Abstract
The specific cellular role of mitochondria is influenced by the surrounding environment as effective mitochondrial function requires delivery of inputs (e.g. oxygen) and export of products (e.g. signaling molecules) to and from other cellular components, respectively. Recent technological developments in mitochondrial imaging have led to a more precise and comprehensive understanding of the spatial relationships governing function of this complex organelle opening a new era of mitochondrial research. Here, I highlight current imaging approaches for visualizing mitochondrial form and function within complex cellular environments. Increasing clarity of mitochondrial behavior within cells will continue to lend mechanistic insights into the role of mitochondria under normal and pathological conditions and point to spatially-regulated processes that can be targeted to improve cellular function.
Keywords: Energy metabolism, super-resolution microscopy, 3D electron microscopy, systems-level imaging
Mitochondria Function as Part of the Cellular Team
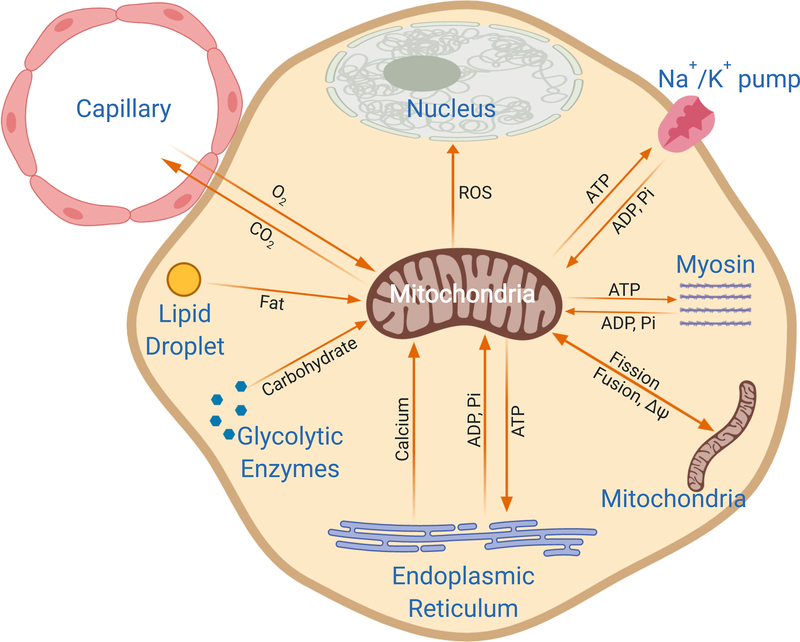
In recent years, it has become increasingly clear that all mitochondria are not the same. Indeed, mitochondrial form, protein and lipid composition (see glossary), and function are all now known to vary according to the type of cells in which they reside [1–7] as mitochondrial function within a given cell appears to be tuned to help achieve the overall functional goals of that particular cell. In other words, mitochondria are team players and adapt to fit the needs of the team. Regardless of whether its major output is ATP, signaling molecules, or other metabolites, how well a mitochondrion is able to perform its required functions within a cell is intrinsically dependent on its spatial relationships with other components within or nearby that cell (Figure 1).
Figure 1: Mitochondrial function relies on cellular interactions.
Depiction of a mitochondrion within a cell highlighting some of the many interactions with other cellular structures that can regulate acute mitochondrial function. Mitochondrial inputs are listed next to arrows pointing toward the mitochondrion and mitochondrial products are listed next to arrows pointing away from the mitochondrion. Interactions involved in assembling and degrading mitochondria are not shown. Structures not drawn to scale.
With the aim of better understanding how mitochondria support cellular functions and how this support may be altered by disease states, biologists have begun to employ a variety of innovative imaging techniques in order to visualize mitochondrial structure and function within the context of the rest of the cell. Here, I discuss the ins and outs of recent advances in mitochondrial imaging that are generating new hypotheses and contributing mechanistic insights into the relationships between mitochondrial form and function within cells.
Mitochondrial Form
The structure of mitochondria within a cell, together with mitochondrial protein and lipid composition, dictates the capacity for mitochondrial function within that cell (Box 1). Cellular conditions at any given time will determine how much of that capacity will actually be used in that moment, but maximal mitochondrial function will always be limited by 1) the amount of mitochondria within a cell, 2) the internal components within mitochondria, and 3) the ability of mitochondria to interact with other cellular structures in order to receive inputs and deliver products as needed. While biochemical methods [8, 9] or protein analyses [10–12] can be used to assess some of the parameters involved in determining mitochondrial functional capacity, microscopic observations of mitochondria can be used to evaluate each aspect within the spatial context of the intact cell. The two primary types of microscopy used for viewing mitochondrial form within cells are light microscopy and electron microscopy (Figure 2), and each hold specific advantages for measuring different structural characteristics (Table 1). Below I discuss recent breakthroughs as well as examine the benefits and tradeoffs associated with currently available options for visualizing mitochondrial structure.
Box 1: Clinician’s Corner.
The function of mitochondria within a cell is regulated by the amount of mitochondria within the cell, their composition, and how well they are able to communicate with other organelles. A better understanding of how each aspect of mitochondrial function contributes to overall cellular function in both health and disease is needed.
Dysfunctional mitochondria are the cause for a very heterogeneous group of devastating human disorders referred to as Mitochondrial Diseases for which no cure currently exists. Novel imaging approaches may help further understand the underlying pathologies.
Mitochondrial structures within cells can be visualized to provide an index of capacity for mitochondrial function. Recent developments in microscopy now allow simultaneous imaging of multiple proteins or molecular targets within mitochondria and how they interact with other cellular structures.
Measurement of mitochondrial structure together with function within the context of the entire cell may help facilitate the identification and/or impact of potential therapeutic targets aimed at improving cellular function.
Application of antibody and fluorescent dye based techniques are well suited for visual evaluation of mitochondrial form and function in patient blood samples and tissue biopsies using relatively few cells compared to biochemical or molecular biology approaches. Genetically encoded indicators of mitochondrial structure and function provide an additional suite of tools compatible with patient derived cell lines.
Figure 2: Recent advances in mitochondrial imaging.
A) Fluorescent antibody labeled mitochondria (magenta) and lysosomes (yellow) within two layer V pyramidal neurons of the mouse somatosensory cortex visualized by expansion lattice light sheet microscopy. Scale bars: 5μm (inset 10μm). B) Mitochondrial cristae structure within live HeLa cells expressing a Cox8a-SNAP fusion protein and labeled with SNAP-Cell SiR dye as imaged by a stimulated emission depletion super-resolution microscope. Scale bar: 1μm. C) 3D rendering of contacts between mouse skeletal muscle mitochondria (cyan), sarcoplasmic reticulum (magenta), and t-tubules (orange) visualized with focused ion beam scanning electron microscopy and machine learning organelle segmentation. Scale bar: 1μm. D) Dynamic interactions between genetically-encoded mitochondria (mEmerald-Tomm20, green), lysosomes (HaloTag-Lamp1, magenta), and microtubules (mCherry-ensconsin, yellow) in COS-7 cells visualized on a grazing incidence structured illumination microscope. Scale bar: 1μm. Reproduced with permission from [23], [36], [2], [40], respectively.
