Abstract
An accurate, rapid, and cost-effective biosensing strategy for the quantification of disease biomarkers is vital for the development of early-diagnostic point-of-care systems, benefiting overall human health. The recent discovery of the trans-cleavage activity of CRISPR type V effectors makes CRISPR a potential high-accuracy sensing system. In this study, we report the first CRISPR Cas12a (cpf1) based electrochemical biosensor (E-CRISPR), which is more cost-effective and portable comparing with optical transduction based biosensing systems. The trans-cleavage activity of Cas12a endonuclease is investigated by ssDNA reporter linked with electrochemical tag on a three-electrode based disposable sensor. To optimize the on-chip trans-cleavage activity of CRISPR in order to boost the detection sensitivity, the activity of different Cas12a analogs (LbCas12a and AsCas12a), the surface density of ssDNA reporter, the length of the ssDNA reporter, the trans-cleavage period, the activated operation concentration of Cas12a-crRNA complex and the concentration of divalent cation were investigated, providing an endonuclease favored condition for enhanced in vitro cleavage activity. To demonstrate the versatility of the system, the developed E-CRIPSR was firstly applied for the detection of nucleic acids, including Human papillomavirus 16 (HPV-16) and Parvovirus B19 (PB-19). Owing to the optimized chemical environment for the in vitro trans-cleavage of Cas12a, without any enzymatic amplification, a detection limit at pico molar level was achieved. We further designed an aptamer based E-CRISPR cascade for the detection of protein target. Quantification of transforming growth factor beta 1 (TGF-β1) protein in clinical samples was demonstrated as an example for protein detection. Owing to the combination of electrochemistry and CRISPR, we believe that the E-CRISPR could be a powerful enabler for wide developments of portable, accurate, and cost-effective point-of-care diagnostic systems.
Keywords: CRISPR Cas12a (cpf1), Trans-cleavage, Electrochemical biosensor, Bioanalytical chemistry, E-CRISPR
Graphical Abstract
Introduction
An accurate, rapid, and cost-effective sensing strategy for the quantification of disease biomarkers is vital for the development of early-diagnostic point-of-care systems, further leading to personalized medicine and benefiting overall human health. Electrochemistry based biosensing platforms have been widely developed, owing to its rapid signal readout, affordable transduction element and simple sensing platform.[1] One of the critical challenges for such sensing system is its accuracy. Recent robust developments of CRISPR (clustered regularly interspaced short palindromic repeats) based gene editing systems demonstrated the accuracy of the CRISPR system in targeting nucleic acids owing to the complementarity dependent CRISPR cleavage event.[2] Utilizing the Cas-crRNA target recognition-and-cleavage event induced collateral (trans) cleavage effect of the nonspecific ssDNA reporter, CRISPR type III, V, VI RNA guided nucleases (Csm6, Cas12a, Cas13) have been applied for the detection of nucleic acid (RNA/DNA) through fluorescence transduction system.[3] Herein, we extend the application of CRISPR-Cas12a (cpf1) system to the development of an electrochemical biosensing system, because of its relative cost-efficiency and portable nature of the transduction system comparing with those of the fluorescent transducer. Moreover, for a comprehensive and robust point-of-care system, a potential universal biosensing platform that can detect different categories of analytes is of high importance for the clinical applications. Therefore, built upon the accurate CRISPR based sensing system, other than detection of nucleic acid, we further design a Cas12a based biosensing cascade as a strategy for the detection of protein. Our study demonstrates a CRISPR-Cas12a (cpf1) based electrochemical biosensing platform (E-CRISPR) for the detection of the major categories of biomolecules, providing a potential deployable point-of-care system for the healthcare industry.
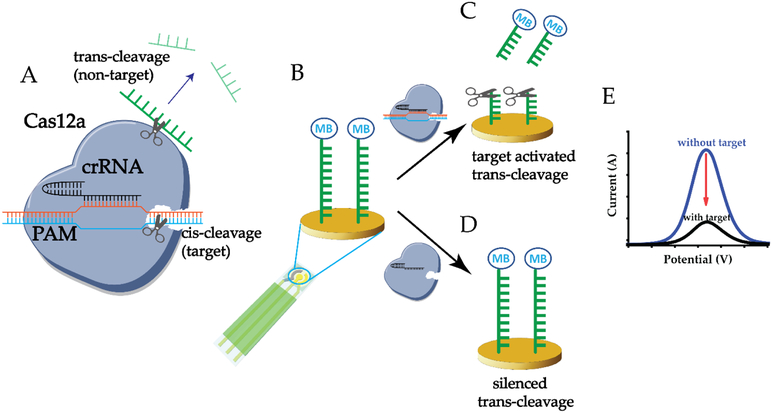
The E-CRISPR introduces a simple transduction method for CRISPR type III, V, VI nucleases based sensing systems and provides a new liberty in the classes of analytes the sensing system can detect. A disposable, micro-fabricated gold based three-electrode sensor with gold as working and counter electrodes and Ag/AgCl as the reference electrode was applied for the development of the electrochemical biosensor (Figure S1).[4] Cas12a-crRNA duplex is designed to specifically recognize and cleave target nucleic acid strand based on the protospacer adjacent motif (PAM) sequence of the target and crRNA sequence (Figure 1A).[5] The PAM recognition depends on the specific 5’ TTTN nucleic acid sequence located at the opposite strand (blue strand) of the recognition strand (orange strand). Only upon the recognition of the PAM sequence by the Cas protein, the Cas protein, acting as a DNA helicase, would unwind the target DNA. After the separation of the target strands, the complementarity (between crRNA and target) dependent cleavage activity can further be activated.[6] To achieve the electrochemical transduction of CRISPR detection signal, the target cis-cleavage initiated trans-cleavage (collateral cutting) effect of Cas12a on the nonspecific ssDNA is probed through an electrochemical method. A nonspecific ssDNA reporter is designed with a methylene blue (MB) electrochemical tag for signal transduction and a thiol moiety to tether on the sensor surface in order to acquire the signal electrically (Figure 1B).[7] Consequently, the electron transfer process between the gold electrode and the redox active species on the ssDNA can be electrochemically initiated and transduced. With the presence of the target, the Cas12a trans-cleavage activity is activated, cleaving the MB-ssDNA reporter off the electrode surface, therefore decreasing the MB signal transduced (Figure 1C). Without the presence of the target, the Cas12a trans-cleavage activity is silenced, retaining the MB-ssDNA reporter on the surface (Figure 1D). A representation of electrochemical signal output based on the conditions without/with target is shown in Figure 1E. The design of the MB-ssDNA reporter covered electrode is generally applicable for any CRISPR type III, V and VI systems as a simple and cost-effective signal transduction strategy. Based on the concept of E-CRISPR, we developed this platform as a universal biosensing strategy for the detection of nucleic acid, protein and small molecule.
