Abstract
Can molecular dynamics simulations predict the mechanical behavior of protein complexes? Can simulations decipher the role of protein domains of unknown function in large macromolecular complexes? Here, we employ a wide-sampling computational approach to demonstrate that molecular dynamics simulations, when carefully performed and combined with single-molecule atomic force spectroscopy experiments, can predict and explain the behavior of highly mechanostable protein complexes. As a test case, we studied a previously unreported homologue from Ruminococcus flavefaciens called X-module-Dockerin (XDoc) bound to its partner Cohesin (Coh). By performing dozens of short simulation replicas near the rupture event, and analyzing dynamic network fluctuations, we were able to generate large simulation statistics and directly compare them with experiments to uncover the mechanisms involved in mechanical stabilization. Our single-molecule force spectroscopy experiments show that the XDoc-Coh homologue complex withstands forces up to 1 nN at loading rates of 105 pN/s. Our simulation results reveal that this remarkable mechanical stability is achieved by a protein architecture that directs molecular deformation along paths that run perpendicular to the pulling axis. The X-module was found to play a crucial role in shielding the adjacent protein complex from mechanical rupture. These mechanisms of protein mechanical stabilization have potential applications in biotechnology for the development of systems exhibiting shear enhanced adhesion or tunable mechanics.
Introduction
Molecular structure and motion can be studied using various techniques, but none offer a level of detail comparable to molecular dynamics (MD) simulations. By depicting atomic-level motions with femtosecond resolution, all-atom MD provides researchers with a unique computational microscope.1 It is crucial, however, to validate simulations by direct comparison with experiments.2,3 Once validated, MD can provide structural and mechanistic insights at extremely high resolution. One field in which insights gleaned from MD simulations can have a major impact is in the study of molecular biomechanics. Molecular biomechanics4−6 is a topic that is challenging to study experimentally because molecular level mechanical properties remain hidden to ensemble averaging methods and bioinformatic methods, and require measurements with high spatial and temporal resolution. MD simulations can therefore aid experimentalists and provide mechanistic insights of molecules under force.
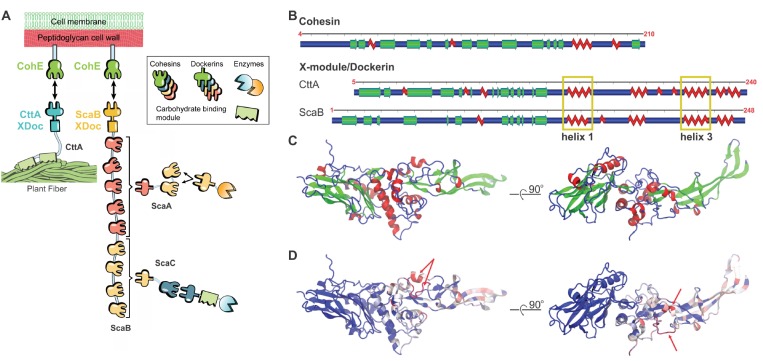
We previously reported on the extreme mechanostability among folded domains7−9 and receptor–ligand complexes10−16 derived from cellulose-degrading enzyme networks known as cellulosomes. The binding partners within these networks belong to grouped families of conserved domains called Cohesin (Coh) and Dockerin (Doc),17 comprising tens of dozens of known homologues that exhibit high affinity and specificity.18 X-modules (Xmod) are Ig-like folds frequently found as tandem dyads adjacent to scaffold-borne Docs that, despite having been heavily studied, have no known function.19 The most mechanostable Coh–Doc interaction reported to date is a complex from Ruminococcus flavefaciens (Rf) formed between the tandem dyad CttA-XDoc and the cell-wall anchored Cohesin E (CohE).12,20 In its native pulling geometry, this complex was capable of resisting forces of 500–800 pN at loading rates ranging from 2 to 300 nN/s.12
Here, we report a related protein complex that is significantly stronger. Depicted schematically in Figure 1A, Rf scaffold B (ScaB) is found within the same cellulosome as the previously reported CttA-XDoc:CohE complex12,15 and contains a C-terminal XDoc (ScaB-XDoc) that shares high sequence homology with CttA-XDoc. The role of ScaB-XDoc is to connect the large protein machinery of the Rf cellulosome to the cell wall via high-affinity binding to CohE.21−23 CohE is itself covalently linked to the peptidoglycan outer cell wall by a sortase-mediated pathway.24,25
Figure 1.
Homology model of ScaB-XDoc:CohE. (A) Schematic of the Rf cellulosome protein network for fiber degradation.20 The two cell-surface anchoring complexes are CttA-XDoc:CohE and ScaB-XDoc:CohE. (B) Representation of secondary structures of Coh (CohE) and XDoc domains from both CttA (PDB ID: 4IU3) and ScaB (model protein). (C) The structural homology model of the ScaB-XDoc:CohE complex colored by secondary structure and (D) colored by sequence similarity (BLOSUM60) to CttA-XDoc:CohE. In (D), regions indicated by red arrows are ScaB-XDoc insert sequences not present in CttA.
To investigate the adhesion forces at play in the ScaB-XDoc:CohE complex, we employed steered molecular dynamics (SMD)26−28 simulations. Using a wide sampling approach, simulations were performed with many replicas for each of the systems, allowing us to analyze experiments and simulations in the same statistical framework. Wide sampling combined with dynamic network analysis29 allowed us to visualize most probable deformation pathways through the protein architecture and understand how resistance to mechanical stress arises at the level of individual complexes. Simulation results were validated using single-molecule force spectroscopy (SMFS) experiments carried out with an atomic force microscope (AFM) over a range of loading rates. By recording the force required to break the complex for thousands of individual interactions, we built up sufficient statistics to analyze the interaction and unfolding pathways. We describe remarkable agreement between simulations and experiments, demonstrating that they probe fundamentally the same molecular process. Such an approach provided a strong basis for the molecular-level mechanistic descriptions that emerged from detailed analysis of MD simulations.
Results and Discussion
Modeling
There were no structural data available on the ScaB-XDoc:CohE complex, so we built a homology model30 based on CttA-XDoc. The primary sequences of CttA-XDoc and ScaB-XDoc are highly similar containing 47% sequence identity.20 ScaB-XDoc is slightly longer (Figure 1B), containing small sequence insertions in the Xmod domain and Doc insert regions. Doc insert regions are sequence additions within the conserved Doc sequence that make contact with Xmod in the CttA-XDoc:CohE crystal structure.20 It was suggested that these Doc inserts provide structural buttresses for the elongated stalk-like conformation of the Xmod.20 We employed MODELLER 9.1731 and used CttA-XDoc (PDB 4IU3)20 as a template to obtain a model of the ScaB-XDoc structure. We then used VMD32 to align helices 1 and 3 of the model structure (ScaB-XDoc) with those contained in the template (CttA-XDoc:CohE complex) to build a homology model of the ScaB-XDoc:CohE complex (see Figure 1C). For the two sequence gaps in the Doc insert loop regions marked by red arrows in Figure 1D, we performed a loop optimization protocol using ROSETTA.33 The obtained structure was further refined with MD simulations. Equilibration for 100 ns was performed using NAMD34,35 through its QwikMD interface.36Figure 1C,D shows the structure obtained after loop optimization and equilibration by MD simulation. The equilibration resulted in a stable complex with no major changes in conformation when compared to the initial structural model.
Mechanical Dissociation in Silico and in Vitro
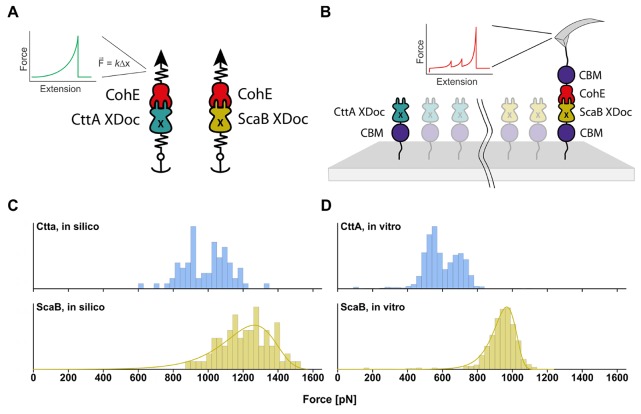
To evaluate the behavior of ScaB-XDoc:CohE under force, we first performed in silico SMD simulations28 employing NAMD34 through the QwikMD36 interface. An atom of the ligand was attached to a dummy atom via a virtual spring of known stiffness. The dummy atom was then moved at constant velocity and the force was determined using Hooke’s law. Simultaneously, an atom of the receptor was anchored to its initial position with another virtual spring of high stiffness. Specifically, the N-terminus of ScaB-XDoc was fixed and the C-terminus of CohE was pulled at a constant velocity of 5.0 Å/ns (Figure 2A). For comparison, CttA-XDoc:CohE was simulated under the same conditions. To capture the stochastic nature of the thermally driven unbinding process, 75 replicas were performed for each complex.