Table 1:
How to Visualize Mitochondrial Form and Function
| Mitochondrial Parameter | Light Microscopy | Electron Microscopy |
|---|---|---|
| Content | Straightforward for detecting large changes in mitochondrial content. Super resolution increases the sensitivity to detect smaller changes. Care must be taken that membrane potential or protein expression changes do not confound results if using dyes or antibodies, respectively. | High resolution permits detection of small differences in content. Must ensure that images adequately represent the entire cell volume. Segmentation of greyscale images can be more difficult than specifically labeled light microscopy images. |
| Shape | Easily used to compare elongated and spherical mitochondrial shapes. May lack resolution to discriminate between individual mitochondrial structures. | Individual mitochondria are easily observed and measured. 2D images may be misleading due to inability to see complete 3D structure. |
| Ultrastructure | Recommended for mtDNA nucleoids which can be labeled transgenically, with antibodies, or dyes. Super resolution microscopy is approaching capacity to resolve inner membrane and matrix spaces. | Recommended for cristae junctions, inner membrane, and matrix spaces due to higher resolution. Often difficult to see mtDNA nucleoids. |
| Protein Localization | Recommended due to ease of specific antibody or transgenic labeling of proteins. | Further developments are needed. Current transgenic approaches have proven difficult to adapt across models, particularly in animals. Immunogold is easier but lacks precision. |
| Organelle Interactions | Recommended for observing dynamic organelle interactions due availability of transgenic and dye labeling approaches for multiple | 3D approaches are ideal for assessing direct organelle interactions due to high resolution and ability to discern up to 10 or more structures per cell |
| structures in live cells. Generally lacks resolution to specifically detect functional contacts (<30 nm) between organelles. The number of organelle types observed can be limited by ability to specifically label different structures. | volume. Accurate and fast image segmentation of greyscale image volumes is current limitation. 2D images are informative but lack complete picture of 3D images. | |
| Energetics | A number of dyes and endogenous markers are available to assess mitochondrial energetic status. Care should be taken that changes in fluorescence are not simply due to differences in mitochondrial content. Membrane potential probes may also be sensitive to changes in pH or ROS. | Not directly assessed by electron microscopy in cells. |
| Signaling | A number of dyes and endogenous markers are available to assess mitochondrial calcium, ROS, and redox status, among others. Care should be taken that changes in fluorescence are not simply due to differences in mitochondrial content. ROS and calcium probes may be sensitive to changes in pH or membrane potential. | Not directly assessed by electron microscopy in cells. |
| Dynamics | Recommended for assessing frequency of fission and fusion events as well as motility due to ability to visualize live cells across time. | Mitochondria fixed during fission and fusion events can be observed but unable to see live events. |
| Turnover | Several recently developed transgenic | Mitochondria engulfed by autophagosomes or within |
| approaches are available for live cell assessment of mitophagy. Antibodies are also available for assessing mitophagy markers in fixed cells. | lysosomes can be observed but can sometimes be difficult to discriminate from other structures. Cannot observe live events. |
Mitochondrial Content
The amount of mitochondria within a cell, or mitochondrial content, is generally considered to be proportional to cellular capacity for mitochondrial function. While there are several methods for measuring cellular mitochondrial content [2, 13–17], imaging is the only way to directly assess the volume of a cell that is occupied by mitochondria. For light microscopy, there are three general approaches to visualizing mitochondrial structures: 1) labeling mitochondria in live cells with fluorescent dyes, 2) immunostaining fixed cells with mitochondria-specific fluorescent antibodies, and 3) using genetically encoded fluorescent mitochondrial tags. Using live cell fluorescent dyes to assess mitochondrial structure is straightforward in theory as all it takes is incubating the cells of interest with the dye for 20–60 minutes and then imaging and analyzing cellular fluorescence. The MitoTracker (MT) series of dyes are commonly used to assess mitochondrial content and work by binding to thiol groups within the mitochondria [18]. MT Orange, Red, and Far Red dyes all load into the mitochondria according to the mitochondrial membrane potential (ΔΨ), thus, differences in mitochondrial fluorescence between cells may be attributable to altered cellular conditions rather than a difference in mitochondrial content. MT Green is considered to load independent of ΔΨ and thus is a more consistent marker of content for most live cells. However, MT Green is not retained well in cells after fixation, so the other MT dyes may become a better option, for example, when combining MT with antibody labeling of specific proteins or when using fixation to stop cellular or mitochondrial movement while imaging. Additionally, MT Green was recently shown to be sent out of mitochondria by xenobiotic efflux pumps in hematopoietic stem cells [13]. The above information, combined with the known toxicity of mitochondrial dyes [19], suggests that while fluorescent dyes can be used to assess mitochondrial content quickly, results should be interpreted with appropriate caution.
Similar in concept to a spatially-resolved western blot, immunofluorescent staining can simultaneously be performed on multiple mitochondrial or other cellular proteins to assess structure and content within fixed cells in process that takes 1–2 days. Antibodies are generally available from several sources and can often be used with a fluorophore of choice leading to great experimental flexibility. The choice of antibody should be for mitochondrial proteins whose expression level per mitochondrion is not expected to be altered between comparison groups in order to separate changes in mitochondrial content from changes in mitochondrial composition. Also, targeting more abundant mitochondrial proteins (e.g. VDAC) should result in stronger antibody labeling, brighter fluorescence, and, thus, easier image analysis and interpretation. One downside of immunofluorescence is potential non-specific antibody labeling of untargeted proteins which can confound structural assessments, so using validated antibodies or performing control experiments is recommended. Additionally, uniform antibody penetration throughout thick cells or tissues has traditionally been a problem, but recently developed tissue clearing techniques such as CLARITY, SWITCH, or iDISCO can be used to overcome this issue [20–22].
Genetically encoded fluorescent mitochondrial tags provide specific and stable labeling of mitochondrial structures within cells and can be targeted either to specific proteins or to the mitochondrial matrix in general using a mitochondrial targeting sequence. Transgenic labeling is often the approach of choice for cell biologists due to the relative ease of transfection of cell lines though transfection efficiency is often lower in primary cell cultures and live animals. Compared to the abundance of antibodies, there are fewer transgenic constructs available to label mitochondria. Additionally, genetically encoded cell cultures and animals can take days to months to develop, respectively, making time a major limitation of this approach.
Mitochondrial Shape
The approaches to visualizing mitochondrial shapes are similar to those described for mitochondrial content, though assessing individual mitochondrial shapes requires higher resolution images. In many cases, the maximum resolution allowed by traditional light microscopy (~200–250nm in x,y, 500nm in z) is insufficient to clearly resolve individual mitochondrial structures necessitating the use of electron microscopy (maximum resolution ~0.2nm). However, earlier this year, the Betzig and Boyden labs reported on a recent collaboration that overcame this resolution limitation and allowed for assessment of thousands of individual mitochondrial shapes with molecular precision in whole tissues [23]. They combined expansion microscopy (ExM) with lattice light sheet microscopy (LLSM), each recent major breakthroughs for imaging cellular structures [24, 25]. ExM, which achieves super-resolution by physically magnifying biological samples rather than optical magnification and thus can be performed with any light microscope, involves embedding fixed samples labeled with either antibodies or transgenic fluorophores in a swellable polymer gel, digestion of unlabeled tissue, and then expanding the gel in a solvent. ExM has been used to increase resolution up to 20-fold [26], but a four-fold increase may be a more realistic goal for uniform expansion without distortion of cellular structures [23]. A disadvantage of ExM is that image acquisition times and file sizes increase proportional to the level of expansion cubed for 3D data (i.e. 4x expansion ➔ 64-fold more data). LLSM together with ExM overcomes the speed issue by sweeping a thin sheet of Bessel beams through the sample perpendicular to the objective with the resulting fluorescence collected by a high speed camera at rates of several hundred images per second and with very little photobleaching. This combined approach will be very useful for future studies evaluating mitochondrial structure in whole tissues or large cells such as cardiac and skeletal muscle or neurons, though the fixation required for ExM does not permit study of live cells.
