Abstract
Inherited retinal dystrophies (IRDs) are characterized by progressive photoreceptor degeneration and vision loss. Usher syndrome (USH) is a syndromic IRD characterized by retinitis pigmentosa (RP) and hearing loss. USH is clinically and genetically heterogeneous, and the most prevalent causative gene is USH2A. USH2A mutations also account for a large number of isolated autosomal recessive RP (arRP) cases. This high prevalence is due to two recurrent USH2A mutations, c.2276G>T and c.2299delG. Due to the large size of the USH2A cDNA, gene augmentation therapy is inaccessible. However, CRISPR/Cas9-mediated genome editing is a viable alternative. We used enhanced specificity Cas9 of Streptococcus pyogenes (eSpCas9) to successfully achieve seamless correction of the two most prevalent USH2A mutations in induced pluripotent stem cells (iPSCs) of patients with USH or arRP. Our results highlight features that promote high target efficacy and specificity of eSpCas9. Consistently, we did not identify any off-target mutagenesis in the corrected iPSCs, which also retained pluripotency and genetic stability. Furthermore, analysis of USH2A expression unexpectedly identified aberrant mRNA levels associated with the c.2276G>T and c.2299delG mutations that were reverted following correction. Taken together, our efficient CRISPR/Cas9-mediated strategy for USH2A mutation correction brings hope for a potential treatment for USH and arRP patients.
Introduction
Inherited retinal dystrophies (IRDs) are a clinically and genetically heterogeneous group of neurodegenerative disorders. They are characterized by progressive vision loss due to degeneration of the light-sensing photoreceptor cells of the retina. IRDs affect approximately 1 in 2,000 individuals worldwide.1 They can be divided into non-syndromic forms, characterized by an isolated retinal phenotype, or syndromic forms, in which another organ in addition to the eye is affected. The most common form of non-syndromic IRD is retinitis pigmentosa (RP), characterized by progressive tunnel vision, which has a prevalence of 1 in 4,000 individuals worldwide.2 The most prevalent form of syndromic IRD is Usher syndrome (USH), which associates RP and hearing loss, and in severe cases, vestibular dysfunction. USH is the most common cause of inherited deaf-blindness and has a prevalence of approximately 1 in 6,000 individuals.3 Three clinical forms can be distinguished according to disease severity and progression: USH type 1 (USH1), USH type 2 (USH2), and USH type 3 (USH3), each of which is further subdivided depending on the causative gene. USH2 is the most frequent form and is characterized by congenital moderate-to-severe hearing loss and post-pubertal onset of RP.4
Up to 85% of USH2 patients have causative mutations in the gene USH2A.5 In addition, USH2A mutations account for 8%6 to 22%7 of non-syndromic autosomal recessive RP (arRP) cases, depending on the origin of the population. Therefore, taken together, USH2A is considered the most prevalent causative gene for both isolated and syndromic arRP.8,9 Over 600 causative USH2A mutations have been identified and are distributed throughout the gene (https://databases.lovd.nl/shared/genes/USH2A). The majority of these are private mutations; however, there do exist recurrent mutations likely because of founder effects.9, 10, 11 The most prevalent mutations are c.2276G>T (p.Cys759Phe) and c.2299delG (p.Glu767Serfs*21). These pathogenic variants are located 22 bp apart in exon 13 and account for approximately half of the cases of USH2 and arRP. Interestingly, when c.2276G>T is present in the homozygous or heterozygous state, it leads to isolated arRP.12 This missense variant is thus considered as a “retinal disease-specific” USH2A allele.13 By contrast, c.2299delG is a severe allele and, unless it is present in the compound heterozygote state with a retinal disease-specific allele, leads to USH2.
USH has an autosomal recessive mode of transmission and thus could be potentially treated by gene augmentation therapy. Gene augmentation using adeno-associated viral (AAV) vectors has proven to be a safe and encouraging treatment for autosomal recessive IRDs.14, 15, 16, 17 However, the major limitation of AAV vectors is their cloning capacity (<4.7 kb), which hinders the transfer of larger cDNAs. This limitation was circumvented for the 7.5-kb (GenBank: NM_000260) cDNA of the USH1B causative gene, MYO7A. MYO7A gene transfer was accomplished using an equine infectious anemia virus (EIAV)-based lentiviral vector, which has a cloning capacity of 9 kb.18 By contrast, the 15.6-kb USH2A cDNA (GenBank: NM_206933.2) makes even EIAV-mediated transfer inaccessible for this gene. A promising alternative is gene correction using genome-editing strategies, such as the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated nuclease (Cas) (CRISPR/Cas system), which has shown positive results for the correction of IRD causative genes.19, 20, 21
The CRISPR/Cas system is a bacterial adaptive immune system,22,23 which has been largely used for in vivo and ex vivo genome-editing therapies.24, 25, 26 The system comprises two primary elements: first, the Cas nuclease, the most commonly used is Cas9 of Streptococcus pyogenes (SpCas9); and second, the single guide RNA molecule (sgRNA). The Cas nuclease is specifically guided to the target locus in the DNA by the sgRNA sequence and a protospacer adjacent motif (PAM), a 3-nt sequence found at the 3′ end of the sgRNA sequence (NGG in the case of SpCas9), where it will induce a double-strand break (DSB).27 After cleavage of the DNA, the target locus will typically undergo one of the two major pathways for DNA repair. In the error-prone non-homologous end-joining (NHEJ) pathway, the two ends of the DSB are randomly re-ligated, leaving insertions and deletions (INDELs) at the desired region. Alternatively, the homology-directed repair (HDR) pathway can be used with a donor template to precisely edit the genome at the desired location.
The coupling of the simple, rapid, and highly efficient CRISPR/Cas9 system together with patient-specific induced pluripotent stem cell (iPSC) technology has opened up a window for regenerative therapy and personalized medicine for IRDs.28 In this vein, as a proof-of-concept study, we developed two CRISPR/Cas9 strategies to target the recurrent USH2A mutations in the iPSCs of patients: one patient carried the homozygous c.2299delG mutation and presented with USH2,29 and the other patient was compound heterozygous for the c.2276G>T mutation and the c.2299delG mutation, and presented with isolated arRP.30 We demonstrate successful correction of the mutant alleles in the iPSCs of both patients. Moreover, we used an enhanced specificity SpCas9 (eSpCas9), which, together with an optimal sgRNA sequence, demonstrated high efficiency and specificity without detectable events in the predicted off-targets regions. In addition, the CRISPR/Cas9-mediated correction of the iPSCs did not disturb their genetic integrity or pluripotency. Lastly, and unexpectedly, our results show aberrant USH2A mRNA levels associated with the c.2299delG mutation, which were rescued following gene correction. Moreover, these levels appeared normalized in the presence of the c.2276G>T mutation. This mRNA expression profile is particularly intriguing and raises considerations for the genotype-phenotype correlation of these two recurrent mutations.
Results
An Optimal CRISPR/Cas9 Design for Correcting the Most Prevalent USH2A Mutations in Exon 13
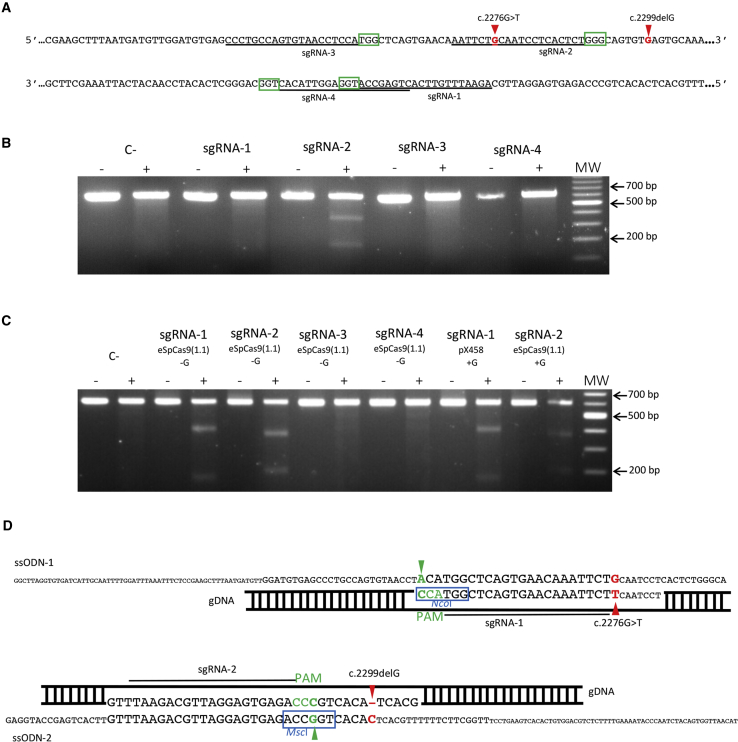
We designed four different sgRNAs in the vicinity of the c.2276G>T and c.2299delG mutations in exon 13. The sgRNAs were chosen according to the presence of the canonical NGG PAM site used by SpCas9 (Figure 1A). All four sgRNAs were individually subcloned into the enhanced specificity Cas9 plasmid [eSpCas9 (1.1), #71814; Addgene]. This plasmid co-expresses the sgRNA and the eSpCas9 with EGFP, which is linked to the C terminus of eSpCas9 by a 2A peptide. This variant of the wild-type SpCas9 has been shown to induce robust on-target activity with significant reduction in off-targets.31 All four sgRNAs were designed and cloned with a mismatched G at the 5′ of the sgRNA sequence because it has been reported that the human U6 promoter, which drives sgRNA expression in this plasmid, prefers a G to start transcription.32
Figure 1.
Design and Selection of the Optimal sgRNA and ssODN
(A) Sequence of exon 13 of USH2A. Red arrowheads indicate the c.2276G>T and c.2299delG mutations. The four different sgRNAs (sgRNA-1 to sgRNA-4) are underlined. The green boxes correspond to the 3′ PAM sequences adjacent to each sgRNA. (B) Representative gel image of a T7E1 assay in HEK293 cells for each sgRNA. Untreated DNA from the same HEK293 cells was used as a negative control (C−). Samples not treated with T7E1 are indicated by a minus sign. Samples treated with T7E1 are indicated by a plus sign. (C) Representative gel image of a T7E1 assay in HEK293 cells for each sgRNA. The absence or presence of the mismatched G in the sgRNA sequence is indicated as “−G” or “+G,” respectively. Untreated DNA from the same HEK293 cells was used as a negative control (C−). Samples not treated with T7E1 are indicated by a minus sign. Samples treated with T7E1 are indicated by a plus sign. (D) Diagram showing the design of the exogenous repair templates ssODN-1 (upper panel) and ssODN-2 (lower panel). The position of the PAM sequences is shown in green, and green arrowheads indicate the PAM silent mutations introduced into the ssODNs. The restriction enzyme digestion sites for NcoI and MscI in ssODN-1 and ssODN-2, respectively, are boxed in blue. In ssODN-1, a red arrowhead indicates the guanine substitution to correct the c.2276G>T mutation. In ssODN-2, a red arrowhead indicates the inclusion of the cytosine (antisense allele) to correct the c.2299delG mutation.
To determine the cleavage efficiency of the selected sgRNAs, each eSpCas9 (1.1) plasmid with the corresponding sgRNA was transfected into HEK293 cells. Forty-eight hours post-transfection, cells were harvested for genomic DNA extraction. The eSpCas9-induced cleavage directed by the sgRNAs was assessed by the T7 endonuclease I (T7E1) assay. Only one of the four sgRNAs (sgRNA-2) resulted in a DSB in the DNA by eSpCas9 (Figure 1B). Recent studies have suggested that the addition of the mismatched G in the sgRNA sequence when using Cas9 variants with enhanced specificity might interfere with the on-target Cas9 activity.33 Thus, we redesigned all four sgRNAs without the mismatched G, subcloned them into the eSpCas9 (1.1) plasmid, and re-assessed cleavage using the T7E1 assay. Cleavage was again induced in the presence of the modified sgRNA-2 but also in the presence of sgRNA-1. There was no detectable cleavage using sgRNA-3 and sgRNA-4 (Figure 1C). To confirm that the cleavage directed by the modified sgRNA-1 in the eSpCas9 plasmid was due to the removal of the mismatched G, we subcloned the original sgRNA-1, with the mismatched G, into a wild-type SpCas9 plasmid (pX458, #106097; Addgene) and re-performed a T7E1 assay. As shown in Figure 1C, sgRNA-1 with the mismatched G permitted cleavage of the targeted DNA by wild-type SpCas9. In view of these results, the modified sgRNA-1 and sgRNA-2 without the mismatched Gs were retained for the correction of the missense variant c.2276G>T and the c.2299delG mutation, respectively.
