During translation, the presence of an mRNA structure prolongs a ribosomal state favorable to spontaneous frameshifting.
Abstract
Maintenance of translational reading frame ensures the fidelity of information transfer during protein synthesis. Yet, programmed ribosomal frameshifting sequences within the coding region promote a high rate of reading frame change at predetermined sites thus enriching genomic information density. Frameshifting is typically stimulated by the presence of 3′ messenger RNA (mRNA) structures, but how these mRNA structures enhance −1 frameshifting remains debatable. Here, we apply single-molecule and ensemble approaches to formulate a mechanistic model of ribosomal −1 frameshifting. Our model suggests that the ribosome is intrinsically susceptible to frameshift before its translocation and this transient state is prolonged by the presence of a precisely positioned downstream mRNA structure. We challenged this model using temperature variation in vivo, which followed the prediction made based on in vitro results. Our results provide a quantitative framework for analyzing other frameshifting enhancers and a potential approach to control gene expression dynamically using programmed frameshifting.
INTRODUCTION
The ribosome faithfully maps amino acids to corresponding three-nucleotide codons to synthesize proteins. Translational reading frame maintenance is an essential aspect of this information transfer process, as a transition to alternative reading frames during translation typically results in premature termination with negative biological impacts. The ribosome normally maintains the reading frame while translating hundreds of codons, with a spontaneous frameshift error estimated to be one in 105 codons (1, 2). Yet, most known organisms have genes whose expression requires programmed ribosomal frameshifting (3, 4), which yields a tuned ratio of multiple protein products from a single mRNA sequence via −2, −1, +1, and/or +2 frameshifting events. Frameshifting is enriched in viral genes and transposon elements (3), increasing the repertoire of protein products from their limited mRNA species. In particular, programmed −1 frameshifting signals are often conserved in retroviral genomes, including human immunodeficiency virus (5, 6), where changes in frameshifting efficiency are deleterious to viral replication. The importance of viral programmed −1 frameshifting is such that a component of interferon-induced proteins, shiftless, has evolved to inhibit this frameshifting as part of the host antiviral strategy (7).
Signals in mRNAs that efficiently induce programmed −1 frameshifting usually require a slippery sequence and additional stimulatory elements, frequently a folded mRNA structure 3′ of the shift site, and in bacteria, an internal Shine-Dalgarno (SD) sequence (Fig. 1A) (3, 4). The precise location and direction of −1 frameshifting are determined by the slippery sequence that permits the realignment of tRNA anticodons from the original frame to the shifted frame. For −1 frameshifting, one of the most efficient −1 frameshifting slippery sequences in Escherichia coli is A-AAA-AAG (a dash indicates the current, or zero, reading frame) (8, 9), which commonly permits realignment of two tRNA anticodons (mnm5s2UUU of E. coli tRNALys) within the ribosome to the −1 frame (AAA-AAA-G). While a slippery sequence is an integral part of frameshifting cassettes, ribosomal frameshifting on the known slippery sequences alone is inefficient, yielding a −1 frameshifting efficiency on the order of 2% (8, 9). More efficient frameshifting systems include a downstream mRNA structure, either an RNA hairpin (10) or an RNA pseudoknot (11, 12), which greatly enhances the frameshifting efficiency. The spacing between the mRNA structure and the slippery sequence is an important factor in determining the efficiency of the frameshifting. The most common spacing between the 5′ start of a stimulatory RNA structure and the 3′ end of the slippery sequence is commonly five to six nucleotides for bacterial ribosomes (five to nine for mammalian ribosomes), where decreasing or increasing the spacer length can result in sharp reduction in −1 frameshifting efficiency (10, 13). This precise spacing requirement argues that the mRNA structure has to be presented at a specific step of translation to enhance frameshifting on the slippery sequence, not sooner or later.
Fig. 1. mRNA structure in −1 frameshifting induces translocation pause.
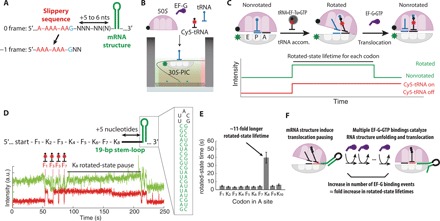
(A) An example of the programmed −1 frameshifting cassette containing two mRNA elements: slippery sequence and 3′ mRNA structure (nts: nucleotides). (B) Schematic of single-molecule in vitro fluorescence assay to monitor translation dynamics. Elongation components—BHQ-2–labeled 50S, elongation factor G (EF-G), and Cy5-labeled and unlabeled tRNA—are delivered to the translation preinitiation complex (30S-PIC) containing Cy3B-labeled 30S tethered to the ZMW nanostructure. (C) Structural changes of the ribosome during translation and corresponding fluorescence signals to measure the rotated-state lifetime (time between tRNA accommodation and translocation) for each codon. (D) The mRNA construct used (F: UUC codon for Phe; K: AAA codon for Lys) and the representative trace. Translocation is severely hindered on codon 8 (K8) when the ribosome encounters mRNA structure. a.u., arbitrary units; bp, base pair. (E) Measured translocation time for each mRNA codon. Translocation into the structured mRNA region occurs after a substantial pause (n = 114 molecules; error bars represent 95% confidence interval from fitting the single-exponential distributions). (F) Model of unfolding mRNA structure during translocation, catalyzed by repeated binding of EF-G•GTP.
Despite the studies cited above, a general quantitative and mechanistic model of −1 frameshifting is still lacking. The presence of a downstream mRNA structure has been previously shown to delay ribosomal translocation on the slippery sequence using ensemble kinetics and single-molecule methods, which was suggested to activate −1 frameshifting pathways that are kinetically unfavorable (13–17) and may involve noncanonical conformations of the ribosome induced by its encounter with an mRNA structure. Yet, it remains unresolved how a delay before translocation and which of its intermediate states lead to a −1 frameshifting event and how mRNA structure and slippery sequences enhance −1 frameshifting. To understand the mechanism of −1 frameshifting, we here applied in vitro single-molecule fluorescence assays that monitor the conformational and compositional changes of individual ribosomes during translation. Using this approach, we first probed the effects of mRNA structure on the translation kinetics and then observed frameshifting on the slippery sequence for single translating ribosomes. This has permitted a quantitative description of the kinetic pathways leading to −1 frameshifting. Our results lead to a mechanistic model for ribosomal frameshifting and its energy landscape, which were used to predict −1 frameshifting efficiencies and changes in response to temperature change. This model was validated by bulk frameshifting assays in vivo.
RESULTS AND DISCUSSION
Unfolding mRNA structure delays translocation of ribosomes catalyzed by elongation factor G
To identify the role of mRNA structure in frameshifting, we applied in vitro single-molecule assays to monitor compositional and conformational changes of actively translating ribosomes. The ribosome cycles through two main global conformations termed nonrotated and rotated states, per each codon during translation elongation. The conformational changes of the ribosome were monitored by observing Förster resonance energy transfer (FRET) between a Cy3B–BHQ-2 donor-quencher pair, site-specifically attached to E. coli small and large ribosomal subunits, respectively (Cy3B-30S and BHQ-50S) (18). While transitions between rotated and nonrotated states have been previously tracked using different fluorescent labeling sites on the ribosome (19), we have used fluorescent probes site-specifically attached to helices 44 and 101 of the 16S and 23S ribosomal RNAs (rRNAs), respectively (20). In previous studies (18, 21), we have used this single-molecule FRET (smFRET) signals to show that each state transition is coupled to a successful enzymatic activity: Transitions from the nonrotated to rotated state are coupled with tRNA decoding catalyzed by elongation factor Tu (EF-Tu) and subsequent peptide bond formation, and transitions from the rotated to nonrotated state are coupled with a translocation event catalyzed by elongation factor G (EF-G) (Fig. 1, B and C). We monitored the rotated-state lifetimes for each translated codon and measured changes in translocation kinetics at codon resolution. The ribosomal intersubunit FRET signal was fortified by the addition of a Cy5-labeled tRNA compositional signal (18); binding of Cy5-labeled tRNA that is correlated with the non–rotated- to rotated-state transitions marks the position of the ribosome on the mRNA transcript (Fig. 1, B and C) (18). To tolerate the high concentration of free Cy5-labeled factors in solution, we performed the experiments using a zero-mode waveguide (ZMW)–based single-molecule fluorescence instrument, as benchmarked previously (18).