Mitochondrial Ultrastructure
The mitochondrial inner membrane contains many folds called cristae which can regulate the organization, and thus function, of the enzymes involved in the energy conversion and reactive oxygen species production processes [27–29]. Also contained within mitochondria are nucleoids which enclose mitochondrial DNA (mtDNA). Due to the small sizes of the internal mitochondrial components and spaces (~100nm or less), visualization of mitochondrial ultrastructure has typically been performed using electron microscopy on fixed cells [30–32]. However, the emergence of and continued improvements in super-resolution light microscopy has now opened a window into mitochondrial ultrastructure within live cells. Super-resolution (stimulated emission depletion or STED) microscopy was recently used to visualize individual ellipsoid shaped mtDNA nucleoids finding that each nucleoid contains a single copy of mtDNA that is compacted by mitochondrial transcription factor A (TFAM) [33]. STED microscopy works by using a second laser to selectively deplete the emission of a fluorophore with the shape of the additional laser resembling a donut. Thus, the remaining non-depleted fluorescence is in the shape of the donut-hole, or smaller (~30–80nm resolution) than without using the depletion laser. STED can be used in tandem with confocal microscopy and is even available as a modular add-on on some commercial systems. This can be useful when imaging multiple fluorophores where not all require super-resolution (e.g. nuclei or lipid droplets) as the depletion laser can be selectively used for some or none of the fluorophores and reduce the potential photodamage from additional laser exposure.
The shape of mitochondrial cristae and how the structures change across time has also been traditionally difficult to capture in live cells [34, 35]. Recently, a STED approach to imaging cristae was developed using a genetically encoded mitochondrial inner membrane SNAP-tag which could then be linked with a fluorophore of choice allowing simultaneous visualization of cristae and mtDNA nucleoids with ~50nm resolution [36]. However, photobleaching limited timecourse imaging to 10–20 frames over 2 minutes. Development of another super-resolution microscopy approach (Hessian structured illumination microscopy or Hessian-SIM) which utilizes a grid illumination pattern together with a Hessian deconvolution algorithm to reconstruct high resolution images at low light doses allowed visualization of individual cristae with ~90nm resolution for up to 800 frames using MT Green dye [37]. Thus, STED approaches currently offer higher resolution which may be important for mitochondria with tightly packed cristae such as in the heart, but SIM can be used to image faster and with less photobleaching and may be better for cells with highly dynamic mitochondria. Application of these and similar types of live cell imaging techniques [38–40] to specific biological questions will likely offer powerful insight into the dynamics of mitochondrial ultrastructure and the potential functional roles during normal and pathological conditions.
Mitochondrial Protein Localization
Approaches for imaging mitochondrial or potential mitochondrial proteins are similar to those for other cellular structures and can be done with either antibodies or transgenic probes as discussed above. Importantly, images should be taken at high enough resolution to clearly discriminate between mitochondria and other closely associated organelles such as the endoplasmic reticulum (ER) [41, 42]. Determining in which mitochondrial compartment or membrane a protein resides can indicate the types of other proteins it may interact with and offer further insight into potential protein function (Box 2). Super-resolution techniques as discussed above can provide enough resolution to discriminate between mitochondrial compartments in some cases [43, 44]. Fluorescence lifetime imaging (FLIM) has also been used to distinguish between different mitochondrial compartments for genetically encoded proteins based on the length (lifetime) of exponential fluorescence decay after excitation using time-correlated single photon counting [45]. One advantage of FLIM is that several different proteins or molecules with the same or similar fluorescence emission wavelengths can be differentiated if the lifetimes can be clearly resolved [46].
Box 2: Tandem Labeling Approaches for Evaluating Sub-mitochondrial Protein Localization.
Visualizing the intra-mitochondrial localization of proteins without the use of specialized super-resolution or FLIM equipment can be done using one of several tandem labeling approaches. The split fluorophore (e.g. split-GFP) approach involves genetically labeling the protein of interest with half of a fluorophore and a potential interacting protein with the other half resulting in fluorescence only where the two proteins are in close enough proximity (~10–50nm depending on length of spacer encoded on one end [111]) to combine into the complete fluorophore [112, 113].
Forster resonance energy transfer (FRET) relies on genetically encoding one protein with a donor fluorophore and the other with an acceptor fluorophore [114, 115]. When the two proteins are in close proximity, the FRET donor transfers energy to the acceptor resulting in an increased fluorescence of the acceptor and a decrease in the donor. Thus, the fluorescence ratio of the two fluorophores is the measured output for FRET. Only specific fluorophore pairs work with FRET (e.g. YFP+CFP) and the use of two fluorophores takes up more of the emission bandwidth than the split-GFP approach, but because the energy transfer efficiency is directly related to distance between fluorophores, FRET can be used to more quantitatively assess the distance between proteins.
Proximity ligation assays (PLA) [116] label both proteins with primary antibodies and secondary antibodies linked to specific short DNA strands such that when the two proteins are in proximity, the two DNA strands form a circular DNA molecule which can be amplified and bound by fluorescently labeled oligonucleotides resulting in fluorescence specific to the sites of protein interaction. PLA does not require transgenic expression of probes and thus is more readily available across model systems compared to the split-GFP or FRET approaches. However, PLA also requires fixation and thus, cannot be used to monitor dynamic events in live cells.
Mitochondrial protein localization can also be assessed by performing immunogold labeling which is similar to immunofluorescence labeling, but instead of a fluorophore, the secondary antibody is labeled with 5–10nm gold particles that appear as electron dense spots in electron microscopy images [47, 48]. Analysis of immunogold labeled images relies on labeling density near the structure of interest and can be confounded by background signal from non-specific antibody labeling. Recently, genetically encoded tags for electron microscopy have been developed (e.g. miniSOG and APEX) [49–52] which can be linked to a protein of interest to provide increased image contrast where the protein resides within mitochondria or the cell. miniSOG relies on exposing an internal flavoprotein to UV light resulting in generation of singlet oxygen which then polymerizes added diaminobenzidine (DAB) leading an increase in electron density after addition of osmium. APEX works somewhat similarly but is a modified ascorbate peroxidase that polymerizes DAB upon exposure to hydrogen peroxide. Use of miniSOG and APEX require careful titration of the UV light dose and hydrogen peroxide exposure, respectively, so as to polymerize enough DAB to increase electron density but not too much that the increased electron density spreads beyond the protein of interest. Compared to the adoption of fluorescently tagged proteins a decade ago, application of genetically encoded electron microscopy tags has moved more slowly perhaps due to the sensitivity of the DAB polymerization step or just the inherently slower nature of electron versus light microscopy. At current rates, pushing the boundaries of super-resolution light microscopy appears to be the more promising avenue to pursue for high resolution assessment of mitochondrial protein localization.
