NACC FTLD = 0 for two years in a row (but images used to create a risk score were always those obtained at the first time point). Family members without mutations, carriers with CDR® plus NACC FTLD = 0.5, or any carrier with more than two time points were left out of the model for use as a predictive data set in the survival analysis described below. We also left out any carrier observed to convert from CDR® plus NACC FTLD 0 or 0.5 to a higher level over time. The classification model was based on a L2-regularized logistic regression algorithm [31] and implemented in a machine learning Python package [32] using the Fast Incremental Gradient Method [33].The targeted probability distribution is
| (1) |
where the w-burden wi, for the participant i, is a vector whose dimensions represent the integral of the local w-score Wij for each ROI j. The label t ∈ {0, 1} indicates a patient’s mental status as healthy or impaired. The parameters vector α is fitted by optimizing the empirical log-logistic loss using an L2-regularization term [31]:
| (2) |
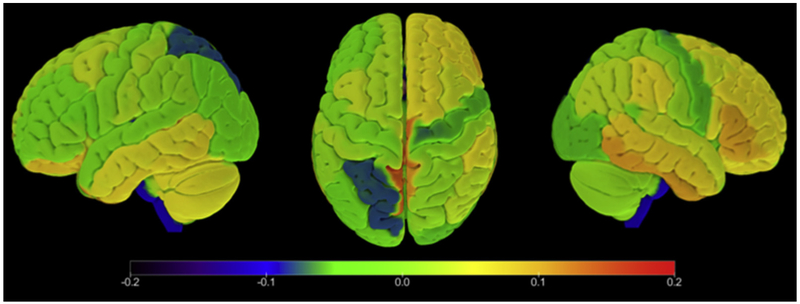
The predicted values from the fitted logistic regression range from 0 to 1 and represent the probability of being assigned to the CDR® plus NACC FTLD ≥ 1 group. Before classification, the W-scores wij in each ROI j were standardized across participants’ predictors such that ∑wij=0 for i = 1, …, n and j = 1, …, p where n is the number of participants and p is the number of ROIs. To estimate model performance, a 5-fold cross-validation scheme was used [34] across a grid of values for the parameter λ, which specifies the regularization strength in equation 2. The optimal λ = 10 value was selected based on the highest mean R-squared over the 5-folds. The optimized weight distribution of the vector α is presented in Fig. 3.
Fig. 3.
Distribution of weights in each ROI resulting from the optimization of the logistic regression model. This figure displays the fitted weights (coefficients) associated with each ROI in the logistic regression model. These weights were subsequently used to calculate prediction scores for the survival analysis. Abbreviation: ROI, Region of interest.
2.6. Analysis
To assess prediction accuracy, the dementia prediction scores were binarized such that regression score values above 0.5 were set to 1 (predicting dementia/CDR® plus NACC FTLD = 1) and those equal to, or below 0.5 were set to 0 (predicting asymptomatic/CDR® plus NACC FTLD = 0). The predicted label was compared with the actual diagnosis, with accuracy being calculated as the proportion of participants correctly classified. 101 nonmutation-carrying family members of f-FTLD patients with the three mutations of interest were used as a validation group. In addition, the risk scores were plotted in 36 f-FTLD mutation carriers with a CDR® plus NACC FTLD = 0.5 and a single visit. Please note that this latter group was not used to create the logistic regression, nor were they included in the accuracy score, but rather plotted for illustrative purposes.
For those participants not included in the model used to derive the dementia risk score (i.e., stable CDR® plus NACC FTLD = 0.5 for at least 2 time points, stable CDR® plus NACC FTLD = 0 with more than 2 time points, and those who converted from CDR® plus NACC FTLD = 0 or 0.5 to CDR® plus NACC FTLD≥ 1) individual dementia risk scores from their MRIs were calculated for all available time points. These mutation carriers with longitudinal data were examined to assess the potential utility of the dementia risk score for predicting development of symptoms (e.g., converting from CDR® plus NACC FTLD = 0 or CDR® plus NACC FTLD = 0.5 to CDR® plus NACC FTLD≥1). This was accomplished using Cox regression models where atrophy score was treated as a time-varying/dependent predictor and CDR® plus NACC FTLD score as the outcome [35].
3. Results
3.1. Participant demographics
The total study group included 268 family members, including 101 nonmutation carriers, 127 mutation carriers were included in the logistic model and accuracy analysis (54 C9orf72, 37 GRN, and 36 MAPT), and 40 in the survival analysis (14 C9orf72, 12 GRN, and 14 MAPT). Table 1 summarizes the age, sex, and symptom severity breakdown for each analysis, including the imaging reference group. Table 2 describes the clinical diagnoses in these groups.
