Abstract
Reliable data on the diversity of the genus Diplostomum (Digenea: Diplostomidae) parasitising freshwater fishes in South Africa, as well as in Africa, is almost non-existent. Most of the morphology-based identifications of species within this genus reported from Africa require critical revision. The aim of the present study was to determine the diversity of Diplostomum metacercariae in South African fishes applying molecular and traditional morphological techniques. To achieve this aim, a total of 216 fishes belonging to 21 species collected in the Rivers Phongolo, Riet, Usuthu and Mooi in three provinces of South Africa were examined. Metacercariae of Diplostomum were recovered from the eye lenses of 38 fishes belonging to five species of the families Anguillidae, Cichilidae and Mochokidae, with an overall low prevalence of infection (18%). Metacercariae were subjected to morphological study and molecular sequencing of the partial mithochondrial cox1 and ribosomal 28S rDNA genes as well as of ribosomal ITS1-5.8S-ITS2 region. Morphological and phylogenetic analyses revealed the presence of three species which matched those previously reported from Nigeria, Iraq and China, therefore those from Tilapia sparrmanii and Synodontis zambezensis were named Diplostomum sp.; those from Anguilla labiata, Oreochromis mossambicus and S. zambezensis were named Diplostomum sp. 14; and those from Pseudocrenilabrus philander were named Diplostomum sp. 16. Geographic distribution of several species of Diplostomum appeared to be wider than expected. Morphological description and novel sequence data generated during this study will contribute to the elucidation of the life cycles of Diplostomum sp., Diplostomum sp. 14 and Diplostomum sp. 16 and advance further research of diplostomids in Africa.
Keywords: Trematoda, Metacercariae, Freshwater fish, Morphology, DNA, South Africa
Graphical abstract
Highlights
-
•
Exploring the diversity of trematodes in freshwater fishes from South Africa.
-
•
Molecular evidence for three species of Diplostomum in the Southern Hemisphere.
-
•
First detailed morphological data for metacercariae of Diplostomum in South Africa.
-
•
First report on the distribution of Diplostomum across the Palaearctic and Afrotropics.
1. Introduction
Trematodes from the genus Diplostomum von Nordmann, 1832 (Digenea: Diplostomidae) are intestinal parasites of fish-eating birds, reported from all continents, but with the majority of species described from the Nearctic and Palaearctic (Shigin, 1986, 1993). For the successful completion of their life cycles, species of Diplostomum utilise freshwater snails and fish as intermediate hosts (Niewiadomska, 2002). Metacercarial stages are regarded as pathogenic for their fish hosts and therefore they remain the focus of numerous ecological, behavioural and evolutionary studies (Ballabeni and Ward, 1993; Owen et al., 1993; Kalbe and Kurtz, 2006; Seppälä et al., 2004, Seppälä et al., 2011; Benesh and Kalbe, 2016; Klemme et al., 2016). A significant amount of research effort has recently been invested in developing the molecular sequence library for species within this genus (Galazzo et al., 2002; Moszczynska et al., 2009; Locke et al., 2010a, 2010b; 2015; Behrmann-Godel, 2013; Georgieva et al., 2013; Pérez-del-Olmo et al., 2014; Blasco-Costa et al., 2014; Selbach et al., 2015; Kuhn et al., 2015; Soldánová et al., 2017; Kudlai et al., 2017; Enabulele et al., 2018). The complete mitochondrial genomes of two closely related species from the Palaearctic, D. spathaceum (Rudolphi, 1819) and D. pseudospathaceum Niewiadomska, 1984 were characterised (Brabec et al., 2015). Currently, molecular data for Diplostomum available in GenBank includes sequences for eight species and 38 unidentified species/species-level genetic lineages from Europe (4 species and 15 unidentified species/species-level genetic lineages), North America (4 and 19, respectively), Asia (1 and 3, respectively) and Africa (1 unidentified species) (see Chibwana et al., 2013; Locke et al., 2015; Kudlai et al., 2017; Soldánová et al., 2017; Gordy and Hanington, 2019). Nevertheless, the utility of available molecular data remains limited due to the heavy bias nature towards larval stages. Among the sequences of Diplostomum spp. currently available in GenBank, only 6% were generated from the adult isolates collected from bird definitive hosts, 19% from the cercarial isolates collected from snails and 75% from the metacercarial isolates collected from fishes. The majority of molecular sequences were derived from the metacercarial stages that, in contrast to adult stages, lack sufficient morphological characteristics used for accurate species identification. Therefore, numerous metacercarial isolates remain unidentified and require the sequences from their adult in bird definitive hosts. To date, sequences for nine species generated from the adult isolates have been published, with only six being identified to species level. Of these, two species are from Europe: D. spathaceum and D. pseudospathaceum; and seven species are from North America: four named species, Diplostomum ardeae Dubois 1969, Diplostomum baeri Dubois, 1937, Diplostomum huronense (La Rue, 1927), Diplostomum indistinctum (Guberlet, 1923); and three unidentified species, Diplostomum sp. 1, 3, 4 sensu Locke et al. (2010a, 2010b).
Studies of the global diversity of Diplostomum are restricted due to sampling insufficiencies. Although numerous research studies have been done in the Northern Hemisphere, there were no comprehensive studies published from the Southern Hemisphere. To date, ten species of Diplostomum were described and reported from Argentina [D. minutum Szidat, 1964], Antarctica [D. antarcticum Freiler, 1986, D. dominicanum Freiler, 1986 and D. minutum Szidat 1964], Australia [D. amygdalum Dubois and Pearson, 1965, D. auriculosum Dubois and Pearson, 1967, D. murrayense (Johnston and Cleland, 1938), D. parvulum Dubois and Angel, 1972 and D. triangulare (S. J. Johnston, 1904)], and Brazil [D. compactum (Lutz, 1928)]. However, no molecular evidence for the distribution of Diplostomum spp. in the Southern Hemisphere exists. In Africa, eight species have been described and reported from freshwater fishes (Kudlai et al., 2018; Hoogendoorn et al., 2019) based on morphological examination. Thus far, molecular data is only available for one unidentified species, Diplostomum sp. from Nigeria (Chibwana et al., 2013).
As part of an ongoing survey of diplostomid trematodes from freshwater fishes in South Africa, we studied the diversity of the genus Diplostomum based on combined morphological and multi-locus molecular analyses involving the partial nuclear 28S rRNA and mitochondrial cox1 genes, as well as the ribosomal internal transcribed spacer region ITS1-5.8S-ITS2. We provide detailed morphological descriptions for the metacercariae of the detected species of Diplostomum, explore their phylogenetic position and relationships within the genus and provide the first molecular evidence for species of Diplostomum distributed in the Southern Hemisphere.
2. Materials and methods
2.1. Sample collection
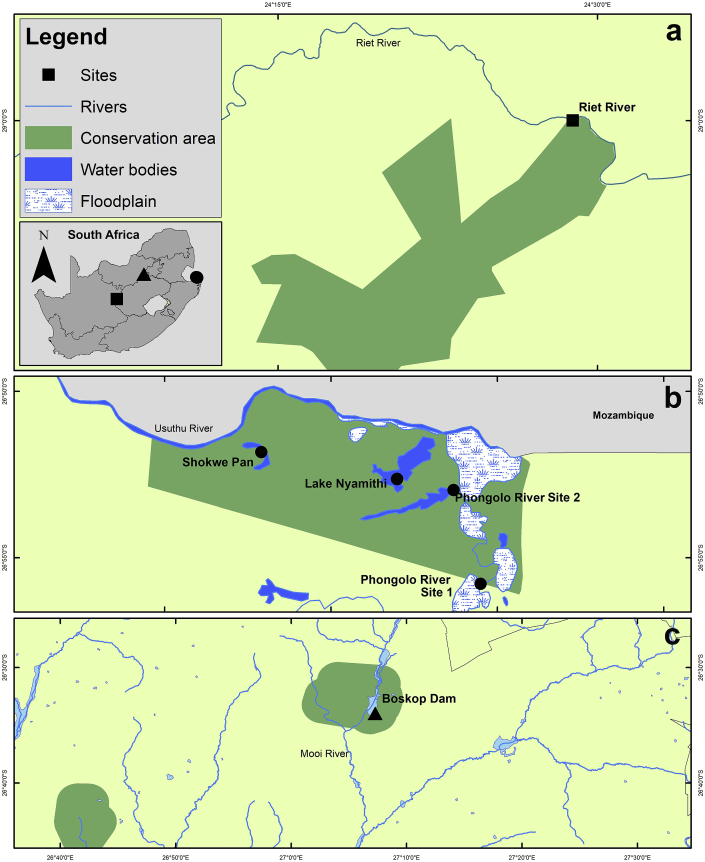
Samples were collected at five localities in three provinces of South Africa: KwaZulu-Natal Province [River Phongolo Site 1 (26°55′47″S, 32°19′30″E), River Phongolo Site 2 (26°52′58″S, 32°184′1″E), Lake Nyamithi of the River Phongolo system (26°53′35″S, 32°17′35″E), and Shokwe Pan of the River Usuthu system (26°51′50″S, 32°12′55″E) within the Ndumo Game Reserve in 2016, 2017 and 2018], Northern Cape Province [River Riet (28°59′60″S, 24°28′50″E) within the Mokala National Park in 2016], and North West Province [River Mooi at Boskop Dam (26°33′58″S, 27°07′16″E) within the Boskop Dam Nature Reserve in 2019] (Fig. 1). Sampling was carried out under the permits OP 1582/2018 (Ezemvelo KZN Wildlife) and NW 8065/03/2019 (Department of Rural, Environmental and Agricultural Development). Fishes were identified using Skelton (2001) and taxonomy followed FishBase (Froese and Pauly, 2019). The vitreous humour, retina, eye lenses and brain of each of the 216 fishes belonging to 21 species from 11 families (Table 1) were examined for the presence of metacercariae of Diplostomum. The metacercariae were examined, counted and preserved directly in 96% molecular grade ethanol. Selected representatives of metacercariae were prepared for morphological analyses, DNA isolation and sequencing. Voucher material was deposited in the Parasite Collection of the National Museum (NMB), Bloemfontein, South Africa.
Fig. 1.
Map illustrating the sampling localities on (a) River Riet in Mokala National Park (b) River Phongolo (Site 1, Site 2 and Nyamithi Lake) and the River Usuthu (Shokwe Pan) in Ndumo Game Reserve and (c) River Mooi (Boskop Dam) in Boskop Dam Nature Reserve, South Africa. The illustration was compiled in ArcGIS 10.6 (Available from https://support.esri.com/en/downloads).
