Abstract
Introduction
Long non-coding RNAs (lncRNAs) have obtained increasing attention due to their regulatory functions in many cancers. This work aimed to investigate the functional roles of lncRNA TTN-AS1 in colorectal cancer (CRC) and to explore the underlying mechanisms.
Methods
The expression profiles of TTN-AS1 and miR-497 in CRC tissues and cell lines were determined by RT-qPCR analysis. MTT assay, transwell assay, western blot analysis, and xenograft tumors in nude mice were employed to analyze the effects of TTN-AS1 on the proliferation, migration, invasion, EMT, and in vivo tumorigenesis of CRC cells. Bioinformatics analysis and dual-luciferase reporter assay determined the direct binding relation between TTN-AS1 and miR-497 in CRC.
Results
We observed a significant increase of TTN-AS1 expression level in CRC tissues and cell lines compared with normal counterparts. High expression of TTN-AS1 predicted a poor prognosis and was correlated with aggressive clinicopathological characteristics in CRC patients. Functionally, gain- and loss-of-function studies indicated that TTN-AS1 knockdown suppressed the proliferation, migration, invasion and epithelial–mesenchymal transition of CRC cells in vitro, whereas TTN-AS1 overexpression showed the complete opposite effects. Mechanistically, we found that TTN-AS1 could directly interact with miR-497, and co-transfection with miR-497 mimics blocked the activation of PI3K/Akt/mTOR signaling, and reversed the effects of TTN-AS1 overexpression in CRC cells.
Conclusion
To conclude, our findings provide novel insight into CRC tumorigenesis and indicate that TTN-AS1 may serve as a potential therapeutic target for CRC treatment.
Keywords: colorectal cancer, long non-coding RNA TTN-AS1, epithelial-mesenchymal transition, microRNA-497, PI3K/Akt/mTOR signaling
Introduction
Colorectal cancer (CRC), a prevailing gastrointestinal malignancy, is also a main cause of cancer-associated deaths globally.1 Despite considerable advancements in the diagnostic approaches and therapeutic strategies, the clinical outcomes of patients with advanced CRC remain unfavorable.2 Accordingly, it is desirable and urgently needed to identify the potential molecular mechanisms underlying CRC progression and thus provide novel therapeutic targets.
The human genome produces coding and non-coding RNAs. Long non-coding RNAs (lncRNAs), a group of non-protein coding RNA transcripts characterized by a length of over 200 nucleotides, have emerged recently as major regulators in multiple biological processes, including tumorigenesis.3 TTN-AS1 is a newly discovered lncRNA located at human chromosomal region 2q31.2. Deregulated expression of TTN-AS1 has been reported in a variety of human malignancies, such as cervical cancer, papillary thyroid cancer, and gastric cancer.4–6 However, at present, little is understood about the role of TTN-AS1 in CRC.
Therefore, in this work, we aimed to explore the expression profile and clinical significance of TTN-AS1 in CRC and revealed the functional effects of TTN-AS1 on the malignant behaviors of CRC cells.
Materials and Methods
Tumor Tissues
95 matched pairs of CRC and pair-matched normal tissues were collected from patients who underwent tumor surgical resection at The Second Hospital of Shandong University (Jinan, China). Before operation, these patients were histologically confirmed to be CRC and did not receive chemotherapy or radiotherapy. After collection, the tissue specimens were immediately frozen in liquid nitrogen and then stored at −80°C. The use of human tissues was approved by the Ethics Committee of The Second Hospital of Shandong University and was conducted in accordance with the Declaration of Helsinki. We obtained written informed consent from every patients.
Cell Culture and Transfection
A series of CRC cell lines (SW480, SW620, HT29, HCT116) and human normal colon epithelial cell (FHC), obtained from American Type Culture Collection (ATCC; Manassas, VA, USA), were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin/streptomycin in a humidified incubator (37°C, 5% CO2).
