Abstract
Background
Long non-coding RNA (lncRNA) DiGeorge syndrome critical region gene 5 (DGCR5) plays different roles in different types of human cancer, but its role in prostate cancer (PC) has not been reported.
Methods
DGCR5 and TGF-β1 expression in paired tumor and adjacent healthy tissues from 64 PC patients was analyzed by performing RT-qPCR. A 5-year follow-up study was performed to analyze the prognostic value of DGCR5 for PC. The interaction between DGCR5 and TGF-β1 was analyzed by overexpression experiments. Cell stemness was analyzed by cell stemness assay.
Results
In our study, we found that DGCR5 was down-regulated in tumor tissues than in adjacent healthy tissues of PC patients, but TGF-β1 was up-regulated in the tumor tissues. DGCR5 expression was not affected by clinical stages, but low DGCR5 level in the tumor was correlated with poor survival. DGCR5 and TGF-β1 were inversely correlated in tumor tissues but not in adjacent healthy tissues. DGCR5 over-expression resulted in down-regulation of TGF-β1, while TGF-β1 treatment did not significantly affect DGCR5 expression. DGCR5 over-expression led to decreased stemness of PC cells, but TGF-β1 treatment played a reverse role and attenuated the effects of DGCR5 over-expression. DGCR5 may decrease the stemness of PC cells by down-regulating TGF-β1.
Keywords: prostate cancer, lncRNA DGCR5, TGF-β1, stemness
Introduction
The incidence of prostate cancer (PC) ranks to be the 1st place among male malignancies in western countries.1 PC is also the 2nd leading cause of deaths among males due to its fast progression and extreme malignant nature.1 With the growth of aging population, incidence of PC is predicted to be continuously and significantly increased.2 The unclear pathogenesis of PC is the major challenge in its clinical treatment.3,4 Studies on molecular pathways involved with PC have revealed that genetic factors are the main contributors to PC.5 However, very limited number of oncogenes and tumor suppressors have been identified during the development and progression of PC. Current evidence is not sufficient to explain all aspects of its molecular pathogenesis.
Long non-coding RNAs (lncRNAs) (>200nt, lncRNAs) have recently been characterized as critical determinants in human diseases,6 such as cancer biology.7,8 The main function of lncRNAs is to regulate gene expression at multiple levels.9 Altered expression of lncRNAs in cancer cells is usually correlated with the dysregulated expression of certain oncogenes or tumor suppressors.10 Besides, the regulation of lncRNAs may help the treatment of cancer by affecting downstream gene expression.7,8 Therefore, characterizations of lncRNAs in cancer development and progression are always necessary. LncRNA DiGeorge syndrome critical region gene 5 (DGCR5) has been proven as an oncogene in lung cancer,11 and a tumor suppressor in liver cancer.12 Our study was performed to explore the function of DGCR5 in PC.
Materials and Methods
Subjects
Our study included 64 PC patients (40 and 69 years old, 55.9 ± 5.1 years old) to serve as research subjects. All these patients were enrolled at the First Peoples’ Hospital of Foshan between Sep 2011 and Sep 2013. Inclusion criteria: 1) first diagnosis and no therapies received; 2) no previous history of malignancies; 3) willing to participate in follow-up. Exclusion criteria: 1) who had been treated within 3 months before admission; 2) combined with other clinical conditions. Based on AJCC staging system, there were 14, 20, 18 and 12 cases at stage I-IV, respectively. All 64 PC patients signed informed consent. Ethics Committee of The First Peoples’ Hospital of Foshan approved this study before the admission of patients.
Follow-Up
All patients were followed up for 5 years old every month throughout patient visit and/or telephone. During follow-up, 2 patients were lost and 5 died of other diseases or accidence. These 7 patients were not included in follow-up study.
Specimen Collection and PC Cell Lines
PC tissues and adjacent (within 2 cm around tumors) non-cancer tissues were obtained from each participant before therapies. All tissues were confirmed by 3 experienced pathologists.
PC cell lines 22Rv1 and DU145 were used in this study to perform cell experiments. Cells of both cell lines were from ATCC (USA). 10% fetal bovine serum (FBS) was added into Eagle’s Minimum Essential Medium to be used as culture medium and cells were cultivated in an incubator with conditions of 37°C and 5% CO2.
