Abstract
Purpose.
In this pilot study, we used shotgun metagenome sequencing (SMS) strategy on bronchoalveolar lavage (BAL) samples from hospitalized patients with suspected ventilate-associated pneumonia (VAP) in order to explore its potential for improving detection of ventilator-associated-pneumonia (VAP) etiology.
Methodology.
In total, 67BAL samples from patients with VAP were tested with SMS strategy for detection of respiratory pathogens. Results of SMS and routine respiratory culture were compared.
Results.
SMS detected all pathogens recovered by cultivation approaches. In addition, putative pathogens other than the organisms recovered by culture were detected by SMS in culture-positive samples. In 40 of 45 (89 %) culture-negative samples, a potential pathogen was detected by SMS.
Conclusion.
This proof-of-concept study demonstrates that SMS is able to detect bacterial, fungal and viral organisms in BAL, including culture-negative cases.
Keywords: shotgun metagenomic sequencing, ventilate-associate pneumonia, respiratory pathogens
Introduction
Culture-independent molecular methods have become part of the standard care for diagnosis and treatment of upper respiratory viral infections. However, diagnosis of pneumonia still relies on culture of bronchoalveolar lavage (BAL), tracheal aspirates and sputum to recover potential pathogens. Quantitative cultivation provides a relative count of respiratory tract bacteria including normal flora and pathogens, assisting clinicians in determining whether the potential pathogen is a colonizer or a causative. In general, BAL colony counts <103 c.f.u. ml−1 suggest colonization, counts of 103 to 104 c.f.u. ml−1 are indeterminant and counts >104 c.f.u. ml−1 indicate both the presence of pneumonia and the etiologic agent in patients with suspected pneumonia [1]. Thresholds for tracheal aspirates are 1–2 log10 higher, while protected specimen brush and mini-BALs are one log10 lower. Routine bacterial cultures of respiratory samples are slow and technically intensive, requiring plating specimens on sheep-blood agar, chocolate agar and MacConkey agar followed by incubate for 48 h. Significant pathogens that are slow-growing, fastidious, inert or unculturable under these conventional media are not recovered by this culture procedure [2]. The inability to cultivate host-associated pathogens is one facet of the ‘great plate count anomaly’ [3], and can have serious implications for proper patient care. For example, one study, employing cultivation approaches and targeted PCR, reported that an etiologic agent could not be identified in approximately 46 % of cases of community-acquired pneumonia [4].
Cultivation-independent molecular approaches offer a potential solution in the search for etiologic agents in pneumonia patients. In particularly, high-throughput next-generation sequencing (NGS) approaches, both targeted and non-targeted, can be deployed in the search for such agents. Direct sequencing of clinical samples detects organisms unable to be recovered with conventional culture-based techniques [5–7]. Microbiome studies of respiratory samples with NGS demonstrate a diverse normal lung microbial community that varies in composition and diversity between and within different stages of chronic lung diseases. The lung microbiome of patients with cystic fibrosis, chronic obstructive pulmonary disorder (COPD) and asthma have been extensively studied using NGS methods [8–10]. In contrast, NGS pathogen detection to improve pneumonia diagnosis has been not well studied.
In many studies, PCR-based deep sequencing of a marker gene such as 16S rRNA of 30S small subunit (SSU) allows detection of a large number of bacterial species [11]. In this approach, amplification products, generally 250–500 bp in length, generated from mixed communities are then sequenced using next-generation sequencers [12]. Data generated from these approach are not strictly quantitative, however. First, the data provide only estimates of relative abundance, but of greater concern is distortion of the underlying community structure through PCR bias [13], variable rRNA gene copy number [14], problematic primer-taxon mismatches [15], and inconsistent resolution at species- and strain-level taxonomy [16]. Finally, it is not within the scope of microbial 16S rRNA gene amplicon to target eukaryotic or viral pathogens. Thus, although amplicon-based NGS approaches are useful in circumventing cultivation limitations and in some cases high-host DNA burden, non-targeted shotgun metagenome sequencing (SMS) is needed to reduce bias associated with PCR, and to expand the target range of cultivation-independent molecular approaches to target pathogens from all domains of life and viruses [17–19]. As long as host genomic DNA does not dominate the sample to be analysed, SMS also provides a mechanism to complement taxonomic data with functional gene information including antibiotic resistance genes and to perform de novo assembly of microbial genomes from mixed communities.
