In this study, Koo et al. set out to identify the key downstream target genes of Hippo pathway effectors YAP/TAZ involved in organ size control. Using a battery of genetic experiments in cultured cells and in vivo assays on liver growth the authors identified that YAP/TAZ are directly involved in immediate early gene induction, including AP-1; and this AP-1 induction by YAP is essential for the functions of YAP in organ size control and cancer cell growth.
Keywords: Hippo, c-fos, liver, bilirubin, hepatomegaly, cancer
Abstract
Yes-associated protein (YAP) and its homolog transcriptional coactivator with PDZ-binding motif (TAZ) are key effectors of the Hippo pathway to control cell growth and organ size, of which dysregulation yields to tumorigenesis or hypertrophy. Upon activation, YAP/TAZ translocate into the nucleus and bind to TEAD transcription factors to promote transcriptional programs for proliferation or cell specification. Immediate early genes, represented by AP-1 complex, are rapidly induced and control later-phase transcriptional program to play key roles in tumorigenesis and organ maintenance. Here, we report that YAP/TAZ directly promote FOS transcription that in turn contributes to the biological function of YAP/TAZ. YAP/TAZ bind to the promoter region of FOS to stimulate its transcription. Deletion of YAP/TAZ blocks the induction of immediate early genes in response to mitogenic stimuli. FOS induction contributes to expression of YAP/TAZ downstream target genes. Genetic deletion or chemical inhibition of AP-1 suppresses growth of YAP-driven cancer cells, such as Lats1/2-deficient cancer cells as well as Gαq/11 mutated uveal melanoma. Furthermore, AP-1 inhibition almost completely abrogates the hepatomegaly induced by YAP overexpression. Our findings reveal a feed-forward interplay between immediate early transcription of AP-1 and Hippo pathway function.
Gene transcription is a fundamental process for cells to change its functional machinery for homeostatic regulation including cell and organ growth. In response to many external signals such as serum, lysophosphatidic acid (LPA), growth factors, developmental cues, phorbol esters, and cellular stress, the very first group of genes, known as “immediate early genes,” are rapidly induced (Herschman 1991; Iyer et al. 1999). But the expressions of many immediately early genes are very transient and last for only a short period of time even in the continuous presence of the stimuli. These genes are particularly important in the overall transcription program because they often participate in the next wave of gene transcription. Indeed, many immediate early gene products are transcription factors or partners of DNA-binding proteins, and play important roles in the temporal regulation of gene induction in response to external stimuli.
One of the best-characterized immediate early gene products is AP-1 transcription factor, which is composed of Fos family proteins (FOS, FOSB, and FRA1 and FRA2) dimerized with Jun family proteins (JUN, JUNB, and JUND) (Eferl and Wagner 2003). As a heterodimer, AP-1 binds to the promoter region of specific target genes, converting extracellular signals into changes in gene expression. Since Fos family proteins are barely expressed at basal state and only FOS and FOSB have transcriptional activation domain, AP-1 activity is determined by de novo transcription of FOS and FOSB (Foletta et al. 1994; Bergers et al. 1995; Eferl and Wagner 2003). Previous studies have shown that FOS induction is one of the most critical events in cellular processes such as proliferation, differentiation, and survival (Vaquerizas et al. 2009). Moreover, studies have revealed that FOS is involved in tumorigenesis in most types of cancers, including uveal melanoma and hepatocellular carcinoma (Liu et al. 2002; Mallikarjuna et al. 2006). Recently it has been also shown that FOS may play a key role in organ size regulation (Bakiri et al. 2017). Ectopic expression of FOS in hepatocytes led to dramatic enlargement of the liver in mice, due to uncontrolled cell growth. While induction of FOS is known to be driven by several transcription factors, SRF has been regarded as the dominant transcription factor to induce FOS and other immediate early genes in response to serum or serum containing factors (Graham and Gilman 1991). However, the role of other serum-induced transcription machinery, such as the recently characterized YAP of the Hippo pathway, in AP-1 induction has not been investigated.
The Hippo pathway has emerged as a central regulator of cell proliferation and tissue homeostasis (Piccolo et al. 2014; Moroishi et al. 2015a; Yu et al. 2015). Core kinase cascade of the Hippo pathway consists of MST1/2, MAP4Ks, and LATS1/2. The Hippo pathway functions to suppress the activity of YAP and TAZ, two transcriptional coactivators as the main functional effectors of the Hippo pathway. When the Hippo pathway is active, MST1/2 and MAP4Ks activate LATS1/2 by phosphorylating their hydrophobic motifs, and LATS kinases then repress YAP/TAZ through phosphorylation on multiple residues. Constitutive inhibition of the Hippo pathway is reported as a driving force in many cancers (Moroishi et al. 2015a). For instance, in uveal melanoma more than 90% of cancers carry activating mutations in either GNAQ or GNA11, which acts by inhibiting the Hippo pathway (Van Raamsdonk et al. 2009, 2010; Yu et al. 2014; Robertson et al. 2017). Similarly, mutations of NF2, which activates the Hippo pathway, are frequently observed in mesothelioma and schwannoma (Murakami et al. 2011; Li et al. 2014). However, the most remarkable and distinct role of Hippo pathway is to limit organ size. Liver mass is tightly regulated to a set point of ∼3% body weight (∼5% in mice) (Michalopoulos and DeFrances 1997). Even after surgical ablation up to 70% in human or mice, the remaining hepatocytes undergo rapid growth, bringing back the organ to its original mass in days. Notably, hepatic overexpression of mutant YAP that is not repressed by the Hippo pathway induces up to five-fold increase in mouse liver mass, due to proliferation of mature hepatocytes (Camargo et al. 2007; Dong et al. 2007). Consistently, deletion of Hippo pathway components Mst1/2, Sav1, or Nf2 also causes liver overgrowth (Zhou et al. 2009; Benhamouche et al. 2010; Lee et al. 2010; Lu et al. 2010; Song et al. 2010; Zhang et al. 2010). Despite these observations, the underlying mechanism underpinning how Hippo pathway controls cell growth, and organ size remains enigmatic.
In response to mitogenic signals, the Hippo pathway is inhibited and YAP/TAZ are released from repression. The active YAP/TAZ translocate into the nucleus to bind TEAD family transcription factors (Zhao et al. 2008). YAP/TAZ–TEAD complex stimulates expression of target genes, such as CTGF, CYR61, AMOTL2, and ANKRD1 (Yu et al. 2015). Although TEAD binding seems to be the most important in YAP/TAZ target gene induction, YAP/TAZ–TEAD complex can further cooperate with other DNA-binding partners (Totaro et al. 2018). One of such factors is AP-1 (Zanconato et al. 2015; Liu et al. 2016). In breast cancer cells, a significant portion of YAP/TAZ-TEAD binding sites are co-occupied with AP-1. AP-1 has been shown to synergize with YAP/TAZ and TEAD to promote mammosphere formation and tumor xenograft growth. It is noteworthy that YAP/TAZ are dephosphorylated by the same upstream signals that also induce AP-1 expression (Yu et al. 2015). Given that YAP/TAZ nuclear localization occurs earlier than FOS induction upon serum or LPA treatment, we speculated that YAP/TAZ may participate in AP-1 regulation.
In this study, we show that AP-1 induction requires the presence of YAP/TAZ with TEAD binding and that AP-1 assembly itself contributes to the functions of YAP, constituting a feed-forward machinery. We discovered that deletion of YAP/TAZ blocks transcription of immediate early genes including AP-1 components. Mechanistically, YAP/TAZ-TEAD complex acts as a direct transcriptional regulator for FOS. In addition, FOS induction contributes to YAP/TAZ-mediated target gene transcription and oncogenic cell growth. Moreover, AP-1 induction plays a key role in the physiological functions of YAP/TAZ in supporting uveal melanoma growth and liver size regulation. Our study uncovers a functional interplay between immediate early gene transcription and Hippo biology.