Following validation of the activity of the two sgRNAs in HEK293 cells, repair templates in the form of a single-stranded oligodeoxynucleotide (ssODN) were designed for the correction of the two mutations via HDR. ssODNs have been shown to be effective and powerful templates for directing HDR upon DSB in the genome.34,35 In addition, a previous study demonstrated that the use of asymmetric ssODN complementary to the non-targeted DNA strand enhanced HDR efficiency.36 Therefore, we designed the ssODNs following this criterion and using the reference sequence for USH2A (GenBank: NM_206933.2). Two ssODNs were designed: ssODN-1, for sgRNA-1, which would convert the mutant T into the wild-type G to correct the c.2276G>T variant, and ssODN-2 for sgRNA-2, which would introduce a C on the antisense allele to correct the c.2299delG variant (Figure 1D). Moreover, to avoid re-cleavage of the target DNA by Cas9 after HDR repair, silent mutations were introduced into the PAM sequences in both ssODNs.37 For ssODN-1, the PAM silent mutation destroyed an NcoI restriction enzyme site present in the USH2A reference sequence. By contrast, the PAM silent mutation for ssODN-2 introduced an MscI site into the target sequence. Thus, these two PAM silent mutations could also be used to facilitate genotyping of HDR events. The effect of the PAM silent mutations on possible splicing alterations was assayed using the computational prediction tools NetGene238 and Human Splicing Finder.39 The synonymous nucleotide changes in ssODN-1 and ssODN-2 did not result in any predicted effects on splicing. Lastly, phosphorothioate-modified terminal bases were added at both ends of the ssODNs to enhance ssODN stability.40
CRISPR/Cas9 Mediates Correction of the c.2299delG Mutation in the iPSCs of an USH2 Patient
We previously generated an iPSC line, INMi002 or USH2A-USH-iPSC, from a patient presenting USH2 caused by the homozygous USH2A mutation c.2299delG29 (Table 1). The USH2A-USH-iPSC line was co-nucleofected with the eSpCas9-sgRNA-2 plasmid and the ssODN-2. Forty-eight hours post-nucleofection, EGFP-positive iPSCs were single-cell sorted by fluorescence-activated cell sorting (FACS) and seeded into three 96-well plates. The surviving iPSC clones (5 out of 288) were then expanded for screening of HDR events.
Table 1.
Patient Samples
| Cell Lines | Clinical Diagnosis | Mutation in A1 | Mutation in A2 | Reference |
|---|---|---|---|---|
| USH2A-USH-iPSC | USH2 | c.2299delG | c.2299delG | 29 |
| USH2A-RP-iPSC | non-syndromic arRP | c.2276G>T | c.2299delG | 28,30 |
A1, allele 1; A2, allele 2.
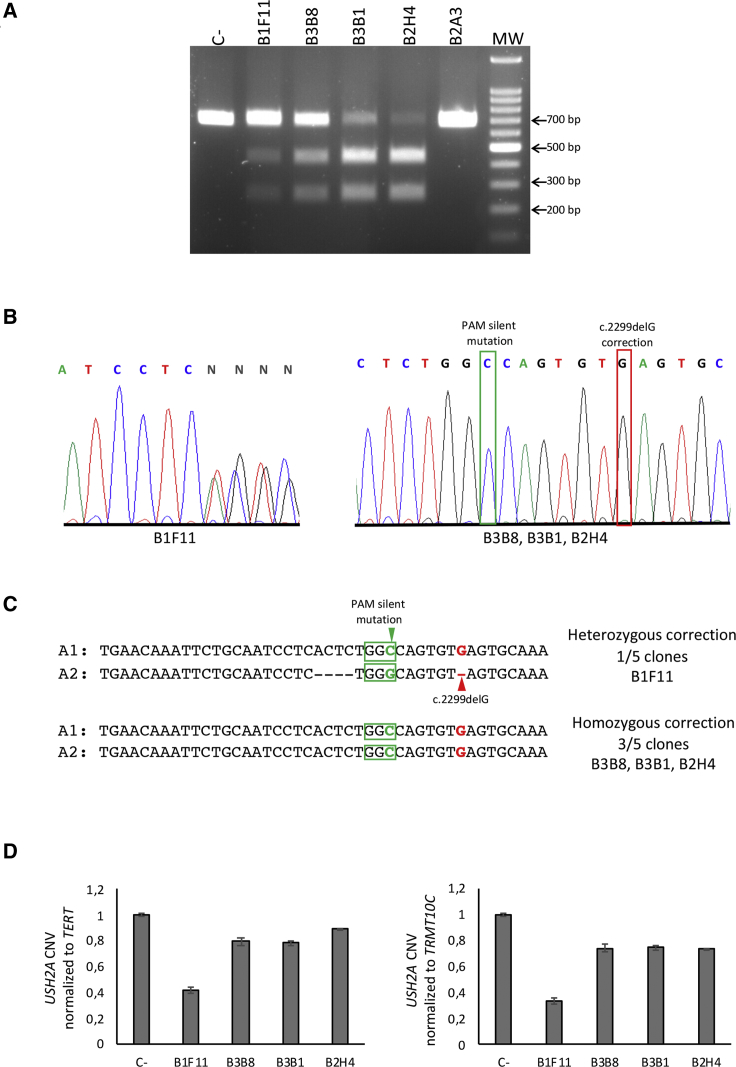
We first screened the clones by PCR amplification followed by restriction enzyme digestion. MscI digestion of the amplicon showed that four of the five clones (B1F11, B3B8, B3B1, and B2H4) had used ssODN-2 to repair the Cas9-induced DSB (Figure 2A). We confirmed HDR events by Sanger sequencing, both before and after subcloning of the PCR products (Figure 2B). Of the four positive clones, one clone (B1F11) showed heterozygous correction (Figure 2B), as determined by the presence of the PAM silent mutation and the corrected c.2299delG mutation in one allele (A1), compared with the second allele (A2) in which Cas9-induced INDELs were observed (Figure 2C). The three remaining positive clones (B3B8, B3B1, and B2H4) demonstrated homozygous correction of the c.2299delG mutation, as well as homozygous introduction of the PAM silent mutation (Figures 2B and 2C). The fifth clone (B2A3) showed no CRISPR/Cas-induced modifications (data not shown).
Figure 2.
Gene Correction of the c.2299delG Mutation in the USH2A-USH-iPSC Cell Line
(A) MscI restriction enzyme digestion of DNA from the surviving USH2A-USH-iPSC clones B1F11, B3B8, B3B1, B2H4, and B2A3. DNA from the parental USH2A-USH-iPSC line was used as a negative control (C−). (B) Electropherograms showing the Sanger sequencing results of CRISPR/Cas9-mediated genome correction of the c.2299delG mutation: (left panel) heterozygous correction; (right panel) homozygous correction (boxed in red). The incorporated PAM silent mutation is boxed in green. (C) Sequence analysis of allele 1 (A1) and allele 2 (A2) from the CRISPR/Cas9-edited region after subcloning. The red arrowhead indicates the c.2299delG mutation. PAM sequences are boxed by green. The green arrowhead shows the PAM silent mutation present in ssODN-2. (D) qPCR analysis of copy number variation (CNV) of USH2A in the corrected clones compared with that of the parental USH2A-USH-iPSC line (C−). The data were normalized to the housekeeping genes TERT (left panel) and TRMT10C (right panel).
We then assayed copy number variation (CNV) of USH2A in the corrected clones, using a quantitative PCR (qPCR) assay, to exclude the presence of large Cas9-induced deletions.41 To this end, we designed a primer set surrounding the c.2299delG mutational site. As an internal control, the forward primer was directly designed to hybridize in the region where the B1F11 clone presented INDELs in A2. The qPCR results showed that all the homozygous clones (B3B8, B3B1, and B2H4) presented similar CNV relative values of USH2A, normalized to two invariable housekeeping genes, TERT and TRMT10C, as compared with the negative control (C−, parental USH2A-USH-iPSC line). By contrast, the heterozygous-corrected B1F11 clone presented half the relative CNV values of USH2A (Figure 2D).
In conclusion, although we have no way to determine whether the non-surviving iPSC clones that were FACS sorted had been genome edited, we did achieve a high (80%, 4/5 clones) efficiency of correction for the prevalent c.2299delG USH2A mutation in the surviving iPSCs from a patient presenting with USH2.
A SNP in an arRP Patient Results in Cas9 Allele-Specific Cleavage In trans to c.2276G>T
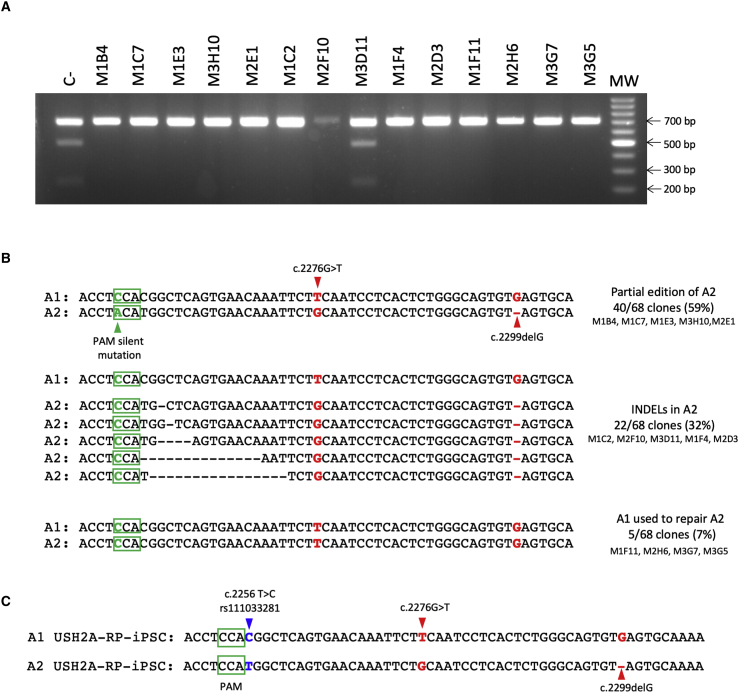
We previously reported an iPSC line, INMi001 or USH2A-RP-iPSC, from a patient presenting with arRP caused by the compound heterozygous mutations c.2276G>T and c.2299delG30 (Table 1). In order to target the c.2276G>T mutation, we nucleofected the USH2A-RP-iPSC line with the eSpCas9-sgRNA-1 plasmid in combination with ssODN-1, and the EGFP-positive cells were single-cell sorted by FACS 48 h post-nucleofection. The target region of the surviving clones (68 out of 288) was PCR amplified and initially screened for HDR events by NcoI digestion. A representative gel of 14 NcoI-digested clones is shown in Figure 3A. Encouragingly, NcoI digested only one clone (M3D11) with a profile identical to that of the parental USH2A-RP-iPSC line (C−), suggesting incorporation of the ssODN-1 into the host DNA of the remaining 13 clones.
Figure 3.
Cas9 Allele-Specific Cleavage In trans to c.2276G>T in the USH2A-RP-iPSC Cell Line
(A) Representative NcoI restriction enzyme digestion of 14 of the surviving USH2A-RP-iPSC clones. DNA from the parental USH2A-RP-iPSC was used as a negative control (C−). (B) Representative sequence analysis of allele 1 (A1) and allele 2 (A2) in the 14 clones shown in (A). Highlighted in red are the positions of the mutations c.2276G>T and c.2299delG. Green boxes represent PAM sequences. The green arrowhead shows the position of the PAM silent mutation present in ssODN-1. Black dashes correspond to the Cas9-induced INDELs, which were determined by comparison with the wild-type USH2A reference sequence. (C) Sequence analysis of A1 and A2 from the parental USH2A-RP-iPSC cell line after subcloning. Red arrowheads indicate the c.2276G>T and c.2299delG mutations. Green boxes represent the PAM sequences. A blue arrowhead indicates the SNP c.2256T>C (rs111033281) present in A1.
