Abstract
MicroRNAs (miRNAs) are endogenous small noncoding RNAs that negatively regulate gene expression by interfering with the translation or stability of target transcripts. Some tumor-suppressive miRNAs can concurrently target multiple cancer-promoting genes and may be useful as therapeutic anticancer agents. However, the development of drug delivery systems is critical for the implementation of miRNA-based therapeutics. We have previously demonstrated that the enforced expression of miR-634 effectively induces apoptosis by concurrently and directly targeting genes associated with mitochondrial homeostasis, antiapoptosis signaling, antioxidant ability, and autophagy in cancer cells. In the current study, we validated the therapeutic potential of lipid nanoparticle (LNP)-mediated delivery of miR-634 for cancer therapy. We confirmed the ability of enforced expression of miR-634 to induce apoptosis in various cancer cell lines, including pancreatic cancer cells. Intravenous administration of LNPs harboring miR-634 significantly reduced the xenograft tumor growth of BxPC-3 pancreatic cancer cells in mice. These findings suggest that LNP-mediated delivery of miR-634 can potentially be used for cancer therapy.
Keywords: microRNA, DDS, cancer therapy, miR-634, LNP
Introduction
MicroRNAs (miRNAs) are endogenous small noncoding RNAs that can negatively regulate gene expression by concurrently interfering with the translation and/or stability of multiple target transcripts by directly binding to the 3′ untranslated region (3′ UTR) or coding region and functioning as a “fine-tuner” of numerous biological processes.1,2 Dysregulated miRNA expression is associated with many human diseases, including cancer, and the downregulation of several tumor-suppressive miRNAs (TS-miRNAs) has been associated with processes involved in tumor progression, including cell proliferation, invasion/metastasis, and chemoresistance.1, 2, 3, 4, 5 Given that miRNAs have the potential to simultaneously regulate multiple targets in entire cancer-related cascades, the therapeutic restoration of TS-miRNAs could represent a novel strategy for the development of more practical strategies for cancer therapy.3, 4, 5, 6, 7 Importantly, the restoration of TS-miRNAs that endogenously alter target pathways is more tolerable in normal cells than conventional therapies, especially because they exert minimal toxic effects.3, 4, 5, 6, 7
We have identified multiple TS-miRNAs related to the inhibition of cell growth, cell invasion, and chemoresistance in human cancer, including endometrial cancer, oral cancer, and esophageal squamous cell carcinoma (ESCC).8, 9, 10, 11, 12, 13, 14 Notably, we reported that the expression of miR-634 was downregulated in ESCC cells, and that the overexpression of miR-634 effectively induced apoptosis in ESCC cells by concurrently and directly targeting multiple genes associated with cytoprotective processes, including those involved in mitochondrial homeostasis (OPA1 and TFAM), antiapoptosis signaling (APIP, XIAP, and BIRC5), redox (NRF2), and autophagy-lysosomal degradation (LAMP2).11 The cytoprotective processes upregulated in chemotherapy-resistant cancer cells seem to be logical targets to overcome chemoresistance in cancer. Indeed, the enforced expression of miR-634 markedly enhanced chemotherapy-induced cytotoxicity in ESCC cells.11 Furthermore, in vivo local administration of synthetic double-strand (ds) miR-634 mimic into xenograft tumors was shown to be therapeutically effective.11 Thus, our previous findings strongly suggest that the ds-miR-634 mimic may be a useful agent for the implementation of miRNA-based cancer therapeutics.
In the present study, we formulated lipid nanoparticles (LNPs)15,16 incorporating ds-miR-634 mimics. Intravenously administered miR-634-LNP effectively delivered ds-miR-634 mimics to tumors in BxPC-3 tumor-bearing mice. The protein expression of miR-634 target genes was significantly downregulated in the tumors, and tumor growth was significantly reduced in mice treated with miR-634-LNP. Taken together, these findings suggest the therapeutic potency of the LNP-mediated delivery of ds-miR-634 mimics for cancer therapy.
Results
Induction of Apoptotic Cell Death by miR-634 Overexpression in Pancreatic Cancer Cells
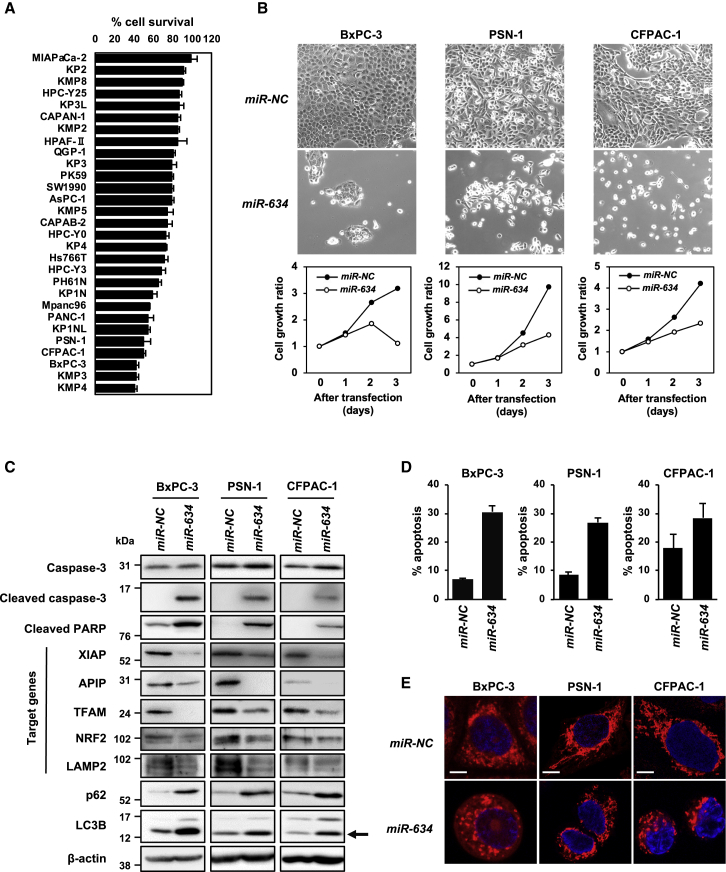
We examined the effect of miR-634 overexpression on cell viability in a total of 117 cell lines of various cancer types, including pancreatic cancer. We found that sensitivity to miR-634 was cell-type dependent and was very high in 38.5% (45 of 117 cell lines) of the cell lines, as indicated by a <50% reduction in the cell survival rate (Figure 1A; Table S1). None of the cancer types showed any specificity to miR-634. Furthermore, we showed that the sensitivity to miR-634 was very low in normal fibroblasts derived from healthy human volunteers (Figure S1).11
Figure 1.
