Abstract
BACKGROUND
Hepatocellular carcinoma (HCC) is a common malignant gastrointestinal tumor. There are currently few clinical diagnostic and prognostic markers for HCC. LncRNA cancer susceptibility candidate 9 (CASC9) is a long-chain non-coding RNA discovered in recent years, and previous studies have found that lncRNA CASC9 participates in the occurrence and development of HCC, but its clinical value remains unclear.
AIM
To determine the expression of lncRNA CASC9 in HCC and its diagnostic and prognostic value.
METHODS
Data on CASC9 expression in patients with HCC were collected from the Cancer Genome Atlas (TCGA) database to analyze the relationship between CASC9 and patient survival. A total of 80 HCC patients treated in The First Affiliated Hospital of Guangxi Medical University from May 2012 to January 2014 were enrolled in the patient group, and 50 healthy subjects were enrolled in the control group during the same period. CASC9 expression in the two groups was determined using quantitative real-time polymerase chain reaction, and its diagnostic and prognostic value was analyzed based on the CASC9 data and pathological data in these HCC patients. The relationship between CASC9 and patient survival was assessed during the 5-year follow-up period.
RESULTS
Analysis of data from TCGA database revealed that control samples showed significantly lower CASC9 expression than carcinoma tissue samples (P < 0.001); the low CASC9 expression group had a higher survival rate than the high CASC9 expression group (P = 0.011), and the patient group showed significantly increased expression of serum CASC9, with the area under the curve (AUC) of 0.933. CASC9 expression was related to tumor size, combined hepatitis, tumor, node, metastasis (TNM) staging, lymph node metastasis, differentiation and alpha fetoprotein, and the high CASC9 expression group showed lower 1-year, 3-year and 5-year survival rates than the low CASC9 expression group (all aP < 0.05). Multivariate Cox regression analysis revealed that TNM staging, lymph node metastasis, differentiation, alpha fetoprotein and CASC9 were independent factors affecting the prognosis of patients. Stage I+II patients with lymph node metastasis, low differentiation, and alpha fetoprotein > 200 ng/mL had a poor 5-year survival rate.
CONCLUSION
High CASC9 expression is beneficial in the prognosis of HCC patients. CASC9 is expected to be a potential diagnostic and prognostic indicator of HCC.
Keywords: LncRNA cancer susceptibility candidate 9, Hepatocellular carcinoma, Prognosis, Kyoto Encyclopedia of Genes and Genomes, Gene Ontology, Competing endogenous RNA
Core tip: Previous studies have found that lncRNA cancer susceptibility candidate 9 (CASC9) can promote the survival of hepatocellular carcinoma (HCC) cells through AKT ions, but it is not clear whether lncRNA CASC9 can be used as a clinical indicator of diagnosis and prognosis in patients with HCC. This study confirmed that lncRNA CASC9 can be used as a potential prognostic and diagnostic indicator in patients with HCC and it may be a potential therapeutic target for HCC.
INTRODUCTION
Liver cancer is a common digestive tract cancer[1]. An epidemiological investigation reported 800 thousand new and dead liver cancer patients in 2018, and liver cancer was ranked the sixth most common cancer worldwide and the second major cause of cancer death in humans[2]. Hepatocellular carcinoma (HCC), the most common liver cancer, is accompanied by insidious clinical manifestations in the early stage; thus, most HCC patients are already in middle or advanced stages when admitted to hospital, and are unsuitable for surgical treatment, which seriously affects patients' quality of life[3]. At present, there are no good diagnostic markers for early HCC, and alpha fetoprotein (AFP) is the most widely used serological marker for HCC diagnosis; however, 30% of patients with HCC do not show increased AFP, and can even be negative, which increases the diagnosis difficulty of HCC[4]. Therefore, it is particularly important to identify a biomarker of HCC with high specificity.
Due to its prominent use in various disciplines, lncRNA has been a hot research topic in recent years[5]. lncRNAs are non-coding RNAs with lengths exceeding 200 nucleotides. Initially, scholars considered that lncRNAs lacked prominent protein coding ability, but recent studies found that lncRNAs have regulatory functions in a variety of mechanisms, including epigenetic modification, transcriptional regulation and post-transcriptional modification[6-8]. A previous study demonstrated that lncRNA ZEB1-AS1 is a potential prognostic indicator of HCC due to its differential expression in this tumor[9]. Another study found that lncRNA MALAT1 promotes the proliferation, migration and invasion of HCC by antagonizing miR-142-3p[10].
Cancer susceptibility candidate 9 (CASC9), located on human chromosome 8q21.13, is a member of the lncRNA family[11]. Previous studies revealed that CASC9 was differentially expressed in esophageal cancer and lung adenocarcinoma, and was closely related to biological functions such as proliferation, invasion and metastasis[12,13]. However, there are few studies on whether CASC9 is expressed in HCC and whether it has clinical diagnostic and prognostic value. Therefore, this study analyzed CASC9 expression in HCC based on the Cancer Genome Atlas (TCGA) database and clinical verification, in order to provide a potential index for clinical diagnosis.
MATERIALS AND METHODS
Acquisition and analysis of TCGA data
Data on gene expression in HCC were downloaded for analysis by logging into TCGA, selecting Access TCGA Data and entering the database. The data were acquired from the core sample database of TCGA, and sequencing and analysis were performed on the acquired data based on a standardized processing scheme (http://can-cergenome.nih.gov/cancergenomics/tissuesamples). The data included a total of 424 patients, involving 374 cancer samples and 50 control samples. Clinical data on HCC patients were acquired from http://gdac.broadinstitute.org for analysis, and excluded patients without detailed data and a survival time less than 30 d. Original data on CASC9 were processed through log (X+1, 2) to analyze the differences between control samples and carcinoma tissues, and a survival curve for high and low expression groups was drawn according to the median value of CASC9.
Bioinformatic analysis of lncRNA
Starbase 3.0 was adopted to predict lncRNA CASC9 targeted microRNA (miR), and miRDB, miRTarBase, and TargetScan were adopted to predict target genes of potential miR on-line. Cytoscape software was adopted to draw the competing endogenous (ce)RNA network, and David software to analyze target genes based on the Kyoto Encyclopedia of Genes and Genomes and Gene Ontology (GO) enrichment analysis.
