Abstract
Food safety has emerged as an important global issue with international trade and public health implications. Bacillus cereus is an important cause of food poisoning worldwide. A total of 200 individual meat samples were collected from meat retail outlets and restaurants and investigated the frequency of B. cereus and hemolysin BL (Hbl), non-hemolytic enterotoxin (Nhe) complex genes. The meat samples were immediately homogenized and cultured on Bacillus cereus selective agar and subjected for confirmatory biochemical tests and molecular detection of gyrB, hblA, hblC, hblD, nheA, nheB and nheC genes. A total of 29 (14.5 %) meat samples were positive for the presence of B. cereus. The frequency of B. cereus in raw meat (14.1 %) was similar to cooked beef samples (15 %) (P > 0.05). Twenty six (89.6 %) isolates carried at least one or more enterotoxin genes. We found nheA (58.6 %) and hblD (51.7 %) genes with higher frequency than others. Hemolysin BL complex genes were found in lower frequency than Nhe complex (P > 0.05). Detection of enterotoxigenic B. cereus in meat samples shows a probable risk for public health. Therefore, the reliable molecular methods for monitoring of potentially pathogenic B. cereus are strongly recommended for the routine food examination.
Keywords: Bacillus cereus, Meat, Hbl complex, Nhe complex, PCR
1. Introduction
Bacillus cereus is one of the most common causes of food borne outbreaks. It is frequently associated with diarrheal and emetic syndromes [1,2]. B. cereus emetic syndrome is related to the consumption of foods containing sufficient amounts of cereulide toxin. Furthermore, diarrheal food poisoning usually occurs as a result of germination and multiplication of surviving spores in cooked or pasteurized foods [3]. B. cereus widely distributed in different environments (water, soil and dust) and it is often present in a variety of foods such as rice, spices, milk and dairy products, vegetables, meat and meat products, cakes and other desserts [3,4].
Bacillus cereus produces emetic toxin and several enterotoxins including non-hemolytic enterotoxin (Nhe), hemolysin BL (Hbl), cytolysin K (CytK), hemolysin II (HlyII), enterotoxin FM (EntFM), and enterotoxin T (bc-D-ENT) [3,4].
Three secreted pore-forming cytotoxins Hbl, Nhe, and CytK are considered as the primary virulence factors in B. cereus diarrhea [5]. Hemolysin BL is a three-component toxin consisting of two lytic proteins, L2 and L1, and a binding component B encoded by hblC, hblD and hblA, respectively [5,6]. The presence of three components Hbl-B, Hbl-L1, and Hbl-L2 is necessary for hemolysin BL activity. Distribution of hbl genes in B. cereus species is quite diverse, and strains show different capability of producing diarrheal toxins. Several factors can group such strains based on growth temperature, food matrix and nutritional availability [5,7,8]. Non-hemolytic enterotoxin also is a pore forming toxin consisting of two lytic elements NheA and NheB, and a protein NheC with unknown function encoded by nheA, nheB, and nheC, respectively. CytK is a single-component toxin that has necrotic and hemolytic effects and is toxic for human intestine epithelium cells [5,9].
Due to the increasing incidence of B. cereus food-borne disease and the wide spread distribution of B. cereus in food, rapid detection methods are required for diagnostic purposes and for the prevention of food contamination and food-borne outbreaks [10]. Risk assessment and microbial monitoring will continue to play important role in quality assurance of meat products [11]. The minimal bacterial count required to provoke emetic and diarrheal syndromes was estimated to be approximately >105 colony-forming units (cfu)/g of ingested food [1,5]. However, there are some reports of emetic syndrome associated with foods containing only 103 cfu/g of food [1]. Methods currently used for detection of B. cereus in food are culture on selective media and biochemical tests, Enzyme-Linked Immunosorbent Assay (ELISA) and polymerase chain reaction (PCR) technique [1,11,12].
In Iran, no information is available on the incidence of enterotoxigenic strains of B. cereus in raw and cooked meat. Based on this fact, the objective of present study was to investigate the frequency of Hbl and Nhe complex genes of B. cereus in meat samples collected from meat retail outlets and restaurants in Zanjan, Iran.
