Abstract
Homologous chromosome segregation during meiosis I (MI) in mammalian oocytes is carried out by the acentrosomal MI spindles. Whereas studies in human oocytes identified Ran GTPase as a crucial regulator of the MI spindle function, experiments in mouse oocytes questioned the generality of this notion. Here, we use live‐cell imaging with fluorescent probes and Förster resonance energy transfer (FRET) biosensors to monitor the changes in Ran and importin β signaling induced by perturbations of Ran in mouse oocytes while examining the MI spindle dynamics. We show that unlike RanT24N employed in previous studies, a RanT24N, T42A double mutant inhibits RanGEF without perturbing cargo binding to importin β and disrupts MI spindle function in chromosome segregation. Roles of Ran and importin β in the coalescence of microtubule organizing centers (MTOCs) and MI spindle assembly are further supported by the use of the chemical inhibitor importazole, whose effects are partially rescued by the GTP hydrolysis‐resistant RanQ69L mutant. These results indicate that RanGTP is essential for MI spindle assembly and function both in humans and mice.
Keywords: importazole, importin β, meiosis I, oocyte, RanGTP
Subject Categories: Cell Adhesion, Polarity & Cytoskeleton; Cell Cycle
Live‐cell imaging with fluorescent probes and FRET biosensors clarify previous discrepancies between findings in human and mouse oocytes, supporting conserved roles of Ran GTPase in acentrosomal meiotic spindle formation in mammals.
Introduction
The formation of female gametes—eggs—depends on the function of the meiosis I and II (MI and MII) spindles in chromosome segregation during the two meiotic divisions. Together with the simultaneous breakdown of the prophase I nuclear envelope (termed germinal vesicle breakdown, GVBD), the onset of the MI spindle assembly marks the re‐entry to meiosis in fully grown oocytes. While mitotic spindles in somatic cells rely on the pair of centrosomes to guide the assembly of their bipolar structure, fully functional centrosomes are eliminated before the start of meiosis in oocytes of mammals and many other animal species (Szollosi et al, 1972, 1986; Schatten & Sun, 2015; Simerly et al, 2018). As a result, the meiotic spindles in mammalian oocytes are more dependent on the self‐assembly of microtubule (MT) structures and the role of chromosomal signaling via the small GTPase Ran (Gruss, 2018).
In cells containing nuclei (including the prophase I, germinal vesicle (GV)‐containing oocytes), Ran regulates the macromolecular translocations between the nucleus and the cytoplasm such that the high RanGTP concentration in the nucleus and low in the cytoplasm dictate the direction of the traffic. The nuclear RanGTP enables the loading of nuclear export cargos on exportins and releases the nuclear localization signal (NLS)‐containing cargos of importins inside the nucleus. The nuclear import and chromatin‐binding activity of the guanine nucleotide exchange factor RCC1, the cytoplasmic localization of the Ran GTPase activating protein RanGAP1 and its cofactors RanBP1/2 are responsible for the nuclear accumulation of RanGTP and its cytoplasmic scarcity (Clarke & Zhang, 2008; Kalab & Heald, 2008; Forbes et al, 2015). Since RCC1 binds chromatin and RanGAP1 remains cytoplasmic after nuclear envelope disassembly, diffusional concentration gradients of RanGTP surround mitotic or meiotic chromosomes (Gorlich et al, 2003; Bastiaens et al, 2006; Kalab & Heald, 2008). Downstream, RanGTP binding to importins dissociates their cargos, inducing spatial activity gradients of NLS‐containing spindle assembly factors (SAFs) that are inhibited by importin binding (Clarke & Zhang, 2008; Kalab & Heald, 2008; Forbes et al, 2015). The abundant importin β is the prominent RanGTP‐regulated inhibitor of several SAFs that bind importin β directly (direct cargos, such as HURP; Sillje et al, 2006; Breuer et al, 2010) or through the importins α (indirect cargos, such as the MT‐nucleating factor TPX2; Gruss et al, 2001; Brunet et al, 2008; Forbes et al, 2015).
Early studies showed that spindle assembly in the X. laevis egg extracts was disrupted by the inhibition of RanGTP and cargo gradients by the GTP binding‐resistant RanT24N (Kalab et al, 2002; Clarke & Zhang, 2008; Kalab & Heald, 2008), which blocks GTP loading on the endogenous Ran by sequestering and inhibiting RCC1 (Dasso et al, 1994; Lounsbury et al, 1996). However, while the expression of RanT24N in mouse oocytes abolished the chromosomal importin β cargo gradients (Dumont et al, 2007), the MI spindles assembled and segregated chromosomes (Dumont et al, 2007), although MT nucleation at MTOCs decreased (Schuh & Ellenberg, 2007) and the MI spindles were misshapen (Bury et al, 2017). In contrast to mouse oocytes, the expression of RanT24N in human oocytes abolished their MI spindle assembly and chromosome segregation (Holubcova et al, 2015), suggesting that species‐specific factors define Ran function in meiotic spindle assembly in mammalian oocytes.
The difference in the role of Ran in human versus mouse oocytes is unlikely involving the variation in Ran or importin β sequences since they are nearly identical (94 and 99% identity for Ran and importin β, respectively). Potentially, the heightened dependence of the MI spindles on Ran in human oocytes could arise from adaptations to longer MI time (16 versus 4‐5 h in mouse oocytes; Holubcova et al 2015) and to the absence of γ‐tubulin or pericentrin‐containing MTOCs in human oocytes (Holubcova et al, 2015), as compared to the MTOC‐dependent MI spindle assembly in mouse oocyte (Breuer et al, 2010; Luksza et al, 2013; Clift & Schuh, 2015; Bennabi et al, 2016). Supporting the potential role of MTOCs in the lower dependence on Ran in the mouse oocytes, transgenic mouse oocytes depleted of pericentrin were more sensitive to RanT24N expression than the wild‐type (WT) oocytes (Baumann et al, 2017).
Two lines of evidence made us wonder if the role of Ran in mouse MI spindle assembly is more pervasive than what the previous research indicated. First, the evidence for the involvement of Ran in the MI spindle assembly (or the lack of it) so far relied on the application of RanT24N and the notion that it acts solely as an inhibitor of RanGTP formation (Dumont et al, 2007; Schuh & Ellenberg, 2007; Holubcova et al, 2015; Bury et al, 2017). However, studies in the meiotic X. laevis egg extracts showed that RanT24N co‐precipitated with importin β (Hughes et al, 1998), which would be impossible if RanT24N acted only as RCC1 inhibitor. Second, we noticed that mouse pericentrin contains NLS sequences that are highly similar to NLS that conferred the binding of human pericentrin to importin β (Liu et al, 2010), suggesting that pericentrin could function as a conserved RanGTP‐ and importin β‐regulated SAF. Moreover, the organization of mouse oocyte MTOCs depends on the MT‐binding protein HURP (Breuer et al, 2010), which is a SAF, which has been shown to be regulated by RanGTP and direct binding to importin β in human cells (Sillje et al, 2006). We suspected that the reliance on RanT24N as a putatively dominant‐negative inhibitor of RanGTP underestimated the importance of RanGTP and that RanGTP and importin β could potentially regulate MTOCs, if not other aspects of MI spindle assembly in the mouse oocytes.
To test these ideas, we initially manipulated Ran and importin β function in mouse oocytes by the expression of RanT24N (Dumont et al, 2007; Schuh & Ellenberg, 2007; Bury et al, 2017), injection of the RanGTP‐resistant cargo domain of importin β (importin β(71–876); Nachury et al, 2001), and the treatments with importazole (IPZ), a chemical inhibitor of RanGTP–importin β interaction (Soderholm et al, 2011). We combined those treatments with analyses of their effects on RanGTP and indirect (importin α/importin β complex‐dependent) importin β cargos in oocytes by applying the fluorescence lifetime imaging microscopy (FLIM) with FRET biosensors RBP‐3 (RanGTP‐binding probe‐3 spectral variant of RBP‐4; Hasegawa et al, 2013) and Rango‐3 (Ran‐regulated importin β cargo‐3; Soderholm et al, 2011), respectively. We found that while RanT24N inhibited the spatial RanGTP and cargo gradients around chromosomes, consistent with previous reports (Dumont et al, 2007; Hasegawa et al, 2013), in a concentration‐dependent manner, RanT24N also increased, rather than decreased the average levels of importin β‐free cargos, as it would be expected from a specific RCC1 inhibitor. Since reduced binding of cargos to importin β could counteract the effects of RCC1 inhibition, these results confirmed our suspicion that previous studies could have underestimated the role of Ran in mouse oocytes by relying on RanT24N to inhibit the RCC1‐dependent RanGTP formation (Dumont et al, 2007; Schuh & Ellenberg, 2007; Bury et al, 2017). To resolve the quandary, we developed and tested RanT24N, T42A double mutant, which acts as an inhibitor of RCC1 that is devoid of the detectable interference with importin β‐cargo interaction and whose expression strongly disrupts the MI spindle assembly and function in mouse oocytes. Together with analyses of the perturbations of MI spindles by the constitutively active importin β and IPZ, our results show that the MI spindle function in mouse oocytes critically depends on the role of the RanGTP and importin β‐regulated mechanisms.
Results
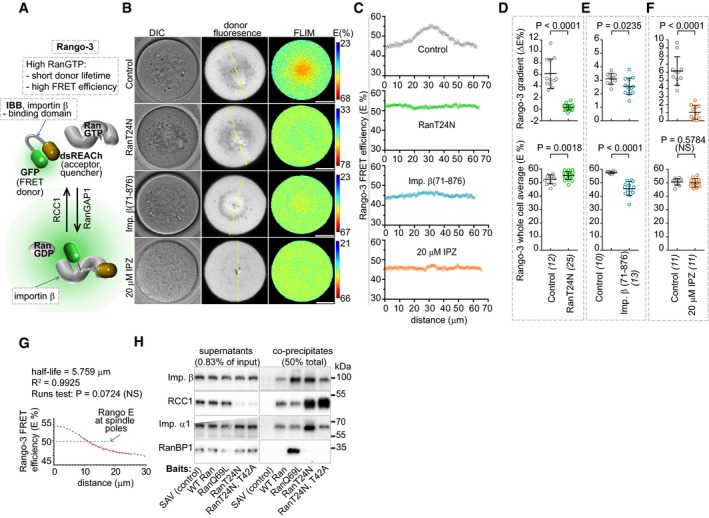
We began by applying the previously described FRET sensor Rango‐3 (Soderholm et al, 2011) to examine the effects of inhibitors on the RanGTP‐ and importin β‐mediated signaling in MI oocytes (Fig 1A). Rango‐3 consists of importin β‐binding domain (IBB) flanked by the GFP FRET donor protein and the non‐fluorescent dsREACh acceptor (Hasegawa et al, 2013). The donor and acceptor are spatially separated in the extended conformation of the importin β‐bound Rango‐3, increasing the lifetime of the donor fluorescence (τdonor). RanGTP binding to importin β releases the free form of Rango‐3 in which the donor and acceptor can interact with each other, resulting in increased FRET and reduced τdonor (Fig 1A). We detected the changes in the τdonor with the use of time‐correlated single‐photon counting FLIM. To compare the effects of various treatments, we used the amplitude‐weighted average τdonorAW values measured by FLIM in oocyte to obtain estimates of the FRET efficiency (E) using the equation E = 1 – (τdonorAW/τdonorREFAW) (Lakowicz, 2006), where the τdonorREFAW corresponds to τdonorAW measured in the absence of the acceptor.
Figure 1. Quantitative FLIM/FRET imaging reveals pleiotropic effects of RanT24N on chromosomal gradients and importin β‐cargo binding.
-
ASchematics of Rango‐3 sensor.
-
BRepresentative images of importin β cargo gradients detected by Rango‐3 FLIM/FRET imaging. Scale bars, 20μm.
-
CLine scans of Rango‐3 FRET efficiency (E) in 10‐μm‐wide areas centered at the dashed lines in (B).
-
D–FQuantification of the Rango‐3 E gradients (top panels) and average cellular Rango‐3 E (bottom) in involving untreated oocytes (controls) or oocytes injected with RanT24N mRNA, importin β(71–876) protein or treated with 20 μM IPZ. Data for each panel are from at least 2 separate experiments. Means ± SDs, t‐test, oocyte numbers indicated in brackets.
-
GNonlinear single‐exponential regression was used to fit the average of radial Rango‐3 τdonor line scans in control oocyte (Fig 1D) with the one‐phase decay model. The dashed line corresponds to the average of spindle pole distance from chromosomes in the same cell.
-
HImmunoblotting in biochemical pulldowns with recombinant Ran proteins added to human DLD1 cell lysates. A representative of at least five repeats.
The signal of importin β cargo gradients reaches beyond the MI spindle poles
Consistent with the previous report (Dumont et al, 2007), chromosomes in MI oocytes were surrounded by the highest cellular concentration of free importin β cargos (Fig 1B). Rango‐3 lifetime increased over the distance from chromosomes along a single‐exponential curve with the mean half‐life 7.7 ± 1.7 μm (R 2 > 0.95; Fig 1G, Appendix Fig S1A), indicating that the corresponding SAF activities are elevated throughout the spindles, including their poles. The mono‐exponential decay profile of the Rango‐3 signal is consistent with the expected behavior of the sensor by indicating the dominant influence of a single Rango‐3 ligand, importin β.
