Abstract
The Hippo pathway, which plays a critical role in organ size control and cancer, features numerous WW domain‐based protein–protein interactions. However, ~100 WW domains and 2,000 PY motif‐containing peptide ligands are found in the human proteome, raising a “WW‐PY” binding specificity issue in the Hippo pathway. In this study, we have established the WW domain binding specificity for Hippo pathway components and uncovered a unique amino acid sequence required for it. By using this criterion, we have identified a WW domain‐containing protein, STXBP4, as a negative regulator of YAP. Mechanistically, STXBP4 assembles a protein complex comprising α‐catenin and a group of Hippo PY motif‐containing components/regulators to inhibit YAP, a process that is regulated by actin cytoskeleton tension. Interestingly, STXBP4 is a potential tumor suppressor for human kidney cancer, whose downregulation is correlated with YAP activation in clear cell renal cell carcinoma. Taken together, our study not only elucidates the WW domain binding specificity for the Hippo pathway, but also reveals STXBP4 as a player in actin cytoskeleton tension‐mediated Hippo pathway regulation.
Keywords: PY motif, STXBP4, the Hippo pathway, WW domain, YAP
Subject Categories: Cancer; Post-translational Modifications, Proteolysis & Proteomics; Signal Transduction
Identification of a conserved WW domain sequence mediating specific interactions among Hippo pathway components uncovers a new regulator of Hippo signaling as potential kidney cancer tumor suppressor.
Introduction
Signaling proteins often entail modular domains that facilitate protein–protein interactions to assemble functional protein complexes, control enzymatic activity and regulate protein cellular localization (Cohen et al, 1995; Pawson & Scott, 1997). Importantly, the recognition between domains and their peptide ligands is usually specific, thus allowing the transduction of unique information through signaling cascades (Das & Smith, 2000; Hu et al, 2004). The WW domain is a small protein module that is defined by the presence of two tryptophan (W) residues separated apart by ~ 25 amino acids (Sudol et al, 1995b). WW domain and its cognate proline‐rich peptide motif have been identified within various protein complexes widely distributed in plasma membrane, cytoplasm, and nucleus. Failure of their recognition is associated with multiple human diseases including Alzheimer's disease (Mandelkow & Mandelkow, 1998; Liu et al, 2007), Huntington's disease (Faber et al, 1998; Passani et al, 2000), Liddle syndrome (Hansson et al, 1995), Golabi‐Ito‐Hall syndrome (Lubs et al, 2006; Tapia et al, 2010), muscular dystrophy (Bork & Sudol, 1994; Rentschler et al, 1999; Ervasti, 2007), and cancers (Chang et al, 2007; Salah & Aqeilan, 2011). These facts highlight a crucial role of the WW domain‐mediated protein–protein interaction in biological processes and tissue homeostasis.
WW domain was initially uncovered by characterizing the protein sequence of YAP, a key transcriptional co‐activator downstream of the Hippo pathway (Sudol et al, 1995a; Pan, 2010; Jiang et al, 2015). The Hippo pathway is a highly conserved signaling pathway involved in tissue homeostasis, organ size control, and cancer development (Pan, 2010; Halder & Johnson, 2011; Piccolo et al, 2014; Jiang et al, 2015; Yu et al, 2015). In mammals, the Hippo pathway is composed of a kinase cascade (two serine/threonine kinases, MST and LATS; and the adaptors SAV1 for MST and MOB1 for LATS), downstream effectors (YAP and TAZ), and nuclear transcriptional factors (TEADs). MST phosphorylates and activates LATS, which in turn phosphorylates YAP and TAZ. The phosphorylated YAP/TAZ can be recognized by 14‐3‐3 proteins, retained in the cytoplasm, and eventually targeted by β‐TRCP E3 ligase complex for degradation. When the Hippo pathway is inactivated, unphosphorylated YAP/TAZ enter into the nucleus, where they associate with TEAD transcriptional factors to promote the transcription of genes that are involved in proliferation and survival.
Notably, many Hippo pathway components and regulators contain either the WW domain or its proline‐rich peptide ligand, mostly “PPxY” motif (P, proline; Y, tyrosine; x, any amino acid; hereafter named as “PY” motif) (Sudol, 2010; Salah & Aqeilan, 2011). YAP, TAZ, SAV1, and KIBRA, an upstream component of the Hippo kinase cascade (Yu et al, 2010), are four known WW domain‐containing components of the Hippo pathway (Salah & Aqeilan, 2011). In the nucleus, the WW domain of YAP/TAZ is a requirement for their association with a group of nuclear transcriptional factors and regulators that contain the PY motif to regulate gene transcription (Strano et al, 2001, 2005; Ferrigno et al, 2002; Zhang et al, 2011; Haskins et al, 2014; Qiao et al, 2016; Chang et al, 2018; Liu et al, 2018). In the cytoplasm, the PY motif of LATS1/2 is involved in the LATS1/2‐mediated YAP/TAZ phosphorylation (Hao et al, 2008; Verma et al, 2018); several PY motif‐containing proteins can physically bind the WW domain of YAP/TAZ and promote YAP/TAZ's cytoplasmic translocation (Espanel & Sudol, 2001; Chan et al, 2011; Wang et al, 2011, 2012b, 2014; Zhao et al, 2011; Liu et al, 2013; Michaloglou et al, 2013; Tavana et al, 2016). Moreover, the phosphorylated YAP/TAZ can negatively regulate Wnt pathway by forming a complex with DVL2, which is mediated by the WW domain of YAP/TAZ and the PY motif of DVL2 (Varelas et al, 2010). As a Hippo upstream component, KIBRA can similarly associate with several Hippo PY motif‐containing proteins and negatively regulate YAP (Wilson et al, 2014; Tavana et al, 2016). On the other hand, several WW domain‐containing proteins have been shown to modulate the Hippo pathway activity by regulating the Hippo PY motif‐containing components and regulators (Salah et al, 2011, 2013; Ulbricht et al, 2013; Yeung et al, 2013; Abu‐Odeh et al, 2014; Wang et al, 2015). Collectively, these facts suggest that the WW domain and PY motif‐mediated protein–protein interaction plays a fundamental role in building up the major framework of the Hippo pathway.
Actually, ~ 100 WW domains and 2,000 PY motif‐containing peptides have been predicted in the human proteome (Tapia et al, 2010), raising an issue of binding specificity for the proteins containing WW domain and PY motif. Indeed, a large scale of WW domain array screen only confirmed 10% of the tested WW domain–ligand interactions (Hu et al, 2004). Several large‐scale proteomic studies exclusively identified a group of PY motif‐containing proteins (e.g., LATS1/2, AMOTs, PTPN14) as the binding partners for the Hippo WW domain‐containing components (Couzens et al, 2013; Hauri et al, 2013; Wang et al, 2014). These facts indicate the binding specificity for the Hippo WW domain‐mediated protein–protein interaction, while the underlying mechanism is still largely unknown.
In this study, we demonstrated the WW domain binding specificity for the Hippo pathway proteins and uncovered a highly conserved amino acid sequence required for it. By using this criterion, we identified STXBP4 as a novel Hippo pathway regulator in human proteome. Mechanistically, STXBP4 assembled a complex with α‐catenin and several Hippo PY motif‐containing components/regulators to negatively regulate YAP when actin cytoskeleton tension is low. Moreover, both TCGA data and tissue array studies suggested STXBP4 as a potential tumor suppressor in human kidney cancer, whose downregulation is significantly correlated with YAP activation in clear cell renal cell carcinoma. Collectively, our study not only elucidated the WW domain binding specificity for the Hippo pathway protein–protein interaction network, but also identified STXBP4 as a Hippo pathway regulator and a potential tumor suppressor in kidney cancer development.
Results
Binding specificity exists for the Hippo WW domain‐containing components
We re‐analyzed our previously published proteomic data (Wang et al, 2014) for four Hippo WW domain‐containing components YAP, TAZ, SAV1, and KIBRA (Fig 1A) and found that most of the known Hippo PY motif‐containing proteins (e.g., AMOT, AMOTL1, AMOTL2, LATS1, LATS2, PTPN14, PTPN21, WBP2) were hardly detected in the SAV1‐associated protein complex (Fig 1B). Moreover, proteomic analysis of the WW domains isolated from these four Hippo components (Fig EV1A) further confirmed this finding, where the WW domain of YAP, TAZ, and KIBRA, but not that of SAV1, retrieved most of these known Hippo PY motif‐containing proteins (Fig 1B). These data suggest that the WW domain of SAV1 is different from that of YAP, TAZ, and KIBRA in associating with the known Hippo PY motif‐containing proteins.
Figure 1. The Hippo WW domain shows binding specificity with the known Hippo PY motif‐containing proteins. (This figure is related to Fig EV1 and Tables EV1, EV2, EV3 and EV4).
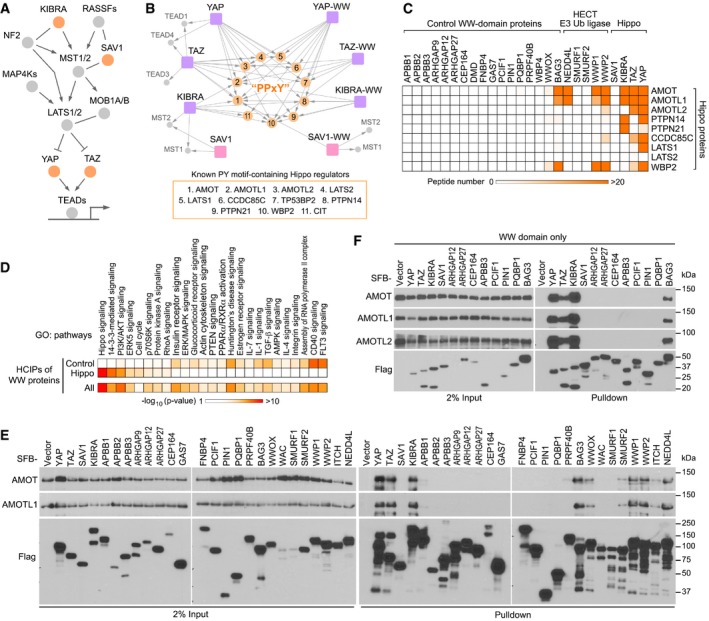
- Schematic illustration of the human Hippo pathway, where the Hippo WW domain‐containing components are highlighted.
- A summary map of cytoscape‐generated merged interaction network for the Hippo WW domain‐containing components and their WW domains.
- The Hippo WW domain‐containing proteins show binding specificity to the known Hippo PY motif‐containing proteins. TAP‐MS analysis of a series of WW domain‐containing proteins were performed, and their binding with the indicated Hippo PY motif‐containing proteins was summarized in a heatmap.
- The HCIPs for the Hippo WW domain‐containing proteins were involved in different signaling pathways compared to those retrieved from the control WW domain‐containing proteins. Gene Ontology analysis was performed.
- Validation of the binding specificity for the Hippo WW domain‐containing proteins. HEK293T cells were transfected with the indicated SFB‐tagged constructs and subjected to the pull‐down assay.
- Validation of the binding specificity for the derived WW domains from the Hippo WW domain‐containing proteins. HEK293T cells were transfected with the indicated SFB‐tagged constructs and subjected to the pull‐down assay.
Source data are available online for this figure.
Figure EV1. Proteomic analysis of the WW‐containing proteins. (This figure is related to Fig 1).
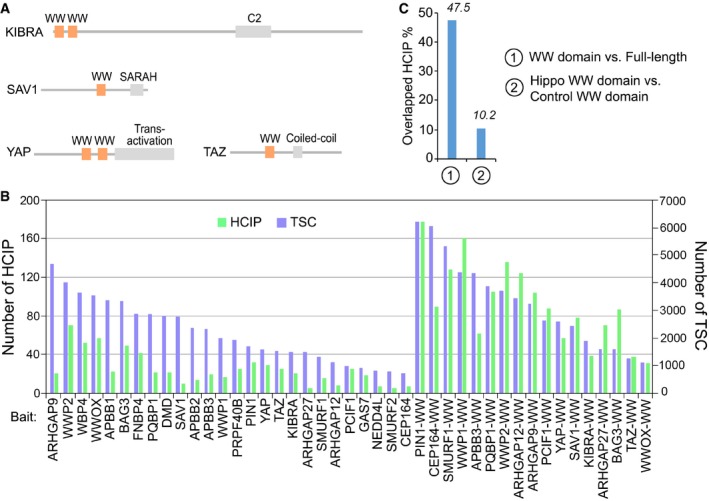
- Schematic illustration of the Hippo WW domain‐containing components.
- The total spectral counts (TSCs) and corresponding numbers of HCIPs for the indicated proteomic experiments were listed.
- The overlapped HCIP rate was compared for the full‐length protein vs. its WW domain, and Hippo WW domains vs. control WW domains, respectively.
