Abstract
Though the role of Estrogen Receptor (ER)α in breast cancer has been studied extensively, there is little consensus about the role of alternative ER isoform ERβ in breast cancer biology. ERβ has significant sequence homology to ERα but is located on a different chromosome and maintains both overlapping and unique functional attributes. Five variants exist, resulting from alternative splicing of the C-terminal region of ERβ. The relevance of ERβ variants in breast cancer outcomes and response to therapy is difficult to assess because of conflicting reports in the literature, likely due to variable methods used to assess ERβ in patient tumors. Here, we quantitatively assess expression of ERβ splice variants on over 2,000 breast cancer patient samples. Antibodies against ERβ variants were validated for staining specificity in cell lines by siRNA knockdown of ESR2 and staining reproducibility on formalin-fixed paraffin-embedded tissue by quantitative immunofluorescence (QIF) using AQUA technology. We found antibodies against splice variants ERβ1 and ERβ5, but not ERβ2/cx, which were sensitive, specific, and reproducible. QIF staining of validated antibodies showed both ERβ1 and ERβ5 QIF scores, which have a normal (bell shaped) distribution on most cohorts assessed, and their expression is significantly associated with each other. Extensive survival analyses show that ERβ1 is not a prognostic or predictive biomarker for breast cancer. ERβ5 appears to be a context-dependent marker of worse outcome in HER2-positive and triple-negative patients, suggesting an unknown biological function in the absence of ERα.
Introduction
Estrogen Receptor (ER) β was first identified in rat prostate in 1996 and, like ERα, is a nuclear hormone receptor that dimerizes in the presence of estrogen, binds sequence-specific estrogen response elements present in DNA, and subsequently recruits transcriptional activators and repressors to nearby genes [1]. The transcriptional profiles of cell lines expressing ERα and/or ERβ in the presence of estrogen have been extensively studied and generally show some overlapping targets of the two ERs, though differences in the DNA binding domain as well as the activation function domains of the two ERs likely account for the different, seemingly antagonistic role of ERβ to ERα signaling [2–6]. Variants of both ERs resulting from alternative splicing have been identified over the years, and their ability to alter cellular response to estrogen in vitro suggests that estrogen signaling is not solely carried out by full-length ERα [7–11]. The ERβ variants ERβ1, ERβ2/cx, and ERβ5 represent proteins that are identical exclusive of the C-terminus. ERβ2/cx and ERβ5 both have truncations that alter ligand binding capabilities though all three ERβ variants can heterodimerize with ERα and potentially alter estrogen signaling.
For decades, ERα has been an essential, though imperfect, breast cancer biomarker in the clinic for initial diagnosis and subsequent therapeutic decision-making. The selective estrogen receptor modulator tamoxifen is given to patients whose tumors express ERα, but not all of these patients will respond initially and many will eventually relapse [12–14]. Despite the discovery of ERβ nearly 10 years ago, the relevance of its expression in breast cancer to prognosis and response to therapy is still uncertain. The literature examining ERβ correlations with clinicopathological tumor characteristics, survival, and response to therapy is astoundingly discordant [15]. Possible explanations for these discrepancies are non-specificity of reagents for the different splice variants of ERβ, small sample sizes, the lack of a standardized method of measurement, or many other issues inherent in small discovery-based studies. One major limitation to evaluating ERβ protein levels is a lack of commercially available antibodies specific to the alternatively spliced forms. As such, there is no consensus on the usefulness of ERβ in clinical diagnosis of breast cancer. While it could potentially provide novel insight in predicting response to endocrine therapy, there is no high level data supporting either clinical validity or clinical utility.
In this study, we have set out to clarify the relevance of ERβ as a breast cancer biomarker for prognosis and response to therapy by rigorously validating specificity and reproducibility of antibodies against the ERβ variants and then quantitatively measuring their expression on thousands of patients from multiple institutions with clinical follow-up and treatment information.
Methods
Cell lines and siRNA knockdown
Cell lines MCF7-ERβ1 and MCF7-ERβ2 are constructs with doxycycline-inducible expression of ERβ1 and ERβ2/cx constructed and shared by LC Murphy and previously described in [16]. Cells were grown on coverslips, and expression of ERβ was induced with 2 μg/ml of doxycycline for 24 h then knocked down with ESR2 Stealth siRNA (Invitrogen, Cat#1299001) for 24 h. For knockdown of ERβ5, A431 cells were grown on coverslips and knocked down with ESR2 siRNA as above. Immunofluourescent staining was performed on coverslips briefly as follows; cells were fixed in 3.7 % formaldehyde, permeabilized in 0.2 % TritonX-100 then blocked in 2 % BSA. To assess staining in cell lines grown on coverslips, primary antibodies were incubated overnight at 4 °C at dilutions of 1:500, 1:100, and 1:250 for ERβ1 (Thermo-Scientific, PPG5/10), ERβ2/cx (Serotec, 57/3), and ERβ5 (Serotec, 5/25), respectively. 1 h secondary antibody Alexa546-conjugated goat anti-mouse (Life Technologies) diluted 1:500 in BSA was followed by DAPI stain.
