Abstract
Transcriptomic and proteomic analyses were performed on three replicates of tomato fruit pericarp samples collected at nine developmental stages, each replicate resulting from the pooling of at least 15 fruits. For transcriptome analysis, Illumina-sequenced libraries were mapped on the tomato genome with the aim to obtain absolute quantification of mRNA abundance. To achieve this, spikes were added at the beginning of the RNA extraction procedure. From 34,725 possible transcripts identified in the tomato, 22,877 were quantified in at least one of the nine developmental stages. For the proteome analysis, label-free liquid chromatography coupled to tandem mass spectrometry (LC-MS/MS) was used. Peptide ions, and subsequently the proteins from which they were derived, were quantified by integrating the signal intensities obtained from extracted ion currents (XIC) with the MassChroQ software. Absolute concentrations of individual proteins were estimated for 2375 proteins by using a mixed effects model from log10-transformed intensities and normalized to the total protein content. Transcriptomics data are available via GEO repository with accession number GSE128739. The raw MS output files and identification data were deposited on-line using the PROTICdb database (http://moulon.inra.fr/protic/tomato_fruit_development) and MS proteomics data have also been deposited to the ProteomeXchange with the dataset identifier PXD012877. The main added value of these quantitative datasets is their use in a mathematical model to estimate protein turnover in developing tomato fruit.
Keywords: Proteomics, Transcriptomics, Tomato fruit development, Pericarp, Time-series, Absolute quantification, Protein turnover
Specifications Table
| Subject | Plant Science |
| Specific subject area | Plant physiology, transcriptomic and proteomic quantitative data, tomato fruit development |
| Type of data | Tables and Figures |
| How data were acquired | Illumina-sequenced libraries for transcriptomics. Label-free LC-MS/MS for proteomics |
| Data format | Raw and transformed in quantitative concentrations (fmol.gFW-1) for both transcripts and proteins. |
| Parameters for data collection | Total proteins and transcripts were extracted from the fleshy part of the tomato fruit pericarp at 9 developmental stages, i.e. at 8, 15, 21, 28, 34, 42, 48, 50 and 53 days post-anthesis. |
| Description of data collection | Tomato plants were grown in a greenhouse under optimal conditions of commercial production. One sample results from the pooling of at least 15 fruits. Replicates 1, 2 and 3 correspond to the 5th, 6th and 7th truss respectively. |
| Data source location | INRA France. |
| Data accessibility | Transcriptomics data are available via Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) an international public repository with accession number GSE128739 For proteomics data, the raw MS output files and identification data were deposited on-line using the PROTICdb database (http://moulon.inra.fr/protic/tomato_fruit_development) The proteomics data have also been deposited to the ProteomeXchange with the dataset identifier PXD128739 (http://proteomecentral.proteomexchange.org) |
| Related research article | [2] Isma Belouah, Christine Nazaret, Pierre Pétriacq, Sylvain Prigent, Camille Bénard, Virginie Mengin, Mélisande Blein-Nicolas, Alisandra K. Denton, Thierry Balliau, Ségolène Augé, Olivier Bouchez, Jean-Pierre Mazat, Mark Stitt, Björn Usadel, Michel Zivy, Bertrand Beauvoit, Yves Gibon, Sophie Colombié Modeling Protein Destiny in Developing Fruit Plant Physiology 2019. DOI: https://doi.org/10.1104/pp.19.00086 |
