Abstract
Many of the protective responses observed for flavonoids in the gastrointestinal track resemble aryl hydrocarbon receptor (AhR)-mediated effects. Therefore, we examined the structure–activity relationships of isoflavones and isomeric flavone and flavanones as AhR ligands on the basis of their induction of CYP1A1, CYP1B1, and UGT1A1 gene expression in colon cancer Caco2 cells and young adult mouse colonocyte (YAMC) cells. Caco2 cells were significantly more Ah-responsive than YAMC cells, and this was due, in part, to flavonoid-induced cytotoxicity in the latter cell lines. The structure–activity relationships for the flavonoids were complex and both response and cell context specific; however, there was significant variability in the AhR activities of the isomeric substituted isoflavones and flavones. For example, 4′,5,7-trihydroxyisoflavone (genistein) was AhR-inactive whereas 4′,5,7-trihydroxyflavone (apigenin) induced CYP1A1, CYP1B1, and UGT1A1 in Caco2 cells. In contrast, both 5,7-dihydroxy-4-methoxy substituted isoflavone (biochanin A) and flavone (acacetin) induced all three AhR-responsive genes; 4′,5,7-trimethoxyisoflavone was a potent AhR agonist, and the isomeric flavone was AhR-inactive. These results coupled with simulation studies modeling flavonoid interaction within the AhR binding pocket demonstrate that the orientation of the substituted phenyl ring at C-2 (flavones) or C-3 (isoflavones) on the common 4-H-chromen-4-one ring strongly influences the activities of isoflavones and flavones as AhR agonists.
Graphical Abstract
INTRODUCTION
The consumption of fruits and vegetables has long been recognized for their health benefits, which are primarily due to diverse phytochemicals and their direct and indirect effects on the host.1–4 Indirect health promoting responses of phytochemicals could be due to metabolically mediated effects or modulation of gut microbial populations and their metabolites.3,4 Among phytochemical compounds, polyphenolics including flavonoids, phenolic acids, stilbenes, and lignans have been frequently linked to health benefits in epidemiologic, human, and laboratory animal studies.5,6 Flavonoids include structurally related flavones, flavanols, anthocyanins, isoflavones, and chalcones, and with the exception of the chalcones, all of these contain a common 4-H-chromen-4-one (chromone) ring substituted in the heterocyclic ring with a (±substituent’s) phenyl group at C-2 or C-3. The potential health benefits of flavonoids are diverse and are associated with their antioxidant/radical scavenging and anti-inflammatory activities and also interactions with multiple target genes/enzymes and receptors.7–11 Epidemiological studies show that either estimated intake or serum levels of flavonoids correlate with a lower risk of colon cancer, type 2 diabetes, and macular degeneration, enhance cardiovascular functions, and protect from intestinal inflammation.10–17 Soy food products primarily contain daidzein and genistein conjugates, and their bioavailability is dependent on intestinal glucuronidases.1–4 Medium isoflavone daily intake in one study was 0.154 mg/d.18 Daidzein is further metabolized to the estrogenic metabolite equol, and levels of this isoflavone are dependent on multiple factors including the availability of specific bacterial species.
A major functional difference between isoflavones and flavones is the estrogenic activity of the former compounds, particularly genistein and daidzein, which bind both estrogen receptorα (ERα) and ERβ with higher binding affinities for ERβ18,19 On the basis of their phytoestrogenic activity, soy isoflavones have been used as alternative therapies for problems associated with estrogen depletion in postmenopausal women.18 Many of the beneficial health effects of flavonoids (flavones and isoflavones) and particularly those associated with intestinal anti-inflammatory activities resemble those observed for compounds that bind the aryl hydrocarbon receptor (AhR).20–23 We have recently characterized the structure–AhR activity relationships among a series of flavones and flavanols, and the results showed that the pentahydroxyflavones such as quercetin taxifolin and robinetin were among the most active compounds.24 With a few exceptions, decreased AhR-dependent activities were observed for hexa- and tetra/trihydroxy flavonoids; however, there were also response-specific differences in terms of activation of different AhR-response genes. Structure–activity relationships for isoflavones as AhR agonists have previously been reported; however, the effects are variable and dependent on the end point and the cell line used.25–30
The commonly occurring soy isoflavones such as genistein contain ≤4 hydroxyl substituents, and on the basis of the results obtained for hydroxyflavones,24 the major soy isoflavones would not be AhR ligands. This study investigates and compares the AhR activity of isomeric hydroxy-, hydroxy/methoxy-, and methoxy- isoflavones and flavones and some corresponding flavanones as inducers of drug-metabolizing enzyme (CYP1A1, CYP1B1, and UGT1A1) gene expression in Caco2 colon cancer cells and young adult mouse colonocyte (YAMC) cells. Some isoflavones are AhR-active; however, their effects are structure-, response-, and cell-context dependent. We also used molecular modeling approaches, including docking and stimulations to understand ligand–AhR interactions and showed that substituted isoflavones and flavones bind with different orientations in the ligand binding pocket of the AhR.
EXPERIMENTAL PROCEDURES
Cells and Reagents.
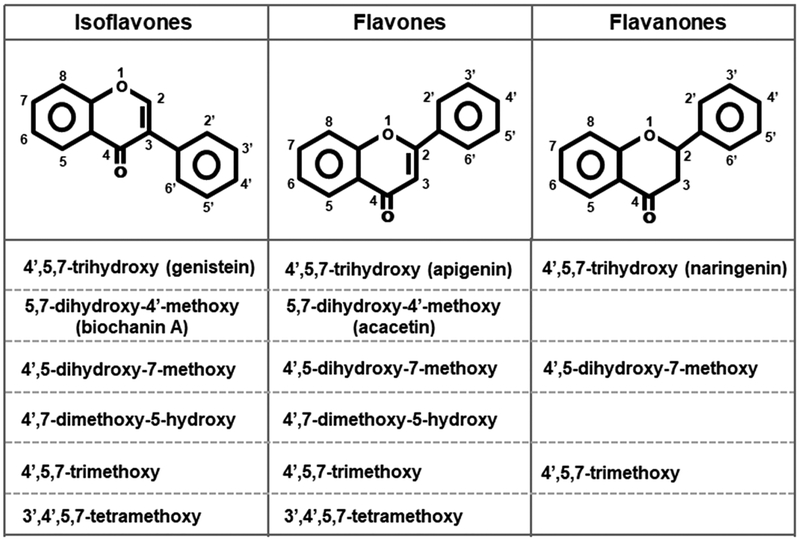
The human Caco2 cell line was purchased from the American Type Culture Collection (ATCC, Manassas VA), and mouse YAMC cells were previously used in our laboratory.24,30 Caco2 cells were maintained in Dulbecco’s modified Eagle medium nutrient mixture supplemented with 20% fetal bovine serum, 10 mL/L 100× MEM nonessential amino acid solution (Gibco), and 10 mL/L 100× antibiotic solution (Sigma-Aldrich, St. Louis MO). The cells were maintained at 33 °C under 5% carbon dioxide. The YAMC cells were maintained in RPMI 1640 medium with 5% fetal bovine serum, 5 units/mL interferon-α (IF005) (EMP Millipore, MA), and 0.1% ITS-minus (insulin, transferrin, selenium) (41-400-045) (Life Technologies, Grant Island, NY), and experiments were carried out at 33 °C. Cells were seeded at 60–70% density and were subconfluent after treatment and subsequent analysis. The Caco2-AhRKO and YAMC-AhRKO cell lines were previously generated by CRISPR/Cas9 and lack expression of the AhR.24,30 All the flavonoids (Figure 1) were purchased from Indofine Chemical Co (Hillsborough, NJ) and were >98% pure as determined by liquid chromatography–mass spectrometry using an Executive Plus Orbitrap Mass Spectrameter (Thermo Scientific, Waltham MA) coupled to a binary pump HPLC (Ultimate 300, Thermo Scientific). Sodium acetate, sodium propionate, and sodium butyrate were purchased from Sigma-Aldrich, and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) was synthesized in our laboratory.
