Abstract
Hematopoiesis is dynamically regulated to maintain blood system function under non-homeostatic conditions such as inflammation and injury. However, common surface marker and HSC reporter systems used for prospective enrichment of HSC have been less rigorously tested under these non-homeostatic conditions. Here, we use two surface markers, EPCR/CD201 and CD34 to re-analyze dynamic changes in the HSC-enriched phenotypic SLAM compartment in a mouse model of chronic IL-1 exposure. EPCR and CD34 coordinately identify four functionally and molecularly distinct compartments within the SLAM fraction, including an EPCR+/CD34− fraction whose long-term serial repopulating activity is only modestly impacted by chronic IL-1 exposure, relative to unfractionated SLAM cells. Notably, the other three fractions expand in frequency upon IL-1 treatment and represent actively proliferating, lineage-primed cell states with limited long-term repopulating potential. Importantly, we find that the Fgd5-ZSGreen HSC reporter mouse enriches for molecularly and functionally intact HSC regardless of IL-1 exposure. Altogether, our findings provide further functional and molecular evidence of dynamic heterogeneity within a commonly used HSC-enriched phenotypic compartment under stress conditions. Importantly, they also show that stringency of prospective isolation approaches can enhance interpretation of findings related to HSC function when studying models of hematopoietic stress.
Introduction
Hematopoietic stem cells (HSC) are tasked with maintaining lifelong blood production, including under non-homeostatic conditions induced by physiological stresses[1]. HSC must therefore cope with demands brought about by infection, injury, aging, inflammatory disease and myeloablative therapeutic interventions such as irradiation (IR) and chemotherapy[2]. Thus, understanding the impact of these stresses on HSC function and the long-term viability of the HSC pool is crucial for identifying mechanisms that drive pathogenic processes such as leukemogenesis, aging and bone marrow (BM) failure.
Prospective identification and isolation of HSC under stress conditions by flow cytometry offers opportunities for detailed studies to address the unique biological features of stress hematopoiesis. Phenotypic definitions such as Lineage (Lin)−/cKit+/Sca-1+/Flk2−/CD48−CD150+ surface marker combination (hereafter referred to as SLAM), are commonly used to enrich for HSC in these studies[3–5]. Surface markers including CD34 and Endothelial Protein C Receptor (EPCR/CD201) have also been used in combination with the SLAM definition to further enrich for HSC activity [6, 7]. Other methods, such as Hoescht side-population (SP) and rhodamine dye have also been used for further enrichment in combination with SLAM [8–10]. In addition, a growing body of work has highlighted functional and molecular heterogeneity at the population and single-cell levels within phenotypic HSC gates [11–16], suggesting that markers such as SLAM contain distinct functional compartments within. Moreover, proteins such as CD41 and interferon-gamma receptor have been used to distinguish unique functional compartments within the SLAM gate under stress conditions [17, 18]. Outcomes common to most studies of inflammatory stress include loss of long-term repopulating activity as measured by transplantation of SLAM HSC, expansion of the phenotypic SLAM HSC compartment, increased cell proliferation, and activation of unique HSC-like subsets, including megakaryocyte (Mk)-biased CD41+ cells, within the SLAM gate[17, 19–23].
However, a wide variety of stressors including IR, chemotherapy agents such as 5-FU, and inflammatory cytokines can induce phenotypic shifts in key surface markers such as c-Kit and Sca-1 that may lead to contamination of the phenotypic HSC gate with non-HSC cell types [19, 24–26], or exclusion of stem/progenitor cells that may transiently express traditionally excluded lineage markers such as Mac-1 under stress conditions [27]. In recent years, numerous reporter mouse lines have also been developed to enrich for HSC, based on expression of fluorescent reporters driven by promoter of genes including Hoxb5, Fgd5, Vwf, Gprc5c and Krt18 [28–32]. Altogether, the capacity of surface markers and reporters to identify functional HSC in non-homeostatic conditions such as inflammation have not been extensively evaluated. Hence, careful evaluation and enhancement of commonly used HSC marker/reporter definitions can ensure correct interpretation of experimental results in studies of stress hematopoiesis.
In the present study, we used a model of chronic IL-1β exposure to evaluate changes within the SLAM gate during inflammatory stress. We use the markers EPCR and CD34 to coordinately identify four distinct SLAM fractions with unique functional and molecular properties. Strikingly, we find that the relative abundance of these fractions changes significantly following IL-1 treatment, with a sharp reduction in the EPCR+/CD34− fraction, which enriches for the vast majority of HSC activity. Importantly, we find that chronic IL-1 exposure does not substantially change the molecular or proliferative properties of each fraction. Likewise, we find that Fgd5-ZSGreen expression identifies HSC with equivalent molecular and functional properties regardless of IL-1 exposure, indicating that a functional long-term HSC pool is retained even under chronic inflammatory stress. Altogether, this work provides critical insights into the dynamic nature of stress hematopoiesis and provides critical insight into improved strategies to identify functional HSC under non-homeostatic conditions.
Materials and methods
Mice
Wild-type C57BL/6, CD45.1+ congenic B6.SJL-PtprcaPepcb/BoyJ (Boy/J) mice and Fgd5-ZSGreen-CreERT mice [30]were obtained from The Jackson Laboratory and bred in-house. Vwf-GFP mice [33] were a kind gift of Dr. C. Nerlov, Oxford University, UK. Both male and female mice were used for experiments. All procedures were performed in accordance with an IACUC-approved University of Colorado Anschutz Medical Campus animal protocol (protocol number 00091).
In vivo experiments
Recombinant murine IL-1β injections were performed as previously described [20]. Mice were injected intraperitoneally with 0.5 μg IL-1β (Peprotech) in 100 μl of 0.22 μm sterile-filtered PBS/0.2% bovine serum albumin (BSA), or 100 μl PBS/0.2% BSA alone. Mice were injected daily on alternating sides for 20 days. Transplantation experiments were performed as previously described [20]. 8–12 week old Boy/J mice were lethally irradiated with 11 Gy in a split dose 3 hours apart using a Cs137 fixed-beam source (J.L. Shepherd) and injected retro-orbitally with 250 donor test cells plus 5×105 Sca-1-depleted Boy/J helper BM cells. Mice were maintained on Bactrim in water for three weeks following transplantation. Secondary transplantations were performed similarly, but with 500 donor test cells. Donor peripheral blood chimerism was analyzed every four weeks by submandibular bleed and collection of blood in 4ml of ACK buffer (150 mM NH4Cl/10 mM KHCO3) for flow cytometry analyses. EdU labeling was performed by injecting 2mg EdU in PBS intraperitoneally at the same time as the final IL-1/PBS injection, and euthanizing mice 24 hours later. At the termination of all experiments, mice were euthanized by CO2 inhalation followed by cervical dislocation according to approved protocols.
Flow cytometry
BM cells were isolated by crushing legs, arms, pelvis and spines from mice in Staining Medium (SM) consisting of Hanks’ Buffered Saline Solution (HBSS) with 2% heat-inactivated FBS. Cells were subsequently incubated on ice with ACK buffer for 3 min to remove erythroid cells, washed with SM and centrifuged on a Histopaque gradient (Histopaque 1119; Sigma-Aldrich). To enrich c-Kit+ cells for sorting, BM cells were incubated for 20 minutes with c-Kit microbeads (Miltenyi Biotec, 130–091-224), washed with SM and enriched on an AutoMACS Pro cell separator (Miltenyi Biotec). For analysis of BM populations, BM cells were flushed with SM from both femurs and tibiae from each mouse, treated with ACK and counted on a ViCell automated counter (Beckman-Coulter) prior to staining. For hematopoietic stem and progenitor cell analyses, 107 cells were blocked with purified rat IgG (Sigma Aldrich) and stained with PE-Cy5-conjugated lineage antibodies against B220, CD3, CD4, CD5, CD8, Gr1 and Ter119, plus CD34-FITC, EPCR-PE, Mac-1-PE/Cy7, Flk2-Biotin, ESAM-APC, CD48-A700, and c-Kit-APC/Cy7 for 30 min on ice, washed with SM, and stained with Sca-1-BV421, CD41-BV510, Strepatavidin-BV605, and CD150-BV785 in a staining buffer comprised of 1:3 vol/vol Brilliant Buffer (BD Biosciences)/SM. Cells were also analyzed for IL-1R (anti-IL-1R-PE) and CD49b (anti-CD49b-PE/Cy7) by substituting EPCR-APC for EPCR-PE. For SLAM chimerism analyses, a variation of this panel was used in which CD41 and Mac-1 were omitted and CD45.1 BV650 and CD45.2-PE/Cy7 were used. For mature BM cell analyses, 5×105 cells were stained with Gr-1-Pacific Blue, B220-BV785, CD4-FITC, CD8-PE, Mac-1-PE/Cy7, IgM-APC, CD3-A700 and CD19-A780. Dead cells were excluded by resuspending cells in SM containing 1 μg/ml propidium idodide (PI). Samples were analyzed on a 12-channel, 3-laser FACSCelesta or a 20-channel, 5-laser Fortessa X-20 analyzer running FACSDiva software (Becton-Dickenson). Data were analysed using FlowJo version 10 (FlowJo). For cell sorting, anti-CD41 antibodies were not included in the staining cocktail. For sorting experiments, cells were double-sorted to purity on a FACSAria IIu or Aria Fusion cell sorter (Becton-Dickenson). For Ki67/DAPI cell cycle analysis, 107 cells were stained as previously described [34] using the 12-color panel above, except using Sca-1-PE/Cy7 and excluding CD41 and Mac-1. After staining, cells were fixed with CytoFix/CytoPerm (BD Biosciences) for 30 minutes at room temperature (RT), washed with 1x PermWash buffer (BD Biosciences), permeablized with Perm Buffer Plus (BD Biosciences), washed with PermWash buffer, re-fixed with CytoFix/CytoPerm for 10 minutes at RT, and stained with anti-Ki67-PerCP-Cy5.5 for 1 hour at RT before washing in PermWash. Cells were subsequently incubated with SM containing 2 μg/ml DAPI. For EdU analysis, we found that EPCR was destroyed by the fix/perm regimen. Hence, we double-sorted SLAM fractions from EdU-injected mice and mixed the cells with 1 × 105 B220+ carrier cells harvested from the spleen, and EdU was visualized using the Click-iT Plus EdU Flow Cytometry Assay Kit (Thermo Fisher Scientific) containing the AlexaFluor 488 picolyl azide fluorochrome and following the kit instructions. Fix, perm/wash and click reagents were diluted according to the manufacturer’s instructions, and sorted cells were fixed for 15 minutes at RT, washed in PBS/1% BSA, permeablized for 15 minutes at RT, washed and incubated with click reagents for 30 minutes at RT. Cells were then washed in perm/wash before analysis. For all flow cytometry applications, cells were analyzed on a four-laser, 20-channel LSRII analyzer running FACSDiva software (Becton-Dickenson) and analyzed using FlowJo. For all procedures listed above, antibody clone, manufacturer, catalog number and dilution information are contained in Supplementary Table 1.