RT-qPCR
RNAzol reagent (Sigma-Aldrich, USA) was used to extract total RNAs from PC tissues, non-cancer tissues as well as cells of both 22Rv1 and DU145 cell lines to perform reverse transcription, which was carried out using AMV Reverse Transcriptase (Promega Corporation). To detect the expression of DGCR5 and TGF-β1, PCR systems were prepared using Applied Biosystems™ PowerUp™ SYBR™ Green Master Mix (Thermo Fisher Scientific, Inc.) with 18S rRNA as the endogenous control of DGCR5 and GAPDH as the endogenous control of TGF-β1. Each PCR experiment was performed 3 times and 2−ΔΔCq method was used to process Ct values.
Transient Cell Transfection
All transient cell transfections in this study were performed using Lipofectamine® 2000 reagent (Thermo Fisher Scientific, Inc.). PcDNA3.1 vector was used to construct DGCR5 and TGF-β1 expression vectors (construction service provided by Sangon, Shanghai, China). 10 nM vector was used in the transfection and an empty vector was used as negative control. The control group was the cells with no transfection but were treated with Lipofectamine® 2000 reagent. Subsequent experiments were carried out at 36 hrs after transfections.
Flow Cytometry for Stemness Evaluation
Cells of 22Rv1 and DU145 cell lines were harvested at 36 hrs after transfection and subjected to trypsinization. After that, cells were subjected to the incubations with IgG1-PE or CD133-PE antibody (130-093-193, Meltenyi Biotec, Germany) for 20 min at 4°C. After that, cells were washed and resuspended in PBS. FACS Aria system (BD Immunocytometry Systems, USA) was used to detect the signals and Cell Quest software (Becton Dickinson Ltd) was used to process the signals. Cell stemness was reflected by the percentage of CD133+ cells.
Western-Blotting
Cells of 22Rv1 and DU145 cell lines were harvested at 36 hrs after transfection and were mixed with RIPA solution (Thermo Fisher Scientific, Inc.) to extract total protein. Protein samples were denatured and electrophoresis was carried out using 10% SDS-PAGE gel. Following gel transfer to PVDF membranes, incubation with primary antibodies of rabbit anti-human TGF-β1 (ab92486, 1:1200; Abcam) and rabbit anti-human GAPDH and GAPDH (ab9485, 1:1200, Abcam) as well as anti-rabbit IgG-HRP secondary antibody (1:1000, MBS435036, MyBioSource) were carried out. After that, signals were produced using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific). Image J v.1.46 software was used for data normalizations.
Statistical Analysis
Three biological replicates were included in each experiment. Difference analysis between PC and non-cancer tissues was carried out using the paired t-test. Differences analysis among different clinical stages or cell transfection groups were performed using one-way ANOVA and Tukey t-test. Correlation analyses were performed using linear regression. 57 PC patients completed follow-up and they were divided into high (n=26) and low (n=31) DGCR5 groups using DGCR5 expression data in PC tissues according to Youden’s index. K-M method and log-rank t-test were used to plot and compare survival curves, respectively. Differences were statistically significant when p<0.05.
Results
DGCR5 and TGF-β1 Were Dysregulated in PC Tissues
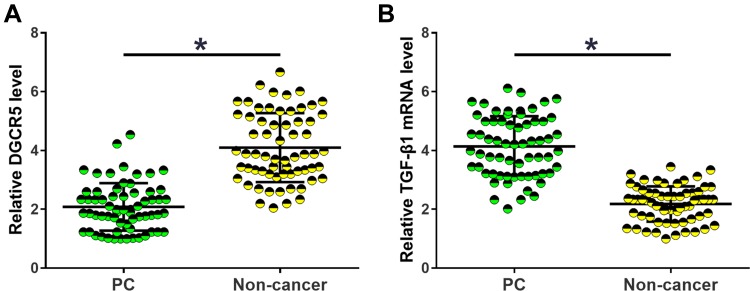
Expression of DGCR5 and TGF-β1 in tissue specimens collected from PC patients was analyzed by RT-qPCR. The sample with the lowest expression level was set to “1”, and all other samples were normalized to this sample. Relative levels of DGCR5 in PC tissues ranged from 1 to 4.78. Relative levels of TGF-β1 mRNA in non-tumor tissues ranged from 2.05 to 7.12. Relative levels of DGCR5 in PC tissues ranged from 1.77 to 6.32. Relative levels of DGCR5 in non-tumor tissues ranged from 1 to 3.54. Comparisons of DGCR5 and TGF-β1 expression levels between PC and non-cancer tissues were performed by paired t-test. The results showed that DGCR5 was significantly down-regulated (Figure 1A), while TGF-β1 was significantly up-regulated (Figure 1B) in PC tissues than in non-cancer tissues of PC patients (p<0.05).
