Abstract
Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) encodes a 403-amino acid protein with an amino-terminal domain that shares sequence homology with the actin-binding protein tensin and the putative tyrosine-protein phosphatase auxilin. Crystal structure analysis of PTEN has revealed a C2 domain that binds to phospholipids in membranes and a phosphatase domain that displays dual-specific activity toward both tyrosine (Y), serine (S)/threonine (T), as well as lipid substrates in vitro. Characterized primarily as a lipid phosphatase, PTEN plays important roles in multiple cellular processes including cell growth/survival as well as metabolism.
PTEN STRUCTURE PREDICTS A DUAL PROTEIN PHOSPHATASE
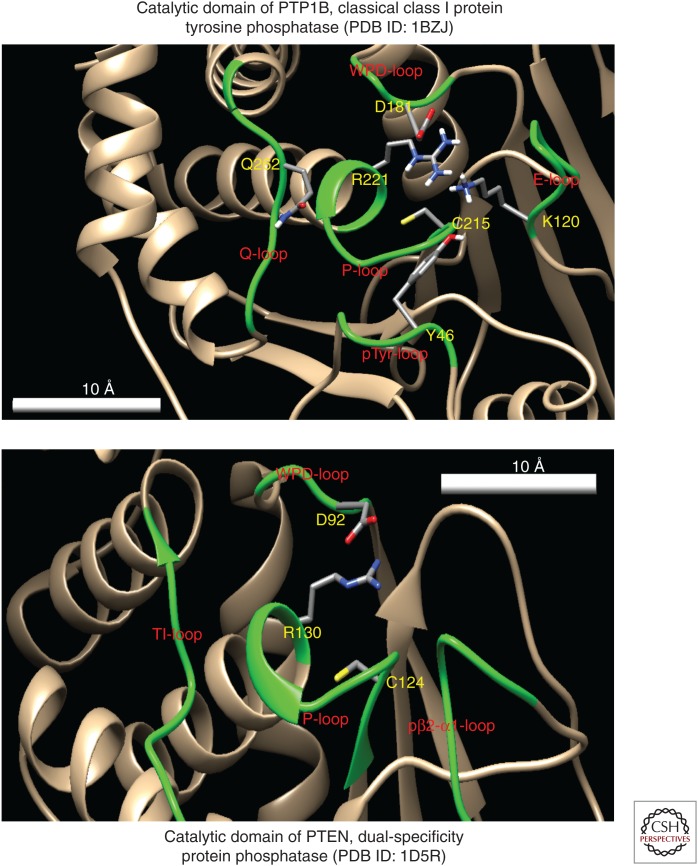
Initial amino acid sequence analysis suggested that PTEN belonged to the protein tyrosine phosphatase family (Nelen et al. 1997). All protein tyrosine phosphatases (e.g., PTP1B) contain a signature CX5R motif that forms the catalytic active pocket termed the P-loop (Fig. 1). The amino acid selectivity of protein phosphatases is determined partially by the accessibility of the catalytic cysteine at the base of the catalytic cleft (Tautz et al. 2013). For protein tyrosine phosphatases, the depth of the cleft matches the length of a phosphotyrosine residue, whereas the shorter phosphoserine/threonine residues are not able to reach the catalytic cysteine. In contrast, typical catalytic loop of dual-specific phosphatases are at the bottom of a much shallower pocket, which gives access to all three phosphorylated hydroxyl amino acids (Myers et al. 1997; Bhattacharyya et al. 2012). The amino-terminal domain of PTEN contains the signature motif HCKAGKGR for protein tyrosine phosphatases, forming the P-loop (residues H123–R130) at the bottom of its active site pocket with cysteine being at the bottom of the catalytic pocket (Lee et al. 1999). Uniquely, the catalytic pocket of PTEN, although it shares a similar depth as PTP1B, shows a much wider opening. This wider opening allows tyrosine and serine/threonine to reach the cysteine even though it is deep in the pocket. Thus, both phosphotyrosine and phosphoserine/threonine have been found to be substrates for PTEN (Myers et al. 1997; Lee et al. 1999).
Figure 1.
Structure of catalytic pocket of PTEN defines it as a dual protein phosphatase and a phosphoinositide phosphatase. The CX5R motif on protein tyrosine phosphatases forms a P-loop that sits at the catalytic active pockets. A cysteine at the base of this pocket is required for the phosphatase to react with the substrate. (Top) Structure of the catalytic pocket of PTP1B, a protein tyrosine phosphatase also contains Q-loop (left), WPD-loop (top), E-loop (right), and pTyr-loop (bottom) (labeled in green). A conserved glutamine on the Q-loop position a water molecule for hydrolysis. E-loop functions to regulate the dynamics of the WPD-loop. pTyr-loop controls specificity of classical PTP substrates. (Bottom) In addition to P-loop and WPD-loop that are also found on PTP1B, PTEN also contains a TI-loop and a pβ2-α1-loop. TI-loop determines the size of the catalytic pocket of PTEN and allowed accommodation of large lipids such as phosphatidylinositol 3,4,5-triphosphate (PIP3). Function of pβ2-α1-loop is not clear but it may have a similar location as pTyr-loop in classical PTPs. (Image generated using data from the Protein Data Bank and analyzed using the UCSF Chimera software.)
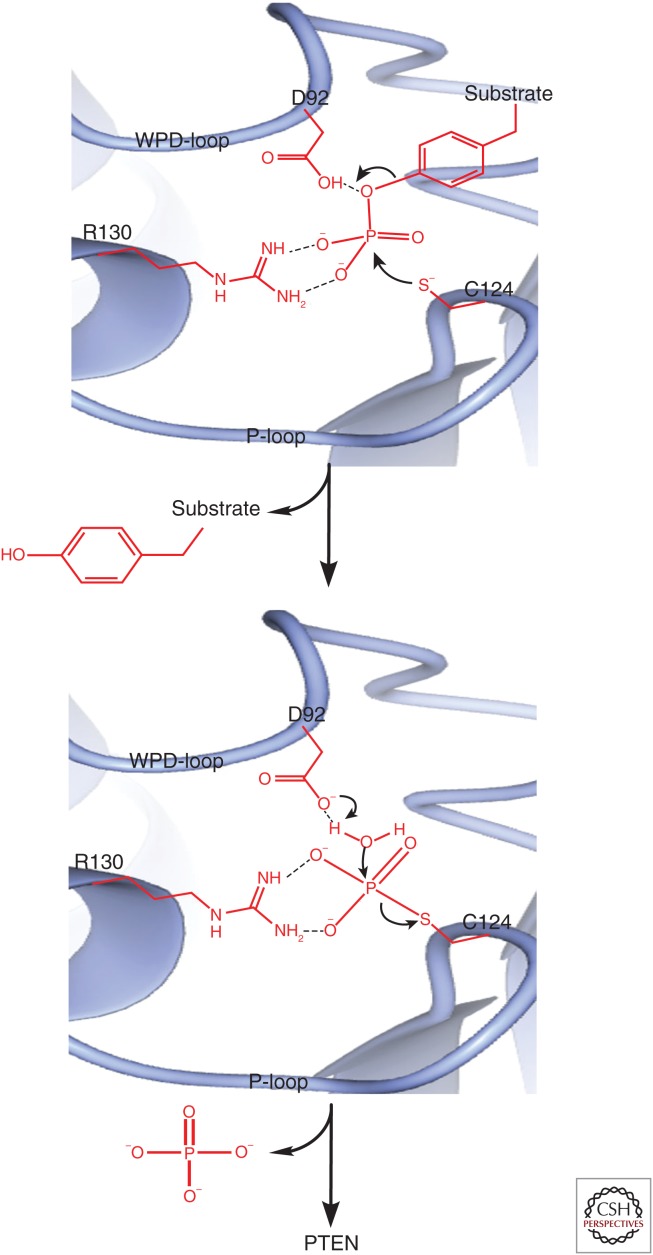
Similar to other protein tyrosine phosphatases, the catalytic process of PTEN is mainly coordinated by three amino acids on the P-loop and WPD-loop (Tautz et al. 2013). The catalytic cysteine (C124) of the P-loop initiates a nucleophilic attack on the phosphorous atom of the phosphopeptide. This results in the breaking of the oxygen–phosphorus bond between the peptide and its phosphate group, leading to the formation of a phosphocysteine through a phosphorous–sulfur bond. This process is facilitated by aspartic acid (D92) of the WPD-loop that donates a proton to the hydroxy leaving group on the substrate peptide. The oxygen atom of aspartic acid (D92) also binds to the hydrogen on the water molecule, allowing the nucleophilic hydroxy group of the water molecule to hydrolyze the phosphorous–sulfur bond and release the free phosphate. A guanidinium group of a conserved arginine (R130) on the P-loop is needed to coordinate the position of the phosphate group during this process (Fig. 2).
Figure 2.
The interaction of PTEN with its protein substrates at the catalytic pocket. The catalytic cysteine (C124) of the P-loop initiates a nucleophilic attack on the phosphorous atom of the phosphopeptide. This nucleophilic attack by C124 results in the breakage of the oxygen–phosphorus bond between the peptide and its phosphate group, leading to the formation of a phosphocysteine through a phosphorous–sulfur bond. This process is facilitated by aspartic acid (D92) of the WPD-loop, which donates a proton to the hydroxy leaving group on the substrate protein. The aspartic acid (D92) that donated a proton is left with a negatively charged oxygen atom, which now binds to the hydrogen on the water molecule. This allows the nucleophilic hydroxy group of the water molecule to hydrolyze the phosphorous–sulfur bond and release the free phosphate. A guanidinium group of a conserved arginine (R130) on the P-loop is needed to coordinate the position of the phosphate group during this process. (Data downloaded from the Protein Data Bank and analyzed using CCP4MG software.)