Figure 1. Principle of E-CRISPR.
A) Cas12a (cpf1) performs crRNA guided cis-cleavage (specific target) initiated trans-cleavage activity (nonspecific ssDNA). B) Nonspecific ssDNA reporter with methylene blue tag immobilized on the gold electrode. C) With the presence of the target, Cas12a-crRNA would initiate the trans-cleavage activity on nonspecific ssDNA reporter, resulting a low electrochemical current of methylene blue. D) Without the presence of the target, Cas12a-crRNA would not initiate the trans-cleavage activity on nonspecific ssDNA reporter, resulting a high electrochemical current of methylene blue. E) A representation of electrochemical current outputs based on the without & with target conditions.
Results and Discussion
Verification of E-CRISPR on Nucleic Acid Detection
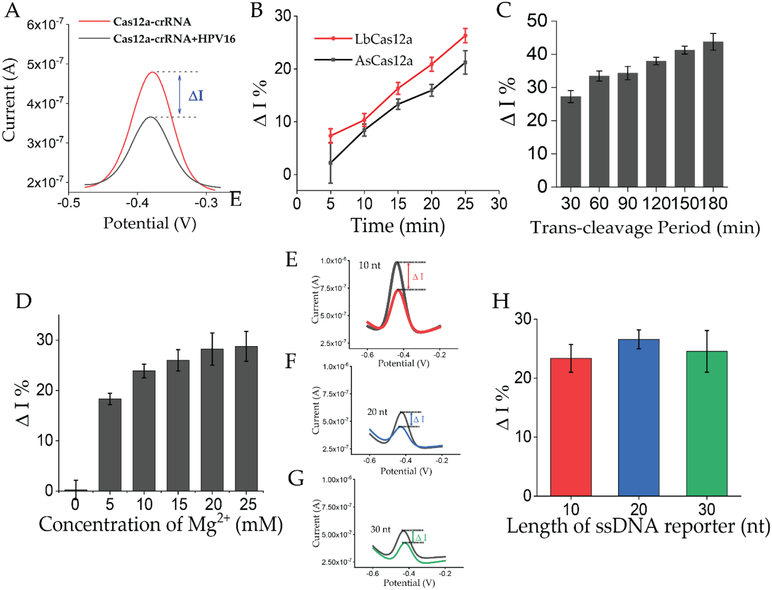
To examine the feasibility of the E-CRISPR on nucleic acid detection, a human papilloma virus (HPV) subtype, HPV-16, which is critical to carcinogenesis[8], was selected as the target. A target sequence in the L1-enconding gene of HPV16 was identified based on the TTTN PAM sequence required by the Cas12a endonuclease.[9] The electrochemical biosensing platform was initially developed based on the Cas12a endonuclease from Acidaminococcus sp (AsCas12a).[10] We first investigated the on-chip collateral cleavage performance based on the AsCas12a-crRNA duplex targeting the HPV-16 sequence. After assembling the HPV-16 and the AsCas12a-crRNA, the triplex complex was directly incubated onto the ssDNA reporter covered electrode. Square wave voltammetry (SWV) was applied to evaluate the MB signal, which was decreased only in the presence of the cognate target with corresponding AsCas12a-crRNA (Figure 2A).
Figure 2. Optimization of on-chip trans-cleavage activity.
A) Representation of square wave voltammetry (SWV) evaluation of E-CRISPR in response to HPV-16. Red curve represents the background signal of 50 nM of Cas12a-crRNA duplex. Black curve represents the signal generated by the 50 nM of Cas12a-crRNA-target induced trans-cleavage activity. B) Evaluation of 50 nM of Cas12a orthologs from Lachnospiraceae bacterium and Acidaminococcus sp on its activity for on-chip trans-cleavage activity based on the change of current between background signal and target-mediated signal. . (Red line- LbCas12a; Black line- AsCas12a) C) Evaluation of trans-cleavage activity using 50 nM of LbCas12a-crRNA-target triplex. D) Evaluation of the effect of the concentration of divalent metal ions on the trans-cleavage activity of RuvC domain based on 50 nM of LbCas12a-crRNA-target triplex. E) & F) & G) SWV graphs of different lengths of surface ssDNA reporters based on 30 nM of LbCas12a-crRNA-target triplex. H) Comparison of signal change from different lengths of surface ssDNA reporters. SWV graphs in these figures present the result of a single test. Error bars in figures present the standard error (SE) based on at least three individual trails using at least three different sensors.