miR-497 mimics and negative mimics control (NC) were purchased from GenePharma Co., Ltd. (Shanghai, China). To overexpress TTN-AS1, the full-length sequence of TTN-AS1 cDNA was cloned into the pcDNA3.1 vector (Invitrogen). An empty pcDNA3.1 vector was used as a negative control. To knockdown TTN-AS1, chemically synthesized siRNA sequence targeting TTN-AS1 was inserted into the shRNA expression vector pGPH1/Neo (GenePharma Co., Ltd.). Cells were cultured to about 80% confluence and then transfected with the vectors or oligonucleotides using Lipofectamine 2000 (Invitrogen). After 48 hrs, the transfection efficacy was verified by RT-qPCR analysis. In some cases, cells were treated with a specific Akt activator, SC79 (5 μM; Invitrogen).7
RT-qPCR Analysis
Total RNA was isolated using TRIzol reagent (Invitrogen), and cDNA synthesis was performed using the PrimeScript RT reagent Kit (TaKaRa, Dalian, China). qPCR reactions were then carried out using a SYBR Green PCR Kit (TaKaRa) on the MyIQ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The relative expression levels of target genes were calculated by the 2−ΔΔCt method,8 with GAPDH or U6 as an internal control. The sequences of the primers were shown as follows: TTN-AS1, forward primer: 5ʹ-GCCAGGTAGAGTTGCAGGTT-3ʹ and reverse primer: 5ʹ-GAAGCTGCTGCGGATGAATG-3ʹ; GAPDH, forward primer: 5ʹ-AGGTCGGTGTGAACGGATTTG-3ʹ and reverse primer: 5ʹ-TGTAGACCATGTAGTTGAGGTCA-3ʹ; miR-497, forward primer: 5ʹ-AGTCCAGTTTTCCCAGGAATCCCT-3ʹ and reverse primer: 5ʹ-ACCAGCAGCACACTGTGGTTTGT-3ʹ; U6, forward primer: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ and reverse primer: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
MTT Assay
Cell proliferation was monitored by MTT assay. Cells were incubated in 96-well-plates at a density of 4 × 103 cells/well. At the indicated time points, 20 μL MTT (5 mg/mL; Beyotime, Shanghai, China) was added to each well. After additional 4 hrs, the formazan crystals were dissolved in 150 μL DMSO, and the absorbance of the plate was then read at 570 nm.
Transwell Assay
Cells resuspended in 200 μL of serum-free medium were added into the upper chambers of Matrigel-uncoated and -coated transwell inserts (8 μm pore size; Corning, Inc., Corning, NY, USA). The lower chambers were loaded with 600 µL of medium containing 10% FBS. After incubation for 24 hrs, the cells that had migrated or invaded to the lower chamber were fixed by 4% paraformaldehyde, stained with 0.1% crystal violet and photographed under an optical microscope.
Western Blot Analysis
Total protein was isolated using RIPA lysis buffer (Beyotime). Equivalent amounts of proteins were fractionated by SDS-PAGE gel and then transferred onto PVDF membranes (Millipore, Billerica, MA, USA). The membranes were then incubated with specific primary antibodies against E-cadherin (1:2000; Abcam, Cambridge, MA, USA), N-cadherin (1:2000; Abcam), vimentin (1:2000; Abcam), p-PI3K (1:1000; Cell Signaling Technology, Inc., Danvers, MA, USA), PI3K (1:1000; Cell Signaling Technology, Inc.), p-Akt (1:1000; Cell Signaling Technology, Inc.), Akt (1:1000; Cell Signaling Technology, Inc.), p-mTOR (1:1000; Cell Signaling Technology, Inc.), mTOR (1:1000; Cell Signaling Technology, Inc.), and GAPDH (1:500; Santa Cruz Biotechnology, Inc., Dallas, TX, USA) overnight at 4°C, followed by incubation with HRP-conjugated secondary antibody (1:10,000; Santa Cruz Biotechnology, Inc.) at room temperature for 1 hr. The immunoreactive bands were detected using the enhanced chemiluminescence detection kit (Millipore). GAPDH was used as the control.
Dual-Luciferase Reporter Assay
The fragment of TTN-AS1 containing predicted miR-497 binding site was amplified by PCR and inserted into the pGL3-basic luciferase vector (Promega, Madison, WI, USA). Site-directed mutagenesis was performed using Quickchange XL site-directed mutagenesis kit (Agilent Technologies, Santa Clara, CA, USA). With Lipofectamine 2000, cells seeded in 24-well plates were co-transfected with the luciferase reporters and miR-497 mimics or NC. After 48 hrs, the luciferase activities were measured using the Dual-Luciferase Reporter Assay System (Promega).
Tumorigenesis in Nude Mice
Twelve BALB/c athymic nude mice (male, 4-week-old), obtained from Shanghai SLAC Laboratory Animal, Co., Ltd. (Shanghai, China), were housed under SPF condition and underwent a reverse dark-light cycle. Standard rodent chow and tap-water were provided ad libitum. The experimental procedures involving animals were performed with the approval of the Ethics Committee of The Second Hospital of Shandong University. All efforts were made to reduce the suffering of animals, according to the Guidelines for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006).
