Abstract
Malformations of cortical development encompass heterogeneous groups of structural brain anomalies, associated with complex neurodevelopmental disorders, and diverse genetic and non-genetic etiologies. Recent progress in understanding the genetic basis of brain malformations has been driven by extraordinary advances in DNA sequencing technologies. For example, somatic mosaic mutations that activate mTOR signaling in cortical progenitor cells during development are now recognized as the cause of hemimegalencephaly and some types of focal cortical dysplasia. Also, research on brain development has begun to reveal the cellular and molecular basis of cortical gyrification and axon pathway formation, so disorders involving these processes may be better understood. New neuroimaging techniques with improved resolution have enhanced our ability to characterize subtle malformations, such as those associated with intellectual disability and autism. In this review, we broadly discuss cortical malformations and focus on several in which genetic etiologies have elucidated pathogenesis.
Keywords: Microcephaly, Hemimegalencephaly, Focal Cortical Dysplasia, Polymicrogyria, Lissencephaly, Hippocampal Dysgenesis
1. INTRODUCTION
The past few years have been marked by unprecedented progress in the rapid identification of genes and signaling pathways that cause different types of cortical malformations. Concurrently, improved resolution and new techniques in neuroimaging have facilitated the recognition of increasingly subtle malformations, associated with disorders such as focal epilepsy. Recent studies have also revealed novel mechanisms of cortical development relating to processes such as brain gyrification, especially in humans. Neuropathological studies have also played an important part in recent progress, as genetic and histological studies of patient brain specimens are critical to diagnosis and research. The purpose of this review is to integrate these advances in brain development, genetics, and malformations in an updated perspective, focusing on disorders (such as cortical overgrowth disorders) where progress has led to the greatest new insights.
2. GENERAL FEATURES OF CORTICAL MALFORMATIONS
2.1. Impact of cortical malformations
Corticogenesis is a complex sequential process that, in humans, leads to the formation of a layered cortex with consistent gyral patterns and axonal connections. Broadly, corticogenesis can be divided into three partially overlapping stages, consisting of cell proliferation, neuronal migration, and postmigrational cortical organization (1). Disruption of any of the steps may result in malformations of cortical development (MCDs), a large spectrum of disorders with varied cortical morphologies, genetic or extrinsic etiologies, related syndromes, and clinical manifestations (2,3). Epilepsy, developmental delay (DD), intellectual disability (ID), autism, and schizophrenia are among the common clinical consequences of MCDs, and they sometimes occur together in various combinations.
The definite incidence of MCDs is unknown. Up to 40% of medically refractory childhood seizures are attributable to MCDs (4), and at least 75% of patients with MCDs develop seizures (5). Some MCDs are discovered incidentally after magnetic resonance imaging (MRI) for other conditions or unrelated events, and some cases are probably never detected (6).
2.2. Causes of cortical malformations; role of de novo mutations
The majority of MCDs are thought to be caused by underlying genetic mutations, which disturb the encoded proteins and associated molecular pathways involved in early and/or later stages of cerebral cortex development (2). Mutations that arise de novo, during gametogenesis or postzygotic development, are increasingly recognized as causes of MCDs, epilepsy, ID, and autism (7–9). In addition, environmental insults, such as infection or ischemia in utero at different stage of brain development, or during the perinatal or postnatal period, may contribute to the development of MCDs (10,11). The time of insult during corticogenesis also seems to impact the severity of cortical malformation, with disruptions of later neurodevelopmental processes proposed to cause more severe network disruptions (12). The severity of MCDs also reflects their focal or widespread impact on cortical structure.
Some MCDs are associated with postzygotic mutations that produce somatic mosaicism, classified as either type 1 (new heterozygous mutation; “first hit”) or type 2 (loss of heterozygosity, on germline heterozygous background; “second hit”) mosaicism (13,14). For example, type 2 TSC2 point mutations have been identified in lesions from tuberous sclerosis patients (15). Type 1 mosaicism is represented by some forms of hemimegalencephaly and focal cortical dysplasia (FCD), caused by activating mutations in genes such as PIK3CA or AKT3 (14).
2.3. Detection of cortical malformations
Some MCDs, such as lissencephaly, polymicrogyria, or large heterotopia (e.g., subependymal nodules in tuberous sclerosis), can be detected in utero by fetal ultrasound or MRI (16). However, many MCDs are detected postnatally or during the first year of life, depending on the severity of the malformation (2). Although neuroimaging is the clinical cornerstone of MCD detection, cytogenetic and genetic studies, including next generation (deep) sequencing, as well as neuropathology, represent essential tools for the modern diagnosis of these disorders.
2.4. Classification of cortical malformations
Since MCDs are complex disorders involving multiple etiologies and neurodevelopmental processes, classification has always been challenging and, to some degree, incomplete. Traditionally, MCDs have been classified into disorders of neuronal and glial proliferation or apoptosis; disorders of cell migration; disorders of postmigrational development; and malformations caused by metabolic disorders, peroxisomal disorders, or sublobar dysplasia (12,17) (Table 1). However, classifications continue to evolve with the identification of new types of MCDs and causative genes, and with ongoing advances in the understanding of brain development (6,18,19).
Table 1.
Simplified classification of genetic MCDs
| MCD group | MCD type | Morphologies | Related pathways |
|---|---|---|---|
| Disorders of proliferation, apoptosis, and/or differentiation | Microcephalies | Microcephaly, microlissencephaly Alobar, lobar, variant holoprosencephaly |
Tubulinopathies, microtubule-associated proteins Decreased RTK-PI3K-AKT-mTOR Sonic hedgehog pathway Midline differentiation |
| Cortical overgrowth disorders (focal and diffuse) | Megalencephaly, hemimegalencephaly, polymicrogyria, FCD-II | Overactive RTK-PI3K-AKT-mTOR | |
| Disorders of neuronal migration | Classic lissencephaly spectrum | Smooth lissencephaly, microlissencephaly, subcortical band heterotopia | Tubulinopathies, microtubule-associated proteins Variant lissencephalies (non-cytoskeletal) |
| Cobblestone malformations | Rough lissencephaly, polymicrogyria, leptomeningeal glioneuronal heterotopia | Dystroglycanopathies Other basement membrane - glia limitans interaction disorders |
|
| Periventricular heterotopia | Nodular or linear periventricular heterotopia | Microtubule-associated proteins | |
| Dyslamination without cytologic dysplasia or growth abnormality | FCD-I | Overactive RTK-PI3K-AKT-mTOR Other rare forms (e.g., variant Rett syndrome) |
|
| Disorders of axon pathway formation | Isolated callosal defects | Agenesis, hypogenesis, dysgenesis of corpus callosum | Axon growth and guidance Midline differentiation |
| Other isolated axon defects (putative) | Unknown | Axon growth and guidance |
Classification schemes increasingly incorporate the genetic basis of MCDs as an important organizing principle (2,6). Human genetic studies have identified hundreds of genes associated with one or more types of MCDs, which are involved in various cellular activities, such as cell-cycle regulation and apoptosis, cytoskeletal structure and function, basement-membrane function, neuronal migration, and intracellular metabolic activities.
For instance, micrencephalies, although sharing a common phenotype, are genetically heterogeneous. Underlying genes are involved in cell cycle progression and checkpoint regulation (MCPH1, CDK5RAP2, CENPJ); mitotic spindle formation (WDR62, NDE1); and centrosome duplication and maturation (CDK5RAP2, NDE1) (20,21). In addition, mutated genes involved in microtubule formation (TUBA1A, TUBB2B, TUBB3, TUBG1) are identified not only in primary microcephalies, but also in other MCDs, including lissencephaly, microlissencephaly, pachygyria, and various examples of polymicrogyria (22). Microtubule-associated proteins (MAPs), including LIS1, DCX, DYNC1H and KIF5C are associated with cytoskeleton development and are identified in patients with severe microcephaly, diverse lissencephalies, and subcortical band heterotopia (23). Postmigrational microcephalies (2), usually developing during the first two postnatal years, are associated with a congenital variant of Rett syndrome caused by FOXG1 mutations (24), and with pontocerebellar hypoplasia secondary to CASK gene deletion (25).
3. DEVELOPMENT OF THE CEREBRAL CORTEX
Research on cortical development has progressed rapidly in the last five years, and in particular, has been marked by the extension of cellular and molecular research from experimental animals (especially mice) to human developing brain. These advances have been especially relevant to understanding the morphogenesis of gyri and sulci, and carry important implications for disorders of gyrus formation, such as pachygyria.
Classic studies of cortical development from previous centuries showed that cell proliferation occurs mainly adjacent to the embryonic ventricles; that new neurons migrate from the periventricular zones towards the pial surface, to form the cortical plate; that cortical networks are organized in columns, derived from corresponding “radial unit” progenitors; that cortical layers are generated “inside-out,” with deep layers formed earlier than superficial layers; that axons and dendrites begin to grow concurrently with cell migration; and that gyri and sulci are initially generated by differential growth of developmental zones in regions of cortex (26,27). Subsequent studies have elucidated the cellular and molecular bases of these processes, and revealed additional complexities, especially in the past five years.
3.1. Projection neurons and interneurons: distinct cell types with separate origins
Until about 20 years ago, all cortical neurons, including projection neurons (long axon, glutamatergic) and interneurons (short axon, GABAergic) were thought to arise from common progenitor cells that divided at the ventricular surface of the embryonic neuroepithelium. A succession of important discoveries dramatically changed this view. The first key discovery was that interneurons are produced outside the cortical neuroepithelium, in basal forebrain compartments known as the ganglionic eminences (28,29). The interneurons, comprising about 15–20% of all cortical neurons, migrate tangentially into all cortical areas, including hippocampus, before integrating radially into cortical columns. This discovery harkened back to the observations of Ramón y Cajal, who was the first to classify cortical neurons into fundamentally different projection neurons and interneurons. The separate origins of interneurons and projection neurons have significant implications for interpreting the effects of mosaic mutations that arise during development.
3.2. Projection neurons are generated from several types of progenitor cells
Studies in the past several years have revealed that multiple types of progenitor cells are present in the developing cortex, where projection neurons are produced (Fig. 1). The primary source of projection neurons and glial cells in mammalian cortex is radial glial cells, long known as scaffolds that guide the migration of new neurons from periventricular zones to the cortical plate. The “radial glial progenitors” (RGPs), as they are now known, have the capacity to produce diverse types of neurons and glia (multipotency), and to proliferate extensively by self-renewal. These properties (neural multipotency and extensive self-renewal) mark RGPs as a special type of neural stem cells (NSCs) in the developing cortex (30,31).
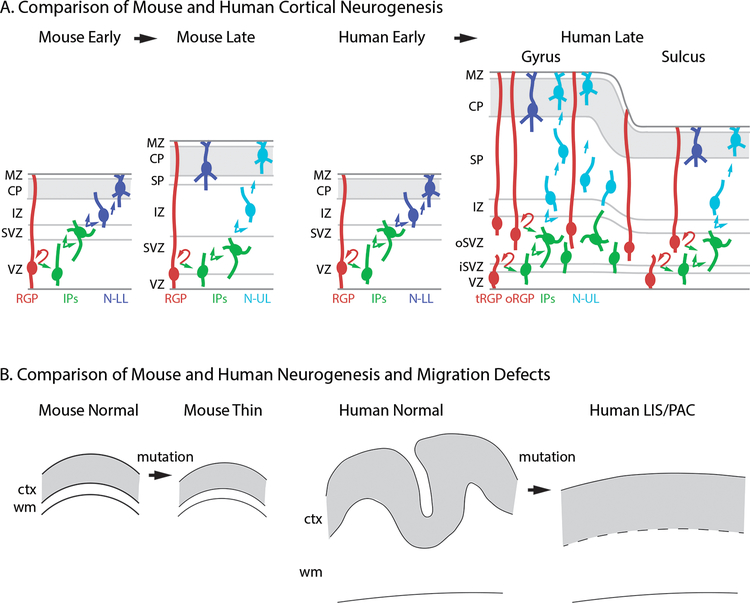
Figure 1.