BIOLOGICAL FUNCTIONS FOR PTEN AS A PROTEIN PHOSPHATASE
As the C124 on the P-loop is critical for the phosphatase function of PTEN, mutations of C124 completely eliminate its catalytic activity toward either protein or lipid substrates (Myers et al. 1998). Other mutations that lead to complete or partial loss-of-function have also been reported (Myers et al. 1997; Herschbein and Liesveld 2018). In particular, studies of mutants with loss of only lipid or protein phosphatase activity but not both have led to some understanding of how the protein versus lipid phosphatase activity of PTEN may play a role in its biological function. One such mutation is that of glycine at the P-loop. PTEN G129E retains the protein phosphatase activity toward polyGlu4Tyr1 (Myers et al. 1998) but results in lost activity toward its lipid substrate (Rodríguez-Escudero et al. 2011). Using this mutant, cell motility was established as the function dependent on PTEN protein tyrosine phosphatase activity (Tamura et al. 1998; Gildea et al. 2004). In U87-MG glioma cells, tyrosine phosphorylation of focal adhesion kinase (FAK) can be induced by spreading of the cells on fibronectin. PTEN G129E was reported to inhibit this phosphorylation as well as cell spreading similar to wild-type PTEN (wtPTEN), whereas PTEN C124S that lacks all phosphatase activity cannot (Tamura et al. 1998). Transforming growth factor β (TGF-β)-induced phosphorylation of FAK during the induction of epithelial-mesenchymal transition (EMT) in H358ON lung cancer cells was also shown to be inhibited by PTEN G129E mutants (Kusunose et al. 2015). The dephosphorylation of FAK resulting from the PTEN protein phosphatase activity was also shown to be responsible for wtPTEN and PTEN G129E to inhibit hyaluronic acid-induced matrix metalloproteinase-9 expression in U87-MG glioblastoma cells (Park et al. 2002). Other support for the role of PTEN protein phosphatase activity in cell movement was observed in chemotaxis of bladder cancer cells where G129E was equally as effective at inhibiting motility of the cells as wtPTEN even though it did not have any effect on colony size (Gildea et al. 2004). Vitronectin-directed migration was also found to be dependent on the protein phosphatase activity of PTEN using G129E (Dey et al. 2008). In chick embryos, EMT needed before primitive streak mesoderm cells migrate away from the streak is inhibited by the protein phosphatase activity of PTEN, whereas directional motility is regulated through its lipid phosphatase activity (Leslie et al. 2007).
Although FAK dephosphorylation is related to PTEN protein phosphatase function and partially responsible for the PTEN phosphatase function-regulated cell motility, specific enzyme–substrate relationship between PTEN and FAK has not yet been established. Recently, Ras related in brain 7 (Rab7) was found to be directly dephosphorylated by PTEN on both serine and tyrosine (Shinde and Maddika 2016). Rab7 is a conserved Rab family small GTPase that plays pivotal roles in late endosome trafficking and lysosome biogenesis. Dephosphorylation of Rab7 by PTEN was shown to regulate endosome maturation and play important roles in epidermal growth factor receptor (EGFR) trafficking between membrane and endosome (Shinde and Maddika 2016). Active FAK has been found to localize with integrins on endosome and is positively regulated by integrin endocytosis (Alanko et al. 2015). Whether the PTEN-regulated endosome maturation through Rab7 plays a role in FAK phosphorylation and activation is unknown.
Although ample evidence has reported the role of AKT in PTEN-mediated cell growth effect, PTEN was also found to regulate cell cycle and apoptosis via its protein phosphatase activity. Using a Tet-off expression system in MCF-7 cells, it was shown that PTEN G129E can partially inhibit the expression of cyclin D1 (Weng et al. 2001). During apoptosis induction, PTEN was found to mobilize the transfer of calcium from the endoplasmic reticulum (ER) to mitochondria. This process is dependent on PTEN protein phosphatase activity as G129E and not C124S mutant of PTEN is capable of evoking such a response (Bononi et al. 2013).
Furthermore, PTEN also participates in maintaining the integrity of the genome using its protein phosphatase activity. When localized in the nucleus, PTEN protects the DNA from genotoxic stress. Expression of G129E PTEN is able to protect U87-MG cells from ionizing radiation-induced DNA damage (Bassi et al. 2013). In addition, PTEN prevents gross chromosome alterations by associating and dephosphorylating Polo-like kinase 1 (PLK1) on T210 to regulate cell mitosis. As a result, loss of PTEN protein phosphatase activity leads to failed cytokinesis and increased polyploidization (Zhang et al. 2016). Moreover, minichromosome maintenance complex component 2 (MCM2), a key replication helicase of the replisome essential for DNA replication, was also found to be a substrate for PTEN (Feng et al. 2015). This function of PTEN was found necessary for preventing chromosomal aberrations under replication stress.
A proteomic analysis of U87-MG cells stably expressing wtPTEN, PTEN C124G (no phosphatase activity) and PTEN G129E has identified ∼30 proteins that are associated with PTEN's protein phosphatase activities (Shim et al. 2006). A number of these proteins are involved in cytoskeletal regulation, consistent with the observation that PTEN protein phosphatase activity is primarily associated with morphological changes. However, these analyses do not distinguish whether these candidates are direct or indirect targets of PTEN's protein phosphatase activity. In fact, PTEN is reported to be autodephosphorylated at T366 (Zhang et al. 2012). Phosphorylation of PTEN has been reported to occur on several residues, mostly located at the C2 domain. Two major sites are S370 and S385 (Torres and Pulido 2001; Miller et al. 2002), although phosphorylation on other sites such as the aforementioned T366 have also been reported (Gericke et al. 2006). It is proposed that the phosphorylated PTEN assumes a more stable conformation (Vazquez et al. 2000). Dephosphorylation results in the opening of PTEN structure, leading to a more active but also less stable PTEN. The autophosphorylated PTEN on its carboxy-terminal T366 was reported to regulate the density of neuronal spines in cultured hippocampal slices, a process that is dependent on the protein phosphatase activity of PTEN (Zhang et al. 2012). An unphosphorylated PTEN at the C2 domain has been linked to TGF-β-induced EMT processes (Kusunose et al. 2015).
In vivo, heterozygous mice carrying G129E mutation displayed gain-of-function phenotype for tumor growth when compared with catalytic domain-defective C124R-mutant mice (Wang et al. 2010). This observation was surprising because the protein and lipid phosphatase activities were found to have additive effects on the cell cycle (Hlobilkova et al. 2000; Weng et al. 2001). Thus, the protein phosphatase mutant of PTEN may have its own activities. The gain-of-function tumor phenotype associated with PTEN protein phosphatase activity is either the result of such additional activity or an outcome of potential interacting with the wtPTEN counterpart. The G129E mutant may attenuate the function of the wtPTEN in these heterozygous mice, which does not occur in C124R mice (Papa et al. 2014).
In general, several putative substrates for PTEN protein phosphatase activity have been identified (Table 1). Besides what was discussed above, PTEN was found to bind and dephosphorylate protein tyrosine kinase 6 (PTK6) (Wozniak et al. 2017). Phosphorylation of Y342 activates PTK6 and facilitates translocation of PTK6 to the plasma membrane, which by itself is sufficient to transform fibroblasts. PTEN negatively regulates PTK6 through dephosphorylation of PTK6 on Y342 and plays a role in prostate cancer invasion. Ptk6 deletion in mice inhibits development of invasive prostate adenocarcinoma following prostate-specific Pten deletion (Wozniak et al. 2017). In addition, PTEN was found to be a phosphatase for insulin receptor substrate 1 (IRS1) (Shi et al. 2014). Dephosphorylation of IRS1 on Y612 and Y989 potentially plays a role in insulin and insulin-like growth factor 1 (IGF-1) signaling. PTEN was also reported to dephosphorylate Dishevelled (DVL), a ciliogenesis regulator involved in cilia formation during early stages of development in Xenopus oocytes and in tracheal of mouse embryo (Shnitsar et al. 2015).
Table 1.
Known substrates regulated by protein phosphatase activity of PTEN
| Putative target (site) | Cell type | Biological function | References |
|---|---|---|---|
| PTEN (S366) | U-87MG | Inhibits cell invasion, changes cell morphology | Tibarewal et al. 2012 |
| Dvl (S143) | hTERT RPE-1 | Stabilizes primary cilia | Shnitsar et al. 2015 |
| Rab7 (S72, Y183) | HeLa | Promotes recruitment of Rab7 to late endosomes and late endosome maturation | Shinde and Maddika (2016) |
| PTK6 (Y342) | PC3 | Inhibits progression of prostate adenocarcinoma | Wozniak et al. 2017 |
| IRS1 (Y612, Y989) | MEF (mouse embryonic fibroblast) | Inhibits IGF and insulin signaling | Shi et al. 2014 |
| FAK (Y397)a | U-87MG, NIH/3T3 | Inhibits cell spreading and migration | Tamura et al. 1998; Cai et al. 2005 |
| CREB (S133) | MEF, PE2 | Regulates CREB transcriptional activity and cell survival | Gu et al. 2011 |
| MCM2 (S41) | HCT116 | Regulates replication fork progression | Feng et al. 2015 |
| PLK1 (T210) | MEF | Regulates cell mitosis and polyploid formation | Zhang et al. 2016 |
IGF, Insulin-like growth factor.
aMultiple studies suggested dephosphorylation of this site is associated with PTEN but has not yet been verified as a direct substrate of PTEN.