In light of the promising results, we sequenced the 68 surviving clones. Unexpectedly, the sequencing results did not correlate with the restriction enzyme digestion profiles. In all the CRISPR/Cas9-modified clones (67/68; 98%), the target allele carrying the c.2276G>T variant (referred to as allele 1 or A1) had not incorporated the ssODN-1, but by contrast, the non-targeted allele (allele 2 or A2) had been modified. We further confirmed these results by subcloning and re-sequencing the target region (Figure 3B). In 59% (40/68) of the clones, the ssODN-1 had been exclusively introduced into A2, as determined by the incorporation of the PAM silent mutation in the host DNA. In 32% (22/68) of the clones, we detected Cas9-induced INDELs only in A2. In 7% (5/68) of the clones, we determined that allele A1 had been used to repair A2 because: (1) the PAM silent mutation was not present in either allele, (2) the c.2276G>T mutation was present in both alleles, and (3) the c.2299delG mutation was absent in both alleles. Lastly, 2% (1/68) of the clones showed no CRISPR/Cas9-induced modifications in the DNA (data not shown).
The sequencing of the 68 clones revealed the presence of a SNP c.2256T>C (rs111033281) in A1, in cis with the c.2276G>T missense variant that we aimed to correct. Similarly, we confirmed the presence of the SNP in the parental USH2A-RP-iPSC line (Figure 3C). Therefore, the USH2A sequence of this patient differed from the reference USH2A sequence used to design the CRISPR/Cas9 strategy. Moreover, the SNP in A1 corresponds to the first nucleotide of the sgRNA-1 sequence, and thus likely prevented sgRNA-1 from binding the target sequence in A1. Furthermore, the SNP interfered with the endogenous NcoI site on A1. This, in addition to the observation that the Cas9-induced modifications destroyed the NcoI recognition site on A2, accounts for the lack of NcoI digestion of the majority of the clones (Figure 3A).
In conclusion, successful correction of the c.2276G>T missense variant was not achieved for this patient because of the previously unidentified SNP in the target allele. Nonetheless, we demonstrated a high (98%, 67/68 clones) rate of CRISPR/Cas9 events in the non-target allele of the surviving clones. Hence we showed the exceptionally high level of fidelity of eSpCas9, as a 1-bp difference in the seed region of the sgRNA sequence resulted in trans allele-specific cleavage.
CRISPR/Cas9 Mediates Correction of the c.2276G>T Mutation in the iPSCs of an arRP Patient
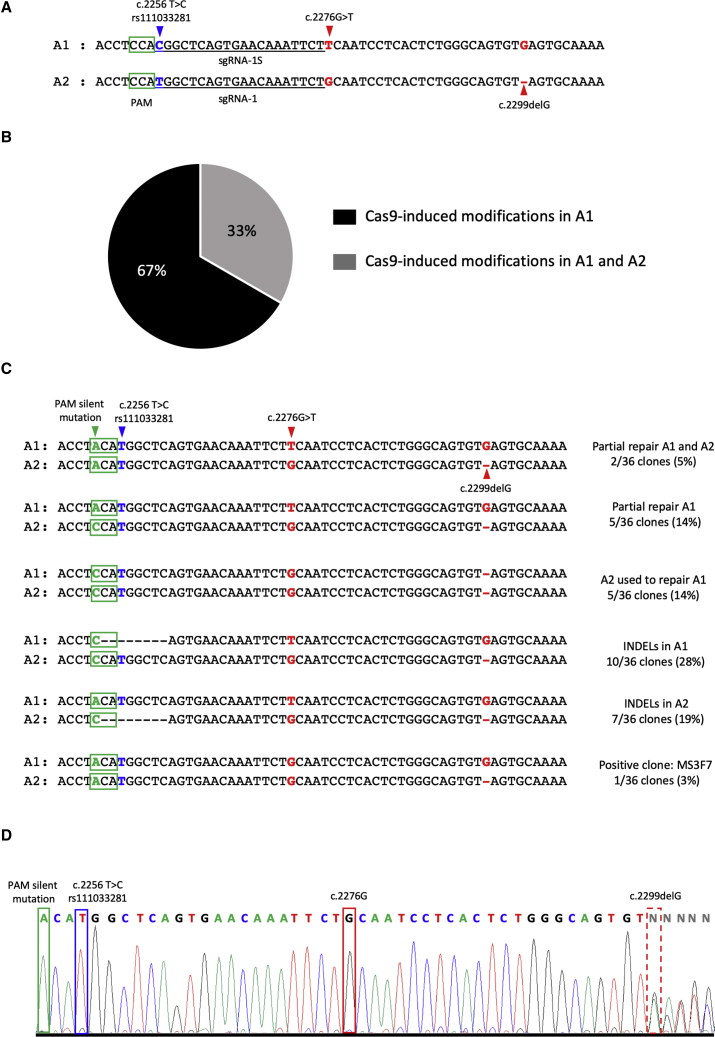
A meta-analysis of our USH2A patient database revealed that 75% of alleles with the missense c.2276G>T mutation also have the c.2256T>C SNP in cis. In order to target the maximum number of patients, we therefore re-designed sgRNA-1 to include the c.2256T>C SNP in the sgRNA sequence (sgRNA-1S; Figure 4A). Following subcloning, we nucleofected the eSpCas9-sgRNA-1S plasmid, together with ssODN-1 (which did not carry the SNP), into the USH2A-RP-iPSC line and single-cell-sorted the EGFP-positive cells. The surviving clones (36/288) were expanded and screened for HDR events.
Figure 4.
Gene Correction of the c.2276G>T Mutation in the USH2A-RP-iPSC Cell Line
(A) Sequence design for sgRNA-1S that targets allele 1 (A1) carrying the SNP c.2256T>C (blue arrowhead), in comparison with sgRNA-1, which targets allele 2 (A2). Green boxes represent the PAM sequences. Red arrowheads indicate the mutations c.2276G>T and c.2299delG in A1 and A2, respectively. (B) Schematic representation of the proportion of CRISPR/Cas9-induced modifications in A1 (black) and in A1 and A2 (gray). (C) Sequence analysis of A1 and A2 in the surviving USH2A-RP-iPSC corrected clones. Green boxes represent PAM sequences, and the green arrowhead indicates the PAM silent mutation present in ssODN-1. The blue arrowhead indicates the SNP present in A1. Red arrowheads indicate the mutations c.2276G>T on A1 and c.2299delG on A2. Black dashes correspond to the Cas9-induced INDELs, which were determined by comparison with the wild-type USH2A reference sequence. (D) Electropherogram showing the results from Sanger sequencing of CRISPR/Cas9-mediated genome correction of the c.2276G>T mutation in the USH2A-RP-iPSC MS3F7 clone. The corrected c.2276G nucleotide is boxed in red. A dashed red box indicates the uncorrected trans allelic c.2299delG mutation. A blue box indicates the SNP, and a green box indicates the PAM silent mutation.
The sequencing results showed that, in contrast to sgRNA-1, sgRNA-1S was able to recognize and induce cutting of not only the target allele A1 containing the SNP, but also A2 (Figure 4B). In 5% (2/36) and 14% (5/36) of clones, we detected partial repair by introduction of the PAM silent mutation in A1 and A2, or in A1 alone, respectively, but without correction of the c.2276G>T variant in A1 (Figure 4C). In 14% (5/36) of the clones, A2 had been used to repair Cas9-induced cleavage of A1, as suggested by the: (1) absence of the PAM silent mutation in A1, (2) correction of the c.2276G>T mutation in A1, and (3) presence of the c.2299delG mutation in both A1 and A2. In 28% (10/36) and 19% (7/36) of the clones, we found INDELs in the target allele A1 or in the non-target allele A2, respectively. In 17% (6/36) of clones, no CRISPR/Cas9-induced modifications could be detected (data not shown). Lastly, in 3% (1/36) of clones, we detected correction of the missense c.2276G>T variant in A1 and the presence of the c.2299delG only in A2 (Figure 4C and 4D). Interestingly, we believe that sgRNA-1S recognized both alleles in this clone, because the PAM silent mutation was also present in A2.
Taken together, we successfully corrected the c.2276G>T mutation in the iPSCs of a patient presenting with arRP. By contrast, a single-nucleotide change in the seed region of the sgRNA sequence caused a dramatic decrease in efficiency and specificity.
CRISPR/Cas9-Mediated Correction Does Not Result in Off-Target Cleavage
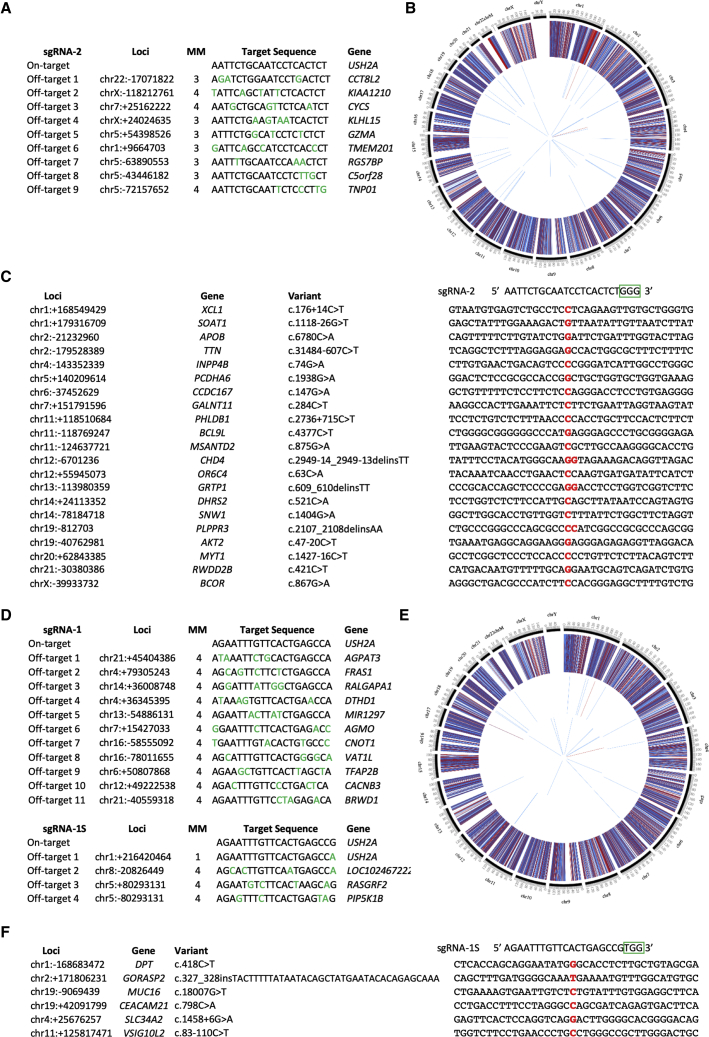
Even though CRISPR/Cas9 has streamlined genome editing, Cas9 has the capacity to cleave DNA in undesired regions in the genome. Thus, despite the added security of using eSpCas9, we initially investigated possible off-target events in the coding regions of the edited iPSCs by whole-exome sequencing (WES). First, we predicted the potential off-target regions for the sgRNAs using the MIT CRISPR Design website (http://www.crispr.mit.edu), which is no longer accessible, and the CRISPOR website tool (http://crispor.tefor.net). For sgRNA-2 (Figure 5A) and sgRNA-1 (Figure 5D), 9 and 11 exonic off-target hits were predicted, respectively. For sgRNA-1S, four exonic off-target sites were predicted (Figure 5D). We then performed WES of the iPSC lines before and after CRISPR/Cas9-mediated gene correction. For this purpose, we selected the corrected USH2A-USH-iPSC clone B3B1 (homozygous correction of c.2299delG) and USH2A-RP-iPSC clone MS3F7 (heterozygous correction of c.2276G>T). The WES results of the clone B3B1 demonstrated that no variants were found in any of the nine predicted off-target regions. Similarly, no variants were found for the clone MS3F7 in the predicted off-target regions for either sgRNA-1 or sgRNA-1S. The WES data also confirmed the effective correction of c.2299delG and c.2276G>T in the B3B1 and MS3F7 clones, respectively, because none of the reads carried the original mutations (with a coverage >100× in both cases; data not shown).
