Abstract
Introduction
Opportunistic bacterial and fungal infections are the major cause of morbidity and mortality among immune suppressed HIV-positive patients. The main objective of this study was to determine bacterial and fungal organisms causing respiratory infections and their susceptibility to commonly prescribed antimicrobials among HIV patients attending a tertiary infectious disease hospital in Kathmandu.
Methods
Sputum samples were collected from the HIV-positive patients attending Sukraraj Tropical and Infectious Disease Hospital (STIDH) from August 2017 to March 2018. A total of 100 sputum samples were cultured on conventional bacterial and fungal culture media. Bacterial and fungal isolates were identified based on their colony characteristics, microscopic morphology and various biochemical tests. Antibiotic susceptibility test (AST) of bacterial isolates was performed by modified Kirby Bauer disc diffusion method.
Results
Out of 100 sputum samples cultured, 24% (n=24) showed bacterial growth, 42% (n=42) showed fungal growth and 10% (n=10) had both bacterial and fungal growth. Among bacteria, 91.6% (n=22) were monomicrobial and 8.4% (n=2) were polymicrobial in growth, of which, Klebsiella pneumoniae (37.5%) were predominant isolates, followed by Pseudomonas aeruginosa (29.2%), and Escherichia coli (16.7%). The antibiotic susceptibility test (AST) showed 68% (17/25) of bacterial isolates were multi-drug resistant (MDR) and among them 41.2% (7/17) were found to be extended spectrum β lactamase (ESBL) producers. Fungal growth was observed in 42% of samples (42/100). A total of six different species of Candida and four different genera of molds were identified. On species differentiation, Candida albicans (20%) were followed by Candida parapsilosis (4%), and Candida dubliniensis (3%); and various molds were Aspergillus fumigatus (4%), Aspergillus flavus (2%), and Penicillium species (5%). CD4 count was inversely associated with bacterial and fungal infections. Fifty percent of the patients with the fungal infections had a CD4 count below 200. No fungal organisms were isolated from HIV-positive patients under antifungal drug treatment.
Conclusion
HIV-positive patients with a CD4 count less than 200 cells/µL are more vulnerable to opportunistic infections of bacterial and fungal origin. Early isolation, identification and appropriate treatment can reduce mortality due to co-infections. Routine screening of opportunistic pathogens is critical to contain the disease progression.
Keywords: HIV, CD4, bacterial and fungal infection, opportunistic infections
Introduction
Globally, an estimated 40 million people were found to be living with Human Immunodeficiency virus (HIV) infections in 2017.1 In South-East Asia, an estimated 3.5 million people were found living with HIV and this area bears the second highest burden after Sub-Saharan Africa.2 Among 11 South-East Asian Countries, Nepal has an estimated 0.2% prevalence rate of HIV with 0.03% incidence per 1000 population. A total of 32,747 HIV cases have been reported in Nepal since the first case of HIV detected in 1988.3
HIV virus infection reduces the number and functionality of CD4 helper lymphocytes that direct and coordinate acquired immunity against most pathogens.4 Once infected, individuals remain infected throughout life. The diminished immunity leads to various opportunistic infections (OIs) in HIV-affected persons.5 Acquired Immune Deficiency Syndrome (AIDS) is a chronic condition that causes the progressive destruction of the immune system leading to recurring OIs, with changes in body composition, adverse nutritional impact, decreased quality of life and mortality.6 Opportunistic infections occur more often than in people with compromised immune systems (CD4 count<500) than in people with normal immune system. A weakened immune system makes it harder for the body to fight off HIV-related OIs.
The majority of HIV-related OIs includes pneumonia, Salmonella infection, toxoplasmosis, candidiasis (thrush) and tuberculosis.7 Lower respiratory tract infections (LRTIs) are the most common respiratory diseases and are frequently the first clinical manifestations of HIV infections.8 It is estimated that 65% of the patients infected with HIV present with pulmonary involvement as their first clinical manifestation, and that approximately 80% of HIV-positive patients develop pulmonary disease over the course of the HIV infection.9 Among various opportunistic infections, respiratory infections account for up to 70% of AIDS-defining illnesses.10
In Nepal, lower respiratory tract infections (LRTIs) are increasingly reported as opportunistic infections. The etiology of bacterial LRTIs among HIV patients was reported to be due to Streptococcus pneumoniae, Hemophilus influenzae, Pseudomonas aeruginosa, Staphylococcus aureus, Klebsiella pneumoniae, Mycobacterium tuberculosis and Acinetobacter species. Bacteria such as Legionella pneumophila, Mycoplasma pneumoniae and Chlamydia pneumoniae have also been found to cause HIV-associated pneumonia.11 Opportunistic fungal infections such as mucocutaneous candidiasis, pneumocystis, cryptococcosis, and histoplasmosis are the most common AIDS-defining conditions. Fungi such as Candida albicans, Cryptococcus neoformans, Histoplasma capsulatum, Coccidioides immitis, Aspergillusspp., Blastomyces dermatitidis, Penicillium marneffei, and Sporothrix schenckii can cause pulmonary infections in persons infected with HIV.12
CD4 count is a marker for the opportunistic infections in AIDS-related conditions.13 The increase in opportunistic infections when the CD4+ T lymphocytes count falls below 500 cells/µL further leads to an increase in morbidity and mortality among HIV-positive individuals.14 The lower the CD4 count, the higher the risk of opportunistic fungal infections. For instance, the risk of developing fungal infections of the lower respiratory tract remains higher in people with a CD4 count less than 200 cells/µL.11 Preventing opportunistic infections by raising the CD4 count through early and sustained adherence to Antiretroviral therapy (ART) is thus critical.15 Over the last decades, ART has greatly reduced the mortality from AIDS-related OIs including LRTIs, by reducing HIV viral load in the blood and by strengthening the immune response.
