Abstract
Hepatitis C virus (HCV) is a major cause of liver disease including metabolic disease, fibrosis, cirrhosis, and hepatocellular carcinoma (HCC). HCV induces and promotes liver disease progression by perturbing a range of survival, proliferative, and metabolic pathways within the proinflammatory cellular microenvironment. The recent breakthrough in antiviral therapy using direct-acting antivirals (DAAs) can cure >90% of HCV patients. However, viral cure cannot fully eliminate the HCC risk, especially in patients with advanced liver disease or comorbidities. HCV induces an epigenetic viral footprint that promotes a pro-oncogenic hepatic signature, which persists after DAA cure. In this review, we summarize the main signaling pathways deregulated by HCV infection, with potential impact on liver pathogenesis. HCV-induced persistent signaling patterns may serve as biomarkers for the stratification of HCV-cured patients at high risk of developing HCC. Moreover, these signaling pathways are potential targets for novel chemopreventive strategies.
Hepatitis C virus (HCV) is a main cause of chronic liver disease worldwide. Chronic HCV infection causes chronic hepatic inflammation, steatosis, and fibrosis, which progresses to cirrhosis and hepatocellular carcinoma (HCC) (Polaris Observatory HCV Collaborators 2017). HCC is the most common type of liver cancer and the second leading cause of cancer-related death on the globe (Baumert and Hoshida 2019). The liver is an extraordinarily resistant organ with a unique regeneration capacity, but the persistent stress induced by chronic inflammation and deregulation of signaling and metabolism culminate in a >10-fold increased HCC risk in HCV-infected patients compared with HCV-negative subjects in cross-sectional and case-control studies (El-Serag 2012). The rate of HCC among HCV-infected persons ranges from 1% to 3% and the interval from infection to HCC has been estimated to be ∼30 years (Thrift et al. 2017). It is believed that a combination of direct (viral proteins) and indirect (chronic inflammation, deregulated signaling) factors are responsible for HCV-induced liver disease development and progression. Because of the absence of a latent phase in the viral life cycle or any DNA integration event, HCV must ensure an optimal condition to maintain its replication (Lupberger et al. 2019) and to escape from the host innate immune response (Gale and Foy 2005). In this review, we summarize the main pathways that are deregulated during chronic HCV infection, which are relevant for the development and progression of HCV-induced liver disease and HCC. Some of these pathways remain deregulated in HCV-cured patients, serving as potential biomarkers for the identification of risk patients and novel drug targets for chemopreventive clinical strategies.
HCV-INDUCED CHRONIC INFLAMMATION, FIBROSIS, AND CIRRHOSIS
Inflammation is a life-preserving process to maintain cellular homeostasis. It is mostly activated in response to pathogens or tissue injury and is part of a physiological recovery response. The liver harbors a large spectrum of immune cells distributed within the hepatic compartments (Freitas-Lopes et al. 2017). This organ is constantly exposed to external signaling from commensal molecules and produces a series of neo-antigens derived by its metabolic activities. This leads to the development of a constant and physiological immunotolerance state in the organ (Jenne and Kubes 2013), which was first recognized by Calne and coworkers in 1969 (Calne et al. 1969). The relative immunotolerance in the liver is necessary to avoid overactivation of the immune system but it also facilitates the adaptation and persistence of different liver pathogens, such as malaria, hepatitis B virus (HBV), and HCV (Horst et al. 2016). HCV has developed several strategies to evade the innate and adaptive antiviral responses to infection (Gale and Foy 2005; Rosen 2013). Consequently, failure of viral clearance promotes a chronically inflamed liver that leads to scarification (fibrosis), cirrhosis, and ultimately provokes the development of HCC. According to the World Health Organization (see who.int), most of the HCV-infected patients do not achieve viral clearance and 60%–80% develop chronic hepatic inflammation. In these patients, the risk of developing cirrhosis is ∼15%–35% after 20–30 years of infection (Thrift et al. 2017). The virus directly accelerates the inflammatory response through a large range of interconnected mechanisms, including pathogen pattern recognition, host–viral protein interactions, activation of inflammasomes, and reactive oxygen species (ROS) production (Gale and Foy 2005; Horner and Gale 2013; Negash et al. 2019). Liver diseases and fibrosis associated with HCV infection evolve in the context of a strong oxidative microenvironment. HCV core, E1, E2, NS3, NS4B, and NS5A are known to encourage the production of ROS (Bureau et al. 2001; Pal et al. 2010; Ivanov et al. 2011). The antioxidant defense machine involves different ROS scavenging enzymes and their synthesis depends on many genes commonly regulated by the transcription factor NF-E2-related factor 2 (Nrf2) (Bureau et al. 2001). Nrf2 expression is inversely correlated with the severity of liver injury in chronic HCV patients and is impaired in end-stage liver disease (Kurzawski et al. 2012; Jiang et al. 2015). In HCV-positive cells, free Nrf2 is trapped at the replicon complexes and is therefore prevented from its entry into the nucleus (Medvedev et al. 2017). This observation is in line with impaired expression levels of antioxidative enzymes like catalase (Lupberger et al. 2019) and superoxide dismutase SOD1 (Levent et al. 2006; Diamond et al. 2012) in infected hepatocytes, which further promote oxidative stress damaging host proteins, lipids, and DNA. This coincides with a perturbed endogenous DNA repair by HCV infection (Nguyen et al. 2018; Lupberger et al. 2019) further contributing to the development of HCC in HCV patients. Because ROS-induced lipid peroxidation hampers viral membrane fusion, HCV has developed strategies to divert oxidative stress, for example, by the modulation of phospholipid hydroperoxide glutathione peroxidase (GPx4) (Brault et al. 2016). Importantly, ROS levels strongly promote liver fibrosis, characterized by an excessive production of extracellular matrix (ECM) and scarring of the tissue (Luangmonkong et al. 2018). At the same time, ROS stimulates pro-oncogenic signaling pathways, promoting cell survival, proliferation, and angiogenesis (Zhang et al. 2016). Chronic inflammation is accompanied by elevated plasma levels of proinflammatory cytokines, such as tumor necrosis factor α (TNF-α), which are further induced by HCV proteins NS3, NS4, and NS5 (Hosomura et al. 2011; Alhetheel et al. 2016). The levels of liver and blood cytokines are associated with HCV microenvironment and liver fibrosis (de Souza-Cruz et al. 2016). In particular, interleukin (IL)-1α is increased in HCV patients and correlates with liver cirrhosis and HCC (Tawfik et al. 2018). Therefore, HCV-induced cytokine signaling increases the oncogenic pressure within the host cell and contributes to a recalibration of hepatocyte functions (Fig. 1).
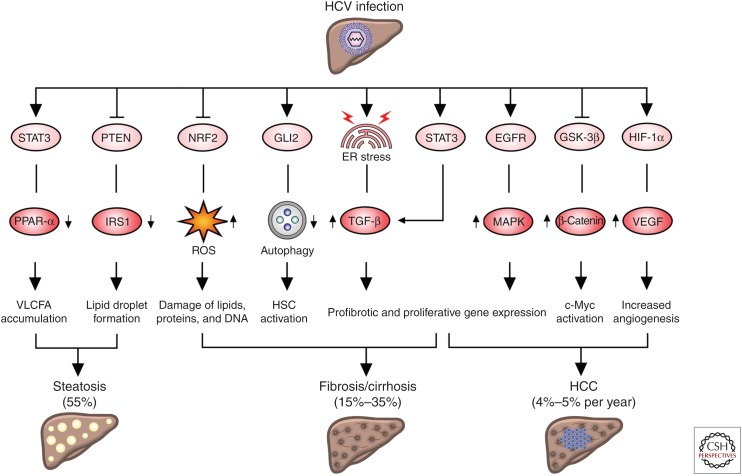
Figure 1.
Hepatitis C virus (HCV) infection alters signaling pathways relevant for liver disease. HCV-mediated activation of signal transducer and activator of transcription 3 (STAT3) causes very long-chain fatty acid (VLCFA) accumulation in the infected hepatocytes via down-regulation of peroxisome proliferator-activated receptor α (PPAR-α) expression. STAT3 activation sustains profibrotic gene expression via up-regulation of transforming growth factor β (TGF-β). Down-regulation of phosphatase and tensin homolog (PTEN) by HCV decreases insulin receptor substrate 1 (IRS1) expression and the formation of large lipid droplets favoring hepatic steatosis. HCV impairs NF-E2-related factor 2 (NRF2) activity and enhances the accumulation of reactive oxygen species (ROS). Activation of the Hedgehog (Hh) pathway via GLI family zinc finger 2 (GLI2) inhibits autophagy in hepatic stellate cells (HSCs), favoring their conversion into myofibroblasts and the development of fibrosis. HCV infection induces endoplasmic reticulum (ER) stress triggering TGF-β expression. Epithelial growth factor receptor (EGFR) is activated by several mechanisms and induces mitogen-activated protein kinase (MAPK) signaling and the expression of genes related to fibrosis and hepatocyte proliferation. Following HCV infection, the Wnt pathway is activated and inhibits the β-catenin destruction complex. As a consequence, β-catenin migrates to the nucleus and activates c-Myc oncogene. HCV sustains vascular endothelial growth factor (VEGF) via the stabilization of hypoxia inducible factor 1 subunit α (HIF1-α), which consequently up-regulates VEGF signaling and increases angiogenesis. The percentage of infected patients developing steatosis, cirrhosis, or the cumulative incidence of hepatocellular carcinoma (HCC) is indicated. GSK-3β, Glycogen synthase kinase 3β.
HCV Sustains Hedgehog Signaling Pathway and Promotes Fibrogenesis
The Hedgehog (Hh) pathway regulates liver development and differentiation and is a critical modulator of adult liver repair (Ingham and McMahon 2001; Machado and Diehl 2018). Interestingly, stimulation of the Hh pathway results in increased permissiveness for HCV replication in cell culture (Choi et al. 2011). HCV activates Hh signaling during fibrogenic repair of liver damage and increases the production of Hh ligands in HCV-infected cells (de Almeida Pereira et al. 2010). Complementary studies confirm that HCV derived from the sera of HCV-infected patients stimulates Hh signaling in human primary fibroblasts via activation of zinc finger protein GLI2 transcription factor. Especially, GLI2 inhibits autophagy in fibroblasts, thus forcing their conversion into myofibroblasts, which promotes fibrogenesis (Granato et al. 2016). The increase in Hh ligands may additionally be sustained by the accumulation of liver damage markers, such as epithelial growth factor (EGF), transforming growth factor β (TGF-β), and platelet-derived growth factor (PDGF) (Stepan et al. 2005; Jung et al. 2008; Omenetti et al. 2008), creating a persistent proliferative and antiapoptotic environment in the infected liver.
