Abstract
Giardia duodenalis (syn. Giardia intestinalis or Giardia lamblia) infSAects over 280 million people each year and numerous animals. G. duodenalis can be subdivided into eight assemblages with different host specificity. Unculturable assemblages have so far resisted genome sequencing efforts. In this study, we isolated single and pooled cysts of assemblages C and D from dog faeces by FACS, and sequenced them using multiple displacement amplification and Illumina paired-end sequencing. The genomes of assemblages C and D were compared with genomes of assemblages A and B from humans and assemblage E from ruminants and pigs. The genomes obtained from the pooled cysts and from the single cysts were considered complete (>99 % marker genes observed) and the allelic sequence heterozygosity (ASH) values of assemblages C and D were 0.89 and 0.74 %, respectively. These ASH values were slightly higher than for assemblage B (>0.43 %) and much higher than for assemblages A and E, which ranged from 0.002 to 0.037 %. The flavohaemoglobin and 4Fe-4S binding domain family encoding genes involved in O2 and NO detoxification were only present in assemblages A, B and E. Cathepsin B orthologs were found in all genomes. Six clades of cathepsin B orthologs contained one gene of each genome, while in three clades not all assemblages were represented. We conclude that whole-genome sequencing from a single Giardia cyst results in complete draft genomes, making the genomes of unculturable Giardia assemblages accessible. Observed differences between the genomes of assemblages C and D on one hand and the assemblages A, B and E on the other hand are possibly associated with host specificity.
Keywords: diplomonad, multiple displacement amplification, synteny, cathepsin, heterozygosity, parasitology
Data Summary
Sequence data were submitted to the European Nucleotide Archive (https://www.ebi.ac.uk/ena) under accession number PRJEB32663: cyste1 (GCA_902209425), cyste2 (GCA_902221465), cyste3 (GCA_902221515), cyste4 (GCA_902221485), pool 5 (GCA_902221535) and pool 8 (GCA_902221545).
Impact Statement.
Giardia duodenalis is a single-celled parasite that can infect many mammalian species, including man. Infection can lead to diarrhoea and weight loss. Each year, 300 million people and countless animals became infected and ill because of this parasite. There are eight sub-species (assemblages) that differ in host specificity and the ability to be cultured in vitro. Knowledge of the DNA sequence of the whole organism (whole-genome sequence) has in recent years became an important tool for studying this parasite. So far, only the assemblages that could be cultured could be sequenced, and these assemblages infect mainly man. These genome sequences gave a lot of information on the biology of the parasite. However, knowledge of the genome sequence of assemblages that infect other mammals is valuable to the understanding of host specificity. Therefore, we isolated individual parasites of two other assemblages from dogs. We multiplied all the DNA from the individual parasites with a new technique and performed whole-genome sequencing. We found that many characteristics of the dog assemblages are shared with the assemblages that infect man, but differences were also found. For example, proteinases (cathepsin B) that are able to digest proteins of the host are different in the different assemblages.
Introduction
Giardia duodenalis (syn. Giardia lamblia, Giardia intestinalis) is a common intestinal parasite of mammals, including man, with diarrhoea being the most common symptom. More than 280 million annual cases of human infections are described [1] and numerous cases in other mammals. In G. duodenalis, eight assemblages (A to H) have been described up till now, with various degree of host specificity [2, 3]. The relative prevalence of an assemblage in a specific host is now regarded as the result of host specificity [2].This host specificity has made a number of researchers suggest that some of the assemblages, such as assemblages A and B, can be considered different species, also because of the large genetic differences between the assemblages [4–6].
The assemblages A and B are primarily found in humans, but have occasionally been found in dogs as well. The assemblages specific for dogs are the assemblages C and D. In humans, only 0.3 % of infections consisted of assemblage C or D [2]. Assemblage A can be divided into sub-assemblages AI, AII and AIII, of which AI occurs both in man and other mammals, such as dogs, while AII occurs almost exclusively in humans [2, 3]. Assemblage B is diverse with no clear sub-assemblages. Assemblages AI, AII and B are assumed to be zoonotic.
The genome of G. duodenalis is a compact genome with few non-coding regions and only four introns [7]. The parasite is supposed to be a clonal reproducing organism with two diploid, almost identical nuclei. Without exchange of DNA between the two nuclei, random mutation will result in increasing sequence differences between the two nuclei and, thus, increasing heterozygosity within the individual. However, the level of heterozygosity in Giardia was lower than expected [7] and the limited exchange of DNA between the nuclei, named diplomixis by Poxleitner et al. [8], could be the cause for this low level of heterozygosity. The allelic sequence heterozygosity (ASH) varies in the different assemblages from very low (<0.0023 %) in assemblage E to much higher (0.53 %) in assemblage B [4]. The causes or the consequences of these differences are unknown.
Of the eight assemblages of G. duodenalis, only three assemblages have their genomes sequenced and published. Two genomes of assemblage A have been sequenced: one from assemblage AI, strain WB [7], and one from assemblage AII, strain DH [9]. From assemblage B, there are also two isolates sequenced, GS [4] and GSB [9]. Of the non-human assemblages, only assemblage E, strain P15, from a pig has been sequenced [10]. This assemblage is associated with artiodactyls (ruminants and pigs). All the genomes are comparable in size, ranging from 10.7 to 12.0 million bp for the haploid genome, and between 5008 and 7477 ORFs [9]. Differences between the assemblages in genome size and number of ORFs can be a biological difference; however, different sequencing methods and subsequent assembly and gene calling can add to these differences as well.
The mechanisms behind the host specificity of the assemblages is not known, nor why some infections are without symptoms, while in other cases severe diarrhoea develops upon infection. Proteins excreted by the trophozoites are likely to play a role in host specificity and immunity. Several proteins from the WB (assemblage A: ATCC_50803) and GS (assemblage B: ATCC_50581) isolates excreted in the absence of host cells were identified by proteomics [11]. The most abundant proteins in both isolates were variant-specific surface proteins (VSPs), cathepsins, high cysteine membrane proteins and tenacins. Differences in proteolytic activity and specificity of the cathepsin paralogs and orthologs of different isolates/assemblages have been described [12, 13]. So far, nothing is known about the cathepsins of the assemblages of the dog.
The whole-genome sequences and the proteomic data so far all have been derived from cultured G. duodenalis trophozoites. Based on whole-genome sequencing (WGS), assemblage-specific proteins, e.g. EF-1α and CxC rich proteins [11], could be identified by proteomics and characterized. Assemblages from the dog (C and D), the cat (F), the rat (G) and from the pinnipeds (H) have resisted culturing so far. Therefore, other methods for obtaining DNA of sufficient quality and quantity for WGS are needed. Purification of cysts from a clinical isolate by immunomagnetic beads is an option. This method has resulted in sufficient pure Giardia DNA for WGS [14], but (unnoticed) mixed-infections will give spurious results. This can be circumvented by starting with the isolation of a single cyst followed by high-quality amplification of the small amount of genomic DNA. Troell et al. [15] were able to isolate individual oocysts of Cryptosporidium parvum prior to whole-genome amplification and sequencing. The coverage of the genome was on average 81 % with a single cell as the template, and by combining 10 individual cells 97.8 % of the whole genome was covered.
The aim of the study was to obtain the whole-genome sequences of the dog-specific assemblages C and D of G. duodenalis, and to perform a comparative analysis with the genomes from assemblages already sequenced. Hereto, the DNA from single cysts and pooled cysts (a control for completeness) was amplified with multiple displacement amplification (MDA). WGS was performed on the resulting MDA product and the genome was assembled and annotated. Finally, the genomes from assemblages C and D obtained from single and pooled cyst were compared with each other and with the already known genomes from the assemblages A, B and E.