Evaluation of the Optimized Condition for On-Chip Trans-Cleavage activity
For biosensing application, the detection sensitivity is critical due to the low abundance of clinically relevant biomarkers in human fluids.[11] For the E-CRISPR detection platform, the trans-cleavage activity is the key for signal transduction, and therefore is critical to the sensitivity performance. We first compared the on-chip trans-cleavage activity of another type of Cas12a protein, Lachnospiraceae bacterium ND2006 Cas12a (LbCas12a),[12] with that of the AsCas12a. LbCas12a demonstrated a more apparent and stable trans-cleavage response within 5 min comparing with that of AsCas12a (Figure 2B). F, LbCas12a presented a more robust trans-cleavage activity within the testing period based on the same experimental condition, therefore LbCas12a was selected for further E-CRISPR development. We further evaluated the possible factors that may affect the trans-cleavage activity for on-chip electrochemical test using HPV-16 as the target. The optimized trans-cleavage period was investigated. The ΔI% continuously increased with the increasing incubation time for the collateral cleavage event (Figure 2c). It is interesting to notice that the trans-cleavage activity is not a simultaneous event of the cis-cleavage activity after the activation of cis-cleavage by the target. Because, the cis-cleavage of target strand is typically finished within 30 min; [3a] however, the trans-cleavage function remained active even after 3 hours (Figure 2C), indicating the target recognition and cis-cleavage activity of Cas12a system is the activator for the trans-cleavage domain of the Cas12a endonuclease.
We further investigated the chemical environment of the Cas12a to optimize the trans-cleavage performance. An important factor that may affect the Cas12a cleavage activity is the divalent cation Mg2+ concentration in the testing solution.[13] Cas12a RuvC domain is known to cleave ssDNA through the two-metal ion mechanism,[14] which involves the Mg2+ ions to induce conformational coordination of the RuvC domain and the ssDNA by shifting the spatial distribution of ssDNA around the RuvC active cutting center. Therefore, we evaluated the effect of concentration of Mg2+ ions in the in vitro cleavage solution on the performance of trans-cleavage activity. The trans-cleavage activity was only activated with the presence of the Mg2+ ions in the testing solution (Figure 2D). Increasing concentration of Mg2+ cations up to 15 mM demonstrated an enhanced trans-cleavage activity. Hence, an optimized Mg2+ concentration of 15 mM was selected for the preparation of Cas12a-crRNA duplex.
In order to perform an efficient surface chemistry based trans-cleavage, the accessibility of Cas12a endonuclease to the nonspecific ssDNA is important. Thus, we evaluated the effect of ssDNA reporter density on the electrode surface on the variation of electrochemical signal before and after trans-cleavage activity. An ideal surface condition can provide an optimized electrostatic environment for charged phosphate backbones and the hydroxyl groups of the passivation agents to ensure an upright ssDNA surface, facilitating the cleavage activity. The surface density of ssDNA reporter was manipulated by the concentration of the ssDNA reporter incubation solution. As shown in Figure S2A, a high surface density of ssDNA reporter significantly decreased the change of signal, because this high surface density decreased the accessibility of Cas endonuclease to the ssDNA reporter, producing a steric hindrance effect, which limited the trans-cleavage activity. An ideal density was prepared by 1 μM of ssDNA reporter and identified as 5.2 × 10−14mol/mm2 (Figure S2B), which created sufficient space for Cas12a to perform collateral cleavage on the electrode surface, providing a sufficient electrochemical signal change and ensuring an excellent detection resolution.
Other than the surface density, the length of the immobilized ssDNA reporter was also evaluated. We hypothesized that ssDNA reporters with different lengths might lead to different cleavage efficiency due to the exposed length difference. Different lengths of ssDNA probes at the same concentration were evaluated based on the same reaction condition of E-CRISPR as investigated previously. Moreover, the effect of passivation agents with different carbon chain lengths may influence the electrostatic interaction between the phosphate backbones of ssDNA probes, therefore was also evaluated for optimized cleavage activity (Figure S3). The selected ssDNA and passivation agent pairs were then compared through the effect of lengths on trans-cleavage activity. However, different lengths of ssDNA reporter only produce a minute variation (< 5%) of signal change (Figure 2H). We observed that for a short ssDNA reporter (10 nt), the electrochemical oxidation current over the background current was larger than that of long ssDNA strand because of its short contact mediated electron tunneling distance to the electrode resulting a faster charge transfer kinetics (Figure 2E). Therefore, the short probe possessed a large baseline current. As for long reporters (20 nt & 30 nt), they gave a relative low background current (Figure 2F&G), but the ΔI% of these long reporters were comparable to that of short reporter. 20 nt ssDNA reporter was selected for further application because of its relative greater degree of signal change and smaller standard error (Figure 2H). With the completion of the surface packing optimization, we tested the storage stability of the optimized packing of ssDNA electrode by storing in 4°C at a humidified environment (Figure S4). A stable SWV signal was retained for around 3 days, which is a sufficient turnaround time for clinical point-of-care routine. If a longer storage stability is needed, a multi-component monolayer system can be applied as demonstrated previously to maintain a high-sensitivity over months.[15] Moreover, evaluation of a multi-component monolayer system (e.g. ternary self-assembled monolayers) might also be a potential solution for future researches seeking for a higher sensitivity through tuning the surface molecular packing condition.[16]
Another interesting finding regarding to the cleavage accessibility is that Cas12a-crRNA based trans-cleavage activity is also significantly concentration dependent as was its analog Cas9.[17] Different concentrations of Cas12a-crRNA in response to a same target concentration were evaluated (Figure S5). A high concentration level (>100 nM) in a 30 uL sample solution significantly decreased the activity of the Cas12a nuclease to nonspecific ssDNA reporter, due to that the large size of the Cas12a probably cause a diffusion hindrance effect in the solution. Hence, a relative minor change of current outputs was observed based on a high concentration level of Cas12a-crRNA. An optimized concentration for Cas12a-crRNA duplex trans-cleavage operation was identified to be 30 nM in a 30 uL solution.
