Abstract
Charcot-Marie-Tooth (CMT) disease is a progressive and heterogeneous inherited peripheral neuropathy. A myriad of genetic factors have been identified that contribute to the degeneration of motor and sensory axons in a length-dependent manner. Emerging biological themes underlying disease include defects in axonal trafficking, dysfunction in RNA metabolism and protein homeostasis, as well deficits in the cellular stress response. Moreover, genetic contributions to CMT can have overlap with other neuropathies, motor neuron diseases (MNDs) and neurodegenerative disorders. Recent progress in understanding the molecular biology of CMT and overlapping syndromes aids in the search for necessary therapeutic targets.
Keywords: CMT, Genetics, Motor Neuron Disease, Neurodegeneration, Disease Spectrum, Ribosome-Associated Quality Control
Introduction
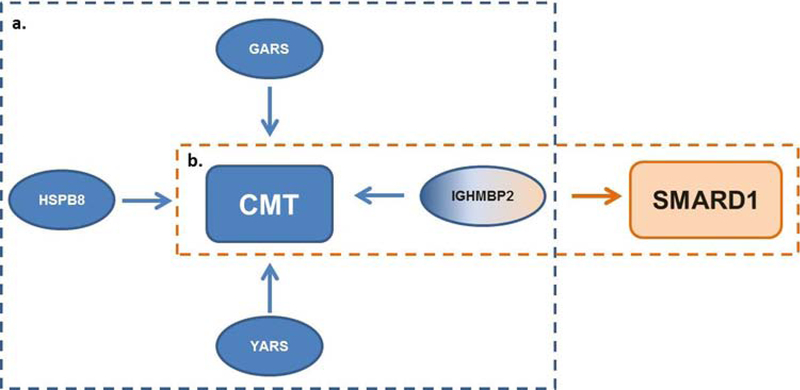
In the mid to late nineteenth century a premier neurologist, Jean-Martin Charcot, set out to correlate clinical findings in neurological patients with anatomical discoveries in the nervous system using a novel method known as the anatomoclinical method. This method provided comprehensive clinical findings coupled with the pathological changes, allowing for specific disease classification. Charcot defined many neurological disorders, chief among them multiple sclerosis (MS) in 1868, amyotrophic lateral sclerosis (ALS) in 1869, and Charcot-Marie-Tooth (CMT) disease in 1886. Although Charcot made great strides in neurology, his work predated modern understandings of inheritance and genetics, and it was not until almost a century later that the first genotype-phenotype correlation was made in CMT (PMP22) in 1991 and ALS (SOD1) in 1993 (Lupski et al., 1991, Timmerman et al., 1992a; Rosen et al., 1993). With the advent of precision genetics, we are now able to examine a disease from its three main faces: clinical presentation, pathological features, and underlying genetics. Variations in different genes can phenotypically converge onto the same clinical classification or disease category; an excellent example of this is Charcot-Marie-Tooth disease (Fig. 1a). Conversely, variants within the same gene can diverge pathogenically and give rise to a spectrum of diseases (Fig. 1b). Among those with clinical and genetic heterogeneity are inherited peripheral neuropathies (IPNs), a group of diseases including CMT and hereditary motor neuropathy (HMN).
Figure 1.
Schematic showing that a. genetic contributions can converge onto a similar clinical phenotype as well as a b. genetic contribution presenting with diverging clinical presentation.
Charcot-Marie-Tooth is characterized by progressive muscular atrophy and loss of sensation in a length-dependent manner and is the most common IPN, with an estimated 1 in 2,500 individuals affected (Skre, 1974). CMT is further clinically subdivided into Types 1, 2, and intermediate. Classically, Type 1 CMTs are demyelinating neuropathies with decreased nerve conduction velocities (NCV), while Type 2 CMTs are axonal neuropathies with normal or near-normal NCVs. More recently, intermediate forms have been recognized with clinical characteristics of both demyelination and axonal degeneration. While CMTs are defined as having both distal sensory and motor involvement, other IPNs can be predominantly motor (hereditary motor neuropathy, HMN) or predominantly sensory (hereditary sensory neuropathy, HSN) (Rossor et al., 2015).
Recent work defining the genetic underpinnings of CMT suggests significant overlap in genes that may also be contributing to motor neuron diseases (MND) and other neurodegenerative diseases (Table 1). Historically, CMT and MNDs have been defined and diagnosed through clinical presentation, but newly-available genetic information can shed new light on these diseases and reshape our understanding of their classification. Genetic factors contributing to one disease presentation in one patient may have a slightly different presentation in another. CMT primarily impacts the most distal axons of a patient, whereas MNDs generally have upper and/or lower motor neuron degeneration as well as central involvement, there can be considerable overlap in the presentation of more severe CMTs and more mild or slowly-progressing MNDs. Common genetic MNDs that will be discussed in this review include: amyotrophic lateral sclerosis (ALS) a later-onset MND resulting in progressive paralysis and mortality, as well hereditary spastic paraplegia (HSP) sometimes referred to as spastic paraplegia (SPG) an inherited MND with bilateral lower-extremity spasticity and weakness. HSP can be defined as uncomplicated (pure) or can be classified as complicated, which is characterized as having additional neurologic findings including, but not limited to, ataxia, intellectual disability, or peripheral neuropathy.
Table 1.
The Broad Genetic Overlap of CMT, MND and Neurodegenerative Diseases.
| Gene Name | Disease-Association | Inheritance | Function | OMIM |
|---|---|---|---|---|
| CMT2E | Dominant | 607684 | ||
| NEFL | CMT1F | Dom/Rec | Neuronal cytoskeleton | 607734 |
| CMT, dominant intermediate G | Dominant | 617882 | ||
| NEFH | CMT2CC | Dominant | Neuronal cytoskeleton | 616924 |
| ALS | Dom/Rec | 105400 | ||
| DYNC1H1 | CMT2OMRD13 | Dominant | 614228 | |
| Dominant | Retrograde transport | 614563 | ||
| SMALED1 | Dominant | 158600 | ||
| DCTN1 | CMT2C | Dominant | 607641 | |
| Perry syndrome | Dominant | Retrograde transport | 168605 | |
| ALS | Dom/Rec | 105400 | ||
| CMT2 | Dominant | |||
| KIF5A | SPG10 | Dominant | Anterograde transport | 604187 |
| ALS | Dominant | 617921 | ||
| NEIMY | Dominant | 617235 | ||
| HSN2C | Recessive | 614213 | ||
| KIF1A | SPG30 | Dom/Rec | Anterograde transport | 610357 |
| MRD9 | Dominant | 614255 | ||
| RTT | Recessive | |||
| CMT4J | Recessive | 611228 | ||
| FIG4 | ALS | Dominant | Vesicle trafficking | 612577 |
| Yunis-Varon syndrome | Recessive | 216340 | ||
| BTOP | Recessive | 612691 | ||
| SPG11 | ALS | Recessive | 616668 | |
| Spastic paraplegia 11 | Recessive | Vesicle trafficking | 602099 | |
| CMT2X | Recessive | 604360 | ||
| IGHMBP2 | SMARD1 | Recessive | RNA/DNA Helicase | 604320 |
| CMT2S | Recessive | 616155 | ||
| CLP1 | PCH10 has lPN component | Recessive | Splicing of intron containing tRNA | 615803 |
| GARS | CMT2D | Dominant | ||
| HMN5A | Dominant | Cytoplasmic and mitochondrial tRNA synthetase | 601472 | |
| blubar ALS | Dominant | 600794 | ||
| AARS | CMT2N | Dominant | Cytoplasmic tRNA synthetase | 616339 |
| MARS | CMT2U | Dominant | Cytoplasmic tRNA synthetase | 616280 |
| HARS | CMT2W | Dominant | Cytoplasmic tRNA synthetase | 616625 |
| KARS | CMTIB | Recessive | Cytoplasmic tRNA synthetase | 613641 |
| YARS | CMT-DIC | Dominant | Cytoplasmic and mitochondrial tRNA synthetase | 608323 |
| GTPBP2 | JABELS | Recessive | Ribosome recycling factor | 617988 |
| NEMF | ID w/hypotonia | Recessive | CATylation Ltn1 recruitment | 608378 |
| LTN1 | Mouse model: locomotor abnormalities neurodegeneration | Recessive | E3 ubiquitin ligase, degradation of aberrant nascent chains | 613083 |
| VCP | CMT2Y | Dominant | Protein degradation | 616687 |
| ALS14 | Dominant | 613954 | ||
| IBMPFD | Dominant | 167320 | ||
| HSPB1 | CMT2F | Dominant | Stress-activated kinase | 606595 |
| dHMN2B | Dominant | 608634 | ||
| HSPB3 | dHMN2C | Dominant | Stress-activated kinase | 613376 |
| CMT2 | Dominant | |||
| HSPB8 | CMT2L | Dominant | Stress-activated kinase, chaperone activity | 608673 |
| dHMN2A | Dominant | 158590 | ||
| DNAJB2 | dSMA5 | Recessive | ||
| dHMN | Recessive | Stress-activated kinase, chaperone activity | 614881 | |
| CMT2-like | Recessive | 614881 |
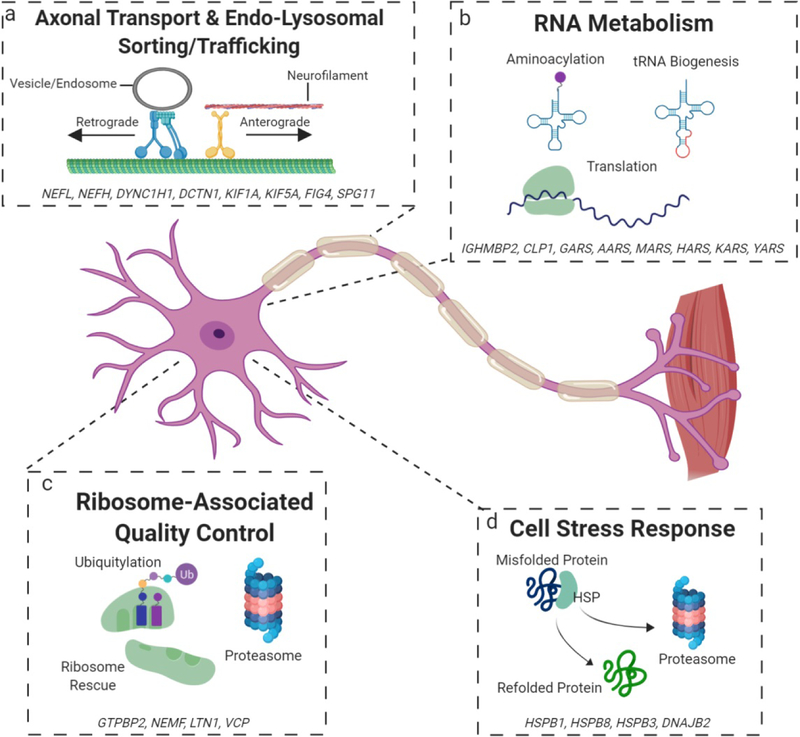
Here we will consider in depth the clinical and genetic features of CMT, as well as a spectrum of MNDs and other neurodegenerative diseases that share overlapping genetic contributions. We highlight emerging themes associated with pathogenic contributions to this spectrum, including disrupted axonal transport (Fig. 2a), dysfunction in RNA metabolism (Fig. 2b), the ribosome-associated quality control pathway (Fig. 2c), and cell stress response (Fig. 2d).
Figure 2. Mechanisms of disease implicated in CMT and a broader neurodegenerative spectrum.
a. Variants in genes involved in the endo-lysosomal sorting/trafficking, cytoskeleton, and axonal transport can disturb cytoskeletal integrity and impair transportation, limiting the movement of necessary organelles between the cell body and distal portions. b. Disruptions in proteins involved in the processing (CLP1) and charging (AARs proteins) of tRNAs, as well as others can impair translation and RNA metabolism, which slows a high translational demand. Additionally, neomorphic activities of AARs proteins can disrupt neuronal homeostasis. c. Mutations in proteins involved in the rescue and recycling of stalled ribosomes can give rise to potentially toxic polypeptides and disrupts the sensitive cellular economy of neurons and distal axons. d. Impairments in heat shock proteins, chaperones for potentially toxic misfolded proteins, can lead to toxic aggregates that further sequester more heat shock proteins, impairing the appropriate cell stress response.
1. Axonal Transport & Endo-Lysosomal Sorting/Trafficking
Endo-Lysosomal sorting and trafficking as well as axonal transport are both critical for axon growth, maintenance, and signaling (Cosker and Segal, 2014; Maday et al., 2014; Kalani and Tyagi, 2015; Mestres and Sung, 2017), so it is not surprising that mutations in the various genes associated with these processes have been tied to a wide variety of neurodegenerative diseases (De Vos et al., 2008; Hinckelmann et al., 2013; Millecamps and Julien, 2013; Neefjes and van der Kant, 2014; Prior et al., 2017; Burk and Pasterkamp, 2019; Vagnozzi and Praticò, 2019). In this section, we will take a closer look at a few of these genes that encode for proteins that make up the axon cytoskeleton, molecular motors, and vesicle trafficking and are implicated in a range of neuropathies (Fig. 2a) (Table 1).
1.1. NEFL
The NEFL gene encodes the neurofilament light chain protein which forms a type IV intermediate filament and plays a major role in the neuronal cytoskeleton (Agrawal et al., 2014). Mutations in this gene are associated with a variety of different CMT subtypes including CMTDIG (dominant intermediate type G) (Züchner et al., 2004), CMT1F (Jordanova et al., 2003), and CMT2E (Georgiou et al., 2002).
CMTDIG is an intermediate, but variable, disease with phenotype onset typically before 20 years of age and a slow progression that results in a spectrum of severity from progressive gait instability, to patients becoming wheelchair bound (Züchner et al., 2004; Berciano et al., 2015). It has been speculated that the demyelinating symptoms observed in patients may actually be secondary to the axon loss (Züchner et al., 2004; Berciano et al., 2017). Patients with autosomal dominant CMT1F typically present between 1–13 years of age with reduced nerve conduction velocity, sensory defects, and pathology consistent with a demyelinating neuropathy (Jordanova et al., 2003; Abe et al., 2009; Yum et al., 2009). One group speculated that a missense mutation in NEFL can also cause a recessive form of CMT1F (Abe et al., 2009) which was later supported by a study using patient-specific derived neurons in culture (Sainio et al., 2018). Dominant CMT2E is typically slower and less severe with onset starting in the 20s or later and pathology showing signs of giant axons indicating an axonopathy (Georgiou et al., 2002; Fabrizi et al., 2004; Miltenberger-Miltenyi et al., 2007).
Cell models carrying CMT2E and CMT1F mutations show that transport and assembly of neurofilament proteins are disrupted causing aggregation (Brownlees et al., 2002; Gentil et al., 2013; Saporta et al., 2015), which may provide a common therapeutic target and is a common feature in many other neurodegenerative diseases, including ALS. The cause of the variability seen in NEFL patients is currently unknown, but one paper notes that mutations in the head domain may slow nerve conduction velocities more than mutations in other regions of the gene (Miltenberger-Miltenyi et al., 2007).
1.2. NEFH
The NEFH gene encodes the heavy chain neurofilament protein which is a major component of the cytoskeleton in large myelinated axons (Elder et al., 1998). A NEFH-null mouse model showed that NEFH protein is not crucial for development but it does plays a critical role in the survival of mature motor and sensory axons (Rao et al., 1998). Overexpression of NEFH results in degeneration of motor neurons (Collard et al., 1995). Mutations in this gene have been associated with both mild ALS and CMT2CC.
While rare, most ALS-associated mutations identified in NEFH change transcription patterns resulting in one long and one short transcript that appear to encode proteins that assemble normally with other neurofilament subunits but may have altered interactions with other cytoskeletal elements (Figlewicz et al., 1994; Tomkins et al., 1998; Al-Chalabi et al., 1999). The dominant mutations in CMT2CC patients produce a frameshift in the last exon of the gene that causes the polymerase to read through the stop codon and add additional amino acids including a cryptic amyloidogenic element (Rebelo et al., 2016; Jacquier et al., 2017). Studies have also shown that the inclusion of these additional elements make the mutant NEFH highly likely to form protein aggregates (Rebelo et al., 2016).
While the mechanism of each of the mutations is slightly different, NEFH protein aggregation occurs in the dominant forms of both NEFH–related ALS and CMT2CC. The age of symptom onset of CMT2CC patients varies highly ranging from the first few years of life to over 50 years old. Aside from heterogeneity between alleles, intrafamilial variability is also observed suggesting that this mutation has that ability to be modulated by genetic background (Rebelo et al., 2016).
