Abstract
Background
Metabolic syndrome (MetS), the clustering of metabolic risk factors, is associated with cardiovascular disease risk. We sought to determine if dysregulation of the lipidome may contribute to metabolic risk factors.
Methods
We measured 154 circulating lipid species in 658 participants from the Framingham Heart Study (FHS) using liquid chromatography-tandem mass spectrometry and tested for associations with obesity, dysglycemia, and dyslipidemia. Independent external validation was sought in three independent cohorts. Follow-up data from the FHS were used to test for lipid metabolites associated with longitudinal changes in metabolic risk factors.
Results
Thirty-nine lipids were associated with obesity and eight with dysglycemia in the FHS. Of 32 lipids that were available for replication for obesity and six for dyslipidemia, 28 (88%) replicated for obesity and five (83%) for dysglycemia. Four lipids were associated with longitudinal changes in body mass index and four were associated with changes in fasting blood glucose in the FHS.
Conclusions
We identified and replicated several novel lipid biomarkers of key metabolic traits. The lipid moieties identified in this study are involved in biological pathways of metabolic risk and can be explored for prognostic and therapeutic utility.
Keywords: Metabolic risk, Metabolic syndrome, Cardiovascular disease, Dysglycemia, Dyslipidemia, Biomarker
Abbreviations: BMI, body mass index; CE, cholesteryl ester; Cer, ceramide; CVD, cardiovascular disease; DAG, diacylglycerol; dhSL, dihydrosphingolipid; ERF, Erasmus Family Study; FDR, false discovery rate; FHS, Framingham Heart Study; HDL-C, High density lipoprotein cholesterol; LC-MS/MS, liquid chromatography-tandem mass spectrometry; LGPL, lysoglycerophospholipid; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; MetS, metabolic syndrome; MRM, multiple reaction monitoring; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PESA, Progression of Early Subclinical Atherosclerosis; PS, phosphatidylserine; SAFHS, San Antonio Family Heart Study; SL, sphingolipid; SM, sphingomyelin; T2DM, type II diabetes mellitus; TAG, triacylglycerol
Research in context
Evidence before this study
Prior metabolomic association studies linking lipids with metabolic risk factors have been limited by the lipid metabolites profiled, lack of independent external replication, and absence of longitudinal association analyses.
Added value of this study
Our global lipidomics approach, along with external replication in three independent cohorts, provides robust cross-sectional associations of several lipid moieties with obesity and dysglycemia. We further strengthen these results by identifying lipids associated with longitudinal changes in metabolic risk factors.
Implications of all the available evidence
Our results, taken together with prior evidence, highlight lysoglycerophospholipids and dihydrosphingolipids as markers of obesity and dysglycemia that should be explored for prognostic and therapeutic utility.
1. Introduction
Cardiovascular disease (CVD), the leading cause of mortality in the United States and worldwide, accounts for roughly 30% of deaths [1] and is closely linked to the burden of metabolic risk factors. Metabolic syndrome (MetS) [2], clinically defined as the simultaneous presence of several metabolic risk factors [3], affects over 20% of American adults and is associated with a doubling in CVD risk [4]. While MetS is treated using lifestyle and drug-based interventions, its pathogenesis is not fully understood. Identifying key mechanistic drivers of metabolic risk factors and the progression from subclinical to overt disease may allow for the development of more targeted and effective interventions. The complexities of the associated metabolic risk factors necessitate global approaches, such as lipidomic profiling, which has advanced in the past decade due to mass spectrometry [5]. Considerable CVD research has shifted towards interrogating the circulating lipidome to further understand the cardiovascular consequences of dysregulated lipid metabolism [6].
Here, we report the results of plasma lipidomic profiling via liquid chromatography-tandem mass spectrometry (LC-MS/MS) in Framingham Heart Study (FHS) participants [7,8] in whom cross-sectional and longitudinal phenotypic characterization are available. Independent external replication of all cross-sectional associations from discovery was sought in the Erasmus Rucphen Family (ERF) study [9], San Antonio Family Heart Study (SAFHS) [10], and Progression of Early Subclinical Atherosclerosis (PESA) study [11]. We posited that the characteristic metabolic traits that define MetS have distinct and shared lipidomic signatures. To that end, we sought to identify lipid metabolomic biomarkers associated with metabolic risk factors to further understand the biology underlying MetS and to highlight high-value targets for the treatment and prevention of cardiometabolic disease.
2. Methods
2.1. Study samples
The discovery study sample included participants from the Offspring (n = 5124) and Third Generation (n = 4095) FHS cohorts (6,7). FHS clinical evaluations, including physical examinations and laboratory tests, were conducted every four to eight years. We sampled FHS Offspring cohort participants evaluated at Examination 8 (2005–2008; n = 3021) and Third Generation cohort participants evaluated at Examination 1 (2002–2005; n = 4095). Exclusion criteria included the following: off-site examination (n = 107), no blood specimen (n = 126), lack of unrestricted consent (n = 335), prevalent CVD at baseline (n = 216), pharmacological treatment for diabetes (n = 257) or dyslipidemia (n = 1201), missing BMI data (n = 4), cigarette smoking at the time of examination (n = 772), and age <50 or >79 years in the Offspring cohort (n = 123) or age <25 or >59 years in the Third Generation cohort (n = 183). The final study sample was selected from 3792 eligible participants.
Independent external replication was conducted in the ERF study, SAFHS, and PESA study. The ERF study is a prospective family-based study consisting of 3465 individuals aged 17–92 years from the southwest region of the Netherlands recruited based on kinship [9,12]. Samples without measures of metabolites or fasting glucose, or available genotype data for family kinship adjustment were excluded. The final study sample consisted of 2564 participants without diabetes. All baseline demographic data was collected between 2002 and 2006, and venous blood samples were collected after at least eight hours of fasting. A more detailed description of the ERF study and baseline characteristics of the study sample have been reported previously [9]
The SAFHS is a population-based, epidemiological study of type II diabetes (T2D) mellitus and CVD in Mexican Americans and non-Hispanic whites [10,13,14]. A total of 1431 individuals comprising 42 extended families who had given informed consent underwent blood sample collection. Of these, plasma samples from 1076 participants had complete data and were used for lipidomic profiling via liquid chromatography electrospray ionization tandem mass spectrometry (LC ESI-MS/MS) [13].
The PESA study is an observational cohort study of subclinical atherosclerosis in 4184 middle-aged (40–54 years) employees at Banco Santander in Madrid, Spain [11]. Recruitment began in 2010, and the baseline examination, including assessment for CVD risk factors and blood sampling, was completed in 2013. PESA participants with prevalent CVD at baseline (n = 34), active treatment for cancer (n = 2), history of transplants requiring active immunotherapy (n = 2), morbid obesity (BMI ≥40 kg/m2) (n = 22), chronic kidney disease (glomerular filtration rate ˂60 mL/min per square meter) (n = 1), an implanted metal device that could produce artifacts in computed tomography (n = 2), and any limitation or disability resulting in failure to comply with the study protocol (n = 1) were excluded. The final PESA sample size included 4120 participants.
2.2. Clinical measures
For the cross-sectional analyses, we evaluated three categorically-defined metabolic risk factors: obesity (BMI ≥30 kg/m2), dysglycemia (fasting blood glucose ≥100 mg/dL [adjusted for diabetes medications as described below]), and dyslipidemia (TAG ≥ 150 mg/dL [adjusted for lipid-lowering treatment as described below] or HDL cholesterol [HDL-C] <40 mg/dL in men or <50 mg/dL in women). All definitions were obtained from the National Cholesterol Education Program's Adult Treatment Panel III report [19]. For the longitudinal analyses, serial changes (from the baseline examination to the follow-up examination) in the continuous measures of the above-mentioned metabolic risk factors were used as the outcomes. The continuous measures for obesity, dysglycemia, and dyslipidemia were BMI [kg/m2], fasting blood glucose [mg/dL], and TAG [mg/dL] and HDL-C [mg/dL], respectively.
