Abstract
Sulfoxide synthases are enzymes involved in the biosynthesis of small sulfur-containing natural products. Their enzymatic activity represents a unique sulfur transfer strategy in nature that is the insertion of a sulfur atom on the imidazole ring of histidine. To date, only two enzymes are known to carry out this function: the sulfoxide synthase EgtB, involved in the biosynthesis of ergothioneine in fungi and bacteria, and the 5-histidylcysteine sulfoxide synthase OvoA, involved in the biosynthesis of ovothiols, found in the eggs and biological fluids of marine invertebrates, some proteobacteria and protists. In particular, ovothiols, thanks to their unique redox properties, are probably the most intriguing marine sulfur-containing molecules. Although they have long been considered as cellular protective molecules, new evidence suggest that their biological activities and ecological role might be more complex than originally thought. Here, we investigate the evolutionary history of OvoA in Metazoa, reporting its monophyletic ancient origins, which could be traced back to the latest common ancestor of Choanozoa. Nevertheless, we show that OvoA is missing in several major extant taxa and we discuss this patchy distribution in the light of the massive genome reduction events documented in Metazoa. We also highlight two interesting cases of secondary acquisition through horizontal gene transfer, which occurred in hydrozoans and bdelloid rotifers. The evolutionary success of this metabolic pathway is probably ascribable to its role in the maintenance of cellular redox homeostasis, which enables organisms to survive in different environmental niches.
Keywords: molecular evolution, sulfoxide synthase, ovothiol, metazoan, invertebrates, phylogenetics
1. Introduction
Small sulfur-containing molecules are endowed with unique and highly specialized features that enable them to serve a variety of different functions in the cell. Diverse redox systems based on low-molecular-weight thiols and their oxidized disulfide have been independently developed along evolution. The careful balance between these highly abundant molecules is fundamental to maintain redox homeostasis in various cellular compartments, to provide protection towards oxidative and xenobiotic stress and to support redox-regulatory and signalling processes [1].
Ovothiols are among the most abundant, yet least investigated marine sulfur-containing metabolites. These π-N-methyl-5-thiohistidines were first isolated from the eggs of marine invertebrates, including sea urchins, sea stars and cephalopods [2]. The unique antioxidant properties and broad distribution of ovothiols among invertebrates, microalgae and proteobacteria suggest that these molecules play important roles in cellular biochemistry [3]. In sea urchins, ovothiols are commonly thought to play a protective role towards the strong oxidative burst occurring at fertilization [4] and in larvae exposed to environmental stressors [5]. Three different forms of ovothiols (A, B and C), differing by the degree of methylation at the lateral amino acidic chain, have been identified so far. Ovothiol A has been found in multiple biological sources, such as the eggs of the sea urchin Paracentrotus lividus, the eggs and biological fluids of some molluscs, the coelomic fluid of marine worms, some microalgae and pathogenic protists [2–7]. On the other hand, ovothiol B and C have been only described in the scallop Chlamys hastata and in the sea urchin Strongylocentrotus purpuratus, respectively [2–4]. The desmethylated form of 5-thiohistidine has been so far identified in larger molecular complexes in cephalopods, in the sea star Dermasterias imbricata and in the sponge Lantruculia brevis [3]. Owing to the presence of a sulfydril group on the imidazole ring of histidine, ovothiols have long been considered as modulators of the cellular redox state, and exhibit a high redox potential compared to other cellular thiols, glutathione, trypanothione and ergothioneine [8,9]. However, these small molecules are likely to act as signals to regulate several biological processes not limited to fertilization and larval development [3], as suggested by their ability to reduce cell proliferation in human liver carcinoma cells [10], to exert an anti-inflammatory activity in endothelial cells from diabetic patients [11] and to ameliorate liver fibrosis [12].
New impetus to the research on ovothiols has recently stemmed from the discovery of their biosynthetic route in bacteria [13–15]. The key enzyme involved in ovothiol biosynthesis is a bifunctional enzyme, which catalyses the formation of the oxidative C–S bond, an unprecedented reaction in nature that results in the net sulfur transfer from cysteine to position 5 of histidine. In detail, the enzyme 5-histidylcysteine sulfoxide synthase (OvoA) catalyses the formation of the 5-histidylcysteine sulfoxide conjugate from cysteine and histidine [14]. Subsequently, a pyridoxal phosphate (PLP)-dependent lyase (OvoB) cleaves this intermediate product to generate 5-thiohistidine [14,15]. Then, OvoA catalyses the methylation at the imidazole ring to produce ovothiol A [14,15]. The only other sulfoxide synthase known in nature is EgtB, which catalyses the insertion of the sulfur atom in position 2 of histidine, producing the trimethyl-histidine ergothioneine in some fungi and bacteria. The primary structures of OvoA and EgtB share a similar central formyl-glycine (FGE)-sulfatase domain, which differs for the regioselectivity of the C2 or C5 position of histidine in sulfur transfer [16]. In addition, OvoA has an S-adenosyl methyl (SAM)-transferase C-terminal domain, which is responsible for the imidazolic methylation of 5-thiohistidine [14]. In ergothioneine biosynthesis, the sulfoxide conjugate intermediate is cleaved by the PLP-dependent C–S lyase EgtE [17]. While the biosynthesis and biological activity of ergothioneine have been widely investigated, ovothiols have only recently attracted the interest of the scientific community because of their therapeutic potential and the possibility of biosynthetic production [3,10–12].
In the present study, we explore the evolutionary history of metazoan OvoA genes by comparative genomics and transcriptomic approaches, extending our investigation to neglected phyla, which have been so far been the target of limited -omic studies. We discuss the phylogenetic placement of these sequences with respect to bacteria and unicellular eukaryotic microorganisms, highlighting the ancestral origins of the metabolic pathway and revealing the frequent occurrence of lineage-specific gene loss and two independent horizontal gene transfer (HGT) events.
2. Methodology
(a). Identification of metazoan OvoA from genomic and transcriptomic resources
The fully sequenced and annotated holozoan genomes available in Ensembl and NCBI genome databases were screened for the presence of OvoA based on homology criteria. In detail, the OvoA protein from the sea urchin P. lividus (PlOvoA) was used as a query for BLASTp searches against the predicted protein collections of each species, using an e-value threshold of 1 × 10−50 for an initial detection of possible matches. Positive hits were further inspected with HMMER v. 3.2.1 [18] to assess the presence of the three expected conserved domains in OvoA, i.e. the DinB-like domain (PF12867), FGE-sulfatase domain (PF03781) and methyltransferase 11 domain (PF08241), pertaining to the SAM-dependent methyltransferase homologous superfamily (SSF53335), based on the e-value threshold of 1 × 10−3. Multiple sequence alignments performed with MUSCLE [19] allowed us to assess the completeness of the inferred encoded protein sequences. Genes suffering from excessive fragmentation and incomplete at the 5′ or 3′-end were discarded, while the full-length OvoA orthologues were selected for further investigation.