Figure 1.
DGCR5 and TGF-β1 were dysregulated in PC tissues. Comparisons of RT-qPCR data by paired t-test revealed that DGCR5 was significantly down-regulated (A), while TGF-β1 was significantly up-regulated (B) in PC tissues than in non-cancer tissues of PC patients. All PCR reactions were repeated 3 times and mean values were presented and compared by paired t-test (*p<0.05).
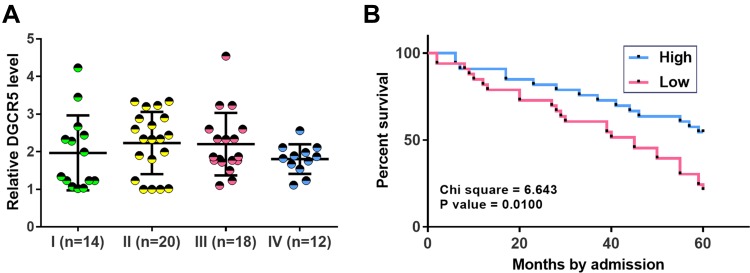
Low DGCR5 Level in PC Tissues Was Correlated with Poor Survival
Comparisons of DGCR5 in PC tissues among patients with different clinical stages were performed by one-way ANOVA and Tukey’s test. No significant differences in DGCR5 expression were found among 4 clinical stages (Figure 2A). In this study, 57 PC patients completed follow-up and they were divided into high (n=26) and low (n=31) DGCR5 groups using DGCR5 expression data in PC tissues according to Youden’s index. K-M method and log-rank t-test were used to plot and compare survival curves, respectively. It was observed that patients in low DGCR5 groups had a significantly lower 5-year overall survival rate (Figure 2B).
Figure 2.
Low DGCR5 level in PC tissues was correlated with poor survival. One-way ANOVA and Tukey’s test were used to compare the expression level of DGCR5 among patients at different clinical stages (A). The 57 PC patients were divided into high (n=26) and low (n=31) DGCR5 groups using DGCR5 expression data in PC tissues according to Youden’s index. K-M method and log-rank t-test were used to plot and compare survival curves, respectively. Survival curve analysis showed that low DGCR5 level in PC tissues was correlated with poor survival (B).
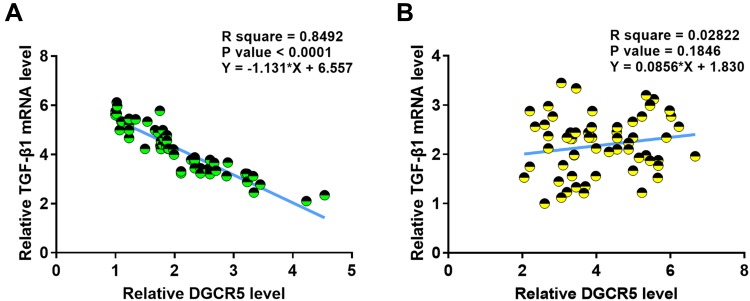
DGCR5 and TGF-β1 Were Inversely Correlated in PC Tissues
Correlations between expression levels of DGCR5 and TGF-β1 in PC tissues were analyzed by linear regression. It was observed that DGCR5 and TGF-β1 were inversely and significantly correlated across cancer tissues (Figure 3A), but not across adjacent non-cancer tissues (Figure 3B).
Figure 3.
DGCR5 and TGF-β1 were inversely correlated in PC tissues. Linear correlation analysis showed that DGCR5 and TGF-β1 were inversely and significantly correlated across PC tissues (A), but not across adjacent non-cancer tissues (B).