Screening of 11 mutants based on amino acid similarities between PTEN and two related lipid phosphatases resulted in the identification of a novel mutant Y138L that lacks the protein but retains the lipid phosphatase activity (Davidson et al. 2010). Unlike the PTEN C124S and G129E mutants, PTEN Y138L inhibited U87-MG cell colony formation (Davidson et al. 2010). Moreover, the same mutant interfered with the directed cell migration in the chicken embryo, corroborating previous observation that the lipid PTEN phosphatase activity also impacts cell movement (Leslie et al. 2007; Davidson et al. 2010). PTEN protein phosphatase activity also appears critical for controlling the neural progenitor cell (NPC) differentiation in the mouse, where the PTEN Y138L mutation, but not the G129E, mimicked the effect of PTEN knockdown on NPC differentiation (Fig. 3; Lyu et al. 2015). One possible mechanistic insight into this function was previously reported, where PTEN was shown to be physically associated with the NMDA receptor and its protein phosphatase activity was shown to act upstream of this key receptor implicated in neuronal development and plasticity (Ning et al. 2004). Protein phosphatase function of PTEN is thought to be important for maintaining the spine density of CA1 pyramidal neurons, as PTEN G129E mutant but not the Y138L and G124S mutants reduced spine density, similar to wild-type PTEN (Zhang et al. 2012). Additionally, PTEN was found to physically associate with drebrin, an actin-binding protein involved in synaptic function (Kreis et al. 2013). Serving as potential substrate for PTEN, dephosphorylation of drebrin mediates the synaptic functions regulated by PTEN.
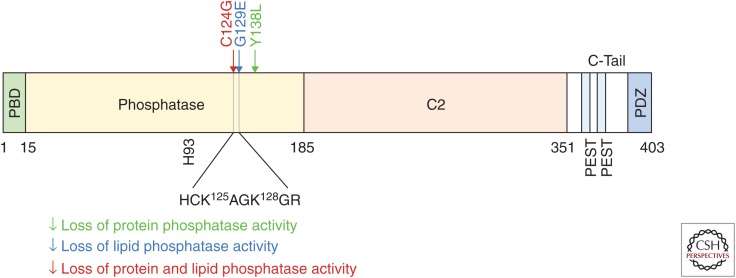
Figure 3.
Domain structure of the PTEN protein. PTEN is composed of five functional domains: (1) a PIP2 (PI(4,5)P2)-binding domain (PBD); (2) a phosphatase domain including the catalytic core (H123–R130) in which cancer-associated mutations are enriched; (3) a C2 domain, which is critical in PTEN subcellular location regulation; its interaction with the membrane-binding domain and the ubiquitination of it have both been found to regulate PTEN translocation; (4) two PEST (proline, glutamic acid, serine, and threonine) domains that are associated with degradation; and (5) a PDZ motif for protein–protein interactions. Critical point mutations that disrupt lipid or protein phosphatase activity or both are indicated with arrows.
UNIQUE FEATURES ENABLING PTEN TO ACT AS A LIPID PHOSPHATASE
In addition to the P-loop and WPD-loop, classical protein tyrosine phosphatases also contain E-loop, Q-loop, and p-Tyr loops within their catalytic domains (Tautz et al. 2013). A pair of residue insertions (residues 42–52 in the pβ2–α1 loop, and residues 163–166 in the “TI” loop) distinguish PTEN from the phosphatases such as PTP1B, a classical type I PTP and VHR (Vaccinia H1-related), a dual specificity protein phosphatase (Lee et al. 1999). These insertions, and in particular the one within the “TI” loop, are key to PTEN function as a lipid phosphatase. The TI-loop, although not forming the critical interactions with the protein substrate, determines the size of PTEN catalytic pocket (Matsuda et al. 2011). PTEN's TI-loop forms a structure that is “displaced away from the center of the pocket” (Lee et al. 1999), creating a catalytic cleft that is even larger than that of the dual specificity phosphatase VHR. As a result, the opening of PTEN's catalytic pocket allows phospholipids to reach the catalytic cysteine (Worby and Dixon 2014). Thus, in addition to being a dual specificity protein phosphatase, PTEN is also capable of accommodating big lipid substrates such as the phosphatidylinositol 3,4, 5-triphosphate (PI(3,4,5)P3) (Maehama and Dixon 1998). Another fundamental characteristic that accounts for PTEN's capability to take in lipid substrates lies within the basic residues located at the active site pocket of PTEN. The two conserved lysine residues (K125 and K128) in the P-loop (HCK125AGK128GR), along with the H93 residing in the “WPD” loop (pβ4–α3 loop, residues 91–94) (Lee et al. 1999), endow PTEN with a positively charged pocket that favors negatively charged substrates. Phosphoinositides (PIPs) carry negative charges under physiological conditions owing to their deprotonated phosphate group (Wu et al. 2014) and thus serve as a preferred substrate for PTEN (Lee et al. 1999).
PTEN crystal structure has also revealed an extensive interface between the phosphatase domain and C2 domain of PTEN (Lee et al. 1999). The interface features a hydrogen-bond network and a buried core formed by the hydrophobic and the aromatic side chains emanating from the two domains (Lee et al. 1999). Disrupting the interdomain integrity significantly impacts the enzymatic activity of PTEN. For example, D252Y, a tumor-derived mutation, disrupts the hydrogen-bond network at the interface and decreases the PI(3,4,5)P3-directed phosphatase activity of PTEN by 85% (Lee et al. 1999). Consistently, most residues that participate in forming the interdomain hydrogen bonds were found to be frequently mutated in cancers, with S170 and R173 among the eight most frequently mutated residues of PTEN (Lee et al. 1999). Based on these observations, the PTEN C2 domain was also proposed to play a role in positioning the catalytic phosphatase domain correctly toward its lipid substrates at the membrane, an observation verified in vitro using artificially membrane targeted PTEN C2 mutants with lipid-binding deficiency (Georgescu et al. 2000).
The C2 domains are membrane-binding modules whose phospholipid-binding ability is originally thought to be regulated by Ca2+ ions, primarily through interaction with the aspartate side chains (Rizo and Südhof 1998). Originally found in PLCδ1, classic PKC isoforms (PKCα, -β, -γ) and cPLA2, the C2 domain, allow these enzymes to access the membrane (Nalefski et al. 1994, 1997; Lomasney et al. 1999; Kohout et al. 2002). The carboxy-terminal domain of PTEN folds into a structure that is similar to the C2 domains found on PLCδ1, PKCδ, and cPLA2, promoting PTEN association with the cell membrane (Lee et al. 1999; Downes et al. 2007). Unlike Ca2+-dependent C2 domains that require cationic Ca2+ ions for them to bind the negatively charged phospholipid phosphoryl groups (Essen et al. 1997), the PTEN C2 domain does not bind Ca2+ and shows a Ca2+-independent characteristic for lipid binding (Lee et al. 1999). Instead of acquiring positive charges through binding to Ca2+, PTEN C2 domain is positively charged as a result of (1) solvent-exposed K260, K263, K266, K267, K269, and the partially hidden H259–D268 pair, in the membrane-interacting portions of PTEN C2 domain (CBR3 loop); and (2) a basic patch resulting from solvent-exposed K327, K330, K332, and R335 on its cα2 helix nearby (Lee et al. 1999). Together, these nine basic residues within the C2 domain help PTEN to associate with the cell membrane anionic lipids, likely through direct contact. In addition, two hydrophobic residues, M264 and L265, at the tip of the CBR3 loop in the C2 domain are thought to facilitate PTEN insertion into the lipid bilayer (Lee et al. 1999). Altering the basic and hydrophobic residues at the CBR3 and the cα2 loops in vitro has been shown to diminish membrane lipid-binding ability of PTEN, as well as its growth-suppressive activity (Lee et al. 1999; Georgescu et al. 2000).
CANONICAL SIGNALS REGULATED BY LIPID PHOSPHATASE PTEN
The unique larger catalytic pocket and C2 domain allow PTEN to accommodate lipids as substrate in which it dephosphorylates PI(3,4,5)P3 at the 3′ position on the inositol ring of PIPs and generates PI(4,5)P2 (Maehama and Dixon 1998; Lee et al. 1999; Worby and Dixon 2014). This enzymatic function of PTEN antagonizes the catalytic reaction performed by class I phosphoinositide 3-kinases (PI3Ks) that add phosphate to the 3′-hydroxyl group on the inositol ring of PI(4,5)P2 to give rise to PI(3,4,5)P3 (Yuan and Cantley 2008). Depending on the receptors from which class I PI3Ks transduce the extracellular stimuli, members of this subclass of PI3Ks are further divided into class IA and class IB, with class IA and IB being activated via receptor tyrosine kinases (RTKs) and G-protein-coupled receptors (GPCRs), respectively (Engelman et al. 2006). The class IA PI3Ks have been intensely studied because of their role in glucose metabolism and human cancer (Engelman et al. 2006; Yuan and Cantley 2008; Chalhoub and Baker 2009). The mechanisms of RTK activation have been previously reviewed (Lemmon and Schlessinger 2010). Upon the activation of RTKs, class IA PI3Ks are recruited to the cell membrane and catalyze the production of a critical lipid second messenger PI(3,4,5)P3 from PI(4,5)P2 to propagate signal transduction (Chalhoub and Baker 2009). Because the enzymatic function of PTEN is to convert PI(3,4,5)P3 back to PI(4,5)P2, PTEN is capable of terminating PI3K/ PI(3,4,5)P3 signaling and thus has been suggested to play a fundamental role in controlling the transient activation of PI3K/PI(3,4,5)P3 signaling in cells (Manning and Toker 2017).
