Abstract
Maternally Expressed Gene 3 (MEG3) is a long noncoding RNA (lncRNA) that coordinates a diverse array of cellular processes by acting as a competitive endogenous (ce)RNA, epigenetic regulation, and interactions with key proteins such as p53. MEG3 expression is affected by epigenetic modifications driven by in utero nutrition and is involved the development of many metabolic dysfunctions. Here, we suggest that these bodies of research are connected and that epigenetic modification of MEG3 expression may assist in adaptation to different metabolic environments. To this end, we discuss how nutritional status either leads to an increase of MEG3 expression that protects against cancer and metabolic dysfunctions, or to its downregulation minimizing its pleiotropic costs of expression. Lastly, we identify research directions that would further shed light on the role of MEG3 within metabolism and its role with larger imprinted gene networks.
Keywords: MEG3, epigenetic imprinting, metabolic programming, miRNA sponge
1. Introduction
Metabolic syndrome, the presentation of a collection of metabolic disorders including insulin resistance and obesity, is a global epidemic that burdens an estimated billion people worldwide [1,2]. Given that metabolic syndrome contributes to the pathogenesis of chronic diseases such as cardiovascular disease and diabetes, it is critical that we identify the factors that underlie its development [1,3]. Recent research has emphasized the impact of in utero exposure to different stressors (e.g., starvation, toxins, or hyperglycemia) leading to lifelong susceptibility to metabolic syndrome through epigenetic modifications [4–7]. However, the epigenetic mechanisms through which the intrauterine environment may predispose infants to metabolic syndrome remain unclear [5]. Nevertheless, epidemiological studies connecting the lifestyles of pregnant mothers to epigenetic alterations in their infants have identified promising candidate genes. The lncRNA Maternally Expressed Gene 3 (MEG3) is noteworthy because its expression is both modulated by the uterine environment and dysregulated in many diseases of metabolic dysfunction [8,9]. Despite this connection, whether epigenetic modification of MEG3 expression participates in metabolic programming remains unexplored. Here, we describe how changes in MEG3 expression may assist in adaptation to the distinct challenges of energy rich and poor environments. In addition, we suggest research directions that would further describe MEG3’s role in metabolism.
2. Location, inheritance, and regulation of MEG3
The advent of next-generation sequencing data has revealed that lncRNAs act as ubiquitous coordinators of diverse biological processes. In this regard, the maternally imprinted lncRNA MEG3 is typical, participating in processes as distinct as cancer, diabetes, and tissue fibrosis [10–12]. MEG3 is located within the DLK1-DIO3 gene cluster, a locus rich in imprinted genes, at human chromosome 14q32.3 [13–15]. The “imprintedness” of this region is the result of the different epigenetic programs that occur on the maternal and paternal chromosomes [15]. On the paternal chromosome, the imprinting control center for the region, the MEG3 intergenetic differentially methylated region (IG-DMR), becomes hypermethylated during spermatogenesis [15]. Conversely, the maternal copy of the MEG3 IG-DMR becomes hypomethylated during oogenesis [15]. Post-fertilization, the methylation patterns at the maternal MEG3 IG-DMR locus are imprinted onto the maternally inherited MEG3 IG-DMR locus of the embryo [16]. As the imprinting control center of the locus, the MEG3 IG-DMR controls expression of MEG3 and the other imprinted genes within the locus through methylation of secondary DMRs. One of the secondary DMRs methylated by the MEG3 IG-DMR is the MEG3 DMR, located on MEG3’s upstream promoter [17]. Although poorly understood, the mechanism by which methylation at the MEG3 DMR regulates MEG3 expression may involve interference in the binding of enhancers to the promoter region [18]. Because both the MEG3 IG-DMR and MEG3 DMR stay hypermethylated on the paternal copy, while remaining comparatively hypomethylated on the maternal copy, MEG3 is maternally expressed [15].
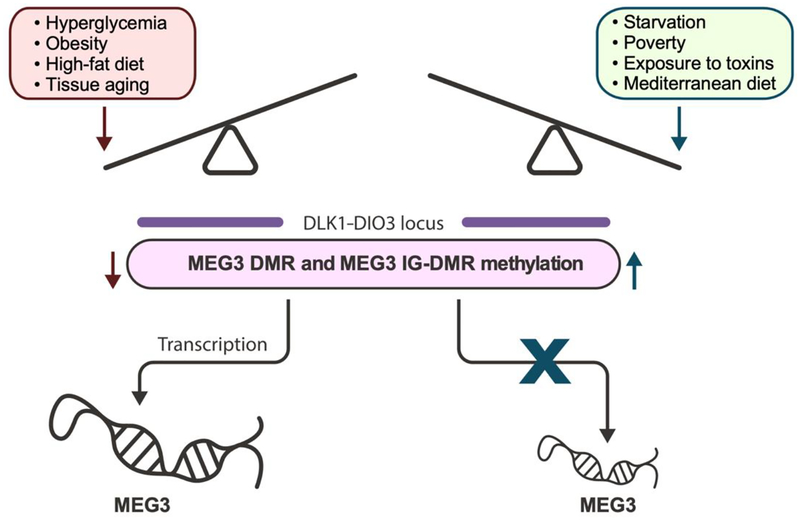
During gestation and embryonic development, maternal nutrition as well as environmental and emotional stressors modulate methylation at the maternal MEG3 IG-DMR and MEG3 DMR. Methylation at the MEG3 IG-DMR and MEG3 DMR is highly sensitive to the uterine environment, so much so that MEG3 expression is highly differential even between identical twins [19]. In response to starvation, consumption of a Mediterranean diet, environmental toxins, or stressors such as maternal depression, these DMRs are hypermethylated [8,20–24]. Conversely, gestational diabetes mellitus, maternal obesity, or a high-fat diet cause them to become hypomethylated [8,25–27]. These methylation changes regulate MEG3 expression from lower to higher levels, respectively. They can also be transmitted intergenerationally, affecting second generation as well as first generation offspring [16]. Interestingly, prenatal exercise improves the metabolic health of pups and is associated with changes in the transcriptome; however, while epigenetic modifications of MEG3 are associated with both muscle development and exercise, there are no MEG3 DMR methylation changes associated with prenatal exercise [28–31]. To conclude, MEG3 is maternally expressed owing to the methylation of the paternal MEG3 IG-DMR. Malnutrition and stress induce epigenetic alterations that reduce MEG3 expression, while overnutrition increases MEG3 expression via promoter hypomethylation (Figure 1).
Figure 1:
Epigenetic regulation of MEG3 expression. Overnutrition and obesity increase MEG3 expression by hypomethylating the MEG3 IG-DMR and MEG3 DMR whereas starvation and other stressors contribute to the decrease in MEG3 expression via hypermethylation. MEG3 IGDMR methylation in most tissues decreases with age, with the notable exception of skeletal muscle [32–35]. Consumption of a Mediterranean diet has been associated with lower MEG3 expression.
3. MEG3-driven metabolic adaptation
The fetal origins hypothesis suggests that epigenetic changes in utero affect susceptibility to future disease [36]. However, whether epigenetic modification of MEG3 expression is adaptive or maladaptive has not been evaluated. Published data suggests that elevated MEG3 induced by in utero hyperglycemia may actually be beneficial, protecting against the development of metabolic diseases such as diabetes [37]. For example, heightened MEG3 expression impedes the development of diabetes by maintaining β-cell mass in the pancreas. Within β-cells, MEG3 is one of the most enriched genes and is essential for insulin production and cell maintenance [38,39]. MEG3 knockdown reduces insulin synthesis and promotes β-cell apoptosis, leading to impaired glucose tolerance and diminished insulin secretion [37]. Mechanistically, this diminishment is the result of reduced MafA expression, a key stimulator of insulin production and a required gene in β-cell maintenance [40–43]. MEG3 knockdown also increases expression of IGF2, which sensitizes β-cells to oxidative damage, inflammation, and is associated with weight gain at middle age [44,45]. Further linking MEG3 expression to diabetes, healthy individuals have higher MEG3 levels in their β-cells, blood serum, and retinal epithelium compared to diabetic individuals [46,47]. While the low levels of MEG3 expression presented by diabetic individuals could be considered solely a symptom of their condition, genome wide association studies, studies of imprinting disorders, and molecular examination of diabetic β-islets suggests MEG3 dysfunction plays a causal role. Susceptibility to both type 1 and 2 diabetes are associated with MEG3 polymorphisms, providing a link between altered MEG3 function and risk of diabetes [48–50]. Further, dysregulated MEG3 expression in the imprinting disorders Temple Syndrome, Silver-Russell Syndrome, and Prader William’s Syndrome explains similarities between the conditions, a clinical overlap that includes obesity, insulin resistance, and other metabolic dysfunctions [51]. Lastly, in diabetic pancreatic islets, MEG3 is not regulated by glucose levels [46]. This suggests that the reduced MEG3 levels in diabetic patients preceded the chronic hyperglycemia caused by their condition [46]. To summarize, MEG3 maintains β-islet function to protect against diabetes.