Collection of patient samples
A total of 80 HCC patients (50 males and 30 females with an average age of 54.6 ± 5.0 years) treated in The First Affiliated Hospital of Guangxi Medical University from May 2012 to January 2014 were enrolled in the patient group, and 50 healthy subjects (29 males and 21 females with an average age of 53.7 ± 4.1 years) were enrolled in the control group. This study was approved by the First Affiliated Hospital of Guangxi Medical University Ethics Committee. The patients meeting the following criteria were included: Patients diagnosed with HCC based on imaging and pathologic biopsy; patients meeting the TNM staging criteria for HCC by the American Joint Committee on Cancer in 2009[14]; patients who had not taken part in any previous targeted tumor research (surgery, radiotherapy and chemotherapy, etc.), understood the study and signed an informed consent form (including their families) and patients whose expected survival time was more than 3 mo. The following patients were excluded: Patients with other combined tumors, renal function diseases, infection before admission, and those unwilling to cooperate.
Sample collection and detection
Peripheral venous blood (5 mL) was obtained from the patients, centrifuged at 3000 rpm for 10 min after 30 min and the supernatant collected. Total RNA in the collected supernatant was extracted with TRIzol reagent (Carlsbad Invitrogen Company, California, United States), and the purity, concentration and integrity of the total RNA were determined using ultraviolet spectrophotometry and agarose gel electrophoresis. Reverse transcription was performed using TransScript® miRNA RT Enzyme Mix and 2×TS miRNA Reaction Mix in the TransScript Green Two-Step quantitative real-time polymerase chain reaction SuperMix kit (Beijing TransGen Biotech Co., Ltd., China) in strict accordance with the original kit instructions. The amplification system of CASC9 consisted of 1 μL of cDNA, 0.4 μL of upstream and downstream primers, respectively, 10 μL of 2X TransScript® Tip Green qPCR SuperMix, Passive Reference Dye (50X), and Nuclease-free water (added to 20 μL in total). The amplification conditions were as follows: Pre-denaturation at 94°C for 30 s, denaturation at 94°C for 5 s and annealing extension at 60°C for 30 s, with 40 cycles in total. Triplicate wells were prepared for each sample, and the experiment was repeated three times. Glyceraldehyde-3-phosphate dehydrogenase was used as an internal reference, and 2-Δct was used to analyze the data. Experiments were carried out using a 7500 PCR instrument (ABI, United States).
Follow-up of patients
The patients were followed-up for 5 years by telephone interview and clinic reexamination. During the 1st year, the patients were followed-up at the 3rd, 6th, 9th and 12th month, respectively, and during the remaining 4 years, they were followed-up once every 4 mo.
Statistical analysis
In this study, the acquired data were statistically analyzed using the SPSS20.0 software package, and figures were drawn using the GraphPad 7 software package. The distribution of measurement data was analyzed using the K-S test, and the data in normal distribution were expressed as mean ± SD. Comparisons between groups were performed using independent-samples T test. Comparisons within groups were analyzed by paired t test, and expressed as t. Ranked data were analyzed using the rank sum test, and expressed as Z. Enumeration data were analyzed using the chi-square test. ROC curves of CASC9 to evaluate its diagnostic value in HCC were drawn, and a K-M curve for 5-year survival of patients was also drawn. The log-rank test was adopted for analysis. Multivariate Cox regression analysis was adopted to analyze independent risk factors for patients. P < 0.05 indicated a significant difference.
RESULTS
CASC9 expression in TCGA
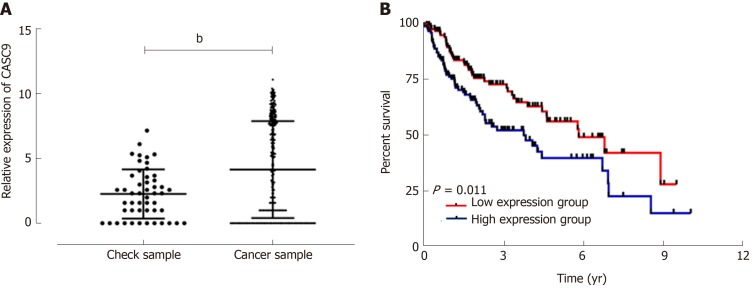
Analysis of data from TCGA database on CASC9 expression in HCC patients showed that control samples had significantly lower CASC9 expression than carcinoma tissue samples (P < 0.001), and the grouping of patients according to median CASC9 expression revealed that the low CASC9 expression group had a higher survival rate than the high CASC9 expression group (P = 0.011, Figure 1).
Figure 1.
Cancer susceptibility candidate 9 expression and survival from TCGA database. A: Cancer susceptibility candidate 9 (CASC9) was highly expressed in cancer patients; B: The survival rate of the high CASC9 expression group was lower than that of the low CASC9 expression group (P = 0.011). bP < 0.01. CASC9: Cancer susceptibility candidate 9.
Comparison of baseline data
Comparisons between the patient group and control group showed that there were no significant differences between the groups in terms of gender, age, body mass index (BMI), past medical history, smoking history, history of alcoholism and place of residence, while a difference in AFP expression was observed between the two groups (P < 0.001, Table 1).
Table 1.
Baseline analysis, n (%)
| Factors | Patient group (n = 80) | Control group (n = 50) | χ2 /t | P value | |
| Gender | 0.264 | 0.609 | |||
| Male | 50 (62.50) | 29(58.00) | |||
| Female | 30 (37.50) | 21 (42.00) | |||
| Age (yr) | 54.6 ± 5.0 | 53.7 ± 4.1 | 1.068 | 0.288 | |
| BMI (kg/m2) | 22.86 ± 1.93 | 23.17 ± 2.07 | 0.866 | 0.388 | |
| Past medical history | |||||
| Hypertension | 25 (31.25) | 10 (20.00) | 1.979 | 0.160 | |
| Hyperlipidemia | 13 (16.25) | 6 (12.00) | 0.445 | 0.505 | |
| Diabetes | 20 (25.00) | 10 (20.00) | 0.433 | 0.510 | |
| Smoking history | 0.081 | 0.776 | |||
| Yes | 50 (62.50) | 30 (60.00) | |||
| No | 30 (37.50) | 20 (40.00) | |||
| History of alcoholism | 0.494 | 0.482 | |||
| Yes | 15 (18.75) | 7 (14.00) | |||
| No | 65 (81.25) | 43 (86.00) | |||
| Place of residence | 0.177 | 0.674 | |||
| Urban area | 45 (56.25) | 30 (60.00) | |||
| Rural area | 35 (43.75) | 20 (40.00) | |||
| Tumor size | |||||
| ≥ 5 cm | 45 (56.25) | ||||
| < 5 cm | 35 (43.75) | ||||
| Combined hepatitis | |||||
| Yes | 70 (87.50) | ||||
| No | 10 (12.50) | ||||
| TNM staging | |||||
| Stage I+II | 33 (41.25) | ||||
| Stage III+IV | 47 (58.75) | ||||
| Lymph node metastasis | |||||
| Yes | 42 (52.50) | ||||
| No | 38 (47.50) | ||||
| Differentiation | |||||
| Low differentiation | 27 (33.75) | ||||
| Moderate + high differentiation | 53 (66.25) | ||||
| AFP (ng/mL) | 59.583 | < 0.001 | |||
| ≤ 200 | 25 (31.25) | 50 (100.00) | |||
| >200 | 55 (68.75) | 0 (0.00) |
BMI: Body mass index; AFP: Alpha fetoprotein; TNM: Tumor, node, metastasis.