Quantitative immunofluorescent staining
Tissue microarrays (TMAs) were deparaffinized for 20 min at 60 °C then soaked in xylene twice for 20 min each. Slides were rehydrated in two 1 min washes in 100 % ethanol followed by one wash in 70 % ethanol and finally rinsed in streaming tap water for 5 min. Antigen retrieval was performed in sodium citrate buffer, pH6 in the PT module from LabVision. Endogenous peroxidases were blocked by 30 min incubation in 2.5 % hydrogen peroxide in methanol. Non-specific antigens were then blocked by 30 min incubation in 0.3 % BSA in TBST. Primary antibodies were prepared diluted to 1:500 (ERβ1; Thermoscientific PPG5/10) and 1:250 (ERβ5; Serotec 5/25) combined with 1:100 pan-cytokeratin (Dako, Cat#Z062201–2) in 0.3 % BSA in TBST and incubated at 4 °C overnight. Primary antibodies were followed by incubation with Alexa 546-conjugated goat anti-rabbit secondary antibody (Life Technologies, Cat#A-11010) diluted 1:100 in mouse EnVision reagent (Dako, Cat#K400111–2) for 1 h. Signal was amplified with Cy5-Tyramide (Perkin Elmer, Cat#SAT705A001EA) for 10 min and then slides were mounted with ProlongGold + DAPI (Life Technologies, Cat#P36931). Immunofluorescence was quantified using AQUA. Briefly, fluorescent images of DAPI, Cy3 (Alexa 546-cytokeratin), and Cy5 (target-ERβ1 or ERβ5) for each TMA spot were collected. Image analysis was carried out using AQUAnalysis software (Genoptix), which uses the cytokeratin stain to generate an epithelial tumor mask. The AQUA score is calculated by dividing the sum of target pixel intensities by the area of the compartments within which they were measured [17, 18].
Patient cohorts
This study includes four cohorts of archived breast cancer cases, constructed into formalin-fixed, paraffin-embedded TMAs. Yale Cohort 1 (n = 649, patients diagnosed 1962–1982) and Yale cohort 2 (n = 398, patients diagnosed 1976–2005) were constructed into TMAs at Yale and represent patients diagnosed at Yale New Haven Hospital. From outside institutions, the Toronto axillary-lymph node-negative cohort consists of 887 patients diagnosed between 1987 and 1996, and the National Cancer Institute Polish Breast Cancer Study (NCI-PBCS) consists of 1,375 patients diagnosed between 2000 and 2003, both constructed into TMAs [19–22]. Clinical variables are shown in Table 1 as reported by the individual institutions from which the cohorts were collected. For Yale Cohort 1, hormone receptor status was determined by immunohistochemistry and HER2 status determined by immunohistochemistry and/or FISH. Since this cohort predates the standardized use of these tests, the TMA was tested for both markers shortly after it was constructed using the same reagents used for IHC testing in our clinical lab. Similarly, FISH was done retrospectively on the TMA. REMARK guidelines were used in the design and evaluation of work presented here [23].
Table 1. Clinicopathological characteristics of the four breast cancer cohorts used in this study.
| Yale Cohort 1 | Yale Cohort 2 | Toronto Cohort | NCI-PBCS | |
|---|---|---|---|---|
| Characteristic | N (%) | |||
| Years diagnosed | 1962–1982 | 1976–2005 | 1987–1999 | 2000–2003 |
| All patients | 649 | 398 | 976 | 1,375 |
| All patients (ERβ5) | 479 (73.8) | 323 (81.2) | 452 (46.3) | 960 (70.0) |
| Age (years) | ||||
| <50 | 122 (25.5) | 102 (31.6) | 177 (39.2) | 283 (29.5) |
| ≥50 | 336 (70.2) | 208 (64.4) | 275 (60.8) | 677 (70.5) |
| Unknown | 21 (4.3) | 13 (4.0) | 0 (0) | 0 (0) |
| Nodal status | ||||
| Positive | 253 (52.8) | 62 (19.2) | 0 (0) | 371 (38.6) |
| Negative | 204 (42.6) | 166 (51.4) | 452 (100) | 566 (59.0) |
| Unknown | 22 (4.6) | 95 (29.4) | 0 (0) | 23 (2.4) |
| Tumor size (cm) | ||||
| ≤2 | 134 (28.0) | 177 (54.8) | 295 (65.3) | 506 (52.7) |
| 2–5 | 236 (49.3) | 105 (32.5) | 142 (31.4) | 406 (42.3) |
| ≥5 | 51 (10.6) | 1 (0.3) | 15 (3.3) | 33 (3.4) |
| Unknown | 58 (12.1) | 40 (12.4) | 0 (0) | 15 (1.6) |
| ER | ||||
| Positive | 246 (51.4) | 158 (48.9) | 288 (63.7) | 656 (68.4) |
| Negative | 208 (43.4) | 102 (31.6) | 118 (26.1) | 271 (28.2) |
| Unknown | 25 (5.2) | 63 (19.5) | 46 (10.2) | 33 (3.4) |
| PR | ||||
| Positive | 232 (48.4) | 137 (42.4) | 227 (50.2) | 524 (54.6) |
| Negative | 201 (41.9) | 114 (35.3) | 185 (40.9) | 401 (41.8) |
| Unknown | 37 (7.7) | 72 (22.3) | 40 (8.9) | 35 (3.6) |
| HER2 | ||||
| Positive | 76 (15.9) | 16 (5.0) | 38 (8.4) | 77 (8.0) |
| Negative | 373 (77.9) | 237 (73.4) | 401 (88.7) | 670 (69.8) |
| Unknown | 30 (6.2) | 70 (12.6) | 13 (2.9) | 213 (22.2) |
| Follow-up (m) | ||||
| Median (range) | 95 (0.4–498) | 123 (7–385) | 98.2 (56.1–158.5) | 116 (4–149) |
| Treatment | ||||
| Chemotherapy | NA | 60 (18.6) | 81 (17.9) | 229 (23.9) |
| Hormonal | NA | 78 (24.2) | 197 (43.6) | 300 (31.2) |
| Both | NA | 41 (12.6) | 17 (3.8) | 308 (32.1) |
| Neither | NA | 71 (22.0) | 156 (34.5) | 123 (12.8) |
| Unknown | 479 (100) | 73 (22.6) | 1 (0.2) | 0 (0) |
Statistical analysis
Statistical analyses for Yale cohorts and the NCI-PBCS cohort were carried out using StatView (SAS Institute Inc.). For the Toronto Cohort, all statistical analyses were preformed using SAS 9.2 software (SAS Inc., Cary, NC, USA), and the Kaplan–Meier curves were generated by R statistical software version 2.3.0 (http://www.r-project.org/). Univariable sample distributions of ERβ are visualized by histograms and fitted by normal distribution. Simple linear regression and Spearman’s rank correlation coefficient were used to estimate the linear relationship and the correlation coefficient between two quantitative scores, respectively. Kaplan–Meier curves were generated to visualize the survival distribution, and the log-rank test was used to compare the survival between different groups. Multivariate Cox Proportional Hazards models included clinical parameters age, node status, tumor size, ERα, PgR, HER2, and ERβ5. Tests with a P-value <0.05 were considered statistically significant.