Mitochondria-Organelle Interactions
Simultaneous imaging of multiple organelles is a powerful tool for assessing how the components of a cell, including mitochondria, work together to achieve functional goals. A few years ago, a novel multispectral imaging approach allowed simultaneous visualization of six different organelles, including mitochondria, across time and in 3D to show that the dynamic organelle interactome was dependent on a functional microtubule system in COS-7 cells [53]. Cells were excited sequentially by six different lasers, emitted light was collected through a series of interference filters, and then a spectral unmixing algorithm was applied to every pixel to separate the CFP, EGFP, YFP, mApple, Texas Red, and Bodipy fluorescent signals corresponding to different organelles. While this multispectral imaging technique can be applied to any cellular structures on any light microscope in theory, they used confocal and light sheet microscopes where the resolution achieved (300+nm) was insufficient to evaluate the specialized functional contacts that occur between organelles when they are within 30nm of each other [54]. Application of multispectral imaging to super-resolution microscopes capable of resolving functional contacts between mitochondria and other organelles would provide a powerful tool for systems level evaluation of dynamic organelle interactions within the cell.
Bleck et al. [2] were able to bypass the resolution limits of light microscopy by using 3D electron microscopy (focused ion beam scanning electron microscopy or FIB-SEM) combined with machine learning image segmentation to show that the intra- and inter-organelle interactions within muscle mitochondrial networks are altered in accordance with the functional goals of different muscle cell types. FIB-SEM images the surface of a fixed sample then uses an ion beam to remove the top layer of the sample and repeats this process many times. Analysis of the resultant 3D volume of hundreds or thousands of greyscale images can be a major limitation of this technique due to the time required to segment different cellular structures [55, 56]. Machine learning techniques in which several training images are used to classify which pixels correspond to mitochondria or other cellular structures and then applied over the entire volume can speed up image segmentation by several thousand-fold with high accuracy [57, 58]. In addition to higher resolution, expression of transgenic probes or use of fluorescent dyes is unnecessary to discriminate between mitochondria, ER, or many other cellular structures with electron microscopy as they can be specified by their characteristic structure in any fixed cell, although the visibility and contrast of different cellular structures is dependent on the heavy metal and other stains used (e.g. osmium, tannic acid, thiocarbazide, etc.). High throughput analysis of functional organelle interactions throughout the cell provides a promising platform for dissecting how cellular structural design translates to cellular function or dysfunction.
Mitochondrial Function
One of the major issues of functional imaging is that, unlike structural imaging where a single snapshot is often sufficient, functional imaging generally requires collection of many images in living cells or tissue across time. Since electron microscopy cannot be performed on living tissue (the electron beams completely destroy live cells) [59], light microscopy is the primary tool for visualizing mitochondrial function within cells. However, while less damaging than electron beams, shining light beams on cells can still cause damage either to the cells or the fluorophores being excited [60]. Thus, the more times a cell or region of a cell is imaged, the greater the likelihood of damage occurring. The issue of photodamage sets off a delicate balance where attempts are made to shine a minimal amount of light on cells for the least amount of time while still shining enough light to get high signal-to-noise ratios and imaging often enough to adequately describe the functional events taking place [60]. This balancing act can vary greatly as the requirements for spatial and temporal resolution as well as signal-to-noise ratio are different for each biological question and can also be dictated by sensitivity and intensity of the fluorophore(s) being imaged. Unfortunately, there is no one-size-fits-all approach to functional mitochondrial imaging. However, faster and gentler microscopes as well as brighter and more stable fluorophores continue to be developed allowing for a clearer window into how mitochondria actually behave within cells. These recent advances and the current state-of-the-art for functional mitochondrial imaging are discussed below.
Mitochondrial Energetics
One of the most important functions of mitochondria is to make ATP. Thus, it is of great interest to visualize how well mitochondria perform this task within cells. Indeed, there are several dyes or genetically encoded probes of varying specificity which enable observation of ATP in cells, [61–63]. One of these methods, ATeam [61], is a genetically encoded FRET probe where instead of using putting the donor and acceptor fluorophores on two different proteins as discussed in Box 2 above, the two fluorophores are linked by a protein which changes conformation upon ATP binding thus bringing the two fluorophores close enough to induce FRET. ATeam is insensitive to pH and can be targeted to either the cytosol or mitochondria making it useful for assessing pathological changes in cellular energy metabolism [64].
Approaches to imaging other components of the mitochondrial energy conversion pathway include observing mitochondrial pyruvate, redox status and ΔΨ [65–67] (Box 3, 4). While each of these approaches provide information on the concentration of energetic markers, concentration of each is a reflection of the balance between its production and utilization within the cell. As a result, if production continues to match utilization during an increase in energy demand, a common occurrence in cells [68–70], concentration of the energetic marker will not indicate that the energetic environment in the cell is different. The ideal marker of mitochondrial energetic function would be a direct measure of how fast mitochondria are making ATP. Unfortunately, a robust, spatially-resolved measure of the rate of ATP production is not currently available. Oxygen consumption is proportional to ATP production, but while methods to image oxygen within cells are improving, current developments have primarily focused on visualizing oxygen content rather than the rate of oxygen consumption [71, 72]. An initial approach to provide a spatially-resolvable measure of the rate of mitochondrial energy conversion was recently published [73] where monitoring of the kinetic response of endogenous mitochondrial NADH fluorescence (see Box 3) to a rapid inhibition of NADH utilization allowed the rate of NADH production to be measured in live mouse skeletal muscle and in cultured neurons under a multiphoton microscope. Further development of methods to quantify rates of mitochondrial ATP production or proportional measures (e.g. oxygen or NADH) would allow for improved testing of how pharmacological interventions affect mitochondria within different regions of the same cell or tissue.
Box 3: Assessing Mitochondrial NADH Redox Status.
The redox status of the mitochondrial NADH pool can be assessed through its endogenous autofluorescence [65, 117] as the NADH molecule itself fluoresces meaning there is no need for additional probes. However, NADH autofluorescence requires ultraviolet (~340–375nm) excitation which is not available on most commercial confocal microscope systems requiring use of more expensive two-photon lasers to excite NADH with higher (double) wavelength (~680–750nm) pulses. Additionally, NADH and another redox molecule, NADPH, have identical optical properties making it difficult to evaluate them separately when present in similar quantities [46], though FLIM offers one approach [46]. For these reasons, transgenic NADH probes have been developed using circularly permuted YFP (cpYFP) linked to a Rex homodimer (Frex-Mit) both of which change conformation upon NADH binding leading to increased fluorescence proportional to NADH concentration [118]. The cpYFP approach is reported to result in a much greater fluorescent dynamic range than FRET probes [119], however, the fluorescent intensity of Frex-Mit is sensitive to both NADH and pH limiting its usefulness to questions where pH can be controlled for. Targeting pH resistant, cytosolic NADH probes, such as SoNar [120], to the mitochondria in a cell-specific manner (e.g. conditional expression through Cre recombinase) would be helpful for evaluating the redox status of small cells with low mitochondrial content (i.e. low endogenous NADH fluorescence) within large tissues such as endothelial or satellite cells within skeletal muscle.
Box 4: Mitochondrial Membrane Potential Probes.
