Abstract
Background
Distinction in the mutational profile between the common histological types, lung adenocarcinoma (LUAD) and squamous cell lung carcinoma (LUSC) has been well‐established. However, comprehensive mutation profiles of the predominant histological subtypes within LUAD and LUSC remains elusive.
Methods
We analyzed the mutational profile of 318 Chinese NSCLC patients of adenocarcinoma and squamous cell carcinoma predominant subtypes from seven hospitals using capture‐based ultra‐deep sequencing of 68 lung cancer‐related genes.
Results
Of the 318 NSCLC patients, 215 were diagnosed with LUAD and 103 with LUSC. Adenocarcinoma in situ and acinar adenocarcinoma were the most predominant subtypes of LUAD. On the other hand, keratinizing squamous cell carcinoma was the most predominant subtype of LUSC. Among the LUAD subtypes, EGFR sensitizing mutations were most prevalent in the invasive lepidic subtype. More than half of the patients with preinvasive adenocarcinoma in situ, minimally invasive, acinar, micropapillary and papillary subtypes were also EGFR‐mutants. Patients with colloidal, invasive mucinous, and fetal subtypes had the least number of EGFR mutations. Moreover, KRAS mutations were prevalent in patients with invasive mucinous, colloid, enteric and solid subtypes. A total of 90% of the LUSC patients harbor mutations in TP53, wherein all patients except five with nonkeratinizing were TP53 mutants. PIK3CA amplifications were most prevalent in keratinizing, followed by basaloid and nonkeratinizing subtypes.
Conclusion
These data suggest that the mutational profiles among the predominant histological subtypes were very distinct, which provided a reliable tool to improve treatment decisions.
Keywords: Adenocarcinoma, histological subtyping, molecular profiling, non‐small cell lung cancer, squamous cell carcinoma
Keypoints
Mutation profiling can aid in distinguishing between the histological subtypes to improve treatment decisions.
Lung adenocarcinoma and squamous cell lung carcinoma are molecularly distinct.
The most predominant lung adenocarcinoma subtypes were adenocarcinoma in situ and acinar adenocarcinoma. The most predominant squamous cell lung carcinoma subtype was keratinizing squamous cell carcinoma.
Introduction
Lung cancer is the leading cause of cancer‐related death in China and around the world.1, 2 Non‐small cell lung cancer (NSCLC) accounts for about 85% of lung cancer cases diagnosed, with two major histological types: adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC) accounting for nearly 50% and 30% of NSCLC, respectively.3 Historically, the histological classification of NSCLC had not been a major determining factor in treatment guidance.4 It is only in the past decade that it has become apparent that LUAD and LUSC have distinctive mutation profiles, responsible for their divergent responses to targeted therapies. Development of targeted therapies such as EGFR‐TKIs has revolutionized the management of EGFR‐mutant LUAD patients.5, 6 Furthermore, pemetrexed7 and bevacizumab8 were approved for nonsquamous NSCLC. On the other hand, nivolumab has recently been approved by the U.S. Food and Drug Administration for metastatic LUSC patients.9 Due to the development of therapeutic agents approved only for particular histological types, the need for histopathological classification grew significantly over the years.
Conventionally, histological classification of NSCLC primarily relied on morphology using light microscopy with hematoxylin‐eosin and mucin stains. However, certain cytology samples obtained from small biopsies are morphologically indistinguishable such as poorly differentiated adenocarcinoma and squamous cell.10 Immunohistochemistry (IHC) markers were introduced in the diagnosis of NSCLC to improve accuracy and reproducibility of histological classification and are now widely used in the subtyping of NSCLC. In line with the growing need for histologic classification of NSCLC, the World Health Organization (WHO) recently revised the guidelines for the classification of lung tumors. Some of the amendments included the emphasis of histology to personalized medicine and modification of the histologic criteria and classifications for both LUAD and LUSC subtypes following the recommendations from the Association for the Study of Lung Cancer, American Thoracic Society and European Respiratory Society (ASLC/ATS/ERS).4 The reclassification in the LUAD and LUSC subtypes were according to the predominant morphologic pattern as well as the general pattern of invasion. LUAD with predominantly lepidic, nonmucinous pattern is characterized as preinvasive adenocarcinoma in situ, minimally invasive adenocarcinoma, or invasive adenocarcinoma with lepidic component depending on the invasion pattern; while other invasive LUAD with identifiable patterns are classified as invasive mucinous, colloid, fetal, enteric, acinar, papillary, micropapillary, and solid subtypes.4 On the other hand, LUSC is further classified as preinvasive squamous cell carcinoma in situ, and invasive squamous cell carcinoma as keratinizing, nonkeratinizing and basaloid subtypes.4
Each cancer histological type and subtype arose from multiple risk factors including genetic and environmental and thus has its own unique genetic mutational profile. Molecular profiling of the individual tumor's genome facilitates the understanding of the distinct molecular mechanism that regulates cancer progression and the discovery of potential therapeutic targets. Due to the advancements in molecular profiling technologies, the mutation profiles of LUAD and LUSC have been well elucidated.11, 12, 13 However, the molecular distinction between each specific histological subtypes within LUAD and LUSC is just beginning to be understood. Among LUAD subtypes, EGFR mutations are more prevalent in lepidic (formerly termed as bronchioalveolar) tumors, while KRAS mutations are more common invasive LUAD subtypes, particularly invasive mucinous and solid subtypes.11, 13, 14, 15, 16, 17, 18, 19 On the other hand, limited information is available on the molecular distinction in LUSC subtypes.13
In this multi‐center comparative study, we aimed to characterize the mutational profiles of the major histological subtypes of adenocarcinoma and squamous cell carcinoma in the Chinese NSCLC patients. This study emphasizes the need for mutational profiling in all NSCLC patients to identify actionable mutations amenable to targeted therapy.