To determine the effect of mRNA structure on translation kinetics, we initially quantified the rotated-state lifetime of the ribosome as it translated an mRNA with a stable RNA structure located five mRNA nucleotides after the eighth codon in the coding region (Fig. 1D). The RNA structure is derived from the pseudoknot used in the insertion sequence 3 (IS3) programmed −1 frameshifting system (see below), where the two pseudoknot helical stems were joined to form a 19–base pair–long RNA hairpin stem-loop [ΔGmfe = −150 kJ mol−1 at 37°C (22); SL2 in fig. S2]. Compared with the rotated-state lifetimes for the first seven codons, the rotated-state lifetime for the eighth codon was 11-fold longer (Fig. 1, D and E). On the basis of the available structures of E. coli ribosomes (23), the distance between the end of the A-site codon and the mRNA entrance (composed of S3, S4, and S5 proteins) that prevents access of any double-stranded mRNA is about five to six nucleotides, suggesting that unfolding the basal part of the mRNA structure before translocation induces pausing in the rotated state (Fig. 2, A and B). Increasing the spacer length from five to seven nucleotides decreased by twofold this increase in the rotated-state lifetimes, possibly suggesting that the magnitude of translocation pause duration may be related to the number of base pairs that must be unfolded before translocation (Fig. 2, C and D). The rotated-state lifetimes for the subsequent 9th and 10th codons were not substantially longer, suggesting either (i) the whole 19 base pairs in the mRNA structure have melted during translocation from the eighth codon or (ii) only the initial encounter of the mRNA structure induces translocation pause on the ribosome, and unfolding of the rest of RNA structure involves a different mechanism (24).
Fig. 2. Characterizing the temperature dependence of translation through the mRNA structure.
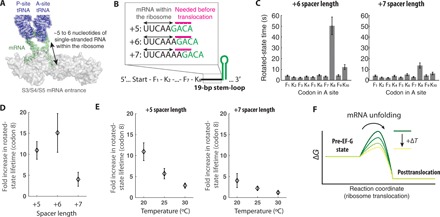
(A) The positioning of mRNA within the ribosome during translation, where any mRNA structures need to be unfolded and threaded through the mRNA entrance channel (composed of ribosomal proteins S3, S4, and S5) five to six nucleotides before decoding [adapted from Protein Data Bank 4V6F (23)]. (B) mRNA constructs with different spacer lengths between the K8 codon and the base of mRNA structure. On the basis of the structure, five to six nucleotides of the mRNA downstream of the A site are expected to be within the ribosome (black arrows), and next single-stranded three nucleotides (magenta) are expected to be available before full translocation step. The structured portion of the mRNA is colored green. (C) Rotated-state lifetimes for codons 1 to 10 for translating mRNA with +6 (left) and +7 spacers (right) (n = 114, 129, and 99 molecules from left to right; error bars represent 95% confidence interval from fitting the single-exponential distribution). (D) The fold increase in rotated-state lifetimes for the K8 codon compared with prior Lys codons (K2, K4, and K6) for mRNAs with three different spacer lengths. (E) The fold increase in rotated-state lifetimes for the K8 codon compared to prior Lys codons (K2, K4, and K6) for +5 spacer mRNA construct (left; n = 136, 100, and 114 molecules from left to right) and for +7 spacer mRNA construct (right; n = 117, 135, and 99 molecules from left to right; error bars represent 95% confidence interval after the error propagation) at different temperatures. (F) A cartoon energy landscape of the downstream mRNA structure unfolding before translocation.
In our assay, the rotated-state lifetimes are rate limited by the EF-G binding event at its low concentration (200 nM) (21), and the fold increase in the rotated-state lifetimes can be interpreted as fold increase in the number of EF-G binding events (Fig. 1F) (18, 25), where multiple samplings of EF-G on the ribosome have been observed previously when ribosomes encounter an mRNA structure in single-molecule assays (16, 17). To confirm that the delay in translocation from the rotated state is due to multiple bindings of EF-G, we directly monitored association and dissociation of Cy5-labeled EF-G to the actively translating ribosome paused in the rotated state (fig. S1). As expected, we observed multiple bindings of Cy5-labeled EF-G to the ribosome paused in the rotated state. This result matches with the previous reports by Kim et al. (14, 16), which used an smFRET signal between the incoming EF-G and either the ribosomal protein or the tRNA to show multiple bindings of EF-G.
To probe the energetic barrier to the unfolding of the downstream mRNA structure by the ribosome before translocation, we measured the fold increase in the rotated-state lifetime (at codon 8) at two additional temperatures (25° and 30°C) and compared them to the measurements made at 20°C. We observed the fold increase in rotated-state lifetime decreasing sharply as the temperature of translation increased (Fig. 2E), suggesting that the underlying rate-limiting process has high activation enthalpy (∆H‡) required to reach the transition state. The observed high activation enthalpy to the transition state is expected not only for unfolding the mRNA structure before the translocation step but also for possible ribosomal conformational change before unfolding the mRNA structure. Together, our results suggest that translocation is slowed upon the first encounter between the mRNA structure and the ribosome and that the magnitude of the delay depends on the reaction temperature.
Multiple GTP hydrolysis events by EF-G may catalyze unfolding of mRNA structure during translocation
Translocation is catalyzed by EF-G, which hydrolyzes guanosine 5′-triphosphate (GTP) on the ribosome and changes its conformation within the ribosome. To probe the relative EF-G position within the ribosome during multiple samplings of EF-G, we used a FRET signal between a Cy3-labeled A-site tRNA and incoming Cy5-labeled EF-G to identify the pretranslocation and posttranslocation states, previously developed by the Blanchard group (Fig. 3, A and B) (16, 26). Translocation may occur in the absence of GTP hydrolysis, but in this case, both tRNA movements and dissociation of EF-G from the posttranslocated ribosome are slowed from the order of 100 ms to seconds (26). Replicating the previously developed tRNA–EF-G FRET signal, we observed that both the FRET efficiency and EF-G binding lifetime on the ribosome change substantially when either GTP hydrolysis or EF-G domain movements are perturbed by substitution of GTP with its nonhydrolyzable analog GTPγS or by the addition of the antibiotic fusidic acid, respectively, compared with that of the normal translocation (Fig. 3, B to E). In stark contrast, we observed that both FRET efficiency and EF-G binding lifetime during the mRNA structure–impeded translocation are indistinguishable from those of unperturbed translocation (Fig. 3, E and F). Our findings thus suggest that the underlying mechanism of translocation perturbation by an mRNA secondary structure differs from those of GTPγS or fusidic acid. The observed fast EF-G dissociation kinetics (lifetime of 0.17 ± 0.01 s) during a prolonged rotated state suggests that each EF-G molecule hydrolyzes GTP (Fig. 3A) in futile translocation cycles and now competes with the delayed pretranslocation rate (proportional to the inverse of the fold increase in the rotated-state lifetimes), resulting in multiple EF-G binding and GTP hydrolysis events attempting to catalyze unfolding the downstream mRNA secondary structure and subsequent translocation.