CASC9 expression in patients and its clinical value
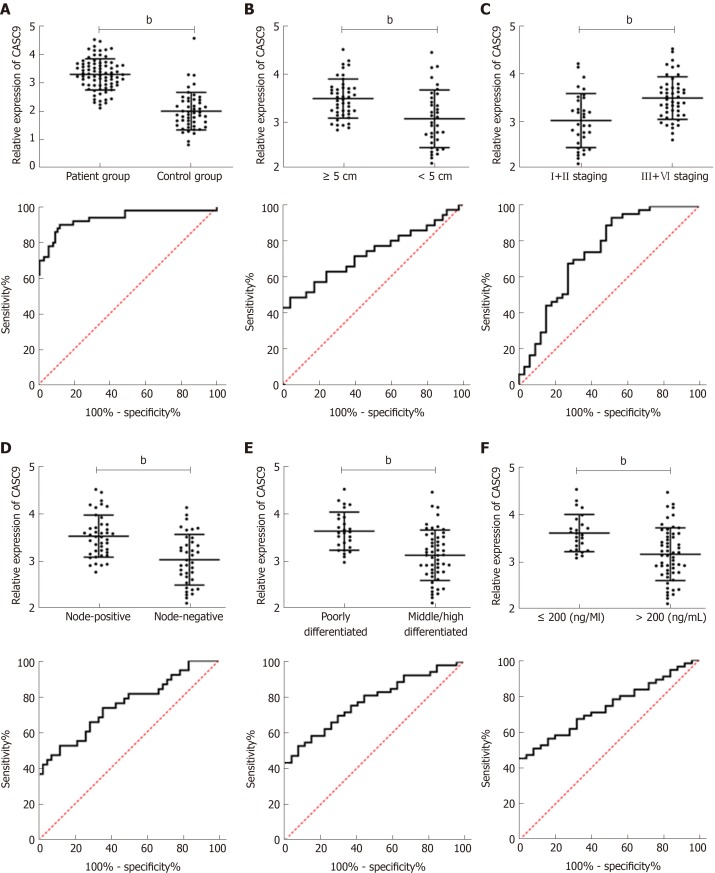
The expression of serum CASC9 in the patient group was significantly higher than that in the control group, and the ROC curve showed that the AUC was 0.933. Further analysis of the relationship between CASC9 and clinical pathological data showed that tumor size, combined hepatitis, TNM staging, lymph node metastasis, differentiation and AFP were closely related to CASC9 expression. In addition, the analysis of ROC curves showed that CASC9 was associated with tumor size, TNM staging, lymph node metastasis, differentiation and AFP (Figure 2 and Tables 2 and 3).
Figure 2.
Cancer susceptibility candidate 9 expression in patients, its relationship with tumor size, Tumor, Node, Metastasis staging, lymph node metastasis, differentiation, and alpha fetoprotein expression, and its diagnostic value. A: Cancer susceptibility candidate 9 (CASC9) was highly expressed in the patient group, and the AUC was 0.933; B: Relationship between CASC9 expression and tumor size. The AUC was 0.726; C: Relationship between CASC9 expression and TNM staging. The AUC was 0.743; D: Relationship between CASC9 expression and lymph node metastasis. The AUC was 0.752; E: Relationship between CASC9 expression and differentiation. The AUC was 0.777; F: Relationship between CASC9 expression and AFP. The AUC was 0.738. bP < 0.01. CASC9: Cancer susceptibility candidate 9; AUC: area under the curve.
Table 2.
Relationship between cancer susceptibility candidate 9 and pathological data
| Factors | Patient group (n = 80) | T | P value | |
| Gender | 0.308 | 0.759 | ||
| Male (n = 50) | 3.287 ± 0.553 | |||
| Female (n = 30) | 3.326 ± 0.553 | |||
| Age (yr) | 0751 | 0.455 | ||
| < 55 (n = 35) | 3.354 ± 0.504 | |||
| ≥ 55 (n = 45) | 3.261 ± 0.585 | |||
| Tumor size | 4.357 | < 0.001 | ||
| ≥ 5 cm (n = 45) | 3.489 ± 0.414 | |||
| < 5 cm (n = 35) | 3.06 ± 0.611 | |||
| Combined hepatitis | 0.575 | 0.567 | ||
| Yes (n = 70) | 3.315 ± 0.540 | |||
| No (n = 10) | 3.208 ± 0.634 | |||
| TNM stage | 4.157 | < 0.001 | ||
| Stage I+II (n = 33) | 3.496 ± 0.448 | |||
| Stage III+IV (n = 47) | 3.024 ± 0.567 | |||
| Lymph node metastasis | 4.557 | < 0.001 | ||
| Yes (n = 42) | 3.54 ± 0.445 | |||
| No (n = 38) | 3.038 ± 0.538 | |||
| Differentiation | 4.387 | < 0.001 | ||
| Low differentiation (n = 27) | 3.642 ± 0.405 | |||
| Moderate + high differentiation (n = 53) | 3.128 ± 0.535 | |||
| AFP (ng/mL) | 3.617 | 0.001 | ||
| ≤ 200 (n = 25) | 3.609 ± 0.393 | |||
| > 200 (n = 55) | 3.162 ± 0.557 |
AFP: Alpha fetoprotein; TNM: Tumor, node, metastasis.
Table 3.