In addition to preserving β-islet function, MEG3 shields epithelial tissue from hyperglycemia induced inflammation, apoptosis, and oxidative stress. Given that epithelial dysfunctions such as retinopathy and atherosclerosis are closely associated with insulin resistance and obesity, this may represent an additional mechanism through which increased MEG3 expression is adaptive in calorie-rich environments [52,53]. As an example, in diabetic retinopathy MEG3 alleviates the endothelial dysfunction caused by chronic hyperglycemia [54]. MEG3 serum levels in diabetic individuals with diabetic retinopathy are reduced compared to diabetic persons without diabetic retinopathy and healthy controls [47]. MEG3 protects against diabetic retinopathy—and endothelial dysfunction generally—through numerous, distinct pathways. These include: i) MEG3-dependent inhibition of the NF-κB pathway, hence reducing inflammation and apoptosis caused by hyperglycemia [55]; ii) downregulation of VEGF and TGF-β1, two critical genes involved in diabetic retinopathy [47]; iii) dampening of inflammation-induced p53 activation, a condition associated with obesity and diabetes, which, in turn, protects endothelial tissue from DNA damage and apoptosis [56]; iv) MEG3-dependent reduction in radical oxygen species production and lipoprotein oxidation in endothelial cells [57]; and v) protection of endothelial progenitor cells through upregulated HDAC7 expression and decreased miR-140–5p expression [58]. In short, MEG3 helps maintain epithelial function under hyperglycemic stress, preventing the development of diabetic retinopathy and endothelial dysfunction in metabolic syndrome.
Further, MEG3 protects against non-alcoholic fatty liver disease, a common manifestation of metabolic syndrome [1,59]. In metabolic syndrome, visceral fat and hyperglycemia induce liver damage via chronic inflammation and ROS production [60–62]. This causes hepatic stellate cells to produce excessive extracellular matrix, leading to liver fibrosis [62,63]. However, MEG3-mediated upregulation of p53, smoothened protein encoded by the SMO gene, and miR-212 can dampen this maladaptive activation of hepatic stellate cells by means of apoptosis [64]. Additionally, MEG3 is progressively downregulated during liver fibrosis progression and is diminished in fibrotic livers compared to healthy controls. Together, this suggests that MEG3 has a role in curbing liver fibrosis, as seen in non-alcoholic fatty liver disease [65,66].
However, the narrative that upregulated MEG3 expression is purely protective against metabolic syndrome is muddled by the fact that MEG3 itself contributes to insulin resistance by stimulating hepatic gluconeogenesis. In the liver, MEG3 is regulated dynamically by glucose and is increased in mice fed a high-fat diet, palmitate, oleate, or linoleate, and in ob/ob mice [9,37]. This increase in MEG3 expression is caused by changes in histone acetylation driven by high-fat-mediated inhibition of HDAC1 and HDAC3 [9]. MEG3 production is also stimulated by glucagon, a key promoter of gluconeogenesis, which induces hepatic MEG3 expression by increasing the levels of CREB, which bind to a CREB response element on its promoter [67]. Subsequently, MEG3 induces gluconeogenesis by upregulating FOXO1, CRTC2, G6PC and PEPCK, all drivers of hepatic gluconeogenesis [9,67]. MEG3 further drives gluconeogenesis by competitively binding to micro (mi)RNAs that otherwise limit gluconeogenesis, such as miR-214 and miR-302a-3p [67,68]. MEG3 upregulation is sufficient to exacerbate hyperglycemic dysfunctions in the liver while its hindered expression attenuates metabolic distortions (e.g., downregulation of glycogen content and upregulation of triglyceride content) in mice fed a high-fat diet [9,67]. Lastly, transient MEG3 overexpression causes cholestatic liver injury by facilitating SHP mRNA decay, resulting in the disruption of bile acid and liver enzyme homeostasis [69]. To conclude, MEG3 may contribute to metabolic syndrome by impairing hepatic insulin sensitivity and altering metabolism in the liver. Why MEG3 promotes gluconeogenesis while protecting against its harmful effects across different organs is an unresolved question.
4. Role of MEG3 in cancer prevention
In addition to its role in metabolism, MEG3 functions as a potent tumor-suppressor, an advantageous quality in cancer-prone, energy-rich environments. To illustrate how energetic excess contributes to cancer development, excess fat is thought to cause as many as 20% of all cancer cases while moderate caloric restriction in adult monkeys reduces cancer risk by 50% [70,71]. MEG3 limits cancer progression through several independent mechanisms, including the upregulation of the canonical tumor suppressor p53, the repression of angiogenesis, and autophagy suppression. On account of this, MEG3 knockdown or downregulation is observed in 25% of neuroblastomas, 81% of hepatocellular cancers, 82% of gliomas, and is associated with poor prognosis [14,72–81].
MEG3 halts cancer progression, in part, by upregulating the canonical tumor suppressor p53. MEG3 induces p53 accumulation by suppressing MDM2, a protein which tags p53 for degradation [82]. In addition, MEG3 directly interacts with p53 to facilitate the activation of downstream targets implicated in the reduction in autophagy, promotion of apoptosis, and cell cycle arrest [76,82–85].
MEG3 also restricts cancer growth by disrupting the PI3K/AKT signaling cascade. Excessive activation of the PI3K pathway is common across cancers and contributes to tumorigenesis by encouraging cell survival and division [86]. In gliomas, MEG3 stops the translocation of AKT to the plasma membrane by downregulating miR-93 [87], thus dampening the activation of downstream target genes. MEG3 further inactivates the PI3K/AKT pathway by upregulating PTEN, which buffers PI3K signaling [88].
Another mechanism through which MEG3 arrests cancer development is by acting as a competitive endogenous RNA (ceRNA) to “sponge” tumorigenic miRNAs. In gastric cancer, MEG3 promotes apoptosis by upregulating Bcl2 through sponging miR-181 [10] while the loss in proliferation of glioma results from the sponging of miR-19a by MEG3 [89]. By competitively binding to miR-21–5p, MEG3 suppresses proliferation and promotes apoptosis of cervical cancer cells [90] while sensitizing non-small cell lung cancer to cisplatin [76,91]. In pancreatic cancer, MEG3 directly targets miR-183 to downregulate p38/ERK/AKT and Wnt/β-catenin signaling, resulting in reduced growth and metastasis [92]. Overall, MEG3 offers anti-cancer protection through interaction with an expansive array of miRNAs, with more interactions surely still undiscovered.
Lastly, MEG3 limits cancer growth by suppressing angiogenesis, a process that is required for the robust metabolic needs associated with tumor development and metastasis [93–95]. In mouse MEG3-null embryos, there is an upregulation of key angiogenic genes in the VEGF pathway, such as VEGFA and VEGRF1, leading to increased microvessel formation [96]. Further, in human umbilical cord, MEG3 knockdown accelerates VEGF-mediated angiogenesis through VEGFR2 upregulation [97]. MEG3 interference stimulates epithelial cell division, sprouting, and tube formation through activation of Notch signaling, which ultimately leads to blood flow improvement [98] and enhanced tumor growth [99]. This may explain why MEG3 levels are reduced in cancer exosomes, which are small secreted vesicles that facilitate intercellular signaling [100,101].
To conclude, MEG3 limits cancer progression by restricting angiogenesis, sponging tumorigenic miRNAs, and interfering in canonical cancer pathways. As calorie-rich environments are tumorigenic, the epigenetic upregulation of MEG3 by caloric excess may be adaptive.
