Summary:
We demonstrate that unconventional ST2- and CD127-negative ILC2 populations are present in mouse lung and are induced by Alternaria, suggesting commonly used ILC2 identification practices do not accurately enumerate the total burden of type 2 cytokine producing ILC2s.
Keywords: asthma, Alternaria, ILC1, ILC2, ILC3, IL-5, IL-13
To the Editor:
In the last decade, group 2 innate lymphoid cells (ILC2s) have been recognized for their ability to drive type 2 responses in experimental animal models in the absence of T cells.1, 2 Mice intranasally challenged with Alternaria alternata, a fungal allergen associated with severe asthma and fatal exacerbations in humans3, potently activates ILC2s via the IL-33/ST2 (IL-33R) axis to robustly secrete type 2 cytokines.4–6 Animal models, including Alternaria-induced lung inflammation in mice, have given us critical insight into pathways of ILC2 regulation that appear to apply to ILC2 responses in human disease.7 Thus, accurate identification of ILC2s in the lungs of mice is critical to future understanding of their biology and contribution to allergic disease. We assessed whether the conventional ILC2 identification markers ST2 and CD127 (IL-7R) sufficiently include all type 2 cytokine-producing ILCs in the mouse lung.
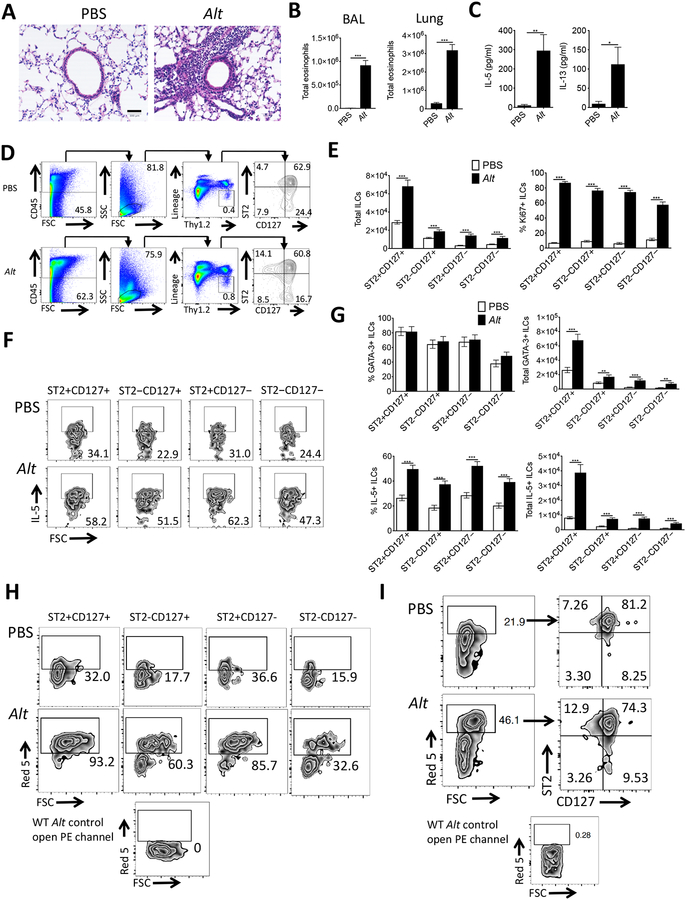
WT mice were challenged intranasally with Alternaria for three consecutive days and 24 hours after the last challenge BAL and lung tissue was harvested. Mice challenged with Alternaria demonstrated increased peri-bronchial inflammation (Fig. 1A), robust BAL and lung eosinophil recruitment (Fig. 1B), as well as elevated BAL IL-5 and IL-13 levels (Fig.1C). Conventional ILC2s were identified as CD45+Lineage−Thy1.2+ST2+CD127+ lymphocyte-sized cells (Fig. 1D & S1A, supplemental methods). As expected, total conventional ILC2s expanded after Alternaria challenge (Fig. 1E). Unexpectedly, ILC populations lacking either or both ST2 and CD127 proliferated significantly, with reduced apoptosis, and represented approximately 40% of the total ILC population (Fig.1E & S1B). All ILC subsets varied within 10% before, during and after Alternaria (Fig. S1C) challenges and also displayed a robust increase in IL-5 (Fig. 1F, 1G, & S1D), IL-13 (Fig. S2A), and GATA-3 (Fig 1G & S2B) expression. Further, all four subpopulations also highly expressed CD25 (IL-2Rα), killer lectin like receptor G1 (KLRG1), inducible T cell costimulator (ICOS), stem cell antigen-1 (Sca-1), c-Kit, CD44, and CD69 and expression of these markers (except c-Kit) was up-regulated by Alternaria (Fig. S3). RAG2 knockout (KO) mice that lack B and T cells but have ILCs showed similar ILC subset activation after Alternaria challenges (Fig. S4). Overall, though conventional ILC2s represented the highest total Th2 cytokine producing ILCs, the sum of the three unconventional populations in WT mice approximated 40% of the total ILC2 burden.