Opportunistic infections caused by both bacterial and fungal origins are life-threatening complications in HIV-infected patients. Management of these opportunistic infections based on the diagnosis is critical. In Nepal, there have been very few studies exploring the prevalence of these opportunistic infections in HIV-infected patients. Most of the studies from Nepal reported only opportunistic parasitic infections.16–18 Few studies have reported the prevalence of bacterial and fungal opportunistic infections in HIV-infected patients from Tribhuvan University Teaching Hospital, Kathmandu,19 National Public Health Laboratory, Kathmandu11 and Manipal Teaching Hospital, Pokhara.20 However, none of these studies included antimicrobial susceptibility of these opportunistic infections.
The current study represents patients attending Sukraraj Tropical and Infectious Disease Hospital (STIDH), a tertiary center specialized for treatment of HIV/AIDS and other tropical infectious diseases in Kathmandu. In addition to exploring the opportunistic bacterial and fungal infections, this study has included the standardized susceptibility test of antimicrobials prescribed for these infections. The main objective of this study was to determine bacterial and fungal organisms causing respiratory infections and their susceptibility to commonly prescribed antimicrobials among HIV patients attending a tertiary infectious disease hospital in Kathmandu. This study provides an update on the use of appropriate and more effective treatment regimens for opportunistic LRTIs and fungal infections.
Materials and Methods
A total of 100 HIV/AIDS patients suspected of LRTIs who attended Sukraraj Tropical and Infectious Disease Hospital (STIDH) between August 2017 and March 2018 were included in the study. All the cases were confirmed HIV/AIDS with clinical diagnosis of LRTIs based on the national guidelines for the treatment of HIV/AIDS.21 A written informed consent was obtained from each participant. Socio-demographic characteristics of patients were collected using a questionnaire form through face-to-face interview with the patients. CD4 counts and the information related to patients were first entered in Microsoft Excel and later analyzed using SPSS version 24.0 (IBM Corporation, Armonk, NY, USA).
Inclusion and Exclusion Criteria
A patient was enrolled in this study if he/she was HIV positive and attended STIDH; was clinically diagnosed to have lower respiratory tract infections based on the national treatment guidelines; and provided written informed consent regardless of their ART status. People living with HIV/AIDS who were below 15 years of age were excluded from the study.
Specimen Collection, Sample Size and Processing
Using Cochrane formula for sample size calculation a minimal sample size required was 94. We collected a total of 100 samples. A total of 100 sputum specimens were collected from HIV patients attending STIDH, Teku from August 2017 to March 2018. Patients were advised to collect 2 mL of early morning sputum specimen in a clean, transparent, plastic container. The sputum samples were delivered to the microbiology laboratory of STIDH, Teku and processed using standard microbiological methods for isolation of bacterial and fungal pathogens. The sputum specimens were observed macroscopically for Gram staining including acid fast staining to identify bacterial pathogens and KOH preparation for fungal pathogens.22,23 Thus, identified bacterial pathogens were subjected to an antibiotic susceptibility test using the Kirby Bauer method to determine multi-drug resistant and ESBL producers. All the fungal isolates were primarily identified at the microbiology laboratory of STIDH, Teku and were further confirmed by an external consultant mycologist for quality control.
Isolation, Identification and Antibiotic Susceptibility Test of Bacterial Isolates
A loop full of mucopurulent part of the sputum sample was selected and inoculated onto blood agar, chocolate agar and MacConkey agar (Hi-Media) and incubated for isolation of bacterial pathogens. Identification of significant bacterial isolates was done using standard microbiological techniques.22 Antimicrobial susceptibility test (AST) was performed using the modified disc diffusion test method (Kirby-Bauer) recommended by the Clinical and Laboratory Standards Institute (CLSI) guidelines.24 The following antimicrobial agents; ampicillin (10 µg), ceftriaxone (30 µg) and cefixime (30 µg), gentamicin (10 µg), amikacin (30 µg), co-trimoxazole (25 µg), tetracycline (10 µg), chloramphenicol (10 µg) ciprofloxacin (5 µg), ofloxacin (5 µg), nalidixic acid (30 µg), amoxycillin clavulanic acid (20/10 µg), imipenem (10 µg), piperacillin (75 µg), piperacillin/tazobactam (100/10 µg) and colistin (10 µg) were used for AST. Isolates suspected as Extended Spectrum ß-lactams (ESBLs) were phenotypically confirmed by combined disk diffusion method using ceftazidime (30 µg) vs ceftazidime clavulanic acid (30/10 µg) based on the CLSI guidelines.24
Identification of Fungal Isolates
For isolation of fungal pathogens, sputum samples were inoculated in duplicate Sabouraud`s Dextrose Agar (SDA) tubes and incubated at room temperatures: 25ºC and 37ºC. The cultures were examined on alternate days for growth and were kept for 21 days before discarding them as negative.25 Fungal isolates were identified macroscopically and microscopically based on the colonial morphology of the isolates on the laboratory media. Mucoid yeast-like growth was processed by carrying out Gram’s staining and capsular staining, germ tube test and urease test. Gram’s staining was done to all the isolates with mucoid- and yeast-like growth and observed for Gram-positive budding yeast cells.5,15
All the isolates with mucoid- and yeast-like growth were tested for capsulated budding yeast cells by performing capsular staining with 10% nigrosine for identification of Cryptococcus neoformans. In addition, the organism was inoculated into bird seed agar; and Christensen’s Urease test was done for confirmation of Cryptococcus neoformans. Upon observing Gram-positive budding yeast on gram staining, Candida was suspected, and all Candida isolates were tested for germ tube formation.4 Different Candida species were identified on a selective and differential chromogenic reaction (ChroMagar Candida). The Candida species were identified as described in the manufacturer’s instruction; Candida albicans (Green), C. tropicalis (Blue), C. glabrata (Pink), C. krusei (Dry pink).5 Identification of mold was done with macroscopic examination of the colony morphology. Microscopic identification of the molds was done by morphology and arrangements of spores and hyphae on the lactophenol cotton blue (LCB) mount.15
Results
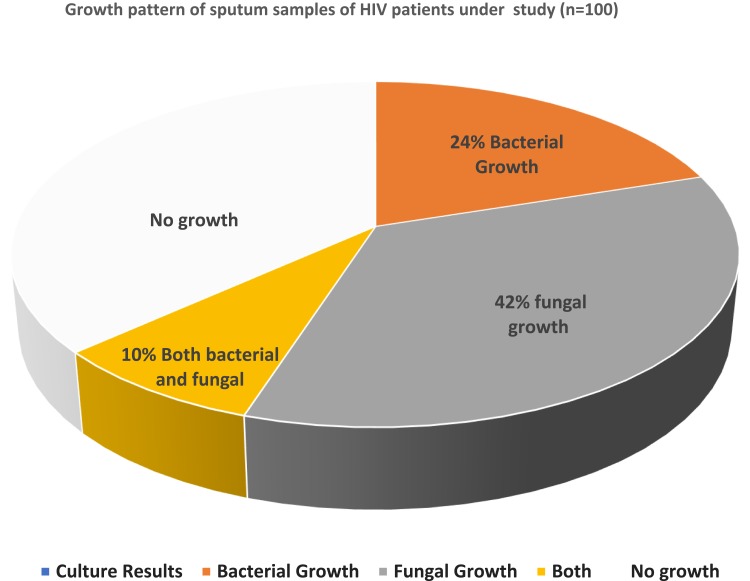
Among 100 sputum samples collected from HIV-positive patients attending STIDH; 24% (n=24) of patients had bacterial infections, 42% (n=42) had fungal infections, and 10% (n=10) had both bacterial and fungal infections (Figure 1).