Results
YAP/TAZ in regulating immediate early genes in response to mitogenic signals
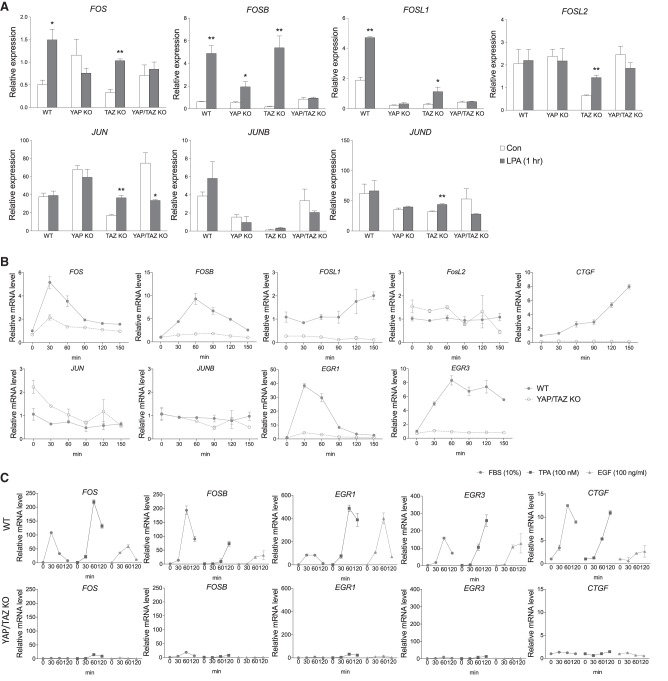
AP-1 has been reported to cooperate with YAP and TEAD in gene expression (Zanconato et al. 2015; Liu et al. 2016). Mitogens, such as serum or LPA are known to activate YAP as well as induce AP-1 expression (Yu et al. 2012). We tested whether YAP/TAZ may play a role in induction of Fos family proteins, which are subunits of AP-1. Cell lines with gene knockout for YAP, TAZ, or both were described previously (Hansen et al. 2015). Since one of the most robust signals to induce AP-1 is LPA, RNA-seq was performed using these cells after short treatment of LPA for 1 h (Plouffe et al. 2018). Among genes comprising AP-1, Fos family are immediately early genes that are rapidly induced transcriptionally, whereas Jun family genes are regulated by phosphorylation in general. As expected, HEK293 cells showed acute induction of FOS, FOSB, and FOSL1 upon LPA treatment. Interestingly, this effect was blunted in YAP knockout cells and was further blocked by additional deletion of TAZ (Fig. 1A). The effect of TAZ single knockout was not as strong as YAP knockout. It has been previously shown that in spite of their functional redundancy, YAP has a stronger influence than TAZ, probably likely due to the higher expression of YAP in HEK293 (Plouffe et al. 2018). Therefore, it can be assumed that YAP and TAZ both play a shared role in early induction of AP-1.
Figure 1.
AP-1 induction requires YAP/TAZ in response to various inputs. (A) Deletion of YAP and TAZ blocks LPA-induced FOS expression. RNA-sequencing results from wild-type and YAP, TAZ, or YAP/TAZ double-knockout HEK293 cells were analyzed for expression of AP-1 components. Cells were serum starved overnight, followed, and treated with LPA for 1 h. (*) P < 0.05; (**) P < 0.01, compared with respective controls, by Student's t-test. (B) Knockout of YAP/TAZ represses transient induction of AP-1. Time-course expression of each immediate early gene in wild-type or YAP/TAZ knockout HEK293 cells were determined by qPCR. LPA was used to stimulate cells for indicated time points. (C) YAP/TAZ are commonly required for induction of immediate early genes by various stimuli. qPCR was done on wild-type and YAP/TAZ knockout cells, which were serum-starved overnight and treated with 10% serum, 100 nM TPA, or 100 ng/mL EGF.
SRF (serum response factor) is the most-studied transcription factor responsible for the induction of immediate early genes, including Fos family (Graham and Gilman 1991). Downstream from mitogenic stimuli the MRTF-A transcription cofactor translocates into the nucleus to form heterocomplex with SRF, which then induces immediate early genes. MRTF is also rapidly regulated by mitogens in a mechanism dependent on G-actin, which binds to and retains MRTF in cytoplasm (Miralles et al. 2003). Mitogen stimulation, such as LPA, induces G-actin polymerization to F-actin, resulting in depletion of G-actin. As a result, MRTF is relieved from inhibition by G-actin (Supplemental Fig. S1A). To confirm the role of SRF in FOS induction, we generated SRF knockout by CRISPR/CAS9. Indeed, SRF knockout strongly blocked the induction of immediate early genes, including FOS (Supplemental Fig. S1B). Notably, deletion of SRF also repressed induction of typical YAP target gene CTGF. This may have resulted from repression of either SRF activity or immediate early gene induction. As SRF is required for FOS induction, our observation is consistent with previous studies that AP-1 plays an important role in expression of YAP target genes.
With a great similarity to MRTF-SRF pathway, nuclear localization of YAP/TAZ is also triggered by the common upstream signals, resulting transcription of their downstream target genes such as CTGF and CYR61. However, previous studies have only focused on SRF on immediate early gene induction, probably because the precise regulatory mechanism of YAP/TAZ was revealed relatively recently. As shown in RNA-seq, qPCR assays confirmed that LPA-induced induction of FOS and FOSB were blunted in the YAP/TAZ KO cells (Fig. 1B). It is noteworthy that without YAP/TAZ, cells failed to induce AP-1 even in the presence of classical regulator SRF. In addition, transcription of other immediate early genes such as EGR1 and EGR3, were also inhibited, implying a general necessity of YAP/TAZ in immediate early gene induction. Other major signals to induce AP-1 include serum, 12-O-Tetradecanoylphorbol 13-acetate (TPA), and epidermal growth factor (EGF). It has been shown that these signals also activate YAP/TAZ (Yu et al. 2015). Similarly, these stimuli commonly required the presence of YAP or TAZ to induce immediate early gene expression (Fig. 1C), indicating a general role of YAP/TAZ in immediate early gene expression. These results reveal a previously unappreciated role of YAP/TAZ in the induction of immediate early genes in response to extracellular stimuli.
Direct association of YAP–TEAD complex on FOS gene promoter for transcriptional activation
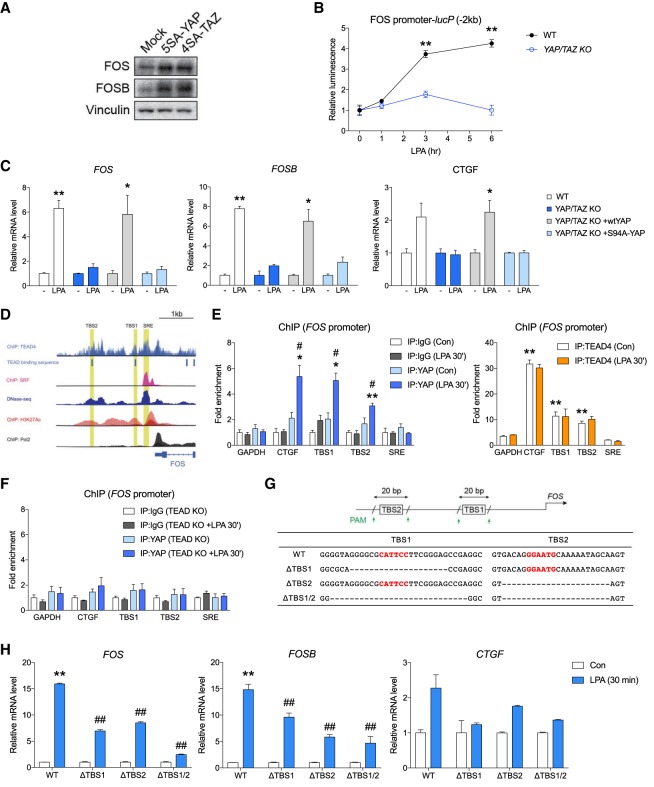
YAP/TAZ activities are tightly controlled by phosphorylation on multiple sites (Zhao et al. 2010). When cells are cultured without serum, LATS1/2 kinases maintain YAP/TAZ in hyperphosphorylated state to sequester them in the cytosol, thereby inhibiting transcriptional activity. To determine whether YAP/TAZ directly affect the expression level of AP-1, we generated HEK293 cells stably overexpressing 5SA-YAP or 4SA-TAZ with all of the LATS1/2 phosphorylation sites mutated to alanine, thereby constitutively active and unresponsive to inhibition by LATS1/2. Although cells were serum-starved for overnight, those expressing active YAP or TAZ displayed higher basal expression of FOS and FOSB (Fig. 2A), indicating that YAP/TAZ activation is sufficient to induce these genes. We next investigated whether YAP/TAZ are required for de novo transcription of FOS. Genomic region comprising a 2-kb promoter of human FOS gene were cloned upstream of firefly luciferase and transfected into wild-type or YAP/TAZ knockout cells. As expected, stimulation with LPA increased promoter activity in a time-dependent manner in wild-type cells, but not in the knockout cells, showing that YAP/TAZ are involved in the transcription of FOS promoter (Fig. 2B). We further examined whether the effect of YAP requires its transcriptional activity. YAP mainly binds to the TEAD transcription factors (TEAD1-4) to induce gene expression, and Ser94 in YAP is required for TEAD binding (Zhao et al. 2008). Wild-type YAP expression in YAP/TAZ knockout cells rescued AP-1 induction. In contrast, Ser94-to-alanine mutant YAP could not rescue AP-1 induction, suggesting the requirement of TEAD binding for YAP to induce AP-1 components (Fig. 2C).
Figure 2.
