Abstract
Circular RNAs (circRNAs) represent a class of noncoding RNAs with a wide expression pattern, and they constitute an important layer of the genome regulatory network. To date, the expression pattern and regulatory potency of circRNAs in the retina, a key part of the central nervous system, are not yet well understood. In this study, RNAs from five stages (E18.5, P1, P7, P14, and P30) of mouse retinal development were sequenced. A total of 9,029 circRNAs were identified. Most circRNAs were expressed in different stages with a specific signature, and their expression patterns were different from those of their host linear transcripts. Some circRNAs could act as sponges for several retinal microRNAs (miRNAs). Furthermore, circTulp4 could function as a competitive endogenous RNA (ceRNA) to regulate target genes. Remarkably, silencing circTulp4 in vivo led to mice having a thin outer nuclear layer (ONL) and defective retinal function. In addition, we found that circRNAs were dysregulated at a much earlier time point than that of disease onset in a retinal degeneration model (rd8 mice). In summary, we provide the first circRNA expression atlas during retinal development and highlight a key biological role for circRNAs in retinal development and degeneration.
Keywords: circular RNA (circRNA), retinal development, retinal degeneration, circTulp4, competing endogenous RNAs (ceRNA), miRNA sponges
Introduction
circRNAs (circular RNAs) are a novel class of noncoding RNAs with tissue- and stage-specific expression patterns, and they are much more stable than linear transcripts.1,2 To date, a large number of circRNAs have been identified and characterized in various cell lines and across different species.3, 4, 5 In addition, the expression patterns and biological functions of circRNAs in the vertebrate brain have been widely studied.1,6, 7, 8, 9, 10 circRNAs are highly abundant in the brain, with specific and dynamic expression patterns during neuronal differentiation.1 Notably, circRNAs may regulate synaptic activity and brain function during development; for example, Cdr1as, which is one component of a regulatory RNA network in the brain, is essential for normal brain function.11,12 These findings show that circRNAs are highly expressed in the central nervous system (CNS) in a spatiotemporal-dependent manner.
Recently, circRNAs were reported to participate in a wide range of biological processes and to be differentially expressed in various diseases, including cancer,13, 14, 15 neurological disorders,16,17 and diabetic retinopathy.18,19 The abundant expression, conservation, specificity, and stability of circRNAs allow them to play potential roles as biomarkers for diagnosis, prognosis, and as predictors of the therapeutic response to treatments.14,19, 20, 21 However, the molecular mechanisms involved in the biological processes of circRNAs and the functions of those circRNAs in vivo remain largely unknown.
As a key but separate part of the CNS, the retina is an essential component of the eye and is responsible for vision production by transducing light signals into neuronal signals. Currently, if and how circRNAs are involved in retinal development and degeneration remains largely unknown. Profiling retinal circRNAs from different stages of development and degeneration would not only help us understand their physiological function, but also provide a circRNA-based therapeutic strategy for retinal diseases. In this study, we attempted to investigate the circRNA expression profile during retinal development and found that circRNAs were abundantly expressed during this process. Several circRNAs were found to serve as microRNA (miRNA) sponges. Strikingly, circTulp4 was validated to be essential for retinal function in vivo. Furthermore, the expression of circRNAs seemed more variable than that of linear transcripts in a retinal degeneration model, implying that circRNAs could act as biomarkers for retinal diseases. Our findings indicate that circRNAs possess distinct signatures and play essential roles in retinal developmental stages and function.
Results
Identification and Characterization of circRNAs in the Mouse Retina
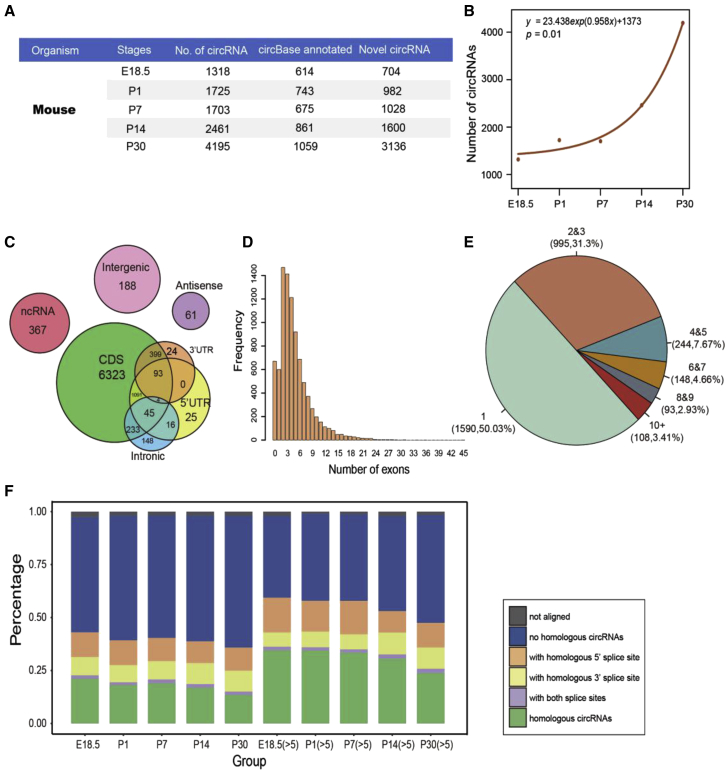
To comprehensively reveal the stage-specific expression patterns of circRNAs in the mouse retina on a genomic scale, we performed transcriptome profiling of circRNAs by deep RNA sequencing (RNA-seq) of ribosomal RNA-depleted total RNA samples. The samples came from five developmental stages, including the embryonic stage (E18.5), early postnatal stage (P1), outer segment of photoreceptor development stage (P7),22 eye opening stage (P14), and maturation (P30). A total of 9,029 unique circRNAs present during retinal development were detected using the find_circ and CIRI computational pipelines (Figure 1A). We found that most circRNAs (5,616, 62.20%) were novel and that only 3413 circRNAs (37.80%) had been annotated in circBase to date.3 Furthermore, the number of circRNAs increased rapidly during retinal development (Figures 1A and 1B). A total of 4,195 circRNAs were detected at P30, accounting for 46.46% of the total circRNAs in the retina.
Figure 1.