These mice were randomized into two groups (n = 6 in each group). SW620 cells (1 × 106 cells in 150 μL of PBS) stably transfected with sh-TTN-AS1 or sh-NC were subcutaneously injected into the flank of each mouse. Tumor volume was calculated every three days using the formula: volume = length × width2/2. After four weeks, the mice were euthanized by a cervical vertebrae luxation, and the tumors were excised for further analysis.
Statistical Analysis
All statistical analyses were performed using GraphPad Prism 6.0 software (GraphPad Software Inc., San Diego, CA, USA) and SPSS 18.0 software (IBM Corporation, Armonk, NY, USA). Continuous data were expressed as mean ± standard deviation (SD) and compared by Student’s t-test or one-way ANOVA as appropriate. Categorical variables were compared by chi-square test. The survival curves were plotted by Kaplan–Meier analysis, and analyzed by log-rank test. Statistical significance was set at P < 0.05.
Results
TTN-AS1 Is Overexpressed in CRC
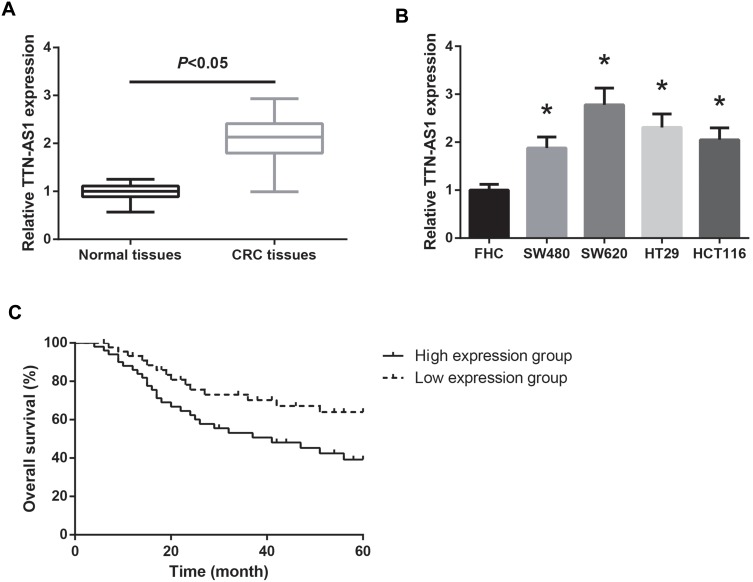
We first determined the expression profile of TTN-AS1 in CRC. As shown in Figure 1A, TTN-AS1 level was markedly increased in CRC tissues compared with paired normal tissues. Moreover, compared with normal colon mucosal epithelial cells (FHC), all of the CRC cell lines (SW480, SW620, HT29, HCT116) exhibited higher TTN-AS1 expression (Figure 1B).
Figure 1.
TTN-AS1 is overexpressed in CRC. (A) RT-qPCR analysis of TTN-AS1 expression in CRC tissues and adjacent normal tissues. (B) RT-qPCR analysis of TTN-AS1 expression in CRC cell lines and normal FHC cells. *P<0.05 vs FHC cells. (C) Association of TTN-AS1 expression with overall survival of CRC patients.
Using the median expression level of TTN-AS1 as a cutoff value, the 95 CRC patients were allocated into low TTN-AS1 expression group (n = 45) and high TTN-AS1 expression group (n = 50). As listed in Table 1, high TTN-AS1 expression was strongly correlated with lymph node metastasis (P = 0.039) and advanced TNM stage (P = 0.015) of CRC patients. We also noticed that CRC patients with higher TTN-AS1 expression had poorer overall survival (Figure 1C).
Table 1.