There are several commonly used ΔΨ sensitive dyes (TMRM, TMRE, Rhod123, JC-1) which are all lipophilic cations and accumulate proportional to the voltage across membranes. Because these dyes accumulate across both the cell membrane and the mitochondrial inner membrane, measures of total cellular fluorescence or only mitochondrial fluorescence can be affected by changes in the plasma membrane potential [121]. Thus, imaging at sufficient resolution to measure the mitochondrial-to-cytosolic fluorescence ratio allows evaluation of ΔΨ independent of changes in plasma membrane voltage. TMRM, TMRE, and Rhod123 each contain one fluorophore whereas JC-1 fluorescence shifts from green to red with increasing aggregation in the matrix and thus is used ratiometrically. However, there are many reported problems with JC-1 including insensitivity to small voltage changes, high photosensitivity, slow equilibration times across membranes, and being prone to artifacts [19] suggesting it should be used with caution if at all. TMRM and TMRE equilibrate more quickly (~15 minutes) than Rhod123, and TMRE and Rhod123 are more toxic than TMRM [19]. Additionally, use of probes above ~100 nM can initiate “quench mode” where the concentration of the probe is so high inside the mitochondria that aggregated dye quenches emitted fluorescence resulting increased signal when ΔΨ goes down (less quenching) and decreased signal when ΔΨ goes up (more quenching). Thus, using the lowest possible concentration of these probes is recommended. Proper imaging of ΔΨ can then be multiplexed with cellular (e.g. calcein-AM) and nuclear (e.g. Hoechst) dyes for high throughput analysis of mitochondrial structure and function within dynamic cell populations under normal or pathological conditions [15].
Mitochondrial Signaling
The most well described mitochondrial signaling molecules are cytochrome c, calcium, reactive oxygen species (ROS), and the redox pairs discussed above, NAD+/NADH and NADP+/NADPH. Several cytochrome c probes have recently been synthesized with the aim of better understanding apoptosis [74–82], however, to date, they have been little used to answer specific questions. Future application of these probes to biological questions will be needed to determine the advantages and disadvantages of each for specific experimental models. Sensors are available in many forms and have been well characterized and discussed extensively elsewhere for both calcium [83–91] and ROS [92–95] and can also be multiplexed with other measures of mitochondrial structure and function [96].
In addition to signaling with other organelles, mitochondria have also been reported to communicate amongst themselves through intermitochondrial junctions or mitochondrial nanotunnels [97–100]. While conduction of ΔΨ among connected mitochondria has been assessed by comparing the tetramethyl rhodamine, methyl ester (TMRM) response of mitochondria adjacent to a region of photoactivatable depolarization [2, 97, 101], propagation of the chemical component of the protonmotive force across the inner mitochondrial membrane, ΔpH, between adjacent mitochondria has also been shown by the Demaurex group [102]. Mitochondrial pH can be visualized ratiometrically with a genetically encoded, pH sensitive cpYFP (MitoSypHer) [103] which is a mutation of the ROS probe, Hyper [104], to remove the sensitivity to hydrogen peroxide. By combining MitoSypHer with a photoactivatable mitochondrial matrix GFP (paGFP), Santo-Domingo et al. [102] showed that rapid electrical coupling could occur between adjacent mitochondria without exchange of matrix contents (i.e. paGFP) and demonstrated the applicability of using multiple mitochondrially targeted probes to elucidate mechanistic information about the nature of ion versus protein exchange between adjacent mitochondria.
Mitochondrial Dynamics and Turnover
Another major function mitochondria are tasked with is to contribute to their own self-maintenance. The removal of damaged mitochondria through mitophagy can now be easily imaged thanks to the development of two recent genetically encoded probes (mt-Keima and mito-QC) [105–107]. Mitochondrial matrix-targeted mt-Keima utilizes a ratiometric, dual excitation system where excitation with 458nm light illuminates mitochondria with a neutral or slightly alkaline pH and 561nm light excites mitochondria in the acidic environment of the lysosome. mito-QC consists of two fluorophores, GFP and mCherry, linked to outer mitochondrial membrane protein Fis1 and detects mitophagy by the loss of GFP signal in the acidic lysosome. While both probes are compatible with live cell imaging, mt-Keima cannot be used in fixed cells precluding its use in tandem with immunofluorescent staining of other proteins. However, the large emission bandwidth used by mito-QC also limits its compatibility with additional indicators to either blue or far red emitting probes.
Maintenance of high quality mitochondria also involves the proper replication and transmission of mtDNA. It has been well known that an exchange of mitochondrial contents can occur when mitochondria undergo fission and fusion [108–110]. However, until recently, it was unknown how or whether this process involved mtDNA. Lewis et al. [42] used three transgenic fluorophores targeted at the same time to the mitochondrial matrix (BFP), mtDNA nucleoids (GFP), and ER (mCherry) to show that replicating mtDNA nucleoids mark the sites of mitochondrial fission which is then mediated by the ER. Thus, simultaneous imaging of two mitochondrial structures and the ER revealed a mechanism of how alterations in mitochondrial shape and ultrastructure can be regulated in coordination with specific interactions between organelles further highlighting the integrated nature of cellular processes and the role of mitochondria within them.
Concluding Remarks
The emergence of mitochondria as multi-faceted organelles tailored to support cellular function has helped drive the development of new imaging approaches aimed at better understanding the mechanisms which guide mitochondrial function within complex cellular environments. Continued improvements in light microscopy, particularly super resolution microscopy, offer a promising path toward visualizing mitochondrial behavior with molecular precision, while increasing accessibility to 3D electron microscopy techniques is expanding our knowledge of how mitochondria interact with other organelles to meet cellular objectives. There remains an unwavering goal to image faster, gentler, with higher resolution, across larger volumes, and a greater number of molecules or structures which continues to guide technological development (see Outstanding Questions). As approaches to imaging mitochondria as part of complex cell systems are realized, the development of big data analytical platforms will be necessary to fully elucidate the principles which govern mitochondrial form and function across health and disease.
Outstanding Questions.
Can super-resolution microscopy meet or surpass the resolution of large-scale, 3D electron microscopy techniques?
Can methods for genetically encoding tags in electron microscopy be improved to facilitate widespread use akin to that of genetically encoded fluorescent tags?
How can the number of mitochondrial proteins or molecules that can be imaged simultaneously by the light microscope be increased? Will multispectral or fluorescent lifetime imaging lead the way?
Can spatially-resolved measures of mitochondrial function reach the quantitative rigor of biochemical methods?
Will a widely accepted, quantitative measure describing the large variety of mitochondrial shapes be developed?
Is the frequency of dynamic mitochondrial cristae events proportional to the frequency of mitochondrial fission and fusion events?
What will it take to measure ATP production rates with subcellular resolution?
Can current and future advancements in cellular imaging be applied to tissues in live animals and humans?
As imaging speed, resolution, volume, and the number of structures imaged all increase, how do we analyze and interpret the very large datasets that will be generated?
Highlights.
Advances in super-resolution microscopy now enable visualization of thousands of individual mitochondria with molecular precision throughout large tissues as well as unprecedented views of the dynamic nature of internal mitochondrial structures.
Expansion of our ability to simultaneously visualize multiple mitochondrial structures and proteins together with other organelles has provided novel mechanistic insights into the intra- and inter-organelle interactions of mitochondrial networks.
Spatially resolved measures of mitochondrial energetic flux provide a promising avenue for evaluating the impact of interventions into cellular energy metabolism within heterogeneous cells and tissues.
Accompaniment of high-throughput image analysis platforms with big data generating microscopy approaches now enables systems level evaluations of how mitochondria behave within the cellular environment.