Results
Identification of specific and reproducible antibodies to ERβ1 and ERβ5
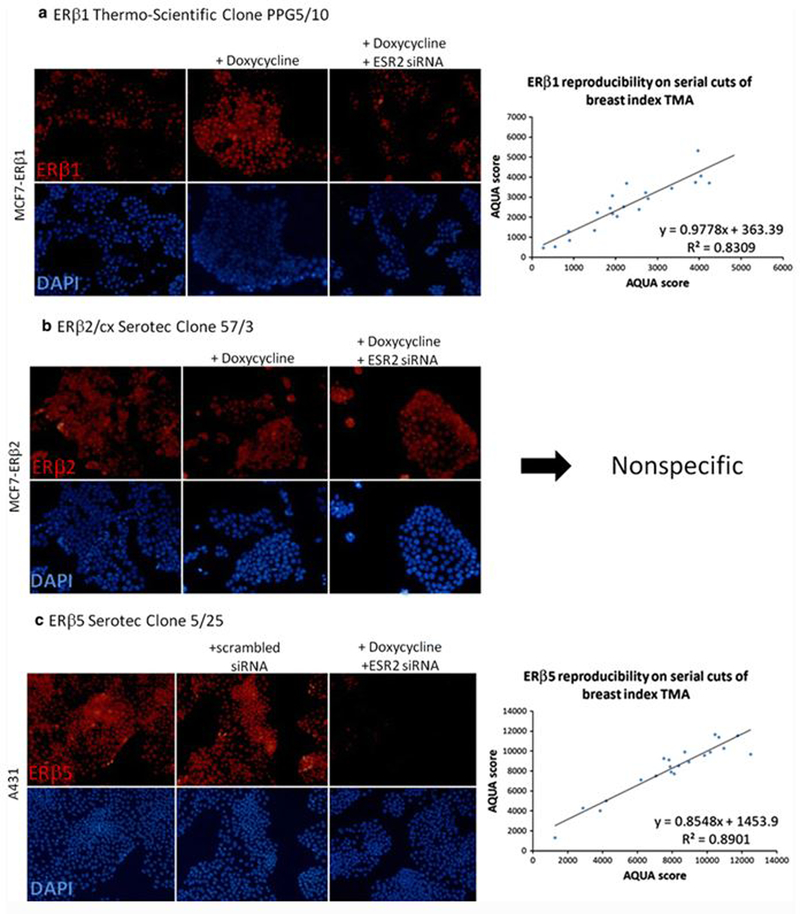
Antibodies against ERβ splice variants were validated by testing their specificity and reproducibility measured by quantitative immunofluorescence (QIF) (Fig. 1). Specificity of ERβ1 and ERβ5 was validated using siRNA knockdown of ESR2 in cell lines. For ERβ1, MCF-7 cells with doxycycline-inducible expression of Xpress-tagged ERβ1 were treated with ESR2 siRNA and staining of ERβ1 assessed on cells grown on coverslips, showing an increase in fluorescence when ERβ1 expression is induced with doxycycline and a substantial decrease when knocked down with ESR2 siRNA (Fig. 1a, left). Similar experiments were done with MCF-7 cells with doxycycline-inducible expression of Xpress-tagged ERβ2/cx, but the antibody used showed no difference in fluorescence in the siRNA knockdown despite successful knockdown of ERβ2/cx, as visualized using an anti-Xpress antibody (data not shown). Thus, the ERβ2 antibody is nonspecific and was not used further (Fig. 1b). To validate an ERβ5 antibody, we used A431 cells, which showed moderate expression of ERβ5 on a TMA with cell line histospots. Staining of ERβ5 on A431 cells grown on coverslips showed corresponding loss of fluorescence in the cells treated with ESR2 siRNA (Fig. 1c, left). Reproducibility of QIF of ERβ1 and ERβ5 antibodies on a set of breast cancer control cases showed reproducible staining, with R 2 > 0.8 on TMAs of serial cut sections (Fig. 1a, c; right).
Fig. 1.