Value of the data
|
1. Data description
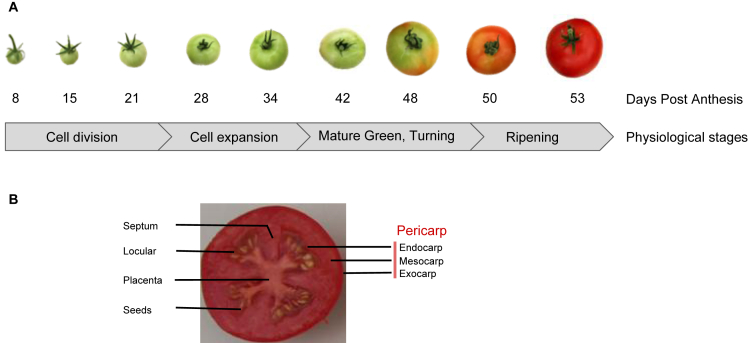
Tomato plants (Solanum lycopersicum cv. Moneymaker) were grown under conditions of commercial production in a greenhouse in the south-west of France. Samples were taken from pericarp of tomato fruits, at nine stages of tomato fruit development, on the 5th, 6th and 7th trusses [1] (Fig. 1). Transcriptomics and proteomics have been performed on these samples. Hierarchical clustering (Fig. 2) and principal component analyzes (Fig. 3) provide an overview of the transcriptome and proteome changes throughout the tomato fruit development. Transcriptomic data are available via GEO with accession number GSE128739. For proteomic, raw files and data are available on-line using the PROTICdb database (http://moulon.inra.fr/protic/tomato_fruit_development) and the ProteomeXchange with identifier PXD012877; quantitative data of proteins are provided in tables. All these data have been used to model protein turnover [2] and to study redox metabolism in the developing tomato fruit [3].
Fig. 1.
Experimental setup. (A) The nine stages of samples with corresponding physiological phases of the tomato fruit development (Solanum lycopersicum cv. ‘Moneymaker’). (B) Description of the analyzed tissue, the pericarp, composed of endocarp, mesocarp and exocarp in tomato fruit at the last stage of development.
Fig. 2.
Hierarchical clustering analysis of (A) transcript and (B) protein concentrations from tomato at nine developmental stages. The hierarchical clustering analysis was performed using Pearson's correlation on mean centered and scaled data. Hierarchical clustering analysis was performed using plyr, gplots and reshape2 packages from R studio (R 3.3.2; http://www.rstudio.com/).
Fig. 3.
Principal component analysis of (A) transcriptomics and (B) proteomics data (fmol.gFW−1). Data were mean centered and scaled. Developmental stages and replicates were distinguished by colors and shapes. Principal component analysis was performed using factoextra and gplots packages from R studio (R 3.3.2; http://www.rstudio.com/).
2. Experimental design, materials, and methods
2.1. Plant material
Tomato plants (Solanum lycopersicum cv. Moneymaker) were cultivated in a greenhouse at Sainte-Livrade (southwest of France, 44° 239 5699 N and 0° 359 2599E) in commercial practice conditions between June and October of 2010. Lateral stems were systematically removed to promote flowering and trusses were pruned to six fruits to limit fruit size heterogeneity. Based on age and color (OECD color gauge), fruits were harvested at nine stages expressed in days post anthesis (DPA), from green/young to red/ripened fruit (8, 15, 21, 28, 34, 42, 48, 50 and 53 DPA; Fig. 1). Each biological replicate was prepared with 15–50 fruits harvested on different plants but on the same truss, which was numbered according to its order of appearance on the plant, i.e. truss 5, 6 or 7. Gel and placenta were quickly removed before 1cm2 of equatorial pericarp zone was quickly cut into small pieces that were immediately shock-frozen in liquid nitrogen. Frozen samples were transported with a dry shipper, then ground into a fine powder with liquid nitrogen using a bead mill and stored at −80 °C. At the end 26 samples were analyzed, with only two biological replicates for the 48 DPA stage.