A characteristic of PIPs is their ability to bind the pleckstrin homology (PH) domains on protein structures. In human, 252 different proteins are suggested to contain at least one PH domain (Lander et al. 2001), which makes the PH domain the 11th most abundant domain structure in human proteome (Lemmon et al. 2002; Yu et al. 2004). High-affinity PH domains have been identified for three PIPs, whereas other PIPs were found to have low affinity for many different PH domain structures (Lemmon 2007). PI(3,4)P2, PI(4,5)P2, and PI(3,4,5)P3 are the three phosphoinositides that can bind with high affinity to specific PH domains because of the basic side chains on these domains that form extensive hydrogen networks with the two adjacent/vicinal phosphate groups on the inositol ring. The 3- and 4-phosphate groups on the inositol ring occupy similar PH domain locations on AKT-, Btk-, or Grp1-PH domains as the 4- and 5-phosphate groups for PLCδ-PH domain with a 180° flip. Dephosphorylation of the 3-phosphate by PTEN reduces levels of PI(3,4)P2 and PI(3,4,5)P3, and results in disruptions of this network (Lemmon 2007).
Using yeast as model organism, the PH domains with high affinity for PI(3,4)P2, PI(4,5)P2, and PI(3,4,5)P3 were shown to possess obvious membrane targeting ability for the proteins. These PH domains provide the proteins with ability to target either plasma or Golgi membranes in studies using GFP fusion proteins (Lemmon 2007). In mammalian systems, AKT (also known as protein kinase B or PKB) is the best-characterized PH domain-containing protein. AKT is a member of the AGC family of protein kinases, highly conserved from primitive metazoans to humans. The three mammalian AKT isoforms are encoded by different genes but share similar structural features. They contain a central catalytic domain related to protein kinases A and C; a carboxy-terminal regulatory domain that contains a phosphorylation site (FXXF(X/T)Y) common to all AGC family of kinases; and an amino-terminal PH domain that allows them to bind to PIPs, particularly PI(3,4)P2 and PI(3,4,5)P3. In the inactive state, the PH domain of AKT folds back and binds intramolecularly to the catalytic domain. This intramolecular interaction stabilizes AKT in an inactive conformation and also prevents phosphorylation of AKT by the upstream kinases (i.e., 3-phosphoinositide-dependent kinase 1 [PDK1]), as the PH domain blocks the phosphorylation site. Thus, in addition to localizing AKT to the membrane, binding of AKT PH domain to the 3-phosphorylated PIPs also releases this intramolecular interaction, allowing its activation by PDK1, another PH domain-containing kinase (Thomas et al. 2002). The PH domain on PDK1 binds to PI(4,5)P2 with high affinity and has little affinity to the 3-phosphorylated PIPs (Komander et al. 2004). PDK1, also an AGC kinase, acts as a master kinase that phosphorylates the activation loop of AKT and many other AGC kinases (Mora et al. 2004). More than 100-PH-domain-containing proteins exist in mammals, and in addition to lipid binding, mediate protein–protein and protein–DNA interactions (Scheffzek and Welti 2012).
Defined primarily through its impact on AKT activity, the lipid phosphatase PTEN is found to regulate diverse signaling and biological processes (Chen et al. 2018). Many of the direct AKT phosphorylation targets reflect the impact of PTEN deregulation of numerous biological processes: (1) cell proliferation and survival via GSK3α (S21) and GSK3β (S9), p21 (T145), p27 (T157), CHK1 (S280), BAD (S99), MDM2 (S166, S186), forkhead transcriptional factors including FOXO1 (T24, S256, S319), FOXO3A (T32, S253, S315), and FOXO4 (T32, S197, S262) (Manning and Cantley 2007); (2) mRNA translation and cell growth via TSC2 (S939, T1462) and PRAS40 (T246), two suppressors of mTORC1 signaling (Inoki et al. 2002; Vander Haar et al. 2007); and (3) glucose metabolism via the AS160 Rab GTPase-activating protein (GAP), to control redistribution of glucose transporter 4 from intracellular compartments to the plasma membrane, as well as forkhead transcription factor to control hepatic gluconeogenesis (Puigserver et al. 2003; Eguez et al. 2005; Mîinea et al. 2005). Independent of AKT, PTEN as a lipid phosphatase regulates (1) cell motility via PI(3,4,5)P3-dependent actin polymerization and Rac-associated GEF and GAP recruitment and activation (Liliental et al. 2000; Campa et al. 2015); and (2) cell polarity via PI(4,5)P2-dependent recruitment of apical lumen formation machinery proteins (Martin-Belmonte et al. 2007).
PI3K signaling network is also subject to extensive feedback regulation. Notably, p70 ribosomal S6 kinase (S6K1), which is phosphorylated and activated by mTORC1 was shown to phosphorylate IRS1 on the phosphotyrosine-binding (PTB) domain and the PI3K-binding domain. These phosphorylation events result in the degradation of IRS1 (Shah and Hunter 2006; Tzatsos and Kandror 2006), and disrupt interaction of IRS1 with insulin receptor (IR) and PI3K (Tanti and Jager 2009), leading to net down-regulation of the pathway. Furthermore, S6K1 phosphorylates Rictor of the mTORC2 complex on T1135. Although this phosphorylation does not affect the activity or localization of mTOR, blocking phosphorylation of this site results in loss of mTORC2-dependent phosphorylation and activation of AKT (Julien et al. 2010). S6K1 also phosphorylates Sin1 at T86 and T398 to reduce mTORC2-dependent activation of AKT by dissociating Sin1 from mTORC2 (Liu et al. 2013). Finally, mTORC1 also phosphorylates growth-receptor-bound protein 10 (Grb10), an adaptor protein for growth factor signaling pathway. This phosphorylation significantly increases Grb10 stability and promotes its suppression of AKT activity (Hsu et al. 2011). Together, the negative feedback loops emanating from activated mTOR effectively serve to temper signaling from the membrane. They also present a major challenge for the therapeutic efforts aimed at inhibiting PI3K/AKT signaling in tumors formed by PTEN loss, as PI3K-directed therapeutics also relieve the inhibitory impact of feedback loops (Herschbein and Liesveld 2018).
LIPID METABOLISM AND STEATOSIS REGULATION BY PTEN
The domain architecture of the AKT family members are generally conserved with the linker region between the PH and catalytic domain showing highest variability (Kumar and Madison 2005). Earlier studies have shown that AKT1 has a function in the regulation of cell-cycle progression (Dudek et al. 1997; Verdu et al. 1999; Stiles et al. 2002; Kumar and Madison 2005; Manning and Cantley 2007; Dillon and Muller 2010; Hers et al. 2011; Song et al. 2019). Mice lacking Akt1 display growth retardation with partial lethality (Chen et al. 2001; Cho et al. 2001b; Yang et al. 2005; Goncalves et al. 2010). AKT2 appears to function predominantly in metabolic regulation in vivo (Cho et al. 2001a; Bae et al. 2003). Nevertheless, AKT2 has also been reported to participate in control of cell-transformation processes, including the EMT (Dillon and Muller 2010). The expression of AKT3 is restricted to the nervous system (Bergeron et al. 2017), consistent with the in vivo phenotypes observed with Akt3 deletion mice that displayed normal glucose metabolism and overall growth but a significantly reduced brain size and weight (Tschopp et al. 2005). Here, we summarize the lipid metabolic regulation by PTEN that is primarily dependent on AKT2.
PTEN as a Metabolic Regulator
In Caenorhabditis elegans and Drosophila, the insulin/PI3K pathway, that is negatively regulated by their respective PTEN homologs, controls dauer formation (worms), metabolism, as well as life span in response to nutrient availability (Ogg and Ruvkun 1998; Goberdhan et al. 1999; Scanga et al. 2000). The mammalian PTEN/PI3K signaling network parallels what was found in those organisms. Ectopic expression of PTEN, by introduction of bacterial artificial chromosomes (BACs) into the mouse genome, led to reduced body size, increased energy expenditure, and low body fat content (Garcia-Cao et al. 2012; Ortega-Molina et al. 2012). Consistent with the observations in C. elegans and Drosophila, these mice also have a longer tumor-free life span. Thus, besides its primary identity as a tumor suppressor, PTEN also acts as a metabolic regulator via the insulin/PI3K/AKT signaling pathway (Ortega-Molina and Serrano 2013; Han et al. 2018).
Lipomas are a frequent component of the tumor spectrum in familial PTEN hamartoma tumor syndrome (PHTS), caused by abnormal overgrowth of adipocytes (Lee et al. 2011). Moreover, an increase of 23% body mass index has been also reported for PHTS (Cowden) patients heterozygous for PTEN mutations, in which PTEN mutations are proposed to cause heightened insulin sensitivity (Pal et al. 2012). Similarly, improved insulin sensitivity, concurrent with increased hepatic steatosis, was observed in mouse models carrying deletion of Pten in the liver (Stiles et al. 2004; Peyrou et al. 2013). These mice also developed liver cancer that appears to be driven by steatosis (Xu et al. 2006; Galicia et al. 2010; He et al. 2016; Debebe et al. 2017; Jia et al. 2017). Blocking steatosis in these mice attenuated tumor growth (Galicia et al. 2010; He et al. 2010; Debebe et al. 2017), suggesting a potential role of PTEN-regulated tumor lipid metabolism in tumorigenesis. Nonetheless, broad metabolic phenotypes induced by PTEN loss in metabolic tissues (Horie et al. 2004; Stiles et al. 2004, 2006; Kurlawalla-Martinez et al. 2005; Wijesekara et al. 2005; Li et al. 2013b; Zeng et al. 2013; Yang et al. 2014) highlight the role of PTEN as a metabolic regulator.