1.3. DYNC1H1
The DYNC1H1 gene codes for dynein heavy chain which assembles with multiple other subunits to form a molecular motor that uses ATP retrograde transport down toward the microtubule minus-ends. In axons, microtubules have uniform “plus-end out” polarity, so dynein is primarily responsible for retrograde transport in this cellular compartment. Mutations in this gene have been associated dominantly with CMT2O (Weedon et al., 2011), lower extremity-predominant spinal muscular atrophy-1 (SMALED1) (Harms et al., 2010, 2012; Tsurusaki et al., 2012), and mental retardation 13 (MRD13) (Vissers et al., 2010). Mouse models have validated these associations showing signs of both a neuropathy (Hafezparast et al., 2003; Chen et al., 2007), as well as neuronal migration defects in the cortex (Ori-McKenney and Vallee, 2011).
Symptom onset for dominant CMT2O appears during childhood before progressing slowly with both motor and sensory impairment as well as indications of learning impairment (Weedon et al., 2011). Interestingly, most of the MRD13 patients also experienced CMT-like symptoms, early-onset MR, seizures, and brain scans of these patients showed cortical malformations, likely due to neuronal migration defects (Vissers et al., 2010; Willemsen et al., 2012; Poirier et al., 2013; Jamuar et al., 2014). SMALED1 is a dominant form of Spinal Muscular Atrophy (SMA) with clinical onset typically in early childhood with predominant paralysis in the lower extremities (Harms et al., 2010, 2012; Tsurusaki et al., 2012). While many patients exhibit severe early phenotypes, one patient’s symptoms were so mild that she was not genetically diagnosed until her children were diagnosed, suggesting that other genetic factors may contribute to susceptibility (Tsurusaki et al., 2012). New work has also shown that some SMALED1 patients may present with a congenital myopathy (Beecroft et al., 2017).
The overlap and variability in phenotype severity illustrate that this may really be a spectrum of overlapping diseases with the potential to be modified by other genetic factors (Lipka et al., 2015). The differences in severity related to DYNC1H1 have been mechanistically associated with both the methylation state of the gene at exon 37 (Maretina et al., 2019) and the change in processivity of dynein to move long distances through its binding to both dynactin and the BICD2 cargo adaptor (Hoang et al., 2017) with the latter possibly explaining the dominance seen in these diseases.
1.4. DCTN1
The DCTN1 gene produces the p150(Glued) protein and is the largest subunit of the dynactin complex which binds to both the dynein motor, as well as acting as a cargo adapter (Holzbaur and Tokito, 1996) and is essential for most dynein function (Gauthier et al., 2004; Schroer, 2013). It normally binds directly with both dynein and microtubules through its N-terminus. Additionally, it plays a role in retrograde transport of autophagosomes (Ikenaka et al., 2013). Mutations in this gene have been associated with Perry Syndrome (Perry et al., 1975; Farrer et al., 2009) and distal hereditary motor neuropathy type VIIB (dHMN7B) (Puls et al., 2003; Nam, 2016), as well as ALS (Munch et al., 2004). While these mutations typically result in impairment of axon transport, it is important to note that this is not always the case (Chevalier-Larsen et al., 2008; Lloyd et al., 2012).
Perry syndrome is the most well studied of the DCTN1-associate diseases. Patients experience an onset of depression later in life followed by Parkinson’s and eventually respiratory failure due to defects in brainstem respiratory neurons (Perry et al., 1975, 2012; Caroppo et al., 2014). There is also substantial neuron loss seen in the substantia nigra and TDP43 pathology (Wider et al., 2010). Mutations associated with Perry syndrome act in a dominant manner and all are located in exon 2 within the microtubule binding domain of the protein (Puls et al., 2003; Farrer et al., 2009). Cell-based assays show that the result of these mutations is decreased microtubule binding (Farrer et al., 2009). dHMN7B is an intermediate motor neuron disease with progressive distal paralysis of the face, hands, and vocal cords with weakness of the lower distal limb progressing later (Puls et al., 2003). Histology shows abnormal dynein-dynactin inclusions within the motor neurons (Puls et al., 2005). While the first dHMN7B mutation was identified in exon 2, others have since been identified in other domains of DCTN1 (Hwang et al., 2016; Konno et al., 2017). Symptoms of dHMN7B overlap with CMT, which may account for some patients being initially diagnosed with CMT (Nam, 2016). As for ALS, while there have been DCTN1 mutations identified in ALS patients, a larger study could not find any conclusive association between DCTN1 mutations and ALS, Parkinson’s disease, or frontotemporal lobar degeneration (Vilariño-Güell et al., 2009). The differences in pathology and affected cell types in both Perry syndrome and dHMN7B indicate that they may have different molecular mechanisms.
1.5. KIF5A
The KIF5A gene encodes Kinesin heavy chain isoform 5A which is a neuron-specific member of the kinesin protein family (Niclas et al., 1994). It is involved in trafficking of a variety of cargoes anterogradely within axons (toward microtubule plus-ends) (Okada et al., 1995; Kanai et al., 2004). Mutations in this gene have been associated with a dominant form of neonatal intractable myoclonus (NEIMY) (Duis et al., 2016), ALS (Nicolas et al., 2018), Spastic paraplegia-10 (SPG10) (Reid et al., 2002a), and CMT2 (Crimella et al., 2012).
NEIMY is a severe neurological disorder in which patients experience myoclonic seizures soon after birth, developmental delays, and mitochondrial dysfunction (DaRe et al., 2013; Duis et al., 2016). The mutations that cause NEIMY are all stop-loss mutations that produce a longer protein that is postulated to alter mitochondrial transport (DaRe et al., 2013; Duis et al., 2016; Rydzanicz et al., 2017). All of the dominant loss-of-function mutations identified in ALS patients were either located near exon 26/27 or the C-terminus and resulted in abnormal splicing (Brenner et al., 2018; Nicolas et al., 2018). SPG10 is a variable disease that classically occurs in childhood or early adulthood with spasticity and hyperreflexia of the lower limbs. Some patients also show involvement of upper limbs and sensory defects. In rare cases, there can also be cognitive decline or Parkinson-like symptoms (Goizet et al., 2009; Crimella et al., 2012). Dominant missense mutations in these patients seem to primarily occur in the motor domain of the gene (Reid et al., 2002b; Crimella et al., 2012). There are some SPG10 patients that present with distal sensory and motor defects in the absence of spasticity which is more a CMT2-like phenotype. There are also patients diagnosed with CMT2 that have KIF5A mutations in the motor domain (Goizet et al., 2009; Crimella et al., 2012; Liu et al., 2014; Nam et al., 2018).
SPG10 and CMT2 share not only overlapping phenotypes, but they also share some overlap in mutations (Nam et al., 2018). This is why it is believed that SPG10 and CMT2 patients represent a spectrum (Crimella et al., 2012; Nam et al., 2018) and indicate that the phenotype can potentially be modified. While NEIMY and KIF5A-associated ALS share different mechanisms of disease from each other and SPG10/CMT2, they do share genetic overlap which can make structure/function assignments difficult.
1.6. KIF1A
KIF1A encodes Kinesin family member 1A which is a kinesin heavy chain protein involved in anterograde transport of synaptic vesicles down microtubules in axons (Rivire et al., 2011). Mutations in this gene have been associated with hereditary sensory neuropathy type IIC (HSN2C) (Rivire et al., 2011), spastic paraplegia 30 (SPG30) (Erlich et al., 2011; Klebe et al., 2012; Pennings et al., 2019), mental retardation autosomal dominant 9 (MRD9) (Hamdan et al., 2011; Esmaeeli Nieh et al., 2015; Lee et al., 2015), and, very recently, Rett Syndrome (RTT) (Wang et al., 2019). At one point, KIF1A was also thought to be a candidate gene for juvenile-onset ALS4, but mapping studies disproved that theory (Keller et al., 1999).
HSN2C is a recessive sensory neuropathy resulting in distal numbness and weakness with a variable but typically young age of onset, and a subset of HSN2C patients also develop mental deficits more like MRD9 (Rivire et al., 2011). SPG30 has a slowly progressive phenotype onset in the first two decades of life and results in unsteady gait and over responsive reflexes (Erlich et al., 2011). Some patients also eventually develop sensory deficits, and SPG30 can be caused by either recessive or dominant loss-of-function mutations (Erlich et al., 2011; Klebe et al., 2012; Pennings et al., 2019). MRD9 is a dominantly inherited form of mental retardation with varying degrees of spasticity, hypotonia, peripheral neuropathy, and cerebellar atrophy (Hamdan et al., 2011; Esmaeeli Nieh et al., 2015; Lee et al., 2015). Recently, one patient with a classical RTT syndrome diagnosis was identified to be compound heterozygous for KIF1A mutations (Wang et al., 2019). She showed signs of delayed development, never achieved independent mobility or the ability speak, and developed classical repetitive behaviors, but more patient data and experiments are needed to see if this association between KIF1A and RTT is causative.
HSN2C patients tend to have truncating mutations where SPG30 patients tend to have missense mutations (Klebe et al., 2012). These missense mutations are commonly found in the motor domain of KIF1A for both forms of SPG30 and MRD9, but some disease associated mutations have been identified outside of this region (Krenn et al., 2017; Pennings et al., 2019). While some mutations slow the transport of synaptic vesicles, several point mutations associated with SPG30 have been shown to produce a hyperactive form of KIF1A (Chiba et al., 2019). While there is some overlap in phenotypes between the various KIF1A-related diseases, the fact that there are both gain and loss-of-function mutations as well as mutations that truncate or alter protein function by either slowing or speeding up transport indicates that the mechanism of disease may not entirely be shared other than altered axon transport and that these diseases are potentially not part of a spectrum.
1.7. SPG11
The SPG11 gene encodes the protein spatacsin. While the exact function of SPG11 is unknown, it has been shown to play a role in anterograde transport, axon maintenance, and lipid turnover (Pérez-Brangulí et al., 2014; Branchu et al., 2017). It has also been shown to localize to axons and dendrites (Branchu et al., 2017). Mutations in this gene have been recessively associated with CMT2X (Montecchiani et al., 2016), autosomal recessive juvenile amyotrophic lateral sclerosis-5 (ALS5) (Hentati et al., 1998; Orlacchio et al., 2010), and spastic paraplegia-11 (SPG11) (Nakamura et al., 1995; Stevanin et al., 2007; Paisan-Ruiz et al., 2008; Guidubaldi et al., 2011; Hinreiner et al., 2016).
CMT2X is a slowly progressive recessive CMT with onset in the first 20 years of life resulting from axonal loss in the distal lower limbs. Almost all of the identified mutations are predicted to produce a non-functional truncated protein (Montecchiani et al., 2016). ALS5 is a recessive and slowly progressive form of ALS with phenotype onset before age 25 due to loss of motor neurons. A majority of the identified mutations implicated in ALS5 are predicted to result in truncated proteins (Orlacchio et al., 2010). SPG11 is a recessive disorder with significant phenotypic variability characterized by spastic movement of the lower limbs, progressive weakness, and a thin corpus callosum (Nakamura et al., 1995). Many patients also experience the development of a sensorimotor peripheral neuropathy and cognitive impairment. SPG11-associated mutations cause a frameshift that is predicted to cause a loss-of-function (Stevanin et al., 2007). There is a strong case for these diseases being modifiable due to the observed phenotypic variability and overlap in clinical features while sharing many of the same mutations (Bauer et al., 2009; Orlacchio et al., 2010; Montecchiani et al., 2016).
1.8. FIG4
The FIG4 gene encodes the Sac3 phosphoinositide phosphatases protein which helps to cleave a phosphate group from phosphoinositides within lysosomal membranes to regulate fission and fusion of vesicles (Sbrissa et al., 2007). Data indicates that Sac3 may also regulate homeostasis of lysosomal membranes in a noncatalytic manner (Bharadwaj et al., 2016), and mutations in FIG4 result in dysfunction in lysosomal storage (Martyn and Li, 2013). Mutations have been associated with CMT4J (Chow et al., 2007; Nicholson et al., 2011), Yunis-Varon syndrome (Campeau et al., 2013; Nakajima et al., 2013; Wiessner et al., 2013), Bilateral Temporooccipital Polymicrogyria (BTOP) (Ouled Amar Ben Cheikh et al., 2009), and ALS (Chow et al., 2009; Osmanovic et al., 2017; Bertolin et al., 2018).
CMT4J is a highly variable recessive CMT with both axonal and demyelinating defects (Nicholson et al., 2011; Yan et al., 2012). Patient mutations are typically compound heterozygous carrying one null allele and one mutant allele (Chow et al., 2007; Nicholson et al., 2011). Patients with recessive Yunis-Varon syndrome have skeletal defects and neuronal loss that is typically lethal in infancy (Campeau et al., 2013). Patients were either homozygous or compound heterozygous for mutations that are predicted to result in a complete loss of protein function (Campeau et al., 2013). BTOP is a rare recessive disorder with a variable onset from childhood to 20’s resulting in seizures as well as psychiatric and behavioral problems. The homozygous mutation identified in these patients results in a partial loss-of-function (Baulac et al., 2014). Dominant ALS-associated mutations in FIG4 are missense, splicing, and truncating mutations that cause either significant or complete loss-of-function (Chow et al., 2009). Not all known missense mutations are pathogenic, and the association of FIG4 with ALS has begun to be questioned (Verdiani et al., 2013).
The wide range of disease severity in CMT4J and FIG4-associated ALS, combined with the fact that different severities have been seen in patients carrying the same mutation combination in CMT4J (Nicholson et al., 2011) and incomplete penetrance of ALS mutations (Osmanovic et al., 2017), indicates that there are likely genetic or environmental factors that modify the phenotype. There may be a case for CMT4J and FIG4-associated ALS being related diseases since they share mutations (Osmanovic et al., 2017), and lysosomal storage defects are a common feature of all of these disorders
2. Translation & tRNA Biogenesis
Dysregulation of protein synthesis is an emerging commonality in hereditary peripheral neuropathies and motor neuron diseases. The process of decoding mRNA into proteins must be tightly regulated for proper cellular function. Neurons and axons are particularly sensitive to deviations in this regulation, as local axonal translation machinery is limited. Variants in genes encoding for proteins involved in translation are associated with a number of neurodegenerative diseases, ranging from infantile-onset Spinal Muscular Atrophy with Respiratory Distress (SMARD) to CMT and ALS. Both autosomal dominant (AD) and recessive (AR) inheritance patterns have been noted, with different diseases represented by AD or AR inheritance patterns within the same gene. Highlighted here are genes encoding proteins that play a role in protein synthesis and are implicated in a broad spectrum of hereditary motor and sensory neuropathies and MNDs (Fig. 2b) (Table 1).
2.1. IGHMBP2
Autosomal recessive mutations in the Immunoglobulin H μ-binding protein 2 (IGHMBP2) gene result in two clinically distinct diseases: Spinal Muscular Atrophy with Respiratory Distress Type 1 (SMARD1) and Charcot-Marie-Tooth Disease Type 2S (CMT2S). IGHMBP2 encodes a ubiquitously expressed dsDNA/dsRNA helicase, found to be localized in the cytoplasm and associated with key translational machinery (Grohmann et al., 2004; De Planell-Saguer et al., 2009, Guenther et al., 2009b). IGHMBP2’s preferred substrate in biochemical assays is dsRNA, highly suggesting that IGHMBP2 plays a role in translation or tRNA processing, as it has also been associated with tRNA fragments (De Planell-Saguer et al., 2009, Guenther et al., 2009b).
Disease-associated Ighmbp2 variants were first identified in a mouse model (nmd2J) (Cox et al., 1998), while a locus containing IGHMBP2 in humans was correlated with patients with early infantile respiratory distress and distal motor impairments (Grohmann et al., 1999). In 2001, Grohmann et al, made the connection between IGHMBP2 mutations and patients of the motor neuron disease, SMARD1, while additionally characterizing the nmd2J mouse model as a SMARD1 model (Grohmann et al., 2001). More recently, Cottenie et al. identified compound heterozygous missense and truncating mutations that were causative of disease symptoms more clinically similar to CMT than SMARD1 (Cottenie et al., 2014; Wagner et al., 2015). Variants in IGHMBP2 have also been linked to cases of Sudden Infant Death Syndrome (SIDS) (Grohmann et al., 1999, 2003; Jackson et al., 2017).
The recessive nature of these diseases, as well as work examining the function and stability of variants reported, suggests that both SMARD1 and CMT2S diseases are caused by loss-of-function alleles (Guenther et al., 2009c, a). Allelic heterogeneity doesn’t solely account for the variability noted in patients, as patients with identical allelic combinations have exhibited phenotypic heterogeneity (Guenther et al., 2009c; Joseph et al., 2009; Hamilton et al., 2015). Some evidence points towards a possible correlation between mutant protein levels and disease severity, as patients with a SMARD1 diagnosis have lower protein levels, while CMT2S patients have reported higher levels (Guenther et al., 2009c). Phenotypic variability may also be driven by genetic modifiers. We have identified a modifier region in the nmd2J mouse model which attenuates the MND, but not a cardiac phenotype (Cox et al., 1998; Maddatu et al., 2005). Interestingly, the modifier region contains a number of tRNAs, strengthening the hypothesis that IGHMBP2 functions in translation or tRNA biogenesis (Maddatu et al., 2004).