Methods
Dogs and parasites
Giardia-positive faecal samples from nine dogs were obtained from the Veterinary Microbiological Diagnostic Centre (VMDC, Faculty of Veterinary Medicine, Utrecht University, The Netherlands).
Isolation of cysts
Faeces were collected and stored at 4 °C for no more than 1 week, before the Giardia cysts were isolated. Isolation of the cysts was done by suspending 1 to 5 g faeces in approximately 25 ml H2O and filtering over an open filter chamber with mesh size of approximately 100 µm (Beldico). The filtrate was centrifuged for 2 min at 2000 g and the pellet was resuspended in 50 ml H2O. Centrifugation and resuspension steps were repeated until the supernatant was clear. Finally, the pellet was resuspended in 25 ml H2O. Ten millilitres sucrose solution (70 g sucrose in 100 ml H2O) was applied at the bottom of the tube without mixing with the cysts suspension. The tube was centrifuged without braking for 15 min at 2000 g. The majority of the Giardia cysts present in the interphase were harvested, washed again with H2O as described and resuspended in 1 ml PBS. Subsequently, the cysts were counted in a modified Fuchs–Rosenthal counting chamber. From each isolate, samples were preserved in 70 % (v/v) ethanol at 4 °C, as well as frozen in 33 µl samples at −20 °C.
Isolation of DNA from cysts
DNA isolation from sucrose-purified cysts was performed with a QIAamp Fast DNA stool mini kit (Qiagen) as described for dog faeces [16] with the following modification: instead of 0.2 g faeces, we used only 33 µl cysts suspension and 167 µl inhibition/extraction buffer.
Determination of assemblage by multilocus genotyping (MLG)
Fragments of the β-giardin gene (bg) and the glutamate dehydrogenase gene (gdh) were amplified by nested PCR. All amplifications were performed with 2.5 µl template and Dreamtag polymerase (Thermo Scientific) supplemented with 0.5 mg BSA ml−1 (Sigma). Nested PCR on the gdh locus was performed with primers described elsewhere [17]. Conditions for primary amplification were: 35 cycles (94 °C for 30 s, 57.5 °C for 30 s and 72 °C for 60 s). The undiluted primary product (2.5 µl) was used as the template for the secondary amplification. Conditions for secondary amplification were: 35 cycles (95 °C for 30 s, 60 °C for 30 s and 72 °C for 60 s). Each amplification started with a denaturation step of 95 °C for 3 min and ended with a final extension step of 72 °C for 7 min. PCR on the bg locus was performed with primers as described elsewhere [18]. Conditions for primary amplifications were: 35 cycles (95 °C for 30 s, 65 °C for 30 s, 72 °C for 60 s). Primary products were 10× diluted before use as the template in secondary amplification. Conditions for secondary amplification were: 35 cycles (95 °C for 30 s, 50 °C for 30 s, 72 °C for 60 s). Each amplification started with a denaturation step of 95 °C for 3 min and ended with a final extension step of 72 °C for 7 min. The amplicons were treated with Exo-SAP-IT (Affymetrics) and Sanger sequencing was performed at BaseClear (Leiden, The Netherlands). The assemblage was determined by aligning the sequences of the amplicons with reference sequences of assemblages A to G [2] with the ClustalW module in DNASTAR Lasergene 14.0 .
Labelling of cysts and FACS
One millilitre ethanol-preserved cysts was added to 100 µl zirconium beads (Biospec products) (diameter 0.5 mm). The cysts were washed twice with 1 ml PBS by centrifugation (2 min, 2000 g) and aspirated without disturbing the beads. PBS (170 µl) and 2 drops detection reagent (Merifluor C/G, Meridian Bioscience), containing FITC-labelled mAb against Giardia and Crytosporidium, were added to the beads–cysts suspension. The suspension was mixed gently end over end, stained for 30 min at room temperature and washed again twice as described. Finally, 200 µl PBS was added to the mixture, mixed gently and the cysts were transferred in the 200 µl (without centrifugation), while taking care not to transfer the beads. The cyst suspension was poured through a 100 µm cell strainer (Greiner Bio-One) and checked for stained Giardia cysts and the absence of Cryptosporidium with fluorescence microscopy. Cysts were counted in a modified Fuchs–Rosenthal counting chamber and adjusted to a concentration of 5 cysts (µl PBS)− 1.
Sorting of Giardia cysts
First, Giardia cysts were identified and gated based on the FACS forward scatter (FSC) and side scatter (SSC) profiles (Fig. S1, available in the online version of this article). Within this gate, doublets were eliminated based on FSC and pulse width parameters. Finally, only the fluorescently labelled particles were sorted based on FSC and FITC-fluorescence parameters. Single cysts and pools of 10 cysts were collected in wells containing 2 µl PBS.
MDA
Individual single Giardia cysts or pools of 10 cysts isolated by FACS were subjected to MDA with the REPLI-g single cell kit (Qiagen). MDA was performed according to the manual, except that all volumes were half of the prescribed volumes. Confirmation of MDA amplification of the right assemblage was achieved by PCR using only the first primer pairs of the gdh and bg loci on the 100× diluted MDA product and subsequent sequencing of the amplicons. Sequences were aligned with the reference sequences [2]. Controls for contamination by small amounts of assemblage C in MDA products of assemblage D, or vice versa, were performed by RFLP on the amplified bg fragment. The bg fragment of assemblage D (AY545647) and assemblage C (AY545646) differ in the number of restriction sites for XhoI. Pooling of cells for MDA reactions [19], as well as pooling of MDA products for sequencing reactions [20], can increase the completeness of the genome. Therefore, 4 MDA products, each obtained from 10 cysts of the same assemblage from the same dog were, when proven to be without contamination, pooled into one vial. This results in pooled MDA products of 4×10 cysts. MDA products obtained from individual cysts were kept separately. All the MDA products were subsequently purified with a QIAampDNA mini kit and eluted with AE buffer. dsDNA content was determined with a Qubit 3.0 fluorometer and all samples were diluted to 200 ng dsDNA in 50 µl AE buffer. This purified dsDNA was used to produce the TruSeq DNA Nano LT libraries.
WGS and genome analysis
WGS was performed on MiSeq platforms (Illumina) using 2×250 bp reads with approximately 230-fold coverage per genome. A total of 85 % of the reads could be aligned to the non-redundant (nr) protein database using diamond [21] and classified using megan [22] to evaluate contamination with bacterial DNA. Approximately 0.25 % of the reads were classified as bacterial and 99.75 % as eukaryotic, of which 99.98 % were assigned to Hexamitidae (Fig. S2). Genomes were assembled with SPAdes v3.10.1 [23] using default settings. Contigs with a size <200 bp and a coverage lower than 10 were removed. All contigs were aligned with the nr protein database. Contigs classified as bacterial by megan were removed (approximately 0.7 % of the contigs). Genes were identified using Prodigal v2.6. with the sequenced Giardia genomes and protein annotations were collected from GiardiaDB (https://giardiadb.org/giardiadb/). An all-versus-all blast was performed for all predicted proteins of the genomes and with Spironucleus salmonicida as the outgroup at an E value cut-off of 1×10−6. To determine the orthologous relationships of all proteins, the blast output was parsed by Orthagogue, v1.02 [24]. Proteins were considered for orthology clustering if the proteins had at least 20 % identity and at least 20 % overlap. To determine the orthologous groups (OGs), Markov clustering (MCL) was performed using MCL-edge, v14-137 [25]. Proteins were aligned with each other within their respective OGs using muscle [26], and the gene names and functions from the reference genomes were added.