PTEN/AKT Signal Coordinately Regulates Glycolysis and Oxidative Phosphorylation
AKT2, the AKT isoform dominantly expressed in metabolic tissues, was found to mediate the effect of insulin on both glucose and lipid metabolism (Cho et al. 2001a; Mîinea et al. 2005; He et al. 2010; Palian et al. 2014; Herschbein and Liesveld 2018). Of note, rate-limiting glycolytic enzymes, hexokinase II (HKII) and phosphofructokinase, are phosphorylated as a result of PI3K/AKT activation (Deprez et al. 1997; Bertrand et al. 1999; Mouton et al. 2010; Roberts et al. 2013). Phosphorylation of HKII by AKT couples glycolysis with oxidative phosphorylation, in which the mitochondrial localized HKII is proposed to mediate apoptosis inhibition caused by AKT activation (Gottlob et al. 2001). This result is supported by the observation that AKT is found in the mitochondria soon after activation of PI3K (Bijur and Jope 2003). In the mitochondria, AKT was found to phosphorylate the mitochondrial pool of GSK3β, as well as pyruvate dehydrogenase (PDH), contributing to increased mitochondrial energetics (Antico Arciuch et al. 2009; Li et al. 2013a).
Additionally, AKT activation controls mitochondrial gene transcription by phosphorylating and activating CREB transcription factor, independent of the cAMP-mediated activation of PKA, a common signal that induces CREB phosphorylation (Li et al. 2013b). Through this mechanism, AKT activation robustly increases the transcriptional activity of estrogen receptor-related receptor (ERR). Acting on the nuclear genome, ERR functions to promote transcription of genes that regulate mitochondrial function (Li et al. 2013b). In hepatocytes, this ERR induction led to increased oxygen consumption and elevated production of reactive oxygen species, contributing to the nonalcoholic fatty liver disease (NAFLD)/nonalcoholic steatohepatitis (NASH) phenotypes found in the Pten-deficient mouse livers (Galicia et al. 2010; Li et al. 2013b).
PTEN/AKT/mTOR Signal-Regulated Lipogenesis
Given the function of PTEN as a negative regulator of PI3K/AKT signaling, PTEN loss-of-function in metabolic tissues is accompanied by augmented response to insulin and increased insulin signaling. In the liver, such signals led to increased de novo lipogenesis (Stiles et al. 2004), primarily owing to the induction of fatty acid synthase (Fasn) via regulation by AKT2 (He et al. 2010). As a result, the liver-specific deletion of Pten led to the development of NAFLD and more severe NASH (Horie et al. 2004; Stiles et al. 2004; Galicia et al. 2010). AKT2 was found to both positively and negatively regulate Fasn expression via sterol regulatory element-binding protein (SREBP) and Maf-1, a transcriptional repressor, respectively (He et al. 2010; Palian et al. 2014). In addition, mTOR also affects the posttranslational processing of SREBP, which involves the SREBP cleavage-activating protein (SCAP) and an insulin-induced gene (Insig) (Du et al. 2006; Yecies et al. 2011).
SUMMARY
In the more than 20 years since the discovery of PTEN, our understanding of its function has been continuously expanding. For many years, the realization of PTEN regulation of the PI3K/AKT signaling pathway has been the main driver for the discovery of PTEN's biological function. In more recent years, compartment-specific roles of PTEN and its regulation are being elucidated. The biological functions of PTEN beyond cell growth and survival are also being recognized, particularly its impact on metabolism. This expanded understanding of PTEN's biological functions broadens the impact of PTEN on not only cancer biology but also other diseases, particularly those related to metabolism and inflammation.
Footnotes
Editors: Charis Eng, Joanne Ngeow, and Vuk Stambolic
Additional Perspectives on The PTEN Family available at www.perspectivesinmedicine.org
REFERENCES
- Alanko J, Mai A, Jacquemet G, Schauer K, Kaukonen R, Saari M, Goud B, Ivaska J. 2015. Integrin endosomal signalling suppresses anoikis. Nat Cell Biol 17: 1412–1421. 10.1038/ncb3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antico Arciuch VG, Galli S, Franco MC, Lam PY, Cadenas E, Carreras MC, Poderoso JJ. 2009. Akt1 intramitochondrial cycling is a crucial step in the redox modulation of cell cycle progression. PLoS ONE 4: e7523 10.1371/journal.pone.0007523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae SS, Cho H, Mu J, Birnbaum MJ. 2003. Isoform-specific regulation of insulin-dependent glucose uptake by Akt/protein kinase B. J Biol Chem 278: 49530–49536. 10.1074/jbc.M306782200 [DOI] [PubMed] [Google Scholar]
- Bassi C, Ho J, Srikumar T, Dowling RJ, Gorrini C, Miller SJ, Mak TW, Neel BG, Raught B, Stambolic V. 2013. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science 341: 395–399. 10.1126/science.1236188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron Y, Bureau G, Laurier-Laurin ME, Asselin E, Massicotte G, Cyr M. 2017. Genetic deletion of Akt3 induces an endophenotype reminiscent of psychiatric manifestations in mice. Front Mol Neurosci 10: 102 10.3389/fnmol.2017.00102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand L, Alessi DR, Deprez J, Deak M, Viaene E, Rider MH, Hue L. 1999. Heart 6-phosphofructo-2-kinase activation by insulin results from Ser-466 and Ser-483 phosphorylation and requires 3-phosphoinositide-dependent kinase-1, but not protein kinase B. J Biol Chem 274: 30927–30933. 10.1074/jbc.274.43.30927 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya S, Dutta D, Saha B, Ghosh AK, Das AK. 2012. Crystal structure of Staphylococcal dual specific inositol monophosphatase/NADP(H) phosphatase (SAS2203) delineates the molecular basis of substrate specificity. Biochimie 94: 879–890. 10.1016/j.biochi.2011.12.007 [DOI] [PubMed] [Google Scholar]
- Bijur GN, Jope RS. 2003. Rapid accumulation of Akt in mitochondria following phosphatidylinositol 3-kinase activation. J Neurochem 87: 1427–1435. 10.1046/j.1471-4159.2003.02113.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bononi A, Bonora M, Marchi S, Missiroli S, Poletti F, Giorgi C, Pandolfi PP, Pinton P. 2013. Identification of PTEN at the ER and MAMs and its regulation of Ca2+ signaling and apoptosis in a protein phosphatase-dependent manner. Cell Death Differ 20: 1631–1643. 10.1038/cdd.2013.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai XM, Tao BB, Wang LY, Liang YL, Jin JW, Yang Y, Hu YL, Zha XL. 2005. Protein phosphatase activity of PTEN inhibited the invasion of glioma cells with epidermal growth factor receptor mutation type III expression. Int J Cancer 117: 905–912. 10.1002/ijc.21251 [DOI] [PubMed] [Google Scholar]
- Campa CC, Ciraolo E, Ghigo A, Germena G, Hirsch E. 2015. Crossroads of PI3K and Rac pathways. Small GTPases 6: 71–80. 10.4161/21541248.2014.989789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub N, Baker SJ. 2009. PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol 4: 127–150. 10.1146/annurev.pathol.4.110807.092311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, et al. 2001. Growth retardation and increased apoptosis in mice with homozygous disruption of the akt1 gene. Genes Dev 15: 2203–2208. 10.1101/gad.913901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chen J, He L, Stiles BL. 2018. PTEN: Tumor suppressor and metabolic regulator. Front Endocrinol (Lausanne) 9: 338 10.3389/fendo.2018.00338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB III, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ. 2001a. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKBβ). Science 292: 1728–1731. 10.1126/science.292.5522.1728 [DOI] [PubMed] [Google Scholar]
- Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. 2001b. Akt1/PKBα is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem 276: 38349–38352. 10.1074/jbc.C100462200 [DOI] [PubMed] [Google Scholar]
- Davidson L, Maccario H, Perera NM, Yang X, Spinelli L, Tibarewal P, Glancy B, Gray A, Weijer CJ, Downes CP, et al. 2010. Suppression of cellular proliferation and invasion by the concerted lipid and protein phosphatase activities of PTEN. Oncogene 29: 687–697. 10.1038/onc.2009.384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debebe A, Medina V, Chen CY, Mahajan IM, Jia C, Fu D, He L, Zeng N, Stiles BW, Chen CL, et al. 2017. Wnt/β-catenin activation and macrophage induction during liver cancer development following steatosis. Oncogene 36: 6020–6029. 10.1038/onc.2017.207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. 1997. Phosphorylation and activation of heart 6-phosphofructo-2-kinase by protein kinase B and other protein kinases of the insulin signaling cascades. J Biol Chem 272: 17269–17275. 10.1074/jbc.272.28.17269 [DOI] [PubMed] [Google Scholar]
- Dey N, Crosswell HE, De P, Parsons R, Peng Q, Su JD, Durden DL. 2008. The protein phosphatase activity of PTEN regulates SRC family kinases and controls glioma migration. Cancer Res 68: 1862–1871. 10.1158/0008-5472.CAN-07-1182 [DOI] [PubMed] [Google Scholar]
