Abstract
Background
Recent studies have shown that circular RNA (circRNA) is rich in microRNA (miRNA) binding sites. We have previously demonstrated that the antidepressant effect of ketamine is related to the abnormal expression of various miRNAs in the brain. This study determined the expression profile of circRNAs in the hippocampus of rats treated with ketamine.
Methods
The aberrantly expressed circRNAs in rat hippocampus after ketamine injection were analyzed by microarray chip, and we further validated these circRNAs by quantitative reverse-transcription PCR (qRT-PCR). The target genes of the different circRNAs were predicted using bioinformatic analyses, and the functions and signal pathways of these target genes were investigated by Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses.
Results
Microarray analysis showed that five circRNAs were aberrantly expressed in rat hippocampus after ketamine injection (fold change > 2.0, p < 0.05). The results from the qRT-PCR showed that one of the circRNAs was significantly increased (rno_circRNA_014900; fold change = 2.37; p = 0.03), while one was significantly reduced (rno_circRNA_005442; fold change = 0.37; p = 0.01). We discovered a significant enrichment in several GO terms and pathways associated with depression.
Conclusion
Our findings showed the abnormal expression of ketamine-induced hippocampal circRNAs in rats.
Keywords: Rat, Ketamine, CircRNA, Hippocampus
Background
The most commonly used antidepressants today effectively improve symptoms of depression, but require at least 2 weeks to take therapeutic effect. In addition, about two-thirds of depressive patients do not respond to the currently available antidepressants and are prone to relapse [1]. Therefore, the development of new, fast-acting antidepressants is particularly important for patients with depression-induced suicidal tendencies [2].
Recent observations suggest that sub-anesthetic doses of ketamine produce rapid therapeutic effects in depressed patients and in animal models of depression [3–6]. These effects are characterized by a rapid onset of action (within hours) after a single dose, a lasting effect (1 week), as well as an efficacy in patients resistant to traditional antidepressant drugs [7, 8].
In a prior investigation, we found that the antidepressant effect of ketamine was related to the regulation of multiple microRNAs (miRNAs) in neurons [9]. Recent studies have shown that circular RNA (circRNA) is rich in miRNA binding sites. CircRNA plays the role of an miRNA sponge in cells, thereby relieving the inhibitory effect of the miRNAs on their target genes and increasing the expression of these genes. The mechanism by which circRNA inhibits miRNAs to increase the expression of their target genes is called the competitive endogenous RNA (ceRNA) mechanism [10, 11].
Based on the previous reports, the aim of this study was to determine the effect of an antidepressant dose of ketamine on the expression of circRNAs in the hippocampus of rats and examine a possible circRNA-mediated mechanism for ketamine’s action. This work may provide new perspectives on the development of circRNA as a possible drug target.
Methods
Animals
These experiments were conducted in male Sprague-Dawley rats (50 days old, 150–200 g), provided by Chengdu Dashuo biological technology Co., Ltd., China (experimental animal production license: SCXK Chengdu 2013–17). These rats were housed 5 per cage in standard cages (42 × 20 × 20 cm) in a room. Animals had access to food and water ad libitum during the experiment. The room was maintained at 25–26 °C with about 65% relative humidity, on a 12-h dark/light cycle (lights on at 7 am). All experiments were performed according to National Institute of Health (NIH) guidelines and approved by the ethics committee of Southwest Medical University (Approval number:20180306038).
Experimental design and procedure
After 1 week of adaptation, the rats were randomly divided into control and experimental group (12 rats/group). The rats in control group were daily injected with 0.9% saline, whereas the rats in experimental group received ketamine (15 mg/kg). All injections were done intraperitoneally for three consecutive days (the volume of injection was 1 ml/kg). The dose of ketamine used in this study was based on our previous study [9]. On day 4, approximately 24 h after the last ketamine or saline injection, the rats were sacrificed by cervical dislocation, and their hippocampus tissues were dissected out for circRNA microarray analysis (n = 3/group) and for qRT-PCR (n = 6 /group). The hippocampus of 3 remaining rats per group were spare specimen according to the quality of hippocampus removed from rats.
CircRNAs analysis from microarray chip
Relevant circRNAs were analyzed according to our previous approach [12]. Hippocampus tissues(n = 3/group) were used for microarray assay to determine differentially expressed circRNA between the two groups. The microarray hybridization was performed based on the Arraystar’s standard protocols, including purifying RNA, transcribing into fluorescent cRNA, and hybridizing onto the rat circRNA Arrays (Arraystar). Finally, the hybridized slides were washed, fixed and scanned to images by the Agilent Scanner G2505C. The Agilent Feature Extraction software (version 11.0.1.1) were used to analyze the acquired array images. The raw data were normalized and data analysis was further performed with the R software Limma package (Agilent Technologies). The statistical significance of differentially regulated circRNAs between the two groups was identified through screening fold change ≥2.0, P < 0.05 and FDR < 0.05.
Quantitative real-time PCR validation
Hippocampus tissues(n = 6/group) were used for qRT-PCR validation. After RNA isolation, M-MLV reverse transcriptase (Invitrogen, USA) was used for synthesizing cDNA according to the manufacturer’s instructions. Subsequently, we performed qRT-PCR using the ViiA 7 Real-time PCR System (Applied Biosystems, Foster City, CA, USA) in a total reaction volume of 10 μl, including 2 μl cDNA, 5 μl 2 × Master Mix, 0.5 μl PCR Forward Primer (10 μM), 0.5 μl PCR Reverse Primer (10 μM) and 2 μl double distilled water. The protocol was initiated at 95 °C for 10 min, then at 95 °C (10 s), 60 °C (60 s) for a total 40 cycles. β-actin was used as a reference. Results were harvested in three independent wells. For quantitative results, the relative expression level of each circRNA was calculated using 2−ΔΔCt method.
Competing endogenous RNA analysis of differentially expressed circRNAs
The candidate miRNA binding sites were searched on the sequences of circRNAs and mRNAs, and the circRNA-miRNA-mRNA interaction were found by the overlapping of the same miRNA seed sequence binding site both on the circRNAs and the mRNA. The miRNA-mRNA interactions were predicted by Targetscan (http://www.targetscan.org/), while the miRNA binding sites were predicted by miRcode (http://www.mircode.org/).
GO and KEGG pathway analysis
Gene Ontology (GO) analysis (http://www.geneontology.org) were conducted to construct meaningful annotation of genes and gene products in the organisms. The ontology includes molecular functions (MFs), biological processes (BPs) and cellular components (CCs). KEGG pathway analysis were also performed to harvest pathway clusters on the molecular interaction and reaction networks in differentially regulated genes. The -log10 (p-value) denotes enrichment score representing the significance of GO term enrichment and pathway correlations among differentially expressed genes.