Association Between TTN-AS1 Expression and Clinicopathological Characteristics of CRC Patients
| Characteristics | Total Number | Low Expression (n=45) | High Expression (n=50) | P value |
|---|---|---|---|---|
| Age (years) | 0.297 | |||
| <55 | 37 | 20 | 17 | |
| ≥55 | 58 | 25 | 33 | |
| Gender | 0.810 | |||
| Male | 54 | 25 | 29 | |
| Female | 41 | 20 | 21 | |
| Tumor size (cm) | 0.183 | |||
| <5 | 61 | 32 | 29 | |
| ≥5 | 34 | 13 | 21 | |
| Differentiation | 0.287 | |||
| Well/Moderately | 58 | 30 | 28 | |
| Poorly | 37 | 15 | 22 | |
| Lymph node metastasis | 0.039 | |||
| Yes | 40 | 14 | 26 | |
| No | 55 | 31 | 24 | |
| TNM stage | 0.015 | |||
| I–II | 53 | 31 | 22 | |
| III–IV | 42 | 14 | 28 |
TTN-AS1 Promotes CRC Cell Proliferation in vitro and CRC Tumor Growth in vivo
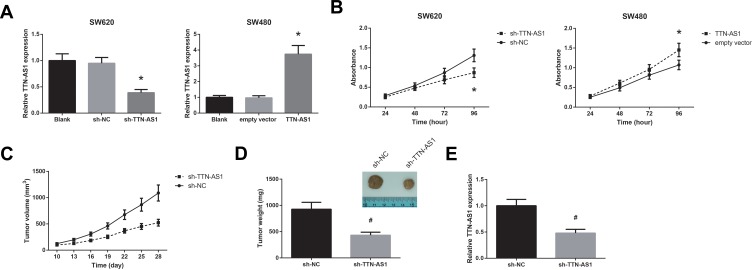
The biological function of TTN-AS1 in CRC was further analyzed. SW620 cells who had highest TTN-AS1 expression were transfected with sh-TTN-AS1, whereas SW480 cells whose TTN-AS1 expression was lowest were transfected with pcDNA3.1-TTN-AS1. The transfection efficiency was validated by RT-qPCR analysis (Figure 2A). MTT assay showed that TTN-AS1 knockdown observably restrained the proliferation ability of SW620 cells, whereas the proliferation rate of SW480 cells was enhanced by TTN-AS1 overexpression (Figure 2B). Moreover, as shown in Figure 2C, the growth of tumors in the sh-TTN-AS1 group was remarkably repressed, and after four weeks of growth, we observed that the average tumor weight in the sh-TTN-AS1 group was markedly decreased (Figure 2D). By RT-qPCR analysis, the downregulation of TTN-AS1 in the tumors of sh-TTN-AS1 group was also confirmed (Figure 2E).
Figure 2.
TTN-AS1 promotes CRC cell proliferation in vitro and CRC tumor growth in vivo. (A) RT-qPCR analysis of TTN-AS1 expression in CRC cells after transfection. (B) Proliferation of CRC cells after transfection, detected by MTT assay. (C) Growth curves of CRC tumors in the nude mice. (D) CRC tumors were extracted and weighed. (E) RT-qPCR analysis of TTN-AS1 expression in the CRC tumors. *P<0.05 vs sh-NC or empty vector-transfected cells; #P<0.05 vs sh-NC group.
TTN-AS1 Promotes CRC Cell Migration, Invasion and EMT in vitro
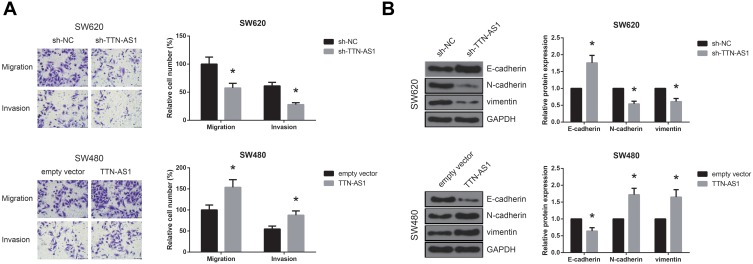
We then performed transwell assay, and the results demonstrated that, following transfection with sh-TTN-AS1, the migratory and invasive capacities of SW620 cells were dramatically suppressed (Figure 3A). On the other hand, TTN-AS1 overexpression remarkably increased the number of migrated and invaded SW480 cells. Moreover, as shown in Figure 3B, TTN-AS1 knockdown notably increased E-cadherin protein level and reduced the levels of N-cadherin and vimentin in SW620 cells, while TTN-AS1 overexpression exerted opposite effects in SW480 cells.
Figure 3.
TTN-AS1 promotes CRC cell migration, invasion, and EMT in vitro. (A) Migration and invasion of CRC cells after transfection, detected by transwell assay. (B) Western blot analysis of EMT-related protein expression in CRC cells after transfection. *P<0.05 vs sh-NC or empty vector-transfected cells.