HCV Modulates Activation of the TGF-β Pathway
TGF-β has a key role in fibrogenesis and it is involved in all stages of liver disease progression (Dooley and ten Dijke 2012; Fabregat et al. 2016). The TGF-β superfamily includes pleiotropic growth factors that are essential for embryonic development and organ homeostasis. TGF-β is responsible for cell proliferation, differentiation, and migration during embryogenesis, while it is involved in tissue regeneration, cell growth control, and remodeling throughout adulthood. Under certain conditions, TGF-β1 is also involved in the induction of apoptotic cell death in the liver (Oberhammer et al. 1992). The TGF-β cytokine is physiologically sequestered in the ECM as part of latent complexes and it is released in response to different environmental perturbations (Xu et al. 2018). This cytokine triggers downstream signaling through the activation of canonical and noncanonical pathways. First, TGF-β mediates the formation of a heterotrimeric complex of type I and type II serine/threonine kinase receptors, which phosphorylate receptor-associated SMAD (R-SMADs) proteins. The trimeric complex formed by R-SMADs (Smad2 and Smad3) and Smad4 enters the nucleus and regulates gene expression (Miyazawa et al. 2002). Second, TGF-β triggers other signaling pathways, such as mitogen-activated protein kinase (MAPK) and transforming protein RhoA cascades, even in absence of SMADs activation (Yu et al. 2002; Derynck and Zhang 2003). In addition, both canonical and noncanonical signaling pathways can be modulated by TGF-β to tightly control epithelial-to-mesenchymal transition (EMT) (Bhowmick et al. 2001; Katsuno et al. 2019), which is a physiopathological program implicated in liver disease progression (Thiery and Sleeman 2006). TGF-β1 triggers hepatic fibrosis and cirrhosis in both animal models and human hepatic disorders (Castilla et al. 1991; Bedossa et al. 1995; Sanderson et al. 1995), and thus most evidently also plays an important role during HCV pathogenesis. Several studies and clinical observations highlighted a clear correlation between TGF-β and chronic HCV infection (Nelson et al. 1997; Grüngreiff et al. 1999; Ray et al. 2003; Chen et al. 2017). TGF-β plasma levels are associated with a high degree of hepatic fibrosis in patients with chronic HCV (Tsushima et al. 1999; Flisiak et al. 2002). Notably, HCV core protein seems to up-regulate the transcription of TGF-β (Taniguchi et al. 2004). HCV induces TGF-β1 via endoplasmic reticulum stress activation and the unfolded protein response (UPR) (Chusri et al. 2016). Additionally, in vitro studies show that HCV-induced oxidative stress indirectly regulates TGF-β1 expression through p38 MAPK, c-jun amino-terminal kinase (JNK), and extracellular signal-regulated kinase (ERK) via nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB) signaling (Erhardt et al. 2002; Lin et al. 2010). More recent studies observed decreased TGF-β1 levels in the serum of chronic HCV-infected patients that achieved sustained virologic response (SVR) after antiviral treatment (Janczewska-Kazek et al. 2006; Kotsiri et al. 2016). Therefore, uncovering the role of HCV proteins in TGF-β signaling pathways may contribute to understanding the mechanisms involved in HCV-induced HCC. Indeed, HCV core and NS3 have been shown to interact with Smad3 in vitro and in vivo (Cheng et al. 2004). Interestingly, some HCV core variants isolated from HCC tissue interact with Smad3 and inhibit TGF-β signaling. According to this study, a possible selection of viral variants during chronic HCV infection gradually promotes antiapoptotic effects in the liver that overcome the initial antiproliferative functions of TGF-β (Cheng et al. 2004). Hence, although TGF-β may have proapoptotic effects during the early stages of chronic liver disease, it probably acquires pro-cancerogenic responses after HCV core variants selection (Pavio et al. 2005; Battaglia et al. 2009).
HCV-Induced IL-6/STAT3 Signaling
Signal transducer and activator of transcription 3 (STAT3) is involved in tissue repair mechanisms by the regulation of proliferative and prosurvival cellular programs. In this context, activation of STAT3 can be induced by a vast number of different cytokines, including IL-6, which sensitizes hepatocytes to regenerative signals (Michalopoulos 2007). Beyond its physiological role, persistent activation of STAT3 induces chronic inflammation and fibrosis, increasing the risk to develop severe pathological conditions (Yu et al. 2014; Kasembeli et al. 2018). HCV requires IL-6/STAT3 signaling to maintain infection (Lupberger et al. 2013; McCartney et al. 2013); therefore, it induces its activation by several mechanisms. HCV core directly binds and sustains STAT3 activation (Yoshida et al. 2002), whereas the expression of NS5A, E1, and NS3 promotes STAT3 signaling indirectly via ROS production (Gong et al. 2001; Machida et al. 2006). The activation of STAT3 is not limited to HCV-infected hepatocytes. miR19a secreted in endosomes from HCV-infected hepatocytes impairs suppressor of cytokine signaling 3 (SOCS3) in hepatic stellate cells (HSCs). As a negative regulator of STAT3, impaired SOCS3 levels cause a subsequent activation of TGF-β in HSCs (Devhare et al. 2017). Therefore, considering the profibrotic role of STAT3 signaling and its strong cooperation with the TGF-β pathway, it has been suggested as a potential target for antifibrotic therapies (Chakraborty et al. 2017).
HCV INCREASES CANCER RISK BY DEREGULATION OF ONCOGENIC SIGNALING PATHWAYS
The liver is a key organ for the detoxification and metabolism of a wide range of potentially harmful substances. Therefore, liver regeneration is a tightly controlled process (Cordero-Espinoza and Huch 2018) that converges in the reconstruction of hepatocyte parenchyma in response to damage. The replacement of the damaged tissue occurs mainly through hepatocyte proliferation and to a lesser extent via an activation of ductal progenitor cells. During regeneration, the HSCs differentiate in myofibroblasts that release ECM within the space of Disse. Under normal conditions, the excess of ECM is promptly degraded by matrix metalloproteinases (MMPs), which restore the original architecture and function of the tissue without scar formation (Kholodenko and Yarygin 2017). During chronic inflammation this balance is perturbed, which leads to a progressive deposition of ECM and the development of liver fibrosis. HCV infection causes oxidative stress, steatohepatitis, and fibrosis, which create a hepatic pro-oncogenic environment. The oncogenic pressure on the diseased liver is further promoted by virus-induced growth factors and signaling pathways such as EGF, vascular endothelial growth factor (VEGF), Wnt/β-catenin, which are strongly implicated in the cirrhotic remodeling of the tissue and hepatocarcinogenesis (Fuchs et al. 2014; Wang et al. 2018a; Moon et al. 2019). As a consequence, patients affected with HCV-associated cirrhosis present a 4% to 5% cumulative annual incidence of HCC (El-Serag 2012).
HCV Up-Regulates EGFR and Stimulates MAPK Signaling
The growing knowledge on the interplay between HCV and epithelial growth factor receptor (EGFR) cascade has markedly contributed to explain the pathologic consequences of the viral infection, such as fibrosis development and HCC (Lupberger et al. 2011, 2013; Fuchs et al. 2014; Roca Suarez et al. 2018). It has been shown that EGFR signaling promotes the formation of the cluster of differentiation 81 (CD81)/claudin1 (CLDN1) coreceptor complex, which is required for HCV entry (Harris et al. 2010; Krieger et al. 2010; Lupberger et al. 2011; Zona et al. 2013). Inhibition of EGFR kinase hampers the CD81/CLDN1 coreceptor association and thus prevents HCV particle entry (Lupberger et al. 2011). The physical link between EGFR kinase and CD81/CLDN1 interaction is mediated by GTPase HRas, activated downstream from the EGFR signaling (Zona et al. 2013). HCV has an interest in maintaining EGFR signaling and elevated EGFR signaling is observed in liver biopsies of HCV patients (Mailly et al. 2015). EGFR signaling is further prolonged by a NS5A-induced retention of activated EGFR in the early endosomal compartment (Mankouri et al. 2008) and by an increasing level of Netrin-1 that impedes EGFR recycling (Plissonnier et al. 2016). Furthermore, NS3/4A protease mediates the down-regulation of T-cell protein tyrosine phosphatase (TC-PTP), which is negative regulator of EGFR and MAPK signaling (Brenndörfer et al. 2009; Stanford et al. 2012). The activation of EGFR during HCV infection induces MAPK signaling (Hayashi et al. 2000; Bürckstümmer et al. 2006; Mankouri et al. 2008; Diao et al. 2012), an evolutionarily conserved mechanism of cellular transduction that regulates many vital cellular functions, such as proliferation, differentiation, survival, and apoptosis (Zhang and Liu 2002; Dhillon et al. 2007). EGFR is overexpressed in ∼50% of patients with chronic HCV and in most patients with cirrhosis and HCC. The extent of EGFR expression is even higher in the advanced stages of HCV-related fibrosis (Badawy et al. 2015). These observations have a potential clinical application because EGF is a major driver of liver disease progression, and inhibition of EGFR signaling using clinical compounds in animal models attenuates the development of liver fibrosis and HCC nodules (Fuchs et al. 2014).
HCV Up-Regulates VEGF and Promotes Angiogenesis
Angiogenesis is a growth factor-dependent program responsible of the formation of new vessels from preexisting ones. It is commonly induced in response to hypoxia-related and inflammatory mechanisms (Paternostro et al. 2010). Hepatic angiogenesis is triggered by HCV via the deregulation of multiple pathways (Hassan et al. 2009). Several studies have shown an up-regulation of VEGF in HCV-related HCC patient tissues (Llovet et al. 2012; Mukozu et al. 2013). The HCV core protein seems to sustain VEGF signaling by several mechanisms. It can lead to hypoxia inducible factor 1 (HIF-1α) stabilization, which consequently up-regulates VEGF expression (Shimoda et al. 1999; Abe et al. 2012; Zhu et al. 2014). Additionally, HCV-mediated VEGF expression seems to also engage Janus kinase (JAK)/STAT signaling. Indeed, the inhibition of the JAK/STAT pathway in cell culture blocks the HCV core protein-mediated activation of the androgen receptor (AR), causing a down-regulation of VEGF (Kanda et al. 2008). HCV core protein potentiates VEGF expression by the activation of activator protein 1 (AP-1) transcription factor, which is binding to the VEGF promoter region (Shao et al. 2017).
HCV Induces β-Catenin Accumulation and Wnt Pathway Activation
Wnt pathway is crucial for embryonic development and cellular differentiation (Kielman et al. 2002; Reya and Clevers 2005; Grigoryan et al. 2008; Bone et al. 2011). When Wnt signaling is active, β-catenin phosphorylation is reduced via the inhibition of the β-catenin destruction complex (Behrens et al. 1998; Amit et al. 2002; Liu et al. 2002). The augmented unphosphorylated β-catenin migrates from the cytoplasm to the nucleus, where it binds to T-cell factor (TCF) and promotes transcription of genes such as Cyclin D1 (Tetsu and McCormick 1999), c-MYC (He et al. 1998), Axin-2 (Jho et al. 2002), and c-Jun (Mann et al. 1999). In cell culture, NS5A triggers the serine/threonine-protein kinase Akt, by interacting with phosphoinositide 3-kinases (PI3K). Consequently, this leads to an inhibition of glycogen synthase kinase (GSK)-3β, which is a key component of the destruction complex (Street et al. 2005). Moreover, NS5A stabilizes β-catenin in the cytoplasm and therefore promotes β-catenin signaling, which is also reflected in elevated β-catenin levels in livers of HCV patients (Park et al. 2009). This is very relevant for liver pathogenesis because β-catenin is most frequently activated in HCC pathogenesis (Khalaf et al. 2018). NS5A-induced stabilization of β-catenin transcription factor stimulates c-Myc expression in cell lines, human liver tissues, and livers from FL-N/35 transgenic mice (Colman et al. 2013; Higgs et al. 2013). c-Myc is an essential regulator of liver regeneration and its perturbation is considered as an early event during HCC development (Colman et al. 2013). Moreover, HCV-induced c-Myc expression drives the metabolic shift from glucose to glutamine dependence, which is a hallmark of cancer cells (Lévy et al. 2017).