2.3. Study design
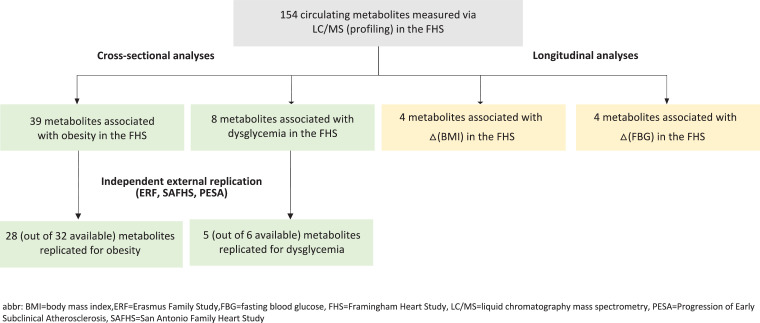
A 2 × 2 × 2 factorial study design was used in the FHS to group samples according to the presence or absence of each of the three central metabolic risk factors. Based on the clinical cut-points described above, eight (2 × 2 × 2 = 8) mutually-exclusive groups were generated from the three dichotomized outcomes. 80–84 individuals were selected for each group while controlling for sex and cohort across all groups. The longitudinal study sample consisted of all FHS participants in the cross-sectional sample who attended the subsequent clinic examination and had complete follow-up data for the traits of interest (n = 554). For those on lipid-lowering (n = 133) or diabetes medications (n = 30) at follow-up, observed values were also adjusted for treatment effects (see Statistical Methods for details). Fig. 1 provides a flow chart so the experimental design can be visualized.
Fig. 1.
Flow chart of experimental design.
2.4. Lipidomic profiling
Lipidomic profiling for replication was carried out via three different platforms – 1) liquid chromatography-mass spectrometry (LC-MS), 2) AbsoluteIDQ p150 Kit of Biocrates Life Sciences AG, and 3) electrospray-ionization mass spectrometry (ESI-MS) – using methods previously described [15], [16], [17], [18]
A detailed description of the LC-MS based lipidomic profiling methods used in the FHS is provided in the Supplementary Text and Table 2a contains the metabolite transitions used in the MRM mass spectrometry.
Table 2.
Cross-sectional lipid associations with obesity in FHS.
| Lipid species | Class | Estimated β* | SE | P-value† | Replication# |
|---|---|---|---|---|---|
| LPC 17:0 | LPC | −0.618 | 0.072 | 1.05E-16 | ✓ |
| LPC 18:2 | LPC | −0.524 | 0.069 | 1.67E-13 | ✓ |
| LPC 18:0p | LPC | −0.503 | 0.070 | 2.28E-12 | ✓ |
| LPC 18:1 | LPC | −0.510 | 0.072 | 3.89E-12 | ✓ |
| LPC 18:0e | LPC | −0.503 | 0.071 | 3.95E-12 | ✓ |
| PC 16:2e | PC | −0.448 | 0.067 | 4.52E-11 | |
| SM 36:0 | SM | 0.493 | 0.075 | 7.81E-11 | |
| LPC 22:5 | LPC | −0.462 | 0.073 | 5.08E-10 | |
| PC 40:8 | PC | −0.460 | 0.074 | 8.80E-10 | |
| LPC 17:1 | LPC | −0.447 | 0.073 | 1.97E-09 | ✓ |
| LPC 15:0 | LPC | −0.439 | 0.076 | 1.06E-08 | ✓ |
| LPC 18:0 | LPC | −0.423 | 0.074 | 1.34E-08 | ✓ |
| LPC 22:6 | LPC | −0.427 | 0.075 | 2.26E-08 | ✓ |
| PC 34:0 | PC | −0.407 | 0.074 | 6.45E-08 | |
| LPC 16:0 | LPC | −0.387 | 0.072 | 1.31E-07 | ✓ |
| LPC 20:4 | LPC | −0.365 | 0.071 | 3.30E-07 | ✓ |
| LPC 18:1e | LPC | −0.373 | 0.073 | 4.35E-07 | ✓ |
| TAG 50:1 | TAG | 0.329 | 0.065 | 6.44E-07 | ✓ |
| TAG 50:2 | TAG | 0.323 | 0.065 | 7.76E-07 | ✓ |
| LPC 18:3 | LPC | −0.369 | 0.076 | 1.40E-06 | |
| PC 38:3 | PC | 0.362 | 0.075 | 1.66E-06 | ✓ |
| SM (d18:0/24:0) | SM | 0.347 | 0.075 | 4.86E-06 | |
| LPE 16:0 | LPE | −0.352 | 0.078 | 7.17E-06 | ✓ |
| TAG 48:0 | TAG | 0.310 | 0.069 | 8.65E-06 | ✓ |
| LPE 18:1 | LPE | −0.343 | 0.077 | 1.04E-05 | ✓ |
| LPC 16:0e | LPC | −0.317 | 0.072 | 1.16E-05 | ✓ |
| LPC 16:1e | LPC | −0.310 | 0.072 | 1.86E-05 | ✓ |
| DAG 32:0 | DAG | 0.280 | 0.067 | 3.04E-05 | |
| LPC 18:2e | LPC | −0.299 | 0.074 | 5.24E-05 | ✓ |
| TAG 48:1 | TAG | 0.276 | 0.069 | 6.40E-05 | ✓ |
| PC 40:7 | PC | −0.302 | 0.075 | 6.85E-05 | ✓ |
| PC 36:3e | PC | −0.283 | 0.072 | 1.01E-04 | ✓ |
| LPC 20:5 | LPC | −0.296 | 0.076 | 1.04E-04 | |
| TAG 52:1 | TAG | 0.251 | 0.066 | 1.51E-04 | ✓ |
| LPE 18:2 | LPE | −0.276 | 0.078 | 4.26E-04 | ✓ |
| PC 34:1e | PC | −0.249 | 0.071 | 4.58E-04 | |
| PC 36:2 | PC | −0.270 | 0.077 | 4.85E-04 | ✓ |
| PC 36:0 | PC | −0.270 | 0.077 | 5.13E-04 | ✓ |
| DAG 34:1 | DAG | 0.221 | 0.064 | 5.48E-04 |
Estimated β coefficients represent the mean differences in standardized lipid measures between participants with and without obesity.
The p-value threshold for significance was determined by the Bonferroni method and based on the number of principal components. In FHS, the p-value threshold was p < 5.7 × 10−4 [0.05/(22×4)].
The p-value threshold for replication in the ERF study, PESA cohort, and SAFHS of the lipid cross-sectional associations with obesity was calculated using Bonferroni correction to be 1.56 × 10−3 (0.05/32).
✓ denotes lipid species that replicated in the ERF study, PESA cohort, or SAFHS.
Abbreviations: DAG=diacylglycerol; LPC=lysophosphatidylcholine; LPE=lysophosphatidylethanolamine; PC=phosphatidylcholine; SM=sphingomyelin; TAG=triacylglycerol.
2.5. Statistical methods
Statistical analyses were performed with SAS software version 9.2 (SAS Institute, Inc., Cary, NC, USA). All lipid measurements were rank-normalized via the Blom method [20]. For the FHS cross-sectional analyses, we used generalized linear mixed models (SAS procedure: GLIMMIX) to identify lipid species that (a) differed across all eight metabolic risk factor groups and (b) were associated with individual risk factors. All analyses were adjusted for the following baseline covariates: age, sex, cohort, and age-cohort interaction. To identify lipid-level associations across all eight groups, each lipid marker was treated as the outcome variable and the eight-level risk group as the main predictor. That is, our models were lipid level ~ cohort age sex cohort*age risk-factor-classes (* levels).