Table 1.
Participant demographics
| Reference group |
Logistic regression model and accuracy calculation by the baseline CDR® plus NACC FTLD score |
Survival analysis by CDR® plus NACC FTLD score |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UCSF | DIAN | PPMI | ADNI | 0 | 0.5* | 1 | 2 | 3 | Stable 0 | Stable 0.5 | Converter | |
| n (visits) | 28 | 143 | 31 | 181 | 35 | 36 | 23 | 26 | 7 | 11 (36) | 15 (33) | 14 (41) |
| Age (SD) | 59.71 (5.86) | 39 (10.3) | 59.0 (11.2) | 73.5 (6.25) | 44.2 (15.67) | 52.2 (10.77) | 58.6 (8.45) | 59.35 (9.9) | 59.86 (10.4) | 43.82 (13.14) | 53.47 (11.61) | 57.86 (11.06) |
| %Male | 48.6 | 42 | 64.5 | 48.6 | 42.86 | 50 | 43.48 | 38.4 | 71.4 | 45.5 | 60 | 25 |
Abbreviations: ADNI, Alzheimer’s Disease Neuroimaging Initiative; FTLD, frontotemporal lobar degeneration; CDR, Clinical Dementia Rating Scale; DIAN, Dominantly Inherited Alzheimer’s Network; PPMI, Parkinson’s Progression Markers Initiative; UCSF, University of California, San Francisco.
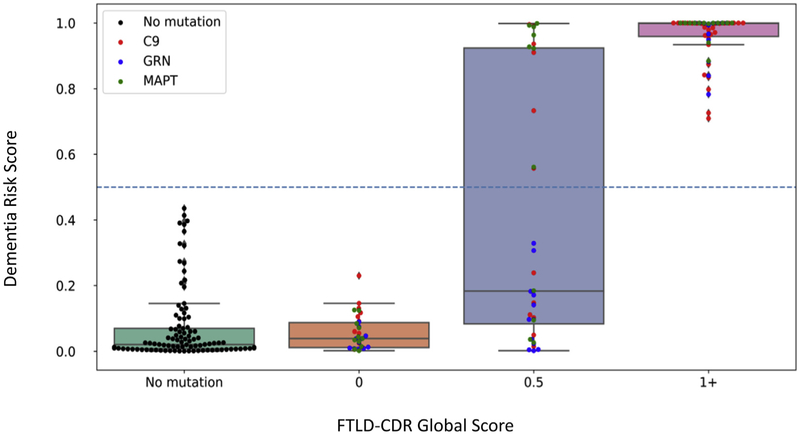
Participants with CDR® plus NACC FTLD = 0.5 are plotted in Fig. 4 for illustrative purposes but were not included in the logistic regression model.
Table 2.
Diagnostic composition at baseline
| Baseline diagnosis | Logistic regression/ accuracy |
Survival analysis |
||
|---|---|---|---|---|
| CDR® plus NACC FTLD = 0.5 |
FTLD CDR ≥1 |
Stable CDR® plus NACC FTLD = 0.5 |
Converters | |
| bvFTD | 2 | 32 | ||
| svPPA | 1 | |||
| nfvPPA | 1 | 1 | ||
| Logopenic variant PPA | 1 | |||
| FTD/ALS | 1 | 6 | ||
| ALS | 1 | |||
| CBS | 1 | 1 | ||
| MCI—Behavior | 6 | 1 | 5 | 2 |
| MCI—Other | 11 | 3 | 6 | 2 |
| AD | 1 | 4 | 1 | |
| AD frontal | 1 | |||
| Primary psychiatric | 2 | 4 | 1 | |
| Clinically normal | 11 | 2 | 7 | |
| DLB | 1 | |||
| Parkinson’s disease | 1 | 1 | ||
Note. MCI other = MCI frontal/exec; MCI mixed/unspecified; MCI–cognitive.
Abbreviations: bvFTD, Behavioral variant frontotemporal dementia; CDR, Clinical Dementia Rating Scale; svPPA, semantic variant primary progressive aphasia; nfvPPA, nonfluent variant PPA; ALS, amyotrophic lateral sclerosis; CBS, corticobasal syndrome; MCI, mild cognitive impairment; AD, Alzheimer’s disease; DLB, dementia with Lewy bodies.