Methods
Patient enrollment
This cohort included 318 Chinese stage I–IV NSCLC patients with either adenocarcinoma or squamous cell types from seven hospitals. All patients underwent complete tumor staging according to the seventh edition tumor, node, and metastasis (TNM) criteria of NSCLC.20 Lung cancer histology was classified according to the predominant subtype following the 2015 World Health Organization (WHO) histopathology classification.4 The patients' clinical data, including demographic information, smoking status and cancer histological subtype, were reviewed. Tumor samples were obtained either by surgical or needle biopsy procedures and sequenced for mutational analysis. The study had been approved by the relevant regulatory and independent ethics committees or institutional review boards of all the participating hospitals. Written informed consent was obtained from each patient for the use of their tissue samples.
Tissue DNA extraction
DNA was extracted from formalin‐fixed, paraffin‐embedded (FFPE) tumor tissues using QIAamp DNA FFPE tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions.
Capture‐based targeted DNA sequencing
A minimum of 50 ng of DNA is required for NGS library construction. Tissue DNA was sheared using Covaris M220, followed by end repair, phosphorylation, and adaptor ligation. Fragments of size 200–400 bp were selected by bead (Agencourt AMPure XP Kit, Beckman Coulter, Brea, CA, USA), followed by hybridization with capture probes baits, hybrid selection with magnetic beads and PCR amplification. Lung Core panel from Burning Rock Biotech (Guangzhou, China) consisting of 68 lung cancer‐related genes spanning 345 kb of the human genome was used. The quality and the size of the fragments were assessed using a Qubit 2.0 Fluorimeter with the dsDNA high sensitivity assay kit (Life Technologies, Carlsbad, CA, USA). Indexed samples were sequenced on Nextseq500 (Illumina, Inc., Madison, WI, USA) with paired‐end reads.
Sequence data analysis
Sequence data were mapped to the reference human genome (hg19) using Burrows‐Wheeler aligner v.0.7.10. Local alignment optimization, variant calling and annotation were performed using Genome Analysis Tool Kit v.3.2, and VarScan. Variants were filtered using the VarScan fpfilter pipeline, loci with depth less than 100 were filtered out. Base calling in tissue samples required at least five and eight supporting reads for small insertion‐deletions (INDELs) and single nucleotide variants (SNVs), respectively. INDELs and SNVs with population frequency over 0.1% in the ExAC, 1000 Genomes, dbSNP or ESP6500SI‐V2 databases were grouped as SNP and excluded from further analysis. Remaining variants were annotated with ANNOVAR and SnpEff v.3.6. Analysis of DNA translocation was performed using Factera v.1.4.3.
Statistical analysis
All the data were analyzed using R software (R version 3.4.0; R: The R Foundation for Statistical Computing, Vienna, Austria). Significance between the groups was calculated using Fisher's exact test with P < 0.05 considered as statistically significant. P‐values were adjusted for variables such as age, gender, smoking history, and pathological stage when applicable.
Results
Patient characteristics
Our cohort consisted of 318 Chinese stage I‐IV NSCLC patients diagnosed with either lung adenocarcinoma (LUAD) or lung squamous cell carcinoma (LUSC) from seven hospitals. Lung cancer histological subtypes other than LUAD and LUSC were excluded from the study cohort. Patient demographics and clinical characteristics are summarized in Table 1. Figure 1 illustrates the distribution of LUAD and LUSC patients according to their histological subtypes. Of the 215 LUAD patients, the most predominant LUAD subtypes were adenocarcinoma in situ and acinar subtypes with 14.4% (31/215) each. Other LUAD subtypes included solid (24/215, 11.2%), enteric (22/215, 10.2%), lepidic (19/215, 8.8%), papillary (19/215, 8.8%), invasive mucinous (14, 6.5%), micropapillary (12/215, 5.6%), colloid (10/215, 4.7%), minimally invasive (8/215, 3.7%) and fetal (2/215, 1%). There were 23 (10.7%) patients with unclassifiable LUAD (Fig 1a). On the other hand, among the 103 LUSC patients, keratinizing subtype (40.8%, 42/103) was the most predominant. Other LUSC subtypes included non‐keratinizing (26/103, 26.2%), basaloid (8/103, 8%), and squamous cell carcinoma in situ (1/103, 1%). There were 25 (24.3%) patients with unclassifiable LUSC (Fig 1b). The pathologic stage distribution of the 215 LUAD and 103 LUSC patients according to predominant histological subtypes were summarized in Tables S1 and S2, respectively.
Table 1.
Patient characteristics
| Clinical characteristics | Total n = 318 | LUAD n = 215 | LUSC n = 103 |
|---|---|---|---|
| Age (years) | |||
| Median, (range) | 61, (29–84) | 61, (29–81) | 61, (36–84) |
| Gender | |||
| Male | 201 (63.2%) | 106 (49.3%) | 95 (92.2%) |
| Female | 117 (36.8%) | 109 (50.7%) | 8 (7.8%) |
| Smoking history | |||
| Smoker | 92 (28.9%) | 40 (18.6%) | 52 (50.5%) |
| Non‐smoker | 166 (52.2%) | 143 (66.5%) | 23 (22.3%) |
| Unknown | 60 (18.9%) | 32 (14.9%) | 28 (27.2%) |
| Clinical stage | |||
| Stage IA | 65 (20.4%) | 50 (23.3%) | 15 (14.6%) |
| Stage IB | 32 (10.1%) | 22 (10.2%) | 10 (9.7%) |
| Stage IIA | 15 (4.7%) | 6 (2.8%) | 9 (8.7%) |
| Stage IIB | 15 (4.7%) | 4 (1.9%) | 11 (10.7%) |
| Stage IIIA | 32 (10.1%) | 24 (11.2%) | 8 (7.8%) |
| Stage IIIB | 6 (1.9%) | 3 (1.4%) | 3 (2.9%) |
| Stage IV | 16 (5.0%) | 13 (6.0%) | 3 (2.9%) |
| NA | 137 (43.1%) | 93 (43.3%) | 44 (42.7%) |
Figure 1.