Induction of Apoptosis by miR-634 in Pancreatic Cancer Cell Lines
(A) Differential sensitivity to miR-634 overexpression in pancreatic cancer cell lines. (B) Phase-contrast images and growth rate of miR-634-transfected cells. Cells were transfected with 20 nmol/L miR-NC or miR-634, and images were obtained at 3 days after transfection (upper panels). Cell growth rate was assessed with the crystal violet (CV) staining assay, and results are reported as the relative ratio compared with day 0 (lower panels). Error bars indicate SD of triplicate experiments. The error bars are not visualized due to being too small. (C) Western blotting analysis of miR-634-transfected cells. Cell lysates were subjected to SDS-PAGE and immunoreacted with the indicated antibodies. For the detection of LAMP2, cell lysates were prepared under nonreducing conditions (without 2-mercaptoethanol [2-ME]). Arrow indicates the band for LC3B form-II, an autophagosome marker. (D) FACS analysis of the apoptotic cell population. FACS analysis was performed at 3 days after transfection. Cells were collected and stained with Annexin V and propidium iodide (PI). Cell population analysis was performed using an Accuri Flow Cytometer. The percentages of apoptotic cells are indicated in each graph. Error bars indicate SD of triplicate experiments. (E) Representative images of mitochondrial staining. After 3 days of transfection, the cells were stained with 100 nmol/L MitoTracker Red CMX ROS for 30 min at 37°C. After fixation, images were obtained by confocal fluorescence microscopy. Scale bars, 10 μm.
It has been recently reported that miR-634 functions as a tumor suppressor in pancreatic cancer.17 We also showed that overexpression of miR-634 induced cell death in three pancreatic cancer cell lines, BxPC-3, PSN-1, and CFPAC-1 (Figures 1A and 1B). Western blotting analysis revealed a marked increase in the expression of the cleaved forms of caspase-3 and poly-ADP ribose polymerase (PARP) in miR-634-expressing cells (Figure 1C). As expected, the expression levels of known target genes of miR-634, such as XIAP, APIP, TFAM, NRF2, and LAMP2, were clearly decreased in miR-634-expressing cells. The fluorescence-activated cell sorting (FACS) analysis showed that the apoptotic population, indicated by the Annexin V and propidium iodide (PI) double-positive fraction, was markedly increased in miR-634-expressing cells compared with miR-NC-expressing cells (Figure 1D). Mitochondrial injury, as indicated by fragmented morphology, was increased in miR-634-expressing cells, but not in miR-NC-expressing cells (Figure 1E). In addition, we showed increased reactive oxygen species (ROS) levels, an indicator of mitochondrial injury, in miR-634-expressing pancreatic cancer cells (Figure S2). Autophagy contributes to mitochondrial quality control through mitophagy, the removal of damaged mitochondria via lysosomal degradation.18 The levels of LC3B-II, an autophagosome indicator, and p62 protein, a substrate of autophagic degradation, were increased in miR-634-expressing cells, suggesting that miR-634 impairs autophagic degradation, possibly by causing lysosomal dysfunction via concurrent downregulation of LAMP2 (Figure 1C).19,20 These findings imply that miR-634 effectively triggers activation of the mitochondrial apoptosis pathway in pancreatic cancer cells, strongly suggesting that synthesized ds-miR-634 mimics may be useful agents for cancer therapy.
Antitumor Effect of Systemic Administration of miR-634-LNP in the BxPC-3 Xenograft Model
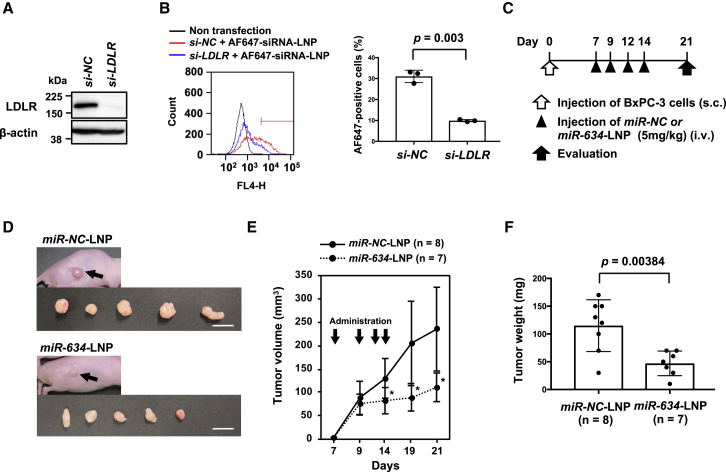
We attempted to validate the therapeutic potential of ds-miR-634 mimics as a cancer therapeutic agent using LNPs as a delivery system to tumors. Cellular uptake of LNPs is known to be attenuated by endocytosis via low-density lipoprotein receptor (LDLR).15 Indeed, when LDLR expression was inhibited in BxPC-3 cells, the cellular uptake of an LNP incorporating Alexa Fluor 647 (AF647)-labeled small interfering RNA (siRNA) was remarkably reduced (Figures 2A and 2B). Next, we formulated an LNP (Table S2) incorporating ds-miR-634 mimic or ds-miR-NC. The average diameter of miR-634-LNPs and miR-NC-LNPs was 48 and 49 nm, respectively. The polydispersity index (PDI) and encapsulation efficiency (EE) of both LNPs were 0.05 and >90%, respectively (Table S2). We first showed that adding miR-634-LNP in the culture medium induced a decreased expression of target proteins and as apoptosis in BxPC-3 cells, suggesting miR-634 incorporating into LNP is functional (Figure S3). The therapeutic effect of miR-634-LNPs was evaluated in BxPC-3 xenografted mice. Seven days after the injection of BxPC-3 cells, 5 mg/kg miR-NC-LNP or miR-634-LNP was intravenously administered through the tail vein into BxPC-3 tumor-bearing mice, with additional treatments on days 9, 12, and 14 (Figure 2C). As a result, tumor growth was significantly reduced in mice treated with miR-634-LNPs compared with mice treated with miR-NC-LNPs (Figures 2D–2F).