2. Materials and methods
2.1. Sampling
From March to June 2015, a total of 200 individual meat samples including 60 raw beef, 60 raw lamb and 80 cooked beef samples were collected from meat retail outlets and restaurants in Zanjan, Iran. Due to the distribution of meat retail outlets and restaurants in Zanjan, the cluster sampling method was used. At first, Zanjan was divided into 5 regions based on the abundance of outlets. Then, a total of 10 meat retail outlets and 10 restaurants were selected. Six samples of raw beef and 6 samples of raw lamb were collected from each meat retail outlet and 8 samples were collected from each restaurant. Twenty five gram of meat samples were packed into a clean polyethylene bag then marked and transported to the laboratory of food microbiology in a cool box for analysis within 1 h.
2.2. Reference strain
Bacillus cereus ATCC 14579 was used as a reference enterotoxin-positive strain for detection of hblACD and nheABC genes in B. cereus isolates using PCR.
2.3. Isolation and identification of B. cereus
The meat samples were transferred in buffered peptone water 0.1 % (PW). Pepton water was used to avoid the strain variation and keep the strain a live as possible.Twenty five gram of meat samples was homogenized for 2 min in a stomacher (Heidolph, Schwabach, Germany) with 225 ml of PW and heated at 90 °C for 10 min to destroy vegetative bacteria and fungi and to make easier the isolation of bacilli from spores that survive the heat treatment [5]. One hundred micro liter of homogenates was streaked onto Bacillus cereus selective agar (Liofilchem, Italy) supplemented with egg yolk and polymyxin B and incubated under aerobic conditions at 37 °C for 24−48 h. The typical pink colonies surrounded by precipitate zone indicating lecithinase production were enumerated as B. cereus and sub-cultured onto blood agar (Merck, Darmstadt, Germany). Bacterial identification was based on morphological and biochemical tests using Gram stain, Shaffer–Fulton stain, catalase, oxidase and urease tests, nitrate reduction, Voges Proskauer reaction, indole production, growth on Simmons citrate agar, sugar fermentation, motility, hydrolysis of starch and gelatin and PCR targeting the B. cereus species specific gyrB gene (B. cereus species specific).
2.4. Genomic DNA extraction
A colony of B. cereus (one colony per sample) was picked from nutrient agar and inoculated into 5 ml of Luria Bertani broth (LB, Merck, Germany) until 2 McFarland standard turbidity (approximate cell density is 6 × 108 CFU/mL) with shaking at 120 rpm at 37 °C. Extraction of genomic DNA was performed according to the protocol provided with the Qiagen Mini Amp kit. The concentration and purity of DNA samples were determined using a NanoDrop Spectrophotometer (ND-1000, Nano-Drop Technologies, Wilmington, DE) at 260 and 260/280 nm, respectively. One microliter of DNA with concentration of 400 ng was used in PCR.
2.5. Detection of hblACD and nheABC in B. cereus isolates by PCR
The presence of B. cereus enterotoxin genes hblA, blC, hblD, nheA, nheB and nheC was assessed using the primers listed in Table 1. Single PCR was performed using DreamTaq PCR Master Mix (Thermo Fisher Scientific), which contains Taq polymerase, dNTPs, MgCl2 and the appropriate buffer. Each PCR tube contained 25 μl reaction mixture composed of 12.5 μl of the master mix, 2.5 μl of each forward and reverse primer solution (in a final concentration of 200 nM), 1 μl of DNA with concentration of 400 ng and nuclease-free water to complete the final volume. PCR was performed using the Gene Atlas 322 system (ASTEC) with the same cycling conditions for hblACD, nheABC genes. Amplification involved an initial denaturation at 94 °C, 5 min followed by 30 cycles of denaturation (94 °C, 1 min), annealing (55 °C, 1.5 min for bal; 54 °C, 1 min for nheB and nheC; 56 °C, 1 min for nheA and hblA; 58 °C, 1 min for hblD and hblC;) and extension (72 °C, 1 min), with a final extension step (72 °C, 8 min). The amplified DNA was separated by submarine gel electrophoresis on 1.5 % agarose, stained with ethidium bromide and visualized under UV transillumination.
Table 1.