5. Pleiotropic costs of MEG3 expression
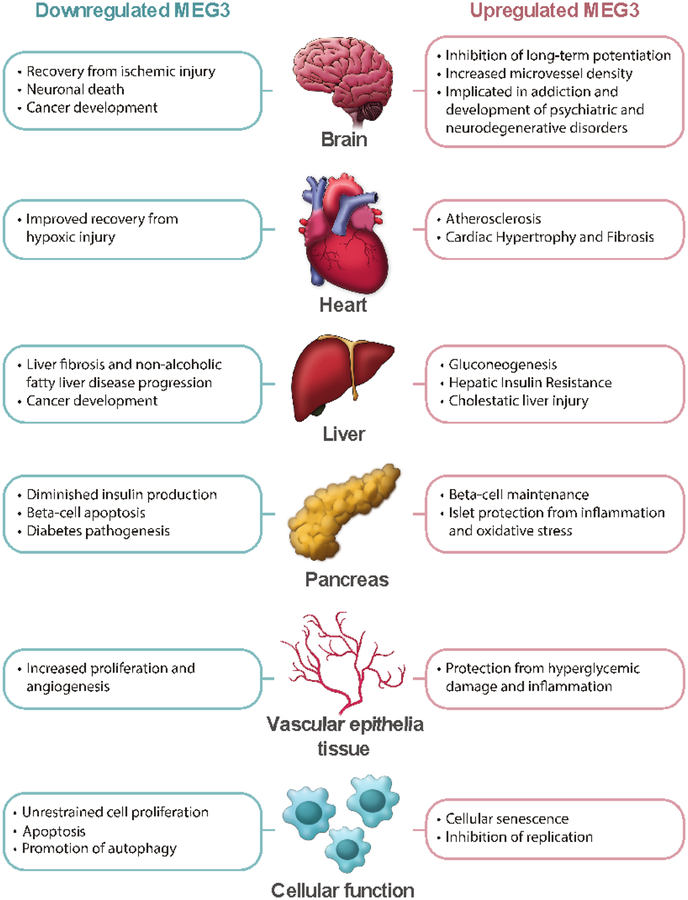
However, increased MEG3 expression also reveals pleiotropic costs, tradeoffs which may hurt more than help in the absence of a nutrient-rich environment. One of these costs is accelerated cellular senescence, a state of permanent growth arrest and altered gene expression associated with tissue dysfunction, cancer, and aging [102]. Through interactions with p53, p21, PI3K/AKT and mTOR, MEG3 potently fights cancer, but can also induce senescence [74,82,103,104]. Expression of recombinant MEG3 gene is sufficient to trigger replicative senescence in cervical cancer lines [103]. Overexpression of MEG3 has been reported in senescent HUVECs, aged liver and muscle, mesenchymal stromal cells, and cardiovascular atrial tissue, thus connecting MEG3 to aging [33,34,105,106]. In aged HUVECS, MEG3 silencing rescues aging-associated changes in sprouting angiogenic activity [107]. In fact, 69% of upregulated mRNAs present in late-passage human adipose-derived stem cells vs. early-passage cells map to the DLK1-DIO3 locus, making the DLK1-DIO3 locus a putative hotspot in aging tissue [35] (Figure 2).
Figure 2:
Schematic representation of the pleiotropic effects of MEG3 expression in various organs and tissues.
In addition to induce cell senescence, MEG3 exacerbates damage from injury by promoting cell death and inhibiting cell growth and angiogenesis. MEG3 has been linked to p53-mediated neuronal cell death following ischemic stroke [108]. Further, inhibition of PI3K/AKT by MEG3 exacerbates neuronal cell injury following subarachnoid hemorrhage [109] and, conversely, MEG3 silencing improves blood flow to tissues following ischemic injury [107]. MEG3 knockdown not only protects PC12 cells against hypoxic injury [110], but it also shields cardiomyocyte-derived H9c2 cells from hypoxic damage by sponging miRNAs that activate the PI3K/AKT/FOXO3a pathway [111]. Similarly, MEG3 silencing accelerates tibia fraction healing by upregulating the Wnt/β-catenin pathway [112]. In summary, increased MEG3 expression reduces the ability for the body to recover from a variety of injuries (Figure 2).
Elevated MEG3 expression may also contribute to the development of neurological disorders. MEG3 is involved in brain development, with its isoforms dynamically expressed across developing regions and its expression levels decreasing as neurons differentiate after birth [113,114]. Within neurons, MEG3 promotes apoptosis, directs cell fate, and limits long-term potentiation by inhibiting PI3K/AKT signaling [96,115–119]. Additionally, MEG3 may affect neural function by regulating angiogenesis within the brain [96]. Epidemiological studies have connected hypomethylation of the MEG3 IG-DMR with childhood maladaptive behavior and lesser social relatability [120,121]. Elevated MEG3 is also implicated in addiction, being upregulated in the nucleus accumbens of heroin users and in mice given methamphetamine [122,123]. However, this correlation may be the result of drug-induced elevation in cAMP levels and subsequent cAMP-response element-mediated MEG3 transcription, instead of MEG3 having a causal role [68,124]. In addition, MEG3 expression is increased in both human and animal models of Huntington’s disease, with its knockdown resulting in a decrease in aggregates caused by mutant huntingtin. Further, aberrant expression of miRNAs, copy number variants, and coding variants from the DLK1-DIO3 locus are correlated with schizophrenia, anxiety, and psychosis [125–129]. Overall, while the mechanisms through which MEG3 affects brain function are still mostly uncharacterized, it appears that elevated expression may result in a variety of dysfunctions (Figure 2).
Additionally, elevated MEG3 expression contributes to cardiac fibrosis. In response to pressure overload, cardiac fibroblasts secrete matrix metalloproteinases that remodel cardiac tissue, leading to heart stiffness, diastolic dysfunction, and heart fibrosis [130]. In cardiac fibroblasts, MEG3 contributes to matrix metalloproteinase-2 (MMP2) signaling by assisting p53 and the fibrotic cytokine TGF-β. Through interaction with MMP2, MEG3 participates in fibrosis progression; conversely, silencing of MEG3 reduces fibrosis and prevents diastolic dysfunction in mouse cardiac tissue following transverse aortic constriction surgery [12,130]. In addition to fibrosis, MEG3 also contributes to heart hypertrophy, a condition where cardiomyocytes are enlarged and heart fibroblasts are overactive, causing an increase in heart rigidity and susceptibility to adverse cardiovascular events [12]. In models of cardiac hypertrophy, MEG3 induces maladaptive cardiomyocyte growth by competitive binding to miR-361–3p, which normally inhibits the pro-hypertrophic factor HDAC9. Silencing MEG3 is sufficient to restore miR-361–3p/HDAC9 axis function, partially reversing the excessive growth of cardiomyocytes that causes cardiac hypertrophy [131]. To conclude, increased MEG3 expression comes at the cost of heart fibrosis and dysfunction (Figure 2).
6. Conclusions and Perspectives
Even as investigation into MEG3 has helped describe its role within metabolism, there remain unanswered questions. One of these is whether MEG3 affects metabolism via the thyroid. The thyroid regulates metabolism by releasing hormones that control basal metabolic rate, appetite, and the breakdown of fat [132–134]. Perhaps not coincidentally, MEG3 is expressed at its highest level in the thyroid compared to all other organs [125]. Further, imprinting disorders of the DLK1-DIO3 locus all share pathologies typical of thyroid dysfunction, including obesity, appetite dysregulation, growth retardation, and disrupted timing of puberty [133,135–138]. All of this would suggest that MEG3 affects metabolism by regulating thyroid hormones. However, investigation thus far into MEG3’s role in the thyroid function has been limited to its contribution to cancer. Thus, experiments probing the relationship between MEG3 and thyroid hormones may reveal an exciting new avenue through which MEG3 may affect metabolism. Further, it is worth investigating how MEG3 coordinates with other imprinted genes within the DLK1-DIO3 region and with other imprinted loci. Because the DLK1-DIO3 locus is regulated as a cohesive unit via the MEG3 IG-DMR, the effects of MEG3 are inherently tethered to that of the other genes within the locus such as DLK1, which also affects insulin resistance and glucose metabolism [139,140]. Further, MEG3 controls expression of other imprinted genes that affect metabolism and growth, such as IGF2, SNURF, and IPW [135,141]. With this in mind, we hypothesize that MEG3 may coordinate with a broader imprinted gene network that affects metabolic imprinting collectively. Studying the function and regulation of other genes within the DLK1-DIO3 locus and within other imprinted regions under different metabolic challenges would help evaluate this hypothesis. Lastly, while the literature suggests that the modulation of MEG3 expression may help mammals adapt to hyperglycemic challenge, experimental validation is necessary. In vivo studies investigating how MEG3 knockdown or overexpression affects the development of disease in animals given a high-fat diet or under caloric restriction would be the most direct approach. While this is complicated by the fact that MEG3 is unique to mammals and that maternal MEG3-knockout causes perinatal death in mice, inducible knockout or transgenic models may provide a solution [142].
Overall, research suggests that MEG3 is involved in mammalian adaptation to the metabolic environment through interactions with p53, PI3K/AKT, and miRNA networks. In an energy abundant environment, elevated MEG3 levels generally protect against many of the most common afflictions associated with hyperglycemia. However, MEG3 expression also carries many pleiotropic costs such as cellular senescence, impaired injury recovery, heart fibrosis, and neurological impairments. These pleiotropic costs may suggest why MEG3 expression is regulated by nutrient availability. In nutrient-rich environments, MEG3-driven prevention of metabolic syndrome and cancer outweighs its costs. However, in nutrient-poor environments, high levels of MEG3 expression may do more harm than good. The cost of a maladapted epigenome is best illustrated by the differing fates of the children exposed to prenatal starvation during The Dutch Hunger Winter compared to The Siege of Leningrad. During The Dutch Hunger Winter, those prepared in utero for scarcity then raised on a normal diet developed cancer and cardiovascular disease at an elevated rate [7]. By comparison, children born during The Siege of Leningrad were starved in utero and endured prolonged starvation, thus conferring protection against these risks [7]. Given that the developing world is undergoing a comparable transition from food scarcity to excess [143], how developmental nutrition may predispose individuals to chronic disease remains a pressing yet unanswered question.