HCV INFECTION ALTERS LIVER METABOLISM
The liver plays an essential role in the metabolic regulation during both the postprandial period and fasting state. The energetic balance of the organism is finely maintained by a series of biochemical reactions involved in metabolism, storing, and redistribution of carbohydrates, proteins, and lipids (Bechmann et al. 2012). HCV circulates in the serum of patients as lipo-viro-particles and interacts with very low-density lipoprotein (VLDL) components of the host. The striking association between the HCV life cycle and the VLDL pathway is not only crucial for HCV entry, maturation, and morphogenesis, but has also an impact on the immune escape capacity of the virus (Miyanari et al. 2007; Gondar et al. 2015). Importantly, the interplay between the virus and metabolic pathways contributes to the pathogenesis of liver disease via deregulation of the host lipid metabolism (Syed et al. 2010). HCV infection is strongly associated with hepatic steatosis and dysmetabolic syndromes, such as hypocholesterolemia, altered body fat distribution, insulin resistance (IR), and hyperuricemia (Kralj et al. 2016). Estimates suggest that ∼55% of HCV-infected patients develop hepatic steatosis, which is defined as an excessive accumulation of triglycerides (TGs) within the hepatocyte cytoplasm (Lonardo et al. 2006; Vilgrain et al. 2013). Although this has been observed for several HCV genotypes, steatosis is most frequent and severe in patients infected with genotype 3 (Leandro et al. 2006), which correlates with the viral load (Rubbia-Brandt et al. 2001). HCV-induced steatosis is triggered by the interaction between HCV proteins and host factors and its development does not require the presence of visceral obesity (Adinolfi et al. 2001). HCV infection deregulates metabolic pathways via miR146a5p expression, probably dependent on NF-κB signaling (Bandiera et al. 2016). In addition, it has been suggested that HCV core protein expression may be sufficient to induce liver fat accumulation and steatosis (Moriya et al. 1997). In particular, core protein 3a induces the activation of miR-21-5p, thereby promoting HCV replication and steatosis (Clément et al. 2019). An important factor in lipid homeostasis is the β-oxidation of fatty acids in mitochondria and the peroxisomal compartment. HCV infection suppresses peroxisomal β-oxidation, which leads to the accumulation of very long-chain fatty acids (VLCFAs) in the infected hepatocytes (Lupberger et al. 2019). This is partially mediated by HCV-induced STAT3 signaling (Van Renne et al. 2018), suppressing the peroxisome proliferator-activated receptor α (PPAR-α) expression (Lupberger et al. 2019). These results are consistent with decreased hepatic PPAR-α levels in HCV-infected patients (Dharancy et al. 2005). Importantly, HCV antiviral therapy can restore lipidic levels in serum (Batsaikhan et al. 2018; Doyle et al. 2019) and attenuate hepatic steatosis after viral clearance (Shimizu et al. 2018). However, many genes relevant for metabolism remain deregulated even after viral cure (Hamdane et al. 2019), including peroxisomal genes. Restoration of peroxisomal function may be therefore a clinical strategy to improve liver function in HCC risk patients. Notably, HCV genotype 3 infection is associated with the down-regulation of phosphatase and tensin homolog deleted on chromosome 10 (PTEN) leading to decreased levels of insulin receptor substrate 1 (IRS1) and the formation of large lipid droplets (Clément et al. 2011). This is relevant for the viral life cycle and liver disease progression because PTEN overexpression has been shown to reduce HCV viral particle secretion (Peyrou et al. 2013), and it is one of the most important tumor suppressors frequently mutated in many tumors, including HCC (Schulze et al. 2015). PTEN is also an important regulator of the insulin pathway and HCV infection perturbs the glucose homeostasis in the liver. Epidemiological studies suggest a link between chronic HCV infection and diabetes type 2 (Shintani et al. 2004; Gastaldi et al. 2017) and HCV core transgenic mice develop IR (Shintani et al. 2004). This is accompanied by a marked reduction in insulin-stimulated Akt phosphorylation without any alterations in MAPK activity in HCV-infected subjects (Aytug et al. 2003). HCV proteins up-regulate the protein phosphatase 2α (PP2A) catalytic subunit and alter signaling pathways controlling hepatic glucose homeostasis by inhibiting Akt and dephosphorylation of FoxO1 (Bernsmeier et al. 2008, 2014). Importantly, DAA treatment improves glycemic control and IR in livers, muscles, and adipose tissues of HCV cured patients (Adinolfi et al. 2018; Lim et al. 2019).
HCV-INDUCED LIVER DISEASE—IS THERE A POINT OF NO RETURN?
Since the discovery of HCV in 1989, there has been a remarkable breakthrough in antiviral therapy using DAAs. Meanwhile, >90% of patients can be cured by interferon-free treatments (Chung and Baumert 2014; Arends et al. 2016). However, in patients with advanced liver disease the risk of mortality and HCC development cannot be fully eliminated (Carrat et al. 2019). It has been estimated that HCV-induced HCC will remain one of the major health burdens for the next decades (Harris et al. 2014; Sievert et al. 2014; Petrick et al. 2016; Baumert et al. 2017). This also raises the question of whether some of the HCV-induced pro-oncogenic signaling pathways remain deregulated after viral cure. Indeed, HCV infection causes epigenetic alterations, which act as genetic circuits that influence gene expression patterns in the long term. DNA hypermethylation has been observed in livers of patients with chronic HCV infection, leading to a silencing of tumor suppressor gene expression (Wijetunga et al. 2017). In addition, HCV induces histone modifications, which also result in persistently altered gene expression patterns (Hamdane et al. 2019; Perez and Gal-Tanamy 2019). Importantly, this epigenetic footprint is still detectable in livers of HCV-cured chimeric mice and patients (Hamdane et al. 2019; Perez and Gal-Tanamy 2019). Associated with this viral footprint, the transcriptional signature reflecting many of the earlier mentioned HCV-induced pro-oncogenic signaling pathways remains deregulated after viral cure (Hamdane et al. 2019). This may partially account for the observed elevated HCC risk. Therefore, a detailed knowledge of these pathways will be potentially useful as biomarkers to identify patients at risk and highlight potential targets for future chemopreventive strategies.
Clinical methods to predict HCV-related fibrosis and cirrhosis and its associated HCC risk are still limited. The clinical outcome also very much depends on comorbidities like human immunodeficiency virus (HIV)/HBV coinfection or alcohol. Hoshida et al. (2008) developed a prognostic liver signature (PLS) from genome-wide transcriptomics of nontumor liver tissues adjacent from HCCs, which correlates to the clinical outcome of the patients. This has been later extended to a composite prognostic model for HCC recurrence (Villanueva et al. 2011). The PLS consists of 186 genes representing a powerful tool to predict the risk for patients to progress to cirrhosis and HCC and help prioritizing those for regular follow-up and HCC surveillance. Importantly, the PLS is induced also by HCV infection (Hoshida et al. 2013; King et al. 2015). PLS components are cytokines and signaling mediators that may be useful as targets for chemoprevention of their biological impact on liver disease development.
Small molecule inhibitors targeting signaling pathways arrived in clinical practice a long time ago, especially in cancer therapy. Some of these inhibitors target pathways that are potentially involved in an HCV-induced signaling pattern and have been tested or are currently in clinical trials for the treatment of liver disease progression. Human fibrosis and HSC activation are regulated by Wnt/β-catenin signaling (Berg et al. 2010; Ye et al. 2013; Lam et al. 2014), which therefore represents a promising target for the treatment of liver fibrosis (Cheng et al. 2008). Proof-of-concept has been provided targeting the interaction of CREB-binding protein (CBP) and β-catenin using the small molecule inhibitor PRI-724. This compound hampers HSC activation and accelerated fibrosis resolution, which seems to be accompanied by an increased expression of MMP2, MMP8, and MMP9 in intrahepatic leukocytes (Osawa et al. 2015). Currently, the safety and tolerability of PRI724 is being evaluated in patients with HCV or HBV-associated cirrhosis (NCT03620474). The Hh pathway is involved in the development of cirrhosis and HCC. Sonidegib (LDE225), a specific inhibitor of Hh is currently being tested in a phase I clinical trial for toxicity in patients with cirrhosis and advanced/metastatic HCC, who are intolerant to sorafenib (NCT02151864). In the last few years, a large number of nonspecific and specific TGF-β inhibitors have been developed (Giannelli et al. 2011; de Gramont et al. 2017). Despite that, galunisertib (LY2157299), a selective ATP-mimetic inhibitor of TGFβRI/ALK5, is the only inhibitor of TGF-β signaling currently under clinical trials in HCC patients (NCT01246986). Moreover, it seems to down-regulate the expression of stemness-related genes (such as CD44 and THY1) in HCC patients (Rani et al. 2018). Receptor tyrosine kinases (RTKs), such as EGFR and vascular endothelial growth factor receptor (VEGFR), have been shown to play crucial roles in fibrogenesis, cirrhosis, and HCC development, highlighting the importance of their therapeutic inhibition (Kömüves et al. 2000; Yoshiji et al. 2003; Fuchs et al. 2014; Badawy et al. 2015). Ramucirumab, a VEGFR-2 inhibitor, was recently evaluated as a second-line treatment for HCC patients previously treated with sorafenib, showing an improved overall survival compared with placebo (Zhu et al. 2019) (NCT02435433). STAT3 signaling pathway has shown to be up-regulated during HCV infection (Yoshida et al. 2002; McCartney et al. 2013; Van Renne et al. 2018) and strong data reveal its role in fibrosis development (Chakraborty et al. 2017). A large spectrum of clinical and preclinical data supports STAT3 as a pharmacological target for different typologies of cancers (Laudisi et al. 2018). This has prompted substantial efforts to design and test different types of STAT3 inhibitors. Some of the potential therapeutic opportunities to target STAT3 pathway are to be found upstream of its activation, at STAT3 SH2 domain and at STAT3 DNA-binding domain levels. AZD1480 (NCT01219543) and AG490 inhibitors belong to the first category and inhibit JAK2 kinase (Meydan et al. 1996; Hedvat et al. 2009). The safety and tolerability of AZD1480 have been tested in a phase I study in patients with solid tumors (including HCC). However, the unusual dose limit toxicity and the lack of clinical activity brought its discontinuation in clinical development (Plimack et al. 2013). OPB-31121, a potent SH2 domain inhibitor exerting also JAK inhibitory activity (Kim et al. 2013; Brambilla et al. 2015), has shown insufficient antitumoral activity and toxicity in patients with advanced HCC (Okusaka et al. 2015). S3I-201 (NSC 74859), discovered by structure-based virtual screening (Siddiquee et al. 2007), seems to suppress HSC activation and proliferation, as well as angiogenesis and fibrogenesis in fibrotic livers (Wang et al. 2018b). A promising therapeutic agent for liver fibrosis can be represented by HJC0123, which inhibits human HSC proliferation and STAT3 dimerization (Chen et al. 2013; Nunez Lopez et al. 2016). Recently, OPB-111077 (NCT01942083) has been shown to be well tolerated in patients with advanced HCC after failure of sorafenib therapy (Yoo et al. 2019). However, the preliminary outcomes of OPB-111077 treatment are still very limited (Yoo et al. 2019), and further investigation of the role of the STAT3 signaling pathway in fibrosis and HCC are required.
CONCLUSION
Studying HCV–host interactions is not only important for the understanding of the viral life cycle but also to answer how the virus manages to tweak its host cell to ensure persistence with all its consequences for liver pathogenesis. The molecular circuits exploited and triggered by HCV strikingly resemble other liver disease etiologies like nonalcoholic fatty liver disease (NAFLD) following a very similar path of disease progression. Studying HCV with all the experimental tools that have been developed during the last 30 years serves here as a powerful model to understand the specific and common mechanisms of liver disease development. This is essential to develop new diagnostic biomarkers and chemopreventive strategies to help HCV cured patients with advanced liver disease to tackle the epigenetic turnouts set by decades of chronic HCV infection. These tools will be potentially very useful also for other liver disease etiologies.