RanT24N overexpression inhibits chromosomal cargo gradients but does not reduce importin β‐free cargos across oocytes
All three inhibitors of importin β signaling that we initially tested (co‐expression of RanT24N, microinjection of importin β(71–876) protein, treatment with 20 μM IPZ) strongly reduced or flattened the MI cargo gradients (Fig 1C–F). While no remaining gradients were detectable in RanT24N‐treated oocytes, a small residual gradient was present in oocytes treated with importin β(71–876), and there was a small local increase in free cargos at the chromosomes compared to the cytoplasm in the IPZ‐treated cells (Fig 1C). Importin β(71–876) sharply reduced the average free cargos in the oocytes, confirming its function as a RanGTP‐resistant, constitutively active importin β (Fig 1E). However, we observed no overall change in importin β‐bound cargos in the IPZ‐treated cells (Fig 1F), indicating that, while the IPZ binds to importin β and inhibits its interaction with RanGTP (Soderholm et al, 2011), the IPZ‐bound importin β also does not stably load its cargos. Unexpectedly, the overall concentration of free cargos slightly but significantly increased in the RanT24N‐treated cells (Fig 1D).
If RanT24N is a specific RCC1 inhibitor, RanT24N expression should lead to lower RanGTP and reduced free importin β cargos across the cell, which is opposite of what we observed (Fig 1D). To understand this discrepancy, we considered that, in meiotic X. laevis egg extracts, RanT24N co‐precipitated with importin β (Hughes et al, 1998). To verify that RanT24N could bind importin β also in mammalian cells, we performed biochemical pulldowns from human tissue culture cell lysates supplemented with recombinant Ran proteins. As expected, the GTP hydrolysis‐resistant RanQ69L co‐precipitated with RanBP1 and importin β (Fig 1H), and RanT24N co‐precipitated RCC1 more efficiently than the WT Ran or RanQ69L, leading to near‐complete depletion of RCC1 from the lysates. However, at the same time, the RanT24N bait also pulled a similar amount of importin β as RanQ69L (Fig 1H), confirming results in meiotic frog egg extracts (Hughes et al, 1998) that oppose the prematurely accepted canon of RanT24N as a specific RCC1 inhibitor. To test how RanT24N interacts with importin β, we targeted threonine 42, which was necessary for WT Ran binding to importin β (Murphy et al, 1997), and substituted it with alanine. Indeed, the RanT24N, T42A double mutant precipitated much less importin β from the cell extracts compared to RanT24N, suggesting that RanT24N directly interacts with importin β through the T42 site. Interestingly, the T42A mutation also reduced the prominent co‐precipitation of importin α1 with RanT24N, indicating that RanT24N could potentially bind importin β without causing the importin α dissociation, and showing that RanT24N does not induce the dissociation of importin α from importin β, as RanGTP does. The composition of the various complexes formed by the RanT24N or RanT24N, T42A proteins will require further investigation. However, together with the FRET imaging data (Fig 1D), the co‐precipitation results indicated that RanT24N expressed in oocytes could sequester the endogenous importin β and globally increase, rather than decrease the fraction of cargos/SAFs that are not inhibited by binding to importin β. Previous studies showed that the overall importin β cargo release but not its spatial regulation is enough to support bipolar spindle assembly in meiotic Xenopus egg extracts or mitotic somatic cells (Maresca et al, 2009; O'Connell et al, 2009). Then, the limited effects of RanT24N on mouse MI spindles (Dumont et al, 2007; Schuh & Ellenberg, 2007; Bury et al, 2017) could have resulted from the incomplete inhibition of importin β targets across the oocytes.
While the expression of RanT24N above a critical threshold fails to further reduce the overall cargo release, a wide range of injected RanT24N, T42A mRNA inhibits cargo gradients and overall cargo release in the oocytes
We wished to verify that the added T42A substitution (Ran double mutant), which presumably blocks the importin β‐binding, would reduce the effects of RanT24N on importin β also in the oocytes. Because the RanT24N binds RCC1 strongly, and RCC1 is less abundant than importin β (Gorlich et al, 2003), we hypothesized that the RanT24N‐induced importin β cargo release could depend on the dose of the overexpressed RanT24N, and the global importin β cargo release should be much lower or non‐detectable in oocytes treated with increasing doses of RanT24N, T42A. Since we obtained the results in Fig 1B–D in oocytes microinjected with 2,500 ng/μl RanT24N mRNA, we tested these ideas by monitoring Rango‐3 FLIM in oocytes treated with the lower range of RanT24N or RanT24N, T42A mRNA (300–1,200 ng/μl). Consistent with our prediction, increasing concentrations of RanT24N mRNA elevated the average free cargo (Rango‐3) levels, while the average free cargos remained lower than in controls and constant in oocytes injected with increasing doses of the double mutant (Fig 2A, Appendix Fig S2A and B). At the same time, the double mutant was less efficient than RanT24N in reducing the chromosomal gradients (Fig 2A–C, Appendix Fig S2A and B). This result suggests that thanks to the reduced or absent interference with importin β, even incomplete inhibition of RCC1 by the Ran double mutant could efficiently deplete oocytes of free importin β cargos. In separate experiments that involved the side‐by‐side comparisons of the two Ran mutants at higher doses (2,000 ng/μl), we observed that RanT24N, T42A reduced cargo binding to importin β, but not the RanT24N mutant (Fig 2D).
Figure 2. The RanT24N, T42A double mutant disrupts chromosomal cargo gradients without perturbing importin β‐cargo binding in oocytes.

- Quantification of chromosomal cargo gradients (top) and average Rango‐3 E (bottom) in oocytes injected with increasing concentrations of Ran mutant mRNA concentrations. Data from oocytes imaged side‐by‐side within one experiment. Means ± SDs, one‐side ANOVA, Tukey's test. Numbers of analyzed oocytes are indicated in the brackets in the top panel and are identical for both panels. Additional analyses of the same data are in Appendix Fig S2.
- Representative images of importin β cargo gradients detected by Rango‐3 FLIM/FRET imaging. Scale bars, 20 μm.
- Line scans of the estimated Rango‐3 FRET efficiency (E) in 10‐μm‐wide areas centered at the dashed lines in (A).
- Quantification of the Rango‐3 E gradients (left panel) and average cellular Rango‐3 E (right panel) oocytes that were treated side‐by‐side by the injection of mRNA for Rango‐3, alongside with buffer (controls) or 2,000 ng/μl mRNA for RanT24N or RanT24N, T42A. Means ± SDs, one‐side ANOVA, Tukey's test, oocyte numbers indicated in brackets.
Inhibition of importin β by IPZ perturbs chromosome condensation and RanGTP gradient
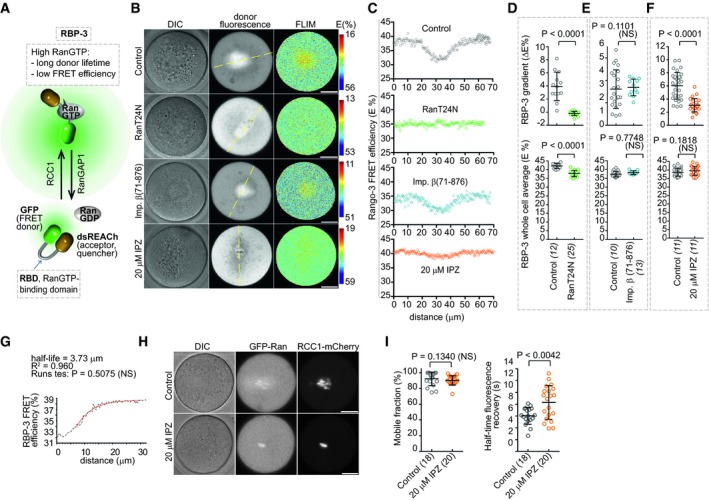
Next, we applied FLIM/FRET imaging with the RBP‐3 sensor (Hasegawa et al, 2013) to examine the RanGTP signaling upstream of the importin β cargo gradient (Kalab et al, 2002). RBP‐3 consists of the RanGTP‐binding domain of the yeast homolog of RanBP1 (Kalab et al, 2002) flanked by the GFP donor and non‐fluorescent dsREACh acceptor. The binding of RBP‐3 to RanGTP is signaled by increased τdonor, corresponding to reduced RBP‐3 FRET efficiency (RBP‐3 E) (Hasegawa et al, 2013) (Fig 3A). The gradients detected by RBP‐3 (Fig 3B and C) reached to a shorter distance from chromosomes compared to the cargo gradients, dissipating with a single‐exponential half‐life 3.6 ± 1.0 μm (Fig 3G, Appendix Fig S1). The shorter range of the RBP‐3 gradients agreed with the predicted steeper decay of the equivalent RanGTP‐RanBP1 gradient (Gorlich et al, 2003; Caudron et al, 2005).
Figure 3. IPZ perturbs the RanGTP gradients through interfering with RCC1 binding to chromatin.
-
ASchematics of RBP‐3 sensor function.
-
BChromosomal gradients detected by RBP3 FLIM/FRET imaging. Scale bars, 20 μm.
-
CLine scans of the estimated RBP‐3 in 10‐μm‐wide areas centered at the dashed lines in (B).
-
D–FQuantification of the RBP‐3 gradients (top panels) and average cellular RBP‐3 E (bottom) in separate experiments involving untreated oocytes (controls) or oocytes injected with RanT24N mRNA, importin β(71–876) protein or treated with 20 μM IPZ. Means ± SDs, t‐test, oocyte numbers indicated in brackets.
-
GNonlinear single‐exponential regression was used to fit the average of radial RBP‐3 τdonor line scans in control oocyte (D) with the one‐phase decay model.
-
HImages of oocytes expressing GFP‐Ran and RCC1‐mCherry from injected mRNAs before FRAP assays. Scale bars, 20 μm.
-
IRCC1‐mCherry half‐time fluorescence recovery (right, means ± SDs, t‐test.) and a mobile fraction (left, means ± SDs, Mann–Whitney test) in control and IPZ‐treated cells, oocyte numbers indicated in brackets.
If RanT24N inhibits GTP loading on Ran by binding to RCC1, the expression of RanT24N should reduce RanGTP levels across the cell, which would be detected as an increase in RBP‐3 E. As expected, RanT24N expression destroyed RBP‐3 gradient; however, it also induced a small but significant drop in the average RBP‐3 E, indicating slightly elevated overall RanGTP levels (Fig 3D). Although this change was significantly smaller compared to the effect of co‐expressed RanQ69L (GTP hydrolysis‐resistant Ran; Appendix Fig S3A), this result indicated that RanT24N expression slightly increased, instead of decreased, RanGTP levels (Fig 1D and H), possibly because RanT24N could compete with RanGTP for access to importin β.
Consistent with data from meiotic X. laevis egg extracts (Kalab et al, 2002), importin β(71–876) did not significantly alter the RBP‐3 chromosomal gradients or the average level of RBP‐3 E (Fig 3B, C and E). Surprisingly, treatment with IPZ inhibited the chromosomal RanGTP gradients, although it caused no significant change in overall RanGTP levels (Fig 3B, C and F). Since chromosomes in IPZ‐treated oocytes appeared hyper‐condensed (Appendix Fig S3B), we hypothesized that RCC1 binding to chromatin was affected. To test this, we performed FRAP (fluorescence recovery after photobleaching) measurements with RCC1‐mCherry expressed in untreated or IPZ‐treated oocytes (Fig 3H and I). Although we have not detected a significant change in the RCC1 mobile fraction, the half‐time of fluorescence recovery of the chromosomal RCC1‐mCherry was significantly increased in the IPZ‐treated cells (Fig 3I). These data suggest that, by inducing changes in chromosome condensation, IPZ indirectly slowed down the dynamics of RCC1 binding to and release from the MI chromatin, which is necessary for the formation of the RanGTP gradient. At the same time, since the overall RanGTP levels in oocytes remained unchanged (Fig 3F), RCC1 activity was unlikely to be altered by the IPZ.
Treatment with IPZ blocks MI spindle formation and function
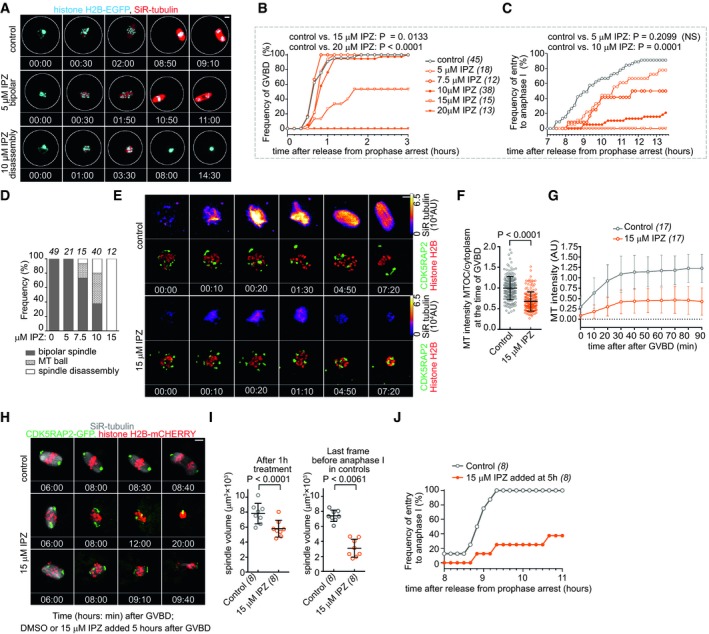
Having confirmed that IPZ disrupts both the chromosomal RanGTP and importin β cargo gradients in oocytes, we were interested in testing how IPZ affects the MI progression in mouse oocytes. To do so, we isolated oocytes from the histone H2B‐EGFP‐expressing transgenic mice, stained the microtubules with the fluorogenic SiR‐tubulin dye (Lukinavicius et al, 2014; Balboula et al, 2016; Courtois et al, 2018), and monitored the meiotic progression in the presence of 5–20 μM IPZ by using confocal live‐cell imaging (Fig 4A, Movie EV1 and Appendix Fig S4A). Although the treatment with 5 μM IPZ did not affect the resumption of meiosis (Fig 4B) or the assembly of a bipolar spindle (Fig 4D), the entry into anaphase I was significantly delayed (Fig 4C). At higher than 7.5 μM IPZ, the frequency of oocytes that entered anaphase I significantly decreased (Fig 4C). At 7.5 and 10 μM IPZ, the proportion of normal bipolar spindles decreased to 73 and 37%, respectively (Fig 4D). In the remaining IPZ‐treated oocytes, only the initial multipolar ball‐like structure of MTs (Schuh & Ellenberg, 2007) was formed, but in all cases, this structure eventually disassembled. Treatment with 15 μM IPZ strongly delayed resumption of meiosis (Fig 4B and Appendix Fig S4B) and induced spindle disassembly in all oocytes (Fig 4D). At 20 μM, IPZ completely blocked the resumption of meiosis (Fig 4B).