Figure 2.
Antitumor Effect of miR-634-LNP Administration in the BxPC-3 Xenograft Model
(A) Western blot analysis of LDLR in BxPC-3 cells. Cells were transfected with 20 nmol/L si-NC (negative control) or si-LDLR. Cell lysates were subjected to SDS-PAGE and immunoreacted with the indicated antibodies. (B) FACS analysis of the cellular uptake of LNP harboring Alexa Fluor 647-labeled siRNA. At 2 days after transfection with siRNA, the cells were incubated with 1 nmol/L LNPs harboring Alexa Fluor 647-labeled siRNA for 60 min at 37°C. Then the cells were collected, and the fluorescence intensity was measured using flow cytometry. The percentage of FL4-Hhigh cells was gated (left panel), and the average is indicated on the graph (right panel). Error bars indicate SD of triplicate experiments. (C) Experimental schedule for the administration of miR-634-LNPs. Tumors were formed by subcutaneous injection of BxPC-3 cells in nude mice. miR-NC-LNPs or miR-634-LNPs were administered intravenously a total of four times (at 7, 9, 12, and 14 days after the injection of cells). At 21 days after the injection of cells, the mice were sacrificed, and the tumors were evaluated. (D) Representative images of resected tumors at day 21. Arrows indicate subcutaneous tumors. Scale bars, 1 cm. (E) Tumor volume in mice treated with miR-NC-LNPs or miR-634-LNPs. Tumor volume in mice treated with miR-NC-LNPs (n = 8) or miR-634-LNPs (n = 7) was calculated. Error bars indicate SD. ∗p values; p = 0.025 at Day 14, p = 0.0067 at Day 19, and p = 0.004 at Day 21. (F) Tumor weight in mice treated with miR-NC-LNPs or miR-634-LNPs. Tumor weight in mice treated with miR-NC-LNPs (n = 8) or miR-634-LNPs (n = 7) is shown. Error bars indicate SD.
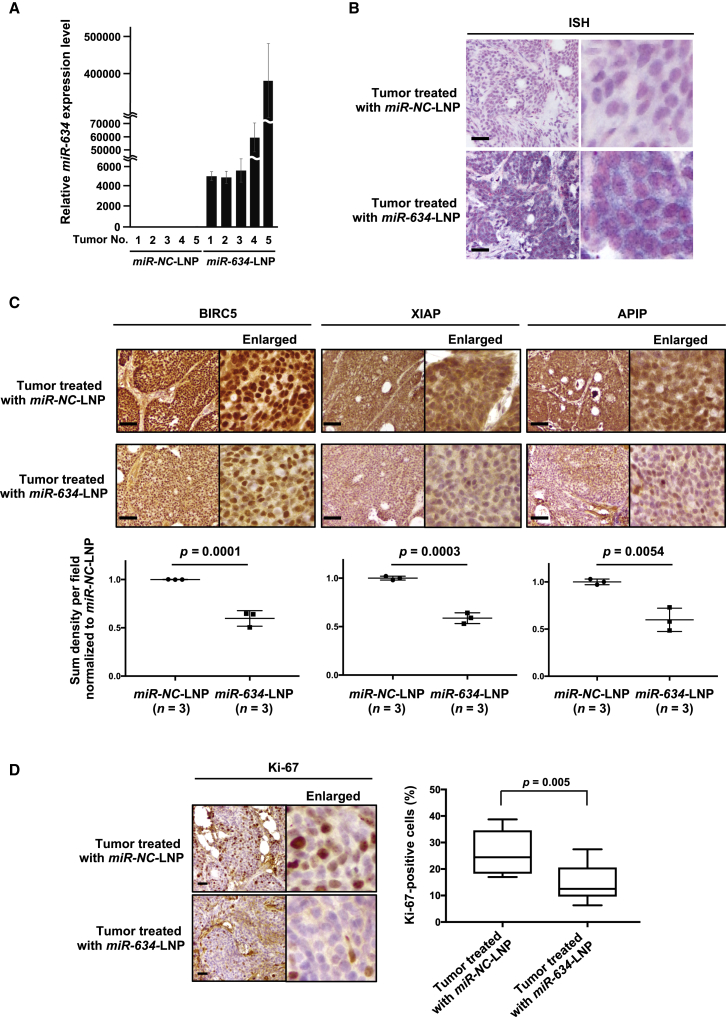
Importantly, expression analysis by quantitative reverse transcriptase PCR (qRT-PCR) showed that miR-634 expression levels were markedly increased in tumor samples resected from mice treated with miR-634-LNPs compared with mice treated with miR-NC-LNPs (Figure 3A). In addition, in situ hybridization (ISH) analysis using a miR-634-specific probe revealed the enforced expression of miR-634, visible as purple in tumor samples resected from mice treated with miR-634-LNP (Figure 3B). Furthermore, immunohistochemistry (IHC) analysis showed that the expression of some miR-634 target genes, such as BIRC5, XIAP, and APIP, was significantly decreased in tumor samples from mice treated with miR-634-LNPs (Figure 3C). Compared with tumors from mice treated with miR-NC-LNPs, those from mice treated with miR-634-LNPs had a significant reduction in the number of Ki67-positive cells, a marker of cell proliferation (Figure 3D). Taken together, these results strongly suggest that LNP-mediated delivery of ds-miR-634 mimics into tumors in BxPC-3 tumor-bearing mice is both successful and effective; therefore, miR-634-LNP can potentially be used in cancer therapy.