3.2. Atrophy-based dementia risk score fitting
The atrophy-based dementia risk score was developed using the 56 mutation carriers with CDR® plus NACC FTLD ≥1 and with 35 mutation carriers with a CDR® plus NACC FTLD = 0 for two years in a row. Fig. 3 shows the weight for each brain region in the final score. Fig. 4 plots the final scores in these individuals after 5-fold cross validation. The model achieved good separation of prediction scores for CDR® plus NACC FTLD = 0 versus CDR® plus NACC FTLD ≥1, achieving an accuracy of 90.0% (SE: 0.06) based on a cutoff score of 0.5. For comparison, scores for the 101 nonmutation carriers and those with CDR® plus NACC FTLD = 0.5 ratings are also plotted in Fig. 4. As expected, nearly all noncarriers have low prediction scores. CDR® plus NACC FTLD = 0.5 cases ranged from very low to very high scores.
Fig. 4.
Prediction scores derived from the logistic regression model. Abbreviations: FTLD, frontotemporal lobar degeneration; CDR, Clinical Dementia Rating Scale.
Participants with CDR® plus NACC FTLD = 0.5 are plotted in Fig. 4 for illustrative purposes but were not included in the logistic regression model. This study used CDR® plus NACC FTLD = 1 as the criterion for the logistic regression, rather than clinical diagnosis. In most cases this corresponds, but as shown in the table, some patients with CDR® plus NACC FTLD = 1 received a clinical diagnosis of MCI rather than dementia.
3.3. Dementia risk score testing
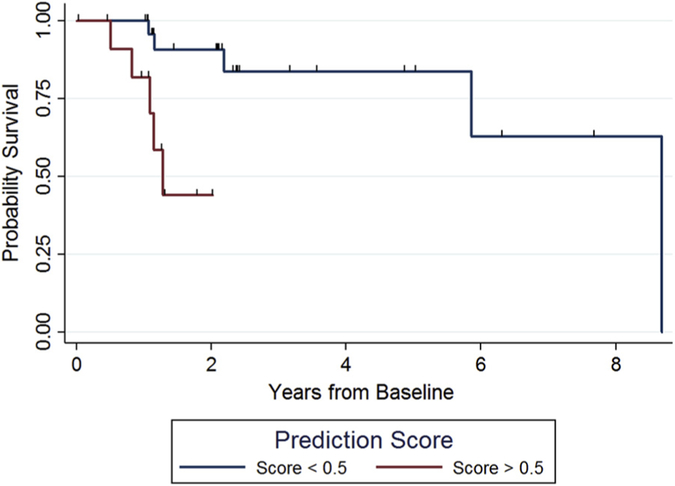
To test whether the model could predict likelihood of conversion, we examined longitudinal data in mutation carriers with stable CDR® plus NACC FTLD = 0 over at least 3 time points, CDR® plus NACC FTLD = 0.5 over at least 2 time points, or conversion from CDR® plus NACC FTLD = 0 to CDR® plus NACC FTLD = 0.5, CDR® plus NACC FTLD = 0 to CDR® plus NACC FTLD ≥1, or CDR® plus NACC FTLD = 0.5 to CDR® plus NACC FTLD ≥1. The numbers of cases in each category and follow-up statistics are summarized in Supplementary Table 1. We fitted a number of Cox regression models to examine conversions between different CDR® plus NACC FTLD levels: model A, 0 to 0.5; model B, from 0.5 to 1 or more (i.e., 0.5+); and model C, from a 0 or 0.5 to 1 or more (1+). The hazard ratios (HR) for a 0.1 unit increase in dementia risk prediction score in those models that included participants enrolled at a CDR® plus NACC FTLD = 0.5 or (0 or 0.5), were 1.28 and 1.51, indicating a shorter time to conversion with increased dementia risk score. Results are displayed in Table 3. Both the model examining conversion from 0.5 to 1+ (HR = 1.28, P = .041), and the model examining conversion from 0 or 0.5 to 1+ (HR = 1.51, P = .001) were statistically significant, but the model examining conversion from 0 to 0.5 + was not statistically significant (HR = 0.98, P = .932). Fig. 5 shows Kaplan-Meier curves for model C (0 or 0.5 to ≥1) after dichotomizing using a baseline cutoff dementia risk score of 0.5. Those with scores above 0.5 all converted to dementia within 2 years, whereas >80% of those with scores below 0.5 still have not converted after 5 years.
Table 3.
Results of survival analyses
| Model parameters |
|||||
|---|---|---|---|---|---|
| Model | n | Number of conversions |
Hazard ratio |
95% CI | P value |
| A. CDR® plus NACC FTLD 0 to 0.5+ | 18 | 7 | 0.98 | 0.64 - 1.5 | .932 |
| B. CDR® plus NACC FTLD 0.5 to 1+ | 25 | 10 | 1.28 | 1.01 - 1.62 | .041 |
| C. CDR® plus NACC FTLD (0 or 0.5) to 1+ | 40 | 10 | 1.51 | 1.17 - 1.94 | .001 |
Abbreviations: CDR, Clinical Dementia Rating Scale; FTLD, frontotemporal lobar degeneration; NACC, National Alzheimer’s Coordinating Center.
