Abstract
Kinetic modeling of the in vivo pyruvate-to-lactate conversion is crucial to investigating aberrant cancer metabolism that demonstrates Warburg effect modifications. Non-invasive detection of alterations to metabolic flux might offer prognostic value and improve the monitoring of response to treatment. In this clinical research project, hyperpolarized [1-13C] pyruvate was intravenously injected in 10 brain tumor patients to measure its rate of conversion to lactate (kPL) and bicarbonate (kPB) via echo-planar imaging. Our aim was to investigate new methods to provide kPL and kPB maps with whole-brain coverage. The approach was data-driven and addressed two main issues: selecting the optimal model for fitting our data and determining an appropriate goodness-of-fit metric. The statistical analysis suggested that an input-less model had the best agreement with the data. It was also found that selecting voxels based on post-fitting error criteria provided improved precision and wider spatial coverage compared to using signal-to-noise cutoffs alone.
Keywords: Brain Cancer, Dissolution Dynamic Nuclear Polarization, Hyperpolarized MRI, Kinetic Modeling, kPL, kPB, Metabolic Imaging
I. Introduction
Dissolution Dynamic Nuclear Polarization (dDNP)1,2 is a powerful technique that enhances nuclear polarization by up to 5 orders of magnitude.
Its chemical3–5, biological6–9, physical10–14, pre-clinical15–21 and clinical22–28 potentials have been investigated in the last two decades. The use of hyperpolarized (HP) substrates has provided important insights into cancer metabolism6,22,29–31 and cardiac imaging23,32–36. Thus far, HP pyruvate is the most common substrate for in-vivo applications of dDNP because of both its relatively long T1(13C), high biological relevance, and rapid uptake and conversion.
The first phase I clinical trial of HP [1-13C]pyruvate was reported in 201322 on patients with prostate cancer and it demonstrated feasibility and safety for this approach to monitoring cancer progression and treatment response by quantifying the Warburg effect37 that is expressed through increased lactate dehydrogenase (LDH) activity. Several institutions around the globe are now performing clinical cancer research of HP pyruvate in prostate, breast38, liver39 and brain28, in order to investigate the clinical value of dDNP HP MRI and its advantages over current molecular imaging methods including Fludeoxyglucose Positron Emission Tomography (FDG-PET)40,41.
FDG-PET is an imaging technique used for the diagnosis of cancer based on the uptake and trapping of the radioactive substrate, but it does not track its kinetic conversion into downstream metabolites. Furthermore, it is of limited use in brain tumors since high glucose uptake is observed not only in cancerous tissues, but also in healthy cortical gray matter42,43, confounding the discrimination of brain tumors. There is an unmet clinical need for a more precise diagnosis of brain tumors which might be overcome by the capability of dDNP to track the pyruvate-to-lactate conversion and the Warburg effect, provided that a robust and reproducible kinetic modeling of the conversion of pyruvate is achieved44–51.
The goal of this work is to develop, implement and test methods for calculating precise kPL and kPB maps describing the enzymatic conversion of [1-13C]pyruvate to lactate and bicarbonate in the human brain. We acquired 22 datasets though a dynamic 2D multislice Echo Planar Imaging (EPI)52 sequence. Dynamic acquisition of MR images has the advantage to provide robust quantification of kinetic processes53, regardless of differences in bolus delivery, which influences, for instance, area-under-curve (AUC) ratios. EPI acquisition of HP substrates offers improved temporal resolution and greater spatial coverage than Echo Planar Spectroscopic Imaging (EPSI) techniques. We present and compare several models and two approaches to cut off voxels with improper fitting, in order to provide precise and spatially-resolved kinetic maps.
II. METHODS
Sample preparation
The sample preparation was carried out in collaboration with UCSF Radiopharmacy. Each sample consisted of a mixture of Good Manufacturing Practices (GMP) [1-13C]pyruvic acid (14.2 M; MilliporeSigma Isotec) and 15 mM Electron Paramagnetic Agent (EPA), (AH111501, GE Healthcare).
Samples were prepared the same morning of each study and they were inserted, under aseptic conditions, into pharmacy kits from GE Healthcare.
Polarization
A SPINlab54 polarizer (GE Healthcare) was used to polarize [1-13C]pyruvate at 5 T and about 0.8 K. The sample was irradiated with microwave at fMW = 140.02 GHz for at least 2 hours, boosting the 13C polarization up to 40% in the liquid state back-calculated to the time of dissolution.
Pre-injection quality check
After the dissolution, the EPA was filtered mechanically and quality control was carried out prior to injection: pH, pyruvate and EPA concentrations, polarization, and temperature were measured and compared to ranges of acceptability. Contemporarily, the integrity of the 0.2 µm sterile filter was tested following manufacturer specifications on its resistance to a pressure-stress of 50 psi. Following the release by the pharmacist, ~250 mM pyruvate was injected at a dose of 0.43 mL/kg and a rate of 5 mL/s, followed by a 20 mL saline flush. A pause of 5 s preceded the acquisition.
Acquisition
All experiments were performed on a 3T clinical MR scanner (MR750, GE Healthcare) with multi-nuclear excitation and 32-channel reception capability. Carbon-13 RF excitation was achieved with a volume transmit coil and detected with custom 13C RF coils with either 8 or 32 channels55.
Images of pyruvate, lactate and bicarbonate were acquired through a 2D EPI dynamic sequence53, capable of exciting multiple 13C frequencies sequentially, and having the following characteristics: 8 slices, 1.5 or 2 cm thick axial slice, TR = 62.5 ms, TE = 21.9 ms, 20 time-points with a 3 s temporal resolution, in-plane resolutions of 1.2 × 1.2, 1.5 × 1.5 or 2 × 2 cm2. In order to improve image quality, we applied noise pre-whitening56 in k-space prior to sum-of-squares coil combination.
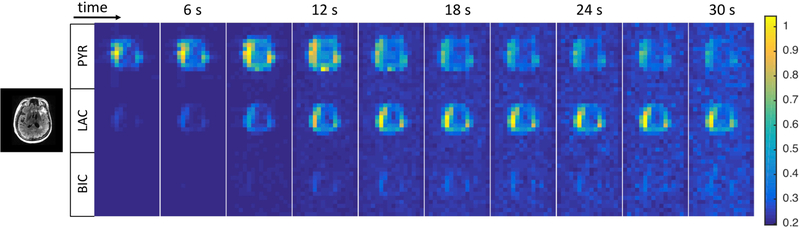
In Fig. 1 we show the temporal trends of the signals of pyruvate, lactate and bicarbonate in a single slice.
Figure 1.
Temporal dynamics of the signals of pyruvate, lactate and bicarbonate in a single slice (data set 3 in Table 1), acquired with the EPI dynamic sequence (see text for details). For anatomic reference, a 1H FLAIR image on the left. Note that data have not been coil corrected and reflect the sensitivity profile of the 8-channel receive-array used in this study.