Table 1.
Number of fish examined during the study.
| Fish species | River Mooi | River Phongolo Site 1 | River Phongolo Site 2 | River Phongolo Lake Nyamithi | River Usuthu | River Riet | Total |
|---|---|---|---|---|---|---|---|
| Alestidae | |||||||
| Brycinus imberi (Peters, 1852) | – | 11 | – | – | – | – | 11 |
| Hydrocynus vittatus Castelnau, 1861 | – | 1 | – | – | – | – | 1 |
| Anguillidae | |||||||
| Anguilla bengalensis labiata (Peters, 1852) | – | 4 | – | – | – | – | 4 |
| Centrarchidae | |||||||
| Micropterus salmoides (Lacepède, 1802) | 6 | – | – | – | – | – | 6 |
| Cichilidae | |||||||
| Coptodon rendalli (Boulenger, 1897) | – | 7 | – | – | – | – | 7 |
| Oreochromis mossambicus (Peters, 1852) | – | 18 | 2 | 9 | – | – | 29 |
| Pseudocrenilabrus philander (Weber, 1897) | 10 | 1 | – | – | – | 4 | 15 |
| Tilapia sparrmanii Smith, 1840 | 11 | 6 | – | – | – | 6 | 23 |
| Clariidae | |||||||
| Clarias gariepinus (Burchell, 1822) | – | 6 | 3 | – | 5 | 5 | 19 |
| Cyprinidae | |||||||
| Cyprinus carpio Linnaeus, 1758 | – | – | – | – | – | 1 | 1 |
| Labeo capensis (Smith, 1841) | – | – | – | – | – | 3 | 3 |
| Labeo congoro Peters, 1852 | – | 1 | – | – | – | – | 1 |
| Labeo cylindricus Peters, 1852 | – | 2 | – | – | – | – | 2 |
| Labeobarbus aeneus (Burchell, 1822) | – | – | – | – | – | 7 | 7 |
| Labeobarbus marequensis (Smith, 1841) | – | 3 | – | – | – | – | 3 |
| Gobiidae | |||||||
| Glossogobius giuris (Hamilton, 1822) | – | 7 | – | – | – | – | 7 |
| Mochokidae | |||||||
| Synodontis zambezensis Peters, 1852 | – | 46 | – | – | – | – | 46 |
| Mormyridae | |||||||
| Marcusenius macrolepidotus (Peters, 1852) | – | 6 | – | – | 5 | – | 11 |
| Petrocephalus wesselsi Kramer and van der Bank, 2000 | – | 2 | – | – | 3 | – | 5 |
| Schilbeidae | |||||||
| Schilbe intermedius Rüppell, 1832 | – | 12 | – | – | – | – | 12 |
| Sparidae | |||||||
| Acanthopagrus berda (Forsskål, 1775) | – | 3 | – | – | – | – | 3 |
2.2. Morphological examination
The selected metacercariae were initially studied live under a Nikon dissecting microscope or Nikon Eclipse Ni compound microscope (when possible), thereafter the parasites were transferred to molecular grade ethanol and re-examined. Photomicrographs of live and fixed metacercariae were captured with a digital camera attached to a Nikon Eclipse Ni microscope using NIS-Elements BR Camera analysis software. All measurements for the representative isolates were taken from the digital images with the aid of ImageJ (Available from https://imagej.nih.gov/ij/download.html). Twenty morphometric variables were measured from the digital images: BL, body length; BW, body width; FL, forebody length; FW, forebody width; HL, hindbody length; HW, hindbody width; OSL, oral sucker length; OSW, oral sucker width; PPHL, prepharynx length; PHL, pharynx length; PHW, pharynx width; PSL, pseudosucker length; PSW, pseudosucker width; VSL, ventral sucker length; VSW, ventral sucker width; HOL, holdfast organ length; HOW, holdfast organ width; VSDAB, distance from ventral sucker to anterior end of body; VSDPFB, distance from ventral sucker to posterior end of forebody; VSDHO, distance between ventral sucker and holdfast organ.
2.3. Generation of sequence data
Total genomic DNA was isolated from a single specimen of metacercaria following manufacturer's protocol of the KAPA Express Extract Kit (Kapa Biosystems, Cape Town, South Africa). DNA amplifications for the partial cytochrome c oxidase subunit 1 (cox1) and 28S rRNA genes, and the entire ITS1-5.8S-ITS2 gene cluster were performed using forward and reverse primers (Table 2) following polymerase chain reaction (PCR) protocols as described in Galazzo et al. (2002), Tkach et al. (2003), Moszczynska et al. (2009), and Van Steenkiste et al. (2015). PCR amplicons were purified and sequenced at Inqaba Biotechnical Industries (Pty) Ltd., Pretoria, South Africa (a commercial sequencing company). The original PCR primers as well as the internal primers (Table 2) were used for sequencing of the 28S rDNA amplicons. Contiguous sequences were assembled and edited using Geneious v. 11 (Biomatters, Auckland, New Zealand). Sequences were deposited in GenBank under the accession numbers MN808616-MN808629; MN813526-MN813549.
Table 2.
Primers used for amplification and sequencing.
| Locus/primer | Sequence | Source |
|---|---|---|
| 28S | ||
| Digl2 (forward) | 5ʹ-AAGCATATCACTAAGCGG-3ʹ | Snyder and Tkach (2001) |
| 1500R (reverse) | 5ʹ-GCTATCCTGAGGGAAACTTCG-3ʹ | Tkach et al. (2003) |
| 300Fa (forward) | 5ʹ-CAAGTACCGTGAGGGAAAGTTG-3ʹ | Littlewood et al. (2000) |
| ECD2a (reverse) | 5ʹ-CCTTGGTCCGTGTTTCAAGACGGG-3ʹ | Littlewood et al. (1997) |
| ITS1-5.8S-ITS2 | ||
| D1 (forward) | 5ʹ-AGGAATTCCTGGTAAGTGCAAG-3ʹ | Galazzo et al. (2002) |
| D2 (reverse) | 5ʹ-CGTTACTGAGGGAATCCTGGT-3ʹ | Galazzo et al. (2002) |
| cox1 | ||
| Plat-diploCOX1F (forward) | 5ʹ-CGTTTRAATTATACGGATCC-3ʹ | Moszczynska et al. (2009) |
| Plat-diploCOX1R (reverse) | 5ʹ-AGCATAGTAATMGCAGCAGC-3ʹ | Moszczynska et al. (2009) |
| Dice1F (forward) | 5ʹ-ATTAACCCTCACTAAATTWCNTTRGATCATAAG-3ʹ | Van Steenkiste et al. (2015) |
| Dice14R (reverse) | 5ʹ-TAATACGACTCACTATACCHACMRTAAACATATGATG-3ʹ | Van Steenkiste et al. (2015) |
Internal primers.
2.4. Phylogenetic analyses
Datasets for the phylogenetic analyses included 38 novel sequences obtained in the present study and 143 sequences for Diplostomum spp. available in GenBank (Supplementary Table 1). Analyses were performed using three alignments that were built according to the gene/region fragment amplified using MUSCLE (Edgar, 2004) implemented in Geneious v. 11 under default parameter values. Sequences of two species of the genus Tylodelphys Diesing, 1850, Tylodelphys mashonensis Berverly-Burton, 1963 (28S, KF189071) and Tylodelphys clavata (von Nordmann, 1832) (ITS, JQ665459; cox1, JX986909) were used as the outgroup based on the results of the phylogenetic analyses of Diplostomum published by Georgieva et al. (2013). The cox1 dataset was aligned with reference to the amino acid translation, using the trematode mitochondrial code (translation table 21; https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi#SG21) (Garey and Wolstenholme, 1989; Ohama et al., 1990). Phylogenetic trees were constructed through Bayesian inference (BI) and maximum likelihood (ML) analyses based on GTR + I + G model for ITS1-5.8S-ITS2 and cox1 datasets and HKY + I + G model for the 28S rDNA dataset. The jModelTest 2.1.2 (Guindon and Gascuel, 2003; Darriba et al., 2012) was used to estimate the best-fitting model of nucleotide substitution based on Akaike information criterion (AIC). Bayesian inference analyses were performed using MrBayes v. 3.2.6 software (Ronquist et al., 2012) run on CIPRES Science Gateway v. 3.3 (Miller et al., 2010). Markov chain Monte Carlo (MCMC) chains were run for 10, 000, 000 generations, log-likelihood scores were plotted and only the final 75% of trees were used to produce the consensus trees by setting the ‘‘burn in’’ parameter at 2,500. Maximum likelihood analyses were performed using PhyML v. 3.0 (Guindon et al., 2010) and run on the ATGC bioinformatics platform (http://www.atgc-montpellier.fr/). Nodal support was estimated using a bootstrap value of 100 pseudoreplicates. Phylogenetic trees were visualised using FigTree v. 1.4 software (Rambaut, 2012). MEGA v. 6 was used to calculate the pairwise genetic distances (p-distance) and number of nucleotide differences between sequences.
3. Results
3.1. General observations
Metacercariae of Diplostomum spp. were found in the eye lenses of 38 fishes belonging to five species: African mottled eel Anguilla labiata (Peters, 1852) (n = 3), Mozambique tilapia Oreochromis mossambicus (Peters, 1852) (n = 1), Southern mouthbrooder Pseudocrenilabrus philander (Weber, 1897) (n = 9), Plain squeaker Synodontis zambezensis Peters, 1852 (n = 24) and Banded tilapia Tilapia sparrmanii Smith, 1840 (n = 1). The overall prevalence of infection with metacercariae of Diplostomum was rather low (18%). The overall intensity of infection appeared to be relatively high in P. philander (3–21 metacercariae per fish) collected in the River Mooi and was low (1–12 metacercariae per fish) in other fish hosts from the Phongolo, Riet and Usuthu rivers.
3.2. Molecular identification of metacercariae
Thirty-eight novel sequences were generated for 16 isolates during this study: eight sequences for the partial 28S rRNA gene, 16 sequences for the ITS1-5.8S-ITS2 region and 14 sequences for the partial cox1 gene.