- Dillon RL, Muller WJ. 2010. Distinct biological roles for the akt family in mammary tumor progression. Cancer Res 70: 4260–4264. 10.1158/0008-5472.CAN-10-0266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downes CP, Ross S, Maccario H, Perera N, Davidson L, Leslie NR. 2007. Stimulation of PI 3-kinase signaling via inhibition of the tumor suppressor phosphatase, PTEN. Adv Enzyme Regul 47: 184–194. 10.1016/j.advenzreg.2006.12.018 [DOI] [PubMed] [Google Scholar]
- Du X, Kristiana I, Wong J, Brown AJ. 2006. Involvement of Akt in ER-to-Golgi transport of SCAP/SREBP: A link between a key cell proliferative pathway and membrane synthesis. Mol Biol Cell 17: 2735–2745. 10.1091/mbc.e05-11-1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek H, Datta SR, Franke TF, Birnbaum MJ, Yao R, Cooper GM, Segal RA, Kaplan DR, Greenberg ME. 1997. Regulation of neuronal survival by the serine-threonine protein kinase Akt. Science 275: 661–665. 10.1126/science.275.5300.661 [DOI] [PubMed] [Google Scholar]
- Eguez L, Lee A, Chavez JA, Miinea CP, Kane S, Lienhard GE, McGraw TE. 2005. Full intracellular retention of GLUT4 requires AS160 Rab GTPase activating protein. Cell Metab 2: 263–272. 10.1016/j.cmet.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Engelman JA, Luo J, Cantley LC. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 7: 606–619. 10.1038/nrg1879 [DOI] [PubMed] [Google Scholar]
- Essen LO, Perisic O, Lynch DE, Katan M, Williams RL. 1997. A ternary metal binding site in the C2 domain of phosphoinositide-specific phospholipase C-δ1. Biochemistry 36: 2753–2762. 10.1021/bi962466t [DOI] [PubMed] [Google Scholar]
- Feng J, Liang J, Li J, Li Y, Liang H, Zhao X, McNutt MA, Yin Y. 2015. PTEN controls the DNA replication process through MCM2 in response to replicative stress. Cell Rep 13: 1295–1303. 10.1016/j.celrep.2015.10.016 [DOI] [PubMed] [Google Scholar]
- Galicia VA, He L, Dang H, Kanel G, Vendryes C, French BA, Zeng N, Bayan JA, Ding W, Wang KS, et al. 2010. Expansion of hepatic tumor progenitor cells in Pten-null mice requires liver injury and is reversed by loss of AKT2. Gastroenterology 139: 2170–2182. 10.1053/j.gastro.2010.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Cao I, Song MS, Hobbs RM, Laurent G, Giorgi C, de Boer VC, Anastasiou D, Ito K, Sasaki AT, Rameh L, et al. 2012. Systemic elevation of PTEN induces a tumor-suppressive metabolic state. Cell 149: 49–62. 10.1016/j.cell.2012.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu MM, Kirsch KH, Kaloudis P, Yang H, Pavletich NP, Hanafusa H. 2000. Stabilization and productive positioning roles of the C2 domain of PTEN tumor suppressor. Cancer Res 60: 7033–7038. [PubMed] [Google Scholar]
- Gericke A, Munson M, Ross AH. 2006. Regulation of the PTEN phosphatase. Gene 374: 1–9. 10.1016/j.gene.2006.02.024 [DOI] [PubMed] [Google Scholar]
- Gildea JJ, Herlevsen M, Harding MA, Gulding KM, Moskaluk CA, Frierson HF, Theodorescu D. 2004. PTEN can inhibit in vitro organotypic and in vivo orthotopic invasion of human bladder cancer cells even in the absence of its lipid phosphatase activity. Oncogene 23: 6788–6797. 10.1038/sj.onc.1207599 [DOI] [PubMed] [Google Scholar]
- Goberdhan DC, Paricio N, Goodman EC, Mlodzik M, Wilson C. 1999. Drosophila tumor suppressor PTEN controls cell size and number by antagonizing the Chico/PI3-kinase signaling pathway. Genes Dev 13: 3244–3258. 10.1101/gad.13.24.3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves MD, Pistilli EE, Balduzzi A, Birnbaum MJ, Lachey J, Khurana TS, Ahima RS. 2010. Akt deficiency attenuates muscle size and function but not the response to ActRIIB inhibition. PLoS ONE 5: e12707 10.1371/journal.pone.0012707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N. 2001. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev 15: 1406–1418. 10.1101/gad.889901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T, Zhang Z, Wang J, Guo J, Shen WH, Yin Y. 2011. CREB is a novel nuclear target of PTEN phosphatase. Cancer Res 71: 2821–2825. 10.1158/0008-5472.CAN-10-3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han CY, Patten DA, Richardson RB, Harper ME, Tsang BK. 2018. Tumor metabolism regulating chemosensitivity in ovarian cancer. Genes Cancer 9: 155–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hou X, Kanel G, Zeng N, Galicia V, Wang Y, Yang J, Wu H, Birnbaum MJ, Stiles BL. 2010. The critical role of AKT2 in hepatic steatosis induced by PTEN loss. Am J Pathol 176: 2302–2308. 10.2353/ajpath.2010.090931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Gubbins J, Peng Z, Medina V, Fei F, Asahina K, Wang J, Kahn M, Rountree CB, Stiles BL. 2016. Activation of hepatic stellate cell in Pten null liver injury model. Fibrogenesis Tissue Repair 9: 8 10.1186/s13069-016-0045-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hers I, Vincent EE, Tavaré JM. 2011. Akt signalling in health and disease. Cell Signal 23: 1515–1527. 10.1016/j.cellsig.2011.05.004 [DOI] [PubMed] [Google Scholar]
- Herschbein L, Liesveld JL. 2018. Dueling for dual inhibition: Means to enhance effectiveness of PI3K/Akt/mTOR inhibitors in AML. Blood Rev 32: 235–248. 10.1016/j.blre.2017.11.006 [DOI] [PubMed] [Google Scholar]
- Hlobilkova A, Guldberg P, Thullberg M, Zeuthen J, Lukas J, Bartek J. 2000. Cell cycle arrest by the PTEN tumor suppressor is target cell specific and may require protein phosphatase activity. Exp Cell Res 256: 571–577. 10.1006/excr.2000.4867 [DOI] [PubMed] [Google Scholar]
- Horie Y, Suzuki A, Kataoka E, Sasaki T, Hamada K, Sasaki J, Mizuno K, Hasegawa G, Kishimoto H, Iizuka M, et al. 2004. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest 113: 1774–1783. 10.1172/JCI20513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. 2011. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332: 1317–1322. 10.1126/science.1199498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Zhu T, Wu J, Guan KL. 2002. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol 4: 648–657. 10.1038/ncb839 [DOI] [PubMed] [Google Scholar]
- Jia C, Medina V, Liu C, He L, Qian D, Taojian T, Okamoto CT, Stiles BL. 2017. Crosstalk of LKB1- and PTEN-regulated signals in liver morphogenesis and tumor development. Hepatol Commun 1: 153–167. 10.1002/hep4.1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien LA, Carriere A, Moreau J, Roux PP. 2010. mTORC1-activated S6K1 phosphorylates Rictor on threonine 1135 and regulates mTORC2 signaling. Mol Cell Biol 30: 908–921. 10.1128/MCB.00601-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohout SC, Corbalán-García S, Torrecillas A, Goméz-Fernandéz JC, Falke JJ. 2002. C2 domains of protein kinase C isoforms α, β, and γ: Activation parameters and calcium stoichiometries of the membrane-bound state. Biochemistry 41: 11411–11424. 10.1021/bi026041k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Fairservice A, Deak M, Kular GS, Prescott AR, Peter Downes C, Safrany ST, Alessi DR, van Aalten DM. 2004. Structural insights into the regulation of PDK1 by phosphoinositides and inositol phosphates. EMBO J 23: 3918–3928. 10.1038/sj.emboj.7600379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreis P, Hendricusdottir R, Kay L, Papageorgiou IE, van Diepen M, Mack T, Ryves J, Harwood A, Leslie NR, Kann O, et al. 2013. Phosphorylation of the actin binding protein Drebrin at S647 is regulated by neuronal activity and PTEN. PLoS ONE 8: e71957 10.1371/journal.pone.0071957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar CC, Madison V. 2005. AKT crystal structure and AKT-specific inhibitors. Oncogene 24: 7493–7501. 10.1038/sj.onc.1209087 [DOI] [PubMed] [Google Scholar]
- Kurlawalla-Martinez C, Stiles B, Wang Y, Devaskar SU, Kahn BB, Wu H. 2005. Insulin hypersensitivity and resistance to streptozotocin-induced diabetes in mice lacking PTEN in adipose tissue. Mol Cell Biol 25: 2498–2510. 10.1128/MCB.25.6.2498-2510.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusunose M, Hashimoto N, Kimura M, Ogata R, Aoyama D, Sakamoto K, Miyazaki S, Ando A, Omote N, Imaizumi K, et al. 2015. Direct regulation of transforming growth factor β-induced epithelial-mesenchymal transition by the protein phosphatase activity of unphosphorylated PTEN in lung cancer cells. Cancer Sci 106: 1693–1704. 10.1111/cas.12831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921. 10.1038/35057062 [DOI] [PubMed] [Google Scholar]
- Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. 1999. Crystal structure of the PTEN tumor suppressor: Implications for its phosphoinositide phosphatase activity and membrane association. Cell 99: 323–334. 10.1016/S0092-8674(00)81663-3 [DOI] [PubMed] [Google Scholar]
- Lee CH, Spence RA, Upadhyaya M, Morrison PJ. 2011. Familial multiple lipomatosis with clear autosomal dominant inheritance and onset in early adolescence. BMJ Case Rep 2011: bcr1020103395 10.1136/bcr.10.2010.3395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA. 2007. Pleckstrin homology (PH) domains and phosphoinositides. Biochem Soc Symp 74: 81–93. 10.1042/BSS2007c08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Schlessinger J. 2010. Cell signaling by receptor tyrosine kinases. Cell 141: 1117–1134. 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM, Abrams CS. 2002. Pleckstrin homology domains and the cytoskeleton. FEBS Lett 513: 71–76. 10.1016/S0014-5793(01)03243-4 [DOI] [PubMed] [Google Scholar]