YAP directly induces FOS in a manner dependent on TEAD. (A) Active YAP or TAZ induces AP-1 protein expression. HEK293A cells transfected with 5SA-YAP, 4SA-TAZ, or control vector were subjected to immunoblot analysis. (B) YAP/TAZ are required for FOS promoter activation by LPA. HEK293A cells were transfected with a luciferase reporter construct that has a 2-kb upstream region from transcription start site of human FOS gene as a promoter. Relative luciferase activities of LPA-treated samples were normalized to nontreated controls. Data are from three independent experiments. (**) P < 0.01 compared with wild-type controls at the same time point, by Student's t test. (C) TEAD binding is required for YAP to support FOS induction by LPA. YAP/TAZ knockout HEK293A cells transfected with wild-type or S94A mutant YAP, which is defective in TEAD binding, were serum-starved for 16 h and then stimulated with LPA for 30 min. (*) P < 0.05; (**) P < 0.01, compared with nontreated controls, by Student's t-test. (D) FOS promoter region contains two TEAD-binding sequences (TBS; CATTCC). Chromatin immunoprecipitation (ChIP) signals for TEAD4, SRF, H3K27Ac, and PolII were imported from ENCODE database. SRE; serum response element, the SRF-binding region. (E) Endogenous YAP and TEAD both bind to the FOS promoter. HEK293 cells starved in serum-free medium for 16 h were stimulated with LPA for 30 min. The cells were then subjected to ChIP with antibodies for endogenous YAP (left) or TEAD4 (right) along with control IgG. The precipitated DNA was quantitated by real-time PCR analysis with primers specific for promoter regions of GAPDH or CTGF, or three different TBS sites in FOS promoter. (*) P < 0.05; (**) P < 0.01; compared with IgG controls; (#) P < 0.05, compared with nontreated samples, by Student's t-test. (F) TEAD is required for YAP to bind FOS promoter. TEAD1/2/4 knockout HEK293 cells were treated as described in E, and analyzed for ChIP using antibody for endogenous YAP. (G) Scheme and sequence of HEK293 cells with TBS deletion. Wild-type, ΔTBS1, ΔTBS2, and ΔTBS1/2 cells were generated using pairs of CRISPR guide RNAs targeting PAM sequences flanking each TBS region. Single clones with indicated sequences were established. TEAD-binding sequences are colored in red. (H) Both TEAD binding sites are required for full induction of FOS. Wild-type, ΔTBS1, ΔTBS2, and ΔTBS1/2 cells were serum-starved for 16 h and treated with LPA for 30 min. Expression levels of FOS, FOSB, and CTGF mRNA were determined by real-time PCR analysis. (*) P < 0.05; (**) P < 0.01, compared with nontreated samples; (##) P < 0.01, compared with wild-type control, by Student's t-test.
Within the 2-kb promoter region in the human FOS gene, there are two consensus TEAD-binding sequences (TBSs) (Fig. 2D). These regions (TBS1 and TBS2) accompanied open chromatin signals (i.e., Acetylated histone or DNase hypersensitivity signals) and were apart from previously identified SRF binding element (SRE). Chromatin immunoprecipitation (ChIP) using anti-YAP antibody revealed that YAP specifically associated with both TBS regions only when cells were stimulated with LPA (Fig. 2E). ChIP using anti-TEAD4 antibody confirmed the binding of TEAD to the TBS sites; however, TEAD binding was present on both sites regardless of the LPA stimulation. These were in line with the current model that YAP shuttles between cytoplasm and nucleus in a stimulation-dependent manner, while TEAD resides in the nucleus but requires YAP for its transcriptional activation. Neither YAP nor TEAD4 bound to the SRF binding site. CTGF, a known direct target gene of YAP-TEAD and GAPDH were used as a positive and a negative control, respectively. Furthermore, additional ChIP assays showed that YAP could not bind to any of the TBS sites when TEAD1/2/4 were knocked out (Fig. 2F). This confirmed that YAP binds to FOS promoter in a manner dependent on TEAD. Taken together, our data suggest that upon LPA stimulation YAP-TEAD directly binds to the promoter of FOS to induce its expression.
To further verify that YAP-TEAD binding sites are responsible for the induction of FOS, the identified TBS regions were eliminated from the genome in HEK293 cells by using CRISPR. Each pair of gRNAs were designed to target the closest PAM sequences that flank respective TBS sites, resulting excision of a ∼20-bp fragment. Single cell clones having deletion of each site (ΔTBS1 or ΔTBS2) or both were selected and verified by genomic DNA sequencing (Fig. 2G). When these cells were stimulated with LPA for 30 min, FOS induction in ΔTBS1 or ΔTBS2 cells were significantly weaker than that in wild-type cells, showing that both sites are functionally involved in FOS expression. Deletion of both sites resulted in more dramatic, although not complete, blockade of FOS induction (Fig. 2H), demonstrating the critical role of YAP-TEAD in FOS transcription. FOSB has been reported to be transcribed by AP-1, which explains why FOSB induction was also dampened (Hong et al. 2011). It is noteworthy that induction of CTGF were also affected by deletion of the TBS cis-regulatory elements within FOS promoter. Although CTGF has an AP-1 binding site on its promoter as well, YAP/TAZ is known to be the dominant transcriptional regulator. This supports a notion that AP-1 might be important for proper function of YAP/TAZ to induce their downstream target genes in certain conditions.
Recently, YAP/TAZ have been shown to bind to distal enhancer regions of genes and recruit Mediator complex to target site for transcriptional activation (Galli et al. 2015). Therefore, putative distal enhancer regions were selected from upstream of FOS promoter, according to histone acetylation and monomethylation pattern (Supplemental Fig. S2A). However, among four putative enhancers (E1–E4) tested, none of them had strong binding with YAP or TEAD by ChIP assays (Supplemental Fig. S2B). Together, our data reveal that two TEAD-binding sites in FOS promoter are functionally indispensable for the proper induction of AP-1 in response to upstream stimulation.
Role of AP-1 induction in YAP/TAZ target gene transcription
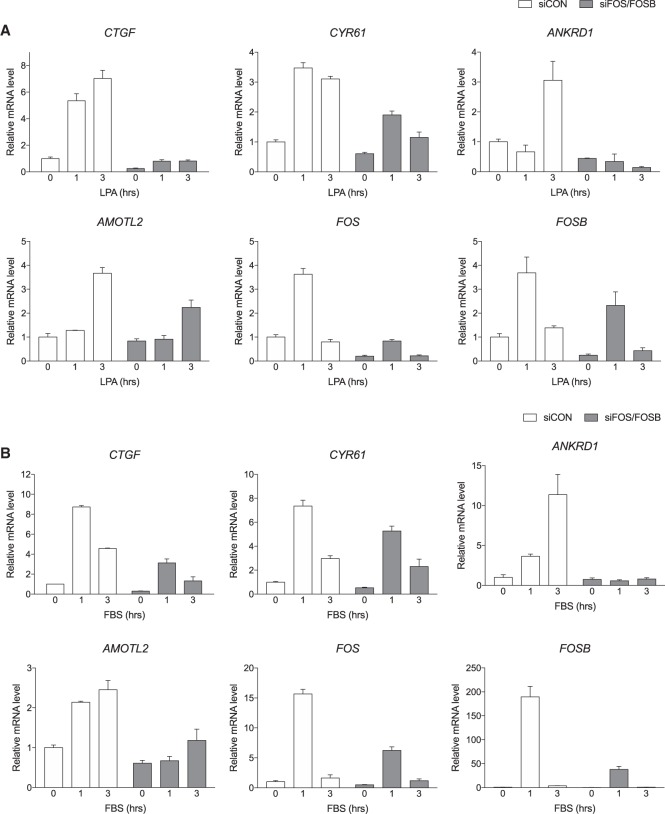
Immediate early genes including FOS are rapidly induced upon extracellular stimuli to drive subsequent transcriptional waves of other genes. Therefore, we hypothesized that YAP/TAZ might first promote the assembly of AP-1 and then cooperate with AP-1 to activate a large transcriptome for cellular function. To this end, we examined induction of known YAP/TAZ target genes in cells with silence of FOS and FOSB. As hypothesized, LPA induced transcription of many well-studied YAP/TAZ targets (i.e., CTGF, CYR61, ANKRD1, and AMOTL2), and expression of these YAP/TAZ target genes were blunted by AP-1 knockdown (Fig. 3A). Although all are affected, there were differences in the extent of suppression among the genes. CTGF and ANKRD1 were more sensitive than CYR61 and AMOTL2 to FOS/FOSB knockdown. Similar results were observed in serum-induced gene expression (Fig. 3B). The effect of AP-1 silencing on YAP target genes was not due to Hippo pathway regulation, since dominant-negative JUN mutant had no effect on the phosphorylation status of YAP (Supplemental Fig. S3). Given that different transcription partners have been reported for YAP/TAZ, the interplays with AP-1 may provide context-dependent transcriptional programs, adding further complexity and specificity to the biological function of YAP/TAZ.
Figure 3.
YAP requires AP-1 for full induction of target genes. (A) AP-1 induction is required for LPA-induced YAP/TAZ target gene expression. HEK293A cells were transfected with siRNAs against FOS and FOSB and incubated for 48 h. The cells were then serum-starved for 16 h and treated with LPA for indicated times. (B) AP-1 induction is required for serum-stimulated YAP/TAZ target gene induction. Experiments were similar to A except cells were stimulated with 10% FBS.