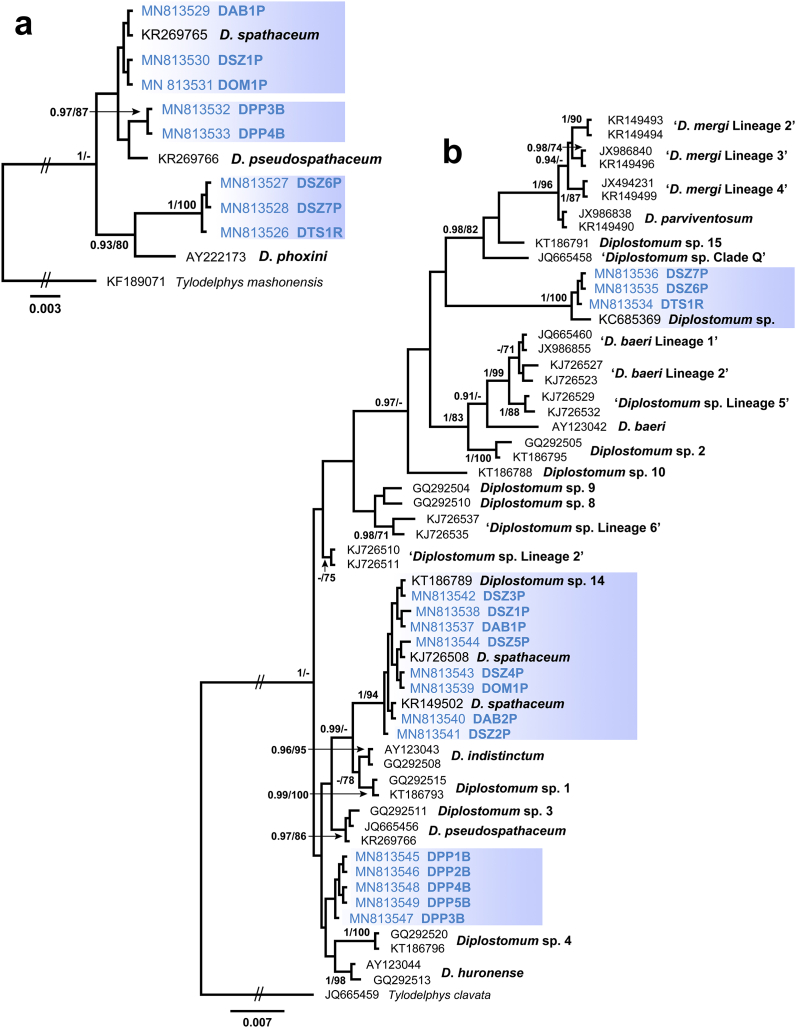
The 28S dataset (1,230 nt positions) consisted of eight sequences obtained in the present study and three sequences for Diplostomum spp. from GenBank (see Supplementary Table 1). Bayesian inference and ML analyses resulted in consensus trees with similar topologies (Fig. 2). The three newly-generated sequences for the isolates DAB1P, DSZ1P and DOM1P (Table 3) clustered with D. spathaceum (KR269765) reported from the Black-headed gull, Larus ridibundus (Linnaeus), from the Czech Republic (Brabec et al., 2015). All four isolates were identical. The sequences of two isolates (DPP3B and DPP4B) obtained from P. philander (Table 3) clustered with the sequence of D. pseudospathaceum (KR269766) also from L. ridibundus collected in the Czech Republic (Brabec et al., 2015) with low support. The genetic divergence between two identical sequences of our isolates and sequence of D. pseudospathaceum was 0.3% (4 nt). Sequences of the three remaining isolates (DSZ6P, DSZ7P and DTS1R) collected from the eye lenses of T. sparrmanii and S. zambezensis clustered with Diplostomum phoxini (Faust, 1918) (AY222173) from the Eurasian minnow, Phoxinus phoxinus (L., 1758) collected in the United Kingdom (Olson et al., 2003). The sequences of our isolates from both hosts were identical, but differed from the sequence of D. phoxini by 1.5% (8 nt).
Fig. 2.
Bayesian inference (BI) and maximum likelihood (ML) phylograms reconstructed using (a) partial 28S rDNA sequences (b) ITS1-5.8S-ITS2 sequences for species of Diplostomum. Nodal support from BI and ML analyses indicated as BI/ML; only values > 0.90 (BI) and >70 (ML) are displayed. Scale-bar indicates the expected number of substitution per site. Sequences generated in this study are in bold and indicated by blue rectangles. Codes with isolate information for newly generated sequences are provided in Table 3. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Table 3.
Summary data for the sequences of Diplostomum spp. obtained during this study.
| Species | Isolate | Host | Locality | GenBank accession numbers |
||
|---|---|---|---|---|---|---|
| 28S | ITS1-5.8S-ITS2 | cox1 | ||||
| Diplostomum sp. | DSZ6P | Synodontis zambezensis | River Phongolo Site 1 | MN813527 | MN813535 | MN808617 |
| Diplostomum sp. | DSZ7P | Synodontis zambezensis | River Phongolo Site 1 | MN813528 | MN813536 | MN808618 |
| Diplostomum sp. | DTS1R | Tilapia sparrmanii | River Riet | MN813526 | MN813534 | MN808616 |
| Diplostomum sp. 14 | DAB1P | Anguilla bengalensis labiata | River Phongolo Site 1 | MN813529 | MN813537 | MN808619 |
| Diplostomum sp. 14 | DAB2P | Anguilla bengalensis labiata | River Phongolo Site 1 | – | MN813540 | – |
| Diplostomum sp. 14 | DOM1P | Oreochromis mossambicus | River Phongolo Site 2 | MN813531 | MN813539 | MN808621 |
| Diplostomum sp. 14 | DSZ1P | Synodontis zambezensis | River Phongolo Site 1 | MN813530 | MN813538 | MN808620 |
| Diplostomum sp. 14 | DSZ2P | Synodontis zambezensis | River Phongolo Site 1 | – | MN813541 | – |
| Diplostomum sp. 14 | DSZ3P | Synodontis zambezensis | River Phongolo Site 1 | – | MN813542 | MN808622 |
| Diplostomum sp. 14 | DSZ4P | Synodontis zambezensis | River Phongolo Site 1 | – | MN813543 | MN808623 |
| Diplostomum sp. 14 | DSZ5P | Synodontis zambezensis | River Phongolo Site 1 | – | MN813544 | MN808624 |
| Diplostomum sp. 16 | DPP1B | Pseudocrenilabrus philander | River Mooi | – | MN813545 | MN808625 |
| Diplostomum sp. 16 | DPP2B | Pseudocrenilabrus philander | River Mooi | – | MN813546 | MN808626 |
| Diplostomum sp. 16 | DPP3B | Pseudocrenilabrus philander | River Mooi | MN813532 | MN813547 | MN808627 |
| Diplostomum sp. 16 | DPP4B | Pseudocrenilabrus philander | River Mooi | MN813533 | MN813548 | MN808628 |
| Diplostomum sp. 16 | DPP5B | Pseudocrenilabrus philander | River Mooi | – | MN813549 | MN808629 |
Sixteen novel sequences and 42 sequences downloaded from GenBank were used in the ITS1-5.8S-ITS2 alignment (963 nt positions). The phylogenetic tree resulted from BI and ML analyses demonstrated that the isolates sequenced in the present study clustered into three clades (Fig. 2b). Sequences of the metacercarial isolates collected from S. zambezensis (DSZ6P and DSZ7P) in the River Phongolo and T. sparrmanii (DTS1R) in the River Riet clustered with the sequence of Diplostomum sp. recorded in the lenses of Synodontis nigrita Valenciennes, 1840 in Nigeria (Chibwana et al., 2013) in a strongly supported clade. The sequence divergence between our isolates and the isolate from S. nigrita was 0.3% (2 nt). Isolates collected from A. labiata (DAB1P and DAB2P), O. mossambicus (DOM1P) and S. zambezensis (DSZ1P–DSZ5P) in the River Phongolo, similarly to the results of 28S rDNA analyses, demonstrated close relationships with the isolates of D. spathaceum (KJ726508; KR149502). This clade also included a sequence of an unidentified species Diplostomum sp. 14 sensu Locke et al. (2015) (KT186789), recently reported by Locke et al. (2015) from the Tench, Tinca tinca (Linnaeus, 1758), in China (Supplementary Table 1). The sequence divergence within the clade was 0.1% (1 nt). The clade consisting of sequences of the isolates recovered from the lenses of P. philander collected in the River Mooi received no support from both BI and ML analyses and was recovered as a sister clade to two species from North America, D. huronense and Diplostomum sp. 4 (Fig. 2b). Sequences of all isolates from P. philander were identical and did not match any sequence of Diplostomum spp. currently available in GenBank.