Publicly available transcriptomic resources were screened to expand the breadth of taxonomical sampling and to cover phyla with no genomic information. Whenever available, assembled transcriptomes were downloaded from the NCBI Transcriptome Shotgun Assembly database or from Dryad (as in the case of the choanoflagellate transcriptomes from the study by Richter et al. [20]). Alternatively, raw sequencing data were downloaded from the NCBI Sequence Read Archive (SRA) database, imported to the CLC Genomics v. 11 environment, trimmed by quality and de novo assembled (setting an automatic selection of word size and bubble size parameters). Transcriptomes were virtually translated using TransDecoder v. 5.3.0 (https://github.com/TransDecoder/), setting the minimal open reading frame length to 100 amino acids. Taking into account the possible inclusion of exogenous sequences (e.g. derived from parasites or microorganisms associated with the target species), positive hits were compared through BLASTn against non-animal sequences deposited in the NCBI nr/nt databases. The detection of positive hits with sequence identity greater than 90% was considered as a strong evidence of exogenous contamination, and led to the exclusion of such sequences from further investigation. Full-length sequences were considered for subsequent analyses, whereas partial matches were only used to mark the presence/absence of OvoA in given taxa. A detailed list of OvoA sequences used in this study is provided in the electronic supplementary material, file S1.
(b). Validation of genomic sequences and assessment of genomic organization
The annotation of OvoA genes was checked by the alignment between genomic DNA and transcriptome-derived complementary DNA (cDNA) sequences of the same species. In the absence of transcriptomes from the same species, the cDNA sequence from a phylogenetically closely related species was used. Annotation errors (i.e. exon skipping, incorrect splicing sites, etc.) were manually corrected, and incomplete sequences, either derived from the miss-assembly of the locus or from the presence of large unresolved gaps, were discarded. Intron and exon boundaries were defined with Splign [21], refining the exact position of donor and acceptor splicing sites with Genie [22]. The possible origin by exogenous genomic DNA contamination of the OvoA sequences found in Hydra vulgaris and Adineta vaga was evaluated by computing the GC content and effective number of codons of all the coding sequences obtained from the two genomes with the EMBOSS geecee and chips tools, respectively [23].
(c). Phylogenetic analysis
The full-length OvoA aa sequences were used to generate two distinct multiple alignments with MUSCLE. The first one (dataset A) included all OvoA sequences identified in UniProtKB; sequences sharing pairwise similarity greater than 55% were clustered with CD-HIT [24]. In this case, the accessory domain of hydrozoan sequences (see the Results section) was removed. The second dataset (B) only contained a selection of the choanozoan OvoA sequences that had been grouped in a monophyletic clade by the previous analysis. These files were processed with GUIDANCE2 [25] to remove highly divergent and phylogenetically poorly informative regions and then evaluated with a ModelTest-NG analysis [26] to detect the best-fitting model of molecular evolution. This was determined as LG + G + I [27] for both datasets, based on the corrected Akaike information criterion [28]. MrBayes v. 3.2 [29] was used to infer the phylogenetic relationships among sequences with a Bayesian approach. Four independent analyses were run in parallel for 3 000 000 generations, for both datasets, i.e. until the observed average standard deviation of split frequencies reached values lower than 0.05, and all estimates for the parameters of the model of molecular evolution reached an effective sample size higher than 200. Convergence of runs was evaluated with Tracer v. 1.6 (http://beast.bio.ed.ac.uk/Tracer). Consensus (50% majority rule) phylogenetic trees were obtained by discarding those obtained during the first 25% of the analysis with the burn-in process. In the graphical representation, poorly supported nodes (posterior probability < 50%) were collapsed. In parallel, maximum-likelihood (ML) inference analyses were performed with the same sequence datasets and model of molecular evolution using RaxML-NG v. 0.9.0 [30].
3. Results
(a). Evolutionary relationship between OvoA from different kingdoms
To shed light on the origin and distribution of OvoA metazoan genes, we performed a comprehensive comparative analysis of all the sequences available in public databases. Besides animals, OvoA with canonical domain organization (electronic supplementary material, figure S1) were found in a large number of prokaryotes (alpha-, beta-, gamma- and delta/epsilon-Proteobacteria, PVC- and FCB-group bacteria), a few cyanobacteria and many unicellular eukaryotes. These included microalgae, such as Haptophyta, Chlorophyta, Cryptophyta and diatoms (Stramenopiles), flagellate protists (Euglenozoa, Parabasalia and Apusozoa), Amoebozoa as well as early-branching holozoan lineages (Ichthyosporea and Choanoflagellatea). No OvoA orthologues were found in Archaea and land plants. In fungi, OvoA was limited to a few species of filamentous ascomycetes.
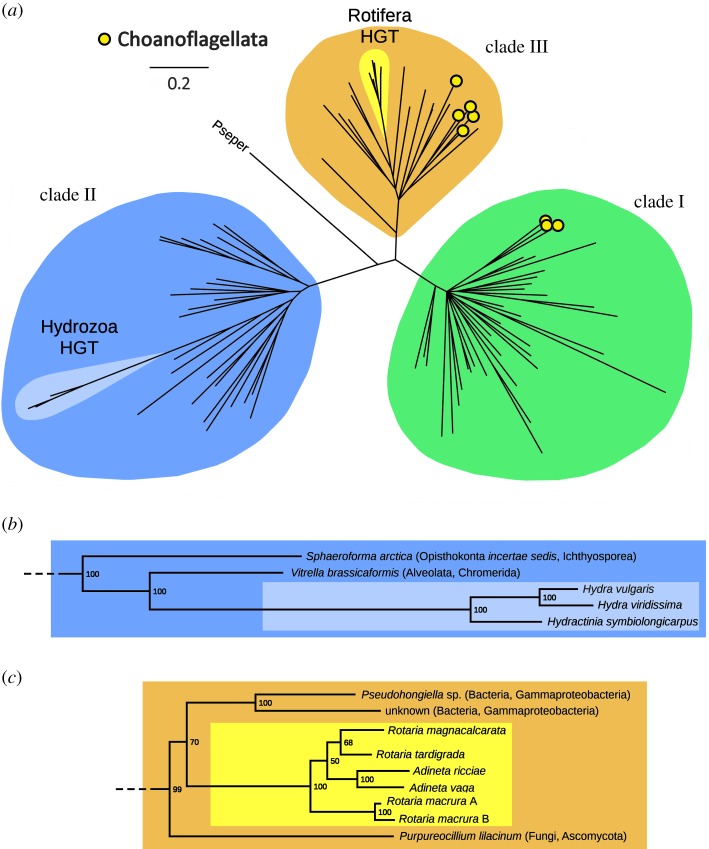
Bayesian and ML inference produced very similar results, placing nearly all metazoan OvoA sequences within a single, highly supported monophyletic group (figure 1a, clade I, 100% posterior probability), well distinct from bacteria and most unicellular eukaryotes. This clade also included all choanoflagellates pertaining to the order Acanthoecida, as well as two species pertaining to the order Craspedida, permitting us to trace back the origins of the prototypical metazoan OvoA gene to the latest common ancestor of Choanozoa. Further attempts made at investigating in more detail the evolutionary history of metazoan sequences produced tree topologies characterized by short basal branches, several polytomies and incongruences with the currently accepted animal phylogeny. These issues most likely reflect a combination between stochastic factors [31] (e.g. lack of phylogenetic signal and inadequate model selection owing to the low number of phylogenetically informative sites present in the multiple sequence alignment), and the rapid radiation of all metazoans which occurred during the Cambrian explosion (electronic supplementary material, figure S2) [32].