Figure 2.
Mechanical dissociation of ScaB-XDoc:CohE and CttA-XDoc:CohE using in silico (left) and in vitro (right) approaches. (A) Schematic illustration of the setup for simulations. Constant velocity SMD simulations were performed at a pulling speed of 5.0 Å/ns. The N-terminus of ScaB-XDoc or CttA-XDoc was restrained in space with a virtual spring, while the C-terminus of CohE was pulled by another spring. The force applied to the spring was monitored during each time step of the simulation. (B) Schematic illustration of the experimental AFM setup, which is analogous to the simulation. Both protein complexes were probed with the same Coh-modified cantilever. (C) Comparison of complex rupture forces arising from mechanical dissociation of ScaB-XDoc:CohE or CttA-XDoc:CohE from in silico MD simulations. Histograms were assembled by taking the highest force reached in each simulation trajectory. (D) Comparison of the forces arising from forced dissociation of ScaB-XDoc:CohE and CttA-XDoc:CohE from in vitro single-molecule AFM at a pulling speed of 1600 nm/s.
For in vitro AFM-SMFS experiments, CttA-XDoc and ScaB-XDoc were produced as purified recombinant fusion proteins in E. coli bearing an N-terminal carbohydrate binding module (CBM) and an N-terminal ybbR-tag. The ybbR tag allowed for covalent and site-specific surface immobilization, catalyzed by 4′-phosphopantetheinyl transferase (Sfp synthase), while the CBM domain served as a fingerprint/marker domain with known unfolding force and contour length increment that was used for filtering large-scale AFM-SMFS data sets as previously described.15 CohE was also produced bearing a C-terminal CBM fusion domain and C-terminal ybbR-tag. These surface anchoring sites (N-terminus for XDoc, C-terminus for Coh) precisely mimic the orientation of these domains within the cellulosome network in vivo. Using microwell silicon masks, ybbR-CBM-ScaB-XDoc and ybbR-CBM-CttA-XDoc were immobilized at spatially separated locations on a single glass slide (Figure 2B). This allowed us to probe both proteins with the same cantilever, which was covalently modified with CohE as described previously.8,13,37,38 In doing so, we could compare the interactions between the two Docs and CohE while circumventing errors that would arise from uncertainty in cantilever calibration, which are known to be on the order of ±10%.39,40
Upon contact between the AFM tip and the surface, an XDoc:CohE complex formed, and the cantilever was retracted at constant speed, resulting in unfolding of two CBM domains in series followed by rupture of the receptor–ligand interaction. This experiment was repeated tens of thousands of times and large-scale data sets of force vs extension curves were obtained and screened for 2× CBM unfolding contour length increments to positively identify single-molecule interactions.10
Force distributions of both in silico (Figure 2C) and in vitro (Figure 2D) experiments consist of the highest forces reached in each experimental single-molecule force–extension curve or simulation trial. In some cases, the Xmod unfolded at high force, followed by complex rupture at low force. For these simulated traces, the Xmod unfolding force was included in the histogram. If the complex dissociated with the Xmod remaining folded, the complex rupture force was included in the histogram. Both simulations and experiments show an unimodal force distribution for ScaB-XDoc:CohE that could be fitted with a two state Bell–Evans model, whereas CttA-XDoc:CohE yielded a bimodal distribution. The simulations predicted that the ScaB-XDoc:CohE interaction should withstand significantly higher forces than those of the CttA-XDoc:CohE interaction. Remarkably, the experiments confirmed this finding, with ScaB-XDoc:CohE ruptures reaching forces of ∼1 nN, ∼50% higher than those observed for CttA-XDoc:CohE at the same loading rate. This finding places ScaB-XDoc:CohE among the most mechanically stable protein receptor–ligand complexes reported to date.
Results from simulations and experiments were found to be in very good agreement despite the 6 orders of magnitude difference in force loading rate. Although the agreement is very good, we have previously reported even better agreements for rupture events in bacteria adhesion complexes.41 Here, our simulation results reveal a rupture force that is approximately 8% lower for the CttA-XDoc:CohE complex and 14% lower for the ScaB-XDoc:CohE complex, when compared to the expected forces based on extrapolations of the experimental results using a Bell–Evans model (see Supporting Information Figure S1). We believe that force field parameters and the use of a homology modeling protocol may be responsible for these small differences.
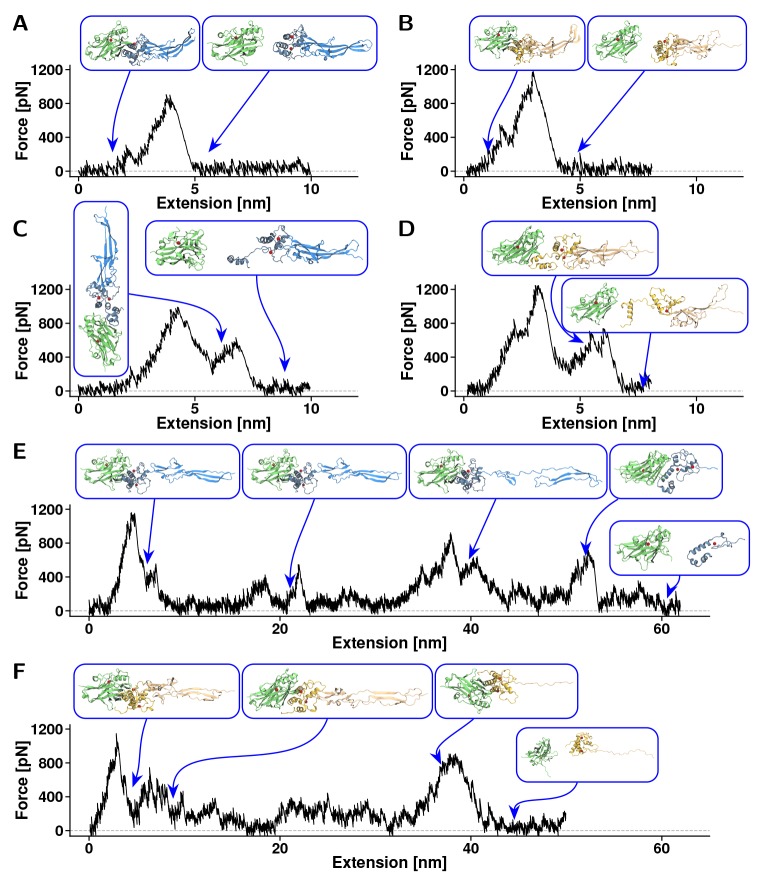
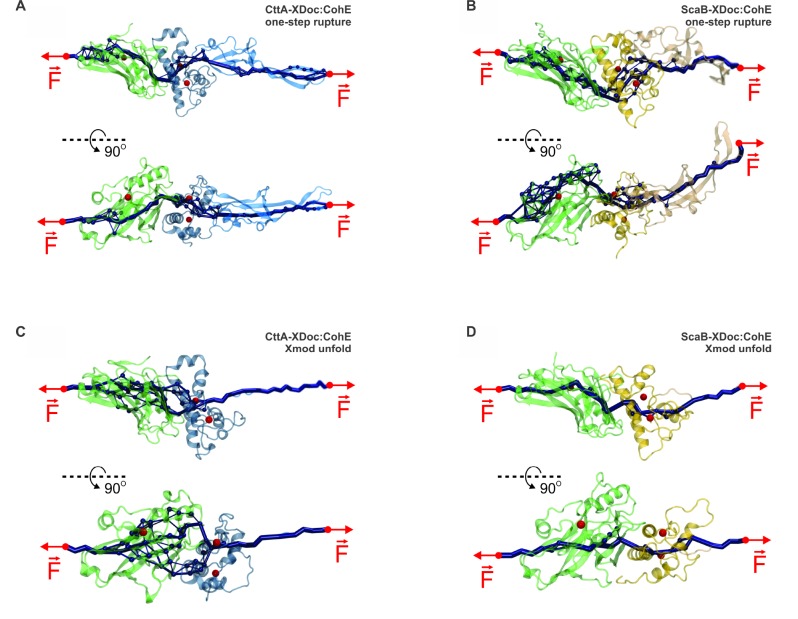