Next, we expanded our proteomic analysis for additional 22 WW domain‐containing proteins (Fig EV1B; [Link], [Link], [Link]) and examined their ability to isolate these known Hippo PY motif‐containing proteins. Consistent with previous reports (Salah et al, 2013; Ulbricht et al, 2013; Yeung et al, 2013; Abu‐Odeh et al, 2014; Wang et al, 2015), WWOX, BAG3, and members of the HECT family of E3 ligases NEDD4L, WWP1, and WWP2 were found to form complexes with the Hippo PY motif‐containing proteins such as AMOT family proteins, CCDC85C, and WBP2 (Fig 1C). However, we failed to identify these Hippo PY motif‐containing proteins as the binding proteins for other tested WW domain‐containing proteins (Fig 1C). Moreover, the high‐confident interacting proteins (HCIPs) of the Hippo WW domain‐containing components were involved in different signaling pathways from those of the control WW domain‐containing proteins (Fig 1D and Table EV4). We also performed proteomic analysis for the WW domains isolated from 13 randomly selected WW domain‐containing proteins and found that only 10.2% of the HCIPs were shared by the Hippo and control WW domains (Fig EV1C). Taken together, these results indicate that the WW domains of the Hippo pathway components YAP, TAZ, and KIBRA possess a binding specificity with the known Hippo PY motif‐containing proteins.
Validation of the Hippo WW domain binding specificity
To validate our proteomic findings, we examined the interaction between a series of WW domain‐containing proteins and AMOT family proteins. Unlike YAP, TAZ, and KIBRA, SAV1 failed to bind AMOT and AMOTL1 (Fig 1E). Consistently, we hardly detected the association between SAV1 and LATS1 in our experimental setting (Appendix Fig S1A). Moreover, BAG3, WWOX, and several members of the HECT family of E3 ligases can interact with AMOT proteins (Fig 1E), which is consistent with our proteomic study (Fig 1C). However, other tested WW domain‐containing proteins as well as their derived WW domains failed to bind AMOT family proteins (Fig 1E and F). These results demonstrate the WW domain binding specificity for the Hippo pathway proteins.
A highly conserved amino acid sequence is required for the Hippo WW domain binding specificity
To further explore the underlying mechanism, we analyzed the WW domain protein sequence for the Hippo pathway components as well as WWOX, BAG3, and several members of the HECT family of E3 ligases, which can bind the known Hippo PY motif‐containing proteins (Fig 2A). Interestingly, in addition to the two tryptophan residues, additional nine amino acids were found to be highly conserved among these WW domains (Fig 2A). We hypothesized that this conserved 9‐amino acid sequence could be required for the specific association with the known Hippo PY motif‐containing proteins.
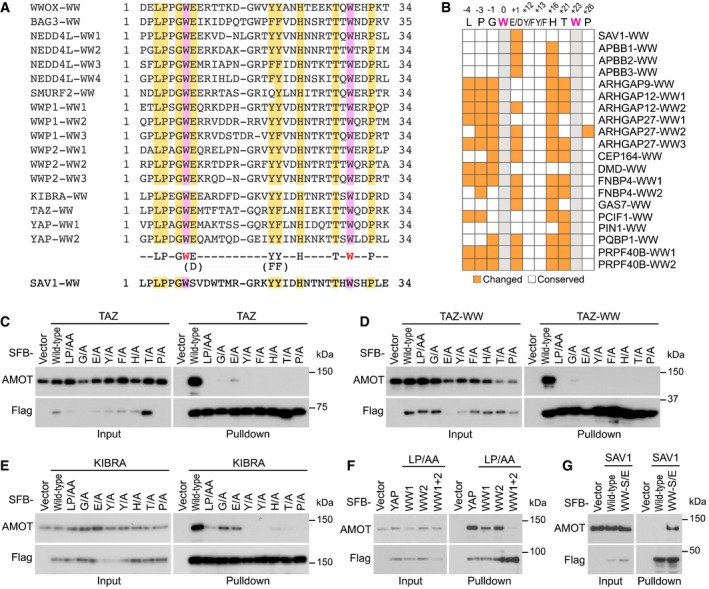
Figure 2. Identification of a conserved 9‐amino acid sequence that determines the Hippo WW domain binding specificity. (This Figure is related to Figs EV2 and EV3; Appendix Figs S1–S3; Table EV5).
-
ASequence alignment of the WW domains derived from the WW domain‐containing proteins that are known to bind the Hippo PY motif‐containing proteins. The two conserved tryptophan restudies were highlighted in purple. Additional conserved amino acid residues were highlighted in yellow.
-
BSummary of the residue difference in the identified 9‐amino acid sequence for the control WW domains. The conserved two tryptophan residues are labeled in gray; the changed residues are labeled in orange; and the unchanged residues are labeled in white.
-
C–GValidation of the identified 9‐amino acid sequence in determining the Hippo WW domain binding specificity. The requirement of the identified 9‐amino acid sequence for AMOT association was, respectively, examined for TAZ (C), TAZ‐WW domain (D), KIBRA (E), YAP (F) and SAV1 (G). HEK293T cells were transfected with the indicated SFB‐tagged constructs and subjected to the pull‐down assay.
Source data are available online for this figure.
To test this hypothesis, we examined the identified 9‐amino acid sequence in the control WW domain‐containing proteins that failed to bind the Hippo PY motif‐containing proteins (Fig 1C) and found that their WW domains have at least one of these nine amino acids replaced by other residues (Figs 2B and EV2A). As for SAV1, the conserved glutamate residue within this 9‐amino acid sequence was found changed to a serine in its WW domain (Fig 2A). Consistently, mutating either of these identified nine amino acids to alanine dramatically disrupted the association of AMOT with TAZ (Fig 2C) or its WW domain (Fig 2D). Similar findings were also observed for both KIBRA (Fig 2E) and YAP (Fig 2F). Notably, mutations of the G and E residues among these identified nine amino acids are less detrimental to the Hippo WW‐PY interaction as compared with other identified sites (Fig 2C–E). We also tested the conservative substitution for the “E/D”, “Y/F”, or “F/Y” of this conserved amino acid sequence and found that the association of AMOT with TAZ and KIBRA was not affected by these substitutions (Appendix Fig S1B). Interestingly, an interaction between SAV1 and AMOT was recovered when the unmatched serine residue was replaced by glutamate, allowing SAV1 WW domain to fit the 9‐amino acid sequence criterion (Fig 2G). Taken together, these results demonstrate that the identified 9‐amino acid sequence determines the WW domain binding specificity for the Hippo pathway proteins.
Figure EV2. Analyses of the identified 9‐amino acid sequence in both control WW domains and evolution. (This figure is related to Fig 2; Appendix Figs S1 and S2; Table EV5).
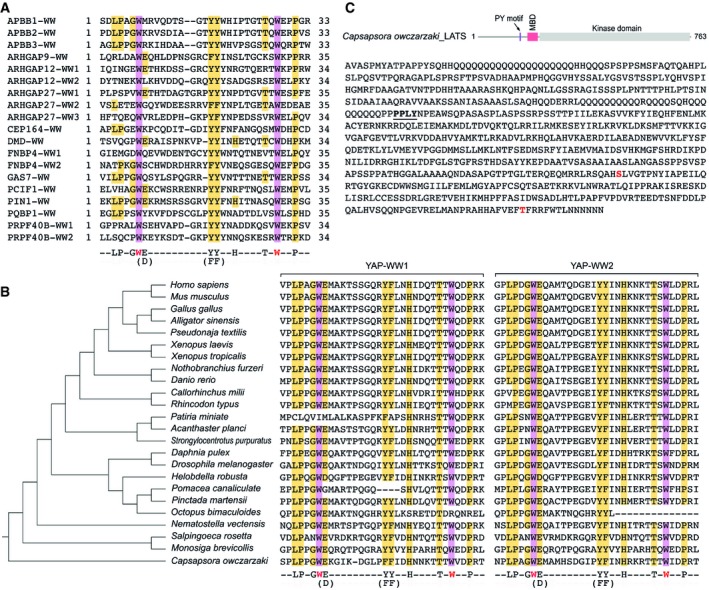
- Sequence alignment of the WW domains derived from the control WW domain‐containing proteins that cannot bind the Hippo PY motif‐containing proteins. The two conserved tryptophan restudies were highlighted in purple, and the identified 9‐amino acid residues were highlighted in yellow.
- Evolutionary analysis of the YAP‐WW domains. The identified 9‐amino acid sequence is highlighted in the two YAP‐WW domains derived from the indicated species.
- A PY motif is identified in Capsapsora owczarzaki LATS. Schematic illustration of the C. owczarzaki LATS protein, where the PY motif is indicated. MBD, MOB1‐binding domain. The PY motif is underlined in the C. owczarzaki LATS protein sequence, where the auto‐phosphorylation site (S586) and the phosphorylation site (T750) in the hydrophobic motif were shown in red.
We also examined the Hippo WW domain‐containing components in Drosophila and found that this 9‐amino acid sequence was highly conserved in the WW domain of Yorkie and Kibra, while Salvador similarly contains a replacement of the conserved glutamate residue by alanine (Appendix Fig S2). By taking YAP as an example, conservation of this 9‐amino acid sequence in the YAP‐WW domains can be even tracked to Capsapsora owczarzaki (Fig EV2B and Table EV5), an unicellular specie that is known to contain the functional Hippo pathway components (Sebe‐Pedros et al, 2012). Interestingly, in C. owczarzaki, a PY motif was also identified in LATS (Fig EV2C), suggesting that this conserved 9‐amino acid sequence may play a crucial role for the Hippo pathway at its premetazoan origin.
Role of the 9‐amino acid sequence in assembly of a specific WW‐PY complex involving the Hippo pathway proteins
Next, we analyzed a NMR solution structure of the YAP‐WW1 domain (the first WW domain of YAP) and SMAD7‐PY motif‐containing peptide complex (Aragon et al, 2012). Interestingly, the identified 9 amino acids form as two functional groups.
First, together with the second tryptophan (W199 of YAP‐WW1), the conserved residues E178, Y188, H192, and T197 were involved in the binding interface with the SMAD7‐PY motif (Fig EV3A). Specifically, hydrogen bond (H‐bond) formation was, respectively, paired between H192 (YAP‐WW1 domain) and Y211 (SMAD7‐PY motif), and T197 (YAP‐WW1 domain) and P209 (SMAD7‐PY motif) (Fig EV3B and C). Hydrophobic contact not only existed within the intramolecular interaction between the W199 and Y188 residues of YAP1‐WW domain, but also mediated their intermolecular interaction with the P208 and P209 residues within SMAD7‐PY motif, respectively (Fig EV3B and C). E178 (YAP‐WW1 domain) functioned in sustaining the intermolecular contact between H192 (YAP‐WW1 domain) and Y211 (SMAD7‐PY motif) by forming both electrostatic and H‐bonding interactions with H192 (Fig EV3B and C).
Figure EV3. Structural analysis of the identified 9‐amino acid sequence. (This figure is related to Fig 2 and Appendix Fig S3).
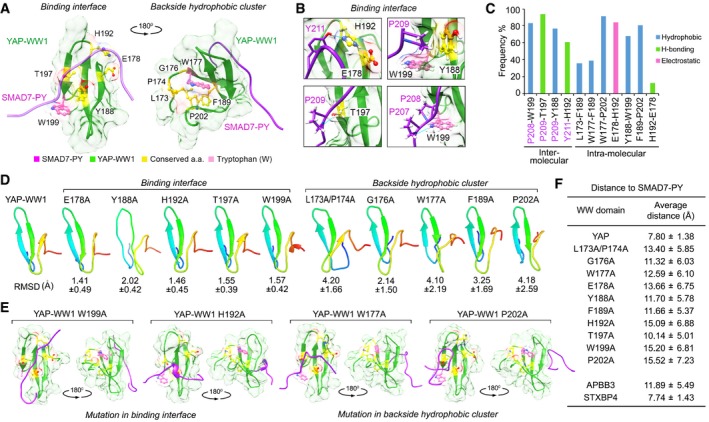
- Illustration of the identified 9‐amino acid residues in the average YAP‐WW1/SMAD7‐PY structure. The initial structure was derived from NMR solution structure (2LTW). SMAD7‐PY peptide was adjusted to 50% transparence to show the residue details on the binding interface.
- Four contact regions within the YAP‐WW1/SMAD7‐PY complex were shown in details from the representative top cluster structures. Residues from SMAD7‐PY motif peptide were labeled in purple. Hydrogen bond is indicated in blue line.
- The binding type and the corresponding frequency rate were shown for the indicated inter‐ and intramolecular residue pairs.
- Simulation analysis of apo YAP‐WW1 domain and its indicated mutants. RMSD value for each mutant simulation (referenced against the average apo YAP‐WW1 domain) was shown.
- Average structures of the indicated YAP‐WW1 mutant/SMAD7‐PY complexes.
- The average distance between SMAD7‐PY motif peptide and the indicated WW domains was summarized in a table.
Second, together with the first tryptophan (W177 of YAP‐WW1 domain), the rest residues L173, P174, G176, F189, and P202 formed a hydrophobic cluster at the backside of the YAP‐WW1/SMAD7‐PY complex (Fig EV3A and C). Although not directly interacted with SMAD7‐PY motif, this hydrophobic cluster may maintain a unique YAP‐WW1 domain structure to facilitate its binding with SMAD7‐PY motif. Since these hydrophobic cluster residues are also frequently replaced by other amino acids in the non‐Hippo WW domains (Figs 2B and EV2A), we consider them as part of the determinants for the specific Hippo WW‐PY recognition.