Function of AP-1 in YAP-mediated anchorage-independent cell growth
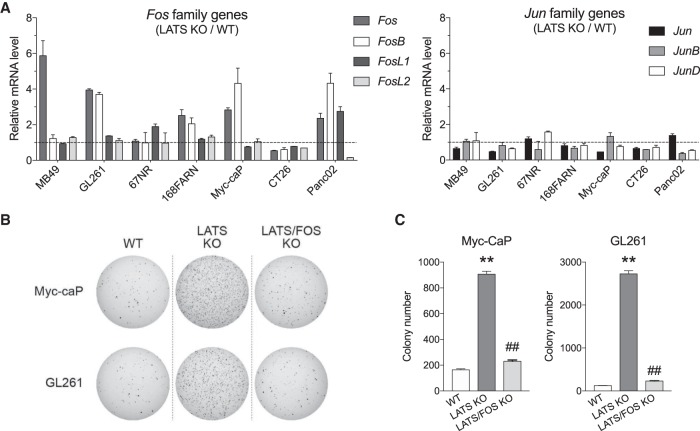
YAP/TAZ are potent regulators of cell proliferation and their hyperactivation is often represented as to drive tumorigenesis in a number of biological contexts. At the same time, AP-1 components are classic proto-oncogenes and play important roles in early tumorigenesis. To investigate the biological significance of our findings, we examined whether YAP/TAZ-mediated tumorigenesis is dependent on AP-1 induction. We have generated Lats1/2 double-knockout cell lines in seven mouse cancer cell lines using CRISPR (Pan et al. 2019). Since LATS1/2 are the inhibitory kinases for YAP/TAZ, endogenous YAP/TAZ were constitutively active in the knockout cells. When compared with their corresponding wild-type cells, those having Lats1/2 knockout showed higher basal expression of Fos family genes in general, particularly for Fos (Fig. 4A). In contrast, Jun family genes were not significantly different.
Figure 4.
YAP-dependent colony formation requires AP-1 activity. (A) Lats1/2 knockout cells show higher basal AP-1 expression compared with wild-type cells. Lats1/2 were knocked out in different cell lines to examine basal mRNA expression for AP-1 components. Cells were maintained in normal growth media with 10% FBS based on either DMEM or RPMI. (B) Deletion of Fos and FosB attenuates anchorage-independent growth of Lats1/2 DKO cells. Fos and FosB were additionally knocked out in Lats1/2 KO Myc-CaP or GL261 cells. Same number (5000 cells) wild-type, Lats1/2 KO, and Lats1/2;Fos/Fosb KO (LATS/FOS KO) cells from single clones were seeded onto a culture medium containing 0.35% agar and incubated for 2 wk at 37°C. The colonies were stained with 0.005% crystal violet and quantified. (C) Quantification of colony formation for images shown in B. (**) P < 0.01 compared with wild-type control; (##) P < 0.01 compared with LATS KO, by Student's t-test.
We next examined the role of AP-1 in oncogenic growth mediated by YAP activation. GL261 and Myc-caP cells were subjected to soft agar colony formation assay because Fos and Fosb were highly elevated when Lats1/2 were deleted. Lats1/2 knockout strongly promoted colony formations of both Myc-caP and GL261 in soft agar when compared with wild-type controls (Fig. 4B,C). Interestingly, additional knockout of Fos and Fosb (Lats/Fos KO) blocked the anchorage-independent growth facilitated by Lats1/2 knockout, suggesting that AP-1 is required for the enhanced oncogenic potential of these cells (Supplemental Fig. S4A,B). These data support a model in which induction of AP-1 by YAP/TAZ is not only involved in their target gene transcription, but also important for biological function to control oncogenic cell growth.
Different roles of AP-1 in YAP-dependent or -independent uveal melanoma
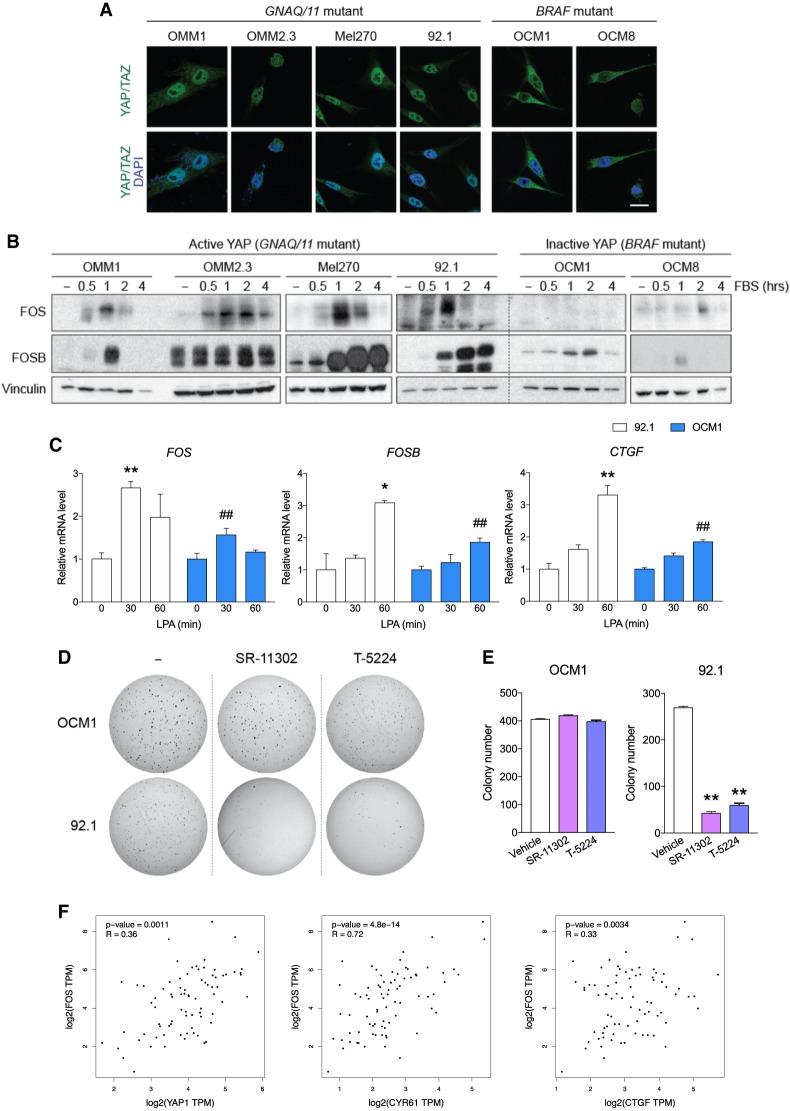
YAP is highly active in many types of cancers, particularly in uveal melanoma (UM), due to mutations in GPCR signaling (Yu et al. 2014). To further examine whether AP-1 induction is selectively associated with YAP-driven cancer cell growth, we compared a series of UM cell lines with activating mutations in either GNAQ/GNA11 or BRAF. The GNAQ/11 mutant UM cells have active nuclear YAP, while the BRAF mutant cells have inactive cytoplasmic YAP (Fig. 5A). The GNAQ mutant UM cells are YAP-dependent, while the BRAF mutant UM cells are YAP-independent as YAP/TAZ knockdown blocks the tumor growth of the GNAQ mutant, but not the BRAF mutant UM cells (Yu et al. 2014). We observed that serum-stimulation evoked strong AP-1 induction across UM cell lines with active YAP, namely OMM1 (GNA11Q209L), OMM2.3, Mel270 (GNAQQ209P), and 92.1(GNAQQ209L), but not in those BRAFV600E OCM1 and OCM8 cell lines that have inactive YAP (Fig. 5B,C). These results further support a role of YAP/TAZ activity in AP-1 induction.
Figure 5.
AP-1 is important for growth of YAP-dependent uveal melanoma cells. (A) YAP/TAZ localization occurs in the uveal melanoma with activating mutation in GNAQ or GNA11, but not in those with BRAF mutation. Different uveal melanoma cells having either GNA11Q209L (OMM1), QNAQQ209P (OMM2.3 and Mel270), or QNAQQ209L (92.1) were compared with those having BRAFV600E (OCM1 and OCM8). Cells were treated with 10% FBS for 1 h and immunostained for YAP/TAZ. Scale bar, 20 µm. (B) Serum induces strong expression of FOS and FOSB only in the YAP active (having mutations in GNAQ or GNA11), but not the YAP-repressed uveal melanoma (having BRAF mutation) cells. Uveal melanoma cells were compared for FOS and FOSB induction after a 16-h serum starvation, followed by stimulation with 10% FBS. (C) LPA-stimulated FOS and FOSB mRNA induction is greater in uveal melanoma harboring active YAP. 92.1 and OCM1 cells were serum-starved for 16 h and stimulated with LPA for qPCR. (*) P < 0.05; (**) P < 0.01, compared with nontreated samples; (##) P < 0.01, compared with 92.1 cells at same time point, by Student's t-test. (D) Inhibition of AP-1 attenuates anchorage-independent growth of the GNAQ mutant 92.1 cells, but not the BRAF mutant OCM1 cells. Same number (5000 cells) of OCM1 or 92.1 cells were seeded onto a culture medium containing 0.35% agar for 2 wk. The medium with AP-1 inhibitor SR-11302 (10 µM), T-5224 (30 µM), or vehicle was changed every 2 d. The colonies were stained with 0.005% crystal violet. (E) Quantification of colony formation for images in D. (**) P < 0.01, compared with vehicle-treated control, by Student's t-test. (F) Dot plots showing the positive correlation between FOS and YAP, CYR61, or CTGF mRNA expression in uveal melanoma from 80 patients. R-values and significance were calculated by Pearson's correlation.