Receiver operating characteristic parameters
| Factors | HCC diagnosis | Tumor size | TNM stage | Lymph node metastasis | Differentiation | AFP |
| AUC | 0.933 | 0.726 | 0.743 | 0.752 | 0.777 | 0.738 |
| SD | 0.026 | 0.060 | 0.059 | 0.055 | 0.051 | 0.054 |
| 95%CI | 0.882-0.983 | 0.608-0.845 | 0.628-0.858 | 0.645-0.859 | 0.678-0.876 | 0.632-0.844 |
| P value | < 0.001 | 0.001 | < 0.001 | < 0.001 | < 0.001 | 0.001 |
| Specificity | 87.50% | 95.56% | 48.48% | 88.10% | 92.59% | 100.00% |
| Sensitivity | 90.00% | 48.57% | 93.62% | 52.63% | 52.83% | 45.45% |
| Youden index | 77.50% | 44.13% | 42.10% | 40.73% | 45.42% | 45.45% |
| Cut-off | < 2.672 | < 2.916 | > 2.919 | < 3.057 | < 3.148 | < 3.057 |
AUC: Area under curve; HCC: Hepatocellular carcinoma; AFP: Alpha fetoprotein.
Relationship between CASC9 and patients' survival
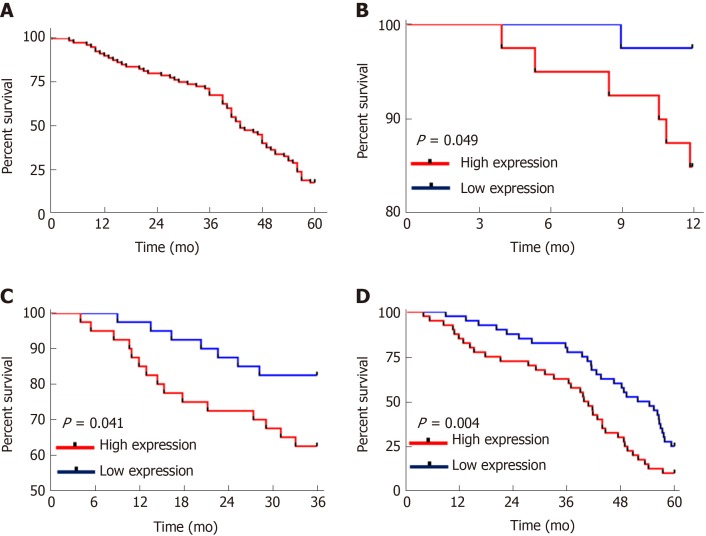
All the patients were successfully followed-up for 5 years in terms of survival. The patient group showed a 5-year survival rate of 17.50% with 14 patients surviving. The patients were grouped into the high CASC9 expression group and the low CASC9 expression group according to the median CASC9 expression (3.305), and survival was compared. The high CASC9 expression group showed significantly lower 1-year, 3-year and 5-year survival rates than the low CASC9 expression group (all P < 0.05, Figure 3).
Figure 3.
Relationship between cancer susceptibility candidate 9 and patients’ survival. A: The overall survival of patients; B: The 1-year survival of patients in the high and low Cancer susceptibility candidate 9 (CASC9) expression groups (P = 0.049); C: The 3-year survival of patients in the high and low CASC9 expression groups (P = 0.041); D: The 5-year survival of patients in the high and low CASC9 expression groups (P = 0.004). CASC9: Cancer susceptibility candidate 9.
Independent factors affecting prognosis of patients
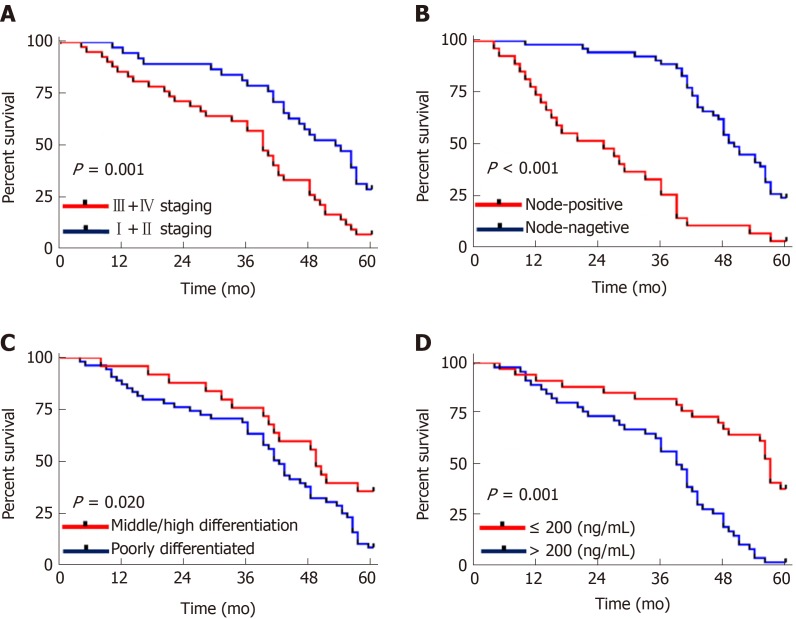
Univariate Cox regression analysis of the pathological data showed that TNM staging, lymph node metastasis, differentiation, AFP and CASC9 were factors which affected the prognosis of patients, and multivariate Cox regression analysis of these factors showed that TNM staging, lymph node metastasis, differentiation, AFP, and CASC9 were independent factors affecting the prognosis of patients (Table 4). In addition, the survival curves in terms of TNM staging, lymph node metastasis, differentiation, AFP and patients' 5-year survival indicated that stage I+II patients with lymph node metastasis, low differentiation, and AFP > 200 ng/mL showed poor 5-year survival (Figure 4).
Table 4.