Figure 1.
Bacterial and fungal growth pattern of sputum samples taken from HIV patients.
Distribution of Bacterial Growth and Fungal Growth in HIV
Among bacterial growth, 91.6% (n=22) were mono-microbial and 8.4% (n=2) were poly-microbial, among which, Klebsiella pneumoniae (37.5%) was predominant, followed by Pseudomonas aeruginosa (29.2%) (Table 1). Among 42 fungal growths, yeast was isolated from 29 samples (69%), only molds were isolated from 12 samples (28.5%); and both yeast and mold were isolated from one sample (2.5%). One patient was found to be co-infected with more than one fungus.
Table 1.
Culture Results of Sputum Samples of HIV Patients (n=100)
| Growth | Number | Percentage | |
|---|---|---|---|
| Bacterial Growth | 24 | 24 | |
| Fungal Growth | 42 | 42 | |
| Both | 10 | 10 | |
| Bacteria (n=24) | |||
| Klebsiella pneumoniae | 9 | 37.5 | |
| Pseudomonas aeruginosa | 7 | 29.2 | |
| Escherichia coli | 4 | 16.6 | |
| Citrobacter freundii | 1 | 4.2 | |
| Acinetobacterspp. | 1 | 4.2 | |
| Acinetobacter spp. + Citrobacter freundii | 1 | 4.2 | |
| Escherichia coli + Pseudomonas aeruginosa | 1 | 4.2 | |
| Fungi (n=42) | |||
| Yeast (n=30) | 29 | 69 | |
| Mold (n=13) | 12 | 28.5 | |
| Both Yeast and Mold(n=1) | 1 | 2.5 | |
| Yeast | |||
| Candida albicans | 20 | 66.7 | |
| Candida parapsilosis | 4 | 13.3 | |
| Candida dubliniensis | 3 | 10.24 | |
| Candida krusei | 1 | 3.3 | |
| Candida tropicalis | 1 | 3.3 | |
| Candida guillermondii | 1 | 3.3 | |
| Mold | |||
| Aspergillus spp. | 6 | 46.15 | |
| Penicillium spp. | 5 | 38.56 | |
| Mucor spp. | 2 | 15.38 | |
Out of a total of 30 isolates of yeast, six different species of yeast were identified. Candida albicans, 20 (66.7%), was the most predominant organism followed by Candida parapsilosis,4 (13.3%). Out of a total of 13 mold isolates, four different types of molds: Aspergillus flavas, Aspergillus fumigatus,Penicillium spp. and Mucorspp. were identified. Aspergillus spp., 6 (46.15%), were the most predominant organism followed by Penicillium spp., 5 (38.56%) and Mucor spp., 2 (15.38%) (Table 1).
Sociodemographic Characteristics, CD4 Count and Their Association with the LRTIs
Among 24 bacterial and 42 fungal culture positive cases, 62.5% (15/24) and 71.4% (30/42) cases of HIV co-infection were reported mostly in the age group 26–49 years (Table 2). The number of bacterial and fungal infections in HIV-positive cases was higher in males (16/65 (24.6%) males had bacterial and 29/65 (44.61%) males had fungal infections) as compared to females (8/35 (22.89% had bacterial infections and 13/35 (37.2%) had fungal infections). There was no significant association between gender of the patients and type of infections among them. Other socio demographic factors such as habit of smoking, habit of taking alcohol were also not significantly associated with development of LRTIs (bacterial and fungal) in HIV patients (Table 2). However, there was a significant association between CD4 count and development of bacterial and fungal infections in HIV patients (p<0.05) (Table 2).
Table 2.