Fig. 5.
Kaplan-Meier curve representing survival from CDR® plus NACC FTLD = (0 or 0.5) to 1+. Note: Hash marks indicate censored observations. Abbreviations: FTLD, Frontotemporal lobar degeneration; CDR, Clinical Dementia Rating Scale; NACC, National Alzheimer’s Coordinating Center.
4. Discussion
The goal of this analysis was to determine whether an atrophy-based risk score could be developed to predict the onset of dementia in asymptomatic or questionably symptomatic individuals who are at high risk due to autosomal dominant mutations. Our results indicate that quantification of each person’s unique pattern of atrophy can separate those with dementia from those with no symptoms with 90% accuracy. Furthermore, in an independent subset of the data composed of longitudinal observations, we were able to show that this atrophy-based risk score increased the risk in a statistically significant manner for progression to dementia from the asymptomatic (CDR® plus NACC FTLD = 0) or questionably symptomatic state (CDR® plus NACC FTLD = 0.5). These results suggest that quantification of individualized atrophy patterns is a promising technique that may be useful for developing new treatments for neurodegenerative disease and for guiding the use of these treatments once they are approved.
This approach for creation of atrophy maps addresses formidable challenges in developing treatments for neurodegenerative disease. Because the age when symptoms develop varies widely across individuals with FTLD and within individual families, drug trials seeking to delay the onset of symptoms must identify biological markers that reliably indicate symptoms will develop within a short time, allowing enrichment of the trial cohort with these participants, or stratification within the study. A similar approach to constructing individualized risk estimates was developed for AD and can significantly predict risk of future decline [36] and conversion from MCI to AD [37]. This approach, however, trained a classifier to produce an atrophy score that reflected similarity with an AD pattern of atrophy. Our approach, which was agnostic to region of atrophy and accounted for age, is necessary in this cohort given the variable age of onset and diversity of brain networks affected in f-FTLD. Another study took a similar approach to distinguish f-FTLD presymptomatic carriers from noncarriers based on their resemblance to bvFTD, again using a trained classifier [38]; this method did not significantly classify presymptomatic carriers from controls. Another study in f-FTLD suggested that volume loss in specific brain regions precedes onset of symptoms [16]. Indeed, our approach can potentially identify brain changes that herald the onset of symptoms regardless of where they occur in the brain, and our validation analysis supports the value of such changes. Similarly, each person’s W-map can potentially be used to define a region of interest for tracking the effect of a drug in slowing atrophy. Once treatments are approved, this type of risk score can be used to avoid potential adverse effects of approved drugs if treatment is delayed until the time when symptoms are more likely to develop. Furthermore, these considerations are not limited to FTLD. For instance, a substantial minority of patients with AD present with symptoms of visuospatial, frontal, or language symptoms [39], and measurements targeted at the hippocampal or entorhinal cortex regions typically affected in AD may miss early brain changes indicative of oncoming symptoms. This idea could be examined in amyloid positive individuals at high risk of developing AD due to genetic risk.
Although this method appears promising, future refinements can be envisioned that will likely improve its utility. Technical factors will need to be addressed, such as the impact of specific scanner and field strength on the estimate. Although this was a multisite study, many of the participants were studied at the University of California, San Francisco, and all participants were scanned at 3T. Replication of these findings in independent data sets will be important. In addition, the regression model was created using patients who had already developed dementia an average of 3 years before the onset of the study. Creation of a model based on patients who were observed to convert to dementia within the last year would likely improve its sensitivity. Such patients will likely be available through projects such as ARTFL and LEFFTDS in the future. Our atrophy-based risk score was derived using 91 mutation carriers in our training group; future iterations will strive to use a larger sample. Furthermore, this model was created by pooling all three types of FTLD mutations as one group to maximize the sample size. The inherent trade-off is that we may have reduced our power to detect mutation-specific atrophy that could have improved model accuracy. With larger numbers of patients, mutation-specific models can be created. For instance, the weighting of each region in our model depicted in Fig. 3 shows that right-sided regions were heavily weighted, and only a limited number of left hemisphere regions were useful in prediction of dementia. In a group with only GRN mutation carriers, there would be a higher likelihood of asymmetric involvement and left-sided involvement, so that the left hemisphere might carry more weight for prediction. Please see the study by Olney et al. (this special issue) for a characterization of brain atrophy patterns by mutation type. In addition, it is likely that additional variables indicative of inflammation or neuronal injury, or other types of imaging or clinical data, when added to this score, could significantly improve prediction in survival analyses. Despite these limitations, these results are an important step toward predicting disease onset in neurodegenerative conditions.