III. KINETIC MODELING
In the last decade, substantial efforts have been dedicated to developing kinetic models of MRI signals of HP metabolites in vivo. Despite several approaches available in literature 14,25–30, no gold-standard is available because strategies may vary according to specific experimental conditions.
Since kinetic modelling might suffer from errors induced by several factors (e.g., variable bolus delivery, acquisition sequence, magnitude images with non-zero mean noise etc), discarding voxels with poor fitting is critical to provide precise kinetic maps.
In this section, we describe kinetic models and fitting criteria to address these issues and to quantify kinetic rate constants in a precise way.
First, suitable models for fitting our data are presented.
Then, we describe two criteria to decide how to discard voxels with suboptimal fitting.
Models
While studying the human brain, in order to obtain insights into complicated metabolic pathways, it is crucial to investigate the kinetics of several metabolites in different cellular environments. Although it is an oversimplification, we consider a three-site system (i.e., pyruvate, lactate and bicarbonate) in a single compartment. In such a system, the magnetization is exchanged and relaxes according to the following equations (single macrons refer to vectors, double macrons to matrixes):
| [1] |
| [2] |
| [3] |
| [4] |
We defined:
and as the rates of labelling exchange from pyruvate to lactate and to bicarbonate (respectively). Note they are first order apparent rate constants and not absolute flux measurements.
as spin longitudinal relaxation rate.
as the experimental temporal trend of the magnetization of pyruvate.
as the experimental magnetization vector at the first time point.
as the experimental magnetization vector at the last time point.
Overall, 5 approaches were tested:
-
, , , , .
This is a model with no input-function57: the fitted signal of pyruvate is bounded to the experimentally acquired signal. The first and last time points are fixed to the experimental values (for all metabolites). Just forward reactions are considered.
Fitted parameters: , , .
Same as A) except for having now (back reaction now allowed with a different rate). Fitted parameters: , , ,
Same as A) except for having now (last time points are now estimated from the fitting). Fitted parameters: , , , .
Same as A) except for having now and (both first and last time points are estimated from the fitting). Fitted parameters: , , , , .
, , . In this model , meaning that there is an input function (defined in Eq. 4) simulating the arrival of the bolus in the region of interest. The magnetization of pyruvate is also fitted. First and last points are set equal to experimental values. Just forward reactions are considered. Fitted parameters: , , , , , , , .
The differential equations 1–4 were solved through a discrete model. The transverse magnetization, , and the residual longitudinal component along the z axis, , were computed in Eq 5 and 6 at each time step (where = TR and N = 1, 2, …, 20), taking into account the excitation flip angles for each metabolite.
The magnetization at time step N+1 was calculated in Eq. 7 by considering the exchange matrix , and .
| [5] |
| [6] |
| [7] |
All models were solved via custom MATLAB routines involving minimization of non-linear least-square problems (i.e., difference between fitted and experimental magnetization). σkPL is half the 95% confidence interval, calculated with MATLAB functions lsqnonlin and nlparci. In all cases, boundary conditions were: 1/80 s−1 < R1 < 1 s−1 and 0 < kPL < 1 s−1.
Experimental T1s of lactate and pyruvate vary significantly in solution. However, a recent work53 has shown that the input-less model is insensitive to errors in pyruvate T1 and that T1 values for pyruvate and lactate are similar in vivo (respectively 30 and 25 s, in patient studies at 3 T). We, therefore, chose to have equal T1 values to improve the stability of the model.
After fitting to experimental data, models were classified according to 3 indicators, Akaike Information Criterion (AIC), and , defined as:
| [8] |
| [9] |
| [10] |
where k is the numbers of parameters estimated in the model, L is the maximum value of the likelihood function for the model (i.e., chi squared in our case), is the number of voxels, in all data sets, that are fitted according to the ith model (with i = A-E), is the corresponding to model A, is the mean kPL, averaged over all data sets, for the ith model and is the corresponding to model A.
The AIC is a measure of the statistical significance of several models with a different number of parameters to be estimated (i.e., the lower the better statistical significance for the model). is informative of how many voxels are fitted if compared to model A (chosen arbitrarily as a standard) and is a measure of how much kPL varies with models.
To assess the reproducibility of the pyruvate bolus delivery, we define a “mean pyruvate time” as the center of mass of the pyruvate signal over time on a voxel-wise basis53:
| [11] |
Criteria to mask voxels on kPL maps
In this paragraph, we introduce two approaches to mask voxels with improper fitting in order to provide “reasonable” kPL maps with an acceptable compromise between precision of the fitting in a voxel and the numbers of voxel for which the fitting itself is retained (i.e. the coverage). Due to low SNR, kPB maps describing the conversion of pyruvate into bicarbonate will not be considered in this analysis. In the “SNR criterion”, prior to fitting temporal trends to a model, we create a mask discarding voxels with peak SNR lower than a set of two SNR thresholds, one for pyruvate and one for lactate, labeled as [, ]. In the “error criterion”, prior to fitting temporal trends to a model, we mask voxels based on [, ], as in the SNR criterion, but, after fitting, we discard all the voxels where is bigger than a fraction of kPL itself.
In this way, the global masking procedure depends on both the set of SNRs and on the statistical outcome of the fitting.
To compare the outcomes of fitting with the two criteria, we will take into account (as in Eq. 9 but now the index refers to the criterion) and R2, defined as:
| [12] |
where yk and fk refer to the experimental and fitted signals at the kth time point and is the average over time of yk. R2 is preferred over AIC or chi squared since these last two vary with different SNR (i.e., if data with lower SNR are filtered, the residuals are bigger).
IV. RESULTS AND DISCUSSION
The subject population comprised 10 patients with recurrent glioma who had received prior surgery and were undergoing standard-of-care treatment, including radiation therapy and temozolomide chemotherapy, along with a variety of adjuvant therapies. A subset of these patients was imaged serially, and all received a single injection per day. Table 1 includes the experimental conditions under which the 22 data sets were acquired (i.e., coil, voxel size, excitation flip angles) and estimation of (the peak SNR in all voxels and slices in the given data set), (mean arrival time of pyruvate) and (mean kPL rate).
Table 1.