Figure 1.
Structures of isoflavones, flavones, and flavanones used in this study.
Quantitative Real-Time Reverse Transcriptase PCR.
Total RNA was extracted using an RNA isolation kit from the cells according to the manufacturer’s protocol. cDNA synthesis was performed from the total RNA of cells using the High Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster City, CA). Real-Time PCR was carried out using Bio-Rad SYSR Universal premix for 1 min at 95 °C for initial denaturing, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min in the Bio-Rad iCycler (MyiQ2) real-time PCR System. Gene expression values were analyzed using the comparative CT method and normalized to expression levels of TATA-binding protein (TBP). The sequences of the primers used for real-time PCR were as follows: CYP1A1 sense 5′-GAC CAC AAC CAC CAA GAA C-3′, antisense 5′-AGC GAA GAA TAG GGA TGA AG-3′; UGT1A1 sense 5′-GAA TCA ACT GCC TTC ACC AAA AT-3′, antisense 5′-AGA GAA AAC CAC AAT TCC ATG TTC T-3′; TBP sense 5′-GAT CAG AAC AAC AGC CTG CC-3′, antisense 5′-TTC TGA ATA GGC TGT GGG GT-3′.
Docking Trimethoxyflavone and Trimethoxyisoflavone to Human AhR.
An in-house docking-refinement protocol was used to identify the most energetically favorable binding mode of isomeric 4′,5,7-trimethoxyflavone and 4′,5,7-trimethoxyisoflavone in complex with human AhR.34,35 The docking-refinement protocol31–35 has been previously used to determine the binding modes of TCDD and 1,4-DHNA to mouse AhR,30 quercetin and apigenin to mouse AhR,24 and other interactions.36–38 Thus, we used the protocol as a tool to refine and predict the structure of a ligand binding to a receptor’s binding pocket through the combination of the following stages: (1) the initial placement of the ligand within the receptor’s binding pocket, (2) the use of multiple short MD docking simulations rotating the ligand within the receptor’s binding pocket using different binding modes while constraining the ligand in the pocket using harmonic and quartic spherical potentials, which collectively enable the nearly exhaustive exploration of a ligand’s binding modes, (3) the use of interaction energy calculations to initially screen for the most probable binding modes generated by the short docking simulations, and (4) the use of multiple explicit-solvent MD simulations and physical–chemistry-based free energy calculations to identify the most favorable binding mode of the ligand–receptor complex.
4′,5,7-Trimethoxyflavone and 4′,5,7-trimethoxyisoflavone were independently placed into the binding site of AhR by structural superposition to the experimentally resolved (5S,7R)-5,7-bis(3-bromophenyl)-4,5,6,7-tetrahydrotetrazolo[1,5-a] pyrimidine39 via the ShaEP algorithm.40 The model of AhR (residues 247 through 406) corresponded to the model used in our previous study investigating the binding of apigenin and quercetin to AhR.24 The structures for both 4′,5,7-trimethoxyflavone and 4′,5,7-trimethoxyisoflavone were built using MarvinSketch, and the compounds were parametrized using CGenFF v1.041 for use in subsequent simulations.
Subsequently, short docking simulations were independently introduced to search and produce binding modes of the flavonoid isomers in complex with AhR using the same parameters as previously described.24 In the docking simulations, 20 independent docking runs were performed. Within each of the 20 docking runs, 200 steps in which the ligand was rotated randomly about a random axis followed by a 2 ps MD simulation and energetic minimization were performed with the structure of the generated binding mode saved for subsequent evaluation. Thus, 4000 saved binding modes of 4′,5,7-trimethoxyflavone and 4′,5,7-trimethoxyisoflavone in complex with AhR, independently, were generated per docking simulation. Throughout the duration of the short docking simulations, ligands were constrained to the binding pocket using harmonic or quartic potential energy functions of the MMFP module of CHARMM,42 and the binding pocket AhR residues were unconstrained and flexible. Six separate sets of docking simulations, or docking systems, were introduced to explore the possible binding modes of the two ligands to AhR. Four of the docking systems utilized harmonic spherical potentials, and two of the docking systems utilized the quartic spherical potentials.30 The independent use of harmonic and spherical potentials constraining the ligands to the binding pocket during the docking procedure energetically allows the ligand to explore binding modes both away from (quartic) and in proximity to (harmonic) their initial positioning. From each of the docking simulation systems, we extracted the three binding modes with the lowest interaction energy for further investigation, resulting in 18 binding modes for each of the two ligands, independently. The 18 binding modes of 4′,5,7-trimethoxyflavone and 4′,5,7-trimethoxyisoflavone in complex with AhR, independently, were investigated using 20 ns explicit-solvent MD simulations to refine the ligand: receptor structures and interactions, determination of the structural stability of the binding modes, and assessment/identification of the most energetically favorable binding modes of the 4′,5,7-trimethoxyflavone–AhR and 4′,5,7-trimethoxyisoflavone–AhR complexes. The structures corresponding to the 18 selected 4′,5,7-trimethoxyisoflavone–AhR docking conformations and the 18 selected 4′,5,7-trimethoxyflavone–AhR docking conformations were used as initial structures for the 36 independent 20 ns explicit-solvent MD simulations (18 runs per molecule under investigation). Upon completion of the 20 ns explicit-solvent MD simulations, the Molecular Mechanics Generalized Born Surface Area (MM-GBSA) approximation43–45 was introduced to identify the most energetically favorable conformation of 4′,5,7-trimethoxyflavone and 4′,5,7-trimethoxyisoflavone in complex with AhR, as previously described.24 We calculated the association-free energy of the 18 complexes per ligand over the 20 ns production runs using snapshots extracted from the simulations every 20 ps. Simulations of the 4′,5,7-trimethoxyisoflavone–AhR and 4′,5,7-trimethoxyflavone–AhR binding modes with the most favorable MM-GBSA association-free energies43–45 (e.g., lowest MM-GBSA association-free energies) were selected as representing the naturally occurring binding conformations of the two ligands in complex with AhR. The simulations comprising the lowest association-free energy simulated binding modes of each of the two ligands were extended for a total simulation duration of 30 ns, independently.
AhR Residue Interaction-Free Energy Analysis.
To determine the key interactions occurring in the lowest association-free energy simulated binding modes, the average per AhR residue interaction-free energies between the AhR receptor and 4′,5,7-trimethoxyflavone or 4′,5,7-trimethoxyisoflavone within the lowest MM-GBSA association-free energy structures were calculated for the final 25 ns of the 30 ns production runs in intervals of 20 ps.24,30,46,47 The first 5 ns of the 30 ns simulations was considered an additional equilibration time as the conformation of both compounds were adjusted during the first few nanoseconds of their respective simulations (see the Results). Thus, the first moments of the simulations allowed for the refinement of the binding poses.
RESULTS
Structure-Dependent Induction of AhR-Responsive Drug Metabolizing Enzymes.