In this pilot study, we used an SMS strategy on BAL samples from hospitalized patients with suspected ventilator-associated pneumonia (VAP) in order to explore its potential for improving detection of VAP etiology. Our study demonstrated that SMS is a valuable tool to detect potential pathogens in culture-negative specimens.
Methods
Case definition and sample collection
BAL was ordered at the discretion of the treating intensivists. Clinical suspicion of pneumonia was generally based on clinical parameters including at least two of the following criteria: (1) fever >37 °C, (2) leukocytosis, (3) elevated procalcitonin, (4) increased respiratory secretions or (5) worsened oxygenation; combined with a new or worsening infiltration on a chest x-ray or computed tomography (CT). VAP was defined as pneumonia acquired more than 48 h after intubation or onset of mechanical ventilation in patients with a chronic tracheostomy.
BAL collection and standard processing
BALs were collected from mechanically ventilated patients in intensive care units (ICUs) in Northwestern Memorial Hospital (NMH). The collection process followed the standard procedure established for either bronchoscopic (Chastre) or non-bronchoscopic BAL (NBBAL) (Kollef). NBBAL was performed by trained respiratory therapists and was available 24 h a day, 7 days a week. Only one sample from each patient was included in the study. Quantitative bacterial culture of BAL was performed by the clinical microbiology laboratory at NMH as part of the standard care. In total, 0.01 ml of each specimen was inoculated to sheep-blood agar, MacConkey agar and chocolate agar plates and incubated for 48 h. A positive culture was reported if at least one potential bacterial pathogen with growth of >102 c.f.u. ml−1. Culture with no growth or growing only normal flora was classified as negative culture. Cell counts and differential were performed manually.
Metagenomic shotgun sequencing
Nucleic acid extraction was performed on 2 ml of residual BAL samples using the Ultra-Deep Microbiome Prep 10 kit (Molzym GmBH and Co KG, Bremen, Germany). The kit is specifically designed for microbial DNA extraction from host-associated clinical specimens for NGS applications. Briefly, human cells were lysed followed by DNase treatment to reduce free-floating DNA, including DNA from lysed human cells and other extracellular DNA. DNA extraction from the microbial cells was carried out with a bind-wash-elute spin column-based process.
Library construction and DNA sequencing
Genomic DNA was prepared for sequencing using the Nextera XT DNA library preparation kit (Illumina, San Diego, CA, USA), according to the manufacturer’s instruction. Some samples did not yield sufficient genomic DNA (gDNA) for the kit requirements, and for these samples gDNA was concentrated using a speed-vac, and half of the total concentrated DNA was added to the preparation protocols. Barcoded libraries were pooled and sequenced using an Illumina NextSeq500 sequencer, employing a high-output kit with paired-end 2×150 base sequencing reads. For libraries with low input DNA, the larger volumes of final library were included in the sequencing pool in an effort to generate additional sequence data from these samples. To evaluate the nucleic acid background of reagent and the contaminated micro-organisms introduced during the nucleic acid extraction and library construction process, negative controls without patient sample were sequenced in the same way as the patient sample. Library preparation and sequencing were performed at the DNA Services (DNAS) facility at the University of Illinois at Chicago (UIC).
Data analysis
The software package One Codex was used for taxonomical classification of sequence data [20]. One Codex is a commercial cloud-based bioinformatics and data platform for microbial genomics. It has a reference database containing the whole genomes of approximately 40 000 bacteria, viruses, protists, archaea and fungi. One Codex classifies unknown nucleotide sequences according to the set of signature sequences within it that are unique to a specific taxonomic group. Each read or contig is then assigned to the microbial clade it most closely resembles. The complete sample is summarized as a collection of these signature sequences indicating the presence of a group of organisms.