To further determine the role of this identified 9‐amino acid sequence from a structure‐based perspective, we mutated each of these conserved residues into alanine in silico and performed root‐mean‐square deviation (RMSD) analyses using the average unbound (apo) structure of YAP‐WW1 domain as a reference. Interestingly, mutating either of the identified residues within the backside hydrophobic cluster significantly altered the YAP‐WW1 protein structure as indicated by their relatively high RMSD values, while this was not the case for the residues within the binding interface with SMAD7‐PY motif (Fig EV3D). These results further confirm the hypothesis that the backside hydrophobic cluster may play a role in maintaining a functional YAP‐WW1 structure. In addition, mutating either of the conserved residues altered the complex structure (Fig EV3E and Appendix Fig S3A) and increased the average distance between YAP‐WW1 domain and SMAD7‐PY motif peptide (Fig EV3F), indicating the intervention of their complex formation. As a control, we analyzed a NMR solution structure of the APBB3‐WW domain (Appendix Fig S3B). The APBB3‐WW domain failed to bind the Hippo PY motif‐containing proteins (Fig 1F), since it contains two unmatched residues (as compared to the identified 9‐amino acid sequence) locating in the PY motif binding interface (Figs 2B and EV2A; Appendix Fig S3B). Consistently, the average distance between APBB3‐WW domain and SMAD7‐PY motif peptide is comparable to that between YAP‐WW1 domain mutants and SMAD7‐PY motif peptide (Fig EV3F), suggesting an unstable complex formation for APBB3‐WW domain and SMAD7‐PY motif. Notably, the standard deviation of average distance value for both YAP‐WW1 domain mutants and APBB3‐WW domain complexes is relatively larger than that of the control YAP‐WW domain complex (Fig EV3F), indicating a substantial movement between SMAD7‐PY motif peptide and the YAP‐WW1 domain mutants as well as APPB3‐WW domain.
Taken together, these simulation analyses suggest that the identified 9‐amino acid sequence is involved in binding PY motif and maintaining a unique WW domain structure, which both determine the Hippo WW domain binding specificity with the known Hippo PY motif‐containing proteins.
Identification of STXBP4, a WW domain‐containing protein, whose WW domain fits the 9‐amino acid sequence criterion
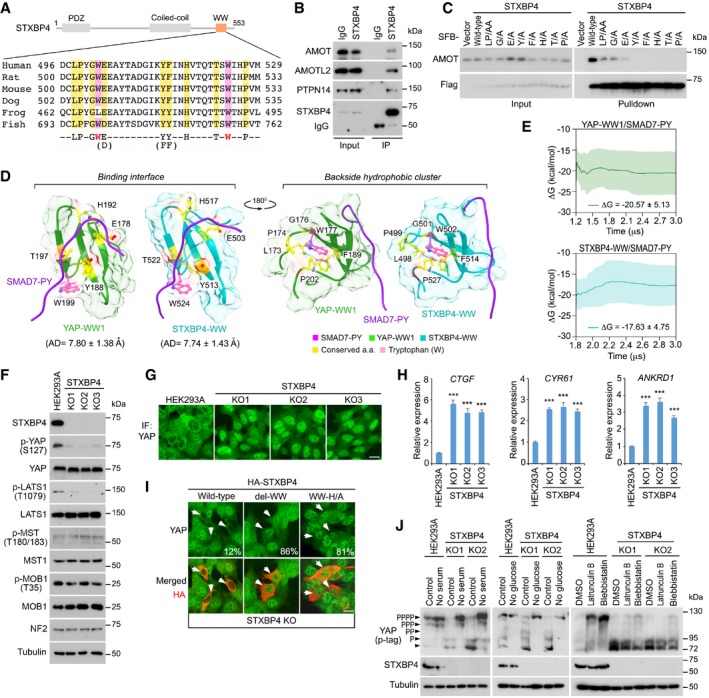
Next, we searched all the WW domain‐containing proteins in the human proteome and identified 12 WW domain‐containing proteins whose WW domains fit such a 9‐amino acid sequence (Fig EV4 and Table EV6). Among them, role of STXBP4 in the Hippo pathway regulation has not been fully characterized (Fig EV4). Although no STXBP4 ortholog is identified in Drosophila, this 9‐amino acid sequence of the STXBP4 WW domain was largely conserved in different species (Fig 3A). Interestingly, STXBP4 can form a complex with several Hippo PY motif‐containing regulators including AMOT, AMOTL2, and PTPN14 (Fig 3B). Mutating either of the conserved 9‐amino acid residues diminished the interaction between STXBP4 and AMOT (Fig 3C). As expected, the association between STXBP4 and these PY motif‐containing Hippo regulators is mediated by the WW domain of STXBP4 (Appendix Fig S4A) and the PY motif of these Hippo regulators (Appendix Fig S4B).
Figure EV4. Schematic illustration of the human proteome search for the WW domain‐containing proteins that fit the Hippo WW domain 9‐amino acid sequence criterion. (This figure is related to Fig 3; Table EV6).
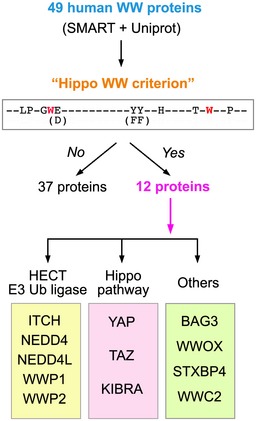
The identified 9‐amino acid sequence was subjected to the 49 WW domain‐containing proteins in human proteome. Total 12 WW domain‐containing proteins were found to contain the WW domains fitting the Hippo WW domain criterion.
Figure 3. STXBP4 is a Hippo pathway regulator, which contains a WW domain that fits the criterion of the Hippo WW domain binding specificity. (This figure is related to Fig EV4, Appendix Figs S4 and S5; Table EV6).
-
ASchematic illustration of STXBP4 protein, where the identified 9‐amino acid sequence of STXBP4‐WW domain was aligned across the indicated species.
-
BSTXBP4 forms a complex with several Hippo PY motif‐containing proteins. Immunoprecipitation was performed with STXBP4 antibody.
-
CThe identified 9‐amino acid sequence is required for the association between STXBP4 and AMOT. HEK293T cells were transfected with the indicated STXBP4 mutants and subjected to the pull‐down assay.
-
DStructural comparison between the YAP‐WW1/SMAD7‐PY and STXBP4‐WW/SMAD7‐PY complexes. The identified 9‐amino acid residues were indicated for both complexes.
-
EThe YAP‐WW1/SMAD7‐PY and STXBP4‐WW/SMAD7‐PY complexes show similar cumulative average trend and average binding free energy (ΔG) within standard deviation (the shaded region) of one another.
-
FLoss of STXBP4 inhibits YAP phosphorylation and LATS activation. Western blotting was performed with the indicated antibodies.
-
G, HLoss of STXBP4 activates YAP. STXBP4 deficiency promotes YAP nuclear translocation (G) and YAP downstream gene transcription (mean ± s.d., n = 3 biological replicates) (H). Scale bar, 20 μm. ***P < 0.001 (Student's t‐test).
-
IWW domain is required for the STXBP4‐mediated YAP cytoplasmic translocation. STXBP4 KO cells were transfected with the indicated STXBP4 constructs, and immunofluorescent staining was performed. HA‐positive cells (arrows) from ˜ 30 different views (˜ 200 cells in total) were randomly selected and quantified for YAP localization. Percentage of HA‐positive cells with nuclear YAP enrichment is shown. Scale bar, 20 μm.
-
JLoss of STXBP4 attenuates YAP phosphorylation as induced by actin cytoskeleton inhibition. The indicated cells were subjected to serum starvation (treatment with no‐serum medium for 12 h), glucose starvation (treatment with no‐glucose medium for 6 h), or actin inhibition (treatment with 0.5 μg/ml latrunculin B or 5 μM blebbistatin for 30 min). YAP phosphorylation was detected using phospho‐tag gel, where the YAP phosphorylation level was indicated.
Source data are available online for this figure.
To gain a structural insight into the STXBP4 WW domain, we compared STXBP4‐WW and YAP‐WW1 through ensemble molecular dynamics simulations and calculating binding free energies (ΔG) using the molecular mechanics Poisson–Boltzmann surface area (MM/PBSA) method. As shown in Appendix Fig S4C, the top 5 predicted clusters for the STXBP4‐WW/SMAD7‐PY complex is similar to those of the YAP‐WW1/SMAD7‐PY complex. By comparing the top one cluster for these two WW‐PY complexes, we found that the identified 9‐amino acid residues as well as the two tryptophan residues are similarly distributed within both the STXBP4‐WW/SMAD7‐PY and YAP‐WW1/SMAD7‐PY complexes, where they form as two groups to, respectively, involve in the binding with SMAD7‐PY motif and assemble a supportive backside hydrophobic cluster for each WW domain (Fig 3D). The average distance between STXBP4‐WW domain and SMAD7‐PY motif is close to that between YAP‐WW1 domain and SMAD7‐PY motif with a similarly low standard deviation value (Fig EV3F). Moreover, binding free energy (ΔG) from MM/PBSA calculations further indicates the similarity between YAP‐WW1 and STXBP4‐WW when they form as a complex with SMAD7‐PY motif peptide (Fig 3E).
Taken together, these data suggest that the STXBP4 WW domain possesses the Hippo WW domain binding specificity, endowing STXBP4 a potential role in the Hippo pathway.
STXBP4 is a negative regulator of YAP
To test the role of STXBP4 in regulation of the Hippo pathway, we examined YAP activation in the STXBP4 knockout (KO) cells (Appendix Fig S5). Interestingly, loss of STXBP4 significantly reduced YAP phosphorylation (Fig 3F), moved YAP into the nucleus (Fig 3G), and activated YAP downstream gene transcription (Fig 3H). Notably, either deleting the WW domain or mutating the histidine residue out of the identified 9‐amino acid sequence to alanine failed to rescue YAP's cytoplasmic localization (Fig 3I), suggesting that the WW domain is required for the STXBP4‐mediated YAP inhibition.
The observation that STXBP4 deficiency reduced YAP phosphorylation at S127 (Fig 3F) suggests that the Hippo pathway is inhibited in the STXBP4 KO cells. Indeed, as shown in Fig 3F, loss of STXBP4 suppressed LATS phosphorylation but did not affect that of MST or its substrate MOB1. These data suggest that STXBP4 is required for LATS activation in the Hippo pathway.
Multiple upstream signaling events have been identified to regulate the Hippo pathway (Yu et al, 2015). Next, we examined the signaling context for the STXBP4‐mediated Hippo pathway regulation. Interestingly, loss of STXBP4 attenuated YAP phosphorylation when actin cytoskeleton was either depolymerized or its tension was inhibited (Fig 3J), whereas YAP was still fully phosphorylated under serum‐ and glucose‐deprived conditions (Fig 3J). These data suggest that STXBP4 is involved in the actin cytoskeleton‐mediated Hippo pathway regulation.
STXBP4 is involved in a protein–protein interaction network comprising multiple Hippo pathway components and regulators
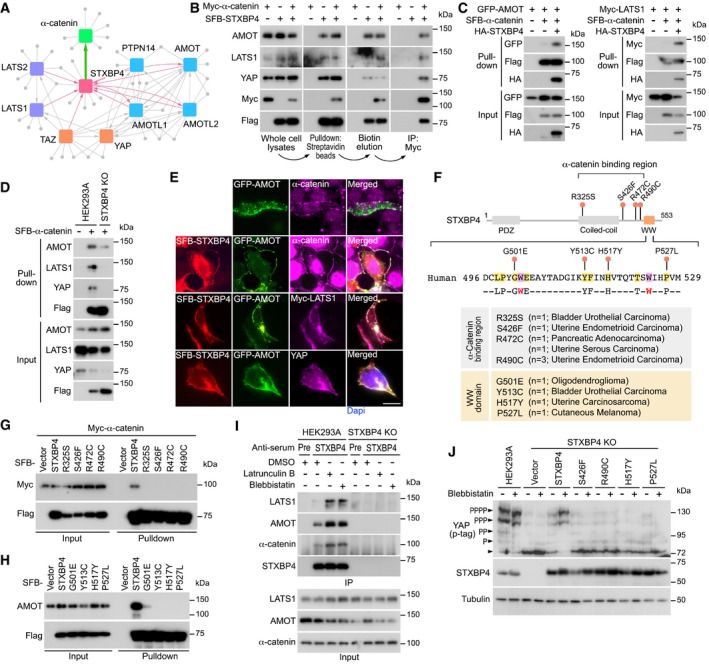
To elucidate the mechanism by which STXBP4 regulates the Hippo pathway, we purified the STXBP4‐associated protein complex and characterized its binding partners by mass spectrometry analysis. As shown in Fig 4A, all the AMOT family proteins were identified to form a complex with STXBP4, which is consistent with our previous findings (Fig 3B and Appendix Fig S4A). Interestingly, we also identified α‐catenin, a known Hippo upstream regulator (Schlegelmilch et al, 2011; Rauskolb et al, 2014; Feng et al, 2016; Vite et al, 2018), as a binding partner for STXBP4 (Fig 4A). STXBP4 was also reciprocally identified as a binding protein for some Hippo pathway components (e.g., LATS1, LATS2, TAZ) and regulators (e.g., AMOT, AMOTL1, AMOTL2, PTPN14) (Couzens et al, 2013; Huttlin et al, 2017; Wang et al, 2014) (Fig 4A). Collectively, these data suggest that STXBP4 involves in a protein–protein interaction network comprising a group of Hippo pathway components and regulators.
Figure 4. STXBP4 functions in the actin cytoskeleton tension‐mediated Hippo pathway regulation by forming a complex with α‐catenin and a group of Hippo PY motif‐containing proteins. (This figure is related to Appendix Figs S6 and S7; Table EV7).
-
AA summary map of cytoscape‐generated protein–protein interaction network for STXBP4, α‐catenin, and a group of Hippo pathway proteins.