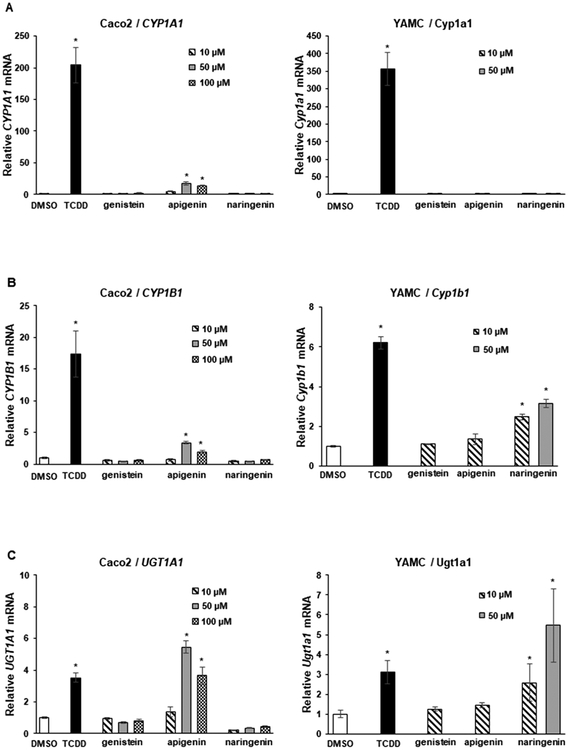
Genistein is a major isoflavone in soy and legumes, and results in Figure 2 compare the pattern of induction of CYP1A1 (Figure 2A), CYP1B1 (Figure 2B), and UGT1A1 (Figure 2C) by genistein and the structurally related 4′,5,7-trihydroxy-flavone (apigenin) and 4′,5,7-trihydroxy flavanone (naringenin) analogs. Genistein (10–100 μM) did not induce CYP1A1; naringenin was inactive, and apigenin induced CYP1A1 in Caco2 cells (<10% of TCDD). Ten and 50 μM, concentrations of the 3 compounds did not induce CYP1A1 in YAMC cells and were cytotoxic at concentrations >50 μM. The pattern of induction of CYP1B1 and UGT1A1 by the 3 compounds in Caco2 cells was similar to that observed for CYP1A1 except that the induction of UGTA1 by 50 and 100 μM apigenin was higher than observed for 10 nM TCDD. Interestingly, only 4′,5,7-trihydroxy flavanone (naringenin) induced CYP1B1 and UGT1A1 in YAMC cells (Figure 2C).
Figure 2.
Induction of drug metabolizing enzymes by trihydroxy compounds. Caco2 and YAMC cells were treated with 10–100 μM flavonoids for 24 h, and induction of CYP1A1 (A), CYP1B1 (B), and UGT1A1 (C) mRNA levels was determined by real time PCR as outlined in the Experimental Procedures. Results are expressed as means ± SD for at least 3 determinations for each treatment, and significant (p < 0.05) induction is indicated (*). The concentration of TCDD was 10 nM.
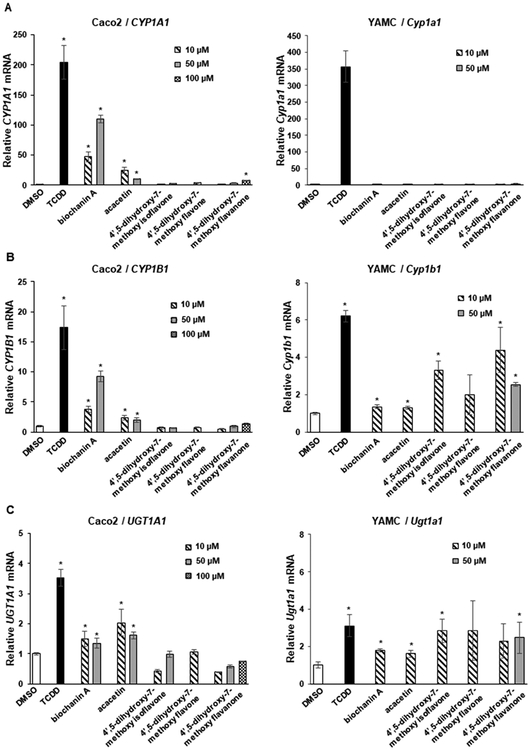
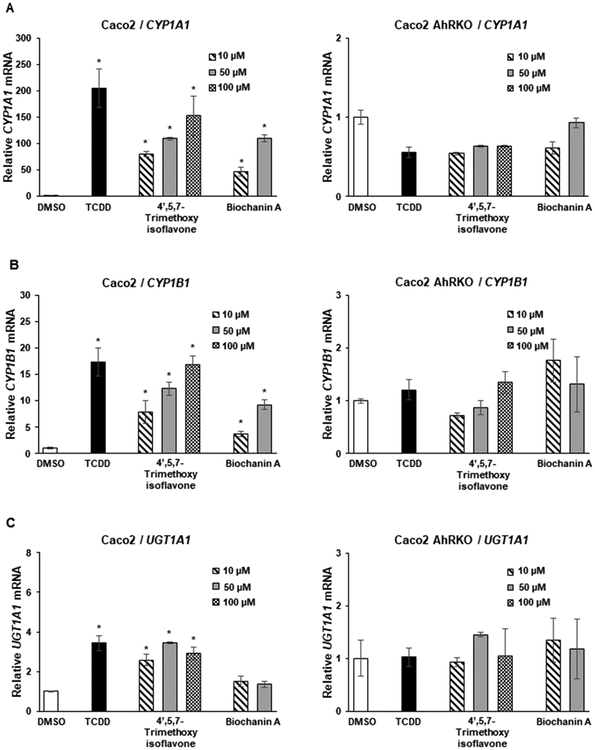
Biochanin A, the 4-methoxy derivative of genistein, induced CYP1A1 in Caco2 but not YAMC cells, whereas the corresponding isoflavone (acacetin) with the same substitution pattern exhibited lower activity in Caco2 and was inactive in YAMC cells (Figure 3A). In contrast, the 4′,5-dihydroxy-7-methoxy-isoflavone, -flavone, and -flavanone analogs were inactive as inducers of CYP1A1 in Caco2 and YAMC cells. Biochanin A and to a lesser extent acacetin induced CYP1B1 in Caco2 but not in YAMC cells, whereas the 4′,5-dihyroxy-7-methoxy analogs were inactive in Caco2 cells and the isoflavone and flavanone analogs induced CYP1B1 in YAMC cells (Figure 3B). Biochanin A and acacetin on the 4′,5-dihydroxy-7-methoxy analogs did not induce UGT1A1 in Caco2 or YAMC cells (Figure 3C). These results indicate that the 4-methoxy analog of genistein was more active than the 4-methoxy analogs of epigenin as an inducer of CYP1A1, CYP1B1, and UGT1A1 in Caco2 cells, whereas the 4,5′-dihydroxy-7-methoxy compounds were inactive in Caco2 cells and only induced (minimal) CYP1B1 and UGT1A1 in YAMC cells.
Figure 3.
Induction of drug-metabolizing enzymes by dihydroxy/methoxy compounds. Caco2 and YAMC cells were treated with 10 nM TCDD and flavonoids for 24 h, and induction of CYP1A1 (A), CYP1B1 (B), and UGT1A1 (C) mRNA levels was determined by real time PCR as outlined in the Experimental Procedures. Results are expressed as means ± SD for at least 3 determinations per treatment group, and significant (p < 0.05) induction is indicated (*).