E-CRISPR on Nucleic Acid Detection
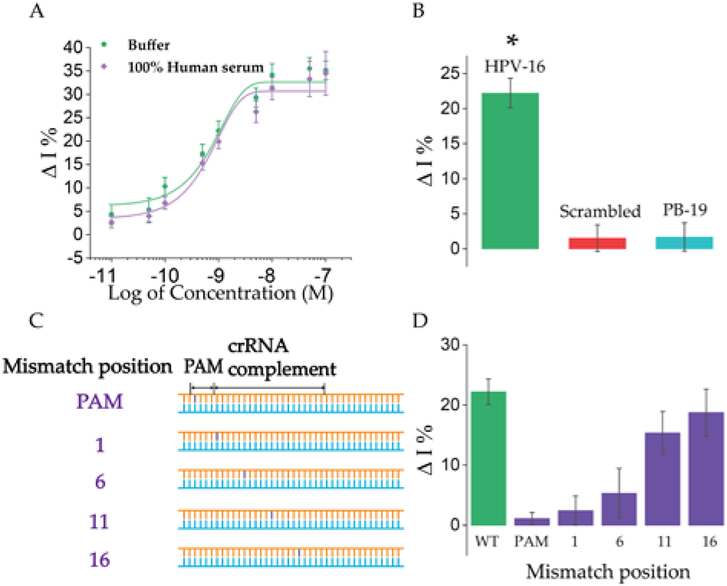
Based on the optimized trans-cleavage condition, we evaluated the E-CRISPR platform on the detection of HPV-16. A broad dynamic range (pM to μM) of more than three orders of magnitude was achieved with a IC50 value of 0.78 nM based on the samples prepared in the buffer solution (Figure 3A). The dose-dependent response curve demonstrated an average standard error (SE) of 2.16% (n=3), indicating a reliable reproducibility. An experimental limit of detection (LOD) at 50 pM was obtained. Worth mentioning, this LOD of optimized trans-cleavage activity based E-CRISPR surpassed previously demonstrated LOD for non-enzymatic amplified nucleic acid detection over two orders of magnitude.[3b] Moreover, the detection performance in complex matrix was also evaluated. A IC50 value in pooled human serum was 0.68 nM, which was comparable with the IC50 value (0.78 nM) in buffer solution, indicating a great potential of E-CRISPR in direct analyzing of biological sample. We also tested the effect of target length on the trans-cleavage signal by increasing the length of HPV-16 targeting sequence from 40-mer to 100-mer in the L1-enconding region.[9] Comparing with the detection performance of 40-mer DNA target, 100-mer DNA target presented a similar IC50 value (0.62 nM) (Figure S6), indicating that the length of the target would not interfere with the in vitro trans-cleavage activity of Cas12a.
Figure 3. E-CRISPR analysis of HPV-16.
A) Dose-response curve of the detection of HPV-16 in different matrixes (green line-10 mM Tris buffer containing 50 mM NaCl and 15 mM MgCl2; purple line-100% human serum). B) Selectivity study through comparison of the signal changes based on non-target nucleic acids (500 nM) with that of 1 nM of HPV-16 (n=3, *P<0.01, target signal vs. non-target signal). C) Target strands with mismatches at different positions, including PAM region and crRNA complement at different positions: 1, 6, 11, 16. D) Evaluation of the influence of mismatches at different positions on the E-CRISPR signal. A target concentration of 1 nM was applied for all the targets (wild type (WT) and mismatched targets).
To evaluate the generality of the detection strategy, we further challenged the E-CRISPR system to detect ssDNA erthrovirus, Parvovirus B19 (PB-19), which is known to cause erythema infectiosum in children and pregnant women.[18] A dynamic detection range from pM to μM was achieved with a IC50 value of 0.60 nM (Figure S7A). The percentage of signal change was similar to that of detection performance by HPV-16, indicating the on-chip trans-cleavage activity would not be affected by different targets.
We further investigated the accuracy of the E-CRISPR platform. A scrambled sequence and PB-19 were applied to evaluate the selectivity for HPV-16 detection. 500 nM of scrambled sequence and PB-19 sequence demonstrated a signal change less than 1.5% and 1.7%, which were lower than the standard error of the signal generated by 1 nM HPV-16 target, indicating a good selectivity of the Cas12a-crRNA duplex on differentiating HPV-16 from non-target (Figure 3B). Selectivity test was also performed for PB-19 detection using HPV-16 and scrambled sequence as interferences (Figure S7B), demonstrating the reliable recognition activity of CRISPR system.