Results
Patient characteristics and culture results
Residual BALs from 67 ventilated patients in any ICU with suspected pneumonia between September 2016 and January 2017 were included. Only one sample from each patient was included in the study.
Table 1 summarizes the patient population and characteristics. The mean age was 61.4±16.4 and 36 (54 %) were male. The duration of ICU stay prior to sampling was 15.4±15.3 days. The average time for a patient on a ventilator was 15.5+21.9 days. Overall, 11 (16 %) patients had a tracheostomy tube when the BAL was collected. Altogether, 54 (87 %) patients received antibiotic treatment prior to the BAL collection.
Table 1.
Patient characteristics
|
Characteristics |
Patients (n=67) |
|---|---|
|
Age in years; mean+sd,(range) |
61.4+16.4, (20–93) |
|
Males |
36 (54 %) |
|
LOS (ICU) in days, mean+sd, median (range) |
15.4+15.3, 11 (2–85) |
|
Days on ventilator, mean+sd, median (range) |
15.5+21.9, 7 (1–101) |
|
No. of patient with trach |
11 (16 %) |
|
Received antibiotics before BAL |
58 (87 %) |
|
Patients with PMN >=80 (n=66) |
31 (47 %) |
|
Patients with positive culture |
16 (24 %) |
BAL, bronchoalveolar lavage; LOS, length of stay; PMN, polymorphonuclear neutrophils.
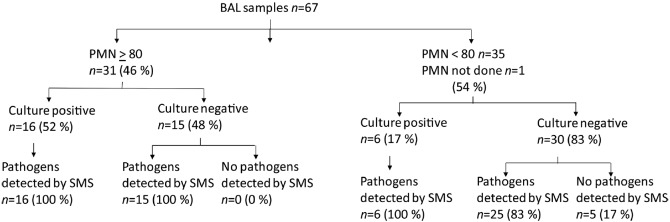
Despite nearly half (46 %) of patients with elevated polymorphonuclear neutrophils (PMNs), cultivation efforts identified at least one respiratory pathogen with colony count ranging from 1×102 c.f.u. ml−1 to greater than 1×106 c.f.u. ml−1 in only 22 samples (33 %) (Table 2). In total, 22 samples had a negative culture, and 23 other samples only grew normal flora. A summary of cultivation and cultivation-independent results is shown in Fig. 1.
Table 2.
Culture results
|
Species |
Positive culture |
Polymicrobial culture |
||
|---|---|---|---|---|
|
PMN>80 |
PMN<80 |
PMN>80 |
PMN<80 |
|
|
3 |
0 |
1 |
0 |
|
|
4 |
3 |
2 |
0 |
|
|
4 |
2 |
2 |
0 |
|
|
3 |
0 |
2 |
0 |
|
|
0 |
1 |
0 |
0 |
|
|
2 |
0 |
1 |
0 |
|
|
1 |
0 |
1 |
0 |
|
|
2 |
0 |
0 |
0 |
|
|
2 |
0 |
1 |
0 |
|
Fig. 1.
Summary of culture and SMS results.
SMS results
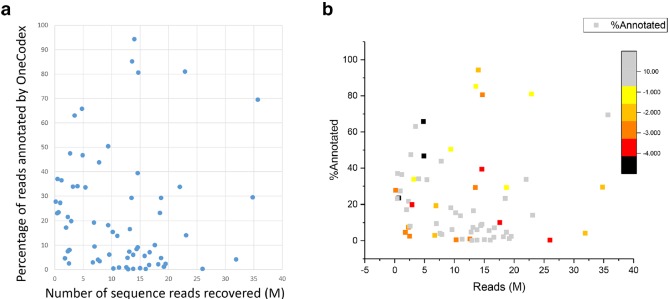
Shotgun sequence data was recovered from all 67 samples, but with highly variable numbers of reads (range 0.17 to 34.8 million clusters per sample) with average reads of 10.9 M per sample. Average reads mapped to micro-organisms per sample, including reads for bacterial, viral and fungal, was 4.6 M. Unmapped reads, including human genome, was detected with an average of 6.4 M per sample. An average of 22 % of the reads from each sample was mapped to micro-organisms in the database.