Identification and Characterization of circRNAs in the Mouse Retina
(A) Summary of circRNAs detected in the mouse retina during five developmental stages. (B) The number of circRNAs across the five developmental stages. (C) Genomic origins of mouse circRNAs. (D) Numbers of circRNAs produced from one gene. (E) Exon number distributions for the mouse circRNAs. (F) Conservation analysis for circRNAs in the retina compared between humans and mice (x axis represents circRNAs in the five stages mouse retinas, all circRNAs [left] and circRNAs ≥5 reads [right]).
We also examined the distribution of the genomic origins of total circRNAs and found that most circRNAs (8,188, 90.69%) originated from coding sequence (CDS) exons, whereas only a small fraction was derived from UTR, intergenic, non-coding RNA (ncRNA), and antisense sequences (Figure 1C). Typically, circRNAs from CDS exons were encoded by three to six exons (Figure 1D). Most of the 3,030 host genes (1,590, 52.48%) produced only one circRNA, while a few genes (108, 3.56%) produced more than 10 circRNAs (Figure 1E). Next, we analyzed circRNA conservation between humans23 and mice using the method previously reported by Rajewsky et al.1,24 More than 13.48% of the circRNAs identified at each developmental stage were expressed in the human retina (Figure 1F). Furthermore, highly expressed mouse circRNAs (>5 reads) were more likely to be conserved in humans at each time point. These results collectively revealed the abundant expression and the spatiotemporal pattern of circRNAs in the retina.
Expression Patterns of circRNAs during Retinal Development
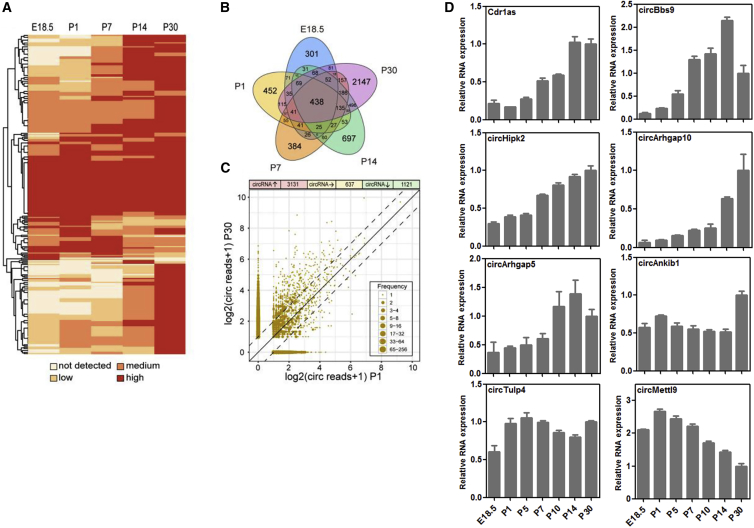
To obtain an overview of the circRNA expression pattern during retinal development, we further profiled the circRNAs at five time points (Data S3). We found that circRNAs were enriched and upregulated during retinal development (Figure 2A). Most circRNAs were specifically expressed at different stages and had a specific signature (Table S1); however, 438 circRNAs were expressed at all tested stages (Figure 2B). Many circRNAs were upregulated during retinal development, especially at P14 and P30 (Figure 2C; Figure S1; Data S4).
Figure 2.
Expression Pattern of circRNAs during Retinal Development
(A) Heatmap comparisons of circRNAs during the five developmental stages. (B) Venn diagram showing the expression patterns of circRNAs across the five stages. (C) circRNAs with log2-fold changes between P30 and P1; the dashed line represents the 2-fold cutoff. (D) qRT-PCR validation of circRNA expression during retinal maturation (n = 5, the normalized values represent the mean ± SEM).
Next, circRNAs with dynamic or high expression patterns during retinal development or those derived from host genes with important roles in the retina were selected for further experimental validation. Seven of the eight selected circRNAs were derived from a protein coding region (CDS), including circBbs9, circHipk2, circArhgap10, circArhgap5, circAnkib1, circTulp4, and circMettl9. The predicted head-to-tail junction sequences of these circRNAs were confirmed by Sanger sequencing (Figure S2). Additionally, the expression patterns of the circRNAs were validated by quantitative RT-PCR (qRT-PCR) (Table S2). Among the eight selected circRNAs, five (Cdr1as, circBbs9, circHipk2, circArhgap10, and circArhgap5) were upregulated during retinal development, one (circMettl9) was downregulated, and two (circAnkib1 and circTulp4) remained unchanged (Figure 2D). Note that the expression patterns of these circRNAs as determined by the qRT-PCR results were not fully consistent with the RNA-seq results (Figure S3), indicating a flaw in the circRNA detection algorithm, which has been described in a previous study.25 In brief, we demonstrated for the first time the expression patterns of retinal circRNAs during development.
circRNA and Linear Transcript Changes Differ during Retinal Development
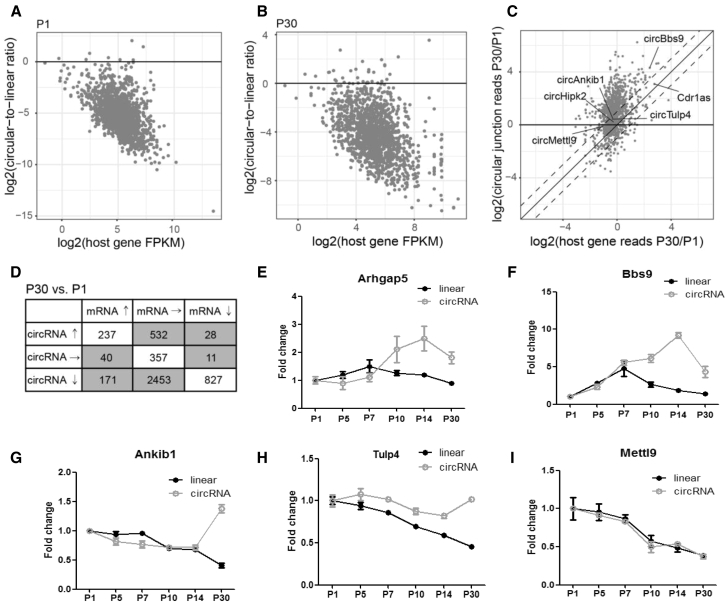
Previous studies have reported that most circRNAs and linear transcripts from the same host gene have distinct expression patterns during development and between cell types.5,26 We combined the circular-to-linear ratio (CLR) with host gene mRNA expression (fragments per kilobase of transcript per million mapped reads [FPKM]),1 and a negative correlation was found between host gene expression and the CLR at P1 and P30 (Figures 3A and 3B), which is inconsistent with the view that circRNAs are a byproduct of occasional aberrant splicing.27,28 In addition, the expression of most circRNAs was lower than that of the corresponding host gene (log2 CLR < 0; most genes were below the black solid line), and highly expressed host genes produced fewer circRNAs (higher FPKM and lower CLR). The changes in circRNAs and their host genes during retinal development were correlated (Figures 3C and 3D; Figures S4A–S4D), and more circRNAs tended to be upregulated (ordinate values greater than 0), indicating the important role of circRNAs in retinal development. Notably, circRNAs levels were more variable than host genes levels. For instance, the expression level of 2,985 circRNAs was changed, while the corresponding host genes were unaltered (P30 versus P1); the expression of 51 host genes was changed, while the corresponding circRNA remained unchanged (Figure 3D). A similar pattern was found at P7 versus P1 and P14 versus P1 (Figure S4).