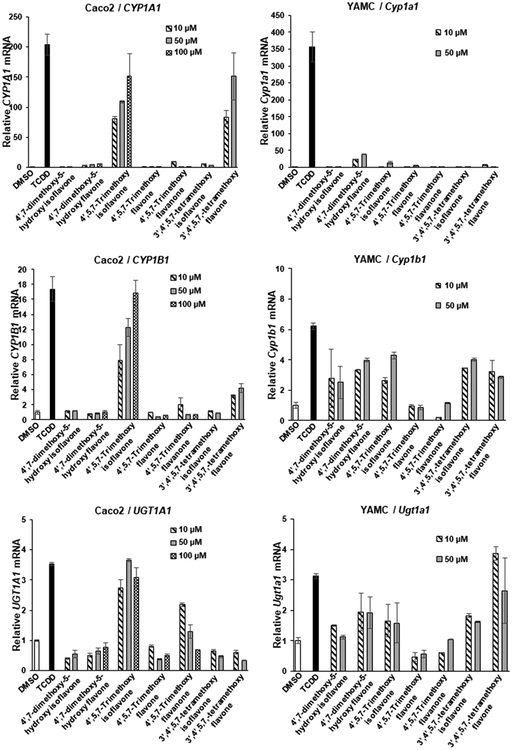
We also examined the AhR activity of 4′,7-dimethoxy-5-hydroxy, 4′,5,7-trimethoxyisoflavone, and 3′,4′,5,7-tetramethoxyisoflavone in Caco2 and YAMC cells (Figure 4). The 4′,7-dimethoxy-5-hydroxy substituted isoflavone and flavone were both inactive as inducers of CYP1A1, CYP1B1, and UGT1A1 in Caco2 cells, whereas 4′,7-dimethoxy-5-hydroxyflavone (but not isoflavone) induced the expression of all three genes in YAMC cells (Figure 4A–C). 4′,5,7-Trimethoxyisoflavone was a potent inducer of CYP1A1, CYP1B1, and UGT1A1 in Caco2 cells, but among the 3 Ah-responsive genes, only CYP1A1 (minimal) and CYP1B1 were significantly induced in YAMC cells. In contrast, the corresponding 4′,5,7-trimethoxyflavone and flavanone were AhR-inactive in both cell lines. The pattern of induction by 3′,4′,5,7-tetramethoxyflavone and isoflavone was also response and cell type dependent; in Caco2 cells, the flavone induced CYP1A1 and CYP1B1, whereas this isoflavone isomer was inactive. In YAMC cells, both 3′,4′,5,7-tetramethoxyisoflavone and flavone isomers induced CYP1B1 and UGT1A1 but not CYP1A1 gene expression. Thus, the introduction of methoxyl groups dramatically altered the AhR-dependent induction responses of both hydroxy-substituted isoflavones and flavones, and induction was flavonoid structure, response, and cell type dependent.
Figure 4.
Induction of drug-metabolizing enzymes by dimethoxy–hydroxy, trimethoxy, and tetramethoxy compounds. Caco2 and YAMC cells were treated with 10 nM TCDD and flavonoids for 24 h, and induction of CYP1A1 (A), CYP1B1 (B), and UGT1A1 (C) mRNA levels was determined by real time PCR as outlined in the Experimental Procedures. Results are expressed as means ± SD for at least 3 determinations per treatment group, and significant (p < 0.05) induction is indicated (*).
Role of the AhR and Isoflavone–SCFA Interactions.
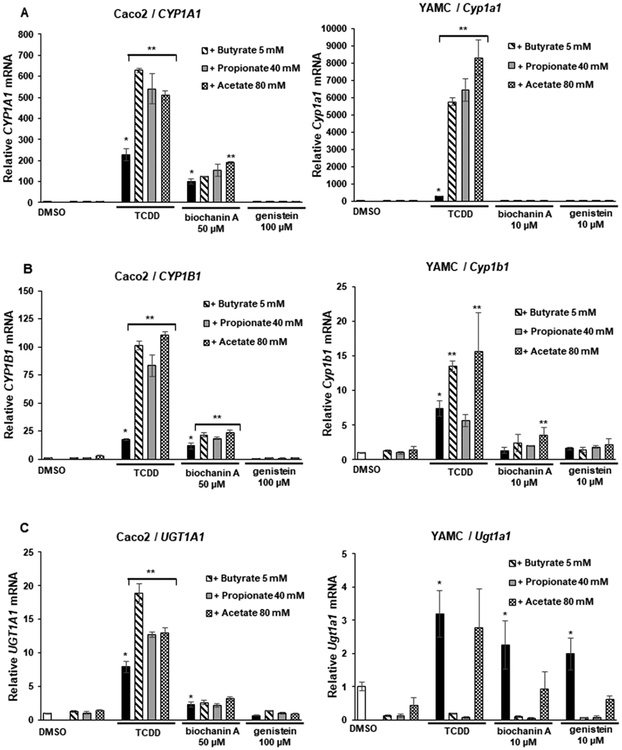
Two of the AhR-active isoflavones, 4′,5,7-trimethoxyisoflavoine and biochanin, were further investigated in Caco2 and Caco2-AhRKO (AhR knockout) cells, which were previously generated by CRISPR/Cas9 silencing of the AhR.30 Both isoflavones and TCDD induced CYP1A1 (Figure 5A), CYP1B1 (Figure 5B), and UGT1A1 (Figure 5C) in wild-type Caco2 cells but not in Caco2-AhRKO cells, thus confirming that gene activation by the isoflavones was AhR dependent. In previous studies, we reported the short chain fatty acids (SCFAs) acetate, propionate, and butyrate synergistically enhanced AhR ligand-induced gene expression in Caco2 cells, and butyrate enhancement of flavones was structure and response dependent.24 In this study, we investigated the effects of SCFAs on the induction of gene expression by biochanin A and genistein, which represent a relatively potent and inactive AhR ligand, respectively. SCFAs increased biochanin-induced CYP1A1 (<2-fold only by acetate) in Caco2 but not YAMC cells, whereas genistein was inactive in the presence or absence of SCFAs in both cell lines (Figure 6A). In contrast, acetate, propionate, and butyrate significantly enhanced TCDD-induced CYP1A1 in Caco2 and YAMC cells. SFCAs only minimally enhanced biochanin-induced (but not genistein) CYP1B1 in Caco2 and YAMC cells (with the exception of propionate), while SCFAs enhanced TCDD-induced CYP1B1 in Caco2 and YAMC cells (Figure 6B). SCFAs had no significant effects on biochanin- or genistein-induced UGT1A1 but enhanced UGT1A1 in Caco2 cells (Figure 6C), whereas the SCFAs either did not affect or inhibited induced UGT1A1 mRNA in YAMC cells. Thus, the SCFA-mediated enhancement of induction of Ah-responsive genes by isoflavones was structure and response dependent and was very different than that observed for TCDD and significantly lower than that previously observed for some flavones.24
Figure 5.
Role of the AhR in the induction response. Caco2 and Caco2-AhRKO (knockout) cells were treated with 10 nM TCDD, biochanin A, or 4′,5,7-trimethoxyisoflavone for 24 h, and the induction of CYP1A1 (A), CYP1B1 (B), and UGT1A1 (C) mRNA levels were determined by real time PCR as outlined in the Experimental Procedures. Results are expressed as means ± SD for at least 3 determinations per treatment group, and significant (p < 0.05) induction is indicated (*).
Figure 6.
Interaction of isoflavones with SCFAs. Caco2 and YAMC cells were treated with 10 nM TCDD, biochanin, and genistein alone or in combination with SCFAs (acetate, propionate, and butyrate), and induction of CYP1A1 (A), CYP1B1 (B), and UGT1A1 (C) mRNA levels was determined by real time PCR as outlined in the Experimental Procedures. Results are expressed as means ± SD for at least 3 determinations per treatment group; significant (p < 0.05) induction is indicated (*), and SCFA-enhanced induction is indicated (**).