Furthermore, as a biosensing platform, discrimination of mismatches in the nucleic acid base pairs is of especially importance for the potential application for the identification of disease related point mutations.[19] Thus, we next challenged the E-CRISPR with artificial mismatched nucleic acid targets (HPV-16). The recognition mechanism of CRISPR-Cas12a involves the identification of PAM region on the target to unwind the DNA target by Cas protein and further hybridization between the crRNA and the target strand.[3a, 14b] Therefore, we designed the mismatches at different positions on the target (Figure 3C). E-CRISPR signal was obtained based on the detection of 1 nM of these artificial targets (Figure 3D). Comparing with the wild type (WT) HPV-16 sequence, mutations in PAM region and PAM-adjacent region (position 1) led to complete diminishment of the trans-cleavage signal. This phenomenon indicates the mandatory requirement of PAM sequence for the Cas12a-crRNA duplex to recognize and cleave the target.[20] Moreover, mismatches in the complementary region of crRNA and target demonstrated retarded trans-cleavage activity, consistent with previous mismatch tolerance study of Cas12a.[3a, 21] The clear differentiable SWV signal between mismatches at different positions also suggests that the trans-cleavage activity of Cas12a might be utilized to identify the position of the mismatched base pairs as a biosensing strategy. Overall, the developed E-CRISPR demonstrates a sensitive, generalized and cost-effective platform for nucleic acid analysis.
Aptamer based E-CRISPR Cascade for Protein Detection
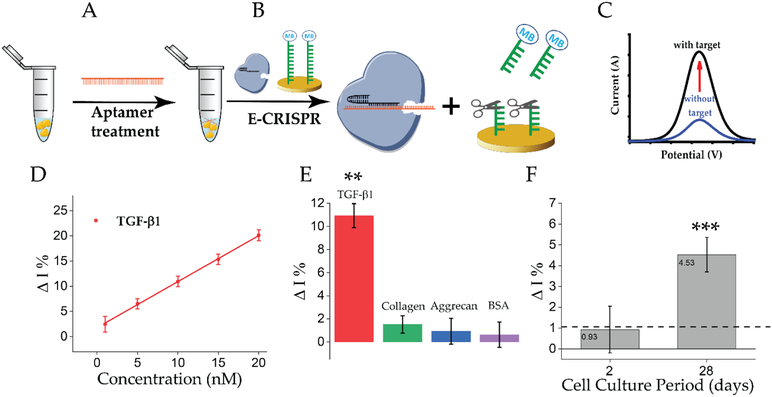
We next explored whether the E-CRISPR could be repurposed as a protein detection platform by utilizing the nucleic acid detection capability of E-CRISPR. For protein detection, ssDNA aptamer was used as the recognition element for a protein of interest. An aptamer based E-CRISPR cascade is designed for protein detection (Figure 4), which allows the direct analysis of complex sample without any time-consuming processing procedures. A fixed concentration of aptamer is firstly applied to treat the sample directly (Figure 4A). Cas12a-crRNA is designed to specifically recognize the aptamer. The E-CRISPR is then applied to determine the remaining concentration of aptamer in the sample (Figure 4B). With the presence of the protein target, less aptamer would be captured and transduced by E-CRISPR, leading to a high electrochemical signal of the methylene blue from the ssDNA reporter. In the absence of the protein target, the electrochemical signal would be lower due to the activation of trans-cleavage activity by the target recognition (Figure 4C).
Figure 4. E-CRISPR cascade for protein detection.
A) Sample containing protein target of interest is firstly treated by a fixed concentration of target specific aptamer (ssDNA). B) A E-CRISPR system is specifically designed for the recognition of the aptamer. The remaining concentration of aptamer is analyzed by E-CRISPR. C) A representation of SWV results based on the with target and without target condition. D) Linear calibration curve of TGF-β1 protein detection with an equation of Y=0.91X+1.79 and R-square value of 0.99 (n=3, SE=1.54%). E) Selectivity study through comparison of the signal outputs based on non-target proteins (10 nM) with that of 10 nM of TGF-β1 (n=3, **P<0.01 versus different interference substances). F) Concentration-dependent signals observed within conditioned medium harvested at two time-points during the chondrogenic differentiation program of human mesenchymal stem cells (hMSCs) containing TGF-β1. The samples were analyzed by three sets of individual experiments using three different sensors (n=3, ***P<0.05, Day 28 vs. Day 2). The horizontal black dashed line represents the average signal variation (n=3) based on the presence of blank conditioned medium.
This designed E-CRISPR array was evaluated for the detection of transforming growth factor beta 1 (TGF-β1) protein, which is a secreted protein contributing to cell proliferation and differentiation,[22] and is also recognized as a biomarker for hepatocellular carcinoma.[23] The dose dependent E-CRISPR for the detection of TGF-β1 aptamer was first evaluated based on the previous established trans-cleavage condition (Figure S8). For proof-of-concept, a fixed concentration of aptamer was first applied to treat sample with and without TGF-β1 protein. E-CRISPR was then applied to analyze the samples with and without TGF-β1 protein demonstrating a clear signal difference (Figure S9). In order to increase the detection resolution for nano molar concentration range, a greater degree of current difference between 1 nM and 50 nM is necessary. Therefore, longer trans-cleavage period was investigated to evaluate whether a higher current difference can be obtained due to that the trans-cleavage activity is multiple-turnover reaction.[3a] Increasing trans-cleavage period indeed leads to a higher detection resolution (Figure S8B), so a trans-cleavage period for protein detection was selected to be 60 min. Therefore, this strategy might be applied to tune the dynamic range and detection limit of the E-CRISPR platform, enhancing the detection performance. An aptamer concentration of 50 nM was selected for protein sample treatment for 30 min. After the treatment, the sample was evaluated by E-CRISPR. A linear detection range was achieved covering three order of magnitudes with an experimental detection limit tested as 0.2 nM (Figure 4D). The detection specificity was investigated using the conditioned medium from hMSCs chonodrogenesis (a complex matrix) biomolecules, including collagen type II, aggrecan protein and bovine serum albumin. The designed strategy defined a good selectivity on target protein over non-specific molecules, indicating an excellent specificity of the applied aptamer in the system (Figure 4E). We further challenged the E-CRISPR platform with samples obtained during the chondrogenic differentiation program of hMSCs, which were cultured in aggregates with complete chondrogenic differentiation medium for 4 weeks.[24] TGF-β1 protein was produced during the chondrogenic differentiation process.[25] A clear difference was identified between the conditioned medium obtained at day 2 and day 28 (Figure 4F). These results are in agreement with the transcriptome analyses performed during Hmsc chondrogenesis of the same analyzed sample (Figure S10), indicating a reliable performance of the designed E-CRISPR array for protein detection. The nucleic acid based receptor is a generalized recognition element for both protein and small molecule.[26] Hence, the designed E-CRISPR array can also be extended to a wide variety of analytes.