Primers used in this study.
| Target | Primer sequences (5'–3') | Amplicon size (bp) | Refs. |
|---|---|---|---|
| gyrB | F-TCATGAAGAGCCTGTGTACG R-CGACGTGTCAATTCACGCGC |
475 | [21] |
| nheA | F-GTTAGGATCACAATCACCGC R-ACGAATGTAATTTGAGTCGC |
755 | [17] |
| nheB | F-TTAGTAGTGGATCTGTACGC R-TTAATGTTCGTTAATCCTCG |
743 | [17] |
| nheC | F-TGGATTCCAAGATGTAACG R-ATTACGACTTCTGCTTGTGC |
683 | [17] |
| hblA | F-AAGCAATGGAATACAATGGG R-AGAATCTAAATCATGCCACTGC |
1154 | [17] |
| hblC | F-GATACTCAATGTGGCAACTGC R-TTGAGAACTGCTCGTCTAGTTG |
740 | [17] |
| hblD | F-ACCGGTAACACTATTCATGC R-GAGTCCATATGCTTAGATGC |
829 | [17] |
2.6. Statistical analysis
The data were analysed with SSPS version 17.0 software (SPSS). A chi-square test was used to determine the statistical significance of the data. A P value of <0.05 was considered significant.
3. Results
3.1. Frequency of B. cereus in meat samples
A total of 200 individual meat samples were studied for the presence of B. cereus. According to conventional cultural method, biochemical tests and molecular analysis of gyrB marker, B. cereus was isolated from 29 (14.5 %) of the samples (Table 2, Fig. 1). B. cereus was isolated from 12 (15 %) of 80 cooked beef samples and from 17 (14.1 %) of 120 raw meat samples.
Table 2.
Frequency of B. cereus in meat samples.
| Meat type (No. of samples examined) | No. (%) of samples containing B. cereus |
|
|---|---|---|
| No. | (%) | |
| Raw lamb (60) | 7 | 3.5 |
| Raw beef (60) | 10 | 5 |
| Cooked beef (80) | 12 | 6 |
| Total (200) | 29 | 14.5 % |
Fig. 1.
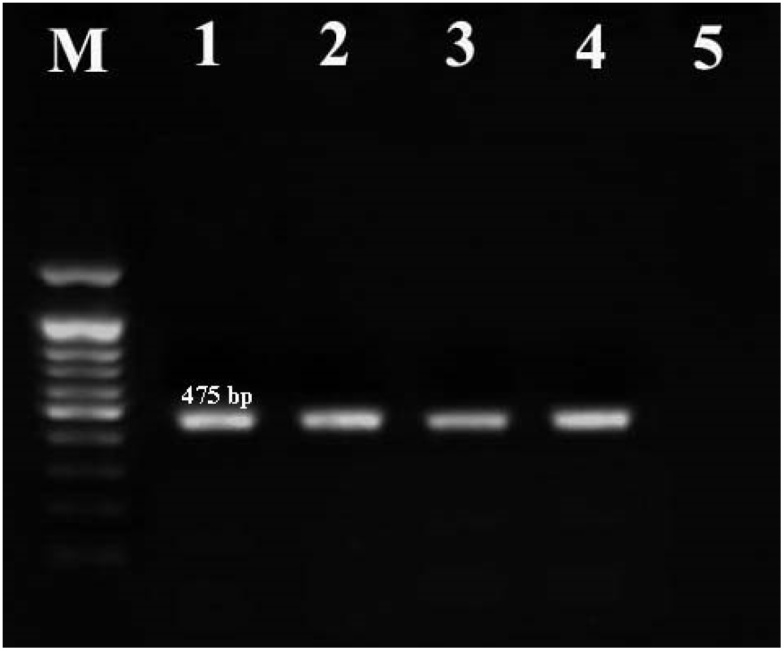
Agarose gel electrophoresis for detection of B. cereus species specific gyrB gene. Lane 1: Positive control, Lanes 2–4: B. cereus isolates, Lane 5: Negative control, Lane M: DNA marker (100 bp).