Specificity and reproducibility of three commercially available ERβ antibodies. a Left, specificity of ERβ1 clone PPG5/10 illustrated by IF of MCF7-ERβ1 cell line with doxycycline-inducible ERβ1 grown on coverslips and ±doxycycline and ±ESR2 siRNA; right, reproducibility of ERβ1 clone PPG5/10 measured by QIF on FFPE TMA consisting of 40 breast cancer histospots. b Specificity of ERβ2 clone 57/3 illustrated by IF of MCF7-ERβ2 cell line with doxycycline-inducible ERβ2 grown on coverslips and ±doxycycline and ±ESR2 siRNA. c Left, specificity of ERβ5 clone 5/25 illustrated by IF of A431 cell line with endogenous expression of ERβ5 and addition of scrambled or ESR2 siRNA; right, reproducibility of ERβ5 clone 5/25 measured by QIF on FFPE TMA consisting of 40 breast cancer histospots
Assessment of ERβ1 and ERβ5 by QIF on four breast cancer cohorts
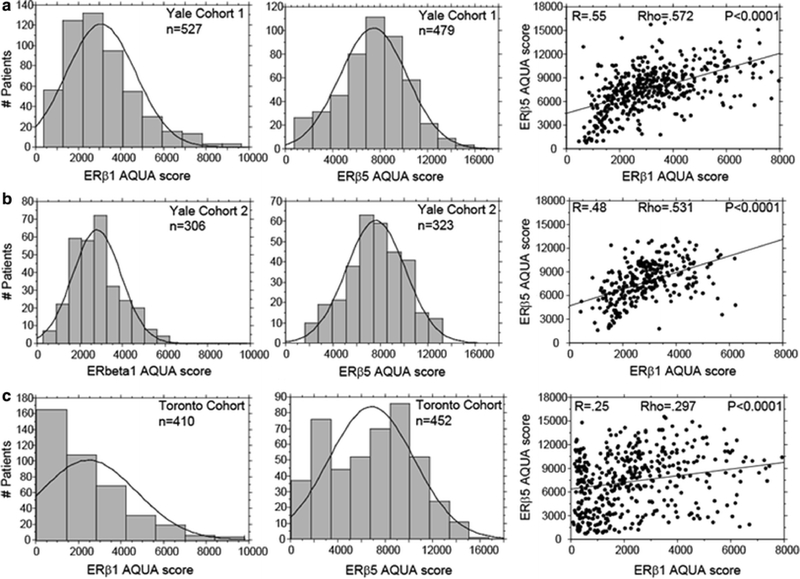
Both ERβ1 and ERβ5 expression by QIF was assessed on 3 breast cancer cohorts (Yale Cohort 1, Yale Cohort 2, and Toronto Cohort) and ERβ5 on one additional cohort to validate findings (NCI-PBCS). See Table 1 for the clinicopathologic characteristics of the four cohorts. With the exception of the NCI-PBCS cohort, each ERβ was assessed in two-fold redundancy, and reproducibility of the QIF assay was assessed with a control TMA consisting of 40 breast cancer histospots with known levels of ERβ1 and ERβ5 expression. The distributions of QIF scores of ERβ1 and ERβ5 on Yale cohort 1, Yale cohort 2, and the Toronto Cohort show similar normal distributions, though for both forms of ERβ, the distribution on the node-negative Toronto Cohort is skewed to the left, showing a greater proportion of lower QIF scores (Fig. 2a–c left and middle panels). Nuclear ERβ5 staining was present in nearly all cases in every cohort, while ERβ1 showed both nuclear and/or cytoplasmic staining in approximately 70 % of cases (data not shown). Linear regression and Spearman-Rank correlation of ERβ1 and ERβ5 on all three cohorts show a tightly correlated expression (Yale Cohort 1 Rho = 0.572; Yale Cohort 2 Rho = 0.531; Toronto Cohort Rho = 0.297), though markedly less so in the Toronto Cohort (Fig. 2a–c, right panels).
Fig. 2.
Comparison of ERβ1 and ERβ5 QIF scores and distributions on three breast cancer cohorts. Histogram distributions of QIF scores of ERβ1 (left) and ERβ5 (middle). Regressions of ERβ1 and ERβ5 QIF scores (right). a Yale Cohort 1. b Yale Cohort 2. c Toronto Cohort
ERβ1 is not prognostic or predictive of response to therapy
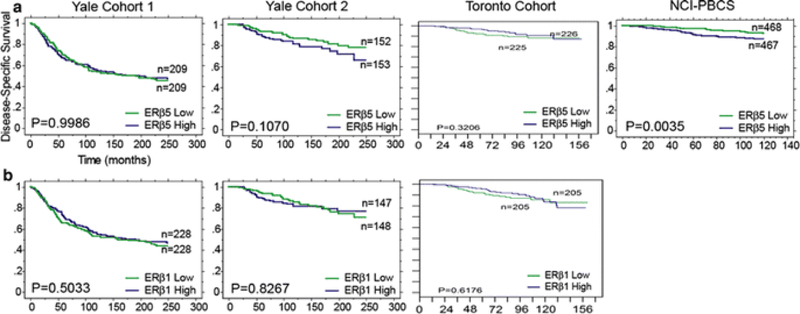
ERβ1 showed no detectable value as a biomarker for patient prognosis or predicting response to therapy in any of the three cohorts examined. Parallel analyses were carried out on the same data set with ERβ1 cut at the estimated visual threshold of detection by immunofluorescence with no significant results. As shown in Fig. 3b, when ERβ1 QIF scores are cut at the median into low and high groups, there is no separation with relation to disease-specific survival. Literature suggests that ERβ1 expression in the nuclear and cytoplasmic cellular compartments may impart differential information with regards to patient prognosis and response to therapy [24], though the data presented here draw from quantitative scores for ERβ1 expression within the entire tumor mask. Individual subcellular compartments were examined alongside total expression within the tumor mask and no differences observed. Extensive exploratory analyses were undertaken to examine ERβ1 in subpopulations of patients, with particular focus on ERα+ and tamoxifen-treated patients, though no associations were observed in any group of patients.
Fig. 3.