Comparison of cortical development and defective neurogenesis in mice and humans. (A) Normal development. In mice, classic RGPs predominate during early (deep-layer) and late (superficial-layer) neurogenesis, and the cortex lacks gyri and sulci. In humans, a transition from classic RGPs to mainly oRGPs occurs between early and late neurogenesis, when gyri and sulci begin to form. The abundance of oRGPs and outer IPs is much higher beneath prospective gyri. (B) Reduced neurogenesis in mice leads to cortical thinning. In humans, reduced neurogenesis is usually associated with impaired gyrification (lissencephaly or pachygyria), and leads to increased cortical thickness. Abbreviations: CP: cortical plate; ctx: cortex; IP: intermediate progenitor; IZ: intermediate zone; LIS: lissencephaly; MZ: marginal zone; N-LL: neuron destined for lower layers; N-UL: neuron destined for upper layers; oRGP: outer radial glial progenitor; PAC: pachygyria; RGP: radial glial progenitor; SVZ: subventricular zone; tRGP: truncated radial glial progenitor; VZ: ventricular zone; wm: white matter.
A second type of cortical progenitor cells, known as intermediate progenitors (IPs), were also found to play a major role in cortical neurogenesis. The IPs are produced from RGPs, but differ from them in several important ways. First, while RGPs undergo M-phase at the ventricular surface, most IPs divide away from the ventricular surface, in not only the ventricular zone (VZ) but also the subventricular zone (SVZ), once thought to harbor only glial progenitors. Second, while RGPs have high proliferative capacity and are multipotent, IPs have low proliferative capacity and are strictly committed to produce projection neurons. Third, while RGPs have long radial processes, IPs have only short processes. Fourth, at the molecular level, RGPs express markers of NSC identity such as Sox2 and Sox9, but IPs express markers of glutamatergic neuronal differentiation, such as Tbr2.
Since the vast majority of cortical projection neurons are generated from IPs (32), they have been proposed to influence evolutionary features of cortical development, such as the growth of cortical upper layers in higher species, or the formation of gyri and sulci by regionally localized IP proliferation (33).
3.3. Basal progenitor cells and gyrification
Most previous studies of cortical development have utilized mice, which have many advantages for research. However, since mice have small, naturally “lissencephalic” brains, their relevance for studying gyrification of the brain is limited. To understand how gyri form, and to correlate human brain malformations with mouse brain defects, studies of larger species are necessary.
Gyrification is an ancient feature of neocortex in large-brained mammals of all orders, including monotremes and marsupials (27,34). In larger mammals such as primates, the developing cortex is histologically more complex than in mice, with a vastly expanded SVZ divided into “inner” and “outer” zones (35). The increased complexity of progenitor zones was an important clue leading to key discoveries concerning the process of gyrification.
More recent studies of gyrencephalic species, including human fetal cortex in slice cultures, revealed a new type of cortical progenitor cells in the outer SVZ, known as “outer” or “basal” RGPs (oRGPs). The oRGPs were distinguished by their location in the outer SVZ of human developing cortex, and by their observed genesis of IPs and neurons (36). Unlike classic ventricular or “apical” RGPs, the oRGPs lack connections to the ventricular surface. Nevertheless, oRGPs retain the capacity to produce IPs and neurons, and they express markers of NSC identity. Also, oRGPs express some specific molecular markers, such as transcription factor HOPX. Significantly, oRGPs are enriched in mTOR signaling activity (37), which has been implicated in human MCDs, such as megalencephaly (see below).
By detaching from the ventricular surface and migrating towards the cortical plate, oRGPs maintain radial columnar organization during neurogenesis, while focally expanding the external surface area of cortex, but not the internal ventricular surface area—a key property required for gyrification (Fig. 1A). While oRGPs appear to be present in all mammals, their increased abundance in larger, gyrencephalic brains provides a putative substrate for the formation of gyri. Consistent with this idea, oRGPs become abundant shortly before the onset of gyrification, during neurogenesis of supragranular (upper-layer) neurons (38). This transition to increased oRGP abundance occurs at about 16.5 gestational weeks in humans. Concurrently, RGPs in the ventricular zone maintain contact with the ventricular surface, but lose contact with the pial surface to become “truncated” RGPs (38).
Significantly, oRGPs, and the outer IPs generated from them, are most abundant beneath cortical regions that form gyri (38,39). The amplification of oRGPs and IPs is driven in part by fibroblast growth factor (FGF) signaling (41). The involvement of specific FGF receptors (FGFRs) in gyrification is directly relevant to disorders of human brain gyrification, such as thanatophoric dysplasia (see below). Furthermore, disorders of human brain gyrification, such as pachygyria and lissencephaly, involve defects of neurogenesis and migration that are not well modeled in mice (Fig. 1B). The cortex is invariably thin in mouse models with reduced neurogenesis, but is abnormally thick in humans with reduced neurogenesis (as occurs in lissencephaly), due to impaired gyrification and decreased cortical surface area (Fig. 1B).
3.4. Prolonged developmental neurogenesis and vulnerability
While it was long believed that human cortical neurogenesis is complete by the middle of gestation, modern research has changed this view. Recent evidence suggests that genesis of projection neurons actually continues until approximately 28 gestational weeks (41). In the dentate gyrus of hippocampus, neurogenesis continues into the postnatal period, albeit at declining levels with increasing age (42). The prolonged genesis of cortical neurons suggests vulnerabilities in fetal and postnatal life that may explain some malformations, as well as susceptibilities to factors that modify neurogenesis, such as radiation and inflammation.
3.5. Cortical area patterning and anomalies
Studies in mice and non-human primates have demonstrated that the parcellation of cortex into distinct motor and sensory regions, and ultimately the definition of functional areas, is driven by gradients of gene expression along rostrocaudal and mediolateral axes (43). Some genes associated with cortical patterning in mice, such as Tbr1 (44) and Fgfr3 (45), are also implicated in human MCDs. While it seems likely that disturbances of cortical arealization occur in human neurodevelopmental disorders, very little is known about these effects.
4. CORTICAL OVERGROWTH AND FOCAL CORTICAL DYSPLASIA
“Cortical overgrowth” encompasses diffuse or partial (e.g., hemispheric) excess of cortical tissue, which may or may not meet strict medical definitions of megalencephaly (>2.5 standard deviations above the mean for age and sex), but usually meets the less stringent definition of brain weight “greater than average for the age and gender of the child” (NINDS Megalencephaly Information Page). Focal cortical dysplasia (FCD) refers to more limited lesions, usually restricted to one or two lobes, in which the cortex is dysplastic, due to abnormal radial and/or tangential organization (FCD type I; FCD-I), in some cases accompanied by neuronal cytomegaly and dysplasia (FCD type II; FCD-II). Additional subtypes of FCD are also described, most relevantly the division of FCD-II into FCD-IIa, in which no balloon cells are present, and FCD-IIb, in which balloon cells are present (46). In children who undergo brain tissue resection for drug treatment-resistant focal epilepsy, FCD is the most common histopathological diagnosis (47).
In recent years, many cortical overgrowth malformations, and some types of FCD (especially FCD-II), have been linked to mutations that cause overactivation of the receptor tyrosine kinase (RTK)→phosphatidylinositol-3-kinase (PI3K)→AKT→mechanistic target of rapamycin (mTOR) signaling pathway (48–50). This signaling cascade mediates progenitor proliferation and cell growth during development. Thus, overactivation due to genetic mutations causes hyperplasia and/or hypertrophy of the brain and sometimes other organs, depending on the specific genes and mutations.
4.1. Outline of the RTK→PI3K→AKT→mTOR signaling pathway
Many growth factors, such as insulin-like growth factor 1 (IGF-1) and most fibroblast growth factors (FGFs), activate cell proliferation and growth by signaling through RTKs. In turn, these RTKs are coupled to downstream signaling pathways, most notably the PI3K→AKT→mTOR pathway, which is critically involved in brain development. Since each kinase in the cascade has multiple downstream targets (for example, dozens of proteins are phosphorylated by activated AKT), the signaling pathway is extremely complex and resembles a network. Besides mTOR, additional downstream pathways activated by growth factor RTKs include Ras-MAPK-Erk, FOXO, primary cilium, and others (51,52). In this review, we focus on elements of this cascade that have been linked to cortical overgrowth, including megalencephaly (MEG), hemimegalencephaly (HME), and FCD (Fig. 2).
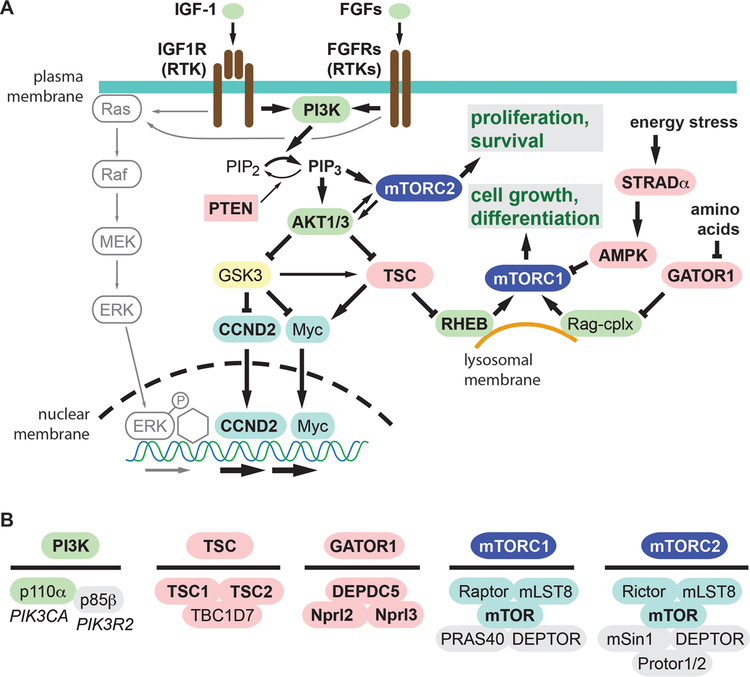
Figure 2.
The RTK-PI3K-AKT-mTOR signaling pathway. (A) The pathways is driven by growth factors interacting with their receptors (RTKs), as well as by intracellular metabolic factors including energy status and amino acids. Activators are colored green, and inhibitors red. Molecules known to be linked to brain overgrowth or FCD are indicated by bold type. (B) Subunit composition of key protein complexes linked to brain overgrowth or FCD. The genes encoding p110α (PIK3CA) and p85β (PIK3R2) are also indicated.