Conclusion
This study introduces a new strategy for the development of electrochemical biosensors by using electrochemistry to probe the CRISPR cleavage activity (E-CRISPR). Owing to the high-specificity of target recognition, other than gene editing tool, we utilized the CRISPR Type V system, Cas12a (cpf1) as an efficient biosensing system, which translates the target recognition activity into a detectable electrochemical signal through an interrogating electrode constructed with non-specific ssDNA. Various factors were investigated to produce an optimized on-chip trans-cleavage activity for a high-sensitivity E-CRISPR detection platform. Moreover, our preliminary implementation illustrates that the E-CRIPSR system can be applied not only for nucleic acid sensing; with the addition of an aptamer based sensing cascade, the E-CRISPR can also be utilized for protein detection, providing a generalizable, robust and cost-effective detection system.
Experimental Section
Fabrication of ssDNA Reporter surface
An array containing twenty sensors was first cleaned through an established procedure using potassium hydroxide, sulfuric acid and nitric acid.[4a] Thiol linked ssDNA reporter was treated with 10 μM of tris(2-carboxyethyl)phosphine (TCEP) to reduce the S-S bond for 10 min in the dark at room temperature. The ssDNA reporter was then diluted to 1 μM using 10 mM Tris buffer containing 10 mM EDTA. 20 μL of the 1 μM ssDNA reporter was directly incubated onto the gold sensor for 1 hr in the dark at room temperature. The ssDNA immobilized sensor array was then cleaned by immersing in 10 mM Tris buffer for 5 min. After cleaning, the sensor array was immersed in 2 mM 6-mercaptoheaxnol (MCH) prepared in 10 mM Tris buffer for 30 min to passivate the surface and replace loosely tethered ssDNA reporter, forming a highly-aligned surface (Operation of MCH related steps should be conducted in a fume hood due to its toxicity). After the MCH treatment, the sensor array was then cleaned by immersing in 10 mM Tris buffer for 5 min. The cleaned sensor array was then dried by nitrogen gas and ready for treatment by CRISPR system. For a short storage period, the cleaned sensor array can be stored in 10 mM Tris buffer (containing 100 mM NaCl) at 4 °C.
In vitro Digestion of Cas12a-crRNA
Cas12a-crRNA duplex was prepared in a buffer prepared by nuclease free water containing 50 mM NaCl, 10 mM Tris-HCl, 15 mM MgCl2, 100 μg/ml BSA with a pH of 7.9. 30 nM of Cas12a-crRNA was assembled and incubated at 25°C for 10 min. Typically, for nucleic acid detection, 4 μL of sample was added into 26 μL of the Cas12a-crRNA duplex to form the Cas12a-crRNA-target triplex and incubated for 10 min at room temperature. 20 μL of the Cas12a-crRNA-target triplex solution was applied to ssDNA reporter covered sensor for trans-cleavage activity at 37°C for 30 min. 80 U/mL of Proteinase k was applied to the CRISPR treated surface at 37°C for 15 min before the electrochemical analysis. For protein detection, 10 μL of 100 nM of aptamer was applied to treat 10 μL of sample (resulting in a 50 nM final concentration of aptamer) and incubated at room temperature for 30 min. E-CRISPR as described above was then applied for protein sample analysis with an elongated trans-cleavage period for 60 min.
On-Chip Electrochemical Analysis
After the on-chip CRISPR reaction, the sensors were cleaned by immersing the sensors into a 10 mM Tris buffer for 5 min. For electrochemical test, a 10 mM Tris buffer containing 100 mM NaCl was applied as the electrolyte. Square wave voltammetry (SWV) was applied before and after the treatment of Cas12a-crRNA-target triplex to obtain the change of current based on a potential range of −0.6V to −0.1V, a frequency of 25 Hz, an amplitude of 25 mV (variation of frequency (15 Hz-120 Hz) and amplitude (25 mV- 50 mV) did not present significant enhancement of the quantity of signal changed or the signal stability).