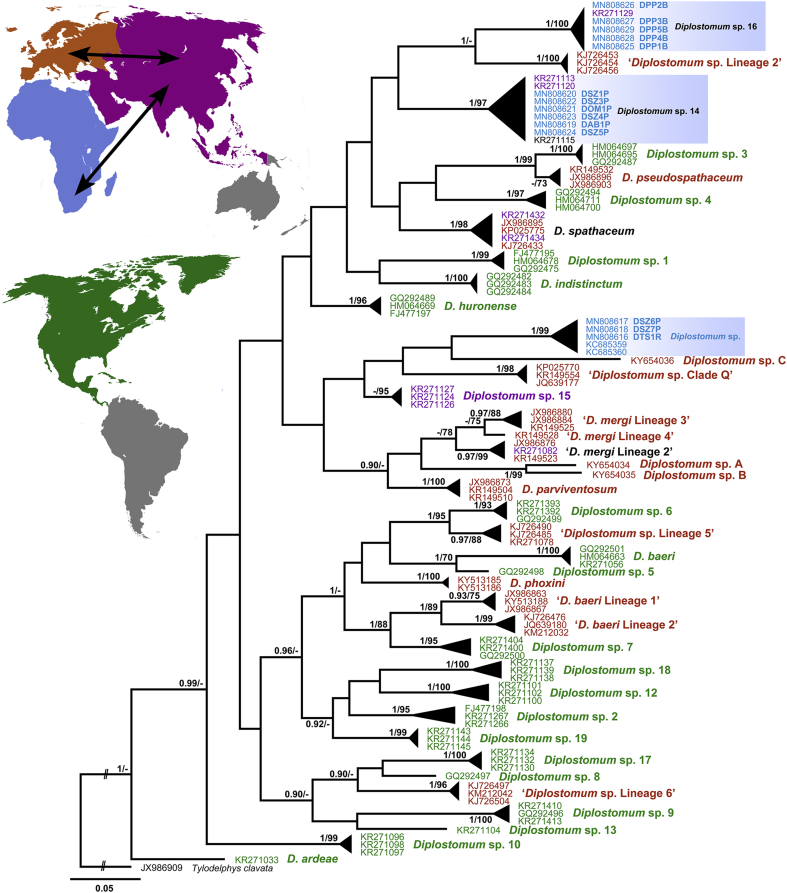
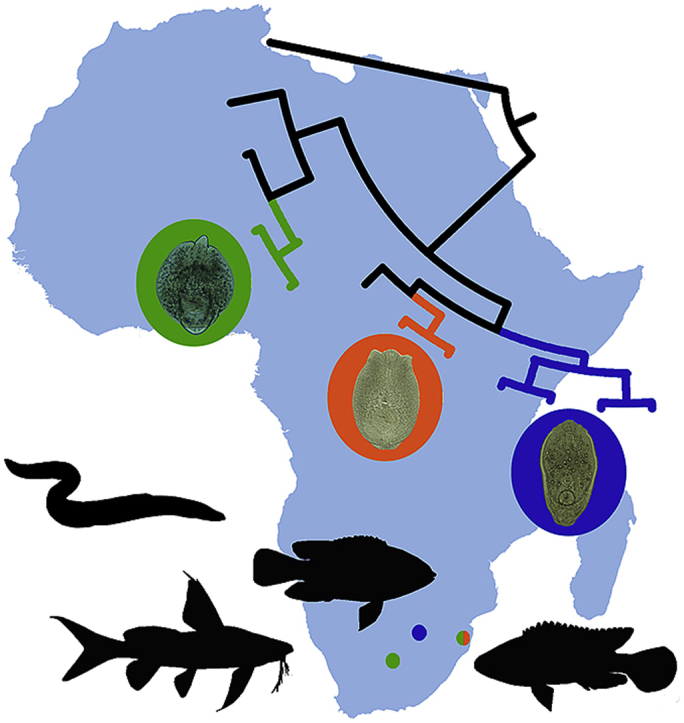
For the cox1 dataset, three sequences per species or per species-level genetic lineages (the longest possible) reported from different countries were selected. The BI and ML analyses based on the cox1 alignment (347 nt positions, 114 sequences) yielded similar phylogenetic trees (Fig. 4), however different from the hypotheses based on nuclear markers, 28S rDNA and ITS1-5.8S-ITS2. Novel sequences of the isolates collected from P. philander in the River Mooi (DPP1B–DPP5B) formed a strongly supported clade with the sequences of an unidentified species of Diplostomum, Diplostomum sp. 16 recently reported from the Tigris bleak, Alburnus caeruleus Heckel, 1843 in Iraq (Locke et al., 2015). No sequence divergence was found between our isolates and the isolate of Diplostomum sp. 16. Sequences of the metacercarial isolates collected from the three fish species in the River Phongolo (DAB1P, DOM1P, DSZ1P and DSZ3P–DSZ5P) (Table 3) clustered together with the two isolates of Diplostomum sp. 14 collected from the Snakehead, Channa argus (Cantor, 1842), and T. tinca in China (Locke et al., 2015) and one isolate of Diplostomum sp. 14 collected from Tigris kingfish, Cyprinion macrostomum Heckel, 1843, in Iraq (Locke et al., 2015) in the strongly supported clade remoted from the clade of D. spathaceum (Fig. 3). The sequence divergence within this clade ranged between 0 and 3.3% (0–9 nt). Sequences of the three metacercarial isolates from S. zambezensis (DSZ6P and DSZ7P) and T. sparrmanii (DTS1R), similarly to the results of the ITS1-5.8S-ITS2 analyses, clustered with the isolates of Diplostomum sp. from S. nigrita in Nigeria (Chibwana et al., 2013). The sequence divergence ranged between 0 and 1.1% (0–3 nt) which is considered as intraspecific.
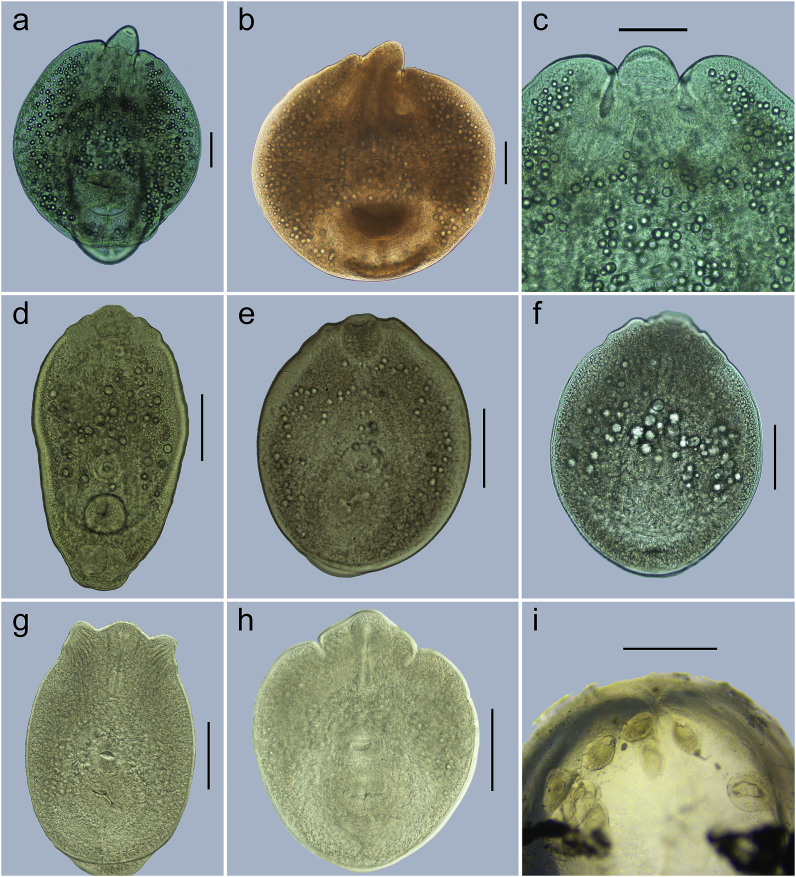
Fig. 4.
Metacercariae of Diplostomum spp. from eye lenses of different fish hosts; (a) Diplostomum sp. from Tilipia sparrmanii, live, ventral view (b) Diplostomum sp. from Tilipia sparrmanii, fixed, ventral view (c) Diplostomum sp. from Tilipia sparrmanii, live, sunken pseudosuckers (arrowhead) (hologenophore, GenBank MN813526, MN813534 and MN808616) (d) Diplostomum sp. 14 sensuLocke et al. (2015) from Synodontis zambezensis, live, ventral view (hologenophore, GenBank MN813541) (e) Diplostomum sp. 14 sensuLocke et al. (2015) from Oreochromis mossambicus, fixed, ventral view, small excretory granules (arrowhead) (hologenophore, GenBank MN813531, MN813539 and MN808621) (f) Diplostomum sp. 14 sensuLocke et al. (2015) from Synodontis zambezensis, fixed, ventral view, large excretory granules (arrowhead) (hologenophore, GenBank MN813541) (g) Diplostomum sp. 16 sensuLocke et al. (2015) from Pseudocrenilabrus philander, fixed, ventral view, everted pseudosuckers (arrowhead) (hologenophore, GenBank MN813532, MN813547 and MN808627) (h) Diplostomum sp. 16 sensuLocke et al. (2015) from Pseudocrenilabrus philander, fixed, ventral view, inverted pseudosuckers (arrowhead) (hologenophore, GenBank MN813533, MN813548 and MN808628) (i) Diplostomum sp. 16 sensuLocke et al. (2015) from Pseudocrenilabrus philander, live metacercariae inside of fish lens. Scale bars: a–h = 100 μm; i = 700 μm.
Fig. 3.
Bayesian inference (BI) and maximum likelihood (ML) phylogram reconstructed using cox1 sequences for species of Diplostomum. Nodal support from BI and ML analyses indicated as BI/ML; only values > 0.90 (BI) and >70 (ML) are displayed. Scale-bar indicates the expected number of substitution per site. Sequences generated in this study are in bold and indicated by blue rectangles. Codes with isolate information for newly generated sequences are provided in Table 3. Sequences derived from Africa are highlighted in blue, from Asia in purple, from Europe in orange, from North America in green (according to the map) and sequences reported from more than one continent are highlighted in black. Black arrows on the map demonstrate distribution of Diplostomum spathaceum and ‘D. mergi Lineage 2’ in both, Asia and Europe, and Diplostomum sp. 14 and Diplostomum sp. 16 in both, Africa and Asia. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
The species identification of the metacercarial isolates recovered from the lenses of the freshwater fishes in South Africa during this study was based on the results of the cox1 gene analyses. A total of three species were identified and they appeared to be conspecific to the three species previously reported from Nigeria (Diplostomum sp.), China (Diplostomum sp. 14) and Iraq (Diplostomum sp. 14 and Diplostomum sp. 16). Therefore, the three species are referred to as Diplostomum sp. sensu Chibwana et al. (2013), Diplostomum sp. 14 sensu Locke et al. (2015) and Diplostomum sp. 16 sensu Locke et al. (2015). The interspecific divergence between Diplostomum sp. and Diplosotmum sp. 14 was 13.2–14.7% (36–40 nt), Diplostomum sp. and Diplostomum sp. 16 was 11.8–12.1% (32–33 nt) and Diplostomum sp. 14 and Diplostomum sp. 16 was 11.8–12.5% (32–34 nt). Although the three species can be well distinguished using molecular sequence data, they also exhibit several prominent characteristics that can be used for the identification based on morphology. The morphological descriptions of the present metacercariae are provided below.
3.3. Morphological descriptions of metacercariae
3.3.1. Diplostomum sp. sensuChibwana et al. (2013)
Description (Fig. 4a−c)
[Based on one live metacercaria]. Body large, oval, 738 × 532, with maximum width at level of ventral sucker. Tegument covered with numerous tiny spines. Forebody subspherical, 590 × 532, larger than hindbody. Hindbody conical, short, 168 × 223, rounded. Forebody/hindbody length ratio 1:0.28, forebody/hindbody width ratio 1:0.42. Pseudosuckers sunken, at level of pharynx (Fig. 4c). Oral sucker subterminal, oval, 63 × 56. Prepharynx long, 21; pharynx muscular, elongate-oval, 68 × 37; oesophagus short; caeca long, reach posterior to holdfast organ. Ventral sucker transversely oval, in forebody, 82 × 102, distinctly larger than oral sucker [sucker width ratio 1:1.82]. Distance from ventral sucker to anterior end of body, 364, and to posterior end of forebody, 237. Holdfast organ large, transversely oval, 103 × 170, in posterior part of forebody. Distance from holdfast organ to ventral sucker, 11. Excretory vesicle large, V-shaped; reserve excretory system of diplostomid type. Excretory granules medium-sized, numerous, scattered throughout forebody. Excretory pore subterminal, oriented ventrally.
