Abstract
Background
Ovarian cancer (OC) is the most frequent aggressive cancer among women worldwide, and chemoresistance is the major challenge in the clinical treatment of OC. Recently, there is evidence that long noncoding RNAs (lncRNAs) are closely related to the regulation of cisplatin (CDDP) resistance in OC cells. However, whether LINC01125, a novel lncRNA, can improve the sensitivity of OC to cisplatin remains unknown.
Material/Methods
In this study, we analyzed aberrantly expressed lncRNAs in miR-200a-overexpressing OC samples by using GSE122123. LINC01125 and miR-1972 expressions were measured by qRT-PCR. The effect of LINC01125 overexpression on cell proliferation was determined by CCK-8 and colony formation assays. The sensitivity of OC cells to cisplatin was determined by CCK-8 assays. The interaction between LINC01125 and miR-1972 was verified through dual-luciferase reporter and RNA immunoprecipitation (RIP) assays, and bioinformatics analysis was performed to predict the target genes of miR-1972.
Results
Our results indicated that LINC01125 expression was significantly downregulated in CDDP-resistant OC tissues and cell lines. Overexpression of LINC01125 inhibited OC cell proliferation and enhanced the cytotoxicity of CDDP in OC cells. Additionally, LINC01125 participated in the apoptosis pathway by directly binding to miR-1972 in OC cells.
Conclusions
Therefore, we suggest that LINC01125 might act as a tumor suppressor in OC and enhances the cisplatin sensitivity of OC cells by binding to miR-1972.
MeSH Keywords: Cisplatin; Ovarian Neoplasms; RNA, Long Noncoding
Background
Ovarian cancer (OC), which ranks seventh in incidence among cancers in women worldwide, is a malignancy with a high mortality rate that claims approximately 150 000 lives every year [1,2]. It was reported that the 5-year survival rate of OC patients could be dramatically improved by diagnosis at early FIGO (The International Federation of Gynecologists and Obstetricians) stages (I and II) [3,4]. However, due to the lack of effective diagnosis measures and biomarkers, as well as the relatively asymptomatic character of OC, the majority of patients are diagnosed at late FIGO stages [2]. Clinical data have revealed that the 5-year survival rate of advanced OC patients is less than 45% [5]. Therefore, it is essential to elucidate the underlying mechanisms of OC and develop a clinically applicable strategy for the early detection of OC. Currently, cytoreductive surgery combined with cisplatin or paclitaxel-based chemotherapy is the most accepted and effective therapeutic strategy for OC patients, especially those with advanced disease [6,7]. Although OC patients initially respond well to this regimen, almost all eventually present with recurrence, and chemoresistance is considered to be the major reason [8,9]. Therefore, it is urgent to confirm novel biomarkers to predict the response of OC patients to available chemotherapy and to select more effective therapeutic strategies.
MicroRNAs (miRNAs), a subtype of noncoding RNAs (ncRNAs) with approximately 22 nucleotides (nt), have been previously reported to participate in multifarious biological processes, including chemoresistance [10,11]. Among miRNAs, miR-200a is well documented to be correlated with cisplatin resistance in OC, indicating a critical role of miR-200a in the regulation of cisplatin resistance in OC [12,13]. However, the molecular mechanisms underlying this role remain largely undetermined. Long noncoding RNAs (lncRNAs) are another important subtype of ncRNAs with 200–100 000 nt [14]. lncRNAs also have important roles in the regulation of multiple biological processes, consisting of cell proliferation, differentiation, cycle, apoptosis, metastasis, and chemoresistance [15]. In addition, there is evidence that lncRNAs can indirectly regulate the expression of genes through binding specific miRNAs to exert biological functions [16]. Therefore, we speculated that miR-200a might play a role in cisplatin resistance by interacting with lncRNAs.
In this study, we analyzed the lncRNA expression profile in GSE122123 upon overexpressing miR-200a. The results showed that LINC01125 was downregulated in miR-200a-overexpressing samples compared to control samples, implying that LINC01125 might also be related to the cisplatin resistance of OC. Previous research showed LINC01125 might be a novel tumor suppressor for breast cancer through actions on the PTEN/AKT/p53 signaling pathway [17]. However, its role in OC remains unclear. Therefore, we aimed to determine the roles and underlying mechanisms of LINC01125 in the pathogenesis and chemoresistance of OC.
Material and Methods
Clinical OC patient samples and cell culture
Twenty-one pairs of OC tissues and corresponding normal tissues were acquired from patients diagnosed with OC in Lanzhou Maternity and Child Health Care Hospital during 2015–2018. Every subject provided written informed consent and all procedures used in this study were approved by the Ethics Committee of Lanzhou Maternity and Child Health Care Hospital. Human CDDP-resistant OC cell lines (SKOV-3/CDDP and A2780/CDDP) and their parental OC lines (SKOV-3 and A2780) were purchased from the Chinese Academy of Sciences (Shanghai, China). All cell lines were maintained in RPMI 1640 (GIBCO-BRL) medium containing 10% fetal bovine serum (FBS) at 37°C with 5% CO2.