-
BSTXBP4 forms a protein complex with α‐catenin and a group of Hippo pathway proteins.
-
CSTXBP4 promotes the association of α‐catenin with AMOT and LATS1. HEK293T cells were transfected with the indicated SFB‐tagged constructs and subjected to the pull‐down assay.
-
DLoss of STXBP4 diminishes the association of α‐catenin with AMOT, LATS1, and YAP. HEK293A and STXBP4 KO cells were transfected with the SFB‐tagged α‐catenin construct and subjected to the pull‐down assay.
-
ESTXBP4 induces the co‐localization between α‐catenin and AMOT as well as LATS1 and YAP. HEK293A cells were transfected with the indicated constructs, and immunofluorescence was performed. Scale bar, 20 μm.
-
F–HIdentification of several STXBP4 missense mutations that disrupt its interaction with α‐catenin and AMOT. The missense mutations within the STXBP4 α‐catenin‐binding region and the 9‐amino acid sequence of the STXBP4 WW domain were indicated and annotated (F). The identified missense mutations, respectively, disrupted the STXBP4‐ α‐catenin (G) and STXBP4‐AMOT (H) complex formation.
-
IInhibition of actin cytoskeleton promotes the STXBP4‐associated protein complex formation. HEK293A and the STXBP4 KO cells were subjected to immunoprecipitation using pre‐immune serum and anti‐STXBP4 serum under the indicated treatments.
-
JThe missense mutations of STXBP4 (F) diminished the ability of STXBP4 to rescue YAP phosphorylation in the STXBP4 KO cells with low actin cytoskeleton tension. YAP phosphorylation was detected using phospho‐tag gel, where the YAP phosphorylation level was indicated.
Source data are available online for this figure.
Notably, most of these STXBP4‐associated proteins are PY motif‐containing proteins (Fig 4A), suggesting that STXBP4 WW domain is required here. Since α‐catenin does not contain a PY motif, we further characterized the α‐catenin‐binding region in STXBP4. To achieve this, a series of STXBP4 truncation and deletion mutants were generated Appendix Fig S6A). As shown in Appendix Fig S6B, deletion of the 300–500 amino acid residues of STXBP4, but not its WW domain, fully abolished its association with α‐catenin. Moreover, we failed to further narrow down the α‐catenin‐binding region in STXBP4 (Appendix Fig S5C), suggesting that this identified 300–500 amino acid sequence region is required for its interaction with α‐catenin.
Taken together, these data indicate that STXBP4 can form a complex with several Hippo PY motif‐containing proteins and α‐catenin through its WW domain and the 300–500 amino acid sequence region, respectively.
STXBP4 functions as a scaffold protein to assemble a protein complex including α‐catenin AMOT, LATS, and YAP
To test this hypothesis, we performed a sequential pull‐down/immunoprecipitation assay using exogenously expressed SFB‐STXBP4 and Myc‐α‐catenin in HEK293T cells. As shown in Fig 4B, we first isolated STXBP4‐associated protein complex using streptavidin beads, eluted the complex with biotin, and purified the α‐catenin‐associated protein complex through immunoprecipitation. This sequential purification approach can help to characterize the proteins within the STXBP4/α‐catenin protein complex. Consistent with our proteomic data (Fig 4A), AMOT, LATS1, and YAP were all identified within the STXBP4/α‐catenin protein complex (Fig 4B).
Next, we examined the role of STXBP4 in this multi‐protein complex. Overexpression of STXBP4 induced the interaction of α‐catenin with both AMOT and LATS1 (Fig 4C), while loss of STXBP4 largely attenuated the association of α‐catenin with AMOT, LATS1, and YAP (Fig 4D). In addition, STXBP4 promoted the co‐localization between AMOT and α‐catenin onto cell adherens junction/membrane region, where both LATS1 and YAP were also identified (Fig 4E). These results suggest a scaffold role of STXBP4 in assembly of a protein complex containing at least α‐catenin, AMOT, LATS, and YAP at adherens junctions.
Both the WW domain and α‐catenin association are required for the STXBP4‐mediated YAP regulation
Given the potential tumor suppressive role of STXBP4 in targeting YAP, we next examined the genetic alteration of STXBP4 in the cBioportal database and found that STXBP4 alleles harbor a series of mutations within cancer patient samples (Appendix Fig S7A and Table EV7). Four missense mutations that are localized in the α‐catenin‐binding region (Fig 4F) disrupted the interaction between STXBP4 and α‐catenin (Fig 4G). As for the STXBP4 WW domain, four out of the identified nine amino acid residues were found mutated in oligodendroglioma (G501E), bladder urothelial carcinoma (Y513C), uterine carcinosarcoma (H517Y), and cutaneous melanoma (P527L), respectively (Fig 4F), and they all diminished the association between STXBP4 and AMOT (Fig 4H). Notably, these cancer‐derived missense mutations in either α‐catenin‐binding region or the WW domain of STXBP4 all failed to rescue YAP's cytoplasmic localization in the STXBP4 KO cells (Appendix Fig S7B), suggesting that association with α‐catenin and the Hippo PY motif‐containing components/regulators is required for the STXBP4‐dependent Hippo pathway regulation.
STXBP4 functions as a potential mechano‐transducer involved in actin cytoskeleton‐mediated Hippo pathway regulation
Notably, α‐catenin is known to play a critical role in mechanotransduction (Yonemura et al, 2010; Charras & Yap, 2018), and loss of STXBP4 significantly attenuated YAP phosphorylation upon disruption of actin cytoskeleton or inhibition of its tension (Fig 3J). Interestingly, depolymerization of actin cytoskeleton by latrunculin B or inhibition of its tension by blebbistatin induced the association of STXBP4 with LATS1, AMOT, and α‐catenin (Fig 4I). Reconstitution of STXBP4, but not its mutants with missense mutations at its α‐catenin‐binding region and WW domain (Fig 4F), significantly rescued YAP phosphorylation when actin cytoskeleton tension was inhibited (Fig 4J). These data indicate that the STXBP4‐mediated protein complex formation with α‐catenin and the Hippo PY motif‐containing proteins plays a role in actin cytoskeleton‐dependent regulation of the Hippo pathway.
STXBP4 is frequently downregulated in kidney cancer and correlated with YAP activation
By analyzing the cancer database, FireBrowse, a platform developed to analyze 14,729 tumor sample data generated by The Cancer Genome Atlas (TCGA), we found that the mRNA level of STXBP4 was downregulated in all the listed kidney cancer subtypes (Fig 5A). This finding was further confirmed through a kidney tissue microarray analysis, where the expression of STXBP4 was found decreased in several types of human kidney cancer: 84.8% clear cell carcinoma, 100% papillary renal cell carcinoma, 50% chromophobe carcinoma, 66.7% carcinoma sarcomatodes, and 50% high‐grade urothelial carcinoma of renal pelvis (Fig 5B). However, downregulation of STXBP4 was only observed in 10% normal kidney tissue (Fig 5B), suggesting an inverse correlation between STXBP4 expression and kidney cancer formation (P = 2.9 × 10−20, R = −0.41). Moreover, our TCGA data analysis indicated that low expression of STXBP4 was significantly correlated with the poor overall survival rate for the cancer patients with clear cell renal cell carcinoma (ccRCC) (Fig 5C), indicating that STXBP4 is a potential tumor suppressor in ccRCC.
Figure 5. STXBP4 is a tumor suppressor in human kidney cancer (This figure is related to Appendix Fig S8).
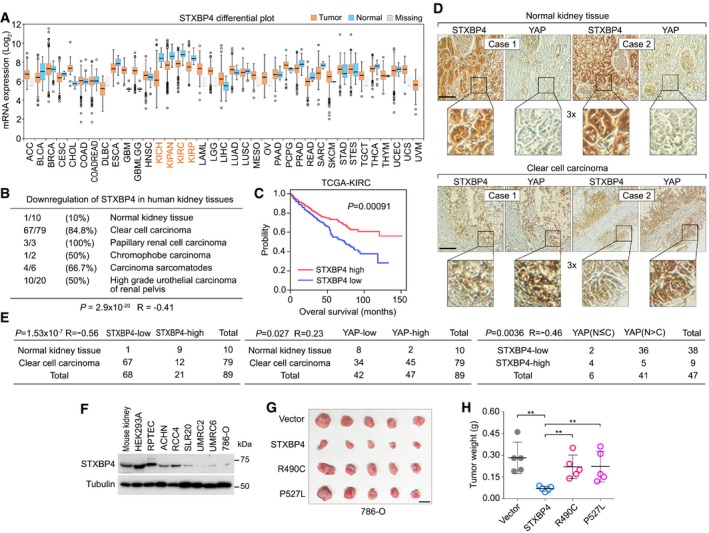
-
A, BSTXBP4 is downregulated in human kidney cancer. The mRNA level of STXBP4 is analyzed in the FireBrowse web database (http://firebrowse.org) (A), where 14,729 tumor sample data generated by TCGA were included. The first quartile, median, and third quartile values were indicated as the boxplots. Outliers were plotted as individual points. Error bars indicated the standard deviation above and below the mean of the data. The expression of STXBP4 was also examined using kidney tissue microarray, where percentage of the indicated tissue samples with downregulated STXBP4 was shown (B). The P value was calculated by using the paired Student's t‐test.
-
CKaplan–Meier curves of overall survival of patients with ccRCC are stratified by STXBP4 expression level. Clinical data of STXBP4 were analyzed in TCGA‐KIRC project containing total 611 patient samples. The P value was calculated by using the Log‐rank (Mantel‐Cox) test.
-
DImmunohistochemical staining of STXBP4 and YAP was performed in a kidney cancer tissue microarray, where the indicated regions in the box were shown three times enlarged. Brown staining indicates positive immunoreactivity. Scale bar, 100 μm.
-
ECorrelation analyses between STXBP4 and YAP in human normal kidney and clear cell carcinoma samples are shown as tables. Statistical significance was determined by chi‐square test. R, correlation coefficient. N, nuclear localization. C, cytoplasmic localization.
-
FSTXBP4 expression is examined in a panel of ccRCC cell lines by Western blotting.
-
G, HBoth the association with α‐catenin and the functional WW domain are required for the STXBP4's tumor suppressive function in 786‐O cells. Overexpression of STXBP4, but not the indicated STXBP4 missense mutants, significantly suppressed the 786‐O cell xenograft tumor formation. Xenograft tumors are shown in (G), and the tumor weight is quantified in (H) (n = 5 mice, mean ± s.d.). **P < 0.01 (Student's t‐test). Scale bar, 1 cm.
Source data are available online for this figure.
YAP is highly expressed and activated in multiple major human cancer types, but genetic mutation for the Hippo pathway components is hardly detected (Jiang et al, 2015), suggesting that additional oncogenic alterations could lead to YAP activation for tumorigenesis. Since loss of STXBP4 activated YAP (Fig 3F–H), we next examined the pathological correlation between STXBP4 and YAP using a kidney cancer tissue microarray. Consistent with previous studies (Cao et al, 2014; Schutte et al, 2014; Godlewski et al, 2018), upregulation of YAP was observed in 57% (45 of 79) of ccRCC tissue samples, while only 20% (2 of 10) of normal kidney tissues showed high YAP expression (Fig 5D and E). Moreover, an inverse correlation between STXBP4 expression and YAP nuclear enrichment was found in the tissue samples with high YAP expression (P = 0.0036, R = −0.46), where 94.7% (36 of 38) of the tested tissue samples with low STXBP4 expression had high nuclear enrichment of YAP (Fig 5D and E). However, there were still 10.6% (5 of 47) of the total tested specimens showing high STXBP4 expression but YAP nuclear enrichment (Fig 5E). These results indicate that downregulation of STXBP4 may contribute to YAP activation in a substantial fraction of ccRCC; however, YAP can still be activated in other tumors via different mechanisms.
Interestingly, although a general low expression of YAP was found in normal kidney tissues, we were still able to observe a relatively high expression of YAP in the podocytes of glomerulus region and partially in the convoluted tubule region (Fig 5D). Even though, these YAP highly expressed normal kidney regions still consistently showed a decreased STXBP4 expression level (Fig 5D), suggesting that their inverse correlation in expression could involve in normal kidney physiology.
Both the α‐catenin association and functional WW domain are required for the STXBP4's tumor suppressive function in kidney cancer
To investigate the role of STXBP4 in kidney cancer, we first determined the STXBP4 expression in normal mouse kidney tissue and a group of human kidney‐related cell lines. Interestingly, STXBP4 had an abundant expression in mouse kidney tissue, an embryonic kidney immortalized cell line HEK293A and an immortalized human renal proximal tubular epithelial cell line RPTEC (Fig 5F). In contrast, STXBP4 showed moderate or low expressions in all the tested ccRCC cell lines (Fig 5F), where YAP was found majorly localized in the nucleus (Appendix Fig S8A). Overexpression of STXBP4, but not its two patient‐derived missense mutants (R490C and P527L) (Fig 4F), in a ccRCC cell line 786‐O (Appendix Fig S8B), significantly suppressed the xenograft tumor formation (Fig 5G and H). Since the R490C and P527L mutations can, respectively, disrupt the STXBP4's interaction with α‐catenin (Fig 4G) and AMOT (Fig 4H), these results indicate that the association with α‐catenin and a functional WW domain are both required for STXBP4's tumor suppressive function.