ACKNOWLEDGMENTS
This work was supported by the European Union (ERC-AdG-2014 HEPCIR to T.F.B., EU H2020 HEPCAR 667273 to T.F.B. and J.L.), the French Cancer Agency (ARC IHU201301187 to T.F.B.), the Agence Nationale de Recherche sur le Sida et les Hépatites Virales (ECTZ4236 to J.L. and ECTZ4446 to A.A.R.S.), Fondation pour la Recherche Médicale en France (FDT201805005763 to A.A.R.S.), the United States Department of Defense (W81XWH-16-1-0363 to T.F.B.), the National Institutes of Health (NIAID R03AI131066, NCI 1R21CA209940, NIAID 5U19AI123862-02 to T.F.B), the Fondation de l'Université de Strasbourg (HEPKIN) (TBA-DON-0002) and the Inserm Plan Cancer 2019-2023 to T.F.B. This work has benefitted from support by the Initiative of Excellence IDEX-Unistra (2018-383i to A.V.) and has been published under the framework of the LABEX ANR-10-LAB-28 (HEPSYS). Inserm Plan Cancer, IDEX, and LABEX are initiatives from the French program “Investments for the Future.”
Footnotes
Editors: Arash Grakoui, Jean-Michel Pawlotsky, and Glenn Randall
Additional Perspectives on Hepatitis C Viruses: The Story of a Scientific and Therapeutic Revolution available at www.perspectivesinmedicine.org
REFERENCES
- Abe M, Koga H, Yoshida T, Masuda H, Iwamoto H, Sakata M, Hanada S, Nakamura T, Taniguchi E, Kawaguchi T, et al. 2012. Hepatitis C virus core protein upregulates the expression of vascular endothelial growth factor via the nuclear factor-κB/hypoxia-inducible factor-1 α axis under hypoxic conditions. Hepatol Res 42: 591–600. 10.1111/j.1872-034X.2011.00953.x [DOI] [PubMed] [Google Scholar]
- Adinolfi LE, Gambardella M, Andreana A, Tripodi MF, Utili R, Ruggiero G. 2001. Steatosis accelerates the progression of liver damage of chronic hepatitis C patients and correlates with specific HCV genotype and visceral obesity. Hepatology 33: 1358–1364. 10.1053/jhep.2001.24432 [DOI] [PubMed] [Google Scholar]
- Adinolfi LE, Nevola R, Guerrera B, D'Alterio G, Marrone A, Giordano M, Rinaldi L. 2018. Hepatitis C virus clearance by direct-acting antiviral treatments and impact on insulin resistance in chronic hepatitis C patients. J Gastroenterol Hepatol 33: 1379–1382. 10.1111/jgh.14067 [DOI] [PubMed] [Google Scholar]
- Alhetheel A, Albarrag A, Shakoor Z, Alswat K, Abdo A, Al-Hamoudi W. 2016. Assessment of pro-inflammatory cytokines in sera of patients with hepatitis C virus infection before and after anti-viral therapy. J Infect Dev Ctries 10: 1093–1098. 10.3855/jidc.7595 [DOI] [PubMed] [Google Scholar]
- Amit S, Hatzubai A, Birman Y, Andersen JS, Ben-Shushan E, Mann M, Ben-Neriah Y, Alkalay I. 2002. Axin-mediated CKI phosphorylation of β-catenin at Ser 45: a molecular switch for the Wnt pathway. Genes Dev 16: 1066–1076. 10.1101/gad.230302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arends JE, Kracht PA, Hoepelman AI; European Study Group for Viral Hepatitits (ESGVH). 2016. Performance of hepatitis C virus (HCV) direct-acting antivirals in clinical trials and daily practice. Clin Microbiol Infect 22: 846–852. 10.1016/j.cmi.2016.05.027 [DOI] [PubMed] [Google Scholar]
- Aytug S, Reich D, Sapiro LE, Bernstein D, Begum N. 2003. Impaired IRS-1/PI3-kinase signaling in patients with HCV: a mechanism for increased prevalence of type 2 diabetes. Hepatology 38: 1384–1392. 10.1053/jhep.2003.09012 [DOI] [PubMed] [Google Scholar]
- Badawy AA, El-Hindawi A, Hammam O, Moussa M, Gabal S, Said N. 2015. Impact of epidermal growth factor receptor and transforming growth factor-α on hepatitis C virus-induced hepatocarcinogenesis. APMIS 123: 823–831. 10.1111/apm.12431 [DOI] [PubMed] [Google Scholar]
- Bandiera S, Pernot S, El Saghire H, Durand SC, Thumann C, Crouchet E, Ye T, Fofana I, Oudot MA, Barths J, et al. 2016. Hepatitis C virus-induced upregulation of microRNA miR-146a-5p in hepatocytes promotes viral infection and deregulates metabolic pathways associated with liver disease pathogenesis. J Virol 90: 6387–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsaikhan B, Huang CI, Yeh ML, Huang CF, Hou NJ, Lin ZY, Chen SC, Huang JF, Yu ML, Chuang WL, et al. 2018. The effect of antiviral therapy on serum lipid profiles in chronic hepatitis C. Oncotarget 9: 21313–21321. 10.18632/oncotarget.25092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia S, Benzoubir N, Nobilet S, Charneau P, Samuel D, Zignego AL, Atfi A, Brechot C, Bourgeade MF. 2009. Liver cancer-derived hepatitis C virus core proteins shift TGF-β responses from tumor suppression to epithelial–mesenchymal transition. PLoS ONE 4: e4355 10.1371/journal.pone.0004355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumert TF, Hoshida Y. 2019. Addressing the challenges of hepatitis C cure and persistent risk of hepatocellular carcinoma. Viruses 11: 441 10.3390/v11050441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumert TF, Jühling F, Ono A, Hoshida Y. 2017. Hepatitis C-related hepatocellular carcinoma in the era of new generation antivirals. BMC Med 15: 52 10.1186/s12916-017-0815-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechmann LP, Hannivoort RA, Gerken G, Hotamisligil GS, Trauner M, Canbay A. 2012. The interaction of hepatic lipid and glucose metabolism in liver diseases. J Hepatol 56: 952–964. 10.1016/j.jhep.2011.08.025 [DOI] [PubMed] [Google Scholar]
- Bedossa P, Peltier E, Terris B, Franco D, Poynard T. 1995. Transforming growth factor-β1 (TGFβ1) and TGF-β1 receptors in normal, cirrhotic, and neoplastic human livers. Hepatology 21: 760–766. [PubMed] [Google Scholar]
- Behrens J, Jerchow BA, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. 1998. Functional interaction of an axin homolog, conductin, with β-catenin, APC, and GSK3β. Science 280: 596–599. 10.1126/science.280.5363.596 [DOI] [PubMed] [Google Scholar]
- Berg T, DeLanghe S, Al Alam D, Utley S, Estrada J, Wang KS. 2010. β-Catenin regulates mesenchymal progenitor cell differentiation during hepatogenesis. J Surg Res 164: 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernsmeier C, Duong FH, Christen V, Pugnale P, Negro F, Terracciano L, Heim MH. 2008. Virus-induced over-expression of protein phosphatase 2A inhibits insulin signalling in chronic hepatitis C. J Hepatol 49: 429–440. 10.1016/j.jhep.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Bernsmeier C, Calabrese D, Heim MH, Duong HT. 2014. Hepatitis C virus dysregulates glucose homeostasis by a dual mechanism involving induction of PGC1α and dephosphorylation of FoxO1. J Viral Hepat 21: 9–18. 10.1111/jvh.12208 [DOI] [PubMed] [Google Scholar]
- Bhowmick NA, Ghiassi M, Bakin A, Aakre M, Lundquist CA, Engel ME, Arteaga CL, Moses HL. 2001. Transforming growth factor-β1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol Biol Cell 12: 27–36. 10.1091/mbc.12.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bone HK, Nelson AS, Goldring CE, Tosh D, Welham MJ. 2011. A novel chemically directed route for the generation of definitive endoderm from human embryonic stem cells based on inhibition of GSK-3. J Cell Sci 124: 1992–2000. 10.1242/jcs.081679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambilla L, Genini D, Laurini E, Merulla J, Perez L, Fermeglia M, Carbone GM, Pricl S, Catapano CV. 2015. Hitting the right spot: Mechanism of action of OPB-31121, a novel and potent inhibitor of the signal transducer and activator of transcription 3 (STAT3). Mol Oncol 9: 1194–1206. 10.1016/j.molonc.2015.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault C, Lévy P, Duponchel S, Michelet M, Sallé A, Pécheur EI, Plissonnier ML, Parent R, Véricel E, Ivanov AV, et al. 2016. Glutathione peroxidase 4 is reversibly induced by HCV to control lipid peroxidation and to increase virion infectivity. Gut 65: 144–154. 10.1136/gutjnl-2014-307904 [DOI] [PubMed] [Google Scholar]
- Brenndörfer ED, Karthe J, Frelin L, Cebula P, Erhardt A, Schulte am Esch J, Hengel H, Bartenschlager R, Sällberg M, Häussinger D, et al. 2009. Nonstructural 3/4A protease of hepatitis C virus activates epithelial growth factor-induced signal transduction by cleavage of the T-cell protein tyrosine phosphatase. Hepatology 49: 1810–1820. 10.1002/hep.22857 [DOI] [PubMed] [Google Scholar]
- Bürckstümmer T, Kriegs M, Lupberger J, Pauli EK, Schmittel S, Hildt E. 2006. Raf-1 kinase associates with hepatitis C virus NS5A and regulates viral replication. FEBS Lett 580: 575–580. 10.1016/j.febslet.2005.12.071 [DOI] [PubMed] [Google Scholar]
- Bureau C, Bérnad J, Chaouche N, Orfila C, Béraud M, Gonindard C, Alric L, Vinel JP, Pipy B. 2001. Nonstructural 3 protein of hepatitis C virus triggers an oxidative burst in human monocytes via activation of NADPH oxidase. J Biol Chem 276: 23077–23083. 10.1074/jbc.M100698200 [DOI] [PubMed] [Google Scholar]
- Calne RY, Sells RA, Pena JR, Davis DR, Millard PR, Herbertson BM, Binns RM, Davies DA. 1969. Induction of immunological tolerance by porcine liver allografts. Nature 223: 472–476. 10.1038/223472a0 [DOI] [PubMed] [Google Scholar]
- Carrat F, Fontaine H, Dorival C, Simony M, Diallo A, Hezode C, De Ledinghen V, Larrey D, Haour G, Bronowicki JP, et al. 2019. Clinical outcomes in patients with chronic hepatitis C after direct-acting antiviral treatment: a prospective cohort study. Lancet 393: 1453–1464. 10.1016/S0140-6736(18)32111-1 [DOI] [PubMed] [Google Scholar]
- Castilla A, Prieto J, Fausto N. 1991. Transforming growth factors β1 and α in chronic liver disease. Effects of interferon alfa therapy. N Engl J Med 324: 933–940. 10.1056/NEJM199104043241401 [DOI] [PubMed] [Google Scholar]
- Chakraborty D, Šumová B, Mallano T, Chen CW, Distler A, Bergmann C, Ludolph I, Horch RE, Gelse K, Ramming A, et al. 2017. Activation of STAT3 integrates common profibrotic pathways to promote fibroblast activation and tissue fibrosis. Nat Commun 8: 1130 10.1038/s41467-017-01236-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yang Z, Ding C, Chu L, Zhang Y, Terry K, Liu H, Shen Q, Zhou J. 2013. Fragment-based drug design and identification of HJC0123, a novel orally bioavailable STAT3 inhibitor for cancer therapy. Eur J Med Chem 62: 498–507. 10.1016/j.ejmech.2013.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Yang W, Wang X, Li X, Qi S, Zhang Y, Gao MQ. 2017. TGF-β1 induces EMT in bovine mammary epithelial cells through the TGFβ1/Smad signaling pathway. Cell Physiol Biochem 43: 82–93. 10.1159/000480321 [DOI] [PubMed] [Google Scholar]