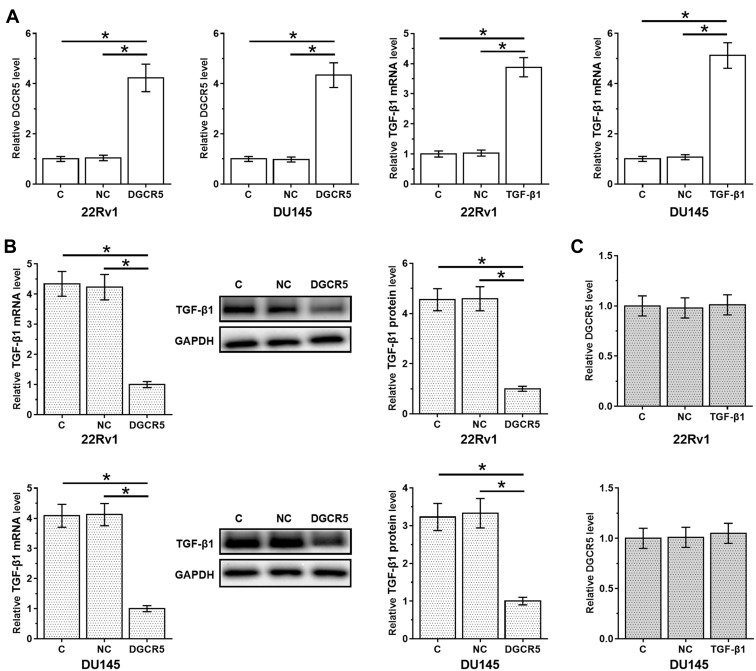
DGCR5 Over-Expression Mediated the Down-Regulation of TGF-β1
DGCR5 and TGF-β1 expression vectors were transfected into cells of both 22Rv1 and DU145 cell lines. At 36 hrs after transfection, compared to two controls (control, C and negative control, NC), expression levels of DGCR5 and TGF-β1 were significantly up-regulated (about 4-fold in all cases, Figure 4A, p<0.05). In addition, DGCR5 over-expression resulted in down-regulation of TGF-β1 at both mRNA and protein levels (3.3- to 4.5-fold, Figure 4B, p<0.05), while TGF-β1 treatment did not significantly affect DGCR5 expression (Figure 4C).
Figure 4.
DGCR5 over-expression mediated the down-regulation of TGF-β1. PC cells were transfected with DGCR5 or TGF-β1 expression vectors to analyze the interaction between them. Comparing to two controls (control, C and negative control, NC), expression levels of DGCR5 and TGF-β1 were significantly up-regulated at 36 hrs after transfection (A), indicating the successful transfections. The effects of DGCR5 over-expression of TGF-β1 at both mRNA and protein levels (B) were analyzed by qPCR and Western blot, respectively. The effects of TGF-β1 treatment on DGCR5 expression were analyzed by qPCR (C). The mean values of 3 biological replicates were presented and compared. (*p<0.05).
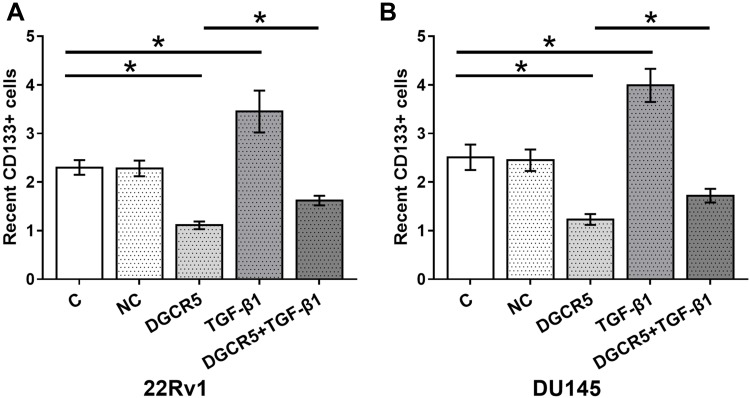
DGCR5 Over-Expression Decreased the Stemness of PC Cells Through TGF-β1
At 36 hrs after transfection, cell stemness was analyzed by performing cell stemness assay. Comparing to two controls (control, C and negative control, NC), DGCR5 over-expression led to significantly decreased stemness (percentage of CD133+ cells) of PC cells, TGF-β1 treatment played a reverse role and attenuated the effects of DGCR5 over-expression on the stemness of both 22Rv1 (Figure 5A) and DU145 (Figure 5B) cells (p<0.05).
Figure 5.