Figure 3.
circRNA and Linear mRNA Expression Changes Differ during Retinal Development
(A–D) Quantification of circRNAs and mRNAs during retinal development. (A and B) Circular-to-linear ratio plotted against the host gene TPM at P1 (A) and P30 (B). (C) circRNA expression changes between the P1 and P30 stages of development are positively correlated with expression changes of the host genes. (D) Summary of circRNA and host gene expression changes. (E–I) qRT-PCR validation of circRNA and host gene expression during retinal development (n = 5, the normalized values represent the mean ± SEM). (E) Arhgap5, (F) Bbs9, (G) Ankib1, (H) Tulp4, and (I) Mettl9.
Next, we validated the expression patterns of selected circRNAs and their host linear transcripts by using qRT-PCR. circArhgap5, circBbs9, and circAnkib1 increased, whereas their linear transcripts decreased during retinal development (Figures 3E–3G). In contrast, circTulp4 had a high expression pattern that remained unchanged during development, whereas the host mRNA continually decreased (Figure 3H). In addition, circMettl9 expression had the same expression tendency between the circRNA and mRNA during retinal development (Figure 3I). Taken together, bioinformatics analysis and quantitative assays indicated that the expression levels of circRNAs and their corresponding linear transcripts differed throughout retinal development.
circTulp4 Functions as a Competitive Endogenous RNA to Regulate Target Genes
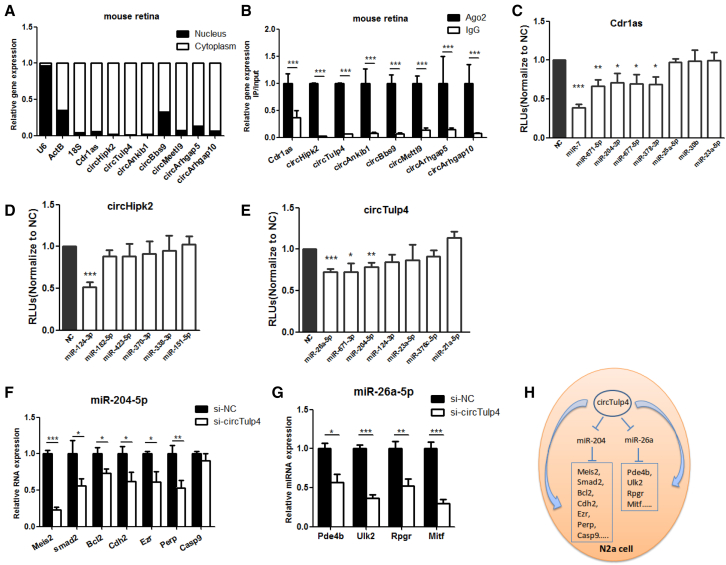
To initially explore the function of the circRNAs identified in the mouse retina, the eight selected circRNAs were studied in a functional investigation. The nuclear versus cytoplasmic enrichment assay showed that the tested circRNAs were predominantly expressed in the cytoplasm (Figure 4A). Because miRNAs exist in the cytoplasm and Ago2 is the core component of the RNA-induced silencing complex (RISC) that binds miRNAs to target mRNAs,29 we attempted to determine whether circRNAs could interact with miRNAs by performing RNA immunoprecipitation (RIP) using an antibody against Ago2. The results showed that all of the studied circRNAs were enriched in the Ago2 protein fraction (Figure 4B), implying that these circRNAs could interact with miRNAs.
Figure 4.
circRNAs Function as ceRNAs to Regulate Target Genes
(A) qRT-PCR detection of circRNAs in the nuclear and cytoplasmic fractions isolated from mouse retinas. U6 is a nuclear transcript used as a control, and ActB and 18S are cytoplasmic transcripts used as controls (n = 3). (B) qRT-PCR detection of immunoprecipitated circRNAs in the mouse retina using Ago2 and IgG antibodies (n = 3). (C–E) Luciferase assay detection of binding between miRNAs and Cdr1as (C), circHipk2 (D), and circTulp4 (E) in 293D cells (n = 3 per group). (F and G) qRT-PCR detection of the miR-204-5p (F) and miR-26a-5p (G) target genes in N2a cells after transfection with si-circTulp4 (n = 3). (H) A model summarizing the circTulp4-miR204/miR-26a-mRNA network in N2a cells. The normalized values represent the mean ± SEM. *p < 0.05, **p < 0.005, ***p < 0.001, Student’s t test.
To further confirm the binding of circRNAs and miRNAs, bioinformatics analysis based on miRanda, RNAhybrid, and TargetScan was performed. We revealed that retina-associated miRNAs could bind the circRNAs studied here (Data S2). Subsequently, Luci-circRNAs were constructed to verify binding between circRNAs and miRNAs. Cdr1as was reported to act as a miR-7 and miR-671-5p sponge, so we performed a luciferase assay for other miRNAs. We found that Cdr1as acting as sponge of miR-204-3p, miR-677-5p, and miR-378-3p (Figure 4C), circHipk2 acting as sponge of miR-124-3p (Figure 4D), and circTulp4 acting as sponge of miR-26a-5p, miR-671-3p, and miR-204-5p (Figure 4E). Other results from the luciferase assay that were performed to detect the sponge activity of circRNAs and miRNAs were summarized in Figure S5. The target sites of miRNAs on corresponding circRNAs were shown in Figure S6. These results demonstrated the potential roles of these circRNAs as competitive endogenous RNAs (ceRNAs) to regulate retinal development.