Figure 5.
Whole-Exome Sequencing of Parental and Corrected iPSC Lines
(A and D) Exonic predicted off-target sites using the MIT portal for sgRNA-2 (A) and sgRNA-1 (D, upper panel) or the CRISPOR website for sgRNA-1S (D, lower panel) are listed. The number of mismatched nucleotides (MM) is shown and represented by green letters. (B and E) Circos plots indicating the variants identified by WES in the parental and corrected USH2A-USH-iPSC (B) and USH2A-RP-iPSC (E) clones. External layer represents the variants (SNVs + INDELs) shared between the parental and corrected iPSCs. The middle layer displays the single-nucleotide variants (SNVs) exclusively identified in the corrected iPSCs. The internal layer shows the INDELs exclusively identified in the corrected iPSCs. The middle and internal layers exclude variants listed in dbSNP151 but include all variants identified prior to manual inspection. Red bars: homozygous variants; blue bars: heterozygous variants. (C and F) Variants retained after filtering in the corrected USH2A-USH-iPSC clone B3B1 (C) and USH2A-RP-iPSC clone MS3F7 (F). Sequences flanking the variants (in red) in comparison with the corresponding sgRNA target sequence; PAM is boxed in green.
Second, to exclude possible off-target effects outside of the predicted regions, we then compared the WES data of the corrected cell lines with that of their respective parental line (Figures 5B and 5E). Divergent variants between the cell lines were filtered out using dbSNP151, leaving approximately 50 variants that were manually inspected. Manual inspection filtered out approximately half of the candidate variants based on: (1) the presence of the candidate variant in the uncorrected cell line but not retained by the variant caller, due to a low coverage or low mapping quality of the WES data; and (2) the poor parameters of the variant itself in the corrected cell line, which are often INDEL artifacts reported by the variant caller. After filtering, 21 variants were retained for the USH2A-USH-iPSC clone B3B1, which were not present in the parental cell line (Figure 5C). For the USH2A-RP-iPSC clone MS3F7, six variants were detected (Figure 5F). All of these variants were heterozygous with a read depth ranging from 40 to 220. Overall, out of the 27 variants identified, the majority (17 variants) were located in genes unknown to cause human pathology. In addition, none of the remaining 10 variants were located in retinal-specific genes. Furthermore, to rule out that these variants were the result of off-target mutagenesis caused by the sgRNAs, we manually compared the surrounding sequences of these variants with the sgRNA sequence used to correct the cell line (Figures 5C and 5F). None of the identified variants shared any sequence homology to their corresponding sgRNA sequence, which is consistent with the fact that they were not listed by the in silico analysis of predicted off-targets. Therefore, this lack of homology excludes the possibility that these variants were introduced by the eSpCas9.
Lastly, we complemented the exonic off-target analysis by assessing the top 10 non-exonic off-target regions for the USH2A-USH-iPSC clone B3B1 (Figure S1) and the USH2A-RP-iPSC clone MS3F7 (Figure S2) by Sanger sequencing. We did not detect sequence alterations indicative of eSpCas9-induced modifications in any of the regions analyzed.
In conclusion, our sgRNA design and eSpCas9-mediated genome editing did not result in off-target modifications, even though exonic variants were detected between parental and corrected cell lines.
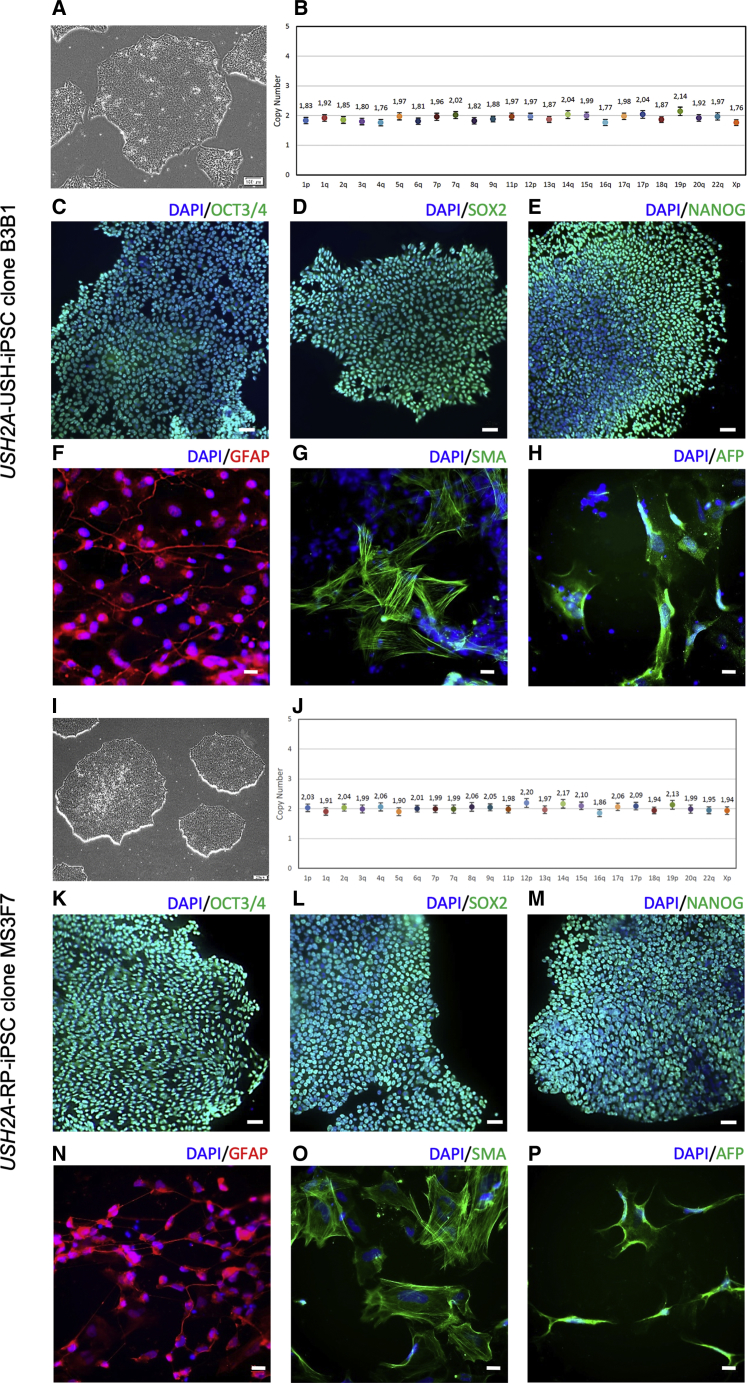
CRISPR/Cas9-Mediated Correction Does Not Affect Genetic Stability or Pluripotency of iPSCs
We then examined whether the CRISPR-corrected iPSC clones maintained the genetic stability and pluripotency characteristics of their parental iPSC lines.29,30 Both USH2A-USH-iPSC clone B3B1 and USH2A-RP-iPSC clone MS3F7 displayed the typical morphology of iPSC clones, composed of tightly packed cells surrounded by a distinct border (Figures 6A and 6I). To rule out any major chromosomal rearrangements that may have been induced by nucleofection or single-cell culture, we assessed the genomic integrity of the corrected iPSCs at mutational hotspots by the iCS-digital Pluri test.42 The results indicated that both corrected lines retained normal genomic stability (Figures 6B and 6J). Furthermore, immunofluorescence analyses showed that the CRISPR/Cas9-corrected clones also retained the expression of the typical pluripotency markers OCT3/4 (Figures 6C and 6K), SOX2 (Figures 6D and 6L), and NANOG (Figures 6E and 6M), as determined by immunofluorescence studies. In addition, we used an embryoid body assay to determine whether the CRISPR/Cas9-corrected lines could differentiate into the three embryonic cell layers. Both B3B1 and MS3F7 differentiated into ectoderm as assessed by immunostaining of glial fibrillary acidic protein (GFAP) expression (Figures 6F and 6N), mesoderm by smooth muscle actin (SMA) expression (Figures 6G and 6O), and endoderm by α-fetoprotein (AFP) expression (Figures 6H and 6P), respectively.
Figure 6.
Genome Stability and Pluripotency of Corrected iPSC Lines
(A and I) Phase-contrast images of USH2A-USH-iPSC clone B3B1 (A) and USH2A-RP-iPSC clone MS3F7 (I). (B and J) Genomic stability of USH2A-USH-iPSC B3B1 (B) and USH2A-RP-iPSC MS3F7 (J) as determined by a digital qPCR analysis of the most commonly rearranged regions reported in iPSCs. The copy number for each chromosomal position is shown with colored dots. (C–E and K–M) Pluripotency of USH2A-USH-iPSC clone B3B1 and USH2A-RP-iPSC clone MS3F7 as determined by immunostaining of the markers OCT3/4 (C and K), SOX2 (D and L), and NANOG (E and M), respectively. Scale bars, 50 μM. (F-H and N-P) Differentiation capacity of USH2A-USH-iPSC clone B3B1 and USH2A-RP-iPSC clone MS3F7 as determined by immunostaining of the germ layer markers GFAP (ectoderm; F and N), SMA (mesoderm; G and O), and AFP (endoderm; H and P), respectively. Scale bars, 20 μM.
Taken together, we successfully generated USH2A-corrected iPSC lines that retained their genetically stable and pluripotent characteristics.
CRISPR/Cas9-Mediated Correction of USH2A in Patient iPSCs Uncovers Abnormal c.2299delG mRNA Levels
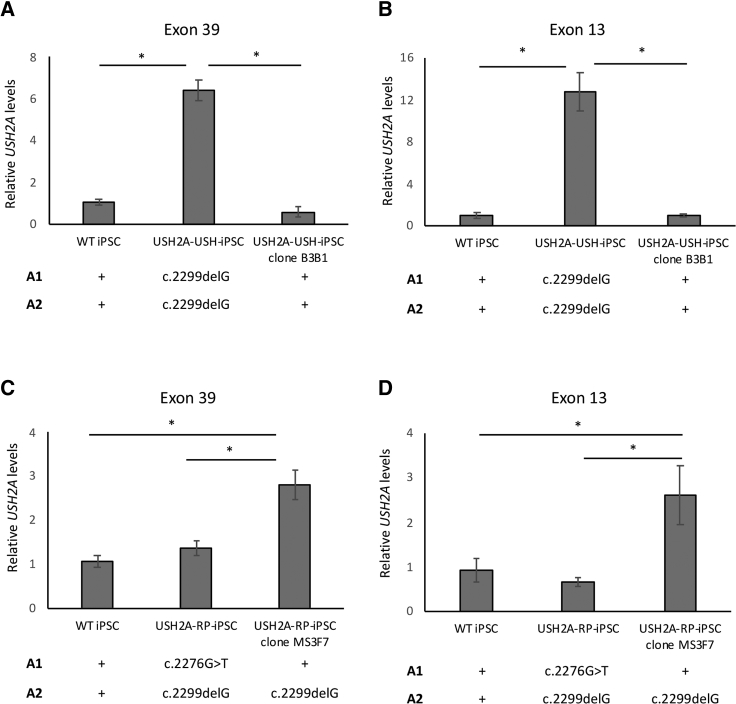
To assess USH2A expression levels in the absence of a functional antibody, we evaluated the expression of USH2A at the mRNA level in parental and CRISPR/Cas9-corrected iPSC lines. USH2A encodes two isoforms, a short isoform with 21 exons and a long isoform with 72 exons.43 Although both isoforms are present in the retina, the long isoform is predominant in photoreceptors.44 For this reason, we designed primers targeting exon 39, which would recognize the long isoform exclusively, as well as exon 13, which would recognize the long and short isoforms, and evaluated USH2A mRNA levels by qPCR.
