In this study, Shyian et al. set out to address mechanistically how the evolutionarily conserved fork pausing complex acts at proteinaceous replication fork barriers (RFBs) to promote fork passage and genome stability. Using several molecular and cell-based assays, the authors propose that forks pause at proteinaceous RFBs through a “sTOP” mechanism (“slowing down with topoisomerases I–II”), which also contributes to protecting cells from topoisomerase-blocking agents.
Keywords: Tof1, Csm3, Mrc1, topoisomerase, Top1, Top2, RFB, replisome, replication fork pausing
Abstract
Replication forks temporarily or terminally pause at hundreds of hard-to-replicate regions around the genome. A conserved pair of budding yeast replisome components Tof1–Csm3 (fission yeast Swi1–Swi3 and human TIMELESS–TIPIN) act as a “molecular brake” and promote fork slowdown at proteinaceous replication fork barriers (RFBs), while the accessory helicase Rrm3 assists the replisome in removing protein obstacles. Here we show that the Tof1–Csm3 complex promotes fork pausing independently of Rrm3 helicase by recruiting topoisomerase I (Top1) to the replisome. Topoisomerase II (Top2) partially compensates for the pausing decrease in cells when Top1 is lost from the replisome. The C terminus of Tof1 is specifically required for Top1 recruitment to the replisome and fork pausing but not for DNA replication checkpoint (DRC) activation. We propose that forks pause at proteinaceous RFBs through a “sTOP” mechanism (“slowing down with topoisomerases I–II”), which we show also contributes to protecting cells from topoisomerase-blocking agents.
The chromosomal DNA of most cells is duplicated once per cell cycle due to the concerted action of DNA helicases unwinding the DNA template, topoisomerases unlinking the parental strands, and DNA polymerases synthesizing the daughter strands in collaboration with a myriad of accessory factors (Bell and Labib 2016). This assembly of proteins on the DNA replication fork is called the “replisome.” In order to achieve complete genome duplication, the replisome should pass through the entirety of all chromosomes. On average budding yeast replisomes move through ∼20 kb of DNA before merging with a converging fork (Pasero et al. 2002). However, in vivo the speed of the replisome is not uniform, as it temporarily or terminally slows/pauses/arrests/stalls at certain locations, called replication fork barriers (RFBs). RFBs are comprised by “unconventional” DNA structures (inverted repeats, trinucleotide repeats, G4 quadruplexes), RNA/DNA hybrids (R-loops), and tight protein/DNA complexes (Gadaleta and Noguchi 2017). Examples in yeast of the latter type of RFB are found at the rDNA repeat array, tRNA genes (tDNA), telomeres, centromeres, silent mating type loci (HML/HMR) silencer elements, and dormant origins of replication (Gadaleta and Noguchi 2017).
Replisome pausing at these protein barriers involves two components: (1) a tight DNA-binding protein block specific for a given locus (e.g., Fob1 [rDNA RFB - rRFB], the RNA polymerase III preinitiation complex, the general regulatory factor Rap1, or the origin recognition complex) and (2) a “fork pausing/protection complex” (FPC)—the evolutionary conserved heterodimer Tof1–Csm3 in budding yeast (Swi1–Swi3 in fission yeast and TIMELESS–TIPIN in humans). Tof1–Csm3 is also found in association with Mrc1 (not itself involved in replication pausing) in a trimeric complex referred to as MTC, which travels with other factors in a still larger assembly on replication forks called the replisome progression complex (RPC) (Gambus et al. 2006). Loss of Tof1–Csm3 leads to a decrease in replisome pausing at many of the studied protein barriers in budding and fission yeast, and human cells (Gadaleta and Noguchi 2017), while increasing blockage at some unconventional DNA structures (Voineagu et al. 2008). Accessory 5′-to-3′ DNA helicase Rrm3 uses its ATPase/helicase activity to assist the main replicative 3′-to-5′ CMG helicase (Cdc45–Mcm2–7–GINS) in progression specifically at protein blocks (Ivessa et al. 2000, 2003; Calzada et al. 2005; Azvolinsky et al. 2006). Replication fork stalling is proposed to promote genome instability, thus fueling tumorigenesis and ageing (Gaillard et al. 2015). However, the molecular mechanism of action of the Tof1–Csm3, Rrm3, and replisome progression through protein blocks is complex and incompletely understood.
In addition to helicases, the replisome must use topoisomerases in order to topologically unlink, or swivel, the two parental DNA strands (Duguet 1997). Topoisomerase I (Top1 in budding yeast) is regarded as the main replicative swivelase, while topoisomerase II (yeast Top2) provides a back-up mechanism when Top1 is not available (Kim and Wang 1989; Bermejo et al. 2007). It was postulated that similarly to helicases, topoisomerase action should be impeded by the presence of tight protein complexes on DNA in front of the fork (Keszthelyi et al. 2016).
We set out here to understand the mechanism of Tof1–Csm3-dependent replisome arrest/pausing at RFBs. We show first that the Tof1–Csm3 fork pausing complex acts independently of the accessory helicase Rrm3. Instead, we found that Tof1–Csm3 engages topoisomerase I at the replisome (and topoisomerase II as a backup) to promote fork pausing at proteinaceous RFBs, which we refer to as the “slowing down with topoisomerases I–II” (sTOP) mechanism. The Tof1 C terminus mediates Top1 association with the replisome and fork pausing but is not required for the DNA replication checkpoint (DRC). sTOP and DRC mechanisms jointly promote cellular resistance to topoisomerase-blocking agents.
Results
Fork pausing complex Tof1–Csm3 acts independently of Rrm3 helicase
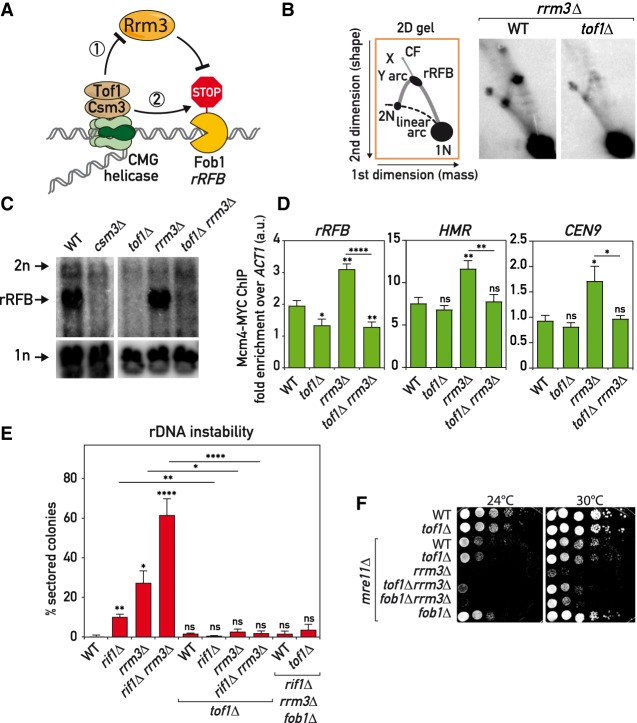
Replication forks slow down at hundreds of tight protein/DNA complexes around the yeast genome (Gadaleta and Noguchi 2017). In a search for the fork pausing mechanism, we started by first confirming with 2D and 1D gels (Brewer and Fangman 1988) that only Tof1–Csm3 but not Mrc1 (Calzada et al. 2005; Tourrière et al. 2005; Hodgson et al. 2007) or other related RPC components are required for fork pausing at the rRFB (Fig. 1A; Supplemental Fig. S1A–C). Accessory helicase Rrm3 helps the replisome to move past protein RFBs throughout the genome (Ivessa et al. 2003). Upon initial characterization of the roles of Tof1 and Csm3 in fork pausing using 2D gels, it was postulated that they work by counteracting the Rrm3 helicase (Fig. 1A, model 1) (Mohanty et al. 2006; Bairwa et al. 2011). If this were true, fork pausing should become completely independent of Tof1–Csm3 in cells lacking Rrm3. However, closer inspection of the 2D gel evidence in the above initial reports suggests that this was not the case.
Figure 1.