Critical outputs of this signaling pathway are mediated by the mTOR protein, a serine/threonine kinase that forms two complexes, mTORC1 and mTORC2, that mainly control cell growth/differentiation and proliferation/survival, respectively (Fig. 2). Importantly, mTORC1 is regulated synergistically by not only growth factor RTK signaling, but also by amino acids (leucine, arginine), as well as overall energy balance. Signaling to mTORC1 from growth factors is mediated through the tuberous sclerosis complex (TSC), which constitutively inhibits mTORC1 activity in the absence of growth factors. In parallel, signaling from amino acids is mediated by pathways that converge on the GATOR1 complex, comprised of DEPDC5, Nprl2, and Nprl3. The GATOR1 complex inhibits mTORC1 activity unless amino acid concentrations are sufficient to support growth. Energy stress (e.g., high AMP levels) also regulates mTORC1 activity, in part via the STRADα complex (composed of STRADα, LKB1, and MO25), which activates AMPK, a direct inhibitor of mTORC1 (53).
Thus, the TSC, GATOR1, and STRADα complexes function as parallel brakes on mTORC1 activity, which must all be released for full activation of mTORC1 (49). Mutations that impair the functions of these complexes all lead to mTORC1 activation, and cortical overgrowth or FCD (Table 2). One of the main biochemical readouts of mTORC1 activation is phosphorylation of ribosomal protein S6 to form phospho-S6 (pS6), a readily detectable marker of mTORC1 activation (Fig. 3).
Table 2.
Genes linked to cortical overgrowth and FCDsa
| Gene | Typical mutations | Syndromes | Brainb | M | H | I | IIa | IIb | Body | References |
|---|---|---|---|---|---|---|---|---|---|---|
| AKT1 | postzygotic mosaic GOF | Proteus | HMEG | ND | + | ND | ND | ND | Asymmetric overgrowth affecting multiple tissues | 54, 62 |
| AKT3 | de novo germline > postzygotic mosaic, GOF; or chr microdup | MPPH | DMEG, HMEG, FCD-Ib, periventricular or subcortical heterotopia | + | + | + | ND | ND | ND: AKT3 is expressed specifically in brain | 56, 57, 61, 62, 64, 65, 127 |
| CCND2 | de novo germline | MPPH | DMEG | + | ND | ND | ND | ND | Postaxial polydactyly | 128 |
| DEPDC5 | heterozygous germline, postzygotic mosaic LOF | BPPc | HMEG, FCD-I,d FCD-IIa/b | ND | + | ND | + | + | ND | 62, 66, 129–133 |
| FGFR3 | de novo constitutional GOF | TD | Occipito-temporal hypergyration | + | ND | ND | ND | ND | Severe dwarfism | 45 |
| MTOR | mosaic, GOF | ND | MEG, HMEG, FCD-IIa/b | + | + | ND | + | + | Pigmentary mosaicism | 127, 129, 134–137 |
| NPRL2 | heterozygous germline, LOF | NDc | FCD-Ia,e FCD-IIa | ND | ND | + | + | ND | ND | 62, 67 |
| NPRL3 | familial, heterozygous germline, LOF | NDc | FCD-IIa | ND | ND | ND | + | ND | ND | 67, 138 |
| PIK3CA | mosaic GOF | CLOVES (severe), MCAP (moderate) | DMEG, HMEG, FCD-IIa | + | + | ND | + | ND | CLOVES: lipomatous overgrowths, vascular malformations, epidermal nevi, spinal/skeletal anomalies MCAP: Asymmetric overgrowth, poly/syndactyly, capillary/lymphatic malformations |
14, 57, 62, 127 |
| PIK3R2 | de novo constitutional > postzygotic mosaic, GOF | MPPH, BPP | DMEG, HMEG | + | + | ND | ND | ND | Polydactyly | 57, 62, 139, 140 |
| PTEN | germline with postzygotic mosaic LOF | Cowden, PTEN-HTSf | DMEG, HMEG, FCD-IIb | + | + | ND | ND | + | Hamartomas (e.g., oral papillomatous papules) | 14, 141 |
| RHEB | de novo constitutional GOF | ND | MEG | + | ND | ND | ND | ND | ND | 59 |
| STRADA | de novo constitutional GOF | PMSE | MEG | + | ND | ND | ND | ND | Polyhydramnios | 53 |
| TSC1 | germline with postzygotic mosaic LOF | TSC | Tubers, subependymal nodules, SEGA, FCD-IIa/b | ND | ND | ND | + | + | Facial angiofibromas, shagreen patch, ungual fibromas, renal angiomyolipoma, cardiac rhabdomyoma | 62, 142–144 |
| TSC2 | germline with postzygotic mosaic LOF | TSC | Tubers, subependymal nodules, SEGA, HMEG, FCD-IIa | ND | + | ND | + | ND | Facial angiofibromas, shagreen patch, ungual fibromas, renal angiomyolipoma, cardiac rhabdomyoma | 62, 80, 142–144 |
Abbreviations: BPP: bilateral perisylvian polymicrogyria; chr: chromosomal; CBTE: cerebellar tonsillar ectopia; CLOVES: congenital lipomatous overgrowth, vascular malformations, epidermal nevi and scoliosis/skeletal/spinal anomalies; DMEG: dysplastic hemimegalencephaly; GOF: gain of function; H: hemimegalencephaly; HMEG: hemimegalencephaly; I: FCD-I; IIa: FCD-IIa; IIb: FCD-IIb; LOF: loss of function; microdup: microduplication; M: megalencephaly; MCAP: megalencephaly-capillary malformation; MEG: megalencephaly; MPPH: megalencephaly-polymicrogyria-polydactyly-hydrocephalus; ND: not detected; PMG: polymicrogyria; PTEN-HTS: PTEN-hamartoma tumor syndrome; SBH: subcortical band heterotopia; SEGA: subependymal giant cell astrocytoma; TD: thanatophoric dysplasia. +: reported in at least one case. Other abbreviations as in text.
Hydrocephalus/ventriculomegaly, and cerebellar tonsillar ectopia/Chiari-I are common findings that accompany cortical overgrowth. Polymicrogyria (often bilateral perisylvian) and/or pachygyria are also frequent findings in megalencephaly or hemimegalencephaly. Bilateral perisylvian polymicrogyria is documented with PIK3R2 (139) and DEPDC5 (133) mutations. Callosal hyperplasia is documented in a minority of megalencephaly and hemimegalencephaly. Periventricular nodular heterotopia were found in patients with germline AKT3 mutations (61), and subcortical nodular heterotopia in AKT3 amplification due to chromosomal microduplication (64).
DEPDC5, NPRL2, and NPRL3 mutations are also associated with temporal lobe epilepsy, febrile seizures, and frontal lobe epilepsy (133).
FCD-I was associated with DEPDC5 mutation, but residual FCD-II was suspected (66).
FCD-Ia has been associated with NPRL2 mutation, but residual FCD-II was considered likely (67).
PTEN-HTS is also known as Bannayan-Riley-Ruvalcaba syndrome.
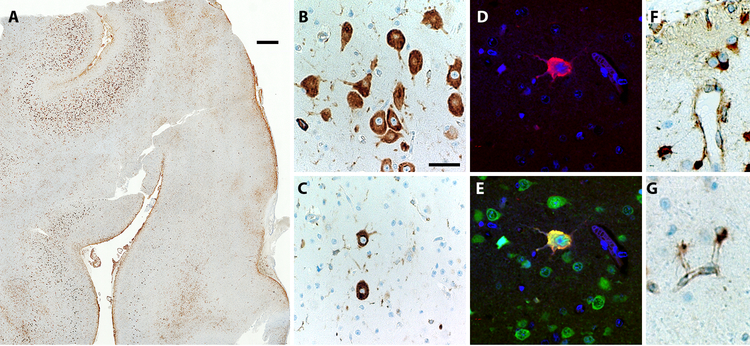
Figure 3.
Patchy mTORC1 activation in a case of FCD-IIa reflects somatic mosaicism. (A) Phospho-S6 immunoreactivity in cortical resection specimen at low magnification. Note local variations in the abundance of immunoreactive cells. (B) Higher magnification shows an area of relatively abundant dysplastic neurons with high phospho-S6 immunoreactivity. (C) An area of sparse dysplastic neurons. (D, E) Two-color immunofluorescence to detect phospho-S6 (red) and NeuN (green) demonstrates that a single dysplastic neuron with high phospho-S6 expression. (F, G) High expression of phospho-S6 in subpial (F) and deep-layer (G) astrocytes is not diagnostically significance, as this may be seen in all forms of epilepsy (Jansen et al., 2015). Scale bar (A): 1 mm. Scale bar (B): 50 µm for B, C; 25 µm for C–F.
Activation of mTORC2 appears to be driven mainly by growth factor RTK signaling (49, 52). Currently, the roles of mTORC2 activation in cortical overgrowth and FCD are not very clear, in part because there is no convenient assay for mTORC2 activation comparable to pS6 for mTORC1.
The association of cortical overgrowth and FCD with overactive RTK→PI3K→AKT→mTOR signaling links these disorders to tuberous sclerosis (TSC), a disorder that shares many clinical and pathologic features with FCD-IIb. Indeed, the cortical tubers in TSC can be considered a syndromic form of FCD-IIb.
4.2. Mutations of the RTK→PI3K→AKT→mTOR pathway in cortical overgrowth and FCD
A landmark in the analysis of brain overgrowth disorders was the linkage of Proteus syndrome, a disorder marked by patchy asymmetric overgrowth of somatic tissues (including hemimegalencephaly in some cases), with activating (“gain-of-function”) mosaic somatic mutations in AKT1 (54). This finding confirmed longstanding hypotheses about the medical significance of mosaic mutations, which arise during development and thus affect only a subset of somatic cells; and set the stage for a new conceptual paradigm of understanding brain overgrowth and FCD, as well as other types of brain malformations, including subcortical band heterotopia (7).
Further studies, focusing on patients with megalencephaly or hemimegalencephaly, quickly identified mutations in AKT3, PIK3CA (p110α catalytic subunit of class IA PI3K), PIK3R2 (p85β regulatory subunit of class IA PI3K), and MTOR, all of which led to overactive RTK→PI3K→AKT→mTOR signaling at some level (55–57).
Indeed, variants in numerous mTOR signaling pathway genes are involved in a range of disorders affecting cortical function—including autism, developmental delay (DD), intellectual disability (ID), and epilepsy—in which sometimes only mild cortical overgrowth is detected (58–60). Depending on the type of mutation and the patient’s genetic background, the degree of cortical overgrowth may range from very mild to extreme, and the degree of functional impairment may likewise vary. For example, RHEB variants were identified as important factors in ID, and in megalencephaly (59). Furthermore, mosaic as well as germline mutations have been detected in many PI3K→AKT→mTOR pathway genes, leading to a spectrum of brain overgrowth and FCD (Table 2).
The specific mutations in each gene are typically diverse, although hotspots are sometimes observed, such as the AKT3 p.E17K activating mutation often found in patients with hemimegalencephaly and other disorders (61). The diversity of mutations is beyond the scope of this review, but it is worth noting that when mosaic mutations occur, different types of mutations and mosaicism are associated with different genes, depending on their functional significance in the signaling pathway. Genes that encode activators of PI3K→AKT→mTOR signaling, such as PIK3CA, are associated with activating (“gain of function”) mutations and type 1 (“first hit”) mosaicism. In contrast, genes that encode inhibitors of PI3K→AKT→mTOR signaling, such as DEPDC5, are associated with inactivating (“loss of function”) mutations and type 2 (“second hit”) mosaicism. This distinction is a reflection of the fact that overactivation of the signaling pathway can be effected by activating mutation of just one allele of an pathway-activating gene, but reduced signaling activity typically requires inactivation of both alleles of pathway-inhibiting genes.