Completeness of the genomes was assessed by calculating the recovery of the single copy genes. There were 2716 single copy core genes present in all the draft genomes of WB (AI), DH (AII), GS (B), GSB (B) and P15 (E). The percentage of those 2716 genes present in the genomes of assemblages C or D was defined as the percentage completeness of the genome.
A protein super alignment of 534 584 positions was created by concatenating the aligned proteins if they were present in all assemblages and the outgroup S. salmonicida as single copies, using the catfasta2phyml script (https://github.com/nylander/catfasta2phyml). Phylogenetic dendrograms were created using iq-tree [27] using the automated model selection method (-m TEST), with 1000 ultrafast bootstrap cycles (-bb 1000) [28], and were visualized using mega 6.6 [29].
Venn diagrams were constructed with an online webtool from the University of Ghent, Belgium (http://bioinformatics.psb.ugent.be/webtools/Venn). Average nucleotide identity (ANI) calculations were performed using FastANI [30] using default settings.
Phylogeny cathepsin B
The OG named the cathepsin-like cysteine proteinase family (OG000011) contained in total 139 sequences from all 11 genomes together. Fourteen sequences with a glycosylphosphatidylinositol (GPI) anchor or truncated sequences with high identity to sequences with a GPI anchor were very dissimilar to the other sequences and, therefore, were removed. From the 125 remaining sequences, those sequences that were not long enough to have a complete peptidase domain (200 aa) were removed together with the sequences that lacked the Gln-Cys-His-Asn residues conserved in cathepsin B [31]. The remaining 85 sequences were aligned and the phylogenetic tree was reconstructed with the maximum-likelihood method in mega 6.6.
The ASH was calculated by scoring all the SNPs with an alternative base calling >14.9 % and a coverage of >10×, according to the definition of ASH [9]. In order to distinguish between real heterozygosity and amplification or sequence errors, SNPs were identified in the genomes of the single cysts. This was done for the common loci for MLG, gdh, bg and triosephophate isomerase (tpi), and for mitotic control protein (dis3), a locus with higher diversity [32]. Shared SNPs between single cysts of the same assemblage indicate real heterozygosity. Synteny of the region around the cathepsin B gene of assemblage C was assessed with mauve (http://www.darlinglab.org/mauve).
Results
Dogs and parasites
Sucrose-purified cysts from nine G. duodenalis-infected dogs were genotyped based on the amplified gdh fragment and the digestion of this fragment by XhoI (Table 1, Figs S3 and S4). The isolates are numbered according the number of the dog they originated from.
Table 1.
Dogs, G. duodenalis isolates and MDA products
|
Dog |
Breed |
Age (months) |
Clinical signs |
Purified cysts |
MDA product |
|
|---|---|---|---|---|---|---|
|
Assemblage* |
Template |
Assemblage† |
||||
|
1 |
Bull terrier |
6 |
Chronic diarrhoea |
C/D |
Single cysts |
Cyst 1 and 3, C; cyst 2 and 4, D |
|
2 |
German shepherd |
10 |
Recurrent diarrhoea |
D |
– |
|
|
3 |
Mixed breed |
6 |
Diarrhoea |
C/D |
– | |
|
4 |
Unknown |
6 |
Diarrhoea |
C/D |
– |
|
|
5 |
French bulldog |
6 |
Watery diarrhoea |
D |
Pooled cysts |
D |
|
6 |
Bullmastiff |
4 |
Diarrhoea |
C |
– |
|
|
7 |
French bulldog |
3 |
Diarrhoea |
C |
– |
|
|
8 |
Galgo Espanol |
7 |
Mild diarrhoea |
C |
Pooled cysts |
C |
|
9 |
Cairn terrier |
2 |
Diarrhoea, abdominal pain |
D |
– |
|
*Based on phylogeny of the gdh locus and on the gdh/XhoI restriction pattern.
†Based on phylogeny of the bg locus and on the bg/XhoI restriction pattern.
FACS and MDA
The individually sorted single cysts from isolate 1 (assemblage C or D), and the cysts sorted in pools of 10 cysts from isolate 5 (assemblage D) and isolate 8 (assemblage C), were used for MDA amplification. The MDA products were subjected to the amplification of the single copy locus bg by PCR. Based on the sequence of the bg fragment and on the digestion with XhoI of this fragment (Figs S5 and S6), it was concluded that pooled MDA products of isolate 5, as well as the cysts 2 and 4 of isolate 1, were assemblage D, without any contamination. Pooled MDA products of isolate 8, as well as of the single cysts 1 and 3 from isolate 1, were assemblage C, also without any contamination.
Characteristics of the genomes
The characteristics of the genomes are given in Table 2 and the presence/absence scores are given in Table S1. The sequencing of the genomes of assemblage C resulted in 3388 to 3917 contigs, with a coverage of 206 to 235. The genome was estimated to be between 11.5 and 12.1 Mbp. The sequencing of the genomes of assemblage D resulted in 2885 to 3489 contigs, with a coverage of 231 to 267. The genome was estimated to be between 11.4 and 11.5 Mbp. All genomes contained about 3900 OGs.
Table 2.
Characteristics of the G. duodenalis genomes
|
Genome |
Assemblage |
No. of reads |
Total bases |
Coverage |
Genome size (bp) |
No. of contigs |
N50 (bp)* |
No. of genes |
No. of OGs |
Completeness (%)† |
ASH (%) |
Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
WB |
AI |
±200 000 |
1.4×108 |
11 |
12 827 416 |
211 |
2 762 469 |
6.027 |
4.199 |
nd |
<0.01 |
[7] |
|
DH |
AII |
4 996 200 |
1.3×109 |
124 |
10 703 894 |
239 |
117 284 |
5.147 |
3.505 |
nd |
0.037 |
[9] |
|
GS |
B |
808 181 |
1.8×108 |
16 |
11 001 532 |
2931 |
36 599 |
4.471 |
3.469 |
nd |
0.53 |
[4] |
|
GSB |
B |
3 019 027 |
6.4×108 |
53 |
12 009 633 |
543 |
58 544 |
6.094 |
3.803 |
nd |
0.43 |
[9] |
|
P15 |
E |
1 639 140 |
5.4×108 |
47 |
11 522 052 |
820 |
71 261 |
5.068 |
3.609 |
nd |
0.0023 |
[10] |
|
SSAL |
2 390 565 |
5.2×108 |
40 |
12 954 588 |
233 |
150 829 |
6.288 |
3.997 |
nd |
nd |
[43] |
|
|
Cyst 1 |
C |
7 903 382 |
2.4×109 |
206 |
11 557 310 |
3388 |
31 204 |
5.550 |
3.993 |
99.3 |
0.85 |
This paper |
|
Cyst 3 |
C |
9 026 378 |
2.7×109 |
235 |
11 550 696 |
3651 |
31 165 |
5.391 |
3.961 |
99.0 |
0.88 |
This paper |
|
Pool 8 |
C |
8 821 718 |
2.7×109 |
220 |
12 061 379 |
3917 |
28 767 |
5.538 |
3.986 |
99.2 |
0.94 |
This paper |
|
Cyst 2 |
D |
8 714 436 |
2.6×109 |
231 |
11 374 926 |
3269 |
31 380 |
5.350 |
3.923 |
99.1 |
0.80 |
This paper |
|
Cyst 4 |
D |
10 000 252 |
3.0×109 |
267 |
11 268 649 |
2885 |
32 917 |
5.460 |
3.974 |
98.6 |
0.63 |
This paper |
|
Pool 5 |
D |
9 496 842 |
2.9×109 |
249 |
11 499 674 |
3489 |
31 255 |
5.549 |
4.016 |
99.0 |
0.78 |
This paper |
nd, Not determined.