Highlights.
MEG3 regulates gene expression through interaction with DNA, RNA and/or proteins.
MEG3 regulates gene expression by sponging miRNAs.
Dysregulation of MEG3 found in several pathophysiological conditions and in aging.
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute on Aging, National Institutes of Health, in Baltimore, Maryland, USA. We wish to thank Marc Raley, Visual Media Specialist, for creating both figures.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest Disclosure:
The authors have no conflicts of interests to declare
References
- [1].Eckel RH, Grundy SM, Zimmet PZ, The metabolic syndrome, Lancet 365 (2005) 1415–1428. doi: 10.1016/S0140-6736(05)66378-7. [DOI] [PubMed] [Google Scholar]
- [2].Saklayen MG, The Global Epidemic of the Metabolic Syndrome, Curr. Hypertens. Rep 20 (2018) 12. doi: 10.1007/s11906-018-0812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Isomaa B, Almgren P, Tuomi T, Forsén B, Lahti K, Nissén M, Taskinen MR, Groop L, Cardiovascular morbidity and mortality associated with the metabolic syndrome, Diabetes Care 24 (2001) 683–689. https://www.ncbi.nlm.nih.gov/pubmed/11315831. [DOI] [PubMed] [Google Scholar]
- [4].Zimmet P, Shi Z, El-Osta A, Ji L, Epidemic T2DM, early development and epigenetics: implications of the Chinese Famine, Nat. Rev. Endocrinol 14 (2018) 738–746. doi: 10.1038/s41574-018-0106-1. [DOI] [PubMed] [Google Scholar]
- [5].Carson C, Lawson HA, Epigenetics of metabolic syndrome, Physiol. Genomics 50 (2018) 947–955. doi: 10.1152/physiolgenomics.00072.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].De Rooij SR, Painter RC, Phillips DIW, Impaired insulin secretion after prenatal exposure to the Dutch famine, Diabetes Care 29 (2006) 1897–1901. [DOI] [PubMed] [Google Scholar]
- [7].Schulz LC, The Dutch Hunger Winter and the developmental origins of health and disease, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 16757–16758. doi: 10.1073/pnas.1012911107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gonzalez-Nahm S, Mendez M, Robinson W, Murphy SK, Hoyo C, Hogan V, Rowley D, Low maternal adherence to a Mediterranean diet is associated with increase in methylation at the MEG3-IG differentially methylated region in female infants, Environ Epigenet 3 (2017). doi: 10.1093/eep/dvx007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhu X, Wu Y-B, Zhou J, Kang D-M, Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via increasing FoxO1 expression, Biochem. Biophys. Res. Commun 469 (2016) 319–325. doi: 10.1016/j.bbrc.2015.11.048. [DOI] [PubMed] [Google Scholar]
- [10].Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, Yu J, Ma R, Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression, J. Exp. Clin. Cancer Res 34 (2015) 79. doi: 10.1186/s13046-015-0197-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Kameswaran V, Golson ML, Ramos-Rodríguez M, Ou K, Wang YJ, Zhang J, Pasquali L, Kaestner KH, The Dysregulation of the DLK1-MEG3 Locus in Islets From Patients With Type 2 Diabetes Is Mimicked by Targeted Epimutation of Its Promoter With TALE-DNMT Constructs, Diabetes 67 (2018) 1807–1815. doi: 10.2337/db17-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Uchida S, Besides Imprinting: Meg3 Regulates Cardiac Remodeling in Cardiac Hypertrophy, Circ. Res 121 (2017) 486–487. doi: 10.1161/CIRCRESAHA.117.311542. [DOI] [PubMed] [Google Scholar]
- [13].Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F, Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q, Genes Cells 5 (2000) 211–220. [DOI] [PubMed] [Google Scholar]
- [14].Zhou Y, Zhang X, Klibanski A, MEG3 noncoding RNA: a tumor suppressor, J. Mol. Endocrinol 48 (2012) R45–53. doi: 10.1530/JME-12-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].da Rocha ST, Edwards CA, Ito M, Ogata T, Ferguson-Smith AC, Genomic imprinting at the mammalian Dlk1-Dio3 domain, Trends Genet 24 (2008) 306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- [16].Jiang Y, Yu Y-C, Ding G-L, Gao Q, Chen F, Luo Q, Intrauterine hyperglycemia induces intergenerational Dlk1-Gtl2 methylation changes in mouse placenta, Oncotarget 9 (2018) 22398–22405. doi: 10.18632/oncotarget.23976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sato S, Yoshida W, Soejima H, Nakabayashi K, Hata K, Methylation dynamics of IG-DMR and Gtl2-DMR during murine embryonic and placental development, Genomics 98 (2011) 120–127. doi: 10.1016/j.ygeno.2011.05.003. [DOI] [PubMed] [Google Scholar]
- [18].Moradi M-T, Fallahi H, Rahimi Z, Interaction of long noncoding RNA MEG3 with miRNAs: A reciprocal regulation, J. Cell. Biochem 120 (2019) 3339–3352. doi: 10.1002/jcb.27604. [DOI] [PubMed] [Google Scholar]
- [19].Gordon L, Joo J-HE, Andronikos R, Ollikainen M, Wallace EM, Umstad MP, Permezel M, Oshlack A, Morley R, Carlin JB, Saffery R, Smyth GK, Craig JM, Expression discordance of monozygotic twins at birth: effect of intrauterine environment and a possible mechanism for fetal programming, Epigenetics 6 (2011) 579–592. [DOI] [PubMed] [Google Scholar]
- [20].Liu Y, Murphy SK, Murtha AP, Fuemmeler BF, Schildkraut J, Huang Z, Overcash F, Kurtzberg J, Jirtle R, Iversen ES, Forman MR, Hoyo C, Depression in pregnancy, infant birth weight and DNA methylation of imprint regulatory elements, Epigenetics 7 (2012) 735–746. doi: 10.4161/epi.20734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Nye MD, King KE, Darrah TH, Maguire R, Jima DD, Huang Z, Mendez MA, Fry RC, Jirtle RL, Murphy SK, Hoyo C, Maternal blood lead concentrations, DNA methylation of MEG3 DMR regulating the DLK1/MEG3 imprinted domain and early growth in a multiethnic cohort, Environ. Epigenet 2 (2016) dvv009. doi: 10.1093/eep/dvv009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Vidal AC, Benjamin Neelon SE, Liu Y, Tuli AM, Fuemmeler BF, Hoyo C, Murtha AP, Huang Z, Schildkraut J, Overcash F, Kurtzberg J, Jirtle RL, Iversen ES, Murphy SK, Maternal stress, preterm birth, and DNA methylation at imprint regulatory sequences in humans, Genet. Epigenet 6 (2014) 37–44. doi: 10.4137/GEG.S18067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Tobi EW, Lumey LH, Talens RP, Kremer D, Putter H, Stein AD, Slagboom PE, Heijmans BT, DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific, Hum. Mol. Genet 18 (2009) 4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Preston JD, Reynolds LJ, Pearson KJ, Developmental Origins of Health Span and Life Span: A Mini-Review, Gerontology 64 (2018) 237–245. doi: 10.1159/000485506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].dos S. Albuquerque D, Study of genetic variants associated with obesity in Portuguese children, eg.uc.pt, 2015. https://eg.uc.pt/handle/10316/28249.