Statistical analysis
The statistical package for the social sciences (SPSS) 11.0 was selected for statistical analysis. All data were expressed as mean ± SEM. The data from the circRNA microarray and qRT-PCR were analyzed by one-way ANOVA or multi-factorial ANOVA followed by Tukey’s post hoc test. P values less than 0.05 with statistically significant differences.
Results
The expression profile of circRNAs in the rat hippocampus using microarray analysis and secondary validation from qRT-PCR
The RNA concentration and purity of all samples meted the requirement (larger than 1.8 and an RNA concentration greater than 30 ng) for subsequent microarray detection of the circRNA expression profile.
As shown in Fig. 1, significantly different circRNAs were selected for hierarchical clustering analysis. The circRNA hierarchical clustering map not only displays circRNA expression, but also exhibits the expression change of a single circRNA from both groups. As shown in Table 1, four circRNAs were upregulated and one was down-regulated in the hippocampus of rats treated with ketamine from circRNA microarray analysis (p < 0.05), but only two of the five circRNAs were confirmed to be differentially expressed from qRT-PCR (Table 3, p < 0.05). As shown in Table 2, the primers for the five different circRNAs were designed using Primer software 5.0.
Fig. 1.
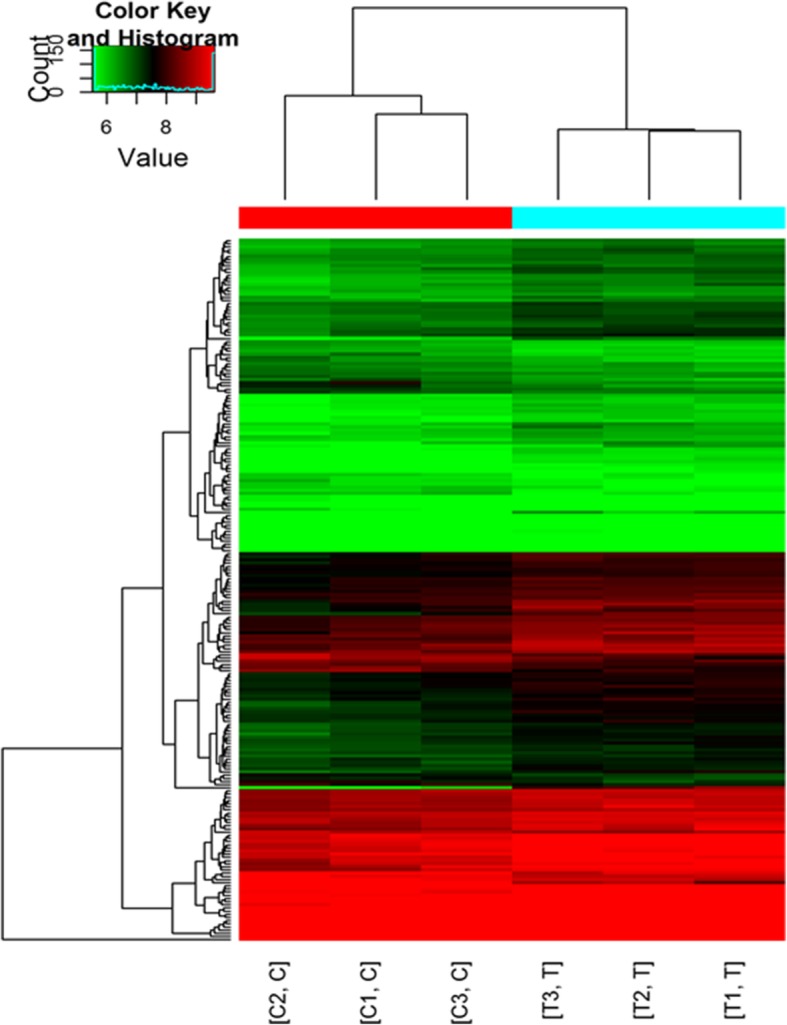
The hierarchical clustering plot of differentially expressed circRNAs (fold change ≥1.5, p < 0.05). Red indicates circRNAs with high expression levels, and green represents circRNAs with low expression levels, the color depth (ranging from black to color) indicates different expression intensities. Each row in the figure indicates a different circRNA, and each column indicates a sample (T is the ketamine group and C is the vehicle group). The left side of the figure shows the circRNA clustering tree, whereas the top shows the hippocampal sample clustering tree
Table 1.
Aberrantly expressed circRNAs in rat hippocampus revealed by microarray analysis (Fold change ≥2.0, p < 0.05)
| circRNA | Fold Change | Regulation | P-value |
|---|---|---|---|
| rno_circRNA_003460 | 5.98 | up | 0.001 |
| rno_circRNA_014900 | 2.28 | up | 0.003 |
| rno_circRNA_006565 | 2.11 | up | 0.010 |
| rno_circRNA_013109 | 2.17 | up | 0.012 |
| rno_circRNA_005442 | 2.01 | down | 0.012 |
Table 3.
qRT-PCR-confirmed expression of circRNAs in rat hippocampus and the predicted target miRNAs
| circRNA | Fold Change | Regulation | P-value | Predicted target miRNAs |
|---|---|---|---|---|
| rno_circRNA_014900 | 2.37 | up | 0.029 |
rno-miR-466b-5p rno-miR-6332 rno-miR-6321 rno-miR-193a-5p rno-miR-1224 |
| rno_circRNA_005442 | 2.72 | down | 0.010 |
rno-miR-323-5p rno-miR-107-5p rno-miR-135a-5p rno-miR-135b-5p rno-miR-344b-5p |
Table 2.