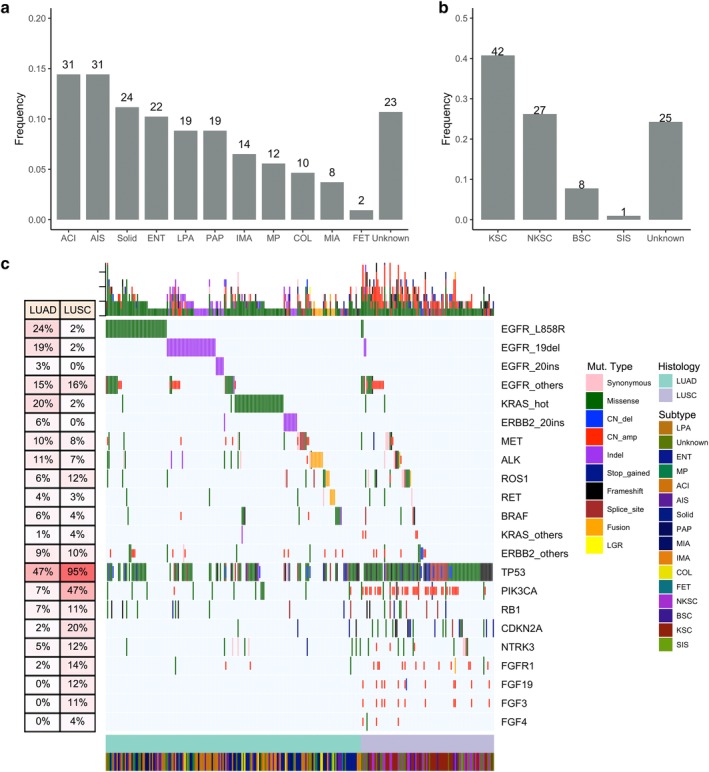
Histological subtype distribution and mutational profile of the lung cancer patients in the cohort. Distribution of (a) LUAD and (b) LUSC patients according to their histological subtypes. X‐axis denotes the histological subtype, Y‐axis, patient frequency. (c) Mutational spectrum of the patients grouped according to histological subtype. Each column represents a patient and each row represents a gene. Table on the left represents the mutation rate of each gene that corresponds to either LUAD or LUSC histological type. Top plot represents the overall number of mutations a patient carried. Different colors denote different types of mutation. ACI, acinar adenocarcinoma; AIS, adenocarcinoma in situ; BSC, basaloid squamous cell carcinoma; COL, colloid adenocarcinoma; ENT, enteric adenocarcinoma; FET, fetal adenocarcinoma; IMA, invasive mucinous adenocarcinoma; KSC, keratinizing squamous cell carcinoma; LPA, lepidic adenocarcinoma; LUAD, adenocarcinoma; LUSC, lung squamous cell carcinoma; MIA, minimally invasive adenocarcinoma; MP, micropapillary adenocarcinoma; NKSC, nonkeratinizing squamous cell carcinoma; PAP, papillary adenocarcinoma; SIS, squamous cell carcinoma in situ; Solid, solid adenocarcinoma.
Comparison of the mutational profile of LUAD and LUSC patients
Of 297 patients, 199 (92.6%) were LUAD and 94 (91.3%) were LUSC. A total of 31 patients (16 LUAD and 15 LUSC) had no mutation detected from our panel.
From the somatic mutational profile of the patients, a distinct pattern between LUAD and LUSC was observed. Mutations in the oncogenic drivers were more prevalent in LUAD than LUSC patients (P < 0.001), with 77.2% (166/215) of the LUAD patients harboring alteration in oncogenic driver mutations. Oncogenic drivers included EGFR L858R and exon 19 deletion (19del), KRAS G12, G13 and Q61, BRAF V600E, MET exon14 skipping and amplification, ERBB2 exon 20 insertion, ALK fusions, RET fusions, ROS1 fusions and NRG1 fusions. Among them, 37.7% (81/215) of the patients carried EGFR L858R and 19del mutations. Mutual exclusivity was observed among driver mutations in LUAD. However, a rare co‐occurrence of EGFR L858R and KRAS Q61H was found in a LUAD patient. Moreover, we also observed concurrent mutations of EGFR L858R and ERBB2 S310F in two adenocarcinoma patients; EGFR L858R and ERBB2 amplification in three LUAD patients (Table S3). In addition to sensitizing mutations in EGFR, we found three rare EGFR mutations including EGFR G719A detected in a patient and EGFR S758I and L838V concurrently detected in a patient (Table S3). A total of 24 patients (11.2%, 24/215) had EGFR compound mutation. KRAS mutations, including G12, G13, Q61, were detected in 17.7% (38/215) of patients. Among the BRAF mutations detected in 5.6% (12/215) of patients, no BRAF V600E was found. A BRAF rare disruptive in‐frame insertion R506_K507insVLR was detected in a LUAD patient (Table S4). Conversely, mutations in ALK, RET and ROS1, including fusions and other types of mutations, were detected in 25 (11.6%), 8 (3.7%) and 12 (5.6%) LUAD patients, respectively. A majority (77.8%, 7/9) of the patients with ALK fusion had EML4 as the fusion partner. Of these nine patients, four had single EML4‐ALK fusion, while three patients had EML4‐ALK and concurrent ALK fusion with previously unreported partners, such as EXOC6B, TTN, ACVR1, and TACR1. The two remaining patients had other unreported ALK fusion partners, including ERBB4 found in a patient and both PRR20A and RHOB detected in another patient. A summary of the ALK fusions detected in our cohort was listed in Table S4. Moreover, a patient with a driver CCDC6‐RET fusion also had a concurrent, previously unreported RET fusion with CCSER2. In addition, CD74‐NRG1 fusions were also detected in three LUAD patients. Rare mutations and concurrent mutations in oncogenic genes were summarized in Table S3.