To identify independent associations between lipid levels and metabolic risk factors, each lipid marker was considered as the outcome variable and each risk factor as the main predictor with the other two risk factors as covariates. That is, our models were lipid level ~ cohort age sex cohort*age obesity dysclycemia dyslipidemia, where cohort, sex, obesity, dysclycemia and dyslipidemia are binary variables.
Secondary analyses of the dyslipidemia axis were conducted using the same models to explore independent associations between lipid species and high versus low TAG and HDL-C. In the PESA samples, generalized linear mixed models were used to identify lipid associations across all eight risk factor groups and with obesity, dysglycemia, and dyslipidemia. All replication models were adjusted for sex and baseline age. In addition, for individuals in the PESA cohort with self-reported use of lipid-lowering or diabetes medications, values were adjusted for treatment effects as follows. HDL-C was lowered by 10% and TAG was raised by 20% for individuals on lipid-lowering treatment, and glucose was raised by 10% or to a maximum of 126 mg/dL for those on diabetes medications [21]. In the ERF, pairwise partial correlation coefficients were generated for each metabolite-trait association, and all analyses were adjusted for age, sex, lipid-lowering medications, and family relationship using polygenic in the GenABEL R package [22]. Bonferroni-corrected p-values for obesity and dysglycemia were individually calculated as 0.05/n where n was equal to the number of metabolites measured in the ERF with available data for the corresponding trait.
For the longitudinal analyses, single marker associations were evaluated using general linear models (SAS procedure: GLM) with each lipid marker as the predictor and longitudinal changes in each metabolic trait (BMI, glucose, TAG, and HDL-C) as the outcome. All models were adjusted for age, sex, cohort, and baseline values of BMI, glucose, TAG, total cholesterol, and HDL-C. For individuals with self-reported use of lipid-lowering or diabetes medications at follow-up, values were adjusted as described above. We evaluated multimarker associations with longitudinal changes using stepwise regression with forward selection. Lipid markers with false discovery rate (FDR) < 0.25 in the single-marker analysis were considered as initial candidates for each trait. A dynamic p-value threshold corresponding to FDR < 0.25 was used for selection at each step [at step i, p < (0.25*i/154)]. For each trait, an aggregate R2 was calculated from the final model after forward selection.
To account for intercorrelations among lipid markers, principal component analyses were performed for the discovery (n = 22 principal components) and PESA replication (n = 13 principal components) sets. The number of principal components with eigenvalues >1 was deemed the number of “independent” tests, and Bonferroni correction was applied to account for multiple testing. For the cross-sectional analyses, the experiment-wide p-value threshold was determined as 0.05/n where n was equal to the number of principal components times four primary analyses (three risk factors plus the eight-group comparison) [23]. This method yielded a threshold of p < 5.7 × 10−4 for the FHS [0.05/(4 × 22)]. The same methodology was used to determine the p-value threshold (p < 5.7 × 10−4) for the longitudinal study, considering the four primary analyses (one per continuous metabolic trait).
2.6. Ethics statement
Informed consent was given by all participants at time of exam cycle from the FHS. The clinical study protocol was approved by the Boston University Medical Campus Institutional Review Board.
3. Results
3.1. Study sample characteristics
Table 1 lists the clinical characteristics of the 658 FHS participants according to the eight prespecified combinatorial groups. By design, mean age and percentage of women were consistent across all eight groups in the FHS, while by design, BMI, glucose, and lipid levels (HDL-C and TAG) varied. Supplementary Table 1 shows the clinical characteristics of the FHS longitudinal sample (n = 554) at the baseline and follow-up examinations. Mean changes, adjusted for lipid-lowering or hyperglycemia medication-use, were determined from baseline to follow-up for the four metabolic traits: 0.6 ± 2.6 kg/m2 for BMI, 0.5 ± 20.4 mg/dL for fasting glucose, 3.6 ± 10.7 mg/dL for HDL-C, and −0.5 ± 84.6 mg/dL for TAG.
Table 1.
Study design and clinical characteristics of FHS samples.
| Clinical characteristics | Group 1 | Group 2 | Group 3 | Group 4 | Group 5 | Group 6 | Group 7 | Group 8 |
|---|---|---|---|---|---|---|---|---|
| Obesity* | – | – | – | – | + | + | + | + |
| Dyslipidemia* | – | – | + | + | – | – | + | + |
| Dysglycemia* | – | + | – | + | – | + | – | + |
| Sample size (n) | 84 | 83 | 84 | 84 | 82 | 84 | 74 | 83 |
| Age (years) | 52 ± 14 | 54 ± 13 | 52 ± 14 | 54 ± 15 | 53 ± 12 | 54 ± 11 | 50 ± 14 | 55 ± 13 |
| Women (%) | 50 | 49 | 50 | 50 | 50 | 50 | 57 | 49 |
| BMI (kg/m2) | 24.5 ± 2.8 | 25.0 ± 2.7 | 25.9 ± 2.7 | 26.6 ± 2.3 | 32.4 ± 2.3 | 35.9 ± 5.1 | 35.4 ± 4.8 | 34.9 ± 4.9 |
| HDL-C (mg/dL) | 65 ± 18 | 62 ± 15 | 45 ± 11 | 44 ± 14 | 61 ± 13 | 58 ± 13 | 42 ± 10 | 42 ± 10 |
| TAG (mg/dL) | 81 ± 26 | 84 ± 26 | 167 ± 115 | 175 ± 91 | 92 ± 28 | 95 ± 29 | 165 ± 77 | 202 ± 148 |
| Glucose (mg/dL) | 92 ± 5 | 107 ± 15 | 92 ± 5 | 107 ± 7 | 93 ± 4 | 109 ± 7 | 93 ± 5 | 122 ± 37 |
(+) and (-) represent the presence and absence, respectively, of obesity, dysglycemia, and dyslipidemia in each of the eight metabolic risk factor groups. Obesity was defined as BMI ≥30 kg/m2. Dysglycemia was defined as fasting blood glucose ≥100 mg/dL. Dyslipidemia was defined as either TAG ≥150 mg/dL or HDL-C <40 mg/dL in men or <50 mg/dL in women.
3.2. Analytical set of potential lipidomic biomarkers
154 distinct metabolites representing ten classes and subclasses were measured in the FHS: four ceramides (Cers), 11 cholesteryl esters (CEs), eight diacylglycerols (DAGs), 21 lysophosphatidylcholines (LPCs), four lysophosphatidylethanolamines (LPEs), 28 phosphatidylcholines (PCs), 11 phosphatidylethanolamines (PEs), one phosphatidylserine (PS), 26 sphingomyelins (SMs), and 40 triacylglycerols (TAGs). A correlation matrix for all 154 lipid species is provided in Supplementary Figure 1. Many metabolite species were positively correlated, with the exception of some LPCs and TAGs, which were negatively correlated. Fig. 1 provides a flow chart of the analytical set and how each marker are used for analysis. .
3.3. FHS cross-sectional associations with metabolic risk factors
Among the aforementioned lipid species, several were cross-sectionally associated with obesity (n = 39; Table 2), dysglycemia (n = 8; Table 3), and dyslipidemia (n = 89; Supplementary Table 4) in the FHS. Many of the lipid markers for obesity were LPCs (n = 18; Table 2). LPCs, PCs, and LPEs were inversely associated, while SMs, TAGs, and DAGs were directly correlated with obesity (Table 2). Eight lipids were associated with dysglycemia; LPCs (n = 5, Table 3) were inversely associated, while SMs and Cer (n = 3, Table 3) were directly correlated with dysglycemia. The SM and Cer species are dihydrosphingolipids (dhSLs), which have backbones comprised of sphinganine (d18:0) rather than sphingosine (d18:1) as is the case for sphingolipids (SLs). Supplementary Table 4 lists all 89 lipid species associated with dyslipidemia (direct = 63, inverse = 26) along with their corresponding replication results. We further analyzed the metabolites by class; results are shown in Supplementary Figures 2, 3, 4, and 5 for obesity, dysglycemia, low vs. high HDL-C, and high vs. low TAG, respectively. Despite directional consistency, the majority of the associations between metabolite classes representing several individual lipid species and metabolic risk factors were not statistically significant, likely due to the broad diversity of individual metabolite species represented.