2.2. CLP1
CLP1, or cleavage and polyadenylation factor I subunit one, encodes for a kinase involved in tRNA splicing and associated with the tRNA splicing endonuclease (TSEN) complex (Vries et al., 2000; Paushkin et al., 2004). The role of TSEN and CLP1 is to edit out the intron that is present within the anticodon loop of select tRNAs (Weitzer and Martinez, 2007; Hanada et al., 2013). This cleavage event generates 5’ and 3’ tRNA exon halves. CLP1 phosphorylates the 3’ tRNA half, a prerequisite for ligation of the two exonic halves, and the production of a mature tRNA (Weitzer and Martinez, 2007). Editing out the intron of tRNAs is critical for translation, as some tRNA families have only intron-containing genes (van Tol and Beier, 1988).
tRNA processing has been found to be particularly sensitive for cerebellar development, as mutations in human CLP1 are associated with the recessively-inherited Pontocerebellar Hypoplasia Type 10 (PCH10) (Karaca et al., 2014; Schaffer et al., 2014; Wafik et al., 2018). PCH10 is characterized by progressive microcephaly, spasticity, and brain atrophy and delayed myelination (Karaca et al., 2014; Schaffer et al., 2014; Wafik et al., 2018). Additionally, a subset of patients present with axonal sensorimotor neuropathy, similar to CMT2 diseases (Karaca et al., 2014). A homozygous missense variant (p.R140H) identified in these families was found to be in a conserved region of the protein, and impaired in its interaction with members of the TSEN complex (Karaca et al., 2014; Schaffer et al., 2014). Changes in tRNA pre-processing were identified in patient neurons and found to have increased sensitivity to oxidative stress-induced death. These changes in tRNA pre-processing and depletion of mature tRNAs could certainly impair the overall translation ability in motor neurons, increasing stalling due to the depletion of available charged tRNAs (Gorochowski et al., 2015a). To further support the importance of CLP1 to neuron viability, CLP1 kinase-dead mice exhibited neo-natal lethality with impaired innervation of the diaphragm leading to respiratory failure, as well as a loss of motor neurons in the cervical and lumbar spinal cord, decreased level of processed tRNAs, and a sensitivity to oxidative stress (Hanada et al., 2013). Like the Ighmbp2 mouse model, CLP1 kinase-dead mutants were severe on the C57BL/6 background, but were found to be less affected on a CBA/J background, suggesting that some modifier locus exists decreasing motor neuron susceptibility to the Clp1 mutations (Hanada et al., 2013). Such differences in genetic background could explain why patients carrying the same homozygous missense mutation (p.R140H) in two different families presented with heterogeneous phenotypes (Karaca et al., 2014; Schaffer et al., 2014).
2.3. GARS.
Aminoacyl-tRNA synthetases (ARSs) are a group of enzymes that catalyze the association of amino acids to the end of their cognate tRNAs. ARSs play a critical role in the interpretation of the genetic code by associating the anticodon sequence of the tRNA to the specific amino acid. Here we discuss the ARS genes that are associated with autosomal dominantly inherited CMTs and distal hereditary motor neuropathies (dHMNs). It should be noted that many of these ARS genes are also associated with multi-systemic autosomal recessive diseases which are not discussed here (Boczonadi et al., 2018).
GARS encodes for both the cytoplasmic and mitochondrial isoforms of glycyl-tRNA synthetases, ensuring that the appropriate tRNAs are charged with glycine. Dominant missense mutations in GARS were first identified in 2003 in families with CMT2D and distal hereditary motor neuropathy type VA (HMN5A) (Antonellis et al., 2003). This has since been supported by a number of familial and individual cases associated with dominant missense variants; however to date no null alleles have been observed (Christodoulou et al., 1995; Ionasescu et al., 1996; Sambuughin et al., 1998; Sivakumar et al., 2005; Del Bo et al., 2006; James et al., 2006; Liao et al., 2015; Sun et al., 2017). The GARS phenotypic spectrum has tentatively been further expanded from inherited peripheral neuropathies (IPNs) to more classical MNDs as a new case study suggests an association with ALS. A patient had developed a late-onset bulbar ALS phenotype and had a familial history of ALS (Corcia et al., 2018). Targeted next-generation sequencing identified a likely pathogenic variant (p.V665L) in a highly conserved region of the anti-codon binding domain of GARS and found no other mutations in known ALS-associated genes (Corcia et al., 2018). Further work determining the pathogenicity of the ALS-associated variant, as well as identifying other cases with GARS variants in bulbar or classical ALS will be informative to strengthen this connection. Overall, these findings indicate that dominant human variants in GARS lead to a spectrum of IPNs such as CMT2D and HMN5A, as well as the potential for later-onset MNDs.
Research determining the pathomechanisms of disease-causing variants has suggested that many are a combination of loss-of-function as well as dominant toxic gain-of-function. Work has suggested that some variants can impair enzymatic activity of GARS, cause localization defects, impair protein translation, and disrupt dimerization through loss-of-function mechanisms (Antonellis et al., 2006; Chihara et al., 2007; Nangle et al., 2007; Griffin et al., 2014; Malissovas et al., 2016). While disruption of the canonical activity and dimerization of GARS has been associated with some disease-associated variants, there is a distinct lack of correlation between canonical loss-of-function and all disease-causing variants. Evidence for a toxic gain-of-function pathomechanism in many of the disease-causing variants has been presented. Work in both mouse and Drosophila models have found that potential neomorphic activity may impair protein translation in motor and sensory neurons, a phenotype which cannot be rescued with overexpression of wildtype protein (Seburn et al., 2006; Motley et al., 2011; He et al., 2015; Niehues et al., 2015). Oprescu et al. highlight a patient with compound heterozygous GARS variants, one which includes a frameshifting putative null allele. The maternal carrier of this null allele was not found to have a CMT2D or HMN5A-like phenotype, further suggesting that haploinsufficiency is not a mechanism of GARS-associated variants. It is plausible that both LOF and GOF variants exist and drive similar phenotypes; however the lack of nonsense and putative null variants in patients with dominant CMT2D and HMN5A, as well as the finding that heterozygous null mice have no discernible phenotype, suggests that a toxic gain-of -function is likely at play.
Such a gain-of-function is likely due to conformational changes of GARS by the disease causing variants, which create novel structures that permit neomorphic binding. Targets for these sites have been identified in a number of studies; observations indicate that mutant GARS can aberrantly associate with Trk receptors, Nrp1, and HDAC6 (He et al., 2015; Sleigh et al., 2017; Mo et al., 2018). Mutant GARS has been found to antagonize the motor neuron VEGF-Nrp1 interaction, which has been implicated in neuronal migration and axonal guidance, however this does not explain why peripheral nerves selectively degenerate (He et al., 2015). Furthermore, mutant GARS aberrantly interact with Trk receptors causing mis-activation of Trk signaling, a process that has proven essential for sensory neuron differentiation and development (Sleigh et al., 2017). This interaction is increased in two of the CMT2D mouse models (GARSC201R/+ and GARSNmf249/+), both of which have sensory involvement. However, what was not tested were variants associated with HMN5A, which may not show spurious mis-activation of Trk. This may be a plausible explanation for the phenotypic heterogeneity of sensory involvement. However, certain variants (p. Ile280Phe and p.Leu129Pro) have been found to segregate with both CMT2D and HMN5A, suggesting that neomorphic Trk receptor binding may not account for all sensory heterogeneity (Sambuughin et al., 1998; James et al., 2006; Liao et al., 2015). Most recently, certain mutant forms of GARS have been shown to aberrantly associate with HDAC6, which impairs α-tubulin acetylation, causing peripheral nerve axonal transport deficits pre-disease onset (Mo et al., 2018). These findings suggest a common, but potentially specific pathomechansim among GARS alleles, by creating stable alternative conformations, which aberrantly interact with various proteins or receptors, causing a cascade of disruption that impacts axonal homeostasis.
Delineating why certain patients present as HMN5A, CMT2D, or ALS, has been difficult as some families present with multiple disease phenotypes (Sivakumar et al., 2005; Del Bo et al., 2006; Corcia et al., 2018). With no apparent allelic segregation, as previously mentioned with the neomorphic Trk receptor binding, it is possible that background genetics increase or decrease susceptibility to certain genetic variants (Sambuughin et al., 1998; Liao et al., 2015). In a CMT2D mouse model, modulating the background strain did alter disease progression and severity (Seburn et al., 2006; He et al., 2015). Likewise, genetic modifiers of disease have been identified in GARS Drosophila models (Ermanoska et al., 2014).
2.4. KARS
KARS, like GARS, is bifunctional in that it encodes for the cytoplasmic and mitochondrial isoforms of the lysyl-tRNA synthetase. However, unlike GARS, only recessive variants have been associated with KARS. The first to be identified was missense variant (p.Leu133His) that significantly impaired enzymatic activity (McLaughlin et al., 2010). This patient presented with an intermediate CMT with axonal and myelination involvement, as well as developmental delay and vestibular Schwannoma. Since this patient diagnosed with CMTRIB, no other KARS-associated CMT patients have been reported. However, there have been multiple reports of patients with neurodegenerative symptoms like leukoencephalopathy, microcephaly, and sensorineural hearing impairment, without peripheral nerve involvement (Santos-Cortez et al., 2013; McMillan et al., 2015; Zhou et al., 2017; Ardissone et al., 2018; Scheidecker et al., 2019).
No models exist to study KARS variants, however the international mouse phenotyping consortium (IMPC) found that the knockout of the mouse KARS is homozygous-lethal, as no homozygous mice were observed post-wean, which is expected as lysine would be rendered useless in protein synthesis if left uncharged (Dickinson et al., 2016).
2.5. AARS
Unlike the aforementioned GARS and KARS, AARS only encodes for the cytosolic isoform of alanyl-tRNA synthetase. Dominant and recessive diseases have both been associated with AARS mutations and variants have been identified in all domains. Phenotypic heterogeneity is noted with some families presenting as CMT2N, characterized by distal sensorimotor neuropathy, while other families present with dHMN, lacking sensory involvement (Latour et al., 2010; Lin et al., 2011; McLaughlin et al., 2012; Zhao et al., 2012; Bansagi et al., 2015; Motley et al., 2015; Weterman et al., 2018). Research regarding the pathomechanisms of the predominantly missense variants has found impaired amino-acetylation and failed complementation in yeast studies (McLaughlin et al., 2012; Motley et al., 2015; Weterman et al., 2018). However, there may be evidence not only for hypomorphic alleles, but also hypermorphic as Weterman et al., 2018, found that p.Glu337Lys increases tRNA charging velocity.
Animal models of AARS variants confirm neural abnormalities in the zebrafish as well as a mouse model, which has degeneration of the cerebellar Purkinje cells (Lee et al., 2006; Weterman et al., 2018). The AARS mouse model has a point mutation in the editing domain of AARS, which results in an increase in the mischarging of ser-tRNAAla. More recent work has found that ANKRD16 is a genetic modifier of the AARS mutant phenotype, as it prevents the charging of serine adenylates to tRNAAla and is a necessary co-regulator of tRNA synthetase editing; such deletion causes widespread protein aggregation and neuron loss (Vo et al., 2018).
2.6. MARS
MARS only encodes for the cytosolic isoform of methionyl-tRNA synthetase and, like other ARSs, has been implicated in a spectrum of dominant neurodegenerative diseases. To further expand the spectrum, dominant variants in MARS have been associated with a late-onset CMT type 2U. (Gonzalez et al., 2013; Hyun et al., 2014; Hirano et al., 2016; Sagi-Dain et al., 2018). Presentation associated with these dominant variants has been heterogeneous, with some families exhibiting incomplete penetrance, and others presenting far earlier than previously reported (Gonzalez et al., 2013; Sagi-Dain et al., 2018). Additionally, research regarding hereditary spastic paraplegia (HSP) candidate genes suggests that MARS variants may be causative of HSP, an upper motor neuron disease (Novarino et al., 2014). Future work to better understand the wide spectrum of disease onset and severity, particularly in dominant variant carriers is needed. Addressing if carriers of loss-of-function alleles, such as parents of patients with recessive interstitial lung and liver disease, have a sub-clinical CMT, or perhaps are not yet symptomatic of the later-onset (6–7th decade) CMT2U, will need to be further explored. Additionally, the potential for modifiers that modulate disease susceptibility also needs to be explored; as such mechanisms have begun to be identified in other ARS-associated diseases.
2.7. HARS
HARS, like previously noted ARSs, encodes for the cytosolic histidyl-tRNA synthetase and has been found to be associated with both dominant and recessive disease. As noted in other ARSs, the CMT-associated form of disease is dominantly inherited.
CMT2W, a dominantly inherited disease, has also been linked to the HARS gene (Vester et al., 2013; Safka Brozkova et al., 2015; Abbott et al., 2018; Royer-Bertrand et al., 2019). Presentation of CMT2W is heterogeneous, with some subsets of HARS variants presenting as pure motor axonal neuropathy (Safka Brozkova et al., 2015). One particularly interesting case found a late-onset demyelinating peripheral neuropathy, comorbid with cerebellar atrophy and cognitive deficit, suggesting that not only the PNS may be sensitive to changes in HARS, but also the CNS (Royer-Bertrand et al., 2019). Many of the CMT2W-causing variants have been studied to determine the functional impact to HARS and it has been found that they are able to form stable dimers, but had reduced overall aminoacylation (Vester et al., 2013; Safka Brozkova et al., 2015; Abbott et al., 2018; Royer-Bertrand et al., 2019). While the enzymatic activity of catalyzing the ligation of histidine to its cognate tRNA is reduced, this impact was not shown to reduce overall protein synthesis, as measured by puromycin click-chemistry assay (Royer-Bertrand et al., 2019). While the global translation level is not specifically impacted, it is plausible that a specific subset of proteins critical to axonal maintenance is impaired.
2.8. YARS
YARS gene encodes for the cytosolic tyrosyl-tRNA synthetase. To date a number of cases have associated YARS variants with DI-CMT (Jordanova et al., 2006; Lin et al., 2011; Hyun et al., 2014). Like many of the other CMT-associated ARSs genes, reduction in aminoacylation was observed in mutant proteins (Jordanova et al., 2006). Work in a Drosophila model expressing mutant YARS found progressive deficits in motor performance and axonal degeneration, with the neuron-specific expression inducing the phenotype, suggesting a cell-autonomous effect (Storkebaum et al., 2009). Furthermore, they found these phenotypes are not due to haploinsufficiency of aminoacylation activity but instead are likely due to a dominant-negative or gain-of-function, as seen in the disrupted localization at the axonal termini (Jordanova et al., 2006; Storkebaum et al., 2009). More recent work identified that mutant GARS and YARS-expressing Drosophila models had similar modifiers for the rough eye phenotype, with some modifiers impacting both ARSs, suggesting common pathomechanisms (Ermanoska et al., 2014). Interestingly, the modifiers were specific to the YARS and GARS mutant models, as models of other neurodegenerative disease Gl-Tau and Gl1 (human ortholog of Dynactin 1) were not modified. The enhancement of the rough eye phenotype (mutant-specific enhancers) was only found in the models of YARS/GARS, suggesting a specific interaction of these proteins with the mutant form rather than additive effects unrelated to the disease pathomechanism.
Commonalities between YARS-related toxicity and GARS-related toxicity may address common pathological findings. Such example is the finding that many disease-associated GARS mutations cause stable alternative conformations, YARS variants have also been found to encode proteins with stable and potentially toxic conformations (Blocquel et al., 2017). While disease-causing variants disrupt the conformation of these proteins, as a common mechanism, what these novel stable conformations bind to may be allele specific. Future work assessing other AARs variants for neomorphic binding partners, as a common pathomechanism in GARS and YARS suggest, could identify easily targetable biological pathways as therapeutic avenues.
3. Ribosome-Associated Protein Quality Control
The Ribosome-associated Quality Control (RQC) pathway responds to ribosomes that stall during translation elongation, facilitating resolution by degrading potentially toxic proteins. Ribosomes can stall during translation for a myriad of reasons; a lack of termination codon (nonstop mRNA), increased mRNA secondary structure, or the deficiency of charged tRNAs (no-go mRNA) (Wallen and Antonellis, 2013, Gorochowski et al., 2015b; Simms et al., 2017; Schuller and Green, 2018). Failure to appropriately respond to a stalled ribosomal complex can deplete the translational machinery, a problem that can be particularly apparent in neurons where machinery is extremely limited and translational demand is elevated. Another detrimental effect of improper RQC is the accumulation of misfolded protein products, which could be potentially toxic. Highlighted here are genes encoding for proteins that play essential roles throughout the RQC pathway and are involved in a range of movement and cognitive neurodegenerative diseases (Fig. 2c) (Table 1).