Identification of the Most Energetically Favorable Binding Modes of 4′,5,7-Trimethoxyflavone and 4′,5,7-Trimethoxyisoflavone in Complex with AhR.
The 4′,5,7-trimethoxyisoflavone–AhR and 4′,5,7-trimethoxyflavone–AhR binding modes acquiring the lowest MM-GBSA association-free energy across all 18 simulated 4′,5,7-trimethoxyisoflavone–AhR and 4′,5,7-trimethoxyflavone–AhR binding modes (see Experimental Procedures), respectively, were selected for further analysis. The binding modes of the flavonoid–AhR complexes originated from the docking simulation system utilizing the quartic and harmonic spherical potentials, respectively. The most energetically favorable binding modes according to average MM-GBSA association-free energy and all other binding modes were far less energetically favorable. Thus, only the most energetically favorable binding modes were selected for further investigation. The selected binding modes of the 4′,5,7-trimethoxyisoflavone–AhR and 4′,5,7-trimethoxyflavone–AhR complex structures were refined during the 30 ns explicit-solvent MD simulations, which is reflected by the high average root-mean-square deviation (RMSD) of the ligands’ heavy atoms and with respect to their initial structure is 4.7 ± 0.3 and 3.8 ± 0.7 Å for 4′,5,7-trimethoxyisoflavone and 4′,5,7-trimethoxyflavone, respectively. The large values are attributed to the fact that, within the first 5 ns of their respective simulations, the position and orientation of the two compounds are stabilized. The average RMSD of the ligands’ heavy atoms with respect to their average structure in the last 25 ns of their respective simulations was 1.2 ± 0.2 and 1.0 ± 0.2 Å for 4′,5,7-trimethoxyisoflavone and 4′,5,7-trimethoxyflavone, respectively. The low average RMSD of both compounds with respect to their average structure within their respective simulations indicates that the compounds are stable within their individual simulations after the first 5 ns.
Interactions between Trimethoxyisoflavone and AhR.
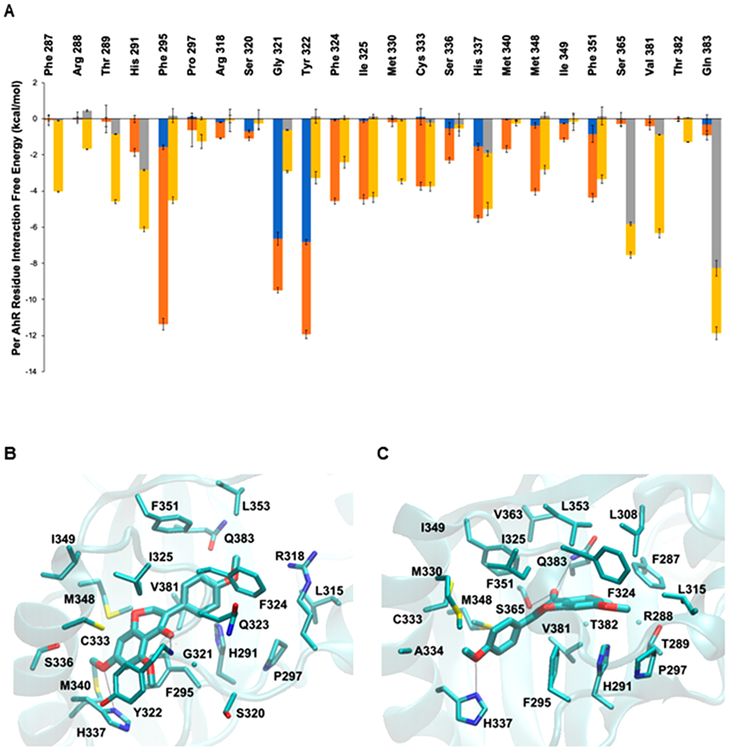
The average per-residue interaction-free energies between AhR residues and 4′,5,7-trimethoxyisoflavone were decomposed into polar and nonpolar contributions, and interactions with average interaction-free energy values less than −1.0 kcal/mol are shown in Figure 7A. In the simulation of the selected 4′,5,7-trimethoxyisoflavone–AhR complex structure, the isoflavone carbonyl group formed a hydrogen bond with the backbone amide group of Gly321 and the methoxy group of the compound formed a hydrogen bond with the ND atom of His337. These hydrogen bonds are indicated with black dotted lines in Figure 7B. As the binding site of AhR is highly hydrophobic, the binding of 4′,5,7-trimethoxyisoflavone to AhR was primarily stabilized by nonpolar interactions. A π–π interaction was formed between the aromatic rings of 4′,5,7-trimethoxyisoflavone and Phe295 as well as Tyr322. In addition, van der Waals interactions were formed between 4′,5,7-trimethoxyisoflavone and the side-chain atoms of His291, Pro297, Ile315, Arg318, Ser320, Phe324, Ile325, Cys333, Ser336, His337, Met340, Met348, Ile349, Phe351, Leu353, Val381, and Gln383 due to their close proximity to the bound compound. These interactions are shown in Figure 7B.
Figure 7.
Average interaction-free energies (kcal/mol) decomposed into polar (blue or gray) and nonpolar (orange or yellow) components for AhR interacting residues and molecular graphics images of the lowest association-free energy binding modes of 4′,5,7-trimethoxyisoflavone and 4′,5,7-trimethoxyflavone in complex with AhR. (A) AhR interacting residues in complex with 4′,5,7-trimethoxyflavone (second bar/residues). The sum of the polar and nonpolar components corresponds to the total average per AhR residue interaction-free energy. Only per residue interactions with total interaction-free energies less than −1.0 kcal/mol are considered in the plot. The average polar and nonpolar interaction-free energy component values and their standard deviation values were calculated over four measurements in which the first, second, third, and fourth measurements correspond to the individual average interaction-free energy component of the first, second, third, and fourth 6.5 ns segment of the last 25 ns of the 30 ns MD simulation production run. Image of the lowest association-free energy binding modes of 4′,5,7 trimethoxyisoflavone (B) and 4′,5,7-trimethoxyflavone (C) in complex with the AhR binding pocket. For both panels, the ligand is shown in licorice representation; AhR is shown in transparent cyan new cartoon representation, and key AhR residues are shown in thin licorice representation.
Interactions between 4′,5,7-Trimethoxyflavone and AhR.
The average per-residue interaction-free energy between AhR residues and 4′,5,7-trimethoxyflavone was decomposed into polar and nonpolar contributions, and interactions with average interaction-free energy values less than −1.0 kcal/mol are presented in Figure 7A. In the simulation of the selected 4′,5,7-trimethoxyflavone–AhR complex structure, the carbonyl group formed hydrogen bonds with the side-chain atoms of both Ser365 and Gln383 and the methoxy group formed a hydrogen bond with the ND atom of His337. These hydrogen bonds are indicated with black dotted lines in Figure 7C. Similarly to 4′,5,7-trimethoxyisoflavone, the binding of 4′,5,7-trimethoxyflavone to AhR was primarily stabilized by nonpolar interactions. van der Waals interactions were formed between 4′,5,7-trimethoxyflavone and the side-chain atoms of Phe287, Thr289, His291, Phe295, Pro297, Tyr322, Phe324, Ile325, Met330, Cys333, Ser336, His337, Met348, Phe351, Ser365, Val381, and Gln383 as well as the backbone atoms of Arg288, Gly321, and T382 due to their close proximity to the bound compound. These interactions are shown in Figure 7C.