In LUSC, only 10 (9.7%, 10/103) patients carried alterations in oncogenic drivers. No mutual exclusivity was observed among driver mutations. Interestingly, four and one LUSC patients carried EGFR sensitizing and KRAS mutations, respectively. Such mutations were believed to occur exclusively in LUAD. On the other hand, BRAF mutations were found in four (3.9%, 4/103) LUSC patients; however, no BRAF V600E mutations were detected. Conversely, EML4‐ALK and CD74‐ROS1 fusions were found in a patient each; while no RET fusion was found in LUSC patients.
On the other hand, mutations in TP53 (P < 0.001) and gene amplifications in particular genes (P < 0.001) were more predominant in LUSC. TP53 mutations were detected in almost all of the LUSC patients (90%, 93/103) but only 41% (88/215) of LUAD patients. In addition, LUSC patients had significantly more mutations in CDKN2 (20/103, P < 0.001), and amplifications in CCND1 (14/103, P < 0.001), PIK3CA (40/103, P < 0.001) and FGFR1 (13/103, P < 0.001) than LUAD patients. Amplifications in FGF19 (10.7%, 11/103), FGF3 (10.7%, 11/103) and FGF4 (2.9%, 3/103) were only found in LUSC patients in our cohort (Fig 1c). Interestingly, a previously unreported FGFR1 fusion with KCNU1 (KCNU1 intergenic: FGFR1 F4) was detected in a patient also harboring concurrent FGFR1 amplification and NTRK1 Q558X stop gain mutation. Moreover, PIK3CA mutation types between LUAD and LUSC were significantly different, with missense mutations more prevalent in LUAD patients, while amplifications were largely found in LUSC patients (P < 0.001, Fig 1c). These data indicate that LUAD and LUSC have very distinct mutational characteristics (P < 0.001).
Mutational profile of major subtypes of lung adenocarcinoma patients (LUAD)
Our data revealed oncogenic mutation detection rate of 80.6% (25/31), 75% (6/8) and 89.5% (137/153) for preinvasive, minimally‐invasive and invasive subtypes, respectively. Patients with invasive subtypes (ie, acinar, colloid, enteric, fetal, lepidic, micropapillary, invasive mucinous, papillary, and solid adenocarcinomas) harbored more total mutations than preinvasive (ie, adenocarcinoma in situ) and minimally‐invasive subtypes (P < 0.001, Fig 2a).
Figure 2.

Mutational profile of LUAD patients based on predominant histological subtype. (a) Total mutation count (including small nucleotide variations, insertion‐deletions, copy number variations and fusions) in LUAD patients. X‐axis denotes the LUAD subtypes. Y‐axis denotes the total mutation count per patient. Each dot represents a patient. (b) Mutational profile in oncogenic gene drivers including ALK fusion, BRAF mutations, EGFR mutations and amplifications, ERBB2 exon20 insertion and amplifications, KRAS G12, G13 and Q61 mutations, MET exon14 skipping and amplifications, RET fusion, ROS1 fusion in LUAD patients. (c) Mutation rate in TP53 in LUAD patients. X‐axis denotes the LUAD subtype. Y‐axis denotes the mutation rate, calculated as a percentage of the ratio of patients positive for the gene mutation against the total number of patients in the particular subtype group. ACI, acinar adenocarcinoma (n = 31); AIS, adenocarcinoma in situ (n = 31); COL, colloid adenocarcinoma (n = 10); ENT, enteric adenocarcinoma (n = 22); FET, fetal adenocarcinoma (n = 2); IMA, invasive mucinous adenocarcinoma (n = 14); LPA, lepidic adenocarcinoma (n = 19); MIA, minimally invasive adenocarcinoma (n = 8); MP, micropapillary adenocarcinoma (n = 12); PAP, papillary adenocarcinoma (n = 19); Solid, solid adenocarcinoma (n = 24); Unknown, unclassified LUAD subtype (n = 23).
In general, EGFR sensitizing mutations were predominant in LUAD (P < 0.001) with relatively even distribution in preinvasive, minimally invasive and invasive LUAD subtypes (P = 0.515, Fig 2b). However, further analysis of the detailed subtypes revealed that among all the subtypes, invasive lepidic adenocarcinoma had the most EGFR‐mutant patients (P = 0.003), followed by acinar and micropapillary subtypes. Meanwhile, both patients with fetal subtype were wild‐type for EGFR (Fig 2b).