Tof1–Csm3 complex functions independently of Rrm3 helicase. (A) Schematics of Rrm3-dependent (1) and -independent (2) mechanisms for Tof1–Csm3 role in replication fork pausing at proteinaceous barriers. (B–D) tof1Δ suppressed fork pausing in rrm3Δ cells. (B) Schematic (left) and images (right) of replication intermediates detected in the asynchronous cultures of strains of indicated genotypes by Southern hybridization with rDNA rRFB probe on BglII-digested DNA separated with 2D gels and blotted to nylon membrane. (C) Same as in B but Southern blot done directly on first-dimension gels. (D) Replisome pausing detection with Mcm4-MYC ChIP-qPCR at several pausing sites in asynchronous cultures of strains of the designated genotypes. (E) tof1Δ suppressed rDNA instability in rrm3Δ and rif1Δ cells—rDNA instability measurement with ADE2 marker loss assay. (F) tof1Δ partially alleviated mre11Δ rrm3Δ synthetic sickness—serial dilution growth assay. (X) X-shaped molecules; (CF) converging forks. Means with SEM are plotted; Welch's t-test was used for quantitative comparisons. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001; (ns) not significant. See also Supplemental Figure S1.
To clarify the Tof1–Csm3 relationship with Rrm3 we used several replication fork pausing and instability assays (Fig. 1). Deletion of TOF1 or CSM3 led to a strong decrease in paused fork signal at Fob1-RFB detected by 1D gels, as expected (Fig. 1C). Significantly, tof1Δ mutation also decreased fork pausing in a rrm3Δ background (Fig. 1B,C), suggesting that in cells lacking Rrm3 helicase, Tof1 still actively promotes replication fork slowdown (Fig. 1A, model 2). Next, we used chromatin immunoprecipitation to probe binding of the replicative helicase components Mcm4 and Cdc45, as it was reported that replisome components are more enriched at pause sites (Azvolinsky et al. 2009). Consistent with the 1D and 2D gel analysis, we detected Tof1-dependent enrichment of Mcm4 and Cdc45 on several pause sites in cells lacking Rrm3 helicase (Fig. 1D; Supplemental Fig. S1D), while pausing at telomeres was less dependent on Tof1.
Lack of the Rrm3 helicase leads to prolonged fork pausing at Fob1-RFB and elevated rDNA instability as a result of fork pausing (Ivessa et al. 2000). Using ADE2 marker loss from the rDNA locus as a measure of ribosomal gene array instability, we found that Tof1 was required for rDNA repeat destabilization in rrm3Δ cells (Fig. 1E). Remarkably, tof1Δ mutation also suppressed the more elevated instability of an rrm3Δ rif1Δ double mutant, which additionally lacks a negative regulator of replication origin firing, Rif1 (Shyian et al. 2016). Viability of rif1Δ cells requires the DSB repair and fork maintenance complex MRX, and the lethality caused by MRX mutations in these cells is suppressed by pausing alleviation through fob1Δ, tof1Δ, or csm3Δ mutations (Shyian et al. 2016). Notably, we observed that tof1Δ partially suppressed synthetic sickness of rrm3Δ and mre11Δ mutations, to an extent slightly stronger than suppression by fob1Δ (Fig. 1F). This difference in suppression by tof1Δ compared with fob1Δ is perhaps related to a more general role of Tof1 in replisome pausing throughout the genome, since Fob1 is thought to act exclusively at rDNA repeats. Altogether, our results show that Tof1 mediates fork pausing, rDNA instability and cellular toxicity in cells lacking Rrm3 helicase. Therefore, it is unlikely that Tof1 promotes fork pausing exclusively by regulating Rrm3 helicase but rather suggests a more direct involvement of Tof1–Csm3 in fork slowdown (Fig. 1A, model 2), albeit through an unknown mechanism.
Tof1–Csm3 complex interacts with Top1
Intrigued by the strong rDNA stabilizing effect of tof1Δ mutation (Fig. 1E), we sought to identify the factor(s) contributing to this stability and regulating replication fork pausing at Fob1-RFB. We carried out an unbiased forward genetic screen for mutants destabilizing the rDNA in either a wild-type (WT) or tof1Δ background, using ADE2 and URA3 loss from the array as a readout (the “cowcatcher” screen) (Materials and Methods; Supplemental Fig. S2A). Mutations in RRM3, SIR2, HST3, CAC1, ORC1, and PSF2 genes were recovered in the WT background but not in tof1Δ. Most of these genes are known to contribute to rDNA stability but do not affect fork pausing (Ide et al. 2007; Saka et al. 2016), except RRM3. Recovery of the above factors in the screen highlighted the viability of our strategy to detect genes involved in rDNA metabolism. Being specifically interested in factors acting in the same pathway as Tof1–Csm3, we anticipated that those will behave differently from rrm3 mutations and will not be suppressed by tof1Δ. Therefore, we next looked at mutations present also (or exclusively) in the tof1Δ background, which could indicate genetic interaction with TOF1. One of the two mutations we discovered specifically in the tof1Δ background was in the TOP1 gene, which encodes topoisomerase I (Supplemental Fig. S2A)—an enzyme required for both DNA replication and stability of rDNA repeats (Christman et al. 1988; Kim and Wang 1989). The highly negative score of this top1-G297D mutation in Protein Variation Effect Analyzer (PROVEAN: −7; cutoff = −2.5) (Choi et al. 2012) implied a deleterious effect of this change on Top1 function. Indeed, complete deletion of the TOP1 ORF led to a strong elevation of rDNA instability (Fig. 2A). In contrast to rrm3Δ and rif1Δ mutations, however, the rDNA instability in top1Δ cells was not suppressed by tof1Δ, suggesting that Top1 and Tof1 may have overlapping roles. This and the fact that TOF1 was originally identified in a yeast two-hybrid screen that used a part of Top1 protein as a bait, as its name implies (“topoisomerase I-interacting factor 1”) (Park and Sternglanz 1999), prompted us to focus further on this factor. The only other mutation recovered in the tof1Δ background was in the leading strand DNA polymerase POL2 gene (Supplemental Fig. S2A). This mutation led to a low expression level of Pol2 protein (Supplemental Fig. S2A′). Since decreasing Pol2 protein level by use of the auxin-inducible degron did not affect fork pausing (Supplemental Fig. S2A′), our phenotype of interest, we did not study pol2 mutants further.
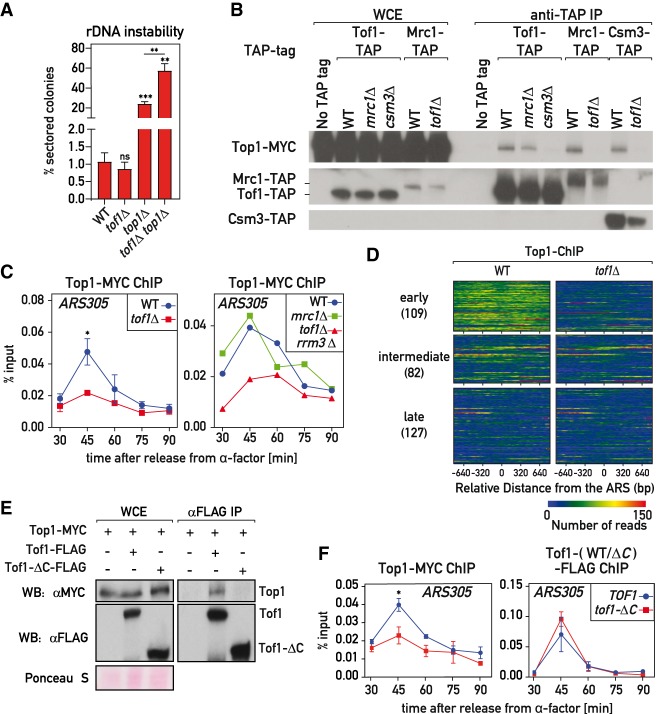
Figure 2.
Tof1–Csm3-dependent recruitment of Top1 to the replisome. (A) tof1Δ did not suppress top1Δ-induced rDNA instability as measured with ADE2 marker loss assay. (B) Top1 was coimmunoprecipitated with MTC complex in a Tof1- and Csm3-dependent but Mrc1-independent manner. (C) Chromatin DNA immunoprecipitated with Top1-MYC from cell cultures synchronously released into S phase from G1 (α-factor) arrest was subjected to qPCR on ARS305. (D) Immunoprecipitated DNA from the 45-min time point (early S phase) was Illumina-sequenced; reads mapping to early, intermediate, and late origins are shown as a heat map. (E,F) Tof1 lacking C terminus (tof1-ΔC strains) did not coimmunoprecipitate Top1-MYC (E) and was defective in Top1-MYC association with ARS305 during S phase (F). Here and on subsequent figures, TOF1 = TOF1-3xFlag; tof1-ΔC = tof1-Δ981-1238-3xFlag. Values plotted and statistics are as in Figure 1. See also Supplemental Figure S2.
