In this study Boskovic et al. set out to elucidate the functions of a small RNA derived from the 5’ end of mature tRNA-Gly-GCC. Using several genomic, biochemical, and molecular methods, the authors reveal a conserved mechanism for 5’ tRNA fragment control of noncoding RNA biogenesis and global chromatin organization.
Keywords: epigenetics, histones, tRNA fragment
Abstract
Small RNAs derived from mature tRNAs, referred to as tRNA fragments or “tRFs,” are an emerging class of regulatory RNAs with poorly understood functions. We recently identified a role for one specific tRF—5′ tRF-Gly-GCC, or tRF-GG—as a repressor of genes associated with the endogenous retroelement MERVL, but the mechanistic basis for this regulation was unknown. Here, we show that tRF-GG plays a role in production of a wide variety of noncoding RNAs—snoRNAs, scaRNAs, and snRNAs—that are dependent on Cajal bodies for stability and activity. Among these noncoding RNAs, regulation of the U7 snRNA by tRF-GG modulates heterochromatin-mediated transcriptional repression of MERVL elements by supporting an adequate supply of histone proteins. Importantly, the effects of inhibiting tRF-GG on histone mRNA levels, on activity of a histone 3′ UTR reporter, and ultimately on MERVL regulation could all be suppressed by manipulating U7 RNA levels. We additionally show that the related RNA-binding proteins hnRNPF and hnRNPH bind directly to tRF-GG, and are required for Cajal body biogenesis, positioning these proteins as strong candidates for effectors of tRF-GG function in vivo. Together, our data reveal a conserved mechanism for 5′ tRNA fragment control of noncoding RNA biogenesis and, consequently, global chromatin organization.
It has been known for some time that mature tRNAs can be cleaved in response to cellular stressors (Lee and Collins 2005), but only recently have the resulting cleavage products—broadly known as tRNA fragments or tRFs—been appreciated as potential regulatory molecules in their own right (Keam and Hutvagner 2015). Although tRNA fragments have in some cases been reported to function in complex with Argonaute proteins and thereby act analogously to microRNAs or endo-siRNAs (Deng et al. 2015; Martinez et al. 2017; Schorn et al. 2017), they have also been reported to have Argonaute-independent regulatory functions ranging from inhibition of translation to control of apoptosis (Elbarbary et al. 2009; Zhang et al. 2009; Ivanov et al. 2011; Couvillion et al. 2012; Gebetsberger et al. 2012; Sobala and Hutvagner 2013; Goodarzi et al. 2015; Molla-Herman et al. 2015; Kim et al. 2017). The diversity of proposed mechanisms for tRF function in part reflects the multitude of types of tRNA fragments that have been identified—22-nt fragments derived from the 3′ ends of mature tRNAs have been found to be associated with Argonaute proteins (Kumar et al. 2014; Kuscu et al. 2018) and have been suggested to direct cleavage of retrotransposon RNAs (Martinez et al. 2017; Schorn et al. 2017), whereas longer (28- to 32-nt) fragments arising from tRNA 5′ ends appear to play more diverse mechanistic roles. For instance, 5′ fragments of valine tRNAs serve as global repressors of translation in archaea, yeast, and mammals, and in some cases appear to act by interfering with translational initiation (Bąkowska-Żywicka et al. 2016; Gebetsberger et al. 2017; Guzzi et al. 2018; Luo et al. 2018).
We previously showed that, both in murine ES cell culture and in preimplantation embryos, interfering with a 5′ fragment of tRNA-Gly-GCC (hereafter, tRF-GG) using an antisense LNA oligonucleotide resulted in derepression of ∼50 genes associated with the long terminal repeat (LTR) of the endogenous retroelement MERVL (Sharma et al. 2016). This functional link between a tRNA fragment and LTR element control is particularly interesting given the ancient and widespread role for tRNAs in LTR element replication—tRNAs almost universally serve as primers for reverse transcription of LTR elements (Marquet et al. 1995)—as well as recent studies reporting that 3′ tRNA fragments can interfere with multiple stages of the LTR element life cycle (Deng et al. 2015; Martinez et al. 2017; Schorn et al. 2017). In the case of tRF-GG-mediated control of MERVL elements, however, we found no identifiable homology between the 5′ tRF-GG and either the LTR or the primer binding sequence of MERVL (which is primed by homology to leucine tRNAs), making it unlikely that MERVL regulation occurs through homology-directed RNA targeting.
Here, we set out to uncover the mechanistic basis for repression of MERVL-associated genes by tRF-GG. To our surprise, we found that control of MERVL elements is a downstream result of an evolutionarily conserved function for tRF-GG in supporting noncoding RNA production. Manipulation of tRF-GG levels in human and mouse ES cells affects the levels of a wide range of noncoding RNAs, including snoRNAs, scaRNAs, and various snRNAs, all of which require a subnuclear organelle known as the Cajal body for stability or function. One such RNA, the U7 noncoding RNA, is essential for 3′ UTR processing of histone pre-mRNAs. Accordingly, we show that tRF-GG control of U7 levels has downstream effects on histone mRNA and protein levels, with concomitant effects on chromatin compaction, and that the effects of tRF-GG on histones and on MERVL target gene transcription can be suppressed by manipulating U7 RNA levels. Finally, we identify the related proteins hnRNP F/H as direct binding partners for tRF-GG, and show that these RNA-binding proteins are required for normal Cajal body biogenesis and for repression of MERVL-driven gene expression. Taken together, our data reveal a novel pathway for tRNA fragment function in mammals, linking tRNA cleavage to regulation of noncoding RNA production.
Results
Chromatin-mediated repression of MERVL target transcription by tRF-GG
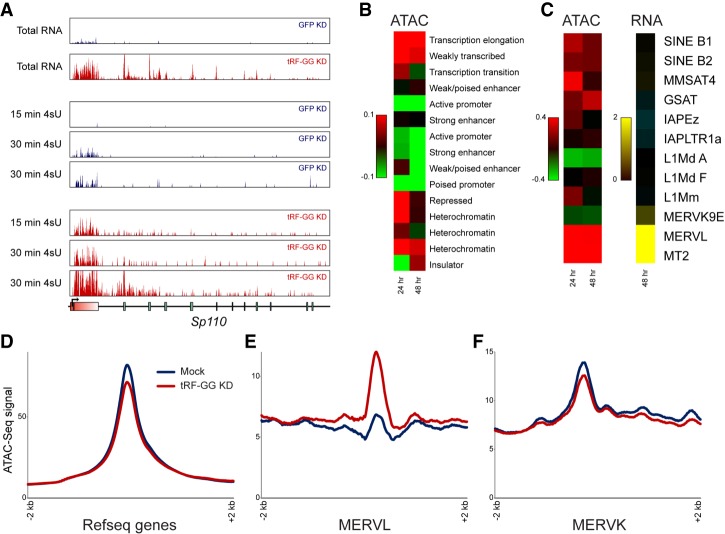
We recently identified a role for a 28-nt RNA derived from the 5′ end of tRNA-Gly-GCC—tRF-Gly-GCC, hereafter referred to as tRF-GG for brevity—as a repressor of genes associated with the long terminal repeat (LTR) of the endogenous retroelement MERVL (Sharma et al. 2016). This repressive activity does not appear to be a consequence of sequence homology between tRF-GG and the MERVL element, based on three observations: (1) There is no significant sequence homology between tRF-GG and MERVL (which uses leucine tRNAs to prime reverse transcriptase), and, in any case, many of the tRF-GG target genes are regulated by “solo” LTRs that have lost the MERVL primer binding sequence. (2) LNAs targeting the 3′ end of tRNA-Gly-GCC have no effect on MERVL target expression (Sharma et al. 2016). (3) Transfection of ES cells with various synthetic 3′ tRNA fragments of potential relevance to either tRF-Gly-GCC or to MERVL has no significant effect on MERVL target gene expression (Supplemental Fig. S1A). Moreover, our initial studies suggested that tRF-GG-mediated repression of MERVL occurs at the level of transcription rather than RNA stability (Sharma et al. 2016). Follow-up metabolic labeling studies using 4-thiouridine (Supplemental Table S1) confirm that tRF-GG inhibition (“tRF-GG KD”) affects target gene synthesis rather than mRNA stability (Fig. 1A; Supplemental Fig. S1B,C).
Figure 1.
tRF-Gly-GCC directs chromatin-mediated repression of MERVL-associated genes. (A) Metabolic labeling reveals transcriptional derepression upon tRF-GG inhibition. Genome browser tracks show total RNA levels, and newly synthesized RNAs obtained after 15 or 30 min of 4-thiouridine (4SU) labeling for ES cells transfected with esiRNAs targeting GFP, or with an LNA oligonucleotide antisense to tRF-GG. Effects of tRF inhibition on previously described MERVL-associated target genes (Macfarlan et al. 2012; Sharma et al. 2016) are nearly identical for total RNA as well as newly synthesized RNA (see also Supplemental Fig. S1B,C). (B) Increased accessibility at heterochromatin and weakly transcribed regions in tRF-GG-inhibited ES cells. Heat map shows log2 fold change in ATAC-seq reads following tRF-GG inhibition, aggregated across the indicated types of chromatin (Bogu et al. 2016). (C) As in B, with tRF-GG effects on ATAC-seq occupancy and RNA abundance averaged across the indicated repeat elements. (D–F) Examples showing average ATAC-seq signal across the indicated genomic elements: RefSeq genes (D), MERVL elements (E), or MERVK elements (F).