Multivariate Cox regression analysis
| Factors |
Univariate Cox |
Multivariate Cox |
||||
| P value | HR | 95%CI | P value | HR | 95%CI | |
| Gender (male vs female) | 0.272 | 1.320 | 1.320-0.804 | |||
| Age (< 55 yr vs ≥ 55 yr) | 0.438 | 0.825 | 0.825-0.508 | |||
| Tumor size (≥ 5 cm vs < 5 cm) | 0.952 | 1.015 | 1.015-0.624 | |||
| Combined hepatitis (yes vs no) | 0.636 | 1.185 | 1.185-0.586 | |||
| TNM staging (stage I+II vs stage III+IV) | 0.000 | 4.271 | 4.271-2.391 | 0.006 | 2.501 | 1.308-4.781 |
| Lymph node metastasis (yes vs no) | 0.001 | 0.428 | 0.428-0.259 | 0.025 | 0.535 | 0.309-0.924 |
| Differentiation (low vs medium + high) | 0.000 | 0.242 | 0.242-0.144 | 0.000 | 0.326 | 0.186-0.569 |
| AFP (≤ 200 vs > 200 ng/mL) | 0.025 | 1.914 | 1.914-1.086 | 0.042 | 1.935 | 1.023-3.662 |
| CASC9 (< 3.305 vs ≥ 3.305) | 0.005 | 2.023 | 2.023-1.235 | 0.021 | 2.024 | 1.112-3.682 |
AFP: Alpha fetoprotein; TNM: Tumor, node, metastasis; CASC9: Cancer susceptibility candidate 9.
Figure 4.
Relationship between tumor, node, metastasis staging, lymph node metastasis, differentiation, alpha fetoprotein and 5-year survival of patients. A: The 5-year survival rate of stage I+II patients was higher than that of stage III+IV patients (P = 0.001); B: The 5-year survival rate of patients without lymph node metastasis was higher than that of patients with lymph node metastasis (cP < 0.001); C: The 5-year survival rate of patients with moderate/high differentiation was higher than that of patients with low differentiation (P = 0.020); D: The 5-year survival rate of patients with AFP ≤ 200 was higher than that of patients with AFP > 200 (P = 0.001).
Bioinformatic analysis
Seventeen CASC9 potential targeted miRs were identified by Starbase 3.0, and 89 mRNAs were found by predicting the downstream mRNA of the 17 targeted miRs with miRDB, miRTarBase, and TargetScan. We used Cytoscape software to construct the interaction map between 1ncRNA-miRNA-mRNAs, and GO enrichment and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis were performed on the 89 mRNAs in the ceRNA network using DAVID and KOBAS, and 18 GO functions with P < 0.05 and 8 signal transduction pathways with P < 0.05 were found (Tables 5 and 6).
Table 5.
The top 10 Gene Ontology enrichment functions
| Term | Count | P value | Genes |
| Negative regulation of transcription from the RNA polymerase II promoter | 9 | 0.001 | PHF19, SQSTM1, CPEB3, E2F7, ESR1, CBX2, SOX6, HMGA2, TWIST1 |
| Positive regulation of transcription regulatory region DNA binding | 3 | 0.002 | WNT3A, HMGA2, TWIST1 |
| Cytoplasm | 27 | 0.003 | CLSPN, CPEB3, TPM2, BDNF, RNF165, MAPT, STRIP2, HOXA10, FASN, PLCB1, CDC37L1, ARL2, IRAK1, SGK1, MAP2K1, KIF5A, DDX39B, PIM1, SOCS6, ESR1, SNAI2, WEE1, CDC25A, ADM, CA8, PSAT1, DUSP6 |
| Positive regulation of transcription, DNA-templated | 6 | 0.009 | RET, MAP2K1, FOXK1, MYRF, ESR1, PLCB1 |
| Osteoblast differentiation | 4 | 0.010 | WNT3A, FASN, SNAI2, TWIST1 |
| Neuron projection morphogenesis | 3 | 0.011 | BDNF, SGK1, WEE1 |
| Transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific | 5 | 0.019 | PLAG1, HOXA10, ESR1, HMGA2, MYBL2 |
| Negative regulation of sequence-specific DNA binding transcription factor activity | 3 | 0.021 | PIM1, ESR1, TWIST1 |
| Neuron projection | 4 | 0.027 | CPEB3, KIF5A, MAPT, SLC6A4 |
PHF19: PHD finger protein 19; SQSTM1: Sequestosome 1; CPEB3: Cytoplasmic polyadenylation element binding protein 3; E2F7: E2F transcription factor 7; ESR1: Estrogen receptor 1; CBX2: Chromobox 2; SOX6: SRY-box transcription factor 6; HMGA2: High mobility group AT-hook 2; TWIST1: Twist family bHLH transcription factor 1; WNT3A: Wnt family member 3A; CLSPN: Claspin; TPM2: Tropomyosin 2; BDNF: Brain derived neurotrophic factor; RNF165: Ring finger protein 165; MAPT: Microtubule associated protein tau; STRIP2: Striatin interacting protein 2; HOXA10: Homeobox A10; FASN: Fatty acid synthase; PLCB1: Phospholipase C beta 1; CDC37L1: Cell division cycle 37 like 1; ARL2: ADP ribosylation factor like GTPase 2; IRAK1: Interleukin 1 receptor associated kinase 1; SGK1: Serum/glucocorticoid regulated kinase 1; MAP2K1: Mitogen-activated protein kinase kinase 1; KIF5A: Kinesin family member 5A; DDX39B: DExD-box helicase 39B; PIM1: Pim-1 proto-oncogene, serine/threonine kinase; SOCS6: Suppressor of cytokine signaling 6; SNAI2: Snail family transcriptional repressor 2; WEE1: WEE1 G2 checkpoint kinase; CDC25A: Cell division cycle 25A; ADM: Adrenomedullin; CA8: Carbonic anhydrase 8; PSAT1: Phosphoserine aminotransferase 1; DUSP6: Dual specificity phosphatase 6; RET: Ret proto-oncogene; FOXK1: Forkhead box K1; MYRF: Myelin regulatory factor; PLAG1: PLAG1 zinc finger; MYBL2: MYB proto-oncogene like 2; SLC6A4: Solute carrier family 6 member 4.
Table 6.