Cell transfection
The LINC01125-overexpression plasmid (control: empty vector) and miR-1972 inhibitor (negative control: scramble miRNA) were designed by and purchased from GenePharma (Shanghai, China). For transfections, OC cells were cultured at 37°C for at least 24 h, followed by transfection with the indicated oligonucleotides using Lipofectamine 3000 (Invitrogen) based on the instructions provided by the supplier.
qRT-PCR assay
TRIzol reagent (Takara, Japan) was used to extract total RNA from OC tissues and cell lines following the protocols provided by the manufacturer. The Bestar™ qPCR RT kit (DBI Bioscience, China) was utilized to synthesize cDNA from extracted RNA following the instructions provided by the supplier. Subsequently, the expression of LINC01125 and miR-1972 in OC tissues and cell lines was quantified using Bestar™ qPCR MasterMix (DBI Bioscience) on the ABI PRISM 7500 Sequence Detection System (Life Technologies, USA). The sequences of the primers are presented in Table 1.
Table 1.
Primer sequences used in this study.
| Gene | Sequence or target sequence |
|---|---|
| LINC01125-F | 5′-TTCTCCATCTGCGCACCACA-3′ |
| LINC01125-R | 5′-GCCAGCCATCGGTGCCATAT-3′ |
| GAPDH-F | 5′-CACCCACTCCTCCACCTTTG-3′ |
| GAPDH-R | 5′-CCACCACCCTGTTGCTGTAG-3′ |
| miR-1972-F | 5′-GGGTCAGGCCAGGCACAGT-3′ |
| miR-1972-R | 5′-CAGTGCGTGTCGTGGAGT-3′ |
| U6-F | 5′-CTCGCTTCGGCAGCACA-3′ |
| U6-R | 5′-AACGCTTCACGAATT TGCGT-3′ |
Cell viability detection (CCK-8 assay)
The viability of treated OC cells was determined through Cell Counting Kit-8 assays (Beyotime Inst Biotech, China) in line with the instructions provided by the manufacturer. In brief, OC cells were seeded into 96-well plates at a concentration of 2×104 cells/well and cultured at 37°C for 24 h, followed by transfection with LINC01125 or vector control and miR-1972 inhibitor or negative control (miR-NC). At 1, 2, 3, 4, and 5 days after transfection, 10 μl of CCK-8 (5 mg/ml) was added to each well, and the plates were maintained at 37°C for 1 h. The absorbance at 450 nm was determined with a microtiter plate reader (SpectraMax, Molecular Devices, USA).
Colony formation assay
After transfection with LINC01125 or miR-1972 inhibitor, OC cells were collected and plated into 6-well plates containing culture medium. Cells were maintained at 37°C in an incubator with 95% O2 and 5% CO2 for 2 weeks. Then, the OC cells were then fixed using methanol and stained with 0.1% crystal violet. The number of visible colonies was counted.
Dual-luciferase reporter assay
The interaction between LINC01125 and miR-1972 was verified by dual-luciferase reporter assays. LINC01125 containing wild-type or mutant miR-1972 binding sites was amplified and inserted into the luciferase vector psi-CHECK2 (Promega, Madison, USA) to generate the recombinant luciferase plasmids psiCHECK2-LINC01125-WT (LINC-WT) and psiCHECK2-LINC01125-WT (LINC-Mut). SKOV-3 and A2780 cells were seeded into 96-well plates at a density of 1×104 cells/well, maintained at 37°C for 24 h, and then transfected with LINC-WT or LINC-Mut and miR-1972 or miR-NC. Subsequently, the luciferase activities of Firefly and Renilla in treated SKOV-3 and A2780 cells were detected by the Dual-Luciferase Assay System (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity.
RNA immunoprecipitation (RIP) assay
RIP assays were applied to further confirm the interaction between miR-1972 and LINC01125 in SKOV-3 and A2780 cells using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA) and the Ago2 antibody (Millipore). In brief, after SKOV-3 and A2780 cells were lysed using RIP lysis buffer, extracts (100 μl) were treated with RIP buffer with magnetic beads conjugated with anti-Ago2 antibody or IgG (negative control). Subsequently, proteinase K was applied to digest proteins, and immunoprecipitated RNAs were isolated and detected by qRT-PCR.
Statistical analysis
All Data are displayed as the mean±SEM. GraphPad software (Ver. Prism 7, GraphPad Prism Software, La Jolla, CA, USA) was utilized to analyze the differences between groups by using the t test or one-way ANOVA. P<0.05 was considered significant.