3.1. GTPBP2
GTP-binding protein 2 (GTPBP2), and its yeast paralogue of Hbs1, function in ribosomal rescue upstream of RQC. Ribosomes that are stalled at the mRNA 3’ end are sensed by GTPBP2 and its binding partner, pelota (PELO; yeast Dom34). Work in the yeast model system has shown that Hbs1 with Dom34 play a role in non-canonical translation termination and recycling of stalled ribosomes in the 3’ region (Doma and Parker, 2006; Shoemaker et al., 2010; Pisareva et al., 2011).
Variants in GTPBP2 have recently been reported to cause the autosomal recessive Jaberi-Elahi syndrome (Jaberi et al., 2015; Bertoli-Avella et al., 2018; Carter et al., 2019). This neurodegenerative syndrome is early-onset and characterized by the presentation of dystonia, motor and sensory neuropathy, ataxia, cognitive dysfunction, and in some cases brain iron accumulation. It is likely that these variants are hypomorphic loss-of-function alleles, as all reported patients have nonsense or splicing variants leading to a putative null state. While mutations in GTPBP2 do not lead exclusively to CMT diseases, many of the patients exhibit progressive peripheral axonal degeneration and demyelination, suggesting pathomechanistic overlap between CMTs and more global neurodegenerative disorders.
Further insight into the pathomechanisms of GTPBP2 variants has come from mouse model work. A homozygous splice site mutation in mouse Gtpbp2 causes neurodegeneration accompanied with truncal ataxia symptoms, apoptosis of inner granular layer and degeneration of retinal neurons, which are only observable on the C57BL/6J background (Ishimura et al., 2014). Unique to the C57BL/6J strain is a mutation in tRNA-Arg-UCU, an isodecoder that is predominantly expressed in brain tissue, but is not critical in other tissue types. Correction of this tRNA mutation, by utilizing a different genetic background, mitigates the neurodegenerative phenotype. The presence of hypomorphic GTPBP2 as well as an imbalance in tRNA abundance increased ribosomal pausing in the cerebellum. These findings suggest that loss-of-function mutations in GTPBP2 are causative of neurodegenerative disease and can be highly modifiable if ribosomal stalling is decreased. More recent work with this model indicates that the kinase GCN2 is activated in the mutant brain, suggesting that activation of the integrated stress response is a regulatory and neuroprotective mechanism (Ishimura et al., 2016).
3.2. NEMF
NEMF (nuclear export mediator factor) was first identified as serologically defined colon cancer antigen 1 gene (SDCCAG1) (Carbonnelle et al., 1999). Work done in the Drosophila model found that the NEMF orthologue, Caliban, mediates nuclear export of fusion proteins, and is critical for neural differentiation and astrocyte development (Bi et al., 2005). However, since then much of the work regarding understanding the function of NEMF has been performed in the yeast model system. Rqc2 (the yeast NEMF orthologue) works in concert with its cofactor Ltn1 (mammalian Listerin). Rqc2 senses and binds the obstructed 60S subunit and the still-attached charged tRNA. After which, Rqc2 performs two critical roles: the first is to recruit and stabilize the binding of its cofactor Ltn1/Listerin to the stalled 60S subunit; the second is to catalyze the C-terminal elongation of the nascent chain with untemplated alanine and threonine tails (CAT tails) (Bengtson and Joazeiro, 2010; Lyumkis et al., 2014; Shao et al., 2015; Shen et al., 2015). The biological role of CAT-tails still remains largely unknown; however, it is thought that CAT-tails play a role in the activation of stress-response pathways to signal the slowing of translation and/or to increase the likelihood that a lysine substrate within the stalled nascent peptide is available for Ltn1 to ubiquitinate (Choe et al., 2016; Frost et al., 2017).
While not much is known about the role of NEMF in mammalian cells, the neuroprotective nature of GTPBP2 would suggest that aberrations in NEMF may also result in neurodegenerative diseases. One study has found NEMF to be a candidate gene for intellectual disability (Anazi et al., 2017). This study identified two families with independent homozygous truncating variants in NEMF. The patients, two sets of siblings from independent families, presented with global developmental delay, intellectual disability, microcephaly, and hypotonia. While it remains unknown if the hypotonia and global developmental delays are neurogenic in nature, the phenotypic overlap between these cases and the patients with variants in GTPBP2 suggest that deficits in RQC can lead to a spectrum of neurodegenerative diseases, including IPNs and ID.
3.3. LTN1
Listerin (LTN1) is an E3 ubiquitin ligase that works in the RQC pathway (Bengtson and Joazeiro, 2010). Ltn1 then ubiquitinates the polypeptide to promote the recruitment of the potentially toxic nascent chain to the proteasome for degradation (Lyumkis et al., 2014; Defenouillère et al., 2016). Failure to ubiquitinate lysine in the nascent chain can cause protein aggregation and signaling of stress-response (Brandman et al., 2012).
While no human diseases are currently associated with mutations in LTN1, a promising mouse model highlights the impact of hypomorphic variants in Ltn1, implicated in peripheral neuropathies and motor neuron diseases (Chu et al., 2009). A splice mutation causes a hypomorphic allele giving rise to a motor neuron disease, while a complete null was found to be embryonic lethal. These findings further support that neurons are vulnerable to disruption in RQC pathways and may suggest that a spectrum of human peripheral and motor neuron disease is associated with pathogenic variants in LTN1.
3.4. VCP
Valosin-containing protein (VCP) is a member of the AAA-ATPase superfamily and plays a role in several cell protein pathways, including protein degradation through autophagic and proteasomal pathways (Hirabayashi et al., 2001; Walther et al., 2001; Rabinovich et al., 2002; Brandman et al., 2012; Defenouillere et al., 2013; van den Boom and Meyer, 2018). In the RQC pathway, VCP is recruited to extract and deliver the ubiquitinated nascent chain to the proteasome for degradation (Defenouillere et al., 2013; Verma et al., 2013). Clearance of the stalled complex is necessary for recycling the ribosome back into the translational pool. Defective ribosome-associated degradation could contribute to the pathophysiology of diseases implicated by variants in VCP.
Variants in VCP are implicated in a spectrum of neurodegenerative and myopathic diseases, predominantly comprised of the autosomal dominantly-inherited inclusion body myopathy with early-onset Paget disease and frontotemporal dementia (IBMPFD), as well as a few cases of sporadic and familial ALS, CMT2, Parkinson’s disease, and HSP. VCP-associated IMBPFD has been found to be highly heterogeneous both intra-familial as well as inter-familial (Haubenberger et al., 2005; van der Zee et al., 2009; Chan et al., 2012; Abrahao et al., 2016). VCP variants alone cannot account for the broad heterogeneity as many of the patients carry p.Arg159His and presentations include FTD alone, IBM with PBD, or any combination of the three (Haubenberger et al., 2005; van der Zee et al., 2009). Further still, this same variant has also been associated with a peripheral neuropathy phenotype, as well as a familial ALS patient, with a strong family history of FTD (Koppers et al., 2012; Segers et al., 2014). Phenotypic overlap is observed in IMBPFD and ALS-FTD, as both can be characterized by TDP-43 positive ubiquitinated inclusions in the muscle and frontal cortex neurons (Cairns et al., 2007; Neumann et al., 2007; Weihl et al., 2008). A review of the genetic spectrum of familial ALS (fALS) found that mutations in VCP may account for 1–2% of all cases, and other work has identified VCP variants associated with a sporadic ALS (sALS) patient (DeJesus-Hernandez et al., 2011; Johnson et al., 2011). To further broaden the phenotypic spectrum, cases of CMT2 and peripheral sensorimotor neuropathy, as well as hereditary spastic paraplegia (HSP) and Parkinson’s Disease have all been associated with variants in VCP (De Bot et al., 2012; Gonzalez et al., 2014; Al-Obeidi et al., 2018).
Pathogenic VCP mutations have been suggested to preferentially disrupt autophagy, leading to cellular degeneration and an accumulation of immature autophagosomes (Ju et al., 2009; Gonzalez et al., 2014). Pathogenic variants were also found to disrupt VCP hexamer formation leading to increased stress-induced toxicity in the Drosophila model (Wang et al., 2016). Mice expressing the p.Arg155His variant exhibited pathology in muscle, brain and bone, recapitulating IBMPFD (Custer et al., 2010). Work from Custer et al., also found that in in vitro studies mutant VCP caused aberrant activation of the NF-kB signaling cascade, which could contribute to the mechanism of pathogenesis in multiple tissues including muscle, bone, and brain. Further studies with the VCP-R155H mouse observed pathology consistent with ALS phenotypes: age-dependent degeneration of the ventral horn motor neurons, TDP-43 positive inclusions, and progressive astrogliosis (Yin et al., 2012). While no patients homozygous for VCP variants have been identified, a homozygous knock-in of the p.Arg155His variant found an earlier-onset, more severe IBMPFD phenotype (Nalbandian et al., 2012). This may suggests that the spectrum of VCP patients may include recessive patients that present at a much earlier time, further expanding the spectrum of overlapping neurodegenerative diseases.
4. Cell Stress Response
Heat shock proteins (HSPs) act as chaperones and monitor for misfolded proteins caused by stressors such as temperature change and UV light. When they detect a misfolded protein, they can either refold the protein into its appropriate tertiary structure or facilitate it towards a proteasome for degradation, promoting cell survival during stressful events (Bakthisaran et al., 2015). If overexpressed, HSPB1, HSPB3, and DNAJB2 have been shown to be neuroprotective in cell cultures modeling Huntington’s, Parkinson’s, amyotrophic lateral sclerosis (ALS), and other neurological diseases associated with deleterious protein folding and aggregation (Carra et al., 2005; Westhoff et al., 2005; Borrell-Pages et al., 2006; Blumen et al., 2012; La Padula et al., 2016; Zarouchlioti et al., 2017). However, when these chaperone proteins are no longer able to properly function proteins aggregate (James and Talbot, 2006; Sanchez et al., 2016). Bound proteins stuck in the aggregates, including HSPs and other homeostatic proteins, are no longer be able to perform their duties. This causes a cascade of other issues to arise, disrupting cell transport, cytoskeletal structure, and ultimately cell death especially for cells that undergo a lot of stress and translation. Neurons are gravely affected when a mutation occurs in fundamental heat shock proteins such as HSPB1, HSPB3, HSPB8, and DNAJB2. The arrest of the homeostatic duties of the heat shock proteins and the number of other processes interrupted by mutations in these genes cause the neurons to degenerate. Though, most mutations in these proteins cause motor neuron death exclusively, it has been seen to sometimes cause sensory neuropathy. Most commonly, mutations in these four genes are associated with an onset of distal motor neuron degeneration. However, in certain cases, both a motor and sensory neuropathy can occur causing an axonal Charcot-Marie-Tooth disorder (Fig. 2d) (Table 1).
4.1. HSPB1
Variants in HSPB1 have been found to have phenotypic heterogeneity and disease overlap, which allele-type alone cannot account for. The S135F variant, found in the α-crystalline domain, was found to cause both dHMN, as well as CMT type 2F (Evgrafov et al., 2004; Chung et al., 2008). Other variants in the same domain also presented as CMT2F or dHMN, but onsets ranged wildly (Kijima et al., 2005; Tang et al., 2005; Houlden et al., 2008). While many of these variants were autosomal dominant dHMN or CMT2F causing, there have been reports of autosomal recessive inheritance of dHMN/CMT2 (Houlden et al., 2008). Phenotypic heterogeneity, especially between patients with the same variant is suggestive that genetic and/or environmental background may impact the severity of the mutation with potential modifiers perhaps mitigating or enhancing the impact of the mutation.
Two transgenic mice have been created based on the human S135F and P182L mutations (D’Ydewalle et al., 2011). These mice had neuron-specific expression of the wildtype or mutated HSPB1 and displayed signs of neuropathy, with differences in onset, severity, and sensory involvement based on allele. The researchers found that these models displayed a decreased α-tubulin acetylation and utilizing pharmacological inhibition of histone deacetylase 6 (HDAC6), mitigated the CMT-like phenotype displayed in the models. The inhibition of HDAC6 has also proven beneficial in motor neurons derived from ALS patients with fused in sarcoma (FUS) mutations (Guo et al., 2017) and in HSPB1-induced CMT2 mouse models in pharmacological trials (Benoy et al., 2017).
A single copy of a mutant HSPB1 expressed specifically in the nerves caused CMT-like phenotypes, suggesting a toxic-gain-of-function (D’Ydewalle et al., 2011). Despite being ubiquitously expressed, HSPB1 seems to have a large impact on peripheral motor and sensory nerves. Further studies assessing the CMT/dHMN variable p.S135F mutation in different mouse backgrounds still need to be conducted in order to determine if the severity and sensory nerve loss can be increased or mitigated by possible mouse strain-specific modifiers.
4.2. HSPB8
Very similarly to HSP1, most mutations are autosomal dominant and found in the α-crystallin domain. Three known mutations all occur at a mutational hot spot, lysine 141, in five independent families. Four of these families that carried the same variant K141N displayed distal Hereditary Motor Neuropathy type 2A (dHMN2A) with presentation in the second and third decades of life and rarely a later onset sensory loss (Timmerman et al., 1992b; Irobi et al., 2004). However a Chinese family, also with p.K141N but a different nucleotide change of c.423G>T, displayed a CMT2L phenotype (Tang et al., 2005). Additionally, at this mutational hot spot, a p.K141T mutation had a CMT2L presentation, while p.K141E presented as dHMN2A (Irobi et al., 2004; Nakhro et al., 2013). Work understanding the cellular impact of variants at this site suggests that HSPB8 has greater binding to its interacting partner HSPB1, promoting formation of intracellular aggregates (Irobi et al., 2004).
In 2018, Bouhy et al., found that HSPB8-null mice have comparable locomotor abilities as wildtype mice, while homozygous HSPB8- K141N mice have impaired locomotion and strength (Bouhy et al., 2018). Interestingly, heterozygous-K141N mice do not show impaired locomotor activity or axon loss, unlike the human patients who are carriers. Similar to the in vivo cellular findings from Irobi et al., mutant mice show increased protein aggregations of myofibrils (Bouhy et al., 2018). This suggests that the mutated HSPB8 takes on a dose responsive gain-of-function with two copies of the K141N HSPB8 allele being necessary to show a phenotype.
Interestingly, human patients have also been found to have myopathies without the nerve degeneration aspect. Patients from independent families presented with the same heterozygous two base-pair deletion in the HSPB8 (c.508_509delCA) and late-onset muscle degeneration leading to impaired locomotor abilities (Echaniz-Laguna et al., 2017). This may suggest an even broader disease spectrum related to HSPB8, depending on variant type.
4.3. HSPB3
Mutations in HSPB3 are not as common as the other aforementioned genes. Kolb et al. in 2010 studied candidate HSP genes in patients presenting with peripheral motor neuropathies with or without sensory involvement that did not have mutations in known neuropathy-associated genes, such as IGHMBP2. They found a heterozygous mutation in HSPB3 in 2 siblings presenting with motor axonal degeneration in distal limbs starting in their 20s. This motor neuropathy was restricted to their legs until about 30 years later when their arms started to become affected as well. Interestingly, the mutation found was not in the α-crystallin region, but in the N-terminal domain. The autosomal dominant mutation, R7S, was found in a highly conserved residue of HSPB3 (Kolb et al., 2010).
More recently, the disease spectrum of HSPB3 has expanded, as a dominant CMT2 has been associated with p.Y118H. These patients presented similarly to the previously reported distal HMN, however also had vibration sensory loss and pes cavus.
4.4. DNAJB2
DNAJB2, or HSJ1, is not as closely related to HSPB1, HSPB3, and HSPB8 as they are with each other, but still retains the same roles as the other heat shock proteins of maintaining homeostasis of proteins despite stressors. What makes DNAJB2 unique from HSPB1, HSPB3, and HSPB8 is the J domain which allows DNAJB2 to interact with Hsp70 and assist in resolving protein mis-folding. DNAJB2 is found to protect neurons in vitro from polyQ induced cell death (Borrell-Pages et al., 2006) as well as reduce aggregation of mutant SOD1 in motor neuron like cells in vitro (Blumen et al., 2012)). This participation of DNAJB2 in the resolution of protein aggregates associated with well-known neurodegenerative diseases highlights the importance of this chaperone in mitigating potentially neurotoxic peptides.
The HSPB mutations are predominantly dominant diseases, while the mutations associated with DNAJB2 have been found to be recessive and typically cause splicing defects. Not only are mutations in DNAJB2 associated with CMT and dHMN, but SMA as well. A splicing mutation in DNAJB2 gave rise to a dHMN phenotype, while a missense gave rise to a more CMT2-like, both however presented in their second-third decade (Gess et al., 2014). One family had a large 3.8kb deletion that removed the J domain associated with an SMA diagnosis; one individual also displayed juvenile Parkinsonism with symptoms of not only muscular atrophy, but tremor, and hypokinesia (Sanchez et al., 2016).