- Leslie NR, Yang X, Downes CP, Weijer CJ. 2007. PtdIns(3,4,5)P3-dependent and -independent roles for PTEN in the control of cell migration. Curr Biol 17: 115–125. 10.1016/j.cub.2006.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Li Y, He L, Agarwal AR, Zeng N, Cadenas E, Stiles BL. 2013a. PI3K/AKT signaling regulates bioenergetics in immortalized hepatocytes. Free Radic Biol Med 60: 29–40. 10.1016/j.freeradbiomed.2013.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, He L, Zeng N, Sahu D, Cadenas E, Shearn C, Li W, Stiles BL. 2013b. Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) signaling regulates mitochondrial biogenesis and respiration via estrogen-related receptor α (ERRα). J Biol Chem 288: 25007–25024. 10.1074/jbc.M113.450353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liliental J, Moon SY, Lesche R, Mamillapalli R, Li D, Zheng Y, Sun H, Wu H. 2000. Genetic deletion of the Pten tumor suppressor gene promotes cell motility by activation of Rac1 and Cdc42 GTPases. Curr Biol 10: 401–404. 10.1016/S0960-9822(00)00417-6 [DOI] [PubMed] [Google Scholar]
- Liu P, Gan W, Inuzuka H, Lazorchak AS, Gao D, Arojo O, Liu D, Wan L, Zhai B, Yu Y, et al. 2013. Sin1 phosphorylation impairs mTORC2 complex integrity and inhibits downstream Akt signalling to suppress tumorigenesis. Nat Cell Biol 15: 1340–1350. 10.1038/ncb2860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomasney JW, Cheng HF, Roffler SR, King K. 1999. Activation of phospholipase C δ1 through C2 domain by a Ca2+-enzyme-phosphatidylserine ternary complex. J Biol Chem 274: 21995–22001. 10.1074/jbc.274.31.21995 [DOI] [PubMed] [Google Scholar]
- Lyu J, Yu X, He L, Cheng T, Zhou J, Cheng C, Chen Z, Cheng G, Qiu Z, Zhou W. 2015. The protein phosphatase activity of PTEN is essential for regulating neural stem cell differentiation. Mol Brain 8: 26 10.1186/s13041-015-0114-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Dixon JE. 1998. The tumor suppressor, PTEN/MMAC1, dephosphorylates the lipid second messenger, phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem 273: 13375–13378. 10.1074/jbc.273.22.13375 [DOI] [PubMed] [Google Scholar]
- Manning BD, Cantley LC. 2007. AKT/PKB signaling: Navigating downstream. Cell 129: 1261–1274. 10.1016/j.cell.2007.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Toker A. 2017. AKT/PKB signaling: Navigating the network. Cell 169: 381–405. 10.1016/j.cell.2017.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Belmonte F, Gassama A, Datta A, Yu W, Rescher U, Gerke V, Mostov K. 2007. PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell 128: 383–397. 10.1016/j.cell.2006.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda M, Takeshita K, Kurokawa T, Sakata S, Suzuki M, Yamashita E, Okamura Y, Nakagawa A. 2011. Crystal structure of the cytoplasmic phosphatase and tensin homolog (PTEN)-like region of Ciona intestinalis voltage-sensing phosphatase provides insight into substrate specificity and redox regulation of the phosphoinositide phosphatase activity. J Biol Chem 286: 23368–23377. 10.1074/jbc.M110.214361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mîinea CP, Sano H, Kane S, Sano E, Fukuda M, Peränen J, Lane WS, Lienhard GE. 2005. AS160, the Akt substrate regulating GLUT4 translocation, has a functional Rab GTPase-activating protein domain. Biochem J 391: 87–93. 10.1042/BJ20050887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SJ, Lou DY, Seldin DC, Lane WS, Neel BG. 2002. Direct identification of PTEN phosphorylation sites. FEBS Lett 528: 145–153. 10.1016/S0014-5793(02)03274-X [DOI] [PubMed] [Google Scholar]
- Mora A, Komander D, van Aalten DM, Alessi DR. 2004. PDK1, the master regulator of AGC kinase signal transduction. Semin Cell Dev Biol 15: 161–170. 10.1016/j.semcdb.2003.12.022 [DOI] [PubMed] [Google Scholar]
- Mouton V, Toussaint L, Vertommen D, Gueuning MA, Maisin L, Havaux X, Sanchez-Canedo C, Bertrand L, Dequiedt F, Hemmings BA, et al. 2010. Heart 6-phosphofructo-2-kinase activation by insulin requires PKB (protein kinase B), but not SGK3 (serum- and glucocorticoid-induced protein kinase 3). Biochem J 431: 267–275. 10.1042/BJ20101089 [DOI] [PubMed] [Google Scholar]
- Myers MP, Stolarov JP, Eng C, Li J, Wang SI, Wigler MH, Parsons R, Tonks NK. 1997. P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci 94: 9052–9057. 10.1073/pnas.94.17.9052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. 1998. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci 95: 13513–13518. 10.1073/pnas.95.23.13513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalefski EA, Sultzman LA, Martin DM, Kriz RW, Towler PS, Knopf JL, Clark JD. 1994. Delineation of two functionally distinct domains of cytosolic phospholipase A2, a regulatory Ca2+-dependent lipid-binding domain and a Ca2+-independent catalytic domain. J Biol Chem 269: 18239–18249. [PubMed] [Google Scholar]
- Nalefski EA, Slazas MM, Falke JJ. 1997. Ca2+-signaling cycle of a membrane-docking C2 domain. Biochemistry 36: 12011–12018. 10.1021/bi9717340 [DOI] [PubMed] [Google Scholar]
- Nelen MR, van Staveren WC, Peeters EA, Hassel MB, Gorlin RJ, Hamm H, Lindboe CF, Fryns JP, Sijmons RH, Woods DG, et al. 1997. Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum Mol Genet 6: 1383–1387. 10.1093/hmg/6.8.1383 [DOI] [PubMed] [Google Scholar]
- Ning K, Pei L, Liao M, Liu B, Zhang Y, Jiang W, Mielke JG, Li L, Chen Y, El-Hayek YH, et al. 2004. Dual neuroprotective signaling mediated by downregulating two distinct phosphatase activities of PTEN. J Neurosci 24: 4052–4060. 10.1523/jneurosci.5449-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogg S, Ruvkun G. 1998. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell 2: 887–893. 10.1016/S1097-2765(00)80303-2 [DOI] [PubMed] [Google Scholar]
- Ortega-Molina A, Serrano M. 2013. PTEN in cancer, metabolism, and aging. Trends Endocrinol Metab 24: 184–189. 10.1016/j.tem.2012.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Molina A, Efeyan A, Lopez-Guadamillas E, Muñoz-Martin M, Gómez-López G, Cañamero M, Mulero F, Pastor J, Martinez S, Romanos E, et al. 2012. Pten positively regulates brown adipose function, energy expenditure, and longevity. Cell Metab 15: 382–394. 10.1016/j.cmet.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Pal A, Barber TM, Van de Bunt M, Rudge SA, Zhang Q, Lachlan KL, Cooper NS, Linden H, Levy JC, Wakelam MJ, et al. 2012. PTEN mutations as a cause of constitutive insulin sensitivity and obesity. N Engl J Med 367: 1002–1011. 10.1056/NEJMoa1113966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palian BM, Rohira AD, Johnson SA, He L, Zheng N, Dubeau L, Stiles BL, Johnson DL. 2014. Maf1 is a novel target of PTEN and PI3K signaling that negatively regulates oncogenesis and lipid metabolism. PLoS Genet 10: e1004789 10.1371/journal.pgen.1004789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa A, Wan L, Bonora M, Salmena L, Song MS, Hobbs RM, Lunardi A, Webster K, Ng C, Newton RH, et al. 2014. Cancer-associated PTEN mutants act in a dominant-negative manner to suppress PTEN protein function. Cell 157: 595–610. 10.1016/j.cell.2014.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MJ, Kim MS, Park IC, Kang HS, Yoo H, Park SH, Rhee CH, Hong SI, Lee SH. 2002. PTEN suppresses hyaluronic acid-induced matrix metalloproteinase-9 expression in U87MG glioblastoma cells through focal adhesion kinase dephosphorylation. Cancer Res 62: 6318–6322. [PubMed] [Google Scholar]
- Peyrou M, Clément S, Maier C, Bourgoin L, Branche E, Conzelmann S, Kaddai V, Foti M, Negro F. 2013. PTEN protein phosphatase activity regulates hepatitis C virus secretion through modulation of cholesterol metabolism. J Hepatol 59: 420–426. 10.1016/j.jhep.2013.04.012 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, et al. 2003. Insulin-regulated hepatic gluconeogenesis through FOXO1–PGC-1α interaction. Nature 423: 550–555. 10.1038/nature01667 [DOI] [PubMed] [Google Scholar]
- Rizo J, Südhof TC. 1998. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem 273: 15879–15882. 10.1074/jbc.273.26.15879 [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Tan-Sah VP, Smith JM, Miyamoto S. 2013. Akt phosphorylates HK-II at Thr-473 and increases mitochondrial HK-II association to protect cardiomyocytes. J Biol Chem 288: 23798–23806. 10.1074/jbc.M113.482026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Escudero I, Oliver MD, Andrés-Pons A, Molina M, Cid VJ, Pulido R. 2011. A comprehensive functional analysis of PTEN mutations: Implications in tumor- and autism-related syndromes. Hum Mol Genet 20: 4132–4142. 10.1093/hmg/ddr337 [DOI] [PubMed] [Google Scholar]
- Scanga SE, Ruel L, Binari RC, Snow B, Stambolic V, Bouchard D, Peters M, Calvieri B, Mak TW, Woodgett JR, et al. 2000. The conserved PI3′K/PTEN/Akt signaling pathway regulates both cell size and survival in Drosophila. Oncogene 19: 3971–3977. 10.1038/sj.onc.1203739 [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Welti S. 2012. Pleckstrin homology (PH) like domain—Versatile modules in protein–protein interaction platforms. FEBS Lett 586: 2662–2673. 10.1016/j.febslet.2012.06.006 [DOI] [Google Scholar]