DISCUSSION
Previous studies in this laboratory systematically investigated hydroxylated flavones as AhR agonists in Caco2 cells, and the results showed that for induction of CYP1A1 and UGT1A1 the order of potency was pentahydroxy- > hexahydroxy- > tetra/trihydroxyflavones.24 Among the exceptions was the low activity of 2′,3′,4′,5,7-pentahydroxyflavone (morin), and this was due to the 2,4′-dihydroxy substitution on the phenyl ring since 3′,4′-(or 4′,5′-)dihydroxy-substituted compounds were among the most AhR-active compounds. The structure–activity relationships for the tetra/trihydroxyflavones in Caco2 cells were also response dependent; although these compounds were inactive as inducers of CYP1A1, both 3′,4′,5,7-tetrahydroxflavone (luteolin) and 4′,5,7-trihydroxyflavone (apigenin) induced UGT1A1 in Caco2 cells.24 Although the biosynthesis of flavones and isoflavones is initiated from common precursors, the hydroxy- and hydroxy/methoxyisoflavones isolated from soy and legumes are primarily trisubstituted compounds typified by genistein and biochanin (Figure 1).48,49 The AhR activity of isoflavones has not been extensively investigated. One report showed that genistein and daidzein activated an AhR-responsive reporter gene in mouse Hepa-1 cancer cells but not in human HepG2 liver or MCF7 breast cancer cell lines.29 In this study, we compared the effects of isoflavones and flavones (and some corresponding flavanones) as inducers of three Ah-responsive genes (CYP1A1, CYP1B1, and UGT1A1) in two different colon-derived cell lines. Most of the isoflavones and flavones were cytotoxic to YAMC cells, and a dose–response (10–100 μM) could not be determined for these compounds. Among the 6 pairs of isoflavone and flavone isomers and 3 flavanones, only minimal (<10% of TCDD) or nondetectable induction of CYP1A1 was observed in YAMC cells even though TCDD was a potent inducer of this gene. In contrast, induction of CYP1B1 and UGT1A1 in YAMC cells by the isoflavones/flavones was observed for some compounds; however, structure–activity effects were not apparent. The differences between the effects of isoflavones in the two cell lines may be due in part to differences in the mouse (YAMC) and human (Caco2) AhR; however, this must be compound specific since TCDD is active in both cell lines. Moreover, previous studies on flavonoids in human (MCF-7 and HepG2) and mouse (Hepa-1) cancer cell lines showed significant differences in the AhR-responsiveness of individual flavonoids in all 3 cell lines.29 Thus, it is likely that ligand binding to the AhR is only one factor that regulates AhR-responsiveness, which includes cell context and cofactors, ligand structure, and response.
Caco2 cells were more responsive than YAMC cells with regard to the induction of drug metabolizing enzymes. This was particularly evident for CYP1A1, which was induced by apigenin, biochanin A, acacetin, 4′,5,7-trimethoxyisoflavone, and 3′,4′,5,7-tetramethoxyflavone. Moreover, with few exceptions, this same set of isoflavones and flavones also induced CYP1B1 and UGT1A1; in contrast, among the 3 flavanones, only naringenin induced the three drug-metabolizing enzymes and all other compounds were inactive.
4′,5,7-Trihydroxyisoflavone (genistein) was inactive as an inducer of Ah-responsive genes in Caco2 cells; however, methylation of the 4′-hydroxyl group (but not the 7-hydroxy group) to generate biochanin A resulted in an AhR active compound. Methylation of both the 4′- and 7-hydroxy groups of genistein resulted in an AhR-inactive compound whereas 100 μM 4′,5,7-trimethoxyisoflavone induced CYP1A1, CYP1B1, and UGT1A1 mRNA levels similar to that observed for 10 nm TCDD. Both 5,7-dihydroxy-4′-methoxyisoflavone and the corresponding flavone (biochanin A and acacetin) induced CYP1A1, CYP1B1, and UGT1A1 mRNA in Caco2 cells. For all other isomer pairs, if the isoflavone was active, then the flavone was inactive and vice versa. This demonstrates the importance of the attachment of the phenol ring to C-2 or C-3 of the chromen-4-one ring.
SCFAs enhance Ah-responsive of several structurally diverse AhR ligands. This is due, in part, to their activity as histone deacetylase inhibitors.50 We have previously demonstrated that acetate, butyrate, and propionate enhanced TCDD-induced CYP1A1 in Caco2 and YAMC cells as indicated in Figure 6. Similar effects were observed with tryptamine, 1,4-dihydroxy-2-naphthoic acid, and the weak AhR agonist indole.50 Butyrate also significantly enhanced hydroxylated flavone-induced CYP1A1 in Caco2 cells,24 whereas interactions of butyrate with biochanin A-or genistein-induced CYP1A1 in Caco2 cells were minimal (Figure 6). This suggests that AhR ligands may also influence interactions with SCFAs. Moreover, since intestinal CYP1A1 plays an important role in regulating formation of AhR-active tryptophan metabolites,51 the magnitude of CYP1A1 expression resulting from SCFA–isoflavonoid interactions will be lower than that observed for SCFA interactions with other microbially- and dietary-derived AhR ligands. These differences may be important if they affect intestinal levels of AhR-active tryptophan metabolites. However, SCFA interactions of isoflavones and flavones with respect to induction of CYP1B1 and particularly UGT1A1 are highly variable (Figure 6B,C). These interactions in terms of regulating other AhR-mediated responses in the intestine and other organs have not been determined and will be essential for understanding the overall functional significance of SCFA–AhR ligand interactions. These studies are now in progress.
Another difference in the AhR activity of two isomeric isoflavone and flavone derivatives was observed for 4′,5,7-trimethoxyisoflavone, a highly potent AhR agonist, and 4′,5,7-trimethoxyflavone, which was essentially AhR-inactive. Despite their structural similarities, the two flavonoids bind to the AhR with different orientations within the binding site (Figure 7). Accordingly, the binding of the two flavonoids to AhR differentially affects CYP1A1 transcription. While the binding of 4′,5,7-trimethoxyisoflavone to AhR results in increased expression of CYP1A1, the binding of 4′,5,7-trimethoxyflavone does not activate this gene. Any similarities of AhR side-chain residues in their interactions with 4′,5,7-trimethoxyisoflavone and other AhR agonists, such as quercetin and TCDD investigated in our previous studies,24,30 can provide insights on amino acids in the binding pocket that may be key for activity. 4′,5,7-Trimethoxyisoflavone, quercetin, and TCDD all interact strongly with (human or corresponding mouse) AhR residues Phe295, Ile325, Cys333, Ser336, Met348, and Phe351;24,30 notably, with the exception of Ser336 and Met348, all the corresponding aforementioned residues were deemed important for TCDD binding to mouse AhR according to previous experiments.52 While 4′,5,7-trimethoxyflavone also interacts favorably with Ile325, Cys333, Met348, and Phe351, interactions with Phe295 and Ser336 are notably weaker than that observed for 4′,5,7-trimethoxyisoflavone, which suggests that such interactions could potentially be important for CYP1A1 agonist activities, at least based on our current and previous studies.24,30 Nevertheless, the signaling activity could be primarily seen as a result of AhR ligand interactions with combinations of amino acids in the AhR ligand binding site rather than single amino acids with different compounds acting as switches for signaling. Thus, as recently reported by Giani Tagliabue et al.,52 it is possible that different classes of compounds could promote activity by interacting with combinations of key amino acids important for signaling. It is worth noting that the recently published study52 exhibited significant overlaps with our previously reported structure of TCDD binding to AhR using our modeling approach,24 and they also predicted specific combinations of residues within the AhR binding cavity that can play a critical role in binding of three distinct structural classes of AhR-binding compounds.