Discussion
In this study, we identified a conserved 9‐amino acid sequence within the WW domain of the Hippo pathway components and regulators (Fig 2), which is required for the specific Hippo WW‐PY complex formation. Notably, this identified 9‐amino acid sequence has at least one residue altered in all the tested control WW domain‐containing proteins (Figs 2B and EV2A), which could help to explain why these control WW domain‐containing proteins fail to interact with the Hippo PY motif‐containing proteins (Fig 1E and F). Since the “WW‐PY” recognition is widely present in the Hippo pathway, manipulation of their recognition is likely to control the outputs of this key signaling pathway in tissue/organ growth and tumorigenesis. Thus, it would be highly exciting if this Hippo WW domain determinants could be utilized for the development of small molecules or peptides to precisely modulate YAP/TAZ activity in cancer therapy and tissue repair.
Mechanistically, the identified 9‐amino acid sequence accounts for both a suitable WW domain structure and the binding interface with the PY motif peptide (Fig EV3A–C), providing a structural basis for the Hippo WW domain binding specificity. Here, our study is only focused on the individual WW domain binding property. Actually, the mechanism underlying the specific “WW‐PY” recognition could be more complicated given the role of WW tandem in mediating PY motif binding (Lin et al, 2019) and the potential homo‐ and hetero‐dimer formations among WW domains (Sudol & Harvey, 2010). Moreover, although our current study mostly focused on the WW domain, it is highly possible that its cognate PY motif ligand could also contribute to the specific Hippo “WW‐PY” recognition. However, the PY motif is relatively short, flexible and could be easily buried into a higher level of protein structure, making it difficult to assess its role at a protein level. Thus, we did not further address this question from the PY motif‐based perspective.
Among the Hippo pathway components, the SAV1 WW domain functions differently from that of YAP, TAZ, and KIBRA to bind Hippo PY motif‐containing proteins (Fig 1). This difference may arise from the change of one conserved glutamate residue in the identified 9‐amino acid sequence for the SAV1 WW domain in both human (Fig 2A and B) and Drosophila (Appendix Fig S2). Based on our E/D substitution data (Appendix Fig S1B) and the structural analysis (Fig EV3C), the negative charge for this residue position could be essential. Interestingly, the substituted serine residue within the human SAV1 WW domain can be phosphorylated in vivo (http://www.phosphosite.org), suggesting that the association between SAV1 and Hippo PY motif‐containing proteins could be regulated through a yet‐to‐be characterized phosphorylation event.
There are only a few WW domain‐containing proteins, whose WW domains fit such 9‐amino acid sequence in human proteome (Fig EV4 and Table EV6). Among them, STXBP4 was found as a negative regulator for YAP (Fig 3F–H) by forming a protein complex with a series of Hippo PY motif‐containing proteins and an adherens junction component, α‐catenin (Fig 4A). Interestingly, STXBP4 serves as a scaffold protein in this network and transduces actin‐based mechanical cues to regulate the Hippo pathway. Since α‐catenin is known to play a role in both cell density and cytoskeleton tension‐dependent regulation of YAP (Schlegelmilch et al, 2011; Rauskolb et al, 2014; Feng et al, 2016; Vite et al, 2018), our findings provided molecular insights into its downstream signaling events. Under the condition with low actin cytoskeleton tension, STXBP4 recruits several Hippo PY motif‐containing proteins including at least AMOT, LATS to form a complex with α‐catenin at adherens junction. YAP/TAZ are also within this complex based on their interaction with AMOT and LATS (Fig EV5). In proximity, LATS phosphorylates and inhibits YAP. When mechanical cues increase actin cytoskeleton tension, both the adherens junction‐associated α‐catenin and the filament actin‐bound AMOT would be affected in their conformation, resulting in the protein complex disassembly and YAP activation (Fig EV5). Exactly how this α‐catenin‐STXBP4‐Hippo PY proteins axis is coordinated with other related signaling events (Low et al, 2014; Qiao et al, 2017; Dutta et al, 2018) in regulating the interplay between actin cytoskeleton and the Hippo‐YAP/TAZ pathway deserves further investigation.
Figure EV5. A proposed model for the STXBP4‐mediated Hippo pathway regulation in response to actin cytoskeleton tension change. (This figure is related to Figs 4 and 5).
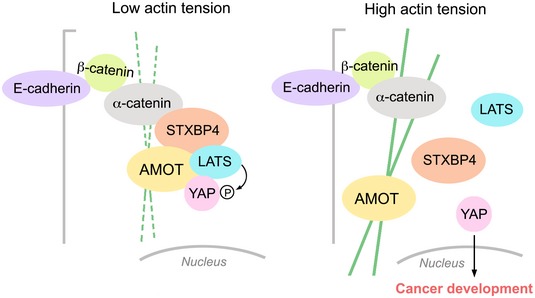
Under low actin tension, STXBP4 assembles a protein complex comprising α‐catenin, AMOT, LATS, and YAP to promote YAP phosphorylation and cytoplasmic retention. When actin cytoskeleton tension increases, the STXBP4‐centered protein complex is dissembled, resulting in YAP's dephosphorylation and nuclear translocation as well as the subsequent cancer development.
Intriguingly, our TCGA database and tissue microarray studies suggested that STXBP4 is a potential tumor suppressor in kidney cancer (Fig 5A–C) and its downregulation is significantly correlated with YAP activation in ccRCC tissues (Fig 5D and E). YAP has been found highly expressed and activated in human kidney cancer including ccRCC (Cao et al, 2014; Schutte et al, 2014; Godlewski et al, 2018). Here, our study identified a pathological relevance between STXBP4 and YAP, providing a potential mechanism for the YAP activation in ccRCC. Notably, a CpG island was identified in the STXBP4 promoter, suggesting that the loss of STXBP4 could occur due to its promoter methylation. In addition, STXBP4 gene alleles harbor a relative high mutation rate (13.45%) including nonsense mutation (6.92%), frameshift deletion (1.92%), in frameshift deletion (0.38%), and gene fusion (4.23%) (Appendix Fig S7A), which could also partially explain the loss of STXBP4 in cancer.
STXBP4 is originally identified as an insulin‐regulated protein involved in GLUT4‐mediated glucose transport in adipocyte (Min et al, 1999) and functions as an inhibitory protein for the SNARE complex‐dependent membrane fusion (Yu et al, 2013). Dysregulated STXBP4 expression was associated with some SNPs in breast cancer (Caswell et al, 2015; Darabi et al, 2016; Masoodi et al, 2017). Recent studies also implicated the role of STXBP4 in squamous cell carcinomas, by regulating N‐terminally truncated isoform of p63 (ΔNp63) (Otaka et al, 2017; Rokudai et al, 2018). Together with these studies, our findings in kidney cancer suggested a complex role of STXBP4 in cancer development, which could depend on tissue context.
Materials and Methods
Antibodies and chemicals
For Western blotting, anti‐α‐tubulin (T6199‐200UL, 1:5,000 dilution), anti‐Flag (M2) (F3165‐5MG, 1:5,000 dilution), and anti‐AMOTL1 (HPA001196, 1:1,000 dilution) antibodies were obtained from Sigma‐Aldrich. Anti‐Myc (sc‐40, 1:500 dilution) and anti‐GFP (sc‐9996, 1:1,000 dilution) antibodies were purchased from Santa Cruz Biotechnology. Anti‐phospho‐YAP (S127) (4911S, 1:1,000 dilution), anti‐phospho‐LATS1 (Thr1079) (8654S, 1:1,000 dilution), anti‐LATS1 (3477S, 1:1,000 dilution), anti‐phospho‐MST (Thr180/Thr183) (3681S, 1:1,000 dilution), anti‐MST1 (3682S, 1:1,000 dilution), anti‐phospho‐MOB1 (Thr35) (8699S, 1:1,000 dilution), anti‐MOB1 (3863S, 1:2,000 dilution), and anti‐NF2 (12896S, 1:2,000 dilution) antibodies were purchased from Cell Signaling Technology. The AMOT, AMOTL2, PTPN14, and YAP polyclonal antibodies were generated as previously described (Wang et al, 2011, 2012b). The STXBP4 antiserum was raised against MBP‐STXBP4 (the 251–553 amino acid residues), and polyclonal antibody was affinity‐purified using an AminoLink Plus Immobilization and Purification Kit (Pierce).
For immunostaining, an anti‐YAP (sc‐101199, 1:200 dilution) monoclonal antibody was purchased from Santa Cruz Biotechnology. Anti‐hemagglutinin (HA) polyclonal antibody (3724S, 1:3,000 dilution) was obtained from Cell Signaling Technology.
For immunohistochemical staining, an anti‐YAP (14074S, 1:15 dilution) monoclonal antibody was purchased from Cell Signaling Technology. The STXBP4 antiserum was raised against MBP‐STXBP4 (the 1–250 amino acid residues), and polyclonal antibody (1:200 dilution) was affinity‐purified using an AminoLink Plus Immobilization and Purification Kit (Pierce).
Latrunculin B and blebbistatin were obtained from Sigma‐Aldrich.
Constructs and viruses
Plasmids encoding the indicated genes were obtained from the Human ORFeome V5.1 library or purchased from Harvard Plasmid DNA Resource Core and Dharmacon. All constructs were generated via polymerase chain reaction (PCR) and subcloned into a pDONOR201 vector using Gateway Technology (Invitrogen) as the entry clones. For tandem affinity purification, all entry clones were subsequently recombined into a lentiviral Gateway‐compatible destination vector for the expression of C‐terminal SFB‐tagged fusion proteins. Gateway‐compatible destination vectors with the indicated SFB tag, HA tag, GFP tag, or Myc tag were used to express various fusion proteins. PCR‐mediated mutagenesis was used to generate all the indicated site mutations and internal region/domain deletion mutations.
All lentiviral supernatants were generated by transient transfection of HEK293T cells with the helper plasmids pSPAX2 and pMD2G (kindly provided by Dr. Zhou Songyang, Baylor College of Medicine) and harvested 48 h later. Supernatants were passed through a 0.45‐μm filter and used to infect cells with the addition of 8 μg/ml hexadimethrine bromide (Polybrene) (Sigma‐Aldrich).
Cell culture and transfection
HEK293T, ACHN, SLR20, and UMRC6 cell lines were purchased from ATCC and kindly provided by Drs. Boyi Gan and Junjie Chen (MD Anderson Cancer Center). HEK293A cells were purchased from Thermo Fisher and kindly provided by Dr. Jae‐Il Park (MD Anderson Cancer Center). RPTEC, 786‐O, RCC4, and UMRC2 cells were purchased from ATCC and kindly provided by Dr. Olga Razorenova (University of California, Irvine). HEK293T, HEK293A, RCC4, UMRC2, and UMRC6 cells were maintained in Dulbecco's modified essential medium (DMEM) supplemented with 10% fetal bovine serum at 37°C in 5% CO2 (v/v). SLR20, and 786‐O cells were grown in RPMI‐1640 medium supplemented with 10% fetal bovine serum at 37°C in 5% CO2 (v/v). RPTEC cells were maintained in DMEM/F12 medium supplemented with 5 pM triiodo‐l‐thyronine, 10 ng/ml epidermal growth factor, 3.5 μg/ml ascorbic acid, 5 μg/ml transferrin, 5 μg/ml insulin, 25 ng/ml prostaglandin E1, 25 ng/ml hydrocortisone, 8.65 ng/ml sodium selenite, and 1.2 mg/ml sodium bicarbonate at 37°C in 5% CO2 (v/v). All the culture media contain 1% penicillin and streptomycin.
Plasmid transfection was performed using a polyethylenimine reagent.
Immunofluorescent staining
Immunofluorescent staining was performed as described previously (Wang et al, 2008) with minor modifications. Briefly, cells cultured on coverslips were fixed with 4% paraformaldehyde for 10 min at room temperature and then extracted with 0.5% Triton X‐100 solution for 5 min. For α‐catenin‐related immunofluorescent staining, cells were pretreated with PBS solution containing 0.5% Trion X‐100 and 1% paraformaldehyde for 4 min, and subjected to 4% paraformaldehyde fixation. After blocking with Tris‐buffered saline with Tween‐20 containing 1% bovine serum albumin, the cells were incubated with the indicated primary antibodies for 1 h at room temperature. After that, the cells were washed and incubated with fluorescein isothiocyanate‐, rhodamine‐, and Cy5‐conjugated secondary antibodies for 1 h. Cells were counterstained with 100 ng/ml 4′,6‐diamidino‐2‐phenylindole (DAPI) for 2 min to visualize nuclear DNA. The coverslips were mounted onto glass slides with an anti‐fade solution and visualized under a Nikon Eclipse Ti spinning‐disk confocal microscope.
Tandem affinity purification (TAP) of SFB‐tagged protein complexes
HEK293T cells stably expressing the indicated SFB‐tagged proteins were selected by culturing in medium containing 2 μg/ml puromycin and confirmed by immunostaining and Western blotting as described previously (Wang et al, 2014). For TAP, HEK293T cells were lysed in NETN buffer (with protease and phosphatase inhibitors) at 4°C for 20 min. The crude lysates were centrifuged at 15,700 g for 15 min at 4°C. The supernatants were incubated with streptavidin‐conjugated beads (GE Healthcare) for 1 h at 4°C. The beads were washed three times with NETN buffer, and bound proteins were eluted with NETN buffer containing 2 mg/ml biotin (Sigma‐Aldrich) for 2 h at 4°C. The elutes were incubated with S protein beads (Novagen) for 1 h. The beads were washed three times with NETN buffer and subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis. Each pull‐down sample was run just into the separation gel so that the whole bands could be excised as one sample and subjected to in‐gel trypsin digestion and MS analysis.