Table 3.
Cross-sectional lipid associations with dysglycemia in FHS.
| Lipid Species | Lipid Class | Estimated β* | SE | P-value† | Replication# |
|---|---|---|---|---|---|
| Cer (d18:0/24:0) | Cer | 0.280 | 0.070 | 6.59E-05 | |
| LPC 18:1e | LPC | −0.279 | 0.073 | 1.50E-04 | ✓ |
| LPC 17:0 | LPC | −0.271 | 0.073 | 1.98E-04 | ✓ |
| SM 36:0 | SM | 0.275 | 0.075 | 2.43E-04 | |
| LPC 18:0e | LPC | −0.260 | 0.071 | 2.80E-04 | ✓ |
| LPC 16:1e | LPC | −0.254 | 0.072 | 4.48E-04 | ✓ |
| SM (d18:0/24:0) | SM | 0.265 | 0.075 | 4.79E-04 | |
| LPC 18:0p | LPC | −0.244 | 0.070 | 5.48E-04 | ✓ |
Estimated β coefficients represent the mean differences in standardized lipid measures between participants with and without dysglycemia.
The p-value threshold for significance in FHS (p < 5.7 × 10−4) was determined by the Bonferroni method [0.05/(22×4)].
The p-value threshold for replication in the ERF study, PESA cohort, and SAFHS of the lipid cross-sectional associations with dysglycemia was calculated using Bonferroni correction to be 8.33 × 10−3 (0.05/6).
✓ denotes lipid species that replicated in the ERF, SAFHS, or PESA.
Abbreviations: Cer = ceramide; LPC = lysophosphatidylcholine; SM = sphingomyelin.
3.4. External replication
Independent external replication of all FHS cross-sectional metabolite associations with obesity and dysglycemia was sought in the ERF study, SAFHS, and PESA. Of the 39 metabolites that were cross-sectionally associated with obesity in the FHS, a total of 32 had available data for replication. Of these 32 metabolites, 28 were significantly associated with obesity at p < 1.56E-03 (0.05/32) (Table 2, Supplementary Table 2). Of the eight metabolites cross-sectionally associated with dysglycemia in the FHS, six had available data for replication. Of these six metabolites, five were associated with dysglycemia at p < 8.3E-03 (0.05/6) (Table 3, Supplementary Table 3).
3.5. Associations with longitudinal changes in metabolic traits
We identified several lipid species associated with longitudinal changes in BMI (n = 4), glucose (n = 4), and HDL-C (n = 1) (Table 4). LPCs 16:0, 18:0, 16:0e, and 22:5 were inversely associated with change in BMI [follow-up minus baseline]. These four LPCs were also inversely associated with BMI in the cross-sectional analyses. LPC 18:1 and PC 36:2 were inversely associated with change in glucose, and SM 36:0 and SM (d18:0/24:0) were directly associated with change in glucose. Both SMs were also directly associated with dysglycemia in the cross-sectional analyses. SM (d18:2/24:1), which was inversely associated with dyslipidemia in cross-sectional analysis, was directly associated with longitudinal change in HDL-C.
Table 4.
Single marker associations with longitudinal changes in metabolic risk factors.
| Change in metabolic trait | Lipid species | Lipid class | Estimated β* | SE | P-value† | FDR | Partial R2 |
|---|---|---|---|---|---|---|---|
| BMI | LPC 16:0 | LPC | −0.57 | 0.14 | 6.7E-05 | 0.01 | 0.029 |
| LPC 18:0 | LPC | −0.50 | 0.13 | 1.7E-04 | 0.01 | 0.026 | |
| LPC 16:0e | LPC | −0.44 | 0.12 | 4.2E-04 | 0.02 | 0.023 | |
| LPC 22:5 | LPC | −0.45 | 0.13 | 4.3E-04 | 0.02 | 0.023 | |
| Glucose | SM 36:0 | SM | 3.71 | 0.91 | 5.0E-05 | 0.01 | 0.030 |
| LPC 18:1 | LPC | −3.37 | 0.93 | 2.9E-04 | 0.02 | 0.024 | |
| SM (d18:0/24:0) | SM | 3.34 | 0.92 | 3.2E-04 | 0.02 | 0.024 | |
| PC 36:2 | PC | −2.97 | 0.85 | 5.2E-04 | 0.02 | 0.022 | |
| HDL-C | SM (d18:2/24:1) | SM | 2.61 | 0.55 | 2.8E-06 | 0.00 | 0.040 |
Estimated β coefficients represent the mean differences in longitudinal change (native units) in the stated metabolic trait per one standard deviation difference in lipid measures.
The p-value threshold for significance (p < 5.7 × 10−4) was determined by the Bonferroni method [0.05/(22×4)].
Abbreviations: LPC = lysophosphatidylcholine; PC = phosphatidylcholine; SM = sphingomyelin.
Lipids also associated in cross-sectional analysis with the corresponding metabolic risk factor are shown in bold.
In multimarker analyses, we identified multimarker panels associated with longitudinal changes in metabolic traits (Table 5). The BMI multimarker consisted of LPC 16:0, which explained 2.9% of the variation in longitudinal change in BMI (baseline BMI explained 2.7%). SM 36:0 and DAG 36:2 were retained in the final glucose model, together accounting for 6.1% of the variation in change in glucose (baseline glucose explained 22.1%). The HDL-C model contained SM (d18:2/24:1), LPC 20:3, and PS 36:0. This multimarker panel explained 8.4% of the variation in change in HDL-C (baseline HDL-C explained 3.2%). LPC 18:3, SM (d18:1/16:0), and Cer (d18:1/24:1) were retained in the final TAG model. These lipids accounted for 8% of the variation in change in TAG (baseline TAG explained 20.7%).
Table 5.
Multimarker associations with longitudinal changes in metabolic risk factors.
| Change in metabolic trait | Parameter | Estimated β* | SE | P-value† | R[2] |
|---|---|---|---|---|---|
| BMI | Baseline BMI | −0.08 | 0.02 | 1E-04 | 0.027 |
| Baseline Covariates‡ | 0.051 | ||||
| LPC 16:0 | −0.56 | 0.14 | <1.0E-04 | 0.029 | |
| Multi-Metabolite Panel | 0.029 | ||||
| Glucose | Baseline Glucose | −0.53 | 0.04 | <1.0E-04 | 0.221 |
| Baseline Covariates‡ | 0.289 | ||||
| SM 36:0 | 4.05 | 0.90 | <1.0E-04 | 0.036 | |
| DAG 36:2 | −4.58 | 1.23 | 2.0E-04 | 0.025 | |
| Multi-Metabolite Panel | 0.061 | ||||
| HDL-C | Baseline HDL-C | −0.15 | 0.04 | <1.0E-04 | 0.032 |
| Baseline Covariates‡ | 0.097 | ||||
| SM (d18:2/24:1) | 2.67 | 0.57 | <1.0E-04 | 0.042 | |
| LPC 20:3 | 1.60 | 0.50 | 1.6E-03 | 0.020 | |
| PS 36:0 | −1.60 | 0.48 | 9.0E-04 | 0.022 | |
| Multi-Metabolite Panel | 0.084 | ||||
| TAG | Baseline TAG | −0.57 | 0.05 | <1.0E-04 | 0.207 |
| Baseline Covariates‡ | 0.260 | ||||
| LPC 18:3 | −12.75 | 3.61 | 4.0E-04 | 0.023 | |
| SM (d18:1/16:0) | −21.18 | 5.05 | <1.0E-04 | 0.032 | |
| Cer (d18:1/24:1) | 15.91 | 4.24 | 2.0E-04 | 0.026 | |
| Multi-Metabolite Panel | 0.080 |
Estimated β coefficients represent the mean differences in longitudinal change (native units) in the stated metabolic trait per one standard deviation difference in lipid measures.