A list of primers used for real-time PCR
| Gene | Bi-directional primer sequence | Annealing temperature (°C) | Primer length (bp) |
|---|---|---|---|
| β-actin (Reference) |
Forward:5′CGAGTACAACCTTCTTGCAGC 3′ Reverse: 5′ ACCCATACCCACCATCACAC 3’ |
60 | 202 |
| rno_circRNA_014900 |
Forward:5′ CTTAGATGACCTGGAGAAGACCT 3′ Reverse: 5′ TGACTTGGTGCTGTTGACTTTAG 3’ |
60 | 124 |
| rno_circRNA_013109 |
Forward:5′ ATTATAGAGCTAATTACAACTTCCG 3′ Reverse:5′ TTATCTGAAGCATGTTAAGACAATA 3’ |
60 | 105 |
| rno_circRNA_006565 |
Forward:5′ CGACTTCAAAAGAGTTGTGGATT 3′ Reverse: 5′ TTCTCCTCGTGAGCTTTTTTCTC 3’ |
60 | 54 |
| rno_circRNA_005442 |
Forward:5′ ACCCCATGAGAAAGACCAGGTC 3′ Reverse:5′ CTGCTCTCTTCAAGTGAAAGACATC 3′ |
60 | 60 |
| rno_circRNA_003460 |
Forward:5′ CGCTAAGCATTTCTTTGGAA 3′ Reverse: 5′ GTAGTGGGTGTAGGGAGGAGA 3′ |
60 | 76 |
Predicted miRNAs sponged by the two circRNAs and their corresponding target genes
As shown in Table 3, the two circRNAs 014900 and 005442 collectively sponged ten miRNAs, namely, rno-miR-466b-5p, rno-miR-6332, rno-miR-6321, rno-miR-193a-5p, rno-miR-1224, rno-miR-323-5p, rno-miR-107-5p, rno-miR-135b-5p, rno-miR-135a-5p, and rno-miR-344b-5p. Each of these miRNAs has target genes that they endogenously regulate, as shown in Table 4, the two circRNAs could indirectly regulate numerous target genes by their endogenous competition mechanism.
Table 4.
Target genes regulated indirectly by the two differentially expressed circRNAs through miRNAs
| circRNA | Sponged miRNAs | Gene Symbol | Gene Description |
|---|---|---|---|
| rno_circRNA_014900 |
rno-miR-193a-5p rno-miR-1224 |
Nova1 | neuro-oncological ventral antigen 1 |
|
rno-miR-193a-5p rno-miR-6332 |
Clasp1 | cytoplasmic linker associated protein 1 | |
|
miR-193a-5p rno-miR-466b-5p |
Rgs4, Mixl1 | regulator of G-protein signaling 4; Mix paired-like homeobox 1 | |
|
rno-miR-466b-5p rno-miR-6332 |
Usp37, RGD1566029, Zfp91, Zmiz1, Pbx1 | ubiquitin specific peptidase 37; similar to mKIAA1644 protein; zinc finger protein 91; zinc finger, MIZ-type containing 1; pre-B-cell leukemia homeobox 1 | |
|
rno-miR-466b-5p rno-miR-1224 |
Calcoco1 | calcium binding and coiled coil domain 1 | |
|
rno-miR-466b-5p rno-miR-6321 |
Brwd3, Nfat5, Cnot7, Camta1 | bromodomain and WD repeat domain containing 3; nuclear factor of activated T-cells 5, tonicity-responsive; CCR4-NOT transcription complex, subunit 7; calmodulin binding transcription activator 1 | |
|
rno-miR-6332 rno-miR-1224 |
Prpf4b, Hgs, Gtdc1, Dgkk | pre-mRNA processing factor 4B;hepatocyte growth factor-regulated tyrosine kinase substrate; glycosyltransferase-like domain containing 1;diacylglycerol kinase kappa | |
|
rno-miR-6332 rno-miR-6321 |
RGD1562037, Usp24,Ssbp2 | similar to OTTHUMP00000046255; ubiquitin specific peptidase 24; single-stranded DNA binding protein 2 | |
|
rno-miR-6332 rno-miR-6321 rno-miR-466b-5p |
Ssbp2, RGD1562037 | singe-stranded DNA binding protein 2; similar to OTTHUMP00000046255 | |
|
rno-miR-6332 rno-miR-466b-5p rno-miR-1224 |
Zfp91 | zinc finger protein 91 | |
| rno_circRNA_005442 |
rno-miR-107-5p rno-miR-323-5p |
Eps8,Rhot1 | epidermal growth factor receptor pathway substrate 8, ras homolog gene family, member T1 |
|
rno-miR-107-5p rno-miR-344b-5p |
Hn1 | hematological and neurological expressed 1 | |
|
rno-miR-107-5p rno-miR-135b-5p |
Ptk2,Tiam1 | protein tyrosine kinase 2;T-cell lymphoma invasion and metastasis 1 | |
|
rno-miR-107-5p rno-miR-135a(b)-5p |
Slc8a1,Man2a1,Tnpo1,Rgl1 | solute carrier family 8 (sodium/calcium exchanger), member 1; mannosidase, alpha, class 2A, member 1;transportin 1; ral guanine nucleotide dissociation stimulator,-like 1 | |
|
rno-miR-323-5p rno-miR-344b-5p |
Tomm6 | translocase of outer mitochondrial membrane 6 homolog (yeast) | |
|
rno-miR-323-5p miR-135a(b)-5p |
Shisa7 | shisa family member 7 | |
|
rno-miR-344b-5p miR-135a(b)-5p |
Arel1 | apoptosis resistant E3 ubiquitin protein ligase 1 | |
|
rno-miR-323-5p rno-miR-107-5p rno-miR-135a(b)-5p |
Rhot1 | ras homolog gene family, member T1 |
GO analysis
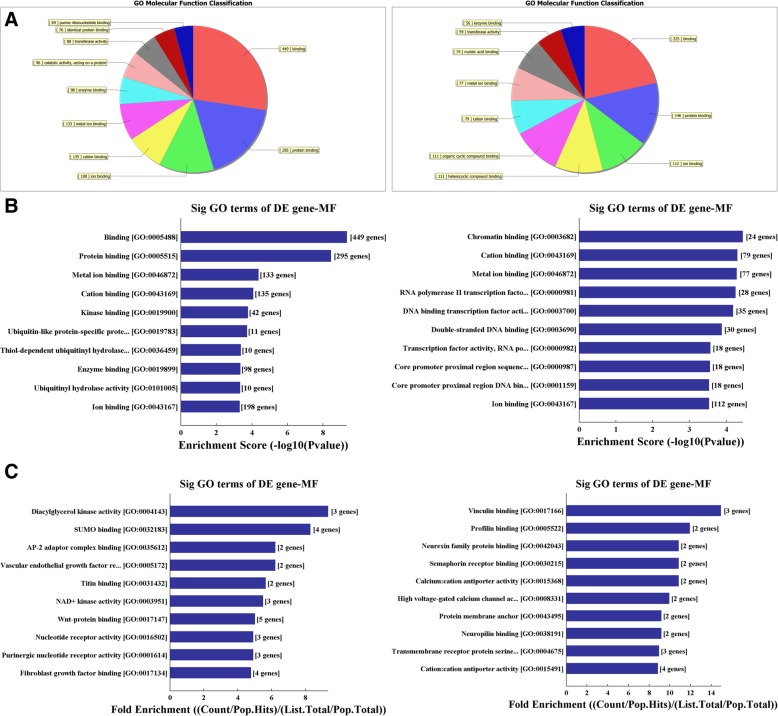
As shown in Fig. 2, the molecular functions (MFs) of neurons that may be regulated by the target genes of the differentially expressed circRNAs (p < 0.05, left: rno_circRNA_014900; right: rno_circRNA_005442). The classification of notable MFs was shown in Fig. 2a and b shows the order of these MFs by their GO analysis enrichment scores. Figure 2c shows the notable activities of neurons using fold enrichment. The detailed list of the genes identified as regulated by circRNA was shown in Additional files 1 and 2.