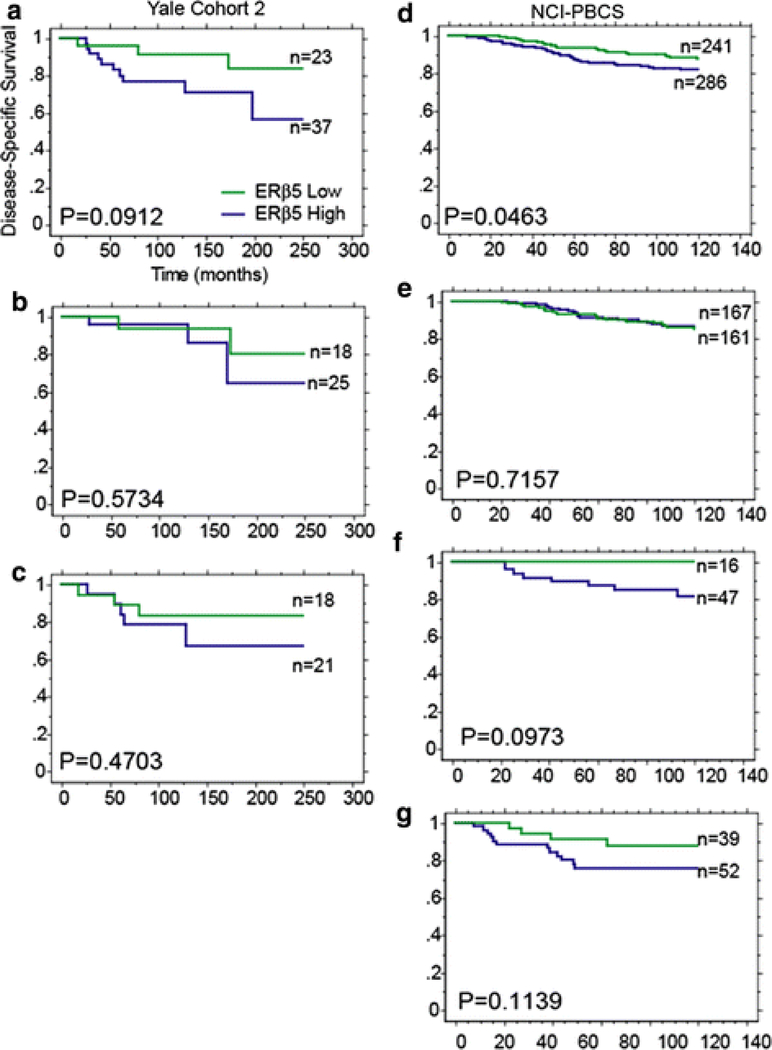
Disease-specific survival stratified by ERβ1 or ERβ5 in breast cancer patients. Kaplan–Meier plots of disease-specific survival stratified by a ERβ5 cut at the median QIF score for Yale Cohort 1, Yale Cohort 2, Toronto Cohort, and NCI-PBCS; b ERβ1 cut at the median QIF score for Yale Cohort 1, Yale Cohort 2, and Toronto Cohort. Note that the only statistically significant finding is ERβ5 low versus high expression on the NCI-PBCS cohort (Log Rank P = 0.0035)
ERβ5 is a marginal marker of worse prognosis in total patient populations
ERβ5, assessed on all four breast cancer cohorts cut at the median QIF score for each patient population, shows a marginal, non-significant potential as a marker of worse prognosis on Yale Cohort 2 (Fig. 3a, middle left). This result is validated on the NCI-PBCS cohort, where ERβ5 is a significant marker of worse prognosis in univariate, but not multivariate, analysis (univariate P = 0.0035, HR 1.931, 95 % CI 1.232–3.029; multivariate P = 0.1951, HR 1.424, 95 % CI 0.834–2.432) (Fig. 3a, far right). No indications of ERβ5 as a prognostic marker were seen on Yale Cohort 1 or the Toronto Cohort in the total patient population.
ERβ5 is prognostic in HER2+ and triple-negative subsets, not luminal
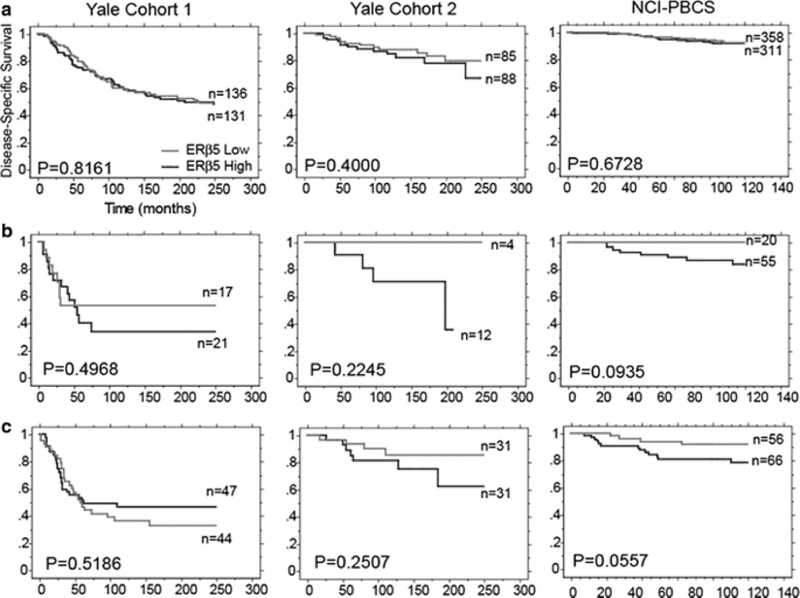
The prognostic implications of ERβ5 in different breast cancer subtypes were examined in Yale Cohort 1, Yale Cohort 2, and the NCI-PBCS cohort. Any hormone receptor positive, HER2-negative patients were classified as luminal, hormone receptor negative, HER2-positive patients classified as HER2-amplified and HER2-negative classified as triple negative. ERβ5 expression cut at the median QIF score of the total patient population within a cohort was then assessed individually in each subtype. ERβ5 shows no prognostic implications in the luminal subtypes of any of the cohorts (Fig. 4a), rather, it appears to be a marker of worse prognosis in HER2-positive and triple-negative subsets, though these results are not significant.
Fig. 4.
Disease-specific survival stratified by ERβ5 in luminal, HER2+, and triple-negative subsets of patients. Kaplan–Meier plots of disease-specific survival stratified by ERβ5 cut at the median QIF score in a luminal, b HER2+, and c triple-negative subsets of patients in Yale Cohort 1, Yale Cohort 2, and NCI-PBCS cohort
In both Yale Cohort 2 and the NCI-PBCS breast cancer cohort, patients received adjuvant chemotherapy, hormonal therapy, both or no adjuvant therapy. ERβ5 expression in treatment subgroups reveals that ERβ5 is a significant marker of worse prognosis only in patients that received adjuvant chemotherapy on the NCI-PBCS cohort, though the same trend is observed in Yale Cohort 2 (Fig. 5a, d). Further analysis of patients that received adjuvant chemotherapy divided into breast cancer subtypes shows results similar to the overall population; ERβ5 correlates with worse prognosis only in HER2+ and triple-negative patients, though these observations are non-significant trends (Fig. 5b–c, e–g).