Summary of the experimental conditions of the 22 human brain data sets. We report coil used, voxel size, flip angles of excitation for each metabolite, peak SNR (along all voxels and slices in the given data set), mean kinetic rate kPL and its standard deviation (calculated with “model A” and the error criterion, described later on in the manuscript) and mean and standard deviation of the mean arrival time of pyruvate tarr.
| Set | Coil | Voxel (cm3) | αpyr(°) | αlac(°) | αbic(°) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient 1 | 1 | 8ch | 2 × 2 × 2 | 33.4 | 33.4 | 33.4 | 393 | 53 | 15 | 0.021±0.008 | 6±2 |
| 2 | 32ch | 1.2 × 1.2 × 2 | 10 | 33.4 | 33.4 | 120 | 22 | 27 | 0.013±0.006 | 7±3 | |
| 3 | 8ch | 2 × 2 × 2 | 33.4 | 33.4 | 33.4 | 437 | 79 | 13 | 0.05±0.02 | 7±4 | |
| 4 | 32ch | 1.2 ×1.2 × 2 | 20 | 30 | 30 | 306 | 22 | 18 | 0.010±0.005 | 6± 3 | |
| Patient 2 | 5 | 8ch | 1.2 ×1.2 × 2 | 33.4 | 33.4 | 33.4 | 306 | 20 | n/a | 0.024±0.009 | 11±4 |
| 6 | 8ch | 2 × 2 × 2 | 20 | 30 | 30 | 170 | 26 | 11 | 0.018±0.007 | 8±5 | |
| 7 | 32ch | 2 × 2 × 2 | 20 | 30 | 30 | 504 | 30 | 15 | 0.016±0.007 | 7±2 | |
| 8 | 8ch | 1.5 ×1.5 × 2 | 20 | 30 | 30 | 403 | 42 | 18 | 0.022±0.009 | 7±2 | |
| Patient 3 | 9 | 8ch | 2 × 2 × 2 | 20 | 30 | 30 | 281 | 41 | 12 | 0.018±0.008 | 7±2 |
| 10 | 32ch | 1.2 ×1.2 × 2 | 20 | 30 | 30 | 576 | 21 | 29 | 0.009±0.004 | 9±3 | |
| Patient 4 | 11 | 8ch | 2 × 2 × 2 | 20 | 30 | 30 | 225 | 43 | 13 | 0.03±0.01 | 10±2 |
| 12 | 32ch | 1 × 1 × 2 | 20 | 30 | 30 | 492 | 25 | 32 | 0.012±0.006 | 8±3 | |
| 13 | 32ch | 1.5 × 1.5 × 1.5 | 20 | 30 | 30 | 181 | 16 | 12 | 0.022±0.008 | 7±3 | |
| 14 | 32ch | 1.5 × 1.5 × 1.5 | 20 | 30 | 30 | 361 | 24 | 14 | 0.02±0.01 | 7±2 | |
| Patient 5 | 15 | 8ch | 1.2 × 1.2 × 2 | 10 | 33.4 | 33.4 | 101 | 22 | 26 | 0.015±0.007 | 6±3 |
| 16 | 32ch | 1.5 × 1.5 × 2 | 20 | 30 | 30 | 392 | 35 | 15 | 0.015±0.008 | 7±2 | |
| 17 | 32ch | 1.5 × 1.5 × 2 | 20 | 30 | 30 | 392 | 35 | 15 | 0.017±0.007 | 11±3 | |
| Patient 6 | 18 | 8ch | 1.2 × 1.2 × 2 | 33.4 | 33.4 | 33.4 | 559 | 22 | 12 | 0.012±0.004 | 10±3 |
| Patient 7 | 19 | 8ch | 2 × 2 × 2 | 20 | 30 | 271 | 41 | 13 | 0.03±0.01 | 9±3 | |
| Patient 8 | 20 | 8ch | 2 × 2 × 2 | 20 | 30 | 568 | 58 | 16 | 0.03±0.01 | 8±2 | |
| Patient 9 | 21 | 8ch | 1.2 × 1.2 × 2 | 10 | 33.4 | 128 | 25 | 13 | 0.02±0.01 | 6±3 | |
| Patient 10 | 22 | 32ch | 1.5 × 1.5 × 2 | 20 | 30 | 30 | 162 | 13 | 13 | 0.009±0.005 | 7±3 |
In Fig. 2, we can see typical temporal trends of the signal-to-noise ratio of pyruvate, lactate and bicarbonate in a single voxel, selected from a slice (also shown for all metabolites). The SNR did not go to zero at later time-points due to the sum-of-squares coil combination used in this work, which results in non-zero mean noise caused by noise rectification of magnitude data.
Figure 2.
[Top] 1H FLAIR and 13C total signal AUC images (data set 3) from the raw data. [Bottom] Temporal dynamics of signals of pyruvate, lactate and bicarbonate in the selected voxel.
While there has been no comprehensive study on the impact of spatial resolution on the measured reaction rate, larger voxels would have a greater risk of partial-volume effects, reducing the measured reaction rate because of the strong vascular pyruvate signal58. Confounding factors such as Rician bias in magnitude data and variations in bolus delivery between patients are global factors that impact quantification. On a voxelwise level, regional variations in perfusion, signal loss from susceptibility/B0 inhomogeneities and reduced SNR in voxels at the center of the brain, farthest from the multichannel receive array, can also impact quantification. The choice of flip angles will also have an impact on the precision of rate constant estimates57,59,60. Using a variable (across frequency) flip angle scheme, with a higher flip angle for downstream metabolites and a lower flip angle for pyruvate, is commonly used to compensate for the lower metabolite concentration and to reduce saturation of pyruvate. This leads to more precise fitting in the presence of noise by increasing the metabolite SNR61. While variable (through time) flip angle schemes can improve SNR, they are also more sensitive to transmit B1 inhomogeneity59,62.
The was excellent for pyruvate and lactate (i.e., greater than 100 and 13, respectively, for all datasets) but greatly variable for bicarbonate.
The mean arrival time of the pyruvate bolus, , was calculated for every data set via Eq. 11. While there is variability throughout the brain due to differences in arrival time between arterial and venous voxels, the mean pyruvate time ranged from 6 s to 11 s post injection, with an average value of 8 ± 2 s across patients. Compared to initial results from the prostate63, the mean arrival time in the brain appears fairly reproducible in our initial cohort.
Mean kPL and its standard deviation were also calculated on each data set, with the approach described in the next paragraphs.
Let us define σkPL as half the 95% confidence interval from non-linear least-square fitting to “model A” with the error criterion with [, ] = [20, 5] and σkPL < 0.6·kPL. We have applied the error criterion with [, ] = [20, 5] and σkPL < 0.6·kPL to compare models A to E. Values of AIC, and are reported in Table 2: model A was the best model at describing data (highest and lowest AIC) and model B the worst (high AIC and negligible ). Model E, which is the only one including an input function, performed worse than model A (the simplest assumption-less model), due to a poor agreement between the input function and our data which may be improved with an earlier start of acquisition.
Table 2.