DGCR5 over-expression decreased the stemness of PC cells through TGF-β1. Cell stemness assay was performed to analyze the effects of DGCR5 and TGF-β1 over-expression on PC cell stemness. DGCR5 over-expression led to significantly decreased stemness (percentage of CD133+ cells) of PC cells, TGF-β1 treatment played a reverse role and attenuated the effects of DGCR5 over-expression on stemness of both 22Rv1 (A) and DU145 (B) cells (p<0.05), (*p<0.05).
Discussion
DGCR5 plays different roles in lung cancer and liver cancer,11,12 while its involvement in PC is not clear. Our study first reported the down-regulation of DGCR5 in PC and demonstrated that DGCR5 may negatively regulate the stemness of PC cells by down-regulating TGF-β1, which contributes to cancer cell stemness.13
TGF-β signaling is a double-edged sword in most types of cancer.14 During the early development of cancer, activation of TGF-β mediates the inhibited proliferation and growth of cancer cells,14 indicating its tumor-suppressive roles, while the activation of TGF-β signaling also promotes the migration and invasion of cancer cells at advanced cancer stages, suggestive of oncogenic role.14 Studies on the role of TGF-β signaling in regulating cancer stemness showed that the promoting and maintenance of cancer cell stemness in diverse types of cancers requires the activation of TGF-β signaling.15,16 Consistent with previous studies, our study showed that over-expression of TGF-β1 also led to the increased stemness of PC cells.
Cancer stemness can also be regulated by certain lncRNAs.17 LncRNA DGCR5 has been proved as an oncogene in lung cancer,11 while a tumor suppressor in liver cancer.12 Our study first reported the down-regulation of DGCR5 in PC tissues and proved that DGCR5 over-expression can lead to the decreased stemness of PC cells, suggestive of its tumor-suppressive role in PC. Therefore, DGCR5 plays different roles in different types of cancer. It has been reported that down-regulation of DGCR5 predicted the poor survival of liver cancer patients.12 In our study, we also showed that patients with lower levels of DGCR5 experienced worse overall survival. Therefore, DGCR5 may also serve as a prognostic factor for PC. It is known that TGF-β signaling in cancer biology can be regulated by lncRNAs.18 For instance, lncRNA NKILA inhibits Epithelial-Mesenchymal Transition in breast cancer inactivating TGF-β.18 Our study reported that DGCR5 is an upstream inhibitor of TGF-β1 in the regulation of PC cell stemness. However, the molecular mechanism remains to be further investigated. It has been reported that DGCR5 can regulate the proliferation of lung cancer cells.11 This study is the first to report the regulatory role of DGCR5 in cancer cell stemness. Therefore, regulating the expression of DGCR5 may serve as a target to treat PC by suppressing cancer stemness.
It was observed that we explored the TCGA dataset and we found that DGCR5 was lowly expressed in PC and its expression level was slightly higher in PC tissues than in non-tumor tissues (0.3 vs 0.24), which is not consistent with our data. So, DGCR5 may be differently expressed in different populations. It is worth noting that, with the development of internet and big data era coming, constructing databases19–21 and establishing a powerful web server22,23 will provide the convenience to most scholars.
In conclusion, lncRNA DGCR5 is down-regulated and plays a tumor-suppressive role in PC. Over-expression of DGCR5 may inhibit the stemness of PC cells by down-regulating TGF-β1.
Acknowledgments
We acknowledge the financial support from Effectiveness and Safety of Abiraterone Combined with Prednisone in the Treatment of mCRPC in Foshan Area (A2018537).