Since UM cells responded differently according to their oncogenic driver mutations and YAP activity, we tested whether the YAP-dependent UM cells are more sensitive to AP-1 inhibition. Interestingly, in a soft agar colony formation assay, chemical AP-1 inhibitors (SR-11302 or T-5224) preferentially suppressed anchorage-independent growth of UM cell line with active YAP (92.1) over cells with inactive YAP (OCM1) (Fig. 5D,E). These results indicate that YAP-driven cell growth is highly dependent on AP-1 and that the inhibiting AP-1 may be a vulnerability of cancers with hyperactive YAP/TAZ signaling.
We additionally analyzed the gene expression in 80 uveal melanoma tissues from The Cancer Genome Atlas (TCGA) database (Robertson et al. 2017). A positive correlation was found between mRNA expression of FOS and YAP or its target genes CYR61 and CTGF (Fig. 5F). These data provide further in vivo evidence supporting that YAP/TAZ contribute to FOS induction.
Essential function of AP-1 in YAP-mediated organ size control
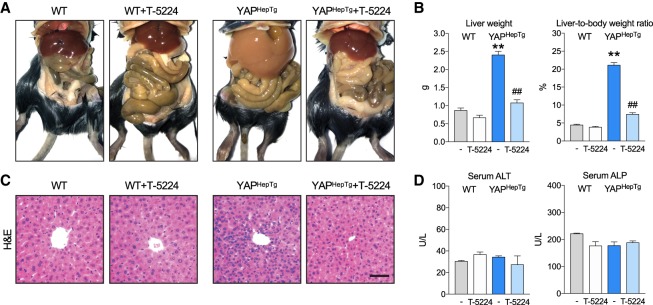
The Hippo pathway has gained great attention owing to its profound effect in organ size control (Yu et al. 2015). To strengthen the physiological significance of our findings, we examined whether AP-1 induction plays a role in YAP-mediated liver size regulation. Transgenic mice expressing hepatocyte-specific, tetracycline-inducible YAP (YAPHepTg) were adopted from a previous study (Yu et al. 2015), which showed a dramatic hepatomegaly upon YAP induction by doxycycline. Either wild-type or YAPHepTg mice were fed doxycycline-containing water, along with daily oral administrations of AP-1 inhibitor T-5224, which blocks the DNA-binding activity of c-Fos/AP-1 (Aikawa et al. 2008). As expected, a massive hepatomegaly was observed in YAPHepTg mice after 17 d of doxycycline treatment. However, the YAPHepTg-induced hepatomegaly was largely suppressed when mice were treated with T-5224, as indicated by gross morphology and tissue weight (Fig. 6A,B). Mice with YAP overexpression showed a slight reduction of body weight. Interestingly, treatment with T-5224 also ameliorated the body weight loss associated with YAP overexpression. Hepatocytes in the YAP-overexpressing livers were more densely packed when compared with wild-type control as an indicative of intense hyperplasia, and again this phenotype was suppressed in the AP-1 inhibitor-treated group (Fig. 6C). The difference in liver size was not due to liver damage since the levels of serum markers for hepatocyte viability, alanine aminotransferase (ALT) or alkaline phosphatase (ALP), were similar across experimental groups (Fig. 6D).
Figure 6.
AP-1 induction is required for YAP-mediated organ size control. (A) Gross appearance of wild-type and hepatocyte-specific tetracycline-inducible YAP transgenic (YAPHepTg) mice. All mice were given doxycycline in drinking water and treated with T-5224 (150 mg/kg) or vehicle for 17 d, starting at 3 wk of age. (B) Quantification of liver weight and liver/body weight ratio. (**) P < 0.01, compared with wild-type control; (##) P < 0.01, compared with YAPHepTg control, by Student's t-test. (C) Representative H&E staining of liver sections. Scale bar, 50 µm. (D) Serum biochemical parameters show that hepatocytes are without significant damage.
AP-1 is required for the majority of YAP-regulated gene expression in the liver
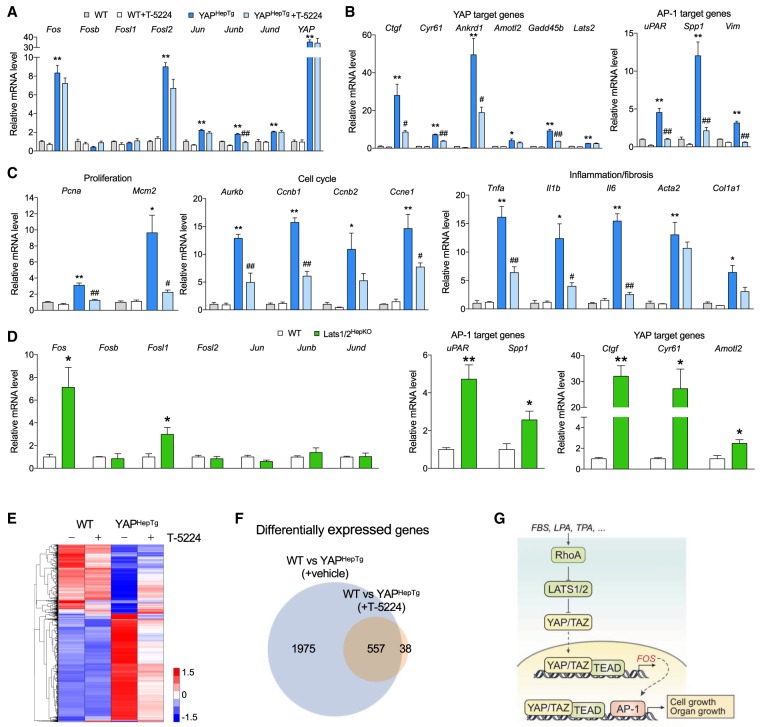
Notably, our finding that YAP drives FOS induction was corroborated in vivo by examining hepatic mRNA expression. Fos mRNA levels were significantly higher in YAPHepTg mice (Fig. 7A). Among AP-1 components, Fos was among the most prominently induced genes. Another finding that YAP and AP-1 form a feed-forward loop was also confirmed in the mouse model. Induction of representative YAP target genes, such as Ctgf, Cyr61, Ankrd1, and Gadd45b, was significantly dampened when AP-1 was inhibited (Fig. 7B). Expression levels of proliferation markers (e.g., Pcna and Mcm2) and cell cycle progression genes (e.g., Aurkb, Ccnb1, Ccnb2, and Ccne1) showed that they were induced by YAP overexpression in a manner dependent on AP-1 activity (Fig. 7C), supporting the macroscopic observation of YAP-induced cell growth and its blockade by AP-1 inhibitor. Representative genes involved in inflammation or fibrosis (e.g., Tnfa, Il1b, Il6, and Col1a1) showed a similar expression pattern, probably since an increased activity of immune cells and hepatic stellate cells are commonly accompanied with intensive replication as seen during liver regeneration (Forbes and Rosenthal 2014).
Figure 7.
AP-1 is required for YAP-regulated gene expression in liver. (A) Elevated FOS mRNA levels after hepatic YAP overexpression. Expression of genes comprising AP-1 were analyzed with qPCR. (B) AP-1 inhibition attenuates YAP target gene expression. Levels of mRNA for YAP/TAZ or AP-1 target genes were quantified. (C) mRNA levels of cell proliferation markers and genes involved in cell cycle progression or inflammation and fibrosis. (D) Elevated mRNA levels of FOS and AP-1 target genes in the livers of hepatocyte-specific Lats1/2 knockout (Lats1/2HepKO) mice. (E) Hierarchically clustered heat map of YAP-responsive genes with or without T-5224 treatment. Total RNAs extracted from three mouse livers of each treatment group were pooled, and then analyzed by RNA-seq. Colors denote genes with high (red) or low (blue) expression levels, which are assigned according to log10(FPKM + 1), ranging from 1.5 to −1.5. (F) Venn diagram comparing genes regulated by YAP and T-5224 treatment. Of the 2532 YAP differentially regulated genes, 1975 genes were blocked by T-5224 treatment. (G) Schematic diagram of AP-1 as downstream effector of YAP in cell and organ growth. Upon signal stimulation, YAP/TAZ are recruited to TEAD on FOS promoter for FOS expression, which forms AP-1 and cooperates with YAP/TAZ for gene expression critical in cell growth and organ size control. For A–D, (*) P < 0.05; (**) P < 0.01, compared to wild-type control; (#) P < 0.05; (##) P < 0.01, compared to YAPHepTg control, by Student's t-test.