Glossary
- Capacity for Mitochondrial Function
The maximal function of mitochondria possible. The capacity for mitochondrial function within a cell is determined by mitochondrial structure, composition, and interactions with other structures
- Electron Microscopy
Microscopy technique using electron beams to illuminate and magnify structures. Electron microscopy can resolve much smaller structures than light microscopy. Greyscale images are generated where contrast depends on the electron density of cellular structures. Many structures can be visualized simultaneously. Metallic stains can be used to increase contrast in specific structures
- Fixed Cells
Cells which have been preserved from biological decay. Several methods of fixation are commonly used such as rapid freezing or chemical fixation by formaldehyde or methanol
- Fluorophore
A fluorescent molecule which emits light when excited by a light beam. Fluorophores come in many varieties including the color of light needed for excitation as well as the color emitted after excitation
- Immunogold
Method for antibody labeling of specific proteins in electron microscopy. Instead of using a fluorescently tagged secondary antibody as with light microscopy, the secondary antibody is conjugated with tiny gold particles which appear as electron dense dots in an electron microscopy image
- Light Microscopy
Microscopy technique using visible light beams to illuminate and magnify structures. For mitochondria, this generally involves excitation of fluorescent molecules and detection of the resultant emitted light
- Mitochondrial Protein and Lipid Composition
The proteins and lipids which a mitochondrion is made of. Proteins can be used to transport molecules, catalyze molecular reactions, and build or breakdown structures among other things. Lipids provide structural support to membranes and some can be synthesized within mitochondria.
- Signal-to-Noise Ratio (SNR)
The ratio between the signal intended to be measured and the background signal. A relatively high background signal, thus low SNR, makes it difficult to clearly distinguish small changes in the desired variable. Conversely, changes in high SNR fluorescent signals can clearly be observed and attributed to the intended biological process
- Super-resolution
A specific type of light microscopy where the achievable resolution is better than the diffraction limit of light (~200 nm). The diffraction limit was thought to set the highest resolution achievable for over 100 years until several recent developments broke through the barrier
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Benador IY et al. (2018) Mitochondria Bound to Lipid Droplets Have Unique Bioenergetics, Composition, and Dynamics that Support Lipid Droplet Expansion. Cell Metab 27 (4), 869–885 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bleck CKE et al. (2018) Subcellular connectomic analyses of energy networks in striated muscle. Nat Commun 9 (1), 5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson DT et al. (2007) Functional consequences of mitochondrial proteome heterogeneity. Am J Physiol Cell Physiol 292 (2), C698–707. [DOI] [PubMed] [Google Scholar]
- 4.Kim Y et al. (2019) Protein composition of the muscle mitochondrial reticulum during postnatal development. J Physiol 597 (10), 2707–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porat-Shliom N et al. (2019) Mitochondrial Populations Exhibit Differential Dynamic Responses to Increased Energy Demand during Exocytosis In Vivo. iScience 11, 440–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vega RB and Kelly DP (2017) Cardiac nuclear receptors: architects of mitochondrial structure and function. J Clin Invest 127 (4), 1155–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fecher C et al. (2019) Cell-type-specific profiling of brain mitochondria reveals functional and molecular diversity. Nature Neuroscience. [DOI] [PubMed] [Google Scholar]
- 8.Zhang H et al. (2016) NAD(+) repletion improves mitochondrial and stem cell function and enhances life span in mice. Science 352 (6292), 1436–43. [DOI] [PubMed] [Google Scholar]
- 9.Kaufmann U et al. (2019) Calcium Signaling Controls Pathogenic Th17 Cell-Mediated Inflammation by Regulating Mitochondrial Function. Cell Metab 29 (5), 1104–1118 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weidberg H and Amon A (2018) MitoCPR-A surveillance pathway that protects mitochondria in response to protein import stress. Science 360 (6385). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richter-Dennerlein R et al. (2016) Mitochondrial Protein Synthesis Adapts to Influx of Nuclear-Encoded Protein. Cell 167 (2), 471–483 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray MW (2015) Mosaic nature of the mitochondrial proteome: Implications for the origin and evolution of mitochondria. Proc Natl Acad Sci U S A 112 (33), 10133–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Almeida MJ et al. (2017) Dye-Independent Methods Reveal Elevated Mitochondrial Mass in Hematopoietic Stem Cells. Cell Stem Cell 21 (6), 725–729 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Z et al. (1999) Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98 (1), 115–24. [DOI] [PubMed] [Google Scholar]
- 15.lannetti EF et al. (2016) Multiplexed high-content analysis of mitochondrial morphofunction using live-cell microscopy. Nat Protoc 11 (9), 1693–710. [DOI] [PubMed] [Google Scholar]
- 16.Wredenberg A et al. (2002) Increased mitochondrial mass in mitochondrial myopathy mice. Proc Natl Acad Sci U S A 99 (23), 15066–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansson A et al. (2004) A switch in metabolism precedes increased mitochondrial biogenesis in respiratory chain-deficient mouse hearts. Proc Natl Acad Sci U S A 101 (9), 3136–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chazotte B (2011) Labeling mitochondria with MitoTracker dyes. Cold Spring Harb Protoc 2011 (8), 990–2. [DOI] [PubMed] [Google Scholar]
- 19.Perry SW et al. (2011) Mitochondrial membrane potential probes and the proton gradient: a practical usage guide. Biotechniques 50 (2), 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murray E et al. (2015) Simple, scalable proteomic imaging for high-dimensional profiling of intact systems. Cell 163 (6), 1500–1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renier N et al. (2014) iDISCO: a simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159 (4), 896–910. [DOI] [PubMed] [Google Scholar]
- 22.Chung K et al. (2013) Structural and molecular interrogation of intact biological systems. Nature 497 (7449), 332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao R et al. (2019) Cortical column and whole-brain imaging with molecular contrast and nanoscale resolution. Science 363 (6424). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen F et al. (2015) Optical imaging. Expansion microscopy. Science 347 (6221), 543–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen BC et al. (2014) Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346 (6208), 1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang J-B et al. (2017) Iterative expansion microscopy. Nature methods 14 (6), 593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cogliati S et al. (2016) Mitochondrial Cristae: Where Beauty Meets Functionality. Trends Biochem Sci 41 (3), 261–273. [DOI] [PubMed] [Google Scholar]
- 28.Quintana-Cabrera R et al. (2018) The cristae modulator Optic atrophy 1 requires mitochondrial ATP synthase oligomers to safeguard mitochondrial function. Nat Commun 9 (1), 3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Friedman JR et al. (2015) MICOS coordinates with respiratory complexes and lipids to establish mitochondrial inner membrane architecture. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brandt T et al. (2017) Changes of mitochondrial ultrastructure and function during ageing in mice and Drosophila. Elife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scorrano L et al. (2002) A distinct pathway remodels mitochondrial cristae and mobilizes cytochrome c during apoptosis. Dev Cell 2 (1), 55–67. [DOI] [PubMed] [Google Scholar]
- 32.Frey TG and Mannella CA (2000) The internal structure of mitochondria. Trends Biochem Sci 25 (7), 319–24. [DOI] [PubMed] [Google Scholar]