Fig. 2.
The molecular functions (MFs) of neurons regulated by the target genes of the differentially expressed circRNAs (p < 0.05, left: rno_circRNA_014900; right: rno_circRNA_005442). a classifies the notable MFs and b shows these same MFs ordered by their GO analysis enrichment scores. c shows the notable activities of neurons that may be regulated by target genes predicted using fold enrichment
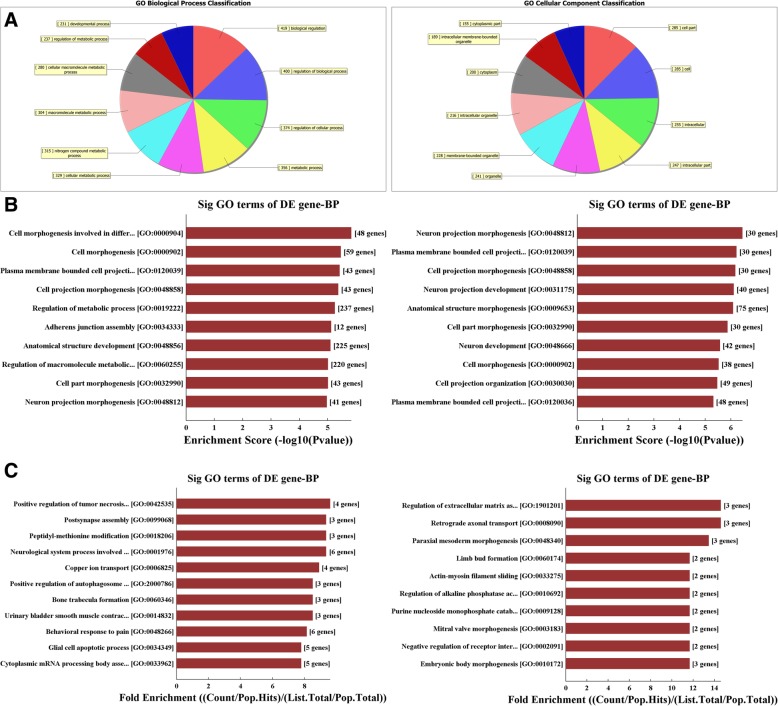
As shown in Fig. 3, the biological processes (BPs) in neurons that may be regulated by target genes of the two circRNAs (p < 0.05, left: rno_circRNA_014900; right: rno_circRNA_005442). The classification of notable BPs was shown in Fig. 3a and b shows the different BPs predicted by enrichment scores. Figure 3c shows the different BPs of neurons predicted using fold enrichment. The detailed list of the genes identified as regulated by circRNA was shown in Additional files 3 and 4.
Fig. 3.
The biological processes (BPs) in neurons regulated by target genes of the two circRNAs (p < 0.05, left: rno_circRNA_014900; right: rno_circRNA_005442). a classifies the predicted BPs and b shows the notable BPs predicted by enrichment scores. c shows the notable BPs of neurons that may be regulated by target genes predicted using fold enrichment
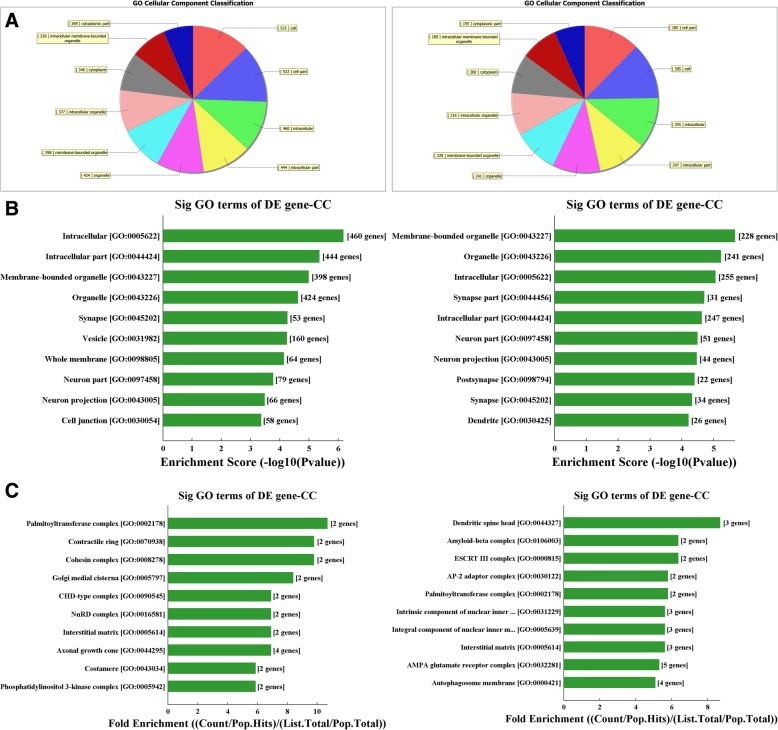
As shown in Fig. 4, the cellular components (CCs) that may be regulated by target genes of the two differentially expressed circRNAs (p < 0.05, left: rno_circRNA_014900; right: rno_circRNA_005442). The classification of notable CCs was shown in Fig. 4a and b shows the different CCs predicted by enrichment scores. Figure 4c shows the different CCs of neurons using fold enrichment. The detailed list of the genes identified as regulated by circRNA was shown in Additional files 5 and 6.
Fig. 4.
The cellular components (CCs) that may be regulated by target genes of the two differentially expressed circRNAs (p < 0.05, left: rno_circRNA_014900; right: rno_circRNA_005442). a classifies the predicted CCs and b shows the notable CCs predicted by enrichment scores. c shows the notable CCs of neurons that may be regulated by target genes predicted using fold enrichment
KEGG pathway analysis
Tables 5 and 6 shows the signaling pathways that may be regulated by target genes of the differentially expressed rno_circRNA_014900 and rno_circRNA_005442 (p < 0.05). The identified pathways were involved in the regulation of central nervous system functions, such as Wnt signaling, long-term depression, PI3K-Akt signaling, dopaminergic synapse activity, mTOR signaling, p53 signaling, apoptosis, TGF-beta signaling, axon guidance, hippo signaling, and MAPK signaling.
Table 5.