Results
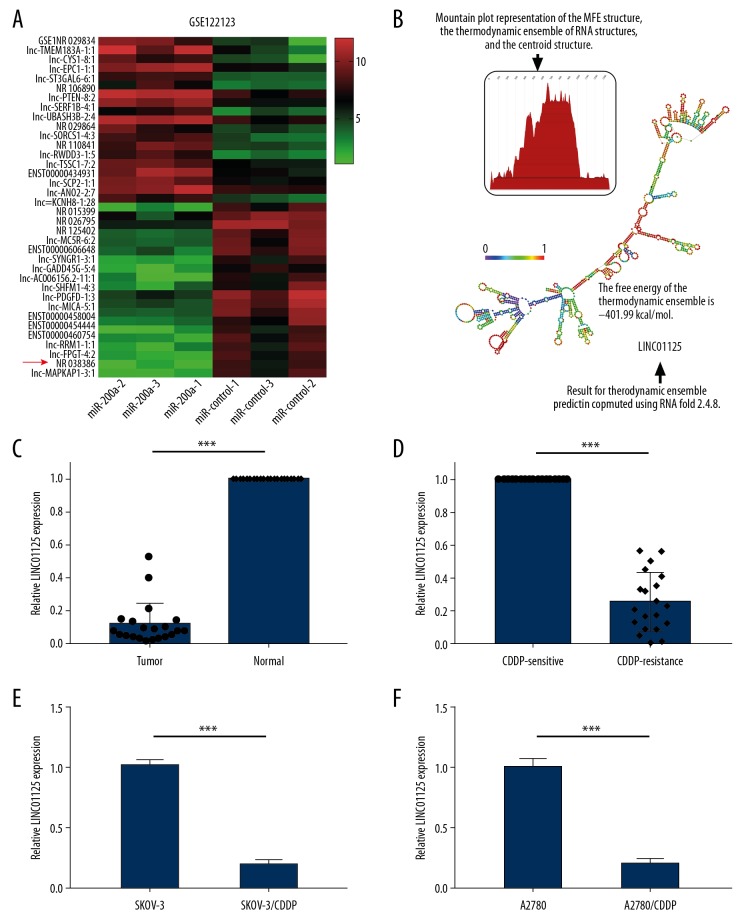
LINC01125 was expressed at a low level in CDDP-resistant OC
As previous studies have suggested, miR-200a contributes to the chemoresistance of OC; however, the mechanisms have been unclear. In this study, we analyzed the lncRNA expression profile in GSE122123 upon overexpressing miR-200a. The results showed that among the top 20 dysregulated lncRNAs, NR_0038386, named LINC01125 in the database, was significantly downregulated in miR-200a-treated samples compared to controls (Figure 1A). RNAfold was used to predict the secondary and MFE structures of LINC01125 (Figure 1B). To investigate whether LINC01125 plays a role in the chemoresistance of OC, we examined its expression in CDDP-resistant OC tissues and cell lines. qRT-PCR assays showed that LINC01125 expression was significantly downregulated in OC tissues compared to corresponding normal tissues (Figure 1C). Compared to CDDP-sensitive OC tissues, CDDP-resistant OC tissues had significantly lower LINC01125 expression (Figure 1D). LINC01125 expression was also markedly decreased in CDDP-resistant OC cell lines (SKOV-3/CDDP and A2780/CDDP) compared to their parental cell lines (SKOV-3 and A2780) (Figure 1E, 1F). This suggested that LINC01125 might play a role in the chemoresistance of OC.
Figure 1.
LINC01125 was expressed at low levels in CDDP-resistant OC. (A) Top 20 dysregulated lncRNAs in GSE122123 after overexpressing miR-200a. (B) The secondary and MFE structures of LINC01125 were predicted by RNAfold. (C) LINC01125 expression in OC tissue samples and corresponding normal samples was measured by qRT-PCR assay (*** P<0.001). (D) Relative LINC01125 expression in CDDP-resistant and CDDP-sensitive OC samples (*** P<0.001). (E, F) Relative LINC01125 expression in CDDP-resistant OC cell lines (SKOV-3/CDDP and A2780/CDDP) and corresponding parental cell lines (SKOV-3 and A2780) (*** P<0.001).
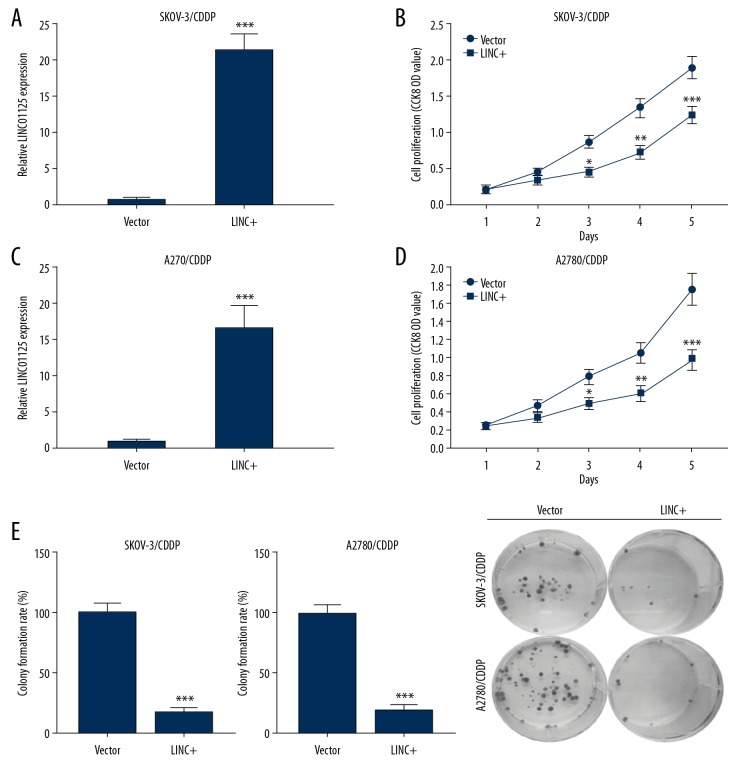
LINC01125 overexpression blocked the proliferation of CDDP-resistant cells
To investigate the biological functions of LINC01125 in OC, we overexpressed this lncRNA in CDDP-resistant OC cells. The efficiency of LINC01125 overexpression in SKOV-3/CDDP and A2780/CDDP cells was examined using qRT-PCR (Figure 2A, 2C). CCK-8 assays showed that the viability of LINC01125-overexpressing SKOV-3/CDDP and A2780/CDDP cells was significantly decreased compared to vector-transfected cells (Figure 2B, 2D). Moreover, in the colony formation assay, we found that the colony number was remarkably decreased in LINC01125-overexpressing SKOV-3/CDDP and A2780/CDDP cells (Figure 2E). These findings indicated that LINC01125 inhibits OC cell proliferation in vitro.