Among the patients with preinvasive adenocarcinoma in situ (AIS) subtype, EGFR mutations were detected in 52% (16/31) of the patients (Fig 2b). No alterations in ALK, ERBB2, MET, and RET were found in AIS patients (Fig 2b). Only an AIS patient harbored ROS1 fusion. Meanwhile, TP53 mutations were found in 19% (6/31) of the AIS patients (Fig 2c). Among the patients with no mutations detected from our panel, five were AIS patients.
In patients with minimally invasive adenocarcinoma subtype (MIA), the only oncogenic driver mutations detected were EGFR, ERBB2 and KRAS (Fig 2b) wherein half (4/8) of the MIA patients were EGFR mutants. ERBB2 and KRAS were found in a patient each. No gene alterations were detected in ALK, BRAF, MET, RET, ROS1 and TP53 in MIA patients (Fig 2b). An MIA patient was negative for mutations in our gene panel.
Among all the patients in the cohort, patients with adenocarcinoma of the invasive type generally had alterations in all oncogenic driver genes. In particular, gene alterations in ALK, MET and RET were only found in these patients (Fig 2b). TP53 mutations were also more prevalent in patients with invasive adenocarcinoma (48%, 74/153, Fig 2c).
Not only among invasive subtypes but among all the LUAD subtypes, lepidic adenocarcinoma subtype (LPA) had the most EGFR mutations, with 78.9% (15/19) EGFR positive patients (P = 0.003, Fig 2b) with the majority being sensitizing mutations. No ALK, BRAF, RET and ROS1 alterations were detected in LPA patients (Fig 2b). Mutations in ERBB2, MET and KRAS were found in 1, 1, and 2 LPA patients (Fig 2b); while mutations in TP53 were detected in three LPA patients (Fig 2c).
Patients with acinar subtype (ACI) had mutations in all oncogenic driver genes (Fig 2b). EGFR mutations were found in 61% (19/31) of these patients (Fig 2b). TP53 mutations were detected in half of the ACI patients (15/31) (Fig 2c). No fusions in ALK, RET and ROS1 were detected in ACI patients.
Solid adenocarcinoma patients had the most TP53 mutations (87.5%, 21/24) among all LUAD subtypes (Fig 2c). Mutations in all oncogenic drivers were detected in these patients except ROS1 fusion (Fig 2b). KRAS mutations were found in 25% (6/24) of the patients with solid subtype; while EGFR mutations were only found in 16.7% (4/24) of the patients with solid subtype.
Patients with enteric adenocarcinoma (ENT) did not have mutations in ALK, BRAF, MET, RET and ROS1. The only mutations found in ENT patients were EGFR (22.7%, 5/22), ERBB2 (27.3%, 6/22) and KRAS (27.3%, 6/22) (Fig 2b). Of the 22 ENT patients, 68% (15/22) had TP53 mutations (Fig 2c). Two ENT patients had no mutation detected from our panel.
Except for RET fusion, patients with papillary subtype (PAP) had mutations in all oncogenic driver genes with 52.6% (10/19) of them positive for EGFR sensitizing mutations (Fig 2b). In addition, half of the PAP patients (9/19) had TP53 mutation (Fig 2c).
Patients with invasive mucinous adenocarcinoma subtype (IMA) patients had the most prevalent KRAS mutations, with 40% (6/14) KRAS‐positive IMA patients (Fig 2b). Instead of sensitizing mutations, only EGFR amplifications were found in two IMA patients (Fig 2b). No BRAF, ERBB2, MET and RET alterations were detected in any of these patients (Fig 2b). A patient with IMA was negative for mutations in our gene panel.
Among the 12 patients with micropapillary adenocarcinoma subtype (MP), 58.3% (7/12) were positive for EGFR sensitizing mutations (Fig 2b). Oncogenic driver mutations in MP patients were only detected in EGFR (7/12), ALK (2/12), ERBB2 (2/12) and RET (2/12) (Fig 2b). The four MP patients with TP53 mutations also coharbored EGFR sensitizing mutations (Figure S1).
Among the 10 colloid adenocarcinoma subtype (COL) patients, 30% (3/10) were positive for KRAS mutations (Fig 2b). Interestingly, two KRAS‐positive patients also had concomitant BRAF mutations (Fig 2b). Moreover, two patients had ALK fusion; one patient had ERBB2 amplification and one had both EGFR 19del and ERBB2 amplification (Fig 2b). Two patients with COL were negative for mutations in our gene panel.
Apart from ERBB2 amplification and KRAS mutation in each of the two fetal adenocarcinoma (FET) patients in the cohort, no other gene alterations in any oncogenic driver and TP53 were detected (Fig 2b,c).
Mutational profile of squamous cell lung carcinoma patients (LUSC) according to subtype
In contrast to LUAD, LUSC had a predominance of mutations in TP53 and copy number variations in particular genes, while oncogenic driver mutations were very few (P < 0.001, Fig 1c, Figure S2). The mutation count among the LUSC histological subtypes were comparable, with the median basaloid squamous cell carcinoma (BSC) patients slightly higher but not statistically different from the other subtypes (P = 0.45, Fig 3a).
Figure 3.