Figure 4. IPZ disrupts meiosis entry, MI spindle assembly, and anaphase I onset.
-
ADose‐dependent effects of IPZ on MI spindle assembly are shown by maximum z‐projection images from confocal time‐lapse imaging of oocytes obtained from transgenic histone H2B‐EGFP mice stained with SiR‐tubulin. White circles indicate oocyte edges; scale bars, 10 μm. All observed phenotype categories after IPZ treatment are shown in Appendix Fig S4A.
-
B, CLive‐cell imaging of oocytes as in (A) was used to quantify the effects of IPZ on GVBD (B) and anaphase I entry (C). Anaphase I was defined as the first time point when chromosome segregation was visible. Fisher's exact test; oocyte numbers are indicated in brackets. The full time course of GVBD kinetics is in Appendix Fig S4B.
-
DFrequency and severity of MI spindle disruption induced by increasing doses of IPZ, as observed by live‐cell imaging in (A–C). Oocyte numbers are shown at the top of the bars.
-
EMI spindle formation in oocytes expressing CDK5RAP2‐EGFP, histone H2B‐mCherry and stained with SiR‐tubulin matured in the presence of 15 μM IPZ. Representative still images of maximum z‐projections are shown. Time in hh:mm is relative to GVBD. Scale bar, 10 μm.
-
FMT intensity at MTOCs at the time of GVBD quantified from SiR‐tubulin signal that was normalized to mean cytoplasmic SiR‐tubulin signal. Mean of MT intensity on MTOCs was arbitrarily centered to 1 in the control group (means ± SDs, Mann–Whitney test). Total of 184 MTOC in 17 control oocytes and a total of 170 MTOCs in 17 IPZ‐treated oocytes were analyzed.
-
GTime course of the MT intensity in the area of spindle after GVBD. Mean of MT intensity was arbitrarily centered to 1 in the control group (means ± SDs). Oocyte numbers are indicated in brackets.
-
HOocytes expressing CDK5RAP2‐EGFP, histone H2B‐mCherry and stained with SiR‐tubulin were treated with 15 μM IPZ after being cultured for 5 h in control media. Maximum intensity z‐projections from confocal live‐cell imaging are shown. Time in hh:mm is relative to GVBD, and DMSO or 15 μM IPZ was added at time 5:00. Scale bars, 10 μm.
-
ISpindle volume at 1 h and metaphase I after IPZ addition. The metaphase time was defined as the last time point before anaphase or, in the IPZ‐treated oocytes that failed anaphase entry, as the average control metaphase time. Means ± SDs, Mann–Whitney test; oocyte numbers are indicated in brackets.
-
JTime of anaphase I entry in oocytes that were treated with 15 μM IPZ after being cultured for 5 h in control media. Oocyte numbers are indicated in brackets.
To examine in more detail how the inhibition of RanGTP signaling by IPZ affects MI spindle assembly, we performed 3D three‐color live imaging (Balboula et al, 2016; Courtois et al, 2018) to simultaneously visualize spindle (SiR‐tubulin), chromosomes (histone H2B‐mCherry), and CDK5RAP2‐EGFP, which co‐localizes with MTOCs, in oocytes treated with 15 μM IPZ or DMSO for control (Fig 4E, Movie EV2). In control oocytes, MTs polymerized around MTOCs already at the time of GVBD (Fig 4F), and about 10 min later, an increase in MT mass occurred in the area defined by condensed chromosomes and perinuclear MTOCs (Fig 4G). Later, multipolar spindles formed an MT ball. Finally, clusters of MTOCs appeared to guide the spindle bipolarization (Fig 4E). Compared to controls, IPZ treatment significantly reduced the MT polymerization at MTOCs (Fig 4F). Although the MT mass increased to a significantly smaller degree in the presence of 15 μM IPZ (Fig 4G), the assembly of multipolar spindle succeeded (Fig 4E). However, instead of subsequent spindle bipolarization, spindles eventually disassembled in all 15 μM IPZ‐treated oocytes, and the perinuclear MTOCs aggregated.
To investigate whether IPZ could affect the already assembled MI spindles, we treated oocytes with 15 μM IPZ starting at 5 h after meiosis resumption, i.e., at the time when bipolar spindles assembled in most control oocytes. The live‐cell imaging showed that all control DMSO‐treated oocytes retained bipolar spindle and entered anaphase I (Fig 4H, Movie EV3). In contrast, the IPZ‐treated oocytes retained significantly smaller spindles (Fig 4I) with unaligned chromosomes (Fig 4H), and only 37% initiated delayed chromosome segregation with small spindles (Fig 4J). Collectively, these data suggest that the RanGTP‐dependent suppression of importin β is essential for the formation, maintenance, and function of the MI spindles in mouse oocytes.
RanGTP but not TPX2 partially rescues the inhibition of MI spindle assembly and anaphase by IPZ
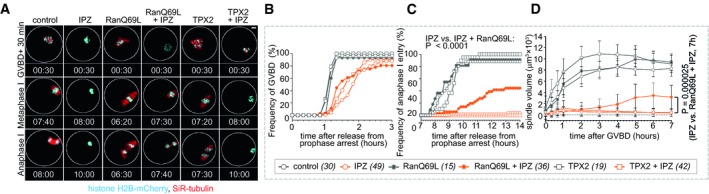
Next, we wished to verify that IPZ disrupts spindle formation through perturbation of RanGTP signaling in oocytes. We tested the effect of IPZ treatment on spindle formation visualized with SiR‐tubulin staining in live oocytes co‐microinjected with histone H2B‐mCherry and the mRNA for the GTP‐locked RanQ69L (Fig 5A, Movie EV4). Although RanQ69L expression induced extended bipolar spindle morphology, the oocytes entered anaphase I entry without delay (Fig 5C, Movie EV4) as previously reported (Dumont et al, 2007). In IPZ‐treated oocytes, the expression of RanQ69L failed to rescue the delayed resumption of meiosis (Fig 5B), consistent with the inhibitory effects of RanQ69L on the nuclear protein import (Palacios et al, 1996) and the requirement for Ran‐regulated nuclear‐cytoplasmic transport during GVBD (Kalab et al, 2011). The RanQ69L overexpression also failed to rescue the assembly of the MT ball‐like structures immediately after GVBD (Fig 5A and D, Movie EV4). However, about 60% of RanQ69L‐expressing and IPZ‐treated oocytes resumed spindle assembly at later stages of meiosis (Fig 5A, Movie EV4), as demonstrated by the significant increase in spindle volume (Fig 5D). Although these partially rescued spindles were significantly smaller than in control, about 40% of the RanQ69L expressing oocytes entered anaphase I in the presence of 15 μM IPZ, in contrast with only 4% of the IPZ‐only treated oocytes (Fig 5C).
Figure 5. RanQ69L partially rescues the IPZ‐induced MI spindle disassembly.
-
AThe rescue of the effects of 15 μM IPZ on MI spindles was examined in mouse oocytes expressing histone H2B‐mCherry from microinjected mRNA, co‐stained with SiR‐tubulin, and noninjected or injected with mRNAs coding for RanQ69L or hTPX2. Maximum intensity z‐stack projections from confocal live‐cell movies 30 min after GVBD, at metaphase I (defined as in Fig 4I) and anaphase I. Time in hh: mm is relative to GVBD. Scale bars, 10 μm.
-
B, CThe frequency of GVBD (B) and anaphase I entry (C) quantified by live confocal imaging of oocytes stained and treated as in (A). Fisher's exact test; oocyte numbers are shown in brackets.
-
DChanges in spindle volume over time after GVBD. Means ± SDs, mixed‐design ANOVA; oocyte numbers are shown in brackets.
Since RanGTP and importin β regulate the TPX2‐dependent MT nucleation and spindle assembly (Gruss et al, 2001; Petry et al, 2013; Zhang et al, 2017), we tested if TPX2 overexpression could restore the spindle assembly in IPZ‐treated oocytes. Interestingly, and in contrast to RanQ69L, overexpression of TPX2 induced significantly increased MT assembly in the IPZ‐treated oocytes in a time window of 0–60 min after GVBD (Fig 5A and D; Appendix Fig S5). However, the initial MT accumulation subsequently reversed (Fig 5D, Appendix Fig S5), and TPX2‐overexpressing and IPZ‐treated oocytes never entered anaphase I (Fig 5C).
We cannot exclude that the only transient effect for TPX2 on spindle recovery or the lack of RanQ69L effect immediately after GVBD resulted from insufficient overexpression levels. On the other hand, the RanQ69L‐induced partial rescue of anaphase I in IPZ‐treated oocytes strongly supports the notion that IPZ disrupts MI spindle assembly by acting as a specific inhibitor of RanGTP function.
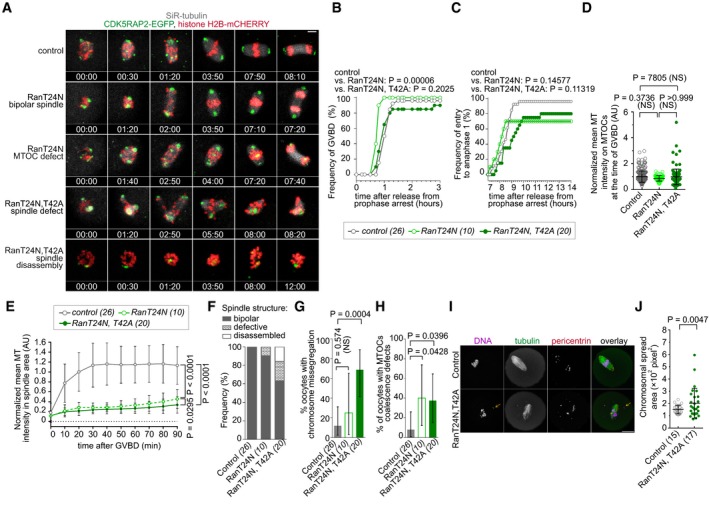
RanT24N, T42A but not RanT24N disrupts anaphase I chromosome segregation
Since biochemical studies and FRET/FLIM imaging indicated that RanT24N, T42A acts as a more specific inhibitor of RanGTP signaling than RanT24N, we expected that RanT24N, T42A would also induce a more profound spindle disruption than RanT24N. To test this prediction, we microinjected oocytes with these mutants and monitored meiosis progression by the 3D three‐color live imaging to visualize chromosomes, microtubules, and CDK5RAP2‐labeled MTOCs (Fig 6A and Movie EV5) as described above. We observed that the expression of RanT24N but not RanT24N, T42A significantly accelerated the resumption of meiosis (Fig 6B). While neither of the Ran mutants had a marked effect on the timing of anaphase I entry, they both reduced the frequency of anaphase entry and polar body extrusion (Fig 6C). Although the Ran mutants did not perturb MT polymerization on MTOC at the time of GVBD (Fig 6D), they both strongly suppressed the rise in MT polymerization around chromosomes and MTOCs within the first 90 min after GVBD, whereby the effect of RanT24N, T42A was more pronounced (mixed ANOVA, P = 0.0295; Fig 6E). While control oocytes formed bipolar spindles, and RanT24N did not induce gross spindle disruptions, RanT24N, T42A increased the frequency of defective spindles, including spindle disassembly (Fig 6A and F; Movie EV5) and chromosome segregation errors (Fig 6G). Both mutants increased the frequency of oocytes with the defective coalescence of MTOCs into MI spindle poles (Fig 6H). In an agreement with increased chromosome segregation errors in RanT24, T42A expressing oocytes observed in movies (Fig 6A, F and G), we found increased chromosomal misalignment in RanT24, T42A expressing oocytes by IF staining in metaphase I (Fig 6I and J). In summary, these data show that while RanT24N perturbs some aspects of the MI spindle assembly the bipolar and functional spindle is eventually formed and correct chromosome segregation ensues, likely owing to the sufficient overall levels of SAFs released from importin β. However, since RanT24N, T42A both disrupt the chromosomal cargo gradients and enable SAF sequestration by importin β, it compromises the formation of the functional bipolar MI spindle and its function in chromosome segregation in anaphase I.
Figure 6. RanT24N, T42A but not RanT24N disrupts MI spindle function in segregating anaphase I chromosomes.
-
AThe effects of RanT24N, T42A, and RanT24N on MI spindle assembly observed in maximum z‐projection images from confocal time‐lapse imaging of oocytes expressing CDK5RAP2‐EGFP, histone H2B‐mCherry and stained with SiR‐tubulin. Time in hh:mm is relative to GVBD. Scale bar, 10 μm.