Sociodemographic Characteristics of Enrolled HIV Cases Under Study (n=100)
| Character | Bacterial Growth | p-value | Total | Fungal Growth | p-value | ||
|---|---|---|---|---|---|---|---|
| Culture Positive | Culture Negative | Culture Positive | Culture Negative | ||||
| N (%) | N (%) | N (%) | N (%) | ||||
| Gender | |||||||
| Male | 16 (66.7%) | 49 (64.5%) | 0.6 | 65 | 29 (69%) | 36 (62.1%) | 0.47 |
| Female | 8 (33.3% | 27 (35.5%) | 35 | 13 (31%) | 22 (37.9%) | ||
| Age | |||||||
| 1625 years | 1 (4.2%) | 12 (15.8%) | 0.26 | 13 | 5 (11.9%) | 8 (13.8%) | 0.48 |
| 26–49 years | 15 (62.5% | 50 (65.8%) | 65 | 30 (71.4%) | 35 (60.3) | ||
| > 50 years | 8 (33.3%) | 14 (18.4%) | 22 | 7 (16.7%) | 15 (25.9%) | ||
| Habit of smoking | |||||||
| Yes | 7 (29.2%) | 28 (36.8%) | 0.3 | 17 (40.5% | 18 (31.0%) | 0.32 | |
| No | 17 (70.8%) | 48 (63.2%) | 25 (59.5%) | 40 (69%) | |||
| Habit of taking alcohol | |||||||
| Yes | 4 (16.7%) | 20 (26.3%) | 0.39 | 24 | 13 (31%) | 11 (19%) | 0.16 |
| No | 20 (83.3%) | 56 (73.7) | 76 | 29 (69%) | 47 (81%) | ||
| CD4 cells per microliter | |||||||
| >500 | 7 (29.2%) | 28 (36.8%) | 0.03 | 35 | 7 (16.7%) | 28 (48.3%) | 0.003 |
| 200–499 | 3 (12.5%) | 27 (35.5%) | 30 | 14 (33.3%) | 16 (27.6%) | ||
| <200 | 14 (58.3%) | 21 (27.6%) | 35 | 21 (50%) | 14(24.1%) | ||
| Intake of antifungal drug | |||||||
| Yes | 8 (33.3%) | 14 (18.4% | 0.26 | 22 | 0 (0%) | 22 (37.9% | 0 |
| No | 16 (66.7%) | 62 (81.6%) | 78 | 42 (100%) | 36 (62.1%) | ||
The majority of infections (14/24 (58.3%) bacterial and 21/42 (50%) fungal infections causing LRTI) was present in the HIV-positive patients with CD4 below 200 cells/µL. A total of 7/24 (29.1%) had bacterial and 7/42 (16.66%) had fungal infections of the LRTI and were associated with a CD4 count below 500 cell/µL. Among LRTIs, Klebsiella pneumoniae infection (29.2%), Candida albicans (33.3%) and Aspergillus fumigatus (15.4%) were major infections in patients with a CD4 count below 200 cells/µL. (Table 3). All the sputum samples were also processed for acid fast staining and GeneXpert analysis. None of the samples were found with acid fast bacilli whereas two cases with CD4 count < 200 cells/µL detected a presence of rifampicin-sensitive Mycobacterium tuberculosis. Candida albicans was isolated from one of the HIV-positive patients infected with Mycobacterium tuberculosis.
Table 3.
Distribution of Bacterial and Fungal Genera on CD4 Count of HIV Patients (n=100)
| Bacterial and Fungal Species | CD4 Counts of the Patient’s Cells Per Microliter | Total | |||||
|---|---|---|---|---|---|---|---|
| >500 | 200–499 | <200 | |||||
| N | % | N | % | N | % | ||
| K. pneumoniae | 0 | 0 | 2 | 8.3 | 7 | 29.2 | 9 |
| E. coli | 3 | 12.5 | 0 | 0 | 1 | 4.2 | 4 |
| P. aeruginosa | 4 | 16.6 | 1 | 4.2 | 2 | 8.3 | 7 |
| C. freundii | 0 | 0 | 0 | 0 | 1 | 4.2 | 1 |
| Acinetobacterspp. | 0 | 0 | 0 | 0 | 1 | 4.2 | 1 |
| Acinetobacterspp. + C. freundii | 0 | 0 | 0 | 0 | 1 | 4.2 | 1 |
| E. coli + P. aeruginosa | 0 | 0 | 0 | 0 | 1 | 4.2 | 1 |
| Total | 7 | 29.2 | 3 | 12.5 | 14 | 58.3 | 24 |
| Candida albicans | 4 | 13.3 | 6 | 20 | 10 | 33.3 | 20 |
| Candida krusei | 0 | 0 | 0 | 0 | 1 | 3.3 | 1 |
| Candida dubliniensis | 2 | 6.7 | 0 | 0 | 1 | 3.3 | 3 |
| Candida parapsilosis | 0 | 0 | 3 | 10 | 1 | 3.3 | 4 |
| Candida guilliermondii | 0 | 0 | 1 | 3.3 | 0 | 0 | 1 |
| Candida tropicalis | 0 | 0 | 0 | 0 | 1 | 3.3 | 1 |
| Total | 6 | 20 | 10 | 33.3 | 14 | 44.7 | 30 |
| Aspergillus fumigatus | 1 | 7.7 | 1 | 7.7 | 2 | 15.4 | 4 |
| Aspergillus flavus | 0 | 0 | 2 | 15.4 | 0 | 0 | 2 |
| Penicillium species | 0 | 0 | 1 | 7.7 | 4 | 30.8 | 5 |
| Mucor species | 0 | 0 | 0 | 0 | 2 | 15.4 | 2 |
| Total (Mold) | 1 | 7.7 | 4 | 30.8 | 8 | 61.5 | 13 |
Antibiotic Susceptibility Pattern of Gram-Negative Bacteria Isolated from Sputum Samples of HIV Patients
Klebsiella pneumoniae isolates were highly sensitive (100%; 9/9) towards gentamicin and imipenem, whereas sensitivity towards ofloxacin and ciprofloxacin was 88.8% (8/9). More than 80% (4/5) of E. coli isolates were sensitive to imipenem, gentamicin, and tetracycline. A total of 60% (3/5) were sensitive to ciprofloxacin, ofloxacin, nalidixic acid and amoxicillin clavulanic acid, 40% (2/5) were sensitive to ampicillin, cefixime, ceftriaxone and chloramphenicol. All Citrobacter freundii isolates were 100% (2/2) sensitive towards imipenem, gentamicin, tetracycline, cotrimoxazole and chloramphenicol. All Pseudomonas aeruginosa isolates were 100% (7/7) sensitive to amikacin, ciprofloxacin and piperacillin-tazobactam, while 85.8% (6/7) were sensitive towards imipenem, gentamycin, and colistin. Only half of the Acinetobacterspp. isolates were sensitive to cefixime, ceftriaxone, imipenem gentamicin, amikacin, and ofloxacin. (Table 4).
Table 4.