Fig. 3. EF-G hydrolyzes multiple GTPs to unfold mRNA structure during translocation.
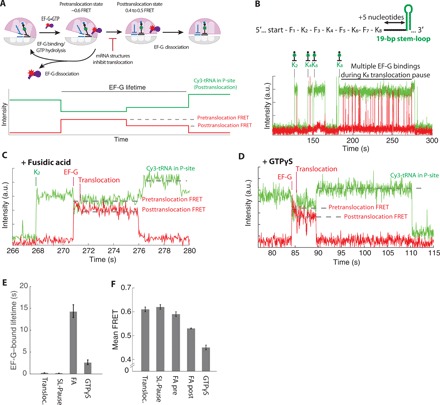
(A) Translocation is monitored via tRNA–EF-G FRET efficiency changes from pretranslocation (Cy3 within the A/P site) to posttranslocation (Cy3 within the P site). Movement of tRNA from the A/P site to the P site induces a slight increase in the Cy3 fluorescence signal, as described previously (26). (B) The representative trace for the normal and delayed (long rotated state induced by mRNA structure) translocation. FRET efficiency change due to translocation may be too fast to detect in the current setup due to rapid EF-G dissociation kinetics after translocation. (C) The representative trace for the fusidic acid (FA) condition. The presence of the FA delays EF-G dissociation after translocation. (D) The representative trace for the GTPγS condition (GTP in the reaction mix is substituted with GTPγS). The presence of the FA delays EF-G dissociation after translocation. (E) Average Cy5-labeled EF-G bound lifetimes at different experimental conditions: Transloc. for normal translocation; SL-Pause for multiple EF-G bindings during translocation through the mRNA structure; FA for translocation in the presence of FA; and GTPγS for translocation in the presence of GTPγS instead of GTP (n = 386, 1211, 364, and 91 molecules; error bars represent 95% confidence interval from fitting the single-exponential distributions). (F) Mean tRNA–EF-G FRET efficiencies measured for (D). First 200 ms (five frames) of each FRET events or the entire FRET event in the FA condition is used for FA pre and for FA post, respectively (error bars represent 95% confidence interval from fitting the normal distribution; fitting of the normal distribution shown is in the fig. S1K).
Increase in the rotated-state pause on a slippery sequence correlates with the frameshifting efficiency
In the efficient −1 programmed ribosomal frameshifting context, the downstream mRNA structure delays translocation on the slippery sequence in the rotated state. To determine the role of the rotated-state delay in frameshifting, we measured both the rotated-state lifetime and the frameshifting efficiency on mRNAs with different downstream mRNA structures (Fig. 4A) and slippery sequences. We designed −1 frameshifting mRNAs based on the bacterial IS3 family (9, 12) (fig. S2), which is a frameshifting system native to E. coli that couples an RNA pseudoknot with the slippery sequence (A-AAA-AAG). mRNA pseudoknots are common 3′-stimulators of eukaryotic −1 frameshifting systems (3). Frameshifting mRNA constructs included a zero-frame stop codon following the slippery sequence, which enabled the classification of single translating ribosomes to either a nonframeshifted population or −1 frameshifted population based on the translation termination at the in-frame stop codon, or continued translation in the −1 frame. Two consecutive binding events of Cy5-labeled tRNALys [Lys-(Cy5)-tRNALys] on the slippery sequence were used as an orthogonal indicator to the ribosomal intersubunit FRET signal monitoring the ribosomal conformation (fig. S3). After classification of ribosomes, the overall frameshifting efficiency was calculated as the fraction of the frameshifted population from the entire population (Fig. 4B). Frameshifting efficiencies measured from the single-molecule assays were compared with in vivo measurements (table S1), showing a quantitative agreement. Thus, our single-molecule in vitro translation system recapitulates the in vivo −1 frameshifting phenomenon.
Fig. 4. mRNA structure enhances frameshifting by lengthening the rotated state.
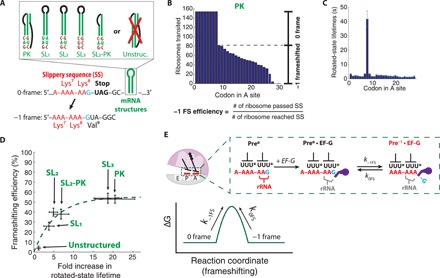
(A) mRNA constructs containing frameshifting cassette and different mRNA structures (SL, stem-loop; PK, pseudoknot; and Unstruc., unstructured) used in the programmed −1 frameshifting. The exact sequence and design of mRNA structures are discussed in fig. S2. (B) Representative translation processivity plot from the PK mRNA construct used to calculate the −1 frameshifting efficiency (percentage of ribosomes that translated codon 9 over ribosomes that reached codon 9; n = 154 molecules). (C) Rotated-state lifetimes for the PK mRNA construct. (D) A plot of the −1 frameshifting efficiency versus the fold increase in the rotated-state lifetime of four mRNA constructs used (n = 108, 126, 195, 137, 101, and 154 molecules from left to right; horizontal error bars represent 95% confidence interval after the error propagation; vertical error bars represent standard error based on the binomial distribution of frameshifting). Dashed line shows the fitting of the equilibrium kinetics model for frameshifting. (E) Proposed model for frameshifting between the 0 frame and −1 frame. Top: The reading frame is maintained via interaction between the codon-anticodon helix and the ribosomal RNA (rRNA) monitoring bases. EF-G disrupts the monitoring bases interaction, leaving the reading frame vulnerable to the spontaneous frame slippage. U* denotes mnm5s2U modification of E. coli tRNALys anticodon. Bottom: A cartoon energy landscape of frameshifting.
In addition to the RNA pseudoknot, we tested various RNA hairpins of different folding stabilities (fig. S2) placed at the same spacer distance (5-nucleotide) from the slippery sequence at the 5′-end. The IS3 pseudoknot construct had two G-C pairs and one A-U pair in the first three base pairs of the RNA hairpin structure. To probe the interplay of terminal base pairing stability, translocation delay, and frameshifting, we designed different mRNA constructs where either the first three base pairs were changed to one G-C pair at the end (SL1) or where all three base pairs were changed to G-C pairs (SL3). To test the effect of the overall stability of the RNA structure in translocation kinetics, we have also designed a stem-loop that shares the first stem and loop of the IS3 pseudoknot but lacks the second stem that base pairs with the RNA loop [SL2–pseudoknot (PK)]. Last, we have designed an mRNA construct that includes the slippery sequence but lacks stable RNA secondary structure in its 3′-end (unstructured). We determined the fold increase in the rotated-state lifetimes (at codon 8) for each mRNA as described above, where each stable mRNA structure delayed translocation on the slippery sequence (Fig. 4C). We observed a positive correlation between the fold increase in rotated-state lifetimes and the frameshifting efficiencies measured in the different mRNA constructs (Fig. 4D). We also observed that the stability of base pairs melted during translocation into the structured RNAs inversely correlated with the rotated-state lifetimes, where a higher G-C content of the first three bases of mRNA stem-loop leads to a longer rotated-state lifetime (Fig. 4D). This indicates that the change in the rotated-state lifetime is highly sensitive to the local stability of the mRNA structures that need to be disrupted before the full translocation.