-
B, CLive‐cell imaging of oocytes as in (A) was used to quantify the effects of Ran mutant. Means ± SDs, Mann–Whitney test; oocyte numbers are indicated in brackets.
-
DMT intensity at MTOCs at the time of GVBD quantified from SiR‐tubulin signal that was normalized to mean cytoplasmic SiR‐tubulin signal. Mean of MT intensity on MTOCs was arbitrarily centered to 1 in the control group. Means ± SDs, Kruskal–Wallis, and Dunn's tests. 25 control oocytes with 264 MTOCs in total, 10 RanT24N‐expression oocytes with 70 MTOCs in total and 19 RanT24N, T42A‐expressing oocytes with 96 MTOCs in total were analyzed.
-
ETime course of the MT intensity in the area of spindle after GVBD. Mean of MT intensity was arbitrarily centered to 1 in the control group. Means ± SDs, mixed‐design ANOVA; oocyte numbers are shown in brackets.
-
FFrequency and severity of MI spindle phenotypes in the oocytes expressing RanT24N or RanT24N, T42A. Oocyte numbers are shown in brackets.
-
G, HQuantification of the percentage of oocytes (G) with chromosome segregation errors in anaphase I and (H) with defective MTOC coalescence during metaphase I. Means and 95% exact confidence interval; oocyte numbers are shown in brackets.
-
IDetection of MTs and pericentrin‐containing MTOCs by IF in control and RanT24N, T42A‐expressing oocytes. Arrow indicates unaligned chromosome.
-
JQuantification of chromosomal spread area in oocytes depicted in (I). Means ± SDs, Mann–Whitney test; oocyte numbers indicated in brackets.
RanGTP and importin β regulate MT assembly and the coalescence of MTOCs into MI spindle poles
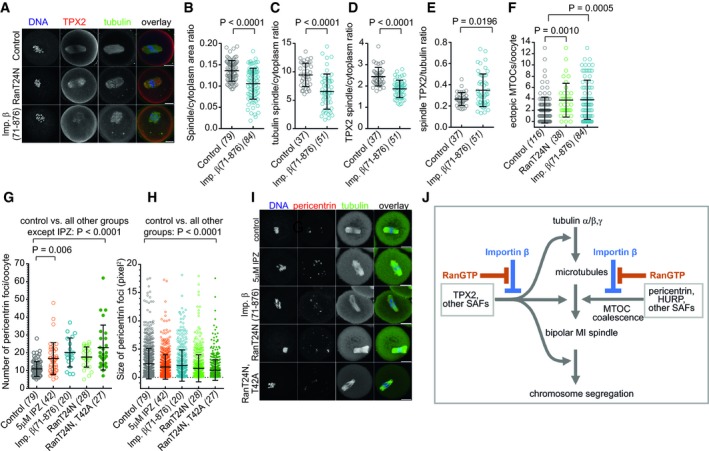
To examine closer how RanGTP and importin β function in MI spindle formation, we followed by analyzing confocal immunofluorescence (IF) images in cells fixed and stained after the injection of importin β(71–876) protein (Fig 7A). The importin β(71–876) significantly reduced the spindle size (Fig 7B), induced multipolar spindles (Fig 7A, Appendix Fig S6B and C), and sharply reduced the spindle‐associated tubulin and TPX2 signals (Fig 7C and D), indicating an overall decrease in spindle MT stability. Interestingly, the importin β(71–876)‐injected oocytes lost more tubulin than TPX2 from their spindles (the ratio of tubulin/TPX2 dropped from 77 to 69%; Fig 7E), suggesting that the MT‐bound TPX2 is less accessible to importin β, which could help to stabilize the already formed spindle MTs. Consistent with previous reports, and in contrast to importin β(71–876), RanT24N caused relatively mild defects in spindle morphology (Bury et al, 2017), including increased spindle width (Appendix Fig S6A). However, in addition to the mild effect on spindle MTs, we noticed that RanT24N markedly increased the number of ectopic MT asters, similar to importin β(71–876) (Fig 7F).
Figure 7. Ran and importin β regulate MT assembly and MTOC coalescence in MI spindles.
-
AMaximum intensity z‐stack projections of confocal IF images (DNA stain, indirect IF for tubulin, and TPX2) in untreated oocytes (control), oocytes expressing RanT24N or injected with importin β(71–876). The representative of at least five experiments is shown.
-
B–DMaximum intensity z‐stack projections of IF images in control and importin β(71–876)‐injected cells were used to quantify the spindle versus whole‐cell area fraction (B) and the ratios of the average signal in the spindle versus cytoplasm for the tubulin (C) and TPX2 (D) staining. Means ± SDs, with t‐test in (B, C) and Mann–Whitney test in (D); oocyte numbers indicated in brackets.
-
ERatios of the spindle‐associated fractions of TPX2 versus tubulin were calculated in maximum intensity z‐stack projections of IF images. Means ± SDs, Mann–Whitney test; oocyte numbers indicated in brackets.
-
FMaximum intensity z‐stack projections of IF images were used to quantify the number of ectopic MT asters. Means ± SDs, ANOVA with Kruskal–Wallis and Dunn's tests; oocyte numbers indicated in brackets.
-
G, HThe number and size of pericentrin foci were quantified in maximum intensity z‐stack projections of IF images stained as in (I) in at least two separate experiments. Means ± SDs, ANOVA with Kruskal–Wallis and Dunn's tests.
-
IMaximum intensity z‐stack projections of confocal IF images stained for DNA, pericentrin, and tubulin (directly labeled primary antibody). A representative example of at least two experiments is shown. Scale bar, 20 μm.
-
JProposed model: The suppression of importin β by RanGTP is required for MT nucleation and polymerization, enabling the assembly and function of the MI spindles. Through separate mechanisms, RanGTP opposes the role of importin β in preventing the coalescence of pericentrin‐containing MTOCs into spindle poles.
We hypothesized that the rise in ectopic MTOCs could result from excessive importin β binding to SAF(s) that are supporting MTOC coalescence during the MI spindle assembly. To test the idea, we used IF staining to analyze the effects of Ran and importin β perturbation on pericentrin, the core scaffolding constituent of MTOCs (Delaval & Doxsey, 2010). The expression of RanT24N or RanT24N, T42A or injection of importin β(71–876) protein significantly increased the number of MTOCs visualized as pericentrin foci throughout the cells (Fig 7G), while at the same time reducing their average size (Fig 7H). To inhibit the RanGTP generation in the oocytes without overexpressing Ran mutants, we adapted the “Trim‐Away” method (Clift et al, 2017) to induce the antibody‐mediated knockdown of endogenous RCC1 in the MI oocytes. Although we could verify only partial RCC1 depletion, the treatment induced a significant increase in the number of ectopic MTOCs (Appendix Fig S7). Finally, since higher than ~10 μM IPZ disrupted spindle MTs, we also used short treatment (< 1 h) with 5 μM IPZ to examine the MT disruption‐independent effects of IPZ on MTOCs. Again, we observed the same overall effect: increased number and the reduced size of MTOCs, indicating their reduced cohesiveness (Fig 7G–I). In summary, our results indicate that, in addition to the critical role of the RanGTP–importin β interaction in activating MI spindle MT nucleation and assembly, the same pathway regulates MI spindle pole formation by targeting SAFs involved in the coalescence of pericentrin‐containing MTOCs at later stages of MI (Fig 7J).
Discussion
We can summarize the main conclusions of this study on the function of Ran in mouse MI oocytes spindle assembly in four points. First, the measurements with FRET biosensors showed that MI chromosomes signal to the meiotic cytoplasm through a short‐ranging spatial gradient of RanGTP and a more extended downstream gradient of importin β cargos. These quantitative live‐cell data could aid mathematical modeling of the spatial regulation by Ran in oocytes. The FRET studies also revealed the unexpected effects of RanT24N on importin β cargo release, indicating that RanT24N does not act solely as RCC1 inhibitor. Second, we designed and tested RanT24N, T42A double mutant, which inhibits the RanGTP‐regulated chromosomal gradients and lacks importin β‐binding activity. We show that the expression of RanT24N, T42A disrupts the MI spindle assembly and anaphase I chromosome segregation, demonstrating the essential role of RanGTP in mouse oocytes. Third, we show that while mouse oocyte MI spindles fail to assemble or remain stable when treated with IPZ, RanQ69L expression partially rescues MI spindle assembly. These results show that the assembly and function of mouse oocyte MI spindles continuously require RanGTP‐dependent negative regulation of importin β, although they do not depend on the distribution of RanGTP in the form of spatial gradients. Fourth, we reveal that in addition to its essential role in MI spindle MT assembly, the RanGTP–importin β pathway contributes to MI spindle function by controlling the coalescence of MTOCs into spindle poles.
Mechanisms of the chromosomal RanGTP and importin β cargo gradients
The importin β cargo gradients were previously detected in maturing mouse oocytes with the use of wide‐field epifluorescence FRET ratio imaging of the Rango‐2 cargo sensor (Dumont et al, 2007). Because the Rango‐2 contains the same importin β‐binding domain as Rango‐3, although the donor–acceptor proteins differ, the results obtained with both should be similar. The confocal FLIM imaging applied here enabled quantification of the FRET signal, and the detection of the exponential decay profile of the gradients (Figs 1G and 3G, Appendix Fig S1), which is expected from mathematical models of the meiotic Ran system and was not detected in the previous study (Dumont et al, 2007). Since the RBP‐3 sensor domain is homologous to the RanGTP domain in RanBP1, the spatial gradients detected by RBP‐3 should mimic the steep decline in RanGTP/RanBP1 complexes that was predicted by mathematical models of Ran signaling in mitotic cytoplasm (Caudron et al, 2005). The significantly steeper decline of the RBP‐3 gradients, compared to the importin β cargo gradients detected by Rango‐3 (Appendix Fig S1), is in line with the model, supporting its validity in mouse oocytes. Further refinement of the model could be achieved by measuring the concentration of Ran system components expressed in the oocytes. The good fit of the FRET signal profiles with mono‐exponential decay pattern (Figs 1G and 3G) also indicates that the signal of both sensors is dominated by binding to a single ligand (importin β for Rango‐3 and RanGTP for RBP‐3), supporting their specificity.
Among the various perturbations of the gradients that we applied, the effects of importin β(71–876) injection were arguably the best aligned with the previously known or expected mechanisms of action. Since the N‐terminal deletion abrogates the importin β binding to RanGTP (Nachury et al, 2001), importin β(71–876) drastically lowered free cargos across the cells (Fig 1B, C and E). Because only a minimal effect on the RanGTP gradient was induced by importin β(71–876), as expected (Fig 3B, C and E), a small residual‐free cargo gradient was still detectable in line scans, indicating that a fraction of the Rango‐3 sensor bound to the endogenous importin β, and was released by the chromosomal RanGTP gradient (Fig 1C).
The role of Ran and importin β in the resumption of meiosis
The strong inhibition of GVBD by > 10 μM IPZ (Fig 4B) hinted that Ran and importin β could be involved in the regulation of meiosis resumption. Although IPZ perturbed RCC1 binding to chromosomes in MI oocytes (Fig 3H and I, Appendix Fig S3B), we have not investigated if IPZ similarly disrupts RCC1 binding to chromosomes inside the GVs, and do not presume that such an effect would be needed to explain how IPZ blocks GVBD. The more likely scenario is that by acting as the specific inhibitor RanGTP‐ and importin β‐dependent nuclear import (Soderholm et al, 2011), IPZ prevented the entry of cell cycle regulator(s) into the GV, blocking the activation of MPF (Kalab et al, 2011). Since the GTP hydrolysis‐resistant RanQ69L inhibits nuclear import (Dickmanns et al, 1996), it is not surprising that it slightly, but not significantly worsened rather than rescued the GVBD resumption (IPZ versus RanQ69L + IPZ: P = 0.4501; Fisher's exact test; Fig 5B). Of note, we performed the experiments involving the rescue of the IPZ effects on MI spindle assembly in oocytes injected with relatively low levels of microinjected RanQ69L mRNA (200 ng/μl) to avoid major disruptions of cell cycle and MI spindles. Future studies will be needed to elucidate the mechanisms responsible for the unexpected, and potentially opposing effects of the RanT24N versus RanT24N, T42A on GVBD (Fig 6B), as well as the identity of the RanGTP and importin β‐regulated cargos involved in GVBD activation.
The mechanisms of RanT24N effects in oocytes
The flattening of both the Rango‐3 and RBP‐3 gradients induced by RanT24N (Figs 1B–D, 2, 3B–D, Appendix Figs S2 and S3B–D) is consistent with the inhibition of the RCC1‐dependent GTP loading on Ran at chromatin. However, at high concentration of the injected RanT24N mRNA (2,500 ng/μl, Figs 1B–D and 3B–D) we detected small but significant changes in the averages of both Rango‐3 and RBP‐3 signals in the opposite direction than expected for a specific RCC1 inhibitor: increase in average free importin β cargos and RanGTP (Figs 1D and 3D). The results of our biochemical pulldowns (Fig 1H) then indicated that these unexpected behaviors of RanT24N could result from its previously documented (Hughes et al, 1998) but so far not thoroughly investigated or understood interactions with importin β.