No significant difference in the number of reads was observed between different groups of samples (i.e. between samples with normal flora, samples with negative culture results and samples with positive culture) (one-way ANOVA; F=0.60, P=0.62) (Fig. 2a). No relationship between the number of reads generated and percent annotation was observed using linear regression analysis (slope =0.0136, r 2=2e-05) (Fig. 2b).
Fig. 2.
Characteristics of sequence reads. (a) One-way ANOVA was used to determine whether there was any statistically significant differences between different groups of samples. (b) Linear regression analysis was used to examine the relationship between the number of reads generated and percent annotation.
In all 22 patients with positive-culture results, at least one of the pathogens was also detected using SMS at the taxonomic level of species. The number of reads derived from the detected pathogen varied widely, representing from 0.0001 to 62.845 % of mapped reads from each sample. In the 15 culture-negative samples from patients with elevated PMN, potential pathogens were detected in all samples by SMS, including six unique bacterial species and one double-stranded DNA virus. The numbers of sequence reads mapped to the pathogen represented 0.0032 to 95.3523 % of classified reads. In the 30 culture-negative samples from patients with PMN <80 %, potential pathogens were detected in 25 samples, including ten different species from six genera. The numbers of sequence reads mapped to the pathogen ranged from 0.0362 to 94.2261 % of classified reads. Tables 3 and 4 listed the most abundant putative pathogens detected by SMS in each sample. In particular, a large number of reads were annotated as Tropheryma whipplei in three samples, and human herpesvirus 1 (HSV1) reads accounted for more than 95 % of classified reads in one sample. No putative pathogens were detected in negative control samples.
Table 3.
Organisms detected by SMS in culture-negative samples from patients with PMN values greater than 80 %
|
SMS identified putative pathogena |
Percentage of mapped reads assigned to the SMS ID (%) |
|---|---|
|
0.0032 |
|
|
0.0042 |
|
|
0.0065 |
|
|
0.0205 |
|
|
0.1015 |
|
|
0.1253 |
|
|
0.1385 |
|
|
0.1504 |
|
|
0.1923 |
|
|
0.5727 |
|
|
0.9313 |
|
|
0.9448 |
|
|
2.4522 |
|
|
9.9639 |
|
|
Human herpesvirus 1 |
95.3523 |
a, species of highest abundance.
Table 4.
Organisms detected by NGS in culture-negative samples from patients with PMN values less than 80 %
|
SMS identified putative pathogena |
Percentage of mapped reads assigned to SMS ID (%) |
|---|---|
|
0.0362 |
|
|
0.0465 |
|
|
0.05 |
|
|
0.1416 |
|
|
0.1535 |
|
|
0.1868 |
|
|
0.1941 |
|
|
0.2105 |
|
|
0.2133 |
|
|
0.2743 |
|
|
0.2884 |
|
|
0.3589 |
|
|
0.3859 |
|
|
0.525 |
|
|
0.5436 |
|
|
0.6431 |
|
|
1.0247 |
|
|
1.1272 |
|
|
2.0776 |
|
|
2.3958 |
|
|
3.4768 |
|
|
8.0532 |
|
|
13.009 |
|
|
52.945 |
|
|
94.2261 |
a, species of highest abundance.
Additional bacterial pathogens detected by SMS in the positive cultures
In 8 of 16 culture-positive samples, in addition to the species recovered by culture other potential pathogens were detected by SMS with the number of reads at least three times more than the culture-recovered species. Five samples grew Gram-negative bacilli while SMS detected S. aureus in two samples, K. oxytoca in one sample, S. pyogenes in one sample and S. pneumoniae in one sample. The reads annotated to the SMS-detected species ranged from 0.3575 to 68.1635 % (Table 5). Besides identifying bacterial pathogens, HSV1 was detected at high levels in three samples, including two samples culture positive for P. aeruginosa and one sample culture positive for S. aureus . In particular, in the sample culture positive for >100 000 c.f.u. ml−1 of P. aeruginosa , the number of reads annotated as HSV1 were 3.6 times greater than those annotated as P. aeruginosa (Table 5).