The top10 Kyoto Encyclopedia of Genes and Genomes signal pathways
| Term | Count | P value | Genes |
| MicroRNAs in cancer | 7 | 0.002 | KIF23, CCNE1, MAP2K1, WNT3A, PIM1, HMGA2, CDC25A |
| Pathways in cancer | 8 | 0.005 | CCNE1, RET, MAP2K1, WNT3A, ITGA2, LAMC1, AXIN2, PLCB1 |
| p53 signaling pathway | 4 | 0.007 | STEAP3, CCNE1, SERPINE1, CHEK1 |
| Chagas disease (American trypanosomiasis) | 4 | 0.024 | GNAL, IRAK1, SERPINE1, PLCB1 |
| Proteoglycans in cancer | 5 | 0.024 | MAP2K1, WNT3A, ESR1, ITGA2, TWIST1 |
| Cell cycle | 4 | 0.030 | CCNE1, CHEK1, WEE1, CDC25A |
| PI3K-Akt signaling pathway | 6 | 0.038 | CCNE1, SGK1, MAP2K1, ITGA2, LAMC1, ANGPT2 |
| Hippo signaling pathway | 4 | 0.048 | WNT3A, SERPINE1, AXIN2, SNAI2 |
| Small cell lung cancer | 3 | 0.081 | CCNE1, ITGA2, LAMC1 |
| Progesterone-mediated oocyte maturation | 3 | 0.082 | MAP2K1, CPEB3, CDC25A |
KIF23: Kinesin family member 23; CCNE1: Cyclin E1; MAP2K1: Mitogen-activated protein kinase kinase 1; WNT3A: Wnt family member 3A; PIM1: Pim-1 proto-oncogene, serine/threonine kinase; HMGA2: High mobility group AT-hook 2; CDC25A: Cell division cycle 25A; RET: Ret proto-oncogene; ITGA2: Integrin subunit alpha 2; LAMC1: Laminin subunit gamma 1; AXIN2: Axin 2; PLCB1: Phospholipase C beta 1; STEAP3: STEAP3 metalloreductase; SERPINE1: Serpin family E member 1; CHEK1: Checkpoint kinase 1; CPEB3: Cytoplasmic polyadenylation element binding protein 3; SGK1: Serum/glucocorticoid regulated kinase 1; ANGPT2: Angiopoietin 2; WEE1: WEE1 G2 checkpoint kinase; ESR1: Estrogen receptor 1; TWIST1: Twist family bHLH transcription factor 1; GNAL: G protein subunit alpha L; IRAK1: Interleukin 1 receptor associated kinase 1; SNAI2: Snail family transcriptional repressor 2.
DISCUSSION
HCC, a digestive system neoplasm with high incidence and mortality, is a common disease in male patients, and morbidity in men compared to women is 2.81:1[15]. A Chinese tumor epidemiology survey reported that more than 400 thousand new and dead patients with HCC were observed in 2015; however, there is no effective diagnosis and treatment plan for this disease[16]. At present, AFP is the main clinical serodiagnostic index for HCC, but relevant studies have found that AFP expression increases in liver diseases such as hepatitis; thus, its specificity is low[17]. Therefore, the key to resolving the problem is to identify biological indices with high sensitivity and specificity.
LncRNA is a long-chain non-coding RNA. A previous study demonstrated that lncRNAs participate in the occurrence and development of various cancers[18]. A study by Ma et al[19] found that the biological function of HCC was inhibited by regulating the expression of miR-122-5p in HCC based on lncRNA ANRIL knockout. CASC9 is a newly discovered tumor susceptibility gene. A study by Luo et al[20] found that CASC9 and CPSF3 could cooperatively regulate TGF-β signaling pathway conduction in colorectal cancer, and a study by Liang et al[21] reported that CASC9 promoted metastasis of esophageal squamous cell carcinoma by up regulating LAMC2 expression through an interaction with CREB binding protein. However, there are few reports on CASC9 in HCC at present. A study by Noh et al[22] showed that CASC9-mediated AKT ions promoted the survival of HCC cells, and there are no other relevant studies on whether CASC9 can be used as a diagnostic and prognostic indicator of HCC. Therefore, this study confirmed the expression and prognostic value of CASC9 in HCC based on the TCGA database, in order to provide a new potential clinical index.
TCGA database, one of the largest Cancer Genome Projects, contains a variety of tumor gene data[23]. In this study, we first extracted data on CASC9 expression in the tissues of HCC patients from TCGA database, analyzed these tissues, and found that CASC9 expression in carcinoma tissue samples was significantly higher than that in adjacent control tissue samples. The patients were divided into the high CASC9 expression group and the low CASC9 expression group according to the median CASC9 expression to determine survival of the patients. It was found that the low CASC9 expression group had a higher survival rate than the high CASC9 expression group. The above results indicated that CASC9 may be a potential diagnostic and prognostic indicator of HCC. Further clinical research demonstrated that the expression of serum CASC9 in HCC patients was consistent with the data from TCGA database. In addition, we found that the AUC of CASC9 expression (> 0.9) had very high diagnostic value based on ROC curves. We further analyzed the relationship between CASC9 and 1-year, 3-year and 5-year survival rates in HCC patients, and differences between the low CASC9 expression group and high CASC9 expression group in 1-year, 3-year and 5-year survival rates were found, which indicated that CASC9 could be adopted as an index for determining the short-term and long-term survival rates of HCC patients. We also analyzed the relationship between CASC9 and pathological data, and found that CASC9 was closely related to tumor size, TNM staging, lymph node metastasis, differentiation, and AFP, which indicated that CASC9 was closely related to the occurrence and development of HCC. In addition, CASC9 had certain diagnostic value in determining tumor size, TNM staging, lymph node metastasis, differentiation and AFP. A study by Gao et al[24] revealed that CASC9 was also highly expressed in esophageal carcinoma, and had relatively high diagnostic value (AUC: 0.814). However, the AUC of CASC9 in our study group was 0.933. This indicated that CASC9 may have higher diagnostic value for HCC than for esophageal carcinoma, but whether it is a diagnostic index still requires further investigation.
Pathological data on HCC patients were obtained to further analyze independent factors affecting the prognosis of patients, and TNM staging, lymph node metastasis, differentiation, AFP and CASC9 were found to be independent prognostic factors for HCC patients based on multivariate analysis. A study by Tan and Huang confirmed that TNM staging, lymph node metastasis, differentiation, and AFP were closely related to the prognosis of patients, which was consistent with the findings in our study[25,26]. However, we found that tumor size was not related to prognosis in this study, which was inconsistent with the previous study findings. We speculated that this may be related to the collected samples. The tumor size data in this study were relatively intensively distributed with an average size of 4.8 ± 0.8 cm, which may be the reason for this difference. Our study confirmed, for the first time, that CASC9 was an independent prognostic factor for HCC patients, indicating that CASC9 may be a prognostic and diagnostic index for HCC.