TTN-AS1 Sponges miR-497 in CRC
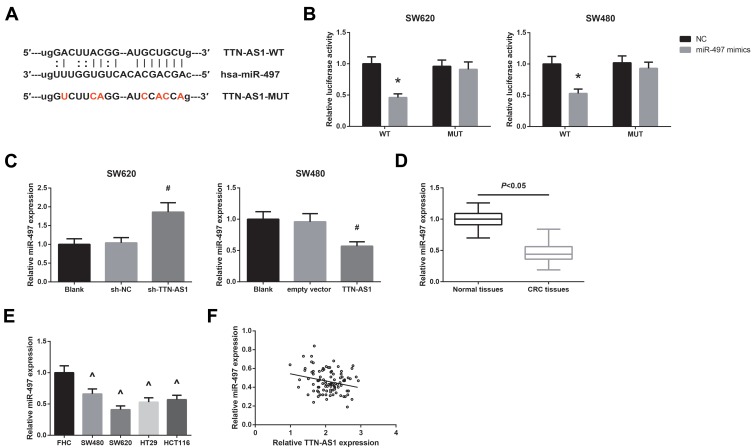
Some lncRNAs can serve as a molecular sponge to sequester miRNAs, resulting in inhibiting the effects of miRNAs. We then predicted the target miRNAs of TTN-AS1 through Starbase database (http://starbase.sysu.edu.cn/index.php), and selected miR-497 as a candidate, with the binding sites shown in Figure 4A. Next, dual-luciferase reporter assay was performed to validate the prediction. As shown in Figure 4B, miR-497 mimics could markedly decrease the luciferase activity of TTN-AS1-WT, while had nearly no effect on TTN-AS1-MUT in both SW620 and SW480 cells. Besides, miR-497 expression was increased in TTN-AS1-silenced SW620 cells and was decreased in SW480 cells with TTN-AS1 overexpression (Figure 4C). We further observed that miR-497 was downregulated in CRC tissues and cell lines (Figure 4D–E). An inverse correlation between TTN-AS1 and miR-497 expression in CRC tissues was also identified (r = −0.248, P = 0.015; Figure 4F).
Figure 4.
TTN-AS1 sponges miR-497 in CRC. (A) The predicted binding sites of miR-497 on TTN-AS1. (B) The binding relation between miR-497 and TTN-AS1 in CRC cells, validated by dual-luciferase reporter assay. (C) RT-qPCR analysis of miR-497 expression in CRC cells after transfection. (D) RT-qPCR analysis of miR-497 expression in CRC tissues and adjacent normal tissues. (E) RT-qPCR analysis of miR-497 expression in CRC cell lines and normal FHC cells. (F) The inverse correlation between miR-497 and TTN-AS1 expression in CRC tissues. *P<0.05 vs NC-transfected cells; #P<0.05 vs sh-NC or empty vector-transfected cells; ^P<0.05 vs FHC cells.
TTN-AS1 Activates PI3K/Akt/mTOR Signaling in CRC
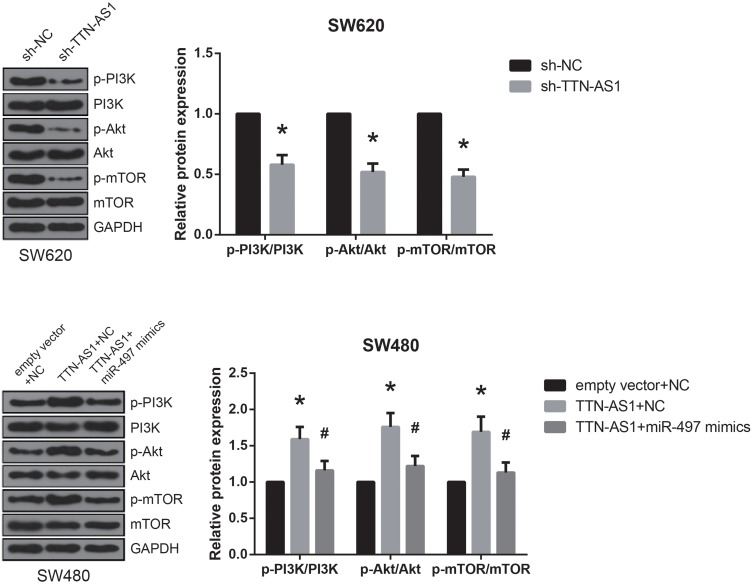
PI3K/Akt/mTOR signaling is one of the key dysregulated pathways in CRC,9 and in this study, through western blot analysis, we found that the expression levels of p-PI3K, p-Akt, and p-mTOR were markedly decreased by TTN-AS1 knockdown in SW620 cells (Figure 5). In addition, in SW480 cells with TTN-AS1 overexpression, co-transfection with miR-497 mimics effectively abrogated the activation of PI3K/Akt/mTOR signaling.