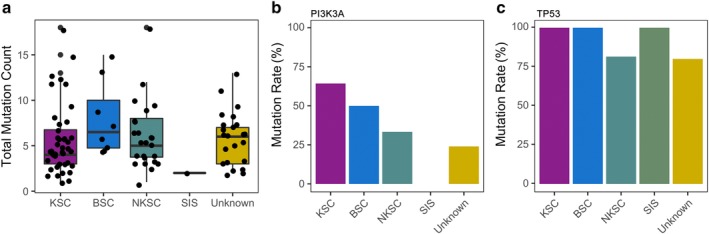
Mutational profile of LUSC patients based on predominant histologic subtype. (a) Total mutation count (including small nucleotide variations, insertion‐deletions, copy number variations and fusions) of LUSC patients. X‐axis denotes the LUSC subtypes. Y‐axis denotes the total mutation count per patient. Each dot represents a patient. Mutation rate in PIK3CA (b) and TP53 (c) in LUSC patients. X‐axis denotes the LUSC subtype. Y‐axis denotes the mutation rate, calculated as the percentage of the ratio of patients positive for the gene mutation against the total number of patients in the particular subtype group. BSC, basaloid squamous cell carcinoma (n = 8); KSC, keratinizing squamous cell carcinoma (n = 42); NKSC, nonkeratinizing squamous cell carcinoma (n = 27); SIS, squamous cell carcinoma in situ (n = 1); Unknown, unclassified LUSC subtype (n = 25).
PIK3CA amplifications were most prevalent in keratinizing squamous cell carcinoma (KSC) patients (64.3%, 27/42, P = 0.015), followed by basaloid squamous cell carcinoma (BSC) (50%, 4/8) and nonkeratinizing squamous cell carcinoma (NKSC) (33.3%, 9/27). No PIK3CA mutation was found in the only squamous cell carcinoma in situ (SIS) patient (Fig 3b).
As expected for LUSC, TP53 mutations were detected in almost all (90%, 93/103) of the LUSC patients, wherein all the patients with KSC (42/42), BSC (8/8) and SIS (1/1) were TP53 mutants. Meanwhile, NKSC patients only had 81% (22/27) TP53 mutants (Fig 3c). Of the five patients with wild‐type TP53, two of them had no mutation detected in any of the genes in our panel (Fig 3b, Figure S2). The other five wild‐type TP53 patients had unclassified subtype (Fig 3c).
The relationship between molecular and clinical features in LUAD and LUSC patients
In LUAD patients, TP53 mutations were associated with older patients. The age of TP53‐positive LUAD patients ranged between 43 to 81 years with a median age of 62 years, while the age of wild‐type TP53 patients ranged between 29 to 79 years with a median age of 59 years (adjusted P < 0.001, Figure S3). KRAS mutations were also found to be associated with smoking status (adjusted P = 0.039). There was no significant correlation in age, gender and genetic alterations among the LUSC patients.
Discussion
The understanding and management of lung cancer has advanced significantly in the past decade. Even with the increasing importance of molecular testing to identify actionable mutations for targeted therapy, histopathological classification of cancer subtypes is still an essential component of clinical diagnosis and making optimal treatment decisions, particularly in patients with no actionable mutations.
To the best of our knowledge, our study is the first to use a unified strategy to compare the mutational profile of LUAD and LUSC predominant histological subtypes in Chinese NSCLC patients.
In our cohort of 215 LUAD NSCLC patients, adenocarcinoma in situ and acinar subtypes were the two most prevalent LUAD histological subtypes. This is in contrast to Caucasian histological prevalence where the top two subtypes were acinar followed by solid subtypes.11, 21 Since previous studies have only used traditional methods of molecular testing, existing literature on the genetic alterations in various histological subtypes are mostly limited to the detection rates of EGFR and KRAS mutations. EGFR mutations are detected in 10%–30% of LUAD patients; however, this prevalence increases to approximately 50% among Chinese LUAD patients.13, 22 Hence, we only considered the reports that included Chinese patients. A previous study reported that among Chinese LUAD patients, lepidic and micropapillary subtypes had the most EGFR mutations with approximately 70% EGFR‐mutant patients from each subtype, while solid subtype had the least number of EGFR mutant patients.18 Meanwhile, another study reported the EGFR mutation detection rates of 68.8%, 70.7%, 69.5%, 22.5%, 80.0%, and 25.0% in Chinese patients with lepidic, papillary, acinar, solid, micropapillary and mucinous subtypes, respectively, with no EGFR mutation detected in the case of fetal adenocarcinoma.23 In contrast, in our cohort, EGFR sensitizing mutations were generally more common in preinvasive and minimally invasive subtypes. Meanwhile, invasive subtypes such as acinar, micropapillary, and papillary also had a substantial number of EGFR mutant patients. However, the least number of EGFR sensitizing mutations were in patients of colloidal subtype (1/10), invasive mucinous (0/14) and fetal (0/2) subtypes. Conversely, KRAS mutations in our cohort were consistent with the reported prevalence.17 In our cohort, KRAS mutations were also more prevalent in invasive mucinous (6/14), colloid (3/10), enteric (4/22) and solid (6/24) subtypes.
Furthermore, we revealed distinct mutation profiles for Chinese LUAD and LUSC patients. In contrast to LUAD patients, the LUSC patients in our cohort had significantly more amplification events and TP53 mutations. Similar to the findings of the TCGA12 and a study among 104 Korean LUSC patients by Kim et al.,24 significantly fewer EGFR and KRAS mutations were detected in Chinese LUSC than LUAD patients. Conversely, another mutational profiling study involving 157 Chinese LUSC patients reported mutation incidence of 56% for TP53, 8.9% for CDKN2, 8.9% for PIK3CA,25 and the incidence rates were significantly lower than those observed in our cohort. Studies involving larger cohorts are necessary to validate the incidence rates.