We extended similar studies in another mouse model of YAP hyperactivation. Livers from hepatocyte-specific Lats1/2 knockout mice (Lats1/2HepKO) were examined for mRNA expression. Interestingly, hepatic Fos mRNA levels were significantly higher in Lats1/2HepKO mice as compared with wild type, with the greatest extent among AP-1 genes (Fig. 7D). Consistently, AP-1 target genes (e.g., uPAR and Spp1) were also higher in Lats1/2HepKO mice.
Given that AP-1 is required for the liver overgrowth caused by Yap overexpression, we sought to examine the function of AP-1 in YAP-regulated transcriptome in the liver. We performed RNA-seq with liver tissues from wild-type and YAPHepTg, treated with or without T-5224 (Fig. 7E). Compared with wild-type animals, 2532 genes were differentially expressed in the livers of YAPHepTg mice. Interestingly, when T-5224 was given to YAPHepTg mice, expression pattern of the YAP-responsive genes was dramatically changed toward the pattern seen in the wild-type livers. On the other hand, only a small portion of genes were affected by T-5224 treatment in wild-type mice (Fig. 7E). Further analysis has shown that 78% (1975 out of 2532 genes) of YAP-regulated genes were blocked by T-5224 treatment. The above results support an essential function of AP-1 in YAP-mediated gene expression as the majority of the YAP-regulated transcriptome requires AP-1 (Fig. 7F).
It is particularly noteworthy that serum bilirubin levels were significantly higher in YAPHepTg mice but not as much in those treated with the AP-1 inhibitor (Supplemental Fig. S5A). In general, hyperbilirubinemia occurs as a result of hepatocyte damage and functional failure (Björnsson and Olsson 2005). Nonetheless, YAPHepTg mice did not display apparent liver damage, as indicated by the relatively normal serum biochemical markers and histology (Fig. 6C,D). Thus, it is likely that YAP may regulate bilirubin metabolism in hepatocytes, and that AP-1 might be involved in this regulation. Indeed, YAPHepTg mice showed lower mRNA expression of Ugt1a1 and Mrp2, the bilirubin-metabolizing enzyme in hepatocytes and the transporter for excretion of metabolized bilirubin to the bile canaliculi, respectively (Supplemental Fig. S5B). Moreover, AP-1 inhibitor prevented these repressions, particularly for Ugt1a1. Furthermore, a dramatic reduction in Ugt1a1 and Mrp2 mRNA levels were also observed in Lats1/2HepKO mice, supporting the notion that YAP and its target AP-1 modulate the expression of bilirubin metabolic enzymes (Supplemental Fig. S5C). Taken together, the above results provide in vivo evidences that AP-1 induction is a critical event for the physiological outcome of YAP activation in the liver, including organ size control as well as functional maintenance (Fig. 7H).
Discussion
The Hippo pathway has gained considerable attention for its role in both physiological and pathological conditions. It has been implicated in organ development, stem cell biology, regeneration, and tumorigenesis (Piccolo et al. 2014; Moroishi et al. 2015a; Yu et al. 2015). Hippo pathway is composed of a core kinase module and a transcription module, the former being the most extensively studied in the past decade. Since YAP/TAZ are the main effectors of Hippo pathway, many studies have focused on how signals affect YAP/TAZ phosphorylation and nuclear location. Relatively recently, there have been increasing attempts to reveal regulatory mechanism of the transcriptional module by which YAP/TAZ can exert selective activity on different cis-regulatory elements (Totaro et al. 2018). However, much less is known about the key downstream target genes that mediate the biological functions of YAP/TAZ. In this study, we uncover a critical layer of regulation on the transcriptional output of YAP/TAZ. We show that YAP/TAZ are indispensable for FOS induction, which is among the best-studied classical transcription factors, and this FOS induction is required for physiological functions of YAP/TAZ.
It is currently understood that nuclear localization is essential for YAP/TAZ activity as they are transcription coactivators. However, recent reports show that other partners have major role in expression of YAP/TAZ target genes, possibly by cooperating with YAP-TEAD in the nucleus (Totaro et al. 2018). So far, SRF has been regarded as the primary factor for transcription of immediate early genes, including AP-1. This report shows an absence of FOS and FOSB induction in YAP/TAZ knockout cells, thus suggests Hippo pathway regulation as another prerequisite for AP-1 induction. Mechanistically, YAP/TAZ seem to act independently of SRF since neither YAP nor TEAD is associated with the SRF-binding region in FOS promoter in ChIP assays, and restricted deletion of TEAD binding motifs was sufficient to block FOS induction. Furthermore, genes of AP-1 complex were not the only immediate early genes of which induction was blocked by YAP/TAZ deletion. Considering that YAP/TAZ translocation occurs rapidly by serum or LPA (Yu et al. 2012), there may be a general role of YAP/TAZ in immediate early gene expression. Our data support a model that YAP/TAZ contribute to AP-1 activation and the active AP-1 then collaborates with YAP/TAZ to regulate gene expression and biological function. Given the enormous volume of investigation on AP-1 regulation and function, our study places AP-1 downstream from YAP/TAZ, and therefore has significantly expanded the potential biology of YAP/TAZ.
The present dogma of the Hippo pathway is that LATS kinase activity is the dominant regulator for downstream function by restricting YAP/TAZ activity through phosphorylation, resulting in 14-3-3 binding and cytoplasmic retention (Zhao et al. 2007). Indeed, LATS knockout cells show nuclear YAP/TAZ under most conditions (Meng et al. 2015). However, we have previously observed that further induction CTGF and CYR61 still occurs when these cells are stimulated with serum (Plouffe et al. 2018), indicating an existence of other factors for full transcriptional activity of YAP/TAZ. By intersecting the mechanism how YAP/TAZ and AP-1 are interwound, we uncovered a critical role of AP-1 in Hippo pathway function. Our data obtained from FOS deletion or chemical inhibition suggest that YAP/TAZ actively promote de novo synthesis of AP-1 to facilitate their downstream function. We have also shown that YAP-mediated anchorage-independent cell growth was dependent on AP-1 activity in cancer cells that acquired malignancy by Lats1/2 deletion. These findings were further expanded to uveal melanoma cells that have constitutively active YAP. AP-1 is essential for growth of YAP-dependent uveal melanoma driven by GNAQ/GNA11 mutation, but not the YAP-independent uveal melanoma driven by BRAF mutation. Chemical inhibition of AP-1 was only effective in repressing GNAQ/GNA11 mutant but not BRAF mutant melanoma cells. Thus, our data suggest a vulnerability of selective AP-1 inhibition for YAP-dependent cancers.
Another important finding of this study is that the cooperation with AP-1 is required for YAP-mediated liver size regulation. Although AP-1 has been extensively studied as to exert prominent effect on cell growth, it is only recently appreciated for its role in organ growth in a transgenic mouse model with liver-specific overexpression of FOS (Bakiri et al. 2017). Hepatocyte-restricted overexpression of FOS caused hepatomegaly at early stage and carcinogenesis at later time period. These phenotypes are very similar to liver-specific YAP overexpression, increased liver size in early time, and tumor development at a late stage. It is noteworthy that the effects on hepatomegaly by FOS induction were much slower than YAP overexpression observed in mouse models (Camargo et al. 2007; Dong et al. 2007). Supporting this notion, AP-1 per se failed to promote anchorage-independent growth in the absence of YAP overexpression in mammary epithelial cells (Zanconato et al. 2015). Nonetheless, we show that AP-1 inhibition strongly represses YAP-mediated hepatomegaly. Therefore, AP-1 is likely to be an important player in the physiological settings accompanied with YAP activation. In addition, our findings add a new insight for bilirubin metabolism, as another physiological role of YAP. Although there was no apparent evidence for hepatocyte damage, YAP-overexpressed mice showed elevated serum bilirubin levels. Moreover, AP-1 was responsible for YAP-mediated repression of bilirubin-removing genes in the liver. Together, our study suggests a model that AP-1 induction plays an important role in YAP-mediated pathophysiology, such as organ size control and tumorigenesis.
The Hippo pathway and YAP/TAZ are identified to play key roles in a wide variety of physiological processes including differentiation, stem cell renewal, and energy metabolism. Future studies are needed to test whether YAP/TAZ require de novo AP-1 induction for all or some of their biological functions and the possibility of targeting AP-1 for intervention of YAP dependent pathophysiology.
Materials and methods
Cell culture
All cell lines were maintained at 37°C with 5% CO2. HEK293A, MB49, GL261, and Myc-caP cells were cultured in DMEM (Invitrogen) and 67NR, 168FARN, CT26, Panc02, and uveal melanoma cell lines were cultured in RPMI (Invitrogen) containing 10% FBS (Gibco) and 50 µg/mL penicillin/streptomycin (Invitrogen). GL261 cells were from DSMZ. MB49 cells were from Millipore. 67NR and 168FARN cells were from Dr. Jing Yang (University of California at San Diego). Uveal melanoma cell lines were provided by Dr. Martine Jager (Leiden University). None of the cell lines in this study was among commonly misidentified cell lines from International Cell Line Authentication Committee and NCBI Biosample. Cells lines were tested and confirmed to be free of mycoplasma.