Figure 5.
TTN-AS1 activates PI3K/Akt/mTOR signaling in CRC. Western blot analysis of PI3K/Akt/mTOR signaling-related protein expression in CRC cells after transfection. *P<0.05 vs sh-NC or empty vector+NC-transfected cells; #P<0.05 vs pcDNA3.1-TTN-AS1+NC-transfected cells.
miR-497 Blocks the Oncogenic Role of TTN-AS1 in CRC
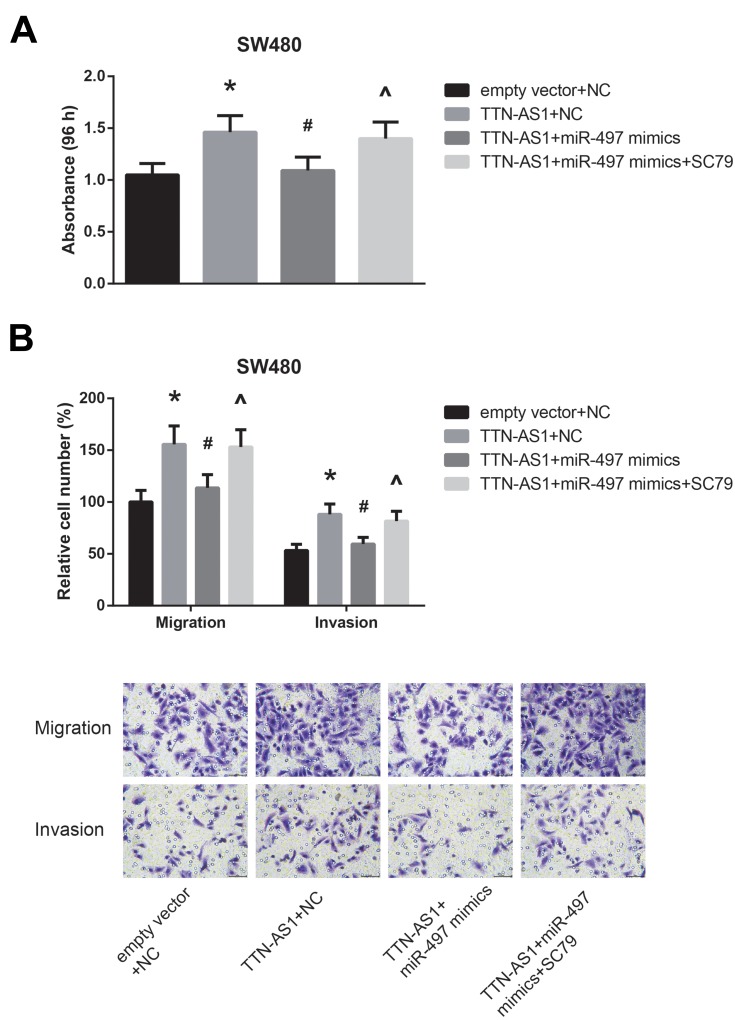
Furthermore, the results of MTT assay and transwell assay revealed that the enhanced proliferation, migration and invasion of TTN-AS1-overexpressing SW480 cells were notably reversed by miR-497 mimics; however, treatment with SC79 obviously diminished the role of miR-497 mimics (Figure 6A–B).
Figure 6.
Mir-497 blocks the oncogenic role of TTN-AS1 in CRC. (A) Proliferation of CRC cells after transfection, detected by MTT assay. (B) Migration and invasion of CRC cells after transfection, detected by transwell assay. *P<0.05 vs empty vector+NC-transfected cells; #P<0.05 vs pcDNA3.1-TTN-AS1+NC-transfected cells; ^P<0.05 vs pcDNA3.1-TTN-AS1+miR-497 mimics-transfected cells without SC79 treatment.
Discussion
The development of CRC is a complex multistep process involving both genetic and epigenetic changes. In recent years, more and more evidence has clearly depicted the critical roles of lncRNAs in CRC tumorigenesis, and a plenty of CRC-related lncRNAs have been identified, such as SNHG16 and DLX6-AS1.10,11 In this work, we focused on the functional role of lncRNA TTN-AS1 in CRC.