Figure 2.
LINC01125 overexpression significantly inhibited the proliferation of CDDP-resistant cells. (A, C) The overexpression efficiency of LINC01125 in SKOV-3/CDDP and A2780/CDDP cells was determined by qRT-PCR assay (*** P<0.001). (B, D) CCK-8 assays were carried out to confirm the effects of LINC01125 overexpression on the viability of SKOV-3/CDDP and A2780/CDDP cells (* P<0.05, ** P<0.01, *** P<0.001). (E) Colony formation assays were conducted to evaluate the proliferation of SKOV-3/CDDP and A2780/CDDP cells transfected with LINC01125 or empty vector.
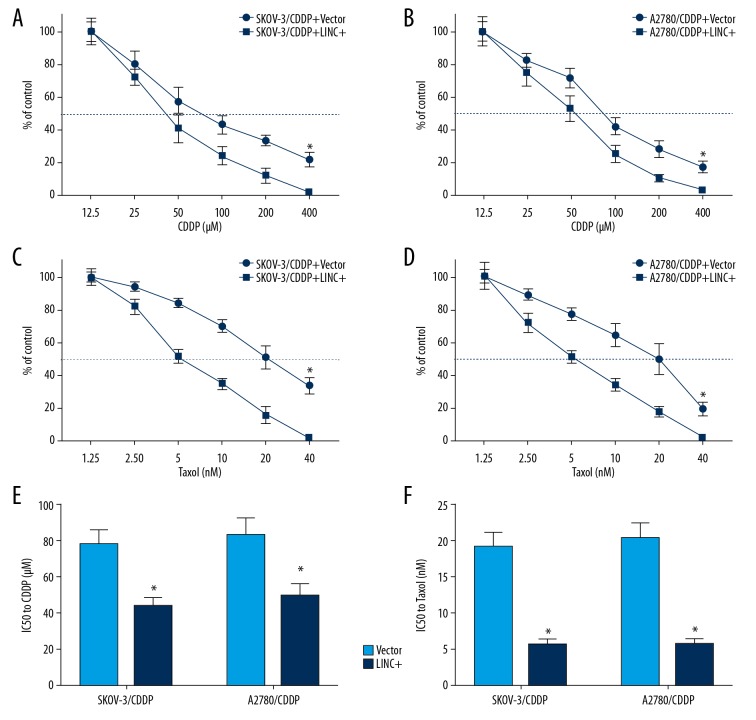
LINC01125 overexpression significantly enhanced the chemosensitivity of CDDP-resistant cells to CDDP and Taxol
We then evaluated the role of LINC01125 in the chemoresistance of OC cells. Upon treating SKOV-3/CDDP and A2780/CDDP cells with different concentrations of CDDP (12.5, 25, 50, 100, 200, and 400 μmol) or Taxol (1.25, 2.50, 5, 10, 20, and 40 nmol), we found that LINC01125 overexpression significantly enhanced the chemosensitivity of CDDP-resistant OC cells to CDDP and Taxol (Figure 3A–3D). Moreover, we demonstrated that LINC01125 overexpression significantly decreased the half maximal inhibitory concentration (IC50) value of CDDP and Taxol in SKOV-3/CDDP and A2780/CDDP cells (Figure 3E, 3F). These findings suggested that LINC01125 overexpression enhances the chemosensitivity of CDDP-resistant cells to CDDP and Taxol.
Figure 3.
LINC01125 overexpression significantly enhanced the chemosensitivity of CDDP-resistant cells to CDDP and Taxol. (A, C) Viability curves of LINC01125-overexpressing SKOV-3/CDDP cells treated with different doses of CDDP (12.5, 25, 50, 100, 200, and 400 μM) and Taxol (1.25, 2.50, 5, 10, 20, and 40 μM) (* P<0.05). (B, D) Viability curves of LINC01125-overexpressing A2780/CDDP cells treated with different doses of CDDP (12.5, 25, 50, 100, 200, and 400 μM) and Taxol (1.25, 2.50, 5, 10, 20, and 40 μM) (* P<0.05). (E, F) The half maximal inhibitory concentration (IC50) of CDDP and Taxol in SKOV-3/CDDP and A2780/CDDP cells treated with LINC01125 (* P<0.05).