The large 3.8kb deletion is not the only allele to show Parkinson’s-like symptoms. One Brazilian family with a splicing mutation shows significant intrafamilial heterogeneity as individuals have presented with cerebellar ataxia, Parkinson’s disease or dHMN. Genetic testing searching for mutations in typical genes associated with parkinsonism, ataxias, and SMA yielded negative results, while whole exome sequencing revealed the 352+1G>A mutation in DNAJB2 (Teive et al., 2016).
5. Conclusions & Perspectives
Unlike Charcot’s anatomoclinical approach, today’s clinicians may turn to genetic screening as the first clinical assessment, particularly with the “1000 dollar” genome nearly in sight. One key to efficiently utilizing a clinicogenetic approach will be determining which variants are pathogenic and which are benign. The current gold standard for determining this includes a multi-faceted approach utilizing population genetics, new powerful modelling and prediction tools as well as functional and segregation data. Furthering the genetic and clinical findings of variants will aid in appropriately interpreting a variant’s potential. Early assessment of the underlying genetic contributions of a given disease is critical as patients, such as those with SMARD, have a very limited window to receive a given therapy prior to irreversible motor neuron death. Therapeutic approaches, like promising gene therapies, are most efficacious when administered at the earliest point. Improved knowledge of the clinical neurogenetics and the spectrum and overlap of inherited axonopathies, MNDs, and other neurodegenerative diseases will aid in the necessary early diagnosis.
Research into the genetics underlying the spectrum of diseases has found that genes contributing to CMTs, HMN, HSPs and MNDs have significantly connected disease modules, forming local connections with each other as they work in related biological pathways (Fig. 1) (Bis-brewer et al., 2019). Among those are biological themes we covered here: axonal trafficking, dysfunction in RNA metabolism and protein homeostasis, and deficits in the cellular stress response (Fig. 2; Table 1). Understanding the molecular pathology of the spectrum and diseases involved in these biological modules will guide research to potential common therapeutic targets and strategies, such as how ALS and SMA have learned from one another (Tosolini and Sleigh, 2017). Cross-over between modules is to be expected, as common pathomechanisms are likely to converge on similar disease phenotypes. Examples of this described here include the changes that variants in GARS and HSPB1 have on HDAC6, causing changes to axonal transport functions. Such findings have observed that for both, HDAC6 inhibitors can attenuate disease severity, indicating a common therapeutic avenue. Furthermore, well-defined family history and advances in genome sequencing technology will improve our knowledge of all genetic contributions to disease, including genetic modifiers that change susceptibility and may potentially shift the spectrum of disease associated with any given variant.
Highlights.
CMT, MNDs and other neurodegenerative diseases share genetic contributions.
Many of these diseases appear to have shared molecular mechanisms.
Clinicogenetics can be useful for the diagnosis of many neurodegenerative diseases.
Acknowledgements
The authors would like to thank Jenn Stauffer for her careful review and insightful editing of the manuscript. We would also like to acknowledge the technical support from BioRender (https://biorender.com/) when producing Figure 2.
Funding Source
This work was supported in part by an NIH R01-NS102414 and the SIMS Family Fund to GAC, and by the University of Maine institutional training grant T32-GM132006 from NIGMS (PIs: L. Liaw and C. Henry) to SEH.
Abbreviations
- AD
Autosomal dominant
- ALS
Amyotrophic lateral sclerosis
- AR
Autosomal recessive
- ARSs
Aminoacyl-tRNA synthetases
- BTOP
Bilateral temporooccipital polymicrogyria
- CMT
Charcot-Marie-Tooth
- FTD
Frontotemporal dementia
- GOF
Gain of function
- HMN
Hereditary motor neuropathy
- HSP
Hereditary spastic paraplegia
- IBM
Inclusion body myopathy
- IBMPFD
Inclusion body myopathy with Paget disease and frontotemporal dementia
- IMPC
International mouse phenotyping consortium
- IPN
Inherited peripheral neuropathy
- LOF
Loss of function
- MND
Motor Neuron Disease
- MS
Multiple sclerosis
- NEIMY
Neonatal intractable myoclonus
- NCV
Nerve conduction velocity
- PBD
Paget’s body disease
- PCH10
Pontocerebellar hypoplasia type 10
- RQC
Ribosome-associated quality control
- SMA
Spinal muscular atrophy
- SMARD
Spinal muscular atrophy with respiratory distress
- SPG
Spastic paraplegia
- TSEN
tRNA splicing endonuclease
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abbott JA, Meyer-Schuman R, Lupo V, Feely S, Mademan I, Oprescu SN, et al. Substrate interaction defects in histidyl-tRNA synthetase linked to dominant axonal peripheral neuropathy. Hum Mutat 2018; 39: 415–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe A, Numakura C, Saito K, Koide H, Oka N, Honma A, et al. Neurofilament light chain polypeptide gene mutations in Charcot-Marie-Tooth disease: Nonsense mutation probably causes a recessive phenotype. J Hum Genet 2009 [DOI] [PubMed] [Google Scholar]
- Abrahao A, Abath Neto O, Kok F, Zanoteli E, Santos B, Pinto WBV de R, et al. One family, one gene and three phenotypes: A novel VCP (valosin-containing protein) mutation associated with myopathy with rimmed vacuoles, amyotrophic lateral sclerosis and frontotemporal dementia. J Neurol Sci 2016; 368: 352–358. [DOI] [PubMed] [Google Scholar]
- Agrawal PB, Joshi M, Marinakis NS, Schmitz-Abe K, Ciarlini PDSC, Sargent JC, et al. Expanding the phenotype associated with the NEFL mutation neuromuscular disease in a family with overlapping myopathic and neurogenic findings. JAMA Neurol 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Chalabi A, Andersen PM, Nilsson P, Chioza B, Andersson JL, Russ C, et al. Deletions of the heavy neurofilament subunit tail in amyotrophic lateral sclerosis. Hum Mol Genet 1999 [DOI] [PubMed] [Google Scholar]
- Al-Obeidi E, Al-Tahan S, Surampalli A, Goyal N, Wang AK, Hermann A, et al. Genotype-phenotype study in patients with valosin-containing protein mutations associated with multisystem proteinopathy. Clin Genet 2018; 93: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anazi S, Maddirevula S, Salpietro V, Asi YT, Alsahli S, Alhashem A. Expanding the genetic heterogeneity of intellectual disability. Hum Genet 2017; 136: 1419–1429. [DOI] [PubMed] [Google Scholar]
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Jordanova A, et al. Glycyl tRNA Synthetase Mutation in Charcot-Marie-Tooth Disease Type 2D and Distal Spinal Muscular Atrophy Type V. Am J Hum Genet 2003: 1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Lee-Lin S-Q, Wasterlain A, Leo P, Quezado M, Goldfarb LG, et al. Functional Analyses of Glycyl-tRNA Synthetase Mutations Suggest a Key Role for tRNA-Charging Enzymes in Peripheral Axons. J Neurosci 2006; 26: 10397–10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardissone A, Tonduti D, Legati A, Lamantea E, Barone R, Dorboz I, et al. KARS-related diseases: Progressive leukoencephalopathy with brainstem and spinal cord calcifications as new phenotype and a review of literature. Orphanet J Rare Dis 2018; 13: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakthisaran R, Tangirala R, Rao CM. Small heat shock proteins: Role in cellular functions and pathology. Biochim Biophys Acta 2015; 1854: 291–319. [DOI] [PubMed] [Google Scholar]
- Bansagi B, Antoniadi T, Burton-Jones S, Murphy SM, McHugh J, Alexander M, et al. Genotype/phenotype correlations in AARS-related neuropathy in a cohort of patients from the United Kingdom and Ireland. J Neurol 2015; 262: 1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer P, Winner B, Schüle R, Bauer C, Häfele V, Hehr U, et al. Identification of a heterozygous genomic deletion in the spatacsin gene in SPG11 patients using high-resolution comparative genomic hybridization. Neurogenetics 2009 [DOI] [PubMed] [Google Scholar]
- Baulac S, Lenk GM, Dufresnois B, Bencheikh BOA, Couarch P, Renard J, et al. Role of the phosphoinositide phosphatase FIG4 gene in familial epilepsy with polymicrogyria. Neurology 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecroft SJ, McLean CA, Delatycki MB, Koshy K, Yiu E, Haliloglu G, et al. Expanding the phenotypic spectrum associated with mutations of DYNC1H1. Neuromuscul Disord 2017 [DOI] [PubMed] [Google Scholar]
- Bengtson MH, Joazeiro CAP. Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 2010; 467: 470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoy V, Vanden Berghe P, Jarpe M, Van Damme P, Robberecht W, Van Den Bosch L. Development of Improved HDAC6 Inhibitors as Pharmacological Therapy for Axonal Charcot-Marie-Tooth Disease. Neurotherapeutics 2017; 14: 417–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berciano J, García A, Gallardo E, Peeters K, Pelayo-Negro AL, Álvarez-Paradelo S, et al. Intermediate Charcot–Marie–Tooth disease: an electrophysiological reappraisal and systematic review. J Neurol 2017 [DOI] [PubMed] [Google Scholar]
- Berciano J, García A, Peeters K, Gallardo E, De Vriendt E, Pelayo-Negro AL, et al. NEFL E396K mutation is associated with a novel dominant intermediate Charcot–Marie–Tooth disease phenotype. J Neurol 2015 [DOI] [PubMed] [Google Scholar]
- Bertoli-Avella AM, Garcia-Aznar JM, Brandau O, Al-Hakami F, Yüksel Z, Marais A, et al. Biallelic inactivating variants in the GTPBP2 gene cause a neurodevelopmental disorder with severe intellectual disability. Eur J Hum Genet 2018; 26: 592–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolin C, Querin G, Bozzoni V, Martinelli I, De Bortoli M, Rampazzo A, et al. New FIG4 gene mutations causing aggressive ALS. Eur J Neurol 2018 [DOI] [PubMed] [Google Scholar]
- Bharadwaj R, Cunningham KM, Zhang K, Lloyd TE. FIG4 regulates lysosome membrane homeostasis independent of phosphatase function. Hum Mol Genet 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi X, Jones T, Abbasi F, Lee H, Stultz B, Hursh DA, et al. Drosophila caliban, a nuclear export mediator, can function as a tumor suppressor in human lung cancer cells. 2005; 1: 8229–8239. [DOI] [PubMed] [Google Scholar]
- Bis-brewer DM, Danzi MC, Wuchty S, Züchner S. OPEN A network biology approach to unraveling inherited axonopathies. 2019: 1–12. [DOI] [PMC free article] [PubMed]
- Blocquel D, Li S, Wei N, Daub H, Sajish M, Erfurth ML, et al. Alternative stable conformation capable of protein misinteraction links tRNA synthetase to peripheral neuropathy. Nucleic Acids Res 2017; 45: 8091–8104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumen SC, Astord S, Robin V, Vignaud L, Toumi N, Cieslik A, et al. A rare recessive distal hereditary motor neuropathy with HSJ1 chaperone mutation. Ann Neurol 2012; 71: 509–519. [DOI] [PubMed] [Google Scholar]
- Del Bo R, Locatelli F, Corti S, Scarlato M, Ghezzi S, Prelle A, et al. Coexistence of CMT-2D and distal SMA-V phenotypes in an Italian family with a GARS gene mutation. Neurology 2006; 66: 752–754. [DOI] [PubMed] [Google Scholar]
- Boczonadi V, Jennings MJ, Horvath R. The role of tRNA synthetases in neurological and neuromuscular disorders. 2018; 592: 703–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Boom J, Meyer H. VCP/p97-Mediated Unfolding as a Principle in Protein Homeostasis and Signaling. Mol Cell 2018; 69: 182–194. [DOI] [PubMed] [Google Scholar]
- Borrell-Pages M, Canals JM, Cordelieres FP, Parker JA, Pineda JR, Grange G, et al. Cystamine and cysteamine increase brain levels of BDNF in Huntington disease via HSJ1b and transglutaminase. J Clin Invest 2006; 116: 1410–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bot ST, Schelhaas HJ, Kamsteeg EJ, Van De Warrenburg BPC. Hereditary spastic paraplegia caused by a mutation in the VCP gene. Brain 2012; 135. [DOI] [PubMed] [Google Scholar]
- Bouhy D, Juneja M, Katona I, Holmgren A, Asselbergh B, De Winter V, et al. A knock - in / knock - out mouse model of HSPB8 - associated distal hereditary motor neuropathy and myopathy reveals toxic gain - of - function of mutant Hspb8. Acta Neuropathol 2018; 135: 131–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchu J, Boutry M, Sourd L, Depp M, Leone C, Corriger A, et al. Loss of spatacsin function alters lysosomal lipid clearance leading to upper and lower motor neuron degeneration. Neurobiol Dis 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li G-W, et al. A Ribosome-Bound Quality Control Complex Triggers Degradation of Nascent Peptides and Signals Translation Stress. Cell 2012; 151: 1042–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D, Yilmaz R, Müller K, Grehl T, Petri S, Meyer T, et al. Hot-spot KIF5A mutations cause familial ALS. Brain 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlees J, Ackerley S, Grierson AJ, Jacobsen NJO, Shea K, Anderton BH, et al. Charcot-Marie-Tooth disease neurofilament mutations disrupt neurofilament assembly and axonal transport. Hum Mol Genet 2002 [DOI] [PubMed] [Google Scholar]
- Burk K, Pasterkamp RJ. Disrupted neuronal trafficking in amyotrophic lateral sclerosis. Acta Neuropathol 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns NJ, Neumann M, Bigio EH, Holm IE, Troost D, Hatanpaa KJ, et al. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am J Pathol 2007; 171: 227–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau PM, Lenk GM, Lu JT, Bae Y, Burrage L, Turnpenny P, et al. Yunis-Varón syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am J Hum Genet 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonnelle D, Liehr T, Jacquot C, Masson D, Lustenberger P, Denis MG, et al. Assignment of the serologically defined colon cancer antigen 1 gene (SDCCAG1) to human chromosome band 14q22 by in situ hybridization. Cytogenet Genome Res 1999; 86: 248–249. [DOI] [PubMed] [Google Scholar]
- Caroppo P, Le Ber I, Clot F, Rivaud-Péchoux S, Camuzat A, De Septenville A, et al. DCTN1 mutation analysis in families with progressive supranuclear palsy-like phenotypes. JAMA Neurol 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carra S, Sivilotti M, Chavez Zobel AT, Lambert H, Landry J. HspB8, a small heat shock protein mutated in human neuromuscular disorders, has in vivo chaperone activity in cultured cells. Hum Mol Genet 2005; 14: 1659–1669. [DOI] [PubMed] [Google Scholar]
- Carter MT, Venkateswaran S, Shapira-Zaltsberg G, Davila J, Humphreys P, Kernohan KD, et al. Clinical delineation of GTPBP2-associated neuro-ectodermal syndrome: Report of two new families and review of the literature. Clin Genet 2019; 95: 601–606. [DOI] [PubMed] [Google Scholar]
- Chan N, Le C, Shieh P, Mozaffar T, Khare M, Bronstein J, et al. Valosin-containing protein mutation and Parkinson’s disease. Park Relat Disord 2012; 18: 107–109. [DOI] [PubMed] [Google Scholar]
- Chen X-J, Levedakou EN, Millen KJ, Wollmann RL, Soliven B, Popko B. Proprioceptive Sensory Neuropathy in Mice with a Mutation in the Cytoplasmic Dynein Heavy Chain 1 Gene. J Neurosci 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier-Larsen ES, Wallace KE, Pennise CR, Holzbaur ELF. Lysosomal proliferation and distal degeneration in motor neurons expressing the G59S mutation in the p150Glued subunit of dynactin. Hum Mol Genet 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba K, Takahashi H, Chen M, Obinata H, Arai S, Hashimoto K, et al. Disease-associated mutations hyperactivate KIF1A motility and anterograde axonal transport of synaptic vesicle precursors. Proc Natl Acad Sci 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara T, Luginbuhl D, Luo L. Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nat Neurosci 2007; 10: 828–837. [DOI] [PubMed] [Google Scholar]
- Choe Y-J, Park S-H, Hassemer T, Körner R, Vincenz-Donnelly L, Hayer-Hartl M, et al. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 2016; 531: 191–5. [DOI] [PubMed] [Google Scholar]