Comparison of models A-E according to Akaike Information Criterion (AIC) and percentage deviation from model A of mean and of number total voxel fitted .
| Model | AIC | ||
|---|---|---|---|
| A: inputless, and bounded, no back reaction | 4 | 0 | 100 |
| B: inputless, and bounded, no back reaction | 11 | −363 | 85 |
| C: inputless, bounded, no back reaction | 6 | −5 | 85 |
| D: inputless, no back reaction | 10 | −7 | 86 |
| E: input function, and bounded, no back reaction. | 13 | −40 | 55 |
Parameters used in both the SNR and the error criterion should be optimized in order to find a compromise between precision and number of voxels fulfilling the statistical requirements based on σkPL (i.e., spatial coverage).
The analysis has been carried out on datasets with peak SNR of lactate lower than 25, by using model A, since it is the one providing the best fit quality.
In the SNR criterion, we have to choose [, ] namely the set of thresholds for masking voxels with low SNR (before fitting). We tested 22 sets obtained by all the possible combinations between = [5, 10, 50, 100] and = [2, 3, 4, 5, 6, 7].
In the error criterion, the SNR mask was computed based on 6 threshold sets, obtained by combining = 5 and = [2, 3, 4, 5, 6, 7]. After fitting, we discarded all the voxels with σkPL < c∙kPL where c = [0.4, 0.6, 0.8, 1.0].
Values of and R2 are reported in Tables 3a and 3b (respectively) for both the SNR criterion and the error criterion. To produce kinetic maps, it is necessary to find a compromise between statistical precision and available number of voxels (or alternatively the voxel count within the FOV) and the final decision may vary from one user to the other.
Table 3a.
Values of for the SNR criterion and the error criterion using model A as standard. Several combinations of and were used for both criteria. In the SNR criterion no conditions are set on σkPL (error of kPL), whereas, in the error criterion, fitting is accepted for σkPL < c·kPL and rejected otherwise.
|
SNR criterion (no check on σkPL after fitting) |
Error criterion:σkPL< c · kPL | |||||||
|---|---|---|---|---|---|---|---|---|
| c =1.0 | c = 0.8 | c = 0.6 | c = 0.4 | |||||
| ╲ | 5 | 10 | 50 | 100 | 5 | 5 | 5 | 5 |
| 2 | 100 | 86 | 26 | 6 | 51 | 45 | 34 | 18 |
| 3 | 50 | 47 | 19 | 5 | 33 | 31 | 27 | 17 |
| 4 | 30 | 29 | 13 | 3 | 21 | 20 | 19 | 14 |
| 5 | 17 | 17 | 9 | 3 | 12 | 12 | 12 | 10 |
| 6 | 11 | 11 | 6 | 3 | 8 | 8 | 8 | 7 |
| 7 | 6 | 6 | 4 | 2 | 4 | 4 | 4 | 4 |
Table 3b.
Values of R2 for the SNR criterion and the error criterion using model A as standard. Several combinations of SNRpyr and SNRlac were used for both criteria. In the SNR criterion no conditions are set on σkPL (error of kPL), whereas, in the error criterion, fitting is accepted for σkPL < c·kPL and rejected otherwise.
| R2 |
SNR criterion (no check on σkPL after fitting) |
Error criterion:σkPL< c · kPL | ||||||
|---|---|---|---|---|---|---|---|---|
| c =1.0 | c = 0.8 | c = 0.6 | c = 0.4 | |||||
| ╲ | 5 | 10 | 50 | 100 | 5 | 5 | 5 | 5 |
| 2 | 0.27 | 0.30 | 0.38 | 0.43 | 0.37 | 0.40 | 0.47 | 0.60 |
| 3 | 0.39 | 0.41 | 0.44 | 0.49 | 0.43 | 0.50 | 0.53 | 0.60 |
| 4 | 0.41 | 0.42 | 0.51 | 0.57 | 0.42 | 0.49 | 0.60 | 0.65 |
| 5 | 0.53 | 0.53 | 0.66 | 0.58 | 0.53 | 0.54 | 0.64 | 0.67 |
| 6 | 0.66 | 0.66 | 0.67 | 0.57 | 0.66 | 0.66 | 0.67 | 0.69 |
| 7 | 0.70 | 0.70 | 0.71 | 0.59 | 0.70 | 0.70 | 0.70 | 0.72 |
Tables 3a and 3b provide insight on how to set limits in our case. In Tab. 3a, drops with higher , higher and lower c, showing that any stricter requirement diminishes the numbers of accepted voxels. For the SNR criterion (Tab. 3b), R2 increases with higher but not necessarily with higher . This suggests that the precision of methods based on the SNR alone depends more critically on lactate rather than on pyruvate. In Tab. 3b, as for the error criterion, R2 increases with both higher and lower c, meaning that both parameters affect the statistical precision.
In Tab. 3b, let us consider the R2 values corresponding to in both criteria (i.e., first column in the SNR criterion and all columns in the error criterion). Values of R2 in case of in the SNR criterion are smaller than any R2 in the error criterion (regardless of c). However, R2 for in the SNR criterion are comparable to those in the error criterion. This means that at low the error criterion performs better than the SNR criterion, while they are comparable at higher .
By comparing Tab. 3a and 3b, we can see that, at similar precisions, the error criterion gives, on average, more voxels with proper fitting (i.e., higher ), yielding to a better spatial coverage in kPL maps. The trend is particularly evident at low .
For example, let us consider two cases with R2 = 0.43 in Tab. 3b: those with [, ] = [100, 2] for the SNR criterion and [, ] = [5, 3] with σkPL < 1.0∙kPL for the error criterion. These two cases exhibit the most dramatic difference since their corresponding values of in Tab. 3a would be 6 and 33, meaning that the error criterion provided 5 times more voxels despite having similar statistical precision.
Furthermore, if one wanted to base the selection on the SNR alone, removing the check on σkPL, in order to choose precise values, it would be necessary to perform many fittings at different SNR thresholds in every dataset and to fix a reasonable threshold.
The error criterion is an easier option since it is sufficient to set and to reasonably low thresholds (high enough to avoid to retain voxels with noisy trends) and leave the main filtering task to the criterion based on σkPL.
We investigated the performances of the error criterion when applied to random noise.
To do so, we generated several sets of lactate signals consisting of Gaussian noise and associated them to the experimental signals of pyruvate from data set 3. Then, we applied the error criterion with [, ] = [5, 3] and σkPL < 0.6∙kPL and calculated the number of fitted voxels, namely those where the fitting fulfils the requirements of the error criterion. We found that, if compared to experimental signals from data set 3, random noise provides, at best, 40 times less fitted voxels.
This suggests that the improvements introduced by the error criterion on and R2 in Tables 3a and 3b are barely influenced by the noise.
All these findings speak in favor of the error criterion, which seems to be more reasonable than the SNR approach at masking voxel to be displayed in kPL maps.
The choice of [, ] and c is arbitrary and looking at the outcome alone might be misleading, as shown in Fig. 3. On the quest to opt for specific values, we started by considering the approach which has been often used in our group so far, namely [, ] = [10, 5]. According to tables 3a and 3b, it has = 17 and R2 = 0.53.