2.2. Transcriptomics
2.2.1. Total RNA extraction
Total RNA was isolated from 100 mg fresh weight aliquots of the frozen powdered samples using Plant RNA Reagent (PureLink kit, Invitrogen™) followed by DNase treatment (DNA-free kit, Invitrogen™), and purification over RNeasy Mini spin columns (RNeasy Plant Mini kit, QIAGEN) following manufacturer's instructions. Total RNA concentration was determined by spectrophotometry (260 nm) considering that an absorbance of 1 unit equals 40 μg of RNA per ml. RNA quality was determined by estimating the RNA integrity number (RIN) with a RNA 6000 Nano kit (Agilent) and an Agilent 2100 Bioanalyzer. A RIN of ‘10’ stands for non-degraded RNA whereas a RIN of ‘1’ stands for a completely degraded RNA. A subsample of at least 5 μg of total RNA from each of 26 RNA extracts was sent to the Get-Plage GenoTOUL facility (Toulouse, France). To determine the absolute concentration of transcript after transcriptome sequencing, eight internal standards (AM 1780, Ambion by Life technologies, Array Control RNA spikes, Invitrogen™) at selected concentrations (in mole, 3.97.10−14 [spike 1], 4.01.10−15 [spike 2], 4.01.10−16 [spike 3], 4.02.10−17 [spike 4], 4.08.10−18 [spike 5], 4.04.10−19 [spike 6], 3.82.10−20 [spike 7], and 3.82.10−21 [spike 8]) were spiked-in the plant extracts at the beginning of the RNA purification process.
2.2.2. Transcript sequencing
RNA-seq libraries were prepared according to Illumina's protocols on a Tecan EVO200 liquid handler using the Illumina TruSeq Stranded mRNA sample prep kit to analyze mRNA. Briefly, mRNA were selected using poly-T beads. Then, mRNA were fragmented to generate double stranded cDNA and adaptors were ligated to be sequenced. Ten cycles of PCR were applied to amplify libraries. Before being quantified by qPCR (Kapa Library Quantification Kit), RNA samples quality was evaluated using an Agilent Bioanalyzer. RNA-seq experiments have been performed on an Illumina HiSeq2000 or HiSeq2500 sequencer using a paired-end read length of 2 × 100 pb with the Illumina TruSeq SBS sequencing kits v3.
2.2.3. Transcriptome analysis and quantification
Genes were mapped to the Solanum lycopersicum HEINZ assembly v2.40, concatenated with the chloroplast (gi|544163592|ref|NC_007898.3|) and mitochondrial genomes (gi|209887431|gb|FJ374974.1|), and an “artificial chromosome” containing the 8 spike sequences (Supplemental Appendix S1). Genome data was downloaded from S. lycopersicum 2.5 and the corresponding ITAG2.4 gene models were downloaded from https://solgenomics.net/(34,725 entries). The quality of library sequencing was checked with FastQC [4]. Quality and adapter trimming was performed with Trimmomatic [5] v0.32. Trimmed reads were mapped to their respective genomes with Star [6] v2.4.2a and the unique counts per locus were quantified with HTSeq [7] v0.6.1. The number of transcripts per million (TPM) was calculated from the unique counts and gene length. The normalized number of fragments per kilobase per million (FPKM) was calculated with cufflinks v2.2.1. Briefly, quantification based on FPKM corresponds to the normalization of data by depth sequencing (summed fragment per sample) divided by one million followed by a normalization by the gene length. Non-default parameters that were used are presented in Supplemental Appendix S1. FPKM were then converted to TPM quantification to get relative transcript abundance among samples. Spikes were quantified as any other transcript. In order to preserve the native dynamic of RNA concentration through tomato fruit development (highest concentration before expansion phase), a standard curve was calculated for each sample. Each standard curve was determined from spiked-in concentrations and corresponding TPM values of the spikes.