3.2. Distribution of enterotoxin genes (hblA, blC, hblD, nheA, nheB and nheC) in B. cereus isolates
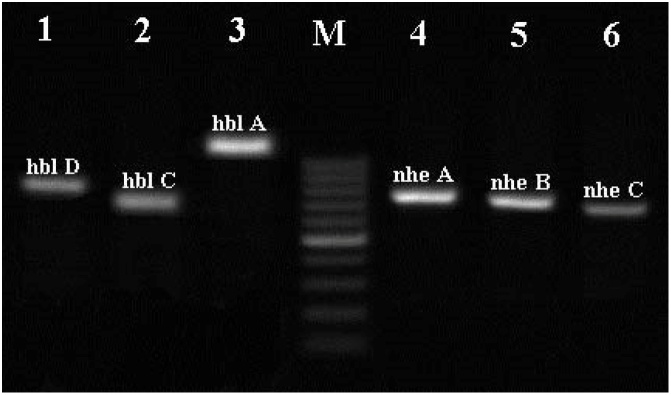
Overall, 89.6 % (26/29) of isolates were positive for the presence of at least one or more enterotoxin genes: 5 isolates (17.2 %) from lamb, 9 isolates (31 %) from beef and 12 (34.5 %) isolates from cooked samples. The frequency of each enterotoxin gene in B. cereus isolates is shown in Table 3 and Fig. 2. The enterotoxigenic profile of the isolates revealed the presence of at least one gene of Hbl complex (hblDAC) in 22 isolates (75.8 %). Furthermore, at least one gene of Nhe complex (nheABC) was detected in 23 isolates (79.3 %) of B. cereus (P > 0.05). Comparison of enterotoxin genes frequency among raw and cooked meat isolates showed different distribution of these genes. The most prevalent enterotoxin gene was nheA (58.6 %), followed by hblD (51.7 %), nheB and nheC (48.3 %).
Table 3.
Distribution of enterotoxin genes (hblA,D,C: hemolysin BL-A,D,C and nheA,B,C: non-hemolytic enterotoxin A,B, C) among 29 B. cereus isolates.
| Enterotoxin | No. (%) of isolates carrying enterotoxin genes |
|||
|---|---|---|---|---|
| Lamb isolates (n = 7) | Beef isolates (n = 10) | Cooked beef isolates (n = 12) | Total (n = 29) | |
| hblA | 3 (10.3 %) | 4 (13.8 %) | 3 (10.3 %) | 10 (34.5 %) |
| hblD | 4 (13.8 %) | 7 (24.1 %) | 4 (13.8 %) | 15 (51.7 %) |
| hblC | 1 (3.4 %) | 4 (13.8 %) | 5 (17.2 %) | 10 (34.5 %) |
| nheA | 4 (13.8 %) | 6 (20.7 %) | 7 (24.1 %) | 17 (58.6 %) |
| nheB | 1 (3.4 %) | 5 (17.2 %) | 8 (27.6 %) | 14 (48.3 %) |
| nheC | 3 (10.3 %) | 5 (17.2 %) | 6 (20.7 %) | 14 (48.3 %) |
Fig. 2.
Agarose gel electrophoresis for detection of B. cereus enterotoxin genes. Lanes 1-6: enterotoxin genes, Lane M: DNA marker (100 bp).
The Hbl and Nhe complex genes were found with different combinations among isolates. Of 26 B. cereus isolates carrying enterotoxin genes, 22 (84.6 %) isolates had simultaneously two or more genes of Hbl or Nhe complex. The number of enterotoxin genes per isolate and their specific combinations are shown in Table 4. The frequent combination of enterotoxin genes was nheA + nheB + nheC + hblA + hblD (11.5 %).
Table 4.
Specific enterotoxin genes (hblA,D,C and nheA,B,C) combinations in B. cereus isolates.