Supplementary Material
RESEARCH IN CONTEXT.
Systematic review: A literature review (PubMed) revealed that although brain volume is reduced in asymptomatic familial frontotemporal lobar degeneration (f-FTLD) mutation carriers, there is no model for using baseline brain volume to predict symptom onset. Previous approaches for calculating atrophy-based risk scores in other dementias have trained classifiers using expected patterns of atrophy. Given the heterogeneity of atrophy in f-FTLD, we developed atrophy-based risk scores that quantify each patients’ atrophy regardless of the pattern.
Interpretation: Individualized atrophy scores separated f-FTLD mutation carriers with dementia from asymptomatic carriers with high accuracy. Moreover, atrophy-based risk scores predicted conversion from asymptomatic or mildly/questionably symptomatic stages to dementia. This biomarker could improve clinical care and enhance the power of clinical trials.
Future directions: This finding should be replicated in other f-FTLD cohorts. Individualized atrophy-based risk scores need to be evaluated as biomarkers of progression in the earliest stages of a variety of neurodegenerative diseases.
Acknowledgments
The authors extend their appreciation to Drs. John Hsiao and Dallas Anderson from the National Institute on Aging, Drs. Marg Sutherland and Codrin Lungu from the National Institute of Neurological Disorders and Stroke, the staff of all centers, and particularly to our patients and their families for their participation in this protocol.
This work is supported by the National Institutes of Health [grants AG045390, NS092089, AG032306, AG021886, AG016976, and K24AG045333] and the Larry L. Hillblom Foundation [2018-A-025-FEL]. Samples from the National Centralized Repository for Alzheimer Disease and Related Dementias (NCRAD), which receives government support under a cooperative agreement grant (U24 AG21886) awarded by the National Institute on Aging (NIA), were used in this study.
Conflict of interest: A.M.S. received research support from the Larry H. Hillblom foundation and support from the NIH. Y.C., S.-Y.M.G., L.B., R.D., K.C., J.B., D.B., C.D., R.D., J.F., D.G., D.H., L.J., A.K., R.K., M.L., P.A.L., M.M., E.R., P.S., J.S., and J.T., had nothing to disclose. J.K. had provided expert witness testimony for Teva Pharmaceuticals in Forest Laboratories Inc. et al. v. Teva Pharmaceuticals USA, Inc., Case Nos. 1:14-cv-00121 and 1:14-cv-00686 (D. Del. filed Jan. 31, 2014 and May 30, 2014) regarding the drug Memantine; for Apotex/HEC/Ezra in Novartis AG et al. v. Apotex Inc., No. 1:15-cv-975 (D. Del. filed Oct. 26, 2015, regarding the drug Fingolimod. He had also given testimony on behalf of Puma Biotechnology in Hsingching Hsu et al., vs. Puma Biotechnology, INC., et al. 2018 regarding the drug neratinib. He received research support from the NIH. B.A. received research support from the CDC. P.B. was employed by the Rainwater Charitable Foundation. G.C., B.C.D., K.F., J.A.F., T.F., L.K.F., R.G., J.G.-R., H.W.H., E.D.H., W.A.K., D.L., B.L.M., L.P., M.P., K.P.R., K.R., L.M.S., S.W., and B.W. received research support from the NIH. J.H.K. receives research support from NIH, the Tau Consortium, and the Larry H. Hillblom Foundation, and provides consultation for Biogen. S.D. was on staff at the Association for Frontotemporal Degeneration and a member of the National Institute for Neurological Disorders and Stroke Advisory Council. N.G. had participated or is currently participating in clinical trials of antidementia drugs sponsored by the following companies: Bristol Myers Squibb, Eli Lilly/Avid Radiopharmaceuticals, Janssen Immunotherapy, Novartis, Pfizer, Wyeth, SNIFF (The Study of Nasal Insulin to Fight Forgetfulness) study, and A4 (The Anti-Amyloid Treatment in Asymptomatic Alzheimer’s Disease) trial. She received research support from Tau Consortium and Association for Frontotemporal Dementia and is funded by the NIH. J.S.G. served as a consultant to the Novartis Alzheimer’s Prevention Advisory Board. She received research support from the NIH, HDSA, New York State Department of Health (RFA # 1510130358). N.G.-R. received royalties from UpToDate and had participated in multicenter therapy studies sponsored by Biogen, TauRx, AbbVie, Novartis, and Lilly. He received research support from the NIH. M.G. received grant support from the NIH, Avid, and Piramal; participated in clinical trials sponsored by Biogen, TauRx, and Alector; served as a consultant to Bracco and UCB; and served on the Editorial Board of Neurology. G.