Figure 3.
Enforced Expression of miR-634 and Downregulation of Target Genes in Resected Tumors
(A) Expression analysis of miR-634 in resected tumors by qRT-PCR. The expression level of miR-634 in tumors from mice treated with miR-NC-LNPs (n = 5) or miR-634-LNPs (n = 5) was measured by qRT-PCR. Error bars indicate SD. (B) In situ hybridization (ISH) analysis of miR-634 in resected tumors. Representative images of ISH in tumors from mice treated with miR-NC-LNPs or miR-634-LNPs. The miR-634-specific probe was visualized in purple in the cytoplasm, and the nucleus was counterstained with nuclear fast red. Scale bars, 100 μm. (C and D) Immunohistochemistry (IHC) analysis of target genes (C) and Ki-67 (D) in resected tumors. Representative IHC images of tumors from mice treated with miR-NC-LNPs or miR-634-LNPs. The sum density was calculated, and results were normalized according to the values of tumor treated with miR-NC-LNP and indicated as the dot plot. The percentage of Ki-67-positive cells is indicated as a boxplot (right panel). Scale bars: 100 μm (upper panels). p values are shown in two-tailed Student’s t test.
Toxicity in Mice Treated with miR-634-LNPs
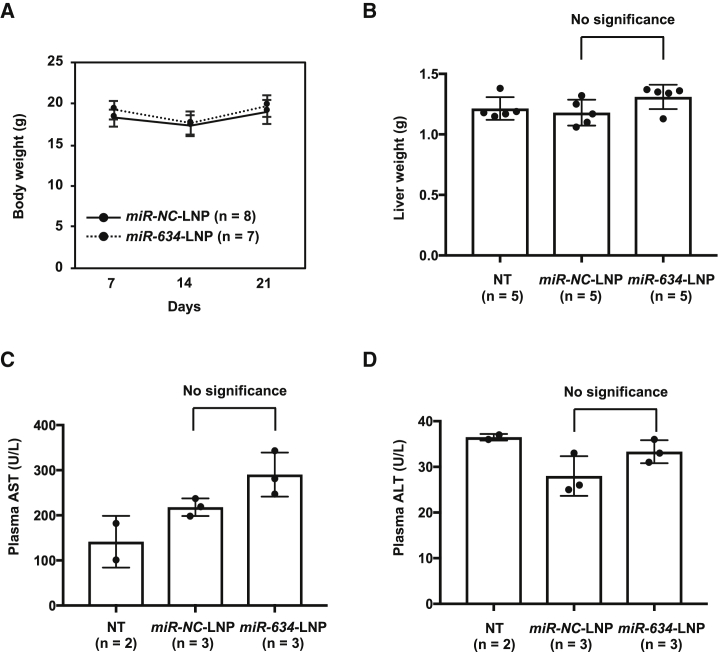
We evaluated the toxicity of miR-634-LNP administration in mice. Mouse body weight tended to decrease slightly at 7 days after treatment with LNPs (14 days after the injection of BxPC-3 cells), but the body weight loss recovered at 14 days after LNP treatment (21 days after the injection of BxPC-3 cells) (Figure 4A). Furthermore, to evaluate hepatotoxicity in mice treated with miR-NC-LNPs and miR-634-LNPs, we measured liver weight and the levels of aspartate aminotransferase (AST) and alanine aminotransferase (ALT) in the plasma of treated mice as markers for liver injury.21 We showed that miR-634 expression was increased in livers resected from mice treated with miR-634-LNPs compared with mice treated with miR-NC-LNPs (Figure S4). Nevertheless, no significant differences in liver weight and the levels of AST and ALT were observed between mice treated with miR-634-LNPs and miR-NC-LNPs, indicating that miR-634-LNP can be safely used in cancer therapy (Figures 4B–4D). However, the levels of AST, but not ALT, were higher in mice treated with either miR-634-LNPs or miR-NC-LNPs than in non-treated mice (Figure 4C). This finding may suggest some hepatotoxicity attributed to the LNPs used in this study.
Figure 4.
No Hepatotoxicity in Mice Treated with miR-634-LNP
(A) Measurement of mouse body weight at 7, 14, and 21 days after the injection of BxPC-3 cells. Error bar, SD from values for miR-NC-LNP (n = 8) or miR-634-LNP (n = 7). (B–D) Liver weight (B), plasma AST levels (C), and plasma ALT levels (D) in mice treated with miR-NC-LNP or miR-634-LNP at 21 days and non-treated mice (NT). Error bar; SD form values for NT (n = 5 in B and n = 2 in C and D), miR-NC-LNP (n = 5 in B and n = 3 in C and D), and miR-634-LNP (n = 5 in B and n = 3 in C and D).
Discussion
To evaluate the therapeutic potential of ds-miR-634 mimics as an anticancer agent, we formulated an LNP harboring ds-miR-634 mimics and showed that systemic administration of miR-634-LNPs inhibited tumor growth in BxPC-3 tumor-bearing mice. Thus, our findings in the current study strongly suggest that the miR-634-LNP formulation may be useful for cancer therapy.
siRNAs and miRNAs are noncoding RNAs that regulate gene expression through RNA interference (RNAi). They have recently been investigated as novel classes of therapeutic agents for the treatment of a wide range of disorders, including cancer. The most attractive aspect of RNAi therapeutics is the ability to target virtually any gene, which may not be possible with small molecules or protein-based drugs, such as monoclonal antibodies. However, there is a major difference between siRNA and miRNA; the former is highly specific with only one mRNA target, whereas the latter has multiple targets.22 Currently, the clinical development of siRNAs is advancing ahead of miRNAs, possibly due to uncertainties regarding the complex roles of miRNAs, which can target multiple genes. However, it is expected that the use of TS-miR, which can concurrently downregulate multiple oncoproteins, is a rational therapeutic strategy for cancers in which multiple cellular cascades are activated to promote tumor progression. Hence miR-634, which was examined in this study, may be a preferred TS-miR and anticancer agent because it can target cytoprotective processes in cancer cells related to cell survival and resistance to chemotherapy, such as mitochondrial homeostasis, antiapoptosis signaling, antioxidation pathways, and autophagy.11 On the other hand, it has been pointed out that somatic mutation on binding site in multiple target genes of TS-miR may reduce the anti-tumor effect by TS-miR.23, 24, 25 A more thorough characterization of miR-634, including further identification of unknown target genes and improved understanding of the physiological function, will be carried out to develop miR-based therapeutics using miR-634.