Figure 1.
(a) Evolutionary relationship of all OvoA sequences detected in UniProtKB, inferred based on Bayesian inference and an LG + I + G model of molecular evolution and four independent Markov chain Monte Carlo chains run for 3 000 000 generations. Posterior probabilities support values are shown close to each major node. Nodes showing posterior probability support lower than 50% were collapsed. Inferred HGT events in Rotifera and Hydrozoa are indicated. Pseper: Pseudocohnilembus persalinus. Choanoflagellate sequences are marked with yellow circles. (b) Detail on tree branches denoting the HGT event inferred in Hydrozoa. (c) Detail of tree branches denoting the HGT event inferred in Rotifera. The scale bar indicates the number of substitutions per site. Green, clade I sequences; blue, clade II sequences; light blue, Hydrozoa horizontally transferred sequences; orange, clade III sequences; yellow, Rotifera horizontally transferred sequences. The multiple sequence alignment, Bayesian and ML tree files are available in the electronic supplementary material, file S2. (Online version in colour.)
With the exception of the sequence from the ciliate Pseudocohnilembus persalinus, all the other sequences were clustered with high confidence in two other clades (figure 1a). Clade II mostly included bacteria, together with a few unicellular eukaryotes (i.e. Ichthyosporea, Alveolata and Stramenopiles). Clade III included some bacterial (including Cyanobacteria), unicellular green algae, most Craspedida choanoflagellates, fungi and a number of other unicellular eukaryotes). Most of these sequences conserve the iron binding motif and the residues known to be involved in the recognition of the substrates [5]. This sequence conservation is predictive of ovothiol production, as confirmed for different bacteria and unicellular eukaryotes [6,7,13,14,16] (electronic supplementary material, figure S1).
Surprisingly, two small groups of metazoan OvoA sequences were placed in clade II (Hydrozoa) and clade III (Rotifera Bdelloidea). The phylogenetic placement (figure 1b,c), unusual gene architecture and unique sequence features of these genes strongly support their exogenous origin by HGT (see Discussion).
(b). OvoA underwent multiple lineage-specific gene-loss events along metazoan evolution
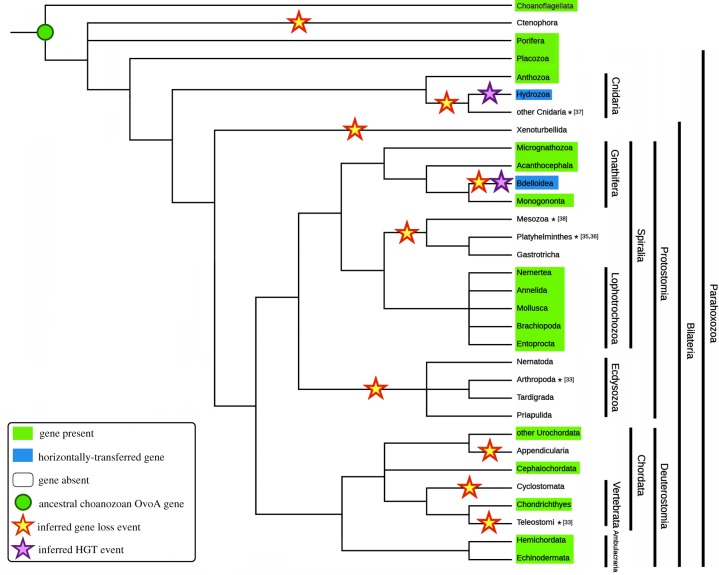
Besides the identification of the ancestral origins of metazoan OvoA genes to the emergence of Choanozoa, our analyses allowed the identification of several independent gene-loss events, often concurrent with well-documented lineage-specific massive orthologous gene-loss events [33,34], or with genome reduction linked with a body plan simplification or with the acquisition of a parasitic lifestyle [35–38] (figure 2). Two major events targeted Ecdysozoa and vertebrates, after the split between Chondrichthyes and Teleostomi, which resulted in the lack of OvoA orthologues in all extant bony fishes and tetrapods [5]. On the other hand, OvoA gene duplication events seem to have only seldom occurred in Metazoa, as most phyla show a single functional OvoA gene (electronic supplementary material, Data note S3).
Figure 2.
Taxonomic spread of OvoA in metazoans. The diagram is a representative of OvoA distribution in the currently accepted metazoan tree of life. Presence, absence, gene loss and HGT events have been inferred through the analysis of genome and transcriptome data. Documented massive gene loss and genome reduction events are marked with an asterisk and the numbers between square brackets indicate the bibliographic reference. (Online version in colour.)
We could confirm the presence of OvoA in Demospongiae and extend its range of taxonomical distribution to two other groups of Porifera i.e. Calcarea and Hexactinellida, whereas no orthologous sequence could be found in Homoscleromorpha. OvoA genes are missing in Ctenophora, one of the most ancient extant animal taxa [39,40], but they are present in Placozoa, another basal metazoan group, included within the group Parahoxozoa, together with Cnidaria and Bilateria [41]. The detailed analysis of genomes and transcriptomes of the phylum Cnidaria highlighted that all anthozoans have OvoA genes, which on the contrary are absent in Myxozoa, Scyphozoa, Cubozoa and Staurozoa. As previously mentioned, Hydrozoa possess largely divergent clade II OvoA-like genes, that were not originated from exogenous DNA contamination (electronic supplementary material, figure S3) and most likely derived from HGT (figure 1b).
We found no OvoA orthologues in the transcriptomes of Xenacoelomorpha (Xenoturbellida + Acoelomorpha), the earliest branching bilaterian phyletic lineage. As previously mentioned, no orthologues were found in Ecdysozoa, confirming previous reports about the absence of OvoA genes in Arthropoda and Nematoda [5] and extending this observation to Priapulida and Tardigrada. On the other hand, the presence of OvoA seems to be a distinctive feature of all Lophotrochozoa, as evidenced by its occurrence in Mollusca, Brachiopoda, Annelida and Nemertea. Transcriptome analysis confirmed the presence of orthologues also in Entoprocta and Phoronida, but not in Bryozoa (probably owing to the limited resources available for this phylum). Although most marginally studied Spiralian species with debated taxonomical placement [42] still lack fully sequenced genomes, we could establish that Platyhelminthes, Mesozoa and Gastrotricha lack OvoA genes. On the contrary, partial sequences were identified in some members of Gnathifera (Acanthocephala, Micrognathozoa and, within Rotifera, Monogononta). As previously mentioned, all bdelloid rotifers display highly divergent, horizontally transferred OvoA genes, which pertain to clade III (figure 1c) and, as in the case of Hydrozoa, were certainly not derived from the assembly of exogenous contaminant genomic DNA (electronic supplementary material, figure S3).