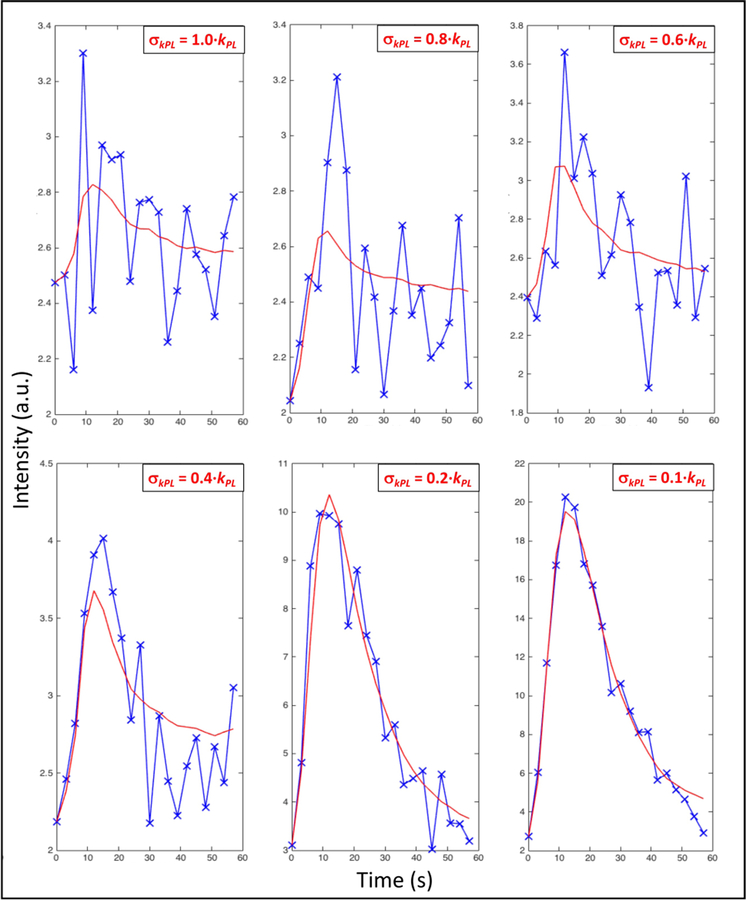
Figure 3.
Temporal dynamics of the signal of lactate in a voxel (blue crosses) and fitting (red line) using model A and the error criterion in 6 cases with different values of the parameter c in the condition σkPL < c kPL (see text for details).
Bearing this in mind, we created kPL and k 64 maps for every data set, by using model A combined with the error criterion with [, ] = [5, 3] and c = 0.6.
These set of values has = 27 and R2 = 0.53, meaning that, if compared to the former approach, it provides about 60% more voxels albeit keeping similar statistical precision.
As for data sets in Table 1, the mean value of the fractional error c = σkPL / kPL in a single data set varies between 0.16 and 0.34, giving an idea of how they link to the graphic outcomes of fitting in Fig. 3.
In Fig. 4 we show examples of 5 kPL maps superimposed to FLAIR images of a patient affected with gliosarcoma (data set 14) and of 5 kPB maps superimposed to T1-weighted images of a patient affected with gliosarcoma (data set 16).
Figure 4.
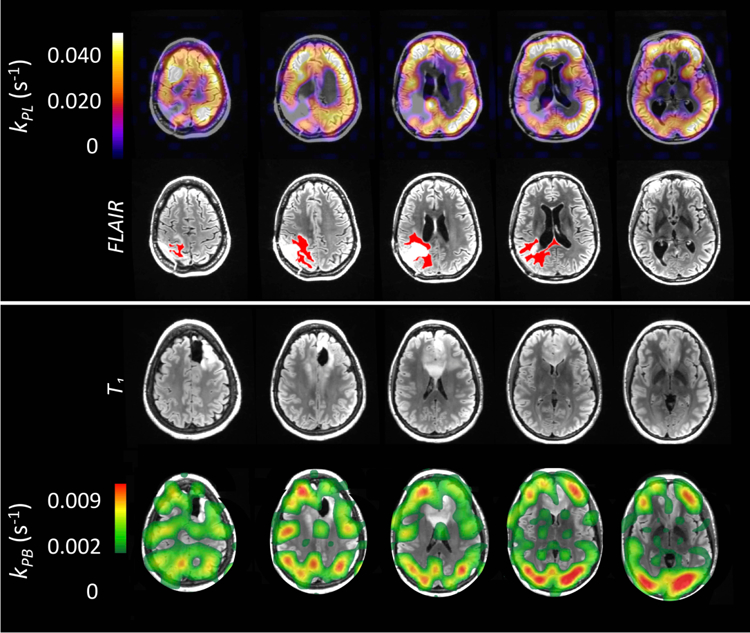
[Top] Examples of kPL maps superimposed to FLAIR images of a patient (data set 14) affected with glioma (active tumor highlighted in red) [Bottom] Examples of kPB maps superimposed to T1-weighted images of a patient (data set 16). Sinc interpolation and the error criterion (σkPL < 0.6∙kPL) were used to generate kinetic maps in both cases (see text for detail).
All the kPL maps have been computed by using the error criterion with [, ] = [5, 3] and σkPL < 0.6∙kPL. The patient affected with gliosarcoma has previously received radiation and temozolomide chemotherapy, along with adjuvant agent pembrolizumab.
The interpretation of the correspondence between higher or lower kPL values and presence of tumor is deferred to future studies since it is beyond the scope of this communication, that is mainly focused on methods to compute kinetic maps for our brain data sets.
V. CONCLUSIONS
We conducted preliminary studies of hyperpolarized [1-13C] pyruvate injected in 10 patients affected with glioma, demonstrating the feasibility for quantifying kinetic rates kPL and kPB, which describe the conversion of pyruvate into lactate and bicarbonate. We compared five kinetic models and two approaches for discarding voxels with improper fitting and we provided quantitative and spatially resolved maps of kinetic rates. Such hyperpolarized MRI experiments enable an unprecedented window into cerebral metabolism, which is not provided by other molecular imaging techniques.
Acknowledgments
We thank the funding sources: NICO and NIH grants P01CA118816, R01EB017449 and P41EB013598. As for the first author, the views expressed are purely those of the writer and may not in any circumstances be regarded as stating an official position of the European Commission.