Both ScaB-XDoc:CohE and CttA-XDoc:CohE interactions were observed to dissociate along at least three different unbinding pathways, which can be seen in Figures 3 and 4A, B. MD simulations revealed the complexes were either broken without prior domain unfolding (one-step event, Figure 3A,B), at nearly the same time as the region connecting the Xmod to the Doc binding helices unfolds (Figure 3C,D), or accompanied by a previous complete Xmod domain unfolding (see Figure 3 E,F). The frequency of occurrence of each of these unbinding pathways is shown in Table 1. In the second class of unfolding trajectories (Figure 3C,D), contact was lost between Doc insert regions and Xmod followed by loss of Doc secondary structure. In these cases, concerted Doc unfolding and complex rupture exhibited highly variable behavior among the various simulation replicas. It is worth emphasizing that the triggering factor for a possible concerted unbinding-unfolding was the loss of contact between Doc inserts and Xmod. As was proposed by Salama-Alber et al.,20 these regions would presumably lend structural reinforcement to the stalk-like conformation of these Xmods. Our results agree with the proposed model, providing dynamical information to the atomic data. The sequence additions within the inset regions of the ScaB Doc domain provide for a stronger contact with XMod and are likely playing a role in the higher mechanical stability of the ScaB-XDoc:CohE complex as compared with the CttA-XDoc:CohE complex. However, we were not able to identify a clear unfolding path in these concerted events, and Figure 3C,D shows exemplary force traces with intermediates that have no statistical significance. These results indicated that, under high-tensile forces, Doc can unfold nearly simultaneously with complex dissociation. Yet some of the Coh:Doc contacts were present for a couple of nanoseconds before the complex broke apart completely (see Figure 3C,D). Thus, complex rupture shows a multistep force–distance trace, with the final separation of Coh and Doc happening as a shielded (lower force) event.
Figure 3.
Exemplary force–extension traces from steered molecular dynamics simulations. Plots represent the three types of rupture events observed in CttA-XDoc:CohE and ScaB-XDoc:CohE simulations. (A) For CttA-XDoc:CohE, one-step rupture events were observed in 57 out of 75 replicas. (B) For ScaB-XDoc:CohE, one step rupture events were observed in 34 out of 75 replica simulations. In the one-step event, all three domains (Coh, Doc, Xmod) remained intact following rupture. (C) For CttA-XDoc:CohE, a destabilization of Doc during breakage was observed in 16 out of 75 replicas. (D) For ScaB-XDoc:CohE, Doc destabilization occurred in 39 out of 75 replicas. Doc partial unfolding led to multipeak force vs extension traces that were highly variable in replica simulations. (E) For CttA-XDoc:CohE, Xmod unfoding was observed in 2 out of 75 replicas. (F) For ScaB-XDoc:CohE, Xmod unfolding was also observed in 2 out of 75 replicas. Xmod unfolding led to multipeak traces that were variable in each trajectory. The first and largest peak in these cases corresponded to the loss of contact between Xmod and Doc insert regions.
Figure 4.
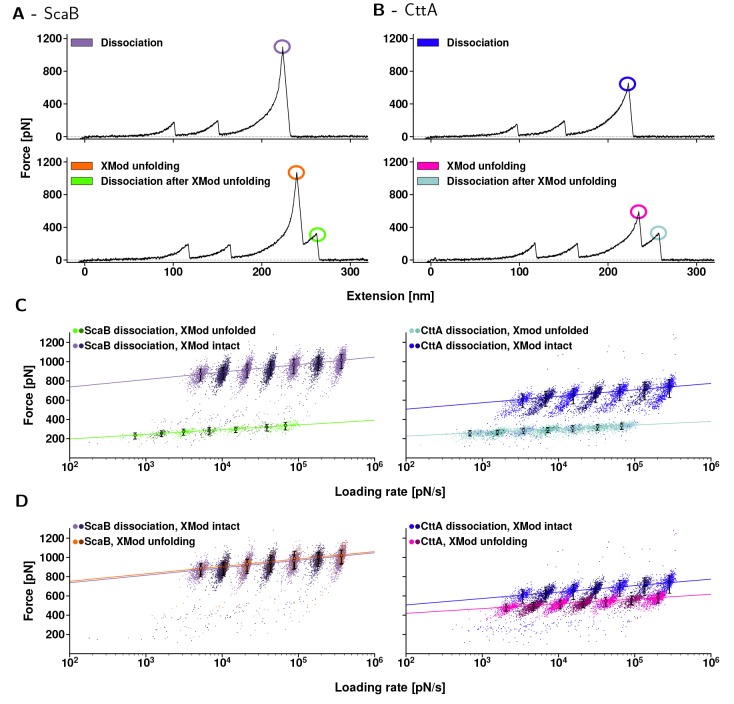
Dynamic force spectra of CttA-XDoc:CohE (left) ScaB-XDoc:CohE (right). (A, B) Typical force–extension traces. First, both CBM-domains unfolded. Their known unfolding behavior served as a fingerprint, indicating that single molecular complexes were probed. Then, for both ScaB-XDoc:CohE and CttA-XDoc:CohE, Xmod remained either folded until complex rupture (upper traces, purple and blue) or unfolded (lower traces, orange and pink) prior to complex rupture. When Xmod unfolding occurred, both complexes ruptured at markedly lower forces (bright green and teal). (C, D) Dynamic force spectra for each class of unfolding or unbinding events that are encircled in (A, B). The colors match the corresponding events in (A, B), and a different color saturation was chosen for every other pulling speed to display the resulting populations more clearly. Data were fitted with the two-state Bell–Evans model. (C) Complex rupture forces. In cases where Xmod remained folded, the ScaB-XDoc:CohE complex ruptured at markedly higher forces than did CttA-XDoc:CohE over the entire range of loading rates tested (purple vs blue). When the Xmod unfolded, the complexes showed nearly identical rupture behavior (bright green vs teal). (D) Comparison of the peak forces reached in both unbinding pathways. The data points either stem from complex rupture events for traces lacking Xmod unfolding, or from unfolding of the Xmod. Interestingly, the most probable unfolding force of ScaB’s Xmod is about the same as the ScaB-XDoc:CohE complex dissociation forces that occur when Xmod remained intact (orange vs purple). The same was not true for CttA-XDoc:CohE, where Xmod unfolding forces were surpassed by complex dissociation forces with no prior Xmod unfolding (blue vs pink). The likelihood of observing Xmod unfolding prior to complex unbinding was only 7% for ScaB-XDoc, as compared to 43% for CttA-XDoc.
Table 1. Occurrence of Each Unbinding Pathway Observed in the SMD Simulations.
| system | one-step rupture | Doc concerted unfolding | Xmod unfolding |
|---|---|---|---|
| ScaB-XDoc:CohE | 34/75 | 39/75 | 2/75 |
| CttA-XDoc:CohE | 57/75 | 16/75 | 2/75 |
The frequency of occurrence of the various unbinding pathways obtained from simulations is shown in Table 1. These data indicate that ScaB-XDoc:CohE unbinding is more often accompanied by Doc unfolding than is CttA-XDoc:CohE unbinding. Since ScaB-XDoc and CttA-XDoc share nearly identical sequence and structure at the Coh:Doc binding interface, we attributed this difference to the higher forces reached for ScaB-XDoc:CohE.