-Y.H. had served as an investigator for clinical trials sponsored by AstraZeneca, Eli Lilly, and Roche/Genentech. He received research support from the Canadian Institutes of Health Research and the Alzheimer Society of British Columbia. D.J.I. received support from the NIH, Brightfocus Foundation, and Penn Institute on Aging. D.T.J. received research support from the NIH and the Minnesota Partnership for Biotechnology and Medical Genomics. K.K. served on the Data Safety Monitoring Board for Takeda Global Research & Development Center, Inc.; data monitoring boards of Pfizer and Janssen Alzheimer Immunotherapy; research support from the Avid Radiopharmaceuticals, Eli Lilly, the Alzheimer’s Drug Discovery Foundation, and the NIH. D.R.K. has served on an Advisory Board for AbbVie and as site PI for studies funded by Roche/Genentech, AbbVie, Avid, Novartis, Eisai, Eli Lilly, and UCSF. D.S.K. served on the DSMB of the DIAN-TU study, is a site PI for clinical trials sponsored by Biogen, Lilly and the University of Southern California, and is funded by the NIH. W.K.K. received research funding from AstraZeneca, Biogen, Roche, DOD, and the NIH. I.L. received research support from the NIH, Parkinson Study Group, Parkinson Foundation, Michael J Fox Foundation, AVID Pharmaceuticals, C2N Diagnostics/Abbvie, and Bristol-Myers Squibb. She was a member of the Biogen and Bristol-Myers Squibb Advisory Boards, Biotie/Parkinson Study Group Medical Advisory Board, and consultant for Toyama Pharmaceuticals. She received salary from the University of California San Diego and as Editor in Frontiers in Neurology. C.L. received honoraria for editorial work from Elsevier, Inc. I.R.M. received research funding from the Canadian Institutes of Health Research. S.M.M. served as an investigator for clinical trials sponsored by AbbVie, Allon Therapeutics, Biogen, Bristol-Myers Squibb, C2N Diagnostics, Eisai Inc., Eli Lilly and Co., Genentech, Janssen Pharmaceuticals, Medivation, Merck, Navidea Biopharmaceuticals, Novartis, Pfizer, and TauRx Therapeutics. He received research support from the NIH. C.O. received research funding from the NIH, the CIHR, and Biogen, Inc. He was also supported by the Jane Tanger Black Fund for Young-Onset Dementias, the Nancy H. Hall Fund for Geriatric Psychiatry, and the gift from Joseph Trovato. R.P. was employed by the Bluefield Project. R.R. received research funding from the NIH and the Bluefield Project to Cure Frontotemporal Dementia. E.D.R. received research support from the NIH, Bluefield Project to Cure Frontotemporal Dementia, Alzheimer’s Association, BrightFocus Foundation, Biogen, Alector, and owns intellectual property related to tau. N.T. was employed by the Association for Frontotemporal Degeneration. A.T. received research support from the NIH and the Alzheimer’s Association. J.Q.T. may accrue revenue in the future on patents submitted by the University of Pennsylvania wherein he is a coinventor and he received revenue from the sale of Avid to Eli Lily as a coinventor on Aβ amyloid imaging–related patents submitted by the University of Pennsylvania. He received research support from the NIH and several nonprofits. Z.W. was supported by the NIH, Mayo Clinic Center for Regenerative Medicine, the gift from Carl Edward Bolch, Jr., and Susan Bass Bolch, the Sol Goldman Charitable Trust, and Donald G. and Jodi P. Heeringa. He has also received grant funding support from Allergan, Inc. (educational grant), and Abbvie (medication trials). A.L.B. received research support from the NIH, the Tau Research Consortium, the Association for Frontotemporal Degeneration, Bluefield Project to Cure Frontotemporal Dementia, Corticobasal Degeneration Solutions, the Alzheimer’s Drug Discovery Foundation, and the Alzheimer’s Association. He served as a consultant for Aeton, Abbvie, Alector, Amgen, Arkuda, Ionis, Iperian, Janssen, Merck, Novartis, Samumed, Toyama and UCB, and received research support from Avid, Biogen, BMS, C2N, Cortice, Eli Lilly, Forum, Genentech, Janssen, Novartis, Pfizer, Roche, and TauRx. B.F.B. served as an investigator for clinical trials sponsored by GE Healthcare and Axovant. He received royalties from the publication of a book entitled Behavioral Neurology of Dementia (Cambridge Medicine, 2009, 2017). He served on the Scientific Advisory Board of the Tau Consortium. He received research support from the NIH, the Mayo Clinic Dorothy and Harry T. Mangurian Jr. Lewy Body Dementia Program and the Little Family Foundation. H.J.R. received research support from Biogen Pharmaceuticals, has consulting agreements with Wave Neuroscience and Ionis Pharmaceuticals, and receives research support from the NIH.