We therefore set out to uncover how tRF-GG directs transcriptional repression of MERVL-driven genes. Like many retroelements, MERVL LTRs are packaged into and repressed by heterochromatin (Macfarlan et al. 2011; Ishiuchi et al. 2015). To investigate the effects of tRF-GG on chromatin architecture genome-wide, we carried out ATAC-seq (Buenrostro et al. 2015) in mouse ES cells upon tRF-GG KD (Supplemental Fig. S1D,E). Consistent with the enhanced transcription observed at MERVL LTRs, we found that inhibition of tRF-GG resulted in a broad increase in chromatin accessibility over MERVL elements and throughout heterochromatin, with minimal changes in ATAC-seq signal over euchromatic transcriptional start sites (Fig. 1B–F; Supplemental Fig. S1D,E). tRF-GG control of chromatin accessibility was also observed in preimplantation embryos following injection with a synthetic tRF-GG oligonucleotide to mimic the process of sperm delivery of tRFs to the zygote (Supplemental Fig. S1F). Thus, tRF-GG manipulation alters chromatin accessibility in both mouse ES cells and in preimplantation embryos.
tRF-GG is a positive regulator of histone genes
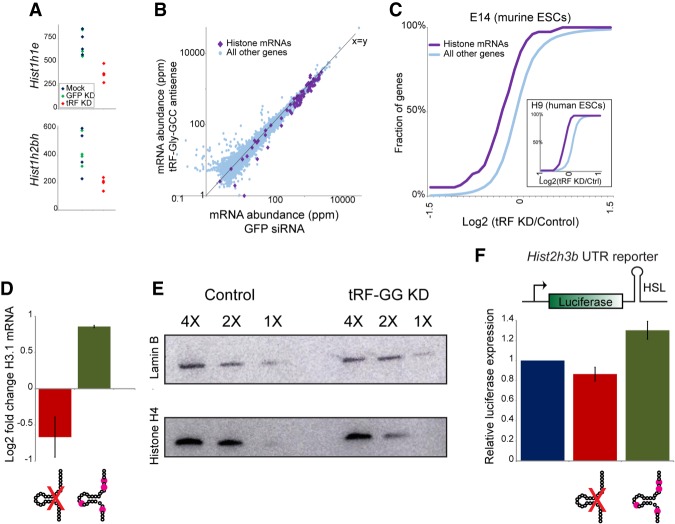
Given the species-specific genomic locations of many ERVs such as MERVL (Franke et al. 2017), we also extended our analyses to H9 human ES cells to identify conserved and divergent transcriptional consequences of tRF-GG function (Supplemental Tables S1, S2). Intriguingly, tRF-GG inhibition in human ESCs had minimal effects on HERV expression, indicating that (under our culture conditions) ERV regulation by this tRNA fragment is confined to mouse ES cells. Instead, we identified two conserved molecular phenotypes resulting from tRF-GG inhibition in both human and mouse ES cells: repression of histone mRNAs (Fig. 2A–C; Supplemental Fig. S2A–C), and decreased expression of a variety of noncoding RNAs such as snoRNAs and scaRNAs (see below). Given our finding of heterochromatin decompaction in tRF-GG-inhibited cells (Fig.1B), and the common derepression of ERV elements in undercompacted genomes (Lenstra et al. 2011; Ishiuchi et al. 2015), we focus first on tRF-GG effects on histone genes. We confirmed by qRT-PCR that tRF-GG inhibition causes a decrease in histone mRNA abundance (Fig. 2D), and that reduced histone mRNA levels are accompanied by a decrease in histone protein levels (Fig. 2E; Supplemental Fig. S2D–E). Importantly, we found that tRF “overexpression” via transfection of ES cells with a synthetic 28-nt tRF-GG also resulted in increased histone mRNA abundance (Fig. 2D), demonstrating that repression of histone genes observed in response to tRF-GG inhibition does not result from an off-target gain of function for our antisense LNA oligo. Together, our gain- and loss-of-function studies demonstrate that tRF-GG plays a conserved role in histone mRNA expression, with the MERVL LTR likely representing a sensitized reporter for chromatin assembly in murine ES cells (with the absence of HERV derepression in hESCs presumably reflecting either lower expression of the transcription factors that target HERV sequences under these growth conditions or the presence of additional redundant silencing mechanisms in hESCs).
Figure 2.
tRF-Gly-GCC represses expression of histone genes via the histone 3′ UTR. (A) mRNA abundance for two example histone genes—Hist1h1e (top) or Hist1h2bh (bottom)—in four replicate RNA-seq libraries from mock-transfected, GFP KD, and tRF-GG KD mES cells, as indicated. (B) Scatter plot comparing RNA abundance for histone genes (purple diamonds) and all other genes in GFP KD ES cells (X-axis) and tRF-GG-inhibited ES cells (Y-axis). Note that nearly all histone genes fall below the X = Y diagonal. (C) Cumulative distribution of the effects of tRF-GG inhibition on histone mRNA expression, with the Y-axis showing cumulative fraction of genes exhibiting any given log2 fold change in expression (X-axis). Main panel shows data from murine ES cells (n = 4 replicates, KS P = 7.7 × 10−5), while inset shows data for human ESCs. See also Supplemental Figure S2. (D) qRT-PCR for Hist2h3b showing effects of transfecting the anti-tRF-GG LNA, or a synthetic tRF-GG oligonucleotide (bearing most of the modified nucleotides expected from human tRNA-Gly-GCC) (Materials and Methods). (E) tRF-GG inhibition leads to decreased histone protein levels. Western blots probed for Histone H4 or loading control Lamin B, as indicated. See also Supplemental Figure S2D,E. (F) tRF-GG regulates histone 3′ UTR-mediated reporter expression. We generated stable ES cell lines carrying a luciferase reporter bearing the 3′ UTR of Hist2h3b (Supplemental Fig. S4A shows data for an independent cell line bearing the Hist1h4j 3′ UTR). Bar graph shows average changes to reporter activity in response to control KD, tRF-GG LNA (14% decrease, P = 0.038), or the modified tRF-GG oligo (30% increase, P = 0.0002).
What is the mechanistic basis for tRF-GG-mediated repression of the histone genes? Although histone expression is largely confined to the S phase of the cell cycle and could thus report on changes in cell cycle profile, FACS analysis of tRF-GG-inhibited ES cells revealed no change in the fraction of cells in S phase (Supplemental Fig. S3), while reanalysis of our RNA-seq data sets confirm that other S-phase-specific genes beyond the histones (Pcna, etc.) are not affected by tRF-GG inhibition or overexpression.
Perhaps the most unique feature of histone expression is the role of several cis-acting RNA elements in the histone 3′ UTR—a short stem loop known as histone stem loop (HSL) that binds to stem loop binding protein (SLBP), and the histone downstream element (HDE) that binds to the U7 noncoding RNA—in regulation of histone pre-mRNA processing (Dominski and Marzluff 1999; Marzluff and Koreski 2017). To separate the effects of tRF-GG manipulation on the histone 3′ UTR from effects on the histone promoter or coding sequence, we generated stable ES cell lines carrying luciferase reporters fused to one of two histone UTRs (Fig. 2F; Supplemental Fig. S4A). Transfection of synthetic tRF-GG drove increased luciferase activity (30%, P = 0.0002), while tRF-GG inhibition resulted in decreased luciferase levels (with values ranging from 14% to 32% in five separate experiments—each in at least triplicate—with P values ranging from 0.038 to 0.000019). tRF-GG inhibition had no effect on a stable ES cell line carrying the wild-type luciferase reporter (data not shown), and minimal effect on a reporter bearing mutations that compromise the histone stem loop (Supplemental Fig. S4A), indicating that a functional histone 3′ UTR is necessary to confer regulation. Moreover, loss of histone 3′ UTR reporter activity was specific to tRF-GG inhibition, as it was not observed in response to four other tRF-directed antisense LNA oligonucleotides (Supplemental Fig. S4B). Finally, consistent with the hypothesis that tRF-GG affects histone 3′ UTR processing, Northern blots in control and tRF-GG-inhibited ESC lysates confirm an increased abundance of misprocessed histone pre-mRNAs (Narita et al. 2007; Sullivan et al. 2009) in response to tRF-GG inhibition (Supplemental Fig. S4C). We conclude from these data that tRF-GG regulates histone mRNA abundance via the histone 3′ UTR.
tRF-GG affects histone expression and MERVL repression via control of U7 noncoding RNA
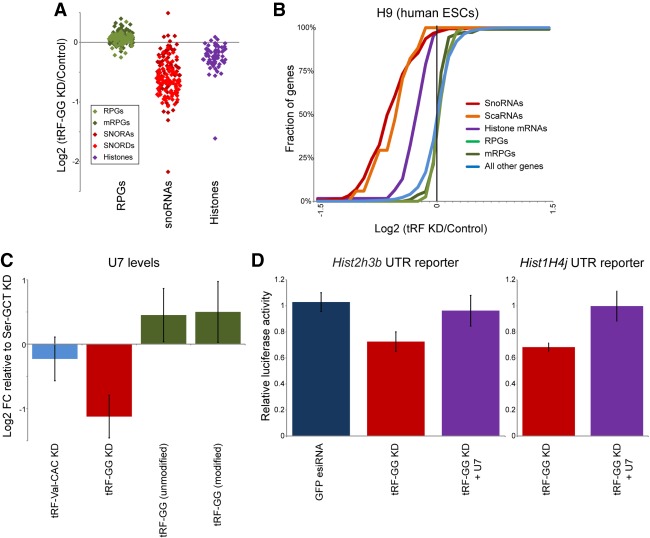
As mentioned, histone mRNA biogenesis involves a complex assembly of 3′ UTR-associated proteins, as well as the noncoding U7 RNA which directs UTR processing via base pairing to the HDE of the histone 3′ UTR (Marzluff and Koreski 2017). Intriguingly, in addition to down-regulation of histone genes, we noted that the other consequence of tRF-GG KD in both human and mouse ES cells was decreased expression of several major classes of noncoding RNA, including snoRNAs, scaRNAs, and, to a lesser extent, various spliceosomal ncRNAs (RNA-seq data shown in Fig. 3A,B; Supplemental Table S2; validation by qRT-PCR and Northern blots shown in Supplemental Fig. S5). Notably, all of these RNAs share a common biogenesis pathway with U7 snRNA, as they all require the subnuclear organelle known as the Cajal body for RNA processing, stability, or function (Wu and Gall 1993; Gall 2000; Machyna et al. 2013). To determine whether tRF-GG also affected levels of U7 RNA, we assayed U7 levels in tRF-GG KD and overexpression cells by Northern blotting (Fig. 3C; Supplemental Fig. S5C) and qRT-PCR (Supplemental Fig. S5B,E). Consistent with the effects of tRF-GG manipulation on other Cajal body RNAs, we found that inhibition of tRF-GG led to reduced U7 expression, while transfecting cells with the synthetic tRF-GG oligo supported higher expression of U7. Together, these findings reveal a conserved role for tRF-GG in promoting noncoding RNA production, and suggest that its effects on the histone 3′ UTR might result from altered U7 levels.