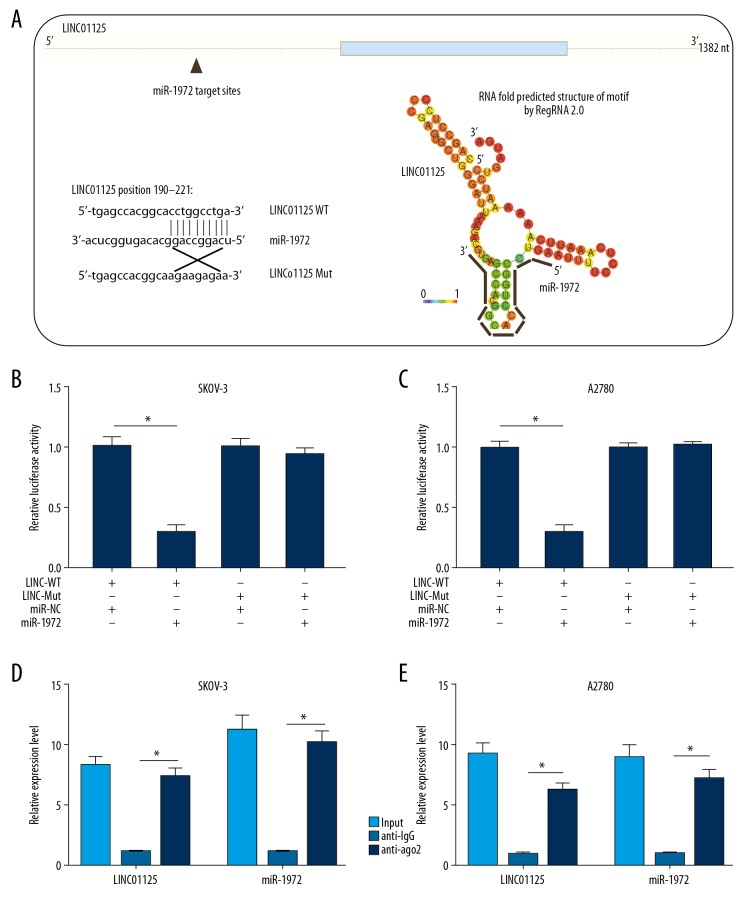
LINC01125 acted as a miRNA sponge of miR-1972 in OC cells
In this study, we predicted the target miRNAs of LINC01125 by bioinformatics analysis. The results showed that miR-1972 might possess a complementary sequence for LINC01125 and physically interact with LINC01125 (Figure 4A). To verify the interaction between LINC01125 and miR-1972, a dual-luciferase reporter assay was performed by cotransfecting SKOV-3 and A2780 cells with LINC01125-WT/Mut plasmid and miR-1972/NC. The results indicated that the luciferase activity of SKOV-3 and A2780 cells driven by LINC01125-WT could be significantly attenuated by miR-1972, but not miR-NC, while the luciferase activity of SKOV-3 and A2780 cells driven by LINC01125-Mut was not affected (Figure 4B, 4C). Moreover, RIP assays were performed to further confirm the interaction between LINC01125 and miR-1972 in SKOV-3 and A2780 cells. The results indicated that LINC01125 and miR-1972 were significantly higher in the anti-AGO2 group than in the anti-IgG group in both SKOV-3 and A2780 cells (Figure 4D, 4E). These results indicated that LINC01125 directly binds to miR-1972 in OC cells.
Figure 4.
LINC01125 acted as a miRNA sponge of miR-1972 in OC cells. (A) Bioinformatics analysis of the interaction between miR-1972 and LINC01125. (B, C) Dual-luciferase reporter assays were utilized to verify the interaction between miR-1972 and LINC01125 in SKOV-3 and A2780 cells (* P<0.05). (D, E) RIP assays were performed in SKOV-3 and A2780 cells to further confirm the interaction between miR-1972 and LINC01125 using input from cell lysate, normal IgG, and anti-AGO2 samples (* P<0.05).
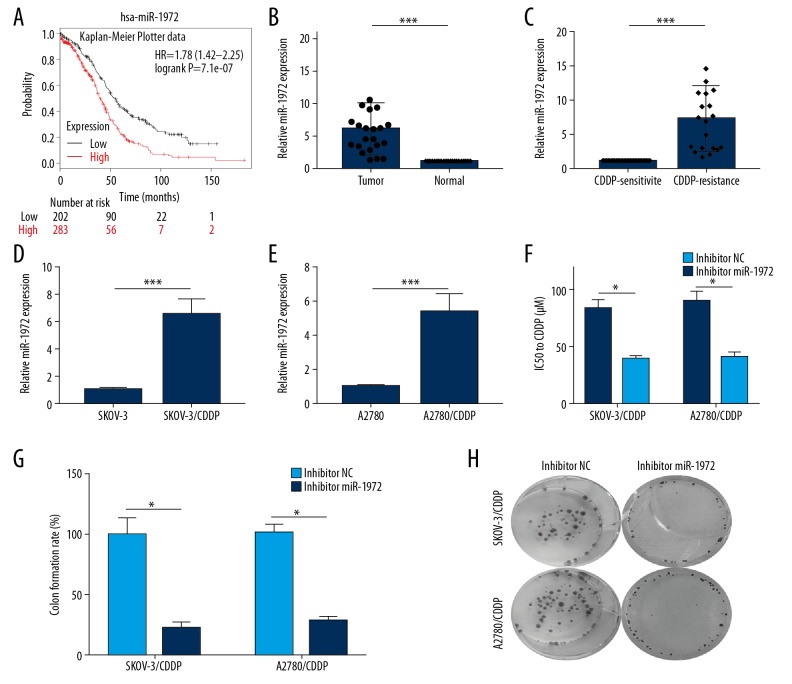
miR-1972 had the function of an oncogene in OC
Using Kaplan-Meier Plotter, we disclosed that OC patients with high miR-1972 expression had a worse prognosis than patients with low miR-1972 expression (Figure 5A). Using qRT-PCR, we found that miR-1972 was upregulated in OC tissues compared to normal tissues (Figure 5B). miR-1972 expression was then examined in CDDP-sensitive or CDDP-resistant OC tissues and cell lines using qRT-PCR. The results indicated that miR-1972 expression was markedly higher in CDDP-resistant OC tissues than in CDDP-sensitive OC tissues (Figure 5C). Relative miR-1972 expression was also considerably higher in SKOV-3/CDDP and A2780/CDDP cells than in SKOV-3 and A2780 cells, respectively (Figure 5D, 5E). We then assessed the effects of miR-1972 inhibition on the resistance of OC cells to CDDP by determining the IC50 value. The IC50 value of CDDP in SKOV-3/CDDP and A2780/CDDP cells treated with the miR-1972 inhibitor was significantly decreased (Figure 5F). In addition, colony formation assay was applied to examine the effects of miR-1972 inhibitor on the proliferation of SKOV-3/CDDP and A2780/CDDP cells. The results indicated that the miR-1972 inhibitor significantly decreased the colony formation rate compared to the NC (Figure 5G, 5H). These findings indicated that miR-1972 inhibition can significantly inhibit the proliferation of CDDP-resistant OC cells and enhance the cytotoxicity of CDDP in CDDP-resistant OC cells.