A key challenge in the implementation of RNAi therapeutics is the development of clinically viable delivery materials.26,27 LNPs have been developed as the most advanced platform for the systemic delivery of siRNAs.28, 29, 30, 31 The LNP delivery mechanism is based on ionizable lipids that facilitate the release of encapsulated siRNA into the cytoplasm.32 It has been demonstrated that LNPs based on an asymmetric ionizable lipid L021 can be systemically delivered to hepatocytes by a mechanism involving apolipoprotein E (apoE) as a targeting ligand and LDLR, which is highly expressed in hepatocytes.15,33 Hence, in this study, we formulated an L021-based LNP (Table S2) harboring ds-miR-634 mimics and showed the successful delivery of ds-miR-634 mimics via L021-LNPs into xenograft tumors upon systemic administration. In parallel with tumor growth inhibition, a slight decrease in body weight during administration and an increase in the levels of AST, but not ALT, were observed in mice treated with either miR-NC- and miR-634-LNP. This mild hepatotoxicity may be attributed to L021, the ionizable lipid, which can be considered the main cause of in vivo hepatotoxicity due to nonspecific adhesion to proteins.15,34 Furthermore, we showed a slight increase, but not significant, of AST level in mice treated with miR-634-LNPs compared with mice treated with miR-NC-LNPs, suggesting that this change may be attributed by an enforced miR-634 expression in livers. L101, the next generation of L021, was recently developed and shown to be rapidly cleared by metabolic degradation in the liver.16 Thus, biodegradable LNPs, coupled with a decreased LNP dosage, will be crucial for improving hepatotoxicity and obtaining a high therapeutic index without causing adverse events.
Accumulating evidence has successfully demonstrated the therapeutic efficacy of TS-miR replacement therapy.5, 6, 7 Despite promising preclinical results, the outcomes of the few translational clinical trials using miRNA replacement therapy have been disappointing, suggesting that several technical barriers still need to be overcome in order to use these RNA molecules clinically. Indeed, the first phase I clinical trial of miRNA replacement therapy involving MRX34, which was designed to restore miR-34 expression in patients with various types of cancer, resulted in an adverse immune response, probably due to the administration of a high dose of miRNA. Furthermore, RNA molecules are unstable due to nuclease degradation within blood circulation and other body fluids.35 Chemical modification of synthesized miRNAs helps overcome the immune responses and poor miRNA stability.36 Finally, in this study, we showed differential sensitivity to miR-634 overexpression in various cancer cell lines, including pancreatic cancer cells, strongly suggesting the importance of stratifying patients by miR-634 responsiveness. Thus, molecular biomarkers for defining sensitivity to miR-634 will be critical for the development of miR-634 therapy based on precision cancer medicine.
Materials and Methods
Cell Culture
HPAF-II, AsPC-1, BxPC-3, CAPAN-1, CAPAN-2, CFPAC-1, Hs766T, MIAPaCa-2, Mpanc96, PANC-1, PSN-1, and SW1990 cells were obtained from ATCC (Manassas, VA, USA). Thirteen cell lines (KMP2, KMP3, KMP4, KMP5, KMP8, KP1L, KP1NL, KP2, KP3, KP3L, KP4, PH61N, and QGP-1) and normal human fibroblasts (WI-38, TIG-1, and IMR-90 cells) were obtained from JCRB cell bank (Tokyo, Japan). HPC-Y0, HPC-Y3, and HPC-Y25 were kindly given to us by Dr. Otsuji E (Kyoto Prefectural University of Medicine). PK-59 cells were obtained from RIKEN Bioresource Research Center (Ibaraki, Japan). All cell lines were cultured in RPMI-1640 medium or DMEM (Wako, Osaka, Japan) containing 10% fetal bovine serum (FBS) at 37°C with 5% CO2.
Antibodies
For western blotting and immunofluorescence analysis, antibodies for caspase-3 (#9662), cleaved caspase-3 (#9661), cleaved PARP (#9541), XIAP (#2042), and BIRC5 (#2808) were purchased from Cell Signaling Technology (Danvers, MA, USA); antibodies for β-actin (A5441), LC3B (L7543), and TFAM (SAB1401383) were from Sigma (St. Louis, MO, USA); antibodies for APIP (ab98153), OPA1 (ab42364), and LAMP2 (ab18529) were from Abcam (Cambridge, UK); antibodies for NRF2 (sc-13032) and p62/SQSTM1 (sc-28359) were from Santa Cruz Biotechnology (Dallas, TX, USA); and the antibody for Ki-67 was from DAKO (#M7240) (Glostrup, Denmark).
Transfection of miRNAs and siRNAs
miRNA or siRNA was transfected into cells using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The miRVana miRNA mimic for miR-634 (5′-AACCAGCACCCCAACUUUGGAC-3′) (PM11538), negative control 1 (miRNA-NC) (4464058), siRNA for LDLR (the pool of four sequences: 1, 5′-GGACAAAUCUGACGAGGAA-3′; 2, 5′-GGACAGAUAUCAUCAACGA-3′; 3, 5′-GAAGUUGGCUGCGUUAAUG-3′; and 4, 5′-GCGAAGAUGCGAAGAUAUC-3′) (M-011073-01), and siRNA negative control (D-001810-05) (siNC) were obtained from Thermo Scientific.