Figure 3.
tRF-Gly-GCC supports production of U7 and other noncoding RNAs. (A) Effects of tRF-GG inhibition on several gene families in human H9 ES cells. Individual dots show individual members of the indicated families, illustrating the widespread down-regulation of histone and snoRNA genes in response to tRF-GG inhibition. Ribosomal protein genes are shown as a representative highly expressed, but tRF-insensitive, gene family for comparison. Effects on several snoRNAs are independently validated by qRT-PCR and Northern blot in Supplemental Figure S5A,D,E. (B) Cumulative distribution plots showing tRF effects on the indicated gene families, as in Figure 2C. (C) Manipulating tRF-GG levels affects U7 snRNA production. ES cells were transfected either with LNA antisense oligos targeting tRF-Ser-GCT, tRF-Val-CAC, or tRF-GG, or with synthetic tRF-GG oligos either bearing appropriate modified nucleotides (modified) or lacking these modifications (unmodified). U7 levels were quantitated by Northern blot (n = 4) and normalized relative to 5S rRNA levels. Change in U7 levels is expressed relative to tRF-Ser-GCT inhibition (as an unrelated LNA control), revealing a significant (P = 0.03) decrease in U7 levels in response to tRF-GG inhibition, as well as modestly increased U7 levels in tRF-GG-supplemented cells. See also Supplemental Figure S5B,C,E. (D) Effects of tRF-GG KD on histone 3′ UTR reporters are suppressed by supplementation with additional U7 snRNA. ES cells were transfected with the LNA antisense to tRF-GG, with or without additional in vitro-synthesized U7 RNA. Effects of tRF-GG KD were significant (P = 0.0039 and 0.00013 for H3 and H4 reporters, respectively), while tRF-GG KD + U7 was statistically indistinguishable from control (P = 0.24 and 0.48, respectively). See also Supplemental Figure S6.
The hypothesis that tRF-GG control of U7 levels is responsible for changes in histone and MERVL expression makes the prediction that manipulating U7 levels should suppress tRF-GG effects on histone expression and on MERVL targets. We therefore transfected our reporter histone 3′ UTR mESC line with the anti-tRF-GG LNA—which results in decreased U7 levels—with or without supplementation of additional U7 RNA, and assayed luciferase activity and MERVL target gene expression. Restoring U7 levels in tRF-GG KD cells reversed the inhibition of histone expression in these cells as assayed by both luciferase reporters (Fig. 3D) and by qRT-PCR (Supplemental Fig. S6A), and this rescue activity was compromised by U7 mutations that interfere with base pairing between U7 snRNA and the histone HDE (Supplemental Fig. S6B). The converse also held true—antisense oligonucleotides directed against U7 were able to reverse the increase in histone levels in ES cells transfected with excess tRF-GG (Supplemental Fig. S6A). Importantly, restoring histone mRNA levels via U7 replenishment in tRF-GG-inhibited cells was able to partially suppress the transcriptional derepression of MERVL-linked genes as assayed both by qRT-PCR and using a MERVL-driven fluorescent reporter cell line (Supplemental Fig. S6C–E). Together, these data illuminate a pathway in which MERVL repression is downstream from tRF-GG-mediated histone expression, rather than being secondary to the tRF's effects on snoRNA or other noncoding RNA production.
tRF-GG is bound by hnRNPF/H in vitro and in vivo
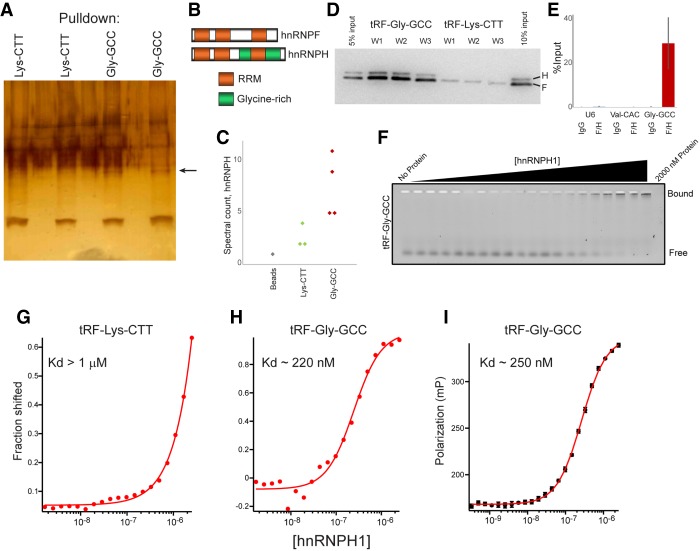
We next turn to the upstream question of how tRF-GG alters noncoding RNA production. To identify direct binding partners of tRF-GG, we used a 3′-biotinylated tRF-GG to isolate candidate tRF-binding proteins from mESC whole-cell extracts. tRF-GG, but not the unrelated tRF-Lys-CTT oligo, pulled down a protein of ∼50 kDa (Fig. 4A). From the potential binding partners enriched in tRF-GG pull-downs relative to the control (Supplemental Table S3), and based on follow-up functional studies of top candidates (below), we focus on the highly homologous pair of heterogeneous nuclear ribonucleoproteins (hnRNP) F and H (Fig. 4B,C). These RNA-binding proteins function redundantly in vivo, and will therefore be referred to below as “hnRNPF/H.” Using an antibody that detects both hnRNPF and hnRNPH, we validated the interaction between tRF-GG and hnRNPF/H by Western blotting—hnRNPF/H was robustly detected in tRF-GG pull-downs, with only modest hnRNPF/H levels detected following tRF-Lys-CTT pull-down (Fig. 4D). In addition to showing that tRF-GG can bind hnRNPF/H in cell extracts, we confirmed that hnRNPF/H binds to tRF-GG in vivo, as immunoprecipitation using the hnRNPF/H antibody revealed specific binding to tRF-GG but not tRF-Val-CAC or U6 snRNA (Fig. 4E).
Figure 4.
tRF-Gly-GCC binds to hnRNPF/H. (A) Biotin-oligo pull-downs from murine ES cell extracts. Silver-stained gel shows two replicates each for pull-downs using biotin-tRF-GG or biotin-tRF-Lys-CTT, as indicated. Arrow indicates ∼50-kDa band enriched in tRF-GG pull-downs. (B) Domain architecture of hnRNPF and hnRNPH1. (C) Mass spec peptide counts for hnRNPH in control, tRF-Lys-CTT, or tRF-GG pull-downs. (D) Western blots show hnRNPF/H recovery following tRF-GG or tRF-Lys-CTT pull-down. Pull-downs were washed four times for 3 min with 50 mM Tris (pH 8.0) supplemented with 100 mM (W1), 250 mM (W2), or 500 mM (W3) NaCl. (E) Enrichment of tRF-GG and two control RNAs in hnRNPF/H immunoprecipitates (and IgG controls) from mESCs, expressed as percent of input. Data show average and standard deviation from six data points: two biological replicates with three technical replicates each. (F) Gel shift analysis of hnRNPH1 binding to tRF-GG. A synthetic oligonucleotide corresponding to tRF-Gly-GCC (GCAJULGUGGUUCAGUGGDAGAAUUCUCGC) was labeled at the 3′ end using fluorescein 5-thiosemicarbazide, then incubated at 3 nM in equilibration buffer (0.01% Igepal, 0.01 mg/mL carrier tRNA, 50 mM Tris at pH 8.0, 100 mM NaCl, 2 mM DTT) for 3 h along with increasing concentrations of purified hnRNPH1 protein from 1.35 nM to 2000 nM. See also Supplemental Figure S7. (G,H) Fit of gel shift binding data for tRF-Lys-CTT and tRF-GG. Fitting the binding data yields an estimated Kd of hnRNPH1 of ∼220 nM for tRF-GG, and >1 μM for tRF-Lys-CTT. (I) Fluorescence polarization data for hnRNPH1 incubations with labeled tRF-GG. Polarization values against the protein concentrations are fit to the Hill equation using Igor Pro software.
To characterize the binding of hnRNPF/H to tRF-GG in more detail, we expressed and purified hnRNPH1 protein (Supplemental Fig. S7A), and carried out quantitative gel shift and fluorescence polarization analyses of hnRNPH1 binding to a fluorescently labeled tRF-GG oligonucleotide. Both assays revealed specific binding between hnRNPH1 and tRF-GG, in contrast to the nearly undetectable binding observed for tRF-Lys-CTT (Fig. 4F–I; Supplemental Fig. S7B). We note that the apparent Kd for hnRNPH1 binding to tRF-GG is roughly threefold weaker (Kd, app ∼250 nM) than that for a positive control—a previously described hnRNPF/H binding site identified in the SV40 pre-mRNA (Kd ∼80 nM) (Alkan et al. 2006). Thus, given that hnRNPF/H are abundant enough to regulate targets bound with 80 nM affinity, the apparent Kd of hnRNPH1 for tRF-GG falls within a plausible physiologically functional range.
hnRNPF/H are robust repressors of the MERVL gene expression program
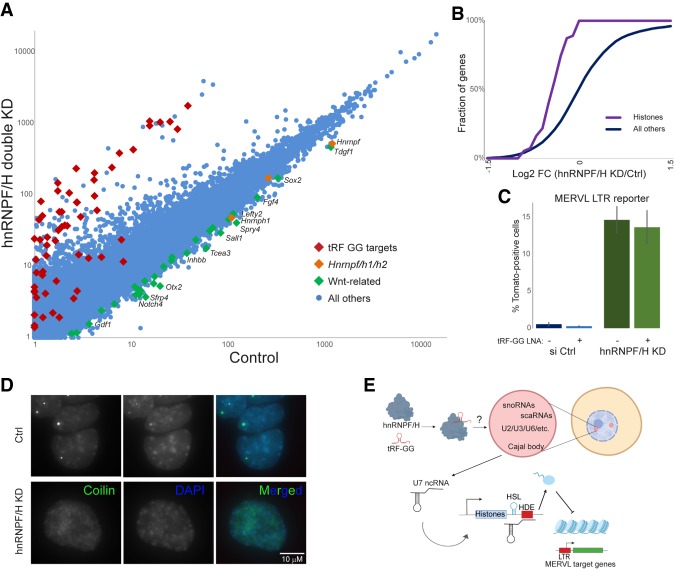
Do hnRNPF/H share any of the in vivo functions we identified for tRF-Gly-GCC? To determine potential roles for hnRNPF/H in histone gene regulation and MERVL repression, we carried out RNA-seq in ES cells subject to double knockdown of both hnRNPF/H. Double knockdown of hnRNPF/H resulted in dramatic alterations in expression of several hundred genes (Fig. 5A; Supplemental Table S4), including significant down-regulation of developmentally relevant genes (Sfrp4, Otx2, Dact2, Spry1/4, Gbx2, Notum, Notch4, Sall1, Fgf15, Tdgf1, Inhbb, Ltbp3, Fgf4, Pou4f2, Prdm14, Lefty2, Bmp4, etc.), consistent with Yamazaki et al. (2018). Importantly, we found that hnRNPF/H also affected the same groups of genes regulated by tRF-GG, with hnRNPF/H knockdown resulting in down-regulation of histone genes and a dramatic derepression of the MERVL program (Fig. 5A,B; Supplemental Fig. S8A). Using a MERVL TdTomato reporter ES cell line (Macfarlan et al. 2012), we found that hnRNPF/H KD led to an ∼30-fold increase in Tomato-positive cells (Fig. 5C; Supplemental Fig. S8B), confirming the derepression of the MERVL program observed in the RNA-seq data set. To our knowledge, hnRNPF/H thus represent the strongest repressors of the MERVL-positive “2C-like” state described to date, as the ∼30-fold derepression of the MERVL reporter in response to hnRNPF/H KD is comparable with, and in fact more dramatic than, that previously observed following Chaf-1 or Ubc9 knockdown (Ishiuchi et al. 2015; Cossec et al. 2018). Importantly, transfection of synthetic tRF-GG had no effect on MERVL repression in the absence of hnRNPF/H (Fig. 5C), consistent with the hypothesis that tRF-GG acts by binding hnRNPF/H.