Table 5.
Additional pathogens detected by SMS in culture-positive samples
|
Culture result |
Culture colony count (c.f.u. ml−1) |
Percentage of mapped reads assigned to the culture isolate (%) |
Addition pathogen detected by SMS |
Percentage of mapped reads assigned to the SMS ID (%) |
|---|---|---|---|---|
|
>100,000 |
0.2327 |
3.7979 |
||
|
Enterobbacter cloacae |
20 000 |
0.0167 |
0.7437 |
|
|
>100 000 |
0.1320 |
6.5403 |
||
|
>100 000 |
0.1724 |
62.8545 |
||
|
>100 000 |
8.5007 |
Human herpesvirus 1 |
30.8683 |
|
|
80 000 |
0.4297 |
Human herpesvirus 1 |
78.7731 |
|
|
Staphyloccus aureus |
>100 000 |
0.3136 |
Human herpesvirus 1 |
68.1635 |
|
Stenotrophomas maltophilia |
30 000 |
0.0651 |
0.3575 |
Discussion
The main purpose of this pilot study is to explore the potential of SMS to detect pathogens in BAL from patients with suspected VAP compared with standard culture results. Overall, 22 of 67 (33 %) samples were culture positive and SMS detected all the putative respiratory pathogens recovered by the culture. The value of SMS was most clearly demonstrated by detecting putative pathogens in 40 of 45 (89 %) culture-negative samples.
Current approaches for VAP patient management relies on clinical criteria and microbiologic data, including gram stain and some degree of quantitation in bacterial culture. The value of quantitative culture for increasing the sensitivity and specificity of clinical diagnosis and identifying the etiologic pathogen has been shown in several studies [21, 22]. However, methodological limitations of culture, including quantitative cultures, are well known. Up to 60 % of clinically diagnosed VAP cannot be confirmed microbiologically [23, 24]. Antibiotics treatment prior to specimen collection, sampling area bias and low reproducibility of quantitative method have been linked to the false negative result [25–28]. Antibiotic inhibition of bacterial growth was clearly demonstrated with our SMS results. In some culture-positive samples, the number of reads mapped to the organism recovered by the culture was much lower than that of the co-infecting pathogen not present in culture of the same specimen. In particular, in three samples culture positive for Gram-negative bacilli with a high colony counts, the reads mapped to the Gram-positive pathogen detected by SMS was at least three times more than the Gram-negative (Table 5). Failure growth of the Gram-positive pathogen may be a result of empiric antibiotic treatment. Incorporation of SMS in patient care will potentially improve VAP diagnosis and targeted treatment.
Compared to culture, SMS non-selectively provides universal detection of bacteria, fungi and viruses with high sensitivity. In three samples, HSV1 was detected as the predominant species in the microbiota with the abundance ranging from 77.8 to 99.5 % of reads (Table 5). HSV bronchopneumonitis in mechanic ventilated patients has been recognized as a distinct clinical entity in critically ill patients. Luyt et al. [29] found that HSV bronchopneumonitis affected 21 % of patients experiencing clinical deterioration in a large cohort of 201 non-immunocompromised patients receiving mechanical ventilation for more than 5 days. Although HSV1 viral shedding has been linked to reduced survival in ventilated patients, the pathogenic role of HSV in VAP remains to be confirmed [30]. In three HSV1-positive samples, two were culture positive for P. aeruginosa , indicating that presence of other respiratory pathogens along with HSV is not uncommon. Treatment of VAP due to Pseudomonas and other serious Gram-negative pathogens has a high clinical failure rate [31]. Occult concomitant HSV1 infection may be a partial explanation. The role that HSV1 played in our patients was unclear because the detected viruses can be from the host cells with latent viral infection. Patient characterization based on comprehensive microbiota analysis of bacteria, fungi and viruses in BAL will be the first step to delineate the contribution of HSV to the disease process. SMS will provide a great value to assist further investigation of HSV in lung infection.