Cell Survival Assay
Cell survival was assessed by the crystal violet staining assay. The cells were washed in PBS and fixed with 0.2% crystal violet (Wako) in 10% formaldehyde in PBS for 3 min. The excess crystal violet solution was discarded, and after the stained cells were completely air-dried, they were lysed with 2% SDS (Wako) by shaking the plates for 1 h. The optical density (OD) was measured at 560 nm using a microplate reader. The percentage absorbance of each well was determined. The OD values of cells in control wells were arbitrarily set at 100% to determine the percentage of viable cells.
Determination of the Apoptotic Cell Population and Intracellular ROS Level by FACS Analysis
Apoptotic cells were stained with the MEBCYTO Apoptosis Kit (MBL, Nagoya, Japan), and cell population analysis was performed using an Accuri Flow Cytometer (BD, San Jose, CA, USA). For the ROS detection assay, cells were incubated with 20 μM 2′,7′ –dichlorofluorescin diacetate (DCFDA) for 30 min at 37°C with 5% CO2. Fluorescence intensity in both procedures was measured using an Accuri Flow Cytometer. For each analysis, median fluorescence intensity was calculated using FlowJo software.
Western Blot Analysis
Whole cell lysates were separated by SDS-PAGE, and the proteins were transferred to polyvinylidene difluoride (PVDF) membranes (GE Healthcare, Chicago, IL, USA). After blocking with TBS containing 0.05% Tween 20 (Sigma) and 5% nonfat dry milk (BD) for 1 h, the membrane was incubated with primary antibodies overnight. The primary antibody dilutions were 1/5,000 for LC3B and β-actin, 1/2,000 for p62, and 1/1,000 for the other antibodies. The membrane was washed and incubated with horseradish peroxidase (HRP)-conjugated anti-mouse or anti-rabbit immunoglobulin G (IgG) secondary antibody (both at 1/4,000) (GE Healthcare) for 1 h. The bound antibodies were visualized in LAS3000 (FUJIFILM, Tokyo, Japan) using a Pierce ECL Western detection kit according to the manufacturer’s instructions (Thermo Scientific).
Immunofluorescence Analysis
Intracellular mitochondria were stained with 100 nmol/L MitoTracker Red CMX ROS (Life Technologies, Carlsbad, CA, USA) for 30 min at 37°C. After fixation with 10% trichloroacetic acid (TCA), images were obtained by confocal fluorescence microscopy (Nikon, Tokyo, Japan).
qRT-PCR
Total RNA was isolated using TRIsure reagent (Nippon Genetics, Tokyo, Japan) according to standard procedures. Real-time qRT-PCR was performed using an ABI PRISM 7500 Fast Real-time PCR System, TaqMan Universal PCR Master Mix, TaqMan Reverse Transcription Kit, and TaqMan MicroRNA Assays (miR-634; Assay ID: 001576) (ABI, Waltham, MA, USA) according to the manufacturer’s instructions. Gene expression values are presented as the ratio (difference in cycle threshold [Ct] values) between miR-634 and an internal reference, RNU6B (for human) (Assay ID: 001093) or snoRNA202 (for mouse) (Assay ID: 001232), respectively.
Cellular Uptake
For the cellular uptake study, cells were seeded in six-well plates and transfected with negative control or LDLR siRNA. After 24 h of transfection, LNPs containing AF647-labeled siRNA (1 nM) were incubated with cells for 1 h. AF647-positive cells were measured by FACS.
Immunohistochemical Analysis
The sections were deparaffinized in xylene and rehydrated using a graded ethanol series (100%, 90%, 80%, 70%, and 50%) to water. After antigen retrieval by boiling in 10 mM citrate buffer (pH 6.0), the sections were treated with 0.3% hydrogen peroxide in methanol to inactivate endogenous peroxidase. Nonspecific binding was blocked by incubation with goat serum in PBS. Next, the slides were incubated with rabbit antibodies against BIRC5, XIAP, APIP, and Ki-67 overnight at room temperature. The bound antibody was visualized using diaminobenzidine as a chromogen (VECTASTAIN Elite ABC kit, Vector Laboratories), and the sections were lightly counterstained with hematoxylin (Wako).
For measurement of the density per field, each of five images on sections from three tumors treated with miR-NC-LNP or miR-634-LNP were manually captured, and a threshold value was defined for each image using the single-point selection tool on the Nikon NIS Elements Br 4.0 software. The sum density was calculated, and results were normalized according to the values of tumors treated with miR-NC-LNP.
ISH Analysis for the Detection of miRNAs
The ISH assay was performed on formalin-fixed and paraffin-embedded (FFPE) tissue sections according to the manufacturer’s instructions (miRCURY LNA microRNA ISH Optimization Kit; Exiqon, Vedbaek, Denmark). In brief, the sections were deparaffinized in xylene, rehydrated with graded ethanol, and incubated with Proteinase K (Exiqon) for 10 min at 37°C. Then the sections were hybridized with digoxigenin (DIG)-labeled miR-634 probes (Exiqon) for 1 h at 55°C, washed stringently, incubated with blocking agent for 15 min, and probed with a specific anti-DIG antibody (Sigma) directly conjugated to alkaline phosphatase (AP) (Roche, Basel, Switzerland). AP converts the soluble substrates 4-nitro-blue tetrazolium (NBT) and 5-bromo-4-chloro-indolyl phosphate (BCIP) into a water- and alcohol-insoluble, dark blue NBT-BCIP precipitate. Finally, the sections were counterstained with nuclear fast red (Vector Laboratories).