A dynamic p-value threshold for entrance into the model was defined based on FDR < 0.25 [at step i, p < (0.25*i)/154].
Baseline covariates were as follows: age; sex; cohort; batch; and baseline values for total cholesterol, HDL-C, glucose, and BMI.
Abbreviations: LPC = lysophosphatidylcholine; DAG = diacylglycerol; PS = phosphatidylserine; SM = sphingomyelin; Cer = ceramide.
Lipids also significant in individual longitudinal associations are shown in italics.
4. Discussion
Associations between plasma lipid classes and metabolic risk factors have been reported [24], [25], [26], [27], [28], [29], but many of these studies were limited to single site studies with small numbers of lipid species or case-control analyses. We used liquid chromatography-tandem mass spectrometry to detect cross-sectional and longitudinal lipidomic signatures of key metabolic risk factors. Our cross-sectional lipidomic signatures of obesity and dysglycemia were highly replicable in external studies and highlight two lipid classes as biomarkers of metabolic risk: lysoglycerophospholipids (LGPLs) and sphingolipids (SLs).
4.1. Lysoglycerophospholipid markers of metabolic traits
Cross-sectional and longitudinal analyses of obesity/BMI and dysglycemia/fasting blood glucose identified significant associations with LGPLs, which are generated along with a free fatty acid by phospholipase-mediated hydrolysis of glycerophospholipids (GPLs; supplementary figure 6c) [30]. The predominant LGPL in our results was lysophosphatidylcholine (LPC). We identified inverse cross-sectional associations between all LPCs and obesity (n = 18; Table 2). Fifteen (83%) of these associations replicated in the ERF and SAFHS (Supplementary Tables 2 and 3). Four of the 18 LPCs cross-sectionally associated with obesity in the FHS were also associated with longitudinal changes in BMI (Table 4). Five of the LPCs associated with obesity cross-sectionally in the FHS were also inversely associated with dysglycemia (Supplementary Table 5), and all five of these associations replicated. Nine phosphatidylcholines (PCs) were associated with obesity (Table 2) – all showing inverse associations, with the exception of PC (38:3), which showed a positive association with obesity (β = 0.36, P = 1.7E-06; Table 2). This association replicated in the ERF, and prior work has shown PC (38:3) to be positively associated with CVD [31] and enriched in atherosclerotic plaques [32]. Of the nine PCs associated with obesity, seven were available for replication and five (71%), including PC (38:3), replicated (Supplementary Table 2). Lysophosphatidylethanolamine (LPE), another LGPL class detected by our lipid-profiling platform, was inversely associated with obesity in the FHS (Table 2); all three LPEs associated with obesity in the FHS replicated (Supplementary Table 2). Overall, LGPLs were inversely associated with metabolic risk factors.
Recent evidence, which has implicated LGPLs in essential signaling processes [33], highlights the potential utility of these lipid classes as novel biomarkers of cardiometabolic risk. LPCs are the most well-characterized and abundant LGPL in human plasma (125–143 µM in healthy individuals) [34], and circulating levels are determined by their generation via two pathways – 1) transfer of a fatty acid from PC to cholesterol by lecithin-cholesterol acyltransferase (LCAT; supplementary Figure 6d), and 2) phospholipase A2 (PLA2)-mediated hydrolysis of PC [35] (Supplementary Figure 6c, bottom) and clearance via transporters such as albumin and alpha-1 acid glycoprotein [34]. While LPCs have been found to be elevated in atherosclerotic plaques [32], and thus might be expected to positively correlate with metabolic risk factors, we found inverse associations between LPCs and obesity and dysglycemia (Tables 2, 3). This seemingly counterintuitive finding for LPCs, however, has been demonstrated for CVD [31,36] and perhaps reflects an increase in LPC catabolism or a decrease in its generation in such physiological states. Indeed, individuals with coronary artery disease have been shown to have lower LCAT activity [37], and three recent studies found reduced plasma LPC levels in obese patients [24,25,13]. Furthermore, higher levels of circulating LPC could reflect a lower rate of atherogenic lysophosphatidic acid production via autotaxin-mediated LPC hydrolysis, thus conferring a net protective effect on CVD [38]. Therefore, in conjunction with recent epidemiological evidence associating dairy consumption with changes in circulating LPC 17:0 [39], an abundance of genetic, epidemiologic, functional, and biochemical data implicating LPCs in CVD suggest that mechanistically understanding the enzyme kinetics and complexities of LPC metabolism may highlight promising therapeutic targets.
4.2. Sphingolipid markers for metabolic traits
Sphingolipids, comprised of Cers, SMs, and their reduced forms (dhCers and dhSMs), were positively associated with obesity (Table 2) and dysglycemia (Table 3). The two dhSMs positively associated with obesity – SM 36:0 and SM (d18:0/24:0) – were also positively associated with dysglycemia, along with Cer (d18:0/24:0). Cer (d18:0/24:0) shares the same sphingosine backbone and fatty acyl side chains as SM (d18:0/24:0) such that the two are inter-convertible via the enzymatic actions of sphingomyelin synthase (SMS; Cer→SM) and sphingomyelinase (SMase; SM→Cer). Of note, SM 36:0 and SM (d18:0/24:0) were also associated with longitudinal change in glucose (Table 4).
Sphingolipids, which are characterized by a sphingosine backbone, are synthesized de novo from the condensation of palmitoyl-CoA and serine by serine palmitoyltransferase [40], with the resulting dhCer being oxidizable by dihydroceramide desaturases (Des) to Cer. Sphingolipids are integral structural constituents of lipid membranes and are involved in cell signaling and apoptosis [41,42], with SMs being the most prevalent SLs in human plasma that can be derived from Cers via SMS, releasing a DAG in the process [41,43] (Supplementary Figure 6a). Sphingomyelin inhibits lipoprotein lipase (LPL)-catalyzed hydrolysis of TAG [44] and LCAT-catalyzed esterification of cholesterol [45] (Supplementary Figure 6b), and individuals with SMase deficiency (Niemann-Pick disease) have low plasma HDL-C levels [46] due to SM-mediated inhibition of cholesterol efflux. Sphingomyelin accumulation has been associated with T2DM [47] and observed in mouse models of insulin resistance [48,49] as well as in the skeletal muscle and serum of obese, insulin-resistant humans [27,50,51].
Prior studies have shown plasma SM levels to predict coronary artery disease risk, possibly due to their strong correlations with apolipoprotein B (apoB) and TAG levels [52]. Indeed, the SM content of apoB-containing or TAG-rich lipoproteins is higher than that in HDL [53], and hydrolysis of LDL-associated SM to Cer causes LDL particle aggregation and macrophage foam cell formation [54,55], which are two hallmarks of atherosclerosis. Recent studies have suggested that Cer provides additional prognostic utility for CVD beyond that from LDL-C [56] – perhaps because it acts as a proxy to standardize changes in LDL-C mass by changes in apoB concentration, which is the biological determinant of atherogenicity of a high LDL-C level [57]. Interestingly, a missense mutation (rs267738) in the CERS2 gene, which encodes the enzyme ceramide synthase that is responsible for biosynthesis of very long-chain Cer (C22/C24/C24:1), is also associated with hemoglobin A1c levels [58], and functional studies have shown that CerS2 haploinsufficiency in mice results in insulin resistance and non-alcoholic fatty liver disease [59], possibly due to a compensatory increase in atherogenic Cer-16. Of note, several dietary intervention studies have demonstrated that improved diet quality blunts the association between Cer and CVD [60], perhaps due to the dependence of Cer synthesis on dietary intake of saturated fats [61]. Nonetheless, the most direct evidence comes from mouse knockout studies in which genetic ablation of key enzymes in Cer synthesis mitigates atherosclerosis [62], insulin resistance [63], and T2DM [64] among other metabolic-related disease phenotypes. Taken together with our results, there is an abundance of evidence for Cers and SMs as markers of obesity and dysglycemia risk.