| enterotoxin combinations | No. (%) of isolates carrying enterotoxin genes combinations |
||
|---|---|---|---|
| Raw meat isolates carrying enterotoxin genes (n = 14) | Cooked meat isolates carrying enterotoxin genes (n = 12) | Total (n = 26) | |
| hblD | 1 (3.8 %) | 0 | 1 (3.8 %) |
| nheA | 0 | 1 (3.8 %) | 1 (3.8 %) |
| nheC | 1 (3.8 %) | 1 (3.8 %) | 2 (7.6 %) |
| nheA + hblC | 1 (3.8 %) | 1 (3.8 %) | 2 (7.6 %) |
| nheA + hblA | 1 (3.8 %) | 0 | 1 (3.8 %) |
| nheA + nheB | 0 | 1 (3.8 %) | 1 (3.8 %) |
| nheB + nheC | 0 | 1 (3.8 %) | 1 (3.8 %) |
| nheB + hblA | 0 | 1 (3.8 %) | 1 (3.8 %) |
| nheA + nheB + hblC | 0 | 1 (3.8 %) | 1 (3.8 %) |
| nheA + nheB + hblD | 0 | 1 (3.8 %) | 1 (3.8 %) |
| nheA + hblC + hblD | 1 (3.8 %) | 0 | 1 (3.8 %) |
| nheA + hblA + hblD | 2 (7.6 %) | 0 | 2 (7.6 %) |
| nheB + hblC + hblD | 1 (3.8 %) | 0 | 1 (3.8 %) |
| nheA + nheC + hblC + hblD | 1 (3.8 %) | 0 | 1 (3.8 %) |
| nheB + nheC + hblC + hblD | 0 | 2 (7.6 %) | 2 (7.6 %) |
| nheA + nheB + nheC + hblA | 0 | 1 (3.8 %) | 1 (3.8 %) |
| nheB + nheC + hblA + hblD | 1 (3.8 %) | 0 | 1 (3.8 %) |
| nheA + nheC + hblA + hblC + hblD | 0 | 1 (3.8 %) | 1 (3.8 %) |
| nheA + nheB + nheC + hblA + hblD | 3 (11.5 %) | 0 | 3 (11.5 %) |
| nheA + nheB + nheC + hblC + hblD | 1 (3.8 %) | 0 | 1 (3.8 %) |
4. Discussion
Food safety has emerged as an important global issue with international trade and public health implications. Bacillus cereus is a most common foodborne pathogen and represents a major public health problem in developing countries [[13], [14], [15]]. Raw meat and meat products reported to be associated with B. cereus food poisoning worldwide [16]. In this study, a total of 29 (14.5 %) meat samples were positive for the presence of B. cereus. The frequency of B. cereus in raw and cooked meat samples was similar (14.1 % and 15 %, respectively). Only a few reports on the frequency of B. cereus in meat samples from Iran have been previously published. According to the previous report from Iran, 15.6 % of the raw chicken meat samples were positive for the presence of this pathogen [16]. Raw meat and meat products contamination with B. cereus has been reported 30.9 % in India yielding positive cultures [5]. According to Das et al. (2009), the frequency of B. cereus in meat products was 23.5 % and 36.7 %, respectively [6]. This variation in B. cereus frequency may be due to differences in the geographical region, reservoir in the various countries, number of samples, seasons of sampling, post-harvest practices and hygienic standards applied during the handling, transport and storage of products, as well as the methods used for isolation and identification of this bacterium. Meat contamination may occur at various stages in preparation including transport, butchering and cut-up in the kitchen and the importance of chopping boards as a source of contamination has been reported [13].
In the present study, 89.6 % of isolates carried at least one or more enterotoxin genes. We found nheA (58.6 %) and hblD (51.7 %) genes with higher frequency than others. At least one gene of NHE complex (nheABC) was detected in 79.3 % of B. cereus isolates, whereas, it was found in 89.7 % and 100 % of isolates in Tewari et al. and van der Voort et al. studies, respectively [5,14]. The enterotoxigenic profile of the isolates revealed the presence of at least one gene of Hbl complex (hblDAC) in 75.8 % of isolates. Similar results were reported by Guinebretiere et al. and Das et al. which found the frequency of 73 % and 71.4 % of Hbl complex in food samples, respectively [6,17]. Lower incidence of Hbl complex (55.2 %) in meat and meat product samples was reported by Tewari et al. [5]. In the present study, the Hbl complex genes were found in lower frequency than Nhe complex. This finding was in accordance with Moravek et al.(2006), Al-khatib et al. (2007) and Tewari et al. reports [5,18,19]. In study conducted by Sadek et al., B. cereus was detected in 21.2 % of milk based fruit, vegetables (15.7 %), honey (17.2 %), rice (14.1 %) and wheat (12 %) and vanished in the infant milk powder samples. Furthermore, 95.5 % and 71.1 % of isolates carried cytK and nheC genes, respectively. The hblA gene was detected only in 11.1 % of B. cereus isolates [20].