Gene expression analysis
For gene expression analyses, pools of 100 cells were sorted into wells of a DNase and RNase-free 96-well plate (Applied Biosystems) containing 5 μl CellsDirect 2x reaction buffer (Invitrogen), centrifuged for 5 minutes at 500×g, snap-frozen on dry ice and stored at −80°C until use. RNA was reverse-transcribed using Superscript III Taq polymerase (Invitrogen) and preamplified for 18 rounds with a custom 96-target DeltaGene (Fluidigm) primer panel on a PCR cycler (Eppendorf). Excess primers were removed from the preamplified product by incubating with Exonuclease-1 (New England Biolabs), and cDNA samples were diluted in DNA buffer. Primers and cDNAs mixed with SsoFast Sybr Green Master Mix (BioRad) were subsequently loaded onto a Fluidigm 96.96 Dynamic Array IFC and run on a BioMark HD system (Fluidigm). Data were subsequently analyzed using Fluidigm Gene Expression Software and normalized to Gusb. Relative changes were subsequently calculated using ΔΔCt approach. Unsupervised clustering of Gusb-normalized delta CT values with Gusb removed along with poorly performing Ebf1 and Hoxa2 primer sets, was performed using average linkage. Clustering and principal component analysis (PCA), and heatmap generation, were performed using ClustVis software (biit.cs.ut.ee/clustvis). PCA and PCA loading plots were generated using Prism 8 (Graphpad) from data generated by ClustVis.
Statistical analysis
Statistical analyses were performed using Prism 8 software (GraphPad). P-values were determined using either a Mann-Whitney u-test for bivariate comparisons or two-way ANOVA for multivariate comparisons. P-values ≤ 0.05 were considered statistically significant.
Results
EPCR and CD34 coordinately enrich for functional HSC during chronic IL-1 exposure
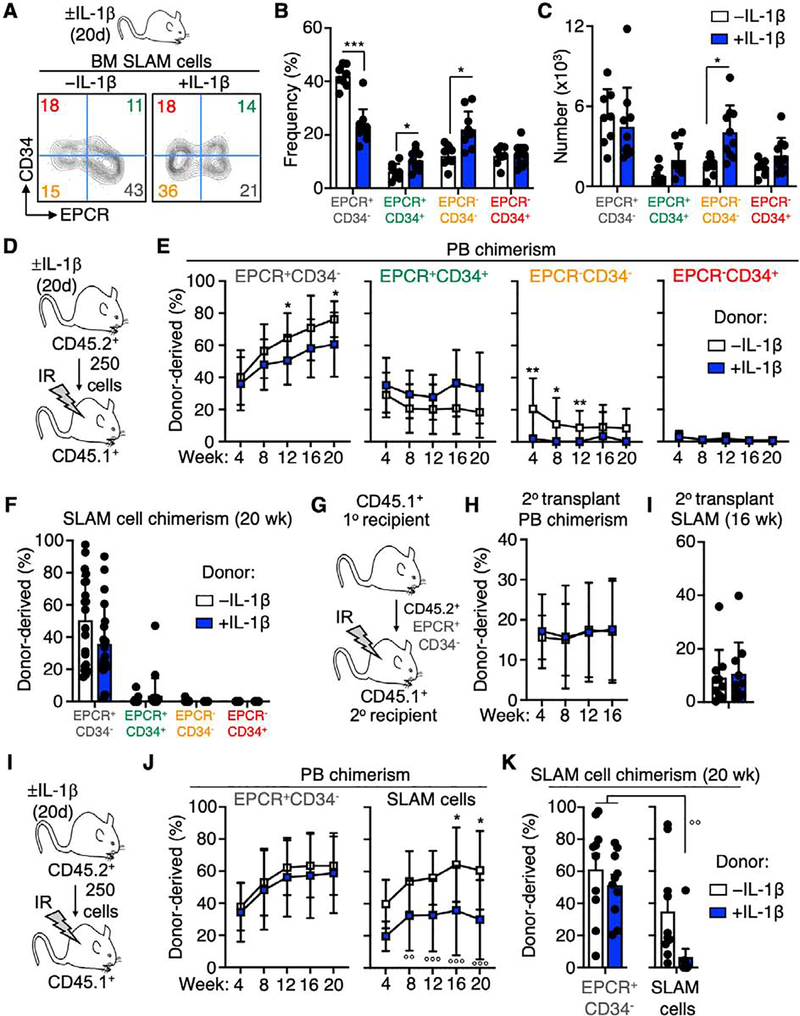
To address the impact of chronic IL-1-driven inflammatory signaling on functional heterogeneity in the HSC-enriched SLAM (LSK/Flk2−/CD48−/CD150+/ESAM+) HSC faction, we injected mice intraperitoneally each day for 20 days ± 0.5 μg of recombinant murine IL-1β. Consistent with our prior published results, chronic IL-1 treatment induced expansion of myeloid cells coincident with contraction of B cell populations in the BM (Fig. S1A–C), as well as expansion of SLAM HSC and multipotent progenitor (MPP)-2 and MPP3 populations [21, 35] (Fig. S1D–E). To study IL-1-induced changes in the SLAM compartment we reanalyzed cells in this compartment using EPCR and CD34, which have been previously used to distinguish functional HSC in the SLAM fraction [7, 36]. Coordinate use of EPCR and CD34 allowed us to prospectively identify four distinct phenotypic compartments within the SLAM gate (Fig. 1A). Strikingly, the relative abundance of these compartments was altered significantly following chronic IL-1 exposure, with a significant decrease in the frequency of EPCR+/CD34− cells, alongside twofold expansion in frequency of the EPCR−/CD34− compartment (Fig. 1B). Despite decreased frequency, the absolute number of EPCR+/CD34− cells in the BM was unchanged (Fig. 1C), whereas increased absolute numbers of the EPCR−/CD34− fraction substantially contributed to the overall numerical expansion of SLAM cells observed following chronic IL-1 exposure (Fig. 1C, Fig. S1D).
Fig. 1. The EPCR+/CD34− SLAM fraction enriches for functional HSC.
(A) Experimental design and representative FACS plots of SLAM cells from mice treated ± IL-1 for 20d fractionated by EPCR and CD34 expression, (B) Frequency and (C) absolute number of EPCR/CD34 SLAM cell fractions (n = 9–10/grp). (D-F) Long-term engraftment of purified SLAM cells fractionated by EPCR and CD34 expression, from mice treated ± IL-1 for 20d. (D) Experimental design; (E) Donor chimerism in peripheral blood (PB) and (F) phenotypic SLAM compartment of recipient mice at the indicated time points (N = 18–20/grp, compiled from two independent experiments). (G) Donor PB and (H) SLAM chimerism following secondary transplant of donor-derived SLAM cells from primary recipient mice in (D) (n = 18–20/grp, compiled from two independent experiments). (I-K) Long-term engraftment of SLAM and EPCR+/CD34− SLAM cells, from mice treated ± IL-1 for 20d. (I) Experimental design; (J) Donor chimerism in peripheral blood (PB) and (K) phenotypic SLAM compartment of recipient mice at the indicated time points (N = 9–10/grp, representative of two independent experiments). ○○ p < 0.01 vs. -IL-1 EPCR+/CD34− SLAM cells by one-way ANOVA; * p < 0.05, ** p < 0.01, *** p < 0.001 vs. -IL-1 condition in each fraction by Mann-Whitney u-test.