Interestingly, we have detected several rare mutations in known oncogenic genes including EGFR and BRAF and unreported fusion partners for RET, FGFR1 and ALK in our cohort. With the increasing use of molecular profiling in the clinical setting, more novel mutations and fusion partner genes are being uncovered. However, the clinical significance of these rare mutations and novel fusion partners requires further studies. Our study is limited by its retrospective nature. Clinical data for some of the patients were incomplete which limits the analysis of clinical features and histological subtypes of the cohort. The limited availability of tissue samples also limits us to explore the clinical significance of rare mutations detected in the study. Future prospective multi‐center studies with a larger cohort are required to further explore the mutation profile of the different subtypes. It would be interesting to explore the stratification of molecular subtypes according to distinct mutation signature and their clinical responses to certain inhibitors.
In summary, this comparative study revealed the mutational distinction between LUAD and LUSC as well as their predominant histological subtypes in the Chinese population. Taking the inherent genetic heterogeneity among tumor subtypes, we further emphasize the need to include comprehensive mutational profiling in the standard management of lung cancer patients of all histological subtypes to understand the genetic landscape of the tumor and further inform clinical decisions.
Disclosure
The authors declare no potential conflicts of interest.
Supporting information
Figure S1 Mutational spectrum of the LUAD patients. Each column represents a patient and each row represents a gene. Top plot represents the overall number of mutations a patient carried. Side bars represent the percentage of patients with a certain mutation. Different colors denote different types of mutation. Negative denotes the absence of any mutation. ACI, acinar adenocarcinoma; AIS, adenocarcinoma in situ; COL, colloid adenocarcinoma; ENT, enteric adenocarcinoma; FET, fetal adenocarcinoma; IMA, invasive mucinous adenocarcinoma; LPA, lepidic adenocarcinoma; MIA, minimally invasive adenocarcinoma; MP, micropapillary adenocarcinoma; PAP, papillary adenocarcinoma; Solid, solid adenocarcinoma; Unknown, LUAD with unclassified subtype.
Figure S2 Mutational profile of LUSC patients according to histological subtypes. Each column represents a patient and each row represents a gene. Top plot represents the overall number of mutations a patient carried. Side bars represent the percentage of patients with a certain mutation. Different colors denote different types of mutation. Negative denotes the absence of any mutation. BSC, basaloid squamous cell carcinoma; KSC, keratinizing squamous cell carcinoma; NKSC, nonkeratinizing squamous cell carcinoma; SIS, squamous cell carcinoma in situ; Unknown, LUSC with unclassified subtype.
Figure S3 The relationship between molecular and clinical features in LUAD patients. Box plot illustrating the relationship between age of the LUAD patients and TP53 mutation. X‐axis denotes the TP53 mutation status, negative for wild‐type. Y‐axis denotes the age of the patients in years.
Table S1 Pathologic stage distribution of the LUAD patients according to the predominant histologic subtypes.
Table S2 Pathologic stage distribution of the LUSC patients according to the predominant histologic subtypes.
Table S3 Summary of concurrent and rare somatic mutations in oncogenic genes found in LUAD patients.
Table S4 ALK fusions detected in 10 patients from our cohort.
Acknowledgments
The authors thank all the patients who participated in this study and their families. We also thank the investigators, study coordinators, operation staff and the whole project team who worked on this study.
This work was supported by the National Natural Science Foundation of China (81773109, 81502089), the Natural Science Foundation of Jiangsu Province (BK20151582, BK20151024, BK20170706), National Key Clinical Specialty Construction Project (2014) and the Fund of the Priority Academic Program Development of Jiangsu Higher Education Institution (JX1023‐1801).
Contributor Information
Ying Ding, Email: dingdongdong815@hotmail.com.
Zhihong Zhang, Email: zhangzh@njmu.edu.cn.
References
- 1. Chen W, Zheng R, Baade PD et al Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66 (2): 115–32. [DOI] [PubMed] [Google Scholar]
- 2. World Health Organization . [Cited 12 Septemebr 2018.] Cancer Fact Sheet No. 297 World Health Organization. Available from URL: http://www.who.int/mediacentre/factsheets/fs297/en/2018.