References and Footnotes
- 1.Ardenkjaer-Larsen JH. On the present and future of dissolution-DNP. Journal of Magnetic Resonance 264, 3–12 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M & Golman K. Increase in signal-to-noise ratio of > 10,000 times in liquid-state NMR. Proceedings of the National Academy of Sciences 100, 10158–10163 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mammoli D, Vuichoud B, Bornet A, Milani J, Dumez J-N, Jannin S & Bodenhausen G. Hyperpolarized para - Ethanol. The Journal of Physical Chemistry B 119, 4048–4052 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Bornet A, Ji X, Mammoli D, Vuichoud B, Milani J, Bodenhausen G & Jannin S. Long-Lived States of Magnetically Equivalent Spins Populated by Dissolution-DNP and Revealed by Enzymatic Reactions. Chemistry - A European Journal 20, 17113–17118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowen S & Hilty C. Rapid sample injection for hyperpolarized NMR spectroscopy. Physical Chemistry Chemical Physics 12, 5766 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Day SE, Kettunen MI, Gallagher FA, Hu D-E, Lerche M, Wolber J, Golman K, Ardenkjaer-Larsen JH & Brindle KM. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nature Medicine 13, 1382–1387 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Ardenkjaer-Larsen J-H, Boebinger GS, Comment A, Duckett S, Edison AS, Engelke F, Griesinger C, Griffin RG, Hilty C, Maeda H, Parigi G, Prisner T, Ravera E, van Bentum J, Vega S, Webb A, Luchinat C, Schwalbe H & Frydman L. Facing and Overcoming Sensitivity Challenges in Biomolecular NMR Spectroscopy. Angewandte Chemie International Edition 54, 9162–9185 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratto R, Bornet A, Milani J, Mammoli D, Vuichoud B, Salvi N, Singh M, Laguerre A, Passemard S, Gerber-Lemaire S, Jannin S & Bodenhausen G. Drug Screening Boosted by Hyperpolarized Long-Lived States in NMR. ChemMedChem 9, 2509–2515 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Zeng H, Ruedisser S, Gossert AD & Hilty C. Nuclear Magnetic Resonance of Hyperpolarized Fluorine for Characterization of Protein–Ligand Interactions. Journal of the American Chemical Society 134, 17448–17451 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Vasos PR, Comment A, Sarkar R, Ahuja P, Jannin S, Ansermet J-P, Konter JA, Hautle P, van den Brandt B & Bodenhausen G. Long-lived states to sustain hyperpolarized magnetization. Proceedings of the National Academy of Sciences 106, 18469–18473 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jannin S, Bornet A, Melzi R & Bodenhausen G. High field dynamic nuclear polarization at 6.7T: Carbon-13 polarization above 70% within 20min. Chemical Physics Letters 549, 99–102 (2012). [Google Scholar]
- 12.Meier B, Dumez J-N, Stevanato G, Hill-Cousins JT, Roy SS, Håkansson P, Mamone S, Brown RCD, Pileio G & Levitt MH. Long-Lived Nuclear Spin States in Methyl Groups and Quantum-Rotor-Induced Polarization. Journal of the American Chemical Society 135, 18746–18749 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Pileio G, Bowen S, Laustsen C, Tayler MCD, Hill-Cousins JT, Brown LJ, Brown RCD, Ardenkjaer-Larsen JH & Levitt MH. Recycling and Imaging of Nuclear Singlet Hyperpolarization. Journal of the American Chemical Society 135, 5084–5088 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Tayler MCD, Marco-Rius I, Kettunen MI, Brindle KM, Levitt MH & Pileio G. Direct Enhancement of Nuclear Singlet Order by Dynamic Nuclear Polarization. Journal of the American Chemical Society 134, 7668–7671 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Gallagher FA, Kettunen MI, Day SE, Hu D-E, Ardenkjær-Larsen JH, Zandt R in ‘t, Jensen PR, Karlsson M, Golman K, Lerche MH & Brindle KM. Magnetic resonance imaging of pH in vivo using hyperpolarized 13C-labelled bicarbonate. Nature 453, 940–943 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Gallagher FA, Kettunen MI & Brindle KM. Biomedical applications of hyperpolarized 13C magnetic resonance imaging. Progress in Nuclear Magnetic Resonance Spectroscopy 55, 285–295 (2009). [Google Scholar]
- 17.Chen AP, Albers MJ, Cunningham CH, Kohler SJ, Yen Y-F, Hurd RE, Tropp J, Bok R, Pauly JM, Nelson SJ, Kurhanewicz J & Vigneron DB. Hyperpolarized C-13 spectroscopic imaging of the TRAMP mouse at 3T—Initial experience. Magnetic Resonance in Medicine 58, 1099–1106 (2007). [DOI] [PubMed] [Google Scholar]
- 18.von Morze C, Larson PEZ, Hu S, Keshari K, Wilson DM, Ardenkjaer-Larsen JH, Goga A, Bok R, Kurhanewicz J & Vigneron DB. Imaging of blood flow using hyperpolarized [13C]Urea in preclinical cancer models. Journal of Magnetic Resonance Imaging 33, 692–697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comment A. Dissolution DNP for in vivo preclinical studies. Journal of Magnetic Resonance 264, 39–48 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Keshari KR, Sriram R, Koelsch BL, Van Criekinge M, Wilson DM, Kurhanewicz J & Wang ZJ. Hyperpolarized 13C-Pyruvate Magnetic Resonance Reveals Rapid Lactate Export in Metastatic Renal Cell Carcinomas. Cancer Research 73, 529–538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marco-Rius I, von Morze C, Sriram R, Cao P, Chang G-Y, Milshteyn E, Bok RA, Ohliger MA, Pearce D, Kurhanewicz J, Larson PEZ, Vigneron DB & Merritt M. Monitoring acute metabolic changes in the liver and kidneys induced by fructose and glucose using hyperpolarized [2–13C]dihydroxyacetone: Monitoring Metabolic