- 33.Kukat C et al. (2015) Cross-strand binding of TFAM to a single mtDNA molecule forms the mitochondrial nucleoid. Proc Natl Acad Sci U S A 112 (36), 11288–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Appelhans T et al. (2012) Nanoscale organization of mitochondrial microcompartments revealed by combining tracking and localization microscopy. Nano Lett 12 (2), 610–6. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt R et al. (2009) Mitochondrial cristae revealed with focused light. Nano Lett 9 (6), 2508–10. [DOI] [PubMed] [Google Scholar]
- 36.Stephan T et al. (2019) Live-cell STED nanoscopy of mitochondrial cristae. Sci Rep 9 (1), 12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang X et al. (2018) Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat Biotechnol 36 (5), 451–459. [DOI] [PubMed] [Google Scholar]
- 38.Shao L et al. (2011) Super-resolution 3D microscopy of live whole cells using structured illumination. Nat Methods 8 (12), 1044–6. [DOI] [PubMed] [Google Scholar]
- 39.Wang C et al. (2019) A photostable fluorescent marker for the superresolution live imaging of the dynamic structure of the mitochondrial cristae. Proc Natl Acad Sci U S A 116 (32), 15817–15822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo Y et al. (2018) Visualizing Intracellular Organelle and Cytoskeletal Interactions at Nanoscale Resolution on Millisecond Timescales. Cell 175 (5), 1430–1442 e17. [DOI] [PubMed] [Google Scholar]
- 41.Phillips MJ and Voeltz GK (2016) Structure and function of ER membrane contact sites with other organelles. Nat Rev Mol Cell Biol 17 (2), 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis SC et al. (2016) ER-mitochondria contacts couple mtDNA synthesis with mitochondrial division in human cells. Science 353 (6296), aaf5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stoldt S et al. (2019) Mic60 exhibits a coordinated clustered distribution along and across yeast and mammalian mitochondria. Proceedings of the National Academy of Sciences 116 (20), 9853–9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Große L et al. (2016) Bax assembles into large ring-like structures remodeling the mitochondrial outer membrane in apoptosis. The EMBO journal 35 (4), 402–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sohnel AC et al. (2016) Probing of protein localization and shuttling in mitochondrial microcompartments by FLIM with sub-diffraction resolution. Biochim Biophys Acta 1857 (8), 1290–1299. [DOI] [PubMed] [Google Scholar]
- 46.Blacker TS et al. (2014) Separating NADH and NADPH fluorescence in live cells and tissues using FLIM. Nat Commun 5, 3936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugiura A et al. (2017) Newly born peroxisomes are a hybrid of mitochondrial and ER-derived pre-peroxisomes. Nature 542 (7640), 251–254. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Q et al. (2016) Mitochondrial endonuclease G mediates breakdown of paternal mitochondria upon fertilization. Science 353 (6297), 394–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shu X et al. (2011) A genetically encoded tag for correlated light and electron microscopy of intact cells, tissues, and organisms. PLoS Biol 9 (4), e1001041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lam SS et al. (2015) Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat Methods 12 (1), 51–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Martell JD et al. (2012) Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat Biotechnol 30 (11), 1143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ariotti N et al. (2018) Ultrastructural localisation of protein interactions using conditionally stable nanobodies. PLoS Biol 16 (4), e2005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Valm AM et al. (2017) Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546 (7656), 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gatta AT and Levine TP (2017) Piecing Together the Patchwork of Contact Sites. Trends Cell Biol 27 (3), 214–229. [DOI] [PubMed] [Google Scholar]
- 55.Helmstaedter M et al. (2013) Connectomic reconstruction of the inner plexiform layer in the mouse retina. Nature 500 (7461), 168–74. [DOI] [PubMed] [Google Scholar]
- 56.Helmstaedter M (2013) Cellular-resolution connectomics: challenges of dense neural circuit reconstruction. Nat Methods 10 (6), 501–7. [DOI] [PubMed] [Google Scholar]
- 57.Beier T et al. (2017) Multicut brings automated neurite segmentation closer to human performance. Nat Methods 14 (2), 101–102. [DOI] [PubMed] [Google Scholar]
- 58.Haberl MG et al. (2018) CDeep3M—Plug-and-Play cloud-based deep learning for image segmentation. Nature methods 15 (9), 677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Jonge N and Peckys DB (2016) Live Cell Electron Microscopy Is Probably Impossible. ACS Nano 10 (10), 9061–9063. [DOI] [PubMed] [Google Scholar]
- 60.Combs CA and Shroff H (2017) Fluorescence Microscopy: A Concise Guide to Current Imaging Methods. Curr Protoc Neurosci 79, 2 1 1–2 1 25. [DOI] [PubMed] [Google Scholar]
- 61.Imamura H et al. (2009) Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci U S A 106 (37), 15651–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang JL et al. (2014) A ratiometric fluorescent probe with unexpected high selectivity for ATP and its application in cell imaging. Chem Commun (Camb) 50 (97), 15411–4. [DOI] [PubMed] [Google Scholar]
- 63.Tantama M et al. (2013) Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat Commun 4, 2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Depaoli MR et al. (2018) Real-Time Imaging of Mitochondrial ATP Dynamics Reveals the Metabolic Setting of Single Cells. Cell Rep 25 (2), 501–512 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rothstein EC et al. (2005) Skeletal muscle NAD(P)H two-photon fluorescence microscopy in vivo: topology and optical inner filters. Biophys J 88 (3), 2165–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sukumar M et al. (2016) Mitochondrial Membrane Potential Identifies Cells with Enhanced Stemness for Cellular Therapy. Cell Metab 23 (1), 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.San Martin A et al. (2014) Imaging mitochondrial flux in single cells with a FRET sensor for pyruvate. PLoS One 9 (1), e85780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balaban RS et al. (1986) Relation between work and phosphate metabolite in the in vivo paced mammalian heart. Science 232 (4754), 1121–3. [DOI] [PubMed] [Google Scholar]
- 69.Chance B et al. (1981) Mitochondrial regulation of phosphocreatine/inorganic phosphate ratios in exercising human muscle: a gated 31P NMR study. Proc Natl Acad Sci U S A 78 (11), 6714–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilson JR et al. (1985) Evaluation of energy metabolism in skeletal muscle of patients with heart failure with gated phosphorus-31 nuclear magnetic resonance. Circulation 71 (1), 57–62. [DOI] [PubMed] [Google Scholar]
- 71.Penjweini R et al. (2018) Intracellular oxygen mapping using a myoglobin-mCherry probe with fluorescence lifetime imaging. J Biomed Opt 23 (10), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Esipova TV et al. (2019) Oxyphor 2P: A High-Performance Probe for Deep-Tissue Longitudinal Oxygen Imaging. Cell Metab 29 (3), 736–744 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Willingham TB et al. (2019) mitoRACE: evaluating mitochondrial function in vivo and in single cells with subcellular resolution using multiphoton NADH autofluorescence. The Journal of Physiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen TT et al. (2015) Fluorescence activation imaging of cytochrome c released from mitochondria using aptameric nanosensor. J Am Chem Soc 137 (2), 982–9. [DOI] [PubMed] [Google Scholar]
- 75.Zhang J et al. (2019) Surface-Enhanced Raman Scattering-Fluorescence Dual-Mode Nanosensors for Quantitative Detection of Cytochrome c in Living Cells. Anal Chem 91 (10), 6600–6607. [DOI] [PubMed] [Google Scholar]
- 76.Liu Y et al. (2016) Upconversion nano-photosensitizer targeting into mitochondria for cancer apoptosis induction and cyt c fluorescence monitoring. Nano Research 9 (11), 3257–3266. [Google Scholar]
- 77.Ma L et al. (2017) A novel upconversion@ polydopamine core@ shell nanoparticle based aptameric biosensor for biosensing and imaging of cytochrome c inside living cells. Biosensors and Bioelectronics 87, 638–645. [DOI] [PubMed] [Google Scholar]