In the AFM-SMFS experiments, we could distinguish only two unbinding behaviors for both ScaB-XDoc:CohE (Figure 4A) and CttA-XDoc:CohE (Figure 4B). One pathway that was consistently observed corresponded to simulation pathway 1 (one-step rupture), where Xmod remained folded prior to rupture of the complex at high force (Figure 4A and B, top). A second experimental pathway corresponding to simulation pathway 3 (Xmod unfolding) was also repeatedly observed in the experiments where Xmod completely unfolded prior to complex dissociation (Figure 4A and B, bottom). Following Xmod unfolding for both CttA-XDoc and ScaB-XDoc, the Doc:CohE complex was significantly destabilized and ruptured at lower forces. We did not detect AFM-SMFS traces corresponding to simulation pathway 2 (Doc concerted unfolding), likely due to the nanosecond lifetime of the intermediate state and the finite response time of the AFM cantilever. The short-lived state likely transitioned to a completely unbound state before the released contour length of the Doc could be resolved by AFM.
From SMFS experiments, we analyzed force-loading rate data of rupture events obtained over a range of loading rates (i.e., dynamic force spectroscopy42), as shown in Figure 4C,D. Out of 16 034 single-molecule traces, unfolding of the Xmod occurred in only 7% of all in vitro traces for ScaB-XDoc, making this unfolding pathway much less frequent, as compared to 43% of all in vitro traces for CttA-XDoc. For both ScaB-XDoc and CttA-XDoc, complex dissociation forces were drastically reduced following Xmod unfolding (Figure 4C). Interestingly, when comparing ScaB-XDoc:CohE vs CttA-XDoc:CohE complexes both with unfolded Xmod domains, higher mechanostability of ScaB-XDoc:CohE was no longer observed. Complex dissociation of ScaB-XDoc:CohE with an unfolded Xmod domain (Figure 4C, bright green) was not stronger than complex dissociation of CttA-XDoc:CohE with its Xmod unfolded (Figure 4C, teal). The similar behavior observed for the two systems when Xmod is unfolded is an indication that the higher forces reached by ScaB-XDoc:CohE stem from differences in the Xmod rather than from differences in the binding interface between Doc and Coh. For ScaB-XDoc:CohE, the dynamic force spectrum of XMod domain unfolding (Figure 4D, orange) was indistinguishable from that of the ScaB-XDoc:CohE complex rupture with an intact Xmod (Figure 4D, purple), indicating that both events may initially arise from a common energy barrier. The same is not true for CttA-XDoc:CohE, where Xmod unfolding events (Figure 4D, blue) occurred at lower forces than the complex dissociation events with no prior Xmod unfolding (Figure 4D, pink). Combining these two events into one histogram yields the bimodal distribution for CttA-XDoc:CohE as displayed in Figure 2D. Rupture force histograms of the data presented in Figure 4 are available in Supporting Information Figures S2–S5. It is worth noting that the exceptionally high rupture forces measured both in silico and in vitro are disproportionate to the equilibrium thermodynamic affinity for both complexes. The KD of CttA-XDoc:CohE obtained by isothermal titration calorimetry (see Supporting Information Figure S6) was 18 nM while the KD of ScaB-XDoc:CohE was 149 nM, both of which are comparable to other Coh:Doc interactions. The discrepancy between the equilibrium analysis, where Ctta-XDoc:CohE exhibits higher affinity, and the mechanical analysis, where ScaB-XDoc:CohE exhibits higher rupture forces, highlights how mechanical dissociation can direct unbinding reactions along pathways that are distinct from those dominant in the absence of force.
High-Force Mechanism
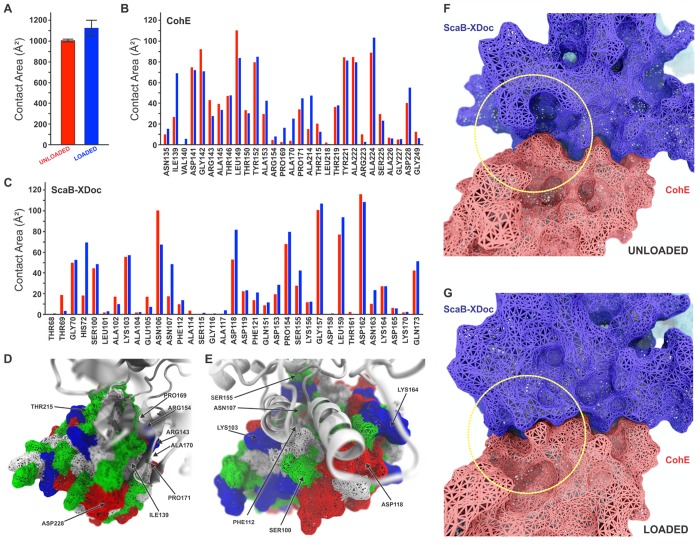
An increase in contact area upon application of force was observed in previous SMD simulations of the CttA-XDoc:CohE complex.12 Taking advantage of the recently developed PyContact,43 we further analyzed our simulation trajectories by quantifying the contact surface area between CohE and ScaB-XDoc in both loaded and unloaded scenarios. For the loaded scenario, we analyzed all trajectories immediately prior to complex rupture over a simulation window of 4.0 ns. For the unloaded scenario, we analyzed equilibrium MD simulations, also over a 4.0 ns window. The total contact area was found to increase upon loading due to a rearrangement of interacting residues, as shown in Figure 5A. We further analyzed the contact surface area per residue (Figure 5B, C), and plotted the contact surface area for main interacting residues as shown in Figure 5D and E.
Figure 5.
Analysis of ScaB-XDoc:CohE binding interface under force from SMD. (A) Average contact surface area between CohE and ScaB-XDoc under unloaded and loaded conditions. The contact area increases under force load condition. The contacts were calculated using PyContact from 4 ns trajectory windows that were combined for all 75 trajectories. Standard deviations were calculated from these combined windows and are presented as error bars. (B) Surface contact area of interacting residues of CohE under unloaded (red) and loaded (blue) conditions. (C) Surface contact area of interacting residues of Doc under unloaded (red) and loaded (blue) conditions. (D,E) Surface of Doc (D) and Coh (E) showing main interface residues. Hydrophobic residues are shown in white, polar residues in green, and negative and positive residues in red and blue, respectively. (F,G) Rearrangement of binding residues of Coh (red) and Doc (blue) under force. The yellow circle shows the region of the binding interface where a rearrangement increases the contact surface from the unloaded (F) to the loaded (G) configurations.
ScaB-XDoc (Figure 5D) and CohE (Figure 5E) both exhibit a binding interface comprising a hydrophobic core (white) surrounded by polar (green) and charged residues (blue, positive; red, negative). This residue pattern suggests that the hydrophilic side chains prevent access of solvent to the interior core. Such hot spots of binding are a common motif in protein–protein binding interfaces, referred to as O-rings.44 Contact area between the two proteins increased upon mechanical loading based on rearrangement of side chains and closing off of the hydrophobic core to solvent (Figure 5F, G).
Exemplary Force Propagation Pathways
To investigate how strain propagates through the protein complex, we employed cross-correlation based network analysis to study one-step rupture events and rupture events following Xmod unfolding. From thermodynamic fluctuation theory, one can infer that paths with high correlation of motion describe the paths along which force propagates through a molecular system.15,45 The relationship between fluctuations of atoms i and j and the force F⃗i on atom i can be described by
| 1 |
where Δr⃗i = r⃗i(t) – ⟨r⃗i(t)⟩ and r⃗i is the position of atom i. If an external force F⃗i acts on atom i and the potential between atoms i and j is steep, the two atoms will move with high positional correlation. By calculating a given element of the correlation matrix Mij = ⟨Δr⃗iΔr⃗jT⟩, we can quantify the strength of an interaction potential between i and j. The deformation response of the molecular complex under applied force will be dominated by the stiff interaction potentials, while the soft potentials become insignificant.15 The propagation of force through the molecules and across the binding interface can then be visualized along the pathways with high correlations of motion. The high-correlation path with the smallest length, namely, the smallest number of amino acid residues, is considered to be the optimal path. When multiple paths of similar length (number of nodes) are found to have high correlation, then these are considered suboptimal paths. It is important to notice that not all residues along these paths need be considered to have the same importance for force propagation. Instead, only residues or interactions that occur in the highest number of suboptimal pathways need to be conserved to guarantee an effective force propagation pathway in the complex.15,29
Although the quasi-harmonic approximation of Pearson correlation is commonly employed in correlation analysis of molecular systems,46−51 it is not a priori justified for complicated biomolecular interactions.52 However, since our analysis relies on the identification of paths of highest correlation through proximate residues, the quasi-harmonic approximation implied using Pearson correlation is justified, particularly for suboptimal pathway analysis.15
In Figure 6, the force propagation pathways through both ScaB-XDoc:CohE (Figure 6A, C) and CttA-XDoc:CohE (Figure 6B, D) complexes are depicted. Figure 6A and B shows correlation networks obtained from one-step rupture event trajectories for ScaB-XDoc:CohE and CttA-XDoc:CohE, respectively. Figure 6C and D shows correlation networks obtained from simulations exhibiting Xmod unfolding prior to rupture. The depicted paths through the system are those along which stress primarily propagates under load, obtained just prior to rupture over a simulation window of 4.0 ns.