Antibiotic Susceptibility Pattern of Bacterial Isolates from HIV Patients (n=100)
| Antibiotics | K. pneumoniae(n=9) | P. aeruginosa(n=7) | E. coli(n=5) | C. freundii (n=2) | Acinetobacter spp (n=2) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | Sensitive | Resistant | ||||||||||||
| N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | N | % | ||
| Amoxycillin clavulanic acid 20/10 µg | 1 | 11.2 | 8 | 88.8 | 3 | 60 | 2 | 40 | 1 | 50 | 1 | 50 | |||||||||
| Cefixime 5 µg | 4 | 44.4 | 5 | 55.5 | 0 | – | 7 | 100 | 2 | 40 | 3 | 60 | 1 | 50 | 1 | 50 | 1 | 50 | 1 | 50 | |
| Ceftriaxone 30 µg | 5 | 55.5 | 4 | 44.4 | 3 | 42.8 | 4 | 57.2 | 2 | 40 | 3 | 60 | 1 | 50 | 1 | 50 | 1 | 50 | 1 | 50 | |
| Imipenem 10 µg | 9 | 100 | 0 | – | 6 | 85.8 | 1 | 14.2 | 4 | 80 | 1 | 20 | 2 | 100 | 0 | – | 1 | 50 | 1 | 50 | |
| Gentamicin 10 µg | 9 | 100 | 0 | – | 6 | 85.8 | 1 | 14.2 | 4 | 80 | 1 | 20 | 2 | 100 | 0 | – | 1 | 50 | 1 | 50 | |
| Tetracyclines 30 µg | 4 | 44.4 | 5 | 55.5 | 4 | 80 | 1 | 20 | 2 | 100 | 0 | – | |||||||||
| Ciprofloxacin 5 µg | 8 | 88.8 | 1 | 11.2 | 7 | 100 | 0 | – | 3 | 60 | 2 | 40 | 1 | 50 | 1 | 50 | 0 | – | 2 | 100 | |
| Ofloxacin 5 µg | 8 | 88.8 | 1 | 11.2 | 6 | 85.8 | 1 | 14.2 | 3 | 60 | 2 | 40 | 1 | 50 | 1 | 50 | 1 | 50 | 1 | 50 | |
| Nalidixic 30 µg | 7 | 77.7 | 2 | 22.2 | 3 | 60 | 2 | 40 | 1 | 50 | 1 | 50 | |||||||||
| Cotrimoxazole 23/23.75 µg | 1 | 11.2 | 8 | 88.8 | 2 | 28.6 | 5 | 71.4 | 1 | 20 | 4 | 80 | 2 | 100 | 0 | – | 0 | – | 2 | 100 | |
| Chloramphenicol 30 µg | 8 | 88.8 | 1 | 11.2 | 2 | 40 | 3 | 60 | 2 | 100 | 0 | – | |||||||||
| Ampicillin 10 µg | 2 | 40 | 3 | 60 | 1 | 50 | 1 | 50 | |||||||||||||
| Amikacin 30 µg | 7 | 100 | 0 | – | 1 | 50 | 1 | 50 | |||||||||||||
| Piperacillin/tazobactum 100/10 µg | 7 | 100 | 0 | – | 0 | – | 2 | 100 | |||||||||||||
| Colistin 10 µg | 6 | 85.8 | 1 | 14.2 | |||||||||||||||||
| Piperacillin 75 µg | 1 | 14.2 | 6 | 85.8 | 0 | – | 2 | 100 | |||||||||||||
Multi-Drug Resistant (MDR) and Extended Spectrum Beta Lactamase (ESBL) in Bacterial Isolates
Among the bacterial isolates from the HIV patients, the prevalence of MDR bacteria were as follows: E. coli 100% (5/5), Acinetobacter species 100% (2/2), Pseudomonas aeruginosa 57.2% (4/7), and Klebsiella pneumoniae 55.6% (5/9). All MDR bacterial isolates were screened for ESBL. Among a total of 17/25 (68%) MDR isolates, 41.2% (7/17) isolates were ESBL producers. Among MDR bacterial isolates, the highest ESBL production was found in Klebsiella pneumoniae (4/9; 80%) and Pseudomonas aeruginosa (2/4; 50%) isolates (Table 5).
Table 5.
Prevalence of MDR and ESBL in Bacterial Isolates Isolated from HIV Patients (n=100)
| Bacterial Pathogens | No. of Isolates | Multi-Drug Resistant | ESBL | ||
|---|---|---|---|---|---|
| N | % | N | % | ||
| Escherichia coli | 5 | 5 | 100 | 1 | 20 |
| Pseudomonas aeruginosa | 7 | 4 | 57.2 | 2 | 50 |
| Klebsiella pneumoniae | 9 | 5 | 55.6 | 4 | 80 |
| Citrobacter freundii | 2 | 1 | 50 | 0 | |
| Acinetobacter spp. | 2 | 2 | 100 | 0 | |
| Total | 25 | 17 | 68 | 7 | 41.2 |
Discussion
Overview of Findings
Opportunistic infections are mostly severe in patients living with HIV infections. Various pathogens such as bacteria, virus, parasites, and fungi are responsible for opportunistic infections in people with HIV.26 In this study, the causative agents of LRTIs were fungi (42%), bacteria (24%), and a mix of bacterial and fungal pathogens (10%). The opportunistic bacterial and fungal infections in HIV patients in our study were lower in comparison to other studies reported from Kathmandu valley (46.6% bacterial infection),11 Hyderabad, India (44.28% bacterial infection),27 and Jaipur, Rajasthan, India (86.1% fungal infection).25 A recent study from Nigeria reported 36% fungal infections in HIV-positive patients.28 The low rate of opportunistic infections in our study might be due to high compliance and adherence to the ART at STIDH. Also, the measures taken at STIDH to ensure compliance and adherence through for example counselling and reminder through mobile text messaging might have been highly effective.