To probe the underlying mechanism of frameshifting, we examined the relationship between the delay in the rotated state and the frameshifting efficiency shown in Fig. 4D. We initially hypothesized that −1 frameshifting was unidirectional—i.e., once a ribosome has −1 frameshifted from Pre0•EF-G state to the Pre−1•EF-G state (“pre” indicates its pretranslocation state bound with EF-G; superscript added to denote the current reading frame), it is unable to return to the original frame. However, this unidirectional model of frameshifting was inconsistent with the observed data, due to the plateau in −1 frameshifting efficiency at 50% (R2 = 0.63) (fig. S4). Instead, our data fit better with a model that allows interconversion between the Pre0•EF-G and Pre−1•EF-G states on the slippery sequence before translocation (R2 = 0.94) (Fig. 4E). Our current model includes one additional fitting parameter (k−1FS and k0FS; forward and backward frameshifting rates, as opposed to the forward frameshifting rate in the unidirectional frameshifting model). This model of frameshifting suggests that the presence of downstream mRNA structures at distances common in productively used −1 frameshifting may not impose directionality of the frameshift event, but instead prolongs the lifetime of a state in which the reading frame can spontaneously and reversibly equilibrate. The rate of the frameshift in both directions (k−1FS and k0FS), which involves rearrangements of base pairing, may be determined primarily by the codon-anticodon interaction in two respective frames, but further influenced by the base context around it (9, 27). This model of reversible slippage of the reading frame during translocation enhances prior models of EF-G–catalyzed translocation, during which contacts between rRNA monitoring bases and the codon-anticodon helices are disrupted (28, 29). Thus, we hypothesize that once the ribosomal contacts to the codon-anticodon helix are disrupted before translocation (possibly aided by EF-G), the frameshifting event may be reversible in both directions as governed by the codon-anticodon pairing of the two tRNAs in the transition state.
Downstream mRNA structure buffers frameshifting efficiency with respect to temperature variation
This model suggests that the frameshifting rate on the slippery sequence might exhibit a strong temperature dependence, because multiple hydrogen bonds and stacking interactions within the codon-anticodon helices need to be disrupted to achieve the transition state for frameshifting. We thus investigated the effect of temperature on frameshifting efficiency. The −1 frameshifting efficiencies were measured at four temperatures ranging from 20° to 35°C by an in vivo translation assay using a dual-tagged reporter with both the frameshift- and nonframeshift-derived products quantified via densitometry of reporters revealed by immunoblotting (Fig. 5) (30). This in vivo translation assay provides the necessary sensitivity to resolve small changes in frameshifting efficiency at different conditions. While the effect of temperature on overall translation may be complex, we observed a clear increase in −1 frameshifting efficiency as the temperature increased in experiments with the frameshifting mRNA without an accompanied mRNA structure near the slippery sequence (Fig. 5A). We also note that the maximal frameshifting efficiency observed (about 12% at 35°C) is still well below the frameshifting efficiency enhanced by the mRNA structure (40 to 50%). Our results indicate that the energetics involved in frameshifting without the mRNA structure can be manipulated by varying the temperature. The directionality of change in the frameshifting efficiency with respect to change in temperature follows the expectations from our model of −1 frameshifting, where reversible codon-anticodon rearrangements may be key processes in frameshifting. However, in the case of frameshifting enhanced by the mRNA structure, the increasing temperature facilitates the unfolding of local mRNA structures before translocation, thereby decreasing the effect of the mRNA structure on prolonging the rotated state (Fig. 2E). As predicted, the strong temperature dependence of −1 frameshifting efficiency was not observed in the mRNA containing an mRNA structure accompanying the slippery sequence (Fig. 5B). The observed insensitivity of frameshifting to changes in temperature on these mRNAs indicates the dual effects of temperature, which may not only affect the kinetics of the codon-anticodon rearrangement leading to the frameshifting but also alter the time window for frameshifting determined by the downstream mRNA structural stability (increased rotated-state lifetimes).
Fig. 5. Characterizing energetics of − 1 frameshifting in vivo.
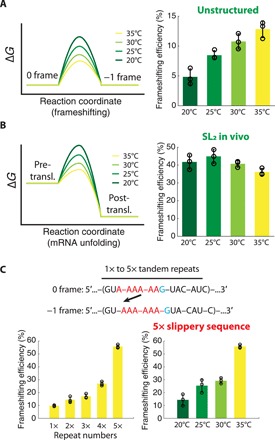
(A) Left: Cartoon energy landscapes of frameshifting on the slippery sequence at different temperatures. Right: The −1 frameshifting efficiency measured on the slippery sequence without mRNA structure at different temperatures. (B) Left: Cartoon energy landscapes of the mRNA structure unfolding before translocation at different temperatures. Right: The −1 frameshifting efficiency measured on the slippery sequence accompanied by the downstream mRNA structure at different temperatures. (C) Top: Frameshifting cassettes composed of tandem repeats of slippery sequences. Bottom left: Frameshifting efficiency of constructs with different repeat numbers measured at 35°C. Bottom right: Frameshifting efficiency of the mRNA construct with five slippery sequence repeats measured at different temperatures (means ± SD are shown from n = 3 technical replicates for all plots in this figure; values are summarized in tables S1 and S2; gel images are present in fig. S5).
To probe the biophysical origin of these in vivo results, we measured both −1 frameshifting efficiency and translocation kinetics on a slippery sequence using our in vitro single-molecule assays. The rotated-state pause induced by the mRNA structure decreased at higher reaction temperatures, while the frameshifting efficiency remained nearly unchanged (fig. S3H). While the protein synthesis response in cells to temperature variation is expected to be complex, the effect of temperature on −1 frameshifting is consistent between the in vitro single-molecule and the in vivo assays and fits our mechanistic model. Our combined results suggest that a stable downstream RNA structure not only stimulates the −1 frameshifting pathway on the slippery sequence but also counterbalances changes in frameshifting kinetics due to external temperature change, resulting in −1 frameshifting efficiency robust to temperature variations.
The temperature dependence of frameshifting, however, could be a desirable feature in controlling the gene expression level in response to an external cue. Therefore, we sought to design an artificial programmed −1 frameshifting system that can be modulated by temperature, while its maximal value for −1 frameshifting efficiency reaches values similar to systems that rely on stimulation by an mRNA structure. Our model of frameshifting shows that the presence of the mRNA structure prolongs the rotated state at the slippery sequence. Alternatively, we reasoned that another way to increase the frameshifting efficiency is via tandem repeats of the slippery sequence across the mRNA, in the absence of a nearby structured region. To limit frameshifting between 0 and −1 frames, nonslippery spacers were included between the repeats of the slippery sequence (Fig. 5C). When tested in the in vivo frameshifting assays, −1 frameshifting efficiency increased with an increase in the number of slippery sequence repeats as expected. All mRNAs with one to five tandem repeats of the slippery sequence showed a strong temperature dependence of frameshifting from 20° to 35°C (Fig. 5C and table S2). At the 35°C condition, the frameshifting efficiency was near 55% in the five-repeat construct, resulting in an overall 15 to 55% tunable range of −1 frameshifting efficiency over the temperature range of 20° to 35°C. These results suggest that our mechanistic model of frameshifting has predictive power and may be used to engineer programmed −1 frameshifting systems in vivo that can tune the relative expression of protein products at translation level using temperature controls.