The abundance of cells carrying mosaic mutations in activating genes such as AKT3 has been found to vary from ~1–40% across brain regions, in disorders such as hemimegalencephaly (61,62). Variable mosaicism also seems to be the rule for inhibitors of mTOR signaling, such as TSC1 (hamartin) and TSC2 (tuberin). While some previous studies have failed to detect loss of heterozygosity in tubers and other tuberous sclerosis lesions (15), recent studies indicate that “second hit” mutations can be detected using deep sequencing approaches (62). The low abundance of mutant cells in some cases has raised the question of how macroscopic lesions, such as tubers and FCD, can arise from such a small proportion of abnormal cells. Possibly, seizures may be driven by a few abnormal neurons; also, secondary effects (e.g., gliosis) and non-autonomous effects (e.g., secreted factors) may contribute. Indeed, AKT3 overactivation results in aberrant secretion of Reelin, an important extracellular regulator of neuron migration (63).
Another important question has been whether lesions arise when mutations involve projection neurons, interneurons, or glial cells. Recent studies in mice, in which Pik3ca was overexpressed selectively in different lineages, indicated that cortical overgrowth occurred after overexpression in projection neurons, but not interneurons (62). Also, in lesions from human cases, mutations were always present in neurons, but only sometimes in glial cells (62). Thus, mutant cortical projection neurons seem to be necessary and sufficient for overgrowth lesions involving the PI3K→AKT→mTOR pathway. Accordingly, deep sequencing has become an important adjunct to neuropathology of resected seizure foci.
In FCD-I, mutations involving the PI3K→AKT→mTOR pathway have been detected much less frequently than in FCD-II. In two cases of FCD-I, mosaic duplications of small chromosomal segments containing AKT3 have been detected (64,65). Also, an activating mutation of AKT3 was documented in a case of extreme megalencephaly (the largest pediatric brain on record), in which no dysplastic cytomegalic neurons or balloon cells were present, and the pathology resembled FCD-I (61). These findings suggest that some AKT3 mutations can cause focal or diffuse brain overgrowth without marked neuronal cytomegaly. Mutations of NPRL2 and DEPDC5 (components of the GATOR1 complex, which inhibits mTORC1; Fig. 2) have also been reported in FCD-I, but in both cases it was suggested that resections of dysplastic cortex were incomplete, and possibly spared regions of FCD-II (66,67).
Other genes rarely implicated in FCD-I include ALDH7A1 (encoding the glial protein Antiquitin), also known as the causative gene of pyridoxine-dependent epilepsy (68); and STXBP1 (syntaxin binding protein 1), a regulator of synaptic vesicle release (69). In one case, the STXBP1 gene was furthermore found to have undergone somatic mosaic mutation (type 2 mosaicism) within the FCD-I lesion (69). However, in most FCD-I cases, mutations have not been found, and the etiology remains puzzling.
While RTKs are major activators of the PI3K→AKT→mTOR pathway, mutations involving RTKs are much less common in brain overgrowth disorders. One exception is a form of severe lethal dwarfism known as thanatophoric dysplasia. In this condition, activating mutations of FGFR3 lead to brain overgrowth with excessive gyrification of the occipitotemporal regions, but no cytomegalic neurons (45). Since FGFR3 activates PI3K→AKT→mTOR signaling (52), this pathway presumably contributes to brain overgrowth. Interestingly, the occipitotemporal overgrowth reflects a gradient of FGFR3 expression during development (45). Another major RTK activator of PI3K→AKT→mTOR signaling is the insulin-like growth factor-1 (IGF-1) and IGF-2 receptor IGF1R, but no mutations in the IGF1R gene have yet been linked to human brain overgrowth. In mice, transgenic overexpression of IGF-1 causes marked cortical overgrowth (70).
4.3. Heterogeneous pathology of FCD, hemimegalencephaly, and megalencephaly
The pathology of FCDs is heterogeneous, as mentioned above, and as encapsulated in the current classification (46,71). The key features include cortical laminar disorganization and, in FCD-II, the presence of cytomegalic dysplastic neurons, either in the absence (FCD-IIa) or presence (FCD-IIb) of eosinophilic “balloon cells.” The significance of balloon cells is unknown, since mutations in the same genes have been found in FCD-IIa and -IIb (Table 2), but no genotype-phenotype correlations have emerged .
Diagnostically, pathologists can screen for mTORC1 activation in FCDs by immunohistochemical detection of pS6, which is elevated in dysplastic neurons (Fig. 3). Since glial cells exhibit elevated levels of pS6 in epilepsy unrelated to FCD or cortical overgrowth (14), and mutant glial cells are not always present in FCD but mutant neurons are (62), pS6 expression in glial cells is not diagnostically significant. Interestingly, studies have also found that FCD-II lesions exhibit abnormal differentiation, including co-expression of neuronal and glial markers in dysplastic neurons, and expression of “primitive” NSC markers, such as Nestin, in balloon cells (72–75). These findings presumably reflect the role of PI3K→AKT→mTOR signaling in regulating neuronal and glial differentiation.
As in FCD, the pathology of hemimegalencephaly and megalencephaly is likewise heterogeneous. Cytomegalic dysplastic neurons may or may not be present, as may various other features such as polymicrogyria, immature cell rests (hamartias), glioneuronal leptomeningeal heterotopia, and subcortical or periventricular heterotopia (61,72,76 ).
4.4. Relation of FCD to tuberous sclerosis
Tuberous sclerosis (TSC) is an autosomal dominant disorder caused by mutations in TSC1 (hamartin) or TSC2 (tuberin). Like forms of brain overgrowth and FCD described above, TSC is classified as an “mTORopathy” (71). Increased mTORC1 signaling, as indicated by markers such as pS6, has been detected in tubers from fetal and adult cortex (77). Accordingly, tubers are now considered to be a special type of FCD-IIb, and indeed the two lesions can be histologically indistinguishable (78,79). Stem cell proteins such as Nestin are also expressed in TSC giant cells (80), as in FCD-IIb balloon cells (72, 74, 75). Moreover, mutations in TSC1 and TSC2 seem to exhibit a spectrum of FCD and hemimegalencephaly (Table 2).
On the other hand, tubers are more often calcified than are FCD-IIb lesions, and more often exhibit inflammatory changes (77,80). Other somatic and brain lesions (such as subependymal nodules) also distinguish TSC from FCD-IIb (71,81).
4.5. Treatment implications of RTK→PI3K→AKT→mTOR pathway overactivation
As a kinase cascade, the RTK→PI3K→AKT→mTOR signaling pathway presents attractive targets for pharmacotherapy, to possibly seizure prevention and counter associated neurodevelopmental problems, such as intellectual disability and autism. This pathway is also important in cancer. Indeed, PI3K inhibitors and mTOR inhibitors have been approved for clinical use (50), and everolimus, an mTOR inhibitor, has been found effective for the treatment of subependymal giant cell astrocytoma in tuberous sclerosis (82,83).
5. LISSENCEPHALY, COBBLESTONE MALFORMATIONS, AND MICRENCEPHALY
Malformations associated with abnormal cell migration are often linked to abnormal cell proliferation as well. For example, brain mass is typically reduced in classic (non-cobblestone) lissencephalies (formerly known as “type I”). Accordingly, disorders of proliferation and migration are considered together in this section. Cobblestone malformations can also cause the appearance of lissencephaly (formerly “type II”), but are mechanistically distinct. Cobblestone malformations result from defects in the pial-limiting membrane, leading to neuronal “overmigration” into the subarachnoid space. The spectrum of cobblestone malformations also includes focal leptomeningeal glioneuronal heterotopia, and more extensive lesions sometimes classified as polymicrogyria, such as bilateral opercular cobblestone cortex caused by GPR56 mutations.
5.1. Classic and variant lissencephalies
Classic lissencephaly (LIS) is a disorder of neuronal proliferation and migration characterized most saliently by absent (agyria) or excessively wide (pachygyria) gyri, along with cortical thickening and disorganization, and misplaced or ectopic neurons in the subcortical white matter, sometimes forming band or nodular heterotopia (2,3,19,84). LIS is uncommon, with an estimated prevalence of about 12 per million births (85). Clinical manifestations are usually severe, with feeding problems, delayed motor milestones, mental retardation, or seizures before 6 months in most cases (84,85). Typical associated abnormalities of classic LIS may include enlarged lateral ventricles, hypoplasia of the corpus callosum, hypoplasia of the pyramidal tracts, and abnormal inferior olives and cerebellum (6,86). Syndromes associated with classic LIS include Miller-Dieker, Norman-Roberts, and X-linked LIS with ambiguous genitalia (XLAG). The brain size and weight are usually well below average; LIS cases that meet the strict definition of micrencephaly (≥2.5 standard deviations below average for age and gender) are designated microlissencephaly.
The cortex in LIS measures 5–10 mm or more in thickness (as compared to normal 3–5 mm), and the boundary with white matter is irregular (5,6,18). Interestingly, different laminar patterns have been observed in LIS related to different causative genes (87). While increased cortical thickness in the face of reduced brain mass may seem paradoxical, the explanation is that cortical surface area is dramatically reduced in LIS (Fig. 1B).
Many genes have been linked to LIS, and diagnosis now depends primarily on genetics along with neuroimaging. (Non-genetic forms of LIS also occur but are less common.) Most classic LIS is caused by mutations in genes encoding cytoskeletal proteins, while “variant” forms are caused by mutations involving other types of proteins, such as the transcription factor ARX, and the secreted protein Reelin (87).
Deletions and mutations in LIS1 (officially known as PAFAH1B1), located on chromosome 17p13.3, account for many cases of LIS, and this was the first LIS gene to be identified (88). Chromosomal microdeletions affecting LIS1 plus other genes cause most cases of Miller-Dieker syndrome (89). The LIS1 gene encodes a 45-kDA protein with dual functions, both as a regulatory subunit of platelet-activating factor acetylhydrolase 1B, and as a component of the neuronal cytoskeleton that interacts with microtubule-associated proteins, especially dynein, and is thus important for cell division and migration (90). Somatic mutations of LIS1, affecting only a subset of cortical neurons, cause subcortical band heterotopia (91). Pafah1b1 mutant mice show dose-dependent disorganization of cortical layers (92).
An X-linked form of LIS in males, or of subcortical heterotopia (“double cortex”) in females, is caused by mutations in the DCX (Doublecortin) gene on Xq22.3-q23. DCX is a microtubule-associated protein that is expressed in IPs and post-mitotic neurons. Most cases of classic LIS are associated with LIS1, while most cases of subcortical band heterotopia (SBH) are associated with DCX mutations. LIS1-related LIS affects posterior brain regions more severely, whereas DCX-related LIS is more severe in anterior cortex (19,84).