- 3. Kim HS, Mitsudomi T, Soo RA, Cho BC. Personalized therapy on the horizon for squamous cell carcinoma of the lung. Lung Cancer 2013; 80 (3): 249–55. [DOI] [PubMed] [Google Scholar]
- 4. Travis WD, Brambilla E, Nicholson AG et al The 2015 World Health Organization classification of lung tumors: Impact of genetic, clinical and radiologic advances since the 2004 classification. J Thorac Oncol 2015; 10 (9): 1243–60. [DOI] [PubMed] [Google Scholar]
- 5. Lindeman NI, Cagle PT, Beasley MB et al Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Thorac Oncol 2013; 8 (7): 823–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Du X, Shao Y, Qin HF, Tai YH, Gao HJ. ALK‐rearrangement in non‐small‐cell lung cancer (NSCLC). Thorac Cancer 2018; 9 (4): 423–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tomasini P, Barlesi F, Mascaux C, Greillier L. Pemetrexed for advanced stage nonsquamous non‐small cell lung cancer: Latest evidence about its extended use and outcomes. Ther Adv Med Oncol 2016; 8 (3): 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Russo AE, Priolo D, Antonelli G, Libra M, McCubrey JA, Ferrau F. Bevacizumab in the treatment of NSCLC: Patient selection and perspectives. Lung Cancer 2017; 8: 259–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Keating GM. Nivolumab: A review in advanced squamous non‐small cell lung cancer. Drugs 2015; 75 (16): 1925–34. [DOI] [PubMed] [Google Scholar]
- 10. Travis WD, Rekhtman N, Riley GJ et al Pathologic diagnosis of advanced lung cancer based on small biopsies and cytology: A paradigm shift. J Thorac Oncol 2010; 5 (4): 411–4. [DOI] [PubMed] [Google Scholar]
- 11. The Cancer Genome Atlas Research Network . Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511: 543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. The Cancer Genome Atlas Research Network . Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012; 489: 519–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Testa U, Castelli G, Pelosi E. Lung cancers: Molecular characterization, clonal heterogeneity and evolution, and cancer stem cells. Cancer 2018; 10 (8): 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li S, Choi YL, Gong Z et al Comprehensive characterization of oncogenic drivers in Asian lung adenocarcinoma. J Thorac Oncol 2016; 11 (12): 2129–40. [DOI] [PubMed] [Google Scholar]
- 15. Finberg KE, Sequist LV, Joshi VA et al Mucinous differentiation correlates with absence of EGFR mutation and presence of KRAS mutation in lung adenocarcinomas with bronchioloalveolar features. J Mol Diagn 2007; 9 (3): 320–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tam IY, Chung LP, Suen WS et al Distinct epidermal growth factor receptor and KRAS mutation patterns in non‐small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res 2006; 12 (5): 1647–53. [DOI] [PubMed] [Google Scholar]
- 17. Yoshizawa A, Sumiyoshi S, Sonobe M et al Validation of the IASLC/ATS/ERS lung adenocarcinoma classification for prognosis and association with EGFR and KRAS gene mutations: Analysis of 440 Japanese patients. J Thorac Oncol 2013; 8 (1): 52–61. [DOI] [PubMed] [Google Scholar]
- 18. Song Z, Zhu H, Guo Z, Wu W, Sun W, Zhang Y. Correlation of EGFR mutation and predominant histologic subtype according to the new lung adenocarcinoma classification in Chinese patients. Med Oncol 2013; 30 (3): 645. [DOI] [PubMed] [Google Scholar]
- 19. Rekhtman N, Ang DC, Riely GJ, Ladanyi M, Moreira AL. KRAS mutations are associated with solid growth pattern and tumor‐infiltrating leukocytes in lung adenocarcinoma. Mod Pathol 2013; 26 (10): 1307–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Edge SB, Compton CC. The American Joint Committee on Cancer: The 7th edition of the AJCC cancer staging manual and the future of TNM. Ann Surg Oncol 2010; 17 (6): 1471–4. [DOI] [PubMed] [Google Scholar]
- 21. Russell PA, Wainer Z, Wright GM, Daniels M, Conron M, Williams RA. Does lung adenocarcinoma subtype predict patient survival?: A clinicopathologic study based on the new International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society international multidisciplinary lung adenocarcinoma classification. J Thorac Oncol 2011; 6 (9): 1496–504. [DOI] [PubMed] [Google Scholar]
- 22. Pi C, Xu CR, Zhang MF et al EGFR mutations in early‐stage and advanced‐stage lung adenocarcinoma: Analysis based on large‐scale data from China. Thorac Cancer 2018; 9 (7): 814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chen Z, Liu X, Zhao J, Yang H, Teng X. Correlation of EGFR mutation and histological subtype according to the IASLC/ATS/ERS classification of lung adenocarcinoma. Int J Clin Exp Pathol 2014; 7 (11): 8039–45. [PMC free article] [PubMed] [Google Scholar]
- 24. Kim Y, Hammerman PS, Kim J et al Integrative and comparative genomic analysis of lung squamous cell carcinomas in East Asian patients. J Clin Oncol 2014; 32 (2): 121–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tao D, Han X, Zhang N et al Genetic alteration profiling of patients with resected squamous cell lung carcinomas. Oncotarget 2016; 7 (24): 36590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Mutational spectrum of the LUAD patients. Each column represents a patient and each row represents a gene. Top plot represents the overall number of mutations a patient carried. Side bars represent the percentage of patients with a certain mutation. Different colors denote different types of mutation. Negative denotes the absence of any mutation. ACI, acinar adenocarcinoma; AIS, adenocarcinoma in situ; COL, colloid adenocarcinoma; ENT, enteric adenocarcinoma; FET, fetal adenocarcinoma; IMA, invasive mucinous adenocarcinoma; LPA, lepidic adenocarcinoma; MIA, minimally invasive adenocarcinoma; MP, micropapillary adenocarcinoma; PAP, papillary adenocarcinoma; Solid, solid adenocarcinoma; Unknown, LUAD with unclassified subtype.
Figure S2 Mutational profile of LUSC patients according to histological subtypes. Each column represents a patient and each row represents a gene. Top plot represents the overall number of mutations a patient carried. Side bars represent the percentage of patients with a certain mutation. Different colors denote different types of mutation. Negative denotes the absence of any mutation. BSC, basaloid squamous cell carcinoma; KSC, keratinizing squamous cell carcinoma; NKSC, nonkeratinizing squamous cell carcinoma; SIS, squamous cell carcinoma in situ; Unknown, LUSC with unclassified subtype.
Figure S3 The relationship between molecular and clinical features in LUAD patients. Box plot illustrating the relationship between age of the LUAD patients and TP53 mutation. X‐axis denotes the TP53 mutation status, negative for wild‐type. Y‐axis denotes the age of the patients in years.
Table S1 Pathologic stage distribution of the LUAD patients according to the predominant histologic subtypes.
Table S2 Pathologic stage distribution of the LUSC patients according to the predominant histologic subtypes.
Table S3 Summary of concurrent and rare somatic mutations in oncogenic genes found in LUAD patients.
Table S4 ALK fusions detected in 10 patients from our cohort.