Next, we studied whether circTulp4 acted as a ceRNA by sponging miR-204-5p and miR-26a-5p. Small interfering RNAs (siRNAs) targeting the backsplice sequence of circTulp4 were designed (Table S3). Transfection of si-circTulp4 into Neuro2a (N2a) cells demonstrated that si-circTulp4 significantly downregulated circTulp4 expression but did not affect expression of the linear Tulp4 transcripts (Figure S7).
Considering that circTulp4 could act as a miR-204-5p and miR-26a-5p sponge, miR-204-5p and miR-26a-5p activity could increase after si-circTulp4 transfection, leading to downregulation of the target genes of miR-204-5p (Mesi2, Bcl2, Cdh2, Ezr, Perp, and Casp9) and miR-26a-5p (Pde4b, Ulk2, and Rpgr). As expected, these genes were decreased significantly after si-circTulp4 transfection in vitro (Figure 4F). Collectively, these results suggested that retinal circRNAs could act as miRNA sponges, and circTulp4 functioned as a ceRNA by acting as sponge for miR-204-5p and miR-26a-5p to regulate their corresponding target genes (Figures 4G and 4H).
circTulp4 Is Essential for Retinal Function in Mice
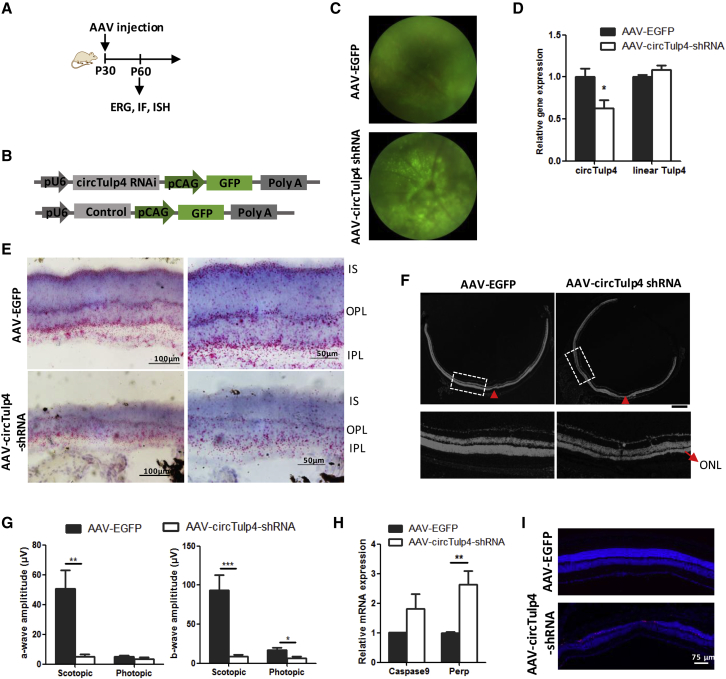
To further determine the function of circTulp4 in the mouse retina, we introduced a GFP-carrying adeno-associated virus (AAV)-circTulp4 short hairpin RNA (shRNA) vector or a control into the subretinal space of mice at P30 (Figures 5A and 5B). Fundus images showed GFP-labeled AAV expressed at P60 (Figure 5C), indicating a successful injection of AAV. As a result, circTulp4 expression decreased to 62.99% in the AAV-shRNA-circTulp4-treated retinas, while linear Tulp4 expression remained unchanged (Figure 5D). An in situ hybridization (ISH) assay displayed circTulp4 localization and expression in the mouse retina. The expression of circTulp4 decreased significantly in the inner segment (IS), outer plexiform layer (OPL), and inner plexiform layer (IPL) (Figure 5E). DAPI staining showed a thinned ONL throughout the retina, indicating that serious photoreceptor degeneration occurred due to the circTulp4 deficiency (Figure 5F). Then, electroretinography (ERG) was performed to evaluate the photoreceptor function. Mouse eyes treated with AAV-circTulp4-shRNA displayed attenuated scotopic and photopic ERG responses. For the scotopic response, the a-wave amplitudes decreased to 9.17%, and the b-wave amplitudes decreased to 9.87% in the shRNA-treated mice (Figure 5G; Figure S8). In addition, several genes promoting apoptosis, such as Perp, were upregulated in the circTulp4 interrupted retina (Figure 5H), which was further confirmed by a terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay. The apoptotic cells that appeared in ONL were consistent with the thinned ONL phenotype (Figure 5I). However, silencing of circTulp4 did not lead to an obvious change in miR-204-5p and miR-26a-5p expression, nor did it result in the downregulation of their target genes, such as Smad2, Mitf, and Cdh2, except for Meis2 (Figures S9A and S9B). As a transcription factor, the target genes of Meis2 were not changed, including Pax6, Tbx5, and Bmp4 (Figure S9C). These results suggested that circTulp4 has functions addition to acting as a miR-204 and miR-26a sponge. The complex microenvironment and compensatory in vivo mechanisms warranted further exploration. Overall, these observations confirmed that circTulp4 is indispensable for proper retinal function.
Figure 5.
circTulp4 Is Indispensable for Retinal Function in Mice
(A) Schematic representation of the experiment. (B) AAV-circTulp4 shRNA and AAV-EGFP expression cassettes driven by the pU6 promoter. (C) Fundus photographs of mice 1 month after AAV-circTulp4 shRNA or AAV-EGFP subretinal injection (n = 5). (D) Quantitative analysis of circTulp4 and linear Tulp4 in the retinas treated with AAV-circTulp4 shRNA and AAV-EGFP (n = 3). (E) Expression and localization of circTulp4 after AAV-circTulp4 shRNA and AAV-EGFP subretinal injection; results are based on the BaseScope assay; red dots shows the sense of ISH image, and the negative control is shown in Figure S10 (n = 3). (F) The thickness of the nucleus in each retinal layer was assessed by DAPI staining (gray) after AAV-circTulp4 shRNA or AAV-EGFP subretinal injection (n = 3). The red arrowheads point to the AAV injection site, the red arrows point to the ONL, and the area in the white dotted box is enlarged at the bottom (n = 3) (G) The a-wave and b-wave amplitudes of the scotopic and photopic response after AAV-circTulp4 shRNA or AAV-EGFP subretinal injection (n = 5). (H) qRT-PCR detection of genes that promote apoptosis in the retina after AAV-circTulp4 shRNA or AAV-EGFP subretinal injection (n = 3). (I) Retinal cell apoptosis was assessed using a TUNEL assay, and TUNEL-positive cells (red) were observed in ONL. The normalized values represent the mean ± SEM. *p < 0.05, **p < 0.005, ***p < 0.001, Student’s t test.