Current studies are focused not only on characterizing the molecular interaction of flavonoids with the AhR but also on correlating the AhR binding and gene activation data with efficacy in ameliorating intestinal inflammation and carcinogenesis. The development of bioassays that can predict AhR-dependent health promoting effects of isoflavones and other phytochemicals will be important for developing food products and nutraceuticals enriched with the appropriate compounds.
Funding
Funding from the National Institutes of Health (R01-AT010282, RO1-CA202697, R35-CA197707, and P30-ES029607), the Syd Kyle Chair, Allen Endowed Chair in Nutrition & Chronic Disease Prevention, and Texas Agrilife is gratefully acknowledged.
ABBREVIATIONS
- AhR
aryl hydrocarbon receptor
- CYP
cytochrome P450
- CHARMM
chemistry at Harvard molecular mechanics
- DHNA
1,4-dihydroxy-2-naphthoic acid
- MMFP
miscellaneous mean-field potential
- SCFA
short chain fatty acids
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- UGT
uridine diphosphate glucuronosyl transferase
- YAMC
young adult mouse colonocyte
Footnotes
The authors declare no competing financial interest.
REFERENCES
- (1).Slavin JL, and Lloyd B (2012) Health benefits of fruits and vegetables. Adv. Nutr 3 (4), 506–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Rodriguez-Casado A (2016) The Health Potential of Fruits and Vegetables Phytochemicals: Notable Examples. Crit. Rev. Food Sci. Nutr 56 (7), 1097–107. [DOI] [PubMed] [Google Scholar]
- (3).David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, and Turnbaugh PJ (2014) Diet rapidly and reproducibly alters the human gut microbiome. Nature 505 (7484), 559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, Keller MP, Attie AD, Rey FE, and Denu JM (2016) Diet-Microbiota Interactions Mediate Global Epigenetic Programming in Multiple Host Tissues. Mol. Cell 64 (5), 982–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Crozier A, Jaganath IB, and Clifford MN (2009) Dietary phenolics: chemistry bioavailability and effects on health. Nat. Prod. Rep 26 (8), 1001–43. [DOI] [PubMed] [Google Scholar]
- (6).Algieri F, Rodriguez-Nogales A, Rodriguez-Cabezas ME, Risco S, Ocete MA, and Galvez J (2015) Botanical Drugs as an Emerging Strategy in Inflammatory Bowel Disease: A Review. Mediators Inflammation 2015, 179616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Havsteen BH (2002) The biochemistry and medical significance of the flavonoids. Pharmacol. Ther 96 (2–3), 67–202. [DOI] [PubMed] [Google Scholar]
- (8).Panche AN, Diwan AD, and Chandra SR (2016) Flavonoids: an overview. J. Nutr. Sci 5, No. e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Salaritabar A, Darvishi B, Hadjiakhoondi F, Manayi A, Sureda A, Nabavi SF, Fitzpatrick LR, Nabavi SM, and Bishayee A (2017) Therapeutic potential of flavonoids in inflammatory bowel disease: A comprehensive review. World J. Gastroenterol 23 (28), 5097–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Vezza T, Rodriguez-Nogales A, Algieri F, Utrilla MP, Rodriguez-Cabezas ME, and Galvez J (2016) Flavonoids in Inflammatory Bowel Disease: A Review. Nutrients 8 (4), 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Mozaffarian D, and Wu JHY (2018) Flavonoids Dairy Foods, and Cardiovascular and Metabolic Health: A Review of Emerging Biologic Pathways. Circ. Res 122 (2), 369–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Sansone R, Rodriguez-Mateos A, Heuel J, Falk D, Schuler D, Wagstaff R, Kuhnle GG, Spencer JP, Schroeter H, Merx MW, Kelm M, Heiss C, and Flaviola Consortium, European Union 7th Framework Program (2015) Cocoa flavanol intake improves endothelial function and Framingham Risk Score in healthy men and women: a randomised, controlled, double-masked trial: the Flaviola Health Study. Br. J. Nutr 114 (8), 1246–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Rienks J, Barbaresko J, Oluwagbemigun K, Schmid M, and Nothlings U (2018) Polyphenol exposure and risk of type 2 diabetes: dose-response meta-analyses and systematic review of prospective cohort studies. Am. J. Clin. Nutr 108 (1), 49–61. [DOI] [PubMed] [Google Scholar]
- (14).Murphy N, Achaintre D, Zamora-Ros R, Jenab M, Boutron-Ruault MC, Carbonnel F, Savoye I, Kaaks R, Kuhn T, Boeing H, Aleksandrova K, Tjonneland A, Kyro C, Overvad K, Quiros JR, Sanchez MJ, Altzibar JM, Maria Huerta J, Barricarte A, Khaw KT, Bradbury KE, Perez-Cornago A, Trichopoulou A, Karakatsani A, Peppa E, Palli D, Grioni S, Tumino R, Sacerdote C, Panico S, Bueno-De-Mesquita HBA, Peeters PH, Rutegard M, Johansson I, Freisling H, Noh H, Cross AJ, Vineis P, Tsilidis K, Gunter MJ, and Scalbert A (2018) A prospective evaluation of plasma polyphenol levels and colon cancer risk. Int. J. Cancer 143, 1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gopinath B, Liew G, Kifley A, Flood VM, Joachim N, Lewis JR, Hodgson JM, and Mitchell P (2018) Dietary flavonoids and the prevalence and 15-y incidence of age-related macular degeneration. Am. J. Clin. Nutr 108 (2), 381–7. [DOI] [PubMed] [Google Scholar]
- (16).Langhorst J, Varnhagen I, Schneider SB, Albrecht U, Rueffer A, Stange R, Michalsen A, and Dobos GJ (2013) Randomised clinical trial: a herbal preparation of myrrh chamomile and coffee charcoal compared with mesalazine in maintaining remission in ulcerative colitis–a double-blind, double-dummy study. Aliment. Pharmacol. Ther 38 (5), 490–500. [DOI] [PubMed] [Google Scholar]
- (17).Oteiza PI, Fraga CG, Mills DA, and Taft DH (2018) Flavonoids and the gastrointestinal tract: Local and systemic effects. Mol. Aspects Med 61, 41–9. [DOI] [PubMed] [Google Scholar]
- (18).Setchell KDR (2017) The history and basic science development of soy isoflavones. Menopause 24 (12), 1338–50. [DOI] [PubMed] [Google Scholar]
- (19).Kuiper GG, Lemmen JG, Carlsson B, Corton JC, Safe SH, van der Saag PT, van der Burg B, and Gustafsson JA (1998) Interaction of estrogenic chemicals and phytoestrogens with estrogen receptor beta. Endocrinology 139 (10), 4252–63. [DOI] [PubMed] [Google Scholar]
- (20).Rothhammer V, and Quintana FJ (2019) The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat. Rev. Immunol 19 (3), 184–97. [DOI] [PubMed] [Google Scholar]
- (21).Esser C, and Rannug A (2015) The aryl hydrocarbon receptor in barrier organ physiology immunology, and toxicology. Pharmacol. Rev 67 (2), 259–79. [DOI] [PubMed] [Google Scholar]
- (22).Stockinger B, Di Meglio P, Gialitakis M, and Duarte JH (2014) The aryl hydrocarbon receptor: multitasking in the immune system. Annu. Rev. Immunol 32, 403–32. [DOI] [PubMed] [Google Scholar]
- (23).Xue Z, Li D, Yu W, Zhang Q, Hou X, He Y, and Kou X (2017) Mechanisms and therapeutic prospects of polyphenols as modulators of the aryl hydrocarbon receptor. Food Funct. 8 (4), 1414–37. [DOI] [PubMed] [Google Scholar]