[Based on three ethanol-fixed metacercariae. Measurements are provided in Table 4]. Body large, subspherical, with maximum width at level of ventral sucker. Tegument covered with numerous tiny spines. Forebody transversely oval, larger than hindbody. Hindbody short, bluntly rounded. Forebody/hindbody length ratio 1:0.13–1:0.16 (1:0.15), forebody/hindbody width ratio 1:0.56–1:0.63 (1:0.60). Pseudosuckers sunken, at level of pharynx. Oral sucker subterminal, subspherical. Prepharynx long; pharynx muscular, elongate-oval; oesophagus short; caeca long, reach posterior to holdfast organ. Ventral sucker transversely oval, distinctly larger than oral sucker [oral/ventral sucker width ratio 1:1.98–1:2.29 (1:2.12)]. Distance from ventral sucker to anterior end of body, 174–298 (256), and to posterior end of forebody, 91–208 (163). Holdfast organ large, transversely oval, in posterior part of forebody, contiguous with ventral sucker. Excretory vesicle large, V-shaped; reserve excretory system of diplostomid type. Excretory granules medium-sized, numerous, scattered throughout forebody. Excretory pore subterminal, oriented ventrally.
Table 4.
Comparative metrical data on Diplostomum spp. (fixed specimens).
| Species |
Diplostomum sp. |
Diplostomum longicollisZhokhov (2014) |
||
|---|---|---|---|---|
| Host |
Synodontis zambezensis, Tilapia sparrmanii |
Enteromius humilis, Garra dembecha |
||
| Country |
South Africa |
Ethiopia |
||
| Source |
Present study |
Zhokhov (2014) |
||
| Character | Range | Mean | Range | Mean |
| BL | 379–615 | 497 | 612–1008 | 748 |
| BW | 456–525 | 491 | 378–576 | 490 |
| FL | 337–568 | 491 | – | – |
| FW | 456–563 | 515 | – | – |
| HL | 55–76 | 66 | – | – |
| HW | 288–296 | 292 | – | – |
| OSL | 38–55 | 48 | 66–72 | 63 |
| OSW | 48–54 | 50 | 66–72 | 65 |
| PPHL | 42–92 | 60 | 72–180 | 123 |
| PHL | 42–65 | 50 | 60–66 | 63 |
| PHW | 31–41 | 35 | 30–42 | 39 |
| VSL | 66–87 | 75 | 72–96 | 89 |
| VSW | 100–112 | 106 | 96–120 | 104 |
| HOL | 104–123 | 115 | 132–180 | 158 |
| HOW | 141–196 | 165 | 150–252 | 183 |
| OSW:VSW | 1:1.98–2.29 | 1:2.12 | 1:1.45–1.67a | 1:1.6a |
Estimated from measurements provided in Zhokhov (2014).
Second intermediate host: Plain squeaker Synodontis zambezensis Peters, 1852 (Siluriformes: Mochokidae); Banded tilapia Tilapia sparrmanii Smith, 1840 (Perciformes: Cichlidae).
Localities: River Phongolo Site 1 (26°55′47″S, 32°19′30″E) and River Riet (28°59′60″S, 24°28′50″E).
Prevalence: 5% (S. zambezensis, River Phongolo Site 1); 1 of 6 (T. sparrmanii, River Riet).
Intensity of infection: 1 metacercaria per fish.
Representative DNA sequences: 28S – 3 sequences (MN813526 – MN813528), ITS1-5.8S-ITS2 – 3 sequences (MN813534 – MN813536), cox1 – 3 sequences (MN808616 – MN808618).
3.3.2. Diplostomum sp. 14 sensuLocke et al. (2015)
Description (Fig. 4d−f)
[Based on 10 live metacercariae]. Body elongate-oval, 300–472 × 175–294 (354 × 229), with maximum width at level of ventral sucker or just anterior to ventral sucker. Tegument covered with numerous tiny spines. Forebody elongate-oval, 277–443 × 178–294 (336 × 235), longer than hindbody. Hindbody rounded, short, 53–84 × 57–100 (68 × 78). Forebody/hindbody length ratio, 1:0.16–1:0.26 (1:0.21). Forebody/hindbody width ratio 1:0.24–1:0.47 (1:0.34). Pseudosuckers elongate-oval, 35–57 × 20–25 (42 × 22). Oral sucker subterminal, subspherical (n = 8), 40–51 × 40–54 (45 × 46) or transversely oval (n = 2), 44–48 × 51–54 (46 × 53). Prepharynx very short, 3–10 (5) or absent; pharynx muscular, elongate-oval, 25–37 × 17–26 (31 × 20); oesophagus short; caeca thick, long, reach posterior to holdfast organ. Ventral sucker transversely oval, postequatorial, 34–46 × 42–53 (39 × 46), smaller or equal to oral sucker [oral/ventral sucker width ratio 1:0.82–1:1.08 (1:0.96)]. Distance from ventral sucker to anterior end of body, 146–224 (177) and to posterior end of forebody, 96–168 (126). Holdfast organ subspherical, 52–67 × 57–78 (61 × 68). Distance from holdfast organ to ventral sucker, 7–15 (11). Excretory granules, medium- or large sized, scattered in forebody, but generally grouped into two lateral extracaecal and one median field. Excretory vesicle V-shaped; reserve excretory system of diplostomid type. Excretory pore subterminal, oriented ventrally.
[Based on 14 ethanol-fixed metacercariae. Measurements are provided in Table 5]. Body oval, with maximum width just anterior to ventral sucker. Tegument covered with numerous tiny spines. Forebody oval, longer than hindbody. Hindbody rounded, short Forebody/hindbody length ratio 1:0.08–1:0.26 (1:0.18), forebody/hindbody width ratio, 1:0.28–1:0.60 (1:0.40). Pseudosuckers elongate-oval. Oral sucker subterminal, subspherical. Prepharynx very short or absent; pharynx muscular, elongate-oval; oesophagus short; caeca long, thick, reach posterior to holdfast organ. Ventral sucker subspherical, postequatorial, smaller to larger than oral sucker [oral/ventral sucker width ratio 1:0.84–1:1.28 (1:1.05)]. Distance from ventral sucker to anterior end of body, 85–187 (133) and to posterior end of forebody, 89–144 (105). Holdfast organ subspherical, in posterior part of forebody. Distance from holdfast organ to ventral sucker, 5–6 (5) or holdfast organ contiguous with ventral sucker. Excretory granules, medium- (Fig. 4e) or large (Fig. 4f) sized, scattered in forebody, but generally grouped into two lateral extracaecal and one median field. Excretory vesicle V-shaped; reserve excretory system of diplostomid type. Excretory pore subterminal, oriented ventrally.
Table 5.
Comparative metrical data on Diplostomum spp. (fixed specimens).
| Species |
Diplostomum sp. 14 |
Diplostomum sp. 16 |
Diplostomum garraeZhokhov (2014) |
Diplostomum montanumZhokhov (2014) |
Diplostomum tilapiaeZhokhov (2014) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| Host |
Anguilla bengalensis labiata, Oreochromis mossambicus, Synodontis zambezensis |
Pseudocrenilabrus philander |
Garra dembecha |
Enteromius humilis, Garra dembecha, Labeobarbus gorgorensis, Varicorhinus beso |
Oreochromis niloticus |
|||||
| Country |
South Africa |
South Africa |
Ethiopia |
Ethiopia |
Ethiopia |
|||||
| Source | Present study |
Present study |
Zhokhov (2014) |
Zhokhov (2014) |
Zhokhov (2014) |
|||||
| Range | Mean | Range | Mean | Range | Mean | Range | Mean | Range | Mean | |
| BL | 237–372 | 302 | 284–434 | 356 | 306–414 | 380 | 432–621 | 552 | 531–828 | 653 |
| BW | 206–271 | 240 | 212–306 | 254 | 252–306 | 283 | 240–372 | 289 | 198–234 | 212 |
| FL | 216–346 | 274 | 293–414 | 346 | – | – | – | – | – | – |
| FW | 206–271 | 237 | 231–277 | 250 | – | – | – | – | – | – |
| HL | 41–91 | 61 | 56–96 | 81 | – | – | – | – | 33–121 | 61 |
| HW | 66–102 | 81 | 72–130 | 104 | – | – | – | – | – | – |
| OSL | 36–55 | 45 | 51–62 | 55 | 54–72 | 65 | 60–78 | 63 | 53–66 | 58 |
| OSW | 35–53 | 43 | 38–59 | 49 | 54–66 | 59 | 48–72 | 67 | 53–57 | 55 |
| PPHL | 3–7 | 5 | 8–22 | 15 | 7–24 | 17 | – | – | – | – |
| PHL | 28–38 | 33 | 31–39 | 36 | 36–54 | 44 | 30–48 | 41 | 29–40 | 33 |
| PHW | 15–27 | 22 | 20–28 | 24 | 24–30 | 29 | 24–36 | 31 | 22–29 | 24 |
| PSL | 30–56 | 41 | 43–63 | 53 | 60–78 | 60 | – | – | 33–66 | 47 |
| PSW | 18–32 | 23 | 28–35 | 32 | – | – | – | – | – | – |
| VSL | 31–49 | 40 | 40–55 | 49 | 42–66 | 56 | 36–80 | 60 | 42–55 | 46 |
| VSW | 34–53 | 44 | 52–68 | 61 | 60–66 | 66 | 48–90 | 99 | 48–57 | 53 |
| HOL | 52–87 | 68 | 77–99 | 91 | 90–120 | 106 | 84–120 | 111 | 88–121 | 98 |
| HOW | 58–91 | 73 | 84–124 | 101 | 90–120 | 112 | 84–120 | 115 | 88–103 | 72 |
| OSW:VSW | 1:0.84–1.28 | 1.05 | 1:0.95–1:1.50 | 1:1.25 | 1:1–1.11a | 1.12a | 1:1–1.26a | 1.48a | 1:0.91–1a | 0.96a |
Estimated from measurements provided in Zhokhov (2014).
Second intermediate host: African mottled eel Anguilla labiata (Peters, 1852) (Anguilliformes: Anguillidae), Plain squeaker Synodontis zambezensis Peters, 1852 (Siluriformes: Mochokidae); Mozambique tilapia Oreochromis mossambicus (Peters, 1852) (Perciformes: Cichlidae).
Localities: River Phongolo Site 1 (26°55′47″S, 32°19′30″E) and River Phongolo Site 2 (26°52′58″S, 32°18′41″E).