According to Ngamwongsatit et al. report, the three genes of Hbl and Nhe complexes occur together in operon [3]. But the present study indicates that the genes in an operon can occur independently of each other and many isolates showed the absence of one or two genes in Hbl and Nhe complexes (Table 4). Furthermore, previous studies reported the variation in genotype and incidence of enterotoxin genes in different geographical locations [5]. In our study, 84.6 % of enterotoxin carrying isolates possessed more than one gene. Seventheen genotypes were observed and the frequent combination was nheA + nheB + nheC + hblA + hblD (11.5 %).
5. Conclusions
Our study revealed that 14.5 % of collected raw and cooked meat samples were infected with B. cereus. Also, 89.6 % of isolates were positive for the presence of at least one or more enterotoxin genes, with the most frequency of nheA. Due to high frequency of enterotoxin genes among isolates, intensive and continuous monitoring of potentially pathogenic B. cereus is strongly recommended in order to evaluate the human health risk arising from food consumption.
CRediT authorship contribution statement
Habib Zeighami: Conceptualization, Methodology, Investigation, Writing - original draft. Gholamreza Nejad-dost: Investigation, Methodology, Resources. Angineh Parsadanians: Methodology, Resources. Shahrzad Daneshamouz: Methodology, Resources. Fakhri Haghi: Conceptualization, Methodology, Investigation, Project administration, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no competing interests.
Acknowledgement
This study as an MSc is in Food Microbiology was supported by the Zanjan University of Medical Sciences, Zanjan, Iran (A-12-535-15, ZUMS.REC.1394.335).
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.toxrep.2019.12.006.
Contributor Information
Habib Zeighami, Email: zeighami@zums.ac.ir.
Fakhri Haghi, Email: haghi@zums.ac.ir.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Zhu L., He J., Cao X., Huang K., Luo Y., Xu W.-T. Development of a double-antibody sandwich ELISA for rapid detection of Bacillus cereus in food. Sci. Rep. 2016;6:16092. doi: 10.1038/srep16092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakshmi S.G., Jayanthi N., Saravanan M., Ratna M.S. Safety assesment of Bacillus clausii UBBC07, a spore forming probiotic. Toxicol. Rep. 2017;5(4):62–71. doi: 10.1016/j.toxrep.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ngamwongsatit P., Buasri W., Pianariyanon P., Pulsrikarn C., Ohba M., Assavanig A., Panbangred W. Broad distribution of enterotoxin genes (hblCDA, nheABC, cytK, and entFM) among Bacillus thuringiensis and Bacillus cereus as shown by novel primers. Int. J. Food Microbiol. 2008;121(3):352–356. doi: 10.1016/j.ijfoodmicro.2007.11.013. [DOI] [PubMed] [Google Scholar]
- 4.Kotiranta A., Lounatmaa K., Haapasalo M. Epidemiology and pathogenesis of Bacillus cereus infections. Microbes Infect. 2000;2(2):189–198. doi: 10.1016/s1286-4579(00)00269-0. [DOI] [PubMed] [Google Scholar]
- 5.Tewari A., Singh S.P., Singh R. Incidence and enterotoxigenic profile of Bacillus cereus in meat and meat products of Uttarakhand, India. J. Food Sci. Technol. 2015;52(3):1796–1801. doi: 10.1007/s13197-013-1162-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Das S., Surendran P.K., Thampuran N.K. PCR-based detection of enterotoxigenic isolates of Bacillus cereus from tropical seafood. Indian J. Med. Res. 2009;129(3):316–320. [PubMed] [Google Scholar]