Although our results indicated that RanT24N could directly interact with importin β in the oocytes, even at the highest concentrations that we tested (2,500 ng/μl injected mRNA), it induced only a relatively small increase in average free importin β cargos (Fig 1B–D) and much smaller increase in RanGTP, compared to the injection of RanQ69L (Appendix Fig S3A). Rather than causing the rapid release of cargos from importin β, similar to RanGTP, it is then more likely that RanT24N competes with both cargo and RanGTP binding to importin β in the oocytes. Because RanT24N binds RCC1 strongly, these effects of RanT24N on importin β start to emerge at RanT24N expression levels are exceeding certain critical threshold, which is likely close to the endogenous RCC1 concentration. Although its concentration in mouse oocytes is unknown, RCC1 is usually a much less abundant protein than Ran or importin β, and its concentration is approximately only 0.24 μM in the meiotic X. laevis egg extracts (Caudron et al, 2005). In comparison, the cytoplasmic concentration of the RanT24N protein injected to mouse oocytes in one of the previous studies was approximately 13 μM (about 50 times over the X. laevis RCC1 level), based on the reported injection conditions (Schuh & Ellenberg, 2007). Interestingly, the robust disruption of the MI spindle assembly by RanT24N in human oocytes (Holubcova et al, 2015) was observed upon injection of nearly four times less concentrated RanT24N protein (75 μM versus 286 μM), suggesting that the potentially lower off‐target effects of RanT24N could have contributed to the much stronger phenotypes.
As indicated by the co‐precipitation of both importin α and importin β with RanT24N in vitro (Fig 1H), the mechanism of RanT24N cellular effects could involve unusual complexes of importin α/β with RanT24N that could potentially interfere with the access of specific types of cargos to importin β. The discrepancy between the moderate effects of RanT24N on spindle MTs (Fig 6A and F; Dumont et al, 2007; Bury et al, 2017), versus strong effect on MTOC dynamics (Figs 6H and 7F–I) possibly reflects different effects of RanT24N on the SAFs that are inhibited by the direct versus indirect (importin α‐mediated) binding to importin β. It is noteworthy that MT nucleation depends on TPX2, an indirect importin β cargo (Gruss et al, 2001; Neumayer et al, 2014), while the MTOC dynamics involves at least two direct importin β cargos: HURP (Sillje et al, 2006; Breuer et al, 2010) and pericentrin (Liu et al, 2010). Our analysis of TPX2 association with MI spindles (Fig 7E) also indicates that the TPX2‐coated MTs are resistant to disassembly by importin β, which could contribute to the low sensitivity of MI spindle MTs to the overexpressed RanT24N.
Overall, our analyses indicated that, in addition to abrogating the RCC1‐dependent chromosomal gradients, RanT24N directly perturbs importin β cargo binding in a manner that could counteract the RCC1 inhibition.
Inhibition of both local and global importin β cargo release by RanT24N, T42A supports the critical role of RanGTP in MI spindle function
We reasoned that we could alleviate the pleiotropic effects of RanT24N on Ran and importin β by mutating the T42 site in Ran that appeared to be essential for the WT Ran‐importin β binding (Murphy et al, 1997). Indeed, we found that, while the injection of increasing concentrations of RanT24N mRNA induced a gradual increase in importin β‐free cargos (Fig 2A), RanT24N, T42A induced a significant overall decrease in importin β‐free cargos across a wide range of microinjected mRNA concentrations (Fig 2, Appendix Fig S2). Although future studies will be needed to characterize its interactions fully, these data indicated that RanT24N, T42A probably mostly lacks the unwanted direct effects on importin β and acts as a more specific inhibitor of RanGTP signaling than RanT24N.
Eager to test this notion, we examined the effects of RanT24N, T42A on MI spindle assembly and meiosis progression by both live‐cell imaging and fixed cell analyses, and compared with the effects of RanT24N (Figs 6 and 7). In agreement with previous reports (Schuh & Ellenberg, 2007), we observed that RanT24N delayed polymerization of MTs within the first 90 min after GVBD (Fig 6E) and increased the MI spindle width (Bury et al, 2017) (Appendix Fig S6A). We also noticed strong effects of RanT24N, T42A on the rise of ectopic MT asters (Fig 7F) and defects in the coalescence of MTOCs into the spindle poles (Figs 6H and 7F–I). The severity and frequency of spindle defects induced by RanT24N, T42A were more pronounced compared to RanT24N (Fig 6A and F). Importantly, live‐cell imaging showed that RanT24N, T42A but not RanT24A induced defective anaphase I chromosome segregation (Movie EV5, Fig 6A and G). By performing IF analyses in MI oocytes fixed and stained after the treatment with the RanT24N, T42A mRNA or buffer, we confirmed defects in chromosome congression in MI spindles (Fig 6I and J). This result also indicates that the RanT24N, T42A‐induced MI defects observed in movies (Movie EV5, Fig 6A) did not stem from sensitizing the oocytes by the application of fluorescent biomarkers and live‐cell imaging. In conclusion, these results show that the mouse MI spindles require RanGTP to accomplish the anaphase I‐mediated reductional diploid chromosome segregation (Fig 6A and F–J), similar to human oocytes (Holubcova et al, 2015). As discussed in the next section, we obtained supporting evidence for the critical role of the global RanGTP‐dependent importin β cargo release in MI spindle function from the analyses of the effects of IPZ on MI oocytes.
The mechanisms of the MI spindle assembly disruption by IPZ, and its partial rescue by RanGTP
The degradation of the RanGTP gradient induced in MI oocyte by the IPZ treatment (Fig 3B, C and F) was unanticipated. The hyper‐condensed appearance of the MI chromosomes (Appendix Fig S3B) and reduced dynamics of RCC1‐mCherry binding to chromosomes (Fig 3H and I) both suggested an intriguing possibility that IPZ blocked a RanGTP‐ and importin β‐dependent regulation of one or more proteins involved in chromosome condensation. One potential mechanism could involve the role of the importin β complex with importin 7 that functions as the chaperone for histone H1 (Jakel et al, 2002), which in turn is required for the organization of meiotic chromosomes in the X. laevis meiotic egg extracts (Freedman et al, 2010). To determine which of the numerous activities controlling chromosome condensation are involved will require future investigations. Because treatments with similar or higher doses of IPZ caused only a partial reduction in the mitotic cargo gradients in HeLa cells (Soderholm et al, 2011), the putative IPZ target(s) in chromosome condensation could be meiotic oocyte‐specific.
While the FRET data showed that the treatment with 20 μM IPZ abolished the RanGTP gradient, the average cell RanGTP concentration remained unaffected (Fig 3F), indicating that RCC1 remained active. Intriguingly, the FLIM images of the IPZ‐treated cells showed a broad zone of increased RanGTP levels that spread > 20 μm away from chromosomes (Fig 3B and C), suggesting that the release of free RanGTP remained only loosely confined to chromosomes. These results suggest that, as a consequence of a slowed‐down binding to and release from the chromosomes, the RCC1‐dependent GDP/GTP exchange on Ran continued in a broader area of cytoplasm surrounding the chromosomes. In the future, it will be essential to clarify how this observation relates to the clear evidence that the phosphorylation‐regulated RanBP1 binding inhibits the ectopic RCC1 activity in mitotic cytoplasm (Zhang et al, 2014).
The IPZ‐induced changes in MI chromosome architecture also suggest that other chromosome‐centered activities involved in MI spindle assembly could have been affected, including the phosphorylation signaling by chromosome passenger complex (CPC)‐associated Aurora B and C kinases (Nguyen et al, 2018). The potential disruption of such pathways could have amplified the effects of IPZ on MI spindles. However, together with the previous evidence documenting the IPZ specificity toward RanGTP–importin β interaction (Soderholm et al, 2011), the partial but significant rescue of the IPZ effect on MI spindle assembly and anaphase by RanQ69L (Fig 5) strongly argues that the RanGTP–importin β complex is the specific target of IPZ also in the oocytes. While previous studies also showed that, in vitro, IPZ directly binds to importin β and not to RanQ69L, transportin 1, and exportin 1 (Soderholm et al, 2011), it remains unknown exactly how the IPZ binding to importin β perturbs its interaction with RanGTP. The lack of detectable changes in the overall Rango‐3 signal indicates that rather than completely blocking RanGTP binding to importin β, IPZ possibly acts by destabilizing or altering the RanGTP–importin β complex in a manner that also precludes the stable release of the importin β cargos.
While only partial, the rescue of the bipolar spindles by RanQ69L also confirmed the previous observation that the mouse MI spindle formation and function do not require RanGTP in the form of the gradient (Dumont et al, 2007). Our results are consistent with the previous conclusion that RanGTP gradient accelerates the initial rise in MT nucleation at chromosomes (Schuh & Ellenberg, 2007), which could be particularly important in human oocytes that do not have pericentrin‐containing MTOCs (Holubcova et al, 2015). In mitotic somatic cells, the spindle assembly is supported by positive feedback between the RanGTP gradients and the MT‐dependent accumulation of active SAFs within the spindle (Oh et al, 2016). Such a mechanism could help to explain why the spatial gradients of SAF activities are not necessary for bipolar MI spindle assembly in meiotic oocytes as well.
Regulation of MTOC dynamics by Ran and importin β
Although centrosomes are missing in the mouse meiotic oocytes, many of the hierarchically organized pericentrosomal constituents are present in the MTOCs and have essential roles in meiotic spindles. These include pericentrin (Carabatsos et al, 2000), γ‐tubulin (Combelles & Albertini, 2001), its interacting partner CDK5RAP2 (also known as Cep215) (Balboula et al, 2016; Baumann et al, 2017), the NEDD1 protein that is required for the recruitment of the MT‐nucleating γ‐tubulin ring complex (γ‐TURC) (Ma et al, 2010), and several kinases with essential roles in MI spindle assembly and MTOC biogenesis (Saskova et al, 2008; Solc et al, 2012, 2015; Bury et al, 2017), in particular the Haspin‐dependent activation of Aurora kinase C (Balboula et al, 2016). As shown by the consequences of pericentrin depletion in transgenic mice (Baumann et al, 2017), pericentrin has a crucial role as a scaffolding protein that stands at the top of the hierarchy of the MTOC assembly. Although the mechanisms of the recombinant constructs, Trim‐Away method or IPZ that we applied in this study differed, they all resulted in the increased number of pericentrin foci dispersed in metaphase I oocytes (Fig 7G, Appendix Fig S7). Since pericentrin functions as a central scaffold which brings together many other MTOC constituents, it remains possible that the correct MTOC function could depend on the precise balance of RanGTP‐regulated local (chromosome‐centered) and global (average across oocytes) activities over time. Disturbances of the balance by different mechanisms then could cause problems in MTOC coalescence.
We summarize that, in agreement with the observations in previous studies (Dumont et al, 2007; Schuh & Ellenberg, 2007; Bury et al, 2017), we show here that RanGTP in the shape of the spatial chromosomal gradients is largely dispensable for at least some aspects of the bipolar MI spindle assembly and anaphase I chromosome segregation in mouse oocytes. However, the present analyses of various perturbations in the same system indicate that the conclusions of studies employing RanT24N as specific RCC1 inhibitor could have been skewed by the counteracting effects of RanT24N on importin β. Our results suggest that the MI spindle assembly and function continuously require RanGTP, which enables the activation of importin β‐regulated SAFs throughout the oocytes. Advances in live‐cell imaging methods, including multi‐color time‐lapse imaging (Courtois et al, 2018) and the quantitative FLIM imaging with optimized FRET biosensors, were important for clarifying the role of RanGTP in mouse MI spindle function.
While there are significant differences between the mouse and human MI oocyte spindles, including their different sizes, composition, and temporal and spatial dynamics of assembly (Holubcova et al, 2015), our results indicate that the dependence of MI spindles on RanGTP is common to both species, supporting the relevance of the mouse model to the mechanisms of human oocyte meiosis. Although species‐specific adaptations likely exist, the high evolutionary conservation of the Ran pathway and importin β predicts that the RanGTP‐ and importin β‐regulated mechanisms are essential for the MI spindle function in other mammalian species.
Materials and Methods
Mouse oocyte preparation and culture
Animals were treated and used according to the guidelines of the Institutional Animal Care and Use Committee of Johns Hopkins University School of Medicine or according to the policies of the Expert Committee for the Approval of Projects of Experiments on Animals of the Academy of Sciences of the Czech Republic. In all experiments, GV oocytes were collected at room temperature from 8‐ to 16‐week‐old female mice 44–48 h after injection with pregnant mare serum gonadotropin (5 IU; Sigma). Live‐cell FLIM imaging in Figs 1, 2, 3 and the IF analyses in fixed cells in Figs 6 and 7 were done with oocytes from the CF1 mice. Oocytes from the CD1 or histone H2B‐EGFP transgenic mice with CD1 genetic background (Hadjantonakis & Papaioannou, 2004; Mayer et al, 2016) were used for live‐cell imaging in Figs 4, 5, 6. The CF1 oocytes were kept in MEM‐PVP containing IMBX (Stein & Schindler, 2011) during collection and microinjection and incubated in the IBMX‐free M16 media (Sigma‐Aldrich). The CD1 and histone H2B‐EGFP transgenic oocytes were collected in M2 media containing 2.5 mM milrinone (Sigma‐Aldrich) and cultured in MEM medium (Sigma‐Aldrich) supplemented with 0.22 mM sodium pyruvate (Sigma‐Aldrich), 4 mg/ml BSA (Sigma‐Aldrich), and penicillin–streptomycin (100 U/ml: 100 mg/ml, Sigma‐Aldrich). The cultures and live imaging were performed at 37°C in a humidified atmosphere with 5% CO2 under paraffin oil. The paraffin oil cover was omitted in all cultures involving the treatment with IPZ.
DNA and tubulin dyes
The GV oocytes were released into the M16 media containing 200 nM SiR700‐DNA (Cytoskeleton) to enable visualization of chromosomes during FLIM measurements. Oocytes were stained with 100 nM SiR‐tubulin (Spirochrome) for MT visualization. SiR‐tubulin was added to both M2 and MEM medium.