Figure 6.
Evolution of force propagation pathways. Force propagation pathways were calculated from correlation-based network analysis carried out using SMD trajectories within 4.0 ns windows just prior to rupture. For one-step rupture of complexes CttA-XDoc:CohE (A) and ScaB-XDoc:CohE (B), force-propagation routes nonparallel to the pulling axis are observed near the Doc:Coh interface. For complex rupture following Xmod unfolding in CttA-XDoc:CohE (C) and ScaB-XDoc:CohE (D) simulation, we again observed redirection of force near XDoc:Coh interface. The thickness of the dark blue tubes represents the number of suboptimal correlation paths connecting two nodes. Thicker tubes correspond to higher likelihood of force propagating through a path. The observed changes in force direction, presented mostly as sharp turns in force propagation routes particularly near the interface, suggest that the ultrastable complexes formed by Coh and Doc achieve mechanostability by directing externally applied forces normal to the pulling axis. Consequently, more force is required to achieve a given separation along the pulling axis.
For the one-step rupture trajectories, it was previously shown for CttA-XDoc:CohE that force propagated through both binding helices of the Doc and along pathways with large normal components with respect to the pulling axis.12 The thickness of the lines in Figure 6 indicates how important these pathways are. That is calculated by considering the multiple possible paths of similar length with high-correlation. Important nodes (amino acid residues) are those that are present in a large number of paths. Likewise, important edges (a line connecting two nodes) are those edges that are part of a large number of paths. Therefore, the most important pathways are those that are shown with thicker lines. Although clear differences between the four force pathways shown in Figure 6 are evident, one can easily observe that these important pathways all have “sharp turns” near the interface. These force-propagation routes nonparallel to the pulling axis for ScaB-XDoc:CohE are advantageous for achieving high dissociation forces.
It was previously shown for another ultrastable protein, namely, silk crystalline units, that curving and branching force paths distributed tension through the entire system.53 A strategy that assumes an indirect path would allow the system to more evenly utilize the interface between binding partners. Such behavior can be simplified in a mechanical picture, where a certain amount of mechanical work dW = F⃗·ds⃗ is required to separate two binding interfaces by a distance Δz and break the interaction. If force F⃗ is locally perpendicular to the direction of the unbinding axis ds⃗, a larger force is required to break the interaction than in a scenario where the force path points along the unbinding axis.
Analysis of the correlation maps indicated that the calcium-binding loops were key in distributing forces through the Coh:Doc binding interface. In all scenarios, force propagated through the calcium loops. Even after unfolding of Xmod, force-propagation through the folded Doc remained largely unchanged. As shown in Figure 4C, complex rupture forces were greatly reduced when the Xmod was unfolded, but remained on the order of 200 pN at the given loading rates. Overall, our correlation analysis supports the view that directing force along a path with significant perpendicular component to the pulling axis leads to high mechanical stability.
Conclusion
The detailed molecular mechanisms of cellulosomes are of broad interest, particularly given their applications in biotechnology54 and their recent discovery as part of the human gut microbiome.55,56 The ScaB-XDoc:CohE receptor–ligand complex is essential for bacterial degradation of cellulose, serving as an adhesion domain and anchoring its organism to a primary carbon source.
The multistep computational pipeline we described to understand the ScaB-XDoc:CohE complex involved homology modeling, wide-sampling of steered molecular dynamics simulations, contact area analysis, and finally dynamic network analysis. We developed this pipeline in order to structurally characterize the ScaB-XDoc:CohE complex, computationally predict its high resistance to applied forces, and understand the molecular mechanisms involved in force resistance. We furthermore were able to confirm the predicted high forces experimentally using parallel high volume acquisition of single-molecule AFM force traces.
The additional strength observed for the ScaB-XDoc:CohE complex is in agreement with its function. The actual load on this complex, with its attached Doc-bearing enzymes and other subunits, would seem to justify the improved mechanical stability of the ScaB-XDoc:CohE complex over the CttA-XDoc:CohE. The primary differences in behavior between ScaB-XDoc and the previously characterized CttA-XDoc arise from the Xmod region, which is significantly longer in ScaB. Unfolding of the Xmod domain prior to complex rupture caused a strong destabilization of the complex, and eliminated the improved mechanical stability of ScaB-XDoc:CohE over CttA-XDoc:CohE complexes. As the ScaB’s Xmod unfolded far less frequently and at higher forces than its CttA analogue, its larger structure rendered it more robust, improving the stabilizing effect it had on the Doc:CohE complex. Taken together, these results depict the Xmod’s function as a structural support of the Doc’s binding helices under high loads, acting in part as a mechanical shield to protect the adjacent Doc from unfolding. Our wide-sampling simulation approach with many simulation replicas was critical for interpreting the mechanical behavior of these receptor–ligand complexes, particularly in describing the mechanisms of mechanical stability. In the future, successful merger of SMD simulations with single-molecule mechanical experiments will benefit from the analysis pipeline presented here in order to gain insight into the fascinating interplay between equilibrium protein binding affinity and adhesion mechanics.
Methods
Structural Model
The structure of the CttA-XDoc:CohE complex has been solved by means of X-ray crystallography at 1.97 Å resolution and is available in the Protein Data Bank (PDB: 4IU3).20 The second system, namely, ScaB-XDoc, had no structure available and was solved by a homology modeling strategy.30 The construction of Coh models was performed using MODELLER 9.1731 software that employs spatial restriction techniques based on the 3D-template structure. The best model was selected by analyzing the stereochemical quality check using PROCHECK57 and overall quality by ERRAT server.58 The structures of two sequence gaps in the loop regions marked by arrows in Figure 1C were resolved by a loop optimization protocol employing ROSETTA.33 Using NAMD34,35 through its QwikMD interface,36 structures were subjected to 100 ns of equilibrium MD to ensure conformational stability. All structures shown are from postequilibration MD simulations.
Molecular Dynamics Simulations
MD simulations were performed employing the NAMD molecular dynamics package.34 The CHARMM36 force field59 along with the TIP3 water model60 was used to describe all systems. Simulations were carried out assuming periodic boundary conditions in the NpT ensemble with temperature maintained at 300 K using Langevin dynamics for pressure, kept at 1 bar, and temperature coupling. A distance cutoff of 14.0 Å was applied to short-range, nonbonded interactions, whereas long-range electrostatic interactions were treated using the particle-mesh Ewald (PME) method. The equations of motion were integrated using the r-RESPA multiple time step scheme34 to update the van der Waals interactions every two steps and electrostatic interactions every four steps. The time step of integration was chosen to be 2 fs for all simulations performed.
In a wide-sampling strategy,61 75 steered molecular dynamics (SMD) runs were carried out for a total of 1.5 μs for each system. To characterize the coupling between dockerin and cohesin, SMD simulations28 with constant velocity stretching (SMD-CV protocol) at 5.0 Å/ns pulling speed were carried out. In all simulations, SMD was employed by restraining the position the N-terminal of ScaB-XDoc or CttA-XDoc, while pulling on the C-terminus of CohE. The procedure is equivalent to attaching one end of a harmonic spring to the end of one domain and pulling on the other domain with a spring, analogous to the experiment. The force applied to the harmonic spring was then monitored during the time of the molecular dynamics simulation.