As mentioned above, Tof1–Csm3 is present in the cell nucleus within the MTC complex; i.e., together with Mrc1 (Bando et al. 2009). Using coimmunoprecipitation experiments, we observed that topoisomerase I was indeed recovered together with all the three components of the MTC complex (Fig. 2B; Supplemental Fig. S2B). This interaction was detected only when whole-cell extracts were treated with benzonase nuclease, which degrades nucleic acids and liberates protein complexes from chromatin (Supplemental Fig. S2C; De Piccoli et al. 2012). Importantly, the MTC–Top1 interaction depended only on Tof1 and Csm3 proteins, but not Mrc1 (Fig. 2B; Supplemental Fig. S2B), suggesting that Mrc1 interacts with Top1 indirectly through a Tof1–Csm3 subcomplex.
Tof1–Csm3 promotes Top1 recruitment to the replisome
Since both Tof1–Csm3 and Top1 are components of the RPC (Gambus et al. 2006) we wondered whether the interaction of Tof1–Csm3 with Top1 occurs in the context of the replisome, which might explain how Top1 is recruited to the replication fork. To investigate this possibility, we conducted chromatin immunoprecipitation (ChIP) experiments to assess Top1 recruitment to origins of replication in cell cultures synchronously released into S phase from α-factor-induced G1 arrest. We detected Top1 association with early origins (ARS305 and ARS607) at the time of their activation (Fig. 2C; Supplemental Fig. S2D,D′) in accordance with a previous study (Bermejo et al. 2007). However, cells lacking Tof1 had much lower levels of Top1 recruitment to these sites (Fig. 2C; Supplemental Fig. S2D). To confirm this result, we analyzed the genome-wide binding of Top1 in early S phase and observed, as expected, that Top1 is enriched at replicating ARSs (Fig. 2D; Supplemental Fig. S2E) and highly transcribed genes (Supplemental Fig. S2E′) that correspond to regions experiencing high helical tension. Remarkably, removing Tof1 abolished the Top1 signal at ARSs, whereas binding at promoters of highly transcribed genes was not affected (Supplemental Fig. S2E,E′). Furthermore, absence of the MTC complex member Mrc1 did not affect Top1 recruitment (Fig. 2C; Supplemental Fig. S2D), which is in line with retention of the Tof1–Top1 interaction in mrc1Δ cells (Fig. 2B; Supplemental Fig. S2B). Moreover, absence of the Rrm3 helicase did not restore the Top1 association with origins in tof1Δ cells (Fig. 2C; Supplemental Fig. S2D).
The last 258 amino acid residues of the C-terminal part of Tof1 were reported to be sufficient for the two-hybrid interaction with Top1 (Park and Sternglanz 1999). Consistent with this part of Tof1 harboring a Top1-interacting domain, we observed a loss of Top1 coimmunoprecipitation and recruitment to origins in cells expressing a Tof1 protein lacking the last 258 amino acids (tof1-ΔC = tof1-Δ981-1238-3xFlag) (Fig. 2E,F; Supplemental Fig. S2F,G). Importantly, recruitment of WT Tof1 and the truncated Tof1-ΔC protein to origins was comparable (Fig. 2F; Supplemental Fig. S2F,G). This suggests that Tof1 promotes Top1 association with origins by directly recruiting Top1 to the replisome.
Top1 positively regulates replication fork pausing at RFBs
As it is not understood how Tof1–Csm3 slows down the replication fork at protein barriers, we wondered whether their interactor Top1 is involved in this process. To assess this putative functional link between Tof1 and topoisomerase I, we evaluated replication pausing at RFBs in asynchronous cultures. Indeed, deletion of TOP1 or dissociation of Top1 from the replisome by tof1-ΔC mutation led to a similar ∼50% decrease in pausing at RFBs both in WT and rrm3Δ backgrounds, as detected by 2D and 1D gels at rRFB (Fig. 3A; Supplemental Fig. S3A,B) or by Mcm4-MYC ChIP at rRFB and tRNA genes (Fig. 3B; Supplemental Fig. S3C). Moreover, the fork pausing decrease in the double mutant tof1-ΔC top1Δ was comparable with that of single tof1-ΔC and top1Δ mutants (Fig. 3B), suggesting that the two factors could act in the same pausing pathway. Consistent with retention of Top1 recruitment to the FPC complex and to the replisome, mrc1Δ had no defect in pausing (Supplemental Fig. S3B), as previously shown (Tourrière et al. 2005; Hodgson et al. 2007). Moreover, as the mrc1Δ mutation is known to decrease fork progression rates even more strongly than does tof1Δ, but has no effect on pausing (Tourrière et al. 2005; Hodgson et al. 2007), it seems unlikely that the decreased fork pausing in tof1-ΔC or top1Δ mutant could be an indirect consequence of any potential change in fork progression rates in these mutants.
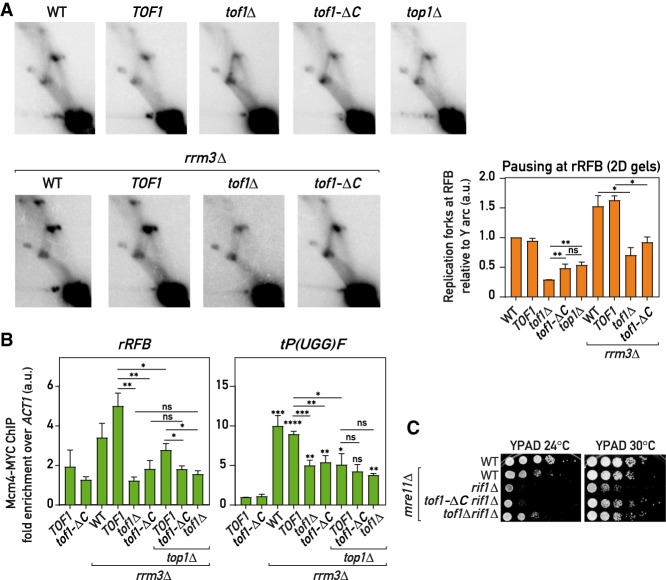
Figure 3.
Tof1-C-dependent recruitment of Top1 to the replisome promotes fork pausing (A) Replication fork pausing at rRFB measured by 2D gels (as in Fig. 1B) in the strains of indicated genotypes: representative gel images and quantification (pausing in WT = 1; n = 4) (see the Materials and Methods). (B) Replisome pausing at rRFB and a tRNA gene (tP(UGG)F) detected with Mcm4-MYC ChIP-qPCR in asynchronous cultures. (C) Tof1-ΔC is less toxic in rif1Δ mre11Δ cells than wild-type Tof1. Alleles: TOF1 = TOF1-3xFlag; tof1-ΔC = tof1-Δ981-1238-3xFlag. Values plotted and statistics are as in Figure 1. See also Supplemental Figure S3.
We had shown previously (Shyian et al. 2016) that rif1Δ leads to increased initiation at the rDNA ARS elements. One consequence of this is increased fork stalling and collapse at the rRFB, which leads to synthetic sickness in combination with mre11Δ. This synthetic growth defect is abolished by deletion of FOB1, confirming its connection to the rDNA fork block. As expected for a pausing defect, we found that tof1-ΔC partially alleviated rif1Δ mre11Δ synthetic sickness (Fig. 3C).
The fact that cells lacking Top1 completely or lacking the Top1-recruiting C terminus of Tof1 still exhibit a pause signal significantly higher than cells lacking the whole of Tof1 protein (Fig. 3A) suggests that some other factor(s) are able to compensate for Top1 loss in a Tof1-dependent way and slow down the replisome in the absence of Top1.
Top1 and Top2 redundantly promote fork pausing at Fob1-RFB
Top1 is believed to be the main replicative swivelase (Kim and Wang 1989), but it is not essential for replication elongation and survival in budding yeast since Top2 is able to compensate for its absence (Kim and Wang 1989; Bermejo et al. 2007). Consistent with this, we also detected Top2 in the immunoprecipitates of Tof1 and Csm3 proteins (Supplemental Fig. S4A) and tof1-ΔC mutation only partially affected this association (Supplemental Fig. S4B). We asked then whether Top2 could compensate for the loss of the Top1 in the replication fork pausing. Indeed, while inactivation of topoisomerase II at elevated temperature in a top2-ts strain or by auxin-induced degradation of the protein had only a small effect on pausing (Fig. 4A,B; Supplemental Fig. S4C,D) doing so in cells lacking Top1 (top1Δ) or in cells with Top1 destabilized from the replisome (tof1-ΔC) led to a dramatic fork pausing loss phenotype similar to the one in tof1Δ cells (Fig. 4A,B; Supplemental Fig. S4C,D,F,G).