Figure 5.
hnRNPF/H represses the MERVL program. (A) Changes in the ES cell transcriptome following hnRNPF/H knockdown. Scatter plot shows mRNA abundance compared between control KD cells (X axis) and hnRNPF/H KD cells (Y axis) (Supplemental Table S4). (B) hnRNPF/H KD results in histone mRNA down-regulation. Cumulative distribution plot shows log2 fold change (hnRNPF/H KD/Ctrl) for histone genes, and all other genes, as indicated. (C) hnRNPF/H suppresses ES cell entry into the MERVL-positive “2C-like state.” ES lines carrying a MERVL LTR-driven tdTomato (Macfarlan et al. 2012) were subject to control or hnRNPF/H KD (with or without synthetic tRF-GG), with bars showing mean ± standard deviation (n = 5 replicates) of the percentage of Tomato-positive cells. (D) hnRNPF/H is required for normal Cajal body morphology and gross chromatin architecture. Panels show typical images for the Cajal body marker coilin (green) and DAPI (blue), in control or hnRNPF/H KD ES cells. See also Supplemental Figure S8B. (E) Schematic of proposed mechanism for tRF-GG function. Our data suggest a model in which 5′ tRF-Gly-GCC supports production of a variety of noncoding RNAs in Cajal bodies, potentially downstream from binding to the hnRNPF/H proteins. Central to the current study is regulation of U7 snRNA production, which controls processing of the histone 3′ UTR via base pairing to the histone downstream element (HDE). Altered expression of histones then leads to downstream effects on the expression of MERVL-associated genes in murine embryonic stem cells and preimplantation embryos. The precise mechanism by which tRF-GG might support hnRNPF/H function remains to be elucidated; tRF-GG could stabilize hnRNPF/H leading to increased functional protein levels, or hnRNPF/H and tRF-GG could function together in a complex as depicted here.
A novel role for hnRNPF/H in Cajal body biogenesis and function
Given the role for tRF-GG in broadly supporting the output of multiple Cajal body products, we examined Cajal body morphology in hnRNPF/H KD cells, using the well-known Cajal body marker coilin (Gall 2000). Remarkably, in contrast to control ES cells that exhibit one or two bright Cajal bodies per nucleus, we found that knockdown of hnRNPF/H leads to more diffuse coilin staining (Fig. 5D; Supplemental Fig. S8B,C). Moreover, DAPI staining was clearly distinctive in hnRNPF/H KD cells, with the typical discrete chromocenters being replaced by more diffuse “lumpy” DAPI staining (Fig. 5D), potentially secondary to altered histone expression in these cells. Our data reveal a novel role for hnRNPF/H in supporting normal Cajal body function in mouse embryonic stem cells, with downstream consequences for histone expression and chromatin-mediated repression of MERVL elements.
How do hnRNPF/H function in Cajal body biogenesis and/or stabilization? hnRNPF/H are well-established splicing regulators, binding to G runs in pre-mRNA and thereby modifying local exon inclusion/skipping as well as more subtle aspects of splicing such as splice site selection (Xiao et al. 2009). To identify direct targets of hnRNPF/H in mouse ES cells, we carried out CLIP-seq (Zarnegar et al. 2016) using the antibody that recognizes both paralogs. Relative to IgG control, the four replicate F/H CLIP data sets exhibited dramatic enrichment for binding to pre-mRNA introns (Supplemental Table S5). F/H targets were enriched for genes encoding a wide range of RNA-binding proteins, with enriched GO annotations including ribosome biogenesis, mRNA binding, and mRNA splicing (Supplemental Fig. S9). Target genes included those encoding several known components of Cajal bodies, including Snrnp70 and Tardbp, as well as a number of other genes recently shown to affect Cajal body formation in a systematic knockdown screen (Berchtold et al. 2018). Consistent with prior studies in human HEK 293T cells (Xiao et al. 2009), analysis of splicing changes in mRNA-seq data from control and hnRNPF/H KD mouse ES cells confirmed that hnRNPF/H influences splicing of many of its directly bound target genes (Supplemental Fig. S10). Taken together, our binding and splicing analyses extend prior studies of hnRNPF/H function and identify a number of candidate target genes with the potential to affect Cajal body formation as a secondary consequence of hnRNPF/H-regulated splicing changes.
Discussion
Together, our studies outline a novel mechanism of action for an abundant tRNA fragment (Fig. 5E).
tRF-Gly-GCC controls histone production via its positive role in Cajal body output
Our data demonstrate that the effects of tRF-GG manipulation on MERVL target genes are mediated by MERVL transcriptional derepression as a result of altered histone gene expression. We document effects of tRF inhibition and tRF “overexpression” on histone expression by qRT-PCR, by deep sequencing, and by quantitative Western blot; we confirm the expected downstream effects on chromatin compaction by ATAC-seq; and finally we show that tRF regulation of histone genes can be recapitulated using two distinct histone 3′ UTR reporters. These data argue that, rather than being directly targeted by tRF-GG via sequence homology between the small RNA and the ERV's primer binding sequence, MERVL instead represents a highly sensitive reporter of global chromatin status in mouse embryonic stem cells. Further supporting this idea is the fact that histone down-regulation, rather than ERV derepression, is a conserved consequence of tRF-GG inhibition in both mouse and human ES cells.
Upstream of the histone 3′ UTR, we document a surprising and conserved role for tRF-GG as a positive regulator of noncoding RNAs that are normally synthesized or processed in the Cajal body. This is demonstrated using both gain- and loss-of-function approaches in both mouse and human ES cells, revealing control of Cajal body output to be a conserved function of tRF-GG in mammals. Although Cajal body-associated ncRNAs play roles in a wide range of biological processes that could be impacted by tRF-GG activity, we show that the key target of tRF-GG in control of MERVL output is the U7 snRNA that is essential for histone 3′ UTR processing. Importantly, not only are U7 levels affected by perturbing tRF-GG activity, but the effects of tRF-GG manipulation on both histone expression and on MERVL derepression can be suppressed by appropriate manipulation of U7 snRNA levels—providing additional U7 to tRF-GG KD cells, which produce less U7, suppresses the decrease in histone mRNA levels, the loss of histone UTR reporter activity, and, most importantly, partially suppresses the up-regulation of MERVL target genes in these cells. Taken together, our results strongly support a mechanism in which tRF-GG support of Cajal body output ensures sufficient U7 production to supply enough histones to fully repress endogenous retroelements in the genome.
hnRNPF/H are novel regulators of the MERVL gene expression program
What is the proximate mechanism by which tRF-GG supports production of Cajal body RNAs? We identify two strong candidates for the relevant effector protein(s), finding that the closely related hnRNPF and H proteins bind directly to tRF-GG both in vitro and in vivo. Moreover, functional studies reveal that hnRNPF/H and tRF-GG exhibit heavily overlapping regulatory roles in vivo, identifying a novel and surprising role for hnRNPF/H in control of the MERVL program in mES cells. Indeed, hnRNPF/H represent the strongest repressors of the so-called “2C-like state” yet observed (Ishiuchi et al. 2015; Cossec et al. 2018), with hnRNPF/H KD leading to a ∼30-fold derepression of a MERVL-driven reporter construct.
hnRNPF/H are RNA-binding proteins with well-characterized roles in mRNA splicing (Wang et al. 2007, 2012; Xiao et al. 2009; Yamazaki et al. 2018), and our RNA-seq analysis of hnRNPF/H KD cells reveals hundreds of altered splicing events (see Supplemental Fig. S10 for examples), consistent with previous studies (Xiao et al. 2009; Yamazaki et al. 2018). How do these changes in RNA splicing—or, alternatively, some unrelated activity of hnRNPF/H—ultimately drive repression of the MERVL program? Given the dramatic changes in Cajal body morphology documented in hnRNPF/H KD cells (Fig. 5D), we favor the hypothesis that one or more hnRNPF/H-regulated transcripts play a key role in Cajal body function. Compromised Cajal body function would affect production of the U7 snRNA, with downstream effects on histone production subsequently driving changes in the highly heterochromatin-sensitive MERVL program. A number of specific target(s) of hnRNPF/H could be responsible for supporting normal Cajal body biogenesis, as a large number of genes encoding chromatin and RNA-binding proteins with potential roles in Cajal bodies (such as Snrnp70, Tardbp, and many other RNA regulators) exhibit altered splicing patterns in hnRNPF/H-depleted cells.