To interrogate the functional properties of SLAM fractions defined by CD34 and EPCR expression during homeostatic and chronic inflammatory conditions, we first isolated each fraction from mice treated ± IL-1 for 20 days, transplanted them into lethally irradiated CD45.1+ recipient mice, and analyzed long-term repopulating activity by bleeding recipient animals every four weeks (Fig. 1D, Fig. S2A). Notably, the EPCR+/CD34− fraction contained the majority of long-term repopulating activity, exhibiting high levels of multilineage donor chimerism in the peripheral blood (PB) out to 20 weeks (Fig. 1E, Fig. S2B). On the other hand, EPCR+/CD34+ cells and both EPCR− fractions generated consistently low to absent donor chimerism by 20 weeks (Fig. 1E). Along these lines, we recovered donor-derived SLAM cells exclusively in mice transplanted with EPCR+/CD34− cells (Fig. 1F, Fig. S2C). Notably, chronic IL-1 exposure moderately decreased donor-derived peripheral blood chimerism in mice transplanted with EPCR+/CD34− cells (Fig. 1E). However, donor-derived SLAM cell chimerism in the BM was unchanged, suggesting that chronic IL-1 exposure has a limited effect on long-term repopulating activity of cells in the EPCR+/CD34− fraction (Fig. 1F). In addition, chronic IL-1 exposure did not impair the capacity of transplanted EPCR+/CD34− cells to give rise to all SLAM fractions including EPCR+/CD34− cells after 20 weeks (Fig. S2C). To address serial repopulating activity of the EPCR+/CD34− fraction, we re-isolated donor-derived EPCR+/CD34− cells from the primary recipient mice and transplanted them into lethally irradiated CD45.1+ secondary recipients (Fig. 1G). In line with our primary transplant data, we observed roughly equal myeloid/lymphoid donor PB chimerism, as well as SLAM HSC chimerism, regardless of chronic IL-1 exposure (Fig. 1H–I, Fig. S2D). Collectively, these experiments establish the EPCR+/CD34− fraction as the major source of long-term HSC activity in the SLAM compartment, consistent with prior studies using this definition [37].
Given the relatively minor impact of chronic IL-1 exposure on EPCR+/CD34− cell repopulating activity, we next assessed whether the EPCR+/CD34− marker definition provides superior enrichment for long-term HSC activity relative to the total SLAM fraction following IL-1 exposure (Fig. 1I). Notably, SLAM cells from IL-1 exposed mice exhibited significantly reduced long-term donor-derived PB and SLAM chimerism relative to controls, as well as relative to EPCR+/CD34− cells in either condition. (Fig. 1J–K), suggesting the EPCR+/CD34− SLAM fraction provides superior enrichment for long-term HSC versus unfractionated SLAM cells. We also analyzed expression of CD49b in each SLAM fraction, since low CD49b expression identifies long-term repopulating cells (LTRC) with chemoresistant ‘reserve HSC’ (R-HSC) properties[38, 39]. We found the EPCR+/CD34− fraction was most highly enriched for CD49blo LTRC (Fig. S3A–B). The non-long-term repopulating EPCR−/CD34− fraction also showed extensive enrichment for CD49blo cells, suggesting that CD49b alone may not completely exclude non-long-term repopulating cells in the SLAM gate. The EPCR+/CD34− fraction also contained CD49bhi cells, indicating this fraction likely contains a mixture of R-HSC and long-term repopulating but chemosensitive ‘primed HSC’ (P-HSC) that can be selectively depleted upon 5-FU stress[40].
Taken together, these findings show that EPCR and CD34 coordinately identify a SLAM fraction that effectively enriches for functional long-term HSC regardless of chronic exposure to IL-1. They also suggest that the impaired long-term repopulating activity of IL-1-exposed SLAM cells we previously reported [20] may be at least partially due to a significantly decreased proportion of EPCR+/CD34− cells occupying the phenotypic SLAM gate following IL-1 treatment, rather than functional impairment of long-term repopulating HSC themselves (Fig. 1A).
EPCR and CD34 coordinately identify distinct molecular phenotypes in the SLAM gate
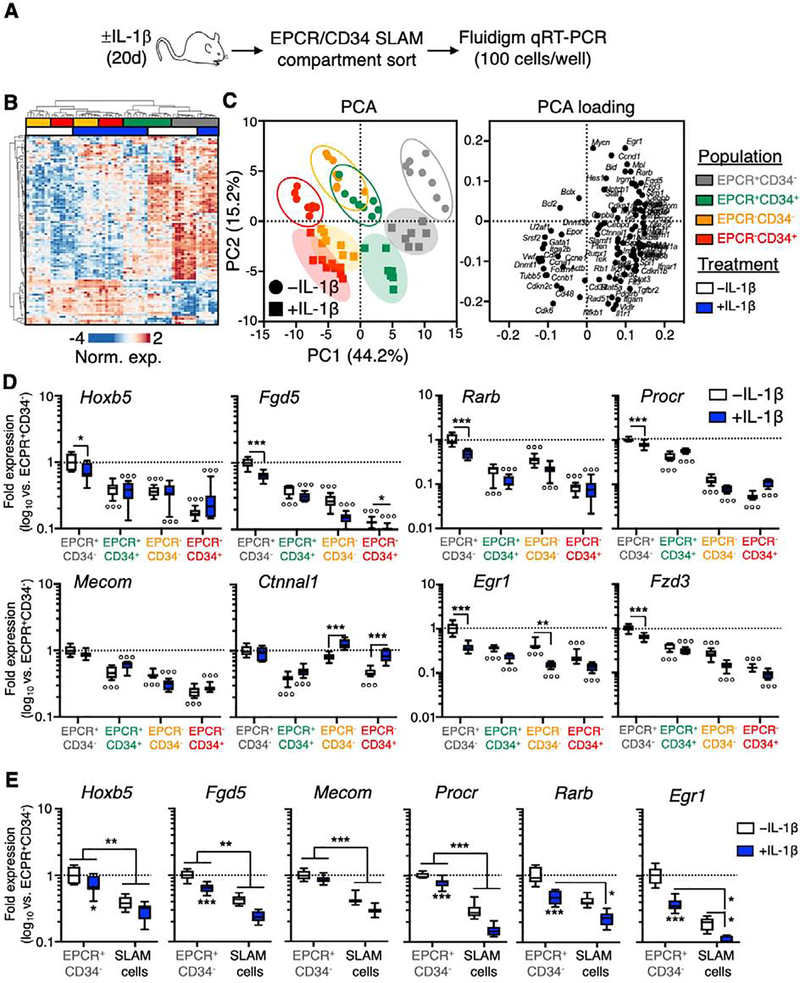
To understand the molecular basis for our transplantation assays, we used a custom 96-gene Fluidigm qRT-PCR array to evaluate the molecular characteristics of each SLAM fraction in mice treated ± IL-1 for 20 days (Fig. 2A). Notably, hierarchical clustering analysis revealed that EPCR expression constitutes the major breakpoint between the four populations, with EPCR+ fractions showing the highest degree of relatedness regardless of IL-1 exposure (Fig. 2B). Along these lines, principal component analysis (PCA) revealed two distinctive molecular phenotypes, with the first principal component (PC1) establishing a linear relationship between SLAM fractions that potentially resembles a differentiation gradient, with HSC activity-enriched EPCR+/CD34− cells and EPCR−/CD34+ cells, which do not exhibit long-term donor engraftment, at the extrema. On the other hand, the second principal component (PC2) separated populations based on IL-1 exposure (Fig. 2C). Interestingly, IL-1 treatment did not substantially alter the order of the fractions in PC space, suggesting that IL-1 does not fundamentally alter the molecular identity of each fraction, but instead activates a discreet gene program(s) within each. Consistent with this model, PC loading analysis identified unique gene sets that define each principal component (Fig. 2C). Along these lines we analyzed expression of genes associated with functional HSC in each SLAM fraction ± chronic IL-1 exposure. As predicted by our transplantation assays, EPCR+/CD34− cells expressed the highest levels of HSC genes including Hoxb5, Fgd5, Rarb and Procr, alongside high surface expression of Sca-1 and EPCR (Fig. 2D, Fig. S3C). Consistently, chronic IL-1 exposure significantly reduced expression of several of these genes in the EPCR−/CD34+ fraction (Fig. 2D, Fig. S3C). On the other hand, expression of HSC genes was lowest in the EPCR−/CD34+ fraction, with EPCR−/CD34+ cells also expressing the highest level of CD48, which serves as a marker of HSC differentiation (Fig. 2D, Fig. S3C–D). Notably, EPCR+/CD34+ cells expressed the highest levels of Flk2, which could be consistent with a proliferative, metabolically active MPP1 phenotype [35]. Moreover, increased CD150 expression, which is associated with hematopoietic stress responses [20, 21], occurred exclusively in EPCR− cell fractions following chronic IL-1 exposure (Fig. S3C–D). Furthermore, we assayed IL-1R expression at the mRNA and protein levels and found detectable levels in each compartment (Fig. S4A–C), with IL-1R surface expression at low levels in at least a fraction of cells, suggesting cells in each compartment can respond to IL-1. While IL-1 treatment led to increased levels of Il1r1 mRNA in all SLAM fractions, this did not translate to increased surface expression (Fig. S4A–C). Lastly, given transplantation of the EPCR+/CD34− SLAM fraction yielded superior enrichment for HSC activity relative to unfractionated SLAM cells following IL-1 treatment, we compared expression of HSC-associated genes between the EPCR+/CD34− fraction and unfractionated SLAM cells. As predicted by our functional analyses, EPCR+/CD34− cells expressed significantly higher levels of Hoxb5, Fgd5, Mecom, Procr, Rarb and Egr1 than unfractionated SLAM cells (Fig. 2E), regardless of IL-1 exposure. Hence, these data indicate that EPCR and CD34 identify molecularly distinct compartments within the SLAM gate, complementing our functional studies. They also provide a molecular basis for the superior repopulating activity in EPCR+/CD34− cells versus unfractionated SLAM cells following IL-1 exposure.