Our findings revealed that the expression of TTN-AS1 was notably ascended in CRC, and its upregulation was associated with lymph node metastasis, advanced TNM stage and poor prognosis in CRC patients. Then, in vitro gain- and loss-of-function experiments were performed using two independent CRC cell lines, and the results verified that loss of TTN-AS1 suppressed, whereas TTN-AS1 overexpression promoted the malignant behaviors, including proliferation, migration, and invasion of CRC cells. Besides, CRC xenograft growth in rodent models was also inhibited by TTN-AS1 knockdown. All these findings indicated that TTN-AS1 might serve as an oncogene in CRC.
Liver metastasis is one of the major causes for the death of CRC patients.12 Epithelial–mesenchymal transition (EMT), a key developmental program in the metastatic cascade, endows epithelial cells with migratory and invasive properties to disseminate from their primary sites.13 EMT-linked loss of basement membranes indicates metastasis and poor survival in CRC patients.14 Mounting evidence highlights a relevant function of lncRNAs on EMT in cancer biology.15 In this study, the activation effect of TTN-AS1 on EMT of CRC cells was proven, consistent with its role in esophageal squamous cell carcinoma and lung adenocarcinoma.16,17
Similar to lncRNAs, miRNAs, a class of small, non-coding RNAs, are also involved in CRC progression.18 Accumulating studies reveal that lncRNAs can serve as a competing endogenous RNA (ceRNA) by sponging miRNAs, thereby causing a loss of miRNA function.19 By bioinformatics prediction, miR-497 was selected as a model miRNA for further study due to its tumor-suppressive role in many types of cancers,20 and our data also indicated a significant downregulation of miR-497 in CRC, in accordance with the previous findings.21,22 Moreover, miR-497 restoration could diminish the effects of TTN-AS1 overexpression on the malignant behaviors of CRC cells, indicating that TTN-AS1 exerted its oncogenic role in CRC partly by regulating miR-497.
PI3K/Akt/mTOR signaling is one of the most critical intracellular pathways in both normal and cancer cells, and inhibition of this signaling has been shown to lead to regression of many tumor types, including CRC.23–25 High TTN-AS1 level in papillary thyroid cancer was closely associated with the activity of PI3K/Akt/mTOR signaling.5 Akt and mTOR are two downstream effectors of PI3K, and Guo et al previously reported that introduction of miR-497 mimics into CRC cells led to inhibition of Akt activation.26 This study verified that TTN-AS1 overexpression activated PI3K/Akt/mTOR signaling in CRC cells partly by regulating miR-497.
To conclude, in this study, we provided the promising evidence showing that TTN-AS1 was overexpressed in CRC, and could promote the proliferation, migration, and invasion of CRC cells by sponging miR-497 and activating PI3K/Akt/mTOR signaling. Along with further investigation, TTN-AS1 may function as a novel therapeutic target for CRC patients.
Acknowledgment
This work was supported by Key Research and Development Project of Shandong Province in 2019 (Grant no. GG201809240047).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi: 10.1016/S0140-6736(13)61649-9 [DOI] [PubMed] [Google Scholar]
- 3.Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–1261. doi: 10.1038/nm.3981 [DOI] [PubMed] [Google Scholar]
- 4.Chen P, Wang R, Yue Q, Hao M. Long non-coding RNA TTN-AS1 promotes cell growth and metastasis in cervical cancer via miR-573/E2F3. Biochem Biophys Res Commun. 2018;503(4):2956–2962. doi: 10.1016/j.bbrc.2018.08.077 [DOI] [PubMed] [Google Scholar]
- 5.Cui Z, Luo Z, Lin Z, Shi L, Hong Y, Yan C. Long non-coding RNA TTN-AS1 facilitates tumorigenesis of papillary thyroid cancer through modulating the miR-153-3p/ZNRF2 axis. J Gene Med. 2019;21(5):e3083. doi: 10.1002/jgm.v21.5 [DOI] [PubMed] [Google Scholar]
- 6.Dong MM, Peng SJ, Yuan YN, Luo HP.LncRNA TTN-AS1 contributes to gastric cancer progression by acting as a competing endogenous RNA of miR-376b-3p. Neoplasma.2019;66:564–575. doi: 10.4149/neo_2018_180927N721 [DOI] [PubMed] [Google Scholar]