A co-expression network of lncRNA-miR-mRNA was constructed and 17 CASC9 targeted miRs, and 89 targeted mRNAs were observed. The top 10 functions in GO enrichment analysis were negative regulation of transcription from the RNA polymerase II promoter, positive regulation of transcription regulatory region DNA binding, cytoplasm, positive regulation of transcription, DNA-templated, osteoblast differentiation, neuron projection morphogenesis, transcriptional activator activity, RNA polymerase II core promoter proximal region sequence-specific, negative regulation of sequence-specific DNA binding transcription factor activity, and neuron projection, respectively, and the 8 different signal transduction pathways in the Kyoto Encyclopedia of Genes and Genomes analysis were microRNAs in cancer, pathways in cancer, p53 signaling pathway, Chagas disease (American trypanosomiasis), proteoglycans in cancer, cell cycle, PI3K-Akt signaling pathway, and Hippo signaling pathway, respectively. According to the above research, CASC9 can participate in various biological pathways by regulating the miR/mRNA axis, and it is noteworthy that signal pathways such as microRNAs in cancer, pathways in cancer, p53 signaling pathway, PI3K-Akt signaling pathway, and Hippo signaling pathway are important cancer-related signal transduction pathways[27-29]. CASC9 can also participate in the occurrence of these pathways by regulating the miR/mRNA axis, which provides an important cornerstone for our subsequent research.
Although this study confirmed the value of CASC9 in HCC, it still has some limitations. Firstly, we only analyzed patients with HCC and healthy subjects, but did not determine CASC9 expression in patients with hepatitis; thus, whether CASC9 can distinguish HCC from hepatitis still requires further research in healthy subjects. Secondly, how CASC9 participates in the occurrence and development of HCC is unclear. Lastly, this study only focused on Chinese patients, and whether CASC9 expression is increased in HCC patients of different races is unclear. Therefore, we hope to carry out bioinformatics and a basic study in the future to further analyze the way that CASC9 affects the occurrence and development of HCC, and obtain different pathological specimens for CASC9 detection in difference races, in order to address the shortcomings in this study. In conclusion, high CASC9 expression is beneficial for the prognosis of HCC patients, and CASC9 is expected to be a potential diagnostic and prognostic indicator of HCC.
ARTICLE HIGHLIGHTS
Research background
Liver cancer, the sixth most common cancer worldwide, is the second leading cause of cancer mortality. A lncRNA is a non-coding RNA with a length exceeding 200 nucleotides. Previous studies have found that lncRNAs are involved in the development and progression of hepatocellular carcinoma (HCC), but whether they can be used as potential diagnostic and prognostic indicators is unclear.
Research motivation
LncRNA is a newly discovered non-coding RNA. Some studies have revealed that lncRNAs are differentially expressed in HCC and are expected to be measures of potential outcome. In this study, we found that lncRNA CASC9 was highly expressed in HCC patients based on the Cancer Genome Atlas (TCGA) database analysis and patients with high expression of CASC9 had a poor prognosis, which indicated that lncRNA CASC9 may be a potential diagnostic and prognostic indicator of HCC.
Research objectives
This study aimed to identify the expression of lncRNA CASC9 in HCC, its diagnostic and prognostic value and to construct ceRNA network maps to further explore its underlying mechanism.
Research methods
Data on the expression of lncRNA CASC9 in TCGA were extracted, clinical samples were collected to further determine the expression of lncRNA CASC9 in HCC, and the correlation between lncRNA CASC9 and pathological data and survival of HCC patients were analyzed. Potential microRNA and target genes of lncRNA CASC9 were analyzed using on-line prediction websites, and ceRNA maps were drawn. In addition, Kyoto Encyclopedia of Genes and Genomes and GO enrichment analysis were employed to analyze the potential mechanisms of lncRNA CASC9 in biological processes.
Research results
Analysis revealed that lncRNA CASC9 was highly expressed in the tissues and serum of HCC patients and high expression of CASC9 was related to tumor size, TNM staging, lymph node metastasis, differentiation, and AFP. Further analysis showed that lncRNA CASC9 could be used as a diagnostic indicator of the above indices. Prognostic analysis revealed that the survival rate of patients with high expression of lncRNA CASC9 decreased, and Cox regression analysis showed that lncRNA CASC9 could be used as an independent prognostic indicator in HCC patients. The bioinformatics analysis revealed that lncRNA CASC9 potentially targeted 17 miRs. A total of 89 mRNAs were found in mRNA prediction. GO enrichment and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis revealed that lncRNA CASC9 participated in 18 GO functions and 8 signal transduction pathways.
Research conclusion
LncRNA CASC9 is highly expressed in HCC patients, and may be a potential diagnostic and prognostic indicator of HCC.
Research perspectives
It is necessary to further explore the value of lncRNA CASC9 in HCC, and prospective experiments and multi-center clinical studies are required to obtain more robust conclusions. In addition, we hope to further verify the relevant mechanisms of lncRNA CASC9 in HCC by carrying out relevant basic experiments, in order to address the deficiencies in this study.
Footnotes
Institutional review board statement: This study was reviewed and approved by the First Affiliated Hospital of Guangxi Medical University Ethics Committee.
Informed consent statement: All patients in our study provided informed consent.
Conflict-of-interest statement: The authors declare no conflict of interest.
STROBE statement: The authors have read the STROBE Statement-checklist of items, and the manuscript was prepared and revised according to the STROBE Statement-checklist of items.
Peer-review started: September 29, 2019
First decision: November 10, 2019
Article in press: December 13, 2019
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Aykan NF, Hann HW S-Editor: Wang J L-Editor: Webster JR E-Editor: Ma YJ
Contributor Information
Yong-Lian Zeng, Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning 530021, Guangxi, China.
Zhen-Ya Guo, Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning 530021, Guangxi, China.
Hui-Zhao Su, Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning 530021, Guangxi, China.
Fu-Di Zhong, Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning 530021, Guangxi, China.
Ke-Qing Jiang, Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning 530021, Guangxi, China.
Guan-Dou Yuan, Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning 530021, Guangxi, China. yuan25958282@163.com.
Data sharing statement
No additional data are available.