Figure 1. Unconventional ILC2 populations are induced by Alternaria.
(A) H&E stained lung sections (scale bar=100mm), (B) eosinophils, and (C) IL-5 and IL-13 from PBS or Alternaria (Alt) challenged mice. Identification, frequency (D), and total numbers (E) of ILCs. Ki67+ frequency within ILC subpopulations (E). Frequency and total number of IL-5+ and GATA-3+ ILCs (F & G). % IL-5+ ILC2s gated from all Thy1.2+ ILCs (H) and ILC ST2/CD127 gating from all IL-5+ ILCs (I) from Red5 mice. Data shown are representative of two to nine independent experiments with three mice per group. *p < 0.05, **p < 0.01, ***p < 0.001, unpaired t test.
To confirm that processing for ILC cytokines using calcium ionophore (supplemental methods) did not influence the presence of IL-5 production by the different ILC subsets, we in performed Alternaria challenges in heterozygous IL-5 reporter (Red5) mice. CD45+Lineage−Thy1.2+ lymphocytes were gated and all subsets (based on ST2 and CD127) showed significant increases in IL-5 expression after Alternaria challenges (Fig. 1H), though the two ST2− populations showed the lowest overall % IL-5 production suggesting a heterogenous mix of ILCs in these subsets. Using the same mice, we found that unconventional ILC2s represented approximately 25% of all IL-5+ CD45+Lineage−Thy1.2+ cells (Fig. 1I). Thus, in WT mice and reporter mice, 25 – 40% of ILC2s are negative for either or both ST2 and/or CD127.
In light of recent reports of ILC plasticity8, we investigated whether Alternaria also induced ILC1 and ILC3 responses. Surprisingly, IFNγ was detected in the BAL and increased significantly after Alternaria challenge (Fig. S5A) though IL-17A was not detected (not shown). IFNγ+ ILCs were increased in percent and total numbers (Fig. S5B), whereas IL-17A+ ILCs were largely increased in total numbers only in challenged mice (Fig. S5C). We found that conventional and unconventional ILC2s expressed similar levels of the ILC1 surface marker IL-18R9, and expression was nearly doubled after Alternaria challenge (Fig. S5D). All four populations also expressed low levels of RORγt (Fig. S5E) and T-bet (Fig. S5F) relative to GATA-3, and expression was not significantly different after Alternaria challenge. To elucidate whether the mixed ILC phenotypes were due to hybrid cells or heterogenous populations, we investigated the simultaneous expression of ILC1, ILC2, and ILC3 cytokine production (Fig. S6A & B). We detected low levels of IL-5+IFNγ+ ILC2s and IL-5+IL-17A+ ILC2s in each population, and the frequency of double-positive cells within each ILC2 subpopulation increased with Alternaria challenge. IL-17+ cells appeared to be distinct from IL-5+ cells though overall IL-5+IFNγ+ and IL-5+IL-17A+ ILC2s were increased following Alternaria challenge (Fig. S6A & B). In aggregate, these results suggest that the ILC populations contains a heterogeneous mixture of hybrid ILCs and distinct ILC1s and ILC3s.