*N50 is the contig length at which 50% of the genome is covered by summed contigs ordered by length.
†Completeness is the percentage recovery of single copy genes.
Completeness of the genomes
Single copy genes are defined here as an OG with exactly one member in all five previously sequenced draft genomes WB, DH, GS, GSB and P15. There were in total 2716 shared single copy genes, and of these genes 99.0–99.3 % and 98.6–99.1 % were present in the genomes of assemblages C and D, respectively (Table 2). No differences in completeness between genomes derived from single cysts or from 40 pooled cysts were found.
Shared and unique OGs in assemblages C and D
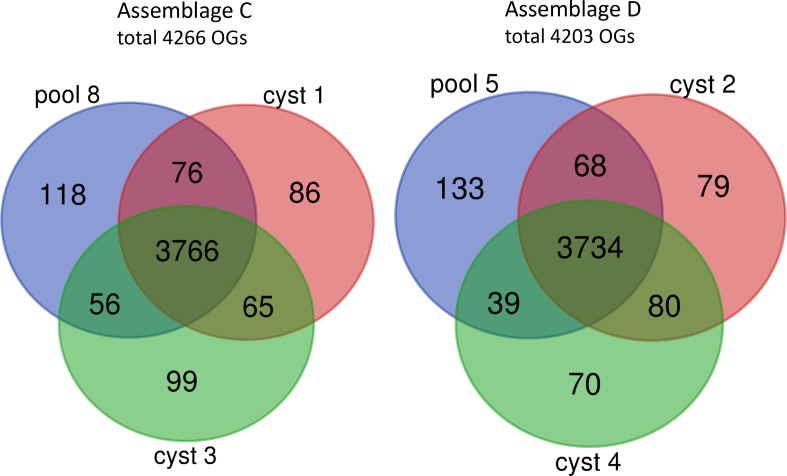
The unique and shared OGs from genomes obtained from individual and pooled cysts were identified for assemblages C and D. In the 3 isolates, there were 4266 and 4203 OGs identified in assemblages C and D, respectively (Fig. 1). The vast majority of OGs (88 –89 %) were shared by all three isolates of the same assemblage and 92.9–93.3 % of the OGs were shared by at least two isolates of the same assemblage. The number of unique OGs was slightly higher in the genomes obtained from the pooled cyst compared to the genomes from the single cysts.
Fig. 1.
Distribution of the OGs of the three genomes within assemblage C (pool 8, cyst 1 and cyst 3) and within assemblage D (pool 5, cyst 2 and cyst 4) shown in a Venn diagram. The genomes from cyst 1, 2, 3 and 4 were obtained from MDA products of single cysts. The genomes from pool 8 and pool 5 were obtained from pooled MDA products of 40 cysts per assemblage.
The proteins in all six genomes from assemblages C and D were compared with each other (Fig. S7). There were in total 4859 OGs, of which 3485 OGs (71.7 %) were shared between all genomes. Furthermore, 243 OGs (5.0 %) and 194 OGs (4.0 %) were specific for assemblages C and D, respectively. Although the sequencing of the PCR products and the RFLP of the MDA product did not reveal cross contamination, the genomes were also inspected for this potential contamination. OGs present in all the genomes from the individual cysts, but absent in one of the genomes from the pooled cysts, is indicative for contamination in the individual cysts. Only 11 (0.23 %) and 10 (0.21 %) OGs were present in all the genomes from the individual cysts, but absent in genomes from pooled cysts of assemblages C and assemblage D, respectively. In genomes of single cyst 1 and cyst 3 from assemblage C there were, respectively, 9 and 21 OGs absent (mean 0.31 %), and from genomes of single cyst 2 and 4 from assemblage D there were, respectively, 2 and 19 OGs (mean 0.22 %) absent. This all indicates that contamination of the genome of one assemblage with that of the other assemblage is, if present, very limited. A direct comparison of unique and shared OGs between the genomes of the single cysts and pooled cysts is also given in Fig. S8.
ASH
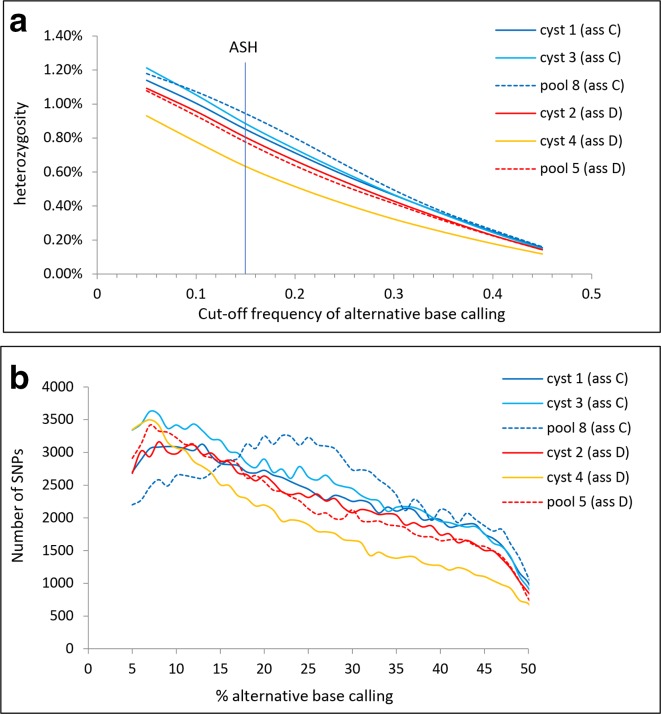
The ASH defined by an alternative base calling of 15 % or higher and a 10-fold coverage or higher [9] resulted in a mean ASH for the three genomes of 0.89 and 0.74 % for assemblages C and D, respectively (Table 2). Shared SNPs from the single cysts from the same assemblages in gdh, tpi, bg and dis3 loci were identified (Fig. S9). The percentage of shared SNPs from single cysts of assemblages C and D was 58 and 80 %, respectively, indicating that the majority of the SNPs were the result of heterozygosity within the cysts and not from amplification or sequence errors. The heterozygosity was similar for the genomes from single cysts and pooled cysts, but was very much dependant on the threshold for alternative base calling (Fig. 2a). The ASH for assemblages C and D was in the same order as that for GS (0.53 %) and much higher than for WB (<0.01 %) and P15 (0.0023 %) [4, 9]. For five out of the six genomes, the number of SNPs decreased linearly with the percentage of alternative base calling (Fig. 2b). Only for the genome from pool 8 (assemblage C), the number of SNPs peaked at an alternative base calling of around 25 %, suggesting the presence of four haploid genomes in this sample.
Fig. 2.
Heterozygosity as a function of the alternative base calling for all three genomes of assemblage C (blue lines) and assemblage D (red and yellow lines). (a) Heterozygosity as a function of the cut-off frequency of alternative base calling. Heterozygosity at alternative base calling of 0.15 is by definition the ASH. (b) Number of SNPs for all three genomes of assemblages C and D as a function of the percentage alternative base calling.