- Chow CY, Landers JE, Bergren SK, Sapp PC, Grant AE, Jones JM, et al. Deleterious Variants of FIG4, a Phosphoinositide Phosphatase, in Patients with ALS. Am J Hum Genet 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CY, Zhang Y, Dowling JJ, Jin N, Adamska M, Shiga K, et al. Mutation of FIG4 causes neurodegeneration in the pale tremor mouse and patients with CMT4J. Nature 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou K, Kyriakides T, Hristova AH, Georgiou DM, Kalaydjieva L, Yshpekova B, et al. Mapping of a distal form of spinal muscular atrophy with upper limb predominance to chromosome 7p. Hum Mol Genet 1995; 4: 1629–1632. [DOI] [PubMed] [Google Scholar]
- Chu J, Hong N a, Masuda C a, Jenkins BV, Nelms K a, Goodnow CC, et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc Natl Acad Sci U S A 2009; 106: 2097–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung KW, Kim S-B, Cho SY, Hwang SJ, Park SW, Kang SH, et al. Distal hereditary motor neuropathy in Korean patients with a small heat shock protein 27 mutation. Exp Mol Med 2008; 40: 304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard JF, Côté F, Julien JP. Defective axonal transport in a transgenic mouse model of amyotrophic lateral sclerosis. Nature 1995 [DOI] [PubMed] [Google Scholar]
- Corcia P, Brulard C, Beltran S, Marouillat S, Bakkouche SE, Andres CR, et al. Typical bulbar ALS can be linked to GARS mutation. Amyotroph Lateral Scler Front Degener 2018; 0: 1–3. [DOI] [PubMed] [Google Scholar]
- Cosker KE, Segal RA. Neuronal signaling through endocytosis. Cold Spring Harb Perspect Biol 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottenie E, Kochanski A, Jordanova A, Bansagi B, Zimon M, Horga A, et al. Truncating and Missense Mutations in IGHMBP2 Cause Charcot-Marie Tooth Disease Type 2. Am J Hum Genet 2014; 95: 590–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox GA, Mahaffey CL, Frankel WN. Identification of the mouse neuromuscular degeneration gene and mapping of a second site suppressor allele. Neuron 1998; 21: 1327–37. [DOI] [PubMed] [Google Scholar]
- Crimella C, Baschirotto C, Arnoldi A, Tonelli A, Tenderini E, Airoldi G, et al. Mutations in the motor and stalk domains of KIF5A in spastic paraplegia type 10 and in axonal Charcot-Marie-Tooth type 2. Clin Genet 2012 [DOI] [PubMed] [Google Scholar]
- Custer SK, Neumann M, Lu H, Wright AC, Taylor JP. Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone. Hum Mol Genet 2010; 19: 1741–1755. [DOI] [PubMed] [Google Scholar]
- D’Ydewalle C, Krishnan J, Chiheb DM, Van Damme P, Irobi J, Kozikowski AP, et al. HDAC6 inhibitors reverse axonal loss in a mouse model of mutant HSPB1-induced Charcot-Marie-Tooth disease. Nat Med 2011; 17: 968–974. [DOI] [PubMed] [Google Scholar]
- DaRe JT, Vasta V, Penn J, Tran NTB, Hahn SH. Targeted exome sequencing for mitochondrial disorders reveals high genetic heterogeneity. BMC Med Genet 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defenouillere Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc Natl Acad Sci 2013; 110: 5046–5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defenouillère Q, Zhang E, Namane A, Mouaikel J, Jacquier A, Fromont-Racine M. Rqc1 and ltn1 prevent c-terminal alanine-threonine tail (cat-tail)-induced protein aggregation by efficient recruitment of cdc48 on stalled 60s subunits. J Biol Chem 2016; 291: 12245–12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Desaro P, Johnston A, Ross OA, Wszolek ZK, Ertekin-Taner N, et al. Novel p.Ile151Val mutation in VCP in a patient of African American descent with sporadic ALS. Neurology 2011; 77: 1102–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, et al. High-throughput discovery of novel developmental phenotypes. Nature 2016; 537: 508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 2006; 440: 561–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duis J, Dean S, Applegate C, Harper A, Xiao R, He W, et al. KIF5A mutations cause an infantile onset phenotype including severe myoclonus with evidence of mitochondrial dysfunction. Ann Neurol 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echaniz-Laguna A, Lornage X, Lannes B, Schneider R, Bierry G, Dondaine N, et al. HSPB8 haploinsufficiency causes dominant adult-onset axial and distal myopathy. Acta Neuropathol 2017; 134: 163–165. [DOI] [PubMed] [Google Scholar]
- Elder GA, Friedrich VL, Kang C, Bosco P, Gourov A, Tu PH, et al. Requirement of heavy neurofilament subunit in the development of axons with large calibers. J Cell Biol 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlich Y, Edvardson S, Hodges E, Zenvirt S, Thekkat P, Shaag A, et al. Exome sequencing and disease-network analysis of a single family implicate a mutation in KIF1A in hereditary spastic paraparesis. Genome Res 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermanoska B, Motley WW, Leitão-Gonçalves R, Asselbergh B, Lee LTH, De Rijk P, et al. CMT-associated mutations in glycyl- and tyrosyl-tRNA synthetases exhibit similar pattern of toxicity and share common genetic modifiers in Drosophila. Neurobiol Dis 2014; 68: 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esmaeeli Nieh S, Madou MRZ, Sirajuddin M, Fregeau B, Mcknight D, Lexa K, et al. De novo mutations in KIF1A cause progressive encephalopathy and brain atrophy. Ann Clin Transl Neurol 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch LV, Dierick I, Leung CL, et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet 2004; 36: 602–606. [DOI] [PubMed] [Google Scholar]
- Fabrizi GM, Cavallaro T, Angiari C, Bertolasi L, Cabrini I, Ferrarini M, et al. Giant axon and neurofilament accumulation in Charcot-Marie-Tooth disease type 2E. Neurology 2004 [DOI] [PubMed] [Google Scholar]
- Farrer MJ, Hulihan MM, Kachergus JM, Dächsel JC, Stoessl AJ, Grantier LL, et al. DCTN1 mutations in Perry syndrome. Nat Genet 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figlewicz DA, Krizus A, Martinoli MG, Meininger V, Dib M, Rouleau GA, et al. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum Mol Genet 1994 [DOI] [PubMed] [Google Scholar]
- Frost A, Weinberg DE, Weissman JS. CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. 2017; 417: 414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier LR, Charrin BC, Borrell-Pagès M, Dompierre JP, Rangone H, Cordelières FP, et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell 2004 [DOI] [PubMed] [Google Scholar]
- Gentil BJ, Mushynski WE, Durham HD. Heterogeneity in the properties of NEFL mutants causing Charcot-Marie-Tooth disease results in differential effects on neurofilament assembly and susceptibility to intervention by the chaperone-inducer, celastrol. Int J Biochem Cell Biol 2013 [DOI] [PubMed] [Google Scholar]
- Georgiou DM, Zidar J, Korošec M, Middleton LT, Kyriakides T, Christodoulou K. A novel NF-L mutation pro22ser is associated with CMT2 in a large Slovenian family. Neurogenetics 2002 [DOI] [PubMed] [Google Scholar]
- Gess B, Auer-Grumbach M, Schirmacher A, Strom T, Zitzelsberger M, Rudnik-Schöneborn S, et al. <em>HSJ1</em>-related hereditary neuropathies. Neurology 2014; 83: 1726 LP–1732. [DOI] [PubMed] [Google Scholar]
- Goizet C, Boukhris A, Mundwiller E, Tallaksen C, Forlani S, Toutain A, et al. Complicated forms of autosomal dominant hereditary spastic paraplegia are frequent in SPG10. Hum Mutat 2009 [DOI] [PubMed] [Google Scholar]
- Gonzalez M, McLaughlin H, Houlden H, Guo M, Liu YT, Hadjivassilious M, et al. Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J Neurol Neurosurg Psychiatry 2013; 84: 1247–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez MA, Feely SM, Speziani F, Strickland AV., Danzi M, Bacon C, et al. A novel mutation in VCP causes Charcot-Marie-Tooth Type 2 disease. Brain 2014; 137: 2897–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorochowski TE, Ignatova Z, Bovenberg RAL, Roubos JA. Trade-offs between tRNA abundance and mRNA secondary structure support smoothing of translation elongation rate. Nucleic Acids Res 2015; 43: 3022–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorochowski TE, Ignatova Z, Bovenberg RAL, Roubos JA. Trade-offs between tRNA abundance and mRNA secondary structure support smoothing of translation elongation rate. Nucleic Acids Res 2015; 43: 3022–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin LB, Sakaguchi R, McGuigan D, Gonzalez MA, Searby C, Züchner S, et al. Impaired Function is a Common Feature of Neuropathy-Associated Glycyl-tRNA Synthetase Mutations. Hum Mutat 2014; 12: n/a–n/a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann K, Rossoll W, Kobsar I, Holtmann B, Jablonka S, Wessig C, et al. Characterization of Ighmbp2 in motor neurons and implications for the pathomechanism in a mouse model of human spinal muscular atrophy with respiratory distress type 1 (SMARD1). Hum Mol Genet 2004; 13: 2031–2042. [DOI] [PubMed] [Google Scholar]
- Grohmann K, Schuelke M, Diers a, Hoffmann K, Lucke B, Adams C, et al. Mutations in the gene encoding immunoglobulin mu-binding protein 2 cause spinal muscular atrophy with respiratory distress type 1. Nat Genet 2001; 29: 75–77. [DOI] [PubMed] [Google Scholar]
- Grohmann K, Varon R, Stolz P, Schuelke M, Janetzki C, Bertini E, et al. Spinal Muscular Atrophy With Respiratory Distress Type 1 (SMARD1). Ann Neurol 2003; 54: 719–724. [DOI] [PubMed] [Google Scholar]
- Grohmann K, Wienker TF, Saar K, Rudnik-Schöneborn S, Stolenburg-Didinger G, Rossi R, et al. Diaphragmatic Spinal Muscular Atrophy with Respiratory Distress Is Heterogeneous, and One Form Is Linked to Chromosome 11q13-q21. Am J Hum Genet 1999: 1457–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther U-P, Handoko L, Laggerbauer B, Jablonka S, Chari A, Alzheimer M, et al. IGHMBP2 is a ribosome-associated helicase inactive in the neuromuscular disorder distal SMA type 1 (DSMA1). Hum Mol Genet 2009; 18: 1288–1300. [DOI] [PubMed] [Google Scholar]
- Guenther UP, Handoko L, Laggerbauer B, Jablonka S, Chari A, Alzheimer M, et al. IGHMBP2 is a ribosome-associated helicase inactive in the neuromuscular disorder distal SMA type 1 (DSMA1). Hum Mol Genet 2009; 18: 1288–1300. [DOI] [PubMed] [Google Scholar]
- Guenther UP, Handoko L, Varon R, Stephani U, Tsao CY, Mendell JR, et al. Clinical variability in distal spinal muscular atrophy type 1 (DSMA1): Determination of steady-state IGHMBP2 protein levels in five patients with infantile and juvenile disease. J Mol Med 2009; 87: 31–41. [DOI] [PubMed] [Google Scholar]
- Guidubaldi A, Piano C, Santorelli FM, Silvestri G, Petracca M, Tessa A, et al. Novel mutations in SPG11 cause hereditary spastic paraplegia associated with early-onset levodopa-responsive Parkinsonism. Mov Disord 2011 [DOI] [PubMed] [Google Scholar]
- Guo W, Naujock M, Fumagalli L, Vandoorne T, Baatsen P, Boon R, et al. HDAC6 inhibition reverses axonal transport defects in motor neurons derived from FUS-ALS patients. Nat Commun 2017; 8: 861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafezparast M, Klocke R, Ruhrberg C, Marquardt A, Ahmad-Annuar A, Bowen S, et al. Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science (80-) 2003 [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, Higashi K, et al. Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MJ, Longman C, O’Hara A, Kirkpatrick M, McWilliam R. Growing up with spinal muscular atrophy with respiratory distress (SMARD1). Neuromuscul Disord 2015; 25: 169–171. [DOI] [PubMed] [Google Scholar]
- Hanada T, Weitzer S, Mair B, Bernreuther C, Wainger BJ, Ichida J, et al. CLP1 links tRNA metabolism to progressive motor-neuron loss. Nature 2013; 495: 474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MB, Allred P, Gardner R, Fernandes Filho JA, Florence J, Pestronk A, et al. Dominant spinal muscular atrophy with lower extremity predominance: Linkage to 14q32. Neurology 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms MB, Ori-McKenney KM, Scoto M, Tuck EP, Bell S, Ma D, et al. Mutations in the tail domain of DYNC1H1 cause dominant spinal muscular atrophy. Neurology 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haubenberger D, Bittner RE, Rauch-Shorny S, Zimprich F, Mannhalter C, Wagner L, et al. Inclusion body myopathy and Paget disease is linked to a novel mutation in the VCP gene. Neurology 2005; 65: 1304–1305. [DOI] [PubMed] [Google Scholar]
- He W, Bai G, Zhou H, Wei N, White NM, Lauer J, et al. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature 2015; 526: 710–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hentati A, Ouahchi K, Pericak-Vance MA, Nijhawan D, Ahmad A, Yang Y, et al. Linkage of a commoner form of recessive amyotrophic lateral sclerosis to chromosome 15q15-q22 markers. Neurogenetics 1998 [DOI] [PubMed] [Google Scholar]
- Hinckelmann MV, Zala D, Saudou F. Releasing the brake: Restoring fast axonal transport in neurodegenerative disorders. Trends Cell Biol 2013 [DOI] [PubMed] [Google Scholar]
- Hinreiner S, Kohl Z, Rödl T, Becher M, Winkler J, Hehr U. Clinical and genetic evaluation of hereditary spastic paraplegia. Medizinische Genet 2016 [Google Scholar]
- Hirabayashi M, Inoue K, Tanaka K, Nakadate K, Ohsawa Y, Kamei Y, et al. VCP/p97 in abnormal protein aggregates, cytoplasmic vacuoles, and cell death, phenotypes relevant to neurodegeneration. Cell Death Differ 2001; 8: 977–984. [DOI] [PubMed] [Google Scholar]
- Hirano M, Oka N, Hashiguchi A, Ueno S, Sakamoto H, Takashima H, et al. Histopathological features of a patient with Charcot-Marie-Tooth disease type 2U/AD-CMTax-MARS. J Peripher Nerv Syst 2016; 21: 370–374. [DOI] [PubMed] [Google Scholar]
- Hoang HT, Schlager MA, Carter AP, Bullock SL. DYNC1H1 mutations associated with neurological diseases compromise processivity of dynein–dynactin–cargo adaptor complexes. Proc Natl Acad Sci 2017; 114: E1597–E1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzbaur ELF, Tokito MK. Localization of the DCTN1 Gene Encoding p150Gluedto Human Chromosome 2p13 by Fluorescencein SituHybridization. Genomics 1996 [DOI] [PubMed] [Google Scholar]
- Houlden H, Laura M, Wavrant–De Vrièze F, Blake J, Wood N, Reilly MM. Mutations in the <em>HSP27</em> (<em>HSPB1</em>) gene cause dominant, recessive, and sporadic distal HMN/CMT type 2. Neurology 2008; 71: 1660 LP–1668. [DOI] [PubMed] [Google Scholar]
- Hwang SH, Kim EJ, Hong Y Bin, Joo J, Kim SM, Nam SH, et al. Distal hereditary motor neuropathy type 7B with Dynactin 1 mutation. Mol Med Rep 2016 [DOI] [PubMed] [Google Scholar]
- Hyun YS, Park HJ, Heo S-H, Yoon BR, Nam SH, Kim S-B, et al. Rare variants in methionyl -and tyrosyl-tRNA synthetase genes in late-onset autosomal dominant Charcot-Marie-Tooth neuropathy. Clin Genet 2014; 86: 592–594. [DOI] [PubMed] [Google Scholar]
- Ikenaka K, Kawai K, Katsuno M, Huang Z, Jiang YM, Iguchi Y, et al. dnc-1/dynactin 1 Knockdown Disrupts Transport of Autophagosomes and Induces Motor Neuron Degeneration. PLoS One 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionasescu V, Searby C, Sheffield VC, Roklina T, Nishimura D, Ionasescu R. Autosomal dominant Charcot-Marie-Tooth axonal neuropathy mapped on chromosome 7p (CMT2D). Hum Mol Genet 1996; 5: 1373–1375. [DOI] [PubMed] [Google Scholar]
- Irobi J, Van Impe K, Seeman P, Jordanova A, Dierick I, Verpoorten N, et al. Hot-spot residue in small heat-shock protein 22 causes distal motor neuropathy. Nat Genet 2004; 36: 597–601. [DOI] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Chuang JH, Ackerman SL. Activation of GCN2 kinase by ribosome stalling links translation elongation with translation initiation. Elife 2016; 5: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishimura R, Nagy G, Dotu I, Zhou H, Yang X-L, Schimmel P, et al. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science (80-) 2014; 345: 455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaberi E, Rohani M, Shahidi GA, Nafissi S, Arefian E, Soleimani M, et al. Identification of mutation in GTPBP2 in patients of a family with neurodegeneration accompanied by iron deposition in the brain. Neurobiol Aging 2015; 38: 216.e11–216.e18. [DOI] [PubMed] [Google Scholar]