First, the USH2A-USH-iPSC line carrying c.2299delG in the homozygous state shows expression levels of the long isoform that were significantly 6-fold higher (p < 0.05; n = 3) than wild-type USH2A levels (Figure 7A). This pattern was confirmed in a second parental iPSC clone (data not shown), indicating the results were not iPSC clone dependent. Furthermore, the homozygous correction of the c.2299delG mutation in the USH2A-USH-iPSC clone B3B1 restored the USH2A mRNA expression levels back to those of wild-type. This same profile was observed following indiscriminate amplification of both the long and short isoforms (p < 0.05; n = 3) (Figure 7B), thus confirming the phenomenon. These results suggest that the c.2299delG mutation has a specific effect on the accumulation of USH2A mRNA levels.
Figure 7.
USH2A mRNA Expression Levels in Parental and Corrected iPSCs
(A and B) USH2A mRNA expression levels in exon 39 (A) and exon 13 (B) for USH2A-USH-iPSC and the corrected USH2A-USH-iPSC clone B3B1. (C and D) USH2A mRNA expression levels in exon 39 (C) and exon 13 (D) for USH2A-RP-iPSC and the corrected USH2A-RP-iPSC clone MS3F7. Wild-type iPSC (WT) was used as a control, and its relative expression was set at 1. The allelic (A1 and A2) mutations carried by each cell line are indicated below the graphs. A plus sign indicates the absence of a mutation. Results were normalized to GAPDH expression, and asterisks represent significant differences (*p < 0.5). Data are expressed as mean ± SEM; n = 3.
Second, the USH2A-RP-iPSC line that carried the c.2276G>T missense variant on A1 and the c.2299delG mutation on A2 showed USH2A mRNA levels of the long isoform that were comparable with those of wild-type cells (Figure 7C). This was also confirmed for a second parental iPSC clone (data not shown). Furthermore, qPCR analysis of mRNA levels in two iPSC clones of another patient compound heterozygous for c.2276G>T and a missense mutation c.7352T>C also showed wild-type levels (Figure S2). By contrast, correction of the c.2276G>T variant, which leaves the c.2299delG mutation in a heterozygous state in A2, results in a significant 3-fold increase (p < 0.05; n = 3) in USH2A mRNA levels when compared with wild-type or non-corrected iPSCs. A similar profile was observed following indiscriminate amplification of both the long and short isoforms (Figure 7D). Together, these results confirmed an accumulation in USH2A mRNA levels associated with the c.2299delG mutation and suggest that they are reduced by the presence of the c.2276G>T variant.
In conclusion, by using a qPCR assay of USH2A expression levels, we were able to validate CRISPR/Cas9-mediated correction of the two most prevalent USH2A mutations. Moreover, this assay revealed an intriguing genotype-related expression of mutant USH2A mRNA levels.
Discussion
There is an unmet need for the development of novel therapies to treat IRDs with mutations in genes that exceed the cloning capacity of AAV vectors, such as USH2A. Along this line, exon skipping of common USH2A splice variants using antisense oligonucleotides (AONs) has been investigated45,46 and has led to an ongoing clinical trial for USH2 (ClinicalTrials.org: NCT03780257). However, a limitation of this strategy is the need for repeated administration due to AON degradation over time. With the recent advances in genome editing, a promising “one-hit” approach is to correct disease-causative mutations directly in the patient’s DNA. This could be an in vivo approach, whereby the disease-causing variant is corrected directly in the DNA of the patient, or an ex vivo approach, in which correction is performed in the patient’s cells prior to transplantation.24 In this study, we demonstrate the feasibility of using CRISPR/eSpCas9-mediated genome editing to correct the two most prevalent USH2A mutations in the iPSCs of patients. To the best of our knowledge, this is the first time eSpCas9 was used to correct disease-causing mutations.
We initially designed four sgRNAs with a mismatched G at the 5′ terminus of the sgRNA sequences to optimize transcription.32 However, the mismatched G appeared to disrupt the interaction of the eSpCas9-sgRNA complex, but not the wild-type SpCas9 complex, for sgRNA-1 in agreement with other studies.33,47,48 These observations confirmed that 5′ G mismatches are largely tolerated by wild-type Cas949 and provide further support for the enhanced specificity of the eSpCas9 variant. By contrast, sgRNA-2 was able to induce a DSB by eSpCas9 into the host DNA regardless of the mismatched G. Interestingly, sgRNA-2 was a highly efficient gRNA, resulting in precise genome editing in 80% of the surviving clones. This suggests that efficient sgRNAs might induce cleavage of the target DNA regardless of the mismatched G when using eSpCas9.
It is widely known that the efficiency of a given sgRNA is determined by its sequence.50,51 Along this line, two of the four sgRNAs tested (sgRNA-3 and sgRNA-4) could not induce Cas9-mediated cleavage either with or without the mismatched G. This may have been inherently because of their specific sequence characteristics or because of other indirect factors, such as sequence accessibility or even structural or epigenetic rearrangements in the DNA that may hamper the attachment of the Cas9-sgRNA complex.52,53 In addition, the cleavage activity of the sgRNAs in the present study was tested using the T7E1 assay, which is based on endonuclease activity and is sensitive only to mismatches above 1%.54 More sensitive methods such as High-Resolution Melt Analysis (HRMA) have recently been reported that can be used to detect CRISPR/Cas9-induced INDELs in cultured cells where endonuclease assays previously failed.55
The eSpCas9 variant was generated by the introduction of several alanine substitutions in the wild-type protein sequence, which weaken the ionic interactions between the protein and the target DNA.31 These substitutions trap the eSpCas9 in an inactive state when it is bound to mismatched targets.56 Consistently, sgRNA-1, which contained a 1-bp mismatch with the target allele A1 due to the SNP, exclusively resulted in cleavage of A2, thus again underlining the enhanced on-target specificity of eSpCas9. This was further supported by the absence of sequence modifications in the predicted off-target regions. Furthermore, the lack of homology between the sgRNA sequences and the sequence surrounding the variants identified by WES suggest that these variants did not arise from the CRIPSR/Cas9-mediated correction itself. It is more likely that they arose from fluctuating temperature and CO2 levels during the manipulation, single-cell sorting, and culture of the iPSCs, consistent with other studies.42,57, 58, 59 A recent study reported that around 10% of all somatic mutations found in iPSCs might arise from handling and manipulation, and that these mutations remain constant between early and later iPSC passages.60 Nevertheless, as promising as these findings are, it is not possible to completely rule out other undesired genomic changes in other regions of the DNA. Empirical assessment of the integrity of the whole genome may be necessary for CRISPR-based genome editing before downstream clinical applications.
In contrast to the highly efficient results obtained with sgRNA-1, we discovered that a single base-pair change (T>C) immediately after the PAM sequence in the sgRNA-1S highly affected the specificity of eSpCas9. It is largely accepted that mismatches in the seed (PAM-proximal) region of the sgRNA sequence are less tolerated compared with mismatches distant to the PAM sequence.22,61 However, Hsu et al.62 demonstrated that there are exceptions to the seed sequence model of SpCas9 specificity. Furthermore, they showed that the degree of tolerance of the mismatch varies with the sequence and should be evaluated individually.62 In addition, the single base-pair substitution created in sgRNA-1S to target A1 creates a GCC motif, previously nonexistent in sgRNA-1. It was recently demonstrated that TT and GCC motifs in the seed region of the sgRNA are enriched in inefficient sgRNAs.63 This finding is in agreement with our results, because sgRNA-1S resulted in 17% of clones without CRISPR/Cas9-induced modifications compared with only 2% of negative clones when using sgRNA-1. Moreover, the authors suggested that the GCC motif in the sgRNA sequence may enhance unspecific binding of the sgRNA to the DNA.63 Consistently, in our study, sgRNA-1S cleaved A1 and A2, compared with sgRNA-1 that cleaved only A2. Although these findings were reported for wild-type SpCas9, we demonstrate that the specificity and efficiency of a given sgRNA is also sequence dependent when using eSpCas9. This should be taken into consideration for future sgRNA design approaches and further evaluated to fully understand DNA targeting specificity associated with eSpCas9.
Genome-editing efficiencies in iPSCs have been reportedly unsatisfactory, and several attempts have been made to improve HDR efficiency, such as the use of small-molecule compounds.64,65 Recently, the transient delivery of BCL-XL in transfected iPSCs reportedly increased cell survival and led to a 20- to 100-fold increase in HDR.66 In addition, marker or drug selection genes have often been used to enrich the transfected cell population. In the current study, we positively selected EGFP-expressing cells, thereby enriching the targeted population and avoiding mixed cell populations. Our HDR efficiency, when using sgRNA-2 and ssODN-2 as a repair template, was 80% (4/5 clones) in the surviving FACS-sorted USH2A-USH-iPSCs without the use of additional compounds. If we take into account all the EGFP-positive single cells that were sorted by FACS, our HDR efficiency would decrease to 1.4% (4/288 clones). However, because the remaining 283 clones did not survive for further screening, there is no way of knowing the nature of the CRISPR/Cas9 genome editing in these cells. We designed and used asymmetric ssODN, with a predefined optimal length (127 bp), and complementary to the non-target strand, because this combination of characteristics has been reported to increase the rate of HDR up to 60% for a single nucleotide substitution.36 By contrast, the HDR efficiency in the surviving USH2A-RP-iPSC clones was 3%. The large difference in HDR efficiency obtained between iPSC lines might be because of the lower efficiency of sgRNA-1S as discussed above. In addition, the predicted cleavage site of sgRNA-2 is 12 bp away from the c.2299delG mutational site. By contrast, the cleavage site of sgRNA-1S is predicted to be 19 bp from the c.2276G>T mutational site. The relative distance of a given sgRNA from the mutational site plays an important role in HDR efficiencies, because mutation correction is distance dependent.37 In summary, by optimizing ssODN design, and when using a sgRNA close to the mutational site coupled to eSpCas9, we report a high rate of HDR efficiency in surviving iPSCs.
Encouraging results were previously reported by another team on a CRISPR/Cas9 genome-editing strategy for USH2A.20 The authors used a shorter (18-nt)67 version of the sgRNA-2 described herein, together with wild-type SpCas9, to correct the c.2299delG mutation in a patient’s fibroblasts. In addition, they used the same sgRNA to successfully introduce the c.2276G>T and c.2299delG mutations into the genome of HEK293 cells. Due to these results and the close proximity of these two USH2A mutations, the authors were tempted to speculate that this particular sgRNA could be used to correct both mutations. However, because this sgRNA sequence spans the wild-type c.2276 site, it would need a 1-bp modification to target the c.2276G>T mutation. Importantly, as we have shown here, a single base-pair change in the sequence of a given sgRNA can drastically interfere with its specificity and efficiency. This underlines the importance of sequencing the target region in the patient’s DNA rather than relying on published reference sequences. Hence, in contrast with this previous study, we have successfully corrected not one, but the two most common USH2A mutations in patients’ cells. Furthermore, by correcting these mutations in iPSCs, rather than fibroblasts, and having isolated a pure, rather than a mixed, cell population, we are one step closer to the translation of this proof-of concept study into a therapy. Moreover, we can now differentiate the genetically stable and pluripotent-corrected iPSCs into retinal organoids, which represent powerful tools to study the molecular mechanisms underlying both USH and arRP pathophysiology.
In order to assess the impact of HDR correction on USH2A expression, and due to the lack of a functional usherin antibody, we assayed the expression levels of USH2A in parental and corrected patients’ iPSCs. The c.2299delG mutation is predicted to result in a premature stop codon, which would be expected to be degraded by the nonsense-mediated mRNA decay (NMD) pathway. In contrast with our expectations, we observed an accumulation of the mutant mRNA in the parental USH2A-USH-iPSC line. Moreover, mRNA levels were reverted to wild-type after CRISPR/Cas9-mediated correction of the mutation. To our knowledge, this is the first time that the effect of the c.2299delG USH2A mutation has been assessed at the mRNA level. These results seem to have uncovered a novel and unreported mechanism for c.2299delG mutation pathogenesis, whereby the mutant transcript evades NMD and accumulates in the cell with possible harmful effects. It has been reported that some premature stop codons do escape NMD; however, the reasons still remain to be elucidated.68,69
Interestingly, when the c.2299delG mutation is present with the c.2276G>T variant in the parental USH2A-RP-iPSC line, there is no mRNA accumulation. It could be argued that c.2276G>T is associated itself with low mRNA expression, which would be compensated by the higher c.2299delG levels, thus appearing to be wild-type. However, the wild-type USH2A mRNA levels detected in a second patient carrying c.2276G>T, in combination with another missense mutation, argue against this possibility. Thus, we hypothesize that when we correct the c.2276G>T variant, leaving the c.2299delG mutation in the heterozygous state, the mRNA levels increase. Furthermore, this increase is equivalent to half the levels observed when c.2299delG is in the homozygous state, at least for the long isoform, further supporting this hypothesis. Therefore, it is tempting to speculate that an intriguing interaction between these two mutations might exist at the mRNA level, which requires further investigation. It is noteworthy that the c.2276G>T allele is associated with a milder phenotype in patients (arRP) compared with the c.2299delG allele (USH2),13 which could suggest a link between the aberrant mRNA levels and pathogenicity. To confirm our observations, we now need to test this hypothesis in iPSCs of patients carrying the c.2276G>T or c.2299delG variants in combination with other USH2A mutations, as well as to validate the results in iPSC-derived retinal organoids. If confirmed, these studies may also help to elucidate the differential pathophysiology of USH2 and RP caused by USH2A mutations.