- 7.Sastalla I., Fattah R., Coppage N., Nandy P., Crown D., Pomerantsev A.P., Leppla S.H. The Bacillus cereus Hbl and Nhe tripartite enterotoxin components assemble sequentially on the surface of target cells and are not interchangeable. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0076955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fagerlund A., Lindback T., Granum P.E. Bacillus cereus cytotoxins Hbl, Nhe and CytK are secreted via the Sec translocation pathway. BMC Microbiol. 2010;10:304. doi: 10.1186/1471-2180-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abbas B., Khudor M., Saeed B. Detection of hbl, nhe and bceT toxin genes in Bacillus cereus isolates by multiplex PCR. Int. J. Curr. Microbiol. Appl. Sci. 2014;3(11):1009–1016. [Google Scholar]
- 10.de Souza C.M.O.C.C., Abrantes S. Detection of enterotoxins produced by B. cereus through PCR analysis of ground and roasted coffee samples in Rio de Janeiro, Brazil. Ciênc. Tecnol. Aliment. Campinas. 2011;31(2):443–449. [Google Scholar]
- 11.Forghani F., Langaee T., Eskandari M., Seo K.H., Chung M.J., Oh D.H. Rapid detection of viable Bacillus cereus emetic and enterotoxic strains in food by coupling propidium monoazide and multiplex PCR (PMA-mPCR) Food Control. 2015;55:151–157. [Google Scholar]
- 12.Lee J., Kwon G.H., Park J.Y., Park C.S., Kwon D.Y., Lim J., Kim J.S., Kim J.H. A RAPD-PCR method for the rapid detection of Bacillus cereus. J. Microbiol. Biotechnol. 2011;21(3):274–276. [PubMed] [Google Scholar]
- 13.Logan N.A. Bacillus and relatives in foodborne illness. J. Appl. Microbiol. 2012;112(3):417–429. doi: 10.1111/j.1365-2672.2011.05204.x. [DOI] [PubMed] [Google Scholar]
- 14.van der Voort M., Abee T. Sporulation environment of emetic toxin-producing Bacillus cereus strains determines spore size, heat resistance and germination capacity. J. Appl. Microbiol. 2013;114(4):1201–1210. doi: 10.1111/jam.12118. [DOI] [PubMed] [Google Scholar]
- 15.Ceuppens S., Uyttendaele M., Drieskens K., Rajkovic A., Boon N., Wiele T.V. Survival of Bacillus cereus vegetative cells and spores during in vitro simulation of gastric passage. J. Food Prot. 2012;75(4):690–694. doi: 10.4315/0362-028X.JFP-11-481. [DOI] [PubMed] [Google Scholar]
- 16.Hanna Tahmasebi R.T., Rahmian Zarif B. Isolated of Bacillus cereus in chicken meat and investigation β-lactamase antibiotic-resistant in Bacillus cereus from chicken meat. Adv. Life Sci. 2014;4(4):7. [Google Scholar]
- 17.Guinebretière M.H., Broussolle V., Nguyen-The C. Enterotoxigenic profiles of food-poisoning and food-borne Bacillus cereus strains. J. Clin. Microbiol. 2002;40(8):3053–3056. doi: 10.1128/JCM.40.8.3053-3056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moravek M., Dietrich R., Buerk C., Broussolle V., Guinebretière M.H., Granum P.E., Nguyen-The C., Märtlbauer E. Determination of the toxic potential of Bacillus cereus isolates by quantitative enterotoxin analyses. FEMS Microbiol. Lett. 2006;257(2):293–298. doi: 10.1111/j.1574-6968.2006.00185.x. [DOI] [PubMed] [Google Scholar]
- 19.Al-Khatib M.S., Khyami-Horani H., Badran E., Shehabi A.A. Incidence and characterization of diarrheal enterotoxins of fecal Bacillus cereus isolates associated with diarrhea. Diagn. Microbiol. Infect. Dis. 2007;59(4):383–387. doi: 10.1016/j.diagmicrobio.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 20.Sadek Z.I., Abdel-Rahman M.A., Azab M.S., Darwesh O.M., Hassan M.S. Microbiological evaluation of infant foods quality and molecular detection of Bacillus cereus toxins relating genes. Toxicol. Rep. 2018;19(5):1–877. doi: 10.1016/j.toxrep.2018.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noor Uddin G.M., Larsen M.H., Christensen H., Aarestrup F.M., Phu T.M., Dalsgaard A. Identification and antimicrobial resistance of bacteria isolated from probiotic products used in Shrimp culture. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0132338. e0132338-e0132338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.