- [26].Soubry A, Murphy SK, Wang F, Huang Z, Vidal AC, Fuemmeler BF, Kurtzberg J, Murtha A, Jirtle RL, Schildkraut JM, Hoyo C, Newborns of obese parents have altered DNA methylation patterns at imprinted genes, Int. J. Obes 39 (2013) 650. doi: 10.1038/ijo.2013.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].El Hajj N, Pliushch G, Schneider E, Dittrich M, Müller T, Korenkov M, Aretz M, Zechner U, Lehnen H, Haaf T, Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus, Diabetes 62 (2013) 1320–1328. doi: 10.2337/db12-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Carter LG, Lewis KN, Wilkerson DC, Tobia CM, Ngo Tenlep SY, Shridas P, Garcia-Cazarin ML, Wolff G, Andrade FH, Charnigo RJ, Esser KA, Egan JM, de Cabo R, Pearson KJ, Perinatal exercise improves glucose homeostasis in adult offspring, Am. J. Physiol. Endocrinol. Metab 303 (2012) E1061–8. doi: 10.1152/ajpendo.00213.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Archer T, Epigenetic changes induced by exercise, J. Reward Defic. Syndr 1 (2015) 71–74. [Google Scholar]
- [30].McCullough LE, Mendez MA, Miller EE, Murtha AP, Murphy SK, Hoyo C, Associations between prenatal physical activity, birth weight, and DNA methylation at genomically imprinted domains in a multiethnic newborn cohort, Epigenetics 10 (2015) 597–606. doi: 10.1080/15592294.2015.1045181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Carter LG, Qi NR, De Cabo R, Pearson KJ, Maternal exercise improves insulin sensitivity in mature rat offspring, Med. Sci. Sports Exerc 45 (2013) 832–840. doi: 10.1249/MSS.0b013e31827de953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Mikovic J, Sadler K, Butchart L, Voisin S, Gerlinger-Romero F, Della Gatta P, Grounds MD, Lamon S, MicroRNA and Long Non-coding RNA Regulation in Skeletal Muscle From Growth to Old Age Shows Striking Dysregulation of the Callipyge Locus, Front. Genet 9 (2018) 548. doi: 10.3389/fgene.2018.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Xing W, Gao W, Mao G, Zhang J, Lv X, Wang G, Yan J, Long non-coding RNA s in aging organs and tissues, Clinical and Experimental Pharmacology and Physiology 44 (2017) 30–37. doi: 10.1111/1440-1681.12795. [DOI] [PubMed] [Google Scholar]
- [34].Wagner W, Bork S, Horn P, Krunic D, Walenda T, Diehlmann A, Benes V, Blake J, Huber F-X, Eckstein V, Boukamp P, Ho AD, Aging and replicative senescence have related effects on human stem and progenitor cells, PLoS One 4 (2009) e5846. doi: 10.1371/journal.pone.0005846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].García-López S, Albo-Castellanos C, Urdinguio RG, Cañón S, Sánchez-Cabo F, Martínez-Serrano A, Fraga MF, Bernad A, Deregulation of the imprinted DLK1-DIO3 locus ncRNAs is associated with replicative senescence of human adipose-derived stem cells, PLoS One 13 (2018) e0206534. doi: 10.1371/journal.pone.0206534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Almond D, Currie J, Killing Me Softly: The Fetal Origins Hypothesis, J. Econ. Perspect 25 (2011) 153–172. doi: 10.1257/jep.25.3.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].You L, Wang N, Yin D, Wang L, Jin F, Zhu Y, Yuan Q, De W, Downregulation of Long Noncoding RNA Meg3 Affects Insulin Synthesis and Secretion in Mouse Pancreatic Beta Cells, J. Cell. Physiol 231 (2016) 852–862. doi: 10.1002/jcp.25175. [DOI] [PubMed] [Google Scholar]
- [38].Segerstolpe Å, Palasantza A, Eliasson P, Andersson E-M, Andréasson A-C, Sun X, Picelli S, Sabirsh A, Clausen M, Bjursell MK, Smith DM, Kasper M, Ämmälä C, Sandberg R, Single-Cell Transcriptome Profiling of Human Pancreatic Islets in Health and Type 2 Diabetes, Cell Metab 24 (2016) 593–607. doi: 10.1016/j.cmet.2016.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Xin Y, Kim J, Okamoto H, Ni M, Wei Y, Adler C, Murphy AJ, Yancopoulos GD, Lin C, Gromada J, RNA Sequencing of Single Human Islet Cells Reveals Type 2 Diabetes Genes, Cell Metab 24 (2016) 608–615. doi: 10.1016/j.cmet.2016.08.018. [DOI] [PubMed] [Google Scholar]
- [40].Zhang C, Moriguchi T, Kajihara M, Esaki R, Harada A, Shimohata H, Oishi H, Hamada M, Morito N, Hasegawa K, Kudo T, Engel JD, Yamamoto M, Takahashi S, MafA is a key regulator of glucose-stimulated insulin secretion, Mol. Cell. Biol 25 (2005) 4969–4976. doi: 10.1128/MCB.25.12.4969-4976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wang N, Zhu Y, Xie M, Wang L, Jin F, Li Y, Yuan Q, De W, Long Noncoding RNA Meg3 Regulates Mafa Expression in Mouse Beta Cells by Inactivating Rad21, Smc3 or Sin3α, Cell. Physiol. Biochem 45 (2018) 2031–2043. doi: 10.1159/000487983. [DOI] [PubMed] [Google Scholar]
- [42].Zhao L, Guo M, Matsuoka T-A, Hagman DK, Parazzoli SD, Poitout V, Stein R, The islet beta cell-enriched MafA activator is a key regulator of insulin gene transcription, J. Biol. Chem 280 (2005) 11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]
- [43].Nishimura W, Takahashi S, Yasuda K, MafA is critical for maintenance of the mature beta cell phenotype in mice, Diabetologia 58 (2015) 566–574. doi: 10.1007/s00125-014-3464-9. [DOI] [PubMed] [Google Scholar]
- [44].O’Dell SD, Miller GJ, Cooper JA, Hindmarsh PC, Pringle PJ, Ford H, Humphries SE, Day INM, Apal polymorphism in insulin-like growth factor II (IGF2) gene and weight in middle-aged males, Int. J. Obes 21 (1997) 822. doi: 10.1038/sj.ijo.0800483. [DOI] [PubMed] [Google Scholar]
- [45].Casellas A, Mallol C, Salavert A, Jimenez V, Garcia M, Agudo J, Obach M, Haurigot V, Vilà L, Molas M, Lage R, Morró M, Casana E, Ruberte J, Bosch F, Insulin-like Growth Factor 2 Overexpression Induces β-Cell Dysfunction and Increases Beta-cell Susceptibility to Damage, J. Biol. Chem 290 (2015) 16772–16785. doi: 10.1074/jbc.M115.642041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kameswaran V, Bramswig NC, McKenna LB, Penn M, Schug J, Hand NJ, Chen Y, Choi I, Vourekas A, Won K-J, Liu C, Vivek K, Naji A, Friedman JR, Kaestner KH, Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets, Cell Metab 19 (2014) 135–145. doi: 10.1016/j.cmet.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhang D, Qin H, Leng Y, Li X, Zhang L, Bai D, Meng Y, Wang J, LncRNA MEG3 overexpression inhibits the development of diabetic retinopathy by regulating TGF-β1 and VEGF, Exp. Ther. Med 16 (2018) 2337–2342. doi: 10.3892/etm.2018.6451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Westra H-J, Martínez-Bonet M, Onengut-Gumuscu S, Lee A, Luo Y, Teslovich N, Worthington J, Martin J, Huizinga T, Klareskog L, Rantapaa-Dahlqvist S, Chen W-M, Quinlan A, Todd JA, Eyre S, Nigrovic PA, Gregersen PK, Rich SS, Raychaudhuri S, Fine-mapping and functional studies highlight potential causal variants for rheumatoid arthritis and type 1 diabetes, Nat. Genet 50 (2018) 1366–1374. doi: 10.1038/s41588-018-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, Clayton DG, The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes, Nat. Genet 42 (2009) 68. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ghaedi H, Zare A, Omrani MD, Doustimotlagh AH, Meshkani R, Alipoor S, Alipoor B, Genetic variants in long noncoding RNA H19 and MEG3 confer risk of type 2 diabetes in an Iranian population, Gene 675 (2018) 265–271. doi: 10.1016/j.gene.2018.07.002. [DOI] [PubMed] [Google Scholar]
- [51].Lokulo-Sodipe K, Ioannides Y, Davies JH, Karen TI, Growth characteristics in children with Temple syndrome: an under-diagnosed imprinting disorder, EJEA (2014). doi: 10.1530/endoabs.36.P66. [DOI] [Google Scholar]
- [52].Ridker PM, On evolutionary biology, inflammation, infection, and the causes of atherosclerosis, Circulation 105 (2002) 2–4. [PubMed] [Google Scholar]
- [53].Barrett-Connor EL, Obesity, atherosclerosis, and coronary artery disease, Ann. Intern. Med 103 (1985) 1010–1019. [DOI] [PubMed] [Google Scholar]