It remains to be determined whether tRF-GG affects Cajal body output as a consequence of binding to hnRNPF/H—although tRF-GG and hnRNPF/H affect overlapping processes, it is clear that tRF-GG only affects a small subset of hnRNPF/H functions as, for example, we did not observe any effect of tRF-GG inhibition on Cajal body morphology or number, in contrast to the dramatic alterations in Cajal bodies seen in response to hnRNPF/H KD (Fig. 5D). Moreover, scrutiny of direct hnRNPF/H binding to target genes following tRF-GG inhibition did not identify reproducible effects of tRF-GG on hnRNPF/H activity in vivo (Supplemental Figs. S9, S10). Thus, although tRF-GG and hnRNPF/H can interact in vivo and in vitro (Fig. 4), and regulate expression of the same target genes downstream from Cajal body ncRNAs (Fig. 5A,B), our only direct evidence that tRF-GG functions through hnRNPF/H is the inability of tRF-GG to affect MERVL targets following hnRNPF/H KD (Fig. 5C). Therefore, at this point we consider hnRNPF/H a promising target, but not a definitive partner, involved in tRF-GG regulation of noncoding RNA production. Ultimately, it may require development of a separation-of-function mutation in hnRNPF/H, preventing tRF-GG binding while still allowing normal Cajal body production, to definitively show that tRF-GG acts through these proteins. Should tRF-GG act through these proteins, it will also be important in future studies to determine the mechanism by which tRF-GG enhances the function of hnRNPF/H—whether tRF-GG stabilizes hnRNPF/H, for example, or whether hnRNPF/H functions in complex with tRF-GG at a subset of targets.
Implications for tRF-Gly-GCC function in the early embryo
We finally turn to the question of the physiological contexts in which tRF-GG is likely to play an important role in cellular function. In typical somatic tissues, tRNAs are cleaved in response to a variety of stress conditions; for example, tRNA cleavage occurs in response to arsenite treatment of neurons to produce tRNA fragments that help to direct global translational down-regulation (Ivanov et al. 2011). Beyond the established case of stress-dependent tRNA fragment production, it is increasingly clear that tRNA cleavage also occurs commonly in the germline of multiple organisms even under apparently stress-free growth conditions (Couvillion et al. 2010; Peng et al. 2012), and tRF-GG is one of the most abundant small RNAs present in mammalian sperm and delivered to the zygote upon fertilization. The control of histone production by tRF-GG is particularly interesting in the context of preimplantation development, as studies in multiple species reveal that the onset of embryonic genome activation is controlled by histone dosage—embryonic transcription is suppressed by an excess of histones, only to be released from repression once enough copies of the genome have been produced to soak up the excess of histones (Amodeo et al. 2015; Wilky et al. 2019). Delivery of tRF-GG by sperm at fertilization therefore has the potential to accelerate or decelerate the initiation of embryonic transcription. It will be interesting in future studies to explore potential long-term consequences of such relatively subtle early changes in development, and to identify any other potential effects of tRF-GG delivery on other cellular processes controlled by Cajal bodies, from ribosome biogenesis (snoRNAs) to splicing (scaRNAs and U RNAs) to global chromatin compaction (U7 RNA).
Together, our findings reveal a novel function for an abundant representative of an understudied class of regulatory RNAs, and suggest that this tRF may have wide-ranging and pleiotropic effects during early development and in other biological contexts.
Materials and methods
Mouse ES cell culture and transfections
All murine ES cell lines were grown in DMEM (Gibco) containing 10% fetal bovine serum and leukemia inhibitory factor (serum + LIF culture conditions) and all transfections were carried out using OptiMEM and Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions at splitting, unless otherwise specified. Inhibition of tRF-Gly-GCC function in mESCs was performed as described in Sharma et al. 2016. Controls included Lipofectamine 2000 only (Mock), anti-GFP esiRNA transfections and/or a scrambled anti-tRF-GG LNA oligo. Transfection of various 3′tRFs (Supplemental Table S6) was performed as for the 5′tRF-GG. Concentrations tested were 5 ng and 100 ng.
U7 rescue experiments were performed by supplementing the tRF-Gly-GCC inhibition transfection reaction with 50 ng of in vitro synthesized U7 snRNA per 3 × 105 cells. Capped mouse U7 small nuclear RNA was generated through in vitro transcription from pGEM-Teasy-U7 plasmid (generous gift from Z. Dominski) using mMessage mMachine T7 kit (Ambion) following plasmid linearization with HindIII restriction enzyme. Different U7 concentrations were tested (ranging from 5 to 500 ng/3 × 105 cells) to determine the physiologically relevant range, which was narrowed to 25–100 ng/3 × 105 cells and the middle value of 50 ng was chosen for subsequent experiments. Human U7 was cloned from hESC cDNA into the pGEM-Teasy plasmid and used as in mouse experiments. Medium was changed after 16 h and cells were allowed to grow for additional 32 h. Cells were processed for various experiments at the end of the 48-h period. U7 knockdown was achieved by transfecting 10 ng of modified antisense oligonucleotides targeting U7 (Supplemental Table S6) synthesized with phosphorothioate linkages.
Double hnRNPF/H knockdown was performed by transient transfection of 20 pmols of siRNAs against mouse hnRNPF and hnRNPH transcripts, respectively (Santa Cruz Biotechnology, sc 37571, sc-38273, and sc-35580) in 12-well format. Forty picomoles of siRNA-A (Santa Cruz Biotechnology, sc-37007) was used as knockdown control. Efficiency of knockdown was validated by Western Blotting and RNA-sequencing. Cells were collected 48 h after transfection for various downstream experiments.
Cell culture (H9)
Line H9 (WA09) human embryonic stem cells were cultured on Matrigel (Corning) in mTESR1 medium (Stem Cell Technologies) in 5% CO2 at 37°C. Nucleofection of oligos was done using human stem cell nucleofector kit 1 (Lonza Bioscience), according to the manufacturer's instructions. Two nanograms of LNA was nucleofected by a 24-well plate. Nucleofection efficiency was checked using pEGFP plasmid control, with successful experiments having more than 80% GFP+ cells. Cells were harvested 12 h after nucleofection for RNA-seq and qRT-PCR.
Metabolic labeling
E14s were labeled in 500 mM 4sU-containing medium for 15 or 30 min, and then RNA was isolated using Trizol and isopropanol precipitation. Fifty micrograms of total RNA was mixed with 0.2 mg/mL EZ-Link Biotin-HPDP (Themo Fisher) in a 500-μL reaction, then incubated for 2 h at 37°C in a shaking thermomixer (750 rpm). Biotinylated RNA was then extracted using phenol:chloroform:isoamyl alcohol (PCI) with phaselock gels and precipitated using isopropanol. RNA pellet was resuspended in 10 μL of water and mixed with 30 μL of washed Dynabeads MyOne Streptavidin C1 beads (Invitrogen) in binding buffer (10 mM Tris-HCl at pH 7.5, 300 mM NaCl, 0.1% Triton-X). The slurry was rotated for 20 min at room temperature to immobilize biotin-tagged RNA, then placed on a magnetic stand and washed with 500 μL of high-salt buffer (50 mM Tris-HCl, 2 M NaCl, 0.5% Triton-X). The supernatant from the first high-salt wash is the unlabeled total RNA. Beads were then stringently washed in high-salt buffer, then two times in binding buffer, and then once in low-salt buffer (5 mM Tris-HCl, 0.1% Triton-X). The biotin-tagged RNA were extracted from the beads with 100 mM DTT twice for 5 min at 65°C. Finally, labeled and unlabeled fractions were PCI-extracted and RNA was isopropanol-precipitated and used to construct RNA-seq libraries.
RNA-seq
Five micrograms of total RNA was depleted of ribosomal RNA using Ribo-Zero rRNA removal kit (human, mouse, rat, Illumina). Less total RNA was used as input from metabolic labeling experiments for both 4sU labeled and unlabeled fractions. Illumina deep sequencing compatible libraries were constructed from rRNA-depleted RNA using an optimized version of a protocol described by Heyer et al. (2015), adding a purification using the RNA Clean and Concentrator (Zymo Research) in between procedures. Ribosome profiling data was published previously (GSE74537), with libraries constructed using the same procedure. Libraries were quantified, multiplexed and either single-end or paired-end sequenced on Illumina NextSeq 500 sequencer.
ATAC-seq
E14 mES cells were transfected with antisense tRF-Gly-GCC LNA or mock-transfected and grown for 24 or 48 h prior to harvesting and counting. ATAC-seq protocol was done essentially the same as described in Buenrostro et al. (2015). Briefly, after titration, 4 μL of TDE1 was determined as sufficient for 50,000 cells. Tagmented DNA was amplified using Kapa HiFi Hotstart polymerase, and libraries were cleaned up using Ampure XT DNA beads. Libraries were quantified, multiplexed, and paired-end sequenced on an Illumina NextSeq 500 sequencer.
Deep sequencing data analysis
RNA-seq libraries were demultiplexed using Novobarcode (v3.02.08). Single end libraries were trimmed of 3′ adapters using Fastx-toolkit (v0.0.14). Quantification was done using RSEM (v1.2.29) to RefSeq GTF annotation, mapped with Bowtie (v1.0.0) to mm10 using default parameters.
ATAC-seq libraries were mapped to mm10 using Bowtie2 (v2.3.2) with the following parameters: -D 15 -R 2 -N 1 -L 20 -i S,1,0.50 –maxins 2000 –no-discordant –no-mixed. Fragment lengths were separated using Python, and coverage of reads in various chromatin states was analyzed in R (v3.4.1) using data from ChromHMM (https://github.com/guifengwei/ChromHMM_mESC_mm10). All coverage data was normalized by global read depth prior to further analysis. Circos plots (v0.69-2) were generated from coverage data calculated by Bedtools (v2.25.0).
irCLIP-seq libraries were demultiplexed and adapters trimmed as in RNA-seq libraries. Reads shorter than 18 nt were excluded from analysis. Reads were mapped to mm10 using the splice-aware STAR-aligner (V2.5.3a) along with the transcriptome GENCODE VM20, and uniquely mapped reads were extracted using samtools (V1.3). FPKM per Refseq gene was counted using RSEM (v1.2.29). Peaks were called using HOMER (V4.9), using “factor” peaking finding algorithm specifying 30-nt peak sizes, with FRD < 0.00001.
qRT-PCR and semiquantitative PCR
For mESCs, RNA was isolated using Trizol (Ambion) according to the standard protocol and the samples were treated with TurboDNase to eliminate genomic DNA contamination. Following TurboDNase treatment, RNA was purified using Zymo RNA Clean and Concentrator 5 kit, quantified on Nanodrop and RT reaction was performed using SuperScript IV RT kit (Invitrogen) according to the manufacturer's protocol. Obtained cDNA was diluted 2× for MERVL target gene qPCR, and 10× for histone and β-actin amplification (based on the standard curves obtained for the primers used) and Kapa SYBR Fast Universal Mastermix was used in all qRT-PCR reactions. The amplification conditions were 3 min at 95°C followed by 40 cycles of 5 sec at 95°C and 20 sec at 60°C (with a plate-reading step between each cycle) based on the previously reported conditions for histone qPCR. All the reactions were read on the BioRad CFX96 qPCR detection machine. The same protocol, with human-specific primers, was performed for H9 human cells.