All analyses of MD trajectories were carried out employing VMD32 and its plugins. Surface contact areas of interacting residues were calculated employing PyContact.43 The Network View plugin29 on VMD was employed to perform dynamical network analysis. A network was defined as a set of nodes comprising all α-carbons connected together by edges. Edges connect pairs of nodes if corresponding monomers are in contact, and two nonconsecutive monomers are said to be in contact if they fulfill a proximity criterion, namely, any heavy atoms (non-hydrogen) from the two monomers are within 4.5 Å of each other for at least 75% of the frames analyzed. As suggested by Sethi et al.,29 nearest neighbors in sequence are not considered to be in contact as they lead to a number of trivial suboptimal paths. Conceptually, suboptimal paths can be understood as allosteric signaling pathways or force propagation pathways. Suboptimal paths are defined as paths that are slightly longer than the optimal path, with a given suboptimal path visiting a node not more than once. These multiple communication paths are nearly equal in length, and not all residues along these paths need be considered important for allostery.
Instead, only residues or interactions that occur in the highest number of suboptimal pathways need to be conserved to guarantee an effective pathway for allosteric communication. The thickness of the edges connecting the nodes reveals the least and most used paths. Allostery can be understood in terms of pathways of residues that efficiently transmit energy, here in the form of mechanical stress,15,62,63 between different binding sites. The dynamical networks were constructed from 4 ns windows of the total trajectories. Using the Floyd–Warshall algorithm, the suboptimal paths were then calculated. The tolerance value used for any path to be included in the suboptimal path was −ln0.5 = 0.69. To calculate the relevance of off-diagonal terms in the correlation matrix, we employed Carma to calculate a correlation matrix where x, y, and z components of each atom were considered independently.
Protein Preparation for Experiments
All proteins were expressed from pET28a vectors using standard induction and expression protocols in NiCo21(DE3)RIPL cells, which were cultivated in ZYM-5052 autoinduction media64 supplemented with kanamycin and chloramphenicol. After pelleting, cells were lysed by sonication and then centrifuged at 4 °C, 39 000 rcf for 60 min. The supernatant was filtered through a 0.22 PES membrane (Carl Roth + Co. KG, Karlsruhe, Germany) and applied to Ni-NTA columns (HisTrap FF, GE Healthcare Europe GmbH, Freiburg, Germany). After washing with 6 column volumes of a buffer containing 25 mM TRIS, pH 8.4, 300 mM NaCl, 20 mM imidazole, and 0.5% (v/v) Triton X-100, the bound fraction was eluted with a buffer containing 25 mM TRIS, pH 8.4, 300 mM NaCl, and 300 mM imidazole. All protein solutions were concentrated using Amicon centrifugal filter units (10K MWCO, Merck KGaA, Darmstadt, Germany), followed by buffer exchange against Ca-TBS (25 mM TRIS, pH 7.2, 75 mM NaCl, and 1 mM CaCl2) buffer using ZebaSpin columns (Zeba spin desalting columns 7K, Thermo Fisher Scientific Inc.). Proteins were stored at −80 °C, with glycerol added to 10% (v/v).
Surface Functionalization
Glass surfaces and silicon nitride cantilevers (BioLever mini BL-AC40TS-C2, Olympus, Tokio, Japan) were silanized with (3-aminopropyl)-dimethyl-ethoxysilane (APDMES, ABCR GmbH, Karlsruhe, Germany). Utilizing silicon masks (CultureWell Reusable Gaskets, Grace Bio-Laboratories, Bend, OR), two spatially separated spots on the silanized glass surfaces were PEGylated with α-maleimindo-hexanoic-ω-NHS polyethylene glycol (NHS-PEG5000-Mal, Rapp Polymere, Tübingen, Germany) dissolved into 25 mM in 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer (HEPES), 50 mM, pH 7.5. Cantilevers were PEGylated using the same conditions. Next, the PEGylated surfaces and cantilevers were coupled to Coenzyme A (CoA, 1 mM) in sodium phosphate buffer, pH 7.2. Finally ybbR-CBM(C63S)-ScaB-XDoc or ybbR-CBM(C63S)-CttA-XDoc was covalently immobilized onto the two spatially separated spots on the glass slide via their ybbR-tags in an Sfp-catalyzed ligation at room temperature for 30 min. Each Dockerin was diluted to 0.5 μM in Ca-TBS supplemented with 20 mM MgCl2, while the Sfp enzyme was added to 1 μM. CohE-CBM(C63S)-ybbR was coupled to cantilevers at a concentration of 20 μM under the same conditions.
Single-Molecule Force Spectroscopy
Measurements were performed in Ca-TBS buffer using custom built AFM instruments (driven vertically by PI-731 piezo actuators and laterally by a 25 × 25 mm piezomotor (U-751) in combination with a 100 × 100 nm (P-734) stage, Physik Instrumente, Germany) in conjunction with MFP-3D AFM controllers (Asylum Research, Santa Barbara, CA). Upon approaching the sample surface with the cantilever tip, the complex between CohE and either CttA-XDoc or ScaB-XDoc was formed and the cantilever was retracted from the surface at constant velocities of 100, 200, 400, 800, 1600, 3200, and 6400 nm/s. After each force–extension curve was acquired, the sample was moved laterally by 100 nm in order to probe a different molecule. Every several hundred measurements, the glass slide was moved laterally between protein spots, such that alternatingly CohE-ScaB-Doc and CohE-CttA-Doc complexes were probed throughout the measurement. In this manner, thousands of force–extension curves were automatically acquired over a measurement time of 24–72 h. Single-molecule interaction traces were identified by filtering the data sets using contour length analysis, and identifying only those traces in which two CBM unfolding events were observed.11 Traces exhibiting two CBM unfolding length increments were then analyzed to create rupture event scatter plots describing the rupture of the XDoc:CohE complexes.
Acknowledgments
We would like to thank Prof. Klaus Schulten (in memoriam), whose contribution to this work, particularly at its early stages, is of inestimable value. Support for this work was provided by the EU seventh Framework Programme NMP4-SL-2013-604530 (CellulosomePlus), the Nanosystems Initiative Munich and the ERC Advanced Grant CelluFuel. M.A.N. acknowledges support from an ERC Starting Grant (#715207). This work was supported by National Institutes of Health (NIH) Grant P41-GM104601, “Center for Macromolecular Modeling and Bioinformatics”. R.C.B. is partially supported by the National Science Foundation (NSF) Grant MCB-1616590, “Molecular Modeling of Bioenergetic Systems”, as well as the Energy Biosciences Institute. Equilibrium molecular dynamics simulations made use of ALCF/Mira and NERSC/Edison supercomputers as part of the DoE ALCC program. This research partially used resources of the Argonne Leadership Computing Facility (ALCF), which is a DOE Office of Science User Facility supported under Contract DE-AC02-06CH11357. This research partially used resources of the National Energy Research Scientific Computing Center (NERSC), a DOE Office of Science User Facility supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Steered molecular dynamics simulations were performed on GPU-accelerated nodes of Blue Waters supercomputer as part of the Petascale Computational Resource (PRAC) Grant The Computational Microscope, which is supported by the National Science Foundation (Award Number ACI-1713784). The state of Illinois and the National Science Foundation (Awards OCI-0725070 and ACI-1238993) support Blue Waters sustained-petascale computing project. We thank Lukas F. Milles for providing the force curve analysis software, as well as Thomas Nicolaus and Angelika Kardinal for laboratory assistance. We thank Marcelo C. R. Melo for the help with SMD analysis and Maximilian Scheurer for the help with PyContact.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.9b06776.
Protein sequences, ITC data, rupture force histograms, dynamic force spectrum combining in vitro and in silico experiments (PDF)
Author Contributions
○ R.C.B. and E.D. contributed equally.
The authors declare no competing financial interest.