A broader taxonomical sampling of deuterostomes compared to previous studies confirmed the loss of OvoA genes in the Teleostomi lineage, and their presence in all the major classes of Ambulacraria (Echinodermata + Hemichordata), Cephalochordata and Urochordata, except for the lineage-specific loss of OvoA in Appendicularia (Oikopleura dioica). While all Chondrichthyes (including both Holocephali and Elasmobranchii) retain OvoA, its lack in lampreys and hagfishes points out that another gene-loss event took place in Cyclostomata.
(c). The structure of OvoA genes is conserved across metazoan evolution
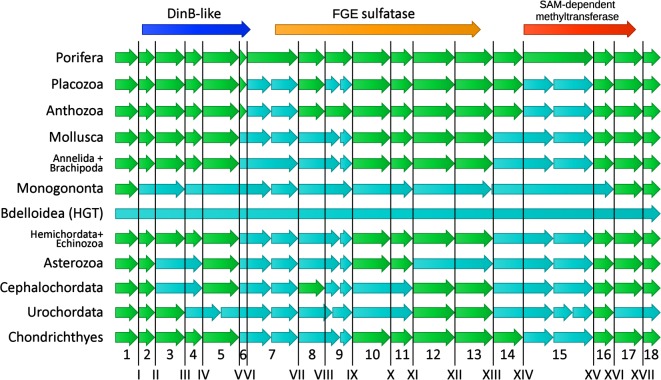
The genomic organization of orthologous genes, in terms of number and position of introns, tends to remain conserved over very long timescales, even in species separated by several hundred million years of divergent evolution [43]. The rates of intron gain have been generally quite low in eukaryotes, while the rate of intron loss has been much higher and variable across lineages, probably owing to the tendency of genome compaction in some phyla (e.g. arthropods and parasitic worms). At the same time, the position of introns appears to be nearly fixed in all eukaryotes, with intron sliding events considered as extremely rare [44]. Therefore, the conservation of intron position is a useful tool for comparative genomics and evolutionary studies and for assessing gene orthology [45]. Owing to the lack of available genomes for Acanthoecida choanoflagellates, we used the gene structure of the Amphimedon queenslandica OvoA as a reference for intron and exon numbering and comparison with other metazoan phyla (figure 3). The OvoA genes of Porifera are characterized by 18 protein-coding exons (plus an additional 5′ exon containing an untranslated region only, disregarded in this study) and by 17 introns.
Figure 3.
Comparative gene architecture of metazoan OvoA. The A. queenslandica gene was used as a reference for exon (Arabic numerals) and intron (Roman numerals) numbering. Exons are represented by arrows and introns are indicated by vertical lines. Exons with boundaries similar to AqOvoA are shown in green, while those with different structure are indicated in light blue. The following representative species were considered: A. queenslandica, T. adherens, Nematostella vectensis, Crassostrea gigas, Octopus bimaculoides, Lottia gigantea, Capitella teleta, Lingula anatina, Brachionus plicatilis, Adineta vaga, S. purpuratus, Saccoglossus kowalevskii, Acanthaster planci, Branchiostoma belcheri, Ciona intestinalis and Callorhinchus milii. (Online version in colour.)
The coding region of OvoA was split among a variable number of exons depending on the phylum, ranging from 10 (in monogonont rotifers, which have extremely compacted genomes [46]) to 21 (in Placozoa) (figure 3). Overall, in spite of several lineage-specific intron loss and intron gain events, common splicing sites were often conserved among largely divergent phyla (e.g. from Porifera to Chordata), consistently with the monophyletic origin of metazoan OvoA genes evidenced by the phylogenetic analysis (figure 1).
The only two exceptions to this general architecture were the genes of bdelloid rotifers and hydrozoans, previously identified as the likely product of HGT (see figures 1b,c and 2). Bdelloid rotifers (i.e. Adineta ssp. and Rotaria spp.) display intron-less genes (figure 3), encoding for OvoA proteins with a canonical size and domain organization. The observed lack of introns, a typical feature of prokaryotic genes, is consistent with the acquisition of nearly 8% of the bdelloid genes by HGT from bacteria in relatively recent times [47]. Hydrozoans, on the other hand, possess highly modified genes, with just five introns, placed in different locations compared to other animals (electronic supplementary material, figure S4). Moreover, the encoded protein contains a large additional N-terminal lyase domain, which is entirely encoded within the first large exon, which may have been recruited through the fusion with another pre-existing gene (see the following section).
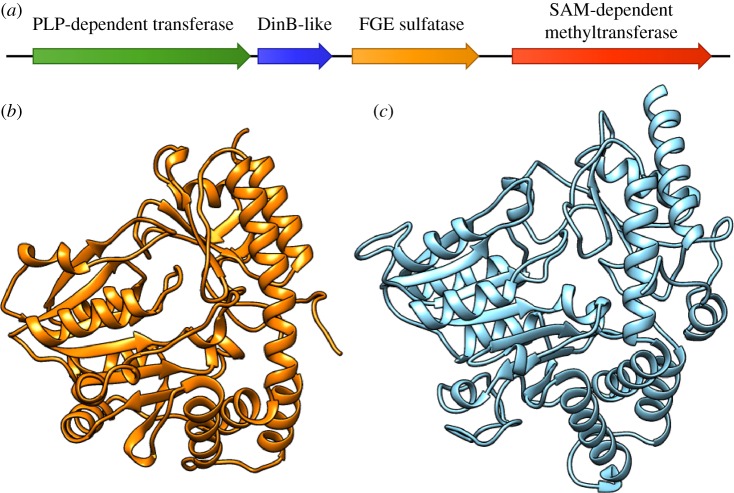
(d). OvoA is fused with a unique sulfide-lyase domain in Hydrozoa
All clade I metazoan and choanoflagellate OvoA proteins display a typical organization, with three conserved domains, i.e. an N-terminal DinB-like domain, a central FGE-sulfatase domain and a C-terminal methyltransferase domain (figure 3; electronic supplementary material, figure S1). While the horizontally transferred clade III OvoA of bdelloid rotifers display the same architecture, the clade II OvoA proteins of hydrozoans include an additional N-terminal PLP-dependent transferase domain (IPR015424) and a much longer C-terminal methyltransferase domain (figure 4a). The additional N-terminal domain is approximately 350-amino acids long and only shows limited homology with a few other metazoan sequences with a much more simple architecture (i.e. containing only an IPR015424 domain), which share similarity with plant mitochondrial cysteine desulfurases. To gather information about the possible function of the N-terminal extension of hydrozoan OvoA, we performed a structural prediction analysis using two different threading/fold-recognition approaches, i.e. Phyre 2 [48] and I-TASSER [49]. The two methods produced similar results, with the former detecting the most significant structural similarities with cyanobacterial l-cystine C–S lyases [50], and the latter finding as the optimal template (confidence > 99.9%) the PLP-dependent C–S lyase Egt2 from Neurospora crassa, which is known to mediate the cleavage of the sulfoxide C–S bond during ergothioneine biosynthesis [51]. The modelled three-dimensional (3D) structure of this domain in H. vulgaris was finally compared with the recently determined crystallographic structure of OvoB, the PLP-dependent C–S lyase involved in ovothiol biosynthesis from Erwinia tasmaniensis (PDB ID: 5Z0Q), which lacked significant primary sequence homology with the Hydra N-terminal domain of OvoA. The superimposition of the two structures through the FATCAT pairwise alignment tool, run in a flexible mode [52], revealed a significant structural match (e-value = 3.87 × 10−6, 296 equivalent positions and an root-mean-square deviation = 3.04 Å). This is consistent with the visible overlap between the backbone structure of both proteins and with the similar arrangement of secondary structural features (figure 4b,c; details of the structural alignment are provided in the electronic supplementary material, File S3). Hence, the hydrozoan OvoA-like gene appears to have been fused with an OvoB-like gene, and may therefore encode a protein that potentially combines three enzymatic activities (sulfoxide synthase and methyl transferase from OvoA and PLP-lyase from OvoB) within the same polypeptidic chain.