- Cheng PL, Chang MH, Chao CH, Lee YH. 2004. Hepatitis C viral proteins interact with Smad3 and differentially regulate TGF-β/Smad3-mediated transcriptional activation. Oncogene 23: 7821–7838. 10.1038/sj.onc.1208066 [DOI] [PubMed] [Google Scholar]
- Cheng JH, She H, Han YP, Wang J, Xiong S, Asahina K, Tsukamoto H. 2008. Wnt antagonism inhibits hepatic stellate cell activation and liver fibrosis. Am J Physiol Gastrointest Liver Physiol 294: G39–G49. [DOI] [PubMed] [Google Scholar]
- Choi SS, Bradrick S, Qiang G, Mostafavi A, Chaturvedi G, Weinman SA, Diehl AM, Jhaveri R. 2011. Up-regulation of Hedgehog pathway is associated with cellular permissiveness for hepatitis C virus replication. Hepatology 54: 1580–1590. 10.1002/hep.24576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung RT, Baumert TF. 2014. Curing chronic hepatitis C—the arc of a medical triumph. N Engl J Med 370: 1576–1578. 10.1056/NEJMp1400986 [DOI] [PubMed] [Google Scholar]
- Chusri P, Kumthip K, Hong J, Zhu C, Duan X, Jilg N, Fusco DN, Brisac C, Schaefer EA, Cai D, et al. 2016. HCV induces transforming growth factor β1 through activation of endoplasmic reticulum stress and the unfolded protein response. Sci Rep 6: 22487 10.1038/srep22487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clément S, Peyrou M, Sanchez-Pareja A, Bourgoin L, Ramadori P, Suter D, Vinciguerra M, Guilloux K, Pascarella S, Rubbia-Brandt L, et al. 2011. Down-regulation of phosphatase and tensin homolog by hepatitis C virus core 3a in hepatocytes triggers the formation of large lipid droplets. Hepatology 54: 38–49. 10.1002/hep.24340 [DOI] [PubMed] [Google Scholar]
- Clément S, Sobolewski C, Gomes D, Rojas A, Goossens N, Conzelmann S, Calo N, Negro F, Foti M. 2019. Activation of the oncogenic miR-21-5p promotes HCV replication and steatosis induced by the viral core 3a protein. Liver Int 10.1111/liv.14112. [DOI] [PubMed] [Google Scholar]
- Colman H, Le Berre-Scoul C, Hernandez C, Pierredon S, Bihouee A, Houlgatte R, Vagner S, Rosenberg AR, Feray C. 2013. Genome-wide analysis of host mRNA translation during hepatitis C virus infection. J Virol 87: 6668–6677. 10.1128/JVI.00538-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero-Espinoza L, Huch M. 2018. The balancing act of the liver: tissue regeneration versus fibrosis. J Clin Invest 128: 85–96. 10.1172/JCI93562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Almeida Pereira T, Witek RP, Syn WK, Choi SS, Bradrick S, Karaca GF, Agboola KM, Jung Y, Omenetti A, Moylan CA, et al. 2010. Viral factors induce Hedgehog pathway activation in humans with viral hepatitis, cirrhosis, and hepatocellular carcinoma. Lab Invest 90: 1690–1703. 10.1038/labinvest.2010.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Gramont A, Faivre S, Raymond E. 2017. Novel TGF-β inhibitors ready for prime time in onco-immunology. Oncoimmunology 6: e1257453 10.1080/2162402X.2016.1257453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. 2003. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425: 577–584. 10.1038/nature02006 [DOI] [PubMed] [Google Scholar]
- de Souza-Cruz S, Victória MB, Tarragô AM, da Costa AG, Pimentel JP, Pires EF, Araújo Lde P, Coelho-dos-Reis JG, Mde S Gomes, Amaral LR, et al. 2016. Liver and blood cytokine microenvironment in HCV patients is associated to liver fibrosis score: a proinflammatory cytokine ensemble orchestrated by TNF and tuned by IL-10. BMC Microbiol 16: 3 10.1186/s12866-015-0610-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devhare PB, Sasaki R, Shrivastava S, Di Bisceglie AM, Ray R, Ray RB. 2017. Exosome-mediated intercellular communication between hepatitis C virus-infected hepatocytes and hepatic stellate cells. J Virol 91: e02225-16 10.1128/JVI.02225-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharancy S, Malapel M, Perlemuter G, Roskams T, Cheng Y, Dubuquoy L, Podevin P, Conti F, Canva V, Philippe D, et al. 2005. Impaired expression of the peroxisome proliferator-activated receptor α during hepatitis C virus infection. Gastroenterology 128: 334–342. 10.1053/j.gastro.2004.11.016 [DOI] [PubMed] [Google Scholar]
- Dhillon AS, Hagan S, Rath O, Kolch W. 2007. MAP kinase signalling pathways in cancer. Oncogene 26: 3279–3290. 10.1038/sj.onc.1210421 [DOI] [PubMed] [Google Scholar]
- Diamond DL, Krasnoselsky AL, Burnum KE, Monroe ME, Webb-Robertson BJ, McDermott JE, Yeh MM, Dzib JF, Susnow N, Strom S, et al. 2012. Proteome and computational analyses reveal new insights into the mechanisms of hepatitis C virus-mediated liver disease posttransplantation. Hepatology 56: 28–38. 10.1002/hep.25649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J, Pantua H, Ngu H, Komuves L, Diehl L, Schaefer G, Kapadia SB. 2012. Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J Virol 86: 10935–10949. 10.1128/JVI.00750-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley S, ten Dijke P. 2012. TGF-β in progression of liver disease. Cell Tissue Res 347: 245–256. 10.1007/s00441-011-1246-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle MA, Galanakis C, Mulvihill E, Crawley A, Cooper CL. 2019. Hepatitis C direct acting antivirals and ribavirin modify lipid but not glucose parameters. Cells 8: 252 10.3390/cells8030252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag HB. 2012. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology 142: 1264–1273e1. 10.1053/j.gastro.2011.12.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erhardt A, Hassan M, Heintges T, Häussinger D. 2002. Hepatitis C virus core protein induces cell proliferation and activates ERK, JNK, and p38 MAP kinases together with the MAP kinase phosphatase MKP-1 in a HepG2 Tet-Off cell line. Virology 292: 272–284. 10.1006/viro.2001.1227 [DOI] [PubMed] [Google Scholar]
- Fabregat I, Moreno-Càceres J, Sánchez A, Dooley S, Dewidar B, Giannelli G, Ten Dijke P, Consortium I-L. 2016. TGF-β signalling and liver disease. FEBS J 283: 2219–2232. 10.1111/febs.13665 [DOI] [PubMed] [Google Scholar]
- Flisiak R, Maxwell P, Prokopowicz D, Timms PM, Panasiuk A. 2002. Plasma tissue inhibitor of metalloproteinases-1 and transforming growth factor β1—possible non-invasive biomarkers of hepatic fibrosis in patients with chronic B and C hepatitis. Hepatogastroenterology 49: 1369–1372. [PubMed] [Google Scholar]
- Freitas-Lopes MA, Mafra K, David BA, Carvalho-Gontijo R, Menezes GB. 2017. Differential location and distribution of hepatic immune cells. Cells 6: 48 10.3390/cells6040048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs BC, Hoshida Y, Fujii T, Wei L, Yamada S, Lauwers GY, McGinn CM, DePeralta DK, Chen X, Kuroda T, et al. 2014. Epidermal growth factor receptor inhibition attenuates liver fibrosis and development of hepatocellular carcinoma. Hepatology 59: 1577–1590. 10.1002/hep.26898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale M Jr, Foy EM. 2005. Evasion of intracellular host defence by hepatitis C virus. Nature 436: 939–945. 10.1038/nature04078 [DOI] [PubMed] [Google Scholar]
- Gastaldi G, Goossens N, Clément S, Negro F. 2017. Current level of evidence on causal association between hepatitis C virus and type 2 diabetes: a review. J Adv Res 8: 149–159. 10.1016/j.jare.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannelli G, Mazzocca A, Fransvea E, Lahn M, Antonaci S. 2011. Inhibiting TGF-β signaling in hepatocellular carcinoma. Biochim Biophys Acta 1815: 214–223. 10.1016/j.bbcan.2010.11.004 [DOI] [PubMed] [Google Scholar]
- Gondar V, Molina-Jiménez F, Hishiki T, García-Buey L, Koutsoudakis G, Shimotohno K, Benedicto I, Majano PL. 2015. Apolipoprotein E, but not apolipoprotein B, is essential for efficient cell-to-cell transmission of hepatitis C virus. J Virol 89: 9962–9973. 10.1128/JVI.00577-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong G, Waris G, Tanveer R, Siddiqui A. 2001. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-κB. Proc Natl Acad Sci 98: 9599–9604. 10.1073/pnas.171311298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granato M, Zompetta C, Vescarelli E, Rizzello C, Cardi A, Valia S, Antonelli G, Marchese C, Torrisi MR, Faggioni A, et al. 2016. HCV derived from sera of HCV-infected patients induces pro-fibrotic effects in human primary fibroblasts by activating GLI2. Sci Rep 6: 30649 10.1038/srep30649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryan T, Wend P, Klaus A, Birchmeier W. 2008. Deciphering the function of canonical Wnt signals in development and disease: conditional loss- and gain-of-function mutations of β-catenin in mice. Genes Dev 22: 2308–2341. 10.1101/gad.1686208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüngreiff K, Reinhold D, Ansorge S. 1999. Serum concentrations of sIL-2R, IL-6, TGF- β1, neopterin, and zinc in chronic hepatitis C patients treated with interferon-α. Cytokine 11: 1076–1080. 10.1006/cyto.1999.0504 [DOI] [PubMed] [Google Scholar]
- Hamdane N, Jühling F, Crouchet E, El Saghire H, Thumann C, Oudot MA, Bandiera S, Saviano A, Ponsolles C, Roca Suarez AA, et al. 2019. HCV-induced epigenetic changes associated with liver cancer risk persist after sustained virologic response. Gastroenterology 156: 2313–2329.e7. 10.1053/j.gastro.2019.02.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris HJ, Davis C, Mullins JG, Hu K, Goodall M, Farquhar MJ, Mee CJ, McCaffrey K, Young S, Drummer H, et al. 2010. Claudin association with CD81 defines hepatitis C virus entry. J Biol Chem 285: 21092–21102. 10.1074/jbc.M110.104836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RJ, Thomas B, Griffiths J, Costella A, Chapman R, Ramsay M, De Angelis D, Harris HE. 2014. Increased uptake and new therapies are needed to avert rising hepatitis C-related end stage liver disease in England: modelling the predicted impact of treatment under different scenarios. J Hepatol 61: 530–537. 10.1016/j.jhep.2014.05.008 [DOI] [PubMed] [Google Scholar]
- Hassan M, Selimovic D, Ghozlan H, Abdel-kader O. 2009. Hepatitis C virus core protein triggers hepatic angiogenesis by a mechanism including multiple pathways. Hepatology 49: 1469–1482. 10.1002/hep.22849 [DOI] [PubMed] [Google Scholar]