Detection of T. whipplei as a predominant species in three samples in our study population raises the possibility that it is a potential respiratory pathogen. T. whipplei has been found in the lung microbiome of uninfected patients, but has also been reported as a causative agent in pneumonia, especially in immunocompetent and immunocompromised patients with ventilated-associated pneumonia and HIV-infected patients [32, 33]. As T. whipplei was only recovered with human fibroblast cell line, further study of the role of the organism in pneumonia pathogenesis can be greatly facilitated by SMS.
This is a proof-of-concept study and therefore has significant limitations. The purpose was to evaluate the feasibility of SMS for direct detection of organisms in BAL without selective amplification. Pathogens detected by SMS only was not confirmed with an additional method. Culture results were generated as a part of standard clinical care; only routine aerobic bacterial culture was performed and cultures were only incubated for 48 h; culture was plated with calibrated loops and cannot detect organisms with colony counts below 100 c.f.u. ml−1. BAL samples were not obtained prior to initiation of new antibiotics in many cases, potentially leading to false negative culture data. Because of the limitations of standard bacterial culture methods, detection of HSV1, T. whipplei , and organisms detected in culture-negative samples was not verified with either an independent method or clinical correlation. Finally, the SMS method used in the study can only detect DNA viral targets and not RNA viruses.
In this study, no genome or targeted amplification process was used to enhance pathogen detection. Sequencing the negative controls demonstrated that the putative pathogens were not present in the reagents, nor were introduced during work process. Therefore, no cut-off for number of species-specific reads as a proportion of classified microbial reads, or minimal genome coverage was used when determining species identification. Shotgun metagenomic strategy has the potential to identify and quantify microbial taxa in a microbial community. The approach used in this study was to classify sequencing reads by using alignment to a reference database genomes. Using the resulting counts of mapped reads to estimate the abundance of taxonomic groups is challenging. Quantitation biases can be introduced during DNA extract and library construction. In addition, reference-based classification cannot account for unknown species, which leaves us with the challenge of converting the remaining reads to a taxon. So comparing species abundance across samples is difficult. Variance in the percentage of reads that map to multiple locations in the same or multiple genomes makes it difficult to compare the relative abundance of species in the same sample. Whether it is possible to correlate relative abundance of mapped reads to bacterial culture colony count requires further investigation.
In summary, a SMS approach to detect respiratory pathogens in BAL from patients with suspected VAP detected all the pathogens recovered by standard respiratory culture. Additional pathogens were detected by SMS in some culture positive samples. In 40 of 45 (89 %) culture-negative samples, putative pathogens were detected by SMS. In addition, SMS results suggest a role for micro-organisms not typically considered to be pneumonia pathogens, including HSV1 and T. whipplei . Further exploration of SMS to assist in diagnosis of pneumonia is warranted.
Funding information
The authors received no specific grant from any funding agency.
Author contributions
J. G., R. W. and C. Q.: conceptualization, methodology, investigation, resources, data curation, and writing – original draft preparation; J. G. and P. S.: formal analysis; P. H., C. P., J. W. and J. K.: writing – review and editing.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Samples used for this study were residual clinical specimens not specifically collected for study purposes. Exemption was granted for patient consent. The human study protocol was approved by the institution review board of Northwestern University
Footnotes
Abbreviations: BAL, bronchoalveolar lavage; COPD, chronic obstructive pulmonary disorder; CT, computed tomography; gDNA, genomic DNA; HSV1, human herpesvirus 1; ICU, intensive care unit; NBBAL, nonbronchoscopic BAL; NGS, next-generation sequencing; PCR, polymerase chain reaction; SMS, shotgun metagenome sequencing; SSU, small subunit; VAP, ventilate-associate pneumonia.