Internal SD sequence may enhance frameshifting by inducing mechanical force
In addition to mRNA structures, cis- and trans-elements (3) that enhance frameshifting efficiency have been identified. Frameshifting systems in bacteria often include an internal SD sequence (3, 4), which base pairs with the 16S rRNA of the small subunit of translating ribosomes. The spacing between the slippery sequence and the frameshift stimulatory internal SD sequence varies according to the frameshifting direction: For a −1 frameshifting system such as dnaX or IS3, the spacing is 10 to 11 nucleotides, whereas the spacing is 3 nucleotides for stimulating +1 frameshifting (4). Recent reports using ribosome profiling assays (31) and bulk kinetic assays (32) have revealed that the internal SD sequence minimally affects the translation rate, suggesting that its frameshifting enhancement mechanism is different from that of the mRNA structure. To understand the role of the internal SD sequence in frameshifting, we tested two mRNAs that contain a strong internal SD sequence, with or without the downstream pseudoknot mRNA structure. Compared with mRNAs that lack an internal SD sequence, mRNAs with an internal SD sequence have higher frameshifting efficiency with indistinguishable rotated-state lifetime (fig. S6). The observed increase in frameshifting efficiency without lengthening of the rotated-state lifetime suggests that the enhancement of −1 frameshifting by internal SD sequence suggests tilting (33) of the frameshifting energy landscape on the slippery sequence, which supports the notion that strain is induced by the internal SD sequence in the transition state for frameshifting and favors shifting in the −1 direction.
Model of the frame maintenance and RNA structure unfolding before translocation
Our findings suggest a model for ribosomal frame maintenance and programmed −1 frameshifting (Fig. 6). During translocation, all the contacts involving the ribosome and codon-anticodon helices that ensure frame maintenance must be disrupted and reformed. These include contacts between the ribosomal monitoring bases and the codon-anticodon helix that have been established during the prior decoding step, as well as contacts that stabilize the P-site tRNA-mRNA interaction. The binding of EF-G may lead to a partial loss of contacts between the ribosome and the codon-anticodon helices during its action to catalyze translocation. In turn, this intermediate translocation state may have limited capability to maintain the reading frame and be prone to spontaneous frameshifting. In the programmed −1 frameshifting cassettes, the slippery sequence imposes frameshift directionality by allowing perfectly matched base pairing in the shifted reading frame. The addition of 3′-stimulatory mRNA structure to the slippery sequence may prolong the translocation intermediate state with limited frame maintenance capability. Our quantitative model expands the previous model of −1 frameshifting (15), highlighting the reversible nature of frameshifting necessary to achieve kinetic selection before translocation. Our model does not rule out the existence of a noncanonical structural state of the ribosome for frameshifting but does not require one as the canonical Pre•EF-G state, an intermediate of normal translocation, may be the frameshifting-prone state. Further studies are needed to reveal the causal relationship between −1 frameshifting and noncanonical states observed by us and others (15, 17, 34, 35). In particular, a recent ribosome structure with disrupted codon-anticodon helices suggested that contacts between tRNA and EF-G may stabilize the codon-anticodon interaction during translocation to enhance reading-frame maintenance (36). This ground-state interaction may be critical to capture the reading frame at a later step, in which EF-G primarily exerts its effect in a transient state. The quantitative agreement of frameshifting efficiencies measured using in vivo and in vitro single-molecule assays, where in the latter the ribosome remains unbound to the EF-G for a prolonged period due to lower EF-G concentration, suggests that frameshifting is less likely to be promoted without EF-G during continued translation, as previously suggested by Kim et al. (14). Increasing the concentration of EF-G from 100 to 1000 nM decreased the rotated-state lifetimes with and without the mRNA secondary structure engaged but had little effect on the relative frameshift efficiencies and the fold increase in the rotated-state lifetime on codon 8 (fig. S7).
Fig. 6. Proposed model of the −1 ribosomal frameshifting.
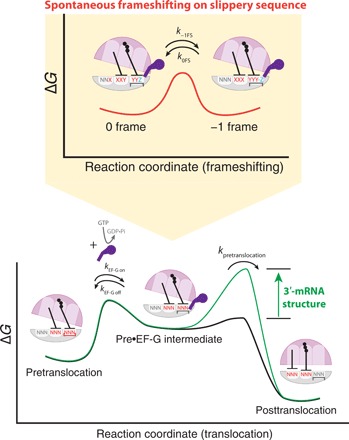
Contacts between the ribosome and the codon-anticodon helices are disrupted by EF-G in the translocation intermediate state (EF-G–bound rotated state or Pre•EF-G), where the reading frame can be spontaneously shifted. The presence of the downstream mRNA structure prolonged the Pre•EF-G state lifetime, thereby increasing the frameshifted population previous to the translocation event.
We note that all four rates involved ( and for frameshifting rates to −1 and 0 frames, respectively, as well as translocation rates and , where superscript denotes respective frames) are important for determining the overall frameshifting efficiency. In our construct design, the spacer length between the slippery sequence and the base of mRNA structure changes from five to six nucleotides upon −1 frameshifting, where a change between translocation rates in respective frames would not be substantial (Fig. 2, C and D). However, with six- or seven-nucleotide spacer length, would be faster than , which may add an additional interpretation of how the −1 frameshifting pathway was observed to be favored in a previous report (15). Furthermore, the spacer length may influence the efficiency of −2 frameshifting. Previous studies have shown that the shorter spacer length between the slippery sequence and the stimulatory mRNA structure enhances the occurrence of −2 frameshifting events over the −1 or +1 frameshifting events (37, 38). We also observe a strong dependence of translocation rate upon small changes in the stability of mRNA structure as well as temperature variations. Increasing the temperature from 20° to 30°C decreased the fold increase in rotated-state lifetimes about 7- to 10-fold (Fig. 2E), which suggests that the pausing during translation due to mRNA structure in vivo at 30° to 37°C may not be significant to trigger other surveillance pathways to degrade abortive translation. The shortened translocation pause at physiological temperature may be counterbalanced with hastened frameshifting kinetics for the A- and P-site codons, showing apparent independence of frameshifting against temperature variation in vivo as observed here (Fig. 5B). Yet, in the case of frameshifting cassettes lacking stable mRNA structures, a slight change in temperature may influence frameshifting efficiency significantly (Fig. 5C). Our results also suggest that mRNA structures could have a substantial impact on the rate of translation during cold shock (39).
Our mechanistic model presents a framework that can be used to incorporate diverse cis- and trans-elements that affect frameshifting efficiency. In addition to the internal SD sequence tested here, distinct elements for stimulating ribosomal frameshifting are being discovered continually, including proteins that bind to mRNA structure (40), the nascent peptide sequence within the ribosomal nascent peptide tunnel (40), and the absence of the ribosomal A-site substrate (41, 42). A systematic study of different frameshifting elements may reveal their effect on the energy landscape of frameshifting delineated here as well as distinctive frameshifting pathways used at other stages of translation. A deep understanding of frameshifting mechanisms may permit its use in artificial regulation of gene expression at the translation level through external cues such as temperature or oligonucleotide signals (43).
MATERIALS AND METHODS
Experimental design
Fluorescence traces from each ZMW nanostructures were selected on the basis of the presence of both fluorophores at different time points (signal from immobile fluorophores on the ribosome was expected to be present at the beginning of the movie, while signal from fluorophores attached to tRNA was not expected to be), and nonrotated and rotated states for 100 to 200 traces were manually assigned following the cross-correlation signals between the intersubunit FRET signal (from Cy3B–BHQ-2 fluorophores) and Cy5-labeled tRNA binding/dissociation signals. In vivo measurements were performed in three technical replicates.