2.3. Proteomics
2.3.1. Total protein extraction
Proteins were extracted by phenol extraction using a modified protocol described by Faurobert et al. [8]. Frozen powder of pericarp tissue (100 mg) was suspended in 10 ml of extraction buffer (0.5 M Tris-HCl pH 7.5, 0.7 M sucrose, 50 mM EDTA, 0.1 M KCl, 10 mM thiourea, 2 mM phenylmethylsulfonyl fluoride, 2% 2-mercaptoethanol). Then an equal volume of water-saturated phenol pH 8 (Ambion) was added and the mixture was incubated with steel beads on a shaker for 30 min and at 4 °C. After 30 min centrifugation (12,000 g at 4 °C), the phenol phase was recovered and transferred into a new tube with 10 ml of extraction buffer followed by shaking without steel beads, and centrifugation (30 min, 12,000 g, 4 °C). The phenol phase was recovered and proteins were precipitated by adding the equivalent of five volumes of cold methanol and 0.1 M acetate ammonium, and incubated overnight at −20 °C. After 30 min centrifugation (10,000 g, 4 °C), the protein pellet was gently washed with methanol and then with cold acetone before being dried in a fume hood. Proteins were then solubilized in 6 M urea, 2 M thiourea, 30 mM Tris HCl pH 8.8, 10 mM dithiotreitol, 0.1% (v/v) zwitterionic acid labile surfactant I (Protea) then quantified using the Plusone 2D Quant kit (GE Healthcare). Proteins were incubated at room temperature for 30 min then alkylated with 50 mM iodoacetamide for 60 min in the dark and at room temperature. Proteins were diluted ten times in 50 mM ammonium bicarbonate buffer to decrease total urea and thiourea concentrations, and then digested overnight at 37 °C with 800 ng trypsin. Trypsin digestion was stopped by acidification with 1% (w/v) trifluoroacetic acid. The resulting peptides were purified by solid phase extraction using a polymeric C18 column (Phenomenex) with a washing solution containing 0.06% (v/v) acetic acid and 3% (v/v) acetonitrile. After elution with 0.06% acetic acid and 40% acetonitrile, peptides were dried under vacuum (Speedvac).
2.3.2. Protein LC-MS/MS analyses
As described in Belouah et al. [2], LC-MS/MS analyses were performed using a NanoLC-Ultra System (nano2DUltra, Eksigent, Les Ulis, France) connected to a Q-Exactive mass spectrometer (Thermo Electron, Waltham, MA, USA). For each sample, about 800 ng of protein digest were loaded onto a Biosphere C18 precolumn (0.1 × 20 mm, 100 Å, 5 μm; Nanoseparation) at 7.5 μl min−1 and desalted with 0.1% formic acid and 2% acetonitrile. After 3 min, the pre-column was connected to a Biosphere C18 nanocolumn (0.075 × 300 mm, 100 Å, 3 μm; Nanoseparation). Electrospray ionization was performed at 1.3 kV with an uncoated capillary probe (10 μm tip inner diameter; New Objective, Woburn, MA, USA). Buffers were 0.1% formic acid in water (A) and 0.1% formic acid and 100% acetonitrile (B). Peptides were separated using a linear gradient from 5 to 35% buffer B for 110 min at 300 nl min−1. One run took 120 min, including the regeneration step at 95% buffer B and the equilibration step at 100% buffer A.
Peptide ions were analyzed using Xcalibur 2.1 (Thermo Electron) with the following data-dependent acquisition steps: (1) MS scan (mass-to-charge ratio (m/z) 300 to 1,400, 70,000 resolution, profile mode), (2) MS/MS (17,500 resolution, normalized collision energy of 30, profile mode). Step 2 was repeated for the eight major ions detected in step (1). Dynamic exclusion was set to 30 seconds. Xcalibur raw datafiles were transformed to mzXML open source format using msconvert software in the ProteoWizard 3.0.3706 package [9]. During conversion, MS and MS/MS data were centroided. The raw MS output files were deposited on-line using PROTICdb database [[10], [11], [12]].
2.3.3. Protein identification
Protein identification was performed using the protein sequence database of S. lycopersicum Heinz assembly v2.40 (ITAG2.4) downloaded from https://solgenomics.net/(34,725 entries). A contaminant database containing the sequences of standard contaminants was also interrogated (58 entries with e.g., trypsin, keratin, and serum albumin). The decoy database comprised the reverse sequences of tomato proteins. Database search was performed with X!Tandem (version 2015.04.01.1; http://www.thegpm.org/TANDEM/) with the following settings. Carboxyamidomethylation of cysteine residues was set to static modification. Oxidation of methionine residues, acetylation or deamination of glutamine and cystein residues were set to possible modifications. Precursor mass precision was set to 10 ppm. Fragment mass tolerance was 0.02 Th. Only peptides with an E-value smaller than 0.05 were reported.