References
- 1.Jorgensen JH, Pfaller MA, Carroll KC, Funke G, Landry ML, et al. editors . Manual of Clinical Microbiology. 11th ed. 2015. [Google Scholar]
- 2.Schlaberg R, Simmon KE, Fisher MA. A systematic approach for discovering novel, clinically relevant bacteria. Emerg Infect Dis. 2012;18:422–430. doi: 10.3201/eid1803.111481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Staley JT, Konopka A. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu Rev Microbiol. 1985;39:321–346. doi: 10.1146/annurev.mi.39.100185.001541. [DOI] [PubMed] [Google Scholar]
- 4.Musher DM, Roig IL, Cazares G, Stager CE, Logan N, et al. Can an etiologic agent be identified in adults who are hospitalized for community-acquired pneumonia: results of a one-year study. J Infect. 2013;67:11–18. doi: 10.1016/j.jinf.2013.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hasman H, Saputra D, Sicheritz-Ponten T, Lund O, Svendsen CA, et al. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J Clin Microbiol. 2014;52:139–146. doi: 10.1128/JCM.02452-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukui Y, Aoki K, Okuma S, Sato T, Ishii Y, et al. Metagenomic analysis for detecting pathogens in culture-negative infective endocarditis. J Infect Chemother. 2015;21:882–884. doi: 10.1016/j.jiac.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Ivy MI, Thoendel MJ, Jeraldo PR, Greenwood-Quaintance KE, Hanssen AD, et al. Direct detection and identification of prosthetic joint infection pathogens in synovial fluid by metagenomic shotgun sequencing. J Clin Microbiol. 2018;56 doi: 10.1128/JCM.00402-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabrera-Rubio R, Garcia-Núñez M, Setó L, Antó JM, Moya A, et al. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J Clin Microbiol. 2012;50:3562–3568. doi: 10.1128/JCM.00767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marri PR, Stern DA, Wright AL, Billheimer D, Martinez FD. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131:346–352. doi: 10.1016/j.jaci.2012.11.013. e1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kramer R, Sauer-Heilborn A, Welte T, Guzman CA, Abraham WR, et al. Cohort study of airway Mycobiome in adult cystic fibrosis patients: differences in community structure between fungi and bacteria reveal predominance of transient fungal elements. J Clin Microbiol. 2015;53:2900–2907. doi: 10.1128/JCM.01094-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69:330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuczynski J, Stombaugh J, Walters WA, González A, Caporaso JG, et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics. 2011;36:10.7.1–10.7.20. doi: 10.1002/0471250953.bi1007s36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green SJ, Venkatramanan R, Naqib A. Deconstructing the polymerase chain reaction: understanding and correcting bias associated with primer degeneracies and primer-template mismatches. PLoS One. 2015;10:e0128122. doi: 10.1371/journal.pone.0128122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klappenbach JA, Saxman PR, Cole JR, Schmidt TM. rrndb: the ribosomal RNA operon copy number database. Nucleic Acids Res. 2001;29:181–184. doi: 10.1093/nar/29.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi H, Sakamoto M, Benno Y. Evaluation of three different forward primers by terminal restriction fragment length polymorphism analysis for determination of fecal B ifidobacterium spp. in healthy subjects. Microbiol Immunol. 2004;48:1–6. doi: 10.1111/j.1348-0421.2004.tb03481.x. [DOI] [PubMed] [Google Scholar]