circRNAs Dysregulated in Retinal Degeneration Mice Model
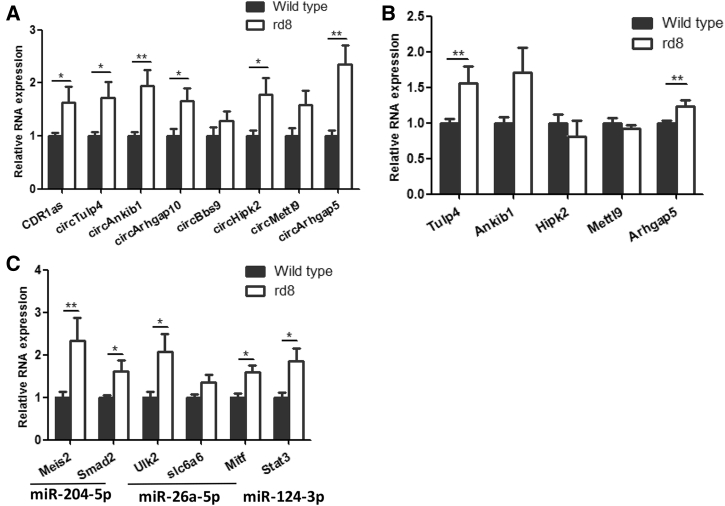
To determine whether circRNAs are dysregulated in a retinal degeneration model, rd8 mice with a crb1 mutation30 were analyzed. Retinal degeneration in rd8 mice started at P90. We investigated whether circRNA changes could be observed before the onset of disease. qRT-PCR was conducted to detect circRNA expression in the retinas of rd8 mice at P30. Remarkably, most of the selected circRNAs were significantly upregulated (Figure 6A); however, their linear transcripts had only minor changes (Figure 6B). These results implied that circRNAs were more responsive than their corresponding linear transcripts to intrinsic retinal damage. Combined with the results shown in Figure 5, we concluded that proper circRNA expression is essential for retinal development and function and that circRNA dysregulation was detrimental to retinal development.
Figure 6.
circRNAs Are Dysregulated in the rd8 Mouse Model
(A and B) qRT-PCR assay detection of circRNAs (A) and host linear mRNA (B) expression in the retinas of wild-type (WT) and rd8 mice. (C) qRT-PCR assay detection of miRNA target gene expression in the retinas of WT and rd8 mice. The normalized values represent the mean ± SEM. *p < 0.05, **p < 0.005, Student’s t test. n = 5 per group.
Our results described in preceding sections showed that circHipk2 and circTulp4 could act as sponges of miR-124-3p (Figure 4C) and miR-204-5p/miR-26a-5p, respectively (Figure 4E). Next, we examined the target genes of these miRNAs in the retinas of rd8 mice and found a significant increase in the expression levels of many target genes (Figure 6C), including Meis2, Smad2, Ulk2, Slc6a6, Stat3, and Mitf. These results further support the hypothesis that circRNAs can function as ceRNAs, and that upregulation of circRNAs can lead to an increase in some miRNA target genes due to the ability of circRNAs to act as sponges for miRNAs.
Discussion
Previous studies have reported that circRNAs possess important regulatory functions,31 especially in the CNS,9,11 which is substantially enriched with circRNAs. However, circRNA expression and function in the retina, which is a key part of the CNS, have not been well profiled, especially at different developmental stages and disease conditions. To address this gap in knowledge, we sequenced and analyzed circRNAs in the mouse retina (Data S4) and found that most circRNAs originated from exons; furthermore, circRNAs showed high expression and distinct signatures at different developmental stages (Figures 2A and 2B), which was consistent with data on circRNAs from the developmental rat retina,2 indicating the important roles of circRNAs at certain retina stages. Additionally, circRNAs showed dynamic expression independent of their linear transcripts (Figure 3). circRNAs changed more dramatically than did their linear transcripts at each critical developmental time point (Figure 3D; Figure S4). Our findings indicated that circRNAs were involved in complex physiologic processes due to their specificity and variability.
By implementing bioinformatics tools and experimental verification, circRNAs acting as ceRNAs during retinal development were identified. Eight circRNAs (Cdr1as, circBbs9, circHipk2, circArhgap10, circArhgap5, circAnkib1, circTulp4, and circMettl9) with a dynamic and high expression pattern during retinal development were selected for further experimental validation. However, only four circRNAs (Cdr1as, circHipk2, circAnkib1, and circTulp4) were confirmed to target miRNAs in the retina. For example, Cdr1as was miR-204/677/378 sponge, circHipk2 was miR-124 sponge, circTulp4 was miR-26a/671/204 sponge, and circAnkib1 was miR-195a sponge. However, while circHipk2 acting as miR-124 sponge has been reported,32 Cdr1as is not expressed in N2a cells. For these reasons, circTulp4 was selected for further functional analysis. A reduction in circTulp4 levels leads to downregulation of miR-204-5p and miR-26a-5p targets, including Meis2, Cdh2, Mitf, and Pde4b in vitro (Figure 4F). Cdh2 may be important for transcriptional control and regeneration of injured retinal ganglion cell axons.33,34 Mitf controls development and function of the retinal pigment epithelium.35 Pde4b is expressed in the inner segment, outer segment, and outer plexiform layer of rod photoreceptors, and it is an important regulator of signal transduction processes.36 These genes were reported to be involved in retinal development and function. The significant phenotype of AAV-circTulp4-shRNA-treated retinas indicated an essential role for circTulp4 in retinal development and function.