Tubulinopathies are another group of cytoskeletal disorders, caused by mutations in tubulin genes, that can cause LIS, microlissencephaly, SBH, and other MCDs (Fig. 4). Mutations in TUBA1A, encoding tubulin α−1A, were the first to be associated with LIS (93). Approximately 1% of patients with classic LIS have a recurrent mutation in the TUBA1A gene, and 30% of patients with LIS and cerebellar hypoplasia also present TUBA1A mutations (94). As well, other isoforms of α-, β-, and γ-tubulin, and various kinesin and dynein isoforms, have been associated with LIS, and with related MCDs including microcephaly, pachygyria, SBH, and polymicrogyria-like cobblestone malformations (6,22,94.95). Microtubules are involved in mitosis, centrosome formation, organization of intracellular structure, axon pathfinding, and protein transport, accounting for the diverse MCDs caused by tubulinopathies. Abnormal brain development in at least some tubulinopathies is caused by a dominant negative effect of heterozygous missense mutations (95).
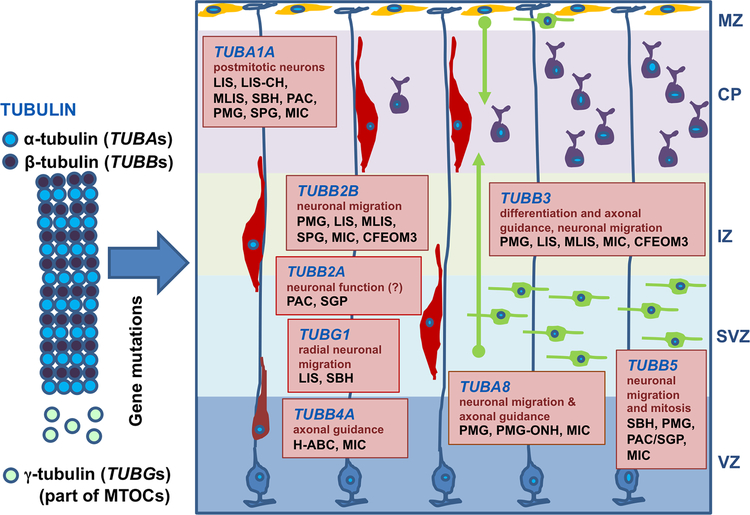
Figure 4.
Tubulinopathies affect multiple processes in cortical development, and cause heterogeneous MCDs. Boxes represent the expression, functions, and MCDs associated with TUBA1A, TUBA8, TUBB2A, TUBB2B, TUBB3, TUBB4A, TUBB5, and TUBG1. Abbreviations: CFEOM3: congenital fibrosis of extraocular muscles type 3; CP: cortical plate; H-ABC: hypomyelination with atrophy of the basal ganglia and cerebellum; IZ: intermediate zone; LIS: lissencephaly; LIS-CH: lissencephaly with cerebellar hypoplasia; MIC: congenital microcephaly; MLIS: microlissencephaly; MTOCs: microtubule organizing centers; MZ: marginal zone (Cajal-Retzius neurons in yellow); PAC: pachygyria; PMG: polymicrogyria; PMG-ONH: polymicrogyria with optic nerve hypoplasia; SBH: subcortical band heterotopia; SGP: simplified gyral pattern; SVZ: subventricular zone; VZ: ventricular zone.
Some forms of genetic LIS are considered “variant” because they do not directly impact cytoskeletal functions. One of these is XLAG, resulting from mutations of a transcription factor gene, ARX, on Xp22.3. Patients with XLAG have occipital-predominant three-layered LIS, agenesis of the corpus callosum, abnormal basal ganglia, thalamocortical tract defects, and hydrocephalus (18,19,87). Also, approximately 5–10% of patients with X-linked ID (but not XLAG) have ARX mutations. ARX mutations cause a reduction of GABAergic neurons in the basal ganglia and cerebral cortex (96), and XLAG has sometimes been considered an “interneuronopathy.” However, ARX also plays important roles in cortical projection neuron development (97).
Another variant LIS is caused by mutations of RELN, encoding the extracellular protein Reelin, which functions in regulating both cell migration and mTOR signaling (98). Patients with autosomal recessive mutations of RELN, or its receptor VLDLR (reelinopathies), are developmentally impaired with global DD and early onset of generalized epilepsy. The MRI phenotype of reelinopathies ranges from simplified gyral pattern with mild cortical thickening, to frank LIS with substantial cortical thickening (6).
Microlissencephaly represents a heterogeneous group of rare disorders (Di Donato et al, 2017). Microlissencephaly can be caused by RELN mutations (Norman-Roberts syndrome). Also, as noted above, some tubulinopathies result in microlissencephaly, most often linked to TUBA1A, less often TUBB2B, TUBB3 or TUBG1 mutations (99). Mutations in CIT (encoding citron kinase, important in mitosis) have also been linked to microlissencephaly (100). Another group includes patients with autosomal recessive microcephalic osteodysplastic primordial dwarfism type 1 (MOPD-1), exhibiting predominantly anterior 3-layered microlissencephaly, glioneuronal heterotopia, hypoplasia of the corpus callosum, and mid- and hindbrain defects (101). Recently, mutations in RNU4ATAC, a noncoding small nuclear RNA gene involved in splicing, have been linked to MOPD-1 (102).
5.2. Cobblestone malformations
Cobblestone LIS, formerly known as LIS type II (103), results from impaired cerebral basement membrane and glia limitans formation, usually attributed to defects in the linkage of radial glia to the basement membrane. Cobblestone LIS represents the severe end of a spectrum, generally characterized by migration of neurons and glia into the subarachnoid space, conferring an irregular “lumpy-bumpy” appearance to the cortical surface (6). Cobblestone malformations are predominantly autosomal recessive disorders with cerebral, ocular and muscular abnormalities.
Different forms of severe cobblestone LIS include Walker-Warburg syndrome (WWS), muscle-eye-brain (MEB) disease, and Fukuyama congenital muscular dystrophy (FCMD). The most severe of these is the rare WWS, with an incidence of 1.2 cases per 100,000 live births (104). In contrast, FCMD, linked to FKTN and FKRP mutations and found mainly in Japanese populations, is typically much less severe clinically (105,106). The FCMD cortical phenotype ranges from nearly normal cerebral cortex with few heterotopia, to cobblestone LIS with large gaps and prominent cortical surface bumps (107).
These related disorders (WWS, MEB, FCMD) are caused by mutations in enzymes that glycosylate α-dystroglycan, encoded by genes including POMT1, POMT2, POMGNT1, FKTN, FKRP, and LARGE. Together, these diseases are known as α-dystroglycanopathies (107). Mutations in TMEM5 and ISPD genes, likewise involved in dystroglycan glycosylation, were also recently detected in severe cobblestone LIS (108). Defective glycosylation of α-dystroglycan on the basal end of radial glia causes defects of radial glia adhesion to the pial basement membrane, leading to aberrant neuronal “overmigration” into the subarachnoid space (109).
Other genes important for basement membrane structure and function have also been linked to cobblestone MCDs. Mutations involving laminins or glycosyltransferases, including LAMB1, LAMB2, LAMC3, and SRD5A3 have been linked to cobblestone malformations (110,111). Mutation of GPR56, encoding a collagen III receptor, is also associated with a cobblestone MCD, manifesting as bilateral frontoparietal polymicrogyria-like dysgenesis (112).
5.3. Micrencephaly
Micrencephaly (referring to small brain) is usually recognized by microcephaly (small head). The most severe forms, the primary micrencephalies, are caused by defects in important genes for cell division, reviewed elsewhere (113). Interestingly, less severe micrencephaly can result from defects in PI3K→AKT→mTOR signaling, such as loss-of-function mutations of AKT3 (114). Non-genetic causes of micrencephaly include toxic, infectious, and ischemic insults, such as fetal alcohol exposure, or Zika virus infection.
6. POLYMICROGYRIA
Like lissencephaly, polymicrogyria (too many, too small gyri) has been variously applied to MCDs of exceedingly diverse etiology and histopathology. Thus, by the broadest definitions, polymicrogyria is observed in many heterogeneous genetic disorders, including brain overgrowth syndromes (Table 2); cobblestone malformations; cortex with excessive but relatively well-formed gyri (polygyria), as in thanatophoric dysplasia; and unique forms of cortical dysgenesis, as in monosomy 1p36 (115). However, definitions of polymicrogyria remain unsettled, even among neuropathologists. For example, one recent study suggested that polymicrogyria is essentially defined by fusion of the molecular layer across gyri (116), while another reported that fusion is present in fewer than one-third of cases, and that disruptions at the brain surface are the most consistent anomaly in polymicrogyria (117).
Given these discrepancies in the definition and features of polymicrogyria, and to avoid redundancy with categories such as brain overgrowth and cobblestone malformations, polymicrogyria may be considered for now as a descriptive grouping that can be applied in many circumstances, rather than as a distinct, independent class of MCDs.
7. AXON PATHWAYS DEFECTS
7.1. Agenesis of the corpus callosum: partial and complete
Corpus callosum abnormalities, which include complete and partial agenesis, as well as dysgenesis, are among the most common brain malformations, observed in isolation or associated with complex malformation syndromes (118). The estimated overall prevalence of callosal abnormalities is 3–7 per 1000 births, and reaches 3–5% in patients with various disorders of brain development (118,119). Most patients with callosal agenesis (~75%) show normal intelligence, with the remainder exhibiting some degree of ID (119).
Callosal anomalies are often diagnosed on neuroimaging studies. Initial formation of the corpus callosum depends on coordination between callosal projection neurons extending axons towards the midline, and multiple midline structures that may guide callosal axons, such as the glial wedge. This conclusion is based on observations from transgenic mice, in which defects of the corpus callosum have been associated with mutations that perturb development of neurons or the glial wedge (120–122).
The etiology of most human callosal abnormalities remains unknown, although genetic and non-genetic factors contribute. It is estimated that up to 45% of callosal abnormalities are associated with genetic defects, including 20% with chromosomal aberrations, and the remainder with genetic syndromes caused by a single or multiple gene variants (118). Indeed, a vast number of MCDs include callosal defects, and thus implicate an equally vast number of genes in callosal development (2,19, 22,123). For example, ciliopathies such as orofaciodigital type 1 syndrome, Meckel-Gruber syndrome, Joubert syndrome, primary ciliary dyskinesia, and Bardet-Biedl syndrome often include callosal defects, and there are dozens of ciliopathy genes (124, 125).
The genetic causes of isolated callosal agenesis appear more elusive, given the lack of strong phenotypes and family studies. Nevertheless, CDK5RAP2 (MDPH3), a gene linked to autosomal recessive microcephaly, was also recently linked to isolated callosal agenesis (126).
7.2. Other axon pathways
Even more elusive are genes linked to the formation or guidance of other axon pathways, such as the corticothalamic pathway. Evaluation of these axon tracts in humans requires sophisticated techniques, such as diffusion tensor imaging (DTI) by neuroradiology, and defects may not have clear phenotypes. In future studies of disorders such as ID and autism that lack definite MCDs but are linked to specific genes, it may be fruitful to study axonal organization by methods such as DTI, since brain wiring may be equally important to cellular structures in such cases.
8. CONCLUSIONS AND FUTURE DIRECTIONS
Among the most important implications of recent research, it has become clear that many genetic MCDs span multiple classification categories based on morphology and primary mechanisms such as proliferation, differentiation, and migration. Polymicrogyria, for example, spans several categories. This reflects the involvement of some genes, such as tubulins, in diverse processes from cell division to axon growth. Thus, genetic diagnoses are an increasingly important adjunct to pathology.