- Jackson L, Madan-Khetarpal S, Naik M, Michaels MG, Riley M. Infant Botulism in the Very Young Neonate: A Case Series. AJP Rep 2017; 7: e163–e166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier A, Delorme C, Belotti E, Juntas-Morales R, Solé G, Dubourg O, et al. Cryptic amyloidogenic elements in mutant NEFH causing Charcot-Marie-Tooth 2 trigger aggresome formation and neuronal death. Acta Neuropathol Commun 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James PA, Cader MZ, Muntoni F, Childs A-M, Crow YJ, Talbot K. Severe childhood SMA and axonal CMT due to anticodon binding domain mutations in the GARS gene. Neurology 2006; 67: 1710–1712. [DOI] [PubMed] [Google Scholar]
- James PA, Talbot K. The molecular genetics of non-ALS motor neuron diseases. Biochim Biophys Acta - Mol Basis Dis 2006; 1762: 986–1000. [DOI] [PubMed] [Google Scholar]
- Jamuar SS, Lam A-TN, Kircher M, D’Gama AM, Wang J, Barry BJ, et al. Somatic Mutations in Cerebral Cortical Malformations. N Engl J Med 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Van Deerlin VM, Trojanowski JQ, Gibbs JR, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron 2011; 68: 857–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordanova A, Irobi J, Thomas FP, Van Dijck P, Meerschaert K, Dewil M, et al. Disrupted function and axonal distribution of mutant tyrosyl-tRNA synthetase in dominant intermediate Charcot-Marie-Tooth neuropathy. Nat Genet 2006; 38: 197–202. [DOI] [PubMed] [Google Scholar]
- Jordanova A, De Jonghe P, Boerkoel CF, Takashima H, De Vriendt E, Ceuterick C, et al. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot-Marie-Tooth disease. Brain 2003 [DOI] [PubMed] [Google Scholar]
- Joseph S, Robb S a, Mohammed S, Lillis S, Simonds a, Manzur a Y, et al. Interfamilial phenotypic heterogeneity in SMARD1. Neuromuscul Disord 2009; 19: 193–5. [DOI] [PubMed] [Google Scholar]
- Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, et al. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J Cell Biol 2009; 187: 875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalani A, Tyagi N. Exosomes in neurological disease, neuroprotection, repair and therapeutics: Problems and perspectives. Neural Regen Res 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron 2004 [DOI] [PubMed] [Google Scholar]
- Karaca E, Weitzer S, Pehlivan D, Shiraishi H, Gogakos T, Hanada T, et al. Human CLP1 Mutations Alter tRNA Biogenesis, Affecting Both Peripheral and Central Nervous System Function. Cell 2014; 157: 636–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller MP, Seifried BA, Rabin BA, Chance PF. Mapping of the kinesin-related gene ATSV to chromosome 2q37. Hum Genet 1999 [DOI] [PubMed] [Google Scholar]
- Kijima K, Numakura C, Goto T, Takahashi T, Otagiri T, Umetsu K, et al. Small heat shock protein 27 mutation in a Japanese patient with distal hereditary motor neuropathy. J Hum Genet 2005; 50: 473–476. [DOI] [PubMed] [Google Scholar]
- Klebe S, Lossos A, Azzedine H, Mundwiller E, Sheffer R, Gaussen M, et al. KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: Distinct phenotypes according to the nature of the mutations. Eur J Hum Genet 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb SJ, Snyder PJ, Poi EJ, Renard EA, Bartlett A, Gu S, et al. Mutant small heat shock protein B3 causes motor neuropathy. Neurology 2010; 74: 502LP–506. [DOI] [PubMed] [Google Scholar]
- Konno T, Ross OA, Teive HAG, Sławek J, Dickson DW, Wszolek ZK. DCTN1-related neurodegeneration: Perry syndrome and beyond. Park Relat Disord 2017; 41: 14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppers M, van Blitterswijk MM, Vlam L, Rowicka PA, van Vught PWJ, Groen EJN, et al. VCP mutations in familial and sporadic amyotrophic lateral sclerosis. Neurobiol Aging 2012; 33: 837.e7–837.e13. [DOI] [PubMed] [Google Scholar]
- Krenn M, Zulehner G, Hotzy C, Rath J, Stogmann E, Wagner M, et al. Hereditary spastic paraplegia caused by compound heterozygous mutations outside the motor domain of the KIF1A gene. Eur J Neurol 2017 [DOI] [PubMed] [Google Scholar]
- Latour P, Thauvin-Robinet C, Baudelet-Méry C, Soichot P, Cusin V, Faivre L, et al. A Major Determinant for Binding and Aminoacylation of tRNAAla in Cytoplasmic Alanyl-tRNA Synthetase Is Mutated in Dominant Axonal Charcot-Marie-Tooth Disease. Am J Hum Genet 2010; 86: 77–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JR, Srour M, Kim D, Hamdan FF, Lim SH, Brunel-Guitton C, et al. De novo mutations in the motor domain of KIF1A cause cognitive impairment, spastic paraparesis, axonal neuropathy, and cerebellar atrophy. Hum Mutat 2015 [DOI] [PubMed] [Google Scholar]
- Lee JW, Beebe K, Nangle LA, Jang J, Longo-Guess CM, Cook SA, et al. Editing-defective tRNA synthetase causes protein misfolding and neurodegeneration. Nature 2006; 443: 50–55. [DOI] [PubMed] [Google Scholar]
- Liao YC, Liu YT, Tsai PC, Chang CC, Huang YH, Soong BW, et al. Two novel de novo GARS mutations cause early-onset axonal Charcot-Marie-Tooth disease. PLoS One 2015; 10: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KP, Soong BW, Yang CC, Huang LW, Chang MH, Lee IH, et al. The mutational spectrum in a cohort of Charcot-Marie-Tooth disease type 2 among the han Chinese in Taiwan. PLoS One 2011; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka J, Kuijpers M, Jaworski J, Hoogenraad CC. Mutations in cytoplasmic dynein and its regulators cause malformations of cortical development and neurodegenerative diseases. Biochem Soc Trans 2015 [DOI] [PubMed] [Google Scholar]
- Liu YT, Laurá M, Hersheson J, Horga A, Jaunmuktane Z, Brandner S, et al. Extended phenotypic spectrum of KIF5A mutations: From spastic paraplegia to axonal neuropathy. Neurology 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd TE, Machamer J, O’Hara K, Kim JH, Collins SE, Wong MY, et al. The p150 Glued CAPGly Domain Regulates Initiation of Retrograde Transport at Synaptic Termini. Neuron 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell 1991; 66: 219–232. [DOI] [PubMed] [Google Scholar]
- Lyumkis D, Oliveira dos Passos D, Tahara EB, Webb K, Bennett EJ, Vinterbo S, et al. Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc Natl Acad Sci 2014; 111: 15981–15986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday S, Twelvetrees AE, Moughamian AJ, Holzbaur ELF. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddatu TP, Garvey SM, Schroeder DG, Hampton TG, Cox G a. Transgenic rescue of neurogenic atrophy in the nmd mouse reveals a role for Ighmbp2 in dilated cardiomyopathy. Hum Mol Genet 2004; 13: 1105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddatu TP, Garvey SM, Schroeder DG, Zhang W, Kim S-Y, Nicholson AI, et al. Dilated cardiomyopathy in the nmd mouse: transgenic rescue and QTLs that improve cardiac function and survival. Hum Mol Genet 2005; 14: 3179–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malissovas N, Griffin LB, Antonellis A, Beis D. Dimerization is required for GARS-mediated neurotoxicity in dominant CMT disease. Hum Mol Genet 2016; 25: 1528–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maretina M, Egorova A, Baranov V, Kiselev A. DYNC1H1 gene methylation correlates with severity of spinal muscular atrophy. Ann Hum Genet 2019; 83: 73–81. [DOI] [PubMed] [Google Scholar]
- Martyn C, Li J. Fig4 deficiency: A newly emerged lysosomal storage disorder? Prog Neurobiol 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin HM, Sakaguchi R, Giblin W, Wilson TE, Biesecker L, Lupski JR, et al. A Recurrent loss-of-function alanyl-tRNA synthetase (AARS) mutation in patients with charcot-marie-tooth disease type 2N (CMT2N). Hum Mutat 2012; 33: 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan HJ, Humphreys P, Smith A, Schwartzentruber J, Chakraborty P, Bulman DE, et al. Congenital Visual Impairment and Progressive Microcephaly Due to Lysyl-Transfer Ribonucleic Acid (RNA) Synthetase (KARS) Mutations: The Expanding Phenotype of Aminoacyl-Transfer RNA Synthetase Mutations in Human Disease. J Child Neurol 2015; 30: 1037–1043. [DOI] [PubMed] [Google Scholar]
- Mestres I, Sung C-H. Nervous system development relies on endosomal trafficking. Neurogenesis 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millecamps S, Julien JP. Axonal transport deficits and neurodegenerative diseases. Nat Rev Neurosci 2013 [DOI] [PubMed] [Google Scholar]
- Miltenberger-Miltenyi G, Janecke AR, Wanschitz JV., Timmerman V, Windpassinger C, Auer-Grumbach M, et al. Clinical and electrophysiological features in Charcot-Marie-Tooth disease with mutations in the NEFL gene. Arch Neurol 2007 [DOI] [PubMed] [Google Scholar]
- Mo Z, Zhao X, Liu H, Hu Q, Chen XQ, Pham J, et al. Aberrant GlyRS-HDAC6 interaction linked to axonal transport deficits in Charcot-Marie-Tooth neuropathy. Nat Commun 2018; 9: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montecchiani C, Pedace L, Lo Giudice T, Casella A, Mearini M, Gaudiello F, et al. ALS5/SPG11/KIAA1840 mutations cause autosomal recessive axonal Charcot-Marie-Tooth disease. Brain 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley WW, Griffin LB, Mademan I, Baets J, De Vriendt E, De Jonghe P, et al. A novel AARS mutation in a family with dominant myeloneuropathy. Neurology 2015; 84: 2040–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley WW, Seburn KL, Nawaz MH, Miers KE, Cheng J, Antonellis A, et al. Charcot-marie-tooth-linked mutant GARS is toxic to peripheral neurons independent of wild-type GARS levels. PLoS Genet 2011; 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munch C, Sedlmeier R, Meyer T, Homberg V, Sperfeld AD, Kurt A, et al. Point mutations of the p150 subunit of dynactin (DCTN1) gene in ALS. Neurology 2004; 63: 724–726. [DOI] [PubMed] [Google Scholar]
- Nakajima J, Okamoto N, Shiraishi J, Nishimura G, Nakashima M, Tsurusaki Y, et al. Novel FIG4 mutations in Yunis-Varon syndrome. J Hum Genet 2013 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Izumi K, Umehara F, Kuriyama M, Hokezu Y, Nakagawa M, et al. Familial spastic paraplegia with mental impairment and thin corpus callosum. J Neurol Sci 1995 [DOI] [PubMed] [Google Scholar]
- Nakhro K, Park J-M, Kim YJ, Yoon BR, Yoo JH, Koo H, et al. A novel Lys141Thr mutation in small heat shock protein 22 (HSPB8) gene in Charcot-Marie-Tooth disease type 2L. Neuromuscul Disord 2013; 23: 656–663. [DOI] [PubMed] [Google Scholar]
- Nalbandian A, Llewellyn KJ, Kitazawa M, Yin HZ, Badadani M, Khanlou N, et al. The Homozygote VCP R155H/R155H Mouse Model Exhibits Accelerated Human VCP-Associated Disease Pathology. PLoS One 2012; 7: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam DE, Yoo DH, Choi SS, Choi BO, Chung KW. Wide phenotypic spectrum in axonal Charcot–Marie–Tooth neuropathy type 2 patients with KIF5A mutations. Genes and Genomics 2018 [DOI] [PubMed] [Google Scholar]
- Nam SH. Identification of Genetic Causes of Inherited Peripheral Neuropathies by Targeted Gene Panel Sequencing. Mol Cells 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangle LA, Zhang W, Xie W, Yang XL, Schimmel P. Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc Natl Acad Sci U S A 2007; 104: 11239–11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neefjes J, van der Kant R. Stuck in traffic: An emerging theme in diseases of the nervous system. Trends Neurosci 2014 [DOI] [PubMed] [Google Scholar]
- Neumann M, Kwong LK, Sampathu DM, Trojanowski JQ, Lee VM-Y. TDP-43 proteinopathy in frontotemporal lobar degeneration and amyotrophic lateral sclerosis: protein misfolding diseases without amyloidosis. Arch Neurol 2007; 64: 1388–1394. [DOI] [PubMed] [Google Scholar]
- Nicholson G, Lenk GM, Reddel SW, Grant AE, Towne CF, Ferguson CJ, et al. Distinctive genetic and clinical features of CMT4J: A severe neuropathy caused by mutations in the PI(3,5)P 2 phosphatase FIG4. Brain 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niclas J, Navone F, Hom-Booker N, Vale RD. Cloning and localization of a conventional kinesin motor expressed exclusively in neurons. Neuron 1994 [DOI] [PubMed] [Google Scholar]
- Nicolas A, Kenna K, Renton AE, Ticozzi N, Faghri F, Chia R, et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehues S, Bussmann J, Steffes G, Erdmann I, Köhrer C, Sun L, et al. Impaired protein translation in Drosophila models for Charcot-Marie-Tooth neuropathy caused by mutant tRNA synthetases. Nat Commun 2015; 6: 7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, et al. Exome Sequencing Links Corticospinal Neurodegenerative Disorders. Science (80-) 2014; 343: 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa N. The neuron-specific kinesin superfamily protein KIF1A is a uniqye monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 1995 [DOI] [PubMed] [Google Scholar]
- Ori-McKenney KM, Vallee RB. Neuronal migration defects in the loa dynein mutant mouse. Neural Dev 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlacchio A, Babalini C, Borreca A, Patrono C, Massa R, Basaran S, et al. SPATACSIN mutations cause autosomal recessive juvenile amyotrophic lateral sclerosis. Brain 2010; 133: 591–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanovic A, Rangnau I, Kosfeld A, Abdulla S, Janssen C, Auber B, et al. FIG4 variants in central European patients with amyotrophic lateral sclerosis: A whole-exome and targeted sequencing study. Eur J Hum Genet 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouled Amar Ben Cheikh B, Baulac S, Lahjouji F, Bouhouche A, Couarch P, Khalili N, et al. A locus for bilateral occipital polymicrogyria maps to chromosome 6q16-q22. Neurogenetics 2009 [DOI] [PubMed] [Google Scholar]
- La Padula V, Staszewski O, Nestel S, Busch H, Boerries M, Roussa E, et al. HSPB3 protein is expressed in motoneurons and induces their survival after lesion-induced degeneration. Exp Neurol 2016; 286: 40–49. [DOI] [PubMed] [Google Scholar]
- Paisan-Ruiz C, Dogu O, Yilmaz A, Houlden H, Singleton A. SPG11 mutations are common in familial cases of complicated hereditary spastic paraplegia. Neurology 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin SV, Patel M, Furia BS, Peltz SW, Trotta CR. Identification of a human endonuclease complex reveals a link between tRNA splicing and pre-mRNA 3′ end formation. Cell 2004; 117: 311–321. [DOI] [PubMed] [Google Scholar]
- Pennings M, Schouten MI, van Gaalen J, Meijer RPP, de Bot ST, Kriek M, et al. KIF1A variants are a frequent cause of autosomal dominant hereditary spastic paraplegia. Eur J Hum Genet 2019: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Brangulí F, Mishra HK, Prots I, Havlicek S, Kohl Z, Saul D, et al. Dysfunction of spatacsin leads to axonal pathology in SPG11-linked hereditary spastic paraplegia. Hum Mol Genet 2014; 23: 4859–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry TL, Bratty PJA, Hansen S, Kennedy J, Urquhart N, Dolman CL. Hereditary Mental Depression and Parkinsonism With Taurine Deficiency. Arch Neurol 1975 [DOI] [PubMed] [Google Scholar]
- Perry TL, Wright JM, Berry K, Hansen S, Perry TL. Dominantly inherited apathy, central hypoventilation, and Parkinson’s syndrome: Clinical, biochemical, and neuropathologic studies of 2 new cases. Neurology 2012 [DOI] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CUT, Pestova TV., Pisarev A V. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J 2011; 30: 1804–1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Planell-Saguer M, Schroeder DG, Rodicio MC, Cox G a., Mourelatos Z. Biochemical and genetic evidence for a role of IGHMBP2 in the translational machinery. Hum Mol Genet 2009; 18: 2115–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier K, Lebrun N, Broix L, Tian G, Saillour Y, Boscheron C, et al. Mutations in TUBG1, DYNC1H1, KIF5C and KIF2A cause malformations of cortical development and microcephaly. Nat Genet 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prior R, Van Helleputte L, Benoy V, Van Den Bosch L. Defective axonal transport: A common pathological mechanism in inherited and acquired peripheral neuropathies. Neurobiol Dis 2017 [DOI] [PubMed] [Google Scholar]
- Puls I, Jonnakuty C, LaMonte BH, Holzbaur ELF, Tokito M, Mann E, et al. Mutant dynactin in motor neuron disease. Nat Genet 2003 [DOI] [PubMed] [Google Scholar]