A key finding of our study was that circRNAs were found to be dysregulated at a much earlier time point than that of disease onset in the rd8 mouse model.30 circRNA levels changed significantly compared to those of linear transcripts, whereas the structure and function of the retina in rd8 mice changed only slightly at P30 (Figures 6A and 6B). circRNAs seemed to be more sensitive and dynamic than linear transcripts in the rd8 model. Furthermore, the upregulation of circTulp4 and circHipk2 led to significant increases in the target genes of their sponged miRNAs (miR-204-5p, miR-26a-5p, and miR-124-3p) (Figure 6C), including genes involved in retinal development and function. These results further validated that circTulp4 functions as a ceRNA in vivo. Our data also implied that certain and precise circRNA levels are required during the normal development of the retina. circRNAs are stable, have tissue- and development-specific expression, and can be found in plasma, cell-free saliva, and exosomes,14,37,38 so we hypothesized that circRNAs might be a potential biomarker for retinal degenerative disease.
In summary, we generated a profile of the entire picture of circRNAs during retinal development and identified circRNA signatures in developing and degenerating retinas. Our findings provided a reference map and new insights into deciphering the biological roles of circRNAs in retinal development and degeneration.
Materials and Methods
Cell Culture and Transfection
Human embryonic kidney (293D) cells and mouse neuroblastoma N2a cells were cultured at 37°C with 5% CO2 in DMEM with 10% fetal bovine serum. Cells were transfected with the described plasmids using Lipofectamine 2000 (Life Technologies) according to the manufacturer’s protocol.
Animals
C57BL/6J and rd8 mice obtained from Charles River (China) were bred in a 12-h light/12-h dark cycle, had free access to food and water, and were maintained in the animal facility of Wenzhou Medical University; the study obtained ethics approval (no. SYXK 2015-0009). The ages of the mice were indicated in the results, and only male mice were used in our study.
RNA Isolation and qRT-PCR
RNAs were extracted from the N2a cells and retinas by using the RNeasy kit (QIAGEN). Nuclear and cytoplasmic fractions were extracted (PARIS kit protein and RNA isolation system, Invitrogen). Complementary DNA (cDNA) was synthesized using a random primer (Promega) and quantified using FastStart Universal SYBR Green Master Mix (Roche). The divergent and convergent primers were designed for circRNAs and linear transcripts, respectively, and GAPDH was used as the reference gene (Table S2).
circRNA Sequencing Analysis
After retinas were dissected and mashed, RNAs were extracted with TRIzol reagent (Life Technologies) and an RNeasy mini kit (QIAGEN). Two samples were prepared from each developmental stage, and each sample contained pooled retinas from multiple mice. RNA purity was detected using a NanoDrop spectrophotometer (Thermo Fisher Scientific), and the RNA concentration was measured with a Qubit RNA HS assay kit in a Qubit 3.0 fluorometer (Life Technologies). RNA integrity was assessed using an Agilent RNA 6000 Pico kit for the Agilent Bioanalyzer 2100 system (Agilent Technologies) (Table S4). After finishing the RNA quality test, 2 μg of total RNA was ribo-depleted using a KAPA RiboErase kit (Kapa Biosystems, USA). Sequencing libraries were generated by using the KAPA stranded RNA-seq kit (Kapa Biosystems, USA) according to the manufacturer’s recommendations, and index codes were added to identify sequences. Libraries were pooled and sequenced on the Illumina X-Ten PE150 platform. The number of reads per sample is listed in Table S5. Raw data were submitted to the NCBI GEO database (GEO: GSE137128). Computational analysis was performed as described in a previous report.1 FASTQ data were mapped to the mouse reference genome mm9. circRNA detection and quantification were performed using the find_circ and CIRI software,26,39 and the circRNA abundance was measured through the total number of reads (head-to-tail junction). To examine circRNA relative expression (CLR), the following equation was used: CLR = no. of reads_circular/maximum (no. of reads_linear_5-prime, no. of reads_linear_3-prime).
circRNA Expression During Retinal Development
To summarize and compare circRNA expression, circRNAs were assigned to four classes: (1) “high,” top 5% most highly expressed circRNAs; (2) “medium,” between the 80th and 95th percentiles of circRNA expression; (3) “low,” all other circRNAs; and (4) “not detected,” circRNAs that were not detected in a particular sample.
Conservation Analysis between Humans and Mice
The conservation of circRNAs between humans and mice was analyzed. circRNA data in the human retina were derived from our published data.23 We used a homologous splice site as a rationale, and the UCSC liftOver tool was used to convert the 5′ and 3′ flank coordinates of mouse circRNA to human genome coordinates.24 Subsequently, circRNAs were classified into the following categories: (1) “not aligned” indicated that sites could not be mapped to the human genome; (2) “no homologous circRNA” indicated that no splice sites were detected within 2 nt of the converted genome coordinates; (3) “with homologous 5′ splice site” indicated that only the 5′ splice site was shared by the circRNAs in humans; (4) “with homologous 3′ splice” indicated that only the 3′ splice site was shared by the circRNAs in humans; (5) “with both splice sites” indicated that both the 3′ and 5′ splice sites were shared by the circRNAs in humans; and (6) “homologous circRNA” indicated that a circRNA spliced from sites within 2 nt of the splice sites predicted by liftOver was found in humans.
Bioinformatics Analysis
The binding patterns between miRNAs and circRNAs were predicted using Miranda (http://miranda.org.uk/),40 RNAhybrid (https://omictools.com/rnahybrid-tool),41 and TargetScan (http://www.targetscan.org/vert_72/).42
Luciferase Assay
The dual-luciferase reporter psi-CHECK2 plasmid was used in our study. The entire mouse circAnkib1, circHipk2, and circTulp4 segments were amplified by PCR using mouse retinal cDNA as a template; the primers are listed in Table S6. The PCR products were inserted into the psi-CHECK2 plasmid to create Luci-circAnkib1, Luci-circHipk2, and Luci-circTulp4, which were cotransfected into 293D cells with a mimic-miRNA. A mimic-negative control (NC) and the psi-CHECK2 empty vector were used as negative controls. After transfection for 48 h, firefly and Renilla substrates were added in sequentially, and the corresponding luminescence was measured on a microplate spectrophotometer.