Figure 2. EPCR and CD34 identify molecularly distinct SLAM compartments.
(A) Experimental design of Fluidigm-based molecular analysis of SLAM cells fractionated by EPCR and CD34 expression from mice treated ± IL-1 for 20d. (B) Heatmap and hierarchical clustering analysis, and (C) principal component analysis (PCA; left) and PCA loading plot (right) of Fluidigm gene expression data (N = 7–8 per group). (D) Expression of HSC-associated genes in SLAM cells fractionated by EPCR and CD34 expression, from mice treated ± IL-1 for 20d. (E) Expression of HSC-associated genes in total SLAM cells and EPCR+/CD34− SLAM cells from mice treated ± IL-1 for 20d. Data in (D-E) are expressed as log10 fold expression vs. -IL-1 EPCR+/CD34− SLAM cells. ○ p < 0.05, ○○ p < 0.01, ○○○ p < 0.001 vs. -IL-1 EPCR+/CD34− SLAM cells; * p < 0.05, ** p < 0.01, *** p < 0.001 vs. -IL-1 condition within each fraction, as determined by two-way ANOVA. Data are representative of two independent experiments.
The EPCR+/CD34− SLAM fraction remains quiescent following chronic IL-1 exposure
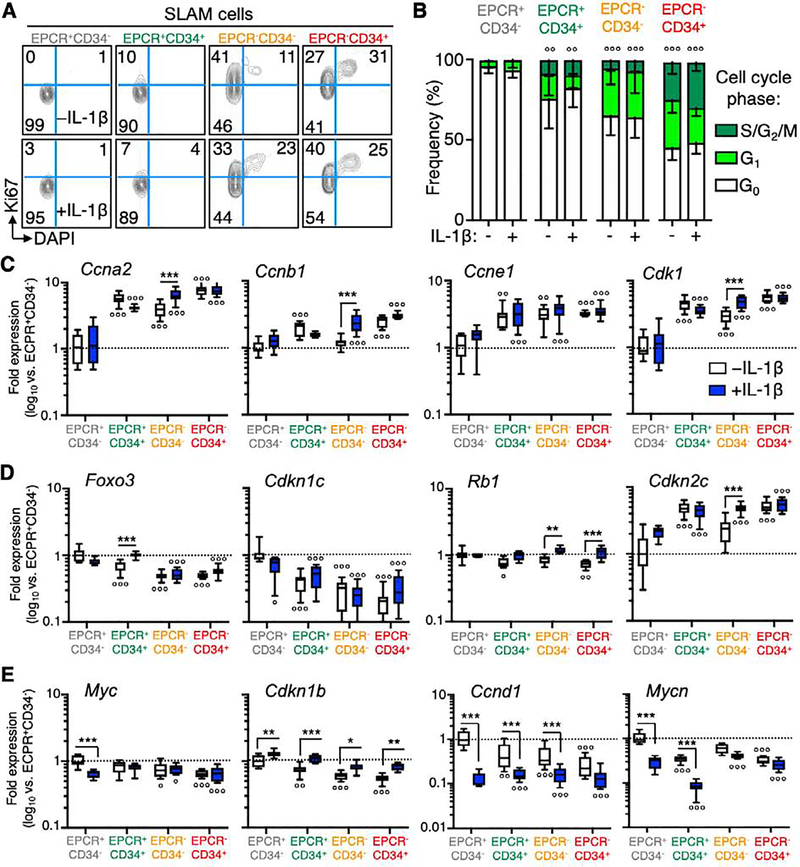
SLAM cells remain largely quiescent during chronic inflammatory conditions such as interferon (IFN) stimulation, obesity and collagen-induced arthritis (CIA)[19, 21, 41, 42]. To better understand the biology of the different SLAM fractions following chronic IL-1 exposure, we analyzed their cell cycle distribution by Ki67/DAPI staining in mice treated ± IL-1 for 20 days. Interestingly, we uncovered a gradient of cell cycle activity in each compartment, with EPCR+/CD34− cells almost entirely in G0, whereas roughly half of EPCR−/CD34+ cells were actively cycling and spread between G1 and S/G2/M phases (Fig. 3A–B). Consistent with the MPP1 phenotype, CD34 expression marked elevated cell cycle activity regardless of EPCR expression level. Strikingly, cell cycle distribution of all SLAM fractions was unchanged following chronic IL-1 exposure, suggesting that chronic IL-1 does not substantially modulate this key cellular characteristic. We confirmed these findings independently using EdU incorporation to measure the proliferative activity in each SLAM fraction (Fig. S5A). Consistent with our Ki67/DAPI analyses we found that EPCR− SLAM fractions had a higher proliferative index than EPCR+ cells. Also similar to our Ki67/DAPI analyses, IL-1 exposure did not significantly alter proliferative activity in any fraction (Fig. S5B–C). Consistent with our cell cycle data, our molecular analyses showed that induction of CD34 expression in EPCR+ SLAM cells, followed by loss of EPCR expression, were associated with increased priming for cell cycle activity, with progressively increasing levels of Ccna2, Ccnb1, Ccne1 and Cdk1 between EPCR+/CD34− and EPCR−/CD34+ fractions (Fig. 3C). On the other hand, consistent with an HSC identity, EPCR+/CD34− cells had the highest expression of cell cycle inhibitor genes including Foxo3, Cdkn1c/p57, and Rb1 (Fig. 3D). Cdkn2c/p18 levels were highest in EPCR−/CD34+, suggesting distinct mechanisms may regulate cell cycle activity in these SLAM fractions. Consistent with minimal induction of cell cycle activity by chronic IL-1, expression levels of cell cycle activator genes did not undergo substantial shifts following IL-1 exposure (Fig. 3D). Interestingly, Rb1 and Cdkn2c/p18 levels increased significantly with IL-1 exposure in both EPCR− fractions, perhaps indicating activation of an Rb-mediated ‘braking’ mechanism unique to these cells (Fig. 3D). On the other hand, while Foxo3 and Cdkn1c/p57 levels decreased in EPCR+/CD34− cells following IL-1 exposure, our analyses identified significant IL-1 treatment-associated decreases in expression of Myc, Mycn and Ccnd1, which are crucial mediators of HSC cell cycle entry, as well as increased levels of the Cdkn1b/p27 (Fig. 3E). Notably, this expression pattern is similar to the cell cycle restriction gene program we recently identified in SLAM cells from mice with collagen-induced arthritis [41], suggesting modulation of these genes is a conserved response to multiple inflammatory stimuli.
Figure 3. EPCR+/CD34− SLAM cells are highly quiescent.
(A) Representative FACS plots and (B) quantification of cell cycle distribution in SLAM cells fractionated by EPCR and CD34 expression, from mice treated ± IL-1 for 20d (n = 5/grp). (C-E) Expression of cell cycle activator and inhibitor genes in SLAM cells fractionated by EPCR and CD34 expression from mice treated ± IL-1 for 20d (n = 7–8/grp). Data in (C-E) are expressed as log10 fold expression vs. -IL-1 EPCR+/CD34− SLAM cells. ○ p < 0.05, ○○ p < 0.01, ○○○ p < 0.001 vs. -IL-1 EPCR+/CD34− SLAM cells; * p < 0.05, ** p < 0.01, *** p < 0.001 vs. -IL-1 condition within each fraction, as determined by two-way ANOVA. Data are representative of two independent experiments.