- [54].Fong DS, Aiello L, Gardner TW, King GL, Blankenship G, Cavallerano JD, Ferris FL 3rd, Klein R, American Diabetes Association, Retinopathy in diabetes, Diabetes Care 27 Suppl 1 (2004) S84–7. [DOI] [PubMed] [Google Scholar]
- [55].Tong P, Peng Q-H, Gu L-M, Xie W-W, Li W-J, LncRNA-MEG3 alleviates high glucose induced inflammation and apoptosis of retina epithelial cells via regulating miR-34a/SIRT1 axis, Exp. Mol. Pathol (2018). doi: 10.1016/j.yexmp.2018.12.003. [DOI] [PubMed] [Google Scholar]
- [56].Shihabudeen Haider Ali MS, Cheng X, Moran M, Haemmig S, Naldrett MJ, Alvarez S, Feinberg MW, Sun X, LncRNA Meg3 protects endothelial function by regulating the DNA damage response, Nucleic Acids Res (2018). doi: 10.1093/nar/gky1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Lan Y, Li Y-J, Li D-J, Li P, Wang J-Y, Diao Y-P, Ye G-D, Li Y-F, Long non-coding RNA MEG3 prevents vascular endothelial cell senescence by impairing miR-128-dependent Girdin down-regulation, Am. J. Physiol. Cell Physiol (2018). doi: 10.1152/ajpcell.00262.2018. [DOI] [PubMed] [Google Scholar]
- [58].Liu HZ, Wang QY, Zhang Y, Qi DT, Li MW, Guo WQ, Ma YH, Wang LY, Chen Y, Gao CY, Pioglitazone up-regulates long non-coding RNA MEG3 to protect endothelial progenitor cells via increasing HDAC7 expression in metabolic syndrome, Biomed. Pharmacother 78 (2016) 101–109. doi: 10.1016/j.biopha.2016.01.001. [DOI] [PubMed] [Google Scholar]
- [59].Wang X, Wang J, High-content hydrogen water-induced downregulation of miR-136 alleviates non-alcoholic fatty liver disease by regulating Nrf2 via targeting MEG3, Biol. Chem 399 (2018) 397–406. doi: 10.1515/hsz-2017-0303. [DOI] [PubMed] [Google Scholar]
- [60].Marchesini G, Brizi M, Bianchi G, Tomassetti S, Bugianesi E, Lenzi M, McCullough AJ, Natale S, Forlani G, Melchionda N, Nonalcoholic fatty liver disease: a feature of the metabolic syndrome, Diabetes 50 (2001) 1844–1850. [DOI] [PubMed] [Google Scholar]
- [61].Almeda-Valdés P, Cuevas-Ramos D, Aguilar-Salinas CA, Metabolic syndrome and non-alcoholic fatty liver disease, Ann. Hepatol 8 Suppl 1 (2009) S18–24. [PubMed] [Google Scholar]
- [62].Malaguarnera M, Di Rosa M, Nicoletti F, Malaguarnera L, Molecular mechanisms involved in NAFLD progression, J. Mol. Med 87 (2009) 679–695. doi: 10.1007/s00109-009-0464-1. [DOI] [PubMed] [Google Scholar]
- [63].Chiang DJ, Pritchard MT, Nagy LE, Obesity, diabetes mellitus, and liver fibrosis, Am. J. Physiol. Gastrointest. Liver Physiol 300 (2011) G697–702. doi: 10.1152/ajpgi.00426.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Yu F, Geng W, Dong P, Huang Z, Zheng J, LncRNA-MEG3 inhibits activation of hepatic stellate cells through SMO protein and miR-212, Cell Death Dis 9 (2018) 1014. doi: 10.1038/s41419-018-1068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].He Y, Wu Y-T, Huang C, Meng X-M, Ma T-T, Wu B-M, Xu F-Y, Zhang L, Lv X-W, Li J, Inhibitory effects of long noncoding RNA MEG3 on hepatic stellate cells activation and liver fibrogenesis, Biochim. Biophys. Acta 1842 (2014) 2204–2215. doi: 10.1016/j.bbadis.2014.08.015. [DOI] [PubMed] [Google Scholar]
- [66].Zhang Y, Luo G, Zhang Y, Zhang M, Zhou J, Gao W, Xuan X, Yang X, Yang D, Tian Z, Ni B, Tang J, Critical effects of long non-coding RNA on fibrosis diseases, Exp. Mol. Med 50 (2018) e428. doi: 10.1038/emm.2017.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zhu X, Li H, Wu Y, Zhou J, Yang G, Wang W, Kang D, Ye S, CREB-upregulated lncRNA MEG3 promotes hepatic gluconeogenesis by regulating miR-302a-3p-CRTC2 axis, J. Cell. Biochem (2018). doi: 10.1002/jcb.27706. [DOI] [PubMed] [Google Scholar]
- [68].Zhu X, Li H, Wu Y, Zhou J, Yang G, Wang W, lncRNA MEG3 promotes hepatic insulin resistance by serving as a competing endogenous RNA of miR-214 to regulate ATF4 expression, Int. J. Mol. Med 43 (2019) 345–357. doi: 10.3892/ijmm.2018.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhang L, Yang Z, Trottier J, Barbier O, Wang L, Long noncoding RNA MEG3 induces cholestatic liver injury by interaction with PTBP1 to facilitate shp mRNA decay, Hepatology 65 (2017) 604–615. doi: 10.1002/hep.28882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Longo VD, Fontana L, Calorie restriction and cancer prevention: metabolic and molecular mechanisms, Trends Pharmacol. Sci 31 (2010) 89–98. doi: 10.1016/j.tips.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Wolin KY, Carson K, Colditz GA, Obesity and cancer, Oncologist 15 (2010) 556–565. doi: 10.1634/theoncologist.2009-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zhao J, Dahle D, Zhou Y, Zhang X, Klibanski A, Hypermethylation of the promoter region is associated with the loss of MEG3 gene expression in human pituitary tumors, J. Clin. Endocrinol. Metab 90 (2005) 2179–2186. doi: 10.1210/jc.2004-1848. [DOI] [PubMed] [Google Scholar]
- [73].Zhang X, Gejman R, Mahta A, Zhong Y, Rice KA, Zhou Y, Cheunsuchon P, Louis DN, Klibanski A, Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression, Cancer Res 70 (2010) 2350–2358. doi: 10.1158/0008-5472.CAN-09-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Lu K-H, Li W, Liu X-H, Sun M, Zhang M-L, Wu W-Q, Xie W-P, Hou Y-Y, Long non-coding RNA MEG3 inhibits NSCLC cells proliferation and induces apoptosis by affecting p53 expression, BMC Cancer 13 (2013) 461. doi: 10.1186/1471-2407-13-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Zhang Y, Wu J, Jing H, Huang G, Sun Z, Xu S, Long noncoding RNA MEG3 inhibits breast cancer growth via upregulating endoplasmic reticulum stress and activating NF-κB and p53, J. Cell. Biochem (2018). doi: 10.1002/jcb.27982. [DOI] [PubMed] [Google Scholar]
- [76].Xia Y, He Z, Liu B, Wang P, Chen Y, Downregulation of Meg3 enhances cisplatin resistance of lung cancer cells through activation of the WNT/β-catenin signaling pathway, Mol. Med. Rep 12 (2015) 4530–4537. doi: 10.3892/mmr.2015.3897. [DOI] [PubMed] [Google Scholar]
- [77].Benetatos L, Vartholomatos G, Hatzimichael E, MEG3 imprinted gene contribution in tumorigenesis, Int. J. Cancer 129 (2011) 773–779. doi: 10.1002/ijc.26052. [DOI] [PubMed] [Google Scholar]
- [78].Tian Z-Z, Guo X-J, Zhao Y-M, Fang Y, Decreased expression of long non-coding RNA MEG3 acts as a potential predictor biomarker in progression and poor prognosis of osteosarcoma, Int. J. Clin. Exp. Pathol 8 (2015) 15138–15142. [PMC free article] [PubMed] [Google Scholar]
- [79].Zhuo H, Tang J, Lin Z, Jiang R, Zhang X, Ji J, Wang P, Sun B, The aberrant expression of MEG3 regulated by UHRF1 predicts the prognosis of hepatocellular carcinoma, Mol. Carcinog 55 (2016) 209–219. doi: 10.1002/mc.22270. [DOI] [PubMed] [Google Scholar]
- [80].Zhang J, Lin Z, Gao Y, Yao T, Downregulation of long noncoding RNA MEG3 is associated with poor prognosis and promoter hypermethylation in cervical cancer, J. Exp. Clin. Cancer Res 36 (2017) 5. doi: 10.1186/s13046-016-0472-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Jia L-F, Wei S-B, Gan Y-H, Guo Y, Gong K, Mitchelson K, Cheng J, Yu G-Y, Expression, regulation and roles of miR-26a and MEG3 in tongue squamous cell carcinoma, Int. J. Cancer 135 (2014) 2282–2293. doi: 10.1002/ijc.28667. [DOI] [PubMed] [Google Scholar]
- [82].Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A, Activation of p53 by MEG3 non-coding RNA, J. Biol. Chem 282 (2007) 24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- [83].Ying L, Huang Y, Chen H, Wang Y, Xia L, Chen Y, Liu Y, Qiu F, Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer, Mol. Biosyst 9 (2013) 407–411. doi: 10.1039/c2mb25386k. [DOI] [PubMed] [Google Scholar]