Fig. 5.
Disease-specific survival stratified by ERβ5 in chemotherapy-treated patients in total, luminal, HER2+, and triple-negative subsets. Kaplan–Meier plots of disease-specific survival in chemotherapy-treated patients stratified by ERβ5. Yale Cohort 2: a all patients, b luminal, c triple negative; NCI-PBCS: d all patients, e luminal, f HER2+ , g triple-negative patients
Discussion
ERβ and its alternatively spliced forms do not have a comprehensively understood role in estrogen signaling. Interest in the clinical relevance of ERβ to breast cancer motivated the numerous accounts of ERβ in retrospectively collected breast cancer tissue. However, discrepant results from these studies have left the question of whether or not ERβ is a clinically relevant breast cancer biomarker unanswered. The presence of various splice variants with unique functionality could be one reason for these conflicting reports [7, 10, 11, 25]. However, even within those studies undertaken that delineate the different ERβ splice variants there are conflicting reports [26, 27]. The general consensus regarding ERβ1, derived most conclusively from cell line studies, is that it opposes ERα signaling and has tumor-suppressor function. ERβ5, on the other hand, appears to oppose both ERα and ERβ1 signaling [7, 25]. This rudimentary biological understanding of ERβ1 and ERβ5 function has not been translated to a clinically relevant tool due to varying results when the proteins are assessed in patient tumors.
We report that rigorous antibody validation is required for accurate assessment of ERβ protein expression. Figure 1 shows the specificity and reproducibility of two commercially available antibodies against ERβ1 and ERβ5 and non-specificity of an antibody against ERβ2/cx. Further, we used a quantitative approach to score staining of ERβ to remove the subjectivity associated with qualitative scoring and to be able to rigorously assess reproducibility of staining to ensure a high level of quality control. Many reports in the literature draw conclusions from small patient samples with few events, resulting, perhaps, in statistically underpowered data that have likely led to much of the confusion in the ERβ field. This study examines ERβ1 and ERβ5 expression in over 1,000 patient tumors on four different patient populations collected from the United States, Canada, and Poland, and, thus, we present the most comprehensive study to date of ERβ in breast cancer patient populations.
ERβ1 expression has no correlation with patient prognosis or response to therapy in any patient population examined. Studies have suggested ERβ1 to act as a marker of good prognosis that can predict response to endocrine therapy in the presence or absence of ERα [24, 28–31]. These results may be highly context dependent. However, this study examined four distinct populations, and ERβ1 was not found to be predictive or prognostic within any of them. Our results imply that ERβ1 will not apply to a broad breast cancer population as a general marker of prognosis or response to endocrine therapy.
A context-dependent, marginal effect of ERβ5 expression on survival was observed. While ERβ5 significantly marks worse prognosis on the NCI-PBCS cohort, the result is neither significant in multivariate analysis nor in any other patient cohort examined in this study. On two of the four cohorts examined, ERβ5 is a marker of worse prognosis in HER2-positive and triple-negative subsets of patients. ERβ5 shows no prognostic or predictive value in the luminal subset of patients in any cohort, a surprising result given that ERβ5 has no known function apart from its ability to interfere with estrogen signaling via interaction with ERα or ERβ1. Known biological functions of ERβ5 require ERβ1 or ERα as a binding partner; ERβ5 on its own has no known ligand. Given its relevance to survival only in ERα-negative patients and lack of any indication that ERβ5 effect on survival is dependent on ERβ1, we speculate that ERβ5 may have yet to be discovered functions in breast cancer cells. Of the two cohorts in which no correlation of ERβ5 expression with survival was seen, Yale Cohort 1 consists of patients diagnosed as early as the 1960s and represents a population of larger, higher grade tumors. The other outlying cohort is the Toronto Cohort and consists only of node-negative breast cancer patients. We speculate that the lack of correlation of ERβ5 with survival on these two cohorts is largely due to the difference in populations and the resulting differences in treatment.
This study has several limitations. We set out to examine ERβ splice variants and were only able to examine two of the three that have been studied in the literature. We were unable to validate an antibody to ERβ2/cx and thus have not performed a fully comprehensive study of the known ERβ splice variants in breast cancer. Secondly, one critique of existing literature is inconsistent results, and we report the same for ERβ5 association with worse outcome among four multi-institutional breast cancer cohorts examined in this study; only two of them show this trend, and, arguably, this trend could represent a random, under-powered result. We believe that the lack of any prognostic implications on the Toronto Cohort and Yale Cohort 1 reflects differences in patient populations; the Toronto Cohort is only node-negative patients, and Yale Cohort 1 contains many patients diagnosed and treated prior to universal use of endocrine therapy for hormone receptor-positive breast cancer. This suggests that the effect of ERβ5 in breast cancer is highly context dependent, only showing prognostic potential in ERα-negative or HER2-amplified populations. Finally, while we have shown that ERβ5 correlates with worse outcome in breast cancer patients while ERβ1 does not, a mechanism of action of ERβ5 in ERα-negative patients remains elusive.
In summary, we report that ERβ splice variants ERβ1 and ERβ5 do not predict response to endocrine therapy, but ERβ5 does indicate worse outcome in patients with ERα-negative and/or HER2-amplified tumors in a population-dependent manner. The results indicate that further investigation of ERβ5 in an ERα-negative context is warranted, but the ultimate clinical utility of ERβ5 is undetermined.