The signaling pathways that may be regulated by targeted genes of the differentially expressed rno_circRNA_014900(p < 0.05)
| PathwayID | Definition | Fisher-Pvalue | Enrichment_Score |
|---|---|---|---|
| rno04510 | Focal adhesion - Rattus norvegicus (rat) | 4.05765E-06 | 5.391726 |
| rno05205 | Proteoglycans in cancer - Rattus norvegicus (rat) | 0.000263739 | 3.578826 |
| rno05200 | Pathways in cancer - Rattus norvegicus (rat) | 0.000839484 | 3.075988 |
| rno04216 | Ferroptosis - Rattus norvegicus (rat) | 0.000947662 | 3.023347 |
| rno04015 | Rap1 signaling pathway - Rattus norvegicus (rat) | 0.001242467 | 2.905715 |
| rno00564 | Glycerophospholipid metabolism - Rattus norvegicus (rat) | 0.001767738 | 2.752582 |
| rno04310 | Wnt signaling pathway - Rattus norvegicus (rat) | 0.002533546 | 2.596271 |
| rno05165 | Human papillomavirus infection - Rattus norvegicus (rat) | 0.003213976 | 2.492957 |
| rno04140 | Autophagy - animal - Rattus norvegicus (rat) | 0.00464105 | 2.333384 |
| rno05222 | Small cell lung cancer - Rattus norvegicus (rat) | 0.005031787 | 2.298278 |
| rno04070 | Phosphatidylinositol signaling system - Rattus norvegicus (rat) | 0.006461245 | 2.189684 |
| rno00561 | Glycerolipid metabolism - Rattus norvegicus (rat) | 0.00791785 | 2.101393 |
| rno04213 | Longevity regulating pathway - multiple species - Rattus norvegicus (rat) | 0.00791785 | 2.101393 |
| rno04730 | Long-term depression - Rattus norvegicus (rat) | 0.00791785 | 2.101393 |
| rno04151 | PI3K-Akt signaling pathway - Rattus norvegicus (rat) | 0.00874211 | 2.058384 |
| rno04512 | ECM-receptor interaction - Rattus norvegicus (rat) | 0.009516369 | 2.021529 |
| rno04137 | Mitophagy - animal - Rattus norvegicus (rat) | 0.009920558 | 2.003464 |
| rno04728 | Dopaminergic synapse - Rattus norvegicus (rat) | 0.01248382 | 1.903653 |
| rno04211 | Longevity regulating pathway - Rattus norvegicus (rat) | 0.01286284 | 1.890663 |
| rno04136 | Autophagy - other - Rattus norvegicus (rat) | 0.01346501 | 1.870793 |
| rno04150 | mTOR signaling pathway - Rattus norvegicus (rat) | 0.01358861 | 1.866825 |
| rno04115 | p53 signaling pathway - Rattus norvegicus (rat) | 0.01402018 | 1.853246 |
| rno04215 | Apoptosis - multiple species - Rattus norvegicus (rat) | 0.01492946 | 1.825956 |
| rno05214 | Glioma - Rattus norvegicus (rat) | 0.01496318 | 1.824976 |
| rno04270 | Vascular smooth muscle contraction - Rattus norvegicus (rat) | 0.02428063 | 1.61474 |
| rno01521 | EGFR tyrosine kinase inhibitor resistance - Rattus norvegicus (rat) | 0.02549639 | 1.593521 |
| rno05219 | Bladder cancer - Rattus norvegicus (rat) | 0.02580699 | 1.588263 |
| rno05224 | Breast cancer - Rattus norvegicus (rat) | 0.02850884 | 1.54502 |
| rno05166 | HTLV-I infection - Rattus norvegicus (rat) | 0.02925304 | 1.533829 |
| rno04066 | HIF-1 signaling pathway - Rattus norvegicus (rat) | 0.03188594 | 1.496401 |
| rno04068 | FoxO signaling pathway - Rattus norvegicus (rat) | 0.03490339 | 1.457132 |
| rno04144 | Endocytosis - Rattus norvegicus (rat) | 0.03599259 | 1.443787 |
| rno04720 | Long-term potentiation - Rattus norvegicus (rat) | 0.03856442 | 1.413813 |
| rno04978 | Mineral absorption - Rattus norvegicus (rat) | 0.04045532 | 1.393024 |
| rno04725 | Cholinergic synapse - Rattus norvegicus (rat) | 0.04272807 | 1.369287 |
| rno05030 | Cocaine addiction - Rattus norvegicus (rat) | 0.04327309 | 1.363782 |
| rno04010 | MAPK signaling pathway - Rattus norvegicus (rat) | 0.04346333 | 1.361877 |
| rno04120 | Ubiquitin mediated proteolysis - Rattus norvegicus (rat) | 0.04667443 | 1.330921 |
| rno00062 | Fatty acid elongation - Rattus norvegicus (rat) | 0.04768946 | 1.321578 |
| rno05211 | Renal cell carcinoma - Rattus norvegicus (rat) | 0.04775832 | 1.320951 |
| rno01522 | Endocrine resistance - Rattus norvegicus (rat) | 0.04798442 | 1.3189 |
Table 6.
The signaling pathways that may be regulated by targeted genes of the differentially expressed rno_circRNA_005442 (p < 0.05)
| PathwayID | Definition | Fisher-Pvalue | Enrichment_Score |
|---|---|---|---|
| rno04350 | TGF-beta signaling pathway - Rattus norvegicus (rat) | 0.002056745 | 2.68682 |
| rno04960 | Aldosterone-regulated sodium reabsorption - Rattus norvegicus (rat) | 0.002806053 | 2.551904 |
| rno05231 | Choline metabolism in cancer - Rattus norvegicus (rat) | 0.0039363 | 2.404912 |
| rno05412 | Arrhythmogenic right ventricular cardiomyopathy (ARVC) - Rattus norvegicus (rat) | 0.005000479 | 2.300988 |
| rno04550 | Signaling pathways regulating pluripotency of stem cells - Rattus norvegicus (rat) | 0.005251424 | 2.279723 |
| rno04218 | Cellular senescence - Rattus norvegicus (rat) | 0.005747859 | 2.240494 |
| rno04360 | Axon guidance - Rattus norvegicus (rat) | 0.005747859 | 2.240494 |
| rno05225 | Hepatocellular carcinoma - Rattus norvegicus (rat) | 0.005747859 | 2.240494 |
| rno04144 | Endocytosis - Rattus norvegicus (rat) | 0.006773032 | 2.169217 |
| rno00510 | N-Glycan biosynthesis - Rattus norvegicus (rat) | 0.007410522 | 2.130151 |
| rno04390 | Hippo signaling pathway - Rattus norvegicus (rat) | 0.009694587 | 2.013471 |
| rno04068 | FoxO signaling pathway - Rattus norvegicus (rat) | 0.01587784 | 1.799209 |
| rno04371 | Apelin signaling pathway - Rattus norvegicus (rat) | 0.02058289 | 1.686494 |
| rno04520 | Adherens junction - Rattus norvegicus (rat) | 0.02501545 | 1.601792 |
| rno05210 | Colorectal cancer - Rattus norvegicus (rat) | 0.02501545 | 1.601792 |
| rno04973 | Carbohydrate digestion and absorption - Rattus norvegicus (rat) | 0.02580168 | 1.588352 |
| rno04010 | MAPK signaling pathway - Rattus norvegicus (rat) | 0.03810185 | 1.419054 |
| rno05410 | Hypertrophic cardiomyopathy (HCM) - Rattus norvegicus (rat) | 0.04204027 | 1.376335 |
| rno05414 | Dilated cardiomyopathy (DCM) - Rattus norvegicus (rat) | 0.04832562 | 1.315823 |
Discussion
There are about 340 million patients suffering from depression around the world, and it is estimated that up to 1 million people die by depression-induced suicide every year. Therefore, depression has become a global public health problem [13]. The major clinical drawback of currently available antidepressants is their slow onset (14 days on average) and poor effects [1]. Therefore, the development of rapid-onset antidepressants is particularly important for patients suffering from major depressive disorder with suicidal tendency. The antidepressant effect of ketamine initiates quickly and is also effective in treating refractory depression [7, 8]. However, the psychosocial side effects induced by ketamine have restricted its use in the treatment of depression [14]. An in-depth understanding of the mechanism of ketamine’s rapid antidepressant effect can provide new targets for similar antidepressants.