Figure 5.
miR-1972 served as an oncogene in OC. (A) Kaplan-Meier Plotter data showed the prognosis of OC patients with low miR-1972 expression. (B) Relative miR-1972 expression in OC and normal tissue samples was examined using qRT-PCR assay (*** P<0.001). (C) Relative miR-1972 expression of CDDP-resistant and CDDP-sensitive OC samples (*** P<0.001). (D, E) Relative miR-1972 expression was measured in SKOV-3 and SKOV-3/CDDP cells, as well as in A2780 and A2780/CDDP cells (*** P<0.001). (F) The IC50 values in SKOV-3/CDDP and A2780/CDDP cells treated with vector and miR-1972 inhibitor (*** P<0.001). (G, H) Colony formation assays were performed in miR-1972 inhibitor- or vector-treated SKOV-3/CDDP and A2780/CDDP cells to evaluate the effects of miR-1972 silencing on cell proliferation (* P<0.05).
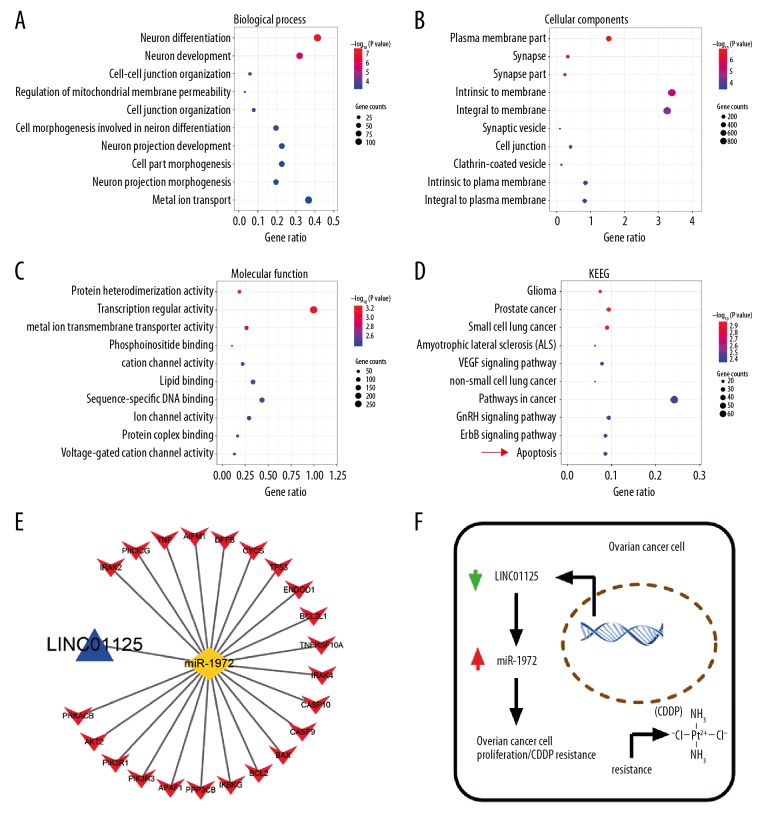
miR-1972 is involved in regulating the apoptosis pathway in OC
The target genes of miR-1972 predicted by TargetScan were analyzed using gene set enrichment analysis to obtain the biological process (BP), cellular component (CC), molecular function (MF), and KEGG pathways. The BP genes exhibited significant enrichment in neuron differentiation, neuron development, and cell-cell junction organization (Figure 6A). Among the CC genes, the top 3 functional items were plasma membrane part, intrinsic to membrane, and integral to membrane (Figure 6B). The MF genes showed significant enrichment in transcription regulator activity, protein heterodimerization activity, and metal ion transmembrane transporter activity (Figure 6C). KEGG pathway enrichment analysis showed the most significant enrichment in glioma, prostate cancer, small cell lung cancer, and apoptosis (Figure 6D). Taken together, our results show that LINC01125 might regulate the proliferation and CDDP sensitivity of OC through the apoptosis pathway by increasing miR-1972 (Figure 6E, 6F).
Figure 6.
miR-1972 was involved in regulating the apoptosis pathway in OC. (A–D) The target genes of miR-1972 predicted by TargetScan were analyzed with gene set enrichment analysis to obtain the biological process, cellular component, molecular function, and KEGG pathways. (E, F) Diagram of the mechanisms underlying the LINC01125/miR-1972 axis in OC.