Time‐lapse confocal imaging
Time‐lapse image acquisitions in Figs 4A and 5A were performed using Leica TCS SP5 with an HCX PL Apo Lambda Blue 40 × 1.25 oil objective with 1× zoom, and 12 confocal 7.5‐μm optical sections were taken with a 1,024 × 1,024 pixel image resolution using 10‐min time intervals as described previously (Mayer et al, 2016). For MTOC imaging (Figs 4E and H, and 6A) in oocytes expressing mEGFP‐mCDK5RAP2, histone H2B‐mCHERRY from microinjected mRNAs and stained with SiR‐tubulin, we used the tracking function of the Matrix Screen module in LAS AF software (Leica Microsystems). 16 confocal 2.5‐μm optical sections with a 7.75× zoom on the area of the chromosomes of individual oocytes in 256 × 256 pixel image resolution by 1,000 Hz speed bi‐directional scan were acquired as described previously (Balboula et al, 2016).
Plasmids
The plasmids used in this study are listed in Table 1. We used previously described plasmids for the in vitro transcription of histone H2B‐mCherry, mEGFP‐CDK5RAP2, YFP‐hTPX2, RanQ69L, and RanT24N (Dumont et al, 2007; Brunet et al, 2008; Kitajima et al, 2011; Balboula et al, 2016) and plasmids for the production of the recombinant importin β(71–876) proteins in E. coli (Nachury et al, 2001). The pEGFP‐N1‐VAPA (1–242) was a gift from Axel Brunger (Addgene plasmid # 18874), and the pET His6 Sumo TEV LIC cloning vector (1S) was a gift from Scott Gradia (Addgene plasmid # 29659). Plasmids for YFP‐hTPX2, RanQ69L, and RanT24N were kindly provided by Marie‐Hélène Verlhac.
Table 1.
Recombinant DNA constructs
| DNA construct name | Reference or source |
|---|---|
| pRN3 RanT24N | Dumont et al (2007) |
| pRN3 RanT24N, T42A (pK1005) | This paper |
| pRN3 RanQ69L | Dumont et al (2007) |
| Histone H2B‐mCherry | Kitajima et al (2011) |
| EGFP‐CDK5RAP2 | Balboula et al (2016) |
| YFP‐hTPX2 | Brunet et al (2008) |
| pGEMHE | Liman et al (1992) |
| pGEMHE2 (pK738) | This paper |
| pGEMHE2 Rango 3 (pK742) | This paper |
| pGEMHE2 RBP3 (pK745) | This paper |
| pGEMHE2 RCC1‐mCherry (human) (pK954) | This paper |
| pGEMHE2 EGFP‐Ran (human) (pK949) | This paper |
| pGEMHE‐V5‐mTrim21 | This paper |
| pGEMHE2 EGFP‐VAPA (pK800) | This paper |
| pAC‐biotin ligase | Avidity Inc. |
| pET30a 6His‐S‐Importin β(71–876) (pKW488) | Nachury et al (2001) |
| pET30a 6His‐SUMO‐ Importin β(71–876) (pK1031) | This paper |
| pRSET AviTag‐WT Ran (human) (pK1087) | This paper |
| pRSET AviTag‐RanQ69L (human) (pK988) | This paper |
| pRSET AviTag‐RanT24N (human) (pK224) | This paper |
| pRSET AviTag‐RanT24N, T42A (human) (pK1007) | This paper |
The pGEMHE2 plasmid was prepared by inserting synthetic oligo with multiple restriction cloning sites between the Nhe1 and Not1 sites in pGEMHE (Liman et al, 1992). Restriction cloning was used to insert the Rango‐3 (Soderholm et al, 2011) between the BamH1 and HindIII in pGEMHE2. RBP‐3 was created by replacing the IBB domain in Rango‐3 with RBD, and cloned between Not1 and Xho1 in pGEMHE2. The WT human RCC1 α‐mCherry (Hasegawa et al, 2013) was cloned between BamH1 and BsrG1 in pGEMHE1‐RBP3 to create the pGEMHE2‐RCC1‐mCherry. The human WT Ran was cloned between BsrG1 and HindIII in pGEMHE2‐RBP3 to create the pGEMHE2‐EGFP‐Ran. The EGFP‐tagged rat VAPA(1–242) (Kaiser et al, 2005) was inserted between Nhe1 and BsrG1 in pGEMHE derivative. The human importin β(71–876) from pET30a 6His‐S‐Importin β(71–876) (Nachury et al, 2001) released by EcoRV and Xho1 was cloned between the SspI and Xho1 sites in pET‐6His‐SUMO TEV LIC (1C). The pGEMHE‐mCherry‐mTrim21 (Clift et al, 2017) was modified by cloning the V5 tag between Age1 and Xho1 sites to replace mCherry, resulting in pGEMHE‐V5‐mTrim21.
The human WT Ran was cloned by PCR into pRSET‐A, followed by the insertion of synthetic oligos coding for the N‐terminal AviTag and a linker (sequence: PSPSTPPTSC), resulting in a construct for the E. coli expression of N‐terminally biotinylated Ran protein. Restriction cloning was used to replace the WT Ran in the above with RanT24N and RanQ69L from pET30a plasmids (Kalab et al, 2002). The Q5 mutagenesis kit (NEB) was used to introduce the T42A mutation into the pRN3 RanT24N (Dumont et al, 2007) and to pRSET AviTag vectors for the WT or T24N Ran described above.
Preparation of mRNA by in vitro transcription
Plasmids were linearized and in vitro transcribed using a mMESSAGE mMACHINE T3 and T7 kits (Ambion) to prepare the mRNAs. All mRNAs, except H2B‐mCherry and mEGFP‐CDK5RAP2, were then polyadenylated, using the Poly(A) Tailing kit (Thermo Fisher Scientific). Before storage at −80°C, all mRNAs were purified using the RNAeasy kit (QIAGEN).
Microinjection of mRNA or proteins
The GV oocytes were microinjected with approximately 5–10 pl of transcribed mRNA solution using micromanipulator (IM 300 microinjector, Narishige or PM 2000B microinjector, MicroData Instrument). The injections contained 700 ng/μl Rango‐3, 400–500 ng/μl RBP‐3, 300–2,500 ng/μl RanT24N (as indicated in the figures), 300–2,000 ng/μl RanT24N, T42A, 200 ng/μl RanQ69L, 50 ng/μl histone H2B‐mCherry, 125 ng/μl mEGFP‐CDK5RAP2, and 200 ng/μl YFP‐hTPX2. Following microinjections, the oocytes were cultured overnight in IBMX (Rango‐3, RBP‐3, RanT24N, RanT24N, T42A) or for 3 h in milrinone (RanQ69L, histone H2B‐mCherry, mEGFP‐CDK5RAP2, YFP‐hTPX2, RanT24N, RanT24N, T42A) to allow for sufficient exogenous protein expression. The recombinant importin β(71–876) proteins were microinjected at 200 μM concentration 2–3 h after GVBD in freshly isolated oocytes or in oocytes that were previously microinjected with the Rango‐3 or RBD‐3 mRNA.
Immunofluorescence
Oocytes were fixed in 4% paraformaldehyde (30 min) in PBS and then transferred to a membrane permeabilization solution (0.5% Triton X‐100 in PBS) for 20 min. After blocking with 1% BSA in PBS for 1 h, the oocytes were incubated with primary antibodies diluted in PBS overnight and then subjected to staining with secondary antibodies (1:100) for 1 h. To enable the simultaneous detection of pericentrin and tubulin with mouse monoclonal antibodies, the FITC‐conjugated mouse anti‐tubulin antibody was applied for 2 h after the indirect immunofluorescence staining with the anti‐pericentrin and Donkey anti‐mouse, DyLight‐560‐conjugated secondary antibodies. Oocytes were washed three times with 0.1% Tween 20 and 0.01% Triton X‐100 in PBS, stained with Hoechst 33342 (10 μg/ml in PBS) for 10 min, and then subjected to confocal microscopy imaging. The Zeiss LSM 800/Axio Observer.Z1/7 microscope equipped with the Plan Apochromat 63×/1.40 oil DIC M27 objective was used for most IF imaging. The Airyscan imaging of the IF in oocytes treated with the RCC1 Trim‐Away (Appendix Fig S7) was performed with Zeiss LSM 880/Axiobserver equipped with the EC Plan‐Neofluar 40×/1.30 Oil DIC M27 lens.
Recombinant protein expression
The WT and mutant Ran proteins containing the N‐terminal 6His and AviTag (biotin‐acceptor peptide) were co‐expressed in the T7‐Shuffle (NEB) or BL21DE3 (Thermo) E. coli cells together with the untagged E. coli biotin ligase (pAC‐6, Avidity Inc). The cell cultures were outgrown in Dynamite media (Taylor et al, 2017) at 37°C to OD(600 nm) = approx. 3.0, then cooled to 30°C, supplemented with 150 μM biotin and 200 μM IPTG, and incubated for another 3 h at 30°C. The centrifuged cell pellets were snap‐frozen in liquid nitrogen and stored at −80°C. Cell pellets were thawed in PBS supplemented with 10 mM imidazole (pH 7.4), 10 μM GTP, 5 mM MgCl2, and protease inhibitors without EDTA (Roche) and lysed by sonication in an ice bath. The lysates were clarified by centrifugation at 15,000 g for 40 min, incubated with the Ni‐NTA affinity resin (Qiagen), followed by washes with 10 mM imidazole in PBS. The proteins were eluted with 0.2 M imidazole/PBS, dialyzed in PBS, and stored at −80°C. The recombinant importin β(71–876) proteins were expressed in T7‐Shuffle (NEB) E. coli cells transfected with either pET30a‐importin β(71–876) (Nachury et al, 2001) or pET‐6His‐TEV‐SUMO‐importin β(71–876). The expression was induced with 300 μM IPTG at 37°C for 3 h, starting with OD600 nm = approximately 1.0. The clarified lysates were incubated with the Ni‐NTA columns; the columns washed with PBS containing 450 mM NaCl and 10 mM imidazole and the proteins eluted with increasing concentrations of imidazole in PBS (25–200 mM). The batches eluting with 150–200 mM imidazole were pooled, dialyzed in PBS, concentrated to 350–400 μM, filtered, and stored in aliquots at −80°C.
Cell lysates
The human colorectal carcinoma cells DLD1 (ATCC; CCL‐21) were grown in RPMI1640 media with 10% FBS, harvested by trypsinization, washed with Dulbecco's PBS (DBPS, no Ca2+, Mg2+, Thermo #14190‐144), snap‐frozen in liquid nitrogen, and stored at −80°C. The frozen pellets were thawed on the ice after the addition of 2 volumes of ice‐cold DPBS containing the complete protease inhibitor cocktail mix without EDTA (Roche) and sonicated on ice (15–20 bursts, Branson sonicator, power output 3.5). The lysates were clarified by centrifugation (4°C, 16,300 g, 10 min), their protein concentration measured with the Bio‐Rad Protein Assay (#500‐0006), adjusted to 5 or 10 μg/μl, snap‐frozen in liquid nitrogen, and stored at −80°C.
Pulldowns and immunoblotting
The DLD1 cell lysates were thawed on ice, and 600 μg aliquots were supplemented with the final concentration of 5 μM biotinylated Ran proteins or DPBS (control). After mixing, the lysates were incubated for 1 h on ice. At the end of the 1‐h incubation, streptavidin (SAV)‐coated magnetic microbeads (Dynabeads MyOne Streptavidin C1, Thermo #650.01) were washed in DPBS with protease inhibitor cocktail mix without EDTA (Roche) and distributed to Eppendorf tubes in aliquots corresponding to 20 μl original volume before removing the DPBPs by using a magnetic stand. The cell lysates that had been incubated with the biotinylated proteins were then transferred to the tubes with the washed SAV bead aliquots, mixed, and returned on ice. The lysates with SAV beads were incubated on ice for another 1 h, with gentle mixing every 5–10 min. The beads were then separated from the lysates on a magnetic stand (Thermo), washed 6× with Tris‐buffered saline (TBS) with 0.005% Tween 20, resuspended in 40 μl SDS–PAGE loading buffer, and heated for 5 min at 100°C. Aliquots of the SAV bead supernatants (5 μg) and of the bead eluates (50%) were separated on 4–20% SDS–PAGE mini gels (Thermo) and blotted to PVDF membranes (Immobilon‐P, Millipore) using the Bio‐Rad TransBlot Turbo apparatus. The blots were incubated for 30 min with 5% non‐fat dried milk in TBS, 0.05% Tween‐20 (TTBS), rinsed in distilled water, and incubated overnight at 4°C with the primary antibodies diluted in TTBS. After incubation with secondary Hpx‐coupled antibodies and the washes in TTBS, the chemiluminescence signal developed with the SuperSignal West Femto kit (Thermo, #34095) or SuperSignal West Pico (Thermo, 1856136) was captured by the Bio‐Rad ChemiDoc XRS+ digital imaging system.