RNA Immunoprecipitation Assay
The retinas were isolated and collected in lysis buffer (Thermo Scientific) containing PMSF and propidium iodide (PI); then, proteins were extracted and quantified using a bicinchoninic acid (BCA) kit (Thermo Scientific). Three hundred micrograms of protein was incubated with 4 μg of Ago2 (Abcam) or immunoglobulin (Ig)G antibodies (Thermo Scientific) and incubated at 4°C overnight with shaking. Fifty microliters of protein A/G magnetic beads (Thermo Scientific) was added and incubated at 4°C for 2–4 h with shaking. The beads were separated using a magnetic strip and then washed four times with 0.2% Nonidet P-40 in PBS. The beads were suspended in TRIzol reagent, and RNA was extracted and subjected to qRT-PCR analysis.
In Situ Hybridization
For ISH, a BaseScope reagent kit-RED and a circTulp4 probe targeting the junction site were designed and supplied by Advanced Cell Diagnostics (ACD). Cryosections of mouse retinas were dried completely at room temperature (RT) and treated with hydrogen peroxide and protease plus; then, probe hybridization was performed strictly according to the manufacturer’s protocol. Images were acquired using a microscope (Nikon Eclipse).
TUNEL Assay
Apoptosis of retinal cells was assessed using a TUNEL assay (Beyotime, China) according to the manufacturer’s protocol.
AAV8-Mediated circTulp4 Knockdown
To generate the AAV8-mmu-circTulp4 shRNA, the target sequence 5′-ACAACAGTGAGAGTTGTAA-3′ was cloned into a GV478 vector containing U6-MCS-CAG-EGFP. The control sequence 5′-CGCTGAGTACTTCGAAATGTC-3′ was cloned into a GV478 vector to serve as the control. The AAV8-mmu-circTulp4-shRNA and AAV-EGFP control titers were 1.27 × 1013 and 1.50 × 1013 transduction units (TU)/mL, respectively. The subretinal injection of AAV was performed as previously described.43 C57BL/6J mice at P30 were anesthetized with a mixture of ketamine and xylazine. A small incision was created in the lens near the sclera with a sharp 30-gauge needle. Then, 1 μL (1.27 × 1013 TU/mL) of AAV8-mmu-circTulp4-shRNA and an equal dose of control vector AAV-EGFP were injected slowly into the subretinal space of the right eye and left eye, respectively, using a blunt 5-μL Hamilton syringe held in a micromanipulator. All injections were performed bilaterally.
Focal ERG
The focal ERG was carried out as described in the instrument manuals (Phoenix Research Laboratories) and as previously described.37 Briefly, mice were dark-adapted overnight and anesthetized as described previously. Before light stimulation, their pupils were dilated, and their corneas were anesthetized with 0.5% phenylephrine and 0.5% tropicamide. A drop of 1% methyl cellulose was applied on the cornea to improve conjunction with a corneal gold wire electrode. Ground electrodes and a referential needle were punctured into the tail and cheek, respectively. The “Aming light” was turned to a high value to ensure the biggest light spot around the optic nerve head; the size of the stimulus region was approximately two-thirds of the entire retina. Scotopic ERG was performed five times with an inter-stimulus interval of 20 s at a 3.0 log cd·s/m2 stimulus intensity. After 10 min of light adaptation with a background illumination of 5.3 cd/m2, the mice were subjected to photopic measurements obtained five times at 3.3 log cd·s/m2 with a 20-s inter-stimulus. The a-wave and b-wave amplitudes of the scotopic and photopic responses from mice treated with AAV-circTulp4 shRNA and AAV-EGFP were recorded and averaged.
Fundus Photography
Mice injected with AAV-circTulp4 shRNA and AAV-EGFP were anesthetized as described previously. Before the examination, 2.5% hydroxypropyl methylcellulose was dropped into the eyes to improve the connection with the machine (Micron IV, Phoenix Research Labs), and then fundus photography was performed.
Author Contributions
X.-J.C., X.-Y.W., H.-Q.Z, C.-J. Z., K.-C.W., M.-L.L., Y.-Y.J., Y.M., L.X., and L.-F.S. performed the research; H.-Q.Z and M.Z. analyzed the data; Z.-B.J. and X.-J.C designed the research; Z.-B.J. and X.-J.C wrote the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81870690 and 81970838); National Key R&D Program of China (2017YFA0105300 and 2017YFB0403700); Zhejiang Provincial Natural Science Foundation of China (LD18H120001); Zhejiang Provincial Key Research and Development Program (2015C03029); and the 111 Project (D16011).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.11.016.
Contributor Information
Meng Zhou, Email: zhoumeng@wmu.edu.cn.
Zi-Bing Jin, Email: jinzb@mail.eye.ac.cn.