Generation of knockout cells
pSpCas9(BB)-2A-Puro (PX459) was a gift from Dr. Feng Zhang (Broad Institute of Massachusetts Institute of Technology and Harvard). The guide RNAs were designed using the CRISPR design tool at http://crispr.mit.edu. Single-guide RNAs (sgRNAs) were cloned into the empty PX459 vector. HEK293A cells were transfected with single or paired CRISPR vectors according to experiments. After 24 h of transfection, cells were selected using puromycin for 3 d. SRF KO cells were used as a pool, shortly after selection. For all other KO lines, cells were single-cell-sorted by flow cytometer (BD Influx) into 96-well plates. Expanded single clones were screened by protein immunoblotting and/or genomic DNA sequencing. YAP/TAZ KO, LATS KO, TEAD KO cells were generated as previously described (Hansen et al. 2015; Moroishi et al. 2015b; Lin et al. 2017; Pan et al. 2019). The guide sequences for knockout cells used in this experiment are provided in Supplemental Table S1.
DNA transfection
Cells were transfected with plasmids using Polyjet transfection reagent (Signagen Laboratories) according to the manufacturer's protocol. Cells were transfected with pQCXIH or pBABE empty vector, pQCXIH-wtYAP (wild type), pQCXIH-S94A-YAP, pQCXIH-5SA-YAP, or pBABE-4SA-TAZ. Experiments were done at least 24–48 h after transfection.
RNA interference
Cells were transfected with siRNAs against different genes using Lipofectamine RNAiMAX transfection reagent (ThermoFisher Scientific). Cells were seeded in 12-well plates and transfected with 10 pmol of siRNA each well, per manufacturer's instructions. After 36 h, cells were used for experiments. The siRNAs were formulated in duplexed Dicer substrate interfering RNA (Integrated DNA Technologies). siRNA sequences were GGGGCAAGGUGGAACAGUUAUCUdCdC (forward) with GGAGAUAACUGUUCCACCUUGCCCCUC (reverse) against human FOS and CCAAGACUUGGAAACUUGAUUGUdTdG (forward) with CAACAAUCAAGUUUCCAAGUCUUGGUU (reverse) against human FOSB.
RNA extraction and quantitative real-time PCR analysis
Cells and mouse tissues were harvested for RNA extraction using the RNeasy Plus mini kit (Qiagen). Reverse transcription was done using iScript reverse transcriptase (Bio-Rad). qRT-PCR was performed using the KAPA SYBR FAST qPCR kit (Kapa Biosystems) with QuantStudio 3 real-time PCR system (Applied Biosystems). All mRNA levels were normalized to GAPDH expression. Primer sequences used for PCR are available in Supplemental Table S1.
Western blot
Cells were harvested using sample buffer containing 10% SDS and β-mercaptoethanol. Samples were boiled for 5 min and total proteins were separated by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Millipore). Proteins of interest were probed with primary antibodies and horseradish peroxidase-linked secondary antibodies. Chemiluminescent signals were developed on X-ray film using HRP substrate luminal reagent (BioLegend). The primary antibodies against FOS, FOSB, and Vinculin are from Cell Signaling.
Luciferase reporter assay
Destabilized luciferase having PEST sequence (luc2P) were used for reporter assay. Cells were transfected with a pGL4.22-Luc reporter having a 2-kb region upstream of human FOS transcription start site. Luciferase activity was assayed using the Neolite reporter gene assay system (PerkinElmer Life Sciences) according to the manufacturer's instructions.
ChIP assay
Cells were cross-linked with 1% formaldehyde for 15 min. After lysis, genomic DNA was digested through incubation with micrococcal nuclease. Immunoprecipitation reactions were done on chromatin extracts overnight at 4°C using 5 µg each of antibodies to YAP, TEAD1, or normal control rabbit IgG. Two percent of the chromatin extract was set aside for input. Precipitated DNA was quantitated by real-time PCR analysis. All ChIP signals were normalized to the input.
Immunofluorescence analysis
Cells were seeded on coverslips pretreated with poly-L-ornithine solution (Sigma). Cells were fixed in 4% paraformaldehyde (Electron Microscopy Sciences) for 15 min, followed by permeabilization with 0.1% Triton-X for 5 min. Cells were blocked in 3% BSA for 1 h and incubated overnight at 4°C in primary antibody against YAP/TAZ (Santa Cruz Biotechnology) diluted in 3% BSA. Secondary antibody was diluted in 3% BSA and incubated for 1 h. Slides were mounted with Prolong gold antifade reagent with DAPI (Invitrogen). Each image is a single Z section at the same cellular level. Images were captured with a Nikon Eclipse Ti confocal microscope.
Soft agar colony formation assay
Each well of the six-well plate was coated with 1.5 mL of bottom agar composed of DMEM containing 10% FBS and 0.5% Difco agar noble. For each well, 5000 cells were suspended in 1.5 mL of top agar composed of DMEM containing 10% FBS and 0.35% Difco agar noble. Cells were incubated for 3 wk with fresh medium replaced every 3 d. Colonies were fixed with 4% formaldehyde and stained using 0.005% crystal violet.
Animal experiments
Mice harboring a hepatocyte-specific tetracycline-inducible human S127A-YAP transgene (YAPHepTg) were previously established (Dong et al. 2007). Three-week-old transgenic and nontransgenic mice were fed 0.2 mg/mL doxycycline (Sigma) in drinking water supplemented with 2.5% sucrose. T-5224 was diluted in polyvinylpyrrolidone solution and given 150 mg/kg body weight per oral, once daily for 17 d. Serum and livers were immediately frozen in dry ice for further evaluation. Hepatocyte-specific LATS1/2 knockout mice (LATS1/2HepKO) were established in a previous study (Lee et al. 2016). Livers isolated at postnatal day 1 were analyzed for mRNA expression.
Statistics
All in vitro assays were performed in triplicates. Data are shown as the mean ± SEM. Groups were compared by two-tailed t-tests. The criterion for statistical significance was set at P-values < 0.05.
Study approval
All animal studies were approved by the Institutional Animal Care and Use Committee of the University of California at San Diego and Korea Advanced Institute of Science and Technology.
Competing interest statement
K.L.G. is a cofounder of and has an equity interest in Vivace Therapeutics, Inc. The terms of this arrangement have been reviewed and approved by the University of California at San Diego in accordance with its conflict of interest policies.
Supplementary Material
Acknowledgments
We thank Dr. Duojia Pan (University of Texas Southwestern Medical Center) for providing the YAPHepTg mice. This work is supported by National Institutes of Health grants CA196878 and CA217642 to K.L.G., and DE015964 to C.-Y.W.
Author contributions: J.H.K., S.W.P., and K.-L.G. conceived the project and designed the research. J.H.K., S.W.P., and Z.M. performed the studies and analyzed the data. D.H.L. and D.S.L. acquired animal tissues and performed experiments. J.H.K. and K.-L.G. wrote the manuscript. C.-Y.W. and K.-L.G. provided administrative support and obtained funding. K.-L.G. provided overall data supervision.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.331546.119.