Fluorescence recovery after photobleaching (FRAP)
The GV oocytes were microinjected with mRNA for EGFP‐Ran and RCC1‐mCherry and incubated overnight at 37°C, 5% CO2 in the presence of IBMX. Four hours after the release from the meiotic arrest, a group of oocytes was left untreated and another group was supplemented with 20 μM IPZ. After an additional 1‐h incubation, RCC1‐mCherry FRAP measurements were performed with the Zeiss 780 confocal microscope and the 40×/1.1 W Korr M27 lens. The entire oocyte was first captured with a 1,024‐pixel square frame, using the 561 nm and 488 nm excitation and while acquiring the emission of mCherry, GFP, and the transmitted light DIC image. Next, the zoom was changed to focus on the chromosomes, and frame size reduced to the 96‐pixel square. A small circular area (15 pixels, corresponding to 1.92 μm) was selected on chromosome arm labeled with the RCC1‐mCherry, and a series of 150 frames (8× averaging; 0.36s/frame) was acquired using the 0.1% power of the 561‐nm laser for mCherry excitation—the series consisted of 15 pre‐bleaching frames, followed by 500 iterations of the 561‐nm laser at 100% within the bleaching area, and 135 post‐bleaching frames. At least three measurements in separate areas on chromosomes were performed in each cell. The time‐lapse intensity profiles in the bleached areas were exported, and the online EasyFRAP (Rapsomaniki et al, 2012) was used to calculate the fluorescence recovery half‐time and the RCC1‐mCherry immobile fraction.
RCC1 Trim‐Away for the analysis of pericentrin‐containing MTOCs
The RCC1 knockdown by the Trim‐Away method was performed as previously described (Clift et al, 2017), with some modifications. The goat polyclonal anti‐RCC1 antibody (Santa Cruz Biotechnology sc‐1161, 0.2 μg/μl) was concentrated 4‐fold, and the buffer was exchanged for PBS with the use of Pierce 0.5 ml 10K MWCO concentrator (Thermo cat. # 88513). GV oocytes were injected with V5‐mTrim21 mRNA (750 ng/μl) and kept 3 h in IBMX‐containing M16 medium. The resumption of meiosis was initiated by washing out the IBMX, and the Trim21‐expressing oocytes were then microinjected with the anti‐RCC1 antibody (approx. 0.8 μg/μl) or buffer after GVBD. After 5 h in culture, the MI oocytes were fixed and processed for immunofluorescence with pericentrin and α‐tubulin antibodies and the Hoechst 33342 dye for DNA.
Validation of the RCC1 Trim‐Away
To validate the antibody for RCC1 Trim‐Away and confirm the RCC1 knockdown, we fluorescently labeled aliquot of the goat polyclonal anti‐RCC1 antibody (Santa Cruz Biotechnology sc‐1161) by using the Alexa Fluor 647 NHS ester (Thermo A37573). First, 100 μl aliquot of the antibody was immobilized on 20 μl (dry bed volume) Protein A/G Plus Sepharose (Santa Cruz Biotechnology, sc‐2003) placed in 1.5 ml disposable column (Bio‐Rad #732‐6204) and washed with ice‐cold 150 mM NaCl, 20 mM HEPES pH 8.2 (HBS). Most of the buffer was then removed from the beads, and the bottom of the column was capped. One 50 μg aliquot of the Alexa Fluor 647 NHS ester was diluted in anhydrous DMSO (Molecular Probes, Thermo D12345), mixed with 18 μl HBS, and immediately applied to the Protein A/G column with the immobilized antibody, thoroughly mixed and incubated for 30 min at room temperature. The buffer with unreacted dye was then removed from the column, and the labeling was repeated with a freshly diluted aliquot of the dye. The column was then washed with HBS to remove all unbound dye. The antibody was eluted by successively applying 20 μl aliquots of 0.2 M acetic acid to the column and collecting the eluates into 5 μl 1 M TrisCl pH9.0 to immediately neutralize the pH. All aliquots containing the AF647 dye were pooled. The Pierce 0.5 ml 10K MWCO concentrator (Thermo # 88513) was then used to concentrate the labeled antibody and exchange buffer for PBS. The yield of the labeling procedure was low (< 10%). The labeled antibody was applied in RCC1 Trim‐Away experiments as described above, except that the concentration of the microinjected antibody was only about 0.2 μg/μl and the results were evaluated by live‐cell imaging of the anti‐RCC1‐AF647 signal instead of by the IF.
Fluorescence lifetime imaging microscopy (FLIM) of Ran FRET sensors
Confocal FLIM of oocytes expressing the Rango‐3 or RBP‐3 sensors was performed with the use of Zeiss LSM 780 microscope and a PicoQuant system consisting of the PicoHarp 300 time‐correlated single‐photon counting (TCSPC) module, two separate hybrid PMA‐04 detectors, and Sepia II laser control module. During imaging, we maintained oocytes in Tecon environmental chamber at 37°C and 5% CO2. The imaging was performed with Apochromat 40×/1.1W Corr 27 lens (Zeiss) and started by focusing on the center of the chromosomes stained with SiR700‐DNA, using the 633 HeNe laser for excitation. Then, a single 16× averaged 1,024‐pixel square confocal scan was performed to simultaneously capture the SiR700‐DNA image and the transmitted light DIC image, using the TPMT detector. The FLIM data were then collected while using no averaging, the same lens, frame size, position and z‐thickness, and the 482‐nm diode laser (PicoQuant) that was pulsed at 32.5 MHz and set to 70% of the maximum output power. The emission light was reflected by the MBS T80/20 mirror (Zeiss) or PicoQuant 485/640 dichroic, passed through a transparent plate and the Picoquant 520 nm ± 35 nm emission filter into PicoQuant PMA‐04 hybrid detector. The pinhole size was individually set to acquire data from 1.5‐ to 5‐μm z‐sections to limit the emission photon count rate below 1% of the laser excitation pulse rate. The single 1,024‐pixel square FLIM images were acquired at zoom 2.7 (78.72 μm2) with pixel dwell time 25.2 μs/pixel (61.96 s total scan time) or 16.4 μs/pixel (40.27 s total), upon update of Zeiss 780 operation system. Noticed no detectable changes in FLIM detection as a result of the update.
FLIM data processing and quantification
FLIM data were processed with SymPhoTime 64 v. 2.3 (PicoQuant) using the customized script for the calculation of the internal response function (IRF) from 100 data points with no smoothing or with SymPhoTime 64 v. 2.4 which includes similar script as a default. The data were binned to assure > 450 photons per binned pixel, and a cell‐specific threshold was applied to eliminate out‐of‐cell fluorescence. The 3‐exponential re‐convolution was used to fit the fluorescence decays into every binned pixel.
The SymPhoTime 64 was used to calculate the amplitude‐weighted fluorescence lifetime averages (τAW) in different areas of the cells. The Magic Wand and Inverse ROI tools in the SymPhoTime were used to select the complete areas of the thresholded oocytes automatically. Regions of interest (ROIs) were manually drawn with the Free ROI tool to select areas corresponding to the position of chromosomes visualized by the emission intensity counts (peaks of the gradients) and the areas of the cytoplasm beyond the reach of the gradient, as visualized by the FLIM pseudocolor images (cytoplasmic average). The SymPhoTime was used to compute the amplitude‐weighted average fluorescence lifetime in the chromosomal, cytoplasmic, and whole‐cell regions, and the data were then exported to Excel for further processing. The average amplitude‐weighted reference EGFP donor‐only fluorescence lifetime (τdonorREFAW) was measured in MI oocytes expressing EGFP‐VAPA from microinjected mRNA, corresponding to 2.252 ns.
Although the detection of FRET by FLIM is relatively insensitive to donor concentration (Kalab et al, 2006), we observed some variation in the average FRET levels in controls that appeared to result from the expression conditions, as dictated by the co‐expressed mRNA or microinjected protein. For that reason, we analyzed the effects of most Ran perturbations in pairwise comparisons, as shown in Figs 1B and D–F, and 2B and D–F. The exceptions are the side‐by side comparisons of RanT24N and RanT24N, T42A (Fig 2 and Appendix Fig S2) containing up to 7 groups analyzed in one experiment.
Pseudocolor FLIM images and line scans
Data for the preparation of FLIM figures were exported as ASCII files and opened using a custom MATLAB application to assign a pseudocolor scale to fluorescence lifetimes. After removing the first three annotation lines in FLIM ASCII files using Notepad (Microsoft), the ASCII files were imported into Fiji as Text Images. A single 10‐μm‐wide line corresponding to the estimated position of the spindle axis was drawn across the oocyte and the profile plot exported to Excel and or GraphPad Prism 6 for figure preparation.
Quantification of Rango‐3 and RBP‐3 gradient half‐lives
The plot profiles of 6–12 radial line scans, 10‐μm‐wide, and originating from the center of the gradients in the symmetric MI oocytes were exported to Excel to calculate a single averaged plot profile for each oocyte, along with the μm distance scale that accounted for the FLIM image binning. The one‐phase decay nonlinear fit of the curve was calculated using GraphPad Prism6. The range of the fit was individually adjusted to start approximately at the edge of the chromosomes and to achieve the highest R 2. Only half‐life values with R 2 > 0.95 and an insignificant deviation from the expected model (the Runs test > 0.05) were included in the average Rango‐3 and RBP‐3 half‐life calculation (Appendix Fig S1).
Analysis and quantification of the time‐lapse and IF imaging
The analysis of the time‐lapse and IF imaging was done with Fiji (Schindelin et al, 2012). SiR‐tubulin on MTOCs (Figs 4F and 6D) was measured from 3D reconstructed images using 3D Object Counter plugin (Bolte & Cordelieres, 2006) when MTOCs were segmented based on the EGFP‐CDK5RAP2 signal. SiR‐tubulin in the area of the whole spindle (Figs 4G and 6E) was measured on maximum z‐projections when spindle area was segmented by intensity thresholding individually in each time point. For spindle volume measurements (Figs 4I and 5D), 3D volumes were reconstructed from confocal sections, and we segmented the spindles by intensity thresholding.
Ectopic MTOCs and pericentrin foci quantification
The number of ectopic MTOCs was manually counted in maximum intensity projection confocal z‐stacks of cells stained for DNA (Hoechst 33342) and with anti‐tubulin antibodies. Two observers independently verified the results.
We quantified the pericentrin foci number in z‐stack maximum intensity projections of IF‐stained oocytes, using custom‐made ImageJ macro. After manually removing the out‐of‐cell nonspecific signal for each cell, the calculations in the macros involved (i) median filter, radius two pixels; (ii) auto‐threshold by moments, dark; (iii) convert to mask; (iv) run watershed (to discern fused pericentrin foci); (v) analyze particles (options from top to bottom: size = 2‐Infinity; pixel; display; clear; include; add in situ); and (vi) display results. The measurements, including foci number and size, were exported to Excel and statistically analyzed and prepared for presentation by GraphPad 6.
Antibodies
| Antibodies | Manufacturer | Catalog number |
|---|---|---|
| Mouse anti‐Pericentrin (IF, 1:100) | BD Transduction | 611814 |
| Rabbit anti‐TPX2 (Thr288) (IF, 1:100) | Novus Biologicals | NB500‐179 |
| Mouse anti‐α‐tubulin, clone DM1A (IF, 1:100) | Sigma | T6199 |
| Mouse anti‐α‐tubulin‐FITC, clone DM1A (IF, 1:100) | Sigma | F2168 |
| Goat anti‐Mouse IgG, Atto 647 conjugate (IF, 1:100) | Sigma | 50185 |
| Donkey anti‐Rabbit IgG, DyLight 560 conjugate (IF, 1:100) | Thermo | SA5‐10039 |
| Donkey anti‐Mouse IgG, Alexa 488 Conjugate (IF, 1:100) | Invitrogen | A21202 |
| Mouse anti‐importin β (IB, 1:500) | BD Biosciences | 610559 |
| Rabbit anti‐RCC1 (IB, 1:2,000) | Abcam | ab109379 |
| Rabbit anti‐RanBP1 (IB. 1:1,000) | Cell Signaling Technology | 6780 |
| Mouse anti‐importin α1 (IB, 1:1,000) | BD Biosciences | 610485 |
| Goat anti‐RCC1 (0.8 μg/μl, Trim‐Away) | Santa Cruz Biotechnology | 1161 |
Reagents
| Reagents | Manufacturer | Catalog number |
|---|---|---|
| SiR700‐DNA Kit | Cytoskeleton | CY‐SC015 |
| SiR‐Tubulin Kit | Spirochrome | SC002 |
| Hoechst 33342 | Sigma | 14533 |
| Importazole | Sigma | SML0341 |
| mMESSAGE mMACHINE SP6 Transcription Kit | Thermo Fisher Scientific | AM1340 |
| mMESSAGE mMACHINE T7 Transcription Kit | Thermo Fisher Scientific | AM1344 |
| mMESSAGE mMACHINE T3 Transcription Kit | Thermo Fisher Scientific | AM1348 |
| Poly(A) Tailing Kit | Thermo Fisher Scientific | AM1350 |
| Alexa Fluor 647 NHS ester | Thermo Fisher Scientific | A37573 |
| Protein A/G Plus Agarose | Santa Cruz Biotech | 2003 |
Author contributions
PK and PS designed experiments and managed collaboration. PK wrote the paper. PS, RL, XD, and DD contributed to writing the manuscript. DD, XD, and PK performed and analyzed experiments. DD, XD, PK, and PS prepared figures. All authors analyzed the data and edited the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Review Process File
Acknowledgements
The US part of this study was supported by grant RO1‐HD086577 from the National Institutes of Health to RL. The Czech part of this study was supported by the Inter‐Excellence Program award LTAUSA17097 to PS and by the LO1609 award from National Sustainability Program of the Czech Ministry of Education, Youth and Sports.
The EMBO Journal (2020) 39: e101689
Contributor Information
Petr Kalab, Email: petr@jhu.edu.
Petr Solc, Email: solc@iapg.cas.cz.