Figure 4.
Schematic domain organization of hydrozoan OvoA proteins (a) and comparison between the three-dimensional structure of E. tasmaniensis OvoB (PDB ID: 5Z0Q), as experimentally determined by X-ray diffraction (b), and the model of the H. vulgaris N-terminal sulfide-lyase domain, obtained through a threading approach with I-TASSER (c). (Online version in colour.)
4. Discussion
Ovothiols are small sulfur-containing natural products that are supposed to have played a pivotal role in the evolution of redox homeostasis, providing an adaptive value for the response of animals to challenges posed by the environment. Little is known about the biological function of these compounds [3]. They are supposed to protect eggs and early embryos from oxidative burst occurring at fertilization in sea urchins [4], thanks to their ability to scavenge peroxides and make redox exchange with glutathione. They have been also suggested to act as redox regulators in chloroplasts of microalgae and to play a defence role from the immune response of the host in Trypanosoma species [6,7]. Although ovothiols are present in a broad range of organisms, their biosynthetic pathways have been so far characterized in detail only in a few microorganisms [14,53,54] and the key enzyme involved in this process, OvoA, has been the subject of limited study in animals [5]. In the present study, through an in-depth genomic investigation, we shed light on previously unknown aspects related to the evolution of ovothiol biosynthesis in Metazoa. We revealed that the taxonomic spread of OvoA genes is much more complex and discontinuous than previously thought, owing to the occurrence of several lineage-specific gene-loss events and two independent HGT events in Bdelloidea and Hydrozoa, paired with the recruitment of an additional functional domain in the latter case.
Overall, both phylogenetic evidence (figure 1) and the remarkable conservation of intron position in the animal tree of life (figure 3) support the monophyly of metazoan OvoA sequences, which probably derive from an ancestral gene present in the latest common ancestor of Choanozoa and subsequently rapidly underwent molecular diversification along with the radiation of Metazoa. In fact, clade I OvoA sequences were identified in some (but not all) choanoflagellates, namely all the members of the Acanthoecida and two Craspedida species (Salpingoeca dolichothecata and Codosiga hollandica). The other chonaoflagellates and another group of early-branching holozoans (Ichthyosporea) possess OvoA genes pertaining clade II and III, respectively (figure 1a).
In spite of the monophyletic origin of all metazoan OvoA genes, their evolution did not proceed linearly. While the ability to produce ovothiols has been previously speculated as a beneficial trait for marine life [5], the numerous gene-loss events that we have evidenced (figure 2) do not coincide with the adaptation to terrestrial life. Indeed, numerous examples of large marine taxa lacking OvoA (e.g. Crustacea), as well as of animals living in terrestrial or freshwater environments (e.g. land snails, freshwater bivalves and earthworms) exist.
One of the most innovative results provided by this study was certainly the identification of two distinct cases where an ancestral loss of the OvoA gene was followed by its secondary re-acquisition by HGT. The first of such events occurred in bdelloid rotifers, unusual pseudocoelomate metazoans. The OvoA genes from Adineta spp. and Rotaria spp. were placed with high confidence within clade III, together with sequences from Bacteria and various unicellular eukaryotes (figure 1a), consistently with the report that rotiferan genomes harbour up to 8% genes acquired through HGT from bacteria, fungi and plants [55]. The intron-less nature of the A. vaga gene matches the typical organization of prokaryotic genes, further supporting its exogenous origins (figure 3). From a phylogenetic perspective, bdelloid OvoA sequences shared the highest similarity with Pseudohongiella spp. and other unclassified Gammaproteobacteria (50% similarity, 58% posterior probability) (figure 1c), coherently with the observation that most horizontally transferred rotiferan genes derive from eubacterial donors [56].
Besides these canonical OvoA genes, bdelloid rotifers also possess a second type of highly divergent intron-less OvoA-like sequences. Compared with canonical OvoA, the encoded proteins possess a different C-terminal domain, i.e. a SAM-dependent histidine-specific methyltransferase (PF10017), shared by bacterial EgtD proteins, which suggests that these organisms may possess two alternative thiol-biosynthetic pathways, a hypothesis which will certainly require additional investigation in the near future (see the electronic supplementary material, Data note S6). Considering the presence of clade I OvoA genes in monogonont rotifers, the HGT event which led to the acquisition of OvoA and OvoA-like genes of bdelloid rotifers needs to be placed before the radiation of this group, estimated at more than 60 Ma [56], but after the split between Bdelloidea and Monogononta (figure 2).
The second case of HGT involves Hydrozoa, that have OvoA genes that clearly pertain to clade II (figure 1), together with OvoA genes from bacteria and some unicellular eukaryotes. Hydrozoan OvoA genes were much larger than those of other metazoans, they had a significantly lower number of exons (i.e. electronic supplementary material, figure S4) and encoded proteins with an additional N-terminal PLP-dependent transferase domain (figure 4a). The relevant length of this region, which is entirely encoded by the first exon, suggests that its acquisition is the result of a gene fusion event. Modelling approaches highlighted a surprising structural similarity between this N-terminal domain and OvoB from E. tasmaniensis, the β-lyase responsible for the cleavage of the sulfoxide conjugate intermediate produced by OvoA [16]. To the best of our knowledge, this is the first report of a single polypeptide chain that combines all the three enzymatic activities (sulfatase, lyase and methyltransferase) responsible for ovothiol biosynthesis in metazoans.