Mass spectrometry (MS) analysis
The mass spectrometry was performed as described previously (Wang et al, 2014; Tavana et al, 2016). Briefly, the excised gel bands described above were cut into approximately 1‐mm3 pieces. The gel pieces were then subjected to in‐gel trypsin digestion (Shevchenko et al, 1996) and dried. Samples were reconstituted in 5 μl of high‐performance liquid chromatography (HPLC) solvent A (2.5% acetonitrile, 0.1% formic acid). A nanoscale reverse‐phase HPLC capillary column was created by packing 5‐μm C18 spherical silica beads into a fused silica capillary (100 μm inner diameter × ~20 cm length) with a flame‐drawn tip. After the column was equilibrated, each sample was loaded onto the column via a Famos autosampler (LC Packings). A gradient was formed, and peptides were eluted with increasing concentrations of solvent B (97.5% acetonitrile, 0.1% formic acid).
As the peptides eluted, they were subjected to electrospray ionization and then entered into an LTQ‐Velos mass spectrometer (Thermo Fisher Scientific). The peptides were detected, isolated, and fragmented to produce a tandem mass spectrum of specific fragment ions for each peptide. Peptide sequences (and hence protein identity) were determined by matching protein databases with the fragmentation pattern acquired by the software program SEQUEST (ver. 28) (Thermo Fisher Scientific). Enzyme specificity was set to partially tryptic with two missed cleavages. Modifications included carboxyamidomethyl (cysteine, fixed) and oxidation (methionine, variable). Mass tolerance was set to 0.5 Da for precursor ions and fragment ions. The database searched was UniProt. Spectral matches were filtered to contain a false discovery rate of less than 1% at the peptide level using the target‐decoy method (Elias & Gygi, 2007), and the protein inference was considered followed the general rules (Nesvizhskii & Aebersold, 2005), with manual annotation based on experiences applied when necessary. This same principle was used for isoforms when they were present in the database. The longest isoform was reported as the match.
Bioinformatic analysis
The full‐length YAP, TAZ, SAV1, and KIBRA dataset was retrieved from a previous study (Wang et al, 2014). The TAP‐MS dataset for a group of full‐length WW domain‐containing proteins randomly selected from human proteome, and the WW domains isolated from these proteins as well as the four Hippo pathway WW components (YAP, TAZ, SAV1, and KIBRA) were newly generated in this study. We combined these two datasets and assigned quality scores to the identified protein–protein interactions using MUSE algorithm as previously described (Li et al, 2016), where a group of unrelated TAP‐MS experiments (1,806 experiments using stably expressed TAP‐tagged protein baits and 20 experiments using empty vector baits) were included as a control group. Through it, we considered any interaction with a MUSE score of at least 0.9 and raw spectra count greater than 1 to be a high‐confident interacting protein (HCIP). The overall HCIP reproducibility rate was close to 85%, which increased when the cutoff peptide number increased. The full‐length WW domain‐containing proteins and their corresponded WW domains shared 47.5% HCIPs, and only 10.2% overlapped HCIPs were identified for the WW domains isolated from the Hippo WW domain‐containing components and the control ones (Fig EV1C).
The WW domain‐containing proteins' interactomes were enriched in signaling pathways, biological processes, and diseases using the HCIPs identified in our studies. The P values were estimated using the Knowledge Base provided by Ingenuity Pathway software (Ingenuity Systems, http://www.ingenuity.com), which contains findings and annotations from multiple sources including the Gene Ontology database, KEGG pathway database, and Panther pathway database. Only statistically significant correlations (P < 0.05) are shown. The –log (P value) for each function and related HCIPs is listed.
Screen of human WW domain‐containing proteins using the identified Hippo WW domain binding criterion
All the WW domain‐containing proteins were retrieved from human proteome using a Simple Modular Architecture Research Tool (SMART) (http://smart.embl-heidelberg.de), and the WW domain‐containing protein list was further refined in UniProt (https://www.uniprot.org). Based on the definition, the WW domain‐containing proteins are defaulted with two tryptophan (W) residues as separated by 20–22 amino acids within the sequence. All the WW domain sequences were downloaded from UniProt and subjected to scan with the identified 9‐amino acid sequence manually. The list of all the human WW domain‐containing proteins and the searching result are listed in Table EV6.
Gene inactivation by CRISPR/Cas9 system
To generate the STXBP4 knockout cells, five distinct single‐guide RNAs (sgRNA) were designed by CHOPCHOP website (https://chopchop.rc.fas.harvard.edu), cloned into lentiGuide‐Puro vector (Addgene plasmid # 52963), and transfected into HEK293A cells with lentiCas9‐Blast construct (Addgene plasmid # 52962). The next day, cells were selected with puromycin (2 μg/ml) for 2 days and subcloned to form single colonies. Knockout cell clones were screened by Western blotting to verify the loss of STXBP4 expression, and their genomic editing was further confirmed by sequencing (Appendix Fig S5).
The sequence information for sgRNAs used for STXBP4 knockout cell generation is as follows:
STXBP4_sgRNA1: AGACTTAATGTTGAGGCTTG;
STXBP4_sgRNA2: GGCTTGGTGTTGTTCCTTTG;
STXBP4_sgRNA3: TGCTTTCACCAAAGTAGCCT;
STXBP4_sgRNA4: GGAAACAGGCCTTGGCCTGA;
STXBP4_sgRNA5: AGGTACTAGGAGGAATTAAC.
RNA extraction, reverse transcription, and real‐time PCR
RNA samples were extracted with TRIzol reagent (Invitrogen). Reverse transcription assay was performed using the Script Reverse Transcription Supermix Kit (Bio‐Rad) according to the manufacturer's instructions. Real‐time PCR was performed using Power SYBR Green PCR master mix (Applied Biosystems). For quantification of gene expression, the 2−ΔΔCt method was used. GAPDH expression was used for normalization.
The sequence information for each primer used for gene expression analysis is as follows:
CTGF‐Forward: 5′‐CCAATGACAACGCCTCCTG‐3′;
CTGF‐Reverse: 5′‐GAGCTTTCTGGCTGCACCA‐3′;
CYR61‐Forward: 5′‐AGCCTCGCATCCTATACAACC‐3′;
CYR61‐Reverse: 5′‐GAGTGCCGCCTTGTGAAAGAA‐3′;
ANKRD1‐Forward: 5′‐CACTTCTAGCCCACCCTGTGA‐3′;
ANKRD1‐Reverse: 5′‐CCACAGGTTCCGTAATGATTT‐3′;
GAPDH‐Forward: 5′‐ATGGGGAAGGTGAAGGTCG‐3′;
GAPDH‐Reverse: 5′‐GGGGTCATTGATGGCAACAATA‐3′.
Molecular dynamics simulations
All simulations were conducted using the AMBER18 molecular dynamics suite (Case et al, 2005, 2017; Gotz et al, 2012; Salomon‐Ferrer et al, 2013). Initial parameterization of complexes and apo conformations was conducted with the LeAP module in AMBER18, using the protein force field ff14SB (Maier et al, 2015). YAP‐WW1 domain bound to SMAD7‐PY motif‐containing peptide was initially parameterized using the PDB structure, 2LTW. The SMAD7‐PY motif‐containing peptide structure was removed from 2LTW and docked to the STXBP4‐WW domain (PDB: 2YSG) to form a complex (STXBP4‐WW/SMAD7‐PY). In the N‐terminal sequence of STXBP4, four non‐native residues (GSSG) were removed prior to docking and formation of the complex STXBP4‐WW/SMAD7‐PY to maintain consistent residue number with the YAP‐WW1 domain. To generate the mutant complexes, all the conserved residues from 2LTW were mutated into alanine using MODELLER v9.21 (Sali & Blundell, 1993; Fiser et al, 2000; Marti‐Renom et al, 2000; Webb & Sali, 2016), and initial docked poses between mutated YAP‐WW1 domains and SMAD7 were generated using the HADDOCK docking program (Dominguez et al, 2003; de Vries et al, 2007) prior to simulation (Appendix Table S1). This docking procedure was also repeated for the APBB3‐WW/SMAD7‐PY simulations. An apo form of SMAD7‐PY and YAP‐WW1 (wild‐type domain mutants: L173A/P174A, G176A, W177A, E178A, Y188A, F189A, H192A, T197A, W199A, P202A) was also derived from PDB structure 2LTW, for simulations (Appendix Table S1).
Neutralized with either Na+ or Cl− counter ions, systems were solvated using a 10 Å buffer of TIP3P waters in a truncated octahedron box. All complexes and apo forms were minimized in a two‐step process using the PMEMD program to remove any steric clashes and overlaps. Complexes were heated to 300K for 100 ps in the canonical (NVT) ensemble and equilibrated for 10 ns at 300K in the isothermal‐isobaric (NPT) ensemble. Production runs were generated using the accelerated CUDA version of PMEMD (Gotz et al, 2012; Salomon‐Ferrer et al, 2013) in the NVT ensemble with 2‐fs time steps at 300K, until MM/PBSA calculations converged. Appendix Table S1 outlines the complete simulation conditions for each complex and apo structure.
The MM/PBSA module in AMBER18 (Wang et al, 2006, 2012a, 2016, 2017; Cai et al, 2010; Wang & Luo, 2010; Miller et al, 2012; Botello‐Smith & Luo, 2015) was employed to calculate the binding free energies (∆G) of wild‐type and mutant complexes. Calculations do not take into consideration entropy; however, all complexes retain SMAD7‐PY as a common binder meeting the necessary requirements for MM/PBSA calculation and comparison. Convergence of both YAP‐WW1 and STXBP4‐WW complex simulations was determined via cumulative average calculations of ∆G values and timeframes for all subsequent analyses (e.g., clustering, averaging, and RMSD) of each complex were determined based on this metric.
Utilizing the AMBER post‐processing program (CPPTRAJ) (Roe & Cheatham, 2013) module in the AMBER18 package, clustering was performed for each complex using only the Cα atoms in SMAD7‐PY motif‐containing peptide. We chose to cluster using SMAD7‐PY motif‐containing peptide that coordinates upon observation of the relative stability of both wild‐type YAP‐WW1 and STXBP4‐WW domains. For wild‐type complexes (YAP‐WW1 or STXBP4‐WW bound to SMAD7‐PY), all frames were incorporated to generate representative clusters, and only the top five clusters are displayed (Appendix Fig S4C). Conformations were clustered using the hierarchical agglomerative clustering algorithm (average‐linkage), with 2.33 Å criteria set as the minimum distance between clusters. Average structures were calculated from only converged timeframes indicated in Appendix Table S1. Using only Cα atoms, the conformation with the smallest RMSD to the average structure was used to represent the average conformation (Figs EV3A and E, and 3D). Hydrogen bonds were quantified using the Baker‐Hubbard (Baker & Hubbard, 1984) criteria and the MDTraj (McGibbon et al, 2015) python module. Ionic salt bridge interactions were determined with a distance criterion (Barlow & Thornton, 1983) (6 Å) between centers of charged groups (positively charged atoms from basic residues Arg, Lys, His: NH*, NZ*, NE2; regions of partial positive charge from His: NE2, HE*, CE1, HD2; negatively charged atoms from acidic residues Glu and Asp: OE*, OD*). Hydrophobic interactions were also measured via a distance criterion of 3.9 Å between carbon atoms. Initially identified in WT YAP‐WW1/SMAD7‐PY simulations, four intermolecular residue pairs (P208‐W199, P209‐T197, Y211‐H192, P209‐Y188) and their Cα atoms were used to calculate the average distance (AD) values in frames outlined in the simulation conditions table (Fig EV3F and Appendix Table S1). This AD calculation procedure was repeated for all complex simulations (SMAD7‐PY bound to YAP‐WW1 mutants, STXBP4‐WW, and APBB3‐WW), with Cα atoms of residues in equivalent positions of YAP‐WW1 residues.
Xenograft assays
Athymic nude (nu/nu) mouse strain was used for the xenograft tumor assay in this study. Four‐week‐old female nude mice were purchased from Jackson Laboratory (002019) and kept in a pathogen‐free environment. The xenograft tumor experiments were followed institutional guidelines, approved by the Institutional Animal Care and Use Committee of the University of California, Irvine, and performed under veterinary supervision. The indicated 786‐O cells (2 × 106) were subcutaneously injected into the nude mice. After 60 days' adaptation, mice were euthanized, and tumor weights were analyzed.
Immunohistochemical analysis
The kidney tissue array (BC07115a) was purchased from US Biomax, Inc. According to the Declaration of Specimen Collection provided by US Biomax, each specimen collected from any clinic was consented by both hospital and individual.
The kidney tissue array was deparaffinized and rehydrated. The antigens were retrieved by applying Unmask Solution (Vector Laboratories) in a steamer for 40 min. To block endogenous peroxidase activity, the sections were treated with 3% hydrogen peroxide for 30 min. After 1 h of pre‐incubation in 10% goat serum to prevent non‐specific staining, the samples were incubated with an antibody at 4°C overnight. The sections were incubated with SignalStain Boost detection reagent at room temperature for 30 min. Color was developed with SignalStain 3,3′‐diaminobenzidine chromogen‐diluted solution (all reagents were obtained from Cell Signaling Technology). Sections were counterstained with Mayer hematoxylin. To quantify the results, a total score of protein expression was calculated from both the percentage of immunopositive cells and immunostaining intensity. High and low protein expressions were defined using the mean score of all samples as a cutoff point. Pearson chi‐square analysis test was used for statistical analysis of the correlation of STXBP4 with tissue type (normal vs. cancer) and the correlation between STXBP4 and YAP.