Acknowledgments
The authors of the study thank Pei Chao and Michael Stagner from Information Management Services (Silver Spring, MD) for data management support; the participants, physicians, pathologists, nurses, and interviewers from participating centers in Poland for their efforts during field-work; and Drs. Montserrat Garcia-Closas, Louise Brinton, Jolanta Lissowska, B. Peplonska for their contributions to the PBCS study design. Funding for this study was partially provided by the Breast Cancer Research Foundation.
Disclaimers Dr. Rimm is a consultant for, and receives laboratory support from Genoptix Inc. References
References
- 1.Kuiper G, Enmark E, PeltoHuikko M, Nilsson S, Gustafsson JA (1996) Cloning of a novel estrogen receptor expressed in rat prostate and ovary. Proc Natl Acad Sci USA 93(12):5925–5930. doi: 10.1073/pnas.93.12.5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu MM, Albanese C, Anderson CM, Hilty K, Webb P, Uht RM, Price RH, Pestell RG, Kushner PJ (2002) Opposing action of estrogen receptors alpha and beta on cyclin D1 gene expression. J Biol Chem 277(27):24353–24360. doi: 10.1074/jbc.M201829200 [DOI] [PubMed] [Google Scholar]
- 3.Monroe DG, Secreto FJ, Subramaniam M, Getz BJ, Khosla S, Spelsberg TC (2005) Estrogen receptor alpha and beta heterodimers exert unique effects on estrogen- and tamoxifen-dependent gene expression in human U2OS osteosarcoma cells. Mol Endocrinol 19(6):1555–1568. doi: 10.1210/me.2004-0381 [DOI] [PubMed] [Google Scholar]
- 4.Ogawa S, Inoue S, Watanabe T, Hiroi H, Orimo A, Hosoi T, Ouchi Y, Muramatsu M (1998) The complete primary structure of human estrogen receptor beta (hER beta) and its heterodimerization with ER alpha in vivo and in vitro. Biochem Biophys Res Commun 243(1):122–126. doi: 10.1006/bbrc.1997.7893 [DOI] [PubMed] [Google Scholar]
- 5.Williams C, Edvardsson K, Lewandowski SA, Strom A, Gustafsson JA (2008) A genome-wide study of the repressive effects of estrogen receptor beta on estrogen receptor alpha signaling in breast cancer cells. Oncogene 27(7):1019–1032. doi: 10.1038/sj.onc.1210712 [DOI] [PubMed] [Google Scholar]
- 6.Charn TH, Liu ETB, Chang EC, Lee YK, Katzenellenbogen JA, Katzenellenbogen BS (2010) Genome-wide dynamics of chromatin binding of estrogen receptors alpha and beta: mutual restriction and competitive site selection. Mol Endocrinol 24(1):47–59. doi: 10.1210/me.2009-0252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poola I, Abraham J, Baldwin K, Saunders A, Bhatnagar R (2005) Estrogen receptors beta4 and Beta5 are full length functionally distinct ER beta isoforms—cloning from human ovary and functional characterization. Endocrine 27(3):227–238. doi: 10.1385/endo:27:3:227 [DOI] [PubMed] [Google Scholar]
- 8.Ogawa S, Inoue S, Watanabe T, Orimo A, Hosoi T, Ouchi Y, Muramatsu M (1998) Molecular cloning and characterization of human estrogen receptor beta cx: a potential inhibitor of estrogen action in human. Nucleic Acids Res 26(15):3505–3512. doi: 10.1093/nar/26.15.3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leung YK, Mak P, Hassan S, Ho SM (2006) Estrogen receptor (ER)-beta isoforms: a key to understanding ER-beta signaling. Proc Natl Acad Sci USA 103(35):13162–13167. doi: 10.1073/pnas.0605676103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Omoto Y, Eguchi H, Yamamoto-Yamaguchi Y, Hayashi S (2003) Estrogen receptor (ER) beta 1 and ER beta cx/beta 2 inhibit ER alpha function differently in breast cancer cell line MCF7. Oncogene 22(32):5011–5020. doi: 10.1038/sj.onc.1206787 [DOI] [PubMed] [Google Scholar]
- 11.Peng B, Lu B, Leygue E, Murphy LC (2003) Putative functional characteristics of human estrogen receptor-beta isoforms. J Mol Endocrinol 30(1):13–29. doi: 10.1677/jme.0.0300013 [DOI] [PubMed] [Google Scholar]
- 12. (EBCTCG) EBCTCG (2005) Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet 365(9472):1687–1717. doi: 10.1016/S0140-6736(05)66544-0 [DOI] [PubMed] [Google Scholar]
- 13.Clarke M, Collins R, Davies C, Godwin J, Gray R, Peto R, Early Breast Cancer Trialists Collaborative G (1998) Tamoxifen for early breast cancer: an overview of the randomised trials. Lancet 351(9114):1451–1467 [PubMed] [Google Scholar]
- 14.Davies C, Godwin J, Gray R, Clarke M, Cutter D, Darby S, McGale P, Pan HC, Taylor C, Wang YC, Dowsett M, Ingle J, Peto R, (EBCTCG) EBCTCG, (2011) Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: patient-level meta-analysis of randomised trials. Lancet 378(9793):771–784. doi: 10.1016/S0140-6736(11)60993-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haldosén LA, Zhao C, Dahlman-Wright K (2014) Estrogen receptor beta in breast cancer. Mol Cell Endocrinol 382(1):665–672. doi: 10.1016/j.mce.2013.08.005 [DOI] [PubMed] [Google Scholar]
- 16.Murphy LC, Peng B, Lewis A, Davie JR, Leygue E, Kemp A, Ung K, Vendetti M, Shiu R (2005) Inducible upregulation of oestrogen receptor-beta 1 affects oestrogen and tamoxifen responsiveness in MCF7 human breast cancer cells. J Mol Endocrinol 34(2):553–566. doi: 10.1677/jme.1.01688 [DOI] [PubMed] [Google Scholar]