Data Sharing Statement
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Consent for Publication
All authors have read and approved the final manuscript.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387 [DOI] [PubMed] [Google Scholar]
- 2.EAU-ESTRO-SIOG guidelines on prostate cancer, Mottet N, Bellmunt J, Bolla M, et al. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2017;71(4):618–629. doi: 10.1016/j.eururo.2016.08.003 [DOI] [PubMed] [Google Scholar]
- 3.Nelson WG, De Marzo AM, DeWeese TL. The molecular pathogenesis of prostate cancer: implications for prostate cancer prevention. Urology. 2001;57(4):39–45. doi: 10.1016/S0090-4295(00)00939-0 [DOI] [PubMed] [Google Scholar]
- 4.Loberg RD, Logothetis CJ, Keller ET, et al. Pathogenesis and treatment of prostate cancer bone metastases: targeting the lethal phenotype. J Clin Oncol. 2005;23(32):8232–8241. doi: 10.1200/JCO.2005.03.0841 [DOI] [PubMed] [Google Scholar]
- 5.Tao ZQ, Shi AM, Wang KX, et al. Epidemiology of prostate cancer: current status. Eur Rev Med Pharmacol Sci. 2015;19(5):805–812. [PubMed] [Google Scholar]
- 6.Chen X, Yan CC, Zhang X, et al. Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief Bioinform. 2016;18(4):558–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang Y, Fullwood MJ. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinf. 2016;14(1):42–54. doi: 10.1016/j.gpb.2015.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ling H, Vincent K, Pichler M, et al. Junk DNA and the long non-coding RNA twist in cancer genetics. Oncogene. 2015;34(39):5003–5011. doi: 10.1038/onc.2014.456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villegas V, Zaphiropoulos P. Neighboring gene regulation by antisense long non-coding RNAs. Int J Mol Sci. 2015;16(2):3251–3266. doi: 10.3390/ijms16023251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–463. doi: 10.1016/j.ccell.2016.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong HX, Wang R, Jin XY, et al. LncRNA DGCR5 promotes lung adenocarcinoma (LUAD) progression via inhibiting hsa-mir-22-3p. J Cell Physiol. 2018;233(5):4126–4136. doi: 10.1002/jcp.26215 [DOI] [PubMed] [Google Scholar]
- 12.Huang R, Wang X, Zhang W, et al. Down-regulation of LncRNA DGCR5 correlates with poor prognosis in hepatocellular carcinoma. Cell Physiol Biochem. 2016;40(3–4):707–715. doi: 10.1159/000452582 [DOI] [PubMed] [Google Scholar]
- 13.Bellomo C, Caja L, Moustakas A. Transforming growth factor β as regulator of cancer stemness and metastasis. Br J Cancer. 2016;115(7):761–769. doi: 10.1038/bjc.2016.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Akhurst RJ, Derynck R. TGF-β signaling in cancer – a double-edged sword. Trends Cell Biol. 2001;11:S44–S51. doi: 10.1016/s0962-8924(01)02130-4 [DOI] [PubMed] [Google Scholar]
- 15.Bruna A, Greenwood W, Le Quesne J, et al. TGFβ induces the formation of tumour-initiating cells in claudin low breast cancer. Nat Commun. 2012;3:1055. [DOI] [PubMed] [Google Scholar]
- 16.Hasegawa T, Yashiro M, Nishii T, et al. Cancer-associated fibroblasts might sustain the stemness of scirrhous gastric cancer cells via transforming growth factor-β signaling. Int J Cancer. 2014;134:1785–1795. doi: 10.1002/ijc.28520 [DOI] [PubMed] [Google Scholar]
- 17.Eades G, Zhang YS, Q L L, et al. Long non-coding RNAs in stem cells and cancer. World J Clin Oncol. 2014;5(2):134–141. doi: 10.5306/wjco.v5.i2.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu W, Chen F, Cui X, et al. LncRNA NKILA suppresses TGF-β-induced epithelial-mesenchymal transition by blocking NF-κB signaling in breast cancer. Int J Cancer. 2018;143(9):2213–2224. doi: 10.1002/ijc.v143.9 [DOI] [PubMed] [Google Scholar]
- 19.Cui T, Zhang L, Huang Y, et al. MNDR v2. 0: an updated resource of ncRNA-disease associations in mammals. Nucleic Acids Res. 2017;46(D1):D371–D374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang ZY, H Y L, Yang H, et al. Pro54DB: a database for experimentally verified sigma-54 promoters. Bioinformatics. 2017;33(3):467–469. doi: 10.1093/bioinformatics/btw630 [DOI] [PubMed] [Google Scholar]
- 21.Hu B, Zheng L, Long C, et al. EmExplorer: a database for exploring time activation of gene expression in mammalian embryos. Open Biol. 2019;9(6):190054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H, Lv H, Ding H, et al. iRNA-2OM: a sequence-based predictor for identifying 2′-O-methylation sites in homo sapiens. J Comput Biol. 2018;25(11):1266–1277. doi: 10.1089/cmb.2018.0004 [DOI] [PubMed] [Google Scholar]
- 23.Zuo Y, Li Y, Chen Y, et al. PseKRAAC: a flexible web server for generating pseudo K-tuple reduced amino acids composition. Bioinformatics. 2016;33(1):122–124. doi: 10.1093/bioinformatics/btw564 [DOI] [PubMed] [Google Scholar]