References
- 1.Ryerson AB, Eheman CR, Altekruse SF, Ward JW, Jemal A, Sherman RL, Henley SJ, Holtzman D, Lake A, Noone AM, Anderson RN, Ma J, Ly KN, Cronin KA, Penberthy L, Kohler BA. Annual Report to the Nation on the Status of Cancer, 1975-2012, featuring the increasing incidence of liver cancer. Cancer. 2016;122:1312–1337. doi: 10.1002/cncr.29936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Zhao X, Chen Q, Liu W, Li Y, Tang H, Liu X, Yang X. Codelivery of doxorubicin and curcumin with lipid nanoparticles results in improved efficacy of chemotherapy in liver cancer. Int J Nanomedicine. 2015;10:257–270. doi: 10.2147/IJN.S73322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sauzay C, Petit A, Bourgeois AM, Barbare JC, Chauffert B, Galmiche A, Houessinon A. Alpha-foetoprotein (AFP): A multi-purpose marker in hepatocellular carcinoma. Clin Chim Acta. 2016;463:39–44. doi: 10.1016/j.cca.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Palazzo AF, Lee ES. Non-coding RNA: what is functional and what is junk? Front Genet. 2015;6:2. doi: 10.3389/fgene.2015.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Sun Y, Cai R, Wang G, Shu X, Pang W. Long noncoding RNA: multiple players in gene expression. BMB Rep. 2018;51:280–289. doi: 10.5483/BMBRep.2018.51.6.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiu HS, Somvanshi S, Patel E, Chen TW, Singh VP, Zorman B, Patil SL, Pan Y, Chatterjee SS Cancer Genome Atlas Research Network, Sood AK, Gunaratne PH, Sumazin P. Pan-Cancer Analysis of lncRNA Regulation Supports Their Targeting of Cancer Genes in Each Tumor Context. Cell Rep. 2018;23:297–312.e12. doi: 10.1016/j.celrep.2018.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klingenberg M, Matsuda A, Diederichs S, Patel T. Non-coding RNA in hepatocellular carcinoma: Mechanisms, biomarkers and therapeutic targets. J Hepatol. 2017;67:603–618. doi: 10.1016/j.jhep.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Li T, Xie J, Shen C, Cheng D, Shi Y, Wu Z, Deng X, Chen H, Shen B, Peng C, Li H, Zhan Q, Zhu Z. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35:1575–1584. doi: 10.1038/onc.2015.223. [DOI] [PubMed] [Google Scholar]
- 10.Yu Q, Xiang L, Chen Z, Liu X, Ou H, Zhou J, Yang D. MALAT1 functions as a competing endogenous RNA to regulate SMAD5 expression by acting as a sponge for miR-142-3p in hepatocellular carcinoma. Cell Biosci. 2019;9:39. doi: 10.1186/s13578-019-0299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shang C, Sun L, Zhang J, Zhao B, Chen X, Xu H, Huang B. Silence of cancer susceptibility candidate 9 inhibits gastric cancer and reverses chemoresistance. Oncotarget. 2017;8:15393–15398. doi: 10.18632/oncotarget.14871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan Z, Mao W, Bao Y, Zhang M, Su X, Xu X. The long noncoding RNA CASC9 regulates migration and invasion in esophageal cancer. Cancer Med. 2016;5:2442–2447. doi: 10.1002/cam4.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, Xiao H, Yang X, Tian H, Xu Z, Zhong Y, Ma L, Zhang W, Qiao G, Liang J. Long noncoding RNA CASC9.5 promotes the proliferation and metastasis of lung adenocarcinoma. Sci Rep. 2018;8:37. doi: 10.1038/s41598-017-18280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kee KM, Wang JH, Lin CY, Wang CC, Cheng YF, Lu SN. Validation of the 7th edition TNM staging system for hepatocellular carcinoma: an analysis of 8,828 patients in a single medical center. Dig Dis Sci. 2013;58:2721–2728. doi: 10.1007/s10620-013-2716-8. [DOI] [PubMed] [Google Scholar]
- 15.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 17.Shu H, Li W, Shang S, Qin X, Zhang S, Liu Y. Diagnosis of AFP-negative early-stage hepatocellular carcinoma using Fuc-PON1. Discov Med. 2017;23:163–168. [PubMed] [Google Scholar]
- 18.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma J, Li T, Han X, Yuan H. Knockdown of LncRNA ANRIL suppresses cell proliferation, metastasis, and invasion via regulating miR-122-5p expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2018;144:205–214. doi: 10.1007/s00432-017-2543-y. [DOI] [PubMed] [Google Scholar]
- 20.Luo K, Geng J, Zhang Q, Xu Y, Zhou X, Huang Z, Shi KQ, Pan C, Wu J. LncRNA CASC9 interacts with CPSF3 to regulate TGF-β signaling in colorectal cancer. J Exp Clin Cancer Res. 2019;38:249. doi: 10.1186/s13046-019-1263-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liang Y, Chen X, Wu Y, Li J, Zhang S, Wang K, Guan X, Yang K, Bai Y. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25:1980–1995. doi: 10.1038/s41418-018-0084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noh JH, Gorospe M. AKTions by Cytoplasmic lncRNA CASC9 Promote Hepatocellular Carcinoma Survival. Hepatology. 2018;68:1675–1677. doi: 10.1002/hep.30165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Computer Science. 2016;2:e67. [Google Scholar]
- 24.Gao GD, Liu XY, Lin Y, Liu HF, Zhang GJ. LncRNA CASC9 promotes tumorigenesis by affecting EMT and predicts poor prognosis in esophageal squamous cell cancer. Eur Rev Med Pharmacol Sci. 2018;22:422–429. doi: 10.26355/eurrev_201801_14191. [DOI] [PubMed] [Google Scholar]
- 25.Tan YL, Bai ZG, Zou WL, Ma XM, Wang TT, Guo W, Liu J, Li JS, Jie-Yin, Zang YJ, Zhang ZT. miR-744 is a potential prognostic marker in patients with hepatocellular carcinoma. Clin Res Hepatol Gastroenterol. 2015;39:359–365. doi: 10.1016/j.clinre.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 26.Huang CS, Yu W, Cui H, Wang YJ, Zhang L, Han F, Huang T. Increased expression of miR-21 predicts poor prognosis in patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:7234–7238. [PMC free article] [PubMed] [Google Scholar]
- 27.Garzon R, Calin GA, Croce CM. MicroRNAs in Cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- 28.Mello SS, Attardi LD. Deciphering p53 signaling in tumor suppression. Curr Opin Cell Biol. 2018;51:65–72. doi: 10.1016/j.ceb.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, Wu C, Chen N, Gu H, Yen A, Cao L, Wang E, Wang L. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget. 2016;7:33440–33450. doi: 10.18632/oncotarget.7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No additional data are available.