- Shah OJ, Hunter T. 2006. Turnover of the active fraction of IRS1 involves raptor-mTOR- and S6K1-dependent serine phosphorylation in cell culture models of tuberous sclerosis. Mol Cell Biol 26: 6425–6434. 10.1128/MCB.01254-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Wang J, Chandarlapaty S, Cross J, Thompson C, Rosen N, Jiang X. 2014. PTEN is a protein tyrosine phosphatase for IRS1. Nat Struct Mol Biol 21: 522–527. 10.1038/nsmb.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim JH, Kim YS, Bahk YY. 2006. Proteome profile changes that are differentially regulated by lipid and protein phosphatase activities of tumor suppressor PTEN in PTEN-expressing U-87 MG human glioblastoma cells. Proteomics 6: 81–93. 10.1002/pmic.200500076 [DOI] [PubMed] [Google Scholar]
- Shinde SR, Maddika S. 2016. PTEN modulates EGFR late endocytic trafficking and degradation by dephosphorylating Rab7. Nat Commun 7: 10689 10.1038/ncomms10689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnitsar I, Bashkurov M, Masson GR, Ogunjimi AA, Mosessian S, Cabeza EA, Hirsch CL, Trcka D, Gish G, Jiao J, et al. 2015. PTEN regulates cilia through Dishevelled. Nat Commun 6: 8388 10.1038/ncomms9388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Bode AM, Dong Z, Lee MH. 2019. AKT as a therapeutic target for cancer. Cancer Res 79: 1019–1031. 10.1158/0008-5472.CAN-18-2738 [DOI] [PubMed] [Google Scholar]
- Stiles B, Gilman V, Khanzenzon N, Lesche R, Li A, Qiao R, Liu X, Wu H. 2002. Essential role of AKT-1/protein kinase Bα in PTEN-controlled tumorigenesis. Mol Cell Biol 22: 3842–3851. 10.1128/MCB.22.11.3842-3851.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles B, Wang Y, Stahl A, Bassilian S, Lee WP, Kim YJ, Sherwin R, Devaskar S, Lesche R, Magnuson MA, et al. 2004. Liver-specific deletion of negative regulator Pten results in fatty liver and insulin hypersensitivity [corrected]. Proc Natl Acad Sci 101: 2082–2087. 10.1073/pnas.0308617100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles BL, Kuralwalla-Martinez C, Guo W, Gregorian C, Wang Y, Tian J, Magnuson MA, Wu H. 2006. Selective deletion of Pten in pancreatic β cells leads to increased islet mass and resistance to STZ-induced diabetes. Mol Cell Biol 26: 2772–2781. 10.1128/MCB.26.7.2772-2781.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. 1998. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science 280: 1614–1617. 10.1126/science.280.5369.1614 [DOI] [PubMed] [Google Scholar]
- Tanti JF, Jager J. 2009. Cellular mechanisms of insulin resistance: Role of stress-regulated serine kinases and insulin receptor substrates (IRS) serine phosphorylation. Curr Opin Pharmacol 9: 753–762. 10.1016/j.coph.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Tautz L, Critton DA, Grotegut S. 2013. Protein tyrosine phosphatases: Structure, function, and implication in human disease. Methods Mol Biol 1053: 179–221. 10.1007/978-1-62703-562-0_13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas CC, Deak M, Alessi DR, van Aalten DM. 2002. High-resolution structure of the pleckstrin homology domain of protein kinase B/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr Biol 12: 1256–1262. 10.1016/S0960-9822(02)00972-7 [DOI] [PubMed] [Google Scholar]
- Tibarewal P, Zilidis G, Spinelli L, Schurch N, Maccario H, Gray A, Perera NM, Davidson L, Barton GJ, Leslie NR. 2012. PTEN protein phosphatase activity correlates with control of gene expression and invasion, a tumor-suppressing phenotype, but not with AKT activity. Sci Signal 5: ra18 10.1126/scisignal.2002138 [DOI] [PubMed] [Google Scholar]
- Torres J, Pulido R. 2001. The tumor suppressor PTEN is phosphorylated by the protein kinase CK2 at its C terminus. Implications for PTEN stability to proteasome-mediated degradation. J Biol Chem 276: 993–998. 10.1074/jbc.M009134200 [DOI] [PubMed] [Google Scholar]
- Tschopp O, Yang ZZ, Brodbeck D, Dummler BA, Hemmings-Mieszczak M, Watanabe T, Michaelis T, Frahm J, Hemmings BA. 2005. Essential role of protein kinase Bγ (PKBγ/Akt3) in postnatal brain development but not in glucose homeostasis. Development 132: 2943–2954. 10.1242/dev.01864 [DOI] [PubMed] [Google Scholar]
- Tzatsos A, Kandror KV. 2006. Nutrients suppress phosphatidylinositol 3-kinase/Akt signaling via raptor-dependent mTOR-mediated insulin receptor substrate 1 phosphorylation. Mol Cell Biol 26: 63–76. 10.1128/MCB.26.1.63-76.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Haar E, Lee SI, Bandhakavi S, Griffin TJ, Kim DH. 2007. Insulin signalling to mTOR mediated by the Akt/PKB substrate PRAS40. Nat Cell Biol 9: 316–323. 10.1038/ncb1547 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Ramaswamy S, Nakamura N, Sellers WR. 2000. Phosphorylation of the PTEN tail regulates protein stability and function. Mol Cell Biol 20: 5010–5018. 10.1128/MCB.20.14.5010-5018.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdu J, Buratovich MA, Wilder EL, Birnbaum MJ. 1999. Cell-autonomous regulation of cell and organ growth in Drosophila by Akt/PKB. Nat Cell Biol 1: 500–506. 10.1038/70293 [DOI] [PubMed] [Google Scholar]
- Wang H, Karikomi M, Naidu S, Rajmohan R, Caserta E, Chen HZ, Rawahneh M, Moffitt J, Stephens JA, Fernandez SA, et al. 2010. Allele-specific tumor spectrum in Pten knockin mice. Proc Natl Acad Sci 107: 5142–5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng LP, Brown JL, Eng C. 2001. PTEN coordinates G1 arrest by down-regulating cyclin D1 via its protein phosphatase activity and up-regulating p27 via its lipid phosphatase activity in a breast cancer model. Hum Mol Genet 10: 599–604. 10.1093/hmg/10.6.599 [DOI] [PubMed] [Google Scholar]
- Wijesekara N, Konrad D, Eweida M, Jefferies C, Liadis N, Giacca A, Crackower M, Suzuki A, Mak TW, Kahn CR, et al. 2005. Muscle-specific Pten deletion protects against insulin resistance and diabetes. Mol Cell Biol 25: 1135–1145. 10.1128/MCB.25.3.1135-1145.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worby CA, Dixon JE. 2014. Pten. Annu Rev Biochem 83: 641–669. [DOI] [PubMed] [Google Scholar]
- Wozniak DJ, Kajdacsy-Balla A, Macias V, Ball-Kell S, Zenner ML, Bie W, Tyner AL. 2017. PTEN is a protein phosphatase that targets active PTK6 and inhibits PTK6 oncogenic signaling in prostate cancer. Nat Commun 8: 1508 10.1038/s41467-017-01574-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu EL, Qi Y, Song KC, Klauda JB, Im W. 2014. Preferred orientations of phosphoinositides in bilayers and their implications in protein recognition mechanisms. J Phys Chem B 118: 4315–4325. 10.1021/jp500610t [DOI] [PubMed] [Google Scholar]
- Xu X, Kobayashi S, Qiao W, Li C, Xiao C, Radaeva S, Stiles B, Wang RH, Ohara N, Yoshino T, et al. 2006. Induction of intrahepatic cholangiocellular carcinoma by liver-specific disruption of Smad4 and Pten in mice. J Clin Invest 116: 1843–1852. 10.1172/JCI27282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZZ, Tschopp O, Di-Poi N, Bruder E, Baudry A, Dummler B, Wahli W, Hemmings BA. 2005. Dosage-dependent effects of Akt1/protein kinase Bα (PKBα) and Akt3/PKBγ on thymus, skin, and cardiovascular and nervous system development in mice. Mol Cell Biol 25: 10407–10418. 10.1128/MCB.25.23.10407-10418.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KT, Bayan JA, Zeng N, Aggarwal R, He L, Peng Z, Kassa A, Kim M, Luo Z, Shi Z, et al. 2014. Adult-onset deletion of Pten increases islet mass and beta cell proliferation in mice. Diabetologia 57: 352–361. 10.1007/s00125-013-3085-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yecies JL, Zhang HH, Menon S, Liu S, Yecies D, Lipovsky AI, Gorgun C, Kwiatkowski DJ, Hotamisligil GS, Lee CH, et al. 2011. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab 14: 21–32. 10.1016/j.cmet.2011.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Mendrola JM, Audhya A, Singh S, Keleti D, DeWald DB, Murray D, Emr SD, Lemmon MA. 2004. Genome-wide analysis of membrane targeting by S. cerevisiae pleckstrin homology domains. Mol Cell 13: 677–688. 10.1016/S1097-2765(04)00083-8 [DOI] [PubMed] [Google Scholar]
- Yuan TL, Cantley LC. 2008. PI3K pathway alterations in cancer: Variations on a theme. Oncogene 27: 5497–5510. 10.1038/onc.2008.245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng N, Yang KT, Bayan JA, He L, Aggarwal R, Stiles JW, Hou X, Medina V, Abad D, Palian BM, et al. 2013. PTEN controls β-cell regeneration in aged mice by regulating cell cycle inhibitor p16ink4a. Aging Cell 12: 1000–1011. 10.1111/acel.12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XC, Piccini A, Myers MP, Van Aelst L, Tonks NK. 2012. Functional analysis of the protein phosphatase activity of PTEN. Biochem J 444: 457–464. 10.1042/BJ20120098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hou SQ, He J, Gu T, Yin Y, Shen WH. 2016. PTEN regulates PLK1 and controls chromosomal stability during cell division. Cell Cycle 15: 2476–2485. 10.1080/15384101.2016.1203493 [DOI] [PMC free article] [PubMed] [Google Scholar]