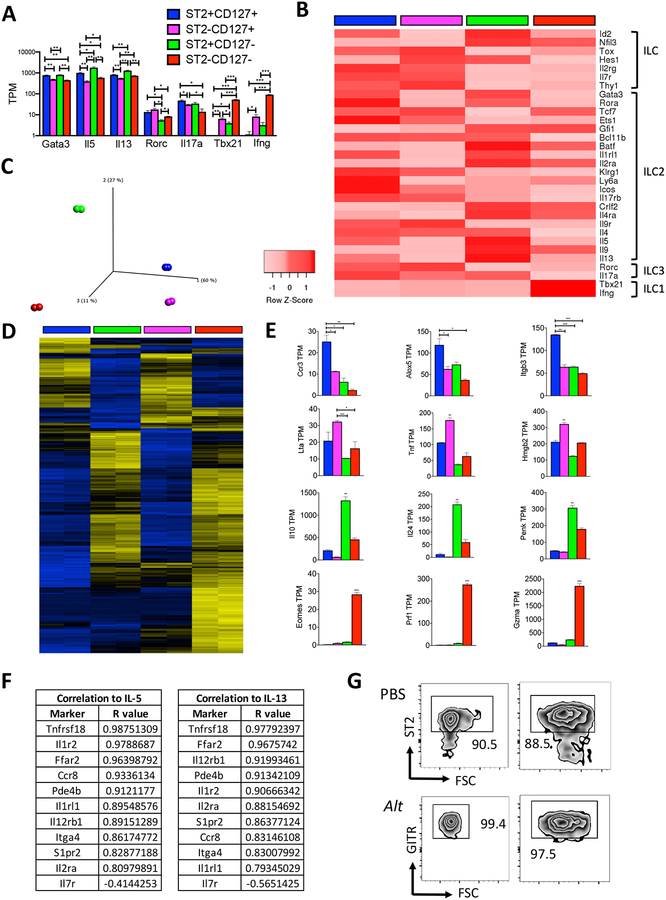
We next compared these ILC populations using RNA sequencing and qPCR with FACS sorted lung ILC subsets (Fig. 7A & supplemental methods) from mice challenged with Alternaria. In agreement with our flow cytometry protein-level data, we found high levels of ILC2-defining surface markers, transcription factors, and cytokine transcripts expressed by all four subpopulations, relative to ILC1 and ILC3 signatures (Fig. 2A & B). Importantly, RNA-Seq and qPCR confirmatory studies demonstrated that the ST2− populations express reduced levels of Il1rl1 transcript (encodes ST2), and the CD127− populations express less Il7ra transcript (Fig. S7B & C). Consistent with the protein data, the ST2+ populations expressed more ILC2-related transcripts (Gata3, Il5, Il9, Il13, and Rora) compared to the ST2− counterparts, and the ST2+CD127− population overall expressed the greatest number of ILC2-related transcripts while the ST2−CD127− population generally expressed the lowest number of ILC2-related transcripts (Fig. 2A & B). The CD127+ populations had the greatest ILC3 signature compared to the CD127− counterparts, whereas the ST2−CD127− population had the greatest ILC1 signature. Further, PCA analysis showed that ST2+CD127+ and ST2−CD127+ populations are more similar, compared with the ST2+CD127− and ST2−CD127− populations, which are individually more unique (Fig. 2C & D). Ccr3, Alox5, and Itgb3 transcripts were increased in the double-positive population and ST2−CD127+ ILCs showed increased expression of Lta, Tnf, and Hmgb2 (Fig. 2E). ST2+CD127− ILCs showed elevated levels of anti-inflammatory cytokines Il10 and Il24 and the ST2−CD127− ILCs had ILC1/NK transcript increases in Eomes, Gzma, and Prf1. These results suggest that all populations contain ILC2s as well as unique transcripts supporting overall heterogeneity in the four subsets.
Figure 2. ILC subpopulations contain ILC2 as well as heterogenous ILC transcriptomes.