Reagents and buffers for single-molecule experiments
Reagents and buffers were prepared as previously reported (18, 21, 20, 44). Briefly, each small and large subunit was mutated to include a weakly forming RNA hairpin at helices 44 and 101, which was used to attach Cy3B/BHQ-2–labeled DNA oligonucleotides via RNA/DNA hybridization [DNA sequences for short oligonucleotides are 5′-GAGGCCGAGAAGTG-(BHQ-2)-(BHQ-2)-3′ and 5′-GGGAGATCAGGATA-(Cy3B)-3′; both purchased from Trilink]. Individual tRNA species used were purchased from Chemical Block Ltd. tRNALys was labeled at the acp3U47 position with Cy5 using N-hydroxysuccinimide (NHS) chemistry as previously described (20, 45), with Cy5-NHS ester purchased from GE Healthcare. tRNAfMet was labeled at the s4U8 position using maleimide chemistry as previously described (20, 45) (Cy3-maleimide was purchased from GE Healthcare). Biotinylated RNAs used for single-molecule translation assays were synthesized in house using T7 RNA polymerase, where the reaction was supplemented with biotin-GMP (Trilink) to introduce biotin on the 5′ of mRNAs. The mRNA sequences used in this study were provided separately (data file S1). Translational factors, ribosomal S1 protein, and aminoacylated tRNAs were prepared as previously reported (21, 44). Labeling of EF-G and purification were prepared as previously reported (46, 47). All single-molecule experiments were conducted in a tris-based polymix buffer consisting of 50 mM tris-acetate (pH 7.5), 100 mM potassium chloride, 5 mM ammonium acetate, 0.5 mM calcium acetate, 5 mM magnesium acetate, 0.5 mM EDTA, 5 mM putrescine-HCl, and 1 mM spermidine, with additional 4 mM GTP.
Single-molecule experiments on the ZMW instrument
Detailed development and specification of the Pacific Bioscience RSII instrument as a platform for single-molecule fluorescence microscope have been published previously (47). Immediately before each experiment, 30S and 50S ribosomal subunits were mixed with respective fluorescently labeled DNA oligonucleotide at 1:1.2 stoichiometric ratio with 1 μM final concentration of the subunit in the polymix buffer and incubated in the 37°C water bath for 10 min, followed by incubation in the 30°C water bath for 20 min. Afterward, the tube containing the 50S subunit was kept on ice, while the 30S subunit was incubated with S1 ribosomal protein at 1:1 stoichiometric ratio with 0.5 μM final concentration in the 37°C water bath for 5 min, as S1 ribosomal proteins may not have been purified with the small subunit.
Using the labeled 30S ribosomal subunit, the preinitiation complex was formed by mixing it with biotinylated mRNA, initiation factor 2, aminoacylated formyl-methionine tRNA at 1:2:13:4 with 150 nM final concentration of the small subunit in the polymix buffer, supplemented with 4 mM GTP, and incubating in the 37°C water bath for 5 min. The formed complex was diluted to 10 nM in the polymix buffer supplemented with 4 mM GTP and the imaging mix [2.5 mM of protocatechuic acid, 2.5 mM of TSY, and 2× PCD (protocatechuate-3,4-dioxygenase), purchased from Pacific Bioscience; PCD was added last], and incubated in the ZMW chip treated with NeutrAvidin at room temperature for 3 min, which was bound to the biotin–polyethylene glycol on the chip. After immobilizing the preinitiation complex, the chip was washed three times using the same buffer without the complex to remove unbound complexes and loaded onto the instrument (RSII, purchased from Pacific Bioscience).
At the same time, the delivery solution, a polymix buffer supplemented with 4 mM GTP, the imaging mix, varying concentrations of tRNA ternary complexes (labeled or unlabeled), varying concentrations of EF-G (labeled or unlabeled), and 200 nM of the BHQ-2–labeled 50S ribosomal subunit was prepared and loaded onto the instrument. For experiments measuring the effect of mRNA structure on translation, purified 20 nM Phe-(Cy5)-tRNAPhe, 20 nM Lys-tRNALys, and 40 nM EF-G were used, as the first 10 codons of mRNA coding regions only included the Phe and Lys codons. For experiments with frameshifting mRNA constructs, 200 nM Lys-tRNALys and 1.6 μM total tRNA mix (Roche) charged with 19 amino acids (excluding lysine) and 100 nM of EF-G were used. A higher concentration of factors was used to monitor events after a long translation pause induced by the presence of an mRNA structure, especially in the presence of an RNA pseudoknot. The tRNA ternary complex was formed by incubating tRNA with 100 μM EF-Tu•GTP within the polymix buffer without the spermidine and the putrescine, supplemented with fresh 1 mM GTP, and incubated in the 37°C water bath for 1 min. In the experiments using Cy5-labeled EF-G, Cy5-labeled tRNA components described above were substituted with the corresponding unlabeled reagent while keeping the final concentration of factors the same. In the tRNA–EF-G FRET experiment, ribosomes were not incubated with their respective labeling DNA oligonucleotide.
At the start of the experiment, the instrument delivered the delivery solution to the chip and recorded an 8-min movie with a frame rate of 10 frames per second, illuminated by 60 mW/mm2 of 532-nm laser and 10 mW/mm2 of 642-nm laser for ribosomal intersubunit FRET experiments with labeled tRNA. For experiments using Cy5-labeled EF-G, an 8-min movie was recorded with a frame rate of 25 frames per second, illuminated by 72 mW/mm2 of 532-nm laser and 24 mW/mm2 of 642-nm laser or by 80 mW/mm2 of 532-nm laser only for measuring tRNA–EF-G FRET. Experiments were performed with the chip temperature clamped to the specified temperature, usually ranging from 20° to 30°C.
The resulting movies were analyzed using in-house–written MATLAB (MathWorks) scripts, as previously described (46, 47). Briefly, traces from each ZMW well were filtered based on the presence of both fluorophores at different time points (signal from immobile fluorophores on the ribosome was expected to be present at the beginning of the movie, while signal from fluorophores attached to tRNA was not expected to be) and a single photobleaching step for each fluorophores. Filtered traces were manually assigned to the rotated state and nonrotated state after the subunit joining event, cross-correlated with the labeled tRNA binding signals. Cross-correlating the intersubunit FRET signal (from Cy3B–BHQ-2 fluorophores) and Cy5-labeled tRNA signals has been powerful in the correct manual state assignments, validated using the ensemble kinetic methods (18, 25). The labeled tRNA signals were placed across the mRNA construct strategically to ensure accurate kinetic measurements at the pause point [Lys-(Cy5)-tRNALys used for the slippery sequence that encodes for two lysine codons] during translation.
From assigned traces, both rotated- and nonrotated-state lifetimes were calculated by fitting a single-exponential distribution to the measured state lifetimes using maximum likelihood estimation in MATLAB. Fold increase in the rotated-state lifetimes was calculated by comparing average rotated-state lifetimes for codons 2 to 6 (before mRNA structure engages the ribosome, with the first codon excluded due to possible effect of transitioning from the initiation to elongation phase) to codon 8 (when translocation is inhibited by the downstream mRNA structure). Fold increase was used, as it was a measurement of kcat/Km change as observed earlier, assuming that EF-G binding kinetics during the rotated state was not perturbed by the presence of the mRNA structure (fig. S1). Resulting fold increase measurements (the kcat/Km change measurements) were interpreted as an increase in the fold increase in the EF-G binding events, following the previous report (18, 25). The equilibrium kinetics equation used was in this form
where k−1FS and k0FS are the −1 frameshifting rate and the reverse frameshifting rate, respectively, and t is the fold increase in the rotated-state lifetime. In the case of the irreversible frameshifting model, k0FS has been changed to 0.
In vivo frameshifting assay: Bacterial strains
The E. coli strains DH5α and BL21 (DE3) were used for plasmid propagation and protein expression, respectively. Strains were grown in LB medium.
In vivo frameshifting assay: Insert construction
Constructs were produced by amplification of complementary oligonucleotides (Integrated DNA Technologies) to produce a full-length sequence containing 5′ Spe I and 3′ Bam HI restriction sites. These were cloned into the vector pJ307 (GST-MBP-His fusion vector) (30). To distinguish between the termination product and the trans-frame product, we present the MBP tag in the alternative −1 frame relative to the GST tag. MBP and GST were in frame for the positive control. Primers used in this study are provided separately in the Supplementary Materials.