Adding to the challenges of classification and diagnosis, it has also become clear that some genetic MCDs, especially FCDs, can arise from type 1 or type 2 somatic mosaic mutations in many different genes. Deep sequencing of resected brain tissues, along with the application of markers such as pS6 to evaluate mTORC1 activation (Fig. 3), can help resolve uncertainties about the involved gene and pathways, as well as the type of mosaicism.
While tremendous progress has been made in studying MCDs, subtle defects, such as disorganization of axon pathways and cortical arealization in disorders such as autism, remain to be explored. While genetic studies of ID and autism have revealed the important role of de novo mutations, more sophisticated techniques in neuroimaging, such as DTI to detect axon defects, and functional MRI to assess cortical arealization, may help advance the phenotypic characterizations. Advances in these and other areas of MCD research will be important to drive future progress in diagnosis and treatment.
ACKNOWLEDGMENTS
Supported by NIH Grants R01 NS092339, and R01 NS085081 to R.F.H. We thank our colleagues in research on cortical malformations, especially Dr. William Dobyns and Dr. Ghayda Mirzaa, for their constant encouragement and stimulating discussions.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, membership, funding, or financial holding that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Bystron I, Blakemore C, Rakic P. 2008. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci 9:110–132 [DOI] [PubMed] [Google Scholar]
- 2.Barkovich AJ, Guerrini R, Kuzniecky RI, Jackson GD, Dobyns WB. 2012. A developmental and genetic classification for malformations of cortical development: update 2012. Brain 135:1348–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerrini R, Dobyns W. 2014. Malformations of cortical development: clinical features and genetic causes. Lancet Neurology 13:710–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuzniecky RI. 1994. Magnetic resonance imaging in developmental disorders of the cerebral cortex. Epilepsia 35 Suppl 6:S44–56 [DOI] [PubMed] [Google Scholar]
- 5.Leventer RJ, Phelan EM, Coleman LT, et al. 1999. Clinical and imaging features of cortical malformations in childhood. Neurology 53:715–722 [DOI] [PubMed] [Google Scholar]
- 6.Barkovich AJ, Dobyns WB, Guerrini R. 2015. Malformations of cortical development and epilepsy. Cold Spring Harb Perspect Med 5:a022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jamuar SS, Lam AT, Kircher M, D’Gama AM, Wang J, et al. 2014. Somatic mutations in cerebral cortical malformations. N Engl J Med 371:733–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Cai T, Jiang Y, Chen H, He X, et al. 2016. Genes with de novo mutations are shared by four neuropsychiatric disorders discovered from NPdenovo database. Mol Psychiatry 21:290–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilfert AB, Sulovari A, Turner TN, Coe BP, Eichler EE. 2017. Recurrent de novo mutations in neurodevelopmental disorders: properties and clinical implications. Genome Med 9:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lombroso CT. 2000. Can early postnatal closed head injury induce cortical dysplasia. Epilepsia. 41:245–253 [DOI] [PubMed] [Google Scholar]
- 11.Sarnat HB. 1987. Disturbance of late neuronal migrations in the perinatal period. Am J Dis Child 141:969–980 [DOI] [PubMed] [Google Scholar]
- 12.Hong SJ, Bernhardt BC, Gill RS, Bernasconi N, Bernasconi A. 2017. The spectrum of structural and functional network alterations in malformations of cortical development. Brain 140:2133–2143 [DOI] [PubMed] [Google Scholar]
- 13.Happle R 1997. A rule concerning the segmental manifestation of autosomal dominant skin disorders. Review of clinical examples providing evidence for dichotomous types of severity. Arch Dermatol 133:1505–9 [PubMed] [Google Scholar]
- 14.Jansen LA, Mirzaa GM, Ishak GE, O’Roak BJ, Hiatt JB, et al. 2015. PI3K/AKT pathway mutations cause a spectrum of brain malformations from megalencephaly to focal cortical dysplasia. Brain 138:1613–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin W, Chan JA, Vinters HV, Mathern GW, Franz DN, et al. 2010. Analysis of TSC cortical tubers by deep sequencing of TSC1, TSC2 and KRAS demonstrates that small second-hit mutations in these genes are rare events. Brain Pathol 20:1096–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallinger G, Kindron D, Schreiber L, Ben-Sira L, Hoffman C, et al. 2007. Prenatal diagnosis of malformations of cortical development by dedicated neurosonography. Ultrasound Obstet Gynecol 29:178–91 [DOI] [PubMed] [Google Scholar]
- 17.Barkovich AJ, Kuzniecky RI, Dobyns WB, Jackson GD, Becker LE, et al. 1996. A classification scheme for malformations of cortical development. Neuropediatrics 27:59–63 [DOI] [PubMed] [Google Scholar]
- 18.Desikan RS, Barkovich AJ. 2016. Malformations of cortical development . Ann Neurol 80:797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero DM, Bahi-Buisson N, Francis F. 2017. Genetics and mechanisms leading to human cortical malformations. Semin Cell Dev Biol [Epub ahead of print] [DOI] [PubMed]
- 20.Thornton GK, Woods CG. 2009. Primary microcephaly: do all roads lead to Rome? Trends Genet 25:501–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu TW, Mochida GH, Tischfield DJ, Sgaier SK, Flores-Sarnat L, et al. 2010. Mutations in WDR62, encoding a centrosome-associated protein, cause microcephaly with simplified gyri and abnormal cortical architecture. Nat Gen 42:1015–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bahi-Buisson N, Poirier K, Fourniol F, Saillaour Y, Valence S, et al. 2014. The wide spectrum of tubulinopathies: what are the key features for the diagnosis? Brain 137:1676–1700 [DOI] [PubMed] [Google Scholar]
- 23.Di Donato N, Chiari S, Mirzaa GM, Aldinger K, Parrini E, et al. 2017. Lissencephaly: expanded imaging and clinical classification. Am J Med Genet 173A:1473–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bahi-Buisson N, Poirier N, Boddaert N, Fallet-Bianco C, Specchio N, et al. 2010. GPR56-related bilaeral frontoparietal polymicrogyria: further evidence for an overlap with the cobblestone complex. Brain 133:3194–209 [DOI] [PubMed] [Google Scholar]
- 25.Rivas L, Blanco O, Torreira C, Repáraz A, Melcón C, Amado A. 2017. Pontocerebellar hypoplasia secondary to CASK gene deletion. Case report. Rev Chil Pediatr 88:529–33 [DOI] [PubMed] [Google Scholar]
- 26.Rakic P 1988. Specification of cerebral cortical areas. Science 241:170–6 [DOI] [PubMed] [Google Scholar]
- 27.Welker W 1990. Why does cerebral cortex fissure and fold? A review of determinants of gyri and sulci in cerebral cortex. In: Cerebral Cortex, Vol 8B (eds. Jones EG & Peters A) Springer, 1990. [Google Scholar]
- 28.Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. 1997. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science 278:474–6 [DOI] [PubMed] [Google Scholar]
- 29.Chu J, Anderson SA. 2015. Development of cortical interneurons. Neuropsychopharmacology 40:16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lui JH, Hansen DV, Kriegstein AR. 2011. Development and evolution of the human neocortex. Cell 146:18–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sun T, Hevner RF. 2014. Growth and folding of the mammalian cerebral cortex: from molecules to malformations. Nat Rev Neurosci 15:217–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kowalczyk T, Pontious A, Englund C, Daza RA, Bedogni F, et al. 2009. Intermediate neuronal progenitors (basal progenitors) produce pyramidal-projection neurons for all layers of cerebral cortex. Cereb Cortex 19:2439–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kriegstein A, Noctor S, Martínez-Cerdeño V. 2006. Patterns of neural stem and progenitor cell division may underlie evolutionary cortical expansion. Nat Rev Neurosci 7:883–90 [DOI] [PubMed] [Google Scholar]
- 34.Hevner RF. 2016. Evolution of the mammalian dentate gyrus. J Comp Neurol 524:578–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smart IH, Dehay C, Giroud P, Berland M, Kennedy H. 2002. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb Cortex 12:37–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hansen DV, Lui JH, Parker PR, Kriegstein AR. 2010. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 464:554–61 [DOI] [PubMed] [Google Scholar]
- 37.Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, et al. 2017. Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358:1318–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nowakowski TJ, Pollen AA, Sandoval-Espinosa C, Kriegstein AR. 2016. Transformation of the radial glia scaffold demarcates two stages of human cerebral cortex development. Neuron 91:1219–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.de Juan Romero C, Bruder C2, Tomasello U1, Sanz-Anquela JM3, Borrell V 2015. Discrete domains of gene expression in germinal layers distinguish the development of gyrencephaly. EMBO J 34:1859–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsumoto N, Shinmyo Y, Ichikawa Y, Kawasaki H. 2017. Gyrification of the cerebral cortex requires FGF signaling in the mammalian brain. Elife 14;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malik S, Vinukonda G, Vose LR, Diamond D, Bhimavarapu BB, et al. 2013. Neurogenesis continues in the third trimester of pregnancy and is suppressed by premature birth. J Neurosci 33:411–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorrells SF, Paredes MF, Cebrian-Silla A, Sandoval K, Qi D, et al. 2018. Human hippocampal neurogenesis drops sharply in children to undetectable levels in adults. Nature [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 43.Greig LC, Woodworth MB, Galazo MJ, Padmanabhan H, Macklis JD. 2013. Molecular logic of neocortical projection neuron specification, development and diversity. Nat Rev Neurosci 14:755–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bedogni F, Hodge RD, Elsen GE, Nelson BR, Daza RA, et al. 2010. Tbr1 regulates regional and laminar identity of postmitotic neurons in developing neocortex. Proc Natl Acad Sci USA 107:13129–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hevner RF. 2005. The cerebral cortex malformation in thanatophoric dysplasia: neuropathology and pathogenesis. Acta Neuropathol 110:208–21 [DOI] [PubMed] [Google Scholar]
- 46.Blümcke I, Thom M, Aronica E, Armstrong DD, Vinters HV, et al. 2011. The clinicopathologic spectrum of focal cortical dysplasias: a consensus classification proposed by an ad hoc Task Force of the ILAE Diagnostic Methods Commission. Epilepsia 52:158–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Blümcke I, Spreafico R, Haaker G, Coras R, Kobow K, et al. 2017. Histopathological findings in brain tissue obtained during epilepsy surgery. N Engl J Med 377:1648–56 [DOI] [PubMed] [Google Scholar]