Footnotes
Supplementary Data
Supplementary data related to this article can be found at https://doi.org/10.1016/j.jalz.2019.04.007.
References
- [1].Reiman EM. Alzheimer disease in 2016: Putting AD treatments and biomarkers to the test. Nat Rev Neurol 2017;13:74–6. [DOI] [PubMed] [Google Scholar]
- [2].Tsai RM, Boxer AL. Therapy and clinical trials in frontotemporal dementia: past, present, and future. J Neurochem 2016;138:211–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bateman RJ, Xiong C, Benzinger TLS, Fagan AM, Goate A, Fox NC, et al. Clinical and biomarker changes in dominantly inherited Alzheimer’s Disease. N Engl J Med 2012;367:795–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Jack CR, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol 2010;9:119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Das P, Verbeeck C, Minter L, Chakrabarty P, Felsenstein K, Kukar T, et al. Transient pharmacologic lowering of Aβ production prior to deposition results in sustained reduction of amyloid plaque pathology. Mol Neurodegener 2012;7:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].McDade E, Bateman RJ. Stop Alzheimer’s before it starts. Nature 2017;547:153–5. [DOI] [PubMed] [Google Scholar]
- [7].Aguirre-Acevedo DC, Lopera F, Henao E, Tirado V, Muñoz C, Giraldo M, et al. Cognitive decline in a Colombian kindred with autosomal dominant Alzheimer disease. JAMA Neurol 2016;73:431–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Carrillo MC, Brashear HR, Logovinsky V, Ryan JM, Feldman HH, Siemers ER, et al. Can we prevent Alzheimer’s disease? Secondary “prevention” trials in Alzheimer’s disease. Alzheimer’s Dement 2013;9:123–131.e1. [DOI] [PubMed] [Google Scholar]
- [9].Coyle-Gilchrist ITS, Dick KM, Patterson K, Vázquez Rodríquez P, Wehmann E, Wilcox A, et al. Prevalence, characteristics, and survival of frontotemporal lobar degeneration syndromes. Neurology 2016; 86:1736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Roberson ED, Hesse JH, Rose KD, Slama H, Johnson JK, Yaffe K, et al. Frontotemporal dementia progresses to death faster than Alzheimer disease. Neurology 2005;65:719–25. [DOI] [PubMed] [Google Scholar]
- [11].Woollacott IOC, Rohrer JD. The clinical spectrum of sporadic and familial forms of frontotemporal dementia. J Neurochem 2016;138:6–31. [DOI] [PubMed] [Google Scholar]
- [12].Leverenz JB, Yu CE, Montine TJ, Steinbart E, Bekris LM, Zabetian C, et al. A novel progranulin mutation associated with variable clinical presentation and tau, TDP43 and alpha-synuclein pathology. Brain 2007;130:1360–74. [DOI] [PubMed] [Google Scholar]
- [13].Ryman DC, Acosta-Baena N, Aisen PS, Bird T, Danek A, Fox NC, et al. Symptom onset in autosomal dominant Alzheimer disease: A systematic review and meta-analysis. Neurology 2014;83:253–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR, International Huntington’s Disease Collaborative Group. A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin Genet 2004;65:267–77. [DOI] [PubMed] [Google Scholar]
- [15].Papma JM, Jiskoot LC, Panman JL, Dopper EG, den Heijer T, Donker Kaat L, et al. Cognition and gray and white matter characteristics of presymptomatic C9orf72 repeat expansion. Neurology 2017; 89:1256–64. [DOI] [PubMed] [Google Scholar]
- [16].Rohrer JD, Nicholas JM, Cash DM, van Swieten J, Dopper E, Jiskoot L, et al. Presymptomatic cognitive and neuroanatomical changes in genetic frontotemporal dementia in the Genetic Frontotemporal dementia Initiative (GENFI) study: a cross-sectional analysis. Lancet Neurol 2015;14:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yokoyama JS, Sirkis DW, Miller BL. C9ORF72 hexanucleotide repeats in behavioral and motor neuron disease: clinical heterogeneity and pathological diversity. Am J Neurodegener Dis 2014;3:1–18. [PMC free article] [PubMed] [Google Scholar]
- [18].Rohrer JD, Rosen HJ. Neuroimaging in frontotemporal dementia. Int Rev Psychiatry 2013;25:221–9. [DOI] [PubMed] [Google Scholar]