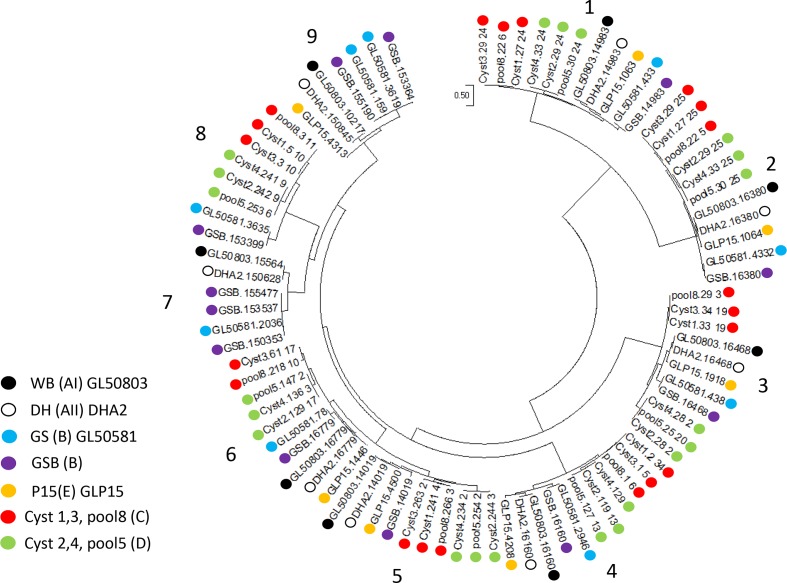
Phylogeny
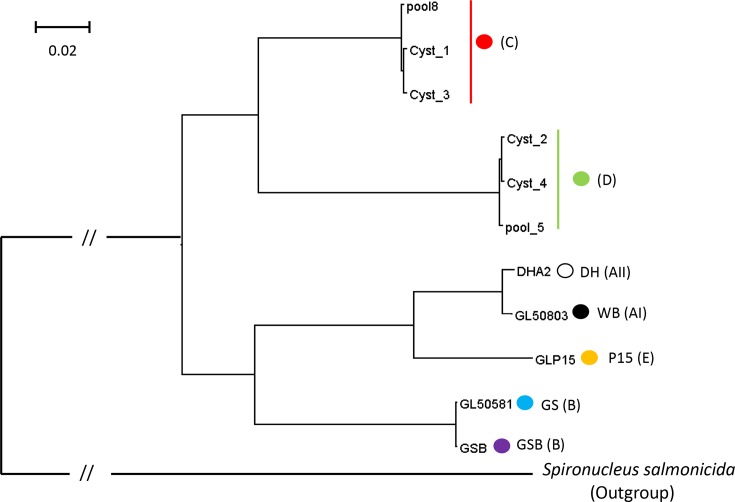
We reconstructed a phylogenetic tree of concatenated alignments of single copy proteins shared by G. duodenalis and S. salmonicida. The phylogenetic tree is given in Fig. 3. As expected, there is very little difference within the three genomes of assemblage C or D (ANI=99.4 to 99.5 and 99.4 to 99.6, respectively). The genomes from assemblages C and D form one clade. This in contrast to the genomes of the human assemblages A and B. Assemblage A is more related to assemblage E than to assemblage B. However, the distance between assemblages C and D (ANI=78.8) is comparable to the distance between the assemblages A and B (ANI=77.8), and is larger than between assemblages A and E (ANI=85.2).
Fig. 3.
Phylogenetic tree of the G. duodenalis genomes of assemblages A (WB and DH), B (GS and GSB), C (cyst 1, cyst 3 and pool 8), D (cyst 2, cyst 4 and pool 5) and E (P15) with S. salmonicida as the outgroup. Analysis was performed with the maximum-likelihood method. The analysis involved 11 genomes and 534 584 positions. Bar indicates the number of amino acid substitutions per site analysed. All branches have a bootstrap value of 100.
Shared and unique OGs of all assemblages
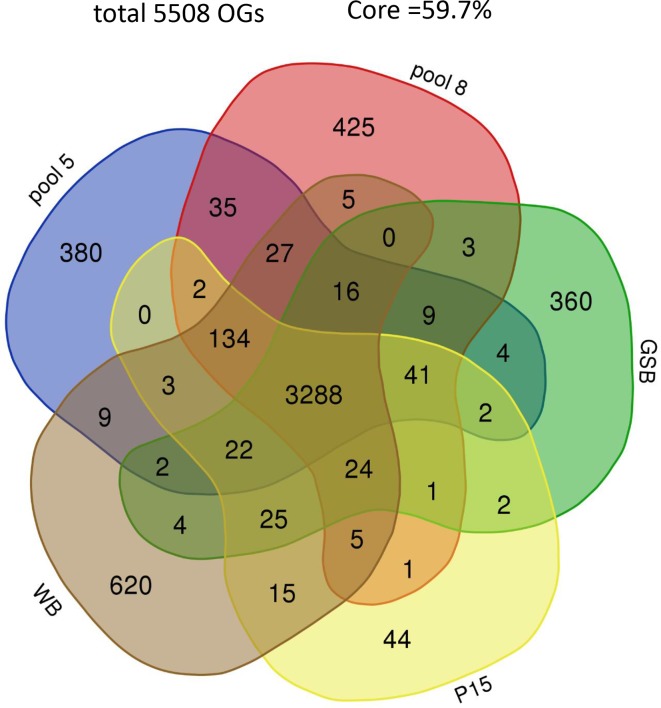
The unique and shared OGs for all assemblages are given in Fig. 4. For reasons of clarity, we used only one genome per assemblage. The GSB genome was preferred over the GS genome, because of the better assembly, resulting in less and longer contigs [9]. For the same reason, the WB genome was preferred over the DH genome (Table 2). All assemblages together counted 5508 OGs, of which 3288 OGs (59.7 %) were shared by all assemblages (core genome). The percentage assemblage-specific OGs for assemblages A, B, C, D and E were 14.7, 9.5, 10.6, 9.6 and 1.2 %, respectively.
Fig. 4.
Distribution of the OGs from five genomes representing the assemblages AI (WB), B (GSB), C (pool 8), D (pool 5) and E (P15) of G. duodenalis shown in a Venn diagram. In total, there were 5508 OGs, and 3288 OGs (59.7 %) were shared by all assemblages.
OGs unique for dog-specific assemblages
OGs present in all the six genomes of the dog-specific assemblages C and D, but in none of the other genomes, are considered dog-specific. Initially, 19 OGs were identified as dog-specific (Table S2). Manual blastx analysis against the nr protein database suggested that 14 of these dog-specific OGs contained gene fragments with a low identity to genes from the non-dog assemblages. Two dog-specific OGs had a high identity with proteins from the non-dog assemblages, but they were annotated in a wrong orientation or frame. Three OGs coded for proteins (two hypothetical proteins and protein 21.1) with a very low identity to proteins from non-dog assemblages and seemed, therefore, dog-specific. Only one OG had no significant hit at all.
OGs unique for assemblages specific for hosts others than dogs
There were 14 OGs present in all genomes except the genomes of assemblages C and D (Table S3). Two of the genes belonging to these fourteen OGs were those encoding flavohaemoglobin and a 4Fe-4S binding domain family protein, one of the five ferredoxins in G. duodenalis [33]. Both genes belong to the oxidative response network of Giardia [34]. Flavohaemoglobin is upregulated by NO in strain WB [35], and by O2 in strains WB and GS [34], and is involved in the detoxification of O2 and NO. Ferredoxin mediates electron transfer in the detoxification of O2 and NO [34]. The 2Fe-2S ferredoxin gene (GL50803.27266) was also missed in the automated annotation of the dog assemblages by Prodigal. That was because this gene contains an intron and genes with introns are not recognized by Prodigal. Manual search identified this gene, including the intron, in assemblage C (not shown).