Supplemental Information
References
- 1.Rybak-Wolf A., Stottmeister C., Glažar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 2.Han J., Gao L., Dong J., Bai J., Zhang M., Zheng J. The expression profile of developmental stage-dependent circular RNA in the immature rat retina. Mol. Vis. 2017;23:457–469. [PMC free article] [PubMed] [Google Scholar]
- 3.Glažar P., Papavasileiou P., Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo J.U., Agarwal V., Guo H., Bartel D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lu C., Sun X., Li N., Wang W., Kuang D., Tong P., Han Y., Dai J. circRNAs in the tree shrew (Tupaia belangeri) brain during postnatal development and aging. Aging (Albany N.Y.) 2018;10:833–852. doi: 10.18632/aging.101437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen B.J., Yang B., Janitz M. Region-specific expression of circular RNAs in the mouse brain. Neurosci. Lett. 2018;666:44–47. doi: 10.1016/j.neulet.2017.12.022. [DOI] [PubMed] [Google Scholar]
- 8.Westholm J.O., Miura P., Olson S., Shenker S., Joseph B., Sanfilippo P., Celniker S.E., Graveley B.R., Lai E.C. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 2014;9:1966–1980. doi: 10.1016/j.celrep.2014.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 10.You X., Vlatkovic I., Babic A., Will T., Epstein I., Tushev G., Akbalik G., Wang M., Glock C., Quedenau C. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 2015;18:603–610. doi: 10.1038/nn.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kleaveland B., Shi C.Y., Stefano J., Bartel D.P. A network of noncoding regulatory RNAs acts in the mammalian brain. Cell. 2018;174:350–362.e17. doi: 10.1016/j.cell.2018.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Piwecka M., Glažar P., Hernandez-Miranda L.R., Memczak S., Wolf S.A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C.A., Fenske P. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science. 2017;357:eaam8526. doi: 10.1126/science.aam8526. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Z.J., Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514–521. doi: 10.1080/15476286.2015.1122162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J., Chen D., Gu J., He X., Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng L., Yuan X.Q., Li G.C. The emerging landscape of circular RNA ciRS-7 in cancer (Review) Oncol. Rep. 2015;33:2669–2674. doi: 10.3892/or.2015.3904. [DOI] [PubMed] [Google Scholar]
- 16.Lukiw W.J. Circular RNA (circRNA) in Alzheimer’s disease (AD) Front. Genet. 2013;4:307. doi: 10.3389/fgene.2013.00307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosal S., Das S., Sen R., Basak P., Chakrabarti J. Circ2Traits: a comprehensive database for circular RNA potentially associated with disease and traits. Front. Genet. 2013;4:283. doi: 10.3389/fgene.2013.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shan K., Liu C., Liu B.H., Chen X., Dong R., Liu X., Zhang Y.Y., Liu B., Zhang S.J., Wang J.J. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136:1629–1642. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S.J., Chen X., Li C.P., Li X.M., Liu C., Liu B.H., Shan K., Jiang Q., Zhao C., Yan B. Identification and characterization of circular RNAs as a new class of putative biomarkers in diabetes retinopathy. Invest. Ophthalmol. Vis. Sci. 2017;58:6500–6509. doi: 10.1167/iovs.17-22698. [DOI] [PubMed] [Google Scholar]
- 20.Chen Y., Li C., Tan C., Liu X. Circular RNAs: a new frontier in the study of human diseases. J. Med. Genet. 2016;53:359–365. doi: 10.1136/jmedgenet-2016-103758. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z., Yang T., Xiao J. Circular RNAs: promising biomarkers for human diseases. EBioMedicine. 2018;34:267–274. doi: 10.1016/j.ebiom.2018.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daum J.M., Keles Ö., Holwerda S.J., Kohler H., Rijli F.M., Stadler M., Roska B. The formation of the light-sensing compartment of cone photoreceptors coincides with a transcriptional switch. eLife. 2017;6:e31437. doi: 10.7554/eLife.31437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun L.F., Zhang B., Chen X.J., Wang X.Y., Zhang B.W., Ji Y.Y., Wu K.C., Wu J., Jin Z.B. Circular RNAs in human and vertebrate neural retinas. RNA Biol. 2019;16:821–829. doi: 10.1080/15476286.2019.1591034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y., Zhang J., Zhao F. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2018;19:803–810. doi: 10.1093/bib/bbx014. [DOI] [PubMed] [Google Scholar]
- 25.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F. The UCSC Genome Browser database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X., Yang L., Chen L.L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell. 2018;71:428–442. doi: 10.1016/j.molcel.2018.06.034. [DOI] [PubMed] [Google Scholar]
- 27.Cocquerelle C., Daubersies P., Majérus M.A., Kerckaert J.P., Bailleul B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992;11:1095–1098. doi: 10.1002/j.1460-2075.1992.tb05148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nigro J.M., Cho K.R., Fearon E.R., Kern S.E., Ruppert J.M., Oliner J.D., Kinzler K.W., Vogelstein B. Scrambled exons. Cell. 1991;64:607–613. doi: 10.1016/0092-8674(91)90244-s. [DOI] [PubMed] [Google Scholar]
- 29.Winter J., Jung S., Keller S., Gregory R.I., Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009;11:228–234. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 30.Chang B., Hawes N.L., Nishina P.M., Smith R.S., Davisson M.T., Heckenlively J.R. Two new mouse models of retinal degeneration (rd8 and Rd9) Invest. Ophthalmol. Vis. Sci. 1999;40(Suppl):S976. [Google Scholar]
- 31.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 32.Huang R., Zhang Y., Han B., Bai Y., Zhou R., Gan G., Chao J., Hu G., Yao H. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy. 2017;13:1722–1741. doi: 10.1080/15548627.2017.1356975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Q., Londraville R.L., Azodi E., Babb S.G., Chiappini-Williamson C., Marrs J.A., Raymond P.A. Up-regulation of cadherin-2 and cadherin-4 in regenerating visual structures of adult zebrafish. Exp. Neurol. 2002;177:396–406. doi: 10.1006/exnr.2002.8008. [DOI] [PubMed] [Google Scholar]
- 34.Li B., Paradies N.E., Brackenbury R.W. Isolation and characterization of the promoter region of the chicken N-cadherin gene. Gene. 1997;191:7–13. doi: 10.1016/s0378-1119(97)00006-1. [DOI] [PubMed] [Google Scholar]
- 35.Saule S. Both PAX6 and MITF are required for pigment epithelium development in vivo. Pigment Cell Melanoma Res. 2012;25:541–543. doi: 10.1111/j.1755-148x.2012.01037.x. [DOI] [PubMed] [Google Scholar]
- 36.Whitaker C.M., Cooper N.G.F. The novel distribution of phosphodiesterase-4 subtypes within the rat retina. Neuroscience. 2009;163:1277–1291. doi: 10.1016/j.neuroscience.2009.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bahn J.H., Zhang Q., Li F., Chan T.M., Lin X., Kim Y., Wong D.T., Xiao X. The landscape of microRNA, Piwi-interacting RNA, and circular RNA in human saliva. Clin. Chem. 2015;61:221–230. doi: 10.1373/clinchem.2014.230433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vo J.N., Cieslik M., Zhang Y., Shukla S., Xiao L., Zhang Y., Wu Y.M., Dhanasekaran S.M., Engelke C.G., Cao X. The candscape of circular RNA in cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y., Wang J., Zhao F. CIRI: an efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015;16:4. doi: 10.1186/s13059-014-0571-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner D. An overview of Miranda. SIGPLAN Not. 1986;21:158–166. [Google Scholar]
- 41.Krüger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34:W451-4. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 43.Xiang L., Chen X.J., Wu K.C., Zhang C.J., Zhou G.H., Lv J.N., Sun L.F., Cheng F.F., Cai X.B., Jin Z.B. miR-183/96 plays a pivotal regulatory role in mouse photoreceptor maturation and maintenance. Proc. Natl. Acad. Sci. USA. 2017;114:6376–6381. doi: 10.1073/pnas.1618757114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.