- (24).Jin UH, Park H, Li X, Davidson LA, Allred C, Patil B, Jayaprakasha G, Orr AA, Mao L, Chapkin RS, Jayaraman A, Tamamis P, and Safe S (2018) Structure-Dependent Modulation of Aryl Hydrocarbon Receptor-Mediated Activities by Flavonoids. Toxicol. Sci 164 (1), 205–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Van der Heiden E, Bechoux N, Muller M, Sergent T, Schneider YJ, Larondelle Y, Maghuin-Rogister G, and Scippo ML (2009) Food flavonoid aryl hydrocarbon receptor-mediated agonistic/antagonistic/synergic activities in human and rat reporter gene assays. Anal. Chim. Acta 637 (1–2), 337–45. [DOI] [PubMed] [Google Scholar]
- (26).Fukuda I, Mukai R, Kawase M, Yoshida K, and Ashida H (2007) Interaction between the aryl hydrocarbon receptor and its antagonists flavonoids. Biochem. Biophys. Res. Commun 359 (3), 822–7. [DOI] [PubMed] [Google Scholar]
- (27).Medjakovic S, and Jungbauer A (2008) Red clover isoflavones biochanin A and formononetin are potent ligands of the human aryl hydrocarbon receptor. J. Steroid Biochem. Mol. Biol 108 (1–2), 171–7. [DOI] [PubMed] [Google Scholar]
- (28).Bialesova L, Novotna A, Macejova D, Brtko J, and Dvorak Z (2015) Agonistic effect of selected isoflavones on arylhydrocarbon receptor in a novel AZ-AhR transgenic gene reporter human cell line. Gen Physiol Biophys 34 (3), 331–4. [DOI] [PubMed] [Google Scholar]
- (29).Zhang S, Qin C, and Safe SH (2003) Flavonoids as aryl hydrocarbon receptor agonists/antagonists: effects of structure and cell context. Environ. Health Perspect 111 (16), 1877–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Cheng Y, Jin UH, Davidson LA, Chapkin RS, Jayaraman A, Tamamis P, Orr A, Allred C, Denison MS, Soshilov A, Weaver E, and Safe S (2017) Editor’s Highlight: Microbial-Derived 1,4-Dihydroxy-2-naphthoic Acid and Related Compounds as Aryl Hydrocarbon Receptor Agonists/Antagonists: Structure-Activity Relationships and Receptor Modeling. Toxicol. Sci 155 (2), 458–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Tamamis P, Kieslich CA, Nikiforovich GV, Woodruff TM, Morikis D, and Archontis G (2014) Insights into the mechanism of C5aR inhibition by PMX53 via implicit solvent molecular dynamics simulations and docking. BMC Biophys. 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Tamamis P, and Floudas CA (2013) Molecular recognition of CXCR4 by a dual tropic HIV-1 gp120 V3 loop. Biophys. J 105 (6), 1502–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Tamamis P, and Floudas CA (2014) Molecular recognition of CCR5 by an HIV-1 gp120 V3 loop. PLoS One 9 (4), No. e95767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Tamamis P, and Floudas CA (2014) Elucidating a key component of cancer metastasis: CXCL12 (SDF-1alpha) binding to CXCR4. J. Chem. Inf. Model 54 (4), 1174–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tamamis P, and Floudas CA (2015) Elucidating a key anti-HIV-1 and cancer-associated axis: the structure of CCL5 (Rantes) in complex with CCR5. Sci. Rep 4, 5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Orr AA, Jayaraman A, and Tamamis P (2018) Molecular Modeling of Chemoreceptor:Ligand Interactions. Methods Mol. Biol 1729, 353–72. [DOI] [PubMed] [Google Scholar]
- (37).Mohan RR, Wilson M, Gorham RD Jr., Harrison RES, Morikis VA, Kieslich CA, Orr AA, Coley AV, Tamamis P, and Morikis D (2018) Virtual Screening of Chemical Compounds for Discovery of Complement C3 Ligands. ACS Omega 3 (6), 6427–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Yoon K, Chen CC, Orr AA, Barreto PN, Tamamis P, and Safe S (2019) Activation of COUP-TFI by a Novel Diindolylmethane Derivative. Cells 8 (3), 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Scheuermann TH, Stroud D, Sleet CE, Bayeh L, Shokri C, Wang H, Caldwell CG, Longgood J, MacMillan JB, Bruick RK, Gardner KH, and Tambar UK (2015) Isoform-Selective and Stereoselective Inhibition of Hypoxia Inducible Factor-2. J. Med. Chem 58 (15), 5930–41. [DOI] [PubMed] [Google Scholar]
- (40).Vainio MJ, Puranen JS, and Johnson MS (2009) ShaEP: molecular overlay based on shape and electrostatic potential. J. Chem. Inf. Model 49 (2), 492–502. [DOI] [PubMed] [Google Scholar]
- (41).Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, and Mackerell AD Jr. (2010) CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem 31 (4), 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Brooks BR, Brooks CL 3rd, Mackerell AD Jr., Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, and Karplus M (2009) CHARMM: the biomolecular simulation program. J. Comput. Chem 30 (10), 1545–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hayes JM, and Archontis G (2012) MM-GB(PB)SA Calculations of Protein-Ligand Binding Free Energies In Molecular Dynamics - Studies of Synthetic and Biological Macromolecules, pp 171–190, InTech,. [Google Scholar]
- (44).Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Cieplak P, Srinivasan J, Case DA, and Cheatham TE 3rd. (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res 33 (12), 889–97. [DOI] [PubMed] [Google Scholar]
- (45).Gohlke H, and Case DA (2004) Converging free energy estimates: MM-PB(GB)SA studies on the protein-protein complex Ras-Raf. J. Comput. Chem 25 (2), 238–50. [DOI] [PubMed] [Google Scholar]
- (46).Tamamis P, Morikis D, Floudas CA, and Archontis G (2010) Species specificity of the complement inhibitor compstatin investigated by all-atom molecular dynamics simulations. Proteins: Struct., Funct., Genet 78 (12), 2655–2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Tamamis P, Lopez de Victoria A, Gorham RD Jr., Bellows-Peterson ML, Pierou P, Floudas CA, Morikis D, and Archontis G (2012) Molecular dynamics in drug design: new generations of compstatin analogs. Chem. Biol. Drug Des 79 (5), 703–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Barnes S (2010) The biochemistry chemistry and physiology of the isoflavones in soybeans and their food products. Lymphatic Res. Biol 8 (1), 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Deavours BE, and Dixon RA (2005) Metabolic engineering of isoflavonoid biosynthesis in alfalfa. Plant Physiol. 138 (4), 2245–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Jin UH, Cheng Y, Park H, Davidson LA, Callaway ES, Chapkin RS, Jayaraman A, Asante A, Allred C, Weaver EA, and Safe S (2017) Short Chain Fatty Acids Enhance Aryl Hydrocarbon (Ah) Responsiveness in Mouse Colonocytes and Caco-2 Human Colon Cancer Cells. Sci. Rep 7 (1), 10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Schiering C, Wincent E, Metidji A, Iseppon A, Li Y, Potocnik AJ, Omenetti S, Henderson CJ, Wolf CR, Nebert DW, and Stockinger B (2017) Feedback control of AHR signalling regulates intestinal immunity. Nature 542 (7640), 242–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Giani Tagliabue S, Faber SC, Motta S, Denison MS, and Bonati L (2019) Modeling the binding of diverse ligands within the Ah receptor ligand binding domain. Sci. Rep 9 (1), 10693. [DOI] [PMC free article] [PubMed] [Google Scholar]