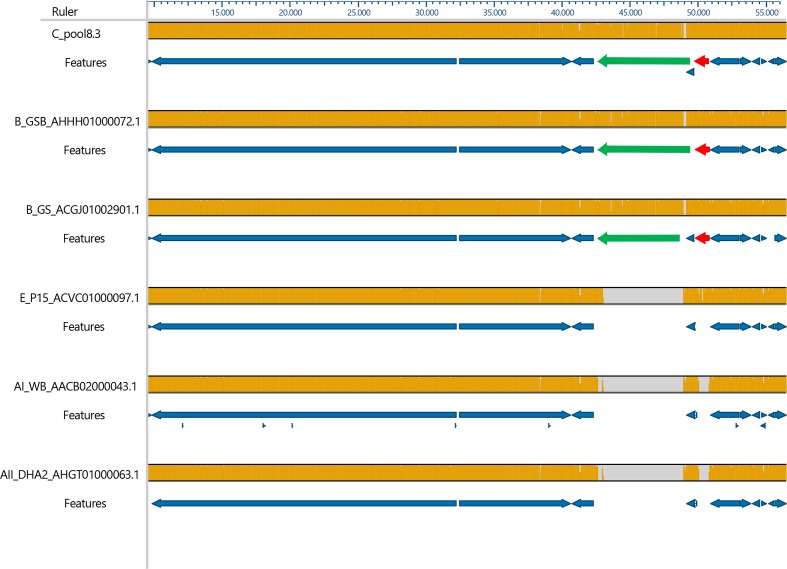
Cathepsin B
The phylogenetic tree of the 85 cathepsin B sequences is shown in Fig. 5. The traditional view of the same phylogenetic tree is shown in Fig. S10. Clades 1 to 6 contained one gene from each genome, except for clade 5 that missed only the gene from the GS genome. Within these six clades, the sequence variation between the orthologs was very low. Clades 7, 8 and 9 were more diverse and closely related to each other. In these three clades, more than one cathepsin gene per genome can be found within one clade, for example three cathepsin paralogs of GSB are found in clade 7. However, not all genomes were represented in all these three clades. Clade 8 was the only clade that contained genes from assemblages C and D, but the genes from assemblages A and E were missing in this clade. Therefore, the synteny of cathepsin B genes of clade 8 was studied by aligning a 47 000 bp fragment with the homologous contigs of the other assemblages (Fig. 6). Two deletions in the genomes of assemblages A and E were found close to each other, one of 6300 bp and another of 670 bp, causing the deletion of the chromosome segregation ATPase encoding gene and the cathepsin B gene, respectively, in the genomes of assemblages A and E.
Fig. 5.
Molecular phylogenetic analysis by the maximum-likelihood method with gamma distribution of the 85 cathepsin B genes from 11 genomes, indicated by coloured dots: WB (black), DH (white with a black outline), GS (blue), GSB (purple), P15 (orange), assemblage C (red) and assemblage D (green). The analysis involved 85 amino acid sequences with a total of 620 positions in the final data set. Bar indicates the number of substitutions per site. Analysis were conducted in mega 6.6.
Fig. 6.
Synteny study of cathepsin B genes from clade 8. Homologous contigs of all genomes were aligned in mauve. Alignment was restricted to a 47 000 bp fragment. Orange in the bar indicates homology, grey indicates deletions. The annotated genes are indicated in the features sections of the figure. The chromosome segregation ATPase encoding gene (green arrow) and the cathepsin B gene (red arrow) were deleted in the genomes from assemblages E, AI and AII. Pool 5 (assemblage D) was completely homologous with pool 8.3, but is not shown, because the 47 000 bp region consisted of three short contigs, hampering automated annotation.
Discussion
Whole-genome sequences were successfully obtained from the dog-specific assemblages C and D of G. duodenalis. After an initial step on a sucrose cushion and fixation in ethanol, it was possible to isolate labelled single cysts by FACS. MDA of the genome of single cysts yielded DNA of sufficient quality and quantity to perform WGS successfully.
The single cysts of assemblages C and D were isolated from the same dog with a mixed infection without contamination with the other assemblage. Only a small number of OGs had to be removed because of bacterial origin, these were all contained on contaminating contigs. This was especially the case in the genome from pool 8 of assemblage C. More extensive washing of the cysts, and further dilution of the cyst suspension before cell sorting, can likely reduce this contamination even further in future experiments.
The whole-genome sequences of assemblages C and D were 99 % complete compared to single copy genes from the other draft genomes of G. duodenalis. The completeness of the genomes derived from single cysts was the same as that from 40 pooled cysts. The chance that more than 1 cyst per well were sorted seems unlikely, because of the diluted suspension of cysts and because of the gating, which selects only single cysts. MDA performed on single cells prior to WGS was described before for prokaryotes [19, 20]. In order to increase the completeness of the genome, Ellegaard et al. [19] suggested pooling of cells prior to MDA amplification, whereas, Lasken [20] suggested pooling the MDA products prior to WGS. We combined this approach by applying the MDA procedure on 10 pooled cysts, followed by pooling 4 MDA products before sequencing, and found very little difference between the genomes obtained from 1 cyst or from 4×10 cysts. MDA of individual single-celled parasites prior to WGS was described before for a single oocyst of C. parvum [15]. There, on average, 81 % of the reference genome could be accounted for in the individual single cell sequence data. The cysts of Giardia are 16 N [36], while the Cryptosporidium oocysts are only 4 N. This is a possible explanation why the completeness of the genome was in the case of Giardia single cell sequencing better than with Cryptosporidium, as more DNA is available per reaction.
The genomes of assemblages C and D were of about the same size and contained the same number of genes. Of the OGs, 71.7 % were shared and about 4 % were unique for each assemblage. The phylogenetic tree of the genomes demonstrated that assemblages C and D belong to the same clade. This is in agreement with the phylogeny based on a single locus (gdh) reported by Feng and Xiao [3]. This is not the case with the two human assemblages A and B. The genomes from assemblage A are found to be more related to assemblage E, than to the other human assemblage B. This has been found before [37]. Nevertheless, the divergence between assemblages A and B is similar to that between assemblages C and D. In a study where the distance between assemblages of Giardia were compared with different species of Leismania or Theileria, [9, 10], they found a shorter distance between species of Leishmania and Theileria, than for assemblages A and B. This justified the suggestion to change the taxon of assemblages A and B into two species, G. duodenalis and G. enterica, respectively [38]. The same authors suggested changing the dog assemblages C and D into a single new species Giardia canis. Other authors [39] suggested not to group the two dog assemblages into one species, but to make two species of it because of the differences in the sequences of the loci used for MLG. The genetic difference between the genomes of assemblages C and D, comparable to the difference between species of Leismania or Theileria, potentially justifying this division into two species. Furthermore, the symptoms seemed to differ between assemblage C and assemblage D infected dogs, with less symptoms in the case of an infection with assemblage C [40]. Interestingly, in our study, the dogs infected with assemblage C also seem to have less symptoms than the dogs infected with assemblage D or with assemblages C and D.