- [84].Fridman JS, Lowe SW, Control of apoptosis by p53, Oncogene 22 (2003) 9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- [85].Chen J, The Cell-Cycle Arrest and Apoptotic Functions of p53 in Tumor Initiation and Progression, Cold Spring Harb. Perspect. Med 6 (2016) a026104. doi: 10.1101/cshperspect.a026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, González-Barón M, PI3K/Akt signalling pathway and cancer, Cancer Treat. Rev 30 (2004) 193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- [87].Zhang L, Liang X, Li Y, Long non-coding RNA MEG3 inhibits cell growth of gliomas by targeting miR-93 and inactivating PI3K/AKT pathway, Oncol. Rep 38 (2017) 2408–2416. doi: 10.3892/or.2017.5871. [DOI] [PubMed] [Google Scholar]
- [88].Georgescu M-M, PTEN Tumor Suppressor Network in PI3K-Akt Pathway Control, Genes Cancer 1 (2010) 1170–1177. doi: 10.1177/1947601911407325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Qin N, Tong G-F, Sun L-W, Xu X-L, Long Noncoding RNA MEG3 Suppresses Glioma Cell Proliferation, Migration, and Invasion by Acting as a Competing Endogenous RNA of miR-19a, Oncol. Res 25 (2017) 1471–1478. doi: 10.3727/096504017X14886689179993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zhang J, Yao T, Wang Y, Yu J, Liu Y, Lin Z, Long noncoding RNA MEG3 is downregulated in cervical cancer and affects cell proliferation and apoptosis by regulating miR-21, Cancer Biol. Ther 17 (2016) 104–113. doi: 10.1080/15384047.2015.1108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Wang P, Chen D, Ma H, Li Y, LncRNA MEG3 enhances cisplatin sensitivity in non-small cell lung cancer by regulating miR-21–5p/SOX7 axis, Onco. Targets. Ther 10 (2017) 5137–5149. doi: 10.2147/OTT.S146423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Zhang Y-Y, Feng H-M, MEG3 Suppresses Human Pancreatic Neuroendocrine Tumor Cells Growth and Metastasis by Down-Regulation of Mir-183, Cell. Physiol. Biochem 44 (2017) 345–356. doi: 10.1159/000484906. [DOI] [PubMed] [Google Scholar]
- [93].Folkman J, Role of angiogenesis in tumor growth and metastasis, Semin. Oncol 29 (2002) 15–18. doi: 10.1053/sonc.2002.37263. [DOI] [PubMed] [Google Scholar]
- [94].Nishida N, Yano H, Nishida T, Kamura T, Kojiro M, Angiogenesis in cancer, Vasc. Health Risk Manag 2 (2006) 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Hanahan D, Weinberg RA, Hallmarks of cancer: the next generation, Cell 144 (2011) 646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- [96].Gordon FE, Nutt CL, Cheunsuchon P, Nakayama Y, Provencher KA, Rice KA, Zhou Y, Zhang X, Klibanski A, Increased expression of angiogenic genes in the brains of mouse meg3-null embryos, Endocrinology 151 (2010) 2443–2452. doi: 10.1210/en.2009-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Ruan W, Zhao F, Zhao S, Zhang L, Shi L, Pang T, Knockdown of long noncoding RNA MEG3 impairs VEGF-stimulated endothelial sprouting angiogenesis via modulating VEGFR2 expression in human umbilical vein endothelial cells, Gene 649 (2018) 32–39. doi: 10.1016/j.gene.2018.01.072. [DOI] [PubMed] [Google Scholar]
- [98].Liu J, Li Q, Zhang K-S, Hu B, Niu X, Zhou S-M, Li S-G, Luo Y-P, Wang Y, Deng Z-F, Downregulation of the Long Non-Coding RNA Meg3 Promotes Angiogenesis After Ischemic Brain Injury by Activating Notch Signaling, Mol. Neurobiol 54 (2017) 8179–8190. doi: 10.1007/s12035-016-0270-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Carmeliet P, Jain RK, Angiogenesis in cancer and other diseases, Nature 407 (2000) 249–257. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- [100].Zhang J, Liu S-C, Luo X-H, Tao G-X, Guan M, Yuan H, Hu D-K, Exosomal Long Noncoding RNA s are Differentially Expressed in the Cervicovaginal Lavage Samples of Cervical Cancer Patients, J. Clin. Lab. Anal 30 (2016) 1116–1121. doi: 10.1002/jcla.21990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Li X-B, Zhang Z-R, Schluesener HJ, Xu S-Q, Role of exosomes in immune regulation, J. Cell. Mol. Med 10 (2006) 364–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Hernandez-Segura A, Nehme J, Demaria M, Hallmarks of Cellular Senescence, Trends Cell Biol 28 (2018) 436–453. doi: 10.1016/j.tcb.2018.02.001. [DOI] [PubMed] [Google Scholar]
- [103].Abdelmohsen K, Gorospe M, Noncoding RNA control of cellular senescence, Wiley Interdiscip. Rev. RNA 6 (2015) 615–629. doi: 10.1002/wrna.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Xiong Y, Liu T, Wang S, Chi H, Chen C, Zheng J, Cyclophosphamide promotes the proliferation inhibition of mouse ovarian granulosa cells and premature ovarian failure by activating the lncRNA-Meg3-p53-p66Shc pathway, Gene 596 (2017) 1–8. doi: 10.1016/j.gene.2016.10.011. [DOI] [PubMed] [Google Scholar]
- [105].Xia X, Chen W, McDermott J, Han J-DJ, Molecular and phenotypic biomarkers of aging, F1000Res 6 (2017) 860. doi: 10.12688/f1000research.10692.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Fuschi P, Carrara M, Voellenkle C, Garcia-Manteiga JM, Righini P, Maimone B, Sangalli E, Villa F, Specchia C, Picozza M, Nano G, Gaetano C, Spinetti G, Puca AA, Magenta A, Martelli F, Central role of the p53 pathway in the noncoding-RNA response to oxidative stress, Aging. 9 (2017) 2559–2586. doi: 10.18632/aging.101341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Boon RA, Hofmann P, Michalik KM, Lozano-Vidal N, Berghäuser D, Fischer A, Knau A, Jaé N, Schürmann C, Dimmeler S, Long Noncoding RNA Meg3 Controls Endothelial Cell Aging and Function, J. Am. Coll. Cardiol 68 (2016) 2589–2591. doi: 10.1016/j.jacc.2016.09.949. [DOI] [PubMed] [Google Scholar]
- [108].Yan H, Yuan J, Gao L, Rao J, Hu J, Long noncoding RNA MEG3 activation of p53 mediates ischemic neuronal death in stroke, Neuroscience 337 (2016) 191–199. doi: 10.1016/j.neuroscience.2016.09.017. [DOI] [PubMed] [Google Scholar]
- [109].Liang Z, Chi Y-J, Lin G-Q, Xiao L-F, Su G-L, Yang L-M, LncRNA MEG3 participates in neuronal cell injury induced by subarachnoid hemorrhage via inhibiting the Pi3k/Akt pathway, Eur. Rev. Med. Pharmacol. Sci 22 (2018) 2824–2831. doi: 10.26355/eurrev_201805_14983. [DOI] [PubMed] [Google Scholar]
- [110].Han L, Dong Z, Liu N, Xie F, Wang N, Maternally Expressed Gene 3 (MEG3) Enhances PC12 Cell Hypoxia Injury by Targeting MiR-147, Cell. Physiol. Biochem (2017). doi: 10.1159/000484452 [DOI] [PubMed] [Google Scholar]
- [111].Gong L, Xu H, Chang H, Tong Y, Zhang T, Guo G, Knockdown of long non-coding RNA MEG3 protects H9c2 cells from hypoxia-induced injury by targeting microRNA-183, J. Cell. Biochem 119 (2018) 1429–1440. doi: 10.1002/jcb.26304. [DOI] [PubMed] [Google Scholar]
- [112].Liu Y-B, Lin L-P, Zou R, Zhao Q-H, Lin F-Q, Silencing long non-coding RNA MEG3 accelerates tibia fraction healing by regulating the Wnt/β-catenin signalling pathway, J. Cell. Mol. Med (2019). doi: 10.1111/jcmm.14229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].McLaughlin D, Vidaki M, Renieri E, Karagogeos D, Expression pattern of the maternally imprinted gene Gtl2 in the forebrain during embryonic development and adulthood, Gene Expr. Patterns 6 (2006) 394–399. doi: 10.1016/j.modgep.2005.09.007. [DOI] [PubMed] [Google Scholar]
- [114].Qu C, Jiang T, Li Y, Wang X, Cao H, Xu H, Qu J, Chen J-G, Gene expression and IG-DMR hypomethylation of maternally expressed gene 3 in developing corticospinal neurons, Gene Expr. Patterns 13 (2013) 51–56. doi: 10.1016/j.gep.2012.11.003. [DOI] [PubMed] [Google Scholar]