The goal of this study was to develop an efficient CRISPR/Cas9-based genome-editing strategy that would allow reliable correction of the two most prevalent USH2A mutations in patients’ iPSCs, as a proof-of-concept for future autologous cell therapy of patients. Over recent years, technological advances in the generation of human stem cell-derived photoreceptors70, 71, 72, 73, 74, 75 have brought hope for future transplantation therapies.76,77 Moreover, iPSC-derived photoreceptor progenitor transplantation in animal models has been shown to restore visual function as assessed by electrophysiology and synapse recordings with the host bipolar cells.78,79 However the fate of the transplanted cells in wild-type versus diseased retinas still needs to be addressed.76,77 An alternative therapeutic option would be in vivo CRISPR/Cas9 mutation correction directly in the patient’s photoreceptors. Our study can also double as a validation of the CRISPR/Cas9 systems described herein for in vivo genome editing in USH2A patients, particularly in the case of the highly efficient sgRNA-2/ssODN-2 combination targeting the c.2299delG mutation.
However, an important issue that still needs to be addressed for in vivo genome editing is the fact that photoreceptors are post-mitotic cells in which the rate for HDR-mediated repair is extremely low.32 To this end, different strategies are being developed to promote in vivo genome editing in photoreceptors cells. A system called homology-independent targeted integration (HITI) was proven to successfully improve visual function in the Royal College of Surgeons rat model of RP.80 Recently, the coupling of the CRISPR/Cas9 system together with the bacterial RecA protein was shown to significantly increase the frequency of HDR in an RP mouse model (rd1).81 These recent advances provide hope for the future implementation of CRISPR/Cas9-based genome-editing strategies in vivo. Along this line, following preclinical studies in mice and non-human primates,82 Editas Medicine is currently preparing a clinical trial (ClinicalTrials.org: NCT03872479) for a CRISPR/Cas9-mediated strategy based on NHEJ to remove a causative intronic variant to treat the IRD Leber congenital amaurosis type 10. An AAV vector will be used to deliver the CRISPR/Cas9 components directly into the retina; however, the constitutive expression of Cas9 raises significant safety concerns.83
In conclusion, we designed a CRISPR/Cas9 strategy that allows correction of the two most prevalent USH2A mutations in patients’ iPSCs, which could result in the treatment of a non-negligible patient population because of the high prevalence of this IRD-causative gene. Importantly, we highlight features that promote high efficiency and specificity for eSpCas9, which, in addition to the absence of off-target events, help to minimize the clinical concerns of translating this proof-of-concept study into a possible future therapy.
Materials and Methods
Patients
The patients from the study were recruited following informed consent under the biomedical research approval number 2014-A00549-38 provided by the French Agency for the Safety of Health Products (ANSM).
Design of sgRNAs and Plasmid Construction
sgRNA-2 and sgRNA-1 were designed using the MIT CRISPR Design website (http://www.crispr.mit.edu), which is no longer accessible, and the online design tool CRISPOR (http://crispor.tefor.net/). sgRNAs were selected according to their proximity to the mutational site and following the rule 5′-(G)N (20 nt) NGG-3′. A pair of oligonucleotides for each sgRNA was purchased from Eurogenetec (Angers, France) and subcloned into the eSpCas9 1.1 (#79145; Addgene) or Px458 (#48138; Addgene) plasmid as described previously.32 In brief, complementary oligonucleotides were annealed by an initial denaturation at 94°C and gradually cooled. Plasmid DNA was digested using the BbsI restriction enzyme and ligated with the annealed oligonucleotides using T4 DNA ligase (Promega, Charbonnières les Bains, France) according to the manufacturer’s instructions.
Design of the ssODNs
Single-stranded ODNs were manually designed to: (1) have 91 nt in the PAM-proximal region and 36 nt in the PAM-distal region, (2) be complementary to the non-targeted strand, and (3) have silent changes in the PAM sequence. They were purchased from GENEWIZ (Leipzig, Germany) with phosphorothioate modifications at both ends.
HEK293 Cell Culture and Transfections
HEK293 cells were maintained in DMEM/F12 (GIBCO, Thermo Fisher Scientific, Villebon sur Yvette, France) supplemented with 10% fetal bovine serum (GIBCO) and 1% penicillin-streptomycin (GIBCO) at 37°C under 5% CO2. Plasmid DNA was purified using the Plasmid Midi Kit (QIAGEN, Villebon sur Yvette, France), and 1.5 μg of each construct was transfected separately into 3 × 105 HEK293 cells using Lipofectamine 3000 (Thermo Fisher Scientific). Forty-eight hours post-transfection, genomic DNA was isolated using the DNeasy Blood & Tissue Kit (QIAGEN) according to the manufacturer’s instructions.
T7Endonuclease I Assay
The target region was amplified from genomic DNA with specific primers (Table S1) targeting exon 13 of USH2A using the high-fidelity TaKaRa polymerase (Thermo Fisher Scientific). To allow heteroduplex formation, the PCR product was denatured by heating at 95°C for 5 min and reannealed by a ramp down to 85°C at −2°C/s followed by a ramp down to 25°C at −0.1°C/s. Heteroduplexes were then treated with 1 μL of T7E1 (New England Biolabs, Evry, France) and incubated at 37°C for 15 min. The reaction was stopped by adding 1 μL of Proteinase K and incubating the mix at 37°C for 5 min. The digested product was migrated on a 2% agarose gel and stained with SYBR safe DNA Gel stain (Thermo Fisher Scientific).
iPSC Culture and Transfection
The previously generated USH2A-USH-iPSC29 and USH2A-RP-iPSC30 cell lines were cultured in mTeSR1 medium (STEMCELL Technologies, Grenoble, France). Large-scale plasmid preparations were performed using the QIAGEN EndoFree Plasmid Maxi kit according to the manufacturer’s instructions. The iPSCs were dissociated with Accutase (STEMCELL Technologies), and 1.5 × 106 cells were electroporated with 5 μg of eSpCas9 (1.1) plasmid, containing either sgRNA-1, sgRNA-1S, or sgRNA-2 and 3 μg of the corresponding ssODN using the Amaxa nucleofector system (Lonza, Levallois-Perret, France). Cells when then recovered in mTeSR1 medium supplemented with 10 mM Rho-associated kinase (ROCK) inhibitor Y-27632 (MACS; Miltenyi Biotec, Paris, France). Forty-eight hours post-transfection, EGFP-positive cells were single cell-sorted by FACS (FACSAria III, Becton Dickinson, San Jose, CA, USA) into 96-well plates. Two to three weeks post-nucleofection, surviving colonies were manually picked and expanded for culture and HDR screening.
HDR Screening
The target region in exon 13 of USH2A was amplified from 150 ng of genomic DNA using the AmpliTaq Gold 360 Master Mix (Thermo Fisher Scientific). The PCR products were then cleaned using the ExoSAP-IT PCR Clean-up Kit (GE Healthcare, Velizy Villacoublay, France) prior to sequencing with the BigDye Terminator Cycle Sequencing Ready Reaction kit V3.1 on an Applied Biosystems 3130xL Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). For subcloning, the amplicons were TA cloned into the pGEM-T Easy Vector System (Promega) according to the manufacturer’s instructions and sequenced as above.
In Vitro Differentiation Assay
iPSCs were dissociated with Accutase and seeded onto ultra-low-attachment dishes for 2 days in Essential 8 medium (GIBCO) containing Y-27632. At day 3, the medium was changed to DMEM/F12 supplemented with 20% Knockout serum replacement (GIBCO), 1% penicillin-streptomycin, 1% GlutaMAX (GIBCO), 55 mM β-mercaptoethanol (GIBCO), and 1% non-essential amino acids (NEAAs) (GIBCO). At day 7, the embryoid bodies were seeded onto Matrigel-coated wells and cultured for a further 10 days before immunofluorescence staining.
Immunofluorescence Staining
iPSC colonies and embryoid bodies were fixed using 4% paraformaldehyde (PFA) and permeabilized with 0.1% Triton X-100 (Sigma-Aldrich, St. Quentin Fallavier, France) in PBS. Non-specific binding was blocked with 1% BSA (Sigma-Aldrich) and 10% donkey serum (Millipore, Saint Quentin en Yvelines, France). Primary antibodies were used at a 1:200 dilution in blocking solution and incubated overnight at 4°C: rabbit anti-NANOG (Abcam, Paris, France), mouse anti-OCT3/4 (Santa Cruz Biotechnology, Heidelberg, Germany), and rabbit anti-SOX2 (Thermo Fisher Scientific) for the iPSCs, and rabbit anti-GFAP (Dako, Les Ulis, France), mouse anti-SMA (Dako), and mouse anti-AFP (Sigma Aldrich) for the embryoid bodies. Fluorescence-conjugated secondary anti-mouse and anti-rabbit antibodies (Jackson ImmunoResearch Laboratories, Suffolk, UK) were used at a 1:500 dilution and incubated for 1 h at room temperature. Nuclei were stained with 0.2 μg/mL bisBenzimide (Sigma-Aldrich). Cells were imaged using a Zeiss ApoTome 2 Upright wide-field microscope.
qPCR Analyses
For the CNV studies, 2.5 ng of genomic DNA was used per reaction, and qPCR amplification was performed using the LightCycler 480 SYBR Green I Master mix on a LightCycler 480 II thermal cycler (Roche, Meylan, France). The results were analyzed using LightCyclerV 480 software and the Microsoft Excel program. Quantification was performed using the ΔΔCt method, and expression levels were normalized to either TERT or TRMT10C levels. For the USH2A expression studies, RNA was extracted using the QiaShredder and RNeasy mini kits (QIAGEN) according to the manufacturer’s instructions. The RNA was treated with RNase-Free DNase (QIAGEN), and 0.5 μg was reverse transcribed using the Superscript III Reverse Transcriptase kit (Thermo Fisher, Waltham, MA, USA). qPCR amplification was performed using 5 ng of cDNA per reaction, and results were normalized to GAPDH expression levels. Experiments were performed in triplicate. Genomic stability in corrected iPSCs was analyzed using digital qPCR by Stem Genomics (Montpellier, France).
WES Analyses
WES of genomic DNA from the USH2A-USH-iPSC, USH2A-USH-iPSC clone B3B1, USH2A-RP-iPSC, and USH2A-RP-iPSC clone MS3F7 cell lines was performed using the Roche NimbleGen Medexome commercial kit on an Illumina NextSeq instrument (Illumina, San Diego, CA, USA). Secondary analysis employed an in-house pipeline named Nenufaar (https://github.com/beboche/nenufaar) based on BWA84 and GATK3.8.85 Variant comparisons were performed using the RTG Tools package (https://github.com/RealTimeGenomics/rtg-tools), and GATK 4 and SnpSift from the SnpEff package.86 In brief, putative off-targets regions that were covered by the WES design were scanned for variants. Then overlapping variants between corrected clones were filtered out, and the remaining were annotated against dbSNP v.151. Non-matching variants were manually assessed using Integrative Genomics Viewer (IGV)87 and annotated with VariantValidator.88 CircosVCF89 was used to generate the Circos plots.