Identified proteins were filtered and sorted by using X!TandemPipeline (version 3.3.4, [13]). Criteria used for protein identification were (1) at least two different peptides identified with an E-value smaller than 0.01, and (2) a protein E-value (product of unique peptide E-values) smaller than 10−5.
2.3.4. Peptide and protein quantification
Peptide ions were quantified using extracted ion chromatograms (XIC) and the MassChroQ software [14] version 2.2 with the following parameters: “ms2_1” alignment method, tendency_halfwindow of 10, MS1 smoothing halfwindow of 0, MS2 smoothing halfwindow of 15, “quant1” quantification method, XIC extraction based on max, min and max ppm range of 10, anti-spike half of 5, mean filter half hedge, minmax_half_edge and maxmin_half_edge respectively set to 2, 4, and 3. Detection thresholds on min and max at 30,000 and 50,000, respectively, peak post-matching mode.
Peptides intensities of each sample were normalized using peptides intensities of a reference sample. In the reference sample, peptide ions extract of the 26 samples were pooled and analyzed (identification, quantification) using the same pipeline used for each sample. After removing shared and dubious peptide ions (standard deviation of retention time higher than 30 seconds), proteins were quantified based on a method named Model [15]. Briefly, peptide ion intensities were log10 transformed and quantified using a mixed effect model. Abundances of proteins are given in Table 1.
Absolute quantification was approximated based on the “Total Protein Amount” approach [16], which is based on the main hypothesis that the sum of MS signal corresponds to the total protein content in the cell. Then the concentration of each protein is determined as a relative abundance of the total protein content (Equation (1)).
| (1) |
With the concentration of each proteini (i = 1:2494) in the sample k (k = 1:26) in fmol gFW-1, n the total number of protein (n = 2494), (Total protein content) k the total amount of proteins in the sample k in g gFW-1 and the molar weight (in g.mol-1) of the proteini.
Total protein quantification is given in Table 2 and the proxy of absolute concentration of proteins is given in Table 3.
Acknowledgments
We acknowledge funding from INRA BAP, University of Bordeaux, and ANR (ANR-15-CE20-0009-01 FRIMOUSS). This work was performed in collaboration with PAPPSO (http://pappso.inra.fr/index.php) and GeT core facility (Toulouse, France, http://get.genotoul.fr).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.dib.2019.105015.
Conflict of Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Abundance of tomato fruit proteins quantified using Model method at nine developmental stages (n = 3 replicates, except for 48 DPA).
Total protein extracted from the pericarp of the tomato fruit at nine developmental stages and quantified using the PlusOne 2-D Quant kit (GE Healthcare). Total protein is expressed in g per gram of fresh weight (g.gFW−1).
Proxy of protein concentration at nine developmental stages throughout tomato fruit development. Concentrations were determined based on abundances (Model method see Table 1) and using the “Total Protein Abundance” approach. Protein concentration is expressed in fmol.gFW−1.
RNASeq parameters and spikes.