- [115].Tan MC, Widagdo J, Chau YQ, Zhu T, Wong JJ-L, Cheung A, Anggono V, The Activity-Induced Long Non-Coding RNA Meg3 Modulates AMPA Receptor Surface Expression in Primary Cortical Neurons, Front. Cell. Neurosci 11 (2017) 124. doi: 10.3389/fncel.2017.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Yen Y-P, Hsieh W-F, Tsai Y-Y, Lu Y-L, Liau ES, Hsu H-C, Chen Y-C, Liu T-C, Chang M, Li J, Lin S-P, Hung J-H, Chen J-A, Dlk1-Dio3 locus-derived lncRNAs perpetuate postmitotic motor neuron cell fate and subtype identity, Elife 7 (2018). doi: 10.7554/eLife.38080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Mercer TR, Qureshi IA, Gokhan S, Dinger ME, Li G, Mattick JS, Mehler MF, Long noncoding RNAs in neuronal-glial fate specification and oligodendrocyte lineage maturation, BMC Neurosci 11 (2010) 14. doi: 10.1186/1471-2202-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Liu X, Hou L, Huang W, Gao Y, Lv X, Tang J, The Mechanism of Long Non-coding RNA MEG3 for Neurons Apoptosis Caused by Hypoxia: Mediated by miR-181b-12/15-LOX Signaling Pathway, Front. Cell. Neurosci 10 (2016) 201. doi: 10.3389/fncel.2016.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Mo C-F, Wu F-C, Tai K-Y, Chang W-C, Chang K-W, Kuo H-C, Ho H-N, Chen H-F, Lin S-P, Loss of non-coding RNA expression from the DLK1-DIO3 imprinted locus correlates with reduced neural differentiation potential in human embryonic stem cell lines, Stem Cell Res. Ther 6 (2015) 1. doi: 10.1186/scrt535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].House JS, Mendez M, Maguire RL, Gonzalez-Nahm S, Huang Z, Daniels J, Murphy SK, Fuemmeler BF, Wright FA, Hoyo C, Periconceptional Maternal Mediterranean Diet Is Associated With Favorable Offspring Behaviors and Altered CpG Methylation of Imprinted Genes, Front Cell Dev Biol 6 (2018) 107. doi: 10.3389/fcell.2018.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Fuemmeler BF, Lee C-T, Soubry A, Iversen ES, Huang Z, Murtha AP, Schildkraut JM, Jirtle RL, Murphy SK, Hoyo C, DNA Methylation of Regulatory Regions of Imprinted Genes at Birth and Its Relation to Infant Temperament, Genet. Epigenet 8 (2016) 59–67. doi: 10.4137/GEG.S40538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Su H, Zhu L, Li J, Wang R, Liu D, Han W, Cadet JL, Chen T, Regulation of microRNA-29c in the nucleus accumbens modulates methamphetamine -induced locomotor sensitization in mice, Neuropharmacology 148 (2019) 160–168. doi: 10.1016/j.neuropharm.2019.01.007. [DOI] [PubMed] [Google Scholar]
- [123].Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ, Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers, J. Neurochem 116 (2011) 459–466. doi: 10.1111/j.1471-4159.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Wu D-Y, Saterlee JS, Decoding drug abuse in noncoding RNA?, ScientificWorldJournal 7 (2007) 142–145. doi: 10.1100/tsw.2007.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Li J, Zhu L, Guan F, Yan Z, Liu D, Han W, Chen T, Relationship between schizophrenia and changes in the expression of the long non-coding RNAs Meg3, Miat, Neat1 and Neat2, J. Psychiatr. Res 106 (2018) 22–30. doi: 10.1016/j.jpsychires.2018.09.005. [DOI] [PubMed] [Google Scholar]
- [126].Gardiner E, Beveridge NJ, Wu JQ, Carr V, Scott RJ, Tooney PA, Cairns MJ, Imprinted DLK1-DIO3 region of 14q32 defines a schizophrenia-associated miRNA signature in peripheral blood mononuclear cells, Mol. Psychiatry 17 (2012) 827–840. doi: 10.1038/mp.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Moon HJ, Yim S-V, Lee WK, Jeon Y-W, Kim YH, Ko YJ, Lee K-S, Lee K-H, Han S-I, Rha HK, Identification of DNA copy-number aberrations by array-comparative genomic hybridization in patients with schizophrenia, Biochem. Biophys. Res. Commun 344 (2006) 531–539. doi: 10.1016/j.bbrc.2006.03.156. [DOI] [PubMed] [Google Scholar]
- [128].Sudhalkar N, Rosen C, Melbourne JK, Park MR, Chase KA, Sharma RP, Long Non-Coding RNAs Associated with Heterochromatin Function in Immune Cells in Psychosis, Noncoding RNA 4 (2018). doi: 10.3390/ncrna4040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Middeldorp CM, Hottenga J-J, Slagboom PE, Sullivan PF, de Geus EJC, Posthuma D, Willemsen G, Boomsma DI, Linkage on chromosome 14 in a genome-wide linkage study of a broad anxiety phenotype, Mol. Psychiatry 13 (2008) 84–89. doi: 10.1038/sj.mp.4002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Piccoli M-T, Gupta SK, Viereck J, Foinquinos A, Samolovac S, Kramer FL, Garg A, Remke J, Zimmer K, Batkai S, Thum T, Inhibition of the Cardiac Fibroblast-Enriched lncRNA Meg3 Prevents Cardiac Fibrosis and Diastolic Dysfunction, Circ. Res 121 (2017) 575–583. doi: 10.1161/CIRCRESAHA.117.310624. [DOI] [PubMed] [Google Scholar]
- [131].Zhang J, Liang Y, Huang X, Guo X, Liu Y, Zhong J, Yuan J, STAT3-induced upregulation of lncRNA MEG3 regulates the growth of cardiac hypertrophy through miR-361–5p/HDAC9 axis, Sci. Rep 9 (2019) 460. doi: 10.1038/s41598-018-36369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Khakisahneh S, Zhang X-Y, Nouri Z, Hao S-Y, Chi Q-S, Wang D-H, Thyroid hormones mediate metabolic rate and oxidative, anti-oxidative balance at different temperatures in Mongolian gerbils (Meriones unguiculatus), Comp. Biochem. Physiol. C. Toxicol. Pharmacol 216 (2019) 101–109. doi: 10.1016/j.cbpc.2018.11.016. [DOI] [PubMed] [Google Scholar]
- [133].Amin A, Dhillo WS, Murphy KG, The central effects of thyroid hormones on appetite, J. Thyroid Res 2011 (2011) 306510. doi: 10.4061/2011/306510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Roncari DA, Murthy VK, Effects of thyroid hormones on enzymes involved in fatty acid and glycerolipid synthesis, J. Biol. Chem 250 (1975) 4134–4138. [PubMed] [Google Scholar]
- [135].Abi Habib W, Brioude F, Azzi S, Rossignol S, Linglart A, Sobrier ML, Giabicani E, Steunou V, Harbison MD, Le Bouc Y, Netchine I, Transcriptional profiling at the DLK1/MEG3 domain explains clinical overlap between imprinting disorders, Sci. Adv 5 (2019) eeau9425. doi: 10.1126/sciadv.aau9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Reinehr T, Obesity and thyroid function, Mol. Cell. Endocrinol 316 (2010) 165–171. doi: 10.1016/j.mce.2009.06.005. [DOI] [PubMed] [Google Scholar]
- [137].Weiss RE, Refetoff S, Thyroid Hormone Resistance Syndromes, The Thyroid and Its Diseases (2019) 741–749. doi: 10.1007/978-3-319-72102-6_49. [DOI] [Google Scholar]
- [138].Uday S, Davies C, Gleeson H, Disorders of the Thyroid in Childhood and Adolescence, in: Llahana S, Follin C, Yedinak C, Grossman A (Eds.), Advanced Practice in Endocrinology Nursing, Springer International Publishing, Cham, 2019: pp. 609–628. doi: 10.1007/978-3-319-99817-6_32. [DOI] [Google Scholar]
- [139].Moon YS, Smas CM, Lee K, Villena JA, Kim K-H, Yun EJ, Sul HS, Mice lacking paternally expressed Pref-1/Dlk1 display growth retardation and accelerated adiposity, Mol. Cell. Biol 22 (2002) 5585–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Abdallah BM, Ditzel N, Laborda J, Karsenty G, Kassem M, DLK1 Regulates Whole-Body Glucose Metabolism: A Negative Feedback Regulation of the Osteocalcin-Insulin Loop, Diabetes 64 (2015) 3069–3080. doi: 10.2337/db14-1642. [DOI] [PubMed] [Google Scholar]
- [141].Calderari S, Gangnerau M-N, Thibault M, Meile M-J, Kassis N, Alvarez C, Portha B, Serradas P, Defective IGF2 and IGF1R protein production in embryonic pancreas precedes beta cell mass anomaly in the Goto–Kakizaki rat model of type 2 diabetes, Diabetologia 50 (2007) 1463–1471. doi: 10.1007/s00125-007-0676-2. [DOI] [PubMed] [Google Scholar]
- [142].Zhou Y, Cheunsuchon P, Nakayama Y, Lawlor MW, Zhong Y, Rice KA, Zhang L, Zhang X, Gordon FE, Lidov HGW, Bronson RT, Klibanski A, Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene, Development 137 (2010) 2643–2652. doi: 10.1242/dev.045724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Swinburn BA, Sacks G, Hall KD, McPherson K, Finegood DT, Moodie ML, Gortmaker SL, The global obesity pandemic: shaped by global drivers and local environments, Lancet 378 (2011) 804–814. doi: 10.1016/S0140-6736(11)60813-1. [DOI] [PubMed] [Google Scholar]