Bacterial and Fungal Opportunistic Infections in HIV Patients
In this study, one fifth of the infections were caused by bacterial pathogens and nearly half of the infections were caused by fungal pathogens. Among bacterial isolates, Klebsiella pneumoniae was the predominant bacterial pathogen followed by Pseudomonas aeruginosa and Escherichia coli. Among fungal isolates, Candidial infection was the major one followed by Aspergillus spp. of which Candida albicans was the predominant one. The findings of our study are in line with the studies reported from within Nepal (National Public Health Laboratory, Teku, Kathmandu)11 and Pokhara, Nepal29 and elsewhere such as Taiwan,30 Ilorin, Nigeria,31 Hyderabad, India,27 Jaipur, Rajasthan, India,25 Amritsar, India,32 Visakhapatnam, India,14 and Kano Metropolis, Northern Nigeria.33
The significant association between lower CD4 count and the infections by bacteria and fungi has been established previously. The lower the CD4 count, the higher the rate of colonization by bacteria and fungi.34 These findings are similar with the other studies conducted in Kathmandu, Nepal,11 Aligarh, India,12 Nnewi, Nigeria,35 Awka, Nigeria36 and Atlanta, USA.37 When the person is infected with HIV virus, they are prone to various opportunistic infections due to diminished immunity that depletes the CD4 T lymphocyte cells.
A number of studies have shown the co-infections with fungal infection in patients with low CD4 count. Mycobacterium species were often found to be the major fungal isolates in HIV patients in Nigeria.35 Candida albicans were the most common pathogens resulting in co-infection with pulmonary tuberculosis in HIV-positive patients in Sweden.38 In this study, of the two co-infected cases with Mycobacterium tuberculosis, one of the patients had Candida albicans. Although in this study, Candida species were isolated from patients with different levels of CD4 counts, the prevalence of Candida species in patients with a CD4 count less than 200 cell/μL was expected to be more. Other studies from Nigeria have reported Candida albicans from patients with CD4+ T lymphocyte cell count <200 cells/µL.35,39
Effective Treatment for Bacteria Isolated from Sputum Samples of HIV Patients
Of all the antibiotics tested, Imipenem and Gentamycin were found to be more effective against bacterial isolates. Most of the bacterial isolates showed variability in antibiotic sensitivity. Among isolated gram-negative bacterial isolates, most of them showed the highest sensitivity (>80%) to imipenem and gentamycin. However, Acinetobacterspp showed only 50% sensitivity to imipenem and gentamicin. The major selective force favoring the emergence of antibiotic resistance is their extensive use. In our study, cotrimoxazole, a WHO recommended drug for the treatment of pneumonia was resistant (>80%) against all the isolates except Citrobacter freundii (100% sensitive). Findings in this study resonate with other similar studies from Ekiti State, Nigeria.40 The resistance to cotrimoxazole has been evolving in recent years as has been the case in Lagos, Nigeria.41
In this study, only Gram-negative bacteria were isolated from the sputum sample and two thirds of them were multidrug resistant and among the MDR bacteria, half of them were ESBL producers. Basal media (blood agar, MacConkey agar) were used for primary screening of bacterial pathogens from sputum samples of HIV patients in this study. However, specialized media were not used for trapping the fastidious bacteria, mostly Streptococcus pneumoniae, a major gram-positive bacteria isolated from sputum. This might be the possible cause of not getting Gram-positive bacteria from the sputum samples.
This study further highlights the emergence of ESBL organisms which have been reported to be due to increasing use of broad-spectrum antibiotics.42 Decrease susceptibility of Gram-negative isolates towards third generation cephalosporins such as cefotaxime, ceftriaxone and ceftazidime, could be attributed to ESBL or Amp C beta-lactamase producers or some other relevant underlying mechanisms.43 ESBL production was most common among K. pneumoniae followed by P .aeruginosa and E. coli. These findings were similar with the latest study reported from Bahir Dar, Amhara Region, Ethiopia.44
Implications for Treatment of Opportunistic Infections
It is necessary to screen all HIV-positive cases with bacterial and fungal pathogens in addition to conducting stool examinations for parasitic tests. This reduces further complications by early diagnosis of opportunistic infections and appropriate treatment with better clinical outcome. Routine screening of opportunistic pathogens is necessary, particularly the cases with CD4 count <500 cells/µL, so that opportunistic infections in HIV-positive patients can be managed in time before the development of complications. Detection and identification of fungal infections should utilize a combination of microbiological, histopathological and serological evidence. Laboratory personnel involved in routine investigation of HIV patients should be trained on standard operating protocol for isolation and identification of bacterial and fungal pathogens of clinical significance. Routine monitoring of diagnostics-based use of antimicrobials through increased reporting of infectious diseases and the prescription can further reduce the burgeoning rise in antimicrobial resistance.45 Further studies are recommended using more sensitive methods such as PCR, to compare and make a logical conclusion in order to establish bacteria and fungi as causative agents of lower respiratory tract infections among HIV-positive patients.
Conclusion
HIV-positive patients with a CD4 count less than 200 cells/µL are more vulnerable to infectious bacterial and fungal opportunistic infections. Invasive bacterial and fungal colonization of the lungs remain important causes of death in immunocompromised patients. An early isolation, identification and appropriate treatment can reduce the mortality due to co-infections. Routine screening of opportunistic pathogens together with AFB staining is necessary to contain the disease progression. As all ESBL isolates were MDR, treatment should be strictly based on antibiotic susceptibility pattern reports.
Acknowledgments
We acknowledge the Sukraraj Tropical and Infectious Disease Hospital, Teku, Kathmandu for support. We thank all the team members, laboratory technicians of the hospital and staff of Central Department of Microbiology, Tribhuvan University, for their strong and generous support throughout the study period. We are grateful to all the HIV patients who cooperated to be enrolled in the study. We are thankful to Prof. Dr Niranjan Nayak from Manipal Teaching Hospital, Pokhara, for his support in identification of fungi.
Funding Statement
This research was supported by Departmental funds of the Central Department of Microbiology, Tribhuvan University, Kirtipur, Kathmandu.
Ethics Approval
The study obtained ethical approval from Nepal Health Research Council (NHRC Registration Number 454/2017 and 456/2017). All aspects of the study were conducted according to Good Clinical Practice (GCP) and Good Laboratory Practice (GLP) guidelines. Written informed consents (thumbprint or signature) were obtained from all of the participants. Participant information was securely stored and identified by Study Number.
Availability of Supporting Data
All data pertaining to this study are within the manuscript.