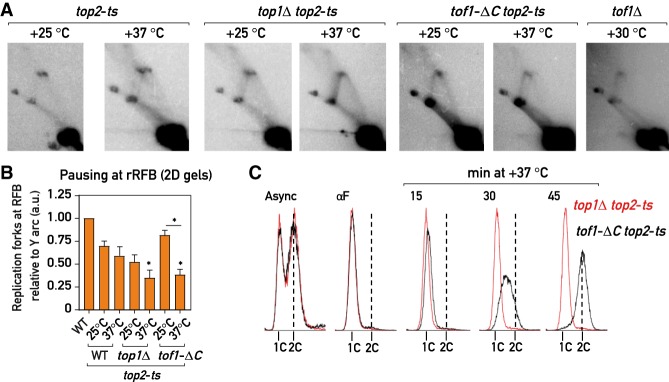
Figure 4.
Top2 partially compensates for the fork pausing upon Top1 loss from the replisome (A,B) 2D agarose gel Southern blots (as in Fig. 1B): representative images (A) and quantification (B); pausing in WT = 1; n = 4; (see the Materials and Methods) of replication intermediates in asynchronous cultures of the strains of the indicated genotypes cultured continuously at +25°C or transferred for 1 h to +37°C. (C) Flow cytometry DNA content profile of the top1Δ top2-ts (red) and tof1-ΔC top2-ts (black) strains upon release in S phase at +37°C from G1 (αF) arrest. Values plotted and statistics as in Figure 1. Asterisks indicate P-values for comparison with top2-ts strain at +25°C. See also Supplemental Figure S4.
We observed a similar loss of fork slowdown when using different means to simultaneously deplete Top1 and Top2: temperature inactivation of Top2 in top1Δ top2-ts and tof1-ΔC top2-ts strains (Fig. 4A,B; Supplemental Fig. S4C), degradation of both proteins (Top1-AID and Top2-AID) or degradation of Top2 in top1Δ and/or tof1-ΔC cells by the auxin-induced degron (Morawska and Ulrich 2013) system (Supplemental Fig. S4D,F,G), and anchoring away (Haruki et al. 2008) of Top2 in a top1Δ TOP2-FRB background (Supplemental Fig. S4H). Depletion of Top3 on its own or in combination with either Top1 or Top2 did not abolish the block (Supplemental Fig. S4D), in accord with a recent study (Mundbjerg et al. 2015) and consistent with Top3 having a role in recombination but not replication (Pommier et al. 2016). Remarkably, replication intermediates in cells lacking both Top1 and Top2 had an appearance very similar to those of tof1Δ strains (Figs. 3A, 4A; Supplemental Fig. S4C–H), in which the loss of the pausing signal at the Fob1-RFB was accompanied by an increase in the intensity of the descending part (left half) of the Y arc. We speculate that the latter might be due to a head-on collision of the replication fork liberated from Fob1-RFB with the RNA polymerase I transcribing the adjacent rRNA gene. Thus, Top1 and Top2 proteins act in parallel to promote replication fork pausing at Fob1-RFB, and the replisome appears to be able to move past the Fob1-RFB in their absence.
Nevertheless, these data have to be interpreted with caution since it was reported that simultaneous inactivation of topoisomerase I and II leads to DNA damage checkpoint activation and to rapid replication cessation (Bermejo et al. 2007), which could in theory contribute to the observed fork pausing phenotypes. However, addressing the checkpoint issue, we found that degradation of both Top1 and Top2 in the checkpoint-deficient backgrounds rad53-K227A (kinase-dead Rad53) or rad9Δ abolished pausing to an extent similar to that in checkpoint-proficient cells (Supplemental Fig. S4H), indicating that checkpoint activation is not necessary for the loss of replication fork slowdown. With regard to replication cessation, when released from G1 arrest into S phase at +37°C, top1Δ top2-ts strains indeed failed to progress through S phase and arrested with close to 1C DNA content (Fig. 4C), consistent with previous findings (Kim and Wang 1989; Bermejo et al. 2007). However, tof1-ΔC top2-ts and tof1Δ top2-ts cells rapidly progressed through the S phase in these conditions (similarly to top2-ts only cells) (Fig. 4C; Supplemental Fig. S4E). Since both top1Δ top2-ts and tof1-ΔC top2-ts cells show a similar decrease of fork pausing at +37°C (Fig. 4A,B; Supplemental Fig. S4C,D), while only the former exhibits an S phase progression defect, we reasoned that the fork slowdown by Top1 and Top2 is not an indirect consequence of their genome-wide replication role but rather an in cis effect of these topoisomerases at the replisome, promoted by the Tof1–Csm3 complex. Moreover, it appears that Top1 (and perhaps Top2) anchoring at the replisome by Tof1 is not essential for general S phase progression, but is specifically important for fork pausing.
tof1-ΔC is a separation of function mutation that leaves replication checkpoint roles intact
Since Tof1–Csm3 is an evolutionary conserved complex performing both fork pausing and replication checkpoint functions at the replisome (McFarlane et al. 2010), we wondered whether the Tof1 C terminus might be specifically involved in only the fork pausing role.
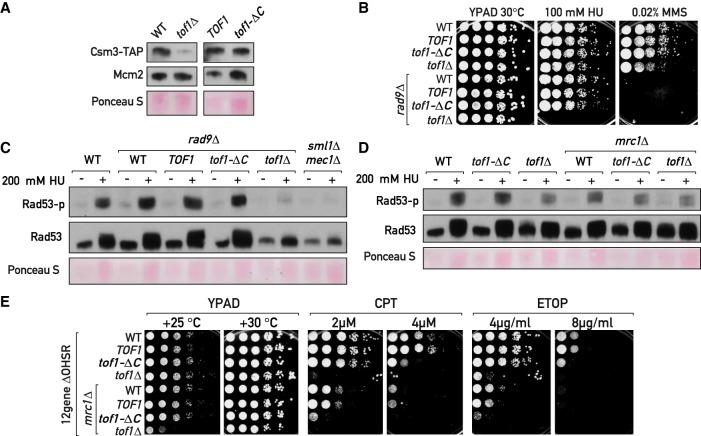
First, similar to the wild-type version, Tof1-ΔC protein appears to protect its partner Csm3 from degradation (Fig. 5A), an evolutionarily conserved Tof1 function (Chou and Elledge 2006; Bando et al. 2009). In addition, Tof1 positively regulates the DNA replication checkpoint (DRC) (Foss 2001), promoting survival of DNA damage response-deficient cells (rad9Δ) subjected to hydroxyurea-induced replication stress. Tof1-ΔC was still able to carry out this function (Fig. 5B), indicating that it is likely checkpoint-proficient. Accordingly, Tof1-ΔC supported DRC activation as measured by Rad53 phosphorylation in both rad9Δ and in WT cells (Fig. 5B,C), while tof1Δ rad9Δ cells had a prominent defect in Rad53 phosphorylation similar to checkpoint-defective mec1Δ sml1Δ cells, as expected (Foss 2001). Furthermore, Tof1 and Mrc1 appear to act in the same DRC pathway, as tof1Δ and tof1Δ mrc1Δ cells showed a similar defect in Rad53 phosphorylation under HU treatment (Fig. 5D). It remains to be seen whether Tof1 directly activates DRC or whether its contribution is completely indirect, for example, via promoting Mrc1 association with replication forks as previously suggested (Tourrière et al. 2005; Bando et al. 2009).
Figure 5.
Fork pausing is a separable function of Tof1–Csm3. (A, C, D) Western blotting of TCA-extracted proteins. (A) In contrast to tof1Δ, tof1-ΔC cells do not degrade Csm3-TAP. (B,E) Serial dilution growth assays. (B) Tof1-ΔC supports viability of rad9Δ cells under hydroxyurea (HU) treatment. (C,D) Tof1-ΔC is proficient in DRC activation under HU treatment. (E) Mrc1 supports tof1-ΔC cells survival under topoisomerase-blocking damage. (CPT) Camptothecin; (ETOP) etoposide; (MMS) methyl methanesulfonate; (12geneΔ0HSR) multidrug sensitive yeast background. See also Supplemental Figure S5.