We acknowledge several limitations of our study. First, the effect sizes and metabolite species differed among the FHS and replication cohorts, despite directional consistency in the replicable metabolites. These discrepancies were in part due to differences in environmental exposures (i.e. smoking) [13,65] as well as discrepancies in lipidomic and analytical techniques across studies. Moreover, some lipids were unavailable for replication. Even considering these limitations, however, consistent relationships were observed, thus reinforcing the robustness of the replicated results. Second, the longitudinal changes in mean TAG levels (<1 mg/dL over six-years of follow up) were small and not of a magnitude to confer clinical consequences. Third, our lipid quantification methods did not allow for the separation of isomeric species and thus multiple isomers could have contributed to each lipid measurement. Fourth, the low replication rate for sphingolipids was partially due to the fact that many of the sphingolipids quantified in the FHS via liquid chromatography tandem mass spectrometry were not measured in the replication cohorts, which used different lipidomic platforms. Finally, larger-scale, multi-site epidemiological studies as well as functional experiments are needed to confirm and extend the findings of this investigation.
5. Conclusions
We identified numerous lipid species that were associated with metabolic risk factors cross-sectionally and with their longitudinal changes. Of note, we demonstrated robust and replicable associations of lysoglycerophospholipids and dihydrosphingolipids with obesity and dysglycemia. Taken together with biological evidence and prior interventional and functional studies, we hypothesize that the lipid species identified in this study may serve as key mechanistic drivers of metabolic dysregulation and can lay the groundwork for future studies to explore their potential therapeutic relevance.
CRediT authorship contribution statement
Xiaoyan Yin: Conceptualization, Writing - original draft. Christine M. Willinger: Conceptualization, Writing - original draft. Joshua Keefe: Data curation, Formal analysis, Writing - review & editing. Jun Liu: Project administration, Validation. Antonio Fernández-Ortiz: Project administration, Validation. Borja Ibáñez: Project administration, Validation. José Peñalvo: Project administration, Validation. Aram Adourian: Project administration, Validation. George Chen: Project administration, Validation. Dolores Corella: Project administration, Validation. Reinald Pamplona: Project administration, Validation. Manuel Portero-Otin: Project administration, Validation. Mariona Jove: Project administration, Validation. Paul Courchesne: Data curation, Project administration. Cornelia M. van Duijn: Project administration, Validation. Valentín Fuster: Project administration, Validation. José M. Ordovás: Data curation, Formal analysis. Ayşe Demirkan: Project administration, Validation. Martin G. Larson: Data curation, Formal analysis. Daniel Levy: Conceptualization, Supervision, Writing - review & editing.
Declaration of Competing Interest
Dr. Adourian contributed to this project while affiliated with BG Medicine, Inc., a biomarker discovery company. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Acknowledgments
Acknowledgements & Funding
The Framingham Heart Study is funded by National Institutes of Health (NIH) contract N01-HC-25195. This study was made possible by a CRADA between BG Medicine, Inc., Boston University, and the NHLBI, and the laboratory work for this research was supported by the Division of Intramural Research of the National Heart, Lung, and Blood Institute (NHLBI). Analytical work was funded by the Division of Intramural Research of NHLBI as well as the Center for Information Technology, NIH, Bethesda, MD. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Heart, Lung, and Blood Institute; the National Institutes of Health; or the U.S. Department of Health and Human Services.
The PESA study is supported by a non-competitive unrestricted grant shared between the National Center for Cardiovascular Research Carlos III (CNIC) and the Bank of Santander. The PESA study is a noncommercial study independent of the health and pharmaceutical industry. The CNIC is supported by the Spanish Ministry of Economy and Competitiveness and the Pro-CNIC Foundation. JMO is supported by the US Department of Agriculture, under agreement no. 8050-51000-098-00D. MPO and MJ acknowledge an Institute of Health Carlos III grant (PI 17-00134). This research was in part funded by the Spanish Ministry of Economy and Competitiveness, Institute of Health Carlos III (PI14/00328), co-financed by FEDER funds from the European Union (‘A way to built Europe’), and the Generalitat of Catalonia, Department of Health(SLT002/16/00250) and Department of Business and Knowledge(2017SGR696) to R.P. MJ is a Serra Húnter Fellow.
Data Sharing
The lipidomic data have been deposited in the NCBI's dbGaP database (phs000007.v30.p11). All Framingham Heart Study data is considered ‘Controlled access’ and accessible with direct application to dbGaP.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2019.10.046.
Appendix. Supplementary materials
References
- 1.Organization WH. Global status report on noncommunicable diseases 2010. 2011; Geneva, Switzerland.
- 2.Patti A.M., Al-Rasadi K., Giglio R.V., Nikolic D., Mannina C., Castellino G. Natural approaches in metabolic syndrome management. Arch Med Sci. 2018;14(2):422–441. doi: 10.5114/aoms.2017.68717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ford ES. Risks for all-cause mortality, cardiovascular disease, and diabetes associated with the metabolic syndrome: a summary of the evidence. Diabetes Care. 2005;28(7):1769–1778. doi: 10.2337/diacare.28.7.1769. [DOI] [PubMed] [Google Scholar]
- 4.Mottillo S., Filion K.B., Genest J., Joseph L., Pilote L., Poirier P. The metabolic syndrome and cardiovascular risk a systematic review and meta-analysis. J Am Coll Cardiol. 2010;56(14):1113–1132. doi: 10.1016/j.jacc.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 5.Murphy S.A., Nicolaou A. Lipidomics applications in health, disease and nutrition research. Mol Nutr Food Res. 2013;57(8):1336–1346. doi: 10.1002/mnfr.201200863. [DOI] [PubMed] [Google Scholar]
- 6.Hoefer I.E., Steffens S., Ala-Korpela M., Back M., Badimon L., Bochaton-Piallat M.L. Novel methodologies for biomarker discovery in atherosclerosis. Eur Heart J. 2015;36(39):2635–2642. doi: 10.1093/eurheartj/ehv236. [DOI] [PubMed] [Google Scholar]
- 7.Feinleib M., Kannel W.B., Garrison R.J., McNamara P.M., Castelli W.P. The Framingham offspring study. Design and preliminary data. Prev Med. 1975;4(4):518–525. doi: 10.1016/0091-7435(75)90037-7. [DOI] [PubMed] [Google Scholar]
- 8.Splansky G.L., Corey D., Yang Q., Atwood L.D., Cupples L.A., Benjamin E.J. The third generation cohort of the national heart, lung, and blood institute's Framingham heart study: design, recruitment, and initial examination. Am J Epidemiol. 2007;165(11):1328–1335. doi: 10.1093/aje/kwm021. [DOI] [PubMed] [Google Scholar]
- 9.Santos RL, Zillikens MC, Rivadeneira FR, Pols HA, Oostra BA, van Duijn CM. Heritability of fasting glucose levels in a young genetically isolated population. Diabetologia. 2006;49(4):667–672. doi: 10.1007/s00125-006-0142-6. [DOI] [PubMed] [Google Scholar]