RNA sequencing-derived transcript levels (A) and heat map (B) of ILC1, ILC2, ILC3 transcripts following Alternaria challenge (B). PCA plot (C), heat map (D), and differentially expressed candidate genes (E) following Alternaria challenge. Transcript correlation with IL-5 and IL-13 (F). (G) % ST2 and GITR of IL-5+ Thy1.2+ ILCs from Fig 1I. (A-F) Data shown are representative of a single experiment with two replicates that include pooled lungs from 9 mice. (G) Representative of 3 mice per group. *p < 0.05, **p < 0.01, ***p < 0.001, unpaired t test. Unless indicated by bars, stats in (E) compared to all other groups.
Finally, we correlated various transcript markers to the type 2 cytokine transcripts Il5 and Il13 across all populations. Tnfrsf18, that encodes glucocorticoid-induced TNFR family related gene (GITR), showed the strongest correlation with IL-5 and IL-13 transcripts, well above Il1rl1 (ST2) and Il7r (CD127) transcripts (Fig. 2F). Mirroring the transcript results, we found that GITR expression more accurately identified IL-5+ ILCs from Red5 mice by FACS compared with ST2 in both PBS and Alternaria challenged mice (Fig. 2G). Thus, our work strongly suggests that current methods of ILC2 identification do not sufficiently include the entire type 2 cytokine producing ILC2 population. Further, our studies highlight the heterogeneity and complexity of ILC responses to Alternaria alternata, a natural allergen associated with severe asthma.
Supplementary Material
Acknowledgements
We would like to thank Cody Fine and Jesus Olvera of the UCSD Human Embryonic Stem Cell Core for help with cell sorting, the LJI RNA Sequencing Core for help with RNA sequencing (NIH Sequencer grant #: S10OD016262), the Histology Core at the University of California, San Diego Moores Cancer Center for help with lung sectioning and staining (NIH grant #: P30CA23100), and Karina N. Roman for help with mouse work.
T.A.D. is supported by NIH grants AI 114585 and AI 070535, D.H.B. is supported by NIH grants AI 070535, AI 107779, AI 242236, and M.C. is supported by NIH grant AI 070535.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest disclosure: The authors declare no relevant conflicts of interest.
Bibliography
- 1.Doherty TA, Broide DH. Airway innate lymphoid cells in the induction and regulation of allergy. Allergol Int 2019; 68:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tait Wojno ED, Artis D. Emerging concepts and future challenges in innate lymphoid cell biology. J Exp Med 2016; 213:2229–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bush RK, Prochnau JJ. Alternaria-induced asthma. J Allergy Clin Immunol 2004; 113:227–34. [DOI] [PubMed] [Google Scholar]
- 4.Doherty TA, Khorram N, Sugimoto K, Sheppard D, Rosenthal P, Cho JY, et al. Alternaria Induces STAT6-Dependent Acute Airway Eosinophilia and Epithelial FIZZ1 Expression That Promotes Airway Fibrosis and Epithelial Thickness. J Immunol 2012; 188:2622–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doherty TA, Khorram N, Chang JE, Kim HK, Rosenthal P, Croft M, et al. STAT6 regulates natural helper cell proliferation during lung inflammation initiated by Alternaria. Am J Physiol Lung Cell Mol Physiol 2012; 303:L577–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartemes KR, Iijima K, Kobayashi T, Kephart GM, McKenzie AN, Kita H. IL-33-responsive lineage- CD25+ CD44(hi) lymphoid cells mediate innate type 2 immunity and allergic inflammation in the lungs. J Immunol 2012; 188:1503–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jurak LM, Xi Y, Landgraf M, Carroll ML, Murray L, Upham JW. Interleukin 33 Selectively Augments Rhinovirus-Induced Type 2 Immune Responses in Asthmatic but not Healthy People. Front Immunol 2018; 9:1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ohne Y, Silver JS, Thompson-Snipes L, Collet MA, Blanck JP, Cantarel BL, et al. IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat Immunol 2016; 17:646–55. [DOI] [PubMed] [Google Scholar]
- 9.Silver JS, Kearley J, Copenhaver AM, Sanden C, Mori M, Yu L, et al. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat Immunol 2016; 17:626–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.