The loss of Tof1–Csm3 complex, but not Mrc1, confers strong sensitivity to the Top1-trapping agent camptothecin (CPT) (Fig. 5E; Supplemental Fig. S5A,B; Redon et al. 2006; Reid et al. 2011). In addition, we found that cells lacking any of the MTC complex components display impaired growth in the presence of etoposide (ETOP) (Fig. 5E; Supplemental Fig. S5A,B), a chemical blocking topoisomerase II, with tof1Δ and csm3Δ again having a greater effect than mrc1Δ. Importantly, tof1Δ and csm3Δ mutations impaired growth specifically in the presence of topoisomerase blocking agents but not upon DNA double-strand break induction by phleomycin or fork stalling and breakage by the alkylating agent MMS (Supplemental Fig. S5A,B). Therefore, the Tof1–Csm3 complex appears to protect cells from blocked topoisomerases. We wondered whether this protection stems from the ability of Tof1–Csm3 to engage with Top1 and Top2. Surprisingly, tof1-ΔC mutant was still significantly resistant to CPT and ETOP (Fig. 5E). We reasoned that the higher sensitivity of tof1Δ to these agents in contrast to tof1-ΔC mutant could be due to the preservation of another function in the Tof1-ΔC protein and, since Tof1-ΔC is proficient in the Mrc1-dependent DRC (Figs. 4D, 5C), speculated that this might also be related to a role shared with Mrc1. We therefore removed Mrc1 from the tof1-ΔC mutant cells and indeed observed an increase in CPT and ETOP sensitivity in the tof1-ΔC mrc1Δ double mutant to an extent comparable with that of tof1Δ cells (Fig. 5E). Interestingly, tof1Δ, but not tof1-ΔC, grew slowly in combination with mrc1Δ (at 25°C) (Fig. 5E) and in spore colonies, suggesting that Tof1-ΔC protein still performs an additional function important for growth in parallel to Mrc1. Thus, the Tof1–Csm3 complex appears to protect the cell from trapped topoisomerases by both interacting with them directly (through the C terminus of Tof1 and perhaps other regions) and by acting together with Mrc1, likely by promoting the DRC and/or stabilizing forks at the topoisomerase-trapping sites (Strumberg et al. 2000).
Discussion
In summary, we showed that Tof1–Csm3 mediates replication fork pausing at proteinaceous RFBs through a pathway independent of Rrm3 helicase. Instead, Tof1–Csm3 complex interacts with topoisomerases I and II and mediates Top1 association with the replisome in normal S phase. Although we did not detect Top2 recruitment to replisomes in unchallenged cells by ChIP, alternative approaches should be used in the future to assess Top2 recruitment and its dependency on FPC. Top1 was previously identified as a part of the RPC (Gambus et al. 2006) and our report pinpoints the precise factor responsible for its engagement and suggests that eukaryotic cells do not rely exclusively on the DNA topology-mediated recruitment of topoisomerases to replicate chromosomes, but rather have an association hub (Tof1–Csm3) to enrich them on the replisome. We imagine that this pathway could serve to prevent buildup of excess torsional stress in the vicinity of the replisome by ensuring topoisomerase presence. This may avert uncontrolled escape of supercoils away from the fork by diffusion, supercoil “hopping” (van Loenhout et al. 2012) or fork rotation (Schalbetter et al. 2015), possibilities that warrant further investigation. It is conceivable that in csm3Δ, tof1Δ, and tof1-ΔC strains topoisomerases I and II do eventually relieve torsional stress in front of the fork (either independently or as part of multiprotein complexes), but perhaps only after the torsion has reached higher levels. Of note, Top2 was recently shown in vitro to strongly favor a single chromatin fiber over two independent fibers as a substrate (Le et al. 2019), a preference that would support its general activity ahead of the fork irrespective of replisome composition.
Our findings indicate that either Top1 or Top2 is able to impose replication fork pausing at the Fob1-RFB, through a mechanism that we dub “sTOP” (“slowing down with topoisomerase I and II”) (Fig. 6). Indeed, it is assumed that in eukaryotes topoisomerase I and II act in front of the replication fork to unlink the parental DNA strands (Duguet 1997), while topoisomerase II acts also behind the fork to remove precatenanes. Either Top1 or Top2 is sufficient to assist in DNA replication elongation (Pommier et al. 2016), explaining why TOP1 is not essential. The essential role of TOP2 stems not from the replication elongation step, but from its crucial role in chromosome segregation during replication termination (Baxter and Diffley 2008). We imagine that the local increase in topoisomerase concentration/activity in the vicinity of replisome afforded by Tof1–Csm3 recruitment might assist general replication elongation by alleviating torsional stress. In cells lacking the Tof1–Csm3 complex topoisomerases would act more distributively but still ensure replisome progression, albeit perhaps less efficiently. We note that recruitment of an essential enzymatic function to the replisome by nonessential RPC factors is not unprecedented, since another RPC component, Ctf4, serves to recruit DNA polymerase α/primase and Mrc1 stimulates the interaction of the leading strand DNA polymerase ε (Bell and Labib 2016).
Figure 6.
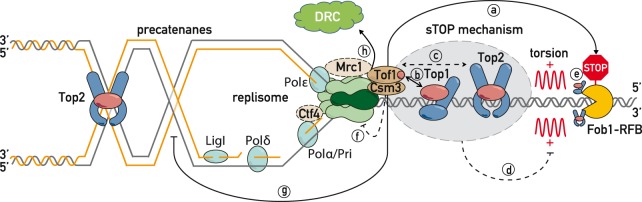
Replisome “sTOP” model (“slowing down with topoisomerases I–II”) Tof1–Csm3 promotes replication fork pausing at proteinaceous barriers (a) via topoisomerase I and II, either by recruiting topoisomerases to the replisome (b,c) to dampen positive torsion in front of the fork (d), or/and by recognizing topoisomerases bound at barriers (e) and potentially regulating CMG helicase activity (f) (Cho et al. 2013). (g) Tof1–Csm3 also suppresses fork rotation (Schalbetter et al. 2015). (h) sTOP function of Tof1–Csm3 is distinct from its Mrc1-shared role in DRC (DNA replication checkpoint). See the text for further details.
In order to assist in DNA replication, the topoisomerase swivelase should be placed in front of the replication fork (Duguet 1997)—a setting where Top1 and Top2 might be the first replisome components to encounter obstacles. The slowing of the replication fork could be either a consequence of an inhibitory signal propagating from stalled topoisomerases through Tof1–Csm3 to the CMG helicase, or a result of topoisomerase activity itself. Consistent with first mode of action, it was reported that Tof1–Csm3 orthologs are able to inhibit the ATPase activity of MCM proteins in vitro (Cho et al. 2013). According to the second model, the absence of Top1 and Top2 at the replisome might promote bypass of barriers by increasing superhelical tension at the fork and simplifying blocking protein dissociation from DNA. In line with this possibility, bacterial topA mutants cause a loss of replication fork pausing at Tus/Ter sites likely by an increase in negative superhelicity, as this effect is suppressed by compensatory gyrB mutations (Valjavec-Gratian et al. 2005). Moreover, it was proposed that topoisomerase inhibition leads to nucleosome destabilization due to increased positive torsion ahead of transcribing RNA polymerase II (Teves and Henikoff 2014). It is thus tempting to speculate that by recruiting topoisomerases to the fork, Tof1–Csm3 precludes torsional stress buildup ahead of the replisome, helping to maintain chromatin integrity (binding of both nonhistone and histone proteins). Further studies, particularly with single-molecule approaches, will help to assess whether this is the case and elucidate the exact molecular details of how Top1 and Top2 promote replication fork pausing at proteinaceous barriers and general fork progression.
Although topoisomerase would still be expected to assist DNA replication elongation by replisomes lacking the Tof1–Csm3 complex (since tof1Δ and csm3Δ cells are viable), the failure to recognize topoisomerases in front of the fork, and perhaps to duly pause until they dissociate from the template, might lead to replisome–topoisomerase collisions. We speculate that collision and replication runoff (Strumberg et al. 2000) with subsequent failure to properly activate the DNA replication checkpoint and repair the collapsed forks might explain the elevated sensitivity of tof1Δ and csm3Δ mutants to topoisomerase blocking conditions (Fig. 5E; Supplemental Fig. S5A,B; Redon et al. 2006; Reid et al. 2011). Accordingly, it was recently proposed that the Csm3 ortholog TIPIN may help to recognize topoisomerase I trapped by CPT and preclude replisome collision with it (Hosono et al. 2014).