Prevalence: 55% (S. zambezensis); 3 of 4 (A. labiata); 1 of 3 (O. mossambicus, River Phongolo Site 2).
Intensity of infection: 1–12 metacercariae per fish (S. zambezensis, River Phongolo Site 1); 2–6 metacercariae per fish (A. labiata, River Phongolo Site 1); 1 metacercaria per fish (O. mossambicus, River Phongolo Site 2).
Voucher material: 14 voucher specimens deposited in NMB P 526–530 [NMB P 526 (2 specimens), NMB P 527 (2 specimens), NMB P 528 (3 specimens), all from S. zambezensis, River Phongolo, KwaZulu-Natal, South Africa; NMB P 529 (3 specimens) and NMB P 530 (4 specimens), both from A. labiata, River Phongolo, KwaZulu-Natal, South Africa].
Representative DNA sequences: 28S – 3 sequences (MN813529 – MN813531), ITS1-5.8S-ITS2 – 8 sequences (MN813537 – MN813544), cox1 – 6 sequences (MN808619 – MN808624).
3.3.3. Diplostomum sp. 16 sensuLocke et al. (2015)
Description (Fig. 4g−i)
[Based on 15 ethanol-fixed metacercariae. Measurements are provided in Table 5]. Body elongate-oval, with maximum width at level of ventral sucker or just anterior to ventral sucker. Tegument covered with numerous tiny spines. Forebody elongate-oval, longer than hindbody. Hindbody rounded, short. Forebody/hindbody length ratio 1:0.19–1:0.27 (1:0.23), forebody/hindbody width ratio 1:0.31–1:0.49 (1:0.41). Pseudosuckers elongate-oval, everted (n = 14; Fig. 4g) or inverted (n = 1; Fig. 4h). Oral sucker subterminal, elongate-oval. Prepharynx short; pharynx muscular, elongate-oval; oesophagus short; caeca long, reach posterior to holdfast organ. Ventral sucker transversely oval, equatorial, equal or larger than oral sucker [oral/ventral sucker width ratio 1:0.95–1:1.50 (1:1.25)]. Distance from ventral sucker to anterior end of body, 128–207 (167) and to posterior end of forebody, 105–158 (133). Holdfast organ transversely oval, in posterior part of forebody, contiguous with ventral sucker. Excretory granules, medium-sized, grouped into two lateral extracaecal and one median field. Excretory vesicle V-shaped; reserve excretory system of diplostomid type. Excretory pore subterminal, oriented ventrally.
Second intermediate host: Southern mouthbrooder Pseudocrenilabrus philander (Weber, 1897) (Perciformes: Cichlidae).
Localities: River Mooi (26°33′58″S, 27°07′16″E).
Prevalence: 9 of 10.
Intensity of infection: 3–21 metacercariae per fish.
Voucher material: 15 voucher specimens deposited in NMB P 531–533 [NMB P 531 (7 specimens), NMB P 532 (5 specimens), NMB P 533 (3 specimens), all from P. philander, River Mooi, North West Province, South Africa].
Representative DNA sequences: 28S – 2 sequences (MN813532; MN813533), ITS1-5.8S-ITS2 – 5 sequences (MN813545 – MN813549), cox1 – 5 sequences (MN808625 – MN808629).
3.4. Remarks
The three species of Diplostomum described above represent species that were previously reported from freshwater fishes in Nigeria (Chibwana et al., 2013), Iraq and China (Locke et al., 2015) based on the analyses of molecular data. The previous reports did not include morphological descriptions of the metacercarial isolates and, thus our study provides the first morphological characterisation of these three species of Diplostomum linked to molecular sequences. Morphologically, metacercariae of the present species are well-distinguishable from each other. The most characteristic feature differentiating metacercariae of Diplostomum sp. from the two other species in our study is the presence of pseudosuckers of the sunken type. In addition the metacercariae of Diplostomum sp. differs from both, Diplostomum sp. 14 and Diplostomum sp. 16 by the shape (subsperical body vs elongate-oval vs elongate-oval, respectively) and size of body [379–615 × 456–525 (497 × 491) vs 237–372 × 206–271 (302 × 240) vs 284–434 × 212–306 (356 × 254)], longer prepharynx [42–92 (60) vs 3–7 (5) vs 8–22 (15)], larger pharynx [42–65 × 31–41 (50 × 35) vs 28–38 × 15–27 (33 × 22) vs 31–39 × 20–28 (36 × 24)], ventral sucker [66–87 × 100–112 (75 × 106) vs 31–49 × 34–53 (40 × 44) vs 40–55 × 52–68 (49 × 61)], oral/ventral suckers ratio [1:1.98–1:2.29 (1:2.12) vs 1:0.84–1:1.28 (1:1.05) vs 1:0.95–1:1.50 (1:1.25)] and holdfast organ [104–123 × 141–196 (115 × 165) vs 52–87 × 58–91 (68 × 73) vs 77–99 × 84–124 (91 × 101)]. Furthermore, the size and distribution of the excretory granules in the metacercariae of Diplostomum sp., i.e. medium-sized and scattered throughout the forebody, differs from the state observed in the two other species where the excretory granules are of small to large size and grouped into two lateral extracaecal and one median field.
Although the body length and width of the metacercariae of Diplostomum sp. 14 and Diplostomum sp. 16 overlap in range [237–372 × 206–271 (302 × 240) vs 284–434 × 212–306 (356 × 254)], the measurements of the metacercariae of Diplostomum sp. 14 is on average smaller than those of Diplostomum sp. 16. Diplostomum sp. 14 further differ from Diplostomum sp. 16 in having a smaller oral sucker [36–55 × 35–53 (45 × 43) vs 51–62 × 38–59 (55 × 49)], shorter prepharynx [3–7 (5) vs 8–22 (15)], smaller ventral sucker [31–49 × 34–53 (40 × 44) vs 40–55 × 52–68 (49 × 61)] and smaller holdfast organ [52–87 × 58–91 (68 × 73) vs 77–99 × 84–124 (91 × 101)].
Morphologically, the metacercariae of Diplostomum sp. strongly resemble the metacercariae of Diplostomum longicollis Zhokhov (2014) reported by Zhokhov (2014) from Enteromius humilis (Boulenger, 1902) and Garra dembecha Getahun and Stiassny, 2007 in Ethiopia in the presence of pseudosuckers of the sunken type. However, morphometric data comparisons of the fixed metacercariae revealed that the specimens in our study exhibit shorter body [379–615 (497) vs 612–1,008 (748)], smaller oral sucker [36–55 × 48–54 (48 × 50) vs 66–72 × 66–72 (63 × 65)], shorter prepharynx (42–92 vs 72–180), lower low limits for pharynx length (42–65 vs 60–66) and ventral sucker length (66–87 vs 72–96), and smaller holdfast organ [104–123 × 141–196 (115 × 165) vs 132–180 × 150–252 (158 × 183)].
The metacercariae of Diplostomum sp. 14 are morphologically most similar to the metacercariae of D. montanum Zhokhov (2014) from the eye lenses of E. humilis, G. dembecha, L. gorgorensis, V. beso and D. tilapiae Zhokhov (2014) from the eye lenses of O. niloticus collected in Ethiopia (Zhokhov, 2014). These similarities include: the shape of the body and pseudosuckers, position and size of the holdfast organ in relation to the ventral sucker and position of the ventral sucker. However, almost all body dimensions of metacercariae in our material are smaller than those of metacercariae of D. montanum and D. tilapiae as described by Zhokhov (2014) (see Table 5 for details).
The metacercariae of Diplostomum sp. 16 possess features similar to metacercariae of Diplostomum garrae Zhokhov (2014) found in the eye lens of G. dembecha in Ethiopia (Zhokhov, 2014). These include shape of the body, pseudosuckers and holdfast organ and the position of the ventral sucker. Metacercariae of Diplostomum sp. 16 can further be distinguished from D. garrae in having a lower low limits for a number of features, including length and width of body, oral sucker, pharynx, ventral sucker, holdfast organ and the length of the prepharynx (see Table 5 for details).
It should be noted that metacercariae reported in the present study were not compared to the four species of Diplostomum, D. heterobranchi Wedl, 1861, D. magnicaudum El-Naffar (1979), Diplostomum sp. type I Prudhoe and Hussey (1977) and Diplostomum sp. type II Prudhoe and Hussey (1977) collected in the brain or encysted in the mesenteries of the North African catfish Clarias gariepinus (Burchell, 1822) in Egypt and South Africa (Prudhoe and Hussey, 1977; Khalil and Polling, 1997), because detailed examination of the descriptions and illustrations of the metacercariae of these species suggested that their placement within the genus Diplostomum was erroneous.
4. Discussion
Freshwater ecosystems in South Africa are characterised by a rich fish diversity with over 180 species currently recognised (Froese and Pauly, 2019). Even though the members of the Cichlidae and Cyprinidae dominate the ichtyofauna, much information concerning diplostomid trematodes originates from studies on C. gariepinus (Siluriformes: Clariidae) (Prudhoe and Hussey, 1977; Khalil and Polling, 1997; Barson and Avenant-Oldewage, 2006; Moema et al., 2013), a species widely used in commercial aquaculture in Africa. Whilst five species of the Diplostomidae known in South Africa have been found parasitising C. gariepinus (Khalil and Polling, 1997; Kudlai et al., 2018; Hoogendoorn et al., 2019), cichlid fishes (T. sparrmanii and P. philander) have been reported as hosts only for a single species (Moema et al., 2013). Recently, Hoogendoorn et al. (2019) examined T. sparrmanii in the North West Province, South Africa and reported four diplostomid species (Bolbophorus sp. 3, Posthodiplostomum sp. 9, Uvulifer sp. 4 and Diplostomidae gen. sp.) that were not previously detected neither in cichlids or cyprinids nor in C. gariepinus. Therefore, it is not surprising that a wider sampling effort for potential hosts, including 21 species from 11 families collected in three provinces of South Africa resulted in the discovery of at least three species of Diplostomum not previously reported from this region. Two out of the three species reported here, Diplostomum sp. and Diplostomum sp. 14 were found to infect more than one host species from different fish families (Anguillidae, Cichilidae and Mochokidae). The wide host range of Diplostomum sp. 14 in South Africa is not surprising as the host range in Iraq and China is even broader and includes members of the Channidae, Cyprinidae, Hemiramphidae, Odontobutidae, Bagridae, Gobiidae, Percichthyidae. However, the wide host range of Diplostomum sp. is interesting and unexpected since it has been previously reported from only a single host species (S. nigrita) in Nigeria (Chibwana et al., 2013). Our data of numerous fish hosts therefore extends its host range. In the present study Diplostomum sp. 16 was only found parasitising a cichlid species, P. philander; which is similar to its previous record where it was found infecting a single host species (A. caeruleus) of cyprinid fish in Iraq (Locke et al., 2015). The fact that metacerariae of all three species in the present study were found in cichlid fishes suggests that their transmission is mainly associated with this group. Further comprehensive assessment of freshwater fishes in other parts of southern Africa may reveal more information on the host ranges of Diplostomum.