Formulation of miR-634-LNPs
LNPs harboring ds-miR-NC or ds-miR-634 were formulated using L021, an ionizable lipid with an asymmetric lipid tail, according to a previously reported procedure.15,16 The miRNAs were dissolved in 10 mM citric acid buffer (pH 4.0). The ionizable lipid, DSPC (1,2-distearoyl-sn-glycero-3-phosphocholine), cholesterol, and distearoyl-rac-glycerol-polyethyleneglycol (DSG-PEG) (55:10:30:5 mol ratios) were dissolved in ethanol. The miRNA/total lipid ratio was 0.05 (wild type [WT]/WT). Using two syringe pumps, we mixed the miRNA and lipid solutions at a flow rate of 3 and 1 mL/min, respectively. These solutions were dialyzed overnight with PBS (pH 7.4) using 100-kDa dialysis tubing. The resulting solution was filtered through a 0.22-μm membrane filter to isolate LNPs, which were used in the subsequent experiments. Zetasizer Nano ZS (Malvern Panalytical, Malvern, UK) was used to determine the particle size and zeta potential. The free and total miRNA concentrations in LNPs were determined using the Quant-iT RiboGreen RNA Assay Kit (Thermo Fisher Scientific, Eugene, OR, USA) as previously described.19 The EE was calculated as follows: EE (%) = (1 − free miRNA concentration/total miRNA concentration) × 100.
In Vivo Tumor Growth Assay and Administration of miR-634-LNPs
Six-week-old female BALB/c nude mice were purchased from Charles River Laboratories (Yokohama, Japan). A total of 1.0 × 107 BxPC-3 cells in 100 μL of PBS were subcutaneously injected into the flanks of the mice. On day 7 after tumor cell inoculation, the administration of miRNA-LNPs (miR-NC or miR-634) was started. miRNA-LNPs (5 mg/kg) were intravenously injected through the tail vein of each mouse on days 7, 9, 12, and 14. At 21 days after the injection of cells, the mice were sacrificed, and tumors were resected. Tumor volume was calculated using the following formula: (shortest diameter)2 × (longest diameter) × 0.5. All experimental protocols involving mice were approved by the Tokyo Medical and Dental University Animal Care and Use Committee.
Measurement of Hepatotoxicity
Upon tumor resection on day 21, liver weight and plasma AST and ALT levels in mice were measured. The levels of AST and ALT were measured using AST/ALT assay kit (Wako) on Hitachi 7180 Biochemical Analyzer (Hitachi, Japan) in Oriental Yeast.
Statistical Analysis
Independent experiments were performed in triplicate. All p values were calculated by the two-tailed Student’s t test, and differences were considered significant at p < 0.05.
Author Contributions
J. Inoue and J. Inazawa designed the experiments. K.G. and J. Inoue performed studies including animal experiments. H.I. and K.K. contributed to the discussion of experimental results. K.G., J. Inoue, and J. Inazawa wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors thank Ayako Takahashi and Rumi Mori (Tokyo Medical and Dental University, Japan) for their technical assistance. This work was supported in part by Grants-in-Aid for Scientific Research (18K06954 to J. Inoue and 16K14630 to J. Inazawa); a Grant-in-Aid for Scientific Research on Innovative Areas “Conquering cancer through NEO-dimensional systems understandings” (15H05908 to J. Inazawa) from JSPS and MEXT; a research program of the Project for Cancer Research and Therapeutic Evolution (P-CREATE); and the Tailor-Made Medical Treatment with the BioBank Japan Project (BBJ) from the Japan Agency for Medical Research and Development (AMED). This study was supported by Nanken-Kyoten, TMDU.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.10.045.
Contributor Information
Jun Inoue, Email: jun.cgen@mri.tmd.ac.jp.
Johji Inazawa, Email: johinaz.cgen@mri.tmd.ac.jp.
Supplemental Information
References
- 1.Gebert L.F.R., MacRae I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Brien J., Hayder H., Zayed Y., Peng C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. (Lausanne) 2018;9:402. doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin S., Gregory R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer. 2015;15:321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peng Y., Croce C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 6.Hosseinahli N., Aghapour M., Duijf P.H.G., Baradaran B. Treating cancer with microRNA replacement therapy: A literature review. J. Cell. Physiol. 2018;233:5574–5588. doi: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 7.Mollaei H., Safaralizadeh R., Rostami Z. MicroRNA replacement therapy in cancer. J. Cell. Physiol. 2019;234:12369–12384. doi: 10.1002/jcp.28058. [DOI] [PubMed] [Google Scholar]
- 8.Kozaki K., Inazawa J. Tumor-suppressive microRNA silenced by tumor-specific DNA hypermethylation in cancer cells. Cancer Sci. 2012;103:837–845. doi: 10.1111/j.1349-7006.2012.02236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsuruta T., Kozaki K., Uesugi A., Furuta M., Hirasawa A., Imoto I., Susumu N., Aoki D., Inazawa J. miR-152 is a tumor suppressor microRNA that is silenced by DNA hypermethylation in endometrial cancer. Cancer Res. 2011;71:6450–6462. doi: 10.1158/0008-5472.CAN-11-0364. [DOI] [PubMed] [Google Scholar]
- 10.Uesugi A., Kozaki K., Tsuruta T., Furuta M., Morita K., Imoto I., Omura K., Inazawa J. The tumor suppressive microRNA miR-218 targets the mTOR component Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res. 2011;71:5765–5778. doi: 10.1158/0008-5472.CAN-11-0368. [DOI] [PubMed] [Google Scholar]
- 11.Fujiwara N., Inoue J., Kawano T., Tanimoto K., Kozaki K., Inazawa J. miR-634 activates the mitochondrial apoptosis pathway and enhances chemotherapy-induced cytotoxicity. Cancer Res. 2015;75:3890–3901. doi: 10.1158/0008-5472.CAN-15-0257. [DOI] [PubMed] [Google Scholar]