Author Contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.UNAIDS. Global HIV & AIDS statistics-2019 fact sheet; 2019. Available from https://www.unaids.org/en/resources/fact-sheet. Accessed July20, 2019.
- 2.Pendse R, Gupta S, Yu D, Sarkar S. HIV/AIDS in the South-East Asia region: progress and challenges. J Virus Erad. 2016;2:1–6. [PMC free article] [PubMed] [Google Scholar]
- 3.National Center for AIDS & STD Control (NCASC), Nepal. Fact sheet 1: HIV epidimic update of Nepal; 2018. Available from http://www.ncasc.gov.np/WAD2018/FACTSHEET-2018-FINAL/Factsheet-2018-final.pdf. Accessed July20, 2019.
- 4.Limper AH. The changing spectrum of fungal infections in pulmonary and critical care practice: clinical approach to diagnosis. Proc Am Thorac Soc. 2010;7:163–168. doi: 10.1513/pats.200906-049AL [DOI] [PubMed] [Google Scholar]
- 5.Koziel H, Eichbaum Q, Kruskal BA, et al. Reduced binding and phagocytosis of Pneumocystis carinii by alveolar macrophages from persons infected with HIV-1 correlates with mannose receptor downregulation. J Clin Invest. 1998;102:1332–1344. doi: 10.1172/JCI560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geraix J, Carvalhaes MABL, Pereira PCM. Different nutritional-state indicators of HIV-positive individuals undergoing antiretroviral therapy. J Venom Anim Toxins Incl Trop Dis. 2008;14:340. doi: 10.1590/S1678-91992008000200011 [DOI] [Google Scholar]
- 7.USA, Department of Health and Human services. United States of America. HIV and opportunistic infection, coinfection and conditions; 2018. Available from https://aidsinfo.nih.gov/understanding-hiv-aids/fact-sheets/26/86/what-is-anopportunistic-infection. Accessed June15, 2018.
- 8.Mayaud C, Parrot A, Cadranel J. Pyogenic bacterial lower respiratory tract infection in human immunodeficiency virus-infected patients. Eur Respir J Suppl. 2002;36:28–39. doi: 10.1183/09031936.02.00400602 [DOI] [PubMed] [Google Scholar]
- 9.Silva R, Teixeira P, Moreira D. Induced sputum for the diagnosis of lung disease in HIV-positive patients. J Bras Pneumol. 2004;30:452–458. doi: 10.1590/S1806-37132004000500009 [DOI] [Google Scholar]
- 10.Walker PA, White DA. Pulmonary disease. Med Clin North Am. 1996;80:1337–1362. doi: 10.1016/S0025-7125(05)70493-4 [DOI] [PubMed] [Google Scholar]
- 11.Ojha CR, Rijal N, Khagendra KC, et al. Lower respiratory tract infections among HIV positive and control group in Nepal. Virus Dis. 2015;26:77–81. doi: 10.1007/s13337-015-0254-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.PA MA K, Fatima N, Shameem M. Profile of fungal lower respiratory tract infections and CD4 counts in HIV positive patients. Virol Mycol. 2013;2:113. [Google Scholar]
- 13.Ratnam M, Nayyar AS, Reddy DS, Ruparani B, Chalapathi KV, Azmi SM. CD4 cell counts and oral manifestations in HIV infected and AIDS patients. J Oral Maxillofac Pathol. 2018;22:282. doi: 10.4103/jomfp.JOMFP_191_17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ying R, Granich RM, Gupta S, Willams BG. CD4 cell count: declining value for antiretrovial therapy eligibility. Clin Infect Dis. 2016;62(8):1022–1028. doi: 10.1093/cid/civ1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.World Health Organization (WHO). Laboratory manual for diagnosis of fungal opportunistic infection in HIV/AIDS patients; 2009. Available from https://apps.who.int/iris/handle/10665/205404. Accessed June21, 2019.
- 16.Adhikari NA, Rai SK, Singh A, Dahal S, Ghimire G. Intestinal parasitic infections among HIV seropositive and high risk group subjects for HIV infection in Nepal. Nepal Med Coll J. 2006;8:166–170. [PubMed] [Google Scholar]
- 17.Sherchan JB, Ohara H, Sakurada S, et al. Enteric opportunistic parasitics infections among HIV seropositive patients in Kathmandu, Nepal. Kathmandu Univ Med J (KUMJ). 2012;11(2):15–17. [DOI] [PubMed] [Google Scholar]
- 18.Ghimire A, Bhandari S, Tandukar S, Amatya J, Bhandari D, Sherchand JB. Enteric parasitic infection among HIV-infected patients visiting Tribhuvan University Teaching Hospital, Nepal. BMC Res Notes. 2016;9:204. doi: 10.1186/s13104-016-1938-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma S, Dungana G, Pokharel BM, Rijal BP. Opportunistic infections in relation to CD4 level among HIV seropositive patients from Central Nepal. Nepal Med Coll J. 2010;12(1):1–4. [PubMed] [Google Scholar]
- 20.Dhungel BA, Dhungel KU, Easow JM, Singh YI. Opportunistic infection among HIV seropositive cases in Manipal Teaching Hospital, Pokhara, Nepal. Kathmandu Univ Med J (KUMJ). 2008;6:335–339. doi: 10.3126/kumj.v6i3.1708 [DOI] [PubMed] [Google Scholar]
- 21.National Center for AIDS & STD Control. National Consolidated Guidelines for Treating and Preventing HIV in Nepal. Goverenment of Nepal. Kathmandu, Nepal: Ministry of Health& Population; 2014. [Google Scholar]
- 22.Cheesbrough M. District Laboratory Practice in Tropical Countries Part II. Cambridge University Press; 2006. [Google Scholar]
- 23.Forbes BASD, Weissfeld AS. Bailey and Scott’s Diagnostic Microbiology. Vol. 11 St. Louis: Mosby; 2002. [Google Scholar]
- 24.Clinical and Laboratory Standards Institute. CLSI Document M100-S25. Performance Standards for Antimicrobial Susceptibility Testing: Twenty Fifth Informational Supplement Ed. Wayne: CLSI; 2015. [Google Scholar]