For semiquantitative PCRs to confirm candidate alternative splicing events, primers were designed in flanking exons. The number of PCR cycles were empirically determined to avoid saturation of PCR amplicons.
Western blotting
Mock or tRF-GG inhibited cells were grown as before and 48-h after transfection, cells were trypsinized and counted. A defined number of cells (usually 50,000) was spun from each group, washed in PBS, pelleted, and lysed directly by boiling at 100°C for 15 min in 2× Laemmli sample buffer containing β-mercaptoethanol. Serial dilutions (2×) of the cell lysates were loaded onto a 15% SDS–polyacrylamide gel. Following protein resolution on the gel, proteins were transferred to a nitrocellulose membrane for 1 h at 100 V through wet transfer. Membrane was blocked in 5% milk in TBSt for at least an hour prior to the addition of primary antibodies overnight at 4°C. Primary antibodies used were antihistone H3 (Millipore, #07-690), antihistone H4 (Millipore, #05-858), anti β-actin (Abcam ab8224), anti-GAPDH (Abcam ab9485), and anti-Lamin B (Santa Cruz Biotechnologies sc-6217). Membranes were then washed 3X in TBSt for 15 min at room temperature. Membrane was incubated with secondary antibody in 5% milk for 1 h at room temperature. Secondary antibodies used were antimouse IgG, HRP-linked (Cell Signaling 7076S) and antirabbit IgG, HRP-linked (Cell Signaling 7074S). Following three additional TBSt washes, the substrate for HRP was added (Amersham ECL Western blotting analysis system, GE) and the blots were exposed on the Amersham Imager 600 machine.
Northern blotting
DNA probes for snoRNAs were ordered from IDT (Supplemental Table S6) and 5′ end-labeled with [γ-32P] using PNK (New England Biolabs). U7 was present in very low abundance and had to be probed using a full-length [γ-32P]C-labeled RNA probe synthesized from a pGEM plasmid containing a cloned mouse U7. Two micrograms of total RNA was separated on a 6% urea PAGE gel. RNA was transferred onto a Hybond-N+ membrane (GE Healthcare) by semidry transfer method (Bio-Rad) in SSC buffer. RNA was then cross-linked to the membrane at 254 nm. Probes were then hybridized to RNA overnight with agitation at 68C. Membranes were then washed and exposed to BioMax film (Kodak) for up to 3 d. Data was quantified using ImageJ.
Histone Northern blots
5′tRF-GG knockdown and RNA isolation from E14 mESCs was performed as above. From total RNA, polyadenylated RNA was isolated using PolyATtract mRNA Isolation System IV (Promega). Denatured poly A (+) RNA (0.4 μg per lane) was resolved on a 6%TBE gel and transferred onto Hybond-N+ membrane. Following transfer, membrane was UV cross-linked (twice at 0.12 J) and prehybridization buffer (7% SDS, 200 mM Na2HPO4 at pH 7.0, 5 μg/mL salmon sperm DNA [SSDNA]) was added to the membrane for a minimum of 2 h at 37°C. The prehybridization buffer was then removed, and hybridization buffer containing 50 ng of biotin-labeled probes against histone H3 coding sequence was added. Probe was generated through asymmetric PCR using biotinylated dCTP in dNTP mix. The biotin-labeled probe did not require denaturation. The membrane was hybridized for 12–16 h at 37°C with gentle shaking and subsequently rinsed with washing buffer (1× SSC, 0.1% SDS). Membrane was then briefly blocked for 15 min in EDTA-containing washing buffer (1× SSC, 0.1% SDS, 1 mM EDTA) followed by a 20-min incubation with washing buffer containing High Sensitivity Streptavidin-HRP (Pierce, 1:10000). Next, the membrane was washed with 10%, 1%, and 0.1% SDS in PBS (10 min/wash). The substrate for the peroxidase was added and biotin-labeled probes were detected using Amersham Imager 680.
Embryo microinjection studies
Zygotes were generated through IVF as described previously (Sharma et al. 2016). After IVF, embryos were washed and placed in KSOM for culturing for 2 h to recover. Following this, zygotes were washed twice in M2 medium and microinjected with (1) control (H3.3-GFP mRNA at 100 ng/μL) and (2) tRF-modified (H3.3-GFP mRNA+ mod-tRF-GG at 200 ng/μL). After micromanipulation, zygotes were placed back in KSOM for overnight culture, and the following day (24 h after IVF) fluorescence was checked in two-cell stage embryos. GFP-positive embryos were washed twice in M2 medium and their zona pellucida was removed by acid tyrode solution. Embryos were then neutralized by two washes in M2, and mouth-pipetted into 5 μL of ATAC-lysis buffer (containing 1% Triton X, without NP-40) on ice. Fifteen microliters of tagmentation reaction mix was added (11.25 μL of TD buffer, 1.5 μL of TDE1, 2.25 μL of H2O), and the reaction was incubated for 30 min at 37C in a thermoblock with intermittent shaking. Tagmentation reaction was stopped by the addition of EDTA (25 mM final) and incubation for 10 min at 50°C. Prior to PCR amplification, reaction was supplemented with MgCl2 and the rest of the protocol was performed as described for ATAC-seq in cultured cells. After amplification, libraries were quantified by Qubit DNA high sensitivity assay, concentration was adjusted between control and experimental groups, and qPCR was performed using Kapa SYBR-FAST Universal qPCR mix on Bio-Rad CFX96 real-time system. Amplification conditions were 3 min at 95°C (initial denaturation), followed by 40 cycles of 10 sec at 95°C, 15 sec at 60°C, and 15 sec at 72°C. The efficiency of primers for MERV-L and tubulin was validated using previously sequenced ATAC-seq libraries generated from mouse embryonic stem cells.
Histone 3′ UTR luciferase reporter assay
Histone H3b (Hist2h3b) and histone H4j (Hist1h4j) 3′ UTR sequences (∼300 nt) were cloned into the PsiCheck 2 vector downstream from the Renilla luciferase coding sequence, between XhoI and NotI restriction sites. As PsiCheck2 vector does not encode for a eukaryotic selectable marker, cells were cotransfected with PsiCheck2-empty or PsiCheck2-Histone3′UTR together with a carrier plasmid pCDNA3.1+ Hygro and stable cell lines were selected. Following 7 d of Hygromycin selection, individual clones were picked, expanded, and tested for luciferase expression. Based on their expression level, one clone from all cell lines was selected for subsequent experiments. Reporter ES cell lines were transfected as previously. Forty-eight hours after transfection, cells were washed twice with PBS and cell lysate for the luminescence reading was prepared as directed by the dual-luciferase assay system (Promega). Firefly and Renilla luminescence was measured using GLOMAX96 Microplate Luminometer, and Renilla luminescence was normalized to the internal control of firefly luminescence.
Cell cycle analysis
E14 mESCs were transfected with antisense 5′tRF-Gly-GCC LNA-containing oligo or mock transfected as described above. Medium was changed 16 h after transfection. After 24 h, cells were synchronized in G1/S phase of the cell cycle by single thymidine block (5 mM thymidine in culture medium) for 16 h. Following treatment, cells were washed twice with PBS, and fresh culture medium without thymidine was added and were allowed to progress through the cell cycle. Cells were collected by trypsinization and 2X PBS washes at time 0 and every 2 h for an 8-h period after removal of the thymidine block. Cold 70% ethanol was added to cells dropwise with light vortexing, and cells were fixed for 30 min at 4°C. Following fixation, cells were pelleted, washed twice with PBS, and treated with RNase A (final concentration 0.2 mg/mL). Cells were stained with propidium iodide solution (final concentration 10 mg/mL) and DNA content was analyzed using FACScan flow cytometer (Becton Dickinson).
Oligonucleotide pull-downs and mass spectrometry
Mouse ES cells were washed twice with PBS and collected by trypsinization. Around 20 million cells were lysed in NP-40 buffer (50 mM Tris at pH 7.5, 0.1% NP-40, 150 mM NaCl, 1 mM CaCl2, 1 mM DTT, 40 units SuperaseIn, 1× Protease inhibitor cocktail) for 15 min on ice. Lysates were cleared by centrifugation for 15 min at 4°C and maximum speed and the supernatant was transferred to a new tube. The lysates were then divided in two and incubated with 500 pmol of tRF-Gly-GCC-biotin or tRF-Lys-CTT-biotin for 1 h at room temperature with end-over-end rotation. Following the incubation, 100 μL of C1 Dynabeads were added to the reactions and incubated with end-over-end rotation for an additional hour. Captured RNPs were then washed three times each with low-salt buffer (30 mM Tris-HCl at pH 7.5, 120 mM KCl, 3.5 mM MgCl2, 0.5 mM DTT), medium salt buffer (30 mM Tris-HCl at pH 7.5, 300 mM KCl, 3.5 mM MgCl2, 0.5 mM DTT) and high-salt buffer (30 mM Tris-HCl at pH 7.5, 0.5 M KCl, 3.5 mM MgCl2, 0.5 mM DTT) for 5 min at room temperature. Following the last wash, elution was performed by disrupting the biotin-streptavidin bond using 95% formamide and 10 mM EDTA at 65°C for 5 min. Eluate was boiled in 2X Laemmli sample buffer and loaded into 4%–20% gradient polyacrylamide gel. Gels were stained by Coomassie Brilliant Blue solution for 1 h at room temperature, then destained overnight in the destaining solution (40% MeOH, 10% acetic acid). Bands were cut from the gel and submitted for mass spectrometry at the University of Massachussetts Medical School Mass Spectrometry Facility.