Recent many researches have shown that many transcriptional products come from the non-coding RNA (ncRNA), including the microRNA (miRNA), long non-coding RNA (lncRNA), and circular RNA (circRNA), these ncRNAs can regulate gene expression at the DNA level, pre-transcriptional level, transcriptional level, post-transcriptional level, translational level, and post-translational level [15]. Previous study from our group found that ketamine induced abnormal expression of miRNAs in the brain, and subsequent target gene and functional analysis revealed that this abnormal expression of miRNAs was closely related to the antidepressant effect of ketamine, but the therapeutic effect of antidepressants focusing on a single mechanism is poor due to the complex mechanism of major depressive disorder [9]. Therefore, we believe that a single miRNA plays relatively weak role in the regulation of a specific target gene. It was hypothesized that a substance in the body that could gather several miRNAs regulated the same specific target gene, then the regulatory action on the specific target gene would be obviously increased. It was interesting that circRNAs, a novel type of non-coding RNAs [15], were perceived as a rare curiosity to having a central regulatory role in RNA metabolism, and accumulating evidences suggested that circRNAs could function as miRNA sponges (the ceRNA mechanism), therefore, circRNA could relieve the inhibitory effect of miRNA on its target gene and increase the expression of the target gene [10–12].
Our results showed that the expression of rno_circRNA_014900 was significantly increased by ketamine, while the expression of rno_circRNA_005442 was obviously decreased. Since these two circRNAs were not investigated in previous studies on depression, we further investigated their regulated target genes and signaling pathways. Competing endogenous RNA analysis found that the rno_circRNA_014900 could sponge rno-miR-466b-5p, rno-miR-6332, rno-miR-6321, rno-miR-193a-5p and rno-miR-1224, whereas the rno_circRNA_005442 had the binding sites in rno-miR-323-5p, rno-miR-107-5p, rno-miR-135a-5p, rno-miR-135b-5p and rno-miR-344b-5p. In a prior investigation, we found that miR-206 was a critical novel gene for the expression of BDNF (brain-derived neurotrophic factor) induced by ketamine [9], our experimental results were expected to be that the circRNA identified in this study would target the miRNA-206, but structural prediction analysis showed that the miRNAs sponged by rno_circRNA_014900 and rno_circRNA_005442 did not include miRNA-206, we thought rno_circRNA_014900 and rno_circRNA_005442 did not regulate the expression of miRNA-206, how ketamine affects the expression of miRNA-206 needs to be further explored in future research. Further prediction analysis revealed that rno_circRNA_014900 and rno_circRNA_005442 may inhibit up to four miRNAs closely related to depression. For example, the predicted target genes related to depression include NOVA1 (regulated by miR-193-5p and miR-1224) [16], Rgs4 (regulated by miR-193-5p and miR-466b-5p) [17–20], zinc-finger protein (regulated by miR-6332, miR-466b-5p, and miR-1224) [21], PBX1 (regulated by miR-466b-5p and miR-6332) [22], SLC8A1 (regulated by miR-107-5p, miR-135a(b)-5p, and miR-135b-5p) [23], PTK2 (regulated by miR-107-5p and miR-135b-5p) [24], and Tiam1 (regulated by miR-107-5p and miR-135b-5p) [25]. Therefore, we believed rno_circRNA_014900 or rno_circRNA_005442 could relieve the inhibitory effect of miRNA on its target gene and increase the expression of the target gene, the phenomenon needs to be further confirmed in future studies.
The results from GO analysis found that the the target genes of the two circRNAs regulated many molecular functions (including protein phosphatase binding, SUMO binding, Wnt-protein binding, etc.), biological processes (including adherens junction assembly, neuron projection morphogenesis, neurological processes, etc.), and cellular components (including dendritic spines, AMPA-glutamate receptor binding, neuronal cell body, etc.). The results from KEGG pathway prediction showed that the signaling pathways regulated by target genes of the two circRNAs included Wnt signaling, long-term depression, PI3K-Akt signaling, dopaminergic synapses, mTOR signaling, p53 signaling, apoptosis, MAPK signaling, TGF-beta signaling, axon guidance, Hippo signaling, etc.. These signaling pathways may be involved in the occurrence and development of depression, because some researches found that the Wnt signaling pathway played important roles in the depression-like behaviors [26–28], the PI3K-Akt signaling pathway was related to the rapid antidepressant-like effects of some drugs [29–33].
As we analyzed in other study about circRNAs [12], novel therapies should include multiple genes and pathways due to the complex mechanisms of major depression disorder. Because many pathophysiological processes of stress-related depression were regulated by several miRNAs, therefore these miRNAs may be commom targets of antidepressant therapies. It was not difficult to understand that the increase in relevant circRNAs expression would enhance the translation of their target genes due to miRNA sponges, a down-expression in relevant circRNAs would result in the obvious silencing of downstream target genes. Therefore, in the future, artificial circRNA drugs will be developed to restore normal transcriptional regulation.