- [19].Whitwell JL, Boeve BF, Weigand SD, Senjem ML, Gunter JL, Baker MC, et al. Brain atrophy over time in genetic and sporadic frontotemporal dementia: a study of 198 serial magnetic resonance images. Eur J Neurol 2015;22:745–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Morris JC, Aisen PS, Bateman RJ, Benzinger TL, Cairns NJ, Fagan AM, et al. Developing an international network for Alzheimer’s research: the Dominantly Inherited Alzheimer Network. Clin Investig (Lond) 2012;2:975–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Marek K, Jennings D, Lasch S, Siderowf A, Tanner C, Simuni T, et al. The Parkinson Progression Marker Initiative (PPMI). Prog Neurobiol 2011;95:629–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Weintraub S, Besser L, Dodge HH, Teylan M, Ferris S, Goldstein FC, et al. Version 3 of the Alzheimer disease centers’ neuropsychological test battery in the Uniform Data Set (UDS). Alzheimer Dis Assoc Disord 2017;32:10–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Knopman DS, Kramer JH, Boeve BF, Caselli RJ, Graff-Radford NR, Mendez MF, et al. Development of methodology for conducting clinical trials in frontotemporal lobar degeneration. Brain 2008; 131:2957–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gefen T, Ahmadian SS, Mao Q, Kim G, Seckin M, Bonakdarpour B, et al. Combined Pathologies in FTLD-TDP Types A and C. J Neuropathol Exp Neurol 2018;77:405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sled JG, Zijdenbos AP, Evans AC. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 1998;17:87–97. [DOI] [PubMed] [Google Scholar]
- [26].Ashburner J A fast diffeomorphic image registration algorithm. Neuroimage 2007;38:95–113. [DOI] [PubMed] [Google Scholar]
- [27].Ashburner J, Ridgway GR. Symmetric diffeomorphic modeling of longitudinal structural MRI. Front Neurosci 2012;6:197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jack CR, Petersen RC, Xu YC, Waring SC, O’Brien PC, Tangalos EG, et al. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer’s disease. Neurology 1997;49:786–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].La Joie R, Bejanin A, Fagan AM, Ayakta N, Baker SL, Bourakova V, et al. Associations between [18F]AV1451 tau PET and CSF measures of tau pathology in a clinical sample. Neurology 2018;90:e282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, et al. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage 2006;31:968–80. [DOI] [PubMed] [Google Scholar]
- [31].Liu J, Chen J, Ye J. Large-scale sparse logistic regression. In: Proceedings of the 15th ACM SIGKDD international conference on Knowledge discovery and data mining - KDD ’09 New York, New York, USA: ACM Press; 2009. p. 547. [Google Scholar]
- [32].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: machine learning in python. J Mach Learn Res 2011;12:2825–30. [Google Scholar]
- [33].Defazio A, Bach F, Lacoste-Julien S. SAGA: A fast incremental gradient method with support for non-strongly convex composite objectives. CoRR 2014. [Google Scholar]
- [34].Fushiki T. Estimation of prediction error by using K-fold cross-validation. Stat Comput 2011;21:137–46. [Google Scholar]
- [35].Liu X. Survival Analysis: Models and Applications 2012. West Sussex, United Kingdom: John Wiley & Sons Ltd; 2012. [Google Scholar]
- [36].McEvoy LK, Fennema-Notestine C, Roddey JC, Hagler DJ, Holland D, Karow DS, et al. Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology 2009;251:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].McEvoy LK, Holland D, Hagler DJ, Fennema-Notestine C, Brewer JB, Dale AM, et al. Mild cognitive impairment: baseline and longitudinal structural MR imaging measures improve predictive prognosis. Radiology 2011;259:834–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Feis RA, Bouts MJRJ, Panman JL, Jiskoot LC, Dopper EGP, Schouten TM, et al. Single-subject classification of presymptomatic frontotemporal dementia mutation carriers using multimodal MRI. Neuroimage Clin 2018;20:188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ossenkoppele R, Cohn-Sheehy BI, La Joie R, Vogel JW, Möller C, Lehmann M, et al. Atrophy patterns in early clinical stages across distinct phenotypes of Alzheimer’s disease. Hum Brain Mapp 2015; 36:4421–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.