In vivo frameshifting assay: Western blot analysis
Overnight cultures of strains containing the appropriate plasmid were diluted 1:100 in LB medium. Each culture was grown in triplicate over a range of temperatures from 20° to 35°C at 200 revolutions per minute (20°, 25°, 30°, and 35°C). Once an optical density of 0.4 to 0.5 was reached, the cultures were induced with 1 mM isopropyl-β,d-thiogalactopyranoside (IPTG) for 1 hour at their respective temperatures. After induction, cultures were incubated on ice for 10 min. Cultures were spun down, resuspended in lysis buffer [50 mM Tris (pH 8.0), 10% glycerol, lysozyme (100 μg/ml), 1 mM phenylmethylsulfonyl fluoride, deoxyribonuclease (1 U/ml), 5 mM dithiothreitol, and 2 mM MgCl2], and incubated for 30 min on ice. Cells were subsequently sonicated four times for 5 s (power 10, 1-min on/off cycles).
Lysates were then centrifuged at 20,000g for 30 min at 4°C. On the basis of the A280 NaNodrop readings, equivalent amounts of proteins (50 μg per lane) were diluted and boiled for 10 min at 95°C in Laemmli sample buffer. Proteins were separated on a 10% SDS–polyacrylamide gel and transferred onto nitrocellulose membrane (Protran). Immunoblots were incubated at 4°C overnight in 5% milk/phosphate-buffered saline–Tween containing a 1:2000 dilution of rabbit anti-GST. Immunoreactive bands were detected on membranes after incubation with appropriate fluorescently labeled secondary antibodies using a LI-COR Odyssey Infrared Imaging Scanner (LI-COR Biosciences). The amounts of termination and frameshift products were quantified using Image Studio Lite (LI-COR Biosciences). The frameshifting efficiency was determined by taking the amount of frameshift product as a ratio of the total amount of termination plus frameshift products.
Statistical analysis
Measurements from single-molecule fluorescence assay resulted from a specified number (n) of molecules from a single experiment, and measurements from individual molecules were used to calculate state lifetimes and FRET distributions using single-exponential and normal distribution fitting, respectively. Measurements from the in vivo Western blot assay resulted from three technical replicates, and individual measurements were used to calculate mean and SDs for each condition.
Supplementary Material
Acknowledgments
We thank O. Fayet for the advice in designing the initial experimental constructs, and past and present members of Puglisi Laboratory for the discussions and input. Funding: This work was supported by the U.S. National Institutes of Health (NIH) grants GM51266 and GM113078 to J.D.P., by a Stanford Bio-X fellowship to J.C., and by the Science Foundation Ireland grant 13/IA/1853 and Irish Research Council grant IRCLA/2019/74 to J.F.A. Author contributions: J.C. performed single-molecule experiments and analyzed data with the guidance of J.D.P. S.O. performed in vivo experiments with the guidance of J.F.A. All authors contributed in writing the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All source data for main figures and extended data figures are available as accompanying Supplementary Experimental Data excel spread sheet. All additional experimental data and MATLAB scripts used are available from the authors upon request.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/1/eaax6969/DC1
Fig. S1. EF-G–bound lifetimes for translocation and mRNA structure unfolding.
Fig. S2. Design of the frameshifting cassette and mRNA structures.
Fig. S3. Assignment of −1 frameshifted population in smFRET assay.
Fig. S4. Simulation of irreversible −1 frameshifting.
Fig. S5. Western blot gel images for Fig. 5.
Fig. S6. Internal SD sequence tilts the energy landscape of the frameshifting.
Fig. S7. The concentration of EF-G does not alter frameshifting efficiency.
Table S1. In vivo measurements for frameshifting constructs.
Table S2. In vivo measurements for frameshifting on slippery sequence repeat constructs.
Data file S1. Nucleic acid sequences for mRNA construct used in the study.
Data file S2. Experimental data presented in the main and supplementary figures.
REFERENCES AND NOTES
- 1.Atkins J. F., Elseviers D., Gorini L., Low activity of β-Galactosidase in frameshift mutants of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 69, 1192–1195 (1972). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jørgensen F., Kurland C. G., Processivity errors of gene expression in Escherichia coli. J. Mol. Biol. 215, 511–521 (1990). [DOI] [PubMed] [Google Scholar]
- 3.Atkins J. F., Loughran G., Bhatt P. R., Firth A. E., Baranov P. V., Ribosomal frameshifting and transcriptional slippage: From genetic steganography and cryptography to adventitious use. Nucleic Acids Res. 44, 7007–7078 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.J. F. Atkins, R. F. Gesteland, Recoding: Expansion of Decoding Rules Enriches Gene Expression (Springer Verlag, New, 2010). [Google Scholar]
- 5.Jacks T., Power M. D., Masiarz F. R., Luciw P. A., Barr P. J., Varmus H. E., Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331, 280–283 (1988). [DOI] [PubMed] [Google Scholar]
- 6.Korniy N., Goyal A., Hoffmann M., Samatova E., Peske F., Pöhlmann S., Rodnina M. V., Modulation of HIV-1 Gag/Gag-Pol frameshifting by tRNA abundance. Nucleic Acids Res. 47, 5210–5222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X., Xuan Y., Han Y., Ding X., Ye K., Yang F., Gao P., Goff S. P., Gao G., Regulation of HIV-1 Gag-Pol Expression by Shiftless, an Inhibitor of Programmed −1 Ribosomal Frameshifting. Cell 176, 625–635.e14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weiss R. B., Dunn D. M., Shuh M., Atkins J. F., Gesteland R. F., E. coli ribosomes re-phase on retroviral frameshift signals at rates ranging from 2 to 50 percent. New Biol. 1, 159–169 (1989). [PubMed] [Google Scholar]
- 9.Sharma V., Prere M.-F., Canal I., Firth A. E., Atkins J. F., Baranov P. V., Fayet O., Analysis of tetra- and hepta-nucleotides motifs promoting −1 ribosomal frameshifting in Escherichia coli. Nucleic Acids Res. 42, 7210–7225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen B., Gesteland R. F., Atkins J. F., Structural probing and mutagenic analysis of the stem-loop required for Escherichia coli dnaX ribosomal frameshifting: programmed efficiency of 50%. J. Mol. Biol. 271, 47–60 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manktelow E., Shigemoto K., Brierley I., Characterization of the frameshift signal of Edr, a mammalian example of programmed −1 ribosomal frameshifting. Nucleic Acids Res. 33, 1553–1563 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazauric M.-H., Licznar P., Prère M.-F., Canal I., Fayet O., Apical loop-internal loop RNA Pseudoknots. J. Biol. Chem. 283, 20421–20432 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Kontos H., Napthine S., Brierley I., Ribosomal pausing at a frameshifter RNA pseudoknot is sensitive to reading phase but shows little correlation with frameshift efficiency. Mol. Cell. Biol. 21, 8657–8670 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim H.-K., Liu F., Fei J., Bustamante C., Gonzalez R. L. Jr., Tinoco I. Jr., A frameshifting stimulatory stem loop destabilizes the hybrid state and impedes ribosomal translocation. Proc. Natl. Acad. Sci. U.S.A. 111, 5538–2243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caliskan N., Katunin V. I., Belardinelli R., Peske F., Rodnina M. V., Programmed −1 frameshifting by kinetic partitioning during impeded translocation. Cell 157, 1619–1631 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H.