Statistical Analyses
Global statistical analyses were performed using a Kruskal-Wallis ANOVA and post hoc 2X2 comparisons using a Mann-Whitney test. A p value <0.5 was considered statistically significant.
Author Contributions
C.S.-S. and V.K. designed the study. C.S.-S., N.E., D.B., and D.M. performed the experiments. C.S.-S., D.B., A.-F.R., and V.K. analyzed the data. C.P.H. and I.M. provided clinical material. C.S.-S. and V.K. wrote the paper. C.P.H. and V.K. provided funding. C.S.-S., A.-F.R., and V.K. provided final approval of the manuscript.
Conflicts of Interest
C.S.-S. and V.K. filed a patent on genome editing of USH2A. V.K. is co-founder of Horama.
Acknowledgments
We thank the patients involved in this study. We are grateful to Christel Vaché and James Poulter for helpful discussions, and Michalitsa Diakatou for technical support. We thank the Neurogenetic facility of the INM for sequencing analysis, and the MRI Imaging facility for FACS and microscopy analyses. This work was supported by the patient associations Aviesan-Unadev, Vaincre Usher 2, and SOS retinite.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.11.016.
Supplemental Information
References
- 1.Berger W., Kloeckener-Gruissem B., Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog. Retin. Eye Res. 2010;29:335–375. doi: 10.1016/j.preteyeres.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 2.McGee T.L., Seyedahmadi B.J., Sweeney M.O., Dryja T.P., Berson E.L. Novel mutations in the long isoform of the USH2A gene in patients with Usher syndrome type II or non-syndromic retinitis pigmentosa. J. Med. Genet. 2010;47:499–506. doi: 10.1136/jmg.2009.075143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kimberling W.J., Hildebrand M.S., Shearer A.E., Jensen M.L., Halder J.A., Trzupek K., Cohn E.S., Weleber R.G., Stone E.M., Smith R.J. Frequency of Usher syndrome in two pediatric populations: Implications for genetic screening of deaf and hard of hearing children. Genet. Med. 2010;12:512–516. doi: 10.1097/GIM.0b013e3181e5afb8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Millán J.M., Aller E., Jaijo T., Blanco-Kelly F., Gimenez-Pardo A., Ayuso C. An update on the genetics of Usher syndrome. J. Ophthalmol. 2011;2011:417217. doi: 10.1155/2011/417217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan D., Liu X.Z. Genetics and pathological mechanisms of Usher syndrome. J. Hum. Genet. 2010;55:327–335. doi: 10.1038/jhg.2010.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eisenberger T., Neuhaus C., Khan A.O., Decker C., Preising M.N., Friedburg C., Bieg A., Gliem M., Charbel Issa P., Holz F.G. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: the example of retinal dystrophies. PLoS ONE. 2013;8:e78496. doi: 10.1371/journal.pone.0078496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge Z., Bowles K., Goetz K., Scholl H.P.N., Wang F., Wang X., Xu S., Wang K., Wang H., Chen R. NGS-based Molecular diagnosis of 105 eyeGENE(®) probands with Retinitis Pigmentosa. Sci. Rep. 2015;5:18287. doi: 10.1038/srep18287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pennings R.J.E., Te Brinke H., Weston M.D., Claassen A., Orten D.J., Weekamp H., Van Aarem A., Huygen P.L., Deutman A.F., Hoefsloot L.H. USH2A mutation analysis in 70 Dutch families with Usher syndrome type II. Hum. Mutat. 2004;24:185. doi: 10.1002/humu.9259. [DOI] [PubMed] [Google Scholar]
- 9.Baux D., Blanchet C., Hamel C., Meunier I., Larrieu L., Faugère V., Vaché C., Castorina P., Puech B., Bonneau D. Enrichment of LOVD-USHbases with 152 USH2A genotypes defines an extensive mutational spectrum and highlights missense hotspots. Hum. Mutat. 2014;35:1179–1186. doi: 10.1002/humu.22608. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Garcia G., Aparisi M.J., Jaijo T., Rodrigo R., Leon A.M., Avila-Fernandez A., Blanco-Kelly F., Bernal S., Navarro R., Diaz-Llopis M. Mutational screening of the USH2A gene in Spanish USH patients reveals 23 novel pathogenic mutations. Orphanet J. Rare Dis. 2011;6:65. doi: 10.1186/1750-1172-6-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aller E., Larrieu L., Jaijo T., Baux D., Espinós C., González-Candelas F., Nájera C., Palau F., Claustres M., Roux A.F., Millán J.M. The USH2A c.2299delG mutation: dating its common origin in a Southern European population. Eur. J. Hum. Genet. 2010;18:788–793. doi: 10.1038/ejhg.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivolta C., Sweklo E.A., Berson E.L., Dryja T.P. Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am. J. Hum. Genet. 2000;66:1975–1978. doi: 10.1086/302926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenassi E., Vincent A., Li Z., Saihan Z., Coffey A.J., Steele-Stallard H.B., Moore A.T., Steel K.P., Luxon L.M., Héon E. A detailed clinical and molecular survey of subjects with nonsyndromic USH2A retinopathy reveals an allelic hierarchy of disease-causing variants. Eur. J. Hum. Genet. 2015;23:1318–1327. doi: 10.1038/ejhg.2014.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Russell S., Bennett J., Wellman J.A., Chung D.C., Yu Z.F., Tillman A., Wittes J., Pappas J., Elci O., McCague S. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390:849–860. doi: 10.1016/S0140-6736(17)31868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.MacLaren R.E., Groppe M., Barnard A.R., Cottriall C.L., Tolmachova T., Seymour L., Clark K.R., During M.J., Cremers F.P.M., Black G.C.M. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014;383:1129–1137. doi: 10.1016/S0140-6736(13)62117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dimopoulos I.S., Chan S., MacLaren R.E., MacDonald I.M. Pathogenic mechanisms and the prospect of gene therapy for choroideremia. Expert Opin. Orphan Drugs. 2015;3:787–798. doi: 10.1517/21678707.2015.1046434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghazi N.G., Abboud E.B., Nowilaty S.R., Alkuraya H., Alhommadi A., Cai H., Hou R., Deng W.T., Boye S.L., Almaghamsi A. Treatment of retinitis pigmentosa due to MERTK mutations by ocular subretinal injection of adeno-associated virus gene vector: results of a phase I trial. Hum. Genet. 2016;135:327–343. doi: 10.1007/s00439-016-1637-y. [DOI] [PubMed] [Google Scholar]
- 18.Zallocchi M., Binley K., Lad Y., Ellis S., Widdowson P., Iqball S., Scripps V., Kelleher M., Loader J., Miskin J. EIAV-based retinal gene therapy in the shaker1 mouse model for usher syndrome type 1B: development of UshStat. PLoS ONE. 2014;9:e94272. doi: 10.1371/journal.pone.0094272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bassuk A.G., Zheng A., Li Y., Tsang S.H., Mahajan V.B. Precision Medicine: Genetic Repair of Retinitis Pigmentosa in Patient-Derived Stem Cells. Sci. Rep. 2016;6:19969. doi: 10.1038/srep19969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuster-García C., García-García G., González-Romero E., Jaijo T., Sequedo M.D., Ayuso C., Vázquez-Manrique R.P., Millán J.M., Aller E. USH2A Gene Editing Using the CRISPR System. Mol. Ther. Nucleic Acids. 2017;8:529–541. doi: 10.1016/j.omtn.2017.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burnight E.R., Gupta M., Wiley L.A., Anfinson K.R., Tran A., Triboulet R., Hoffmann J.M., Klaahsen D.L., Andorf J.L., Jiao C. Using CRISPR-Cas9 to Generate Gene-Corrected Autologous iPSCs for the Treatment of Inherited Retinal Degeneration. Mol. Ther. 2017;25:1999–2013. doi: 10.1016/j.ymthe.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339:823–827. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanjurjo-Soriano C., Kalatzis V. Guiding Lights in Genome Editing for Inherited Retinal Disorders: Implications for Gene and Cell Therapy. Neural Plast. 2018;2018:5056279. doi: 10.1155/2018/5056279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu P., Wu F., Mosenson J., Zhang H., He T.C., Wu W.S. CRISPR/Cas9-Mediated Genome Editing Corrects Dystrophin Mutation in Skeletal Muscle Stem Cells in a Mouse Model of Muscle Dystrophy. Mol. Ther. Nucleic Acids. 2017;7:31–41. doi: 10.1016/j.omtn.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu W., Mookherjee S., Chaitankar V., Hiriyanna S., Kim J.-W., Brooks M., Ataeijannati Y., Sun X., Dong L., Li T. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat. Commun. 2017;8:14716. doi: 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doudna J.A., Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 28.Zheng A., Li Y., Tsang S.H. Personalized therapeutic strategies for patients with retinitis pigmentosa. Expert Opin. Biol. Ther. 2015;15:391–402. doi: 10.1517/14712598.2015.1006192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanjurjo-Soriano C., Erkilic N., Manes G., Dubois G., Hamel C.P., Meunier I. Generation of a human iPSC line, INMi002-A, carrying the most prevalent USH2A variant associated with Usher syndrome type 2. Stem Cell Res. 2018;33:247–250. doi: 10.1016/j.scr.2018.11.007. [DOI] [PubMed] [Google Scholar]
- 30.Sanjurjo-Soriano C., Erkilic N., Manes G., Dubois G., Hamel C.P., Meunier I. Generation of an iPSC line, INMi001-A, carrying the two most common USH2A mutations from a compound heterozygote with non-syndromic retinitis pigmentosa. Stem Cell Res. 2018;33:228–232. doi: 10.1016/j.scr.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Slaymaker I.M., Gao L., Zetsche B., Scott D.A., Yan W.X., Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kato-Inui T., Takahashi G., Hsu S., Miyaoka Y. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 with improved proof-reading enhances homology-directed repair. Nucleic Acids Res. 2018;46:4677–4688. doi: 10.1093/nar/gky264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Strouse B., Bialk P., Niamat R.A., Rivera-Torres N., Kmiec E.B. Combinatorial gene editing in mammalian cells using ssODNs and TALENs. Sci. Rep. 2014;4:3791. doi: 10.1038/srep03791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang L., Guell M., Byrne S., Yang J.L., De Los Angeles A., Mali P., Aach J., Kim-Kiselak C., Briggs A.W., Rios X. Optimization of scarless human stem cell genome editing. Nucleic Acids Res. 2013;41:9049–9061. doi: 10.1093/nar/gkt555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richardson C.D., Ray G.J., DeWitt M.A., Curie G.L., Corn J.E. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat. Biotechnol. 2016;34:339–344. doi: 10.1038/nbt.3481. [DOI] [PubMed] [Google Scholar]
- 37.Paquet D., Kwart D., Chen A., Sproul A., Jacob S., Teo S., Olsen K.M., Gregg A., Noggle S., Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 38.Brunak S., Engelbrecht J., Knudsen S. Prediction of human mRNA donor and acceptor sites from the DNA sequence. J. Mol. Biol. 1991;220:49–65. doi: 10.1016/0022-2836(91)90380-o. [DOI] [PubMed] [Google Scholar]
- 39.Desmet F.O., Hamroun D., Lalande M., Collod-Béroud G., Claustres M., Béroud C. Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res. 2009;37:e67. doi: 10.1093/nar/gkp215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Renaud J.B., Boix C., Charpentier M., De Cian A., Cochennec J., Duvernois-Berthet E., Perrouault L., Tesson L., Edouard J., Thinard R. Improved Genome Editing Efficiency and Flexibility Using Modified Oligonucleotides with TALEN and CRISPR-Cas9 Nucleases. Cell Rep. 2016;14:2263–2272. doi: 10.1016/j.celrep.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 41.Adikusuma F., Plitz S., Corbett M.A., Turvey M., McColl S.R., Helbig K.J. Large deletions induced by Cas9 cleavage. Nature. 2018;560:E8–E9. doi: 10.1038/s41586-018-0380-z. [DOI] [PubMed] [Google Scholar]