We also analyzed our gene expression data to uncover potential regulatory mechanisms promoting quiescence and long-term repopulation capacity in EPCR+/CD34− cells during chronic IL-1. We found that EPCR+/CD34− SLAM cells maintained high relative levels of Nrf2- and Hif1α-regulated genes versus the remaining SLAM fractions. Both pathways are known to be highly active in HSC, and deletion of key components such as Nrf2 and Hif1a leads to excessive HSC proliferation and/or mobilization and loss of self-renewal[43–45]. We also observed high levels of target genes that regulate redox state (Gstt3) and promote quiescence-associated metabolic pathways including fatty acid metabolism (Cpt1a) and glycolysis (Gapdh) [46, 47] (Fig. S6A–B). We also observed robust induction of the Hif1α target genes Vldlr and Pdgfrb by IL-1 in multiple SLAM fractions, which could reflect metabolic adaptation to increased hypoxia in the BM or changes in energy source abundance. Hence, EPCR+/CD34− cells retain key metabolic properties associated with their quiescence and self-renewal under chronic IL-1 conditions. Altogether, these data show that the SLAM compartment is comprised of cell populations with heterogenous cell cycle states whose distinct proliferative and metabolic activities are maintained during chronic inflammation. They also identify IL-1-driven modulations in key cell cycle and metabolic regulators that may account for minimal cell cycle changes.
EPCR expression resolves lineage priming in SLAM compartments
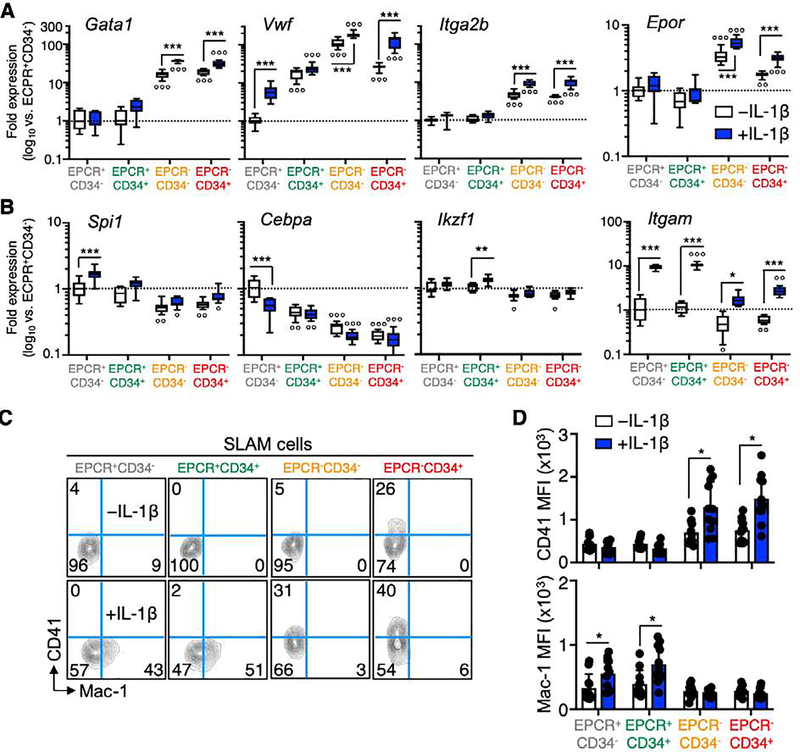
In addition to changes in cell cycle gene expression, we investigated expression of key lineage determinant genes in SLAM fractions. Our PC loading analyses identified the megakaryocyte/erythroid (Mk/E)-lineage genes Gata1, Vwf and Itga2b (Cd41) and EpoR as key markers distinguishing EPCR+ and EPCR− SLAM fractions (Fig. 2C). Along these lines, we observed significant upregulation of these four genes in both EPCR− cell fractions, suggesting they may represent Mk/E-biased populations. (Fig. 4A). Expression of these genes also increased significantly in EPCR− fractions following chronic IL-1 exposure. On the other hand, both EPCR+ fractions were enriched for expression of the myeloid lineage determinants Spi1/Pu.1 and Cebpa, as well as the lymphoid gene Ikzf1, consistent with a more extensive myeloid/lymphoid repopulating activity in these fractions relative to EPCR− cells (Fig. 1E). Notably, the chronic inflammation-mediated increase in Spi1 expression we had previously documented in unfractionated SLAM cells [20, 41] was particularly robust in the EPCR+/CD34− SLAM fraction (Fig. 4B). Interestingly, expression of Itgam (Mac-1), normally considered a myeloid lineage marker, increased significantly in all SLAM fractions after chronic IL-1 exposure, though expression levels were nearly 10-fold higher in the EPCR+ fractions (Fig. 4B). Given our molecular findings, we assessed CD41 and Mac-1 levels on the surface of each SLAM fraction. In line with previous work identifying an inflammation-induced Mk-biased CD41+ SLAM compartment [17, 20], we found that CD41 expression was induced by IL-1 exclusively in EPCR− SLAM fractions. (Fig. 4C, D). Strikingly, on the other hand Mac-1 surface expression was induced exclusively on EPCR+ SLAM fractions following IL-1 exposure (Fig. 4C, D). Collectively, these data suggest EPCR− and EPCR+ SLAM fractions have distinct lineage priming, with EPCR− fractions exhibiting a strong Mk/E signature. On the other hand, EPCR+ fractions express myeloid and lymphoid gene markers consistent with robust myeloid/lymphoid repopulating activity, particularly in the EPCR+/CD34− fraction. Our data also indicate that Mac-1, which is often excluded as a mature myeloid lineage marker in HSC staining panels, can be expressed at low levels in SLAM fractions enriched for long-term HSC activity, following chronic inflammatory stimulation.
Figure 4. EPCR expression distinguishes Mk/E-primed SLAM cells.
(A-B) Expression of (A) Mk/E and (B) myeloid/lymphoid lineage genes in SLAM cells fractionated by EPCR and CD34 expression, from mice treated ± IL-1 for 20d (n = 7–8/grp). Data in (A-B) are expressed as log10 fold expression vs. -IL-1 EPCR+/CD34− SLAM cells. (C) Representative FACS plots and (D) geometric mean fluorescence intensity (MFI) quantification of CD41 and Mac-1 expression in cells fractionated by EPCR and CD34 expression, from mice treated ± IL-1 for 20d (n = 10–11/grp). ○ p < 0.05, ○○ p < 0.01, ○○○ p < 0.001 vs. -IL-1 EPCR+/CD34− SLAM cells; * p < 0.05, ** p < 0.01, *** p < 0.001 vs. -IL-1 condition within each fraction, as determined by two-way ANOVA. Data are representative of two independent experiments.
Fgd5 enriches for functional HSC in homeostatic and inflammatory conditions
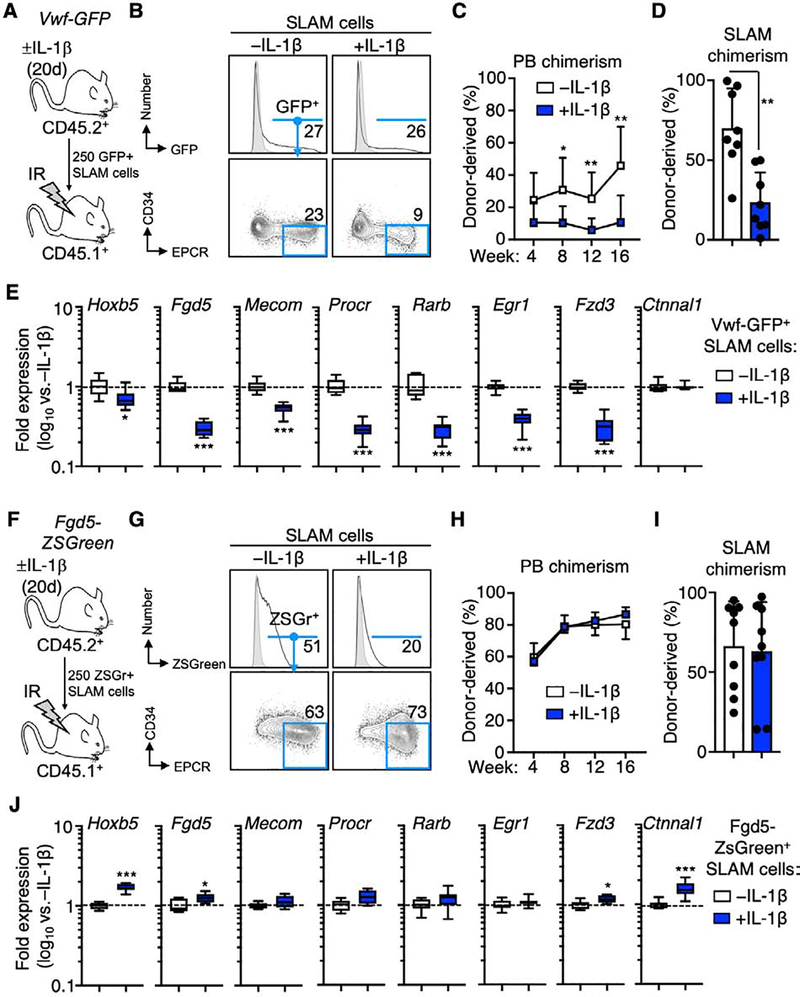
We next assessed whether two reporter strains shown to identify HSC, Vwf-eGFP[33] and Fgd5-ZSGreen[30], faithfully enrich for HSC activity under chronic inflammatory stress. We therefore transplanted reporter-positive SLAM cells from mice treated ± IL-1β for 20 days into lethally irradiated CD45.1+ recipient mice (Fig. 5A, F). Interestingly, while the proportion of Vwf-eGFP+ SLAM cells remained roughly equal following chronic IL-1 exposure, the proportion of EPCR+/CD34− cells in the Vwf-eGFP+ SLAM gate decreased following chronic IL-1 exposure (Fig. 5B). Consistently, donor chimerism as measured by CD45.2 expression in the peripheral blood and BM SLAM compartment was significantly lower in mice transplanted with Vwf-GFP+ SLAM cells following chronic IL-1 exposure (Fig. 5C–D). Consistent with decreased abundance of EPCR+/CD34− cells in the Vwf-eGFP+ SLAM gate, levels of HSC genes, with exception of Ctnnal1, were significantly lower in Vwf-eGFP+ SLAM cells from chronic IL-1-treated mice (Fig. 5E). Likewise, this phenotypic change was reflected in the decreased proportion of Vwf-eGFP+ cells in the EPCR+/CD34− SLAM fraction itself, whereas the abundance of Vwf-eGFP+ cells increased in EPCR− SLAM fractions (Fig. S7A). This suggests that the Vwf-eGFP+ SLAM gate becomes contaminated with non-long term engrafting cells following chronic exposure to IL-1.