Supplementary Material
References
- Perilla J. R.; Goh B. C.; Cassidy C. K.; Liu B.; Bernardi R. C.; Rudack T.; Yu H.; Wu Z.; Schulten K. Molecular dynamics simulations of large macromolecular complexes. Curr. Opin. Struct. Biol. 2015, 31, 64–74. 10.1016/j.sbi.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson S.; Wu H.; Paul F.; Clementi C.; Noé F. Combining experimental and simulation data of molecular processes via augmented Markov models. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 8265–8270. 10.1073/pnas.1704803114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottaro S.; Lindorff-Larsen K. Biophysical experiments and biomolecular simulations: A perfect match?. Science 2018, 361, 355–360. 10.1126/science.aat4010. [DOI] [PubMed] [Google Scholar]
- Schönfelder J.; Alonso-Caballero A.; De Sancho D.; Perez-Jimenez R. The life of proteins under mechanical force. Chem. Soc. Rev. 2018, 47, 3558–3573. 10.1039/C7CS00820A. [DOI] [PubMed] [Google Scholar]
- Alonso-Caballero A.; Schönfelder J.; Poly S.; Corsetti F.; De Sancho D.; Artacho E.; Perez-Jimenez R. Mechanical architecture and folding of E. coli type 1 pilus domains. Nat. Commun. 2018, 9, 2758. 10.1038/s41467-018-05107-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milles L. F.; Unterauer E. M.; Nicolaus T.; Gaub H. E. Calcium stabilizes the strongest protein fold. Nat. Commun. 2018, 9, 4764. 10.1038/s41467-018-07145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnoo M.; Cazade P.-A.; Galera-Prat A.; Nash M. A.; Czjzek M.; Cieplak M.; Alvarez B.; Aguilar M.; Karpol A.; Gaub H.; Carrin-Vzquez M.; Bayer E. A.; Thompson D. Nanoscale Engineering of Designer Cellulosomes. Adv. Mater. 2016, 28, 5619–5647. 10.1002/adma.201503948. [DOI] [PubMed] [Google Scholar]
- Verdorfer T.; Bernardi R. C.; Meinhold A.; Ott W.; Luthey-Schulten Z.; Nash M. A.; Gaub H. E. Combining in Vitro and in Silico Single-Molecule Force Spectroscopy to Characterize and Tune Cellulosomal Scaffoldin Mechanics. J. Am. Chem. Soc. 2017, 139, 17841–17852. 10.1021/jacs.7b07574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnoo M.; Cazade P.-A.; Orlowski A.; Chwastyk M.; Liu H.; Ta D. T.; Cieplak M.; Nash M.; Thompson D. Steered molecular dynamics simulations reveal the role of Ca2+ in regulating mechanostability of cellulose-binding proteins. Phys. Chem. Chem. Phys. 2018, 20, 22674–22680. 10.1039/C8CP00925B. [DOI] [PubMed] [Google Scholar]
- Stahl S. W.; Nash M. A.; Fried D. B.; Slutzki M.; Barak Y.; Bayer E. A.; Gaub H. E. Single-molecule dissection of the high-affinity cohesin–dockerin complex. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 20431–20436. 10.1073/pnas.1211929109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobst M. A.; Schoeler C.; Malinowska K.; Nash M. A. Investigating receptor-ligand systems of the cellulosome with AFM-based single-molecule force spectroscopy. J. Visualized Exp. 2013, e50950 10.3791/50950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeler C.; Malinowska K. H.; Bernardi R. C.; Milles L. F.; Jobst M. a.; Durner E.; Ott W.; Fried D. B.; Bayer E. a.; Schulten K.; Gaub H. E.; Nash M. a. Ultrastable cellulosome-adhesion complex tightens under load. Nat. Commun. 2014, 5, 5635. 10.1038/ncomms6635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten M.; Ott W.; Jobst M. A.; Milles L. F.; Verdorfer T.; Pippig D. A.; Nash M. A.; Gaub H. E. From genes to protein mechanics on a chip. Nat. Methods 2014, 11, 1127–1130. 10.1038/nmeth.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobst M. A.; Milles L. F.; Schoeler C.; Ott W.; Fried D. B.; Bayer E. A.; Gaub H. E.; Nash M. A. Resolving dual binding conformations of cellulosome cohesin-dockerin complexes using single-molecule force spectroscopy. eLife 2015, 4, e10319 10.7554/eLife.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeler C.; Bernardi R. C.; Malinowska K. H.; Durner E.; Ott W.; Bayer E. A.; Schulten K.; Nash M. A.; Gaub H. E. Mapping mechanical force propagation through biomolecular complexes. Nano Lett. 2015, 15, 7370–7376. 10.1021/acs.nanolett.5b02727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott W.; Jobst M. A.; Schoeler C.; Gaub H. E.; Nash M. A. Single-molecule force spectroscopy on polyproteins and receptor–ligand complexes: The current toolbox. J. Struct. Biol. 2017, 197, 3–12. 10.1016/j.jsb.2016.02.011. [DOI] [PubMed] [Google Scholar]
- Smith S. P.; Bayer E. A. Insights into cellulosome assembly and dynamics: from dissection to reconstruction of the supramolecular enzyme complex. Curr. Opin. Struct. Biol. 2013, 23, 686–694. 10.1016/j.sbi.2013.09.002. [DOI] [PubMed] [Google Scholar]
- Mechaly A.; Yaron S.; Lamed R.; Fierobe H.-P.; Belaich A.; Belaich J.-P.; Shoham Y.; Bayer E. A. Cohesin-dockerin recognition in cellulosome assembly: Experiment versus hypothesis. Proteins: Struct., Funct., Genet. 2000, 39, 170–177. . [DOI] [PubMed] [Google Scholar]
- Fontes C. M.; Gilbert H. J. Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 2010, 79, 655–681. 10.1146/annurev-biochem-091208-085603. [DOI] [PubMed] [Google Scholar]
- Salama-Alber O.; Jobby M. K.; Chitayat S.; Smith S. P.; White B. A.; Shimon L. J.; Lamed R.; Frolow F.; Bayer E. A. Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES. J. Biol. Chem. 2013, 288, 16827–16838. 10.1074/jbc.M113.466672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditto I.; et al. Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 7136–7141. 10.1073/pnas.1601558113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bule P.; Alves V. D.; Israeli-Ruimy V.; Carvalho A. L.; Ferreira L. M. A.; Smith S. P.; Gilbert H. J.; Najmudin S.; Bayer E. A.; Fontes C. M. G. A. Assembly of Ruminococcus flavefaciens cellulosome revealed by structures of two cohesin-dockerin complexes. Sci. Rep. 2017, 7, 759–759. 10.1038/s41598-017-00919-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israeli-Ruimy V.; et al. Complexity of the Ruminococcus flavefaciens FD-1 cellulosome reflects an expansion of family-related protein-protein interactions. Sci. Rep. 2017, 7, 42355. 10.1038/srep42355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon M. T.; Čepeljnik T.; Martin J. C.; Lamed R.; Barak Y.; Bayer E. A.; Flint H. J. Unconventional mode of attachment of the Ruminococcus flavefaciens cellulosome to the cell surface. Journal of bacteriology 2005, 187, 7569–7578. 10.1128/JB.187.22.7569-7578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rincon M. T.; Dassa B.; Flint H. J.; Travis A. J.; Jindou S.; Borovok I.; Lamed R.; Bayer E. A.; Henrissat B.; Coutinho P. M.; Antonopoulos D. A.; Miller M. E. B.; White B. A. Abundance and diversity of dockerin-containing proteins in the fiber-degrading rumen bacterium, Ruminococcus flavefaciens FD-1. PLoS One 2010, 5, e12476 10.1371/journal.pone.0012476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izrailev S.; Stepaniants S.; Balsera M.; Oono Y.; Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 1997, 72, 1568–1581. 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubmüller H.; Heymann B.; Tavan P. Ligand binding: molecular mechanics calculation of the streptavidin-biotin rupture force. Science 1996, 271, 997–999. 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- Isralewitz B.; Gao M.; Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 2001, 11, 224–230. 10.1016/S0959-440X(00)00194-9. [DOI] [PubMed] [Google Scholar]