This novel replication fork “sTOP” mechanism offers a solution to an unresolved problem of how Tof1–Csm3 manages to recognize molecularly distinct RFBs: The Top1 and Top2 topological (or physical) interaction with RFBs might serve as a unifying common feature of different barriers. We also note that catalytically engaged Top1 is present at the Fob1-RFB (due to an interaction with Tof2) (Krawczyk et al. 2014) throughout the cell cycle (Di Felice et al. 2005) and Top1 assists in progression of RNA polymerase II complexes (Teves and Henikoff 2014). Therefore, an intriguing question would be whether Tof1–Csm3 could mediate recognition by the replisome of Top1 and Top2 present as a part of these and other chromatin complexes in the path of a replication fork. However, since pausing at some protein barriers does not depend on Tof1 (Larsen et al. 2014), while DNA structural impediments actually slow forks more in the absence of Tof1–Csm3 (Voineagu et al. 2008), the pausing mechanisms in these and other contexts and the potential contribution of topoisomerases remain to be determined.
Recently, DNA replication elongation reactions (Yeeles et al. 2017) and fork pausing at Fob1 barriers were successfully reconstituted in vitro (Hizume et al. 2018), where Tof1–Csm3 supported high elongation rates and mediated pausing, respectively. It will be of interest to test whether these in vitro phenotypes of Tof1–Csm3 are mediated via recruitment of Top1 and Top2 to the replisome. Another fascinating question is whether the role of Tof1–Csm3 orthologs in other systems, such as replication pausing and imprinting control by Swi1–Swi3 at the mat locus in fission yeast (Dalgaard and Klar 2000), circadian clock regulation in metazoans (McFarlane et al. 2010), and survival in the face of replication stress (Bianco et al. 2019) are mediated via interactions with topoisomerases.
Materials and methods
Yeast strains, genetics, and growth conditions
Standard genetic methods for budding yeast strain construction and crossing were used (Shyian et al. 2016). Stains used in this study are listed in the Supplemental Table S1. Genotoxic agent sensitivity was assessed in multidrug-sensitive yeast background (Chinen et al. 2011). For growth assays, saturated cultures of the respective genotypes were serially diluted (1:10) and spotted onto YPAD plates or YPAD plates supplemented with genotoxic agents. The plates were imaged following 2–4 d of incubation at 30°C or 25°C. ADE2 marker loss assays were performed essentially as in Shyian et al. (2016). Degradation of AID-tagged proteins (Morawska and Ulrich 2013) and cytoplasmic anchoring of the FRB-tagged proteins (Haruki et al. 2008) was achieved by addition of 1 mM IAA (Indole-3-acetic acid) for 60 min or 1 μg/mL RAPA (Rapamycin) for 45 min to the exponentially growing cultures. Heat inactivation of the Top2-ts protein was achieved by shifting exponentially growing yeast cultures for 60 min at +25°C to +37°C. For the cell cycle progression analysis in top2-ts background, the exponentially growing cells were arrested in G1 with αF treatment for 2.5 h at +25°C, transferred for an additional 1 h to +37°C, washed twice with H2O, and released from the G1 arrest at +37°C in pronase-containing medium (Mattarocci et al. 2014).
rDNA instability (ADE2 loss) assay
rDNA instability was assessed by the ADE2 maker loss assay (Kaeberlein et al. 1999; Shyian et al. 2016). Saturated yeast cultures were diluted in water to around 400 cells per volume and plated onto YPD plates supplemented with 5 mg/mL adenine or onto SC plates (with or without 5-FOA). Plates were incubated for 3 d at 30°C, then for 2 d at 4°C, and subsequently for 1 d at 25°C. The colonies were counted using ImageJ software Colony Counter plugin and the marker loss was plotted as the percentage of white colonies having red sectors to all the colonies except completely red colonies (where ADE2 marker was lost in previous cell divisions).
1D and 2D gels and Southern blot
2D gels were performed essentially as in Shyian et al. (2016) using BglII enzyme for genomic DNA digestion and Fob1-RFB Southern blot hybridization probe. The images were acquired with Typhoon FLA 9500 (GE Healthcare Life Sciences) and the intensity of signals quantified with ImageQuant TL 8.1 software (GE Healthcare Life Sciences). The ratio of the signals at the rRFB spot to the remainder of Y arc of a given mutant was normalized to the respective ratio in WT present on the same 2D gel blot membrane and reported as “replication forks at RFB relative to Y arc” value; this value in all the WT samples therefore equals 1. For 1D gels, the first dimension gel was stained with EtBr, directly transferred to nylon membrane, and probed with a radioactively labeled probe specific to Fob1-RFB site (Brewer and Fangman 1988). The membranes were exposed to K-screens (Bio-Rad) for 6 h to 7 d before phosophorimaging.
Chromatin immunoprecipitation (ChIP)
Mcm4-13MYC, Cdc45-13MYC, Top1-13MYC anti-Myc, and Tof1-3Flag, Tof1-980aa-3Flag anti-Flag ChIP assays were performed essentially as in Mattarocci et al. (2014). Mcm4-13MYC and Cdc45-13MYC ChIP experiments were done using asynchronously growing cultures. Cell synchronization and flow cytometry for Top1-13xMYC, Tof1-3xFlag, Tof1-Δ981-1238-3Flag, and RPA ChIP experiments were performed essentially as described in Mattarocci et al. (2014). Briefly, exponentially growing yeast cell cultures were arrested in G1 phase by treating with 12.5 ng/mL α-factor (BACHEM AG) for 2.5 h at +30°C, washed twice with H2O, and synchronously released into S phase at +18°C by suspending the pellet in fresh YPAD media supplemented with 0.1 mg/mL pronase (Calbiochem). Where indicated, precipitated DNA was used to prepare sequencing libraries with TruSeq (Illumina) and sequenced on iGE3 Genomics Platform of University of Geneva. FASTQ files were mapped to S. cerevisiae genome with Mapping tool of “HTSstation” (David et al. 2014). All ChIP-seq sequencing data sets generated in this study were submitted to NCBI GEO database under the accession number GSE140034.
‘Cowcatcher’ screen
Strains containing single copy of ADE2 and URA3 genes inserted into rDNA array were used for mutagenesis with EMS at 50% survival. EMS-treated cultures were split in 10 separate tubes, inoculated into SC-ADE-URA liquid medium, and grown overnight (to counterselect mutations in ADE2 and URA3). Next, aliquots were inoculated into YPAD and grown overnight to allow for marker loss from the rDNA. Dilutions were plated on 5-FOA plates (selection for URA3 loss) and incubated as in ADE2 loss assay above. After visual inspection, red sectored colonies from 5-FOA plates were manually selected and their white sectors were streaked sequentially twice onto SC plates. Of ∼50,000 colonies from 5-FOA plates, 30 independent, reproducibly high-sectoring isolates were chosen. These were back-crossed, sporulated, dissected and assessed for segregation of the high sectoring phenotype. Isolates showing 2:2 segregation for sectoring (consistent with Mendelian monoallelic mutations) were subjected to causative mutation identification using Pooled Linkage Analysis (as in Birkeland et al. 2010; Lang et al. 2015). Briefly, 20 spore colonies with a sectoring phenotype were pooled (+phenotype) and 20 white spore colonies were pooled (−phenotype) and their genomic DNA was isolated with a Qiagen genomic-tip kit. Total genomic DNA of the two pools was submitted to iGE3 Genomics Platform of University of Geneva for fragmentation, library preparation, and whole genome deep sequencing. The resulting FASTQ files were mapped to the S. cerevisiae genome with the mapping tool of “HTSstation” (David et al. 2014). The SNPs were identified with the SNP tool of “HTSstation.” The SNPs unique/over-represented in the plus-phenotype pool compared with the minus-phenotype pool were identified in Excel.