Figure 5. Fgd5 enriches for functional EPCR+/CD34− HSC under inflammatory conditions.
(A) Experimental design; (B) representative FACS plots showing frequency of Vwf-GFP+ SLAM cells and EPCR+/CD34− cells within the Vwf-GFP+ gate; (C) donor-derived PB and (D) BM SLAM cell chimerism following transplantation into recipient mice (n = 8/grp). (E) Expression of HSC-associated genes in Vwf-GFP+ SLAM cells from mice treated ± IL-1 for 20d. (F) Experimental design; (G) representative FACS plots showing frequency of ZSGreen+ SLAM cells and EPCR+/CD34− cells within the Fgd5-ZSGreen+ gate; (H) donor-derived PB and (I) BM SLAM cell chimerism following transplantation into recipient mice (n = 9/grp). (J) Expression of HSC-associated genes in Fgd5-ZSGreen+ SLAM cells from mice treated ± IL-1 for 20d. Data are representative of two independent experiments. Data in (E) and (J) are expressed as log10 fold expression vs. -IL-1 condition. * p < 0.05, ** p < 0.01, *** p < 0.001 vs. -IL-1 condition, as determined by Mann-Whitney u-test.
On the other hand, while the Fgd5-ZSGreen reporter marked a smaller proportion of SLAM cells in chronic IL-1-exposed Fgd5-ZSGreen mice, the Fgd5-ZSGreen+ gate contained roughly equal proportions of EPCR+/CD34− cells regardless of IL-1 exposure (Fig. 5G). Consistent with this observation, donor chimerism in the PB and BM SLAM compartments was essentially equivalent (Fig. 5H–I). Likewise, expression of HSC genes was equal or even higher in Fgd5-ZSGreen+ SLAM cells from IL-1-treated mice (Fig. 5J). On the other hand, while the abundance of EPCR+/CD34− cells in the Fgd5+ SLAM compartment was essentially unchanged, the fraction of Fgd5+ cells in the EPCR+/CD34− SLAM fraction decreased (Fig. S7B). This is consistent with decreased Fgd5 mRNA levels (Fig. 2D) in the EPCR+/CD34− SLAM fraction following IL-1 treatment and the moderate decrease in their engraftment following IL-1 exposure (Fig. 1E). These data suggest that Fgd5 expression can identify a subset of EPCR+/CD34− SLAM cells with essentially unimpaired functional properties during inflammatory challenge. Altogether, these data indicate that the Fgd5-ZSGreen reporter faithfully marks phenotypic and functional HSC following chronic IL-1 exposure. These findings are consistent with a report that Fgd5 reporter expression enriches for functional HSC following acute poly I:C stimulation [48]. These data also suggest that the EPCR+/CD34− surface marker combination may have predictive value in assessing the degree to which HSC-specific reporter strains faithfully mark functional HSC in inflammatory conditions.
Discussion
In the present study, we show that the surface markers CD34 and EPCR can be used to prospectively identify a continuum of molecular and functional cell states within the phenotypic SLAM compartment, that include induction of cell cycle activity and distinct lineage programs. Using transplantation, we find these cell states are associated with varying degrees of serial myeloid/lymphoid repopulating activity, with EPCR+/CD34− SLAM cells enriching for a highly potent HSC activity under normal and inflammatory conditions. We show that EPCR expression distinguishes CD41+ Mk/E-primed SLAM cells from long-term repopulating HSC, which express Mac-1 following IL-1 treatment. Lastly, we find that the Fgd5-ZSGreen reporter strain can be used to prospectively enrich for EPCR+/CD34− SLAM cells with equivalent long-term repopulating activity following IL-1 exposure. Altogether, these data indicate that the phenotypic SLAM compartment is dynamically regulated under non-homeostatic conditions, while still maintaining a viable long-term repopulating HSC compartment. They also suggest that careful validation of prospective isolation strategies is required to interpret models of stress hematopoiesis, and that different HSC definitions (e.g., unfractionated SLAM versus Fgd5+) can enhance studies addressing the functional status of HSC following inflammatory stress.
Numerous markers have been demonstrated to enrich for functional long-term HSC activity. The extent to which they faithfully enrich for long-term HSC activity under non-homeostatic conditions has not been extensively studied. Here, we show that EPCR and CD34 mark a continuum of cell states, with EPCR+/CD34− representing the most immature, quiescent cellular fraction. Interestingly, our data show that acquisition of CD34 by EPCR+ cells is associated with induction of cell cycle genes and loss of repopulating activity, consistent with prior work showing that CD34+ SLAM cells, also termed MPP1, represent a metabolically activated SLAM fraction with poor reconstitution activity[35]. Likewise, other studies have demonstrated equivalent levels of HSC reconstitution activity shortly after 5-FU treatment or IR, based on marker systems that exclude c-Kit and Sca-1 while incorporating EPCR and other markers such as CD48/CD150 or CD27[26, 49]. Along these lines, we find that the EPCR+/CD34− fraction is highly enriched for CD49blo cells, which were recently defined as chemoresistant R-HSC[40]. Hence, it is likely there is a high degree of phenotypic and functional overlap between these two definitions, with the intersection between both markers likely representing a highly enriched, chemoresistant R-HSC fraction. Along these lines, our data suggest that a viable, functional HSC pool enriched by EPCR+/CD34− is maintained in the BM despite ongoing hematopoietic stress. Combinatorial use of markers such as CD49b and EPCR may also provide highly stringent enrichment for functional HSC under stress conditions and should be investigated further. As EPCR is also expressed on human HSC[50], similar systems may also be applicable for enriching human HSC in disease settings. Hence, future studies can determine the extent to which the EPCR+/CD34− definition identifies functional HSC under multiple stress conditions. Moreover, studies to assess HSC responses to stress stimuli at the single-cell level may provide insight into how the HSC pool is maintained under stress conditions and which gene(s) most faithfully report functional long-term repopulating HSC in such contexts.
EPCR is an anti-coagulant, anti-inflammatory endothelial surface protein that has also been characterized based on its capacity to identify long-term HSC with quiescent cell cycle activity and chemoresistant properties[6, 51]. Notably, maintenance of EPCR expression and retention in the BM niche has been shown to require a close partnership between HSC and endothelial cells. Indeed, these cells facilitate activated protein C (aPC) processing and binding to EPCR, which induces cleavage of protease-activated receptor 1 (PAR1) to promote HSC retention in the BM, via a mechanism of nitric oxide (NO) restriction[51, 52]. Interestingly, activation of the protease thrombin by inflammation and other insults leads to PAR1 cleavage at a distinct site that drives increased NO production, EPCR shedding and HSC mobilization from the BM niche[51, 52]. Here, while we found a small but significant decrease in EPCR surface expression on EPCR+/CD34− cells that could be related to thrombin activity and EPCR shedding, it does not appear sufficient to mobilize large numbers of HSC as we do not observe decreased HSC numbers in the BM. Hence, our data imply activation of homeostatic mechanism(s) that enforce HSC retention and quiescence despite ongoing inflammatory stress.