A high ASH of 0.89 and 0.74 % was found for assemblages C and D, respectively. Shared SNPs between single cysts from the same assemblage indicates that at least 58 to 80 % of the SNPs for assemblages C and D, respectively, were the result of heterozygosity within the cysts and not due to amplification or sequence errors. Taking this into account, the ASH for assemblages C and D will be 0.52 and 0.58 %, respectively. This is in the same range as that for genome GSB and GS, 0.43 and 0.53 %. respectively, both of assemblage B. Although it is difficult to compare the ASH of the different genomes, because different sequencing methods are used, the difference in ASH between A and E on one side and B, C and D on the other side is striking. The same division of the assemblages can be seen in the phylogenetic tree with assemblages E and A forming one clade. It suggests that having a low ASH is a derived character from a predecessor with a high ASH as found in assemblages B, C and D. The ASH of assemblages F, G and H are not known. Given the position of assemblage F between assemblages A and E in the phylogenetic tree [3], it is predicted that it will have also a low ASH. Likewise, assemblages G and H located on the base of the rooted phylogenetic tree are predicted to have a high ASH.
In the present study, the genome obtained from the pooled cysts of pool 8 (assemblage C) showed a peak in the number of SNPs at 25 % alternative base calling. This suggests the presence of four haploid genomes with equal abundancy, like the four haploid genomes present in a trophozoite [36]. Unexpectedly, a linear decrease in heterozygosity was seen as a function of the cut-off frequency of alternative base calling in the other five genomes. This is expected if a cyst consists of 16 independent haploid genomes with random SNPs. However, the 16 N stage in the cyst is reach by a repeated duplication of the 4 N trophozoite and, therefore, a peak at 25 % alternative base calling seemed more likely. Why this peak is only seen in the genome of pool 8 is difficult to explain; however, it could be that much higher sequencing depths are needed to distinguish alternative base calling frequencies.
After a manual blastx search, four OGs were identified as dog-specific. Three of these OGs encoded a hypothetical protein. One OG encoded protein 21.1. This protein is characterized by the presence of ankyrin repeats [9], but these were not present in this specific protein sequence.
There were 14 OGs found in all genomes of assemblages A, B and E, but lacking in the dog assemblages C and D. Two genes of this group are those encoding flavohaemoglobin and 4Fe-4S binding domain family protein, although a gene fragment of the 4Fe-4S binding domain was found in dog-specific OGs after blastx. Both proteins are part of the oxidative response network [34]. Flavohaemoglobin and ferredoxins play a role in detoxification of O2 and NO. Flavoprotein (OG000298) and other ferredoxins were present in all genomes and possibly this makes flavohaemoglobin and 4Fe-4S binding domain family protein redundant in the dog assemblages C and D.
Cathepsin B from Giardia are known to play a role in host–parasite interaction. Several cathepsin B from different assemblages have been demonstrated to be excreted [11], and digestion by cathepsins B of host IL-8 [41] and of proteins involved in the tight junction of host epithelial cells and several chemokines [13] have been described. Different cathepsin B paralogs within the same assemblages can have different substrate specificity and pH optimum as demonstrated for GL50803.16160, GL50803.14019 and GL50803.16779 from assemblage AI [13] (these paralogs belong in our phylogenetic tree to clades 4, 5 and 6, respectively). Differences in specificity between paralogs may not be surprising, because paralogs, even when identical just after duplication, will become different in the course of evolution or will be lost. Perhaps more interesting for our study is the fact that cathepsin B orthologs can have different characteristics. GL50803.16779 and GL50581.78 are orthologs (clade 6) from assemblage AI and B, respectively, but the first one is much more secreted than the second one [42]. In another orthologous pair (GL50803.15564 and GL50581.2036, clade 7), very strong positive selection (dN/dS >26) is found and, indeed, this suggests these cathepsins play a different role in each assemblage [11]. This is also interesting for our study, because this clade 7 has no orthologs of assemblage C, D or E. Furthermore, the adjacent clade 8 is missing cathepsin B from assemblages A and E, and clade 9 is missing cathepsin B genes from assemblages C, D and E. This suggests that the presence or absence of cathepsin B genes in the different assemblages can also be the result of selection. Because the different assemblages have different host specificities and because of the interaction of cathepsin B with the host, the selection on cathepsin B can be related with host specificity. In the future, it would be interesting to study cathepsin B further, especially those from the clades 7, 8 and 9.
Conclusions
We demonstrated that a 99 % complete genome sequence can be obtained from single G. duodenalis cysts of assemblages C and D from clinical samples. This method would be useful for generating whole-genome sequences from other single-celled parasites that cannot be cultured. The number of genes (±5300) and the size of the genome (±11.5 Mbp) were similar to those of the already known genomes of other assemblages of G. duodenalis, and the genetic distance between assemblages C and D was similar to that of assemblages A and B. Cathepsin B is a cysteine protease present in all assemblages and known to effect the host's cells. It is a large gene family; however, some clades of the family did not contain genes from all assemblages. These clades can be of special interest because of the differences between assemblages, which are possibly related with host specificity.
Data bibliography
GiardiaDB (https:// giardiadb.org/giardiadb) December 2017.
Supplementary Data
Funding information
This work received no specific grant from any funding agency.
Acknowledgements
We are grateful to the Veterinary Microbiological Diagnostic Centre (VMDC, Faculty of Veterinary Medicine, Utrecht University, The Netherlands) for the samples of Giardia-positive dogs. Kristel van Rooijen is thanked for her technical assistance with the isolation of the individual cysts. Dr Ger Arkesteijn is thanked for the FACS analysis.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
All faecal samples were from clinical samples without need for approval by the ethics committee.
Footnotes
Abbreviations: ANI, average nucleotide identity; ASH, allelic sequence heterozygosity; FSC, forward scatter; MDA, multiple displacement amplification; MLG, multilocus genotyping; nr, non-redundant; OG, orthologous group; WGS, whole-genome sequencing.