- 48.Vanhaesebroeck B, Stephens L, Hawkins P. 2012. PI3K signalling: the path to discovery and understanding. Nat Rev Mol Cell Biol 13:195–203 [DOI] [PubMed] [Google Scholar]
- 49.Saxton RA, Sabatini DM. 2017. mTOR signaling in growth, metabolism, and disease. Cell 168:960–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Janku F, Yap TA, Meric-Bernstam F. 2018. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol [Epub ahead of print] [DOI] [PubMed]
- 51.Iwata T, Hevner RF. 2009. Fibroblast growth factor signalling in development of the cerebral cortex. Dev Growth Differ 51:299–323 [DOI] [PubMed] [Google Scholar]
- 52.Kunova Bosakova M, Varecha M, Hampl M, Duran I, et al. 2018. Regulation of ciliary function by fibroblast growth factor signaling identifies FGFR3-related disorders achondroplasia and thanatophoric dysplasia as ciliopathies. Hum Mol Genet [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 53.Puffenberger EG, Strauss KA, Ramsey KE, Craig DW, Stephan DA, et al. 2007. Polyhydramnios, megalencephaly and symptomatic epilepsy caused by a homozygous 7-kilobase deletion in LYK5. Brain 130:1929–41 [DOI] [PubMed] [Google Scholar]
- 54.Lindhurst MJ, Sapp JC, Teer JK, Johnston JJ, Finn EM, et al. 2011. A mosaic activating mutation in AKT1 associated with the Proteus syndrome. N Engl J Med 365:611–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, et al. 2012. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet 44:941–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Poduri A, Evrony GD, Cai X, Elhosary PC, Beroukhim R, et al. 2012. Somatic activation of AKT3 causes hemispheric developmental brain malformations. Neuron 74:41–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rivière JB, Mirzaa GM, O’Roak BJ, Beddaoui M, Alcantara D, et al. 2012. De novo germline and postzygotic mutations in AKT3, PIK3R2 and PIK3CA cause a spectrum of related megalencephaly syndromes. Nat Genet 44:934–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borrie SC, Brems H, Legius E, Bagni C. 2017. Cognitive dysfunctions in intellectual disabilities: the contributions of the Ras-MAPK and PI3K-AKT-mTOR pathways. Annu Rev Genomics Hum Genet 18:115–42 [DOI] [PubMed] [Google Scholar]
- 59.Reijnders MRF, Kousi M, van Woerden GM, Klein M, Bralten J, et al. 2017. Variation in a range of mTOR-related genes associates with intracranial volume and intellectual disability. Nat Commun 8:1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yeung KS, Tso WWY, Ip JJK, Mak CCY, Leung GKC, et al. 2017. Identification of mutations in the PI3K-AKT-mTOR signalling pathway in patients with macrocephaly and developmental delay and/or autism. Mol Autism [Epub ahead of print] [DOI] [PMC free article] [PubMed]
- 61.Alcantara D, Timms AE, Gripp K, Baker L, Park K, et al. 2017. Mutations of AKT3 are associated with a wide spectrum of developmental disorders including extreme megalencephaly. Brain 140:2610–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.D’Gama AM, Woodworth MB, Hossain AA, Bizzotto S, Hatem NE, et al. 2017. Somatic mutations activating the mTOR Pathway in dorsal telencephalic progenitors cause a continuum of cortical dysplasia. Cell Rep 21:3754–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baek ST, Copeland B, Yun EJ, Kwon SK, Guemez-Gamboa A, et al. 2015. AKT3-FOXG1-reelin network underlies defective migration in human focal malformations of cortical development. Nat Med 21:1445–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conti V, Pantaleo M, Barba C1, Baroni G, Mei D, et al. 2015. Focal dysplasia of the cerebral cortex and infantile spasms associated with somatic 1q21.1-q44 duplication including the AKT3 gene. Clin Genet 88:241–7 [DOI] [PubMed] [Google Scholar]
- 65.Milone R, Valetto A, Battini R, Bertini V, Valvo G, et al. 2016. Focal cortical dysplasia, microcephaly and epilepsy in a boy with 1q21.1-q21.3 duplication. Eur J Med Genet 59:278–82 [DOI] [PubMed] [Google Scholar]
- 66.Baulac S, Ishida S, Marsan E, Miquel C, Biraben A, et al. 2015. Familial focal epilepsy with focal cortical dysplasia due to DEPDC5 mutations. Ann Neurol 77:675–83 [DOI] [PubMed] [Google Scholar]
- 67.Weckhuysen S, Marsan E, Lambrecq V, Marchal C, Morin-Brureau M, et al. 2016. Involvement of GATOR complex genes in familial focal epilepsies and focal cortical dysplasia. Epilepsia 57:994–1003 [DOI] [PubMed] [Google Scholar]
- 68.Jansen LA, Hevner RF, Roden WH, Hahn SH, Jung S, Gospe SM Jr. 2014. Glial localization of antiquitin: implications for pyridoxine-dependent epilepsy. Ann Neurol 75:22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Uddin M, Woodbury-Smith M, Chan A, Brunga L, Lamoureux S, et al. 2017. Germline and somatic mutations in STXBP1 with diverse neurodevelopmental phenotypes. Neurol Genet 3(6):e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.O’Kusky J, Ye P. 2012. Neurodevelopmental effects of insulin-like growth factor signaling. Front Neuroendocrinol 33:230–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Najm IM, Sarnat HB, Blümcke I. 2018. Review: The international consensus classification of Focal Cortical Dysplasia - a critical update 2018. Neuropathol Appl Neurobiol 44:18–31 [DOI] [PubMed] [Google Scholar]
- 72.Flores-Sarnat L, Sarnat HB, Dávila-Gutiérrez G, Alvarez A. 2003. Hemimegalencephaly: part 2. Neuropathology suggests a disorder of cellular lineage. J Child Neurol 18:776–85 [DOI] [PubMed] [Google Scholar]
- 73.Englund C, Folkerth RD, Born D, Lacy JM, Hevner RF. 2005. Aberrant neuronal-glial differentiation in Taylor-type focal cortical dysplasia (type IIA/B). Acta Neuropathol 109:519–33 [DOI] [PubMed] [Google Scholar]
- 74.Lamparello P, Baybis M, Pollard J, Hol EM, Eisenstat DD, et al. 2007. Developmental lineage of cell types in cortical dysplasia with balloon cells. Brain 130:2267–76 [DOI] [PubMed] [Google Scholar]
- 75.Orlova KA, Tsai V, Baybis M, Heuer GG, Sisodiya S, et al. 2010. Early progenitor cell marker expression distinguishes type II from type I focal cortical dysplasias. J Neuropathol Exp Neurol 69:850–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salamon N, Andres M, Chute DJ, Nguyen ST, Chang JW, et al. 2006. Contralateral hemimicrencephaly and clinical-pathological correlations in children with hemimegalencephaly. Brain 129:352–65 [DOI] [PubMed] [Google Scholar]
- 77.Tsai V, Parker WE, Orlova KA, Baybis M, Chi AW, et al. 2014. Fetal brain mTOR signaling activation in tuberous sclerosis complex. Cereb Cortex 24:315–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Boer K, Crino PB, Gorter JA, Nellist M, Jansen FE, et al. 2010. Gene expression analysis of tuberous sclerosis complex cortical tubers reveals increased expression of adhesion and inflammatory factors. Brain Pathol 20:704–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mühlebner A, van Scheppingen J, Hulshof HM, Scholl T, Iyer AM, et al. 2016. Novel histopathological patterns in cortical tubers of epilepsy surgery patients with tuberous sclerosis complex. PLoS One 11(6):e0157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Prabowo AS, Anink JJ, Lammens M, Nellist M, van den Ouweland AM, et al. 2013. Fetal brain lesions in tuberous sclerosis complex: TORC1 activation and inflammation. Brain Pathol 23:45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Taylor DC, Falconer MA, Bruton CJ, Corsellis JA. 1971. Focal dysplasia of the cerebral cortex in epilepsy. J Neurol Neurosurg Psychiatry 34:369–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Krueger DA, Care MM, Holland K, Agricola K, Tudor C, et al. 2011. Everolimus for subependymal giant-cell astrocytomas in tuberous sclerosis. N Engl J Med 363:1801–11 [DOI] [PubMed] [Google Scholar]
- 83.Franz DN, Belousova E, Sparagana S, Bebin EM, Frost M, et al. 2014. Everolimus for subependymal giant cell astrocytoma in patients with tuberous sclerosis complex: 2-year open-label extension of the randomised EXIST-1 study. Lancet Oncol 15:1513–20 [DOI] [PubMed] [Google Scholar]
- 84.Parrini E, Conti V, Dobyns WB, Guerrini R. 2016. Genetic basis of brain malformations. Mol Syndromol 7:220–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guerrini R, Filippi T. 2005. Neuronal migration disorders, genetics, and epileptogenesis. J Child Neurol 20:287–99 [DOI] [PubMed] [Google Scholar]
- 86.Leventer RJ, Guerrini R, Dobyns WB. 2008. Malformations of cortical development and epilepsy. Dialogues Clin Neurosci 10:47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Forman MS, Squier W, Dobyns WB, Golden JA. 2005. Genotypically defined lissencephalies show distinct pathologies. J Neuropathol Exp Neurol 64:847–57 [DOI] [PubMed] [Google Scholar]
- 88.Reiner O, Carrozzo R, Shen Y, Wehnert M, Faustinella F, et al. 1993. Isolation of a Miller-Dieker lissencephaly gene containing G protein beta-subunit-like repeats. Nature 364:717–721. [DOI] [PubMed] [Google Scholar]
- 89.Cardoso C, Leventer RJ, Ward HL, Toyo-Oka K, Chung J, et al. 2003. Refinement of a 400-kb critical region allows genotypic differentiation between isolated lissencephaly, Miller-Dieker syndrome, and other phenotypes secondary to deletions of 17p13.3. Am J Hum Genet 72:918–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Reiner O1, Sapir T. 2013. LIS1 functions in normal development and disease. Curr Opin Neurobiol 23:951–6 [DOI] [PubMed] [Google Scholar]
- 91.Sicca F, Kelemen A, Genton P, Das S, Mei D, et al. 2003. Mosaic mutations of the LIS1 gene cause subcortical band heterotopia. Neurology 61:1042–6 [DOI] [PubMed] [Google Scholar]
- 92.Hirotsune S, Fleck MW, Gambello MJ, Bix GJ, Chen A, et al. 1998. Graded reduction of Pafah1b1 (Lis1) activity results in neuronal migration defects and early embryonic lethality. Nat Genet 19:333–9 [DOI] [PubMed] [Google Scholar]
- 93.Keays DA, Tian G, Poirier K, Huang GJ, Siebold C, et al. 2007. Mutations in alpha-tubulin cause abnormal neuronal migration in mice and lissencephaly in humans. Cell 128:45–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fallet-Bianco C, Loeuillet L, Poirier K, Loget P, Chapon F, et al. 2008. Neuropathological phenotype of a distinct form of lissencephaly associated with mutations in TUBA1A. Brain 131:2304–20 [DOI] [PubMed] [Google Scholar]
- 95.Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, et al. 2013. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet 45:639–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Colasante G, Collombat P, Raimondi V, Bonanomi D, Ferrai C, et al. 2008. Arx is a direct target of Dix2 and thereby contributes to the tangential migration of GABAergic interneurons. J Neurosci 28:10674–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Simonet JC, Sunnen CN, Wu J, Golden JA, Marsh ED. 2015. Conditional loss of Arx from the developing dorsal telencephalon results in behavioral phenotypes resembling mild human ARX mutations. Cereb Cortex 25:2939–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ventruti A, Kazdoba TM, Niu S, D’Arcangelo G. 2011. Reelin deficiency causes specific defects in the molecular composition of the synapses in the adult brain. Neuroscience 189:32–42 [DOI] [PubMed] [Google Scholar]