Coimmunoprecipitation, SDS-PAGE, and Western blot
Coimmunoprecipitation was performed as in Gambus et al. (2006) and De Piccoli et al. (2012). Briefly, 50 mL of exponentially growing cells at OD600 = 0.6 was pelleted, washed twice with cold H2O, suspended in 1 mL of lysis buffer (100 mM HEPES-KOH pH 7.9, 100 mM potassium acetate, 10 mM magnesium acetate, 10% glycerol, 0.1% NP-40, 2 mM EDTA, 2 mM glycerol 2-phosphate, and freshly added 2 mM sodium fluoride, 1 mM DTT, 1 mM PMSF, Roche protease inhibitor cocktail and PhosStop), and transferred into a cryotube with 500 μL of zirconia/silica beads. The cells where homogenized in a Minibeadbeater at max power twice for 1.5 min with a 1-min interval. The lysed cells were recovered by centrifugation through a hole in the bottom of a cryotube and treated with 100 U of benzonase (Millipore) for 40 min at +4°C with rotation. The whole-cell extract (WCE) was obtained as supernatant after centrifugation at 13,000 rpm for 30 min at +4°C. Thirty microliters of IgG Sepharose beads (GE Healthcare) or anti-Flag M2 beads (Sigma) prewashed four times with lysis luffer was used for immunoprecipitation of the TAP-tagged or Flag-tagged proteins, respectively, from the WCE for 2 h at +4°C with rotation. The beads were washed three times with lysis buffer at +4°C with rotation and boiled for 10 min with 50 μL of 2× Laemmli buffer. The proteins were resolved on 8% iD PAGE GELS (Eurogentec) and transferred onto nitrocellulose membrane (Amersham). The proteins were detected with anti-TAP (Thermo Fisher), anti-MYC (Cell Signaling), or anti-Flag (Sigma) antibodies. For Csm3-TAP protein level detection (Fig. 4A) and Rad53 phosphorylation detection (Fig. 4C,D), total cellular proteins were isolated using the TCA-urea method (Mattarocci et al. 2014). Total and active autophosphorylated Rad53 were detected with anti-Rad53 antibodies (Mab clone EL7) and (Mab clone F9), respectively, provided by A. Pellicioli (University of Milan) (Fiorani et al. 2008).
Statistical methods
Welch's t-test (two-tailed, unpaired t-test with Welch's correction) was used to assess statistical significance of differences in all the quantitative comparisons ( P < 0.05 [*], P < 0.01 [**], P < 0.001 [***], and P < 0.0001 [****]). Mean values ± SEM (standard error of the mean) are reported on graphs. GraphPad Prism 8 (GraphPad Software, Inc.) was used to prepare the graphs and perform statistical comparisons.
Supplementary Material
Acknowledgments
We thank Nataliia Serbyn for discussions, advice, and assistance; Jessica Bruzzone for assistance with sequencing library preparation; Slawomir Kubik for advice on data analysis; and all members of the Shore laboratory for critical comments. We thank iGE3 Genomics Platform of the University of Geneva for sequencing library preparation and high-throughput sequencing experiments. We thank Benoit Kornmann for advice on the pooled linkage analysis, Usui Takeo for sharing 12geneΔ0HSR background strains, and Francoise Stutz for the BY deletion and DAmP strains. We thank Nicolas Roggli for expert graphical work, and Pascal Damay for ordering and maintaining reagent stocks.
Author contributions: M.S. conceived the project. M.S., B.A., and D.S. designed all the experiments. M.S., B.A., A.M.Z., V.I., G.C., and D.D. performed the experiments and analyzed the data. B.A. and V.I. contributed strains and reagents. M.S. wrote the manuscript, which was revised by M.S. and D.S. with input from B.A.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.331868.119.
References
- Azvolinsky A, Dunaway S, Torres JZ, Bessler JB, Zakian VA. 2006. The S. cerevisiae Rrm3p DNA helicase moves with the replication fork and affects replication of all yeast chromosomes. Genes Dev 20: 3104–3116. 10.1101/gad.1478906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azvolinsky A, Giresi PG, Lieb JD, Zakian VA. 2009. Highly transcribed RNA polymerase II genes are impediments to replication fork progression in Saccharomyces cerevisiae. Mol Cell 34: 722–734. 10.1016/j.molcel.2009.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairwa NK, Mohanty BK, Stamenova R, Curcio MJ, Bastia D. 2011. The intra-S phase checkpoint protein Tof1 collaborates with the helicase Rrm3 and the F-box protein Dia2 to maintain genome stability in Saccharomyces cerevisiae. J Biol Chem 286: 2445–2454. 10.1074/jbc.M110.189456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bando M, Katou Y, Komata M, Tanaka H, Itoh T, Sutani T, Shirahige K. 2009. Csm3, Tof1, and Mrc1 form a heterotrimeric mediator complex that associates with DNA replication forks. J Biol Chem 284: 34355–34365. 10.1074/jbc.M109.065730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter J, Diffley JF. 2008. Topoisomerase II inactivation prevents the completion of DNA replication in budding yeast. Mol Cell 30: 790–802. 10.1016/j.molcel.2008.04.019 [DOI] [PubMed] [Google Scholar]
- Bell SP, Labib K. 2016. Chromosome duplication in Saccharomyces cerevisiae. Genetics 203: 1027–1067. 10.1534/genetics.115.186452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo R, Doksani Y, Capra T, Katou YM, Tanaka H, Shirahige K, Foiani M. 2007. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev 21: 1921–1936. 10.1101/gad.432107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco JN, Bergoglio V, Lin YL, Pillaire MJ, Schmitz AL, Gilhodes J, Lusque A, Mazières J, Lacroix-Triki M, Roumeliotis TI, et al. 2019. Overexpression of Claspin and Timeless protects cancer cells from replication stress in a checkpoint-independent manner. Nat Commun 10 10.1038/s41467-019-08886-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkeland SR, Jin N, Ozdemir AC, Lyons RH Jr, Weisman LS, Wilson TE. 2010. Discovery of mutations in Saccharomyces cerevisiae by pooled linkage analysis and whole-genome sequencing. Genetics 186: 1127–1137. 10.1534/genetics.110.123232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer BJ, Fangman WL. 1988. A replication fork barrier at the 3′ end of yeast ribosomal RNA genes. Cell 55: 637–643. 10.1016/0092-8674(88)90222-X [DOI] [PubMed] [Google Scholar]
- Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. 2005. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev 19: 1905–1919. 10.1101/gad.337205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinen T, Ota Y, Nagumo Y, Masumoto H, Usui T. 2011. Construction of multidrug-sensitive yeast with high sporulation efficiency. Biosci Biotechnol Biochem 75: 1588–1593. 10.1271/bbb.110311 [DOI] [PubMed] [Google Scholar]
- Cho WH, Kang YH, An YY, Tappin I, Hurwitz J, Lee JK. 2013. Human Tim–Tipin complex affects the biochemical properties of the replicative DNA helicase and DNA polymerases. Proc Natl Acad Sci 110: 2523–2527. 10.1073/pnas.1222494110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Sims GE, Murphy S, Miller JR, Chan AP. 2012. Predicting the functional effect of amino acid substitutions and indels. PLoS One 7: e46688 10.1371/journal.pone.0046688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou DM, Elledge SJ. 2006. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci 103: 18143–18147. 10.1073/pnas.0609251103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christman MF, Dietrich FS, Fink GR. 1988. Mitotic recombination in the rDNA of S. cerevisiae is suppressed by the combined action of DNA topoisomerases I and II. Cell 55: 413–425. 10.1016/0092-8674(88)90027-X [DOI] [PubMed] [Google Scholar]
- Dalgaard JZ, Klar AJ. 2000. swi1 and swi3 perform imprinting, pausing, and termination of DNA replication in S. pombe. Cell 102: 745–751. 10.1016/S0092-8674(00)00063-5 [DOI] [PubMed] [Google Scholar]
- David FP, Delafontaine J, Carat S, Ross FJ, Lefebvre G, Jarosz Y, Sinclair L, Noordermeer D, Rougemont J, Leleu M. 2014. HTSstation: a Web application and open-access libraries for high-throughput sequencing data analysis. PLoS One 9: e85879 10.1371/journal.pone.0085879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Piccoli G, Katou Y, Itoh T, Nakato R, Shirahige K, Labib K. 2012. Replisome stability at defective DNA replication forks is independent of S phase checkpoint kinases. Mol Cell 45: 696–704. 10.1016/j.molcel.2012.01.007 [DOI] [PubMed] [Google Scholar]
- Di Felice F, Cioci F, Camilloni G. 2005. FOB1 affects DNA topoisomerase I in vivo cleavages in the enhancer region of the Saccharomyces cerevisiae ribosomal DNA locus. Nucleic Acids Res 33: 6327–6337. 10.1093/nar/gki950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duguet M. 1997. When helicase and topoisomerase meet! J Cell Sci 110: 1345–1350. [DOI] [PubMed] [Google Scholar]
- Fiorani S, Mimun G, Caleca L, Piccini D, Pellicioli A. 2008. Characterization of the activation domain of the Rad53 checkpoint kinase. Cell Cycle 7: 493–499. 10.4161/cc.7.4.5323 [DOI] [PubMed] [Google Scholar]