Conclusions
In summary, we found that ketamine treatment resulted in the abnormal expression of the two circRNAs in the hippocampus of rats, and these two circRNAs may be associated with stress-related depression disorders. CircRNAs should remain the focus of researches investigating antidepressant targets because they have considerable potential in the clinical treatment of stress-related depression. As an invaluable topic for future biomedical studies, we plan to screen for specific circRNAs in the context of depression and to examine their potential value in the diagnosis and treatment of this debilitating disorder.
Supplementary information
Additional file 1. MF results for circRNA01490. The detailed list of the genes identified as regulated by the circRNA 01490, MF: molecular function.
Additional file 2. MF results for circRNA005442. The detailed list of the genes identified as regulated by the circRNA005442, MF: molecular function.
Additional file 3. BP results for circRNA01490. The detailed list of the genes identified as regulated by the circRNA 01490, BP: biological process.
Additional file 4. BP results for circRNA005442. The detailed list of the genes identified as regulated by the circRNA005442, BP: biological process.
Additional file 5. CC results for circRNA01490. The detailed list of the genes identified as regulated by the circRNA 01490, CC: cellular component.
Additional file 6. CC results for circRNA005442. The detailed list of the genes identified as regulated by the circRNA005442, CC: cellular component.
Acknowledgments
We would like to thank LetPub (www.letpub.com) for providing linguistic assistance during the preparation of this manuscript.
Abbreviations
- BPs
Biological processes
- CCs
Cellular components
- ceRNA
Competitive endogenous RNA
- circRNA
Circular RNA
- GO
Gene Ontology
- i.p.
Intraperitoneal
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- lncRNA
Long non-coding RNA
- MFs
Molecular functions
- miRNA
MicroRNA
- ncRNA
Non-coding RNA
- qRT-PCR
Quantitative reverse-transcription PCR
- SEM
Standard error of the mean
Authors’ contributions
JM, TL, DF, HZ, JF, LL and XW made substantial contribution to conception of the intervention and design of the study and/or were involved in acquisition of data. JM, TL, DF and HZ made the first draft of the manuscript and LL, CZ and XW critically revised the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the Projects of the National Natural Science Foundation of China (Grant No. 81271478), Department of Science &Technology of Sichuan Province (Grant No. 14JC0093), Department of Education of Sichuan Province (Grant No. 14ZA0142) and Department of Science &Technology of Luzhou City (Grant No. 2015-S-46). The funding bodies had no role in the design of the study or collection, analysis, and interpretation of data or in writing the manuscript.
Availability of data and materials
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
All experimental procedures involving animals were performed in accordance with the NIH Guide for the Care and Use of Laboratory Animals and the Chinese Society for Neuroscience and Behavior recommendations for animal care. All of the experiments involving surgeries and treatments were approved by the ethics committee of Southwest Medical University (Ethics number: 20180306038).
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Jing Mao and Tianmei Li contributed equally to this work.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12888-019-2374-2.
References
- 1.Duman RS, Aghajanian GK. Synaptic dysfunction in depression: potential therapeutic targets. Science. 2012;338(6103):68–72. doi: 10.1126/science.1222939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Machado-Vieira R, Salvadore G, Diazgranados N, Zarate CA. Ketamine and the next generation of antidepressants with a rapid onset of action. Pharmacol Ther. 2009;123(2):143–150. doi: 10.1016/j.pharmthera.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman RM, Cappiello A, Anand A, Oren DA, Heninger GR, Charney DS, et al. Antidepressant effects of ketamine in depressed patients. Biol Psychiatry. 2000;47(4):351–354. doi: 10.1016/S0006-3223(99)00230-9. [DOI] [PubMed] [Google Scholar]
- 4.Wang XB, Chen YL, Zhou X, Liu F, Zhang T, Zhang C. Effects of propofol and ketamine as the combined anesthesia for electroconvulsive therapy in patients with depressive disorder. J ECT. 2012;28(2):128–132. doi: 10.1097/YCT.0b013e31824d1d02. [DOI] [PubMed] [Google Scholar]
- 5.Garcia LS, Comim CM, Valvassori SS, Réus GZ, Barbosa LM, Andreazza AC, et al. Acute administration of ketamine induces antidepressant-like effects in the forced swimming test and increases BDNF levels in the rat hippocampus. Prog Neuropsychopharmacol. 2008;32(1):140–144. doi: 10.1016/j.pnpbp.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 6.Sun HL, Zhou ZQ, Zhang GF, Yang C, Wang XM, Shen JC, et al. Role of hippocampal p11 in the sustained antidepressant effect of ketamine in the chronic unpredictable mild stress model. Transl Psychiatry. 2016;6:e741. doi: 10.1038/tp.2016.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang JJ, Wang N, Yang C, Shi JY, Yu HY, Hashimoto K. Serum interleukin-6 is a predictive biomarker for ketamine’s antidepressant effect in treatment-resistant patients with major depression. Biol Psychiatry. 2015;77(3):e19–e20. doi: 10.1016/j.biopsych.2014.06.021. [DOI] [PubMed] [Google Scholar]
- 8.Murrough JW, Iosifescu DV, Chang LC, Jurdi RK, Green CE, Perez AM, et al. Antidepressant efficacy of ketamine in treatment resistant major depression: a two-site randomized controlledtrial. Am J Psychiatry. 2013;170(10):1134–1142. doi: 10.1176/appi.ajp.2013.13030392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang X, Yang Q, Wang X, Luo C, Wan Y, Li J, et al. MicroRNA expression profile and functional analysis reveal that miR-206 is a critical novel gene for the expression of BDNF induced by ketamine. NeuroMolecular Med. 2014;16(3):594–605. doi: 10.1007/s12017-014-8312-z. [DOI] [PubMed] [Google Scholar]
- 10.Van Rossum D, Verheijen BM, Pasterkamp RJ. Circular RNAs: novel regulators of neuronal development. Front Mol Neurosci. 2016;9:74. doi: 10.3389/fnmol.2016.00074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang MZ, Su PX, Liu Y, Zhang XT, Yan J, An XG, et al. Abnormal expression of circRNA_089763 in the plasma exosomes of patients with post operative cognitive dysfunction after coronary artery bypass grafting. Mol Med Rep. 2019;20:2549–2562. doi: 10.3892/mmr.2019.10521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferrari AJ, Charlson FJ, Norman RE, Patten SB, Freedman G, Murray CJ, et al. Burden of depressive disorders by country, sex, age, and year: findings from the global burden of disease study. PLoS Med. 2010;10(11):e1001547. doi: 10.1371/journal.pmed.1001547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newcomer JW, Farber NB, Jevtovic-Todorovic V, Selke G, Melson AK, Hershey T, et al. Ketamine-induced NMDA receptor hypofunction as a model of memory impairment and psychosis. Neuropsychopharmacology. 1999;20(2):106–118. doi: 10.1016/S0893-133X(98)00067-0. [DOI] [PubMed] [Google Scholar]