- 12.Endo H., Muramatsu T., Furuta M., Uzawa N., Pimkhaokham A., Amagasa T., Inazawa J., Kozaki K. Potential of tumor-suppressive miR-596 targeting LGALS3BP as a therapeutic agent in oral cancer. Carcinogenesis. 2013;34:560–569. doi: 10.1093/carcin/bgs376. [DOI] [PubMed] [Google Scholar]
- 13.Harazono Y., Muramatsu T., Endo H., Uzawa N., Kawano T., Harada K., Inazawa J., Kozaki K. miR-655 Is an EMT-suppressive microRNA targeting ZEB1 and TGFBR2. PLoS ONE. 2013;8:e62757. doi: 10.1371/journal.pone.0062757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamamoto S., Inoue J., Kawano T., Kozaki K., Omura K., Inazawa J. The impact of miRNA-based molecular diagnostics and treatment of NRF2-stabilized tumors. Mol. Cancer Res. 2014;12:58–68. doi: 10.1158/1541-7786.MCR-13-0246-T. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki Y., Ishihara H. Structure, activity and uptake mechanism of siRNA-lipid nanoparticles with an asymmetric ionizable lipid. Int. J. Pharm. 2016;510:350–358. doi: 10.1016/j.ijpharm.2016.06.124. [DOI] [PubMed] [Google Scholar]
- 16.Suzuki Y., Hyodo K., Suzuki T., Tanaka Y., Kikuchi H., Ishihara H. Biodegradable lipid nanoparticles induce a prolonged RNA interference-mediated protein knockdown and show rapid hepatic clearance in mice and nonhuman primates. Int. J. Pharm. 2017;519:34–43. doi: 10.1016/j.ijpharm.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 17.Chen D., Wu X., Zhao J., Zhao X. MicroRNA-634 functions as a tumor suppressor in pancreatic cancer via directly targeting heat shock-related 70-kDa protein 2. Exp. Ther. Med. 2019;17:3949–3956. doi: 10.3892/etm.2019.7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green D.R., Levine B. To be or not to be? How selective autophagy and cell death govern cell fate. Cell. 2014;157:65–75. doi: 10.1016/j.cell.2014.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klionsky D.J., Abdalla F.C., Abeliovich H., Abraham R.T., Acevedo-Arozena A., Adeli K., Agholme L., Agnello M., Agostinis P., Aguirre-Ghiso J.A. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozer J., Ratner M., Shaw M., Bailey W., Schomaker S. The current state of serum biomarkers of hepatotoxicity. Toxicology. 2008;245:194–205. doi: 10.1016/j.tox.2007.11.021. [DOI] [PubMed] [Google Scholar]
- 22.Lam J.K., Chow M.Y., Zhang Y., Leung S.W. siRNA Versus miRNA as Therapeutics for Gene Silencing. Mol. Ther. Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziebarth J.D., Bhattacharya A., Cui Y. Integrative analysis of somatic mutations altering microRNA targeting in cancer genomes. PLoS ONE. 2012;7:e47137. doi: 10.1371/journal.pone.0047137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhattacharya A., Ziebarth J.D., Cui Y. SomamiR: a database for somatic mutations impacting microRNA function in cancer. Nucleic Acids Res. 2013;41:D977–D982. doi: 10.1093/nar/gks1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lopes-Ramos C.M., Barros B.P., Koyama F.C., Carpinetti P.A., Pezuk J., Doimo N.T.S., Habr-Gama A., Perez R.O., Parmigiani R.B. E2F1 somatic mutation within miRNA target site impairs gene regulation in colorectal cancer. PLoS ONE. 2017;12:e0181153. doi: 10.1371/journal.pone.0181153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitehead K.A., Langer R., Anderson D.G. Knocking down barriers: advances in siRNA delivery. Nat. Rev. Drug Discov. 2009;8:129–138. doi: 10.1038/nrd2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haussecker D. Current issues of RNAi therapeutics delivery and development. J. Control. Release. 2014;195:49–54. doi: 10.1016/j.jconrel.2014.07.056. [DOI] [PubMed] [Google Scholar]
- 28.Walsh C., Ou K., Belliveau N.M., Leaver T.J., Wild A.W., Huft J., Lin P.J., Chen S., Leung A.K., Lee J.B. Microfluidic-based manufacture of siRNA-lipid nanoparticles for therapeutic applications. Methods Mol. Biol. 2014;1141:109–120. doi: 10.1007/978-1-4939-0363-4_6. [DOI] [PubMed] [Google Scholar]
- 29.Zuckerman J.E., Davis M.E. Clinical experiences with systemically administered siRNA-based therapeutics in cancer. Nat. Rev. Drug Discov. 2015;14:843–856. doi: 10.1038/nrd4685. [DOI] [PubMed] [Google Scholar]
- 30.Coelho T., Adams D., Silva A., Lozeron P., Hawkins P.N., Mant T., Perez J., Chiesa J., Warrington S., Tranter E. Safety and efficacy of RNAi therapy for transthyretin amyloidosis. N. Engl. J. Med. 2013;369:819–829. doi: 10.1056/NEJMoa1208760. [DOI] [PubMed] [Google Scholar]
- 31.Fitzgerald K., Frank-Kamenetsky M., Shulga-Morskaya S., Liebow A., Bettencourt B.R., Sutherland J.E., Hutabarat R.M., Clausen V.A., Karsten V., Cehelsky J. Effect of an RNA interference drug on the synthesis of proprotein convertase subtilisin/kexin type 9 (PCSK9) and the concentration of serum LDL cholesterol in healthy volunteers: a randomised, single-blind, placebo-controlled, phase 1 trial. Lancet. 2014;383:60–68. doi: 10.1016/S0140-6736(13)61914-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allen T.M., Cullis P.R. Liposomal drug delivery systems: from concept to clinical applications. Adv. Drug Deliv. Rev. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki Y., Hyodo K., Tanaka Y., Ishihara H. siRNA-lipid nanoparticles with long-term storage stability facilitate potent gene-silencing in vivo. J. Control. Release. 2015;220(Pt A):44–50. doi: 10.1016/j.jconrel.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 34.Barros S.A., Gollob J.A. Safety profile of RNAi nanomedicines. Adv. Drug Deliv. Rev. 2012;64:1730–1737. doi: 10.1016/j.addr.2012.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Davis S., Lollo B., Freier S., Esau C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006;34:2294–2304. doi: 10.1093/nar/gkl183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lennox K.A., Behlke M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011;18:1111–1120. doi: 10.1038/gt.2011.100. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.