Changes with [2–13C]dihydroxyacetone. Magnetic Resonance in Medicine 77, 65–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson SJ, Kurhanewicz J, Vigneron DB, Larson PEZ, Harzstark AL, Ferrone M, van Criekinge M, Chang JW, Bok R, Park I, Reed G, Carvajal L, Small EJ, Munster P, Weinberg VK, Ardenkjaer-Larsen JH, Chen AP, Hurd RE, Odegardstuen L-I, Robb FJ, Tropp J & Murray JA. Metabolic Imaging of Patients with Prostate Cancer Using Hyperpolarized [1–13C]Pyruvate. Science Translational Medicine 5, 198ra108–198ra108 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunningham CH, Lau JYC, Chen AP, Geraghty BJ, Perks WJ, Roifman I, Wright GA & Connelly KA. Hyperpolarized 13C Metabolic MRI of the Human HeartNovelty and Significance: Initial Experience. Circulation Research 119, 1177–1182 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aggarwal R, Vigneron DB & Kurhanewicz J. Hyperpolarized [1–13C]-Pyruvate Magnetic Resonance Imaging Detects an Early Metabolic Response to Androgen Ablation Therapy in Prostate Cancer. European Urology 72, 1028–1029 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park I, Larson PEZ, Gordon JW, Carvajal L, Chen H-Y, Bok R, Van Criekinge M, Ferrone M, Slater JB, Xu D, Kurhanewicz J, Vigneron DB, Chang S & Nelson SJ. Development of methods and feasibility of using hyperpolarized carbon-13 imaging data for evaluating brain metabolism in patient studies: Hyperpolarized Carbon-13 Metabolic Imaging of Patients With Brain Tumors. Magnetic Resonance in Medicine 80, 864–873 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H-Y, Larson PEZ, Gordon JW, Bok RA, Ferrone M, van Criekinge M, Carvajal L, Cao P, Pauly JM, Kerr AB, Park I, Slater JB, Nelson SJ, Munster PN, Aggarwal R, Kurhanewicz J & Vigneron DB. Technique development of 3D dynamic CS-EPSI for hyperpolarized 13C pyruvate MR molecular imaging of human prostate cancer. Magnetic Resonance in Medicine (2018). doi: 10.1002/mrm.27179 [DOI] [PMC free article] [PubMed]
- 27.Gordon JW, Hansen RB, Shin PJ, Feng Y, Vigneron DB & Larson PEZ. 3D hyperpolarized C-13 EPI with calibrationless parallel imaging. Journal of Magnetic Resonance 289, 92–99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miloushev VZ, Granlund KL, Boltyanskiy R, Lyashchenko SK, DeAngelis LM, Mellinghoff IK, Brennan CW, Tabar V, Yang TJ, Holodny AI, Sosa RE, Guo YW, Chen AP, Tropp J, Robb F & Keshari KR. Metabolic Imaging of the Human Brain with Hyperpolarized 13C Pyruvate Demonstrates 13C Lactate Production in Brain Tumor Patients. Cancer Research 78, 3755–3760 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albers MJ, Bok R, Chen AP, Cunningham CH, Zierhut ML, Zhang VY, Kohler SJ, Tropp J, Hurd RE, Yen Y-F, Nelson SJ, Vigneron DB & Kurhanewicz J. Hyperpolarized 13C Lactate, Pyruvate, and Alanine: Noninvasive Biomarkers for Prostate Cancer Detection and Grading. Cancer Research 68, 8607–8615 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brindle KM, Bohndiek SE, Gallagher FA & Kettunen MI. Tumor imaging using hyperpolarized 13C magnetic resonance spectroscopy. Magnetic Resonance in Medicine 66, 505–519 (2011). [DOI] [PubMed] [Google Scholar]
- 31.Golman K, Zandt R. i., Lerche M, Pehrson R & Ardenkjaer-Larsen JH. Metabolic Imaging by Hyperpolarized 13C Magnetic Resonance Imaging for In vivo Tumor Diagnosis. Cancer Research 66, 10855–10860 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Lau AZ, Chen AP, Ghugre NR, Ramanan V, Lam WW, Connelly KA, Wright GA & Cunningham CH. Rapid multislice imaging of hyperpolarized 13C pyruvate and bicarbonate in the heart. Magnetic Resonance in Medicine 64, 1323–1331 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Merritt ME, Harrison C, Storey C, Jeffrey FM, Sherry AD & Malloy CR. Hyperpolarized 13C allows a direct measure of flux through a single enzyme-catalyzed step by NMR. Proceedings of the National Academy of Sciences 104, 19773–19777 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schroeder MA, Cochlin LE, Heather LC, Clarke K, Radda GK & Tyler DJ. In vivo assessment of pyruvate dehydrogenase flux in the heart using hyperpolarized carbon-13 magnetic resonance. Proceedings of the National Academy of Sciences 105, 12051–12056 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golman K, Petersson JS, Magnusson P, Johansson E, Åkeson P, Chai C-M, Hansson G & Månsson S. Cardiac metabolism measured noninvasively by hyperpolarized13C MRI. Magnetic Resonance in Medicine 59, 1005–1013 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Fuetterer M, Busch J, Peereboom SM, von Deuster C, Wissmann L, Lipiski M, Fleischmann T, Cesarovic N, Stoeck CT & Kozerke S. Hyperpolarized 13C urea myocardial first-pass perfusion imaging using velocity-selective excitation. Journal of Cardiovascular Magnetic Resonance 19, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warburg O. On the origin of cancer cells. Science 123, 309–314 (1956). [DOI] [PubMed] [Google Scholar]
- 38.McLean MA, Daniels C, Grist J, Schulte R, Lanz T, Chhabra A, Earl HM, Basu B, Wilkinson IB, Lomas D, Caldas C, Abraham JE, Graves M, Gilbert F, Brindle KM & Gallagher FA Feasibility of metabolic imaging of hyperpolarized 13C-pyruvate in human breast cancer. (2018). doi: 10.17863/CAM.21154 [DOI]
- 39.Zhu Z & et al. Development of Hyperpolarized 13C Dynamic Breath-held MR Techniques and Initial Feasibility Studies for Metabolic Imaging of Liver Metastases. Proceeding of Joint Annual Meeting ISMRM-ESMRMB (2018).