- 78.Zhang H et al. (2018) Label-free fluorescence imaging of cytochrome c in living systems and anti-cancer drug screening with nitrogen doped carbon quantum dots. Nanoscale 10 (11), 5342–5349. [DOI] [PubMed] [Google Scholar]
- 79.Cai M et al. (2019) Label-free fluorometric assay for cytochrome c in apoptotic cells based on near infrared Ag2S quantum dots. Analytica chimica acta 1056, 153–160. [DOI] [PubMed] [Google Scholar]
- 80.Shamsipur M et al. (2016) Detection of early stage apoptotic cells based on label-free cytochrome c assay using bioconjugated metal nanoclusters as fluorescent probes. Analytical chemistry 88 (4), 2188–2197. [DOI] [PubMed] [Google Scholar]
- 81.Tang J et al. (2018) Azoreductase and target simultaneously activated fluorescent monitoring for cytochrome c release under hypoxia. Analytical chemistry 90 (9), 5865–5872. [DOI] [PubMed] [Google Scholar]
- 82.Qi G et al. (2018) Smart Plasmonic Nanorobot for Real-Time Monitoring Cytochrome c Release and Cell Acidification in Apoptosis during Electrostimulation. Analytical chemistry 91 (2), 1408–1415. [DOI] [PubMed] [Google Scholar]
- 83.Smith NA et al. (2018) Fluorescent Ca2+ indicators directly inhibit the Na, K-ATPase and disrupt cellular functions. Sci. Signal 11 (515), eaal2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lock JT et al. (2015) A comparison of fluorescent Ca2+ indicators for imaging local Ca2+ signals in cultured cells. Cell calcium 58 (6), 638–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Akerboom J et al. (2013) Genetically encoded calcium indicators for multi-color neural activity imaging and combination with optogenetics. Frontiers in molecular neuroscience 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Davidson SM and Duchen MR (2018) Imaging Mitochondrial Calcium Fluxes with Fluorescent Probes and Single-or Two-Photon Confocal Microscopy In Mitochondrial Bioenergetics, pp. 171–186, Springer. [DOI] [PubMed] [Google Scholar]
- 87.Whitaker M (2010) Genetically encoded probes for measurement of intracellular calcium In Methods in cell biology, pp. 153–182, Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.De Michele R et al. (2014) Mitochondrial biosensors. The international journal of biochemistry & cell biology 48, 39–44. [DOI] [PubMed] [Google Scholar]
- 89.Wu J et al. (2014) Red fluorescent genetically encoded Ca2+ indicators for use in mitochondria and endoplasmic reticulum. Biochemical Journal 464 (1), 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Contreras L et al. (2010) Mitochondria: the calcium connection. Biochimica et Biophysica Acta (BBA)-Bioenergetics 1797 (6–7), 607–618. [DOI] [PubMed] [Google Scholar]
- 91.Pozzan T and Rudolf R (2009) Measurements of mitochondrial calcium in vivo. Biochimica et Biophysica Acta (BBA)-Bioenergetics 1787 (11), 1317–1323. [DOI] [PubMed] [Google Scholar]
- 92.Zhang X and Gao F (2015) Imaging mitochondrial reactive oxygen species with fluorescent probes: current applications and challenges. Free radical research 49 (4), 374–382. [DOI] [PubMed] [Google Scholar]
- 93.Dikalov SI and Harrison DG (2014) Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxidants & redox signaling 20 (2), 372–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Woolley J et al. (2013) Recent advances in reactive oxygen species measurement in biological systems. Trends in biochemical sciences 38 (11), 556–565. [DOI] [PubMed] [Google Scholar]
- 95.Wang X et al. (2013) Imaging ROS signaling in cells and animals. Journal of molecular medicine 91 (8), 917–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sieprath T et al. (2016) Integrated high-content quantification of intracellular ROS levels and mitochondrial morphofunction In Focus on Bio-Image Informatics, pp. 149–177, Springer. [DOI] [PubMed] [Google Scholar]
- 97.Glancy B et al. (2015) Mitochondrial reticulum for cellular energy distribution in muscle. Nature 523 (7562), 617–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Picard M et al. (2015) Trans-mitochondrial coordination of cristae at regulated membrane junctions. Nat Commun 6, 6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vincent AE et al. (2017) Mitochondrial Nanotunnels. Trends Cell Biol 27 (11), 787–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Patel KD et al. (2016) The electrochemical transmission in I-Band segments of the mitochondrial reticulum. Biochim Biophys Acta 1857 (8), 1284–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Glancy B et al. (2017) Power Grid Protection of the Muscle Mitochondrial Reticulum. Cell Rep 19 (3), 487–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Santo-Domingo J et al. (2013) OPA1 promotes pH flashes that spread between contiguous mitochondria without matrix protein exchange. EMBO J 32 (13), 1927–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Poburko D et al. (2011) Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. J Biol Chem 286 (13), 11672–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Belousov VV et al. (2006) Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods 3 (4), 281–6. [DOI] [PubMed] [Google Scholar]
- 105.McWilliams TG et al. (2016) mito-QC illuminates mitophagy and mitochondrial architecture in vivo. J Cell Biol 214 (3), 333–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sun N et al. (2015) Measuring In Vivo Mitophagy. Mol Cell 60 (4), 685–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.McWilliams TG et al. (2018) Basal Mitophagy Occurs Independently of PINK1 in Mouse Tissues of High Metabolic Demand. Cell Metab 27 (2), 439–449 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Szabadkai G et al. (2004) Drp-1-dependent division of the mitochondrial network blocks intraorganellar Ca2+ waves and protects against Ca2+-mediated apoptosis. Mol Cell 16 (1), 59–68. [DOI] [PubMed] [Google Scholar]
- 109.Molina AJ et al. (2009) Mitochondrial networking protects beta-cells from nutrient-induced apoptosis. Diabetes 58 (10), 2303–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bleazard W et al. (1999) The dynamin-related GTPase Dnm1 regulates mitochondrial fission in yeast. Nat Cell Biol 1 (5), 298–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Cieri D et al. (2018) SPLICS: a split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death Differ 25 (6), 1131–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang F et al. (2015) The cAMP phosphodiesterase Prune localizes to the mitochondrial matrix and promotes mtDNA replication by stabilizing TFAM. EMBO Rep 16 (4), 520–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ruan L et al. (2017) Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543 (7645), 443–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mahajan NP et al. (1998) Bcl-2 and Bax interactions in mitochondria probed with green fluorescent protein and fluorescence resonance energy transfer. Nat Biotechnol 16 (6), 547–52. [DOI] [PubMed] [Google Scholar]
- 115.Sekar RB and Periasamy A (2003) Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J Cell Biol 160 (5), 629–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Misawa T et al. (2013) Microtubule-driven spatial arrangement of mitochondria promotes activation of the NLRP3 inflammasome. Nat Immunol 14 (5), 454–60. [DOI] [PubMed] [Google Scholar]
- 117.Glancy B et al. (2014) In vivo microscopy reveals extensive embedding of capillaries within the sarcolemma of skeletal muscle fibers. Microcirculation 21 (2), 131–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhao Y et al. (2011) Genetically encoded fluorescent sensors for intracellular NADH detection. Cell Metab 14 (4), 555–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hu H et al. (2018) Glucose monitoring in living cells with single fluorescent protein-based sensors. RSC advances 8 (5), 2485–2489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhao Y et al. (2015) SoNar, a Highly Responsive NAD+/NADH Sensor, Allows High-Throughput Metabolic Screening of Anti-tumor Agents. Cell Metab 21 (5), 777–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nicholls DG (2006) Simultaneous monitoring of ionophore- and inhibitor-mediated plasma and mitochondrial membrane potential changes in cultured neurons. J Biol Chem 281 (21), 14864–74. [DOI] [PubMed] [Google Scholar]