Discussion
OC is one of the most lethal gynecologic malignancies in the world, especially at the advanced stages, posing a serious threat to the health of women [18]. Exploring preventive and therapeutic strategies for OC has been the focus of modern fundamental research [19]. Currently, the main therapeutic strategy for OC patients with early FIGO stage disease is surgical resection; however, due to the high risk of recurrence (25–30%), the majority of patients receive concurrent adjuvant chemotherapy [2,20]. Usually, surgical resection is not curative for patients with advanced stage OC, and postoperative chemotherapy has been widely accepted as the standard for the treatment of advanced OC [21]. Cisplatin, a well-known antitumor agent that was first synthesized in 1844 by M. Peyrone, has been proven to combat miscellaneous tumors, such as breast cancer, lung cancer, and liver cancer, significantly improving the prognosis of cancer patients [22]. Recently, accumulating evidence has shown that cisplatin also has significant anticancer activity in patients with advanced OC [23]. However, despite initial efficacy, more than 70% of OC patients treated with cisplatin relapse within 2–3 years and generally require second-line therapy [24]. Therefore, it is urgent to improve the efficacy of cisplatin treatment in OC patients.
The expression of lncRNAs was proved to be dysregulated in cancers, such as OC, lung cancer, and breast cancer, suggesting that lncRNAs has significant effects in cancer progression [25,26]. Although the exact molecular mechanisms of OC chemoresistance remain largely unclear, emerging evidence has shown that lncRNAs might be involved [27]. For example, Wang et al. reported that the lncRNA EBIC promotes cell proliferation, metastasis, and cisplatin resistance of OC and is associated with poor prognosis of OC patients [28]. In our study, we analyzed the expression profile of lncRNAs from control and miR-200a-overexpressing samples. LINC01125, which is expressed at low levels in both cisplatin-resistant tissues and cell lines, was selected for further study. Because miR-200a was previously shown to be relevant to the initiation, progression, and chemoresistance of OC, LINC01125, which was downregulated in the miR-200a-overexpressing samples, was also considered and verified to be an enhancer of cisplatin cytotoxicity.
Studies have concluded that the biological effects of lncRNAs on tumor progression are mainly mediated by specific miRNAs, which further regulate tumor suppressor genes or oncogenes [29,30]. Therefore, in this study, we predicted the target miRNAs of LINC01125 by TargetScan and identified miR-1972. miR-1972 was previously reported to mediate the promotive effects of the lncRNA DANCR on osteosarcoma [31]. However, its role in OC remains undetermined. In this study, we demonstrated miR-1972 was highly expressed in cisplatin-resistant OC tissues and cells, and inhibition of miR-1972 significantly enhanced the sensitivity of OC cells to cisplatin. In the following gene set enrichment analysis of miR-1972 target genes, we revealed that miR-1972 was relevant to the apoptosis pathway.
Conclusions
Our findings indicated that LINC01125 was expressed at a low level in cisplatin-resistant OC tissues and cells, while miR-1972 was highly expressed. Both LINC01125 overexpression and miR-1972 knockdown enhanced the sensitivity of OC cells to cisplatin. LINC01125 might exert its regulatory effects on the cisplatin resistance of OC by directly binding to miR-1972 to regulate apoptosis pathway. Taken together, the data suggest that LINC01125 affects the sensitivity of OC to cisplatin by miR-1972.
Footnotes
Source of support: Departmental sources
Conflict of interest
None.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Di Lorenzo G, Ricci G, Severini GM, et al. Imaging and therapy of ovarian cancer: Clinical application of nanoparticles and future perspectives. Theranostics. 2018;8(16):4279–94. doi: 10.7150/thno.26345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koshiyama M, Matsumura N, Konishi I. Subtypes of ovarian cancer and ovarian cancer screening. Diagnostics (Basel) 2017;7(1) doi: 10.3390/diagnostics7010012. pii: E12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathieu KB, Bedi DG, Thrower SL, et al. Screening for ovarian cancer: Imaging challenges and opportunities for improvement. Ultrasound Obstet Gynecol. 2018;51(3):293–303. doi: 10.1002/uog.17557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart SL, Harewood R, Matz M, et al. Disparities in ovarian cancer survival in the United States (2001–2009): Findings from the CONCORD-2 study. Cancer. 2017;123(Suppl 24):5138–59. doi: 10.1002/cncr.31027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright AA, Bohlke K, Armstrong DK, et al. Neoadjuvant chemotherapy for newly diagnosed, advanced ovarian cancer: Society of Gynecologic Oncology and American Society of Clinical Oncology Clinical Practice Guideline. Gynecol Oncol. 2016;143(1):3–15. doi: 10.1016/j.ygyno.2016.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang C, Wang M, Shi C, et al. Long non-coding RNA Linc00312 modulates the sensitivity of ovarian cancer to cisplatin via the Bcl-2/Caspase-3 signaling pathway. Biosci Trends. 2018;12(3):309–16. doi: 10.5582/bst.2018.01052. [DOI] [PubMed] [Google Scholar]