References
- Aikawa Y, Morimoto K, Yamamoto T, Chaki H, Hashiramoto A, Narita H, Hirono S, Shiozawa S. 2008. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat Biotechnol 26: 817–823. 10.1038/nbt1412 [DOI] [PubMed] [Google Scholar]
- Bakiri L, Hamacher R, Graña O, Guío-Carrion A, Campos-Olivas R, Martinez L, Dienes HP, Thomsen MK, Hasenfuss SC, Wagner EF. 2017. Liver carcinogenesis by FOS-dependent inflammation and cholesterol dysregulation. J Exp Med 214: 1387–1409. 10.1084/jem.20160935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamouche S, Curto M, Saotome I, Gladden AB, Liu CH, Giovannini M, McClatchey AI. 2010. Nf2/Merlin controls progenitor homeostasis and tumorigenesis in the liver. Genes Dev 24: 1718–1730. 10.1101/gad.1938710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergers G, Graninger P, Braselmann S, Wrighton C, Busslinger M. 1995. Transcriptional activation of the fra-1 gene by AP-1 is mediated by regulatory sequences in the first intron. Mol Cell Biol 15: 3748–3758. 10.1128/MCB.15.7.3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Björnsson E, Olsson R. 2005. Outcome and prognostic markers in severe drug-induced liver disease. Hepatology 42: 481–489. 10.1002/hep.20800 [DOI] [PubMed] [Google Scholar]
- Camargo FD, Gokhale S, Johnnidis JB, Fu D, Bell GW, Jaenisch R, Brummelkamp TR. 2007. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol 17: 2054–2060. 10.1016/j.cub.2007.10.039 [DOI] [PubMed] [Google Scholar]
- Dong J, Feldmann G, Huang J, Wu S, Zhang N, Comerford SA, Gayyed MF, Anders RA, Maitra A, Pan D. 2007. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130: 1120–1133. 10.1016/j.cell.2007.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. 2003. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3: 859–868. 10.1038/nrc1209 [DOI] [PubMed] [Google Scholar]
- Foletta VC, Sonobe MH, Suzuki T, Endo T, Iba H, Cohen DR. 1994. Cloning and characterisation of the mouse fra-2 gene. Oncogene 9: 3305–3311. [PubMed] [Google Scholar]
- Forbes SJ, Rosenthal N. 2014. Preparing the ground for tissue regeneration: from mechanism to therapy. Nat Med 20: 857–869. 10.1038/nm.3653 [DOI] [PubMed] [Google Scholar]
- Galli GG, Carrara M, Yuan WC, Valdes-Quezada C, Gurung B, Pepe-Mooney B, Zhang T, Geeven G, Gray NS, de Laat W, et al. 2015. YAP drives growth by controlling transcriptional pause release from dynamic enhancers. Mol Cell 60: 328–337. 10.1016/j.molcel.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham R, Gilman M. 1991. Distinct protein targets for signals acting at the c-fos serum response element. Science 251: 189–192. 10.1126/science.1898992 [DOI] [PubMed] [Google Scholar]
- Hansen CG, Ng YL, Lam WL, Plouffe SW, Guan KL. 2015. The Hippo pathway effectors YAP and TAZ promote cell growth by modulating amino acid signaling to mTORC1. Cell Res 25: 1299–1313. 10.1038/cr.2015.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschman HR. 1991. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem 60: 281–319. 10.1146/annurev.bi.60.070191.001433 [DOI] [PubMed] [Google Scholar]
- Hong S, Skaist AM, Wheelan SJ, Friedman AD. 2011. AP-1 protein induction during monopoiesis favors C/EBP: AP-1 heterodimers over C/EBP homodimerization and stimulates FosB transcription. J Leukoc Biol 90: 643–651. 10.1189/jlb.0111043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer VR, Eisen MB, Ross DT, Schuler G, Moore T, Lee JC, Trent JM, Staudt LM, Hudson J Jr., Boguski MS, et al. 1999. The transcriptional program in the response of human fibroblasts to serum. Science 283: 83–87. 10.1126/science.283.5398.83 [DOI] [PubMed] [Google Scholar]
- Lee KP, Lee JH, Kim TS, Kim TH, Park HD, Byun JS, Kim MC, Jeong WI, Calvisi DF, Kim JM, et al. 2010. The Hippo-Salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci 107: 8248–8253. 10.1073/pnas.0912203107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Park JO, Kim TS, Kim SK, Kim TH, Kim MC, Park GS, Kim JH, Kuninaka S, Olson EN, et al. 2016. LATS-YAP/TAZ controls lineage specification by regulating TGFβ signaling and Hnf4α expression during liver development. Nat Commun 7: 11961 10.1038/ncomms11961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Cooper J, Zhou L, Yang C, Erdjument-Bromage H, Zagzag D, Snuderl M, Ladanyi M, Hanemann CO, Zhou P, et al. 2014. Merlin/NF2 loss-driven tumorigenesis linked to CRL4(DCAF1)-mediated inhibition of the hippo pathway kinases Lats1 and 2 in the nucleus. Cancer Cell 26: 48–60. 10.1016/j.ccr.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KC, Moroishi T, Meng Z, Jeong HS, Plouffe SW, Sekido Y, Han J, Park HW, Guan KL. 2017. Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. Nat Cell Biol 19: 996–1002. 10.1038/ncb3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Kimmoun E, Legrand A, Sauvanet A, Degott C, Lardeux B, Bernuau D. 2002. Activation of NF-κ B, AP-1 and STAT transcription factors is a frequent and early event in human hepatocellular carcinomas. J Hepatol 37: 63–71. 10.1016/S0168-8278(02)00064-8 [DOI] [PubMed] [Google Scholar]
- Liu X, Li H, Rajurkar M, Li Q, Cotton JL, Ou J, Zhu LJ, Goel HL, Mercurio AM, Park JS, et al. 2016. Tead and AP1 coordinate transcription and motility. Cell Rep 14: 1169–1180. 10.1016/j.celrep.2015.12.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Li Y, Kim SM, Bossuyt W, Liu P, Qiu Q, Wang Y, Halder G, Finegold MJ, Lee JS, et al. 2010. Hippo signaling is a potent in vivo growth and tumor suppressor pathway in the mammalian liver. Proc Natl Acad Sci 107: 1437–1442. 10.1073/pnas.0911427107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallikarjuna K, Pushparaj V, Biswas J, Krishnakumar S. 2006. Expression of insulin-like growth factor receptor (IGF-1R), c-Fos, and c-Jun in uveal melanoma: an immunohistochemical study. Curr Eye Res 31: 875–883. 10.1080/02713680600878790 [DOI] [PubMed] [Google Scholar]
- Meng Z, Moroishi T, Mottier-Pavie V, Plouffe SW, Hansen CG, Hong AW, Park HW, Mo JS, Lu W, Lu S, et al. 2015. MAP4K family kinases act in parallel to MST1/2 to activate LATS1/2 in the Hippo pathway. Nat Commun 6: 8357 10.1038/ncomms9357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos GK, DeFrances MC. 1997. Liver regeneration. Science 276: 60–66. 10.1126/science.276.5309.60 [DOI] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113: 329–342. 10.1016/S0092-8674(03)00278-2 [DOI] [PubMed] [Google Scholar]
- Moroishi T, Hansen CG, Guan KL. 2015a. The emerging roles of YAP and TAZ in cancer. Nat Rev Cancer 15: 73–79. 10.1038/nrc3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T, Park HW, Qin B, Chen Q, Meng Z, Plouffe SW, Taniguchi K, Yu FX, Karin M, Pan D, et al. 2015b. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev 29: 1271–1284. 10.1101/gad.262816.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Mizuno T, Taniguchi T, Fujii M, Ishiguro F, Fukui T, Akatsuka S, Horio Y, Hida T, Kondo Y, et al. 2011. LATS2 is a tumor suppressor gene of malignant mesothelioma. Cancer Res 71: 873–883. 10.1158/0008-5472.CAN-10-2164 [DOI] [PubMed] [Google Scholar]
- Pan WW, Moroishi T, Koo JH, Guan KL. 2019. Cell type-dependent function of LATS1/2 in cancer cell growth. Oncogene 38: 2595–2610. 10.1038/s41388-018-0610-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S, Dupont S, Cordenonsi M. 2014. The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev 94: 1287–1312. 10.1152/physrev.00005.2014 [DOI] [PubMed] [Google Scholar]
- Plouffe SW, Lin KC, Moore JL 3rd, Tan FE, Ma S, Ye Z, Qiu Y, Ren B, Guan KL. 2018. The Hippo pathway effector proteins YAP and TAZ have both distinct and overlapping functions in the cell. J Biol Chem 293: 11230–11240. 10.1074/jbc.RA118.002715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, Hess JM, Uzunangelov V, Walter V, Danilova L, et al. 2017. Integrative analysis identifies four molecular and clinical subsets in Uveal Melanoma. Cancer Cell 32: 204–220 e215. 10.1016/j.ccell.2017.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, Chen Y, Park O, Chang J, Simpson RM, et al. 2010. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci 107: 1431–1436. 10.1073/pnas.0911409107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totaro A, Panciera T, Piccolo S. 2018. YAP/TAZ upstream signals and downstream responses. Nat Cell Biol 20: 888–899. 10.1038/s41556-018-0142-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, Simpson EM, Barsh GS, Bastian BC. 2009. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457: 599–602. 10.1038/nature07586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, et al. 2010. Mutations in GNA11 in uveal melanoma. N Engl J Med 363: 2191–2199. 10.1056/NEJMoa1000584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. 2009. A census of human transcription factors: function, expression and evolution. Nat Rev Genet 10: 252–263. 10.1038/nrg2538 [DOI] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, et al. 2012. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell 150: 780–791. 10.1016/j.cell.2012.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Luo J, Mo JS, Liu G, Kim YC, Meng Z, Zhao L, Peyman G, Ouyang H, Jiang W, et al. 2014. Mutant Gq/11 promote uveal melanoma tumorigenesis by activating YAP. Cancer Cell 25: 822–830. 10.1016/j.ccr.2014.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Guan KL. 2015. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163: 811–828. 10.1016/j.cell.2015.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F, Forcato M, Battilana G, Azzolin L, Quaranta E, Bodega B, Rosato A, Bicciato S, Cordenonsi M, Piccolo S. 2015. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat Cell Biol 17: 1218–1227. 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Bai H, David KK, Dong J, Zheng Y, Cai J, Giovannini M, Liu P, Anders RA, Pan D. 2010. The Merlin/NF2 tumor suppressor functions through the YAP oncoprotein to regulate tissue homeostasis in mammals. Dev Cell 19: 27–38. 10.1016/j.devcel.2010.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971. 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. 2010. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(β-TRCP). Genes Dev 24: 72–85. 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D, Conrad C, Xia F, Park JS, Payer B, Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al. 2009. Mst1 and Mst2 maintain hepatocyte quiescence and suppress hepatocellular carcinoma development through inactivation of the Yap1 oncogene. Cancer Cell 16: 425–438. 10.1016/j.ccr.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.