References
- Balboula AZ, Nguyen AL, Gentilello AS, Quartuccio SM, Drutovic D, Solc P, Schindler K (2016) Haspin kinase regulates microtubule‐organizing center clustering and stability through Aurora kinase C in mouse oocytes. J Cell Sci 129: 3648–3660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastiaens P, Caudron M, Niethammer P, Karsenti E (2006) Gradients in the self‐organization of the mitotic spindle. Trends Cell Biol 16: 125–134 [DOI] [PubMed] [Google Scholar]
- Baumann C, Wang X, Yang L, Viveiros MM (2017) Error‐prone meiotic division and subfertility in mice with oocyte‐conditional knockdown of pericentrin. J Cell Sci 130: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennabi I, Terret ME, Verlhac MH (2016) Meiotic spindle assembly and chromosome segregation in oocytes. J Cell Biol 215: 611–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelieres FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsci 224: 213–232 [DOI] [PubMed] [Google Scholar]
- Breuer M, Kolano A, Kwon M, Li CC, Tsai TF, Pellman D, Brunet S, Verlhac MH (2010) HURP permits MTOC sorting for robust meiotic spindle bipolarity, similar to extra centrosome clustering in cancer cells. J Cell Biol 191: 1251–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S, Dumont J, Lee KW, Kinoshita K, Hikal P, Gruss OJ, Maro B, Verlhac MH (2008) Meiotic regulation of TPX2 protein levels governs cell cycle progression in mouse oocytes. PLoS ONE 3: e3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bury L, Coelho PA, Simeone A, Ferries S, Eyers CE, Eyers PA, Zernicka‐Goetz M, Glover DM (2017) Plk4 and Aurora A cooperate in the initiation of acentriolar spindle assembly in mammalian oocytes. J Cell Biol 216: 3571–3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabatsos MJ, Combelles CM, Messinger SM, Albertini DF (2000) Sorting and reorganization of centrosomes during oocyte maturation in the mouse. Microsci Res Tech 49: 435–444 [DOI] [PubMed] [Google Scholar]
- Caudron M, Bunt G, Bastiaens P, Karsenti E (2005) Spatial coordination of spindle assembly by chromosome‐mediated signaling gradients. Science 309: 1373–1376 [DOI] [PubMed] [Google Scholar]
- Clarke PR, Zhang C (2008) Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol 9: 464–477 [DOI] [PubMed] [Google Scholar]
- Clift D, Schuh M (2015) A three‐step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat Commun 6: 7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D, McEwan WA, Labzin LI, Konieczny V, Mogessie B, James LC, Schuh M (2017) A method for the acute and rapid degradation of endogenous proteins. Cell 171: 1692–1706 e1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combelles CM, Albertini DF (2001) Microtubule patterning during meiotic maturation in mouse oocytes is determined by cell cycle‐specific sorting and redistribution of gamma‐tubulin. Dev Biol 239: 281–294 [DOI] [PubMed] [Google Scholar]
- Courtois A, Solc P, Kitajima TS (2018) Triple‐color live imaging of mouse oocytes. Methods Mol Biol 1818: 89–97 [DOI] [PubMed] [Google Scholar]
- Dasso M, Seki T, Azuma Y, Ohba T, Nishimoto T (1994) A mutant form of the Ran/TC4 protein disrupts nuclear function in Xenopus laevis egg extracts by inhibiting the RCC1 protein, a regulator of chromosome condensation. EMBO J 13: 5732–5744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval B, Doxsey SJ (2010) Pericentrin in cellular function and disease. J Cell Biol 188: 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickmanns A, Bischoff FR, Marshallsay C, Luhrmann R, Ponstingl H, Fanning E (1996) The thermolability of nuclear protein import in tsBN2 cells is suppressed by microinjected Ran‐GTP or Ran‐GDP, but not by RanQ69L or RanT24N. J Cell Sci 109(Pt 6): 1449–1457 [DOI] [PubMed] [Google Scholar]
- Dumont J, Petri S, Pellegrin F, Terret ME, Bohnsack MT, Rassinier P, Georget V, Kalab P, Gruss OJ, Verlhac MH (2007) A centriole‐ and RanGTP‐independent spindle assembly pathway in meiosis I of vertebrate oocytes. J Cell Biol 176: 295–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes DJ, Travesa A, Nord MS, Bernis C (2015) Nuclear transport factors: global regulation of mitosis. Curr Opin Cell Biol 35: 78–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman BS, Miller KE, Heald R (2010) Xenopus egg extracts increase dynamics of histone H1 on sperm chromatin. PLoS ONE 5: e13111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D, Seewald MJ, Ribbeck K (2003) Characterization of Ran‐driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J 22: 1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss OJ, Carazo‐Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW (2001) Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell 104: 83–93 [DOI] [PubMed] [Google Scholar]
- Gruss OJ (2018) Animal female meiosis: the challenges of eliminating centrosomes. Cells 7: E73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjantonakis AK, Papaioannou VE (2004) Dynamic in vivo imaging and cell tracking using a histone fluorescent protein fusion in mice. BMC Biotechnol 4: 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa K, Ryu SJ, Kalab P (2013) Chromosomal gain promotes formation of a steep RanGTP gradient that drives mitosis in aneuploid cells. J Cell Biol 200: 151–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holubcova Z, Blayney M, Elder K, Schuh M (2015) Human oocytes. Error‐prone chromosome‐mediated spindle assembly favors chromosome segregation defects in human oocytes. Science 348: 1143–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes M, Zhang C, Avis JM, Hutchison CJ, Clarke PR (1998) The role of the ran GTPase in nuclear assembly and DNA replication: characterisation of the effects of Ran mutants. J Cell Sci 111(Pt 20): 3017–3026 [DOI] [PubMed] [Google Scholar]
- Jakel S, Mingot JM, Schwarzmaier P, Hartmann E, Gorlich D (2002) Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J 21: 377–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser SE, Brickner JH, Reilein AR, Fenn TD, Walter P, Brunger AT (2005) Structural basis of FFAT motif‐mediated ER targeting. Structure 13: 1035–1045 [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R (2002) Visualization of a Ran‐GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295: 2452–2456 [DOI] [PubMed] [Google Scholar]
- Kalab P, Pralle A, Isacoff EY, Heald R, Weis K (2006) Analysis of a RanGTP‐regulated gradient in mitotic somatic cells. Nature 440: 697–701 [DOI] [PubMed] [Google Scholar]
- Kalab P, Heald R (2008) The RanGTP gradient ‐ a GPS for the mitotic spindle. J Cell Sci 121: 1577–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P, Solc P, Motlik J (2011) The role of RanGTP gradient in vertebrate oocyte maturation. Results Probl Cell Differ 53: 235–267 [DOI] [PubMed] [Google Scholar]
- Kitajima TS, Ohsugi M, Ellenberg J (2011) Complete kinetochore tracking reveals error‐prone homologous chromosome biorientation in mammalian oocytes. Cell 146: 568–581 [DOI] [PubMed] [Google Scholar]
- Lakowicz JR (2006) Principles of fluorescence spectroscopy. New York, NY: Springer; xxvi, 954 pp. [Google Scholar]
- Liman ER, Tytgat J, Hess P (1992) Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron 9: 861–871 [DOI] [PubMed] [Google Scholar]
- Liu Q, Yu J, Zhuo X, Jiang Q, Zhang C (2010) Pericentrin contains five NESs and an NLS essential for its nucleocytoplasmic trafficking during the cell cycle. Cell Res 20: 948–962 [DOI] [PubMed] [Google Scholar]
- Lounsbury KM, Richards SA, Carey KL, Macara IG (1996) Mutations within the Ran/TC4 GTPase. Effects on regulatory factor interactions and subcellular localization. J Biol Chem 271: 32834–32841 [DOI] [PubMed] [Google Scholar]
- Lukinavicius G, Reymond L, D'Este E, Masharina A, Gottfert F, Ta H, Guther A, Fournier M, Rizzo S, Waldmann H et al (2014) Fluorogenic probes for live‐cell imaging of the cytoskeleton. Nat Methods 11: 731–733 [DOI] [PubMed] [Google Scholar]
- Luksza M, Queguigner I, Verlhac MH, Brunet S (2013) Rebuilding MTOCs upon centriole loss during mouse oogenesis. Dev Biol 382: 48–56 [DOI] [PubMed] [Google Scholar]
- Ma W, Baumann C, Viveiros MM (2010) NEDD1 is crucial for meiotic spindle stability and accurate chromosome segregation in mammalian oocytes. Dev Biol 339: 439–450 [DOI] [PubMed] [Google Scholar]
- Maresca TJ, Groen AC, Gatlin JC, Ohi R, Mitchison TJ, Salmon ED (2009) Spindle assembly in the absence of a RanGTP gradient requires localized CPC activity. Curr Biol 19: 1210–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, Baran V, Sakakibara Y, Brzakova A, Ferencova I, Motlik J, Kitajima TS, Schultz RM, Solc P (2016) DNA damage response during mouse oocyte maturation. Cell Cycle 15: 546–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GA, Moore MS, Drivas G, Perez de la Ossa P, Villamarin A, D'Eustachio P, Rush MG (1997) A T42A Ran mutation: differential interactions with effectors and regulators, and defect in nuclear protein import. Mol Biol Cell 8: 2591–2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Maresca TJ, Salmon WC, Waterman‐Storer CM, Heald R, Weis K (2001) Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell 104: 95–106 [DOI] [PubMed] [Google Scholar]
- Neumayer G, Belzil C, Gruss OJ, Nguyen MD (2014) TPX2: of spindle assembly, DNA damage response, and cancer. Cell Mol Life Sci 71: 3027–3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AL, Drutovic D, Vazquez BN, El Yakoubi W, Gentilello AS, Malumbres M, Solc P, Schindler K (2018) Genetic Interactions between the aurora kinases reveal new requirements for AURKB and AURKC during oocyte meiosis. Curr Biol 28: 3458–3468 e3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell CB, Loncarek J, Kalab P, Khodjakov A (2009) Relative contributions of chromatin and kinetochores to mitotic spindle assembly. J Cell Biol 187: 43–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D, Yu CH, Needleman DJ (2016) Spatial organization of the Ran pathway by microtubules in mitosis. Proc Natl Acad Sci USA 113: 8729–8734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I, Weis K, Klebe C, Mattaj IW, Dingwall C (1996) RAN/TC4 mutants identify a common requirement for snRNP and protein import into the nucleus. J Cell Biol 133: 485–494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S, Groen AC, Ishihara K, Mitchison TJ, Vale RD (2013) Branching microtubule nucleation in Xenopus egg extracts mediated by augmin and TPX2. Cell 152: 768–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapsomaniki MA, Kotsantis P, Symeonidou IE, Giakoumakis NN, Taraviras S, Lygerou Z (2012) easyFRAP: an interactive, easy‐to‐use tool for qualitative and quantitative analysis of FRAP data. Bioinformatics 28: 1800–1801 [DOI] [PubMed] [Google Scholar]
- Saskova A, Solc P, Baran V, Kubelka M, Schultz RM, Motlik J (2008) Aurora kinase A controls meiosis I progression in mouse oocytes. Cell Cycle 7: 2368–2376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten H, Sun QY (2015) Centrosome and microtubule functions and dysfunctions in meiosis: implications for age‐related infertility and developmental disorders. Reprod Fertil Dev 27: 934–943 [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B et al (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J (2007) Self‐organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell 130: 484–498 [DOI] [PubMed] [Google Scholar]
- Sillje HH, Nagel S, Korner R, Nigg EA (2006) HURP is a Ran‐importin beta‐regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr Biol 16: 731–742 [DOI] [PubMed] [Google Scholar]
- Simerly C, Manil‐Segalen M, Castro C, Hartnett C, Kong D, Verlhac MH, Loncarek J, Schatten G (2018) Separation and loss of centrioles from primordidal germ cells to mature oocytes in the mouse. Sci Rep 8: 12791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderholm JF, Bird SL, Kalab P, Sampathkumar Y, Hasegawa K, Uehara‐Bingen M, Weis K, Heald R (2011) Importazole, a small molecule inhibitor of the transport receptor importin‐beta. ACS Chem Biol 6: 700–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solc P, Baran V, Mayer A, Bohmova T, Panenkova‐Havlova G, Saskova A, Schultz RM, Motlik J (2012) Aurora kinase A drives MTOC biogenesis but does not trigger resumption of meiosis in mouse oocytes matured in vivo . Biol Reprod 87: 85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solc P, Kitajima TS, Yoshida S, Brzakova A, Kaido M, Baran V, Mayer A, Samalova P, Motlik J, Ellenberg J (2015) Multiple requirements of PLK1 during mouse oocyte maturation. PLoS ONE 10: e0116783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein P, Schindler K (2011) Mouse oocyte microinjection, maturation and ploidy assessment. J Vis Exp 53: e2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi D, Calarco P, Donahue RP (1972) Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J Cell Sci 11: 521–541 [DOI] [PubMed] [Google Scholar]
- Szollosi D, Mandelbaum J, Plachot M, Salat‐Baroux J, Cohen J (1986) Ultrastructure of the human preovulatory oocyte. J In Vitro Fert Embryo Transf 3: 232–242 [DOI] [PubMed] [Google Scholar]
- Taylor T, Denson JP, Esposito D (2017) Optimizing expression and solubility of proteins in E. coli using modified media and induction parameters. Methods Mol Biol 1586: 65–82 [DOI] [PubMed] [Google Scholar]
- Zhang MS, Arnaoutov A, Dasso M (2014) RanBP1 governs spindle assembly by defining mitotic Ran‐GTP production. Dev Cell 31: 393–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Roostalu J, Surrey T, Nogales E (2017) Structural insight into TPX2‐stimulated microtubule assembly. Elife 6: e30959 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Review Process File