- 10.Lorenzo C, Okoloise M, Williams K, Stern MP, Haffner SM, San Antonio Heart S. The metabolic syndrome as predictor of type 2 diabetes: the San Antonio heart study. Diabetes Care. 2003;26(11):3153–3159. doi: 10.2337/diacare.26.11.3153. [DOI] [PubMed] [Google Scholar]
- 11.Fernandez-Ortiz A., Jimenez-Borreguero L.J., Penalvo J.L., Ordovas J.M., Mocoroa A., Fernandez-Friera L. The progression and early detection of subclinical atherosclerosis (PESA) study: rationale and design. Am Heart J. 2013;166(6):990–998. doi: 10.1016/j.ahj.2013.08.024. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, van Klinken JB, Semiz S, van Dijk KW, Verhoeven A, Hankemeier T. A mendelian randomization study of metabolite profiles, fasting glucose, and type 2 diabetes. Diabetes. 2017;66(11):2915–2926. doi: 10.2337/db17-0199. [DOI] [PubMed] [Google Scholar]
- 13.Weir J.M., Wong G., Barlow C.K., Greeve M.A., Kowalczyk A., Almasy L. Plasma lipid profiling in a large population-based cohort. J Lipid Res. 2013;54(10):2898–2908. doi: 10.1194/jlr.P035808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haffner S.M., Miettinen H., Gaskill S.P., Stern M.P. Metabolic precursors of hypertension. the San Antonio heart study. Arch Intern Med. 1996;156(17):1994–2001. [PubMed] [Google Scholar]
- 15.Gonzalez-Covarrubias V, Beekman M, Uh HW, Dane A, Troost J, Paliukhovich I. Lipidomics of familial longevity. Aging Cell. 2013;12(3):426–434. doi: 10.1111/acel.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demirkan A., van Duijn C.M., Ugocsai P., Isaacs A., Pramstaller P.P., Liebisch G. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 2012;8(2) doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Draisma H.H.M., Pool R., Kobl M., Jansen R., Petersen A.K., Vaarhorst A.A.M. Genome-wide association study identifies novel genetic variants contributing to variation in blood metabolite levels. Nat Commun. 2015;6:7208. doi: 10.1038/ncomms8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pizarro C., Arenzana-Ramila I., Perez-del-Notario N., Perez-Matute P., Gonzalez-Saiz J.M. Plasma lipidomic profiling method based on ultrasound extraction and liquid chromatography mass spectrometry. Anal Chem. 2013;85(24):12085–12092. doi: 10.1021/ac403181c. [DOI] [PubMed] [Google Scholar]
- 19.National Cholesterol Education Program Expert Panel on Detection E Treatment of high blood cholesterol in A. third report of the National Cholesterol Education Program (NCEP) expert panel on detection, evaluation, and treatment of high blood cholesterol in adults (Adult treatment panel III) final report. Circulation. 2002;106(25):3143–3421. [PubMed] [Google Scholar]
- 20.Blom G. John Wiley & Sons, Inc.; New York: 1958. Statistical estimates and transformed beta variables. [Google Scholar]
- 21.Levy D., DeStefano A.L., Larson M.G., O'Donnell C.J., Lifton R.P., Gavras H. Evidence for a gene influencing blood pressure on chromosome 17. Genome scan linkage results for longitudinal blood pressure phenotypes in subjects from the framingham heart study. Hypertension. 2000;36(4):477–483. doi: 10.1161/01.hyp.36.4.477. [DOI] [PubMed] [Google Scholar]
- 22.Team RDC . R Foundation for Statistical Computing; 2008. R: a language and environment for statistical computing. ISBN 3-900051-07-0. [Google Scholar]
- 23.Li M.X., Yeung J.M., Cherny S.S., Sham P.C. Evaluating the effective numbers of independent tests and significant p-value thresholds in commercial genotyping arrays and public imputation reference datasets. Hum Genet. 2012;131(5):747–756. doi: 10.1007/s00439-011-1118-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graessler J, Schwudke D, Schwarz PE, Herzog R, Shevchenko A, Bornstein SR. Top-down lipidomics reveals ether lipid deficiency in blood plasma of hypertensive patients. PLoS ONE. 2009;4(7):e6261. doi: 10.1371/journal.pone.0006261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barber MN, Risis S, Yang C, Meikle PJ, Staples M, Febbraio MA. Plasma lysophosphatidylcholine levels are reduced in obesity and type 2 diabetes. PLoS ONE. 2012;7(7):e41456. doi: 10.1371/journal.pone.0041456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grapov D, Adams SH, Pedersen TL, Garvey WT, Newman JW. Type 2 diabetes associated changes in the plasma non-esterified fatty acids, oxylipins and endocannabinoids. PLoS ONE. 2012;7(11):e48852. doi: 10.1371/journal.pone.0048852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haus JM, Kashyap SR, Kasumov T, Zhang R, Kelly KR, Defronzo RA. Plasma ceramides are elevated in obese subjects with type 2 diabetes and correlate with the severity of insulin resistance. Diabetes. 2009;58(2):337–343. doi: 10.2337/db08-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kulkarni H, Meikle PJ, Mamtani M, Weir JM, Barlow CK, Jowett JB. Plasma lipidomic profile signature of hypertension in Mexican American families: specific role of diacylglycerols. Hypertension. 2013;62(3):621–626. doi: 10.1161/HYPERTENSIONAHA.113.01396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhee E.P., Cheng S., Larson M.G., Walford G.A., Lewis G.D., McCabe E. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121(4):1402–1411. doi: 10.1172/JCI44442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farooqui A.A., Horrocks L.A., Farooqui T. Interactions between neural membrane glycerophospholipid and sphingolipid mediators: a recipe for neural cell survival or suicide. J Neurosci Res. 2007;85(9):1834–1850. doi: 10.1002/jnr.21268. [DOI] [PubMed] [Google Scholar]
- 31.Stegemann C, Pechlaner R, Willeit P, Langley SR, Mangino M, Mayr U. Lipidomics profiling and risk of cardiovascular disease in the prospective population-based bruneck study. Circulation. 2014;129(18):1821–1831. doi: 10.1161/CIRCULATIONAHA.113.002500. [DOI] [PubMed] [Google Scholar]
- 32.Stegemann C., Drozdov I., Shalhoub J., Humphries J., Ladroue C., Didangelos A. Comparative lipidomics profiling of human atherosclerotic plaques. Circ Cardiovasc Genet. 2011;4(3):232–242. doi: 10.1161/CIRCGENETICS.110.959098. [DOI] [PubMed] [Google Scholar]
- 33.Engelking L.R. Textbook of veterinary physiological chemistry (Third edition) 2015. Chapter 53 - Overview of lipid metabolism; pp. 340–344. [Google Scholar]
- 34.Law S.H., Chan M.L., Marathe G.K., Parveen F., Chen C.H., Ke L.Y. An updated review of lysophosphatidylcholine metabolism in human diseases. Int J Mol Sci. 2019;20(5) doi: 10.3390/ijms20051149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto T., Kobayashi T., Kamata K. Role of lysophosphatidylcholine (LPC) in atherosclerosis. Curr Med Chem. 2007;14(30):3209–3220. doi: 10.2174/092986707782793899. [DOI] [PubMed] [Google Scholar]
- 36.Croset M., Brossard N., Polette A., Lagarde M. Characterization of plasma unsaturated lysophosphatidylcholines in human and rat. Biochem J. 2000;345(Pt 1):61–67. [PMC free article] [PubMed] [Google Scholar]
- 37.Duivenvoorden R., Holleboom A.G., van den Bogaard B., Nederveen A.J., de Groot E., Hutten B.A. Carriers of lecithin cholesterol acyltransferase gene mutations have accelerated atherogenesis as assessed by carotid 3.0-T magnetic resonance imaging [corrected] J Am Coll Cardiol. 2011;58(24):2481–2487. doi: 10.1016/j.jacc.2010.11.092. [DOI] [PubMed] [Google Scholar]