- 40.Gallagher FA, Bohndiek SE, Kettunen MI, Lewis DY, Soloviev D & Brindle KM. Hyperpolarized 13C MRI and PET: In Vivo Tumor Biochemistry. Journal of Nuclear Medicine 52, 1333–1336 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Keshari KR, Wilson DM, Van Criekinge M, Sriram R, Koelsch BL, Wang ZJ, VanBrocklin HF, Peehl DM, O’Brien T, Sampath D, Carano RAD & Kurhanewicz J. Metabolic response of prostate cancer to nicotinamide phophoribosyltransferase inhibition in a hyperpolarized MR/PET compatible bioreactor: Metabolic Dynamics in MR/PET Bioreactor. The Prostate 75, 1601–1609 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Logothetis NK, Pauls J, Augath M, Trinath T & Oeltermann A. Neurophysiological investigation of the basis of the fMRI signal. Nature 412, 150–157 (2001). [DOI] [PubMed] [Google Scholar]
- 43.Bélanger M, Allaman I & Magistretti PJ. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab. 14, 724–738 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Zierhut ML, Yen Y-F, Chen AP, Bok R, Albers MJ, Zhang V, Tropp J, Park I, Vigneron DB, Kurhanewicz J, Hurd RE & Nelson SJ. Kinetic modeling of hyperpolarized 13C1-pyruvate metabolism in normal rats and TRAMP mice. Journal of Magnetic Resonance 202, 85–92 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bankson JA, Walker CM, Ramirez MS, Stefan W, Fuentes D, Merritt ME, Lee J, Sandulache VC, Chen Y, Phan L, Chou P-C, Rao A, Yeung S-CJ, Lee M-H, Schellingerhout D, Conrad CA, Malloy C, Sherry AD, Lai SY & Hazle JD. Kinetic Modeling and Constrained Reconstruction of Hyperpolarized [1–13C]-Pyruvate Offers Improved Metabolic Imaging of Tumors. Cancer Research 75, 4708–4717 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harrison C, Yang C, Jindal A, DeBerardinis RJ, Hooshyar MA, Merritt M, Dean Sherry A & Malloy CR. Comparison of kinetic models for analysis of pyruvate-to-lactate exchange by hyperpolarized 13C NMR: comparison of models of LDH exchange for 13C hyperpolarized puryvate. NMR in Biomedicine 25, 1286–1294 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hill DK, Orton MR, Mariotti E, Boult JKR, Panek R, Jafar M, Parkes HG, Jamin Y, Miniotis MF, Al-Saffar NMS, Beloueche-Babari M, Robinson SP, Leach MO, Chung Y-L & Eykyn TR. Model Free Approach to Kinetic Analysis of Real-Time Hyperpolarized 13C Magnetic Resonance Spectroscopy Data. PLoS ONE 8, e71996 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kazan SM, Reynolds S, Kennerley A, Wholey E, Bluff JE, Berwick J, Cunningham VJ, Paley MN & Tozer GM. Kinetic modeling of hyperpolarized 13C pyruvate metabolism in tumors using a measured arterial input function: Kinetic Modeling of 13C Pyruvate Metabolism. Magnetic Resonance in Medicine 70, 943–953 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Mariotti E, Orton MR, Eerbeek O, Ashruf JF, Zuurbier CJ, Southworth R & Eykyn TR. Modeling non-linear kinetics of hyperpolarized [1–13C] pyruvate in the crystalloid-perfused rat heart: Modelling non-linear kinetics of hyperpolarized pyruvate. NMR in Biomedicine 29, 377–386 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park JM, Josan S, Jang T, Merchant M, Yen Y-F, Hurd RE, Recht L, Spielman DM & Mayer D. Metabolite kinetics in C6 rat glioma model using magnetic resonance spectroscopic imaging of hyperpolarized [1–13C]pyruvate. Magnetic Resonance in Medicine 68, 1886–1893 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Witney TH, Kettunen MI & Brindle KM. Kinetic Modeling of Hyperpolarized 13C Label Exchange between Pyruvate and Lactate in Tumor Cells. Journal of Biological Chemistry 286, 24572–24580 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gordon JW, Chen H-Y, Autry A, Park I, Van Criekinge M, Mammoli D, Milshteyn E, Bok R, Xu D, Li Y, Aggarwal R, Chang S, Slater JB, Ferrone M, Nelson S, Kurhanewicz J, Larson PEZ & Vigneron DB. Translation of Carbon-13 EPI for hyperpolarized MR molecular imaging of prostate and brain cancer patients. Magnetic Resonance in Medicine 81, 2702–2709 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larson PEZ, Chen H-Y, Gordon JW, Korn N, Maidens J, Arcak M, Tang S, Criekinge M, Carvajal L, Mammoli D, Bok R, Aggarwal R, Ferrone M, Slater JB, Nelson SJ, Kurhanewicz J & Vigneron DB. Investigation of analysis methods for hyperpolarized 13C-pyruvate metabolic MRI in prostate cancer patients: Hyperpolarized Pyruvate Prostate Cancer Analysis Methods. NMR in Biomedicine 31, e3997 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ardenkjaer-Larsen JH, Leach AM, Clarke N, Urbahn J, Anderson D & Skloss TW. Dynamic nuclear polarization polarizer for sterile use intent. NMR in Biomedicine 24, 927–932 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Azema M, Carvajal L, Xu D, Gordon JW, Park I, Vigneron DB, Nelson SJ, Stockmann JP, Keil B & Wald LL. 31-Channel Brain Array for Hyperpolarized 13C Imaging at 3T. ISMRM Abstract#1225 (2017).
- 56.Pruessmann KP, Weiger M, Börnert P & Boesiger P. Advances in sensitivity encoding with arbitrary k -space trajectories: SENSE With Arbitrary k -Space Trajectories. Magnetic Resonance in Medicine 46, 638–651 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Maidens J, Gordon JW, Chen H-Y, Park I, Van Criekinge M, Milshteyn E, Bok R, Aggarwal R, Ferrone M, Slater JB, Kurhanewicz J, Vigneron DB, Arcak M & Larson PEZ. Spatio-temporally constrained reconstruction for hyperpolarized carbon-13 MRI using kinetic models. IEEE Transactions on Medical Imaging 1–1 (2018). doi: 10.1109/TMI.2018.2844246 [DOI] [PMC free article] [PubMed]
- 58.Gordon JW, Niles DJ, Adamson EB, Johnson KM & Fain SB. Application of flow sensitive gradients for improved measures of metabolism using hyperpolarized 13C MRI: Improved Measures of Metabolism with 13C MRI. Magnetic Resonance in Medicine 75, 1242–1248 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maidens J, Gordon JW, Arcak M & Larson PEZ. Optimizing Flip Angles for Metabolic Rate Estimation in Hyperpolarized Carbon-13 MRI. IEEE Transactions on Medical Imaging 35, 2403–2412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xing Y, Reed GD, Pauly JM, Kerr AB & Larson PEZ. Optimal variable flip angle schemes for dynamic acquisition of exchanging hyperpolarized substrates. Journal of Magnetic Resonance 234, 75–81 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Santarelli MF, Positano V, Giovannetti G, Frijia F, Menichetti L, Ardenkjaer-Larsen J-H, De Marchi D, Lionetti V, Aquaro G, Lombardi M & Landini L. How the signal-to-noise ratio influences hyperpolarized 13C dynamic MRS data fitting and parameter estimation: SNR ON 13C DYNAMIC MRS. NMR in Biomedicine 25, 925–934 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Sun C, Walker CM, Michel KA, Venkatesan AM, Lai SY & Bankson JA. Influence of parameter accuracy on pharmacokinetic analysis of hyperpolarized pyruvate: Pharmacokinetic Analysis of Hyperpolarized Pyruvate. Magnetic Resonance in Medicine 79, 3239–3248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Granlund KL & et al. Utilizing hyperpolarized MRI in prostate cancer to assess metabolic dynamics and histopathologic grade. Proceeding of Joint Annual Meeting ISMRM-ESMRMB (2017).
- 64.Grist JT, McLean MA, Riemer F, Schulte RF, Deen SS, Zaccagna F, Woitek R, Daniels CJ, Kaggie JD, Matys T, Patterson I, Slough R, Gill AB, Chhabra A, Eichenberger R, Laurent M-C, Comment A, Gillard JH, Coles AJ, Tyler DJ, Wilkinson I, Basu B, Lomas DJ, Graves MJ, Brindle KM & Gallagher FA. Quantifying normal human brain metabolism using hyperpolarized [1– 13C]pyruvate and magnetic resonance imaging. NeuroImage 189, 171–179 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]