- 16.Poretsky R, Rodriguez-R LM, Luo C, Tsementzi D, Konstantinidis KT. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One. 2014;9:e93827. doi: 10.1371/journal.pone.0093827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 18.Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 19.Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minot SS, Krumm N, Greenfield NB. One codex: a sensitive and accurate data platform for genomic microbial identification. bioRxiv. 2015 [Google Scholar]
- 21.Cook D, Mandell L. Endotracheal aspiration in the diagnosis of ventilator-associated pneumonia. Chest. 2000;117:195S–197S. doi: 10.1378/chest.117.4_suppl_2.195S. [DOI] [PubMed] [Google Scholar]
- 22.Woske HJ, Röding T, Schulz I, Lode H. Ventilator-associated pneumonia in a surgical intensive care unit: epidemiology, etiology and comparison of three bronchoscopic methods for microbiological specimen sampling. Crit Care. 2001;5:167–173. doi: 10.1186/cc1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chastre J, Fagon JY. Ventilator-associated pneumonia. Am J Respir Crit Care Med. 2002;165:867–903. doi: 10.1164/ajrccm.165.7.2105078. [DOI] [PubMed] [Google Scholar]
- 24.Kollef MH. Diagnosis of ventilator-associated pneumonia. N Engl J Med. 2006;355:2691–2693. doi: 10.1056/NEJMe068231. [DOI] [PubMed] [Google Scholar]
- 25.Rouby JJ, Martin De Lassale E, Poete P, Nicolas MH, Bodin L, et al. Nosocomial bronchopneumonia in the critically ill. Histologic and bacteriologic aspects. Am Rev Respir Dis. 1992;146:1059–1066. doi: 10.1164/ajrccm/146.4.1059. [DOI] [PubMed] [Google Scholar]
- 26.Marquette CH, Herengt F, Mathieu D, Saulnier F, Courcol R, et al. Diagnosis of pneumonia in mechanically ventilated patients. Repeatability of the protected specimen brush. Am Rev Respir Dis. 1993;147:211–214. doi: 10.1164/ajrccm/147.1.211. [DOI] [PubMed] [Google Scholar]
- 27.Wermert D, Marquette CH, Copin MC, Wallet F, Fraticelli A, et al. Influence of pulmonary bacteriology and histology on the yield of diagnostic procedures in ventilator-acquired pneumonia. Am J Respir Crit Care Med. 1998;158:139–147. doi: 10.1164/ajrccm.158.1.9710061. [DOI] [PubMed] [Google Scholar]
- 28.Gerbeaux P, Ledoray V, Boussuges A, Molenat F, Jean P, et al. Diagnosis of nosocomial pneumonia in mechanically ventilated patients: repeatability of the bronchoalveolar lavage. Am J Respir Crit Care Med. 1998;157:76–80. doi: 10.1164/ajrccm.157.1.9604070. [DOI] [PubMed] [Google Scholar]
- 29.Luyt CE, Combes A, Deback C, Aubriot-Lorton MH, Nieszkowska A, et al. Herpes simplex virus lung infection in patients undergoing prolonged mechanical ventilation. Am J Respir Crit Care Med. 2007;175:935–942. doi: 10.1164/rccm.200609-1322OC. [DOI] [PubMed] [Google Scholar]
- 30.Ong GM, Lowry K, Mahajan S, Wyatt DE, Simpson C, et al. Herpes simplex type 1 shedding is associated with reduced hospital survival in patients receiving assisted ventilation in a tertiary referral intensive care unit. J Med Virol. 2004;72:121–125. doi: 10.1002/jmv.10524. [DOI] [PubMed] [Google Scholar]
- 31.Planquette B, Timsit JF, Misset BY, Schwebel C, Azoulay E, et al. Pseudomonas aeruginosa ventilator-associated pneumonia. predictive factors of treatment failure. Am J Respir Crit Care Med. 2013;188:69–76. doi: 10.1164/rccm.201210-1897OC. [DOI] [PubMed] [Google Scholar]
- 32.Bousbia S, Papazian L, Auffray JP, Fenollar F, Martin C, et al. Tropheryma whipplei in patients with pneumonia. Emerg Infect Dis. 2010;16:258–263. doi: 10.3201/eid1602.090610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lagier JC, Papazian L, Fenollar F, Edouard S, Melenotte C, et al. Tropheryma whipplei DNA in bronchoalveolar lavage samples: a case control study. Clin Microbiol Infect. 2016;22:875–879. doi: 10.1016/j.cmi.2016.07.010. [DOI] [PubMed] [Google Scholar]