- 99.Chang BS. 2015. Tubulinopathies and their brain malformation syndromes: every TUB on its own bottom. Epilepsy Currents 15:65–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Harding BN, Moccia A, Drunat S, Soukarieh O, Tubeuf H, et al. 2016. Mutations in citron kinase cause recessive microlissencephaly with multinucleated neurons. Am J Human Genet 99:511–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Juric-Sekhar G, Kapur RP, Glass IA, Murray ML, Parnell SE, Hevner RF. 2011. Neuronal migration of disorders in microcephalic osteodysplastic primordial dwarfism type I/III. Acta Neuropathol 121:545–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang Y, Wu X, Du L, Zheng J, Deng S, et al. 2018. Identification of compound heterozygous variants in the noncoding RNU4ATAC gene in a Chinese family with two successive foetuses with severe microcephaly. Hum Genomics 12:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dobyns WB, Kirkpatrick JB, Hittner HM, Roberts RM, Kretzer FL. 1985. Syndromes with lissencephaly. II: Walker-Warburg and cerebro-oculo-muscular syndromes and a new syndrome with type II lissencephaly. Am J Med Genet 22:157–95 [DOI] [PubMed] [Google Scholar]
- 104.Verrotti A, Spalice A, Ursitti F, Papetti, Mariani R, et al. 2010. New trends in neuronal migration disorders. Eur J Paediatr Neurol 14:1–12 [DOI] [PubMed] [Google Scholar]
- 105.Ishigaki K, Ihara C, Mori-Yashimura M, Murakami T, Sato T, et al. 2016. Japanese nationwide registry for Fukuyama congenital muscular dystrophy patients. Neuromuscul Disord 26:suppl2 S164 [Abstract] [DOI] [PubMed] [Google Scholar]
- 106.Kobayashi K, Kato R, Kondo-Iida E, Taniguchi-Ikeda M, Osawa M, et al. 2017. Deep-intronic variant of fukutin in the most prevalent point mutation of Fukuyama congenital muscular dystrophy in Japan. J Hum Genet 62:945–48 [DOI] [PubMed] [Google Scholar]
- 107.Devisme L, Bouchet C, Gonzalés M, Alanio E, Bazin A, et al. 2012. Cobblestone lissencephaly: Neuropathological subtypes and correlations with genes of dystroglycanopathies. Brain 135:469–82 [DOI] [PubMed] [Google Scholar]
- 108.Vuillaumier-Barrot S, Bouchet-Séraphin C, Chelbi M, Devisme L, Quentin S, et al. 2012. Identification of mutations in TMEM5 and ISPD as a cause of severe cobblestone lissencephaly. Am J Hum Genet 91:1135–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Myshrall TD, Moore SA, Ostendorf AP, Satz JS, Kowalczyk T, et al. 2012. Dystroglycan on radial glia end feet is required for pial basement membrane integrity and columnar organization of the developing cerebral cortex. J Neuropath Exp Neurol 71:1047–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Radmanesh F, Caglayan AO, Silhavy JL, Yilmaz C, Cantagrel V, et al. 2013. Mutations in LAMB1 cause cobblestone brain malformation without muscular or ocular abnormalities. Am J Hum Genet 92:468–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Radner S, Banos C, Bachay G, Li YN, Hunter DD, et al. 2013. β2 and γ3 laminins are critical cortical basement membrane components: ablation of Lamb2 and Lamc3 genes disrupts cortical lamination and produces dysplasia. Dev Neurobiol 73:209–29 [DOI] [PubMed] [Google Scholar]
- 112.Bahi-Buisson N, Nectoux J, Girard B, Van Esch H, De Ravel T, et al. 2010. Revisiting the phenotype associated with FOXG1 mutations: two novel cases of congenital Rett variant. Neurogenetics 11:241–49 [DOI] [PubMed] [Google Scholar]
- 113.Duerinckx S, Abramowicz M. 2017. The genetics of congenitally small brains. Semin Cell Dev Biol [Epub ahead of print] [DOI] [PubMed]
- 114.Boland E, Clayton-Smith J, Woo VG, McKee S, Manson FD, et al. 2007. Mapping of deletion and translocation breakpoints in 1q44 implicates the serine/threonine kinase AKT3 in postnatal microcephaly and agenesis of the corpus callosum. Am J Hum Genet 81:292–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shiba N, Daza RA, Shaffer LG, Barkovich AJ, Dobyns WB, Hevner RF. 2013. Neuropathology of brain and spinal malformations in a case of monosomy 1p36. Acta Neuropathol Commun 1:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Judkins AR, Martinez D, Ferreira P, Dobyns WB, Golden JA. 2011. Polymicrogyria includes fusion of the molecular layer and decreased neuronal populations but normal cortical laminar organization. J Neuropathol Exp Neurol 70:438–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jansen AC, Robitaille Y, Honavar M, Mullatti N, Leventer RJ, et al. 2016. The histopathology of polymicrogyria: a series of 71 brain autopsy studies. Dev Med Child Neurol 58:39–48 [DOI] [PubMed] [Google Scholar]
- 118.Edwards TJ, Sherr EH, Barkovich AJ, Richards LJ. 2014. Clinical, genetic and imaging findings identify new causes for corpus callosum development syndromes. Brain 137:1579–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sotiriadis A, Makrydimas G. 2012. Neurodevelopment after prenatal diagnosis of isolated agenesis of the corpus callosum: an integrative review. Am J Obstet Gynecol 206:e331–e335 [DOI] [PubMed] [Google Scholar]
- 120.Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, et al. 2001. Tbr1 regulates differentiation of the preplate and layer 6. Neuron 29:353–66 [DOI] [PubMed] [Google Scholar]
- 121.Donahoo AL, Richards LJ. 2009. Understanding of callosal development through the use of transgenic mouse modes. Semin Pediatr Neurol 16:127–42 [DOI] [PubMed] [Google Scholar]
- 122.Chinn GA, Hirokawa KE, Chuang TM, Urbina C, Patel F, et al. 2015. Agenesis of the corpus callosum due to defective glial wedge formation in Lhx2 mutant mice. Cereb Cortex 25:2707–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.O’Driscoll MC, Black GC, Clayton-Smith J, Sherr EH, Dobyns WB. 2010. Identification of genomic loci contributing to agenesis of the corpus callosum. Am J Med Genet A 152A:2145–59 [DOI] [PubMed] [Google Scholar]
- 124.Badano JL, Mitsuma, Beales PL, Katsanis N. 2006. The ciliopathies: an emerging class of the human genetic disorders. Annu Rev Genomics Hum Genet 7:125–48 [DOI] [PubMed] [Google Scholar]
- 125.Alby C, Boutaud L, Bonniére M, Collardeau-Frachon S, Guibaud L, et al. 2018. In utero ultrasound diagnosis of corpus callosum agenesis leading to the identification of orofaciodigital type 1 syndrome in female fetuses. Birth Defects Res 110:382–389 [DOI] [PubMed] [Google Scholar]
- 126.Jouan L, Bencheikh BOA, Daoud H, Dionne-Laporte A, Dobrzeniecka S, et al. 2016. Exome sequencing identified recessive CDK5RAP2 variants in patients with isolated agenesis of corpus callosum. Eur J Hum Genet 24:607–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lee JH, Huynh M, Silhavy JL, Kim S, Dixon-Salazar T, et al. 2012. De novo somatic mutations in components of the PI3K-AKT3-mTOR pathway cause hemimegalencephaly. Nat Genet 44:941–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mirzaa G, Parry DA, Fry AE, Giamanco KA, Schwartzentruber J, et al. 2014. De novo CCND2 mutations leading to stabilization of cyclin D2 cause megalencephaly-polymicrogyria-polydactyly-hydrocephalus syndrome. Nat Genet 46:510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.D’Gama AM, Geng Y, Couto JA, Martin B, Boyle EA, et al. 2015. Mammalian target of rapamycin pathway mutations cause hemimegalencephaly and focal cortical dysplasia. Ann Neurol 77:720–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Scheffer IE, Heron SE, Regan BM, Mandelstam S, Crompton DE, et al. 2014. Mutations in mammalian target of rapamycin regulator DEPDC5 cause focal epilepsy with brain malformations. Ann Neurol 75:782–7 [DOI] [PubMed] [Google Scholar]
- 131.Scerri T, Riseley JR, Gillies G, Pope K, Burgess R, et al. 2015. Familiar cortical dysplasia type IIA caused by a germline mutation in DEPDC5. Ann Clin Transl Neurol 2:575–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Cen Z, Guo Y, Lou Y, Jiang B, Wang J, Feng J. 2017. De novo mutation in DEPDC5 associated with unilateral pachygyria and intractable epilepsy. Seizure 50:1–3 [DOI] [PubMed] [Google Scholar]
- 133.Ricos MG, Hodgson BL, Pippucci T, Saidin A, Ong YS, et al. 2016. Mutations in the mammalian target of rapamycin pathway regulators NPRL2 and NPRL3 cause focal epilepsy. Ann Neurol 79:120–31 [DOI] [PubMed] [Google Scholar]
- 134.Lim JS, Kim WI, Kang HC, Kim SH, et al. 2015. Brain somatic mutations in MTOR cause focal cortical dysplasia type II leading to intractable epilepsy. Nat Med 21:395–400 [DOI] [PubMed] [Google Scholar]
- 135.Nakashima M, Saitsu H, Takei N, Tohyama J, Kato M, et al. 2015. Somatic mutations in the MTOR gene cause focal cortical dysplasia type IIb. Ann Neurol 78:375–86 [DOI] [PubMed] [Google Scholar]
- 136.Leventer RJ, Scerri T, Marsh AP, Pope K, Gillies G, et al. 2015. Hemispheric cortical dysplasia secondary to a mosaic somatic mutation in MTOR. Neurology 84:2029–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mirzaa GM, Campbell CD, Solovieff N, Goold C, Jansen LA, et al. 2016. Association of MTOR mutations with developmental brain disorders, including megalencephaly, focal cortical dysplasia, and pigmentary mosaicism. JAMA Neurol 73:836–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sim JC, Scerri T, Fanjul-Fernández M, Riseley JR, Gillies G, et al. 2016. Familiar cortical dysplasia caused by mutation in the mammalian target of rapamycin regulator NPRL3. Ann Neurol 79:132–7 [DOI] [PubMed] [Google Scholar]
- 139.Mirzaa GM, Conti V, Timms AE, Smyser CD, Ahmed S, et al. 2015. Characterisation of mutations of the phosphoinositide-3-kinase regulatory subunit, PIK3R2, in perisylvian polymicrogyria: a next generation sequencing study. Lancet Neurol 14:1182–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Terrone G, Voisin N, Abdullah Alfaiz A, Cappuccio G, et al. 2016. De novo PIK3R2 variant causes polymicrogyria, corpus callosum hyperplasia and focal cortical dysplasia. Eur J Hum Genet 24:1359–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shick V, Majores M, Engels G, Spitoni S, et al. 2006. Activation of Akt independent of PTEN and CTMP tumor-suppressor gene mutations in epilepsy-associated Taylor-type focal cortical dysplasia. Acta Neuropathol 112:715–25 [DOI] [PubMed] [Google Scholar]
- 142.Crino PB, Nathanson KL, Henske EP. 2006. The tuberous sclerosis complex. N Engl J Med 35:1345–56 [DOI] [PubMed] [Google Scholar]
- 143.Lim JS, Gopalappa R, Kim SH, Ramakrishna S, Lee M, et al. 2017. Somatic mutations in TSC1 and TSC2 cause focal cortical dysplasia. Am J Hum Genet 100:454–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Martin KR, Zhou W, Bowman MJ, Shih J, Au KS, et al. 2017. The genomic landscape of tuberous sclerosis complex. Nat Commun 8:15816. [DOI] [PMC free article] [PubMed] [Google Scholar]