Streptavidin RNA pull-down assay
For each assay, 2.5 μM of biotin-labeled RNA was incubated with streptavidin beads (Invitrogen) according to the manufacturer's instructions. Beads were then incubated for 2 h with cellular lysate in binding buffer (0.01 mg/mL tRNA, 0.01% NP-40, 0.1 mg/mL BSA, 50 mM Tris-Cl at pH 8.0, 100 mM NaCl, 1/20 SuperaseIn [Ambion]). After 2 h of rotation at room temperature, the beads were washed with 200 μL of wash buffer (100 mM NaCl, 50 mM Tris-Cl at pH 8.0, 0.01% NP-40, 0.01 mg/mL tRNA) four times. Proteins were eluted from beads with sample buffer for 5 min at 95°C and equal amounts are run on an SDS–polyacrylamide gel and analyzed by Western analysis.
hnRNPH1 purification
The sequence encoding amino acids 1–449 of mouse hnRNPH1 was cloned into pMal-ac (New England Biolabs) downstream from an N-terminal maltose-binding protein (MBP) tag and the cloned construct was transformed into BL21(DE3) cells. The cells were induced with 1 mM isopropyl 1-thio-β-D-galactopyranoside for 3 h at 37°C. to express the protein with an N-terminal MBP tag. The cells were lysed in 200 mM NaCl, 50 mM Tris (pH 8.8), 2 mM DTT, and EDTA-free protease inhibitor tablet. Amylose (New England Biolabs) affinity column was used for the first step of purification of hnRNPH1. Protein fractions were eluted in lysis buffer supplemented with 10 mM maltose. Fractions containing the protein were pooled and dialyzed into an S-column buffer (20 mM NaCl, 50 mM MOPS at pH 6.0, 2 mM DTT). Purification was followed by HiTrap S at 4°C. Elution of the protein fractions was achieved by a salt gradient ranging from a low-salt buffer (20 mM NaCl, 50 mM Tris MOPS at pH 6.0, 2 mM DTT) to a high-salt buffer (1 M NaCl, 50 mM MOPS at pH 6.0, 2 mM DTT). Pure fractions were dialyzed in a Q-column buffer (20 mM NaCl, 50 mM Tris at pH 8.8, 2 mM DTT). Final purification was done using a HiTrap Q ion exchange column at 4°C. Protein fractions were eluted by a salt gradient ranging from a low-salt buffer (20 mM NaCl, 50 mM Tris at, pH 8.8, 2 mM DTT) to a high-salt buffer (1 M NaCl, 50 mM Tris at pH 8.8, 2 mM DTT). Pure fractions were determined by Coomassie-stained SDS-PAGE and purified hnRNPH1 was dialyzed into storage buffer (25 mM Tris at pH 8.0, 25 mM NaCl, 2 mM DTT) and stored at 4°C. Pure fractions were concentrated using an Amicon spin concentrator.
Preparation of fluorescently labeled RNA
RNA oligonucleotides were 3′ end-labeled with fluorescein 5-thiosemicarbazide as previously described (Pagano et al. 2011). Briefly, RNA is first oxidized with sodium periodate and then reacted it with fluorescein 5-thiosemicarbazide to form a covalent bond. Labeled RNA is then purified over a Sephadex G25 column.
Electrophoretic mobility shift assay
Electrophoretic mobility shift experiments and data analysis were carried out as previously described with a few modifications (Pagano et al. 2011). Briefly, 3 nM of labeled RNA was incubated with a gradient of hnRNPH1 concentration in equilibration buffer (0.01% Igepal, 0.01 mg/mL tRNA, 50 mM Tris at pH 8.0, 100 mM NaCl, 2 mM DTT) for 3 h. After equilibration, polarization readings were taken in a Victor plate reader. The samples were then mixed with bromocrescol green loading dye and loaded on a 5% native, slab polyacrylamide gel in 1× TBE buffer. The gels were run in 1× TBE buffer for 120 min at 120 V and at 4°C and then scanned using a fluor imager (Fujifilm FLA-5000) with a blue laser at 473 nm. The fraction of bound protein against the protein was fit to the Hill equation using Igor Pro software.
TaqMan assay for tRF-GG and hnRNP F/H association
RNA immunoprecipitations were done using a 10-cm dish of mouse E14 ES cells. Five-hundred micrograms of lysate was incubated with either 10 μg of hnRNP F/H (Santa Cruz Biotechnology) antibody or 10 μg of mouse IgG-conjugated Protein G Dynabeads overnight in the cold room. The beads were then washed sequentially for five washes with Millipore EZ-Magna-RIP wash buffer (#17-701). RNA was then isolated from the beads using Trizol and precipitated using isopropanol following standard procedures. Purified RNA quality was checked using bioanalyzer. tRF and U7 snRNA quantification was performed using custom designed TaqMan microRNA assays according to manufacturer's recommended protocols (Applied Biosystems). Ten nanograms of input RNA, RNA-IP, and control IP RNA was reverse transcribed using the TaqMan microRNA reverse transcription kit. qRT-PCR was performed in 15-µL reactions using TaqMan Universal PCR Master Mix, following standard program (10 min at 95°C, then 15 sec at 95°C, and 1 min at 60°C for 40 cycles).
Immunofluorescence
E14 mouse ES cells were grown and transfected as described above and plated onto gelatinized coverslips. For immunofluorescence, cells were washed twice in PBS and fixed in 4% paraformaldehyde for 20 min at room temperature with mild agitation. Fixed cells were permeabilized by 0.5% Triton-X solution for 20 min at room temperature followed by three washes in PBS containing 0.05% Tween 20 (PBS-Tween). Blocking was performed using 5% milk in TBSt for 1 h at 37°C. Cells were incubated in primary antibody in 3% BSA for 1 h at 37°C, followed by three washes with PBS-Tween. Primary antibodies used were anticoilin (1:50 dilution; Abcam, ab210785) and anti-hnRNPF/H (1:500 dilution; Santa Cruz Biotechnology, sc-32310). Following the washes, cells were incubated in secondary antibody conjugated with fluorophores for 45 min to 1 h at room temperature, in the dark. Secondary antibodies used were Alexa fluor 488 goat antimouse (1:1000 dilution), Alexa flour 488 goat antirabbit (1:500 dilution). Following three additional washes with PBS-Tween, cells were mounted in VectaShield mounting medium containing DAPI for DNA visualization. Microscopy was performed on AxioObserver.Z1/7 microscope using 63×/1.4 NA oil objective. Images were analyzed using ImageJ software.
irCLIP-seq
CLIP-seq was performed by following the irCLIP protocol (Zarnegar et al. 2016). Briefly, 80% confluent mouse ES cells (E14) were cross-linked on ice with 254 nM UV-C at 0.3 J/cm2 and then incubated with ice-cold PBS/10 mM EDTA for 5 min, collected, and pelleted. For a 10-cm dish, 500 μL of SDS lysis buffer (1% SDS, 50 mM Tris at pH 7.5, 1 mM EDTA) was used for cell lysis. Lysates were briefly sonicated using a probe sonicator before clarification by centrifugation at maximum speed for 15 min. Two volumes of IP dilution buffer (1.1% Triton X-100, 50 mM Tris at pH 7.5, 1 mM EDTA, 450 mM NaCl) were added and the lysates were quantitated. Five-hundred nanograms of lysates was incubated with antibody-conjugated Protein G Dynabeads overnight at 4°C. Washes were performed sequentially using 1 mL of high-stringency buffer (20 mM Tris at pH 7.5, 120 mM NaCl, 25 mM KCl, 5 mM EDTA, 1% Triton X-100, 1% Na-deoxycholate), 1 mL of high-salt buffer (20 mM Tris at pH 7.5, 1 Μ NaCl, 5 mM EDTA, 1% Triton X-100, 1% Na-deoxycholate, 0.001% SDS), 1 mL of low-salt buffer (20 mM Tris at pH 7.5, 5 mM EDTA) at 4°C. On-bead nuclease digestions were performed using S1 nuclease (Clontech, cat# 2410) at a final concentration of 2 units/μL. This reaction was then followed up by 3′ ssRNA ligation with biotin and IRDye conjugated adapter as per the published protocol. Immunoprecipitates were then transferred to a nitrocellulose membrane and regions corresponding to the nuclease digestions were cut and the membrane was treated with Proteinase K (Thermo Fisher Scientific, #AM2546) for 1 h at 50°C. RNA is isolated from Proteinase K-treated membranes using Trizol extraction and overnight precipitation at −20°C. The cross-linked and immunoprecipitated RNA is then used for library preparation using the Fast-iCLIP method with the irCLIP modifications (Zarnegar et al. 2016).
Data availability
Raw sequencing data are available at GEO, series GSE127247.
Supplementary Material
Acknowledgments
We thank Zbig Dominski for the U7 expression vector, Jack Huey for help building mutant histone UTR reporter lines, and the Benanti laboratory for help with FACS analyses. We thank Sean Ryder and members of the Rando laboratory for critical reading of the manuscript and insightful discussions. This work was supported by National Institutes of Health grant R01HD080224. A.B. was supported by a fellowship from the Human Frontier Science Program (LT000857/2015-L).
Author contributions: The study was conceived by A.B., X.Y.B., E.K., and O.J.R. A.B. performed all experiments in embryos, made ES reporter lines, carried out the majority of tRF knockdowns and transfections and most qRT-PCR, characterized Cajal bodies by IF, and generated mass spectrometry data for tRF-binding proteins. X.Y.B. carried out metabolic labeling and RNA-seq, carried out tRF perturbations in human ESCs, carried out qRT-PCR and Northerns for various ncRNAs, and was responsible for most bioinformatic analyses along with O.J.R. E.K. carried out hnRNPF/H binding studies, and hnRNPF/H CLIP-seq. The manuscript was written and edited by all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.332783.119.