- Hayashi J, Aoki H, Kajino K, Moriyama M, Arakawa Y, Hino O. 2000. Hepatitis C virus core protein activates the MAPK/ERK cascade synergistically with tumor promoter TPA, but not with epidermal growth factor or transforming growth factor α. Hepatology 32: 958–961. 10.1053/jhep.2000.19343 [DOI] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. 1998. Identification of c-MYC as a target of the APC pathway. Science 281: 1509–1512. 10.1126/science.281.5382.1509 [DOI] [PubMed] [Google Scholar]
- Hedvat M, Huszar D, Herrmann A, Gozgit JM, Schroeder A, Sheehy A, Buettner R, Proia D, Kowolik CM, Xin H, et al. 2009. The JAK2 inhibitor AZD1480 potently blocks Stat3 signaling and oncogenesis in solid tumors. Cancer Cell 16: 487–497. 10.1016/j.ccr.2009.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs MR, Lerat H, Pawlotsky JM. 2013. Hepatitis C virus-induced activation of β-catenin promotes c-Myc expression and a cascade of pro-carcinogenetic events. Oncogene 32: 4683–4693. 10.1038/onc.2012.484 [DOI] [PubMed] [Google Scholar]
- Horner SM, Gale M Jr. 2013. Regulation of hepatic innate immunity by hepatitis C virus. Nat Med 19: 879–888. 10.1038/nm.3253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst AK, Neumann K, Diehl L, Tiegs G. 2016. Modulation of liver tolerance by conventional and nonconventional antigen-presenting cells and regulatory immune cells. Cell Mol Immunol 13: 277–292. 10.1038/cmi.2015.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida Y, Villanueva A, Kobayashi M, Peix J, Chiang DY, Camargo A, Gupta S, Moore J, Wrobel MJ, Lerner J, et al. 2008. Gene expression in fixed tissues and outcome in hepatocellular carcinoma. N Engl J Med 359: 1995–2004. 10.1056/NEJMoa0804525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshida Y, Villanueva A, Sangiovanni A, Sole M, Hur C, Andersson KL, Chung RT, Gould J, Kojima K, Gupta S, et al. 2013. Prognostic gene expression signature for patients with hepatitis C-related early-stage cirrhosis. Gastroenterology 144: 1024–1030. 10.1053/j.gastro.2013.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosomura N, Kono H, Tsuchiya M, Ishii K, Ogiku M, Matsuda M, Fujii H. 2011. HCV-related proteins activate Kupffer cells isolated from human liver tissues. Dig Dis Sci 56: 1057–1064. 10.1007/s10620-010-1395-y [DOI] [PubMed] [Google Scholar]
- Ingham PW, McMahon AP. 2001. Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15: 3059–3087. 10.1101/gad.938601 [DOI] [PubMed] [Google Scholar]
- Ivanov AV, Smirnova OA, Ivanova ON, Masalova OV, Kochetkov SN, Isaguliants MG. 2011. Hepatitis C virus proteins activate NRF2/ARE pathway by distinct ROS-dependent and independent mechanisms in HUH7 cells. PLoS ONE 6: e24957 10.1371/journal.pone.0024957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janczewska-Kazek E, Marek B, Kajdaniuk D, Borgiel-Marek H. 2006. Effect of interferon α and ribavirin treatment on serum levels of transforming growth factor-β1, vascular endothelial growth factor, and basic fibroblast growth factor in patients with chronic hepatitis C. World J Gastroenterol 12: 961–965. 10.3748/wjg.v12.i6.961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenne CN, Kubes P. 2013. Immune surveillance by the liver. Nat Immunol 14: 996–1006. 10.1038/ni.2691 [DOI] [PubMed] [Google Scholar]
- Jho EH, Zhang T, Domon C, Joo CK, Freund JN, Costantini F. 2002. Wnt/β-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol 22: 1172–1183. 10.1128/MCB.22.4.1172-1183.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Bao H, Ge Y, Tang W, Cheng D, Luo K, Gong G, Gong R. 2015. Therapeutic targeting of GSK3β enhances the Nrf2 antioxidant response and confers hepatic cytoprotection in hepatitis C. Gut 64: 168–179. 10.1136/gutjnl-2013-306043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y, Brown KD, Witek RP, Omenetti A, Yang L, Vandongen M, Milton RJ, Hines IN, Rippe RA, Spahr L, et al. 2008. Accumulation of hedgehog-responsive progenitors parallels alcoholic liver disease severity in mice and humans. Gastroenterology 134: 1532–1543.e3. 10.1053/j.gastro.2008.02.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda T, Steele R, Ray R, Ray RB. 2008. Hepatitis C virus core protein augments androgen receptor-mediated signaling. J Virol 82: 11066–11072. 10.1128/JVI.01300-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasembeli MM, Bharadwaj U, Robinson P, Tweardy DJ. 2018. Contribution of STAT3 to inflammatory and fibrotic diseases and prospects for its targeting for treatment. Int J Mol Sci 19: 2299 10.3390/ijms19082299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuno Y, Meyer DS, Zhang Z, Shokat KM, Akhurst RJ, Miyazono K, Derynck R. 2019. Chronic TGF-β exposure drives stabilized EMT, tumor stemness, and cancer drug resistance with vulnerability to bitopic mTOR inhibition. Sci Signal 12: eaau8544 10.1126/scisignal.aau8544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaf AM, Fuentes D, Morshid AI, Burke MR, Kaseb AO, Hassan M, Hazle JD, Elsayes KM. 2018. Role of Wnt/β-catenin signaling in hepatocellular carcinoma, pathogenesis, and clinical significance. J Hepatocell Carcinoma 5: 61–73. 10.2147/JHC.S156701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholodenko IV, Yarygin KN. 2017. Cellular mechanisms of liver regeneration and cell-based therapies of liver diseases. Biomed Res Int 2017: 8910821 10.1155/2017/8910821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielman MF, Rindapää M, Gaspar C, van Poppel N, Breukel C, van Leeuwen S, Taketo MM, Roberts S, Smits R, Fodde R. 2002. Apc modulates embryonic stem-cell differentiation by controlling the dosage of β-catenin signaling. Nat Genet 32: 594–605. 10.1038/ng1045 [DOI] [PubMed] [Google Scholar]
- Kim MJ, Nam HJ, Kim HP, Han SW, Im SA, Kim TY, Oh DY, Bang YJ. 2013. OPB-31121, a novel small molecular inhibitor, disrupts the JAK2/STAT3 pathway and exhibits an antitumor activity in gastric cancer cells. Cancer Lett 335: 145–152. 10.1016/j.canlet.2013.02.010 [DOI] [PubMed] [Google Scholar]
- King LY, Canasto-Chibuque C, Johnson KB, Yip S, Chen X, Kojima K, Deshmukh M, Venkatesh A, Tan PS, Sun X, et al. 2015. A genomic and clinical prognostic index for hepatitis C-related early-stage cirrhosis that predicts clinical deterioration. Gut 64: 1296–1302. 10.1136/gutjnl-2014-307862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kömüves LG, Feren A, Jones AL, Fodor E. 2000. Expression of epidermal growth factor and its receptor in cirrhotic liver disease. J Histochem Cytochem 48: 821–830. 10.1177/002215540004800610 [DOI] [PubMed] [Google Scholar]
- Kotsiri I, Hadziyannis E, Georgiou A, Papageorgiou MV, Vlachogiannakos I, Papatheodoridis G. 2016. Changes in serum transforming growth factor-β1 levels in chronic hepatitis C patients under antiviral therapy. Ann Gastroenterol 29: 79–84. [PMC free article] [PubMed] [Google Scholar]
- Kralj D, Virovic Jukic L, Stojsavljevic S, Duvnjak M, Smolic M, Curcic IB. 2016. Hepatitis C virus, insulin resistance, and steatosis. J Clin Transl Hepatol 4: 66–75. 10.14218/JCTH.2015.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger SE, Zeisel MB, Davis C, Thumann C, Harris HJ, Schnober EK, Mee C, Soulier E, Royer C, Lambotin M, et al. 2010. Inhibition of hepatitis C virus infection by anti-claudin-1 antibodies is mediated by neutralization of E2-CD81-claudin-1 associations. Hepatology 51: 1144–1157. 10.1002/hep.23445 [DOI] [PubMed] [Google Scholar]
- Kurzawski M, Dziedziejko V, Urasińska E, Post M, Wójcicki M, Miętkiewski J, Droździk M. 2012. Nuclear factor erythroid 2-like 2 (Nrf2) expression in end-stage liver disease. Environ Toxicol Pharmacol 34: 87–95. 10.1016/j.etap.2012.03.001 [DOI] [PubMed] [Google Scholar]
- Lam AP, Herazo-Maya JD, Sennello JA, Flozak AS, Russell S, Mutlu GM, Budinger GR, DasGupta R, Varga J, Kaminski N, et al. 2014. Wnt coreceptor Lrp5 is a driver of idiopathic pulmonary fibrosis. Am J Respir Crit Care Med 190: 185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laudisi F, Cherubini F, Monteleone G, Stolfi C. 2018. STAT3 interactors as potential therapeutic targets for cancer treatment. Int J Mol Sci 19: 1797 10.3390/ijms19061787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leandro G, Mangia A, Hui J, Fabris P, Rubbia-Brandt L, Colloredo G, Adinolfi LE, Asselah T, Jonsson JR, Smedile A, et al. 2006. Relationship between steatosis, inflammation, and fibrosis in chronic hepatitis C: a meta-analysis of individual patient data. Gastroenterology 130: 1636–1642. 10.1053/j.gastro.2006.03.014 [DOI] [PubMed] [Google Scholar]
- Levent G, Ali A, Ahmet A, Polat EC, Aytaç C, Ayşe E, Ahmet S. 2006. Oxidative stress and antioxidant defense in patients with chronic hepatitis C patients before and after pegylated interferon α-2b plus ribavirin therapy. J Transl Med 4 10.1186/1479-5876-4-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lévy PL, Duponchel S, Eischeid H, Molle J, Michelet M, Diserens G, Vermathen M, Vermathen P, Dufour JF, Dienes HP, et al. 2017. Hepatitis C virus infection triggers a tumor-like glutamine metabolism. Hepatology 65: 789–803. 10.1002/hep.28949 [DOI] [PubMed] [Google Scholar]
- Lim TR, Hazlehurst JM, Oprescu AI, Armstrong MJ, Abdullah SF, Davies NP, Flintham R, Balfe P, Mutimer DJ, McKeating JA, et al. 2019. Hepatitis C virus infection is associated with hepatic and adipose tissue insulin resistance that improves after viral cure. Clin Endocrinol (Oxf) 90: 440–448. 10.1111/cen.13924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Tsai WL, Shao RX, Wu G, Peng LF, Barlow LL, Chung WJ, Zhang L, Zhao H, Jang JY, et al. 2010. Hepatitis C virus regulates transforming growth factor β1 production through the generation of reactive oxygen species in a nuclear factor κB-dependent manner. Gastroenterology 138: 2509–2518.e1. 2518 e2501 10.1053/j.gastro.2010.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. 2002. Control of β-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell 108: 837–847. 10.1016/S0092-8674(02)00685-2 [DOI] [PubMed] [Google Scholar]