- 8.Damia G, Broggini M. Platinum resistance in ovarian cancer: Role of DNA repair. Cancers (Basel) 2019;11(1) doi: 10.3390/cancers11010119. pii: E119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Markowska A, Sajdak S. Role of cancer stem cells and microRNA in resistance to chemotherapy in patients with ovarian cancer. Eur J Gynaecol Oncol. 2017;38(2):181–83. [PubMed] [Google Scholar]
- 10.Guo L, Zhao Y, Yang S, et al. An integrated analysis of miRNA, lncRNA, and mRNA expression profiles. Biomed Res Int. 2014;2014 doi: 10.1155/2014/345605. 345605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuel P, Pink RC, Brooks SA, Carter DR. miRNAs and ovarian cancer: A miRiad of mechanisms to induce cisplatin drug resistance. Expert Rev Anticancer Ther. 2016;16(1):57–70. doi: 10.1586/14737140.2016.1121107. [DOI] [PubMed] [Google Scholar]
- 12.Liu N, Zhong L, Zeng J, et al. Upregulation of microRNA-200a associates with tumor proliferation, CSCs phenotype and chemosensitivity in ovarian cancer. Neoplasma. 2015;62(4):550–59. doi: 10.4149/neo_2015_066. [DOI] [PubMed] [Google Scholar]
- 13.Liu N, Zeng J, Zhang X, et al. [Involvement of miR-200a in chemosensitivity regulation of ovarian cancer]. Zhonghua Yi Xue Za Zhi. 2014;94(27):2148–51. [in Chinese] [PubMed] [Google Scholar]
- 14.Weidle UH, Birzele F, Kollmorgen G, Ruger R. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics. 2017;14(3):143–60. doi: 10.21873/cgp.20027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kondo Y, Shinjo K, Katsushima K. Long non-coding RNAs as an epigenetic regulator in human cancers. Cancer Sci. 2017;108(10):1927–33. doi: 10.1111/cas.13342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rotini A, Martinez-Sarra E, Pozzo E, Sampaolesi M. Interactions between microRNAs and long non-coding RNAs in cardiac development and repair. Pharmacol Res. 2018;127:58–66. doi: 10.1016/j.phrs.2017.05.029. [DOI] [PubMed] [Google Scholar]
- 17.Wan W, Hou Y, Wang K, et al. The LXR-623-induced long non-coding RNA LINC01125 suppresses the proliferation of breast cancer cells via PTEN/AKT/p53 signaling pathway. Cell Death Dis. 2019;10(3):248. doi: 10.1038/s41419-019-1440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Webb PM, Jordan SJ. Epidemiology of epithelial ovarian cancer. Best Pract Res Clin Obstet Gynaecol. 2017;41:3–14. doi: 10.1016/j.bpobgyn.2016.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Dong X, Men X, Zhang W, Lei P. Advances in tumor markers of ovarian cancer for early diagnosis. Indian J Cancer. 2014;51(Suppl 3):e72–76. doi: 10.4103/0019-509X.154049. [DOI] [PubMed] [Google Scholar]
- 20.Trimbos JB, Vergote I, Bolis G, et al. Impact of adjuvant chemotherapy and surgical staging in early-stage ovarian carcinoma: European Organisation for Research and Treatment of Cancer-Adjuvant ChemoTherapy in Ovarian Neoplasm trial. J Natl Cancer Inst. 2003;95(2):113–25. [PubMed] [Google Scholar]
- 21.Chung YS, Kim YJ, Lee I, et al. Impact of neoadjuvant chemotherapy and postoperative adjuvant chemotherapy cycles on survival of patients with advanced-stage ovarian cancer. PLoS One. 2017;12(9):e0183754. doi: 10.1371/journal.pone.0183754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: Molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–78. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikula-Pietrasik J, Witucka A, Pakula M, et al. Comprehensive review on how platinum- and taxane-based chemotherapy of ovarian cancer affects biology of normal cells. Cell Mol Life Sci. 2019;76(4):681–97. doi: 10.1007/s00018-018-2954-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piccart MJ, Du Bois A, Gore ME, et al. A new standard of care for treatment of ovarian cancer. Eur J Cancer. 2000;36(1):10–12. doi: 10.1016/s0959-8049(99)00210-5. [DOI] [PubMed] [Google Scholar]
- 25.Renganathan A, Felley-Bosco E. Long noncoding RNAs in cancer and therapeutic potential. Adv Exp Med Biol. 2017;1008:199–222. doi: 10.1007/978-981-10-5203-3_7. [DOI] [PubMed] [Google Scholar]
- 26.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77(15):3965–81. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu LL, Li CJ, Xu Y, et al. Role of lncRNAs as novel biomarkers and therapeutic targets in ovarian cancer. Crit Rev Eukaryot Gene Expr. 2017;27(2):183–95. doi: 10.1615/CritRevEukaryotGeneExpr.2017019244. [DOI] [PubMed] [Google Scholar]
- 28.Xu QF, Tang YX, Wang X. LncRNA EBIC promoted proliferation, metastasis and cisplatin resistance of ovarian cancer cells and predicted poor survival in ovarian cancer patients. Eur Rev Med Pharmacol Sci. 2018;22(14):4440–47. doi: 10.26355/eurrev_201807_15495. [DOI] [PubMed] [Google Scholar]
- 29.Xie Y, Dang W, Zhang S, et al. The role of exosomal noncoding RNAs in cancer. Mol Cancer. 2019;18(1):37. doi: 10.1186/s12943-019-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abdollahzadeh R, Daraei A, Mansoori Y, et al. Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: A new look at hallmarks of breast cancer. J Cell Physiol. 2019;234(7):10080–100. doi: 10.1002/jcp.27941. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Zeng X, Wang N, et al. Long noncoding RNA DANCR, working as a competitive endogenous RNA, promotes ROCK1-mediated proliferation and metastasis via decoying of miR-335-5p and miR-1972 in osteosarcoma. Mol Cancer. 2018;17(1):89. doi: 10.1186/s12943-018-0837-6. [DOI] [PMC free article] [PubMed] [Google Scholar]