- Sethi A.; Eargle J.; Black A. A.; Luthey-Schulten Z. Dynamical networks in tRNA: protein complexes. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 6620–6625. 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh B. C.; Hadden J. A.; Bernardi R. C.; Singharoy A.; McGreevy R.; Rudack T.; Cassidy C. K.; Schulten K. Computational methodologies for real-space structural refinement of large macromolecular complexes. Annu. Rev. Biophys. 2016, 45, 253–278. 10.1146/annurev-biophys-062215-011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B.; Sali A. Protein structure modeling with MODELLER. Methods Mol. Biol. 2014, 1137, 1–15. 10.1007/978-1-4939-0366-5_1. [DOI] [PubMed] [Google Scholar]
- Humphrey W.; Dalke A.; Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics 1996, 14, 33–38. 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Mandell D. J.; Coutsias E. A.; Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods 2009, 6, 551–552. 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kale L.; Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo M. C.; Bernardi R. C.; Rudack T.; Scheurer M.; Riplinger C.; Phillips J. C.; Maia J. D.; Rocha G. B.; Ribeiro J. V.; Stone J. E.; Neese F.; Schulten K.; Luthey-Schulten Z. NAMD goes quantum: an integrative suite for hybrid simulations. Nat. Methods 2018, 15, 351. 10.1038/nmeth.4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro J. V.; Bernardi R. C.; Rudack T.; Stone J. E.; Phillips J. C.; Freddolino P. L.; Schulten K. QwikMD—Integrative Molecular Dynamics Toolkit for Novices and Experts. Sci. Rep. 2016, 6, 26536. 10.1038/srep26536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durner E.; Ott W.; Nash M. A.; Gaub H. E. Post-Translational Sortase-Mediated Attachment of High-Strength Force Spectroscopy Handles. ACS Omega 2017, 2, 3064–3069. 10.1021/acsomega.7b00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.; Ta D. T.; Nash M. A. Mechanical Polyprotein Assembly Using Sfp and Sortase-Mediated Domain Oligomerization for Single-Molecule Studies. Small Methods 2018, 2, 1800039. 10.1002/smtd.201800039. [DOI] [Google Scholar]
- Slattery A. D.; Blanch A. J.; Ejov V.; Quinton J. S.; Gibson C. T. Spring constant calibration techniques for next-generation fast-scanning atomic force microscope cantilevers. Nanotechnology 2014, 25, 335705. 10.1088/0957-4484/25/33/335705. [DOI] [PubMed] [Google Scholar]
- Clifford C. A.; Seah M. P. The determination of atomic force microscope cantilever spring constants via dimensional methods for nanomechanical analysis. Nanotechnology 2005, 16, 1666. 10.1088/0957-4484/16/9/044. [DOI] [Google Scholar]
- Milles L. F.; Schulten K.; Gaub H. E.; Bernardi R. C. Molecular mechanism of extreme mechanostability in a pathogen adhesin. Science 2018, 359, 1527–1533. 10.1126/science.aar2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkel R.; Nassoy P.; Leung A.; Ritchie K.; Evans E. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature 1999, 397, 50–53. 10.1038/16219. [DOI] [PubMed] [Google Scholar]
- Scheurer M.; Rodenkirch P.; Siggel M.; Bernardi R. C.; Schulten K.; Tajkhorshid E.; Rudack T. PyContact: Rapid, customizable, and visual analysis of noncovalent interactions in MD simulations. Biophys. J. 2018, 114, 577–583. 10.1016/j.bpj.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan A. A.; Thorn K. S. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998, 280, 1–9. 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- Greene R. F.; Callen H. B. On the formalism of thermodynamic fluctuation theory. Phys. Rev. 1951, 83, 1231. 10.1103/PhysRev.83.1231. [DOI] [Google Scholar]
- Karplus M.; Kushick J. N. Method for estimating the configurational entropy of macromolecules. Macromolecules 1981, 14, 325–332. 10.1021/ma50003a019. [DOI] [Google Scholar]
- Levy R. M.; Karplus M.; Kushick J.; Perahia D. Evaluation of the Configurational entropy for Proteins: Application to Molecular Dynamics Simulations of an α-Helix. Macromolecules 1984, 17, 1370–1374. 10.1021/ma00137a013. [DOI] [Google Scholar]
- Amaro R. E.; Sethi A.; Myers R. S.; Davisson V. J.; Luthey-Schulten Z. A. A network of conserved interactions regulates the allosteric signal in a glutamine amidotransferase. Biochemistry 2007, 46, 2156–2173. 10.1021/bi061708e. [DOI] [PubMed] [Google Scholar]
- Alexander R. W.; Eargle J.; Luthey-Schulten Z. Experimental and computational determination of tRNA dynamics. FEBS Lett. 2010, 584, 376–386. 10.1016/j.febslet.2009.11.061. [DOI] [PubMed] [Google Scholar]
- Eargle J.; Luthey-Schulten Z. A. NetworkView: 3D display and analysis of protein-RNA interaction networks. Bioinformatics 2012, 28, 3000–3001. 10.1093/bioinformatics/bts546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R. C.; Cann I.; Schulten K. Molecular dynamics study of enhanced Man5B enzymatic activity. Biotechnol. Biofuels 2014, 7, 83. 10.1186/1754-6834-7-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange O. F.; Grubmüller H. J. Generalized correlation for biomolecular dynamics. Proteins: Struct., Funct., Genet. 2006, 62, 1053–1061. 10.1002/prot.20784. [DOI] [PubMed] [Google Scholar]
- Xiao S.; Stacklies W.; Cetinkaya M.; Markert B.; Gräter F. Mechanical Response of Silk Crystalline Units from Force-Distribution Analysis. Biophys. J. 2009, 96, 3997–4005. 10.1016/j.bpj.2009.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artzi L.; Bayer E. A.; Moraïs S. Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 2017, 15, 83. 10.1038/nrmicro.2016.164. [DOI] [PubMed] [Google Scholar]
- Ben David Y.; Dassa B.; Borovok I.; Lamed R.; Koropatkin N. M.; Martens E. C.; White B. A.; Bernalier-Donadille A.; Duncan S. H.; Flint H. J.; Bayer E. A.; Moraïs S. Ruminococcal cellulosome systems from rumen to human. Environ. Microbiol. 2015, 17, 3407–3426. 10.1111/1462-2920.12868. [DOI] [PubMed] [Google Scholar]
- Cann I.; Bernardi R. C.; Mackie R. I. Cellulose degradation in the human gut: Ruminococcus champanellensis expands the cellulosome paradigm. Environ. Microbiol. 2016, 18, 307–310. 10.1111/1462-2920.13152. [DOI] [PubMed] [Google Scholar]
- Laskowski R. A.; MacArthur M. W.; Moss D. S.; Thornton J. M. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993, 26, 283–291. 10.1107/S0021889892009944. [DOI] [Google Scholar]
- MacArthur M. W.; Laskowski R. A.; Thornton J. M. Knowledge-based validation of protein structure coordinates derived by X-ray crystallography and NMR spectroscopy. Curr. Opin. Struct. Biol. 1994, 4, 731–737. 10.1016/S0959-440X(94)90172-4. [DOI] [Google Scholar]
- Best R. B.; Zhu X.; Shim J.; Lopes P. E.; Mittal J.; Feig M.; MacKerell A. D. Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257. 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W. L.; Chandrasekhar J.; Madura J. D.; Impey R. W.; Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. 10.1063/1.445869. [DOI] [Google Scholar]
- Bernardi R. C.; Melo M. C.; Schulten K. Enhanced sampling techniques in molecular dynamics simulations of biological systems. Biochim. Biophys. Acta, Gen. Subj. 2015, 1850, 872–877. 10.1016/j.bbagen.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppälä J.; Bernardi R. C.; Haataja T. J. K.; Hellman M.; Pentikäinen O. T.; Schulten K.; Permi P.; Ylänne J.; Pentikäinen U. Skeletal Dysplasia Mutations Effect on Human Filamins’ Structure and Mechanosensing. Sci. Rep. 2017, 7, 4218. 10.1038/s41598-017-04441-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haataja T. J.; Bernardi R. C.; Lecointe S.; Capoulade R.; Merot J.; Pentikäinen U. Non-syndromic mitral valve dysplasia mutation changes the force resilience and interaction of human Filamin A. Structure 2019, 27, 102–112. 10.1016/j.str.2018.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W. Protein production by auto-induction in high-density shaking cultures. Protein Expression Purif. 2005, 41, 207–234. 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.