- 17.Camp RL, Chung GG, Rimm DL (2002) Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med 8(11):1323–1327. doi: 10.1038/nm791 [DOI] [PubMed] [Google Scholar]
- 18.Moeder CB, Giltnane JM, Moulis SP, Rimm DL (2009) Quantitative, fluorescence-based in situ assessment of protein expression. Methods Mol Biol 520:163–175. doi: 10.1007/978-1-60327-811-9_12 [DOI] [PubMed] [Google Scholar]
- 19.Andrulis IL, Bull SB, Blackstein ME, Sutherland D, Mak C, Sidlofsky S, Pritzker KP, Hartwick RW, Hanna W, Lickley L, Wilkinson R, Qizilbash A, Ambus U, Lipa M, Weizel H, Katz A, Baida M, Mariz S, Stoik G, Dacamara P, Strongitharm D, Geddie W, McCready D, Toronto Breast Cancer Study Group (1998) neu/erbB-2 amplification identifies a poor-prognosis group of women with node-negative breast cancer. J Clin Oncol 16(4):1340–1349 [DOI] [PubMed] [Google Scholar]
- 20.Garcıá-Closas M, Brinton LA, Lissowska J, Chatterjee N, Peplonska B, Anderson WF, Szeszenia-Dabrowska N, Bardin-Mikolajczak A, Zatonski W, Blair A, Kalaylioglu Z, Rymkiewicz G, Mazepa-Sikora D, Kordek R, Lukaszek S, Sherman ME (2006) Established breast cancer risk factors by clinically important tumour characteristics. Br J Cancer 95(1):123–129. doi: 10.1038/sj.bjc.6603207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horne HN, Sherman ME, Garcia-Closas M, Pharoah PD, Blows FM, Yang XR, Hewitt SM, Conway CM, Lissowska J, Brinton LA, Prokunina-Olsson L, Dawson SJ, Caldas C, Easton DF, Chanock SJ, Figueroa JD (2014) Breast cancer susceptibility risk associations and heterogeneity by E-cadherin tumor tissue expression. Breast Cancer Res Treat 143(1):181–187. doi: 10.1007/s10549-013-2771-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulligan AM, Pinnaduwage D, Bull SB, O’Malley FP, Andrulis IL (2008) Prognostic effect of basal-like breast cancers is time dependent: evidence from tissue microarray studies on a lymph node-negative cohort. Clin Cancer Res 14(13):4168–4174. doi: 10.1158/1078-0432.CCR-07-4543 [DOI] [PubMed] [Google Scholar]
- 23.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM, Diagnostics SSoN-EWGoC (2006) REporting recommendations for tumor MARKer prognostic studies (REMARK). Breast Cancer Res Treat 100(2):229–235. doi: 10.1007/s10549-006-9242-8 [DOI] [PubMed] [Google Scholar]
- 24.Shaaban AM, Green AR, Karthik S, Alizadeh Y, Hughes TA, Harkins L, Ellis IO, Robertson JF, Paish EC, Saunders PTK, Groome NP, Speirs V (2008) Nuclear and cytoplasmic expression of ER beta 1, ER beta 2, and ER beta 5 identifies distinct prognostic outcome for breast cancer patients. Clin Cancer Res 14(16):5228–5235. doi: 10.1158/1078-0432.ccr-07-4528 [DOI] [PubMed] [Google Scholar]
- 25.Leung YK, Lam HM, Wu SL, Song D, Levin L, Cheng LA, Wu CL, Ho SM (2010) Estrogen receptor beta 2 and beta 5 are associated with poor prognosis in prostate cancer, and promote cancer cell migration and invasion. Endocr Relat Cancer 17(3):675–689. doi: 10.1677/erc-09-0294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fox EM, Davis RJ, Shupnik MA (2008) ER beta in breast cancer—onlooker, passive player, or active protector? Steroids 73(11):1039–1051. doi: 10.1016/j.steroids.2008.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu X, Subramaniam M, Negron V, Cicek M, Reynolds C, Lingle WL, Goetz MP, Ingle JN, Spelsberg TC, Hawse JR (2012) Development, characterization, and applications of a novel estrogen receptor beta monoclonal antibody. J Cell Biochem 113(2):711–723. doi: 10.1002/jcb.23443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hopp TA, Weiss HL, Parra IS, Cui Y, Osborne CK, Fuqua SAW (2004) Low levels of estrogen receptor beta protein predict resistance to tamoxifen therapy in breast cancer. Clin Cancer Res 10(22):7490–7499 [DOI] [PubMed] [Google Scholar]
- 29.Honma N, Horii R, Iwase T, Saji S, Younes M, Takubo K, Matsuura M, Ito Y, Akiyama F, Sakamoto G (2008) Clinical importance of estrogen receptor-beta evaluation in breast cancer patients treated with adjuvant tamoxifen therapy. J Clin Oncol 26(22):3727–3734. doi: 10.1200/jco.2007.14.2968 [DOI] [PubMed] [Google Scholar]
- 30.Nakopoulou L, Lazaris AC, Panayotopoulou EG, Giannopoulou I, Givalos N, Markaki S, Keramopoulos A (2004) The favourable prognostic value of oestrogen receptor beta immunohistochemical expression in breast cancer. J Clin Pathol 57(5):523–528. doi: 10.1136/jcp.2003.008599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esslimani-Sahla M, Simony-Lafontaine J, Kramar A, Lavaill R, Mollevi C, Warner M, Gustallsson JA, Rochefort H (2004) Estrogen receptor beta (ER beta) level but not its ER beta cx variant helps to predict tamoxifen resistance in breast cancer. Clin Cancer Res 10(17):5769–5776. doi: 10.1158/1078-0432.ccr-04-0389 [DOI] [PubMed] [Google Scholar]