- Llovet JM, Pena CE, Lathia CD, Shan M, Meinhardt G, Bruix J, Group SIS. 2012. Plasma biomarkers as predictors of outcome in patients with advanced hepatocellular carcinoma. Clin Cancer Res 18: 2290–2300. 10.1158/1078-0432.CCR-11-2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonardo A, Loria P, Adinolfi LE, Carulli N, Ruggiero G. 2006. Hepatitis C and steatosis: a reappraisal. J Viral Hepat 13: 73–80. 10.1111/j.1365-2893.2005.00669.x [DOI] [PubMed] [Google Scholar]
- Luangmonkong T, Suriguga S, Mutsaers HAM, Groothuis GMM, Olinga P, Boersema M. 2018. Targeting oxidative stress for the treatment of liver fibrosis. Rev Physiol Biochem Pharmacol 175: 71–102. 10.1007/112_2018_10 [DOI] [PubMed] [Google Scholar]
- Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, et al. 2011. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med 17: 589–595. 10.1038/nm.2341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupberger J, Duong FH, Fofana I, Zona L, Xiao F, Thumann C, Durand SC, Pessaux P, Zeisel MB, Heim MH, et al. 2013. Epidermal growth factor receptor signaling impairs the antiviral activity of interferon-α. Hepatology 58: 1225–1235. 10.1002/hep.26404 [DOI] [PubMed] [Google Scholar]
- Lupberger J, Croonenborghs T, Roca Suarez AA, Van Renne N, Juhling F, Oudot MA, Virzi A, Bandiera S, Jamey C, Meszaros G, et al. 2019. Combined analysis of metabolomes, proteomes, and transcriptomes of hepatitis C virus-infected cells and liver to identify pathways associated with disease development. Gastroenterology 10.1053/j.gastro.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado MV, Diehl AM. 2018. Hedgehog signalling in liver pathophysiology. J Hepatol 68: 550–562. 10.1016/j.jhep.2017.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machida K, Cheng KT, Lai CK, Jeng KS, Sung VM, Lai MM. 2006. Hepatitis C virus triggers mitochondrial permeability transition with production of reactive oxygen species, leading to DNA damage and STAT3 activation. J Virol 80: 7199–7207. 10.1128/JVI.00321-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailly L, Xiao F, Lupberger J, Wilson GK, Aubert P, Duong FHT, Calabrese D, Leboeuf C, Fofana I, Thumann C, et al. 2015. Clearance of persistent hepatitis C virus infection in humanized mice using a claudin-1-targeting monoclonal antibody. Nat Biotechnol 33: 549–554. 10.1038/nbt.3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankouri J, Griffin S, Harris M. 2008. The hepatitis C virus non-structural protein NS5A alters the trafficking profile of the epidermal growth factor receptor. Traffic 9: 1497–1509. 10.1111/j.1600-0854.2008.00779.x [DOI] [PubMed] [Google Scholar]
- Mann B, Gelos M, Siedow A, Hanski ML, Gratchev A, Ilyas M, Bodmer WF, Moyer MP, Riecken EO, Buhr HJ, et al. 1999. Target genes of β-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc Natl Acad Sci 96: 1603–1608. 10.1073/pnas.96.4.1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney EM, Helbig KJ, Narayana SK, Eyre NS, Aloia AL, Beard MR. 2013. Signal transducer and activator of transcription 3 is a proviral host factor for hepatitis C virus. Hepatology 58: 1558–1568. 10.1002/hep.26496 [DOI] [PubMed] [Google Scholar]
- Medvedev R, Ploen D, Spengler C, Elgner F, Ren H, Bunten S, Hildt E. 2017. HCV-induced oxidative stress by inhibition of Nrf2 triggers autophagy and favors release of viral particles. Free Radic Biol Med 110: 300–315. 10.1016/j.freeradbiomed.2017.06.021 [DOI] [PubMed] [Google Scholar]
- Meydan N, Grunberger T, Dadi H, Shahar M, Arpaia E, Lapidot Z, Leeder JS, Freedman M, Cohen A, Gazit A, et al. 1996. Inhibition of acute lymphoblastic leukaemia by a Jak-2 inhibitor. Nature 379: 645–648. 10.1038/379645a0 [DOI] [PubMed] [Google Scholar]
- Michalopoulos GK. 2007. Liver regeneration. J Cell Physiol 213: 286–300. 10.1002/jcp.21172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat Cell Biol 9: 1089–1097. 10.1038/ncb1631 [DOI] [PubMed] [Google Scholar]
- Miyazawa K, Shinozaki M, Hara T, Furuya T, Miyazono K. 2002. Two major Smad pathways in TGF-β superfamily signalling. Genes Cells 7: 1191–1204. 10.1046/j.1365-2443.2002.00599.x [DOI] [PubMed] [Google Scholar]
- Moon H, Cho K, Shin S, Kim DY, Han KH, Ro SW. 2019. High risk of hepatocellular carcinoma development in fibrotic liver: role of the Hippo-YAP/TAZ signaling pathway. Int J Mol Sci 20: 581 10.3390/ijms20030581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya K, Yotsuyanagi H, Shintani Y, Fujie H, Ishibashi K, Matsuura Y, Miyamura T, Koike K. 1997. Hepatitis C virus core protein induces hepatic steatosis in transgenic mice. J Gen Virol 78: 1527–1531. 10.1099/0022-1317-78-7-1527 [DOI] [PubMed] [Google Scholar]
- Mukozu T, Nagai H, Matsui D, Kanekawa T, Sumino Y. 2013. Serum VEGF as a tumor marker in patients with HCV-related liver cirrhosis and hepatocellular carcinoma. Anticancer Res 33: 1013–1021. [PubMed] [Google Scholar]
- Negash AA, Olson RM, Griffin S, Gale M Jr. 2019. Modulation of calcium signaling pathway by hepatitis C virus core protein stimulates NLRP3 inflammasome activation. PLoS Pathog 15: e1007593 10.1371/journal.ppat.1007593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Gonzalez-Peralta RP, Qian K, Xu Y, Marousis CG, Davis GL, Lau JY. 1997. Transforming growth factor-β1 in chronic hepatitis C. J Viral Hepat 4: 29–35. 10.1046/j.1365-2893.1997.00124.x [DOI] [PubMed] [Google Scholar]
- Nguyen TTT, Park EM, Lim YS, Hwang SB. 2018. Nonstructural protein 5A impairs DNA damage repair: implications for hepatitis C virus-mediated hepatocarcinogenesis. J Virol 92: e00178-18 10.1128/JVI.00178-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunez Lopez O, Bohanon FJ, Wang X, Ye N, Corsello T, Rojas-Khalil Y, Chen H, Chen H, Zhou J, Radhakrishnan RS. 2016. STAT3 inhibition suppresses hepatic stellate cell fibrogenesis: HJC0123, a potential therapeutic agent for liver fibrosis. RSC Adv 6: 100652–100663. 10.1039/C6RA17459K [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberhammer FA, Pavelka M, Sharma S, Tiefenbacher R, Purchio AF, Bursch W, Schulte-Hermann R. 1992. Induction of apoptosis in cultured hepatocytes and in regressing liver by transforming growth factor β1. Proc Natl Acad Sci 89: 5408–5412. 10.1073/pnas.89.12.5408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okusaka T, Ueno H, Ikeda M, Mitsunaga S, Ozaka M, Ishii H, Yokosuka O, Ooka Y, Yoshimoto R, Yanagihara Y, et al. 2015. Phase 1 and pharmacological trial of OPB-31121, a signal transducer and activator of transcription-3 inhibitor, in patients with advanced hepatocellular carcinoma. Hepatol Res 45: 1283–1291. 10.1111/hepr.12504 [DOI] [PubMed] [Google Scholar]
- Omenetti A, Popov Y, Jung Y, Choi SS, Witek RP, Yang L, Brown KD, Schuppan D, Diehl AM. 2008. The Hedgehog pathway regulates remodelling responses to biliary obstruction in rats. Gut 57: 1275–1282. 10.1136/gut.2008.148619 [DOI] [PubMed] [Google Scholar]
- Osawa Y, Oboki K, Imamura J, Kojika E, Hayashi Y, Hishima T, Saibara T, Shibasaki F, Kohara M, Kimura K. 2015. Inhibition of cyclic adenosine monophosphate (cAMP)-response element-binding protein (CREB)-binding protein (CBP)/β-catenin reduces liver fibrosis in mice. EBioMedicine 2: 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Polyak SJ, Bano N, Qiu WC, Carithers RL, Shuhart M, Gretch DR, Das A. 2010. Hepatitis C virus induces oxidative stress, DNA damage and modulates the DNA repair enzyme NEIL1. J Gastroenterol Hepatol 25: 627–634. 10.1111/j.1440-1746.2009.06128.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Choi SH, Kang SM, Kang JI, Ahn BY, Kim H, Jung G, Choi KY, Hwang SB. 2009. Nonstructural 5A protein activates β-catenin signaling cascades: implication of hepatitis C virus-induced liver pathogenesis. J Hepatol 51: 853–864. 10.1016/j.jhep.2009.06.026 [DOI] [PubMed] [Google Scholar]
- Paternostro C, David E, Novo E, Parola M. 2010. Hypoxia, angiogenesis and liver fibrogenesis in the progression of chronic liver diseases. World J Gastroenterol 16: 281–288. 10.3748/wjg.v16.i3.281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavio N, Battaglia S, Boucreux D, Arnulf B, Sobesky R, Hermine O, Brechot C. 2005. Hepatitis C virus core variants isolated from liver tumor but not from adjacent non-tumor tissue interact with Smad3 and inhibit the TGF-β pathway. Oncogene 24: 6119–6132. 10.1038/sj.onc.1208749 [DOI] [PubMed] [Google Scholar]
- Perez S, Gal-Tanamy M. 2019. Studying the hepatitis C virus-induced epigenetic signature after cure with direct-acting antivirals. Methods Mol Biol 1911: 191–207. 10.1007/978-1-4939-8976-8_13 [DOI] [PubMed] [Google Scholar]
- Petrick JL, Kelly SP, Altekruse SF, McGlynn KA, Rosenberg PS. 2016. Future of hepatocellular carcinoma incidence in the United States forecast through 2030. J Clin Oncol 34: 1787–1794. 10.1200/JCO.2015.64.7412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyrou M, Clément S, Maier C, Bourgoin L, Branche E, Conzelmann S, Kaddai V, Foti M, Negro F. 2013. PTEN protein phosphatase activity regulates hepatitis C virus secretion through modulation of cholesterol metabolism. J Hepatol 59: 420–426. 10.1016/j.jhep.2013.04.012 [DOI] [PubMed] [Google Scholar]
- Plimack ER, Lorusso PM, McCoon P, Tang W, Krebs AD, Curt G, Eckhardt SG. 2013. AZD1480: a phase I study of a novel JAK2 inhibitor in solid tumors. Oncologist 18: 819–820. 10.1634/theoncologist.2013-0198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plissonnier ML, Lahlali T, Michelet M, Lebossé F, Cottarel J, Beer M, Neveu G, Durantel D, Bartosch B, Accardi R, et al. 2016. Epidermal growth factor receptor-dependent mutual amplification between netrin-1 and the hepatitis C virus. PLoS Biol 14: e1002421 10.1371/journal.pbio.1002421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polaris Observatory HCV Collaborators. 2017. Global prevalence and genotype distribution of hepatitis C virus infection in 2015: a modelling study. Lancet Gastroenterol Hepatol 2: 161–176. 10.1016/S2468-1253(16)30181-9 [DOI] [PubMed] [Google Scholar]
- Rani B, Malfettone A, Dituri F, Soukupova J, Lupo L, Mancarella S, Fabregat I, Giannelli G. 2018. Galunisertib suppresses the staminal phenotype in hepatocellular carcinoma by modulating CD44 expression. Cell Death Dis 9: 373 10.1038/s41419-018-0384-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Broor SL, Vaishnav Y, Sarkar C, Girish R, Dar L, Seth P, Broor S. 2003. Transforming growth factor β in hepatitis C virus infection: in vivo and in vitro findings. J Gastroenterol Hepatol 18: 393–403. 10.1046/j.1440-1746.2003.02985.x [DOI] [PubMed] [Google Scholar]