- 7.Jo H, Mondal S, Tan D, et al. Small molecule-induced cytosolic activation of protein kinase Akt rescues ischemia-elicited neuronal death. Proc Natl Acad Sci U S A. 2012;109(26):10581–10586. doi: 10.1073/pnas.1202810109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 9.Bahrami A, Khazaei M, Hasanzadeh M, et al. Therapeutic potential of targeting PI3K/AKT pathway in treatment of colorectal cancer: rational and progress. J Cell Biochem. 2018;119(3):2460–2469. doi: 10.1002/jcb.25950 [DOI] [PubMed] [Google Scholar]
- 10.Li Y, Lu Y, Chen Y. Long non-coding RNA SNHG16 affects cell proliferation and predicts a poor prognosis in patients with colorectal cancer via sponging miR-200a-3p. Biosci Rep. 2019;39(5):pii: BSR20182498 Epub 2019 Apr 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou FR, Pan ZP, Shen F, et al. Long noncoding RNA DLX6-AS1 functions as a competing endogenous RNA for miR-577 to promote malignant development of colorectal cancer. Eur Rev Med Pharmacol Sci. 2019;23(9):3742–3748. doi: 10.26355/eurrev_201905_17800 [DOI] [PubMed] [Google Scholar]
- 12.Wang Y, Liu YF, Cheng Y, et al. Prognosis of colorectal cancer with liver metastasis: value of a prognostic index. Braz J Med Biol Res. 2010;43(11):1116–1122. doi: 10.1590/s0100-879x2010007500103 [DOI] [PubMed] [Google Scholar]
- 13.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007 [DOI] [PubMed] [Google Scholar]
- 14.Spaderna S, Schmalhofer O, Hlubek F, et al. A transient, EMT-linked loss of basement membranes indicates metastasis and poor survival in colorectal cancer. Gastroenterology. 2006;131(3):830–840. doi: 10.1053/j.gastro.2006.06.016 [DOI] [PubMed] [Google Scholar]
- 15.Gugnoni M, Ciarrocchi A. Long noncoding RNA and epithelial mesenchymal transition in cancer. Int J Mol Sci. 2019;20(8):1924. doi: 10.3390/ijms20081924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin C, Zhang S, Wang Y, et al. Functional role of a novel long noncoding RNA TTN-AS1 in esophageal squamous cell carcinoma progression and metastasis. Clin Cancer Res. 2018;24(2):486–498. doi: 10.1158/1078-0432.CCR-17-1851 [DOI] [PubMed] [Google Scholar]
- 17.Jia Y, Duan Y, Liu T, et al. LncRNA TTN-AS1 promotes migration, invasion, and epithelial mesenchymal transition of lung adenocarcinoma via sponging miR-142-5p to regulate CDK5. Cell Death Dis. 2019;10(8):573. doi: 10.1038/s41419-019-1811-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding L, Lan Z, Xiong X, et al. The dual role of MicroRNAs in colorectal cancer progression. Int J Mol Sci. 2018;19(9):2791. doi: 10.3390/ijms19092791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi: 10.1016/j.cell.2011.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang G, Xiong G, Cao Z, et al. miR-497 expression, function and clinical application in cancer. Oncotarget. 2016;7(34):55900–55911. doi: 10.18632/oncotarget.10152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang N, Shen Q, Zhang P.miR-497 suppresses epithelial-mesenchymal transition and metastasis in colorectal cancer cells by targeting fos-related antigen-1. Onco Targets Ther.2016;9:6597–6604. doi: 10.2147/OTT.S114609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Chen J, Gao C, et al. MicroRNA-497 inhibits tumor growth through targeting insulin receptor substrate 1 in colorectal cancer. Oncol Lett. 2017;14(6):6379–6386. doi: 10.3892/ol.2017.7033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polivka J Jr., Janku F. Molecular targets for cancer therapy in the PI3K/AKT/mTOR pathway. Pharmacol Ther. 2014;142(2):164–175. doi: 10.1016/j.pharmthera.2013.12.004 [DOI] [PubMed] [Google Scholar]
- 24.Alzahrani AS.PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin Cancer Biol.2019;59:125–132. doi: 10.1016/j.semcancer.2019.07.009 [DOI] [PubMed] [Google Scholar]
- 25.Pandurangan AK. Potential targets for prevention of colorectal cancer: a focus on PI3K/Akt/mTOR and Wnt pathways. Asian Pac J Cancer Prev. 2013;14(4):2201–2205. doi: 10.7314/APJCP.2013.14.4.2201 [DOI] [PubMed] [Google Scholar]
- 26.Guo ST, Jiang CC, Wang GP, et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene. 2013;32(15):1910–1920. doi: 10.1038/onc.2012.214 [DOI] [PMC free article] [PubMed] [Google Scholar]