- 25.Chandwani J, Vyas N, Hooja S, Sharma B, Maheshwari R. Mycological profile of sputum of HIV positive patients with lower respiratory tract infection and its correlation with CD4+ T lymphocyte count. J Clin Diagn Res. 2016;10:28–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirschtick RE, Glassroth J, Jordan MC, et al. Bacterial pneumonia in persons infected with the human immunodeficiency virus. Pulmonary complications of HIV Infection Study Group. N Engl J Med. 1995;333:845–851. doi: 10.1056/NEJM199509283331305 [DOI] [PubMed] [Google Scholar]
- 27.Shailaja VV, Pai LA, Mathur DR, Lakshmi V. Prevalence of bacterial and fungal agents causing lower respiratory tract infections in patients with human immunodeficiency virus infection. Indian J Med Microbiol. 2004;22:28–33. [PubMed] [Google Scholar]
- 28.Ogba OM, Abia-Bassey LN, Epoke J. The relationship between opportunistic pulmonary fungal infections and CD4 count levels among HIV-seropositive patients in Calabar, Nigeria. Trans R Soc Trop Med Hyg. 2013;107:170–175. doi: 10.1093/trstmh/trs025 [DOI] [PubMed] [Google Scholar]
- 29.Jha BJ, Dey S, Tamang MD, Joshy ME, Shivananda PG, Brahmadatan KN. Characterization of Candida species isolated from cases of lower respiratory tract infection. Kathmandu Univ Med J (KUMJ). 2006;4:290–294. [PubMed] [Google Scholar]
- 30.Lin SH, Kuo PH, Hsueh PR, Yang PC, SH K. Sputum bacteriology in hospitalized patients with acute exacerbation of chronic obstructive pulmonary disease in Taiwan with an emphasis on Klebsiella pneumoniae and Pseudomonas aeruginosa. Respirol. 2007;12:81–87. doi: 10.1111/j.1440-1843.2006.00999.x [DOI] [PubMed] [Google Scholar]
- 31.Salami AK, Olatunji PO, Oluboyo PO, Akanbi AA, 2nd, Fawibe, EA Bacterial pneumonia in the AIDS patients. West Af J Med. 2006;25:1–5. [DOI] [PubMed] [Google Scholar]
- 32.Aggarwal A, Arora U, Bajaj R, Kumari K. Clinico-microbiological study in HIV seropositive patients. J Indian Acad Clin Med. 2005;6:142–145. [Google Scholar]
- 33.Usman AD, Uba A. A survey of bacterial and fungal oppertunistic infection among HIV clients in Kano metropolis. Bajopas. 2011;4(1):148–152. [Google Scholar]
- 34.Sogaard OS, Lohse N, Gerstoft J, et al. Hospitalization for pneumonia among individuals with and without HIV infection, 1995-2007: a Danish population-based, nationwide cohort study. Clin Infect Dis. 2008;47:1345–1353. doi: 10.1086/595712 [DOI] [PubMed] [Google Scholar]
- 35.Ochiabuto MTB, Nwankwo A, Enweani IB, et al. Fungal isolation in HIV patients and CD4 count. International STD Res Rev. 2014;2(2):101–112. doi: 10.9734/ISRR/2014/10408 [DOI] [Google Scholar]
- 36.EU EI U, Oshim IO, Okeke-Nwolisa BC, John GU. Microbiological profile of respiratory tract infections among HIV sero-positive subjects attending Nnamdi Azikiwe University Teaching Hospital Nnewi, Nigeria. Am J Med and Med Sci. 2018;8:37–42. [Google Scholar]
- 37.Rimland D, Navin TR, Lennox JL, et al. Prospective study of etiologic agents of community-acquired pneumonia in patients with HIV infection. AIDS. 2002;16:85–95. doi: 10.1097/00002030-200201040-00011 [DOI] [PubMed] [Google Scholar]
- 38.Mardh PA, Novikova N, Stukalova E. Colonisation of extragenital sites by Candida in women with recurrent vulvovaginal candidosis. BJOG. 2003;110:934–937. doi: 10.1111/j.1471-0528.2003.01445.x [DOI] [PubMed] [Google Scholar]
- 39.Peter YJ, Isa AH, Anzaku AS, Builders MI. Pulmonary candidiasis and CD4 count in HIV positive patients seen in Jos, north central Nigeria. Afr J Cln Exper Microbio. 2016;17:46–52. doi: 10.4314/ajcem.v17i1.6 [DOI] [Google Scholar]
- 40.Ojo-Bolo O, Oluyege AO. Antibiotics resistance of bacteria associated with pneumonia in HIV/AIDS patients in Nigeria. Am J Infect Dis Microbiol. 2014;2:138–144. [Google Scholar]
- 41.Adeleye A, Uju L, Idika N, Sobande O. Cotrimoxazole resistance in Streptococcus pneumoniae isolated from sputum of HIV-positive patients. West Indian Med J. 2008;57:497–499. [PubMed] [Google Scholar]
- 42.Raut S, Gokhale S, Adhikari B. Prevalence of extended spectrum beta-lactamases among E. coli and Klebsiella spp isolates in Manipal, Teaching Hospital, Pokhara, Nepal. JMID. 2015;5(2):69–75. doi: 10.5799/ahinjs.02.2015.02.0179 [DOI] [Google Scholar]
- 43.Bradford PA. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin Microbiol Rev. 2001;14:933–951. doi: 10.1128/CMR.14.4.933-951.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moges F, Eshetie S, Abebe W, et al. High prevalence of extended-spectrum beta-lactamase-producing gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS One. 2019;14:e0215177. doi: 10.1371/journal.pone.0215177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pokharel S, Raut S, Adhikari B. Tackling antimicrobial resistance in low-income and middle-income countries. BMJ Global Health. 2019;4(6):e002104. doi: 10.1136/bmjgh-2019-002104 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- UNAIDS. Global HIV & AIDS statistics-2019 fact sheet; 2019. Available from https://www.unaids.org/en/resources/fact-sheet. Accessed July20, 2019.
Data Availability Statement
All data pertaining to this study are within the manuscript.