- 42.Assou S., Bouckenheimer J., De Vos J. Concise Review: Assessing the Genome Integrity of Human Induced Pluripotent Stem Cells: What Quality Control Metrics? Stem Cells. 2018;36:814–821. doi: 10.1002/stem.2797. [DOI] [PubMed] [Google Scholar]
- 43.van Wijk E., Pennings R.J., te Brinke H., Claassen A., Yntema H.G., Hoefsloot L.H., Cremers F.P., Cremers C.W., Kremer H. Identification of 51 novel exons of the Usher syndrome type 2A (USH2A) gene that encode multiple conserved functional domains and that are mutated in patients with Usher syndrome type II. Am. J. Hum. Genet. 2004;74:738–744. doi: 10.1086/383096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu X., Bulgakov O.V., Darrow K.N., Pawlyk B., Adamian M., Liberman M.C., Li T. Usherin is required for maintenance of retinal photoreceptors and normal development of cochlear hair cells. Proc. Natl. Acad. Sci. USA. 2007;104:4413–4418. doi: 10.1073/pnas.0610950104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liquori A., Vaché C., Baux D., Blanchet C., Hamel C., Malcolm S., Koenig M., Claustres M., Roux A.F. Whole USH2A gene sequencing identifies several new deep intronic mutations. Hum. Mutat. 2016;37:184–193. doi: 10.1002/humu.22926. [DOI] [PubMed] [Google Scholar]
- 46.Slijkerman R.W., Vaché C., Dona M., García-García G., Claustres M., Hetterschijt L., Peters T.A., Hartel B.P., Pennings R.J., Millan J.M. Antisense Oligonucleotide-based Splice Correction for USH2A-associated Retinal Degeneration Caused by a Frequent Deep-intronic Mutation. Mol. Ther. Nucleic Acids. 2016;5:e381. doi: 10.1038/mtna.2016.89. [DOI] [PubMed] [Google Scholar]
- 47.Kulcsár P.I., Tálas A., Huszár K., Ligeti Z., Tóth E., Weinhardt N., Fodor E., Welker E. Crossing enhanced and high fidelity SpCas9 nucleases to optimize specificity and cleavage. Genome Biol. 2017;18:190. doi: 10.1186/s13059-017-1318-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang D., Zhang H., Li T., Chen K., Qiu J.-L., Gao C. Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases. Genome Biol. 2017;18:191. doi: 10.1186/s13059-017-1325-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim S., Bae T., Hwang J., Kim J.S. Rescue of high-specificity Cas9 variants using sgRNAs with matched 5′ nucleotides. Genome Biol. 2017;18:218. doi: 10.1186/s13059-017-1355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu H., Xiao T., Chen C.-H., Li W., Meyer C.A., Wu Q., Wu D., Cong L., Zhang F., Liu J.S. Sequence determinants of improved CRISPR sgRNA design. Genome Res. 2015;25:1147–1157. doi: 10.1101/gr.191452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng T., Hou Y., Zhang P., Zhang Z., Xu Y., Zhang L., Niu L., Yang Y., Liang D., Yi F. Profiling single-guide RNA specificity reveals a mismatch sensitive core sequence. Sci. Rep. 2017;7:40638. doi: 10.1038/srep40638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen X., Rinsma M., Janssen J.M., Liu J., Maggio I., Gonçalves M.A.F.V. Probing the impact of chromatin conformation on genome editing tools. Nucleic Acids Res. 2016;44:6482–6492. doi: 10.1093/nar/gkw524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vojta A., Dobrinić P., Tadić V., Bočkor L., Korać P., Julg B., Klasić M., Zoldoš V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim H.J., Lee H.J., Kim H., Cho S.W., Kim J.S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Housden B.E., Perrimon N. Detection of indel mutations in Drosophila by high-resolution melt analysis (HRMA) Cold Spring Harb. Protoc. 2016;2016:794–801. doi: 10.1101/pdb.prot090795. [DOI] [PubMed] [Google Scholar]
- 56.Chen J.S., Dagdas Y.S., Kleinstiver B.P., Welch M.M., Sousa A.A., Harrington L.B., Sternberg S.H., Joung J.K., Yildiz A., Doudna J.A. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bai Q., Ramirez J.-M., Becker F., Pantesco V., Lavabre-Bertrand T., Hovatta O., Lemaître J.-M., Pellestor F., De Vos J. Temporal analysis of genome alterations induced by single-cell passaging in human embryonic stem cells. Stem Cells Dev. 2015;24:653–662. doi: 10.1089/scd.2014.0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y., Yang Y., Kang X., Lin B., Yu Q., Song B., Gao G., Chen Y., Sun X., Li X. One-Step Biallelic and Scarless Correction of a β-Thalassemia Mutation in Patient-Specific iPSCs without Drug Selection. Mol. Ther. Nucleic Acids. 2017;6:57–67. doi: 10.1016/j.omtn.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kyriakides O., Halliwell J.A., Andrews P.W. Acquired Genetic and Epigenetic Variation in Human Pluripotent Stem Cells. Adv. Biochem. Eng. Biotechnol. 2018;163:187–206. doi: 10.1007/10_2017_22. [DOI] [PubMed] [Google Scholar]
- 60.D’Antonio M., Benaglio P., Jakubosky D., Greenwald W.W., Matsui H., Donovan M.K.R., Li H., Smith E.N., D’Antonio-Chronowska A., Frazer K.A. Insights into the Mutational Burden of Human Induced Pluripotent Stem Cells from an Integrative Multi-Omics Approach. Cell Rep. 2018;24:883–894. doi: 10.1016/j.celrep.2018.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graf R., Li X., Chu V.T., Rajewsky K. sgRNA Sequence Motifs Blocking Efficient CRISPR/Cas9-Mediated Gene Editing. Cell Rep. 2019;26:1098–1103.e3. doi: 10.1016/j.celrep.2019.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu C., Liu Y., Ma T., Liu K., Xu S., Zhang Y., Liu H., La Russa M., Xie M., Ding S., Qi L.S. Small molecules enhance CRISPR genome editing in pluripotent stem cells. Cell Stem Cell. 2015;16:142–147. doi: 10.1016/j.stem.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li G., Zhang X., Zhong C., Mo J., Quan R., Yang J., Liu D., Li Z., Yang H., Wu Z. Small molecules enhance CRISPR/Cas9-mediated homology-directed genome editing in primary cells. Sci. Rep. 2017;7:8943. doi: 10.1038/s41598-017-09306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li X.L., Li G.H., Fu J., Fu Y.W., Zhang L., Chen W., Arakaki C., Zhang J.P., Wen W., Zhao M. Highly efficient genome editing via CRISPR-Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res. 2018;46:10195–10215. doi: 10.1093/nar/gky804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu Y., Sander J.D., Reyon D., Cascio V.M., Joung J.K. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Danckwardt S., Neu-Yilik G., Thermann R., Frede U., Hentze M.W., Kulozik A.E. Abnormally spliced beta-globin mRNAs: a single point mutation generates transcripts sensitive and insensitive to nonsense-mediated mRNA decay. Blood. 2002;99:1811–1816. doi: 10.1182/blood.v99.5.1811. [DOI] [PubMed] [Google Scholar]
- 69.Romão L., Inácio A., Santos S., Avila M., Faustino P., Pacheco P., Lavinha J. Nonsense mutations in the human beta-globin gene lead to unexpected levels of cytoplasmic mRNA accumulation. Blood. 2000;96:2895–2901. [PubMed] [Google Scholar]
- 70.Hirami Y., Osakada F., Takahashi K., Okita K., Yamanaka S., Ikeda H., Yoshimura N., Takahashi M. Generation of retinal cells from mouse and human induced pluripotent stem cells. Neurosci. Lett. 2009;458:126–131. doi: 10.1016/j.neulet.2009.04.035. [DOI] [PubMed] [Google Scholar]
- 71.Meyer J.S., Shearer R.L., Capowski E.E., Wright L.S., Wallace K.A., McMillan E.L., Zhang S.C., Gamm D.M. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 2009;106:16698–16703. doi: 10.1073/pnas.0905245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tucker B.A., Park I.H., Qi S.D., Klassen H.J., Jiang C., Yao J., Redenti S., Daley G.Q., Young M.J. Transplantation of adult mouse iPS cell-derived photoreceptor precursors restores retinal structure and function in degenerative mice. PLoS ONE. 2011;6:e18992. doi: 10.1371/journal.pone.0018992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhong X., Gutierrez C., Xue T., Hampton C., Vergara M.N., Cao L.H., Peters A., Park T.S., Zambidis E.T., Meyer J.S. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 2014;5:4047. doi: 10.1038/ncomms5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reichman S., Terray A., Slembrouck A., Nanteau C., Orieux G., Habeler W., Nandrot E.F., Sahel J.-A., Monville C., Goureau O. From confluent human iPS cells to self-forming neural retina and retinal pigmented epithelium. Proc. Natl. Acad. Sci. USA. 2014;111:8518–8523. doi: 10.1073/pnas.1324212111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reichman S., Slembrouck A., Gagliardi G., Chaffiol A., Terray A., Nanteau C., Potey A., Belle M., Rabesandratana O., Duebel J. Generation of Storable Retinal Organoids and Retinal Pigmented Epithelium from Adherent Human iPS Cells in Xeno-Free and Feeder-Free Conditions. Stem Cells. 2017;35:1176–1188. doi: 10.1002/stem.2586. [DOI] [PubMed] [Google Scholar]
- 76.Pearson R.A., Gonzalez-Cordero A., West E.L., Ribeiro J.R., Aghaizu N., Goh D., Sampson R.D., Georgiadis A., Waldron P.V., Duran Y. Donor and host photoreceptors engage in material transfer following transplantation of post-mitotic photoreceptor precursors. Nat. Commun. 2016;7:13029. doi: 10.1038/ncomms13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Santos-Ferreira T., Llonch S., Borsch O., Postel K., Haas J., Ader M. Retinal transplantation of photoreceptors results in donor-host cytoplasmic exchange. Nat. Commun. 2016;7:13028. doi: 10.1038/ncomms13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barnea-Cramer A.O., Wang W., Lu S.J., Singh M.S., Luo C., Huo H., McClements M.E., Barnard A.R., MacLaren R.E., Lanza R. Function of human pluripotent stem cell-derived photoreceptor progenitors in blind mice. Sci. Rep. 2016;6:29784. doi: 10.1038/srep29784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Homma K., Okamoto S., Mandai M., Gotoh N., Rajasimha H.K., Chang Y.S., Chen S., Li W., Cogliati T., Swaroop A., Takahashi M. Developing rods transplanted into the degenerating retina of Crx-knockout mice exhibit neural activity similar to native photoreceptors. Stem Cells. 2013;31:1149–1159. doi: 10.1002/stem.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Suzuki K., Tsunekawa Y., Hernandez-Benitez R., Wu J., Zhu J., Kim E.J., Hatanaka F., Yamamoto M., Araoka T., Li Z. In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature. 2016;540:144–149. doi: 10.1038/nature20565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cai Y., Cheng T., Yao Y., Li X., Ma Y., Li L., Zhao H., Bao J., Zhang M., Qiu Z., Xue T. In vivo genome editing rescues photoreceptor degeneration via a Cas9/RecA-mediated homology-directed repair pathway. Sci. Adv. 2019;5:eaav3335. doi: 10.1126/sciadv.aav3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Maeder M.L., Stefanidakis M., Wilson C.J., Baral R., Barrera L.A., Bounoutas G.S., Bumcrot D., Chao H., Ciulla D.M., DaSilva J.A. Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat. Med. 2019;25:229–233. doi: 10.1038/s41591-018-0327-9. [DOI] [PubMed] [Google Scholar]
- 83.Fu Y., Foden J.A., Khayter C., Maeder M.L., Reyon D., Joung J.K., Sander J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., Del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics. 2013;43:1–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thorvaldsdóttir H., Robinson J.T., Mesirov J.P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Freeman P.J., Hart R.K., Gretton L.J., Brookes A.J., Dalgleish R. VariantValidator: Accurate validation, mapping, and formatting of sequence variation descriptions. Hum. Mutat. 2018;39:61–68. doi: 10.1002/humu.23348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Drori E., Levy D., Smirin-Yosef P., Rahimi O., Salmon-Divon M. CircosVCF: circos visualization of whole-genome sequence variations stored in VCF files. Bioinformatics. 2017;33:1392–1393. doi: 10.1093/bioinformatics/btw834. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.