- Puls I, Oh SJ, Sumner CJ, Wallace KE, Floeter MK, Mann EA, et al. Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann Neurol 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich K-U, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a Cytosolic Chaperone Required for Endoplasmic Reticulum-Associated Protein Degradation. Mol Cell Biol 2002; 22: 626–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao MV, Houseweart MK, Williamson TL, Crawford TO, Folmer J, Cleveland DW. Neurofilament-dependent radial growth of motor axons and axonal organization of neurofilaments does not require the neurofilament heavy subunit (NF-H) or its phosphorylation. J Cell Biol 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebelo AP, Abrams AJ, Cottenie E, Horga A, Gonzalez M, Bis DM, et al. Cryptic Amyloidogenic Elements in the 3′ UTRs of Neurofilament Genes Trigger Axonal Neuropathy. Am J Hum Genet 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E, Dearlove AM, Rhodes M, Rubinsztein DC. A New Locus for Autosomal Dominant “Pure” Hereditary Spastic Paraplegia Mapping to Chromosome 12q13, and Evidence for Further Genetic Heterogeneity. Am J Hum Genet 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E, Kloos M, Ashley-Koch A, Hughes L, Bevan S, Svenson IK, et al. A Kinesin Heavy Chain (KIF5A) Mutation in Hereditary Spastic Paraplegia (SPG10). Am J Hum Genet 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivire JB, Ramalingam S, Lavastre V, Shekarabi M, Holbert S, Lafontaine J, et al. KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am J Hum Genet 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993; 362: 59–62. [DOI] [PubMed] [Google Scholar]
- Rossor AM, Evans MRB, Reilly MM. A practical approach to the genetic neuropathies. Pract Neurol 2015; 15: 187–198. [DOI] [PubMed] [Google Scholar]
- Royer-Bertrand B, Tsouni P, Mullen P, Campos Xavier B, Mittaz Crettol L, Lobrinus AJ, et al. Peripheral neuropathy and cognitive impairment associated with a novel monoallelic HARS variant. Ann Clin Transl Neurol 2019: acn3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydzanicz M, Jagła M, Kosinska J, Tomasik T, Sobczak A, Pollak A, et al. KIF5A de novo mutation associated with myoclonic seizures and neonatal onset progressive leukoencephalopathy. Clin Genet 2017 [DOI] [PubMed] [Google Scholar]
- Safka Brozkova D, Deconinck T, Beth Griffin L, Ferbert A, Haberlova J, Mazanec R, et al. Loss of function mutations in HARS cause a spectrum of inherited peripheral neuropathies. Brain 2015; 138: 2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagi-Dain L, Shemer L, Zelnik N, Zoabi Y, Orit S, Adir V, et al. Whole-exome sequencing reveals a novel missense mutation in the MARS gene related to a rare Charcot-Marie-Tooth neuropathy type 2U. J Peripher Nerv Syst 2018; 23: 138–142. [DOI] [PubMed] [Google Scholar]
- Sainio MT, Ylikallio E, Mäenpää L, Lahtela J, Mattila P, Auranen M, et al. Absence of NEFL in patient-specific neurons in early-onset Charcot-Marie-Tooth neuropathy. Neurol Genet 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambuughin N, Sivakumar K, Selenge B, Suk Lee H, Friedlich D, Baasanjav D, et al. Autosomal dominant distal spinal muscular atrophy type V (dSMA-V) and Charcot-Marie-Tooth disease type 2D (CMT2D) segregate within a single large kindred and map to a refined region on chromosome 7p15. J Neurol Sci 1998; 161: 23–28. [DOI] [PubMed] [Google Scholar]
- Sanchez E, Darvish H, Mesias R, Taghavi S, Firouzabadi SG, Walker RH, et al. Identification of a Large DNAJB2 Deletion in a Family with Spinal Muscular Atrophy and Parkinsonism. Hum Mutat 2016; 37: 1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Cortez RLP, Lee K, Azeem Z, Antonellis PJ, Pollock LM, Khan S, et al. Mutations in KARS, encoding Lysyl-tRNA synthetase, cause autosomal-recessive nonsyndromic hearing impairment DFNB89. Am J Hum Genet 2013; 93: 132–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saporta MA, Dang V, Volfson D, Zou B, Xie XS, Adebola A, et al. Axonal Charcot-Marie-Tooth disease patient-derived motor neurons demonstrate disease-specific phenotypes including abnormal electrophysiological properties. Exp Neurol 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sbrissa D, Ikonomov OC, Fu Z, Ijuin T, Gruenberg J, Takenawa T, et al. Core protein machinery for mammalian phosphatidylinositol 3,5-bisphosphate synthesis and turnover that regulates the progression of endosomal transport: Novel Sac phosphatase joins the ArPIKfyve-PIKfyve complex. J Biol Chem 2007 [DOI] [PubMed] [Google Scholar]
- Schaffer AE, Eggens VRC, Caglayan AO, Reuter MS, Scott E, Coufal NG, et al. CLP1 founder mutation links tRNA splicing and maturation to cerebellar development and neurodegeneration. Cell 2014; 157: 651–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheidecker S, Bär S, Stoetzel C, Geoffroy V, Lannes B, Rinaldi B, et al. Mutations in KARS cause a severe neurological and neurosensory disease with optic neuropathy [Internet]. 2019. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/humu.23799 [DOI] [PubMed]
- Schroer TA. Dynactin. Encycl Biol Chem Second Ed 2013: 174–177. [Google Scholar]
- Schuller AP, Green R. Roadblocks and resolutions in eukaryotic translation. Nat Rev Mol Cell Biol 2018; 19: 526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron 2006; 51: 715–726. [DOI] [PubMed] [Google Scholar]
- Segers K, Glibert G, Callebaut J, Kevers L, Alcan I, Dachy B. Involvement of peripheral and central nervous systems in a valosin-containing protein mutation. J Clin Neurol 2014; 10: 166–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao S, Brown A, Santhanam B, Hegde RS. Structure and Assembly Pathway of the Ribosome Quality Control Complex. Mol Cell 2015; 57: 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen PS, Park J, Qin Y, Li X, Parsawar K, Larson MH, et al. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of Nascent Chains. Science (80-) 2015; 1: 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyltRNA drop-off to initiate no-go decay. Science (80-) 2010; 330: 369–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms CL, Yan LL, Zaher HS, Simms CL, Yan LL, Zaher HS. Ribosome Collision Is Critical for Quality Control during No-Go Decay Article Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol Cell 2017; 68: 361–373.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivakumar K, Kyriakides T, Puls I, Nicholson GA, Funalot B, Antonellis A, et al. Phenotypic spectrum of disorders associated with glycyl-tRNA synthetase mutations. Brain 2005; 128: 2304–2314. [DOI] [PubMed] [Google Scholar]
- Genetic Skre H. and clinical aspects of Charcot-Marie-Tooth’s disease. Clin Genet 1974; 6: 98–118. [DOI] [PubMed] [Google Scholar]
- Sleigh JN, Dawes JM, West SJ, Wei N, Spaulding EL, Gómez-Martín A, et al. Trk receptor signaling and sensory neuron fate are perturbed in human neuropathy caused by Gars mutations. Proc Natl Acad Sci U S A 2017; 114: E3324–E3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevanin G, Santorelli FM, Azzedine H, Coutinho P, Chomilier J, Denora PS, et al. Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nat Genet 2007; 39: 366–372. [DOI] [PubMed] [Google Scholar]
- Storkebaum E, Leitão-Gonçalves R, Godenschwege T, Nangle L, Mejia M, Bosmans I, et al. Dominant mutations in the tyrosyl-tRNA synthetase gene recapitulate in Drosophila features of human Charcot–Marie–Tooth neuropathy. Proc Natl Acad Sci 2009; 106: 11782–11787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B, Chen Z, Ling L, Yang F, Huang X. Clinical and genetic spectra of Charcot-Marie-Tooth disease in Chinese Han patients. J Peripher Nerv Syst 2017; 22: 13–18. [DOI] [PubMed] [Google Scholar]
- Tang BS, Zhao G hua, Luo W, Xia K, Cai F, Pan Q, et al. Small heat-shock protein 22 mutated in autosomal dominant Charcot-Marie-Tooth disease type 2L. Hum Genet 2005; 116: 222–224. [DOI] [PubMed] [Google Scholar]
- Teive H, Kok F, Raskin S, Arruda W. Distal hereditary motor neuropathy with HSJ1 chaperone mutation, presenting with peripheral motor neuropathy, associated to parkinsonism, and cerebellar ataxia. Case report. Parkinsonism Relat Disord 2016; 22: e154. [Google Scholar]
- Timmerman V, Nelis E, Van Hul W, Nieuwenhuijsen BW, Chen KL, Wang S, et al. The periphereal myelin gene PMP-22 is contained within the Charcot-Marie-Tooth disease type 1A duplication. Nat Genet 1992; 1: 171–175. [DOI] [PubMed] [Google Scholar]
- Timmerman V, Raeymaekers P, Nelis E, De Jonghe P, Muylle L, Ceuterick C, et al. Linkage analysis of distal hereditary motor neuropathy type II (distal HMN II) in a single pedigree. J Neurol Sci 1992; 109: 41–48. [DOI] [PubMed] [Google Scholar]
- van Tol H, Beier H. All human tRNA Tyr genes contain introns as a prerequisite for pseudouridine biosynthesis in the anticodon. Nucleic Acids Res 1988; 16: 1951–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkins J, Usher P, Slade JY, Ince PG, Curtis A, Bushby K, et al. Novel insertion in the KSP region of the neurofilament heavy gene in amyotrophic lateral sclerosis (ALS). Neuroreport 1998 [DOI] [PubMed] [Google Scholar]
- Tosolini AP, Sleigh JN. Motor neuron gene therapy: Lessons from spinal muscular atrophy for amyotrophic lateral sclerosis. Front Mol Neurosci 2017; 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurusaki Y, Saitoh S, Tomizawa K, Sudo A, Asahina N, Shiraishi H, et al. A DYNC1H1 mutation causes a dominant spinal muscular atrophy with lower extremity predominance. Neurogenetics 2012 [DOI] [PubMed] [Google Scholar]
- Vagnozzi AN, Praticò D. Endosomal sorting and trafficking, the retromer complex and neurodegeneration. Mol Psychiatry 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdiani S, Origone P, Geroldi A, Bandettini Di Poggio M, Mantero V, Bellone E, et al. The FIG4 gene does not play a major role in causing ALS in Italian patients. Amyotroph Lateral Scler Front Degener 2013 [DOI] [PubMed] [Google Scholar]
- Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife 2013; 2013: 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester A, Velez-Ruiz G, McLaughlin HM, Nisc Comparative Sequencing Program, Lupski JR, Talbot K, et al. A Loss-of-Function Variant in the Human Histidyl-tRNA Synthetase (HARS) Gene is Neurotoxic In Vivo. Hum Mutat 2013; 34: 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilariño-Güell C, Wider C, Soto-Ortolaza AI, Cobb SA, Kachergus JM, Keeling BH, et al. Characterization of DCTN1 genetic variability in neurodegeneration. Neurology 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vissers LELM, De Ligt J, Gilissen C, Janssen I, Steehouwer M, De Vries P, et al. A de novo paradigm for mental retardation. Nat Genet 2010 [DOI] [PubMed] [Google Scholar]
- Vo MN, Terrey M, Lee JW, Roy B, Moresco JJ, Sun L, et al. ANKRD16 prevents neuron loss caused by an editing-defective tRNA synthetase. Nature 2018; 557: 510–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos KJ, Grierson AJ, Ackerley S, Miller CCJ. Role of Axonal Transport in Neurodegenerative Diseases. Annu Rev Neurosci 2008 [DOI] [PubMed] [Google Scholar]
- De Vries H, Ru U, Hu W, Friedlein A. Human pre-mRNA cleavage factor II m contains homologs of yeast proteins and bridges two other cleavage factors. 2000; 19: 5895–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wafik M, Taylor J, Lester T, Gibbons RJ, Shears DJ. 2 new cases of pontocerebellar hypoplasia type 10 identified by whole exome sequencing in a Turkish family. Eur J Med Genet 2018; 61: 273–279. [DOI] [PubMed] [Google Scholar]
- Wagner JD, Huang L, Tetreault M, Majewski J, Boycott KM, Bulman DE, et al. Autosomal recessive axonal polyneuropathy in a sibling pair due to a novel homozygous mutation in IGHMBP2. Neuromuscul Disord 2015: 1–6. [DOI] [PubMed] [Google Scholar]
- Wallen RC, Antonellis A. To charge or not to charge: Mechanistic insights into neuropathy-associated tRNA synthetase mutations. Curr Opin Genet Dev 2013; 23: 302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Mattaj IW, Warren G, Meyer HH, Bilbao-Cortes D, Hetzer M. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol 2001; 3: 1086–1091. [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang Q, Chen Y, Yu S, Wu X, Bao X. Rett and Rett-like syndrome: Expanding the genetic spectrum to KIF1A and GRIN1 gene. Mol Genet Genomic Med 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Xu W, Qin M, Yang Y, Bao P, Shen F, et al. Pathogenic mutations in the valosin-containing protein/p97(VCP) N-domain inhibit the SUMOylation of VCP and lead to impaired stress response. J Biol Chem 2016; 291: 14373–14384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weedon MN, Hastings R, Caswell R, Xie W, Paszkiewicz K, Antoniadi T, et al. Exome sequencing identifies a DYNC1H1 mutation in a large pedigree with dominant Axonal Charcot-Marie-Tooth disease. Am J Hum Genet 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihl CC, Temiz P, Miller SE, Watts G, Smith C, Forman M, et al. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J Neurol Neurosurg Psychiatry 2008; 79: 1186–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzer S, Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature 2007; 447: 222–226. [DOI] [PubMed] [Google Scholar]
- Westhoff B, Chapple JP, van der Spuy J, Hohfeld J, Cheetham ME. HSJ1 is a neuronal shuttling factor for the sorting of chaperone clients to the proteasome. Curr Biol 2005; 15: 1058–1064. [DOI] [PubMed] [Google Scholar]
- Weterman MAJ, Kuo M, Kenter SB, Gordillo S, Karjosukarso DW, Takase R, et al. Hypermorphic and hypomorphic AARS alleles in patients with CMT2N expand clinical and molecular heterogeneities. Hum Mol Genet 2018; 27: 4036–4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wider C, Dachsel JC, Farrer MJ, Dickson DW, Tsuboi Y, Wszolek ZK. Elucidating the genetics and pathology of Perry syndrome. J Neurol Sci 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiessner M, Roos A, Munn CJ, Viswanathan R, Whyte T, Cox D, et al. Yunis-Varón syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am J Hum Genet 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen MH, Vissers LEL, Willemsen MAAP, van Bon BWM, Kroes T, de Ligt J, et al. Mutations in DYNC1H1 cause severe intellectual disability with neuronal migration defects. J Med Genet 2012 [DOI] [PubMed] [Google Scholar]
- Yan Q, Martyn C, Miller L, Shy M, Li J. Demyelination as an Early Pathophysiological Feature in CMT4J (P05.157). Neurology 2012 [Google Scholar]
- Yin HZ, Nalbandian A, Hsu CI, Li S, Llewellyn KJ, Mozaffar T, et al. Slow development of ALS-like spinal cord pathology in mutant valosin-containing protein gene knock-in mice. Cell Death Dis 2012; 3: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yum SW, Zhang J, Mo K, Li J, Scherer SS. A novel recessive NEFL mutation causes a severe, early-onset axonal neuropathy. Ann Neurol 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarouchlioti C, Parfitt DA, Li W, Gittings LM, Cheetham ME. DNAJ Proteins in neurodegeneration : essential and protective factors. 2017 [DOI] [PMC free article] [PubMed]
- van der Zee J, Pirici D, Van Langenhove T, Engelborghs S, Vandenberghe R, Hoffmann M, et al. Clinical heterogeneity in 3 unrelated families linked to VCP p.Arg159His. Neurology 2009; 73: 626–632. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Hashiguchi A, Hu J, Sakiyama Y, Okamoto Y, Tokunaga S, et al. Alanyl-tRNA synthetase mutation in a family with dominant distal hereditary motor neuropathy. Neurology 2012; 78: 1644–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XL, He LX, Yu LJ, Wang Y, Wang XJ, Wang ED, et al. Mutations in KARS cause early-onset hearing loss and leukoencephalopathy: Potential pathogenic mechanism. Hum Mutat 2017; 38: 1740–1750. [DOI] [PubMed] [Google Scholar]
- Züchner S, Vorgerd M, Sindern E, Schröder JM. The novel neurofilament light (NEFL) mutation Glu397Lys is associated with a clinically and morphologically heterogeneous type of Charcot-Marie-Tooth neuropathy. Neuromuscul Disord 2004 [DOI] [PubMed] [Google Scholar]