References
- 1.Biais B., Benard C., Beauvoit B., Colombié S., Prodhomme D., Menard G., Bernillon S., Gehl B., Gautier H., Ballias P., Mazat J.-P., Sweetlove L., Genard M., Gibon Y. Remarkable reproducibility of enzyme activity profiles in tomato fruits grown under contrasting environments provides a roadmap for studies of fruit metabolism. Plant Physiol. 2014;164:1204–1221. doi: 10.1104/pp.113.231241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belouah I., Nazaret C., Pétriacq P., Prigent S., Bénard C., Mengin V., Blein-Nicolas M., Denton A.K., Balliau T., Augé S., Bouchez O., Mazat J.-P., Stitt M., Usadel B., Zivy M., Beauvoit B., Gibon Y., Colombié S. Modeling protein destiny in developing fruit. Plant Physiol. 2019 doi: 10.1104/pp.19.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Decros G., Beauvoit B., Colombié S., Cabasson C., Bernillon S., Arrivault S., Guenther M., Prigent S., Gibon Y., Pétriacq P. Regulation of pyridine nucleotides metabolism along tomato fruit development through transcript and protein profiling. Front. Plant Sci. 2019 doi: 10.3389/fpls.2019.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc [Google Scholar]
- 5.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dobin A., Gingeras T.R., Spring C. Mapping RNA-seq reads with STAR. Curr. Protoc. Bioinforma. 2015:11.14.1–11.14.19. doi: 10.1002/0471250953.bi1114s51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anders S., Pyl P.T., Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faurobert M., Pelpoir E., Chaïb J. Phenol extraction of proteins for proteomic studies of recalcitrant plant tissues. Methods Mol. Biol. 2007;355:9–14. doi: 10.1385/1-59745-227-0:9. [DOI] [PubMed] [Google Scholar]
- 9.Kessner D., Chambers M., Burke R., Agus D., Mallick P. ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferry-Dumazet H., Houel G., Montalent P., Moreau L., Langella O., Negroni L., Vincent D., Lalanne C., de Daruvar A., Plomion C., Zivy M., Joets J., PROTICdb A web-based application to store, track, query, and compare plant proteome data. Proteomics. 2005;5:2069–2081. doi: 10.1002/pmic.200401111. [DOI] [PubMed] [Google Scholar]
- 11.Langella O., Zivy M., Joets J. Plant Proteomics. Humana Press; New Jersey: 2007. The PROTICdb database for 2-DE proteomics; pp. 279–304. [DOI] [PubMed] [Google Scholar]
- 12.Langella O., Valot B., Jacob D., Balliau T., Flores R., Hoogland C., Joets J., Zivy M. Management and dissemination of MS proteomic data with PROTICdb: example of a quantitative comparison between methods of protein extraction. Proteomics. 2013;13:1457–1466. doi: 10.1002/pmic.201200564. [DOI] [PubMed] [Google Scholar]
- 13.Langella O., Valot B., Balliau T., Blein-Nicolas M., Bonhomme L., Zivy M. X!TandemPipeline: a tool to manage sequence redundancy for protein inference and phosphosite identification. J. Proteome Res. 2017;2(16):494–503. doi: 10.1021/acs.jproteome.6b00632. [DOI] [PubMed] [Google Scholar]
- 14.Valot B., Langella O., Nano E., Zivy M. MassChroQ: a versatile tool for mass spectrometry quantification. Proteomics. 2011;11:3572–3577. doi: 10.1002/pmic.201100120. [DOI] [PubMed] [Google Scholar]
- 15.Belouah I., Blein-Nicolas M., Balliau T., Gibon Y., Zivy M., Colombié S. Peptide filtering differently affects the performances of XIC-based quantification methods. J. Proteomics. 2019;193:131–141. doi: 10.1016/j.jprot.2018.10.003. [DOI] [PubMed] [Google Scholar]
- 16.Wiśniewski J.R., Hein M.Y., Cox J., Mann M. A “proteomic ruler” for protein copy number and concentration estimation without spike-in standards. Mol. Cell. Proteom. 2014;13:3497–3506. doi: 10.1074/mcp.M113.037309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Abundance of tomato fruit proteins quantified using Model method at nine developmental stages (n = 3 replicates, except for 48 DPA).
Total protein extracted from the pericarp of the tomato fruit at nine developmental stages and quantified using the PlusOne 2-D Quant kit (GE Healthcare). Total protein is expressed in g per gram of fresh weight (g.gFW−1).
Proxy of protein concentration at nine developmental stages throughout tomato fruit development. Concentrations were determined based on abundances (Model method see Table 1) and using the “Total Protein Abundance” approach. Protein concentration is expressed in fmol.gFW−1.
RNASeq parameters and spikes.