- Foss EJ. 2001. Toflp regulates DNA damage responses during S phase in Saccharomyces cerevisiae. Genetics 157: 567–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadaleta MC, Noguchi E. 2017. Regulation of DNA replication through natural impediments in the eukaryotic genome. Genes (Basel) 8: 98 10.3390/genes8030098 [DOI] [Google Scholar]
- Gaillard H, García-Muse T, Aguilera A. 2015. Replication stress and cancer. Nat Rev Cancer 15: 276–289. 10.1038/nrc3916 [DOI] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. 2006. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol 8: 358–366. 10.1038/ncb1382 [DOI] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31: 925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Hizume K, Endo S, Muramatsu S, Kobayashi T, Araki H. 2018. DNA polymerase ε-dependent modulation of the pausing property of the CMG helicase at the barrier. Genes Dev 32: 1315–1320. 10.1101/gad.317073.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgson B, Calzada A, Labib K. 2007. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell 18: 3894–3902. 10.1091/mbc.e07-05-0500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosono Y, Abe T, Higuchi M, Kajii K, Sakuraba S, Tada S, Enomoto T, Seki M. 2014. Tipin functions in the protection against topoisomerase I inhibitor. J Biol Chem 289: 11374–11384. 10.1074/jbc.M113.531707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide S, Watanabe K, Watanabe H, Shirahige K, Kobayashi T, Maki H. 2007. Abnormality in initiation program of DNA replication is monitored by the highly repetitive rRNA gene array on chromosome XII in budding yeast. Mol Cell Biol 27: 568–578. 10.1128/MCB.00731-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivessa AS, Zhou JQ, Zakian VA. 2000. The Saccharomyces Pif1p DNA helicase and the highly related Rrm3p have opposite effects on replication fork progression in ribosomal DNA. Cell 100: 479–489. 10.1016/S0092-8674(00)80683-2 [DOI] [PubMed] [Google Scholar]
- Ivessa AS, Lenzmeier BA, Bessler JB, Goudsouzian LK, Schnakenberg SL, Zakian VA. 2003. The Saccharomyces cerevisiae helicase Rrm3p facilitates replication past nonhistone protein-DNA complexes. Mol Cell 12: 1525–1536. 10.1016/S1097-2765(03)00456-8 [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. 1999. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13: 2570–2580. 10.1101/gad.13.19.2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keszthelyi A, Minchell NE, Baxter J. 2016. The causes and consequences of topological stress during DNA replication. Genes (Basel) 7: 134 10.3390/genes7120134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim RA, Wang JC. 1989. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae. J Mol Biol 208: 257–267. 10.1016/0022-2836(89)90387-2 [DOI] [PubMed] [Google Scholar]
- Krawczyk C, Dion V, Schär P, Fritsch O. 2014. Reversible Top1 cleavage complexes are stabilized strand-specifically at the ribosomal replication fork barrier and contribute to ribosomal DNA stability. Nucleic Acids Res 42: 4985–4995. 10.1093/nar/gku148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang AB, John Peter AT, Walter P, Kornmann B. 2015. ER-mitochondrial junctions can be bypassed by dominant mutations in the endosomal protein Vps13. J Cell Biol 210: 883–890. 10.1083/jcb.201502105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NB, Sass E, Suski C, Mankouri HW, Hickson ID. 2014. The Escherichia coli Tus-Ter replication fork barrier causes site-specific DNA replication perturbation in yeast. Nat Commun 5: 3574 10.1038/ncomms4574 [DOI] [PubMed] [Google Scholar]
- Le TT, Gao X, Park SH, Lee J, Inman JT, Lee JH, Killian JL, Badman RP, Berger JM, Wang MD. 2019. Synergistic coordination of chromatin torsional mechanics and topoisomerase activity. Cell 179: 619–631.e15. 10.1016/j.cell.2019.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattarocci S, Shyian M, Lemmens L, Damay P, Altintas DM, Shi T, Bartholomew CR, Thomä NH, Hardy CF, Shore D. 2014. Rif1 controls DNA replication timing in yeast through the PP1 phosphatase Glc7. Cell Rep 7: 62–69. 10.1016/j.celrep.2014.03.010 [DOI] [PubMed] [Google Scholar]
- McFarlane RJ, Mian S, Dalgaard JZ. 2010. The many facets of the Tim–Tipin protein families’ roles in chromosome biology. Cell Cycle 9: 700–705. 10.4161/cc.9.4.10676 [DOI] [PubMed] [Google Scholar]
- Mohanty BK, Bairwa NK, Bastia D. 2006. The Tof1p–Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci 103: 897–902. 10.1073/pnas.0506540103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morawska M, Ulrich HD. 2013. An expanded tool kit for the auxin-inducible degron system in budding yeast. Yeast 30: 341–351. 10.1002/yea.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundbjerg K, Jørgensen SW, Fredsøe J, Nielsen I, Pedersen JM, Bentsen IB, Lisby M, Bjergbaek L, Andersen AH. 2015. Top2 and Sgs1–Top3 act redundantly to ensure rDNA replication termination. PLoS Genet 11: e1005697 10.1371/journal.pgen.1005697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H, Sternglanz R. 1999. Identification and characterization of the genes for two topoisomerase I-interacting proteins from Saccharomyces cerevisiae. Yeast 15: 35–41. [DOI] [PubMed] [Google Scholar]
- Pasero P, Bensimon A, Schwob E. 2002. Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev 16: 2479–2484. 10.1101/gad.232902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Sun Y, Huang SN, Nitiss JL. 2016. Roles of eukaryotic topoisomerases in transcription, replication and genomic stability. Nat Rev Mol Cell Biol 17: 703–721. 10.1038/nrm.2016.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon C, Pilch DR, Bonner WM. 2006. Genetic analysis of Saccharomyces cerevisiae H2A serine 129 mutant suggests a functional relationship between H2A and the sister-chromatid cohesion partners Csm3–Tof1 for the repair of topoisomerase I-induced DNA damage. Genetics 172: 67–76. 10.1534/genetics.105.046128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RJ, Gonzalez-Barrera S, Sunjevaric I, Alvaro D, Ciccone S, Wagner M, Rothstein R. 2011. Selective ploidy ablation, a high-throughput plasmid transfer protocol, identifies new genes affecting topoisomerase I-induced DNA damage. Genome Res 21: 477–486. 10.1101/gr.109033.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka K, Takahashi A, Sasaki M, Kobayashi T. 2016. More than 10% of yeast genes are related to genome stability and influence cellular senescence via rDNA maintenance. Nucleic Acids Res 44: 4211–4221. 10.1093/nar/gkw110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalbetter SA, Mansoubi S, Chambers AL, Downs JA, Baxter J. 2015. Fork rotation and DNA precatenation are restricted during DNA replication to prevent chromosomal instability. Proc Natl Acad Sci 112: E4565–E4570. 10.1073/pnas.1505356112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyian M, Mattarocci S, Albert B, Hafner L, Lezaja A, Costanzo M, Boone C, Shore D. 2016. Budding yeast Rif1 controls genome integrity by inhibiting rDNA replication. PLoS Genet 12: e1006414 10.1371/journal.pgen.1006414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumberg D, Pilon AA, Smith M, Hickey R, Malkas L, Pommier Y. 2000. Conversion of topoisomerase I cleavage complexes on the leading strand of ribosomal DNA into 5′-phosphorylated DNA double-strand breaks by replication runoff. Mol Cell Biol 20: 3977–3987. 10.1128/MCB.20.11.3977-3987.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves SS, Henikoff S. 2014. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol 21: 88–94. 10.1038/nsmb.2723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourrière H, Versini G, Cordón-Preciado V, Alabert C, Pasero P. 2005. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell 19: 699–706. 10.1016/j.molcel.2005.07.028 [DOI] [PubMed] [Google Scholar]
- Valjavec-Gratian M, Henderson TA, Hill TM. 2005. Tus-mediated arrest of DNA replication in Escherichia coli is modulated by DNA supercoiling. Mol Microbiol 58: 758–773. 10.1111/j.1365-2958.2005.04860.x [DOI] [PubMed] [Google Scholar]
- van Loenhout MT, de Grunt MV, Dekker C. 2012. Dynamics of DNA supercoils. Science 338: 94–97. 10.1126/science.1225810 [DOI] [PubMed] [Google Scholar]
- Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. 2008. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci 105: 9936–9941. 10.1073/pnas.0804510105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Janska A, Early A, Diffley JF. 2017. How the eukaryotic replisome achieves rapid and efficient DNA replication. Mol Cell 65: 105–116. 10.1016/j.molcel.2016.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.