-K., Tinoco I. Jr., EF-G catalyzed translocation dynamics in the presence of ribosomal frameshifting stimulatory signals. Nucleic Acids Res. 45, 2865–2874 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J., Petrov A., Johansson M., Tsai A., O’Leary S. E., Puglisi J. D., Dynamic pathways of −1 translational frameshifting. Nature 512, 328–332 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi J., Ieong K.-W., Demirci H., Chen J., Petrov A., Prabhakar A., O'Leary S. E., Dominissini D., Rechavi G., Soltis S. M., Ehrenberg M., Puglisi J. D., N6-methyladenosine in mRNA disrupts tRNA selection and translation-elongation dynamics. Nat. Struct. Mol. Biol. 23, 110–115 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cornish P. V., Ermolenko D. N., Noller H. F., Ha T., Spontaneous Intersubunit Rotation in Single Ribosomes. Mol. Cell 30, 578–588 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dorywalska M., Blanchard S. C., Gonzalez R. L., Kim H. D., Chu S., Puglisi J. D., Site-specific labeling of the ribosome for single-molecule spectroscopy. Nucleic Acids Res. 33, 182–189 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aitken C. E., Puglisi J. D., Following the intersubunit conformation of the ribosome during translation in real time. Nat. Struct. Mol. Biol. 17, 793–800 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M., Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenner L. B., Demeshkina N., Yusupova G., Yusupov M., Structural aspects of messenger RNA reading frame maintenance by the ribosome. Nat. Struct. Mol. Biol. 17, 555–560 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Amiri H., Noller H. F., A tandem active site model for the ribosomal helicase. FEBS Lett. 593, 1009–1019 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Choi J., Indrisiunaite G., DeMirci H., Ieong K.-W., Wang J., Petrov A., Prabhakar A., Rechavi G., Dominissini D., He C., Ehrenberg M., Puglisi J. D., 2′-O-methylation in mRNA disrupts tRNA decoding during translation elongation. Nat. Struct. Mol. Biol. 25, 208–216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Munro J. B., Wasserman M. R., Altman R. B., Wang L., Blanchard S. C., Correlated conformational events in EF-G and the ribosome regulate translocation. Nat. Struct. Mol. Biol. 17, 1470–1477 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bertrand C., Prère M. F., Gesteland R. F., Atkins J. F., Fayet O., Influence of the stacking potential of the base 3′ of tandem shift codons on −1 ribosomal frameshifting used for gene expression. RNA 8, 16–28 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G., Song G., Zhang D., Zhang D., Li Z., Lyu Z., Dong J., Achenbach J., Gong W., Zhao X. S., Nierhaus K. H., Qin Y., EF-G catalyzes tRNA translocation by disrupting interactions between decoding center and codon–anticodon duplex. Nat. Struct. Mol. Biol. 21, 817–824 (2014). [DOI] [PubMed] [Google Scholar]
- 29.Noller H. F., Lancaster L., Zhou J., Mohan S., The ribosome moves: RNA mechanics and translocation. Nat. Struct. Mol. Biol. 24, 1021–1027 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonov I., Coakley A., Atkins J. F., Baranov P. V., Borodovsky M., Identification of the nature of reading frame transitions observed in prokaryotic genomes. Nucleic Acids Res. 41, 6514–6530 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buskirk A. R., Green R., Ribosome pausing , arrest and rescue in bacteria and eukaryotes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borg A., Ehrenberg M., Determinants of the rate of mRNA translocation in bacterial protein synthesis. J. Mol. Biol. 427, 1835–1847 (2015). [DOI] [PubMed] [Google Scholar]
- 33.Liu T., Kaplan A., Alexander L., Yan S., Wen J.-D., Lancaster L., Wickersham C. E., Fredrick K., Noller H., Tinoco I. Jr., Bustamante C. J., Direct measurement of the mechanical work during translocation by the ribosome. eLife 3, e03406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin P., Yu D., Zuo X., Cornish P. V., Structured mRNA induces the ribosome into a hyper-rotated state. EMBO Rep. 15, 185–190 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Namy O., Moran S. J., Stuart D. I., Gilbert R. J. C., Brierley I., A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature 441, 244–247 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou J., Lancaster L., Donohue J. P., Noller H. F., Spontaneous ribosomal translocation of mRNA and tRNAs into a chimeric hybrid state. Proc. Natl. Acad. Sci. U.S.A. 116, 7813–7818 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsufuji S., Matsufuji T., Wills N. M., Gesteland R. F., Atkins J. F., Reading two bases twice: mammalian antizyme frameshifting in yeast. EMBO J. 15, 1360–1370 (1996). [PMC free article] [PubMed] [Google Scholar]
- 38.Lin Z., Gilbert R. J. C., Brierley I., Spacer-length dependence of programmed −1 or −2 ribosomal frameshifting on a U6 A heptamer supports a role for messenger RNA (mRNA) tension in frameshifting. Nucleic Acids Res. 40, 8674–8689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Burkhardt D. H., Rouskin S., Li G.-W., Weissman J. S., Gross C. A., A Stress Response that Monitors and Regulates mRNA Structure Is Central to Cold Shock Adaptation. Mol. Cell 70, 274–286.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Napthine S., Ling R., Finch L. K., Jones J. D., Bell S., Brierley I., Firth A. E., Protein-directed ribosomal frameshifting temporally regulates gene expression. Nat. Commun. 8, 15582 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang R., Xiong J., Wang W., Miao W., Liang A., High frequency of +1 programmed ribosomal frameshifting in Euplotes octocarinatus. Sci. Rep. 6, 21139 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lobanov A. V., et al. , Position-dependent termination and widespread obligatory frameshifting in Euplotes translation. Nat. Struct. Mol. Biol. 24, 585–597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Omabegho T., Gurel P. S., Cheng C. Y., Kim L. Y., Ruijgrok P. V., Das R., Alushin G. M., Bryant Z., Controllable molecular motors engineered from myosin and RNA. Nat. Nanotechnol. 13, 34–40 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blanchard S. C., Gonzalez R. L. Jr., Kim H. D., Chu S., Puglisi J. D., tRNA selection and kinetic proofreading in translation. Nat. Struct. Mol. Biol. 11, 1008–1014 (2004). [DOI] [PubMed] [Google Scholar]
- 45.Marshall R. A., Dorywalska M., Puglisi J. D., Irreversible chemical steps control intersubunit dynamics during translation. Proc. Natl. Acad. Sci. U.S.A. 105, 15364–15369 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J., Petrov A., Tsai A., O’Leary S. E., Puglisi J. D., Coordinated conformational and compositional dynamics drive ribosome translocation. Nat. Struct. Mol. Biol. 20, 718–727 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen J., Dalal R. V., Petrov A. N., Tsai A., O'Leary S. E., Chapin K., Cheng J., Ewan M., Hsiung P.-L., Lundquist P., Turner S. W., Hsu D. R., Puglisi J. D., High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc. Natl. Acad. Sci. U.S.A. 111, 664–669 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/6/1/eaax6969/DC1
Fig. S1. EF-G–bound lifetimes for translocation and mRNA structure unfolding.
Fig. S2. Design of the frameshifting cassette and mRNA structures.
Fig. S3. Assignment of −1 frameshifted population in smFRET assay.
Fig. S4. Simulation of irreversible −1 frameshifting.
Fig. S5. Western blot gel images for Fig. 5.
Fig. S6. Internal SD sequence tilts the energy landscape of the frameshifting.
Fig. S7. The concentration of EF-G does not alter frameshifting efficiency.
Table S1. In vivo measurements for frameshifting constructs.
Table S2. In vivo measurements for frameshifting on slippery sequence repeat constructs.
Data file S1. Nucleic acid sequences for mRNA construct used in the study.
Data file S2. Experimental data presented in the main and supplementary figures.