Clinical Sample-Mesenchymal stem cell (MSCs) culture and differentiation:
Cultures of human bone marrow-derived MSCs from healthy de-identified adult volunteer donors were established as previously described.[27] The bone marrow was collected using a procedure reviewed and approved by the University Hospitals of Cleveland Institutional Review Board; informed consent was obtained from all de-identified donors. Cells were expanded in DMEM-LG supplemented with 10% fetal bovine serum, supplemented with FGF2 (10 ng/ml of) for 14 days. Cells were trypsinized and then resuspended in chondrogenic differentiation medium consisting of DMEM-high glucose supplemented with 1% ITS+,10−7 M dexamethasone, 1mM sodium pyruvate, 120 mM ascorbic acid-2 phosphate, 100 mM nonessential amino acids, and 10 ng/mL TGF-β1 protein. Two hundred microliters of this cell suspension containing 250,000 cells was added per well of a 96-well polypropylene V-bottom, multi-well dish (Phenix Research). The multi-well plates were centrifuged at 500 g for 5 min and then incubated at 37 °C. The differentiation medium was changed every other day. Conditioned medium from these pellets was collected at different time points. Days 2 and 28 were chosen to use in the biosensor platform based on previous transcriptome data (RNAseq) showing a greater difference in TGF-β1 protein expression between days 2 and 28 (Figure S10). To activate the latent secreted TGF-β1 protein to the detectable form, 20 μL of 1 M HCl were added to 100 μL of conditioned medium and incubated for 10 minutes and then neutralized with 20 μL of 1.2 M NaOH/0.5 M HEPES. The samples were assayed immediately. This procedure ensures that only the secreted version of TGF-β1 protein assayed.
Supplementary Material
Acknowledgements
The authors acknowledge the staff of the Electronics Design Center for the experimental support and Xintong Cao for the art work. We acknowledge the funding supports from National Institute of Health under the grant number, NIBIB: 1P41EB021911 and R01DK113185. We also acknowledge the funding supports from Wallace R. Persons Research Fund from Case Alumni Association.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Publisher's Disclaimer: This manuscript has been accepted after peer review and appears as an Accepted Article online prior to editing, proofing, and formal publication of the final Version of Record (VoR). This work is currently citable by using the Digital Object Identifier (DOI) given below. The VoR will be published online in Early View as soon as possible and may be different to this Accepted Article as a result of editing. Readers should obtain the VoR from the journal website shown below when it is published to ensure accuracy of information. The authors are responsible for the content of this Accepted Article.
References
- [1] a.Kelley SO, Mirkin CA, Walt DR, Ismagilov RF, Toner M, Sargent EH, Nature nanotechnology 2014, 9, 969; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Yang Y, Gao W, Chemical Society Reviews 2019, 48, 1465–1491; [DOI] [PubMed] [Google Scholar]; c Kim J, Campbell AS, de Ávila BE-F, Wang J, Nature Biotechnology 2019, 37, 389–406; [DOI] [PMC free article] [PubMed] [Google Scholar]; d Dai Y, Liu CC, Angewandte Chemie International Edition 2019, 58, 12355–12368; [DOI] [PubMed] [Google Scholar]; e Wu Y, Tilley RD, Gooding JJ, Journal of the American Chemical Society 2019, 141, 1162–1170; [DOI] [PubMed] [Google Scholar]; f Furst AL, Francis MB, Chemical Reviews 2019, 119, 700–726; [DOI] [PubMed] [Google Scholar]; g Zwang TJ, Tse ECM, Barton JK, ACS Chemical Biology 2018, 13, 1799–1809; [DOI] [PMC free article] [PubMed] [Google Scholar]; h Lubin AA, Plaxco KW, Accounts of Chemical Research 2010, 43, 496–505; [DOI] [PMC free article] [PubMed] [Google Scholar]; i Yang F, Li Q, Wang L, Zhang G-J, Fan C, ACS Sensors 2018, 3, 903–919. [DOI] [PubMed] [Google Scholar]
- [2] a.Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, Science 2017, 356, 438–442; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Li Y, Li S, Wang J, Liu G, Trends Biotechnol 2019, 37, 730–743; [DOI] [PubMed] [Google Scholar]; c Leung K, Krishnan Y, ACS Central Science 2019, 5, 1111–1113; [DOI] [PMC free article] [PubMed] [Google Scholar]; d Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Science 2013, 339, 819–823; [DOI] [PMC free article] [PubMed] [Google Scholar]; e Huang TP, Zhao KT, Miller SM, Gaudelli NM, Oakes BL, Fellmann C, Savage DF, Liu DR, Nature Biotechnology 2019, 37, 626–631; [DOI] [PMC free article] [PubMed] [Google Scholar]; f Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liu DR, Nature 2018, 556, 57; [DOI] [PMC free article] [PubMed] [Google Scholar]; g Yin H, Song C-Q, Suresh S, Kwan S-Y, Wu Q, Walsh S, Ding J, Bogorad RL, Zhu LJ, Wolfe SA, Koteliansky V, Xue W, Langer R, Anderson DG, Nature Chemical Biology 2018, 14, 311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3] a.Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA, Science 2018, 360, 436–439; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F, Science 2018, 360, 439–444; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Li S-Y, Cheng Q-X, Wang J-M, Li X-Y, Zhang Z-L, Gao S, Cao R-B, Zhao G-P, Wang J, Cell Discovery 2018, 4, 20; [DOI] [PMC free article] [PubMed] [Google Scholar]; d Li S-Y, Cheng Q-X, Liu J-K, Nie X-Q, Zhao G-P, Wang J, Cell research 2018, 28, 491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4] a.Dai Y, Wang C, Chiu L-Y, Abbasi K, Tolbert BS, Sauvé G, Yen Y, Liu C-C, Biosensors and Bioelectronics 2018, 117, 60–67; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Dai Y, Abbasi K, Bandyopadhyay S, Liu CC, ACS Sensors 2019, 4, 1980–1985. [DOI] [PubMed] [Google Scholar]