In support of such a model, we find that activation of Nrf2- and Hif-driven gene programs are maintained in EPCR+/CD34− SLAM cells. Notably, Nrf2 and Hif pathways are also required for HSC quiescence, and Nrf2 deletion also leads to HSC mobilization from the BM[43, 53]. Thus, further studies can assess the degree to which EPCR cooperates and/or converges with metabolic control mechanisms like Nrf2 and Hif to enforce quiescence and localization of EPCR+ HSC in the BM niche, particularly under inflammatory stress conditions. Along these lines, our data raise an intriguing possibility that increased Mac-1 expression exclusively in EPCR+ SLAM fractions following IL-1 exposure could be a mechanism to enforce retention of these cells in the BM. This could serve as a stress-inducible counterpart to VLA-4 expression, which maintains EPCR+ HSC localization in the BM under homeostatic conditions, and is similar to prior work showing increased Mac-1 expression anchors hematopoietic progenitors in the BM following G-CSF administration[27]. The extent to which EPCR regulates surface expression of Mac-1 and other adhesion molecules, either directly or indirectly, under inflammatory stress conditions should therefore be more thoroughly investigated. Lastly, since quiescence and BM niche localization also underwrite HSC chemoresistance, such studies will shed further light on the capacity of phenotypic CD49blo R-HSC within the EPCR+/CD34− SLAM fraction to persist under myeloablative stressors such as 5-FU or IR.
Interestingly, we find that loss of EPCR expression is associated with high levels of cell cycle and Mk/E lineage genes including Gata1 and surface expression of CD41. This is accompanied by minimal long-term myeloid/lymphoid engraftment activity, consistent with prior work showing Gata1 expression anti-correlates with long-term HSC repopulating activity[54]. Our findings are also consistent with work using H2B-GFP transgenic reporter systems to assess divisional history within the SLAM gate[55–57]. These studies showed that divisional history is associated with loss of EPCR expression, induction of Mk/E genes and limited to no long-term myeloid/lymphoid reconstitution activity[55–57]. Likewise, our molecular analyses order SLAM fractions in a linear fashion associated with EPCR and CD34 levels, with acquisition of CD34 in each compartment perhaps representing an activated cellular state within the EPCR+ and EPCR− fractions, with EPCR+/CD34+ cells perhaps marking a ‘decision point’ between self-renewal, myeloid/lymphoid differentiation and/or acquisition of Mk/E priming. As inflammation has been shown to induce ‘emergency’ megakaryopoiesis[17], analyses of platelet and erythrocyte reconstitution kinetics using the Vwf-eGFP system can identify whether EPCR− SLAM cells are indeed primed to generate Mk and/or erythroid cells, and the impact of inflammatory stress on their activity. Additional detailed in vivo lineage tracing studies using single-cell transplants, reporter mice and/or native barcoding systems will be needed to ascertain how fate(s) and localization in the BM of cells within each fraction are regulated.
We show that more stringent approaches to enrich for functional HSC, including the EPCR+/CD34− SLAM and Fgd5-ZsGreen+ SLAM definitions, provided superior enrichment for HSC activity and molecular phenotype relative to unfractionated SLAM cells following chronic IL-1 exposure. Similarly, a recent study showed long-term myeloid/lymphoid reconstitution activity in Fgd5-mCherry+ SLAM cells from poly I:C-treated mice was essentially identical to control mice[48], whereas prior work had shown that unfractionated IFN-exposed SLAM cells exhibited impaired long-term reconstitution activity[19, 23]. As our current study and previous work both show Fgd5+ SLAM cells are highly enriched for EPCR+/CD34− cells[30], we anticipate the EPCR+/CD34− definition may be useful for enriching HSC under multiple inflammatory contexts.
Our data suggest that inflammation-induced changes in the relative abundance of SLAM fractions is likely a factor in the reduced reconstitution capacity of unfractionated SLAM cells from IL-1-exposed mice, as the frequency of long-term engrafting EPCR+/CD34− decreases by nearly twofold following IL-1 exposure, replaced by EPCR− cells with limited to no reconstitution activity. Along similar lines, while the Vwf-eGFP+ SLAM compartment includes abundant long-term HSC in homeostatic conditions [33], we found that IL-1 exposure leads to contamination of the Vwf-eGFP+ SLAM compartment with non-reconstituting EPCR− cells. Thus, when transplantation assays are used to assess the long-term reconstitution capacity of enriched HSC under non-homeostatic conditions, changes in the cellular composition of HSC-enriched compartments such as SLAM can potentially lead to ‘false positive’ readouts of impaired HSC function due to inclusion of non-HSC cell types. Likewise, selection of lineage markers may need to be reconsidered, as long-term HSC can express markers such as Mac-1[58]. Hence, it may be important to experimentally re-evaluate long-term HSC reconstitution activity under stress using stringent approaches.
It is also possible that changes in HSC engraftment represent discreet and transient cellular states within the HSC pool. For instance, this context, transplantation as an assay of self-renewal may provide an incomplete picture of HSC activity, as changes in cell cycle and other parameters may impact functions such as homing, lodging, or engraftment that would not otherwise be impaired if the cells were left in the primary animal. Furthermore, the effects of different inflammatory pathways on HSC biology must also be considered. For instance, exposure to bacterial LPS, double-stranded RNA and live infection with pathogens such as Salmonella and Mycobacterium can induce excessive proliferation, DNA damage and/or attrition in the HSC pool, largely through TRIF and/or interferon-related mechanisms[19, 22, 59–61]. On the other hand, MyD88-dependent signaling, which is conserved between IL-1 signaling and several TLRs, does not contribute to these effects following LPS or Salmonella infection[59]. Hence, the dose, timing and signaling pathway(s) activated by inflammatory challenge may dictate the impact on HSC function. Lastly, changes in HSC activity may be transient effects that cease upon resolution of the insult, suggesting that inflammation may not always result in HSC ‘damage,’ but instead may play a reparative role in the hematopoietic system. Altogether, our study underscores the molecular and phenotypic heterogeneity within the SLAM gate and provides a relevant example of how chronic inflammation can dynamically alter the cellular subsets therein. These data suggest that continual re-evaluation of HSC definitions and the use of prospective and retrospective functional assays is crucial for accurately reporting HSC function in non-homeostatic conditions. Such studies will continue to fuel improved approaches for defining HSC activity and cellular identity, particularly in dynamic stress situations.
Supplementary Material
Highlights.
EPCR and CD34 enriches for long-term repopulating SLAM cells in chronic IL-1 conditions
EPCR and CD34 identify molecularly distinct SLAM fractions
EPCR+/CD34− SLAM cells are highly quiescent regardless of IL-1 exposure
EPCR- SLAM cells possess limited repopulating activity and exhibit Mk priming
Fgd5 expression marks long-term HSC minimally impacted by chronic IL-1
Acknowledgements
This work was supported by K01 DK098315, R01 DK119394 and the Cleo Meador and George Ryland Scott Chair of Medicine in Hematology (to E.M.P.), F31 HL138754 (to J.L.R.) and a National Science Foundation Graduate Research Fellowship Program (to T. S. M.). This work was supported in part by the University of Colorado Cancer Center Flow Cytometry Shared Resource, funded by NCI grant P30 CA046934.