- 38.Morris A.J., Smyth S.S. Lysophosphatidic acid and cardiovascular disease: seeing is believing. J Lipid Res. 2013;54(5):1153–1155. doi: 10.1194/jlr.E037887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Petersen K.S., Keogh J.B., Lister N., Weir J.M., Meikle P.J., Clifton P.M. Association between dairy intake, lipids and vascular structure and function in diabetes. World J Diabetes. 2017;8(5):202–212. doi: 10.4239/wjd.v8.i5.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Magaye R.R., Savira F., Hua Y., Kelly D.J., Reid C., Flynn B. The role of dihydrosphingolipids in disease. Cell Mol Life Sci. 2018 doi: 10.1007/s00018-018-2984-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.AOCS Lipid Library; 2013. An introduction to sphingolipids and membrane rafts. [Google Scholar]
- 42.Merrill A.H., Jr. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem Rev. 2011;111(10):6387–6422. doi: 10.1021/cr2002917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quehenberger O., Armando A.M., Brown A.H., Milne S.B., Myers D.S., Merrill A.H. Lipidomics reveals a remarkable diversity of lipids in human plasma. J Lipid Res. 2010;51(11):3299–3305. doi: 10.1194/jlr.M009449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuksis A., Breckenridge W.C., Myher J.J., Kakis G. Replacement of endogenous phospholipids in rat plasma lipoproteins during intravenous infusion of an artificial lipid emulsion. Can J Biochem. 1978;56(6):630–639. doi: 10.1139/o78-095. [DOI] [PubMed] [Google Scholar]
- 45.Rye K.A., Hime N.J., Barter P.J. The influence of sphingomyelin on the structure and function of reconstituted high density lipoproteins. J Biol Chem. 1996;271(8):4243–4250. doi: 10.1074/jbc.271.8.4243. [DOI] [PubMed] [Google Scholar]
- 46.Viana M.B., Giugliani R., Leite V.H., Barth M.L., Lekhwani C., Slade C.M. Very low levels of high density lipoprotein cholesterol in four sibs of a family with non-neuropathic niemann-pick disease and sea-blue histiocytosis. J Med Genet. 1990;27(8):499–504. doi: 10.1136/jmg.27.8.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hannun Y.A., Obeid L.M. Many ceramides. J Biol Chem. 2011;286(32):27855–27862. doi: 10.1074/jbc.R111.254359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Holland W.L., Brozinick J.T., Wang L.P., Hawkins E.D., Sargent K.M., Liu Y. Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab. 2007;5(3):167–179. doi: 10.1016/j.cmet.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 49.Samad F, Hester KD, Yang G, Hannun YA, Bielawski J. Altered adipose and plasma sphingolipid metabolism in obesity: a potential mechanism for cardiovascular and metabolic risk. Diabetes. 2006;55(9):2579–2587. doi: 10.2337/db06-0330. [DOI] [PubMed] [Google Scholar]
- 50.Straczkowski M, Kowalska I, Nikolajuk A, Dzienis-Straczkowska S, Kinalska I, Baranowski M. Relationship between insulin sensitivity and sphingomyelin signaling pathway in human skeletal muscle. Diabetes. 2004;53(5):1215–1221. doi: 10.2337/diabetes.53.5.1215. [DOI] [PubMed] [Google Scholar]
- 51.Adams JM, 2nd, Pratipanawatr T, Berria R, Wang E, DeFronzo RA, Sullards MC. Ceramide content is increased in skeletal muscle from obese insulin-resistant humans. Diabetes. 2004;53(1):25–31. doi: 10.2337/diabetes.53.1.25. [DOI] [PubMed] [Google Scholar]
- 52.Schlitt A., Blankenberg S., Yan D., von Gizycki H., Buerke M., Werdan K. Further evaluation of plasma sphingomyelin levels as a risk factor for coronary artery disease. Nutr Metab (Lond) 2006;3:5. doi: 10.1186/1743-7075-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chapman M.J. Comparative analysis of mammalian plasma lipoproteins. Methods Enzymol. 1986;128:70–143. doi: 10.1016/0076-6879(86)28063-5. [DOI] [PubMed] [Google Scholar]
- 54.Pentikainen M.O., Lehtonen E.M., Kovanen P.T. Aggregation and fusion of modified low density lipoprotein. J Lipid Res. 1996;37(12):2638–2649. [PubMed] [Google Scholar]
- 55.Tabas I., Li Y., Brocia R.W., Xu S.W., Swenson T.L., Williams K.J. Lipoprotein lipase and sphingomyelinase synergistically enhance the association of atherogenic lipoproteins with smooth muscle cells and extracellular matrix. A possible mechanism for low density lipoprotein and lipoprotein(a) retention and macrophage foam cell formation. J Biol Chem. 1993;268(27):20419–20432. [PubMed] [Google Scholar]
- 56.Cheng JM, Suoniemi M, Kardys I, Vihervaara T, de Boer SP, Akkerhuis KM. Plasma concentrations of molecular lipid species in relation to coronary plaque characteristics and cardiovascular outcome: results of the Atheroremo-ivus study. Atherosclerosis. 2015;243(2):560–566. doi: 10.1016/j.atherosclerosis.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 57.Olofsson S.O., Boren J. Apolipoprotein B: a clinically important apolipoprotein which assembles atherogenic lipoproteins and promotes the development of atherosclerosis. J Intern Med. 2005;258(5):395–410. doi: 10.1111/j.1365-2796.2005.01556.x. [DOI] [PubMed] [Google Scholar]
- 58.Wheeler E, Leong A, Liu CT, Hivert MF, Strawbridge RJ, Podmore C. Impact of common genetic determinants of hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: a transethnic genome-wide meta-analysis. PLoS Med. 2017;14(9) doi: 10.1371/journal.pmed.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Raichur S., Wang S.T., Chan P.W., Li Y., Ching J., Chaurasia B. CerS2 haploinsufficiency inhibits beta-Oxidation and confers susceptibility to diet-induced steatohepatitis and insulin resistance. Cell Metab. 2014;20(5):919. doi: 10.1016/j.cmet.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 60.Estruch R., Ros E., Salas-Salvado J., Covas M.I., Corella D., Aros F. Primary prevention of cardiovascular disease with a mediterranean diet supplemented with extra-virgin Olive oil or Nuts. N Engl J Med. 2018;378(25):e34. doi: 10.1056/NEJMoa1800389. [DOI] [PubMed] [Google Scholar]
- 61.Chavez J.A., Summers S.A. Characterizing the effects of saturated fatty acids on insulin signaling and ceramide and diacylglycerol accumulation in 3T3-L1 adipocytes and C2C12 myotubes. Arch Biochem Biophys. 2003;419(2):101–109. doi: 10.1016/j.abb.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 62.Hojjati M.R., Li Z., Zhou H., Tang S., Huan C., Ooi E. Effect of myriocin on plasma sphingolipid metabolism and atherosclerosis in apoE-deficient mice. J Biol Chem. 2005;280(11):10284–10289. doi: 10.1074/jbc.M412348200. [DOI] [PubMed] [Google Scholar]
- 63.Bikman B.T., Guan Y., Shui G., Siddique M.M., Holland W.L., Kim J.Y. Fenretinide prevents lipid-induced insulin resistance by blocking ceramide biosynthesis. J Biol Chem. 2012;287(21):17426–17437. doi: 10.1074/jbc.M112.359950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ussher JR, Koves TR, Cadete VJ, Zhang L, Jaswal JS, Swyrd SJ. Inhibition of de novo ceramide synthesis reverses diet-induced insulin resistance and enhances whole-body oxygen consumption. Diabetes. 2010;59(10):2453–2464. doi: 10.2337/db09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szymanska E., Bouwman J., Strassburg K., Vervoort J., Kangas A.J., Soininen P. Gender-dependent associations of metabolite profiles and body fat distribution in a healthy population with central obesity: towards metabolomics diagnostics. OMICS. 2012;16(12):652–667. doi: 10.1089/omi.2012.0062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.