- [5] a.Chen JS, Doudna JA, Nature Reviews Chemistry 2017, 1, 0078; [Google Scholar]; b Murugan K, Babu K, Sundaresan R, Rajan R, Sashital DG, Molecular cell 2017, 68, 15–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sundaresan R, Parameshwaran HP, Yogesha S, Keilbarth MW, Rajan R, Cell reports 2017, 21, 3728–3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].O’Brien E, Holt ME, Thompson MK, Salay LE, Ehlinger AC, Chazin WJ, Barton JK, Science 2017, 355, eaag1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Mirabello L, Yeager M, Yu K, Clifford GM, Xiao Y, Zhu B, Cullen M, Boland JF, Wentzensen N, Nelson CW, Raine-Bennett T, Chen Z, Bass S, Song L, Yang Q, Steinberg M, Burdett L, Dean M, Roberson D, Mitchell J, Lorey T, Franceschi S, Castle PE, Walker J, Zuna R, Kreimer AR, Beachler DC, Hildesheim A, Gonzalez P, Porras C, Burk RD, Schiffman M, Cell 2017, 170, 1164–1174.e1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Seedorf K, Krämmer G, Dürst M, Suhai S, Röwekamp WG, Virology 1985, 145, 181–185. [DOI] [PubMed] [Google Scholar]
- [10].Maule G, Casini A, Montagna C, Ramalho AS, De Boeck K, Debyser Z, Carlon MS, Petris G, Cereseto A, Nature communications 2019, 10, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11] a.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S, Cell research 2015, 25, 981; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Tabernero J, Lenz H-J, Siena S, Sobrero A, Falcone A, Ychou M, Humblet Y, Bouché O, Mineur L, Barone C, The Lancet Oncology 2015, 16, 937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Breinig M, Schweitzer AY, Herianto AM, Revia S, Schaefer L, Wendler L, Galvez AC, Tschaharganeh DF, Nature methods 2019, 16, 51. [DOI] [PubMed] [Google Scholar]
- [13].Zetsche B, Gootenberg Jonathan S., Abudayyeh Omar O., Slaymaker Ian M., Makarova Kira S., Essletzbichler P, Volz Sara E., Joung J, van der Oost J, Regev A, Koonin Eugene V., Zhang F, Cell 2015, 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14] a.Yang W, Nature Structural &Amp; Molecular Biology 2008, 15, 1228. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Swarts DC, van der Oost J, Jinek M, Molecular Cell 2017, 66, 221–233.e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kuralay F, Campuzano S, Wang J, Talanta 2012, 99, 155–160. [DOI] [PubMed] [Google Scholar]
- [16] a.Wu J, Campuzano S, Halford C, Haake DA, Wang J, Analytical Chemistry 2010, 82, 8830–8837; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Campuzano S, Kuralay F, Lobo-Castañón MJ, Bartošík M, Vyavahare K, Paleček E, Haake DA, Wang J, Biosensors and Bioelectronics 2011, 26, 3577–3583; [DOI] [PMC free article] [PubMed] [Google Scholar]; c Campuzano S, Kuralay F, Wang J, Electroanalysis 2012, 24, 483–493. [Google Scholar]
- [17].Maji B, Moore CL, Zetsche B, Volz SE, Zhang F, Shoulders MD, Choudhary A, Nature chemical biology 2017, 13, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Young NS, Brown KE, New England Journal of Medicine 2004, 350, 586–597. [DOI] [PubMed] [Google Scholar]
- [19] a.Wang J, Analytica chimica acta 2002, 469, 63–71; [Google Scholar]; b Schwarzenbach H, Hoon DS, Pantel K, Nature Reviews Cancer 2011, 11, 426. [DOI] [PubMed] [Google Scholar]
- [20] a.Kim D, Kim J, Hur JK, Been KW, Yoon S.-h., Kim J-S, Nature Biotechnology 2016, 34, 863; [DOI] [PubMed] [Google Scholar]; b Gao L, Cox DBT, Yan WX, Manteiga JC, Schneider MW, Yamano T, Nishimasu H, Nureki O, Crosetto N, Zhang F, Nature Biotechnology 2017, 35, 789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kim HK, Song M, Lee J, Menon AV, Jung S, Kang Y-M, Choi JW, Woo E, Koh HC, Nam J-W, Kim H, Nature Methods 2016, 14, 153. [DOI] [PubMed] [Google Scholar]
- [22].Xu X, Zheng L, Yuan Q, Zhen G, Crane JL, Zhou X, Cao X, Bone Research 2018, 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23] a.Iwasaki M, Nakata K, Nakahara H, Nakase T, Kimura T, Kimata K, Caplan A, Ono K, Endocrinology 1993, 132, 1603–1608; [DOI] [PubMed] [Google Scholar]; b Coulouarn C, Factor VM, Thorgeirsson SS, Hepatology 2008, 47, 2059–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Somoza RA, Correa D, Labat I, Sternberg H, Forrest ME, Khalil AM, West MD, Tesar P, Caplan AI, Tissue Engineering Part A 2018, 24, 335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25] a.Li Z, Kupcsik L, Yao SJ, Alini M, Stoddart MJ, Journal of cellular and molecular medicine 2010, 14, 1338–1346; [DOI] [PMC free article] [PubMed] [Google Scholar]; b Shinojima N, Hossain A, Takezaki T, Fueyo J, Gumin J, Gao F, Nwajei F, Marini FC, Andreeff M, Kuratsu J-I, Cancer research 2013, 73, 2333–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nakatsuka N, Yang K-A, Abendroth JM, Cheung KM, Xu X, Yang H, Zhao C, Zhu B, Rim YS, Yang Y, Weiss PS, Stojanović MN, Andrews AM, Science 2018, 362, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lennon DP, Caplan AI, Experimental Hematology 2006, 34, 1604–1605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.