TCGA database analysis
Dataset for STXBP4 was downloaded from the Cancer Genome Atlas (TCGA) data portal (https://portal.gdc.cancer.gov/). The mRNA expression and clinical data of STXBP4 were analyzed in TCGA‐KIRC project. The mRNA levels of STXBP4 were categorized into high and low expression groups based on the median value. The correlation between STXBP4 expression and patient survival rate was analyzed. Total 611 patient samples were analyzed.
Quantification and statistical analysis
Each experiment was repeated twice or more, unless otherwise noted. There were no samples or animals excluded for the analyses in this study. As for the mouse experiments, there was no statistical method used to predetermine sample size. We assigned the animals randomly to different groups. A laboratory technician was blinded to the group allocation and tumor collections during the animal experiments as well as the data analyses. Student's t‐test was used to analyze the differences between groups. Data were analyzed by Student's t‐test or Pearson chi‐square analysis. SD was used for error estimation. A P value < 0.05 was considered statistically significant.
Author contributions
WW conceived and supervised the study. RL designed and supervised the simulation analyses. RV, HH, APT, SZ, BY, GS, SO, KC, and WW performed the experiments. VTD, AEA, and RL performed the simulation analyses. YC performed TCGA data and evolutionary analyses. JC, XL, and WW performed the proteomic and bioinformatic analyses. OR provided key reagents and revised the manuscript. VTD, JC, XL, RL, and WW wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Table EV7
Source Data for Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Acknowledgements
We thank Drs. Steven Gygi and Ross Tomaino (Taplin Mass Spectrometry Facility, Harvard Medical School) for help with the mass spectrometry analysis and Dr. Chao Wang (MD Anderson Cancer Center) for the insightful discussion. This work was supported in part by a NIH grant (GM126048), an American Cancer Society Research Scholar grant (RSG‐18‐009‐01‐CCG), and an Anti‐Cancer Challenge pilot project from the Chao Family Comprehensive Cancer Center (P30 CA062203) to W.W.; a NIH grant to R. L. (GM130367); and a Department of Defense Era of Hope Research Scholar Award to J.C. (W81XWH‐09‐1‐0409). R.V. is supported by a NIH Initiative for Maximizing Student Development (IMSD) Fellowship (GM055246). V.T.D is supported by a Mathematical, Computational and Systems Biology Predoctoral NIH Training Grant (T32 EB009418‐08).
The EMBO Journal (2020) 39: e102406
Contributor Information
Junjie Chen, Email: jchen8@mdandersonorg.org.
Ray Luo, Email: rluo@uci.edu.
Xu Li, Email: lixu@westlake.edu.cn.
Wenqi Wang, Email: wenqiw6@uci.edu.
Data availability
The MS proteomic data have been deposited in the ProteomeXchange Consortium database (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaino et al, 2013) with the dataset identifier PXD004649. The detailed project information is as follows:
Project Name: Human WW domain‐containing proteins TAP‐LC‐MSMS
Project accession: PXD004649
References
- Abu‐Odeh M, Bar‐Mag T, Huang H, Kim T, Salah Z, Abdeen SK, Sudol M, Reichmann D, Sidhu S, Kim PM et al (2014) Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multiprotein networks. J Biol Chem 289: 8865–8880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragon E, Goerner N, Xi Q, Gomes T, Gao S, Massague J, Macias MJ (2012) Structural basis for the versatile interactions of Smad7 with regulator WW domains in TGF‐beta Pathways. Structure 20: 1726–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker EN, Hubbard RE (1984) Hydrogen bonding in globular proteins. Prog Biophys Mol Biol 44: 97–179 [DOI] [PubMed] [Google Scholar]
- Barlow DJ, Thornton JM (1983) Ion‐pairs in proteins. J Mol Biol 168: 867–885 [DOI] [PubMed] [Google Scholar]
- Bork P, Sudol M (1994) The WW domain: a signalling site in dystrophin? Trends Biochem Sci 19: 531–533 [DOI] [PubMed] [Google Scholar]
- Botello‐Smith WM, Luo R (2015) Applications of MMPBSA to membrane proteins i: efficient numerical solutions of periodic Poisson‐Boltzmann equation. J Chem Inf Model 55: 2187–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Hsieh MJ, Wang J, Luo R (2010) Performance of nonlinear finite‐difference Poisson‐Boltzmann solvers. J Chem Theory Comput 6: 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao JJ, Zhao XM, Wang DL, Chen KH, Sheng X, Li WB, Li MC, Liu WJ, He J (2014) YAP is overexpressed in clear cell renal cell carcinoma and its knockdown reduces cell proliferation and induces cell cycle arrest and apoptosis. Oncol Rep 32: 1594–1600 [DOI] [PubMed] [Google Scholar]
- Case DA, Cheatham TE III, Darden T, Gohlke H, Luo R, Merz KM Jr, Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26: 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, Cerutti DS, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W et al (2017) AMBER 2017 reference manual. University of California, San Francisco. [Google Scholar]
- Caswell JL, Camarda R, Zhou AY, Huntsman S, Hu D, Brenner SE, Zaitlen N, Goga A, Ziv E (2015) Multiple breast cancer risk variants are associated with differential transcript isoform expression in tumors. Hum Mol Genet 24: 7421–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SW, Lim CJ, Chong YF, Pobbati AV, Huang C, Hong W (2011) Hippo pathway‐independent restriction of TAZ and YAP by angiomotin. J Biol Chem 286: 7018–7026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang NS, Hsu LJ, Lin YS, Lai FJ, Sheu HM (2007) WW domain‐containing oxidoreductase: a candidate tumor suppressor. Trends Mol Med 13: 12–22 [DOI] [PubMed] [Google Scholar]
- Chang L, Azzolin L, Di Biagio D, Zanconato F, Battilana G, Lucon Xiccato R, Aragona M, Giulitti S, Panciera T, Gandin A et al (2018) The SWI/SNF complex is a mechanoregulated inhibitor of YAP and TAZ. Nature 563: 265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charras G, Yap AS (2018) Tensile forces and mechanotransduction at cell‐cell junctions. Curr Biol 28: R445–R457 [DOI] [PubMed] [Google Scholar]
- Cohen GB, Ren R, Baltimore D (1995) Modular binding domains in signal transduction proteins. Cell 80: 237–248 [DOI] [PubMed] [Google Scholar]
- Couzens AL, Knight JD, Kean MJ, Teo G, Weiss A, Dunham WH, Lin ZY, Bagshaw RD, Sicheri F, Pawson T et al (2013) Protein interaction network of the mammalian Hippo pathway reveals mechanisms of kinase‐phosphatase interactions. Sci Signal 6: rs15 [DOI] [PubMed] [Google Scholar]
- Darabi H, Beesley J, Droit A, Kar S, Nord S, Moradi Marjaneh M, Soucy P, Michailidou K, Ghoussaini M, Fues Wahl H et al (2016) Fine scale mapping of the 17q22 breast cancer locus using dense SNPs, genotyped within the Collaborative Oncological Gene‐Environment Study (COGs). Sci Rep 6: 32512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Smith TF (2000) Identifying nature's protein Lego set. Adv Protein Chem 54: 159–183 [DOI] [PubMed] [Google Scholar]
- Dominguez C, Boelens R, Bonvin AM (2003) HADDOCK: a protein‐protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125: 1731–1737 [DOI] [PubMed] [Google Scholar]
- Dutta S, Mana‐Capelli S, Paramasivam M, Dasgupta I, Cirka H, Billiar K, McCollum D (2018) TRIP6 inhibits Hippo signaling in response to tension at adherens junctions. EMBO Rep 19: 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elias JE, Gygi SP (2007) Target‐decoy search strategy for increased confidence in large‐scale protein identifications by mass spectrometry. Nat Methods 4: 207–214 [DOI] [PubMed] [Google Scholar]
- Ervasti JM (2007) Dystrophin, its interactions with other proteins, and implications for muscular dystrophy. Biochim Biophys Acta 1772: 108–117 [DOI] [PubMed] [Google Scholar]
- Espanel X, Sudol M (2001) Yes‐associated protein and p53‐binding protein‐2 interact through their WW and SH3 domains. J Biol Chem 276: 14514–14523 [DOI] [PubMed] [Google Scholar]
- Faber PW, Barnes GT, Srinidhi J, Chen J, Gusella JF, MacDonald ME (1998) Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet 7: 1463–1474 [DOI] [PubMed] [Google Scholar]
- Feng X, Liu P, Zhou X, Li M‐T, Li F‐L, Wang Z, Meng Z, Sun Y‐P, Yu Y, Xiong Y et al (2016) Thromboxane A2 activates YAP/TAZ protein to induce vascular smooth muscle cell proliferation and migration. J Biol Chem 291: 18947–18958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrigno O, Lallemand F, Verrecchia F, L'Hoste S, Camonis J, Atfi A, Mauviel A (2002) Yes‐associated protein (YAP65) interacts with Smad7 and potentiates its inhibitory activity against TGF‐beta/Smad signaling. Oncogene 21: 4879–4884 [DOI] [PubMed] [Google Scholar]
- Fiser A, Do RK, Sali A (2000) Modeling of loops in protein structures. Protein Sci 9: 1753–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godlewski J, Kiezun J, Krazinski BE, Kozielec Z, Wierzbicki PM, Kmiec Z (2018) The immunoexpression of YAP1 and LATS1 proteins in clear cell renal cell carcinoma: impact on patients' survival. Biomed Res Int 2018: 2653623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC (2012) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized born. J Chem Theory Comput 8: 1542–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halder G, Johnson RL (2011) Hippo signaling: growth control and beyond. Development 138: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson JH, Schild L, Lu Y, Wilson TA, Gautschi I, Shimkets R, Nelson‐Williams C, Rossier BC, Lifton RP (1995) A de novo missense mutation of the beta subunit of the epithelial sodium channel causes hypertension and Liddle syndrome, identifying a proline‐rich segment critical for regulation of channel activity. Proc Natl Acad Sci USA 92: 11495–11499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Chun A, Cheung K, Rashidi B, Yang X (2008) Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J Biol Chem 283: 5496–5509 [DOI] [PubMed] [Google Scholar]
- Haskins JW, Nguyen DX, Stern DF (2014) Neuregulin 1‐activated ERBB4 interacts with YAP to induce Hippo pathway target genes and promote cell migration. Sci Signal 7: ra116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri S, Wepf A, van Drogen A, Varjosalo M, Tapon N, Aebersold R, Gstaiger M (2013) Interaction proteome of human Hippo signaling: modular control of the co‐activator YAP1. Mol Syst Biol 9: 713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Columbus J, Zhang Y, Wu D, Lian L, Yang S, Goodwin J, Luczak C, Carter M, Chen L et al (2004) A map of WW domain family interactions. Proteomics 4: 643–655 [DOI] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H et al (2017) Architecture of the human interactome defines protein communities and disease networks. Nature 545: 505–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L, Kon N, Li T, Wang S‐J, Su T, Hibshoosh H, Baer R, Gu W (2015) Ferroptosis as a p53‐mediated activity during tumour suppression. Nature 520: 57–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tran KM, Aziz KE, Sorokin AV, Chen J, Wang W (2016) Defining the protein‐protein interaction network of the human protein tyrosine phosphatase family. Mol Cell Proteomics 15: 3030–3044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Yang Z, Xie R, Ji Z, Guan K, Zhang M (2019) Decoding WW domain tandem‐mediated target recognitions in tissue growth and cell polarity. Elife 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Li B, Tung EJ, Grundke‐Iqbal I, Iqbal K, Gong CX (2007) Site‐specific effects of tau phosphorylation on its microtubule assembly activity and self‐aggregation. Eur J Neuorsci 26: 3429–3436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang N, Figel SA, Wilson KE, Morrison CD, Gelman IH, Zhang J (2013) PTPN14 interacts with and negatively regulates the oncogenic function of YAP. Oncogene 32: 1266–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Dai X, Cao X, Yan H, Ji X, Zhang H, Shen S, Si Y, Zhang H, Chen J et al (2018) PRDM4 mediates YAP‐induced cell invasion by activating leukocyte‐specific integrin beta2 expression. EMBO Rep 19: e45180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low BC, Pan CQ, Shivashankar GV, Bershadsky A, Sudol M, Sheetz M (2014) YAP/TAZ as mechanosensors and mechanotransducers in regulating organ size and tumor growth. FEBS Lett 588: 2663–2670 [DOI] [PubMed] [Google Scholar]
- Lubs H, Abidi FE, Echeverri R, Holloway L, Meindl A, Stevenson RE, Schwartz CE (2006) Golabi‐Ito‐Hall syndrome results from a missense mutation in the WW domain of the PQBP1 gene. J Med Genet 43: e30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C (2015) ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput 11: 3696–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow EM, Mandelkow E (1998) Tau in Alzheimer's disease. Trends Cell Biol 8: 425–427 [DOI] [PubMed] [Google Scholar]
- Marti‐Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A (2000) Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 29: 291–325 [DOI] [PubMed] [Google Scholar]