In stark contrast with bdelloid rotifers, the most obvious source of exogenous genetic material for HGT in Hydrozoa would not be bacteria, but endosymbiotic dinoflagellates, which live in close association with many type of corals [57,58]. While just a limited number of HGT events has been reported in Hydrozoa [59], the acquisition of genetic material from algal and bacterial endosymbionts provides a significant contribution to the genomes of other cnidarians, such as stony corals [60,61]. The in-depth phylogenetic analysis of clade II OvoA sequences revealed that the closest relative to the hydrozoan genes was a sequence from the chromerid Vitrella brassicaformis (100% posterior probability, figure 1b), a species of photosynthetic protists associated with the corals of the Great Barrier Reef [62]. Altogether, these pieces of evidence suggest that the highly modified hydrozoan OvoA genes have been horizontally transferred from an ancestral chromerid or apicomplexan symbiont [63], which might have lived in close association with an ancestor of the hydrozoan lineage. This HGT event needs to be placed, hundreds of million years ago, before the split between Trachylinae and Hydroidoline, because both groups possess OvoA, but after the separation between Hydrozoa and all the other major cnidarian lineages (figure 2). Considering the fact that no significant homology could be detected between the hydrozoan OvoB-like domain and sequences encoded by the genome of V. brassicaformis and other alveolates, this secondary acquisition event has probably been followed by the fusion with a second gene encoding a β-lyase, possibly pre-existing in the ancestral hydrozoan genome.
Overall, the reasons behind the broad taxonomic spread of OvoA genes, their loss in some phyla and secondary re-acquisition by HGT in others are the likely product of multiple independent factors, which may have acted with different relative pressure in different metazoan lineages. First, the evolutionary success of OvoA may be related to its ability to provide an efficient regulation of redox-sensitive pathways, whose modulation is fundamental in the rearrangements of body plan during larval development and key life cycle transitions [64,65]. On the other hand, genome reduction and simplification are now well recognized as prevailing processes in genome evolution [34]. For example, the lack of OvoA in organisms that underwent genome reduction along with the simplification of the body plan (e.g. Mesozoa [38]) or in obligated parasites which lost primary metabolic pathways provided by the host (e.g. Platyhelminthes [35,36] and Myxozoa [37]), is perhaps not surprising. Similarly, OvoA might have been lost along with several thousand other genes in taxa such as Ecdysozoa, where massive lineage-specific gene-loss events have been documented [33]. In other cases, the loss of the ability to produce ovothiols may be related to the independent evolution of alternative and more sophisticated mechanisms for controlling redox homeostasis. These might involve glutathione metabolism, which in mammals can be affected by exogenous ovothiols produced by invertebrates [66]. Additional in silico analyses aimed at studying the coevolution between OvoA and other pathways involved in the biosynthesis of thiols may shed light on these aspects in the near future. The appearance of molecular systems to acquire ovothiol from diet might be considered as another plausible cause of OvoA gene loss, as it would have made the presence of enzymes involved in the biosynthesis of these molecules unnecessary. Interestingly, some metazoans, including humans, can accumulate ovothiol-related molecules (i.e. ergothioneine) thanks to the activity of a specific membrane transporter [67].
5. Conclusion
Although the ability to synthesize ovothiols is a biological trait shared by several diverse groups of prokaryotes and eukaryotes, we highlight here for the first time, to our knowledge, the monophyly of metazoan OvoA genes, which appear to share distinctive molecular and phylogenetic signatures compared with the OvoA sequences from bacteria and most unicellular eukaryotes. We revealed that the origins of the prototypical metazoan OvoA gene can be traced back to the latest common ancestor of Choanozoa, and that OvoA genes have been lost on multiple occasions along evolution. While the loss of OvoA is often paired with well-known genome reduction events, the possibility that this trait might have been lost in parallel with the development of alternative redox homeostasis control mechanisms, or with the ability to acquire ovothiol-related molecules from the diet remains to be investigated. Finally, our -omic approach enabled us to pinpoint two independent HGT events that have targeted bdelloid rotifers and hydrozoans. In particular, the chimeric hydrozoan genes hold interesting premises for acquiring new knowledge about alternative ovothiol biosynthetic pathways, owing to the combination of OvoA and an OvoB-like lyase domain within the same protein. We expect that the extension of genomic studies to additional animal taxa, together with the elucidation of the crystal structure of OvoA will provide a striking contribution towards an improved understanding of the evolutionary and ecological significance of this metabolic pathway in the near future.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Prof. Sandie Degnan for interesting discussions on OvoA from A. queenslandica and Dr Sergio Palmitessa for technical help. We also acknowledge anonymous reviewers for their valuable comments.
Data accessibility
All new data are available in the electronic supplementary material.
Authors' contributions
M.G. and M.S. performed in silico analysis and analysed data. A.P. and I.C. analysed data and designed the work. I.C. and M.G. conceived and drafted the manuscript. All authors read and approved the final manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by Stazione Zoologica Anton Dohrn.