One of the major impediments for accurate species delineation and identification of Diplostomum based on their metacercariae is the simple morphology of larval stages (Shigin, 1986; Georgieva et al., 2013; Kudlai et al., 2017). However, the initial delineation of metacercariae in our material into three species based on morphological and morphometric characteristics was further supported by sequence data analyses. It is worth mentioning that one of the most noticeable and rather rare characteristic in metacercariae of Diplostomum sp. that we found in T. sparmanii and S. zambesensis is the pseudosuckers of the sunken type. To the best of our knowledge, this type of pseudosuckers was previously documented in metacercariae for only four species, Diplostomum gobiorum Shigin, 1965, D. pungiti Shigin, 1965, D. volvens von Nordmann, 1832 and D. longicollis (Shigin, 1986; Zhokhov, 2014).
Following the methodological recommendations in previous studies that focused on Diplostomum (Georgieva et al., 2013; Brabec et al., 2015; Locke et al., 2015), molecular identification of our material was based on multiple molecular markers including the partial 28S rRNA and cox1 genes, and ITS1-5.8S-ITS2 gene cluster. The results of both analyses that employed ribosomal markers suggested that specimens collected from A. labiata, O. mosambicus and S. zambezensis in Ndumo Game Reserve was conspecific with D. spathaceum (Fig. 2). In contrast, analyses based on the mitochondrial cox1 gene, that is considered to be more effective for species identification, demonstrated remarkable differences between these species (Fig. 3) suggesting that the specimens isolated from above mentioned hosts is in fact conspecific with Diplostomum sp. 14 reported from Iraq and China (Locke et al., 2015). This observation has once again proved the importance of the application of multiple genetic markers for accurate species delineation and identification with mitochondrial genes being a priority choice.
The results of our study not only improve the knowledge on the species diversity within Diplostomum in South Africa, but also uncover their geographical range and provide the first molecular evidence for the distribution of Diplostomum spp. in the Southern Hemisphere. The analysis of the data available for Diplostomum spp. from four continents (Africa, Asia, Europe and North America) (see Fig. 3), demonstrated a very broad distribution of several species. Diplostomum sp. recently reported from S. nigrita in Nigeria (Chibwana et al., 2013) appeared to also parasitise S. zambezensis and T. sparrmanii in South Africa, thus this species has a much wider geographical distribution within the Afrotropical region than previously recorded. To date, both Diplostomum sp. 14 and Diplostomum sp. 16 were only known from the Asian continent (Iraq and China) and the presence of these species in South Africa was rather unexpected, especially since most species of Diplostomum have a relatively restricted geographical distribution and have been reported from only one zoogeographical region. Prior to our study, based on molecular data, the only two species D. spathaceum and D. mergi Lineage 2 were known to be distributed across two continents, Europe and Asia within the Palaearctic region (Locke et al., 2015). Our results provide evidence for species within Diplostomum to have a much broader geographical distribution by being common in both Northern (within Palaearctic region) and Southern (within Afrotropical region) hemispheres. The transmission between the continents is primarily associated with the migratory patterns of their definitive hosts – piscivorous birds. Four out of six sampling localities of our survey were situated within the Ndumo Game Reserve, an area known for its high diversity of resident and migratory birds. It accommodates 430 bird species that is 19% of all species present on the African continent (Marnewick et al., 2015). Moreover, this area is located within the African-Eurasian flyways for the migratory birds that has previously been shown to be involved in the transmission of numerous bird parasites.
5. Conclusions
Based on an intensive review of the literature and data obtained in the present study, there are currently seven species of Diplostomum known to exploit freshwater fishes in Africa, namely Diplostomum garrae Zhokhov (2014), Diplostomum longicollis Zhokhov (2014), Diplostomum montanum Zhokhov (2014), Diplostomum tilapiae Zhokhov (2014), Diplostomum sp., Diplostomum sp. 14 and Diplostomum sp. 16. Of these, three species are distributed in South Africa. The discovery of these three species strongly suggests that the diversity of this genus is highly underestimated and understudied in the Southern Hemisphere. Moreover, our findings of these species from five fishes belonging to the families Anguillidae, Cichilidae and Mochokidae, significantly contributes to our knowledge on the host ranges and parasite-host associations in South Africa. In this study, we provide the first detailed morphological descriptions together with molecular data evidence for the three newly discovered species in South Africa, revealing a broader geographical distribution of Diplostomum spp. across the Palaearctic and Afrotropic regions. The expansion of the geographical range of Diplostomum between the Northern and Southern hemispheres based on molecular data evidence in particular, demonstrates the importance of the integration of morphological and molecular characterisation of these parasites. However, detailed descriptions of adults from piscivorous birds are still required for species identification and it is advised to follow a cautionary approach in the identification of new species. The development of a much needed baseline will be beneficial for future studies on the diversity, distribution and life histories of this genus, particularly in the Southern Hemisphere. A comprehensive study on the migratory patterns of piscivorous birds of the Palaerctic and Afrotropics should be considered for the study on the transmission and distribution of Diplostomum between these zoogeographical regions.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgements
We are grateful to Ezemvelo KZN Wildlife and to the Department of Rural, Environmental and Agricultural Development, North-West Provincial Government for providing the permits: OP 1582/2018 for sampling in KwaZulu-Natal and NW 8065/03/2019 for sampling in the North-West Province. We thank our Water Research Group team for their assistance during fish sampling and Dr. Wynand Malherbe for help with the design of the map. CH was partially funded by North-West University and received funding from the National Research Foundation (NRF) [DST-NRF Innovation Master's Scholarship grant 117028]. This study was supported by a Postdoctoral Fellowship from the North-West University, South Africa and a Claude Leon Foundation Postdoctoral Fellowship (2017–2018) to OK. This work is based on research supported in part by the National Research Foundation (NRF) of South Africa [NRF project CPRR160429163437 grant 105979, NJ Smit, PI]. Opinions expressed, and conclusions arrived at, are those of the authors and are not necessarily those of the NRF. This is contribution number 358 of the North-West University Water Research Group.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2019.12.003.
Policy and ethics
Animal handling was approved by the North-West University Animal Care, Research Ethics Committee (Ethics number: NWU-00160-18-S5).
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Ballabeni P., Ward P.I. Local adaptation of the tremadote Diplostomum phoxini to the European minnow Phoxinus phoxinus, its second intermediate host. Funct. Ecol. 1993;7:84–90. [Google Scholar]
- Barson M., Avenant-Oldewage A. On cestode and digenean parasites of Clarias gariepinus (Burchell, 1822) from the Rietvlei Dam, South Africa. Onderstepoort J. Vet. Res. 2006;73:101–110. doi: 10.4102/ojvr.v73i2.154. [DOI] [PubMed] [Google Scholar]
- Behrmann-Godel J. Parasite identification, succession and infection pathways in perch fry (Perca fluviatilis): new insights through a combined morphological and genetic approach. Parasitology. 2013;140:509–520. doi: 10.1017/S0031182012001989. [DOI] [PubMed] [Google Scholar]
- Benesh D.P., Kalbe M. Experimental parasite community ecology: intraspecific variation in a large tapeworm affects community assembly. J. Anim. Ecol. 2016;85:1004–1013. doi: 10.1111/1365-2656.12527. [DOI] [PubMed] [Google Scholar]
- Blasco-Costa I., Faltýnková A., Georgieva S., Skírnisson K., Scholz T., Kostadinova A. Fish pathogens near the Arctic Circle: molecular, morphological and ecological evidence for unexpected diversity of Diplostomum (Digenea: Diplostomidae) in Iceland. Int. J. Parasitol. 2014;44:703–715. doi: 10.1016/j.ijpara.2014.04.009. [DOI] [PubMed] [Google Scholar]
- Brabec J., Kostadinova A., Scholz T., Littlewood D.T. Complete mitochondrial genomes and nuclear ribosomal RNA operons of two species of Diplostomum (Platyhelminthes: trematoda): a molecular resource for taxonomy and molecular epidemiology of important fish pathogens. Parasites Vectors. 2015;8:336. doi: 10.1186/s13071-015-0949-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibwana F.D., Blasco-Costa I., Georgieva S., Hosea K.M., Nkwengulila G., Scholz T., Kostadinova A. A first insight into the barcodes for African diplostomids (Digenea: Diplostomidae): brain parasites in Clarias gariepinus (Siluriformes: Clariidae) Infect. Genet. Evol. 2013;17:62–70. doi: 10.1016/j.meegid.2013.03.037. [DOI] [PubMed] [Google Scholar]
- Darriba D., Taboada G.L., Doallo R., Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Naffar M.K. Parasites of the Egyptian moorhens. I: Diplostomum magnicaudum sp. nov. with part of its life cycle. J. Egypt. Soc. Parasitol. 1979;9:349–358. [Google Scholar]