- 15.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19(2):141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malki K, Tosto MG, Jumabhoy I, Lourdusamy A, Sluyter F, Craig I, et al. Integrative mouse and human mRNA studies using WGCNA nominates novel candidate genes involved in the pathogenesis of major depressive disorder. Pharmacogenomics. 2013;14(16):1979–1990. doi: 10.2217/pgs.13.154. [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Lee S, Kang S, Jeon TI, Kang MJ, Lee TH, et al. Regulator of G-protein signaling 4 (RGS4) controls morphine reward by glutamate receptor activation in the nucleus Accumbens of mouse brain. Mol Cells. 2018;41(5):454–464. doi: 10.14348/molcells.2018.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng D, He S, Yu S, Li G, Ma C, Wen Y, et al. Analysis of the association of MIR124-1 and its target gene RGS4 polymorphisms with major depressive disorder and antidepressant response. Neuropsychiatr Dis Treat. 2018;14:715–723. doi: 10.2147/NDT.S155076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stratinaki M, Varidaki A, Mitsi V, Ghose S, Magida J, Dias C, et al. Regulator of G protein signaling 4 [corrected] is a crucial modulator of antidepressant drug action in depression and neuropathic pain models. Proc Natl Acad Sci U S A. 2013;110(20):8254–8259. doi: 10.1073/pnas.1214696110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P. Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry. 2001;6(3):293–301. doi: 10.1038/sj.mp.4000866. [DOI] [PubMed] [Google Scholar]
- 21.Nätt D, Johansson I, Faresjö T, Ludvigsson J, Thorsell A. High cortisol in 5-year-old children causes loss of DNA methylation in SINE retrotransposons: a possible role for ZNF263 in stress-related diseases. Clin Epigenetics. 2015;7:91. doi: 10.1186/s13148-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grados MA, Specht MW, Sung HM, Fortune D. Glutamate drugs and pharmacogenetics of OCD: a pathway-based exploratory approach. Expert Opin Drug Discov. 2013;8(12):1515–1527. doi: 10.1517/17460441.2013.845553. [DOI] [PubMed] [Google Scholar]
- 23.Douglas LN, McGuire AB, Manzardo AM, Butler MG. High-resolution chromosome ideogram representation of recognized genes for bipolar disorder. Gene. 2016;586(1):136–147. doi: 10.1016/j.gene.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao L, Gao Y, Xu E, Xie J. Microarray analysis of the major depressive disorder mRNA profile data. Psychiatry Investig. 2015;12(3):388–396. doi: 10.4306/pi.2015.12.3.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benoist M, Palenzuela R, Rozas C, Rojas P, Tortosa E, Morales B, et al. MAP 1B- dependent Rac activation is required for AMPA receptor endocytosis during long-term depression. EMBO J. 2013;32(16):2287–2299. doi: 10.1038/emboj.2013.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ni X, Liao Y, Li L, Zhang X, Wu Z. Therapeutic role of long non-coding RNA TCONS_00019174 in depressive disorders is dependent on Wnt/β-catenin signaling pathway. J Integr Neurosci. 2018;17(2):203–215. doi: 10.3233/JIN-170052. [DOI] [PubMed] [Google Scholar]
- 27.Martin PM, Stanley RE, Ross AP, Freitas AE, Moyer CE, Brumback AC, et al. DIXDC1 contributes to psychiatric susceptibility by regulating dendritic spine and glutamatergic synapse density via GSK3 and Wnt/β-catenin signaling. Mol Psychiatry. 2018;23(2):467–475. doi: 10.1038/mp.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou WJ, Xu N, Kong L, Sun SC, Xu XF, Jia MZ, et al. The antidepressant roles of Wnt2 and Wnt3 in stress-induced depression-like behaviors. Transl Psychiatry. 2016;6(9):e892. doi: 10.1038/tp.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cunha MP, Budni J, Ludka FK, Pazini FL, Rosa JM, Oliveira Á, Lopes MW, et al. Involvement of PI3K/Akt signaling pathway and its downstream intracellular targets in the antidepressant-like effect of Creatine. Mol Neurobiol. 2016;53(5):2954–2968. doi: 10.1007/s12035-015-9192-4. [DOI] [PubMed] [Google Scholar]
- 30.Shi HS, Zhu WL, Liu JF, Luo YX, Si JJ, Wang SJ, et al. PI3K/Akt signaling pathway in the basolateral amygdala mediates the rapid antidepressant-like effects of trefoil factor 3. Neuropsychopharmacology. 2012;37(12):2671–2683. doi: 10.1038/npp.2012.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lima IVA, Almeida-Santos AF, Ferreira-Vieira TH, Aguiar DC, Ribeiro FM, Campos AC, et al. Antidepressant-like effect of valproic acid-possible involvement of PI3K/Akt/mTOR pathway. Behav Brain Res. 2017;329:166–171. doi: 10.1016/j.bbr.2017.04.015. [DOI] [PubMed] [Google Scholar]
- 32.Pazini FL, Cunha MP, Rosa JM, Colla AR, Lieberknecht V, et al. Creatine, similar to ketamine, counteracts depressive-like behavior induced by Corticosterone via PI3K/Akt/mTOR pathway. Mol Neurobiol. 2016;53(10):6818–6834. doi: 10.1007/s12035-015-9580-9. [DOI] [PubMed] [Google Scholar]
- 33.Zhou W, Dong L, Wang N, Shi JY, Yang JJ, Zuo ZY, et al. Akt mediates GSK-3β phosphorylation in the rat prefrontal cortex during the process of ketamine exerting rapid antidepressant actions. Neuroimmunomodulation. 2014;21(4):183–188. doi: 10.1159/000356517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. MF results for circRNA01490. The detailed list of the genes identified as regulated by the circRNA 01490, MF: molecular function.
Additional file 2. MF results for circRNA005442. The detailed list of the genes identified as regulated by the circRNA005442, MF: molecular function.
Additional file 3. BP results for circRNA01490. The detailed list of the genes identified as regulated by the circRNA 01490, BP: biological process.
Additional file 4. BP results for circRNA005442. The detailed list of the genes identified as regulated by the circRNA005442, BP: biological process.
Additional file 5. CC results for circRNA01490. The detailed list of the genes identified as regulated by the circRNA 01490, CC: cellular component.
Additional file 6. CC results for circRNA005442. The detailed list of the genes identified as regulated by the circRNA005442, CC: cellular component.
Data Availability Statement
The data sets used and/or analyzed during the current study are available from the corresponding author on reasonable request.