References
- 1.Krauth-Siegel RL, Leroux AE. 2012. Low-molecular-mass antioxidants in parasites. Antioxid. Redox Signal. 17, 583–607. ( 10.1089/ars.2011.4392) [DOI] [PubMed] [Google Scholar]
- 2.Palumbo A, Castellano I, Napolitano A. 2018. Ovothiol: a potent natural antioxidant from marine organisms. In Blue biotechnology: production and use of marine molecules (eds S La Barre, SS Bates), pp. 583–610. Weinheim, Germany: Wiley; ( 10.1002/9783527801718.ch18) [DOI] [Google Scholar]
- 3.Castellano I, Seebeck FP. 2018. On ovothiol biosynthesis and biological roles: from life in the ocean to therapeutic potential. Nat. Prod. Rep. 35, 1241–1250. ( 10.1039/C8NP00045J) [DOI] [PubMed] [Google Scholar]
- 4.Shapiro BM. 1991. The control of oxidant stress at fertilization. Science 252, 533–536. ( 10.1126/science.1850548) [DOI] [PubMed] [Google Scholar]
- 5.Castellano I, Migliaccio O, D'Aniello S, Merlino A, Napolitano A, Palumbo A. 2016. Shedding light on ovothiol biosynthesis in marine metazoans. Sci. Rep. 6, 21506 ( 10.1038/srep21506) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ariyanayagam MR, Fairlamb AH. 2001. Ovothiol and trypanothione as antioxidants in trypanosomatids. Mol. Biochem. Parasitol. 115, 189–198. ( 10.1016/S0166-6851(01)00285-7) [DOI] [PubMed] [Google Scholar]
- 7.O'Neill EC, Trick M, Hill L, Rejzek M, Dusi RG, Hamilton CJ, Zimba PV, Henrissat B, Field RA. 2015. The transcriptome of Euglena gracilis reveals unexpected metabolic capabilities for carbohydrate and natural product biochemistry. Mol. Biosyst. 11, 2808–2820. ( 10.1039/c5mb00319a) [DOI] [PubMed] [Google Scholar]
- 8.Holler TP, Hopkins PB. 1988. Ovothiols as biological antioxidants. The thiol groups of ovothiol and glutathione are chemically distinct. J. Am. Chem. Soc. 110, 4837–4838. ( 10.1021/ja00222a057) [DOI] [Google Scholar]
- 9.Marjanovic B, Simic MG, Jovanovic SV. 1995. Heterocyclic thiols as antioxidants: why ovothiol C is a better antioxidant than ergothioneine. Free Radic. Biol. Med. 18, 679–685. ( 10.1016/0891-5849(94)00186-N) [DOI] [PubMed] [Google Scholar]
- 10.Russo GL, Russo M, Castellano I, Napolitano A, Palumbo A. 2014. Ovothiol isolated from sea urchin oocytes induces autophagy in the Hep-G2 cell line. Mar. Drugs 12, 4069–4085. ( 10.3390/md12074069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castellano I, Di Tomo P, Di Pietro N, Mandatori D, Pipino C, Formoso G, Napolitano A, Palumbo A, Pandolfi A. 2018. Anti-inflammatory activity of marine ovothiol A in an in vitro model of endothelial dysfunction induced by hyperglycemia. Oxid. Med. Cell. Longev. 2018, 2087373 ( 10.1155/2018/2087373) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brancaccio M, D'Argenio G, Lembo V, Palumbo A, Castellano I. 2018. Antifibrotic effect of marine ovothiol in an in vivo model of liver fibrosis. Oxid. Med. Cell. Longev. 2018, 5045734 ( 10.1155/2018/5045734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vogt RN, Spies HS, Steenkamp DJ. 2001. The biosynthesis of ovothiol A (N-methyl-4-mercaptohistidine). Identification of S-(4′-l-histidyl)-l-cysteine sulfoxide as an intermediate and the products of the sulfoxide lyase reaction. Eur. J. Biochem. 268, 5229–5241. ( 10.1046/j.0014-2956.2001.02444.x) [DOI] [PubMed] [Google Scholar]
- 14.Braunshausen A, Seebeck FP. 2011. Identification and characterization of the first ovothiol biosynthetic enzyme. J. Am. Chem. Soc. 133, 1757–1759. ( 10.1021/ja109378e) [DOI] [PubMed] [Google Scholar]
- 15.Naowarojna N, et al. 2018. In vitro reconstitution of the remaining steps in ovothiol A biosynthesis: C-S lyase and methyltransferase reactions. Org. Lett. 20, 5427–5430. ( 10.1021/acs.orglett.8b02332) [DOI] [PubMed] [Google Scholar]
- 16.Seebeck FP. 2013. Thiohistidine biosynthesis. Chimia 67, 333–336. ( 10.2533/chimia.2013.333) [DOI] [PubMed] [Google Scholar]
- 17.Liao C, Seebeck FP. 2017. Convergent evolution of ergothioneine biosynthesis in cyanobacteria. Chembiochem Eur. J. Chem. Biol. 18, 2115–2118. ( 10.1002/cbic.201700354) [DOI] [PubMed] [Google Scholar]
- 18.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, W29–W37. ( 10.1093/nar/gkr367) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. ( 10.1093/nar/gkh340) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Richter DJ, Fozouni P, Eisen MB, King N. 2018. Gene family innovation, conservation and loss on the animal stem lineage. eLife 7, e34226 ( 10.7554/eLife.34226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kapustin Y, Souvorov A, Tatusova T, Lipman D. 2008. Splign: algorithms for computing spliced alignments with identification of paralogs. Biol. Direct 3, 20 ( 10.1186/1745-6150-3-20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reese MG, Eeckman FH, Kulp D, Haussler D. 1997. Improved splice site detection in Genie. J. Comput. Biol. 4, 311–323. ( 10.1089/cmb.1997.4.311) [DOI] [PubMed] [Google Scholar]
- 23.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software suite. Trends Genet. 16, 276–277. ( 10.1016/S0168-9525(00)02024-2) [DOI] [PubMed] [Google Scholar]
- 24.Li W, Jaroszewski L, Godzik A. 2001. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics 17, 282–283. ( 10.1093/bioinformatics/17.3.282) [DOI] [PubMed] [Google Scholar]
- 25.Sela I, Ashkenazy H, Katoh K, Pupko T. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 43, W7–W14. ( 10.1093/nar/gkv318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14, 817–818. ( 10.1093/bioinformatics/14.9.817) [DOI] [PubMed] [Google Scholar]
- 27.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. ( 10.1093/molbev/msn067) [DOI] [PubMed] [Google Scholar]
- 28.Hurvich CM, Tsai C-L. 1989. Regression and time series model selection in small samples. Biometrika 76, 297–307. ( 10.1093/biomet/76.2.297) [DOI] [Google Scholar]
- 29.Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755. ( 10.1093/bioinformatics/17.8.754) [DOI] [PubMed] [Google Scholar]
- 30.Kozlov AM, Darriba D, Flouri T, Morel B, Stamatakis A. 2019. RAxML-NG: a fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35, 4453–4455. ( 10.1093/bioinformatics/btz305) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jeffroy O, Brinkmann H, Delsuc F, Philippe H. 2006. Phylogenomics: the beginning of incongruence? Trends Genet. 22, 225–231. ( 10.1016/j.tig.2006.02.003) [DOI] [PubMed] [Google Scholar]
- 32.Maloof AC, Porter SM, Moore JL, Dudás FÖ, Bowring SA, Higgins JA, Fike DA, Eddy MP. 2010. The earliest Cambrian record of animals and ocean geochemical change. GSA Bull. 122, 1731–1774. ( 10.1130/B30346.1) [DOI] [Google Scholar]