- Enabulele E.E., Awharitoma A.O., Lawton S.P., Kirk R.S. First molecular identification of an agent of diplostomiasis, Diplostomum pseudospathaceum (Niewiadomska 1984) in the United Kingdom and its genetic relationship with populations in Europe. Acta Parasitol. 2018;63:444–453. doi: 10.1515/ap-2018-0054. [DOI] [PubMed] [Google Scholar]
- Froese R., Pauly D., editors. FishBase. World Wide Web electronic publication; 2019. www.fishbase.org version (02/2019) [Google Scholar]
- Galazzo D.E., Dayanandan S., Marcogliese D.J., McLaughlin J.D. Molecular systematics of some North American species of Diplostomum (Digenea) based on rDNA-sequence data and comparisons with European congeners. Can. J. Zool. 2002;80:2207–2217. [Google Scholar]
- Garey J.R., Wolstenholme D.R. Platyhelminth mitochondrial DNA: evidence for early evolutionary origin of a tRNA ser AGN that contains a dihydrouridine arm replacement loop, and of serine-specifying AGA and AGG codons. J. Mol. Evol. 1989;28:374–387. doi: 10.1007/BF02603072. [DOI] [PubMed] [Google Scholar]
- Georgieva S., Soldánová M., Pérez-del-Olmo A., Dangel R.D., Sitko J., Sures B., Kostadinova A. Molecular prospecting for European Diplostomum (Digenea: Diplostomidae) reveals cryptic diversity. Int. J. Parasitol. 2013;43:57–72. doi: 10.1016/j.ijpara.2012.10.019. [DOI] [PubMed] [Google Scholar]
- Gordy M.A., Hanington P.C. A fine‐scale phylogenetic assessment of digenean trematodes in central Alberta reveals we have yet to uncover their total diversity. Ecol. Evol. 2019;9:3153–3238. doi: 10.1002/ece3.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hoogendoorn C., Smit N.J., Kudlai O. Molecular and morphological characterisation of four diplostomid metacercariae infecting Tilapia sparrmanii (Perciformes: Cichlidae) in the North West Province, South Africa. Parasitol. Res. 2019;118:1403–1416. doi: 10.1007/s00436-019-06285-y. [DOI] [PubMed] [Google Scholar]
- Kalbe M., Kurtz J. Local differences in immunocompetence reflect resistance of sticklebacks against the eye fluke Diplostomum pseudospathaceum. Parasitology. 2006;132:105–116. doi: 10.1017/S0031182005008681. [DOI] [PubMed] [Google Scholar]
- Khalil L.F., Polling L. University of the North; Pietersburg, Republic of South Africa: 1997. Check List of the Helminth Parasites of African Freshwater Fishes; pp. 36–38. [Google Scholar]
- Klemme I., Louhi K.R., Karvonen A. Host infection history modifies co‐infection success of multiple parasite genotypes. J. Anim. Ecol. 2016;85:591–597. doi: 10.1111/1365-2656.12472. [DOI] [PubMed] [Google Scholar]
- Kudlai O., Oros M., Kostadinova A., Georgieva S. Exploring the diversity of Diplostomum (digenea: Diplostomidae) in fishes from the River danube using mitochondrial DNA barcodes. Parasites Vectors. 2017;10:592. doi: 10.1186/s13071-017-2518-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudlai O., Scholz T., Smit N.J. Trematoda. In: Scholz T., Vanhove M.P.M., Smit N., Jayasundera Z., Gelnar M.A., editors. vol. 18. ABC Taxa; 2018. pp. 245–268. (A Guide to the Parasites of African Freshwater Fishes). [Google Scholar]
- Kuhn J.A., Kristoffersen R., Knudsen R., Jakobsen J., Marcogliese D.J., Locke S.A., Amundsen P. Parasite communities of two three-spined stickleback populations in subarctic Norway - effects of a small spatial-scale host introduction. Parasitol. Res. 2015;114:1327–1339. doi: 10.1007/s00436-015-4309-2. [DOI] [PubMed] [Google Scholar]
- Littlewood D.T., Curini-Galletti M., Herniou E.A. The interrelationships of Proseriata (Platyhelminthes: seriata) tested with molecules and morphology. Mol. Phylogenetics Evol. 2000;16:449–466. doi: 10.1006/mpev.2000.0802. [DOI] [PubMed] [Google Scholar]
- Littlewood D.T., Rohde K., Clough K.A. Parasite speciation within or between host species? –Phylogenetic evidence from site-specific polystome monogeneans. Int. J. Parasitol. 1997;27:1289–1297. doi: 10.1016/s0020-7519(97)00086-6. [DOI] [PubMed] [Google Scholar]
- Locke S.A., McLaughlin J.D., Dayanandan S., Marcogliese D.J. Diversity and specificity in Diplostomum spp. metacercariae in freshwater fishes revealed by cytochrome c oxidase I and internal transcribed spacer sequences. Int. J. Parasitol. 2010;40:333–343. doi: 10.1016/j.ijpara.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Locke S.A., McLaughlin J.D., Marcogliese D.J. DNA barcodes show cryptic diversity and a potential physiological basis for host specificity among Diplostomoidea (Platyhelminthes: digenea) parasitizing freshwater fishes in the St. Lawrence River, Canada. Mol. Ecol. 2010;19:2813–2827. doi: 10.1111/j.1365-294X.2010.04713.x. [DOI] [PubMed] [Google Scholar]
- Locke S.A., Al-Nasiri F.S., Caffara M., Drago F., Kalbe M., Lapierre A.R., Marcogliese D.J. Diversity, specificity and speciation in larval Diplostomidae (Platyhelminthes: digenea) in the eyes of freshwater fish, as revealed by DNA barcodes. Int. J. Parasitol. 2015;45:841–855. doi: 10.1016/j.ijpara.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Marnewick M.D., Retief E.F., Theron N.T., Wright D.R., Anderson T.A. BirdLife South Africa; Johannesburg: 2015. Important Bird and Biodiversity Areas of South Africa. [Google Scholar]
- Miller M.A., Pfeiffer W., Schwartz T. Gateway Computing Environments Workshop (GCE) IEEE; 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees; pp. 1–8. [Google Scholar]
- Moema E.B., King P.H., Rakgole J.N., Baker C. Descriptions of diplostomid metacercariae (Digenea: Diplostomidae) from freshwater fishes in the Tshwane area. Onderstepoort J. Vet. Res. 2013;80:1–7. doi: 10.4102/ojvr.v80i1.611. [DOI] [PubMed] [Google Scholar]
- Moszczynska A., Locke S.A., McLaughlin J.D., Marcogliese D.J., Crease T.J. Development of primers for the mitochondrial cytochrome c oxidase I gene in digenetic trematodes illustrates the challenge of barcoding parasitic helminths. Mol. Ecol. Resour. 2009;9:75–82. doi: 10.1111/j.1755-0998.2009.02634.x. [DOI] [PubMed] [Google Scholar]
- Niewiadomska K. Family Diplostomidae poirier, 1886. In: Gibson D.I., Jones A., Bray R.A., editors. vol. 1. CAB International and The Natural History Museum; Wallingford - London: 2002. pp. 167–196. (Keys to the Trematoda). [Google Scholar]
- Ohama T., Osawa S., Watanabe K., Jukes T.H. Evolution of the mitochondrial genetic code IV. AAA as an asparagine codon in some animal mitochondria. J. Mol. Evol. 1990;30:329–332. doi: 10.1007/BF02101887. [DOI] [PubMed] [Google Scholar]
- Olson P.D., Cribb T.H., Tkach V.V., Bray R.A., Littlewood D.T. Phylogeny and classification of the digenea (Platyhelminthes: trematoda) Int. J. Parasitol. 2003;33:733–755. doi: 10.1016/s0020-7519(03)00049-3. [DOI] [PubMed] [Google Scholar]
- Owen S.F., Barber I., Hart P.J. Low level infection by eye fluke, Diplostomum spp., affects the vision of three‐spined sticklebacks, Gasterosteus aculeatus. J. Fish Biol. 1993;42:803–806. [Google Scholar]
- Pérez-del-Olmo A., Georgieva S., Pula H.J., Kostadinova A. Molecular and morphological evidence for three species of Diplostomum (Digenea: Diplostomidae), parasites of fishes and fish-eating birds in Spain. Parasites Vectors. 2014;7:502. doi: 10.1186/s13071-014-0502-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudhoe S., Hussey C.G. Some parasitic worms in freshwater fishes and fish-predators from the Transvaal, South Africa. Afr. Zool. 1977;12:113–147. [Google Scholar]
- Rambaut A. University of Edinburgh, Institute of Evolutionary Biology; Edinburgh: 2012. FigTree V1. 4. Molecular Evolution, Phylogenetics and Epidemiology.http://tree.bio.ed.ac.uk/software/figtree Retrieved from. [Google Scholar]
- Ronquist F., Teslenko M., Van Der Mark P., Ayres D.L., Darling A., Höhna S. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach C., Soldánová M., Georgieva S., Kostadinova A., Sures B. Integrative taxonomic approach to the cryptic diversity of Diplostomum spp. in lymnaeid snails from Europe with a focus on the ‘Diplostomum mergi’species complex. Parasites Vectors. 2015;8:300. doi: 10.1186/s13071-015-0904-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppälä O., Karvonen A., Valtonen E.T. Parasite-induced change in host behaviour and susceptibility to predation in an eye fluke–fish interaction. Anim. Behav. 2004;68:257–263. [Google Scholar]
- Seppälä O., Karvonen A., Valtonen E.T. Eye fluke-induced cataracts in natural fish populations: is there potential for host manipulation? Parasitology. 2011;138:209–214. doi: 10.1017/S0031182010001228. [DOI] [PubMed] [Google Scholar]
- Shigin A.A. Nauka; Moscow: 1986. Trematode Fauna of the USSR. Genus Diplostomum. Metacercariae; p. 253. (In Russian) [Google Scholar]
- Shigin A.A. Nauka; Moscow: 1993. Trematodes of the Fauna of Russia and Neighbouring Regions. Genus Diplostomum. Adults; p. 206. (In Russian) [Google Scholar]
- Skelton P.H. Struik Publishers; Cape Town: 2001. A Complete Guide to the Freshwater Fishes of Southern Africa. 395 pp. [Google Scholar]
- Snyder S.D., Tkach V.V. Phylogenetic and biogeographical relationships among some holarctic frog lung flukes (Digenea: haematoloechidae) J. Parasitol. 2001;87:1433–1440. doi: 10.1645/0022-3395(2001)087[1433:PABRAS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Soldánová M., Georgieva S., Roháčová J., Knudsen R., Kuhn J.A., Henriksen E.H., Kostadinova A. Molecular analyses reveal high species diversity of trematodes in a sub-Arctic lake. Int. J. Parasitol. 2017;47:327–345. doi: 10.1016/j.ijpara.2016.12.008. [DOI] [PubMed] [Google Scholar]
- Tkach V.V., Littlewood D.T., Olson P.D., Kinsella J.M., Swiderski Z. Molecular phylogenetic analysis of the microphalloidea ward, 1901 (trematoda: digenea) Syst. Parasitol. 2003;56:1–15. doi: 10.1023/a:1025546001611. [DOI] [PubMed] [Google Scholar]
- Van Steenkiste N., Locke S.A., Castelin M., Marcogliese D.J., Abbott C.L. New primers for DNA barcoding of digeneans and cestodes (Platyhelminthes) Mol. Ecol. Resour. 2015;15:945–952. doi: 10.1111/1755-0998.12358. [DOI] [PubMed] [Google Scholar]
- Zhokhov A.E. Metacercariae of new trematode species of the genus Diplostomum (trematoda, Diplostomidae) from fishes of lake tana, Ethiopia. Inland Water Biol. 2014;7:15–24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.