Sequence data were submitted to European Nucleotide Archive (https://www.ebi.ac.uk/ena) under accession number PRJEB32663.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
References
- 1.Lane S, Lloyd D. Current trends in research into the waterborne parasite Giardia. Crit Rev Microbiol. 2002;28:123–147. doi: 10.1080/1040-840291046713. [DOI] [PubMed] [Google Scholar]
- 2.Sprong H, Cacciò SM, van der Giessen JWB, ZOOPNET Network and Partners Identification of zoonotic genotypes of Giardia duodenalis . PLoS Negl Trop Dis. 2009;3:e558. doi: 10.1371/journal.pntd.0000558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24:110–140. doi: 10.1128/CMR.00033-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Franzén O, Jerlström-Hultqvist J, Castro E, Sherwood E, Ankarklev J, et al. Draft genome sequencing of Giardia intestinalis assemblage B isolate GS: is human giardiasis caused by two different species? PLoS Pathog. 2009;5:e1000560. doi: 10.1371/journal.ppat.1000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jerlström-Hultqvist J, Ankarklev J, Svärd SG. Is human giardiasis caused by two different Giardia species? Gut Microbes. 2010;1:379–382. doi: 10.4161/gmic.1.6.13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Monis PT, Andrews RH, Mayrhofer G, Ey PL. Genetic diversity within the morphological species Giardia intestinalis and its relationship to host origin. Infect Genet Evol. 2003;3:29–38. doi: 10.1016/S1567-1348(02)00149-1. [DOI] [PubMed] [Google Scholar]
- 7.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia . Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 8.Poxleitner MK, Carpenter ML, Mancuso JJ, Wang C-JR, Dawson SC, et al. Evidence for karyogamy and exchange of genetic material in the binucleate intestinal parasite Giardia intestinalis . Science. 2008;319:1530–1533. doi: 10.1126/science.1153752. [DOI] [PubMed] [Google Scholar]
- 9.Adam RD, Dahlstrom EW, Martens CA, Bruno DP, Barbian KD, et al. Genome sequencing of Giardia lamblia genotypes A2 and B isolates (DH and Gs) and comparative analysis with the genomes of genotypes A1 and E (WB and pig) Genome Biol Evol. 2013;5:2498–2511. doi: 10.1093/gbe/evt197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jerlström-Hultqvist J, Franzén O, Ankarklev J, Xu F, Nohýnková E, et al. Genome analysis and comparative genomics of a Giardia intestinalis assemblage E isolate. BMC Genomics. 2010;11:e543. doi: 10.1186/1471-2164-11-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubourg A, Xia D, Winpenny JP, Al Naimi S, Bouzid M, et al. Giardia secretome highlights secreted tenascins as a key component of pathogenesis. Gigascience. 2018;7:giy003. doi: 10.1093/gigascience/giy003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhargava A, Cotton JA, Dixon BR, Gedamu L, Yates RM, et al. Giardia duodenalis surface cysteine proteases induce cleavage of the intestinal epithelial cytoskeletal protein villin via myosin light chain kinase. PLoS One. 2015;10:e0136102. doi: 10.1371/journal.pone.0136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu J, Ma'ayeh S, Peirasmaki D, Lundström-Stadelmann B, Hellman L, et al. Secreted Giardia intestinalis cysteine proteases disrupt intestinal epithelial cell junctional complexes and degrade chemokines. Virulence. 2018;9:879–894. doi: 10.1080/21505594.2018.1451284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanevik K, Bakken R, Brattbakk HR, Saghaug CS, Langeland N. Whole genome sequencing of clinical isolates of Giardia lamblia . Clin Microbiol Infect. 2015;21:192.e1–192.e3. doi: 10.1016/j.cmi.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Troell K, Hallström B, Divne A-M, Alsmark C, Arrighi R, et al. Cryptosporidium as a testbed for single cell genome characterization of unicellular eukaryotes. BMC Genomics. 2016;17:471. doi: 10.1186/s12864-016-2815-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uiterwijk M, Nijsse R, Kooyman FNJ, Wagenaar JA, Mughini-Gras L, et al. Comparing four diagnostic tests for Giardia duodenalis in dogs using latent class analysis. Parasit Vectors. 2018;11:439. doi: 10.1186/s13071-018-3014-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cacciò SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008;38:1523–1531. doi: 10.1016/j.ijpara.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 18.Tseng YC, Ho GD, Chen T TW, Huang BF, Cheng PC, et al. Prevalence and genotype of Giardia duodenalis from faecal samples of stray dogs in Hualien city of eastern Taiwan. Trop Biomed. 2014;31:305–311. [PubMed] [Google Scholar]
- 19.Ellegaard KM, Klasson L, Andersson SGE. Testing the reproducibility of multiple displacement amplification on genomes of clonal endosymbiont populations. PLoS One. 2013;8:e82319. doi: 10.1371/journal.pone.0082319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lasken RS. Single-cell genomic sequencing using multiple displacement amplification. Curr Opin Microbiol. 2007;10:510–516. doi: 10.1016/j.mib.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using diamond. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 22.Huson DH, Beier S, Flade I, Górska A, El-Hadidi M, et al. MEGAN community edition - interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput Biol. 2016;12:e1004957. doi: 10.1371/journal.pcbi.1004957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekseth OK, Kuiper M, Mironov V. orthAgogue: an agile tool for the rapid prediction of orthology relations. Bioinformatics. 2014;30:734–736. doi: 10.1093/bioinformatics/btt582. [DOI] [PubMed] [Google Scholar]
- 25.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 2018;35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ani analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sajid M, McKerrow JH. Cysteine proteases of parasitic organisms. Mol Biochem Parasitol. 2002;120:1–21. doi: 10.1016/S0166-6851(01)00438-8. [DOI] [PubMed] [Google Scholar]
- 32.Ankarklev J, Lebbad M, Einarsson E, Franzén O, Ahola H, et al. A novel high-resolution multilocus sequence typing of Giardia intestinalis assemblage A isolates reveals zoonotic transmission, clonal outbreaks and recombination. Infect Genet Evol. 2018;60:7–16. doi: 10.1016/j.meegid.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 33.Ansell BRE, Baker L, Emery SJ, McConville MJ, Svärd SG, et al. Transcriptomics indicates active and passive metronidazole resistance mechanisms in three seminal Giardia lines. Front Microbiol. 2017;8:e00398. doi: 10.3389/fmicb.2017.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma'ayeh SY, Knörr L, Svärd SG. Transcriptional profiling of Giardia intestinalis in response to oxidative stress. Int J Parasitol. 2015;45:925–938. doi: 10.1016/j.ijpara.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 35.Mastronicola D, Testa F, Forte E, Bordi E, Pucillo LP, et al. Flavohemoglobin and nitric oxide detoxification in the human protozoan parasite Giardia intestinalis . Biochem Biophys Res Commun. 2010;399:654–658. doi: 10.1016/j.bbrc.2010.07.137. [DOI] [PubMed] [Google Scholar]
- 36.Bernander R, Palm JED, Svärd SG. Genome ploidy in different stages of the Giardia lamblia life cycle. Cell Microbiol. 2001;3:55–62. doi: 10.1046/j.1462-5822.2001.00094.x. [DOI] [PubMed] [Google Scholar]
- 37.Monis PT, Andrews RH, Mayrhofer G, Ey PL. Molecular systematics of the parasitic protozoan Giardia intestinalis . Mol Biol Evol. 1999;16:1135–1144. doi: 10.1093/oxfordjournals.molbev.a026204. [DOI] [PubMed] [Google Scholar]
- 38.Monis P, Thompson RCA. Giardia – from genome to proteome. In: Rollinson D, Hay SI, editors. Advances in Parasitology. Vol. 78. London and Amsterdam: Elsevier; 2012. pp. 57–95. [DOI] [PubMed] [Google Scholar]
- 39.Ryan U, Cacciò SM. Zoonotic potential of Giardia . Int J Parasitol. 2013;43:943–956. doi: 10.1016/j.ijpara.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 40.Pallant L, Barutzki D, Schaper R, Thompson RCA. The epidemiology of infections with Giardia species and genotypes in well cared for dogs and cats in Germany. Parasit Vectors. 2015;8:2. doi: 10.1186/s13071-014-0615-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cotton JA, Bhargava A, Ferraz JG, Yates RM, Beck PL, et al. Giardia duodenalis cathepsin B proteases degrade intestinal epithelial interleukin-8 and attenuate interleukin-8-induced neutrophil chemotaxis. Infect Immun. 2014;82:2772–2787. doi: 10.1128/IAI.01771-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emery SJ, Mirzaei M, Vuong D, Pascovici D, Chick JM, et al. Induction of virulence factors in Giardia duodenalis independent of host attachment. Sci Rep. 2016;6:20765. doi: 10.1038/srep20765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu F, Jerlström-Hultqvist J, Einarsson E, Ástvaldsson A, Svärd SG, et al. The genome of Spironucleus salmonicida highlights a fish pathogen adapted to fluctuating environments. PLoS Genet. 2014;10:e1004053. doi: 10.1371/journal.pgen.1004053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.