References
- Alkan SA, Martincic K, Milcarek C. 2006. The hnRNPs F and H2 bind to similar sequences to influence gene expression. Biochem J 393: 361–371. 10.1042/BJ20050538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodeo AA, Jukam D, Straight AF, Skotheim JM. 2015. Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc Natl Acad Sci 112: E1086–E1095. 10.1073/pnas.1413990112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bąkowska-Żywicka K, Kasprzyk M, Twardowski T. 2016. tRNA-derived short RNAs bind to Saccharomyces cerevisiae ribosomes in a stress-dependent manner and inhibit protein synthesis in vitro. FEMS Yeast Res 16: fow077 10.1093/femsyr/fow077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchtold D, Battich N, Pelkmans L. 2018. A systems-level study reveals regulators of membrane-less organelles in human cells. Mol Cell 72: 1035–1049.e5. 10.1016/j.molcel.2018.10.036 [DOI] [PubMed] [Google Scholar]
- Bogu GK, Vizán P, Stanton LW, Beato M, Di Croce L, Marti-Renom MA. 2016. Chromatin and RNA maps reveal regulatory long noncoding RNAs in mouse. Mol Cell Biol 36: 809–819. 10.1128/MCB.00955-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, Greenleaf WJ. 2015. ATAC-seq: a method for assaying chromatin accessibility genome-wide. Curr Protoc Mol Biol 109: 21.29.1–21.29.9. 10.1002/0471142727.mb2129s109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossec JC, Theurillat I, Chica C, Búa Aguín S, Gaume X, Andrieux A, Iturbide A, Jouvion G, Li H, Bossis G, et al. 2018. SUMO safeguards somatic and pluripotent cell identities by enforcing distinct chromatin states. Cell Stem Cell 23: 742–757.e8. 10.1016/j.stem.2018.10.001 [DOI] [PubMed] [Google Scholar]
- Couvillion MT, Sachidanandam R, Collins K. 2010. A growth-essential Tetrahymena Piwi protein carries tRNA fragment cargo. Genes Dev 24: 2742–2747. 10.1101/gad.1996210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvillion MT, Bounova G, Purdom E, Speed TP, Collins K. 2012. A Tetrahymena Piwi bound to mature tRNA 3′ fragments activates the exonuclease Xrn2 for RNA processing in the nucleus. Mol Cell 48: 509–520. 10.1016/j.molcel.2012.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T, Deng X, Zhou J, Lee I, Lee YS, et al. 2015. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol Ther 23: 1622–1629. 10.1038/mt.2015.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. 1999. Formation of the 3′ end of histone mRNA. Gene 239: 1–14. 10.1016/S0378-1119(99)00367-4 [DOI] [PubMed] [Google Scholar]
- Elbarbary RA, Takaku H, Uchiumi N, Tamiya H, Abe M, Takahashi M, Nishida H, Nashimoto M. 2009. Modulation of gene expression by human cytosolic tRNase ZL through 5′-half-tRNA. PLoS One 4: e5908 10.1371/journal.pone.0005908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke V, Ganesh S, Karlic R, Malik R, Pasulka J, Horvat F, Kuzman M, Fulka H, Cernohorska M, Urbanova J, et al. 2017. Long terminal repeats power evolution of genes and gene expression programs in mammalian oocytes and zygotes. Genome Res 27: 1384–1394. 10.1101/gr.216150.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall JG. 2000. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol 16: 273–300. 10.1146/annurev.cellbio.16.1.273 [DOI] [PubMed] [Google Scholar]
- Gebetsberger J, Zywicki M, Künzi A, Polacek N. 2012. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea 2012: 260909 10.1155/2012/260909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebetsberger J, Wyss L, Mleczko AM, Reuther J, Polacek N. 2017. A tRNA-derived fragment competes with mRNA for ribosome binding and regulates translation during stress. RNA Biol 14: 1364–1373. 10.1080/15476286.2016.1257470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodarzi H, Liu X, Nguyen HC, Zhang S, Fish L, Tavazoie SF. 2015. Endogenous tRNA-derived fragments suppress breast cancer progression via YBX1 displacement. Cell 161: 790–802. 10.1016/j.cell.2015.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzzi N, Cieśla M, Ngoc PCT, Lang S, Arora S, Dimitriou M, Pimková K, Sommarin MNE, Munita R, Lubas M, et al. 2018. Pseudouridylation of tRNA-derived fragments steers translational control in stem cells. Cell 173: 1204–1216.e26. 10.1016/j.cell.2018.03.008 [DOI] [PubMed] [Google Scholar]
- Heyer EE, Ozadam H, Ricci EP, Cenik C, Moore MJ. 2015. An optimized kit-free method for making strand-specific deep sequencing libraries from RNA fragments. Nucleic Acids Res 43: e2 10.1093/nar/gku1235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiuchi T, Enriquez-Gasca R, Mizutani E, Bošković A, Ziegler-Birling C, Rodriguez-Terrones D, Wakayama T, Vaquerizas JM, Torres-Padilla ME. 2015. Early embryonic-like cells are induced by downregulating replication-dependent chromatin assembly. Nat Struct Mol Biol 22: 662–671. 10.1038/nsmb.3066 [DOI] [PubMed] [Google Scholar]
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. 2011. Angiogenin-induced tRNA fragments inhibit translation initiation. Mol Cell 43: 613–623. 10.1016/j.molcel.2011.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keam SP, Hutvagner G. 2015. tRNA-Derived fragments (tRFs): emerging new roles for an ancient RNA in the regulation of gene expression. Life (Basel) 5: 1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HK, Fuchs G, Wang S, Wei W, Zhang Y, Park H, Roy-Chaudhuri B, Li P, Xu J, Chu K, et al. 2017. A transfer-RNA-derived small RNA regulates ribosome biogenesis. Nature 552: 57–62. 10.1038/nature25005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Anaya J, Mudunuri SB, Dutta A. 2014. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12: 78 10.1186/s12915-014-0078-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuscu C, Kumar P, Kiran M, Su Z, Malik A, Dutta A. 2018. tRNA fragments (tRFs) guide Ago to regulate gene expression post-transcriptionally in a Dicer-independent manner. RNA 24: 1093–1105. 10.1261/rna.066126.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. 2005. Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 280: 42744–42749. 10.1074/jbc.M510356200 [DOI] [PubMed] [Google Scholar]
- Lenstra TL, Benschop JJ, Kim T, Schulze JM, Brabers NA, Margaritis T, van de Pasch LA, van Heesch SA, Brok MO, Groot Koerkamp MJ, et al. 2011. The specificity and topology of chromatin interaction pathways in yeast. Mol Cell 42: 536–549. 10.1016/j.molcel.2011.03.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S, He F, Luo J, Dou S, Wang Y, Guo A, Lu J. 2018. Drosophila tsRNAs preferentially suppress general translation machinery via antisense pairing and participate in cellular starvation response. Nucleic Acids Res 46: 5250–5268. 10.1093/nar/gky189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Agarwal S, Driscoll S, Lettieri K, Wang J, Andrews SE, Franco L, Rosenfeld MG, Ren B, et al. 2011. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Genes Dev 25: 594–607. 10.1101/gad.2008511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlan TS, Gifford WD, Driscoll S, Lettieri K, Rowe HM, Bonanomi D, Firth A, Singer O, Trono D, Pfaff SL. 2012. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature 487: 57–63. 10.1038/nature11244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machyna M, Heyn P, Neugebauer KM. 2013. Cajal bodies: where form meets function. Wiley Interdiscip Rev RNA 4: 17–34. 10.1002/wrna.1139 [DOI] [PubMed] [Google Scholar]
- Marquet R, Isel C, Ehresmann C, Ehresmann B. 1995. tRNAs as primer of reverse transcriptases. Biochimie 77: 113–124. 10.1016/0300-9084(96)88114-4 [DOI] [PubMed] [Google Scholar]
- Martinez G, Choudury SG, Slotkin RK. 2017. tRNA-derived small RNAs target transposable element transcripts. Nucleic Acids Res 45: 5142–5152. 10.1093/nar/gkx103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF, Koreski KP. 2017. Birth and death of histone mRNAs. Trends Genet 33: 745–759. 10.1016/j.tig.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molla-Herman A, Vallés AM, Ganem-Elbaz C, Antoniewski C, Huynh JR. 2015. tRNA processing defects induce replication stress and Chk2-dependent disruption of piRNA transcription. EMBO J 34: 3009–3027. 10.15252/embj.201591006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita T, Yung TM, Yamamoto J, Tsuboi Y, Tanabe H, Tanaka K, Yamaguchi Y, Handa H. 2007. NELF interacts with CBC and participates in 3′ end processing of replication-dependent histone mRNAs. Mol Cell 26: 349–365. 10.1016/j.molcel.2007.04.011 [DOI] [PubMed] [Google Scholar]
- Pagano JM, Clingman CC, Ryder SP. 2011. Quantitative approaches to monitor protein–nucleic acid interactions using fluorescent probes. RNA 17: 14–20. 10.1261/rna.2428111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L, Han C, Ning L, Cao Y, et al. 2012. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res 22: 1609–1612. 10.1038/cr.2012.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn AJ, Gutbrod MJ, LeBlanc C, Martienssen R. 2017. LTR-retrotransposon control by tRNA-derived small RNAs. Cell 170: 61–71.e11. 10.1016/j.cell.2017.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C, Kucukural A, Serra RW, Sun F, et al. 2016. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science 351: 391–396. 10.1126/science.aad6780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobala A, Hutvagner G. 2013. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biol 10: 553–563. 10.4161/rna.24285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Steiniger M, Marzluff WF. 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol Cell 34: 322–332. 10.1016/j.molcel.2009.04.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Dimova N, Cambi F. 2007. PLP/DM20 ratio is regulated by hnRNPH and F and a novel G-rich enhancer in oligodendrocytes. Nucleic Acids Res 35: 4164–4178. 10.1093/nar/gkm387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Aslanzadeh V, Papa F, Zhu H, de la Grange P, Cambi F. 2012. Global profiling of alternative splicing events and gene expression regulated by hnRNPH/F. PLoS One 7: e51266 10.1371/journal.pone.0051266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilky H, Chari S, Govindan J, Amodeo AA. 2019. Histone concentration regulates the cell cycle and transcription in early development. Development 146: dev177402 10.1242/dev.177402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Gall JG. 1993. U7 small nuclear RNA in C snurposomes of the Xenopus germinal vesicle. Proc Natl Acad Sci 90: 6257–6259. 10.1073/pnas.90.13.6257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X, Wang Z, Jang M, Nutiu R, Wang ET, Burge CB. 2009. Splice site strength-dependent activity and genetic buffering by poly-G runs. Nat Struct Mol Biol 16: 1094–1100. 10.1038/nsmb.1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Liu L, Lazarev D, Al-Zain A, Fomin V, Yeung PL, Chambers SM, Lu CW, Studer L, Manley JL. 2018. TCF3 alternative splicing controlled by hnRNP H/F regulates E-cadherin expression and hESC pluripotency. Genes Dev 32: 1161–1174. 10.1101/gad.316984.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar BJ, Flynn RA, Shen Y, Do BT, Chang HY, Khavari PA. 2016. irCLIP platform for efficient characterization of protein–RNA interactions. Nat Methods 13: 489–492. 10.1038/nmeth.3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Sun L, Kragler F. 2009. The phloem-delivered RNA pool contains small noncoding RNAs and interferes with translation. Plant Physiol 150: 378–387. 10.1104/pp.108.134767 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing data are available at GEO, series GSE127247.