- 33.Wyder S, Kriventseva EV, Schröder R, Kadowaki T, Zdobnov EM. 2007. Quantification of ortholog losses in insects and vertebrates. Genome Biol. 8, R242 ( 10.1186/gb-2007-8-11-r242) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolf YI, Koonin EV. 2013. Genome reduction as the dominant mode of evolution. BioEssays 35, 829–837. ( 10.1002/bies.201300037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jackson AP. 2015. The evolution of parasite genomes and the origins of parasitism. Parasitology 142, S1–S5. ( 10.1017/S0031182014001516) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zarowiecki M, Berriman M. 2015. What helminth genomes have taught us about parasite evolution. Parasitology 142(Suppl. 1), S85–S97. ( 10.1017/S0031182014001449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang ES, Neuhof M, Rubinstein ND, Diamant A, Philippe H, Huchon D, Cartwright P. 2015. Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc. Natl Acad. Sci. USA 112, 14 912–14 917. ( 10.1073/pnas.1511468112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu T-M, Kanda M, Furuya H, Satoh N. 2019. Dicyemid Mesozoans: a unique parasitic lifestyle and a reduced genome. Genome Biol. Evol. 11, 2232–2243. ( 10.1093/gbe/evz157) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simion P, et al. 2017. A large and consistent phylogenomic dataset supports sponges as the sister group to all other animals. Curr. Biol. 27, 958–967. ( 10.1016/j.cub.2017.02.031) [DOI] [PubMed] [Google Scholar]
- 40.Whelan NV, Kocot KM, Moroz LL, Halanych KM. 2015. Error, signal, and the placement of Ctenophora sister to all other animals. Proc. Natl Acad. Sci. USA 112, 5773–5778. ( 10.1073/pnas.1503453112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava M, et al. 2008. The Trichoplax genome and the nature of placozoans. Nature 454, 955–960. ( 10.1038/nature07191) [DOI] [PubMed] [Google Scholar]
- 42.Marlétaz F, Peijnenburg KTCA, Goto T, Satoh N, Rokhsar DS. 2019. A new Spiralian phylogeny places the enigmatic arrow worms among gnathiferans. Curr. Biol. 29, 312–318. ( 10.1016/j.cub.2018.11.042) [DOI] [PubMed] [Google Scholar]
- 43.Irimia M, Roy SW. 2008. Spliceosomal introns as tools for genomic and evolutionary analysis. Nucleic Acids Res. 36, 1703–1712. ( 10.1093/nar/gkn012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. 2003. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr. Biol. 13, 1512–1517. ( 10.1016/S0960-9822(03)00558-X) [DOI] [PubMed] [Google Scholar]
- 45.Henricson A, Forslund K, Sonnhammer EL. 2010. Orthology confers intron position conservation. BMC Genomics 11, 412 ( 10.1186/1471-2164-11-412) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blommaert J, Riss S, Hecox-Lea B, Mark Welch DB, Stelzer CP. 2019. Small, but surprisingly repetitive genomes: transposon expansion and not polyploidy has driven a doubling in genome size in a metazoan species complex. BMC Genomics 20, 466 ( 10.1186/s12864-019-5859-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gladyshev EA, Meselson M, Arkhipova IR. 2008. Massive horizontal gene transfer in bdelloid rotifers. Science 320, 1210–1213. ( 10.1126/science.1156407) [DOI] [PubMed] [Google Scholar]
- 48.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10, 845–858. ( 10.1038/nprot.2015.053) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. ( 10.1038/nprot.2010.5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Campanini B, Schiaretti F, Abbruzzetti S, Kessler D, Mozzarelli A. 2006. Sulfur mobilization in cyanobacteria: the catalytic mechanism of L-cystine C-S lyase (C-DES) from synechocystis. J. Biol. Chem. 281, 38 769–38 780. ( 10.1074/jbc.M607098200) [DOI] [PubMed] [Google Scholar]
- 51.Irani S, et al. 2018. Snapshots of C-S cleavage in Egt2 reveals substrate specificity and reaction mechanism. Cell Chem. Biol. 25, 519–529. ( 10.1016/j.chembiol.2018.02.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ye Y, Godzik A. 2004. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 32, W582–W585. ( 10.1093/nar/gkh430) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mashabela GTM, Seebeck FP. 2013. Substrate specificity of an oxygen dependent sulfoxide synthase in ovothiol biosynthesis. Chem. Commun. 49, 7714–7716. ( 10.1039/C3CC42594K) [DOI] [PubMed] [Google Scholar]
- 54.Chen L, Naowarojna N, Song H, Wang S, Wang J, Deng Z, Zhao C, Liu P. 2018. Use of a tyrosine analogue to modulate the two activities of a nonheme iron enzyme OvoA in ovothiol biosynthesis, cysteine oxidation versus oxidative C-S bond formation. J. Am. Chem. Soc. 140, 4604–4612. ( 10.1021/jacs.7b13628) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flot J-F, et al. 2013. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature 500, 453–457. ( 10.1038/nature12326) [DOI] [PubMed] [Google Scholar]
- 56.Eyres I, Boschetti C, Crisp A, Smith TP, Fontaneto D, Tunnacliffe A, Barraclough TG. 2015. Horizontal gene transfer in bdelloid rotifers is ancient, ongoing and more frequent in species from desiccating habitats. BMC Biol. 13, 90 ( 10.1186/s12915-015-0202-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aranda M, et al. 2016. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6, 39734 ( 10.1038/srep39734) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Davy SK, Allemand D, Weis VM. 2012. Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol. Mol. Biol. Rev. 76, 229–261. ( 10.1128/MMBR.05014-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chapman JA, et al. 2010. The dynamic genome of Hydra. Nature 464, 592–596. ( 10.1038/nature08830) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Baumgarten S, et al. 2015. The genome of Aiptasia, a sea anemone model for coral symbiosis. Proc. Natl Acad. Sci. USA 112, 11 893–11 898. ( 10.1073/pnas.1513318112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhattacharya D, et al. 2016. Comparative genomics explains the evolutionary success of reef-forming corals. eLife 5, e13288 ( 10.7554/eLife.13288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oborník M, et al. 2012. Morphology, ultrastructure and life cycle of Vitrella brassicaformis n. sp., n. gen., a novel chromerid from the Great Barrier Reef. Protist 163, 306–323. ( 10.1016/j.protis.2011.09.001) [DOI] [PubMed] [Google Scholar]
- 63.Janouškovec J, Tikhonenkov DV, Burki F, Howe AT, Kolísko M, Mylnikov AP, Keeling PJ. 2015. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc. Natl Acad. Sci. USA 112, 10 200–10 207. ( 10.1073/pnas.1423790112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Castellano I, Ercolesi E, Palumbo A. 2014. Nitric oxide affects ERK signaling through down-regulation of MAP kinase phosphatase levels during larval development of the ascidian Ciona intestinalis. PLoS ONE 9, e102907 ( 10.1371/journal.pone.0102907) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Castellano I, Ercolesi E, Romano G, Ianora A, Palumbo A. 2015. The diatom-derived aldehyde decadienal affects life cycle transition in the ascidian Ciona intestinalis through nitric oxide/ERK signalling. Open Biol. 5, 140182 ( 10.1098/rsob.140182) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brancaccio M, Russo M, Masullo M, Palumbo A, Russo GL, Castellano I. 2019. Sulfur-containing histidine compounds inhibit γ-glutamyl transpeptidase activity in human cancer cells. J. Biol. Chem. 294, 14 603–14 614. ( 10.1074/jbc.RA119.009304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gründemann D, Harlfinger S, Golz S, Geerts A, Lazar A, Berkels R, Jung N, Rubbert A, Schömig E. 2005. Discovery of the ergothioneine transporter. Proc. Natl Acad. Sci. USA 102, 5256–5261. ( 10.1073/pnas.0408624102) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All new data are available in the electronic supplementary material.