- Reya T, Clevers H. 2005. Wnt signalling in stem cells and cancer. Nature 434: 843–850. 10.1038/nature03319 [DOI] [PubMed] [Google Scholar]
- Roca Suarez AA, Baumert TF, Lupberger J. 2018. Beyond viral dependence: the pathological consequences of HCV-induced EGF signaling. J Hepatol 69: 564–566. 10.1016/j.jhep.2018.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen HR. 2013. Emerging concepts in immunity to hepatitis C virus infection. J Clin Invest 123: 4121–4130. 10.1172/JCI67714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbia-Brandt L, Giostra E, Mentha G, Quadri R, Negro F. 2001. Expression of liver steatosis in hepatitis C virus infection and pattern of response to α-interferon. J Hepatol 35: 307 10.1016/S0168-8278(01)00087-3 [DOI] [PubMed] [Google Scholar]
- Sanderson N, Factor V, Nagy P, Kopp J, Kondaiah P, Wakefield L, Roberts AB, Sporn MB, Thorgeirsson SS. 1995. Hepatic expression of mature transforming growth factor β1 in transgenic mice results in multiple tissue lesions. Proc Natl Acad Sci 92: 2572–2576. 10.1073/pnas.92.7.2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, et al. 2015. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 47: 505–511. 10.1038/ng.3252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao YY, Hsieh MS, Wang HY, Li YS, Lin H, Hsu HW, Huang CY, Hsu CH, Cheng AL. 2017. Hepatitis C virus core protein potentiates proangiogenic activity of hepatocellular carcinoma cells. Oncotarget 8: 86681–86692. 10.18632/oncotarget.21407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu K, Soroida Y, Sato M, Hikita H, Kobayashi T, Endo M, Sato M, Gotoh H, Iwai T, Tateishi R, et al. 2018. Eradication of hepatitis C virus is associated with the attenuation of steatosis as evaluated using a controlled attenuation parameter. Sci Rep 8: 7845 10.1038/s41598-018-26293-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda K, Mori M, Shibuta K, Banner BF, Barnard GF. 1999. Vascular endothelial growth factor/vascular permeability factor mRNA expression in patients with chronic hepatitis C and hepatocellular carcinoma. Int J Oncol 14: 353–359. 10.3892/ijo.14.2.353 [DOI] [PubMed] [Google Scholar]
- Shintani Y, Fujie H, Miyoshi H, Tsutsumi T, Tsukamoto K, Kimura S, Moriya K, Koike K. 2004. Hepatitis C virus infection and diabetes: direct involvement of the virus in the development of insulin resistance. Gastroenterology 126: 840–848. 10.1053/j.gastro.2003.11.056 [DOI] [PubMed] [Google Scholar]
- Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, Yip ML, Jove R, McLaughlin MM, Lawrence NJ, et al. 2007. Selective chemical probe inhibitor of Stat3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci 104: 7391–7396. 10.1073/pnas.0609757104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert W, Razavi H, Estes C, Thompson AJ, Zekry A, Roberts SK, Dore GJ. 2014. Enhanced antiviral treatment efficacy and uptake in preventing the rising burden of hepatitis C-related liver disease and costs in Australia. J Gastroenterol Hepatol 29(Suppl. 1): 1–9. 10.1111/jgh.12677 [DOI] [PubMed] [Google Scholar]
- Stanford SM, Rapini N, Bottini N. 2012. Regulation of TCR signalling by tyrosine phosphatases: from immune homeostasis to autoimmunity. Immunology 137: 1–19. 10.1111/j.1365-2567.2012.03591.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepan V, Ramamoorthy S, Nitsche H, Zavros Y, Merchant JL, Todisco A. 2005. Regulation and function of the sonic hedgehog signal transduction pathway in isolated gastric parietal cells. J Biol Chem 280: 15700–15708. 10.1074/jbc.M413037200 [DOI] [PubMed] [Google Scholar]
- Street A, Macdonald A, McCormick C, Harris M. 2005. Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular β-catenin and stimulation of β-catenin-responsive transcription. J Virol 79: 5006–5016. 10.1128/JVI.79.8.5006-5016.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed GH, Amako Y, Siddiqui A. 2010. Hepatitis C virus hijacks host lipid metabolism. Trends Endocrinol Metab 21: 33–40. 10.1016/j.tem.2009.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi H, Kato N, Otsuka M, Goto T, Yoshida H, Shiratori Y, Omata M. 2004. Hepatitis C virus core protein upregulates transforming growth factor-β1 transcription. J Med Virol 72: 52–59. 10.1002/jmv.10545 [DOI] [PubMed] [Google Scholar]
- Tawfik AK, Amin AM, Yousef M, El-Sayd NM, Elashry H, Elkadeem M, Abd-Elsalam S. 2018. IL-1α correlates with severity of hepatitis C virus-related liver diseases. J Inflamm Res 11: 289–295. 10.2147/JIR.S166564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. 1999. β-Catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 398: 422–426. 10.1038/18884 [DOI] [PubMed] [Google Scholar]
- Thiery JP, Sleeman JP. 2006. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 7: 131–142. 10.1038/nrm1835 [DOI] [PubMed] [Google Scholar]
- Thrift AP, El-Serag HB, Kanwal F. 2017. Global epidemiology and burden of HCV infection and HCV-related disease. Nat Rev Gastroenterol Hepatol 14: 122–132. 10.1038/nrgastro.2016.176 [DOI] [PubMed] [Google Scholar]
- Tsushima H, Kawata S, Tamura S, Ito N, Shirai Y, Kiso S, Doi Y, Yamada A, Oshikawa O, Matsuzawa Y. 1999. Reduced plasma transforming growth factor-β1 levels in patients with chronic hepatitis C after interferon-α therapy: association with regression of hepatic fibrosis. J Hepatol 30: 1–7. 10.1016/S0168-8278(99)80001-4 [DOI] [PubMed] [Google Scholar]
- Van Renne N, Roca Suarez AA, Duong FHT, Gondeau C, Calabrese D, Fontaine N, Ababsa A, Bandiera S, Croonenborghs T, Pochet N, et al. 2018. miR-135a-5p-mediated downregulation of protein tyrosine phosphatase receptor delta is a candidate driver of HCV-associated hepatocarcinogenesis. Gut 67: 953–962. 10.1136/gutjnl-2016-312270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilgrain V, Ronot M, Abdel-Rehim M, Zappa M, d'Assignies G, Bruno O, Vullierme MP. 2013. Hepatic steatosis: a major trap in liver imaging. Diagn Interv Imaging 94: 713–727. 10.1016/j.diii.2013.03.010 [DOI] [PubMed] [Google Scholar]
- Villanueva A, Hoshida Y, Battiston C, Tovar V, Sia D, Alsinet C, Cornella H, Liberzon A, Kobayashi M, Kumada H, et al. 2011. Combining clinical, pathology, and gene expression data to predict recurrence of hepatocellular carcinoma. Gastroenterology 140: 1501–1512.e2. 10.1053/j.gastro.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JN, Li L, Li LY, Yan Q, Li J, Xu T. 2018a. Emerging role and therapeutic implication of Wnt signaling pathways in liver fibrosis. Gene 674: 57–69. 10.1016/j.gene.2018.06.053 [DOI] [PubMed] [Google Scholar]
- Wang Z, Li J, Xiao W, Long J, Zhang H. 2018b. The STAT3 inhibitor S3I-201 suppresses fibrogenesis and angiogenesis in liver fibrosis. Lab Invest 98: 1600–1613. 10.1038/s41374-018-0127-3 [DOI] [PubMed] [Google Scholar]
- Wijetunga NA, Pascual M, Tozour J, Delahaye F, Alani M, Adeyeye M, Wolkoff AW, Verma A, Greally JM. 2017. A pre-neoplastic epigenetic field defect in HCV-infected liver at transcription factor binding sites and polycomb targets. Oncogene 36: 2030–2044. 10.1038/onc.2016.340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zheng L, Yuan Q, Zhen G, Crane JL, Zhou X, Cao X. 2018. Transforming growth factor-β in stem cells and tissue homeostasis. Bone Res 6: 2 10.1038/s41413-017-0005-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B, Ge Y, Perens G, Hong L, Xu H, Fishbein MC, Li F. 2013. Canonical Wnt/β-catenin signaling in epicardial fibrosis of failed pediatric heart allografts with diastolic dysfunction. Cardiovasc Pathol 22: 54–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C, Kang J, Lim HY, Kim JH, Lee MA, Lee KH, Kim TY, Ryoo BY. 2019. Phase I dose-finding study of OPB-111077, a novel STAT3 inhibitor, in patients with advanced hepatocellular carcinoma. Cancer Res Treat 51: 510–518. 10.4143/crt.2018.226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Hanada T, Tokuhisa T, Kosai K, Sata M, Kohara M, Yoshimura A. 2002. Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J Exp Med 196: 641–653. 10.1084/jem.20012127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiji H, Kuriyama S, Yoshii J, Ikenaka Y, Noguchi R, Hicklin DJ, Wu Y, Yanase K, Namisaki T, Yamazaki M, et al. 2003. Vascular endothelial growth factor and receptor interaction is a prerequisite for murine hepatic fibrogenesis. Gut 52: 1347–1354. 10.1136/gut.52.9.1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Hebert MC, Zhang YE. 2002. TGF-β receptor-activated p38 MAP kinase mediates Smad-independent TGF-β responses. EMBO J 21: 3749–3759. 10.1093/emboj/cdf366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Lee H, Herrmann A, Buettner R, Jove R. 2014. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer 14: 736–746. 10.1038/nrc3818 [DOI] [PubMed] [Google Scholar]
- Zhang W, Liu HT. 2002. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res 12: 9–18. 10.1038/sj.cr.7290105 [DOI] [PubMed] [Google Scholar]
- Zhang J, Wang X, Vikash V, Ye Q, Wu D, Liu Y, Dong W. 2016. ROS and ROS-mediated cellular signaling. Oxid Med Cell Longev 2016: 4350965 10.1155/2016/4350965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Liu X, Wang S, Yan X, Tang Z, Wu K, Li Y, Liu F. 2014. Hepatitis C virus core protein induces hypoxia-inducible factor 1α-mediated vascular endothelial growth factor expression in Huh7.5.1 cells. Mol Med Rep 9: 2010–2014. 10.3892/mmr.2014.2039 [DOI] [PubMed] [Google Scholar]
- Zhu AX, Kang YK, Yen CJ, Finn RS, Galle PR, Llovet JM, Assenat E, Brandi G, Pracht M, Lim HY, et al. 2019. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased α-fetoprotein concentrations (REACH-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol 20: 282–296. 10.1016/S1470-2045(18)30937-9 [DOI] [PubMed] [Google Scholar]
- Zona L, Lupberger J, Sidahmed-Adrar N, Thumann C, Harris HJ, Barnes A, Florentin J, Tawar RG, Xiao F, Turek M, et al. 2013. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe 13: 302–313. 10.1016/j.chom.2013.02.006 [DOI] [PubMed] [Google Scholar]