Footnotes
Competing Interests Statement
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.King KY and Goodell MA, Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat Rev Immunol, 2011. 11(10): p. 685–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pietras EM, Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood, 2017. 130(15): p. 1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warr MR, Pietras EM, and Passegue E, Mechanisms controlling hematopoietic stem cell functions during normal hematopoiesis and hematological malignancies. Wiley Interdiscip Rev Syst Biol Med, 2011. 3(6): p. 681–701. [DOI] [PubMed] [Google Scholar]
- 4.Kiel MJ, et al. , SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell, 2005. 121(7): p. 1109–21. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, et al. , Enrichment of hematopoietic stem cells with SLAM and LSK markers for the detection of hematopoietic stem cell function in normal and Trp53 null mice. Exp Hematol, 2008. 36(10): p. 1236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balazs AB, et al. , Endothelial protein C receptor (CD201) explicitly identifies hematopoietic stem cells in murine bone marrow. Blood, 2006. 107(6): p. 2317–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson A, et al. , Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell, 2008. 135(6): p. 1118–29. [DOI] [PubMed] [Google Scholar]
- 8.Winkler IG, et al. , Positioning of bone marrow hematopoietic and stromal cells relative to blood flow in vivo: serially reconstituting hematopoietic stem cells reside in distinct nonperfused niches. Blood, 2010. 116(3): p. 375–85. [DOI] [PubMed] [Google Scholar]
- 9.Shin JY, et al. , High c-Kit expression identifies hematopoietic stem cells with impaired self-renewal and megakaryocytic bias. J Exp Med, 2014. 211(2): p. 217–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Challen GA, et al. , Mouse hematopoietic stem cell identification and analysis. Cytometry A, 2009. 75(1): p. 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.MacLean AL, et al. , Single 0Cell Phenotyping Reveals Heterogeneity Among Hematopoietic Stem Cells Following Infection. Stem Cells, 2017. 35(11): p. 2292–2304. [DOI] [PubMed] [Google Scholar]
- 12.Haas S, Trumpp A, and Milsom MD, Causes and Consequences of Hematopoietic Stem Cell Heterogeneity. Cell Stem Cell, 2018. 22(5): p. 627–638. [DOI] [PubMed] [Google Scholar]
- 13.Morita Y, Ema H, and Nakauchi H, Heterogeneity and hierarchy within the most primitive hematopoietic stem cell compartment. J Exp Med, 2010. 207(6): p. 1173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nestorowa S, et al. , A single-cell resolution map of mouse hematopoietic stem and progenitor cell differentiation. Blood, 2016. 128(8): p. e20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto R, et al. , Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell, 2013. 154(5): p. 1112–1126. [DOI] [PubMed] [Google Scholar]
- 16.Carrelha J, et al. , Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature, 2018. 554(7690): p. 106–111. [DOI] [PubMed] [Google Scholar]
- 17.Haas S, et al. , Inflammation-Induced Emergency Megakaryopoiesis Driven by Hematopoietic Stem Cell-like Megakaryocyte Progenitors. Cell Stem Cell, 2015. 17(4): p. 422–34. [DOI] [PubMed] [Google Scholar]
- 18.Matatall KA, et al. , Type II interferon promotes differentiation of myeloid-biased hematopoietic stem cells. Stem Cells, 2014. 32(11): p. 3023–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pietras EM, et al. , Re-entry into quiescence protects hematopoietic stem cells from the killing effect of chronic exposure to type I interferons. J Exp Med, 2014. 211(2): p. 245–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pietras EM, et al. , Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol, 2016. 18(6): p. 607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pietras EM, et al. , Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell, 2015. 17(1): p. 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walter D, et al. , Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature, 2015. 520(7548): p. 549–52. [DOI] [PubMed] [Google Scholar]
- 23.Essers MA, et al. , IFNalpha activates dormant haematopoietic stem cells in vivo. Nature, 2009. 458(7240): p. 904–8. [DOI] [PubMed] [Google Scholar]
- 24.Domen J and Weissman IL, Hematopoietic stem cells and other hematopoietic cells show broad resistance to chemotherapeutic agents in vivo when overexpressing bcl-2. Exp Hematol, 2003. 31(7): p. 631–9. [DOI] [PubMed] [Google Scholar]
- 25.Herault A, et al. , Myeloid progenitor cluster formation drives emergency and leukaemic myelopoiesis. Nature, 2017. 544(7648): p. 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vazquez SE, Inlay MA, and Serwold T, CD201 and CD27 identify hematopoietic stem and progenitor cells across multiple murine strains independently of Kit and Sca-1. Exp Hematol, 2015. 43(7): p. 578–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hidalgo A, et al. , The integrin alphaMbeta2 anchors hematopoietic progenitors in the bone marrow during enforced mobilization. Blood, 2004. 104(4): p. 993–1001. [DOI] [PubMed] [Google Scholar]
- 28.Chapple RH, et al. , Lineage tracing of murine adult hematopoietic stem cells reveals active contribution to steady-state hematopoiesis. Blood Adv, 2018. 2(11): p. 1220–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabezas-Wallscheid N, et al. , Vitamin A-Retinoic Acid Signaling Regulates Hematopoietic Stem Cell Dormancy. Cell, 2017. 169(5): p. 807–823 e19. [DOI] [PubMed] [Google Scholar]
- 30.Gazit R, et al. , Fgd5 identifies hematopoietic stem cells in the murine bone marrow. J Exp Med, 2014. 211(7): p. 1315–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen JY, et al. , Hoxb5 marks long-term haematopoietic stem cells and reveals a homogenous perivascular niche. Nature, 2016. 530(7589): p. 223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pinho S, et al. , Lineage-Biased Hematopoietic Stem Cells Are Regulated by Distinct Niches. Dev Cell, 2018. 44(5): p. 634–641 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanjuan-Pla A, et al. , Platelet-biased stem cells reside at the apex of the haematopoietic stem-cell hierarchy. Nature, 2013. 502(7470): p. 232–6. [DOI] [PubMed] [Google Scholar]
- 34.Jalbert E and Pietras EM, Analysis of Murine Hematopoietic Stem Cell Proliferation During Inflammation. Methods Mol Biol, 2018. 1686: p. 183–200. [DOI] [PubMed] [Google Scholar]
- 35.Cabezas-Wallscheid N, et al. , Identification of regulatory networks in HSCs and their immediate progeny via integrated proteome, transcriptome, and DNA methylome analysis. Cell Stem Cell, 2014. 15(4): p. 507–522. [DOI] [PubMed] [Google Scholar]
- 36.Kent DG, et al. , Prospective isolation and molecular characterization of hematopoietic stem cells with durable self-renewal potential. Blood, 2009. 113(25): p. 6342–50. [DOI] [PubMed] [Google Scholar]
- 37.Lazare S, et al. , Lifelong dietary intervention does not affect hematopoietic stem cell function. Exp Hematol, 2017. 53: p. 26–30. [DOI] [PubMed] [Google Scholar]
- 38.Kiel MJ, Radice GL, and Morrison SJ, Lack of evidence that hematopoietic stem cells depend on N-cadherin-mediated adhesion to osteoblasts for their maintenance. Cell Stem Cell, 2007. 1(2): p. 204–17. [DOI] [PubMed] [Google Scholar]
- 39.Benveniste P, et al. , Intermediate-term hematopoietic stem cells with extended but time-limited reconstitution potential. Cell Stem Cell, 2010. 6(1): p. 48–58. [DOI] [PubMed] [Google Scholar]
- 40.Zhao M, et al. , N-Cadherin-Expressing Bone and Marrow Stromal Progenitor Cells Maintain Reserve Hematopoietic Stem Cells. Cell Rep, 2019. 26(3): p. 652–669 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hernandez G, et al. , Pro-inflammatory cytokine blockade attenuates myeloid expansion in a murine model of rheumatoid arthritis. Haematologica, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JM, et al. , Obesity alters the long-term fitness of the hematopoietic stem cell compartment through modulation of Gfi1 expression. J Exp Med, 2018. 215(2): p. 627–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai JJ, et al. , Nrf2 regulates haematopoietic stem cell function. Nat Cell Biol, 2013. 15(3): p. 309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singh RP, et al. , HIF prolyl hydroxylase 2 (PHD2) is a critical regulator of hematopoietic stem cell maintenance during steady-state and stress. Blood, 2013. 121(26): p. 5158–66. [DOI] [PubMed] [Google Scholar]
- 45.Ramasz B, et al. , Hematopoietic stem cell response to acute thrombocytopenia requires signaling through distinct receptor tyrosine kinases. Blood, 2019. 134(13): p. 1046–1058. [DOI] [PubMed] [Google Scholar]
- 46.Ito K, et al. , A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med, 2012. 18(9): p. 1350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takubo K, et al. , Regulation of glycolysis by Pdk functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell, 2013. 12(1): p. 49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bujanover N, et al. , Identification of immune-activated hematopoietic stem cells. Leukemia, 2018. 32(9): p. 2016–2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Umemoto T, et al. , Ca(2+)-mitochondria axis drives cell division in hematopoietic stem cells. J Exp Med, 2018. 215(8): p. 2097–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fares I, et al. , EPCR expression marks UM171-expanded CD34(+) cord blood stem cells. Blood, 2017. 129(25): p. 3344–3351. [DOI] [PubMed] [Google Scholar]
- 51.Gur-Cohen S, et al. , PAR1 signaling regulates the retention and recruitment of EPCR-expressing bone marrow hematopoietic stem cells. Nat Med, 2015. 21(11): p. 1307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gur-Cohen S, et al. , Regulation of long-term repopulating hematopoietic stem cells by EPCR/PAR1 signaling. Ann N Y Acad Sci, 2016. 1370(1): p. 65–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wielockx B, et al. , Hypoxia Pathway Proteins in Normal and Malignant Hematopoiesis. Cells, 2019. 8(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Drissen R, et al. , Distinct myeloid progenitor-differentiation pathways identified through single-cell RNA sequencing. Nat Immunol, 2016. 17(6): p. 666–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foudi A, et al. , Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat Biotechnol, 2009. 27(1): p. 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qiu J, et al. , Divisional history and hematopoietic stem cell function during homeostasis. Stem Cell Reports, 2014. 2(4): p. 473–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bernitz JM, et al. , Hematopoietic Stem Cells Count and Remember Self-Renewal Divisions. Cell, 2016. 167(5): p. 1296–1309 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang J, et al. , Lineage marker expression on mouse hematopoietic stem cells. Exp Hematol, 2019. 76: p. 13–23 e2. [DOI] [PubMed] [Google Scholar]
- 59.Takizawa H, et al. , Pathogen-Induced TLR4-TRIF Innate Immune Signaling in Hematopoietic Stem Cells Promotes Proliferation but Reduces Competitive Fitness. Cell Stem Cell, 2017. 21(2): p. 225–240 e5. [DOI] [PubMed] [Google Scholar]
- 60.Matatall KA, et al. , Chronic Infection Depletes Hematopoietic Stem Cells through Stress-Induced Terminal Differentiation. Cell Rep, 2016. 17(10): p. 2584–2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baldridge MT, et al. , Quiescent haematopoietic stem cells are activated by IFN-gamma in response to chronic infection. Nature, 2010. 465(7299): p. 793–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.