- Masoodi TA, Banaganapalli B, Vaidyanathan V, Talluri VR, Shaik NA (2017) Computational analysis of breast cancer GWAS loci identifies the putative deleterious effect of STXBP4 and ZNF404 gene variants. J Cell Biochem 118: 4296–4307 [DOI] [PubMed] [Google Scholar]
- McGibbon RT, Beauchamp KA, Harrigan MP, Klein C, Swails JM, Hernandez CX, Schwantes CR, Wang LP, Lane TJ, Pande VS (2015) MDTraj: a modern open library for the analysis of molecular dynamics trajectories. Biophys J 109: 1528–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaloglou C, Lehmann W, Martin T, Delaunay C, Hueber A, Barys L, Niu H, Billy E, Wartmann M, Ito M et al (2013) The tyrosine phosphatase PTPN14 is a negative regulator of YAP activity. PLoS ONE 8: e61916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR III, McGee TD Jr, Swails JM, Homeyer N, Gohlke H, Roitberg AE (2012) MMPBSA.py: an efficient program for end‐state free energy calculations. J Chem Theory Comput 8: 3314–3321 [DOI] [PubMed] [Google Scholar]
- Min J, Okada S, Kanzaki M, Elmendorf JS, Coker KJ, Ceresa BP, Syu LJ, Noda Y, Saltiel AR, Pessin JE (1999) Synip: a novel insulin‐regulated syntaxin 4‐binding protein mediating GLUT4 translocation in adipocytes. Mol Cell 3: 751–760 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Aebersold R (2005) Interpretation of shotgun proteomic data: the protein inference problem. Mol Cell Proteomics 4: 1419–1440 [DOI] [PubMed] [Google Scholar]
- Otaka Y, Rokudai S, Kaira K, Fujieda M, Horikoshi I, Iwakawa‐Kawabata R, Yoshiyama S, Yokobori T, Ohtaki Y, Shimizu K et al (2017) STXBP4 drives tumor growth and is associated with poor prognosis through PDGF receptor signaling in lung squamous cell carcinoma. Clin Cancer Res 23: 3442–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D (2010) The hippo signaling pathway in development and cancer. Dev Cell 19: 491–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passani LA, Bedford MT, Faber PW, McGinnis KM, Sharp AH, Gusella JF, Vonsattel JP, MacDonald ME (2000) Huntingtin's WW domain partners in Huntington's disease post‐mortem brain fulfill genetic criteria for direct involvement in Huntington's disease pathogenesis. Hum Mol Genet 9: 2175–2182 [DOI] [PubMed] [Google Scholar]
- Pawson T, Scott JD (1997) Signaling through scaffold, anchoring, and adaptor proteins. Science 278: 2075–2080 [DOI] [PubMed] [Google Scholar]
- Piccolo S, Dupont S, Cordenonsi M (2014) The biology of YAP/TAZ: hippo signaling and beyond. Physiol Rev 94: 1287–1312 [DOI] [PubMed] [Google Scholar]
- Qiao Y, Lin SJ, Chen Y, Voon DC, Zhu F, Chuang LS, Wang T, Tan P, Lee SC, Yeoh KG et al (2016) RUNX3 is a novel negative regulator of oncogenic TEAD‐YAP complex in gastric cancer. Oncogene 35: 2664–2674 [DOI] [PubMed] [Google Scholar]
- Qiao Y, Chen J, Lim YB, Finch‐Edmondson ML, Seshachalam VP, Qin L, Jiang T, Low BC, Singh H, Lim CT et al (2017) YAP regulates actin dynamics through ARHGAP29 and promotes metastasis. Cell Rep 19: 1495–1502 [DOI] [PubMed] [Google Scholar]
- Rauskolb C, Sun S, Sun G, Pan Y, Irvine KD (2014) Cytoskeletal tension inhibits Hippo signaling through an Ajuba‐Warts complex. Cell 158: 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentschler S, Linn H, Deininger K, Bedford MT, Espanel X, Sudol M (1999) The WW domain of dystrophin requires EF‐hands region to interact with beta‐dystroglycan. Biol Chem 380: 431–442 [DOI] [PubMed] [Google Scholar]
- Roe DR, Cheatham TE III (2013) PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput 9: 3084–3095 [DOI] [PubMed] [Google Scholar]
- Rokudai S, Li Y, Otaka Y, Fujieda M, Owens DM, Christiano AM, Nishiyama M, Prives C (2018) STXBP4 regulates APC/C‐mediated p63 turnover and drives squamous cell carcinogenesis. Proc Natl Acad Sci USA 115: E4806–E4814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah Z, Aqeilan RI (2011) WW domain interactions regulate the Hippo tumor suppressor pathway. Cell Death Dis 2: e172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salah Z, Melino G, Aqeilan RI (2011) Negative regulation of the Hippo pathway by E3 ubiquitin ligase ITCH is sufficient to promote tumorigenicity. Cancer Res 71: 2010–2020 [DOI] [PubMed] [Google Scholar]
- Salah Z, Cohen S, Itzhaki E, Aqeilan RI (2013) NEDD4 E3 ligase inhibits the activity of the Hippo pathway by targeting LATS1 for degradation. Cell Cycle 12: 3817–3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234: 779–815 [DOI] [PubMed] [Google Scholar]
- Salomon‐Ferrer R, Gotz AW, Poole D, Le Grand S, Walker RC (2013) Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. Explicit solvent particle mesh ewald. J Chem Theory Comput 9: 3878–3888 [DOI] [PubMed] [Google Scholar]
- Schlegelmilch K, Mohseni M, Kirak O, Pruszak J, Rodriguez JR, Zhou D, Kreger BT, Vasioukhin V, Avruch J, Brummelkamp TR et al (2011) Yap1 acts downstream of alpha‐catenin to control epidermal proliferation. Cell 144: 782–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutte U, Bisht S, Heukamp LC, Kebschull M, Florin A, Haarmann J, Hoffmann P, Bendas G, Buettner R, Brossart P et al (2014) Hippo signaling mediates proliferation, invasiveness, and metastatic potential of clear cell renal cell carcinoma. Transl Oncol 7: 309–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe‐Pedros A, Zheng Y, Ruiz‐Trillo I, Pan D (2012) Premetazoan origin of the hippo signaling pathway. Cell Rep 1: 13–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M (1996) Mass spectrometric sequencing of proteins silver‐stained polyacrylamide gels. Anal Chem 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G (2001) Physical interaction with Yes‐associated protein enhances p73 transcriptional activity. J Biol Chem 276: 15164–15173 [DOI] [PubMed] [Google Scholar]
- Strano S, Monti O, Pediconi N, Baccarini A, Fontemaggi G, Lapi E, Mantovani F, Damalas A, Citro G, Sacchi A et al (2005) The transcriptional coactivator Yes‐associated protein drives p73 gene‐target specificity in response to DNA Damage. Mol Cell 18: 447–459 [DOI] [PubMed] [Google Scholar]
- Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D (1995a) Characterization of the mammalian YAP (Yes‐associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem 270: 14733–14741 [DOI] [PubMed] [Google Scholar]
- Sudol M, Chen HI, Bougeret C, Einbond A, Bork P (1995b) Characterization of a novel protein‐binding module–the WW domain. FEBS Lett 369: 67–71 [DOI] [PubMed] [Google Scholar]
- Sudol M (2010) Newcomers to the WW domain‐mediated network of the hippo tumor suppressor pathway. Genes Cancer 1: 1115–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M, Harvey KF (2010) Modularity in the Hippo signaling pathway. Trends Biochem Sci 35: 627–633 [DOI] [PubMed] [Google Scholar]
- Tapia VE, Nicolaescu E, McDonald CB, Musi V, Oka T, Inayoshi Y, Satteson AC, Mazack V, Humbert J, Gaffney CJ et al (2010) Y65C missense mutation in the WW domain of the Golabi‐Ito‐Hall syndrome protein PQBP1 affects its binding activity and deregulates pre‐mRNA splicing. J Biol Chem 285: 19391–19401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavana O, Li D, Dai C, Lopez G, Banerjee D, Kon N, Chen C, Califano A, Yamashiro DJ, Sun H et al (2016) HAUSP deubiquitinates and stabilizes N‐Myc in neuroblastoma. Nat Med 22: 1180–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbricht A, Eppler FJ, Tapia VE, van der Ven PF, Hampe N, Hersch N, Vakeel P, Stadel D, Haas A, Saftig P et al (2013) Cellular mechanotransduction relies on tension‐induced and chaperone‐assisted autophagy. Curr Biol 23: 430–435 [DOI] [PubMed] [Google Scholar]
- Varelas X, Miller BW, Sopko R, Song S, Gregorieff A, Fellouse FA, Sakuma R, Pawson T, Hunziker W, McNeill H et al (2010) The Hippo pathway regulates Wnt/beta‐catenin signaling. Dev Cell 18: 579–591 [DOI] [PubMed] [Google Scholar]
- Verma A, Jing‐Song F, Finch‐Edmondson ML, Velazquez‐Campoy A, Balasegaran S, Sudol M, Sivaraman J (2018) Biophysical studies and NMR structure of YAP2 WW domain ‐ LATS1 PPxY motif complexes reveal the basis of their interaction. Oncotarget 9: 8068–8080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vite A, Zhang C, Yi R, Emms S, Radice GL (2018) alpha‐Catenin‐dependent cytoskeletal tension controls Yap activity in the heart. Development 145: dev149823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaino JA, Cote RG, Csordas A, Dianes JA, Fabregat A, Foster JM, Griss J, Alpi E, Birim M, Contell J et al (2013) The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res 41: D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries SJ, van Dijk AD, Krzeminski M, van Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM (2007) HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins 69: 726–733 [DOI] [PubMed] [Google Scholar]
- Wang J, Wang W, Kollman PA, Case DA (2006) Automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graph Model 25: 247–260 [DOI] [PubMed] [Google Scholar]
- Wang W, Chen L, Ding Y, Jin J, Liao K (2008) Centrosome separation driven by actin‐microfilaments during mitosis is mediated by centrosome‐associated tyrosine‐phosphorylated cortactin. J Cell Sci 121: 1334–1343 [DOI] [PubMed] [Google Scholar]
- Wang J, Luo R (2010) Assessment of linear finite‐difference Poisson‐Boltzmann solvers. J Comput Chem 31: 1689–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Huang J, Chen J (2011) Angiomotin‐like proteins associate with and negatively regulate YAP1. J Biol Chem 286: 4364–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Cai Q, Xiang Y, Luo R (2012a) Reducing grid‐dependence in finite‐difference Poisson‐Boltzmann calculations. J Chem Theory Comput 8: 2741–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Huang J, Wang X, Yuan J, Li X, Feng L, Park JI, Chen J (2012b) PTPN14 is required for the density‐dependent control of YAP1. Genes Dev 26: 1959–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li X, Huang J, Feng L, Dolinta KG, Chen J (2014) Defining the protein‐protein interaction network of the human hippo pathway. Mol Cell Proteomics 13: 119–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Zhang W, Yin MX, Hu L, Li P, Xu J, Huang H, Wang S, Lu Y, Wu W et al (2015) Suppressor of Deltex mediates Pez degradation and modulates Drosophila midgut homeostasis. Nat Commun 6: 6607 [DOI] [PubMed] [Google Scholar]
- Wang C, Nguyen PH, Pham K, Huynh D, Le TB, Wang H, Ren P, Luo R (2016) Calculating protein‐ligand binding affinities with MMPBSA: method and error analysis. J Comput Chem 37: 2436–2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Greene D, Xiao L, Qi R, Luo R (2017) Recent developments and applications of the MMPBSA method. Front Mol Biosci 4: 87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B, Sali A (2016) Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci 86: 2.9.1–2.9.37 [DOI] [PubMed] [Google Scholar]
- Wilson KE, Li YW, Yang N, Shen H, Orillion AR, Zhang J (2014) PTPN14 forms a complex with Kibra and LATS1 proteins and negatively regulates the YAP oncogenic function. J Biol Chem 289: 23693–23700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung B, Ho KC, Yang X (2013) WWP1 E3 ligase targets LATS1 for ubiquitin‐mediated degradation in breast cancer cells. PLoS ONE 8: e61027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Wada Y, Watanabe T, Nagafuchi A, Shibata M (2010) alpha‐Catenin as a tension transducer that induces adherens junction development. Nat Cell Biol 12: 533–542 [DOI] [PubMed] [Google Scholar]
- Yu J, Zheng Y, Dong J, Klusza S, Deng WM, Pan D (2010) Kibra functions as a tumor suppressor protein that regulates Hippo signaling in conjunction with Merlin and Expanded. Dev Cell 18: 288–299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Rathore SS, Shen J (2013) Synip arrests soluble N‐ethylmaleimide‐sensitive factor attachment protein receptor (SNARE)‐dependent membrane fusion as a selective target membrane SNARE‐binding inhibitor. J Biol Chem 288: 18885–18893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Guan KL (2015) Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell 163: 811–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Milton CC, Poon CL, Hong W, Harvey KF (2011) Wbp2 cooperates with Yorkie to drive tissue growth downstream of the Salvador‐Warts‐Hippo pathway. Cell Death Differ 18: 1346–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Lu Q, Wang LH, Liu CY, Lei Q, Guan KL (2011) Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev 25: 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Table EV2
Table EV3
Table EV4
Table EV5
Table EV6
Table EV7
Source Data for Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Data Availability Statement
The MS proteomic data have been deposited in the ProteomeXchange Consortium database (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaino et al, 2013) with the dataset identifier PXD004649. The detailed project information is as follows:
Project Name: Human WW domain‐containing proteins TAP‐LC‐MSMS
Project accession: PXD004649


